Abstract
Cytosine or adenine base editors (CBEs or ABEs) can introduce specific DNA C-to-T or A-to-G alterations1–4. However, we recently demonstrated that they can also induce widespread guide RNA-independent RNA base edits5 and created SElective Curbing of Unwanted RNA Editing (SECURE)-BE3 variants that have reduced unwanted RNA editing activity5. Here, we describe structure-guided engineering of SECURE-ABE variants with reduced off-target RNA editing and comparable on-target DNA activities that are also among the smallest Streptococcus pyogenes Cas9 (SpCas9) base editors described to date. We also tested CBEs with cytidine deaminases other than APOBEC1 and found that the human APOBEC3A (hA3A)-based CBE induces substantial RNA base edits, whereas an enhanced A3A (eA3A)-CBE6, human activation-induced cytidine deaminase (hAID)-CBE7, and the petromyzon marinus cytidine deaminase (pmCDA1)-based CBE Target-AID4 induce reduced RNA edits. Finally, we found that CBEs and ABEs that exhibit RNA off-target editing activity can also self-edit their own transcripts, thereby leading to heterogeneity in base editor coding sequences.
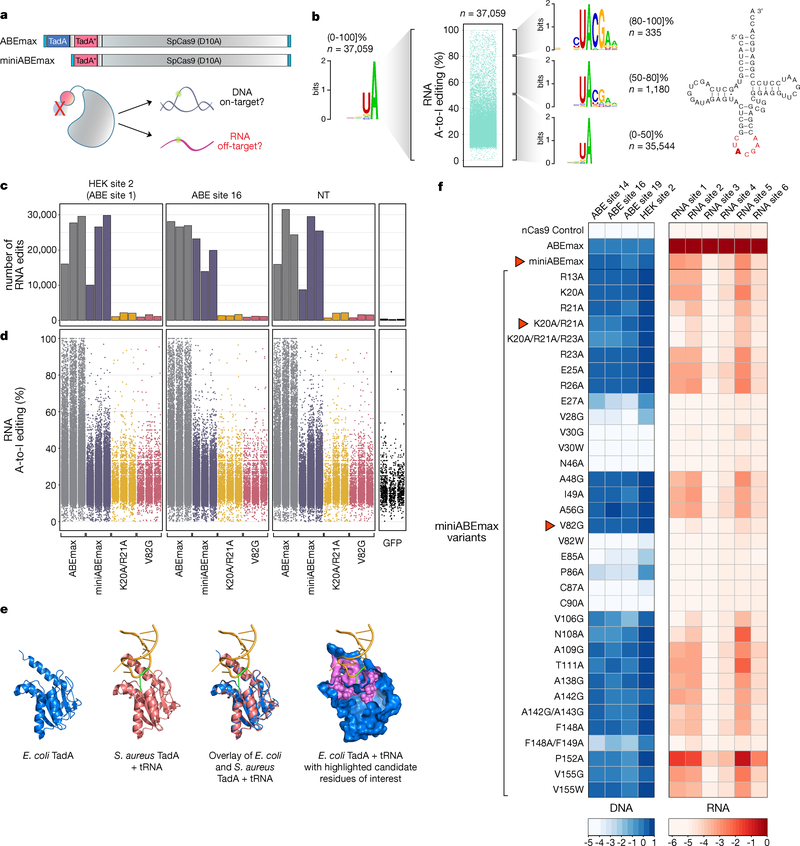
To engineer SECURE-ABE variants, we first used a protein truncation strategy to reduce the RNA recognition capability of the optimized ABEmax fusion. ABEmax harbors a single-chain heterodimer of the wild type (WT) E. coli TadA adenosine deaminase monomer (which deaminates adenines on tRNA) fused to an engineered E. coli TadA monomer that was modified by directed evolution to deaminate DNA adenines3,8,9 (Fig. 1a). Because the WT TadA monomer should still be capable of recognizing its tRNA substrate, one can envision that this domain might recruit ABEmax to deaminate RNA adenines that lie in the same or a similar sequence motif to that present in the tRNA. Consistent with this idea, a re-analysis of our previously published RNA-seq data5 revealed that adenines edited with the highest efficiencies (80–100%) are embedded in a more extended CUACGAA motif, which contrasts to the shorter UA sequence observed across all edits (Fig. 1b). Importantly, the CUACGAA motif matches the sequence surrounding the adenine deaminated in the tRNA substrate of the WT E. coli TadA enzyme (Fig. 1b)8. Therefore, removing the WT TadA domain from ABEmax might reduce its RNA editing activity and doing so might not have a dramatic impact on its on-target DNA editing function (Supplementary Note 1). To test this hypothesis, we generated a smaller ABEmax variant lacking this domain that we refer to as miniABEmax (Fig. 1a).
Figure 1. Engineering of SECURE-ABE variants with reduced off-target RNA editing activities.
(a) Schematic illustration of ABEmax and miniABEmax architectures and overview of experimental testing of miniABEmax for on-target DNA and off-target RNA editing. Light blue boxes = bipartite NLS at N- and C-termini, TadA* = mutant TadA 7.103, and small grey boxes = 32AA flanked XTEN linkers. nCas9 (SpCas9 D10A) = grey shape, TadA WT and mutant monomers = blue and red circles. Green halo = sites of potential adenine deamination on DNA and RNA. (b) Unstratified sequence logo (left) and stratified sequence logos for RNA adenines edited with high (80–100]%, middle (50–80]%, and low (0–50]% efficiencies (right) by ABEmax. n= number of modified adenines. RNA-seq data shown in the Jitter plot was obtained from HEK293T cells in an earlier published study5. Cloverleaf structure of E. coli tRNAArg2 (Ref. 8), illustration adapted from Fig. 1a of Ref. 11. Anticodon loop highlighted in red (Ref. 8). TadA target adenine 34 is highlighted in bold. (c) Bar plots showing the number of RNA A-to-I edits observed in RNA-seq experiments in HEK293T cells with expression of ABEmax, miniABEmax, miniABEmax-K20A/R21A, or miniABEmax-V82G each with three different gRNAs (HEK site 2, ABE site 16, and non-targeting (NT)) and performed in independent replicates (n = 3). Exact number of edits are shown in Supplementary Table 1. GFP negative controls performed as independent replicates (n = 3) are also shown. (d) Jitter plots showing the efficiencies of RNA A-to-I edits from the RNA-seq experiments shown in c. Each dot represents an edited adenine position in RNA. (e) Structural representations of E. coli TadA (PDB 1Z3A), structural representation of S. aureus TadA in complex with tRNA (PDB 2B3J), overlaid structures from E. coli TadA and S. aureus TadA, and surface representation of E. coli TadA in blue with backbone carbons of amino acid positions proximal to the predicted deaminase catalytic site highlighted in pink. Target adenine on tRNA (A34) marked in green. All graphical representations generated with PyMol (Methods). (f) Testing of 34 miniABEmax variants for their on-target DNA editing (A-to-G) and off-target RNA editing (A-to-I) activities. On-target DNA editing was assessed with four different gRNAs and off-target RNA alterations were screened on six RNA adenines previously identified as being efficiently modified by ABEmax5. Efficiencies are shown in heat map format (log2-fold changes), with each box representing the mean of four independent replicates normalized to the editing efficiency observed with ABEmax for each target DNA or RNA off-target site. Red arrows indicate three variants that were chosen for further analysis. Amino acid abbreviations are according to IUPAC nomenclature and residue numbering is based on the amino acid position in E. coli TadA. A = adenine; I = inosine. ABEmax = codon optimized adenine base editor. miniABEmax = ABEmax without N-terminal wild type TadA domain and the proximal 32AA linker.
We used RNA-seq to compare the transcriptome-wide off-target RNA editing activities of miniABEmax to ABEmax in HEK293T cells. Each of these editors and a nickase Cas9 (nCas9) control were assayed with three different gRNAs: two targeted to endogenous human gene sites (HEK site 2 and ABE site 16)3 and one to a site that does not occur in the human genome (NT)5. We performed these studies in triplicate and sorted for GFP-positive cells (each editor or nCas9 was expressed as a P2A-EGFP fusion (Methods)). As an internal control, we confirmed that ABEmax and miniABEmax induced comparable on-target DNA editing with HEK site 2 and ABE site 16 gRNAs (Supplementary Fig. 1a). Edited RNA adenines were identified from RNA-seq experiments as previously described5 by filtering out background editing observed with read-count-matched nCas9 negative controls (Methods). Surprisingly, the total number of edited adenines induced with miniABEmax expression was not consistently lower than what we observed with ABEmax -- the two editors induced on average 80-fold and 54-fold more edited adenines relative to background (determined with a GFP-only negative control) (Fig. 1c and Supplementary Table 1). However, the overall distribution of individual RNA adenine editing efficiencies induced by miniABEmax were generally shifted to somewhat lower values (Fig. 1d and Supplementary Fig. 1b). In addition, the sequence logos of adenines (stratified by editing efficiencies) edited by miniABEmax only yielded shorter GUA or UA motifs, in contrast to the more extended CUACGAA motif observed with ABEmax (Supplementary Figs. 2a and 2b).
We reasoned we might further reduce the off-target RNA editing activity of miniABEmax by altering amino acid residues within its remaining engineered E. coli TadA domain that could potentially mediate RNA recognition. However, although a crystal structure of isolated E. coli TadA has previously been solved10 (PDB 1Z3A; Fig. 1e), no structural information was available to delineate how this protein might recognize its RNA substrate. To overcome this, we exploited the availability of a S. aureus TadA-tRNA co-crystal structure11 (PDB 2B3J) (Fig. 1e and Methods). Although E. coli and S. aureus TadA share only partial amino acid sequence homology (39.5% identity; data not shown), these two proteins share a high degree of structural homology (Fig. 1e). This similarity enabled us to overlay the two structures and thereby to infer 26 amino acid residue positions in E. coli TadA that likely lie near the enzymatic pocket around the substrate tRNA (Fig. 1e). In addition, we mutated three positively charged residues (R13, K20, and R21) in TadA* that we hypothesized might make contacts to the phosphate backbone of a nucleic acid molecule. We reasoned that reducing the potentially non-specifc affinity of miniABEmax in this way might preferentially reduce its Cas9-independent RNA editing activity while preserving its Cas9-assisted on-target DNA editing activity.
We generated 34 miniABEmax variants bearing various substitutions at the amino acid positions described above and screened each editor for on-target DNA editing and off-target RNA editing activities in HEK293T cells. To assess on-target DNA editing, we examined the efficiencies of A-to-G edits induced with four gRNAs targeted to different endogenous gene sequences and found that 23 of the 34 variants induced editing comparable to that observed with miniABEmax and ABEmax (Fig. 1f). To screen for off-target RNA editing activities (using standard transfection conditions, i.e., without sorting for GFP expression; see Methods), we quantified editing by each of the 34 variants at six RNA adenines previously identified as being highly edited with ABEmax overexpression in HEK293T cells5. 14 of the 34 variants showed reduced editing activities on at least three of the six RNA adenines we examined relative to miniABEmax (Fig. 1f). Based on their DNA/RNA editing profiles, we chose to carry forward two miniABEmax variants (K20A/R21A and V82G) for more extensive characterization.
To characterize the transcriptome-wide off-target RNA editing profiles of miniABEmax-K20A/R21A and -V82G, we performed RNA-seq with each of these variants and the HEK site 2, ABE site 16, and NT gRNAs. In contrast to what we observed with miniABEmax, the K20A/R21A and V82G variants both induced substantially reduced numbers of edited adenines relative to ABEmax but still approximately four-fold and three-fold higher numbers, respectively, than background (determined with the GFP-only negative control) (Fig. 1c and Supplementary Table 1). In addition, the distribution of individual RNA adenine editing efficiencies for the two variants was shifted predominantly lower with both variants relative to ABEmax and miniABEmax (Fig. 1d and Supplementary Fig. 1b). The sequence logos of the edited RNA adenines that we derived from these experiments showed that miniABEmax-K20A/R21A and -V82G maintained a UA motif (Supplementary Fig. 2c).
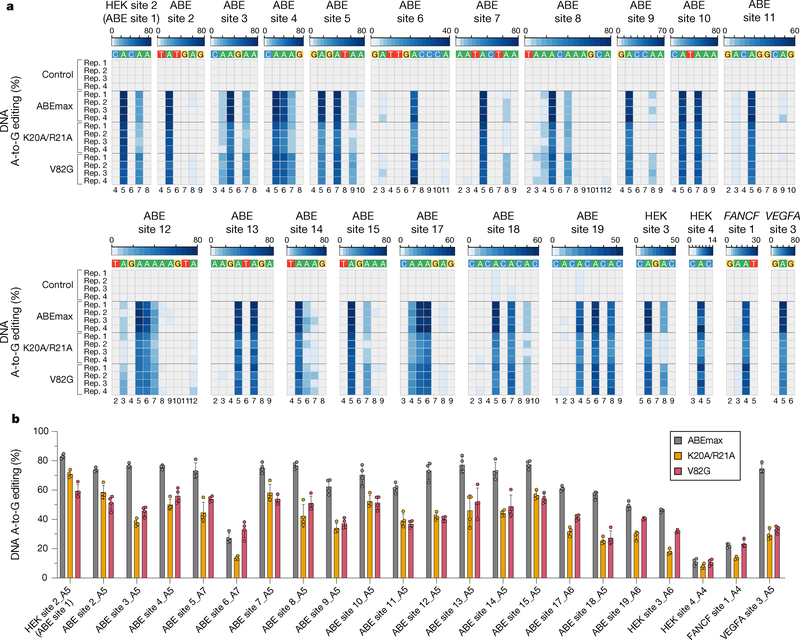
To more fully characterize the on-target editing efficiencies of miniABEmax-K20A/R21A and -V82G, we tested each (without sorting cells) in a variety of different sequence contexts with gRNAs for 22 genomic sites in HEK293T cells3. miniABEmax-K20A/R21A and -V82G retained efficient absolute on-target modification activities (ranges of mean efficiencies of 7.9–70.9% and 10.6–59.4%, respectively; Fig. 2a); however, these efficiencies were typically reduced compared to ABEmax with relative activities across the 22 sites ranging from 38.8 to 85.5% and 44.3 to 121.3% for the most highly edited base in the editing window with miniABEmax-K20A/R21A and -V82G, respectively (Fig. 2b). (The relative activity reductions with the variants may be more apparent here because of the higher on-target editing activities achieved compared with our earlier screening results (Fig. 1f), presumably due to higher transfection efficiencies achieved with a change in the protocol used (Methods)). Neither of the variants showed an apparent preference for a particular sequence context adjacent to the edited adenines (Fig. 2a).
Figure 2. On-target DNA editing activities of ABEmax, miniABEmax-K20A/R21A, and miniABEmax-V82G in HEK293T cells.
Heat maps (a) and bar plots (b) showing the on-target DNA A-to-G editing efficiencies of nCas9 (Control), ABEmax, miniABEmax-K20A/R21A, and miniABEmax-V82G with 22 gRNAs (n=4 independent replicates). For (a), editing window shown includes only the most highly edited adenines and not the entire spacer sequence. A-to-G editing efficiencies are shown in heatmap format. Numbering at the bottom represents spacer position with 1 being the most PAM-distal location. For (b), A-to-G editing efficiencies for only the most highly edited adenine for each gRNA on-target site are reported; error bars represent standard deviation (SD).
Our analysis of ABE activities with 22 gRNAs also identified a new and unexpected imprecise C-to-G base editing activity within the editing windows of some DNA on-target sites. This C-to-G on-target DNA editing was observed with ABEmax and miniABEmax-V82G using the HEK site 2, ABE site 7, and FANCF site 1 gRNAs (Supplementary Fig. 3a). This unwanted editing was consistent across replicates, reached frequencies as high as 14.6% with the FANCF site 1 gRNA, and was not observed with the nCas9 control (Supplementary Fig. 3a). Interestingly, for all three sites, the C showing this unexpected editing was present at position 6 of the spacer and was preceded by a T at FANCF site 1 and by an A at HEK site 2 and ABE site 7 (Supplementary Fig. 3a). Notably, for FANCF site 1, consistent C-to-T and C-to-A edits were also observed at the position C6 (Supplementary Figs. 3b and 3c). Additional studies will be needed to clarify the mechanism by which ABEs can induce this new type of imprecise base edit and to define the positions and sequence contexts that dictate whether a C within the editing window is subject to this alteration.
We also sought to compare the off-target DNA activities of miniABEmax-K20A/R21A and -V82G with that of ABEmax. To do this, we used targeted amplicon sequencing to quantify editing events at ten previously defined potential off-target sites of three gRNAs (targeted to HEK site 2, HEK site 3, and HEK site 4)3,12. We found that ABEmax and miniABEmax-K20A/R21A induced comparable editing patterns and efficiencies on all 10 potential off-target sites (including no detectable mutations on some sites) (Supplementary Fig. 4). miniABEmax-V82G also exhibited comparable editing efficiencies to ABEmax for eight of the ten potential off-target sites examined but did induce some consistent but very low efficiency edits (range of 0.14 – 0.21%) on two sites, both of which are potential off-target sites for the HEK site 3 gRNA (Supplementary Fig. 4). Although additional experiments will be required to more fully define the genome-wide off-target profiles of miniABEmax-K20A/R21A and -V82G, these initial studies suggest that the two variants do not exhibit dramatic alterations in their off-target DNA mutation activities relative to ABEmax.
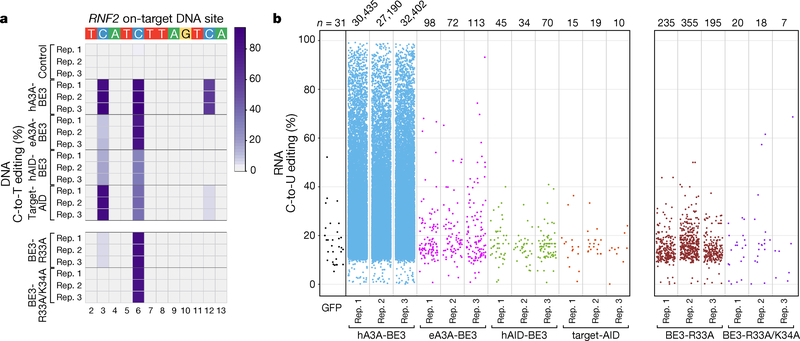
Having previously shown that off-target RNA editing occurs with a CBE harboring the rAPOBEC1 enzyme (BE3) 5, we wanted to determine whether CBEs harboring other cytidine deaminases such as hA3A13, eA3A6 (an engineered A3A with more precise and specific DNA editing activities), hAID7, or a sea lamprey CDA1 (pmCDA1)4 might also induce unwanted edits. To do this, we transfected HEK293T cells in triplicate with plasmids expressing each of these CBEs and a guide RNA (gRNA) targeting a site in the RNF2 gene. We then sorted cells with high CBE expression (top 5% of GFP signal) for isolation of genomic DNA (for on-target DNA amplicon sequencing) and total RNA (for RNA-seq) (Methods). At the RNF2 on-target site, hA3A-BE3, eA3A-BE3, and hAID-BE3 induced mean editing efficiencies of 91%, 82%, and 32%, respectively, at position C6, and Target-AID (with a pmCDA1 deaminase at its C-terminal end) showed a mean editing efficiency of 87.1% at position C3 (Fig. 3a). RNA-seq experiments revealed that hA3A-BE3 induced tens of thousands of C-to-U edits (Fig. 3b and Supplementary Table 1) distributed throughout the transcriptome (Supplementary Fig. 5a). A number of these Cs were edited with very high (>80%) efficiencies (Supplementary Fig. 5b). Sequence logos derived from all Cs edited by hA3A-BE3 show a consensus UC motif (Supplementary Fig. 5a). However, sequence logos from subsets of Cs stratified by editing efficiencies reveal a more extended consensus sequence of CCAUCR for those Cs edited at higher efficiencies (Supplementary Fig. 5a), a motif consistent with a previous study that characterized RNA cytidines edited by the hA3A enzyme14. By contrast, eA3A-BE3 showed a dramatically reduced number of RNA edits relative to hA3A-BE3 but still slightly more (average of approximately three-fold) than what was observed with background in the GFP-only negative control (Fig. 3b, Supplementary Fig. 5b and Supplementary Table 1). Interestingly, hAID-BE3 and Target-AID induced numbers of RNA C-to-U edits comparable to what was observed in the negative control (Fig. 3b, Supplementary Fig. 5b and Supplementary Table 1). The absence of detectable RNA editing in the hAID-BE3 experiments is consistent with a previous study that showed overexpression of isolated AID enzyme in activated B cells did not yield evidence for RNA editing15. By comparison, our two previously described SECURE-BE3 variants5 induced numbers of RNA C-to-U edits slightly higher (BE3-R33A) than eA3A-BE3, hAID-BE3, and Target-AID or comparable to that observed with background (BE3-R33A/K34A)(Fig. 3b).
Figure 3. Transcriptome-wide off-target RNA editing activities of CBEs with non-APOBEC1 cytidine deaminases in HEK293T cells.
(a) Heat map showing the on-target DNA editing efficiencies of nCas9-UGI (Control), hA3A-BE3, eA3A-BE3, hAID-BE3, Target-AID, SECURE BE3-R33A and BE3-R33A/K34A with a gRNA targeted to the RNF2 gene (n= 3 independent replicates). Editing window shown includes only the most highly edited cytosines and not the entire spacer sequence. Numbering at the bottom represents spacer position with 1 being the most PAM distal location. (b) Jitter plots showing transcriptome-wide RNA C-to-U edits observed with a GFP negative control (single replicate) and hA3A-BE3, eA3A-BE3, hAID-BE3, Target-AID, SECURE BE3-R33A and BE3-R33A/K34A (each n= 3 independent replicates). Each dot represents a single edited cytosine. All experiments (except for the GFP control) were performed with co-expression of a gRNA targeting a site in the RNF2 gene and in all experiments the cells were sorted for top 5% of GFP signal except for the GFP control which was sorted for equivalent MFI of top 5% BE3 (Methods)). All CBEs (except Target-AID) used nCas9-UGI as negative control for RNA variant calling; Target-AID used NLS-nCas9-NLS-SH3-UGI (Target-AID without pmCDA1) as a negative control (Supplementary Table 3; Methods). n = total number of modified cytosines. DNA on-target data and RNA-seq data for BE3-R33A and BE3-R33A/K34A presented are from an earlier published study5 (performed using the same experimental conditions) and are shown here to facilitate direct comparison with other CBEs.
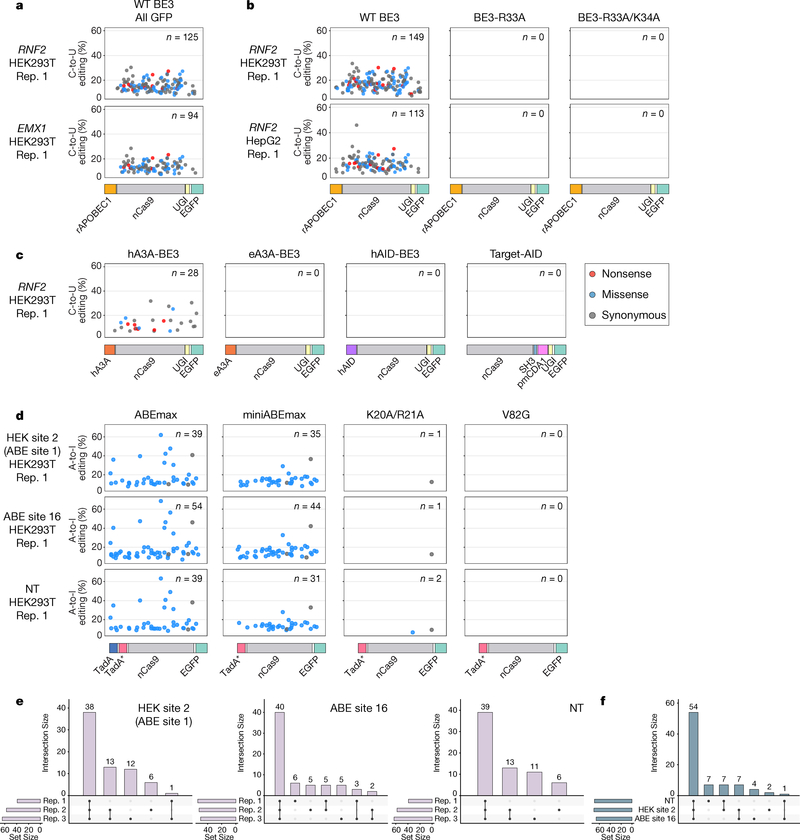
Given their abilities to edit the endogenous human cell transcriptome, we wondered whether CBEs and ABEs might also self-edit their own transcripts, thereby potentially generating sets of heterogeneous base editor proteins. To assess this, we used our analysis pipeline to quantify self-editing events in our previously published RNA-seq data5 performed with BE3 expressed at standard or overexpression levels in HEK293T cells. We observed C-to-U edits at 83 – 125 and 149 – 177 different C positions distributed throughout the BE3 transcript with standard expression and overexpression of BE3, respectively (Fig. 4a and b; Supplementary Fig. 6a and b; Supplementary Table 2); efficiencies of C-to-U editing among replicates ranged from 7.3% - 30.4% with standard BE3 expression and 7.1% - 46% with overexpression. Absolute numbers of missense mutations created by these edits ranged from 25 – 44 and 55 – 64 among replicates with BE3 standard expression and overexpression, respectively (Supplementary Table 2). Importantly, even when overexpressed, the two SECURE-BE3 variants (BE3-R33A and BE3-R33A/K34A) did not induce any detectable C-to-U edits on their own transcripts (Fig. 4b; Supplementary Fig. 6b; Supplementary Table 2). We observed similar results with BE3 and SECURE-BE3 variants expressed in HepG2 cells (Fig. 4b; Supplementary Fig. 6b; Supplementary Table 2). In addition, self-editing was observed with hA3A-BE3 overexpression in HEK293T cells (28 – 31 cytosine positions edited with efficiencies ranging from 4.5% to 33.4% among the replicates) (Fig. 4c; Supplementary Fig. 6c; Supplementary Table 2). As expected, overexpression of eA3A-BE3, hAID-BE3, and Target-AID in HEK293T cells showed no detectable evidence of self-editing of their respective transcripts (Fig. 4c; Supplementary Fig. 6c; Supplementary Table 2). Similarly, ABEmax and miniABEmax both induced of A-to-I changes at dozens (range of 31 – 68) of positions throughout their own transcripts with editing efficiencies ranging from 7% to 69.8% among replicates performed with three different gRNAs (Fig. 4d; Supplementary Fig. 6d; Supplementary Table 2). Nearly all of the edits induced by the ABEs are expected to induce missense mutations (Supplementary Table 2). On average, 57% of adenine positions self-edited by ABEmax appeared to be edited across all three replicates (Fig. 4e). Comparing the unions of self-edits from different gRNAs shows 65.85% of overlap between edits across the three gRNAs, suggesting that self-editing is independent of the gRNA with which the ABE was co-expressed (Fig. 4f). Notably, the two miniABEmax variants showed substantially reduced self-editing activities: K20A/R21A induced only small numbers (range 1 to 3) of self-edits and V82G did not induce any detectable self-edits (Fig. 4d; Supplementary Fig. 6d; Supplementary Table 2).
Figure 4. Self-editing generates a diverse range of heterogeneously edited CBE and ABE transcript sequences in HEK293T and HepG2 cells.
(a) Scatterplots showing C-to-U self-editing of the BE3-encoding RNA transcript observed with WT BE3 (with rAPOBEC1) expression in HEK293T cells (sorted for all GFP-positive cells) with two different gRNAs targeting sites in RNF2 and EMX1. Each dot represents an edited C and the color of the dot indicates the predicted type of mutation caused by a C-to-U edit at that position (Methods). The y-axis shows editing efficiencies for each C-to-U modification and the x-axis represents the position of each C within the BE3 coding sequence (with the architecture of the editor shown schematically below but not displaying the NLS and linkers). Data were obtained by analyzing previously published RNA-seq experiments5. n = total number of modified Cs. (b) Scatterplots illustrating C-to-U self-editing observed with wild-type (WT) BE3 (with rAPOBEC1), SECURE-BE3 (R33A) and SECURE-BE3 (R33A/K34A) in HEK293T and HepG2 cells sorted for top 5% GFP signal with co-expression of the RNF2 gRNA. Data are shown as described in a. Data were obtained by analyzing previously published RNA-seq experiments5. (c) Scatterplots depicting C-to-U self-editing observed in HEK293T cells expressing hA3A-BE3, eA3A-BE3, hAID-BE3, and Target-AID (sorted for top 5% GFP signal). Data are shown as described in a and were obtained using the RNA-seq experiments shown in Fig. 3b. (d) Scatterplots showing A-to-I self-editing induced by expression of ABEmax, miniABEmax, miniABEmax-K20A/R21A, and miniABEmax-V82G (sorted for all GFP-positive cells) with gRNAs targeting HEK site 2, ABE site 16, and a non-targeting gRNA (NT) in HEK293T cells. Data are shown as described in a and were obtained using the RNA-seq experiments shown in Figs. 1c and 1d. n = total number of modified As. (e) UpSet plots showing the intersections of RNA A-to-I self-edits induced by ABEmax on its own transcript across three replicates (data from Fig. 4d and Supplementary Fig. 6d). Each plot shows data from co-expression of ABEmax with one of three different gRNAs. (f) UpSet plots showing the intersection of RNA A-to-I self-edits induced by ABEmax across three different gRNAs using the data shown in Fig. 4d and Supplementary Fig. 6d. For each gRNA, we used A-to-I edits that represent the union of all such edits across the three replicates.
In light of our observation of self-editing, we wondered whether CBEs and ABEs might also be able to edit gRNAs. Although our RNA-seq experiments used RNA extracted from cells by methods optimized for isolation of fragments >200 bases in length, we nonetheless were able to observe thousands of gRNA reads in each of our sequencing data replicates. Therefore, we used our analysis pipeline (Methods) to assess gRNA edits in our RNA-seq data. We did not detect any C-to-U editing of the gRNAs in RNA-seq experiments performed with any of the various CBEs (BE3, BE3-R33A, BE3-R33A/K34A, hA3A-BE3, eA3A-BE3, hAID-BE3, or Target-AID (Supplementary Fig. 7a–c). Analysis of RNA-seq data from our ABE experiments revealed reproducible editing of an A that resides in the loop of stem-loop 2 of the tracrRNA (Supplementary Fig. 7d). Edits at this position were present at frequencies of 4.5 to 19.9% and most consistently observed with miniABEmax and miniABEmax-V82G although edits could also be observed in some replicates with ABEmax and miniABEmax-K20A/R21A (Supplementary Fig. 7d). Given the location and low frequency of this edit, we would not expect it to have a major impact on either activity or specificity of the gRNA-ABE complex.
The work described here extends our understanding of the off-target RNA editing activities of DNA base editors, expands the options available to minimize these unwanted effects, and provides novel SECURE base editor architectures with other desirable properties. The successful engineering of SECURE-ABE variants shows that, as we previously found with the BE3 CBE5, it is possible to minimize unwanted RNA editing while retaining reasonably efficient on-target DNA editing for an ABE. In addition, our characterization of additional CBEs with deaminases other than APOBEC1 further expands the toolbox of base editors that can be used without inducing high-level RNA editing. Recent studies published by others while this work was in preparation have described additional CBE and ABE variants with reduced RNA editing activities16,17. It will be interesting to directly compare all of these variants and perhaps to combine mutations from them to create base editors with even more optimized on-target DNA, off-target DNA, and off-target RNA editing profiles.
Our description of self-editing by DNA base editors provides yet another strong motivation to avoid the use of base editors that possess off-target RNA editing activities and to use expression and/or delivery strategies that limit the duration of activity (e.g., using ribonucleoprotein (RNP) complexes). Self-editing by both CBEs and ABEs potentially creates a heterogeneous population of base editor-encoding transcripts in human cells including missense mutations that might lead to the generation of novel epitopes or other gain/loss-of-function effects. The potential impacts of creating diverse mutated forms of base editor proteins in cells will be particularly important to consider because these fusions will be highly overexpressed for most applications. For CBEs, self-edits also include nonsense mutations that could impact deaminase or Cas9 activities. In addition, because the deaminase is located at the amino-terminal end of most CBEs, the introduction of nonsense mutations into the nCas9 part of the fusion (where the majority of edits occur) could result in truncated proteins that will presumably possess intact deaminase activities. One possibility is that these truncated forms might preferentially increase RNA editing activity levels because these proteins would still be expected to induce off-target RNA editing but not on-target DNA editing. Thus, the existence of self-editing further underscores the importance of using DNA base editors with reduced RNA editing activities for both research and therapeutic applications.
Online Methods
PyMOL Analysis of TadA structures
Escherichia coli tRNA-specific adenosine deaminase (TadA, PDB 1Z3A) and Staphylococcus aureus TadA with tRNA (PDB 2B3J) structures were downloaded from the Protein Data Bank and visualized with PyMOL version 2.2.2. Subunit A (monomer) of S. aureus TadA with tRNA was superimposed with subunit A of E. coli TadA using the “super” command. All related illustrations (Fig. 1e) were generated with PyMOL (Schrödinger).
Plasmid cloning
All ABE constructs (reported in Supplementary Table 3) were cloned using the backbone and the P2A-EGFP-NLS fragment of ABEmax-P2A-EGFP-NLS (AgeI/NotI digest; Addgene ID 112101). ABEmax and variants were expressed under a CMV promoter. Control experiments were performed with a nCas9 negative control that doesn’t contain any TadA domains. All CBE constructs (reported in Supplementary Table 3) were cloned using the backbone of SQT817 and expressed under a CAG promoter (AgeI-NotI-EcoRV digest, Addgene ID 53373). For the P2A-EGFP fragments in these constructs, we used BPK4335 (pCMV-BE3-P2A-EGFP) as a template. APOBEC3A constructs were cloned using JMG5377 (pCAG-hA3A-BE3) as a template. hAID-BE3 was obtained from Addgene (ID 100803). For all CBE plasmids based on the BE3 architecture, nCas9-UGI-NLS-P2A-EGFP (pJUL1001, Addgene ID 123611) was used as a negative control. For Target-AID4, we used NLS-nCas9-NLS-SH3-3xFLAG-NLS-UGI-P2A-EGFP as a separate negative control. Compared to the reference sequence of pmCDA1 from NCBI (ABO15149.1), the pmCDA1 used in Target-AID (as supplied by Addgene, ID 79620) has a R187W single residue modification. This amino acid alteration is also present in other Target-AID derivatives, such as e.g. Target-AID-NG18 (Addgene ID 119861). Guide RNA (gRNA) plasmids were cloned using the SpCas9 gRNA entry vector BPK1520 (pUC19 backbone; BsmbI cassette, Addgene ID 65777). All remaining constructs were generated using isothermal amplification (Gibson assembly, NEB). All gRNA and ABE plasmids were midi or maxi prepped using the Qiagen Midi/Maxi Plus kits.
Cell culture
HEK293T cells (CRL-3216) and HepG2 cells (HB-8065; data from Ref. 5) were purchased from and STR-authenticated by ATCC. Cells were cultured in Dulbecco’s Modified Eagle Medium (DMEM, Gibco) supplemented with 10% (v/v) fetal bovine serum (FBS, Gibco) and 1% (v/v) penicillin-streptomycin (Gibco) for HEK293T or Eagle’s Minimum Essential Medium with 10% (v/v) FBS and 0.5% (v/v) penicillin for HepG2. Cells were passaged every 2–3 days when reaching around 80–90% confluency. HEK293T cells were used only until passage 20 for all experiments, and HepG2 cells until passage 12, and the media was tested every two weeks for mycoplasma.
Transfections
For ABE DNA on-target screening experiments (Fig. 1f), 2×104 HEK293T cells were seeded into 96-well Flat Bottom Cell Culture plates (Corning), transfected 24h post seeding with 165ng base editor or negative control (bpNLS-32AA linker-nCas9(D10A)-bpNLS), 55ng guide RNA expression plasmid, and 0.66μL TransIT-293 (Mirus), and harvested 72h after transfection to obtain genomic DNA. For ABE RNA off-target screening experiments (Fig. 1f), 2×105 HEK293T cells were seeded into 12-well Cell Culture plates (Corning), transfected 24h post seeding with 1.65μg base editor or negative control, 0.55μg guide RNA, and 6.6μL TransIT-293, and harvested 36h after transfection to obtain RNA. For ABE DNA off-target experiments (Supplementary Fig. 4), 3×105 HEK293T cells were seeded into 6-well plates (Corning), transfected 24h post seeding with 825ng base editor or control, 275ng gRNA, and 7.5μL TransIT-X2 (Mirus), and harvested 72h after transfection for DNA. For ABE DNA on-target experiments with 22 gRNAs (Fig. 2 and Supplementary Fig. 3), 1.25×104 HEK293T cells were seeded into 96-well plates, transfected 24h post seeding with 30ng base editor or control, 10ng gRNA, and 0.3μL TransIT-X2, and harvested 72h after transfection to obtain genomic DNA. For experiments with FACS-sorted cells, 6–7×106 HEK293T cells were seeded into 150mm Cell Culture dishes (Corning), transfected 24h post seeding with 37.5μg base editor or an appropriate negative control fused to P2A-EGFP, 12.5μg guide RNA, and 150μL TransIT-293. Sorting was performed 36–40h post transfection.
Fluorescence-activated cell sorting (FACS)
Cells were prepared for sorting by diluting to 1×107 cells per ml with 1X Phosphate Buffer Saline (PBS, Corning) supplemented with 10% FBS and filtering through 35μm cell strainer caps (Corning). Cells were sorted on a FACSAria II (BD Biosciences) using FACSDiva version 6.1.3 (BD Biosciences) after gating for single live cells (Supplementary Note 2). Cells treated with base editor were sorted for either all GFP signal (standard expression) or top 5% of cells with the highest GFP (FITC) signal (overexpression) into FBS; cells treated with nCas9 negative controls were sorted for either all GFP positive cells or the 5% of cells with a mean fluorescence intensity (MFI) matching that of the top 5% of cells treated with base editor. The GFP control shown in Fig. 3b was sorted to match the top 5% GFP signal of BE3-transfected control cells from the same day.
DNA extraction
For ABE DNA on-target experiments in 96-well plates, after washed with PBS, cells were lysed for DNA 72h post-transfection with freshly prepared 43.5μL DNA lysis buffer (50mM Tris HCl pH 8.0, 100mM NaCl, 5mM EDTA, 0.05% SDS, adapted from ref. 19), 5.25μL Proteinase K (NEB), and 1.25μL 1M DTT (Sigma). For experiments with sorted cells, cells were centrifuged (200g, 8 min) and lysed with 174μL DNA lysis buffer, 21μL Proteinase K, and 5μL 1M DTT. Lysates were incubated at 55°C on a plate shaker overnight, then gDNA were extracted with 2x paramagnetic beads (as described in ref. 20), washed 3 times with 70% EtOH, and eluted in 30μL 0.1X EB buffer (Qiagen). For ABE DNA off-target experiments in 6-well plates, cells were washed with PBS, trypsinized, and centrifuged, and gDNA was extracted with QIAmp DNA Mini Kit (Qiagen).
RNA extraction & reverse transcription
Cells were lysed to extract RNA 36h-40h post-transfection with 350μL RNA lysis buffer LBP (Macherey-Nagel), and RNA was extracted with the NucleoSpin RNA Plus kit (Macherey-Nagel) following the manufacturer’s instructions. RNA was reverse transcribed to generate cDNA with the High Capacity RNA-to-cDNA kit (Thermo Fisher) following the manufacturer’s instructions.
Library preparation for DNA or cDNA targeted amplicon sequencing
Next-generation sequencing (NGS) of DNA or cDNA was performed as previously described5. In summary, the first PCR was performed to amplify genomic or transcriptomic sites of interest with primers containing Illumina forward and reverse adapter sequences (see Supplementary Table 4 for primers and amplicons used in this study), using Phusion High-Fidelity DNA Polymerase (NEB). The first PCR products were cleaned with a 0.7x paramagnetic bead clean-up, then the second PCR was performed to add barcodes with primers containing unique sets of p5/p7 Illumina barcodes (analogous to TruSeq CD indexes). The second PCR products were again cleaned with a 0.7x paramagnetic bead clean-up. The libraries were then pooled based on concentrations measured with the QuantiFluor dsDNA System (Promega) and Synergy HT microplate reader (BioTek) at 485/528nm. The final pool was quantified by Qubit or qPCR with the NEBNext Library Quant Kit for Illumina (NEB) and sequenced paired-end (PE) 2×150 on the Illumina MiSeq machine using 300-cycle MiSeq Reagent Kit v2 or Micro Kit v2 (Illumina). FASTQs (post-demultiplexing) were downloaded from Illumina BaseSpace and analyzed using a batch version of CRISPResso 2.
RNA library preparation & sequencing
RNA-seq experiments were performed as previously described5. Briefly, RNA libraries were prepared with the TruSeq Stranded Total RNA Library Prep Gold kit (Illumina) following the manufacturer’s instructions. SuperScript III (Invitrogen) was used for first-strand synthesis, and IDT for Illumina TruSeq RNA unique dual indexes (96 indexes) were used to avoid index hopping. The libraries were pooled based on qPCR measurements with the NEBNext Library Quant Kit for Illumina. The final pool was sequenced PE 2×76 on the Illumina HiSeq2500 machine (for all CBE experiments and one ABE experiment from Ref. 5 shown in Fig. 1b) or PE 2×100 on the NovaSeq6000 machine (for all remaining ABE experiments) at the Broad Institute of Harvard and MIT (Cambridge, MA). To account for variable sequencing depths, all RNA-seq libraries sequenced on the NovaSeq were uniformly downsampled to 100 million reads per library using seqtk version 1.0-r82-dirty (https://github.com/lh3/seqtk).
Amplicon sequencing analysis
Amplicon sequencing data was analyzed with CRISPResso2 v.2.0.2721. The heat maps for the SECURE-ABE screening in Fig. 1f display the highest edited adenine at the target (DNA) or off-target (RNA) sites. Editing efficiency values were averaged over quadruplicates, log2 transformed with a pseudocount of 1, and normalized to ABEmax. Heat maps showing ABE or CBE on-target DNA editing (Figs. 2a and 3a, and Supplementary Fig. 1a) show an editing window that includes the edited As or Cs, respectively, and a grey background for editing efficiencies smaller than 2%. This background cut-off was relaxed for the heat maps showing ABE-induced C-to-N DNA on-target editing (Supplementary Fig. 3) and DNA off-target editing (Supplementary Fig. 4).
RNA variant calling pipeline
All bioinformatic analysis was performed in concordance with GATK Best Practices22,23 for RNA-seq mutation calling as we have previously described5. Briefly, raw sequencing reads were two-pass aligned to the reference hg38 reference genome with STAR24 with parameters to discard multi-mapping reads. After PCR duplicate removal and base recalibration, mutations in RNA-seq libraries were called using GATK HaplotypeCaller. RNA edits in CBE and ABE overexpression experiments were identified using a downstream modification of the GATK pipeline output as we have previously described5. Specifically, mutation positions called by HaplotypeCaller were further filtered to include only those satisfying the following criteria with reference to the corresponding control experiments: (1) Read coverage for a given edit in control experiment should be greater than the 90th percentile of read coverage across all edits in the overexpression experiment. (2) 99% of reads covering each edit in the control experiment were required to contain the reference allele. Edits were further filtered to exclude those with fewer than 10 reads or 0% alternate allele frequencies. A-G edits include A-G edits identified on the positive strand as well as T-C edits identified on the negative strand. For CBE overexpression experiments, C-T edits include C-T edits identified on the positive strand as well as G-A edits derived from the negative strand.
Six A-to-I edits identified from the above pipeline were chosen to test SECURE ABE variants based on the following criteria. These were sites that had (1) read coverage of at least 50 in all replicates of control and overexpression experiments, (2) 99% reads in all control experiments containing reference allele and (3) at least 60% alternate allele frequencies in all replicates. From this list, primers were tested for the top 15 edited sites that were also within 150 bases of an exon-exon junction and the 6 highest edited sites with robust amplification from cDNA were chosen.
To identify self-edits occurring on the base-editing construct, we generated a modified hg38 reference genome with additional contigs for the gRNA and base editor constructs. These additional contigs were appended to the reference genome, and each library was re-processed using GATK best practices, including variant calling with HaplotypeCaller. Variants were then further filtered using a similar process as described above for the transcriptome (i.e. filtering for no more than 1% editing in the negative control) with the exception that positions poorly covered in the control due to differences in the construct design (i.e. the deaminase domain) were not filtered out. We note that since both control and BE constructs were expressed from plasmids, the overall expression of these transcripts is much higher than most detected genes which supersedes the control of coverage between control and BE expression in this analysis (see part 1 of transcriptome variant calling above). Editing efficiencies per position were computed based on the abundance of Gs (ABE) or Ts (CBE) over total coverage from bam-readcount estimated on the PCR deduplicated .bam files. Edits were further filtered to exclude those with fewer than 50 reads or 0% alternate allele frequencies. The stringency of our variant calling pipeline might result in the underestimation of the numbers of CBE or ABE-induced cellular RNA edits and self-edits of BE and gRNA transcripts.
Statistics & Data Reporting
No specific statistical tests were used. Statistical values include mean and median RNA editing efficiencies. Error bars (Fig. 2b) depict the standard deviation (SD) and were plotted using GraphPad Prism 8.1.2. Sample sizes were not predetermined with statistical methods. Investigators were not blinded to experimental conditions or outcome assessments.
Data availability
Plasmids encoding the SECURE-CBE and SECURE-ABE constructs shown in this work are available on Addgene. The RNA-sequencing data used in this study have been deposited in the Gene Expression Omnibus (GEO) repository (National Center for Biotechnology Information). The files are accessible through the GEO Series accession number GSE129894.
Targeted amplicon sequencing data have been deposited at the SRA repository under bioproject accession number PRJNA553185. All other relevant data are available from the corresponding author on request.
Code availability
The authors will make all previously unreported custom computer code used in this work available upon reasonable request.
Life Sciences Reporting Summary
Details regarding statistical tests and experimental design can be found also in the Nature Research Reporting Summary that is attached to this article.
Supplementary Material
Acknowledgements
J.K.J., J.G., and R.Z. are supported by the Defense Advanced Research Projects Agency (HR0011-17-2-0042). Support was also provided by the National Institutes of Health (RM1 HG009490 to J.K.J. and J.G. and R35 GM118158 to J.K.J. and M.J.A.). J.G. was supported by a research fellowship (GR 5129/1-1) of the German Research Foundation (DFG). J.K.J. is additionally supported by the Desmond and Ann Heathwood MGH Research Scholar Award. We thank G. Ciaramella of Beam Therapeutics for the suggestion to delete the wild-type TadA monomer from ABEmax. We thank A. Lapinaite of the Doudna Lab for suggesting the overlay of E. coli and S. aureus TadA structures and S.J. Lee for technical assistance.
Footnotes
Competing Financial Interests Statement
J.K.J. has financial interests in Beam Therapeutics, Editas Medicine, Pairwise Plants, Poseida Therapeutics, Transposagen Biopharmaceuticals, and Verve Therapeutics. J.K.J.’s interests were reviewed and are managed by Massachusetts General Hospital and Partners HealthCare in accordance with their conflict of interest policies. J.K.J. and M.J.A. hold equity in Excelsior Genomics. J.K.J. is a member of the Board of Directors of the American Society of Gene and Cell Therapy. J.G., R.Z., and J.K.J. are co-inventors on patent applications that have been filed by Partners Healthcare/Massachusetts General Hospital on engineered base editor architectures that reduce RNA editing activities and increase their precision.
References
- 1.Rees HA & Liu DR Base editing: precision chemistry on the genome and transcriptome of living cells. Nat Rev Genet 19, 770–788, doi: 10.1038/s41576-018-0059-1 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Komor AC, Kim YB, Packer MS, Zuris JA & Liu DR Programmable editing of a target base in genomic DNA without double-stranded DNA cleavage. Nature 533, 420–424, doi: 10.1038/nature17946 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gaudelli NM et al. Programmable base editing of A*T to G*C in genomic DNA without DNA cleavage. Nature 551, 464–471, doi: 10.1038/nature24644 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nishida K et al. Targeted nucleotide editing using hybrid prokaryotic and vertebrate adaptive immune systems. Science 353, doi: 10.1126/science.aaf8729 (2016). [DOI] [PubMed] [Google Scholar]
- 5.Grunewald J et al. Transcriptome-wide off-target RNA editing induced by CRISPR-guided DNA base editors. Nature 569, 433–437, doi: 10.1038/s41586-019-1161-z (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gehrke JM et al. An APOBEC3A-Cas9 base editor with minimized bystander and off-target activities. Nat Biotechnol 36, 977–982, doi: 10.1038/nbt.4199 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Komor AC et al. Improved base excision repair inhibition and bacteriophage Mu Gam protein yields C:G-to-T:A base editors with higher efficiency and product purity. Sci Adv 3, eaao4774, doi: 10.1126/sciadv.aao4774 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wolf J, Gerber AP & Keller W tadA, an essential tRNA-specific adenosine deaminase from Escherichia coli. EMBO J 21, 3841–3851, doi: 10.1093/emboj/cdf362 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Koblan LW et al. Improving cytidine and adenine base editors by expression optimization and ancestral reconstruction. Nat Biotechnol 36, 843–846, doi: 10.1038/nbt.4172 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kim J et al. Structural and kinetic characterization of Escherichia coli TadA, the wobble-specific tRNA deaminase. Biochemistry 45, 6407–6416, doi: 10.1021/bi0522394 (2006). [DOI] [PubMed] [Google Scholar]
- 11.Losey HC, Ruthenburg AJ & Verdine GL Crystal structure of Staphylococcus aureus tRNA adenosine deaminase TadA in complex with RNA. Nat Struct Mol Biol 13, 153–159, doi: 10.1038/nsmb1047 (2006). [DOI] [PubMed] [Google Scholar]
- 12.Tsai SQ et al. GUIDE-seq enables genome-wide profiling of off-target cleavage by CRISPR-Cas nucleases. Nat Biotechnol 33, 187–197, doi: 10.1038/nbt.3117, nbt.3117 [pii] (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang X et al. Efficient base editing in methylated regions with a human APOBEC3A-Cas9 fusion. Nat Biotechnol, doi: 10.1038/nbt.4198 (2018). [DOI] [PubMed] [Google Scholar]
- 14.Sharma S, Patnaik SK, Kemer Z & Baysal BE Transient overexpression of exogenous APOBEC3A causes C-to-U RNA editing of thousands of genes. RNA Biol 14, 603–610, doi: 10.1080/15476286.2016.1184387 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fritz EL et al. A comprehensive analysis of the effects of the deaminase AID on the transcriptome and methylome of activated B cells. Nat Immunol 14, 749–755, doi: 10.1038/ni.2616 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhou C et al. Off-target RNA mutation induced by DNA base editing and its elimination by mutagenesis. Nature, doi: 10.1038/s41586-019-1314-0 (2019). [DOI] [PubMed] [Google Scholar]
- 17.Rees HA, Wilson C, Doman JL & Liu DR Analysis and minimization of cellular RNA editing by DNA adenine base editors. Sci Adv 5, eaax5717, doi: 10.1126/sciadv.aax5717 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
Methods—only References
- 18.Nishimasu H et al. Engineered CRISPR-Cas9 nuclease with expanded targeting space. Science 361, 1259–1262, doi: 10.1126/science.aas9129 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Laird PW et al. Simplified mammalian DNA isolation procedure. Nucleic Acids Res 19, 4293 (1991). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rohland N & Reich D Cost-effective, high-throughput DNA sequencing libraries for multiplexed target capture. Genome Res 22, 939–946, doi: 10.1101/gr.128124.111 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Clement K et al. CRISPResso2 provides accurate and rapid genome editing sequence analysis. Nat Biotechnol 37, 224–226, doi: 10.1038/s41587-019-0032-3 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McKenna A et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res 20, 1297–1303, doi: 10.1101/gr.107524.110 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.DePristo MA et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet 43, 491–498, doi: 10.1038/ng.806 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dobin A et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21, doi: 10.1093/bioinformatics/bts635 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Plasmids encoding the SECURE-CBE and SECURE-ABE constructs shown in this work are available on Addgene. The RNA-sequencing data used in this study have been deposited in the Gene Expression Omnibus (GEO) repository (National Center for Biotechnology Information). The files are accessible through the GEO Series accession number GSE129894.
Targeted amplicon sequencing data have been deposited at the SRA repository under bioproject accession number PRJNA553185. All other relevant data are available from the corresponding author on request.