Abstract
Patatin-Like Phospholipase Domain Containing 2 (PNPLA2)/Adipose Triglyceride Lipase (ATGL) and PNPLA3/Adiponutrin are close paralogs that appear to have opposite functions on triacylglycerol (TAG) mobilization and storage. PNPLA2/ATGL is a major triglyceride lipase in adipose tissue and liver, whereas a common human variant of PNPLA3, I148M, greatly increases risk of hepatosteatosis. Nonetheless, the function of PNPLA3 and the mechanism by which the I148M variant promotes TAG accumulation are poorly understood. Here we demonstrate that PNPLA3 strongly interacts with α/β hydrolase domain-containing 5 (ABHD5/CGI-58), an essential co-activator of PNPLA2/ATGL. Molecular imaging experiments demonstrate that PNPLA3 effectively competes with PNPLA2/ATGL for ABHD5, and that PNPLA3 I148M is more effective in this regard. Inducible overexpression of PNPLA3 I148M greatly suppressed PNPLA2/ATGL-dependent lipolysis and triggered massive TAG accumulation in brown adipocytes, and these effects were dependent on ABHD5. The interaction of PNPLA3 and ABHD5 can be regulated by fatty acid supplementation and synthetic ABHD5 ligands, raising the possibility that this interaction might be targeted for treatment of fatty liver disease.
Triacylglycerol (TAG) storage and mobilization in adipose tissue and liver are critical processes for systemic energy homeostasis that involve dynamic interactions among evolutionarily conserved proteins. Patatin-Like Phospholipase Domain Containing 2 (PNPLA2), widely known as Adipose Triglyceride Lipase (ATGL), is a member of the Patatin family of lipases and is the rate-limiting step in TAG hydrolysis in fat, muscle, and liver1. While PNPLA2/ATGL exhibits intrinsic TAG lipase activity, this activity is greatly elevated through its interaction with ABHD52 (also known as CGI-58), an ancient protein in the α-β hydrolase family that lacks intrinsic hydrolase activity. The dynamic interaction between ABHD5 and PNPLA2/ATGL is regulated in part by tissue-specific perilipins that function as scaffolds to orchestrate this interaction on the surface or lipid droplets for fatty acid efflux and oxidation3. For example, in adipocytes perilipin 1 (PLIN1) sequesters ABHD5 in the basal state and extracellular signals that lead to PLIN1 phosphorylation release ABHD5 and activate PNPLA2/ATGL; however, additional levels of regulation are likely to exist, especially under conditions that favor TAG storage. The functional interaction between PNPLA2/ATGL and ABHD5 is critical in regulating lipid mobilization, as loss of either PNPLA2 or ABHD5 results in ectopic accumulation of neutral lipids in several tissues4. Nonetheless, null mutations of ABHD5 do not exactly phenocopy loss of PNPLA2/ATGL, suggesting that ABHD5 might have functions that are distinct from lipase activation of PNPLA2/ATGL5,6.
PNPLA3 (also known as adiponutrin) is a close paralog of PNPLA2/ATGL that arose by gene duplication in vertebrates7. Although PNPLA2/ATGL and PNPLA3 are both expressed in fat and liver, PNPLA2 is upregulated by fasting, whereas PNPLA3 is upregulated by feeding8,9, suggesting these proteins serve opposite roles in TAG metabolism. Nonetheless, the precise function of PNPLA3 in neutral lipid metabolism is controversial. In vitro experiments suggest that PNPLA3 has TAG hydrolase, acyltransferase, and transacylase activities10–12. Importantly, a common human genetic variant of PNPLA3 (rs738409), encoding I148M substitution, greatly increases susceptibility to fatty liver disease (FLD)13, likely through mechanisms that involve accumulation of the mutant protein on intracellular lipid droplets (LDs)14,15. Since complete loss of PNPLA3 does not promote FLD16,17, whereas expression of catalytically-inactive (S47A) PNPLA3 phenocopies the I148M variant15, it appears that the ability of PNPLA3 to promote hepatosteatosis might be related in part to non-catalytic interactions with an intersecting pathway. Interestingly, overexpression of PNPLA3 I148M in hepatocytes has been shown to suppress lipolysis, but the molecular mechanism for this suppression is not known18,19.
The fact that certain invertebrate orthologs of PNPLA2/ATGL and ABHD5 interact6 suggests that the ancestral gene of PNPLA2/3 likely functioned as a TAG lipase that was activated by ABHD5. However, biochemical and genetic evidence indicates that PNPLA3 has low functional lipase activity that is not directly affected by ABHD511,12. Nonetheless, if PNPLA3 retained the ability to bind ABHD5 without triggering significant TAG hydrolysis, then such an interaction could sequester ABHD5 from PNPLA2/ATGL, thereby suppressing lipolysis indirectly. Such an interaction would be conceptually similar to the regulation of PNPLA2/ATGL activity by PLIN120, which binds ABHD5 and suppresses lipolysis21. In experiments detailed below we found that PNPLA3 strongly binds ABHD5 and that this interaction is rapidly increased by conditions that promote neutral lipid sequestration and by synthetic ligands of ABHD5. Furthermore, PNPLA3 and PNPLA2/ATGL compete for ABHD5 in transfected cells, and inducible expression of PNPLA3 suppresses ABHD5-dependent lipolysis in brown adipocytes. We found that PNPLA3 I148M reduces ABHD5-dependent lipolysis to a greater degree than the WT protein and thus represents a gain of function with respect to ABHD5 binding and lipolysis suppression. Our results indicate that the interaction of PNPLA3 and ABHD5 can be modulated by both endogenous and synthetic ligands of ABHD5, raising the possibility that this interaction might be targeted for treatment of diseases involving excessive triglyceride storage, such as FLD.
Results
ABHD5 and PNPLA3 co-traffic to Endoplasmic Reticulum
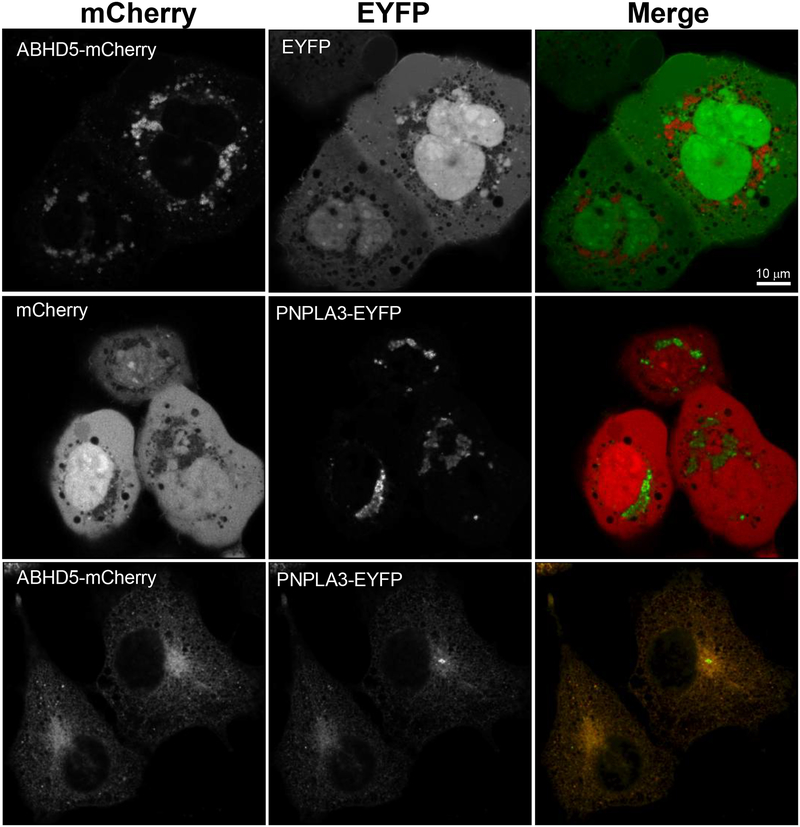
We first asked whether ABHD5 and PNPLA3 traffic to the same subcellular compartments by examining the localization of fluorescently tagged ABHD5 and PNPLA3 individually, and in co-transfected, lipid-loaded COS-7 cells. As previously reported, mCherry tagged ABHD522 mostly localized to lipid droplets (LDs) that were dispersed throughout the cell (Fig. 1), whereas PNPLA3-EYFP localized to clustered structures near the nucleus that appeared to be Golgi (Fig. 1). Immunostaining of cells expressing PNPLA3 with the Golgi marker Receptor-Binding Cancer-Associated Surface antigen-1 (RCAS1) confirmed that PNPLA3 was mostly localized within this subcellular compartment (Supplementary Fig. 1a). In contrast, in cells expressing ABHD5 and PNPLA3, both exhibited strong endoplasmic reticulum (ER) localization (Fig. 1), with some cells showing LD targeting (arrows) as well. (Supplementary Fig. 1b). This ER targeting was verified by co-staining cells with ER marker protein disulfide isomerase (PDI) (Supplementary Fig. 1c). These results demonstrate that ABHD5 and PNPLA3 traffic together to the ER, the main site of TAG synthesis, where they might interact.
Fig. 1. ABHD5 and PNPLA3 co-traffic to endoplasmic reticulum.
COS-7 cells on 25 mm coverslips were co-transfected with pairs of ABHD5-mCherry and EYFP; mCherry and PNPLA3-EYFP; or ABHD5-mCherry with PNPLA3-EYFP and loaded with OA (200 μM) overnight. Cells were fixed with 4% paraformaldehyde and imaged on a Zeiss LSM 800 laser scanning microscope equipped with an Airyscan detector. mCherry was pseudocoloured red and EYFP pseudocoloured green in merged images to show co-localization (yellow). Results are from one experiment and are representative of four independent experiments. Scale bar denotes 10 μm.
PNPLA3 strongly interacts with ABHD5 in vitro and in vivo
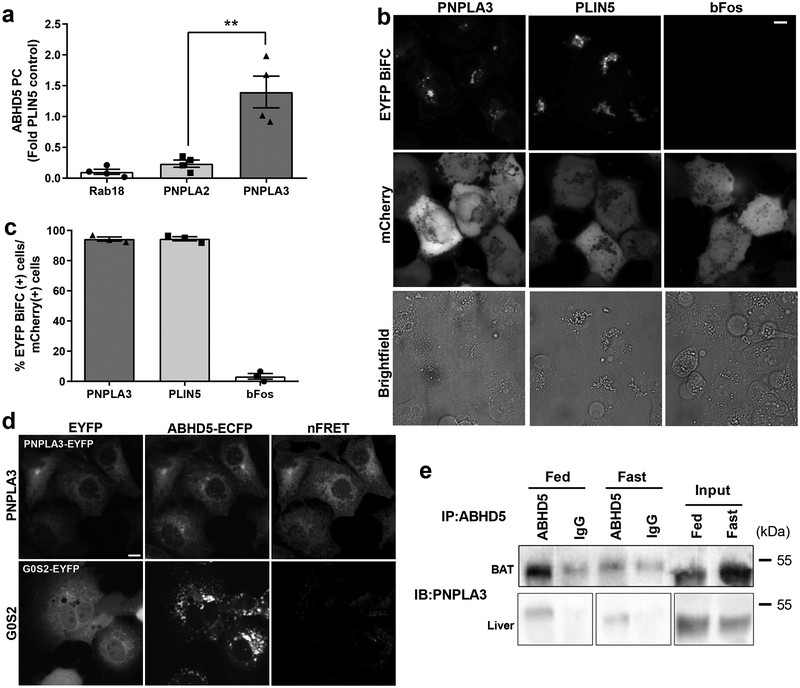
To test whether ABHD5 interacts with PNPLA3, we performed protein complementation (PC) assays in 293A cells, which provide a robust system for transient expression of exogenous proteins (Fig. 2). In these assays, reporter molecules are split into two fragments and fused to test proteins of interest. The interaction of test proteins brings the complementary protein fragments together and reconstitutes the signals generated by the reporter molecule. The first set of experiments used luciferase complementation to test the interaction of ABHD5 with PNPLA3 and PNPLA2/ATGL. The interaction of ABHD5 with lipid droplet (LD) proteins Perilipin 5 (PLIN5) and Rab18 served as positive and negative controls, respectively. As shown in Fig. 2a, ABHD5 strongly interacted with PNPLA3. Indeed, the interaction of ABHD5 with PNPLA3 was as strong as that with PLIN5 (depicted on the y-axis) and was much stronger than that with PNPLA2/ATGL.
Fig. 2. ABHD5 and PNPLA3 interact on ER structures and lipid droplets.
(a) Gaussia luciferase (Gluc) protein complementation (PC) assay in HEK293A cells transfected with complementary fragments of luciferase GlucN-ABHD5 and either Rab18-GlucC, PNPLA2-GlucC, PLIN5-GlucC or PNPLA3-GlucC. Values shown are a fold of PLIN5-GLucC activity and are the averages from three independent experiments performed in quadruplicate. **p=0.0014 for PNPLA3 vs. PNPLA2 as determined by one-way ANOVA with Bonferroni’s post t-test. (b) Bimolecular fluorescence complementation (BiFC) assay performed in COS-7 cells expressing complementary fragments of EYFP Yn-ABHD5 and either PNPLA3-Yc, PLIN5-Yc, or bFos-Yc and loaded with 200 μM oleic acid (OA) overnight. mCherry was used to identify transfected cells and brightfield imaging was used to identify lipid droplets. (c) Quantification of BiFC signal in mCherry+ cells from b and are the averages of biological quadruplicates from three independent experiments. (d) FRET assay in COS-7 cells transfected with ECFP-ABHD5 and either PNPLA3-EYFP or G0S2-EYFP, loaded with OA (200 μM) overnight. Images are from one experiment and are representative of four independent experiments. nFRET, net FRET signal. (e) Immunoprecipitation (IP) of ABHD5/PNPLA3 complex from solubilized lipid droplets from liver of fasted or fed 8 to 12-week-old C57L/B6 male mice. PNPLA3 and ABHD5 proteins were detected by western blotting (WB). Non-specific band (ns). Size markers (in kDa) are depicted on western blot panels. IP results are from one experiment and representative of three independent experiments. Data are expressed as means +/− SEM. Scale bar denotes 10 μm.
To localize the subcellular sites of the interaction between ABHD5 and PNPLA3 we performed Bimolecular Fluorescence Complementation (BiFC) in lipid-loaded COS-7 cells using split fragments of EYFP23,24, with PLIN5 and bFos serving as positive and negative controls, respectively (Fig. 2b and 2c). ABHD5 produced strong BiFC with PNPLA3 and PLIN5, but not with bFos (Fig. 2b). ABHD5-PNPLA3 BiFC was mainly observed on perinuclear ER-like structures and some lipid droplets (LDs), whereas ABHD5-PLIN5 BiFC was observed mostly on LDs. ABHD5-PNPLA3 BiFC was observed in more than 90% of transfected cells, indicating an efficiency of interaction similar to that of ABHD5 with PLIN5 (Fig. 2c). As additional confirmation, we performed Förster Resonance Energy Transfer (FRET) analysis between the ECFP-ABHD5 and PNPLA3-EYFP in lipid-loaded COS-7 cells. G0/G1 Switch 2 (G0S2), which also targets ER membranes, was used as a negative control. ABHD5 and PNPLA3 were targeted to ER membranes, where they exhibited strong energy transfer (Fig. 2d). By contrast, ABHD5 was mainly targeted to LDs in cells expressing G0S2 and exhibited little, if any, energy transfer. Together, these results demonstrate that PNPLA3 strongly interacts with ABHD5 on ER and LD membranes, which are well known sites of TAG synthesis and storage, respectively.
PNPLA3 expression is regulated by nutritional and environmental stimuli25,26 and is highest in mouse brown adipose tissue (BAT)8,9, where its function is largely unknown. In liver, PNPLA3 expression is upregulated by sucrose feeding where expression of I148M variant greatly promotes TAG accumulation. To test whether endogenous ABHD5 and PNPLA3 interact in vivo, we performed immunoprecipitation assays in BAT and liver of mice that were fasted or refed a high sucrose diet. As shown in Fig. 2e, antibodies to ABHD5 immunoprecipitated PNPLA3 from solubilized BAT LDs. Whereas ABHD5 antibodies immunoprecipitated PNPLA3 mostly under fed conditions in BAT, ABHD5/PNPLA3 immunocomplexes were observed in liver LDs from both the fed and fasted mice (Fig 2e). Re-blotting confirmed that the immunoprecipitation of ABHD5 was specific to ABHD5 antibodies (Supplementary Fig. 2).
Endogenous and synthetic ABHD5 ligands promote its interaction with PNPLA3
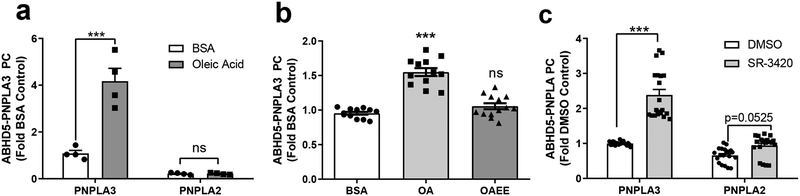
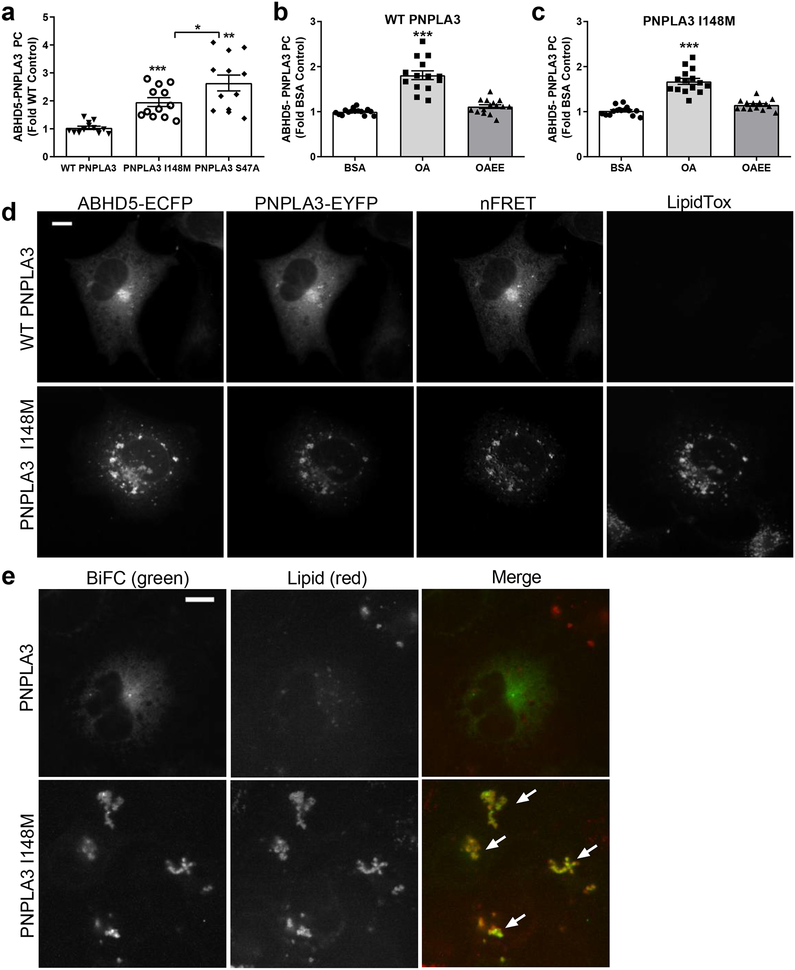
We previously reported that oleoyl-CoA, generated intracellularly from exogenously supplied oleic acid (OA), is an allosteric regulator of ABHD5 that rapidly promotes its interaction with PLIN5 and PLIN123,27. As shown in Fig. 3a, exposing cells to OA/BSA complexes (200 μM) for 3 hr increased the interaction of ABHD5 and PNPLA3 in luciferase complementation assays by approximately four-fold, but did not alter the relatively weak interaction of ABHD5 and PNPLA2/ATGL. Because OA treatment can increase PNPLA3 levels by promoting its stability25, we examined whether the increase in ABHD5/PNPLA3 protein complementation was associated with changes in PNPLA3 protein levels. As shown in Supplementary Fig. 3a, 3 hr OA treatment increased protein levels of PNPLA3, but not ABHD5. We also tested the effects of OA after a 30 minute exposure, when changes in PNPLA3 protein levels were not observed (Supplementary Fig. 3b). Brief OA treatment increased the interaction of ABHD5 and PNPLA3 by 62% ± 5.74 (p<0.001, Fig. 3b). Importantly, the interaction of ABHD5 with PNPLA3 was not promoted by the ethyl ester of OA (OAEE), which cannot form oleoyl-CoA (Fig. 3b).
Fig. 3. The interaction between ABHD5 and PNPLA3 is regulated by endogenous and synthetic ligands of ABHD5.
(a) Gluc protein complementation (PC) assay of HEK293A cells transfected with GlucN-ABHD5 and either PNPLA3-GlucC or PNPLA2-GlucC and treated with BSA or 200 μM oleic acid (OA) the following day for three hours. Data are biological quadruplicates from four independent experiments. ***p=0.0001 effect of OA as determined by two-way ANOVA with Bonferroni post t-test. (b) HEK293A cells expressing GlucN-ABHD5 and PNPLA3-GlucC were treated with either BSA, 200 μM OA, or 200 μM oleic acid ethyl ester (OAEE) for 30 minutes. Data are biological quadruplicates from three independent experiments. ***p=0.0001 indicates a significant difference between BSA and OA as determined by one-way ANOVA with Bonferroni post t-test. (c) Gluc PC assay was performed in HEK293A cells as in a, except cells were treated for one hour with SR-3420 (20 μM). Data are biological quadruplicates from five independent experiments. ***p=0.0001 indicates a significant effect of SR-3420 as determined by two-way ANOVA with Bonferroni post t-test. Data are expressed as means +/− SEM. ns, not significant.
We recently reported the discovery and development of highly-selective synthetic ABHD5 ligands that stimulate adipocyte lipolysis by dissociating ABHD5 from PLIN1/5, thereby promoting the interaction of ABHD5 with ATGL/PNPLA227,28. We examined whether the most potent of these, SR-3420, could promote the interaction with PNPLA3 in the absence of PLIN1/5 in 293A cells. As shown in Fig. 3c, SR-3420 increased the interaction between ABHD5 and PNPLA3 by greater than two-fold (p=0.0001), whereas the interaction between ABHD5 and PNPLA2/ATGL was increased less than 50% (p=0.0525) (Fig. 3c). The effect of SR-3420 occurred without changes in PNPLA3 protein levels (Supplementary Fig. 3c). Overall, these data demonstrate that the interaction between PNPLA3 and ABHD5 can be rapidly regulated by endogenous and synthetic ABHD5 ligands.
PNPLA3 competes with PNPLA2 for binding to ABHD5 and suppresses ABHD5-dependent lipolysis
The above data indicate that the interaction of ABHD5 with PNPLA3 is much stronger than its interaction with PNPLA2/ATGL and is similar to the interaction of ABHD5 with PLIN1 or PLIN5, which are bona fide suppressors of the interaction of ABHD5 with PNPLA2/ATGL20,23. These observations raised the possibility that PNPLA3 and PNPLA2/ATGL compete for ABHD5 and that overexpression of PNPLA3 would suppress ABHD5-dependent lipolysis.
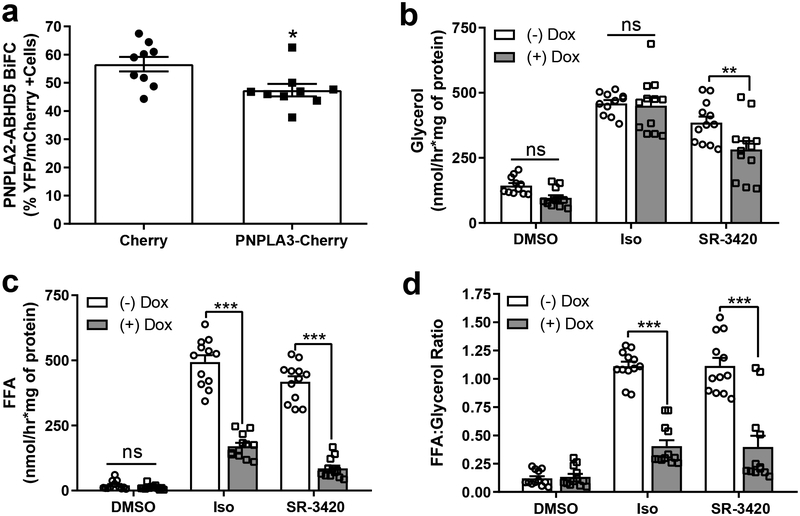
To initially test this hypothesis, we first determined whether PNPLA3 would inhibit the interaction of ABHD5 with PNPLA2/ATGL in lipid-loaded COS-7 cells, as assessed by competitive BiFC. Compared to mCherry negative control, expression of mCherry-tagged PNPLA3 significantly decreased fluorescence complementation between ABHD5 and PNPLA2/ATGL (Fig. 4a). Next we examined the effect of PNPLA3 expression in brown adipocytes (BAs), which provides an excellent model system to test the effect of PNPLA3 on PNPLA2/ATGL-dependent cellular lipolysis,1,27 since these cells lack detectable endogenous PNPLA3 (Supplementary Fig. 4a). To do so, we created a BA cell line in which the levels of PNPLA3 could be tightly controlled with doxycycline induction. Using glycerol release as an index, we found no significant effect of expression of PNPLA3-mCherry (+Dox) on complete lipolysis activated by isoproterenol (Fig. 4b), which triggers lipolysis indirectly by releasing ABHD5 from PLIN1 and directly by phosphorylating hormone sensitive lipase (HSL)21. In contrast, PNPLA3 expression reduced complete lipolysis induced by SR-3420 (Fig. 4b), a synthetic ABHD5 ligand that directly releases ABHD5 from PLIN complexes27 and promotes its interaction with PNPLA3 (Fig. 3c). Interestingly, PNPLA3 expression strongly reduced FFA efflux to both lipolysis activators (Fig. 4c) and suppressed the ratio of fatty acid to glycerol released for both isoproterenol and ABHD5-dependent lipolysis (Fig. 4d), indicating that PNPLA3 promotes FFA re-esterification and/or reduces complete hydrolysis of TAG (Fig. 4c). Doxycycline treatment alone did not affect lipolysis in a control cell line (Supplementary Fig. 4b). We confirmed previous work that ABHD5 promotes the TAG lipase activity of PNPLA2, but not PNPLA3, in an assay using artificial lipid droplets11,12 (Supplementary Fig. 4c). Overall, these data demonstrate that PNPLA3 can compete PNPLA2/ATGL for ABHD5 to suppress lipolysis.
Fig. 4. PNPLA3 competes for ABHD5 and suppresses PNPLA2-dependent lipolysis.
(a) Competitive BiFC assay in COS-7 cells transfected with Yn-ABHD5 and PNPLA2-Yc along with either mCherry or PNPLA3-mCherry and lipid-loaded overnight. The percentage of EYFP+ signal was determined in mCherry+ cells from three independent experiments performed with biological triplicates. *p=0.0145 for PNPLA3 vs. mCherry as determined by two-tailed t-test. (b and c) Control BA (-Dox) or BA expressing WT PNPLA3-mCherry (+Dox) were treated with DMSO, isoproterenol (Iso; 10 nM) or ABHD5 ligand (SR-3420, 20 μM) for one hour and glycerol (b) or free fatty acids (FFA; c) in the media were quantified. Data are biological quadruplicates from three independent experiments. (d) FFA to glycerol ratio as calculated from values in Fig. 4b and c **p=0.0044 (for 4b), and ***p<0.001 indicates a significant effect of PNPLA3 as determined by two-way ANOVA with Bonferroni post t-test. ns, not significant. Data are expressed as means +/− SEM.
PNPLA3 I148M is a gain-of-function mutation for the interaction with ABHD5
PNPLA3 I148M is a common genetic variant in humans13 that greatly increases the risk of developing FLD and its pathological sequelae29,30. Because total loss of PNPLA3 does not elevate hepatic TAG accumulation16, whereas overexpression of the I148M variant or catalytically-inactive S47A each promote hepatic steatosis15, the effect of the I148M mutation on TG accumulation likely involves a non-catalytic gain-of-function.
Both the I148M and S47A mutations increased the interaction of PNPLA3 with ABHD5 compared to the WT protein (Fig. 5a). We next tested whether the interaction between PNPLA3 and ABHD5 was conserved in humans. In luciferase protein complementation assays, human ABHD5 interacted with human PNPLA3, but not PNPLA4, a closely related PNPLA family member, and this interaction was increased with the PNPLA3 I148M mutation (Supplementary Fig. 5a). Interestingly, the increased interaction of mouse PNPLA3 I148M and S47A with ABHD5 was associated with greater cellular levels of the mutant proteins (Supplementary Fig. 5b), suggesting that LD targeting protects these mutants from proteosomal degradation14 and facilitates their interaction with ABHD5. Like WT PNPLA3 (Fig. 5b), acute (30 min) OA treatment increased the interaction of PNPLA3 I148M with ABHD5 (Fig. 5c) without altering expression levels (Supplementary Fig. 5c). Analysis of FRET and BiFC in transfected COS-7 cells confirmed that the interaction between PNPLA3 and ABHD5 occurred mostly on ER, whereas the interaction of ABHD5 with PNPLA3 I148M showed some localization to ER, with greater interaction occurring on LDs (Fig. 5d, e), (Fig. 5e; white arrows). Similar results were found in AML12 hepatocyte cells, where the interaction of PNPLA3 I148M with ABHD5, as detected by FRET, strongly localized to LDs (Supplementary Fig. 5d).
Fig. 5. The PNPLA3 I148M variant enhances the interaction between ABHD5 and PNPLA3 on lipid droplets.
(a) Gluc complementation assay in lipid-loaded HEK293A cells expressing GlucN-ABHD5 and either WT PNPLA3-GlucC, PNPLA3-I148M-GlucC, or PNPLA3 S47A-GlucC. Data are biological quadruplicates from three independent experiments. ***p=0.0001 and **p=0.0051 indicate a significant difference compared to WT PNPLA3, and *p=0.0479 indicates a significant difference between PNPLA3 I148M and S47A as determined by one-way ANOVA with Bonferroni post t-test. (b) and (c) HEK293A cells expressing GlucN-ABHD5 and PNPLA3-GlucC (b) or PNPLA3-I148M-GlucC (c) were treated as in Fig. 3b. Data are biological quadruplicates from three independent experiments. ***p=0.0001 indicates a significant difference between BSA and OA as determined by one-way ANOVA with Bonferroni post t-test. (d) FRET assay in COS-7 cells expressing ECFP-ABHD5 and either WT PNPLA3-EYFP or PNPLA3-I148M-EYFP and loaded with 200 μM OA overnight. Result are from one experiment and are representative of four independent experiments. (e) BiFC assay in COS-7 cells expressing Yn-ABHD5 and either PNPLA3-Yc or PNPLA3 I148M-Yc and loaded with 200 μM oleic acid overnight. Arrows indicate co-localization of BiFC (pseudocoloured green) signal with lipid droplets (pseudocoloured red). Lipid droplets were stained with LipidTox Red. Images are representative of three independent experiments. Data are expressed as means +/− SEM. Scale bars denotes 10 μm.
PNPLA3 I148M is a gain-of-function for suppressing ABHD5-dependent lipolysis
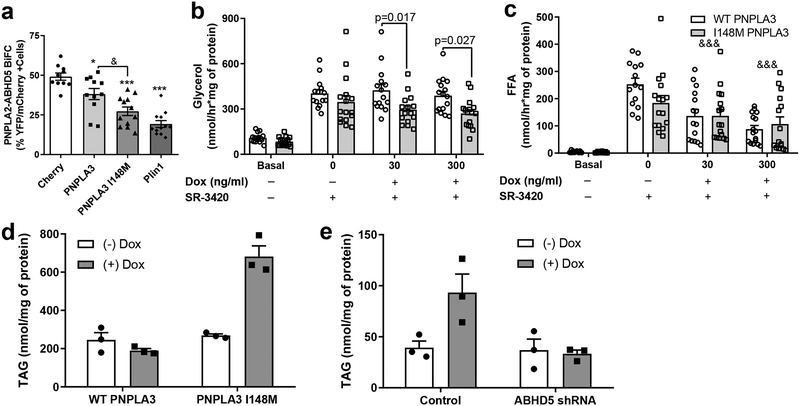
The greater interaction of the I148M variant with ABHD5 suggested that this mutation might suppress the interaction of ABHD5 with PNPLA2 more than WT PNPLA3, and thus represents a gain-of-function. Consistent with this expectation, we found that the I148M variant suppressed the interaction of ABHD5 with PNPLA2/ATGL in BiFC assays more than WT PNPLA3 (Fig. 6a). Furthermore, PNPLA3 I148M was nearly as effective as PLIN1, an established suppressor of ABHD5-ATGL interactions20 (Fig. 6a). We next examined the dose-responsive effects of WT PNPLA3 and PNPLA3 I148M overexpression on ABHD5-driven lipolysis in BA. Direct activation of ABHD5 (no Dox, + SR-3420) increased glycerol release (as a measure of complete lipolysis) by four fold in the absence of PNPLA3 expression (Basal; Fig. 6b). Submaximal (30 ng/ml) and maximal (300 ng/ml) induction of PNPLA3 expression by Dox produced greater suppression of glycerol release by the PNPLA3 I148M variant compared to WT PNPLA3 (Fig. 6b). Similarly, SR-3420 (no Dox, + SR-3420) stimulated fatty acid release by greater than 50-fold over basal in the absence of Dox-induced exogenous PNPLA3 expression in both cell lines (no Dox, no SR-3420). In contrast, Dox-induced expression of exogenous WT and PNPLA3 I148M reduced fatty acid release at both doses of Dox (&&&; main effect of doxycycline observed), however no significant difference was observed between WT and PNPLA3 I148M (Fig. 6c). The greater ability of the PNPLA3 I148M variant to reduce glycerol compared to FFA release was reflected in an increase in the molar ratio of FFA : glycerol when compared to WT PNPLA3 (Supplementary Fig. 6a). As expected from transfection experiments, the greater suppression of lipolysis by PNPLA3 I148M was associated with elevated levels of PNPLA3 I148M-mCherry (Supplementary Fig. 6b).
Fig. 6. PNPLA3 I148M variant is a gain of function for suppressing ABHD5-dependent lipolysis.
(a) Competitive BiFC assay in lipid-loaded COS-7 cells transfected with Yn-ABHD5 and PNPLA2-Yc along with either mCherry, WT PNPLA3-mCherry, PNPLA3 I148M-mCherry, or Plin1-mCherry. Data are biological quadruplicates from four independent experiments. *p=0.0397 and ***p=0.0001 indicates a difference compared to mCherry control, and &p<0.0312 indicates a significant difference between WT PNPLA3 and I148M as determined by one-way ANOVA with Tukey’s post t-test. Glycerol (b) and free fatty acid (FFA; c) release from Basal (no Dox, DMSO) or SR-3420 (20 μM, 1 hour) stimulated WT PNPLA3-mCherry or PNPLA3 I148M-mCherry brown adipocytes (BA) treated with differing doses of doxycycline (Dox; 0, 30, and 300 ng/ml). p values indicate a significant difference in glycerol release between WT PNPLA3 and PNPLA3 I148M as determined by two-way ANOVA with Bonferroni post t-test. &&&p=0.0001 indicates a main effect of doxycycline on FFA release as determined by two-way ANOVA. Data are biological quadruplicates from four independent experiments (d) Triacylglycerol (TAG) levels from control (-Dox) and doxycycline (+Dox) treated wildtype (WT) PNPLA3-mCherry and PNPLA3 I148M-mCherry BAs. (e) TAG levels from Control BA and BA cells with stable knockdown of ABHD5 (ABHD5 shRNA) expressing PNPLA3 I148M (+Dox). (d and e) Data are from one experiment with biological triplicates and are representative of three independent experiments. Data are expressed as means +/− SEM.
Expression of WT PNPLA3 did not have any obvious effect on BA morphology, whereas expression of PNPLA3 I148M promoted the appearance of large LDs (Supplementary Fig. 6c). Consistent with imaging results, PNPLA3 I148M, but not WT PNPLA3, greatly promoted cellular triglyceride accumulation (Fig. 6d). Stable shRNA knockdown of ABHD5 effectively reduced ABHD5 protein levels (Supplementary Fig. 6d) and prevented the ability of the PNPLA3 I148M to increase TAG levels (Fig. 6e). Unexpectedly, knockdown of ABHD5 itself did not increase steady state TAG levels (Fig. 6e), suggesting possible synergistic interactions beyond simple suppression of lipolysis. One possible synergistic mechanism that we considered was lysophosphatidic acid acyltransferase (LPAAT) activity, which has been attributed to both proteins12,31. We observed that expression of PNPLA3 slightly increased LPAAT activity of LDs from BA cells (ANOVA main effect of Dox-induced expression, p = 0.032). However, this overall effect was not significantly modified by the I148M variant or by deficiency of ABHD5 (shABHD5) (Supplementary Fig. 6e).
Discussion
PNPLA2/ATGL and PNPLA3/Adiponutrin arose from the duplication of an ancestral lipase that was likely activated by ABHD5/CGI-587. Interestingly, the differential nutritional regulation suggests these PNPLA paralogs have opposing functions on TAG metabolism. Thus, PNPLA2 is well-established as a critical lipase that mobilizes fatty acids during fasting and is under the control of the fasting-induced transcription factors like FOXO132. In contrast, PNPLA3 is induced by feeding and is upregulated by lipogenic transcription factors like SREBP125,26,33 and ChREBP19,33, suggesting that it might function to promote neutral lipid synthesis and retention.
While the role of PNPLA2/ATGL as a critical intracellular lipase is well established, the biochemical function(s) of PNPLA3 remain poorly understood and controversial. An important insight into the biological role of PNPLA3 was provided in a landmark association study from the Cohen and Hobbs lab demonstrating that a common variant, I148M, greatly increases the risk of FLD13. One pathway by which PNPLA3 I148M promotes TAG accumulation is likely through suppression of lipolysis19,34; however, the molecular mechanism for this suppression was not determined. Because expression of the catalytically inactive S47A mutant phenocopies the I148M variant15, but complete absence of PNPLA3 does not16, PNPLA3 could promote FLD in part by sequestering an upstream activator of lipolysis10,34. A potential role of ABHD5 as such an activator was first suggested by Smagris et al.15 who noted that ABHD5 accumulates on hepatic LDs in knock-in mice expressing the S47A or I148M variant. Chamoun et al. suggested that PNPLA2, PNPLA3 and ABHD5 traffic together to allow targeting of PNPLA2/ATGL to LDs35, however, the potential interaction between PNPLA3 and ABHD5 was not tested.
Our experiments demonstrate that PNPLA3 tightly binds ABHD5, and that this interaction is greatly facilitated by conditions that promote triglyceride accumulation. Furthermore, the interaction of ABHD5 with PNPLA3 was as strong as its interaction with PLIN1 or PLIN5, which are well-established suppressors of PNPLA2/ATGL-dependent lipolysis, and was much stronger than the interaction of ABHD5 with PNPLA2/ATGL. As predicted from these observations, PNPLA3 effectively competed with PNPLA2/ATGL for the interaction with ABHD5. Furthermore, the PNPLA3 I148M variant was more effective in this competition, owing in part to its greater level of expression and accumulation on LD surfaces. Indeed, Donati et al.36 suggested the elevated levels of I148M are an important causative factor in FLD, as a protective variant of PNPLA3 (E434K) reduces PNPLA3 I148M protein levels and decreases the risk of hepatosteatosis. The increased stability and accumulation of the I148M variant14 and its targeting to LDs led to greater sequestration of ABHD5 and suppression of PNPLA2/ATGL-dependent lipolysis, representing a gain of an inhibitory function. This enhanced indirect suppression of PNPLA2, combined with loss of intrinsic lipase activity10,11 likely explains the pronounced accumulation of TAG produced by PNPLA3 I148M expression in BAs. Surprisingly, whole body deletion of PNPLA3 was not found to increase lipolysis in isolated white adipocytes16, which might be explained by a compensatory upregulation of other proteins, including PLIN5, PLIN1 and PNPLA516,17, that might bind ABHD5.
Suppression of lipolysis by PNPLA3 is one mechanism for promoting cellular TAG accumulation; however, it seems reasonable to speculate that the interaction of ABHD5 with PNPLA3 might facilitate TAG synthesis as well, for example, by facilitating subcellular targeting to LD and/or promoting additional enzymatic activities of PNPLA312,37,38, ABHD531,39, or potentially other proteins. Complete TAG hydrolysis involves breakdown to fatty acids and glycerol, however, as much as two-thirds of mobilized fatty acids can be re-esterified40,41. Our results indicate that PNPLA3 greatly suppresses FFA efflux by combined suppression of TAG hydrolysis and promotion of FFA re-esterification. The mechanism for promoting re-esterification are presently unclear; however this could involve transacylase38,42 or acyltransferase activities that have been attributed to PNPLA3. In this regard, we were able to confirm that PNPLA3 expression modestly elevates LPAAT activity; however, this effect was not significantly influenced by the I148M variant or ABHD5 expression.
The degree to which the interaction of PNPLA3 with ABHD5 contributes to hepatosteatosis was not assessed by our experiments. Nonetheless, we found that expression of the I148M variant in BAs promoted TAG accumulation, and this effect required ABHD5. It is likely that the role of ABHD5-PNPLA3 interactions in TAG metabolism will vary among cell types depending on subcellular targeting (ER and LD), the specific complement of interacting proteins (e.g., PLIN1/5, PNPLA1/2/3) and levels of endogenous ligands/substrates (acyl-CoAs). In this regard, minimal artificial assays that are often used to define enzymatic activities might lack the needed cellular context for fully assessing the dynamic role of PNPLA3 in lipolysis regulation and acyl-CoA metabolism.
Our results also have implications regarding functions of ABHD5 that are independent of PNPLA2/ATGL. The tissue expression pattern of ABHD5 only partially overlaps with that of PNPLA2/ATGL and it is now clear that certain metabolic effects of ABHD5 can be fully dissociated from activation of PNPLA2/ATGL5,6. Indeed, recent work identified that PNPLA1 and ABHD5 interact and function in concert in the production of acyl-ceramides, critical in maintaining proper skin barrier43,44. The present results demonstrate that ABHD5 interacts with PNPLA3 and thus might operate more broadly as an allosteric regulator of certain PNPLA family members. Our data also potentially explain the interaction between a genetic pathway (rs738409, PNPLA3 I148M) and diet in promoting FLD45, as the greater availability of acyl-CoAs from diets high in fat and sugar would promote the interaction between ABHD5 and PNPLA3 I148M to channel fatty acids into storage. Finally, the demonstration that highly-selective synthetic ABHD5 ligands27 can directly and indirectly regulate interactions with PNPLA family members raises the possibility that ABHD5-dependent processes might be targeted for treatment of metabolic diseases.
Methods
Mice.
C57BL/6J mice were purchased from Jackson Laboratory, Stock Number 00064 (Bar Harbor, ME). All mice were housed at 24 °C +/− 2 °C with a 12:12 light-dark cycle in an AALAC-approved animal facility at Wayne State University (Detroit, MI). All protocols involving animals were approved by the Institutional Animal Care and Use Committee of Wayne State University and followed the National Institutes of Health Guide for the care and use of Laboratory Animals. Male mice between 8 and 12 weeks of age were fed a high sucrose diet (74% kCal from sucrose, MP Biomedicals, Santa Ana, CA) for one week followed by an overnight fast (7PM-7AM) or fed ad libitum prior to sacrifice.
Cell Lines.
HEK293A, AML12 and COS-7 cells were purchased from ATCC. Mouse brown preadipocyte cells were a gift from Dr. Bruce Spiegelman (Harvard University) and confirmed by their ability to accumulate lipid droplets in response to a differentiation protocol (see below). All cells were maintained in DMEM High Glucose (Hyclone GE Lifesciences) with 10% FCS (Atlanta Biologicals) and Penicillin/Streptomycin (Hyclone, GE Lifesciences). Preadipocytes were induced by treating cells with induction medium (20 nM insulin, 1 nM T3, 0.5 mM isobutylmethylxanthine, 0.5 μM dexamethasone, and 0.125 mM indomethacin). After induction for 48 hours, cells were maintained in differentiation media (20 nM insulin, 1 nM T3) until complete differentiation at 7 days.
Generation of Fluorescent Fusion Proteins and Protein Complementation Constructs.
Mouse PNPLA3 cDNA was amplified from brown adipose tissue using primers 5’-CGC CCA AGC TTC GGC CAC CAT GTA TGA CCC AGA GCG CC-3’ and 5’-CGC CCA CCG GTG GCC TGG TAG AGG GGA GCA GG-3’. The I148M variant was generated using PCR primers 5’-GAT GCC CTG GTG TGT TCC TGC TTC ATG CCC TCT CT-3’ and 5’-GGA TTA GGC CAG GAA GAG GGC CAT GAA GCA GGA ACA C-3’. The S47A mutation was generated using the PCR primers 5’-GAT GCG CGC ACT TTC TTT GGC TGC GCG GCC GGC TGC GCG GCC GGT GCA CTG C-3’ and 5’-GAC CGG TGC GTG CAC CGG CCG CGC AGC CAA AGA AAG TG-3’. After amplification, the PCR products were cloned in frame into EYFP-N1, CFP-N1, and mCherry-N1 (Clontech). The human WT PNPLA3 cDNA and I148M variant were PCR amplified from plasmids provided by Dr. Wanqing Liu (Wayne State University). All sequences were confirmed by DNA sequencing. Gaussia Princeps luciferase complementation constructs were created by substituting split luciferase fragments for the full-length fluorescent proteins above. The N (amino acids 1–92)- and C (amino acids 93–187)-terminal fragments, designated GlucN and GlucC respectively. GlucN was cloned in-frame onto the N terminus of ABHD5 while GlucC was cloned in-frame onto the C termini of the PNPLA3 proteins. Bimolecular fluorescence complementation (BiFC) constructs were created by substituting split fragments of EYFP for the full-length fluorescent proteins. The N-terminal fragment, designated Yn, contained amino acids 1–158, whereas the C-terminal fragment, designated Yc, contained amino acids 155–239. The Yn was cloned in-frame onto the N terminus of ABHD5 while the Yc was cloned in-frame on the C termini of the PNPLA3 proteins.
Co-localization and Immunofluorescence.
COS-7 cells were transfected on 25 mm coverslips with indicated constructs using Lipofectamine LTX (ThermoFisher) and were lipid-loaded overnight with 200 μM oleic acid/BSA complexes. The following day cells were washed with PBS and fixed with 4% paraformaldehyde in PBS. Cells were then permeabilized and blocked with 5% normal goat serum, 0.1% Triton-X 100 for 30 min, and stained with antibodies against RCAS1 (Golgi marker; 1:100; Cell Signaling Technology) or PDI (ER marker; 1:100; Cell Signaling Technology) for 1 hr. Coverslips were washed three times for 10 min. with the same buffer and stained for 45 min with Alexa Fluor 647 (ThermoFisher) or DyLight 649-conjugated (Jackson ImmunoResearch) goat anti-rabbit secondary. Coverslips were washed three times 10 min in block buffer and imaged with a Zeiss LSM 800 laser scanning microscope equipped with an Airyscan detector using a 63× Plan-Apochromat oil immersion lens (1.4NA DIC M27) in the Microscopy, Imaging & Cytometry Resources (MICR) Core at Wayne State University, School of Medicine. EYFP was captured with a 488 nm laser (525 Excitation, 538 Emission), mCherry with a 561 nm laser (587 Excitation, 610 Emission) and AF-647/DyLight-649 with a 640 nm laser (653 Excitation, 668 Emission).
Lentiviral production and Generation of Stable Cells.
For construction of lentiviral vectors, mouse WT PNPLA3-mCherry and PNPLA3 I148M-mCherry were cloned into pINDUCER20 using Gateway LR Clonase II (Invitrogen). 100 ng of either WT-PNPLA3-mCherry or PNPLA3 I148M-mCherry were incubated with 150 ng of pINDUCER20 and LR clonase II enzyme for 3 hours at room temperature. For lentiviral packaging, pINDUCER20 was co-transfected with pMD2.G and psPAX2 packaging vectors into HEK293T cells using lipofectamine LTX and plus reagent (Invitrogen). 48 h and 72 h post transfection, virus-containing culture media was harvested and passed through 0.45 μM filters to remove debris. To collect the virus, the media was centrifuged at 48,000 × g for 2 hours 4°C in a Beckman 25.50 fixed angle rotor, and the virus pellet was resuspended in OPTIMEM. For stable overexpression of WT PNPLA3-mCherry and PNPLA3 I148M-mCherry, mouse brown adipocytes were infected and with pINDUCER20-WT PNPLA3-mCherry or pINDUCER20-PNPLA3 I148M-mCherry virus for 24 hours followed by G418 selection (500 μg/mL) for one week and further selected for mCherry expression by fluorescence-activated cell sorting (FACS) at the Microscopy, Imaging and Cytometry Resources Core at Wayne State School of Medicine.
Gaussia Luciferase (Gluc) Protein Complementation Assay.
HEK293A or AML12 cells in 24 well plates were transfected with Lipofectamine LTX (ThermoFisher) overnight in quadruplicate with 250 ng GlucN-ABHD5 and either 250 ng WT PNPLA3-GlucC, PNPLA3 I148M-GlucC, PNPLA2-GlucC, PLIN5-GlucC, or Rab18- GlucC as specified in the figure legends. To determine luciferase activity, cells were washed once with PBS and suspended in intracellular buffer (IB: 10 mM HEPES (pH 7.3), 140 mM KCl, 6 mM NaCl, 1 mM MgCl2, 2 mM EGTA; 150 μL/ well) followed by repeated freezing at −80°C and thawing at room temperature three times. Lysates (100 μL) were transferred to 96 well white plates, and luminescence was read for 2 seconds after the addition of 100 μL of 20 μM coelenterazine substrate (Nanolight) using a Clariostar luminometer.
Fluorescence Energy Transfer (FRET).
COS-7 cells in 6 well plates with coverslips were transfected using Lipofectamine LTX (ThermoFisher) with 1.25 μg CFP-ABHD5 along with 1.25 μg WT PNPLA3-EYFP, PNPLA3 I148M-EYFP, or G0S2-EYFP. Cells were incubated with 200 μM oleic acid/BSA (OA) overnight to promote lipid droplet formation. The next day, the cells were imaged in Krebs-Ringer Bicarbonate Buffer containing HEPES (H-KRBB) supplemented with 1% Bovine Serum Albumin (BSA) (Alkali Scientific Inc.). Images for FRET were acquired using an Olympus IX-81 microscope equipped with a spinning disc confocal unit. Microscope control and data acquisition were performed using CellSens Dimensions (Olympus) software. FRET was performed using the three-filter method and the net FRET (nFRET) was calculated using the FRET extension of the CellSens Dimensions software. Lipid droplets were stained with LipidTox Red (1:2500; ThermoFisher) in DMEM/F12.
Bimolecular Fluorescence Complementation (BiFC).
COS-7 cells were transfected in 12-well plates with coverslips using Lipofectamine LTX (ThermoFisher) with 100 ng mCherry tracer to identify transfected cells, 500 ng Yn-ABHD5, and either 500 ng WT PNPLA3-Yc, PNPLA3 I148M-Yc, PLIN5-Yc, or bFOS-Yc as specified in the figure legend. Cells were incubated at 30°C with 200 μM OA overnight to promote lipid droplet formation. The next day, the cells were fixed with 4% paraformaldehyde before being imaged in phosphate buffered saline (PBS). Reconstituted EYFP signal in mCherry+ cells were acquired using an Olympus IX-81 microscope equipped with a spinning disc confocal unit. Competitive BiFC20 was performed in COS-7 cells seeded in 12-well plates with coverslips. Cells were co-transfected with 400 ng Yn-ABHD5 and 400 ng PNPLA2-Yc, and either 200 ng mCherry, PNPLA3-mCherry, PNPLA3 I148M-mCherry, or Plin1-mCherry (4:4:2 ratio of Yn:Yc:mCherry). Cells were incubated with 400 μM OA overnight to promote lipid droplet formation. Reconstituted EYFP signal in mCherry+ cells were acquired using an Olympus IX-81 microscope equipped with a spinning disc confocal unit in a blind manner. Five random 40× fields were captured for each coverslip, and cells were scored for the presence of EYFP fluorescence in mCherry+ cells by an analyst who was blinded to the experimental conditions.
Western Blot Analysis.
Total cell proteins were extracted with RIPA buffer (Teknova) supplemented with protease inhibitor tablet (Pierce). Samples were run on 10% Mini-Protean TGX Gels (BioRad), transferred to PVDF membranes and blocked in 5% nonfat milk (Cell Signaling Technology) for 1 hour. The membranes were incubated with rabbit anti-Gaussia luciferase (Nanolight, Cat#401P, 1:5000) at room temperature for 1 hour. The membranes were washed three times over 20 minutes with PBST (PBS containing 0.1% Tween 20) before being incubated with horseradish peroxidase (HRP) -coupled anti-rabbit immunoglobulin G (Cell Signaling Technology, Cat#7470, 1:10,000) for 45 minutes. After four washes over 30 minutes with PBST, the immunolabeled protein bands were detected by enhanced chemiluminescence (ECL) method (Thermo Scientific). Membranes were then stripped with Restore stripping buffer (Thermo Scientific) and re-probed with rabbit anti-α/β tubulin (Cell Signaling Technology, Cat #2148S, 1:1000) and subsequently HRP-coupled anti-rabbit immunoglobulin G (Cell Signaling Technology, Cat #7470, 1:10,000). For the detection of endogenous and exogenous PNPLA3 in the BA cell line, samples were processed as above, and blots incubated with rabbit anti-PNPLA3 (Abcam, Cat#ab81874, 1:1000) overnight at 4°C and detected as above.
Co-Immunoprecipitation of ABHD5 and PNPLA3.
Mice were fed a high sucrose diet as described above and lipid droplets were purified from brown adipose tissue (BAT) and liver15. BAT and liver were excised and washed in ice cold PBS, homogenized in buffer A (250 mM sucrose, 20 mM HEPES, pH 7.8), spun at 100 × g and the supernatant was transferred to an ultracentrifuge tube. Buffer B (100 mM KCl, 2 mM MgCl2, 20 mM HEPES, pH 7.4) was overlaid and the tubes were centrifuged at 18,000 × g for 60 minutes at 4°C in a swinging bucket rotor (SW40). The purified lipid droplets resuspended with Buffer C (20 mM HEPES, 150 mM NaCl, pH 7.4) and then solubilized in immunoprecipitation (IP) Buffer (50 mM Tris, pH 7.4, 150 mM NaCl, 0.5% Nonidet P-40, 1 mM EDTA, 1 mM EGTA, 2 mM NaF, 5 mM sodium glycerophosphate, plus protease inhibitor tablet). A total of 30 μg of protein was incubated with 4 μg rabbit anti ABHD5 (Proteintech) or rabbit IgG (Vector Labs) overnight at 4°C. 25 μL protein A/G agarose (Pierce) beads was added to the protein/antibody mixture and allowed to incubate at 4°C for 2 hours. Beads were washed four times with IP Buffer, boiled, and immunoblotting was performed. For PNPLA3 immunoblotting, membranes were incubated for 1 hour at room temperature with goat anti-PNPLA3 antibody diluted in blocking solution (Everest, Cat#EB08402, 1:10,000). To avoid interference with the immunoglobulin heavy chain, light chain specific mouse anti-goat secondary antibody (Jackson ImmunoResearch, Cat#205-032-176, 1:10,000) was used for detection. The blot was stripped with Restore (ThermoFisher) stripping buffer and re-probed with mouse anti-ABHD5 (Abnova, Cat#H00051099-M01, 1:1000) and subsequently peroxidase linked donkey light chain specific anti mouse secondary (Jackson ImmunoResearch, 1:10,000).
Triglyceride Quantification.
Doxycycline inducible WT PNPLA3-cherry and PNPLA3 I148M-cherry mouse brown adipocytes were seeded on 12 well plates and induced to differentiate as described above. After 2 days, cells were maintained in differentiation media containing no doxycycline or 2 μg/mL of doxycycline for 3 days. Cells were homogenized in distilled water and neutral lipids were extracted three times with isooctane: ethyl-acetate (1:9; Fisher Scientific). The organic layer containing neutral lipids was collected and dried under N2. Triglycerides were resuspended in 5% NP-40, and triglyceride content was measured using a triglyceride determination kit (Sigma). For determining the effect of PNPLA3 I148M-mCherry on triacylglycerol content between control BA cells and those with stable knockdown of ABHD5, the cells were treated with 100 ng/mL and 300 ng/mL of doxycycline, respectively, in order to match expression levels of PNPLA3 I148M-mCherry.
Brown Adipocyte Lipolysis Assays.
Doxycycline inducible WT PNPLA3-cherry and PNPLA3 I148M-cherry mouse brown adipocytes (BAs) were seeded on either 12 well or 24 well plates as noted. After 48 hours of induction medium, the cells were grown in differentiation media for 24 hours, followed by media containing no doxycycline or amount of doxycycline specified for a total of 4 days. After one week of differentiation, the cells were stimulated with specified treatment of either vehicle, SR-3420, or isoproterenol at the indicated concentration in H-KRBB buffer for 1 hour. Free fatty acid release was determined using WAKO kit (Wako Pure Chemicals Industries) adapted to fluorescence detection with Amplex Red (Cayman Chemical). Glycerol release was determined using glycerol reagent from triglyceride determination kit (Sigma).
In vitro lipase assay of triolein substrate.
COS-7 cells in 10 cm plates were transfected (Lipofectamine LTX; ThermoFisher) individually with mCherry, ABHD5-mCherry, PNPLA2(ATGL)-EYFP or PNPLA3-EYFP. The next day, cells were washed and scraped in PBS and lysed in Solution A (20 mM Hepes, 0.25 M sucrose, 1 mM EDTA, 1 mM DTT, pH 7.3, 1 μg/ml pepstatin, 20 μg/ml leupeptin; 1 ml per plate for untransfected COS-7, PNPLA2-EYFP, mCherry, and ABHD5-mCherry; and 0.5 ml per plate for PNPLA3-EYFP) with sonication on ice for 10 sec. Lysates were clarified by centrifugation at 15,000 × g for 20 min and were read for fluorescent intensity on a Clariostar plate reader and normalized to equal fluorescent levels for EYFP and mCherry and with COS-7 control lysate. Artificial lipid droplets were prepared as described46 and consisted of 200 μM phosphatidylcholine:phosphatidylinositol (3:1; Sigma-Aldrich) in chloroform and 500 μM Triolein as substrate (Sigma-Aldrich). Artificial lipid droplets were prepared in 0.1 M potassium phosphate buffer pH 7.0 containing 5% BSA with sonication on ice for 2 times 10 sec bursts using a Heat Systems Sonicator set at 10% output power (Ultrasonics Inc.). 500 μl of artificial lipid droplets were first incubated with mCherry or ABHD5-mCherry for 30 min, spun at 12,000 × g for 5 min to float lipid droplets, and 400 ul of the lower buffer was removed. The 100 ul of concentrated lipid droplets containing mCherry or ABHD5 was incubated with 100 ul of PNPLA2-EYFP, PNPLA3-EYFP, or COS-7 control lysate in a shaking water bath for 1 hour at 37°C. Free fatty acids in the buffer were quantified as above for BA lipolysis assays.
Liquid chromatography–mass spectrometry (LC-MS) analysis of lysophosphatidic acyltransferase (LPAAT) activity.
LDs were purified from BA cell lines as above for BAT and liver except 50 mM Tris pH 7.5 was used instead of Hepes buffer and LDs were centrifuged for 30 min in a Beckman S241.5 swinging-bucket rotor. 10 μg of purified LDs were incubated with 100 μM 1-Oleoyl Lysophosphatidic Acid (LPA, Cayman Chemical), 50 μM oleoyl-CoA (Avanti Lipids) and 10 μM 13C18 Oleoyl-CoA (Sigma-Isotec) for 15 min at 37°C in a 200 μl reaction. The reaction was stopped by adding 0.5ml of iso-octatane/ethyl acetate (9:1) containing 100 ng of C17 phosphatidic acid (PA) as an internal standard. Neutral lipids were removed by performing three extractions with of iso-octatane/ethyl acetate on the aqueous phase. The aqueous phase containing phosphatidic acid was extracted with an acidified Bligh-Dyer47 method with 2% formic acid. The organic phase was dried down with nitrogen gas and resuspended in 500 μl of chloroform: methanol (1:2) and transferred to MS auto-sampling vials.
Extracted samples were dried under nitrogen and reconstituted in methanol-0.1% aqueous ammonium bicarbonate (8:2). The reconstituted samples were subjected to LC-MS analysis to quantify [13C18]-oleic acid containing PA. Mass spectrometric survey scan of the assay extract showed PA (18:1/[13C18]18:1) as the product of the enzymatic reaction with m/z 717 in the negative ion mode. Hence, the samples were monitored by MRM 717→153 and 717→299. PA (17:0/17:0) used as internal standard was monitored by MRM 675→153 and 675→269. HPLC was performed on a Targa C8 (2×20 mm, 5μ) column using methanol-0.1% aqueous ammonium bicarbonate as mobile phase (solvent A: water-methanol (90:10); solvent B: water-methanol (5:95), both containing 0.1% ammonium bicarbonate). The column was eluted with a gradient as follows: 0 min – 80%B, 2 min – 100%B, 6 min – 100%B, at 0.5 ml/min. The column eluent was directly introduced to the TurboV ion source of QTRAP5500 mass analyzer (Sciex). The mass analyzer parameters were Ionization potential: −4500V, Curtain gas, GS1 & GS2: 35 psi, source temperature: 600 °C, CAD gas: low, Declustering potential: −150V, Collision energy: −42V, Entrance and Collision cell exit potentials: −10V. The retention times of PA (17:0/17:0) and PA (18:1/[13C18]18:1) were 2.36 and 2.26 min, respectively.
Quantification and Statistical Analysis.
Results are presented as the Mean +/− SEM (standard error of the mean) for experiments with 3–4 independent biological repeats. Statistical tests were two-sided. Unpaired two-tailed Student’s t-test were used for pairwise comparisons, one-way ANOVA was used for multiple comparison with Bonferroni’s or Tukey’s post t-test for selected comparisons or all pairwise comparison respectively, and two-way ANOVA with Bonferroni’s post t-test for multiple comparisons with two groups. The statistical parameters used for the specific sets of data are further described in the figure legends. Statistical analysis was performed using the Prism 6 or 8 (Graphpad) software.
Reporting Summary.
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Data Availability.
All data generated or analyzed during this study are included in this article and its Supplementary Information files or are available from the corresponding authors upon reasonable request.
Supplementary Material
Acknowledgements
We thank Drs. Jian Wang and Matt Sanders for critical discussions and Dr. Wanqing Liu for providing plasmids containing human PNPLA3. This work was supported by NIH grants, F30-DK116529–01A1 to A.Y., K99-DK114471 to E.P.M, and DK76629 and DK105963 to J.G.G. The Microscopy, Imaging and Cytometry Resources Core is supported, in part, by NIH Center grant P30 CA022453 to the Karmanos Cancer Institute at Wayne State University, and the Perinatology Research Branch of the National Institutes of Child Health and Development at Wayne State University. The Lipidomics Core Facility at Wayne State University was supported in part by National Center for Research Resources, National Institutes of Health Grant S10RR027926.
Footnotes
Competing Interests Statement
The authors declare no competing interests.
References
- 1.Haemmerle G et al. Defective lipolysis and altered energy metabolism in mice lacking adipose triglyceride lipase. Science 312, 734–737 (2006). [DOI] [PubMed] [Google Scholar]
- 2.Lass A et al. Adipose triglyceride lipase-mediated lipolysis of cellular fat stores is activated by CGI-58 and defective in Chanarin-Dorfman Syndrome. Cell Metab 3, 309–319 (2006). [DOI] [PubMed] [Google Scholar]
- 3.Sztalryd C & Brasaemle DL The perilipin family of lipid droplet proteins: Gatekeepers of intracellular lipolysis. Biochim. Biophys. Acta 1862, 1221–1232 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schweiger M, Lass A, Zimmermann R, Eichmann TO & Zechner R Neutral lipid storage disease: genetic disorders caused by mutations in adipose triglyceride lipase/PNPLA2 or CGI-58/ABHD5. Am. J. Physiol. - Endocrinol. Metab 297, E289 (2009). [DOI] [PubMed] [Google Scholar]
- 5.Lord CC et al. Regulation of Hepatic Triacylglycerol Metabolism by CGI-58 Does Not Require ATGL Co-activation. Cell Rep 16, 939–949 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Xie M & Roy R The Causative Gene in Chanarian Dorfman Syndrome Regulates Lipid Droplet Homeostasis in C. elegans. PLoS Genet 11, e1005284 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Holmes R Comparative studies of adipose triglyceride lipase genes and proteins: an ancient gene in vertebrate evolution. Open Access Bioinforma 15 (2012). doi: 10.2147/OAB.S27508 [DOI] [Google Scholar]
- 8.Baulande S, Lasnier F, Lucas M & Pairault J Adiponutrin, a Transmembrane Protein Corresponding to a Novel Dietary- and Obesity-linked mRNA Specifically Expressed in the Adipose Lineage. J. Biol. Chem 276, 33336–33344 (2001). [DOI] [PubMed] [Google Scholar]
- 9.Kershaw EE et al. Adipose triglyceride lipase: function, regulation by insulin, and comparison with adiponutrin. Diabetes 55, 148–157 (2006). [PMC free article] [PubMed] [Google Scholar]
- 10.He S et al. A Sequence Variation (I148M) in PNPLA3 Associated with Nonalcoholic Fatty Liver Disease Disrupts Triglyceride Hydrolysis. J. Biol. Chem 285, 6706–6715 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huang Y, Cohen JC & Hobbs HH Expression and Characterization of a PNPLA3 Protein Isoform (I148M) Associated with Nonalcoholic Fatty Liver Disease. J. Biol. Chem 286, 37085–37093 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kumari M et al. Adiponutrin Functions as a Nutritionally Regulated Lysophosphatidic Acid Acyltransferase. Cell Metab 15, 691–702 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Romeo S et al. Genetic variation in PNPLA3 confers susceptibility to nonalcoholic fatty liver disease. Nat. Genet 40, 1461–1465 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.BasuRay S, Smagris E, Cohen JC & Hobbs HH The PNPLA3 variant associated with fatty liver disease (I148M) accumulates on lipid droplets by evading ubiquitylation: Basuray et al. Hepatology 66, 1111–1124 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Smagris E et al. Pnpla3I148M knockin mice accumulate PNPLA3 on lipid droplets and develop hepatic steatosis. Hepatology 61, 108–118 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Basantani MK et al. Pnpla3/Adiponutrin deficiency in mice does not contribute to fatty liver disease or metabolic syndrome. J. Lipid Res 52, 318–329 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen W, Chang B, Li L & Chan L Patatin-like phospholipase domain-containing 3/adiponutrin deficiency in mice is not associated with fatty liver disease. Hepatology 52, 1134–1142 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li JZ et al. Chronic overexpression of PNPLA3I148M in mouse liver causes hepatic steatosis. J. Clin. Invest 122, 4130–4144 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Perttila J et al. PNPLA3 is regulated by glucose in human hepatocytes, and its I148M mutant slows down triglyceride hydrolysis. AJP Endocrinol. Metab 302, E1063–E1069 (2012). [DOI] [PubMed] [Google Scholar]
- 20.Patel S, Yang W, Kozusko K, Saudek V & Savage DB Perilipins 2 and 3 lack a carboxy-terminal domain present in perilipin 1 involved in sequestering ABHD5 and suppressing basal lipolysis. Proc. Natl. Acad. Sci. U. S. A 111, 9163–9168 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Granneman JG & Moore H-PH Location, location: protein trafficking and lipolysis in adipocytes. Trends Endocrinol. Metab 19, 3–9 (2008). [DOI] [PubMed] [Google Scholar]
- 22.Yamaguchi T, Omatsu N, Matsushita S & Osumi T CGI-58 Interacts with Perilipin and Is Localized to Lipid Droplets: POSSIBLE INVOLVEMENT OF CGI-58 MISLOCALIZATION IN CHANARIN-DORFMAN SYNDROME. J. Biol. Chem 279, 30490–30497 (2004). [DOI] [PubMed] [Google Scholar]
- 23.Granneman JG, Moore H-PH, Mottillo EP & Zhu Z Functional interactions between Mldp (LSDP5) and Abhd5 in the control of intracellular lipid accumulation. J. Biol. Chem 284, 3049–3057 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kerppola TK Design and implementation of bimolecular fluorescence complementation (BiFC) assays for the visualization of protein interactions in living cells. Nat. Protoc 1, 1278–1286 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Huang Y et al. A feed-forward loop amplifies nutritional regulation of PNPLA3. Proc. Natl. Acad. Sci 107, 7892–7897 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Qiao A et al. Mouse patatin-like phospholipase domain-containing 3 influences systemic lipid and glucose homeostasis. Hepatol. Baltim. Md 54, 509–521 (2011). [DOI] [PubMed] [Google Scholar]
- 27.Sanders MA et al. Endogenous and Synthetic ABHD5 Ligands Regulate ABHD5-Perilipin Interactions and Lipolysis in Fat and Muscle. Cell Metab 22, 851–60 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rondini EA et al. Novel Pharmacological Probes Reveal ABHD5 as a Locus of Lipolysis Control in White and Brown Adipocytes. J. Pharmacol. Exp. Ther 363, 367–376 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cohen JC, Horton JD & Hobbs HH Human fatty liver disease: old questions and new insights. Science 332, 1519–1523 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dongiovanni P et al. PNPLA3 I148M polymorphism and progressive liver disease. World J. Gastroenterol 19, 6969–6978 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ghosh AK, Ramakrishnan G, Chandramohan C & Rajasekharan R CGI-58, the Causative Gene for Chanarin-Dorfman Syndrome, Mediates Acylation of Lysophosphatidic Acid. J. Biol. Chem 283, 24525–24533 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chakrabarti P et al. SIRT1 controls lipolysis in adipocytes via FOXO1-mediated expression of ATGL. J. Lipid Res 52, 1693–1701 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dubuquoy C et al. Distinct regulation of adiponutrin/PNPLA3 gene expression by the transcription factors ChREBP and SREBP1c in mouse and human hepatocytes. J. Hepatol 55, 145–153 (2011). [DOI] [PubMed] [Google Scholar]
- 34.Li JZ et al. Chronic overexpression of PNPLA3I148M in mouse liver causes hepatic steatosis. J. Clin. Invest 122, 4130–4144 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chamoun Z, Vacca F, Parton RG & Gruenberg J PNPLA3/adiponutrin functions in lipid droplet formation. Biol. Cell 105, 219–233 (2013). [DOI] [PubMed] [Google Scholar]
- 36.Donati B et al. The rs2294918 E434K variant modulates patatin-like phospholipase domain-containing 3 expression and liver damage. Hepatology 63, 787–798 (2016). [DOI] [PubMed] [Google Scholar]
- 37.Kumashiro N et al. Role of patatin-like phospholipase domain-containing 3 on lipid-induced hepatic steatosis and insulin resistance in rats. Hepatology 57, 1763–1772 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mitsche MA, Hobbs HH & Cohen JC Patatin-like phospholipase domain–containing protein 3 promotes transfers of essential fatty acids from triglycerides to phospholipids in hepatic lipid droplets. J. Biol. Chem 293, 6958–6968 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Montero-Moran G et al. CGI-58/ABHD5 is a coenzyme A-dependent lysophosphatidic acid acyltransferase. J. Lipid Res 51, 709–719 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Edens NK, Leibel RL & Hirsch J Mechanism of free fatty acid re-esterification in human adipocytes in vitro. J. Lipid Res 31, 1423–1431 (1990). [PubMed] [Google Scholar]
- 41.Leibel RL, Hirsch J, Berry EM & Gruen RK Alterations in adipocyte free fatty acid re-esterification associated with obesity and weight reduction in man. Am. J. Clin. Nutr 42, 198–206 (1985). [DOI] [PubMed] [Google Scholar]
- 42.Jenkins CM et al. Identification, Cloning, Expression, and Purification of Three Novel Human Calcium-independent Phospholipase A2 Family Members Possessing Triacylglycerol Lipase and Acylglycerol Transacylase Activities. J. Biol. Chem 279, 48968–48975 (2004). [DOI] [PubMed] [Google Scholar]
- 43.Ohno Y, Nara A, Nakamichi S & Kihara A Molecular mechanism of the ichthyosis pathology of Chanarin–Dorfman syndrome: Stimulation of PNPLA1-catalyzed ω-O-acylceramide production by ABHD5. J. Dermatol. Sci 92, 245–253 (2018). [DOI] [PubMed] [Google Scholar]
- 44.Kien B et al. ABHD5 stimulates PNPLA1-mediated ω- O -acylceramide biosynthesis essential for a functional skin permeability barrier. J. Lipid Res 59, 2360–2367 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Stender S et al. Adiposity amplifies the genetic risk of fatty liver disease conferred by multiple loci. Nat. Genet 49, 842–847 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schweiger M et al. Measurement of Lipolysis in Methods in Enzymology 538, 171–193 (Elsevier, 2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.BLIGH EG & DYER WJ A rapid method of total lipid extraction and purification. Can. J. Biochem. Physiol 37, 911–917 (1959). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated or analyzed during this study are included in this article and its Supplementary Information files or are available from the corresponding authors upon reasonable request.