Abstract
Fanconi anemia (FA) is a genomic instability syndrome with predisposition to congenital abnormalities, bone marrow failure, and cancer. Classical and most frequent congenital abnormalities include all those seen in VACTERL-H association and those described under the PHENOS acronym. Pathogenic variants in at least 22 genes are associated with FA, which code for proteins that comprise the FA/BRCA DNA repair pathway. We reviewed 187 publications and 1101 cases of FA in which the gene or complementation group was identified and analyzed those in whom physical findings were sought. We conducted genotype-phenotype analyses considering the specific gene, the location in the FA/BRCA DNA repair pathway, and the type of variant (null or hypomorphic) as exposures. The outcomes were the presence of any physical abnormality or specific categories of abnormalities. Seventy-nine percent of the patients had at least one physical abnormality. Pathogenic variants in FANCB, FANCD2, the ID complex and downstream genes were associated with several specific anomalies. Patients with biallelic or hemizygous null variants had a higher proportion of at least one abnormality, renal malformations, microcephaly, short stature and the combination of VACTERL-H compared with those with hypomorphic genotypes. VACTERL-H alone or in combination with PHENOS is highly associated with FA, but the absence of those features does not rule out the diagnosis of FA.
Keywords: Congenital Abnormality, Fanconi Anemia, Gene, Genotype-Phenotype Association, PHENOS, VACTERL-H
1. Introduction
Fanconi anemia (FA) is a genomic instability syndrome associated with congenital abnormalities, bone marrow failure (BMF) and cancer predisposition (1, 2). The first cases of FA were reported by Guido Fanconi, a Swiss pediatrician, who in 1927 described three brothers with short stature, physical abnormalities and anemia (3). Cytopenias due to BMF are the most common presentation of FA. Hematological manifestations include aplastic anemia, myelodysplastic syndrome and leukemia (1, 4). The median age at diagnosis of FA is 7 years (2) although symptomatic and asymptomatic (family members) cases have been described from birth to more than 50 years of age. Patients with FA have an extremely high risk of developing malignancies at an early age; the most common are acute myeloid leukemia and squamous cell carcinomas of head and neck and of female genitalia (1).
The physical phenotype in patients with FA is extremely heterogenous and can affect multiple systems (2). Classical congenital abnormalities include those described in the VACTERL-H (Vertebral, Anal, Cardiac, Tracheo-esophageal fistula, Esophageal atresia, Renal, upper Limb and Hydrocephalus) association (OMIM 192350) (5). From 5 to 30% of patients with FA were reported to meet VACTERL-H criteria (presence of at least any 3 of 8 features) (6, 7). Other abnormalities common to FA but not part of VACTERL-H were recently grouped by us as PHENOS (skin Pigmentation, small Head, small Eyes, Nervous system, Otology, Short stature). At least four of the six PHENOS features were more frequent in the patients with FA who also had VACTERL-H (7).
Pathogenic variants in at least 22 genes (FANCA, FANCB, FANCC, FANCD1/BRCA2, FANCD2, FANCE, FANCF, FANCG, FANCI, FANCJ/BRIP1, FANCL, FANCM, FANCN/PALB2, FANCO/RAD51C, FANCP/SLX4, FANCQ/ERCC4/XPF, FANCR/RAD51, FANCS/BRCA1, FANCT/UBE2T, FANCU/XRCC2, FANCV/REV7, and FANCW/RFWD3) have been identified in patients with FA (8). All are autosomal recessive except FANCB which is X-linked, and FANCR which is autosomal dominant. The FA genes code for proteins that comprise a complex network for DNA damage repair (FA/BRCA DNA repair pathway) and other cellular processes (9, 10). The main function of the pathway is the removal of DNA interstrand crosslinks, which interfere with DNA replication and gene transcription (10). The genes are grouped according to their function in the FA/BRCA DNA repair pathway as upstream genes (FANCA, B, C, E, F, G, L, M, and T), ID complex (FANCD2 and I), and downstream genes (FANCD1, J, N, O, P, Q, R, S, U, V, and W). The upstream gene products form the FA core complex upon DNA damage. This leads to monoubiquitination of the ID complex, which then activates the downstream genes, resulting in DNA repair.
Genotype-phenotype associations previously reported in FA include: 1) poor hematological outcomes due to severe cytopenia and increased frequency of leukemia in patients with FANCG and in those with biallelic null variants in FANCA compared with FANCC (11); 2) less frequent congenital abnormalities in patients with FANCC compared with FANCA and G (11); and 3) high risk and early age of cancer in patients with pathogenic variants in the downstream genes FANCD1/BRCA2 and FANCN/PALB2 (12–14).
We revisited the genotype-phenotype relationships in published cases of FA and proposed several questions: What is the association of at least one abnormality or specific groups of abnormalities (such as VACTERL-H or PHENOS) with the genotype? Does it depend on the gene, the location in the FA/BRCA DNA repair pathway, or the type of FA gene pathogenic variant?
2. Patients and Methods
We searched PubMed for publications limited to human subjects from October 1982 through September 2017 using the terms “Fanconi” and “anemia”. We began with 1982 because this was the year of the first patient report was associated with a specific FA complementation group (15, 16). We identified all cases that reported the FA gene or complementation group. We selected cases that had clinical information on the presence or absence of physical abnormalities. We sought genotype-phenotype associations between three types of exposures and several outcomes as shown in Table 1. The exposures were: gene, location in the FA/BRCA DNA repair pathway (upstream, ID complex, or downstream), and type of pathogenic variant (null/no protein production or hypomorphic/some protein production). The outcomes were: presence or absence of at least one physical abnormality of any type, presence or absence of each specific anomaly, presence or absence of VACTERL-H association (≥3 of 8 VACTERL-H features) (5), and presence or absence of PHENOS (≥4 of 6 PHENOS features) (7). Other abnormalities included those that are not part of VACTERL-H and PHENOS; these are listed in detail in Supplemental Table S1. The genotypes with less than 10 patients in each were combined together as “Others” (Supplemental Table S2) and were not considered for specific analyses.
Table 1:
Exposures and Outcomes.
| A) Exposure (independent variables) | B) Outcome (dependent variables) |
|---|---|
| 1. Gene Genes with <10 patients were pooled as Others: FANCE (2 patients), F (7), L (5), M (2), Q (4), R (1), S (1), T (4), U (1), V (1), and W (1). |
1. At least one abnormality of any type |
| 2. Specific congenital abnormalities | |
| 2. Location in the FA/BRCA DNA repair pathway Upstream (FANCA, B, C, E, F, G, L, M, and T) ID (D2 and I) Downstream (D1, J, N, O, P, Q, R, S, U, V, and W) |
3. VACTERL-H, ≥3 of 8 features Vertebral anomalies Anal anomalies Cardiac structural anomalies Tracheal-esophageal fistula Esophageal or duodenal atresia Renal malformations Upper Limb anomalies (radial ray) Hydrocephalus |
| 3. Type of pathogenic variant Null: no protein production (null allele + null allele or FANCB or R null allele) Hypomorphic: some protein production (null allele + hypomorphic allele, both alleles hypomorphic, or FANCB or R hypomorphic allele) |
4. PHENOS, ≥4 of 6 features Pigmentation anomalies, including café au lait macules Small Head circumference Small Eyes Central Nervous system structural anomalies Otological alterations, structural and/or hearing loss Short stature |
| A) Bold means different categories | B) Bold means the letter in the acronym |
We classified pathogenic variants as hypomorphic or null for all the cases for whom molecular data were available. The data were collated into a single file of cDNA positions, which were converted to genomic positions using Mutalyzer (https://mutalyzer.nl). Genomic coordinates were confirmed by visual inspection using the UCSC genome browser (http://genome.ucsc.edu). Variants were annotated using SnpEff (17) and ANNOVAR (18). Variants were then classified according to the following rules: 1) all variants had to be <1% minor allele frequency in the Exome Aggregation Consortium database; if the frequency was >1% the variant was marked as a single nucleotide polymorphism; 2) the variants with loss of function (stop gains or deletions ≥4 nucleotides) or splice donor/acceptor sites were classified as null; missense variants were classified as hypomorphic (19, 20).
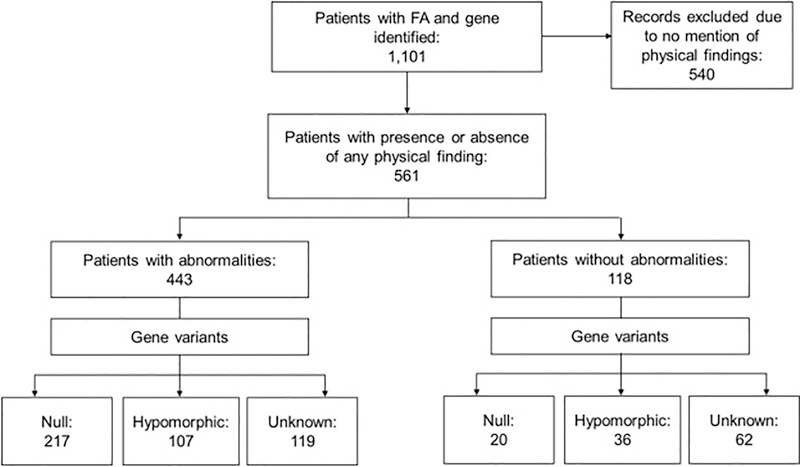
The variants were grouped by classification. The first group was considered null genotype and contained biallelic (homozygous or compound heterozygous) and hemizygous null variants. The second group was hypomorphic genotype and included patients with at least one allele with some protein production. Patients in this group could have either a hypomorphic or null second allele. The third group included patients without information about protein production from any allele, these were called unknown and not considered for the mutational analysis (Fig. 1).
Fig. 1.
Study Flow Chart. We identified 1101 reports of individual patients in which the complementation group or gene related to FA was reported from 1982 through September 2017. Phenotype information was provided for 561 patients (51%); 443 (79%) of these had a physical abnormality and 118 (21%) did not. The type of pathogenic variant (null or hypomorphic) could be classified in 380 patients (68%) with phenotype information; 237 (62%) of these had a null genotype and 143 (38%) a hypomorphic genotype. For the rest of the patients, the authors did not mention molecular details enabling variant classification.
The statistical analyses were performed using Microsoft Excel Office 365 (Microsoft, Redmond, WA, USA) and STATA 14 (StataCorp LP, College Station, TX, USA). We used Fisher’s Exact; p-values <0.05 were significant. The Bonferroni correction was applied for multiple comparisons.
3. Results
3.1. Cases Analyzed
We identified 1101 reports of individual patients in which the complementation group or gene related to FA was reported from a review of 187 articles from 26 countries (Fig. 1). The male to female ratio was 1:0.91 (p=0.5). We excluded 540 patients for whom data regarding the presence or absence of any phenotypic abnormality were not stated. Phenotype information was available for 561 patients (51%), of whom 443 (79%) had at least one abnormality and 118 were stated to have none. Two hundred seventeen of the patients with abnormalities belonged to the null group (46%), 107 to the hypomorphic group (27%), and 119 lacked molecular details for classification (27%). The respective numbers among those reported to have no physical findings were 20 (17%), 36 (30%), and 62 (53%).
The gene distribution among all 1101 patients, in those with phenotype data (561), and in those without phenotype data (540), varied as shown in Supplemental Table S2. The five most frequent genes among the 561 cases with phenotype information were FANCA (40.5%), G (13%), D1 (12%), C (8%), and D2 (7.5%); the frequencies of FANCB, I, J, N, and P were between 1 and 2%; all other genes were each ≤1%.
3.2. Types of Abnormalities
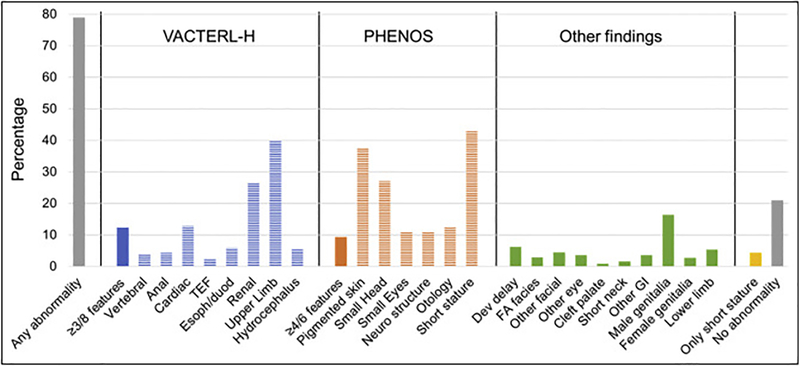
One or more abnormalities were present in 79% of the 561 patients with physical data mentioned. The type and frequency of phenotypic anomalies are shown in Fig. 2. VACTERL-H association (≥3/8 features) was present in 12% of patients, PHENOS (≥4/6 features) in 9%, and both together in 4%. The most frequent individual features were short stature (43%), upper limb (radial ray) abnormalities (40%), skin pigmentation changes including café au lait macules (37%), renal malformations (27%), and microcephaly (27%). The rest of the abnormalities were present in <20% of patients. Four percent of patients had short stature as an isolated finding and 21% had no physical anomalies. The detailed description of the other abnormalities not included in VACTERL-H or PHENOS is provided in Supplemental Table S1.
Fig. 2.
Types of Abnormalities. TEF, trachea-esophageal fistula; Esoph/duod, esophageal or duodenal atresia; neuro, neurological; dev, developmental; FA, Fanconi anemia; GI, gastrointestinal. Upper limb includes abnormal thumb +/− abnormal radius. Neuro structure includes structural brain malformations other than hydrocephalus. Otology comprises ear malformations and/or hearing loss. The heading “Other findings” includes anomalies not contained in VACTERL-H or PHENOS. Gray: any or no abnormality; solid blue or orange: VACTERL-H or PHENOS; horizontal stripes: individual findings. Horizontal axis: abnormalities analyzed; vertical axis: percent of total cases with that abnormality. VACTERL-H association (≥3/8 features) was present in 12% and PHENOS (≥4/6 features) in 9% of patients. The most frequent features (unrelated to criteria for VACTERL-H or PHENOS) were: short stature (43%), upper limb (radial ray) abnormalities (40%), skin pigmentary changes (37%), renal malformations (27%), and small head (27%). The rest of the abnormalities were less than 20%.
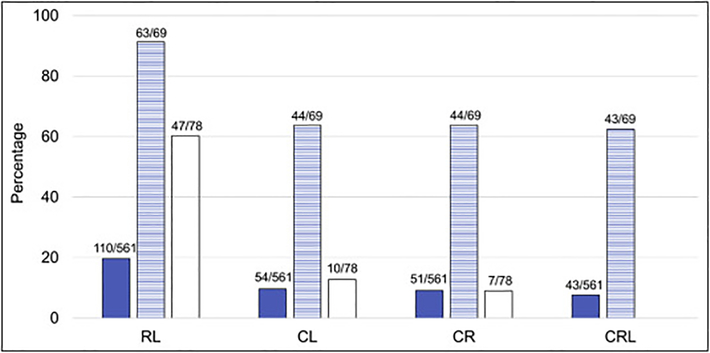
The combinations of two or three most common VACTERL-H features (C: cardiac, R: renal, and L: upper limb [radial ray]) among the 561 cases with phenotype information were: RL 19.6%, CL 9.6%, CR 9.1%, and CRL 7.7%. These combinations were 91.3%, 63.8%, 63.8%, 62.3%, respectively in the 69 patients who met criteria for VACTERL-H (Fig. 3). Among the 492 patients who did not meet criteria for VACTERL-H, 78 patients had only 2 features (60.3% RL, 12.8% CL, and 9% CR; the dual combinations between the other VACTERL-H features [V, A, T, E, and H; Table 1B] were seen in less than 20% of patients).
Fig 3.
Combinations of the most common VACTERL-H features. C: cardiac structural anomalies; R: renal malformations; L: upper limb anomalies (radial ray). Solid: patients with gene and phenotype described (n=561); horizontal stripes: patients who had ≥3/8 VACTERL-H features (n=69); white: patients who had only 2/8 VACTERL-H features (n=78). Horizontal axis: combinations; vertical axis: percent of cases with each combination. The frequencies of the combinations in the patients with gene and phenotype described were RL 19.6%, CL 9.6%, CR 9.1%, and CRL 7.7%; whereas in those who met criteria for VACTERL-H the frequencies were 91.3%, 63.8%, 63.8%, 62.3%, respectively. Eighty-two percent of patients who did not met criteria for VACTERL-H (with only 2 features) had the RL (60.3%), CL (12.8%), or CR (9%) combinations.
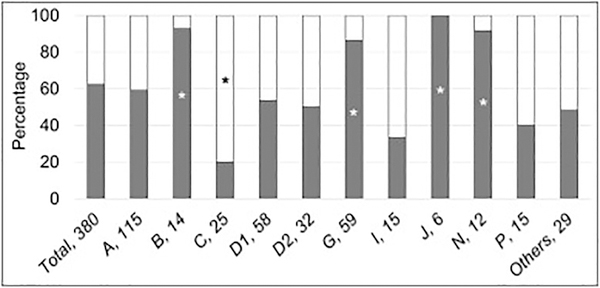
3.3. Types of FA Gene Variants
The type of pathogenic variant could be classified in 380 of the 561 patients (68%) with phenotype data (Fig. 1 and Table 2). Two hundred thirty-seven patients (62%) had biallelic or hemizygous null variants. The distribution of type of pathogenic variants varied across the genes (Fig. 4). Both null and hypomorphic genotypes were equally distributed in patients with pathogenic variants in FANCA, D1, D2, I, and P. The null genotype was significantly more frequent in patients with FANCB, G, J, and N (p≤0.03), whereas 80% of patients with FANCC had a hypomorphic genotype (p=0.004).
Table 2:
Type of Mutation associated with the Presence or Absence of Abnormalities.
| Total number of patients (%)a | Null (%)a | Hypomorphic (%)a | p value | |
|---|---|---|---|---|
| Number | 380 | 237 | 143 | |
| At least one abnormality of any type | 324 (85.2) | 217 (91.5) | 107 (74.8) | <0.0001 |
| No abnormality | 56 (14.8) | 20 (8.5) | 36 (25.2) |
The denominator for percentage is the total number of patients in the column
The bold numbers indicate the significantly higher proportion
Fig 4.
Type of Pathogenic Variants according to Gene. Gray: null genotype; white: hypomorphic genotype. Horizontal axis: gene, number of patients; vertical axis: percent of cases within each gene. *p<0.05. The type of variant could be determined in 380 out of 561 cases (68%) based on available information; 62% of patients had null variants. The frequency of null and hypomorphic variants varied among the genes. FANCB, G, J, and N were more frequently associated with null variants (p≤0.03); FANCC with hypomorphic variants (p=0.004); and, null and hypomorphic variants were equally distributed in the rest of the genes.
3.4. Presence or Absence of at Least One Abnormality According to the Gene, FA/BRCA DNA Repair Pathway, and Type of Pathogenic Variant
Gene
The majority of the patients in each of the FA gene groups had at least one abnormality. FANCB, D1, D2, J, and N were associated with abnormalities in nearly 90% of patients (Fig. 5A). A higher proportion of patients with FANCD2 had at least one abnormality compared with all other gene groups combined (p=0.01), in contrast, a smaller proportion of patients with FANCA had at least one abnormality (p=0.01) (Fig. 5A).
Fig. 5.
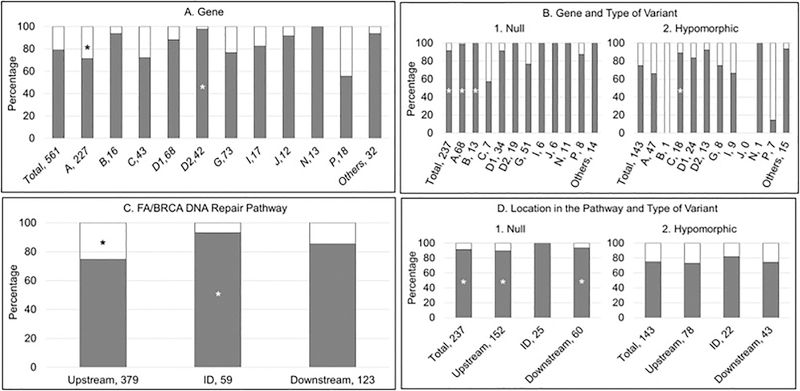
Presence or Absence of at least one Abnormality. Gray: abnormality; white: no abnormality. A and B) Horizontal axis: gene, number of patients; vertical axis: percent of cases within each gene. C and D) Horizontal axis: location in the pathway, number of patients; vertical axis: percent of cases within each location in the pathway. *p<0.05. (A) More than 70% of patients in each gene group had at least one of the abnormalities reported in Figure 2, except for FANCP (56%). Approximately 90% of patients with FANCB, D1, D2, J and N had at least one abnormality. A higher proportion of patients with FANCD2 had at least one abnormality compared with all other genes combined (p=0.01), in contrast with a smaller proportion of patients with FANCA (p=0.01). (B) More than 75% of the patients with a null genotype and more than 65% of the patients with a hypomorphic genotype had at least one abnormality. A higher proportion of patients with a null genotype compared with those with a hypomorphic genotype in FANCA and B had at least one abnormality (p≤0.0001); whereas for FANCC a higher proportion of patients with a hypomorphic genotype compared with biallelic null variants had any abnormality (p=0.01). (C) Presence of any abnormality was more frequent in the ID complex, followed by downstream and then upstream genes (p<0.05). (D) Patients with biallelic or hemizygous null variants were more likely to have any abnormality than those with hypomorphic variants independent of the location in the pathway (p<0.05).
Gene and type of pathogenic variant
A higher proportion of patients with a null genotype had at least one abnormality compared with those with a hypomorphic genotype (p<0.0001) (Table 2 and Fig. 5B). More than 75% of the patients with a null genotype within each individual gene had at least one abnormality, except for FANCC (2 of 5 patients; 40%); and more than 65% of patients with a hypomorphic genotype had at least one abnormality, except for FANCB (0 of 1 patient; 0%) and FANCP (3 of 9 patients; 33%) (Fig. 5B). A higher proportion of patients with null genotype compared with those with hypomorphic genotype in FANCA and B had at least one abnormality (p<0.0001, p=0.0001 respectively); whereas for FANCC, a higher proportion of patients with a hypomorphic genotype compared with a null genotype had any abnormality (p=0.01) (Fig. 5B).
Pathway
The presence of any abnormality was most frequent in the patients with pathogenic variants in the ID complex (93%), followed by downstream (85%) and then by upstream genes (75%) (Fig. 5C). A lower proportion of patients with pathogenic variants in upstream genes had at least one abnormality (p<0.001) compared with the ID complex and downstream genes, whereas a higher proportion of those with pathogenic variants in the ID complex genes had any abnormality (p=0.01) (Fig. 5C).
Pathway and type of pathogenic variant
Eighty-nine percent, 100% and 93% of the patients with null genotype in the upstream, ID complex and downstream genes, respectively, had at least one abnormality compared with 73%, 81% and 74% of patients with hypomorphic genotype (Fig. 5D). These differences were significant for upstream (p=0.003) and downstream (p=0.02) genes.
3.5. VACTERL-H and PHENOS According to the Gene, FA/BRCA DNA Repair Pathway, and Type of Pathogenic Variant
Nine percent of patients had VACTERL-H alone (without PHENOS), 5% had PHENOS alone (without VACTERL-H), and 4% had VACTERL-H plus PHENOS, while 82% had neither (Fig. 6A). Thirty percent of the 69 patients who met criteria for VACTERL-H also had 4 out of 6 PHENOS features, whereas only 6% of the 492 patients who did not meet the criteria for VACTERL-H had PHENOS (p<0.0001). We then examined the distribution of patients with VACTERL-H (≥3/8 features), PHENOS (≥4/6 features), both VACTERL-H plus PHENOS or neither within each FA gene, as well as according to the location in the FA/BRCA DNA repair pathway, and the type of pathogenic variant.
Fig. 6.
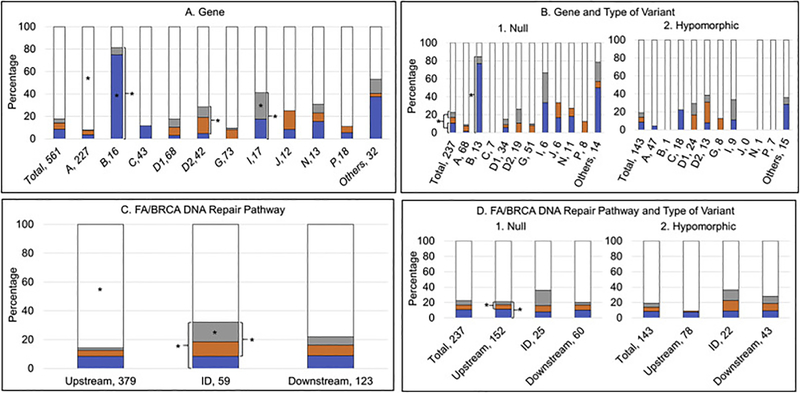
Presence or absence of VACTERL-H and/or PHENOS. Blue, VACTERL-H alone; orange, PHENOS alone; gray, VACTERL-H plus PHENOS; white, neither VACTERL-H nor PHENOS. A and B) Horizontal axis: gene, number of patients; vertical axis: percent of cases within each gene. C and D) Horizontal axis: location in the pathway, number of patients; vertical axis: percent of cases within each location in the pathway. *p<0.05. (A) 9% of all patients had VACTERL-H alone, 5% had PHENOS alone, 4% had VACTERL-H plus PHENOS, and 82% had neither VACTERL-H nor PHENOS. A higher proportion of patients with FANCB and I had VACTERL-H (irrespective of PHENOS) (p<0.001 and p=0.02, respectively). A higher proportion of patients with FANCD2 and I had PHENOS (irrespective of VACTERL-H) (p=0.01 for FANCD2). A higher proportion of patients with FANCB had VACTERL-H alone (without PHENOS) (p<0.001). VACTERL-H plus PHENOS was more frequent in patients with FANCI (p=0.03). VACTERL-H and/or PHENOS was rare in patients with FANCA (p<0.0001). (B) Overall, VACTERL-H (irrespective of PHENOS) was more frequent in patients with biallelic or hemizygous null variants than in those with at least one hypomorphic allele (p=0.01). Gene by gene, VACTERL-H (irrespective of PHENOS) and at least one of the two (VACTERL-H or PHENOS) were more frequent in patients with biallelic or hemizygous null variants in FANCB (p<0.05) and I. (C) VACTERL-H and/or PHENOS were less frequent in patients with pathogenic variants in upstream genes (p<0.0001). A higher proportion of patients with pathogenic variants in ID complex genes had PHENOS (irrespective of VACTERL-H) (p=0.001), both VACTERL-H plus PHENOS (p=0.003), or at least one of the two (p=0.01) compared with patients with pathogenic variants in upstream and downstream genes. (D) Patients with biallelic or hemizygous null variants in upstream genes had a higher proportion of PHENOS (irrespective of VACTERL-H) and at least one of the two (VACTERL-H or PHENOS) than those with hypomorphic genotype (p<0.02); these associations were not seen in ID or downstream genes.
Gene
A higher proportion of patients with FANCB (81%) and I (41%) had VACTERL-H (irrespective of PHENOS) (p<0.001 and p=0.02, respectively), while fewer patients with FANCA and G had this phenotype (p<0.001 and p=0.01, respectively) (Fig. 6A). A higher proportion of patients with FANCD2 (24%) and I (24%) had PHENOS (irrespective of VACTERL-H) (p=0.01 for FANCD2), while this phenotype was rare in FANCA and C (p=0.01 and p=0.02, respectively); FANCC was the only gene in which no patients had PHENOS (Fig. 6A). A higher proportion of patients with FANCB had VACTERL-H alone (without PHENOS) (p<0.001), whereas it was less common in FANCA and G (p=0.003 and p=0.01, respectively) (Fig. 6A). PHENOS alone (without VACTERL-H) was not associated with any gene. VACTERL-H plus PHENOS was more frequent in patients with FANCI (p=0.03), whereas the combination was uncommon in FANCA (p<0.0001) (Fig. 6A).
Gene and type of pathogenic variant
Overall, VACTERL-H (irrespective of PHENOS) was more frequent in patients with null genotype than in those with at hypomorphic genotype (p=0.01) (Fig. 6B). Gene by gene, VACTERL-H (irrespective of PHENOS) and at least one of the two (VACTERL-H or PHENOS) were more frequent in patients with a null genotype in FANCB (p<0.05) and I compared with a hypomorphic genotype (Fig. 6B).
Pathway
VACTERL-H and/or PHENOS were less frequent in patients with pathogenic variants in upstream genes (p<0.0001). A higher proportion of patients with pathogenic variants in ID complex genes had PHENOS (irrespective of VACTERL-H) (p=0.001), both VACTERL-H plus PHENOS (p=0.003), or at least one of the two (p=0.01) compared with patients with pathogenic variants in upstream and downstream genes (Fig. 6C).
Pathway and type of pathogenic variant
A higher proportion of patients with null genotype in upstream genes had PHENOS (irrespective of VACTERL-H) and at least one of the two (VACTERL-H or PHENOS) than those with hypomorphic genotype (p<0.02); these associations were not seen in ID or downstream genes (Fig. 6D).
3.6. Presence or Absence of Specific Type of Abnormality According to the Gene, FA/BRCA DNA Repair Pathway, and Type of Pathogenic Variant
Multiple comparison analyses identified associations between specific congenital abnormalities and the genes, location in the FA/BRCA DNA repair pathway, and type of pathogenic variant. Gene by gene, pathogenic variants in FANCA, C, G, and P did not have any association with the presence of any specific abnormality, whereas FANCB and D2 had the highest number of associations (several abnormalities were associated with those genes) (Table 3). Pathogenic variants in upstream genes did not have any specific association while those with ID complex genes had the highest number of associations followed by downstream genes. A null genotype was significantly associated with renal malformations, microcephaly, short stature and VACTERL-H (irrespective of PHENOS).
Table 3:
Association of Specific Abnormalities with the Gene, FA/BRCA DNA Repair Pathway, and Type of Pathogenic Variant.
| Gene | Pathway | Genotype | ||||||||||||||
| Abnormality | A | B | C | D1 | D2 | G | I | J | N | P | Upstream | ID | Downstream | Null | Hypomorphic | |
| At least one abnormality | + | + | + | |||||||||||||
| VACTERL-H | Vertebral | + | ||||||||||||||
| Anal | + | + | + | |||||||||||||
| Cardiac | + | + | + | |||||||||||||
| Tracheo-esophageal fistula | + | |||||||||||||||
| Esophageal/duodenal atresia | + | |||||||||||||||
| Renal | + | + | + | |||||||||||||
| upper Limb | + | + | + | |||||||||||||
| Hydrocephalus | + | |||||||||||||||
| PHENOS | Pigmented skin | + | + | + | ||||||||||||
| small Head | + | + | + | + | + | + | ||||||||||
| small Eyes | + | + | ||||||||||||||
| Neurologic structure | + | |||||||||||||||
| Otology | + | + | + | |||||||||||||
| Short stature | + | + | ||||||||||||||
| Others | Developmental delay | + | + | |||||||||||||
| Any facial abnormality | + | + | ||||||||||||||
| Lower limb | + | + | ||||||||||||||
| VACTERL-Ha | + | + | + | |||||||||||||
| VACTERL-H alone | + | |||||||||||||||
| PHENOSb | + | + | ||||||||||||||
| VACTERL-H plus PHENOS | + | + | ||||||||||||||
| At least one (VACTERL-H or PHENOS) | + | + | ||||||||||||||
| Neither VACTERL-H nor PHENOS | + | + | ||||||||||||||
| *p<0.005 | *p<0.02 | p<0.05 | ||||||||||||||
+ positive association between the abnormality and the gene, FA/BRCA DNA repair pathway or type or variant.
corrected p-values (Bonferroni)
VACTERL-H (Vertebral, Anal, Cardiac, Tracheo-esophageal fistula, Esophageal atresia, Renal, upper Limb and Hydrocephalus)
PHENOS (skin Pigmentation, small Head, small Eyes, Neurologic structure, Otology, Short stature)
4. Discussion
This is the largest comprehensive review of the literature describing the association of congenital abnormalities with the gene, location in the FA/BRCA DNA repair pathway, and the type of pathogenic variant in patients with FA. The lack of information about physical findings in nearly half of the patients with reported genotype data (mostly patients with FANCA, C and G), may be due to a recent bias towards describing phenotypes in patients with rare genotypes.
The percentage of patients with at least one abnormality of any type (79%) was similar to other reports (60% to 90%) (2, 21). The genotypes that have been consistently associated with abnormalities in the literature, FANCB (22), D1 (13), D2 (23, 24), I (25–28), J (25, 29), and N (14), had the highest frequency of at least one abnormality (more than 80% of patients) in our review. The most frequent findings (seen in 27–43% of patients) were: short stature, radial ray defects, skin pigmentary changes, renal malformations, and microcephaly; all part of VACTERL-H or PHENOS and classically associated with FA (2, 7).
The proportion of patients with FA who met criteria for VACTERL-H association (12%) in this comprehensive review was similar to other reports (5% to 33%) (6, 7). Nine percent of the entire group of patients had ≥4/6 features of PHENOS. We had previously described the association between ≥4/6 findings of PHENOS and VACTERL-H in patients with FA (7). Consistent with the earlier report, patients in this review with VACTERL-H had a higher frequency of ≥4/6 PHENOS features than those without VACTERL-H, highlighting the need to look for PHENOS in patients with VACTERL-H. The presence of VACTERL-H alone or associated with PHENOS suggests the diagnosis of FA, while the lack of these does not rule out the diagnosis, because most patients with FA do not meet criteria for either (82%). The exception is FANCB, the gene most frequently associated with VACTERL-H in this review (81%) and in previous reports (22, 30–32). The observation of at least two out of the three most common VACTERL-H features (cardiac, renal, and upper limb malformations) could guide the clinician to consider FA. Since 5% of patients with FA may have ≥4/6 PHENOS features without meeting criteria for VACTERL-H, the possibility of FA should be considered during the evaluation of patients with PHENOS features, especially if they also have hematological and/or oncological anomalies.
The percentage of male patients with abnormalities of the genitalia was 16%. This could be an important clue during the clinical evaluation of patients suspected to have FA, based on other findings. The lower frequency of females with genitalia abnormalities may be explained by the lack of invasive examination that is necessary to assess for an anomaly.
Associations of specific abnormalities with the gene, location in the FA/BRCA DNA repair pathway, and the type of pathogenic variant were established. The genes associated with a higher number of specific abnormalities were FANCB followed by D2; in the pathway, the ID complex driven mainly by FANCD2 pathogenic variants, followed by downstream genes; and null compared with hypomorphic variants. We were able to delineate a phenotype (specific cluster of congenital abnormalities) according to the gene, location in the FA/BRCA DNA repair pathway, and the type of variant. The reason why some genes are more associated with the presence of congenital abnormalities despite all gene products involved in the DNA repair pathway, could be due to non-canonical gene functions; this hypothesis requires further studies.
5. Strengths and Limitations
There are some limitations to this study. Nearly half of the reports in the literature lacked phenotype information. Approximately one third of the reports did not provide sufficient details to classify the type of variant. Patients who were stated to lack physical abnormalities were assumed to have no anomalies. We did not analyze hematological and oncological features. Strengths of this study include that the genotype-phenotype analysis included 561 patients. In addition to gene by gene analysis, we examined the location in the FA/BRCA DNA repair pathway and the type of variants (null and hypomorphic genotypes).
6. Conclusions and Future Directions
Congenital abnormalities in patients with FA are variable and multisystemic. The majority of patients in whom the genotype and phenotype are described have at least one abnormality. Patients with pathogenic variants in FANCB, D2, ID complex, and downstream genes as well as biallelic or hemizygous null variants have the highest frequency of associations with congenital abnormalities. The most common abnormalities are part of VACTERL-H and PHENOS. The presence of VACTERL-H alone or with PHENOS is highly associated with FA; however, the absence of either or both does not rule out the diagnosis of FA.
The thorough description of phenotype may be used to identify specific groups of findings, such as FA facies, which might facilitate early diagnoses. A more detailed description of the specific congenital abnormalities in patients with FA, including laterality in paired organs, may guide future management. A molecular explanation for the differences between characteristic physical findings among the various inherited bone marrow failure syndromes may contribute to making correct diagnoses in patients who appear to belong to more than one syndrome (2).
Supplementary Material
7. Practice Points.
The spectrum of physical abnormalities in patients with Fanconi anemia is variable and multisystemic.
The most frequent anomalies are: short stature, upper limb structural abnormalities, pigmentary skin changes, renal malformations, and microcephaly; all are included in either VACTERL-H or PHENOS.
FA should be considered in any patient with one or more of the congenital abnormalities listed above.
The clinician should suspect FA even if the patient does not meet criteria for VACTERL-H or PHENOS.
The physical phenotype may guide the genotype.
8. Research Agenda.
Explore the relation between canonical and non-canonical gene functions and the variability of congenital abnormalities (33, 34).
Extend the genotype-phenotype analyses to include hematologic and oncologic outcomes.
Compare the various physical scoring systems for the strength of their association with proven cases with FA: i.e. adding anomalies (35), the CABS (congenital abnormality score) (36), VACTERL-H, PHENOS (7).
Determine the frequency of FA among patients with VACTERL-H.
Acknowledgments
Funding
This research was funded in part by the Intramural Research Program of the Division of Cancer Epidemiology and Genetics of the National Cancer Institute, and a grant from the Fanconi Anemia Research Fund, Inc.
Footnotes
Conflict of Interest
The authors declare no conflicts of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Alter BP, Giri N, Savage SA, Rosenberg PS. Cancer in the National Cancer Institute inherited bone marrow failure syndrome cohort after fifteen years of follow-up. Haematologica. 2018;103(1):30–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shimamura A, Alter BP. Pathophysiology and management of inherited bone marrow failure syndromes. Blood Rev. 2010;24(3):101–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fanconi G Familiäre infantile perniziosaartige Anämie (pernizioses Blutbild und Konstitution). Jahrbuch für Kinderheilkunde. 1927;117:257–89. [Google Scholar]
- 4.Alter BP. Fanconi anemia and the development of leukemia. Best Pract Res Clin Haematol. 2014;27(3–4):214–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Solomon BD, Bear KA, Kimonis V, de Klein A, Scott DA, Shaw-Smith C, et al. Clinical geneticists’ views of VACTERL/VATER association. Am J Med Genet A. 2012;158a(12):3087–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Faivre L, Portnoi MF, Pals G, Stoppa-Lyonnet D, Le Merrer M, Thauvin-Robinet C, et al. Should chromosome breakage studies be performed in patients with VACTERL association? Am J Med Genet A. 2005;137(1):55–8. [DOI] [PubMed] [Google Scholar]
- 7.Alter BP, Giri N. Thinking of VACTERL-H? Rule out Fanconi Anemia according to PHENOS. Am J Med Genet A. 2016;170(6):1520–4. [DOI] [PubMed] [Google Scholar]
- 8.Knies K, Inano S, Ramirez MJ, Ishiai M, Surralles J, Takata M, et al. Biallelic mutations in the ubiquitin ligase RFWD3 cause Fanconi anemia. J Clin Invest. 2017;127(8):3013–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang AT, Smogorzewska A. SnapShot: Fanconi anemia and associated proteins. Cell. 2015;160(1–2):354–354.e1. [DOI] [PubMed] [Google Scholar]
- 10.Rodriguez A, D’Andrea A. Fanconi anemia pathway. Curr Biol. 2017;27(18):R986–r8. [DOI] [PubMed] [Google Scholar]
- 11.Faivre L, Guardiola P, Lewis C, Dokal I, Ebell W, Zatterale A, et al. Association of complementation group and mutation type with clinical outcome in fanconi anemia. European Fanconi Anemia Research Group. Blood. 2000;96(13):4064–70. [PubMed] [Google Scholar]
- 12.Myers K, Davies SM, Harris RE, Spunt SL, Smolarek T, Zimmerman S, et al. The clinical phenotype of children with Fanconi anemia caused by biallelic FANCD1/BRCA2 mutations. Pediatr Blood Cancer. 2012;58(3):462–5. [DOI] [PubMed] [Google Scholar]
- 13.Alter BP, Rosenberg PS, Brody LC. Clinical and molecular features associated with biallelic mutations in FANCD1/BRCA2. J Med Genet. 2007;44(1):1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Reid S, Schindler D, Hanenberg H, Barker K, Hanks S, Kalb R, et al. Biallelic mutations in PALB2 cause Fanconi anemia subtype FA-N and predispose to childhood cancer. Nat Genet. 2007;39(2):162–4. [DOI] [PubMed] [Google Scholar]
- 15.Ishida R, Buchwald M. Susceptibility of Fanconi’s anemia lymphoblasts to DNA-crosslinking and alkylating agents. Cancer Res. 1982;42(10):4000–6. [PubMed] [Google Scholar]
- 16.Pulsipher M, Kupfer GM, Naf D, Suliman A, Lee JS, Jakobs P, et al. Subtyping analysis of Fanconi anemia by immunoblotting and retroviral gene transfer. Mol Med. 1998;4(7):468–79. [PMC free article] [PubMed] [Google Scholar]
- 17.Cingolani P, Platts A, Wang le L, Coon M, Nguyen T, Wang L, et al. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly (Austin). 2012;6(2):80–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang K, Li M, Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010;38(16):e164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kim S, Jhong JH, Lee J, Koo JY. Meta-analytic support vector machine for integrating multiple omics data. BioData Min. 2017;10:2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ioannidis NM, Rothstein JH, Pejaver V, Middha S, McDonnell SK, Baheti S, et al. REVEL: An Ensemble Method for Predicting the Pathogenicity of Rare Missense Variants. Am J Hum Genet. 2016;99(4):877–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Risitano AM, Marotta S, Calzone R, Grimaldi F, Zatterale A. Twenty years of the Italian Fanconi Anemia Registry: where we stand and what remains to be learned. Haematologica. 2016;101(3):319–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McCauley J, Masand N, McGowan R, Rajagopalan S, Hunter A, Michaud JL, et al. X-linked VACTERL with hydrocephalus syndrome: further delineation of the phenotype caused by FANCB mutations. Am J Med Genet A. 2011;155a(10):2370–80. [DOI] [PubMed] [Google Scholar]
- 23.Dragana V, Sandra P, Emilija L, Milos K, Andreja L, Ivana J, et al. Prevalence of FA-D2 rare complementation group of Fanconi anemia in Serbia. Indian J Pediatr. 2014;81(3):260–5. [DOI] [PubMed] [Google Scholar]
- 24.Kalb R, Neveling K, Hoehn H, Schneider H, Linka Y, Batish SD, et al. Hypomorphic mutations in the gene encoding a key Fanconi anemia protein, FANCD2, sustain a significant group of FA-D2 patients with severe phenotype. Am J Hum Genet. 2007;80(5):895–910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Levitus M, Rooimans MA, Steltenpool J, Cool NF, Oostra AB, Mathew CG, et al. Heterogeneity in Fanconi anemia: evidence for 2 new genetic subtypes. Blood. 2004;103(7):2498–503. [DOI] [PubMed] [Google Scholar]
- 26.Dorsman JC, Levitus M, Rockx D, Rooimans MA, Oostra AB, Haitjema A, et al. Identification of the Fanconi anemia complementation group I gene, FANCI. Cell Oncol. 2007;29(3):211–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sims AE, Spiteri E, Sims RJ 3rd, Arita AG, Lach FP, Landers T, et al. FANCI is a second monoubiquitinated member of the Fanconi anemia pathway. Nat Struct Mol Biol. 2007;14(6):564–7. [DOI] [PubMed] [Google Scholar]
- 28.Savage SA, Ballew BJ, Giri N, Chandrasekharappa SC, Ameziane N, de Winter J, et al. Novel FANCI mutations in Fanconi anemia with VACTERL association. Am J Med Genet A. 2016;170a(2):386–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ghazwani Y, AlBalwi M, Al-Abdulkareem I, Al-Dress M, Alharbi T, Alsudairy R, et al. Clinical characteristics and genetic subtypes of Fanconi anemia in Saudi patients. Cancer Genet. 2016;209(4):171–6. [DOI] [PubMed] [Google Scholar]
- 30.Winberg J, Gustavsson P, Papadogiannakis N, Sahlin E, Bradley F, Nordenskjold E, et al. Mutation screening and array comparative genomic hybridization using a 180K oligonucleotide array in VACTERL association. PLoS One. 2014;9(1):e85313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mikat B, Roll C, Schindler D, Gembruch U, Klempert I, Buiting K, et al. X-linked recessive VACTERL-H due to a mutation in FANCB in a preterm boy. Clin Dysmorphol. 2016;25(2):73–6. [DOI] [PubMed] [Google Scholar]
- 32.Umana LA, Magoulas P, Bi W, Bacino CA. A male newborn with VACTERL association and Fanconi anemia with a FANCB deletion detected by array comparative genomic hybridization (aCGH). Am J Med Genet A. 2011;155a(12):3071–4. [DOI] [PubMed] [Google Scholar]
- 33.Cheung RS, Taniguchi T. Recent insights into the molecular basis of Fanconi anemia: genes, modifiers, and drivers. Int J Hematol. 2017;106(3):335–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sumpter R Jr., Levine B. Emerging functions of the Fanconi anemia pathway at a glance. J Cell Sci. 2017;130(16):2657–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Giampietro PF, Verlander PC, Davis JG, Auerbach AD. Diagnosis of Fanconi anemia in patients without congenital malformations: an international Fanconi Anemia Registry Study. Am J Med Genet. 1997;68(1):58–61. [PubMed] [Google Scholar]
- 36.Rosenberg PS, Huang Y, Alter BP. Individualized risks of first adverse events in patients with Fanconi anemia. Blood. 2004;104(2):350–5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.