Abstract
Eukaryotes control nearly every cellular processes in part by modulating the transcription of genes encoded by their nuclear genome. However, these cells are faced with the added complexity of possessing a second genome, within the mitochondria, which encodes critical components of several essential processes, including energy metabolism and macromolecule biosynthesis. As these cellular processes require gene products encoded by both genomes, cells have adopted strategies for linking mitochondrial gene expression to nuclear gene expression and other dynamic cellular events. Here we discuss examples of several mechanisms that have been identified, by which eukaryotic cells link extramitochondrial signals to dynamic alterations in mitochondrial transcription.
1. Introduction
Mitochondria are involved in many diverse metabolic processes. While they possess their own genome, emerging evidence suggests that cellular events outside the mitochondria are tightly coupled to gene expression patterns within the organelle. Coupled with recent pre-clinical success using strategies that modulate mitochondrial translation to inhibit cancer cell growth [1], it has become clear that understanding the mechanisms by which cellular signaling pathways impinge on mitochondrial gene expression will be of significant benefit. Perhaps the best characterized of mitochondrial functions is participation in oxidative phosphorylation via the electron transport chain. Each organelle can contain several copies of the mitochondrial genome, which is comprised of a circular unit of dsDNA approximately 16,600 base pairs in length [2]. This double stranded molecule contains a heavy strand and a light strand, which are distinguished by their relative sedimentation density.
Functionally, the mitochondrial genome encodes 37 known genes: 22 tRNA, 2 rRNA and 13 polypeptides that are part of the oxidative phosphorylation system [2]. The remaining protein components of the mitochondria are encoded and transcribed from the nuclear genome and subsequently imported into the mitochondria [3]. In addition to its transcribed regions, the mitochondrial genome possesses a non-coding control region approximately 1.1 kb in length [2]. The function of this region is not completely characterized, but it contains the origin of replication and transcriptional promoter regions [4, 5]. Transcription begins at one of two promoter sites in the heavy strand (HSP1 and HSP2) and one promoter in the light strand (LSP) [6].
Like the majority of mitochondrial components, the proteins necessary for transcription and replication of the mitochondrial genome are not encoded within the organelle itself, but rather are encoded in the nucleus. Mitochondrial transcription is carried out by the single subunit polymerase POLRMT/mtRNAP, which displays high sequence similarity to the polymerases of T-odd lineages of bacteriophages [4, 7, 8]. Additionally, at least three critical transcription factors have been identified-TFAM, mTERF and TFB1M (or its paralog TFB2M) [9, 10]. TFAM contains two DNA-binding HMG-boxes separated by a linker region and a C-terminal tail involved in promoter-specific transcription [11, 12]. There are several TFAM binding sites in the control region of the mitochondrial genome upstream of transcriptional initiation sites [13]. Functionally, recent work has shown that TFAM introduces a “U-turn” in mtDNA at the LSP, which mediates transcriptional activation [14, 15]. TFB1M and TFB2M are transcription factors that are similar to rRNA methyltransferases [16]. Although TFB2M is two orders of magnitude more efficient than TFB1M, both are able to form heterodimers with POLRMT/mtRNAP and activate transcription [9]. However there is significant evidence since their discovery to support non-redundant functions for TFB1M and TFB2M [17-19]. While TFB2M is required for transcription as a transcription factor, TFB1M has been shown to be involved in translation through its role in rRNA modification via methyltransferase activity. Furthermore, loss of TFB1M does not lead to a decrease in mitochondrial transcription. Additionally, despite being negative results, it has been shown multiple times that TFB1M did not support mitochondrial transcription in highly purified in vitro systems. mTERF, a transcriptional termination factor, is structurally composed of three leucine zippers and two separate basic DNA binding domains [10, 20, 21]. Interestingly, mTERF binding sites on the mitochondrial genome are both upstream and downstream from transcriptional units. This has led to the proposal that transcriptional termination and initiation are functionally linked, with mTERF facilitating the transfer of POLRMT/mtRNAP from the terminated site to the initiation site [22].
The fact that these vital components of mitochondrial transcription are encoded in the nucleus provides an opportunity for regulation and synchronization with other important aspects of cellular physiology. For example, nuclear regulation of mitochondrial transcription may allow the cell to coordinate the production of the mitochondrial and nuclear-encoded subunits of the oxidative phosphorylation system at levels that achieve the appropriate stoichiometry. Another example might include increased biosynthetic reactions in the mitochondria to provide the cell with the building blocks needed for increased cell cycle progression. Here we explore several examples in which nuclear-encoded factors and other extra-mitochondrial aspects of cellular physiology influence mitochondrial transcription. Ultimately we discuss how these factors allow the cell to maintain a tight link between metabolic/biosynthetic demands and mitochondrial function.
Accepting the principle that individual cells might rely on altered mitochondrial transcription as a mechanism for adapting to a changing environment, we are left with the question of how a cell might accomplish this. Among the seemingly simplest mechanisms might be the utilization of nuclear signaling pathways to regulate the expression of genes encoding essential, rate-limiting components of the mitochondrial transcription apparatus. Alternatively, components of the mitochondrial transcriptional apparatus might respond directly to signaling pathways. For example, phosphorylation of the mitochondrial RNA polymerase POLRMT/mtRNAP by growth factor activated kinases could alter its catalytic activity, as it does for RNA pol-II within the nucleus [23, 24]. Another potential mechanism for the dynamic regulation of mitochondrial transcription could be the ability of transcriptional regulators that normally function on the nuclear genome, to transiently translocate into the mitochondria to regulate transcription there. Examples of many of these types of dynamic regulatory mechanisms have come to light in recent years. Here we will provide a handful of examples to illustrate these phenomena.
2. Control of mitochondrial transcription by the MYC and NRF transcription factors
As discussed above, transcription of the mitochondrial genome must respond in a dynamic fashion to changes in the cellular requirement for mitochondrial function and actual numbers of mitochondria, as energy and biosynthetic demands change with alterations in cell cycle progression and other events [25, 26]. For example, as cells receive signals to increase their cell cycle rate, they must increase mitochondrial biogenesis so that progeny cells receive the appropriate complement of these organelles during mitosis. The MYC oncoprotein is among the most potent activators of cell cycle progression in both normal and malignant human cells. Consistent with mitochondrial genome transcription and biogenesis being linked to pro-proliferative signaling pathways, MYC directly controls these processes [27, 28]. Recent studies also implicate MYC in the reprogramming of cellular metabolism that occurs in cancer [29, 30], a process likely dependent on altered mitochondrial transcription.
Interestingly, the landmark study which demonstrated MYC’s role in mitochondrial genome replication and function, identified 281 mitochondrial ontology genes among a total of 2679 MYC responsive, nuclear-encoded genes [27]. Subsequent binding analysis of MYC on the nuclear genome supported the link between MYC and nuclear-encoded mitochondrial genes by showing that MYC directly occupies 107 nuclear genes encoding mitochondrial proteins. In the initial expression profiling study, the best characterized of these was the mitochondrial transcription factor TFAM. Empirical analysis demonstrated direct MYC binding to the TFAM locus and a tight correlation between MYC levels and TFAM transcription. While clearly a central player in the ability of MYC to regulate mitochondrial transcription, these authors suggested that TFAM induction may not explain the entirety of MYC’s effects and others genes among the 281 mitochondrial-ontology targets of MYC almost certainly contribute. For example, the RNA polymerase POLRMT/mtRNAP was among the MYC-regulated genes and MYC’s ability to control the expression of this rate-limiting enzyme may allow it to further modulate mitochondrial transcription. Quantitative proteomic analysis demonstrates that TFAM is present in human mitochondria at levels exceeding those needed for stoichiometric association with the POLRMT/mtRNAP transcription complex [31].
MYC also has other tricks in its arsenal that allow it to control mitochondrial function in general. For example, MYC regulates this nuclear-encoded mitochondrial ontology program largely via a functional interaction with the NRF-1 transcription factor [32], and by increasing expression of the PGC-iβ cofactor [33]. NRF-1 and PGC-1β are among the most potent regulators of the nuclear-encoded mitochondrial ontology program. Coupled with these indirect mechanisms, by directly regulating levels of critical components of the mitochondrial transcription apparatus, such as TFAM and POLRMT/mtRNAP, MYC is able to link its potent cell cycle and metabolic activities to altered mitochondrial function.
A second example of a highly regulated transcription factor that controls transcription of genes encoding the mitochondrial transcription apparatus is the Ets family protein NRF-2 [34]. In response to oxidative stress, NRF-2 regulates the transcription of many cytoprotective genes [35, 36]. However, NRF-2 has recently been demonstrated to directly bind and activate the transcription of several of the genes encoding key effectors of mitochondrial transcription (and mitochondrial genome replication) [34]. Among the recently characterized NRF-2 target genes involved in transcription of the mitochondrial genome are those encoding the transcription factors TFAM and mTERF, while target genes products involved in replication of the mitochondrial genome are the DNA helicase TWINKLE, the single-stranded binding protein mtSSB as well as the essential B subunit of the DNA-γ polymerase itself. Thus, MYC and NRF-2 respond to growth and stress signaling pathways in part by directly modulating the levels of important players in the mitochondrial transcription cycle.
3. Direct roles for traditional nuclear transcription factors in regulating gene expression within the mitochondria.
Glucocorticoids, thyroid hormone and other steroid hormones have long been known to be involved in many key physiologic processes, most notably metabolism. These hormones exert their effects via receptors that belong to the superfamily of nuclear receptors. These receptors function by modulating nuclear gene expression. Upon ligand binding, they dimerize and enter the nucleus where they bind to hormone responsive elements (HREs) in nuclear genome and influence gene expression.
Given the diverse role that these hormones play and since the mitochondria lie at the center of cellular metabolic processes, it is not surprising that they have been shown to modulate mitochondrial functions. The effects that steroid and thyroid hormones have on mitochondrial biogenesis are well-studied in a variety of tissues. Additionally, components of the oxidative phosphorylation (OXPHOS) complex have been shown to be under the control of these hormones [37]. One possible mechanism for this coordination is through indirect transcriptional action in the mitochondria. Indeed, these hormones have been shown to modulate expression of key nuclear-encoded mitochondrial transcription factors, including TFAM, TFB1M and TFB2M [38]. However, the isolation of these nuclear receptors in the mitochondria as well as the identification of HREs in the mitochondrial genome itself suggest that alternate, direct transcriptional effects also occur [39]. The first nuclear receptor identified in mitochondria was the Glucocorticoid Receptor (GR), followed shortly thereafter by Estrogen Receptor (ER) and Thyroid Receptor (TR) [40-42].
Functionally, the evidence suggests that these receptors bind to response elements in the mitochondrial genome and modulate transcription. Early gel shift assays identified interactions between DNA response elements and GR [39]. Subsequently, Psarra and Sekeris showed via chromatin immunoprecipitation that GR binds to the regulatory D-loop of mitochondrial DNA in HepG2 cells. Furthermore they showed that this mitochondrial GR was sufficient to increase levels of the OXPHOS component COX I. This study provided some of the first insights into the molecular mechanisms underlying the direct action that GR (and possibly other “nuclear” receptors) has on mitochondrial transcription.
These observations about the direct action of GR on mitochondrial transcription were later strengthened by experiments involving α-amanitin, an inhibitor of nuclear RNA pol-II function [38]. If indirect effects were all that was involved in the observed increase in transcription following glucocorticoid treatment, then treatment with α-amanitin would ablate this effect. On the contrary, the observed increase in mitochondrial transcription was preserved even in the presence of α-amanitin, strongly suggesting a direct effect of the receptor on mitochondrial transcription.
In addition to GR, multiple in organelle studies have elucidated a similar role for thyroid hormone receptor in mitochondria. Casas et al. studied a TR that was specifically targeted to the mitochondria and found that it increased transcription in a thyroid hormone-dependent manner [43]. Additionally, evidence was presented that showed a significant thyroid hormone dependent increase in mitochondrial transcriptional activity in hypothyroid rat mitochondria, and similar data have been reported in skeletal muscle [44, 45]. While controversial, there is also evidence of the Androgen Receptor having mitochondrial localization and altering transcription there [46].
The reason for the dual effect that these receptors have in modulating both nuclear and mitochondrial transcription may relate to the function of these hormones. These hormones are involved in regulating metabolic activity, a process in which mitochondria and the OXPHOS system lie at the center. Furthermore since the OXPHOS components are encoded by both the nucleus and the mitochondria, it makes intuitive sense that the cell would coordinate gene expression at both sites in response to hormonal stimulation. Presumably, this allows the mitochondria to directly sense the metabolic environment and then synchronize transcription of the OXPHOS system and other metabolic factors with their nuclear counterparts.
The nuclear hormone receptor family transcription factors have an unusual life-cycle and one might imagine that their ability to function on the mitochondrial genome is not shared with other, more classical transcription factors. However, there are now additional examples of traditional nuclear transcription factors that translocate to the mitochondria under some conditions, and directly regulate transcription. NFkB function within the cell is tightly regulated and highly responsive to numerous different cellular stimuli [47]. The NFkB family member RelA has recently been shown to be present within the mitochondria [48], and this localization depends largely on its interaction with the heat shock protein Mortalin [49]. Further analysis using chromatin immunoprecipitation showed that once inside the mitochondria, RelA directly binds to the mitochondrial genome and represses transcription [49]. The ability of RelA to repress transcription appears to involve displacement of the POLRMT/mtRNAP complex from mitochondrial promoters [49]. Like most of NFkB’s functions, its ability to regulate mitochondrial transcription is likely to be cell type and stimulus-specific. Interestingly, this ability of NFkB to regulate mitochondrial transcription is controlled by the DNA damage responsive transcription factor and tumor suppressor p53 [49]. Mechanistically, p53 regulates NFkB function by controlling its importation into the mitochondria. Thus, potentially contributing to the metabolic reprogramming that accompanies p53 mutation in cancer cells.
The phenomenon of nuclear transcription factors working directly at the mitochondrial genome is more widespread than the examples discussed above. As examples, CREB, AP1 and even p53 itself have been documented to function as direct regulators of mitochondrial transcription [50-53].
4. Direct sensing of cellular metabolic state by the mitochondrial transcription machinery
Perhaps the simplest model for establishing a dynamic system linking cellular metabolic state and the transcription of critical metabolic genes within the mitochondria, would be to have components of the mitochondrial transcription apparatus directly sense some central metabolite and response with altered activity. In fact, it has been known for many years that mitochondrial transcription in yeast can be correlated with cellular ATP levels [54], and similar links have been suggested in mammals [55]. More recent mechanistic studies shed additional light on how this mode of regulation might be working. Amiott and Jaehning, using yeast as a model system, showed that mitochondrial transcription is directly linked to changes in mitochondrial ATP levels [56]. In mitochondrial promoters where the initiating nucleotide is ATP, transcription is highly sensitive to mitochondrial ATP levels. This allows differential regulation of the distinct mitochondrial transcripts, despite their being transcribed by apparently identical machinery. (The mitochondrial transcription apparatus is far less sensitive to nucleotide content within the transcript [57].) Additional work has shown similar ATP-mediated regulation of human mitochondrial transcription in in vitro models [58]. These findings may suggest a conserved mechanism of metabolite ‘sensing’ in the regulation of mitochondrial transcription. Additional analysis revealed that the yeast mtRNAP accessory factor Mtf1 is responsible for controlling this heightened sensitivity to the +1 nucleotide. Mtf1 is the yeast ortholog of the human mtTFB protein. Interestingly, human TFB2M has now been shown to also modulate the affinity of the mitochondrial transcription complex for the initiating nucleotide [59], suggesting that this mechanism might be conserved among eukaryotes.
5. Conclusions and future perspective
The examples discussed here provide a broad sense of the widely different avenues by which transcription of the compact and compartmentalized mitochondrial genome might be controlled. Dynamic regulation of the expression of the 13 essential mitochondrial gene products is likely to be of significant benefit to organisms that must adapt to rapid changes in nutrient availability and growth rates. As our understanding of the biochemical events and individual proteins involved in the mitochondrial transcription cycle improves, we will no doubt identify additional mechanism by which mitochondrial transcription can be controlled in response to extra-mitochondrial signals. From the perspective of human health, understanding the mechanisms linking mitochondrial transcription to extramitochondrial events may eventually allow the development of therapeutic strategies for manipulating this process in a rational manner, with the ultimate goal of treating patients with cancer and other diseases where mitochondrial function is abnormal.
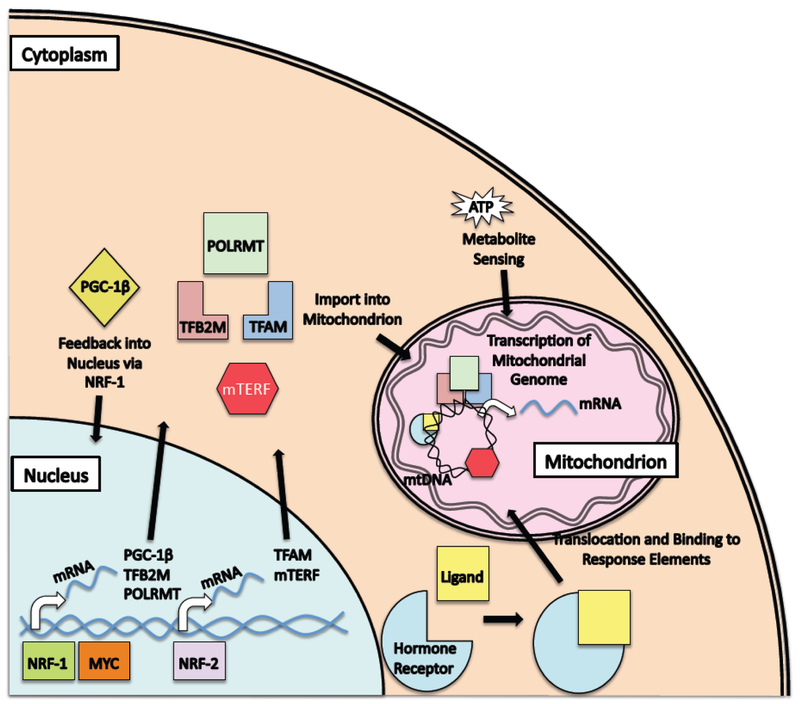
Fig. 1. Mechanisms for dynamic regulation of mitochondrial transcription.
Mitochondrial transcription can be modulated by nuclear events, cytoplasmic signals or direct sensing of metabolites, among other mechanisms. Nuclear events include activation by the MYC and NRF-2 transcription factors of broad programs of nuclear genes encoding mitochondrial proteins. For MYC, part of this regulation is due to its ability to control the expression and/or function of other transcriptional regulators, such as PGC-1β and NRF-1. As part of this process, nuclear encoded component of the mitochondrial transcription apparatus are induced (e.g. TFAM, TFB2M, POLRMT and mTERF). For the nuclear hormone receptor family of transcription factors, control of mitochondrial gene expression is more complicated. Members of this family implicated in mitochondrial gene expression include the Glucocorticoid Receptor, Thyroid Hormone Receptor, Androgen Receptor and Estrogen Receptor. These receptors not only perform their well-characterized role in binding and regulating genes in the nuclear genome, but recent evidence suggests also enter mitochondria and bind and regulate response elements in the mitochondrial genome. As a more direct mechanism for dynamic regulation of mitochondrial gene expression, the mitochondrial transcriptional machinery is responsive to cellular levels of ATP, thus coupling metabolic status with transcriptional regulation. Taken together, these and other mechanisms cooperate to provide the cell with a variety of strategies for dynamically regulating mitochondrial function in response to changes in the cellular environment.
Highlights.
OXPHOS and key biosynthetic reactions require synchronization of nuclear and mitochondrial transcription.
The mitochondrial transcription complex responds directly to changes in cellular energy pools.
An growing number of nuclear transcription factors also function within the mitochondria.
Production of the nuclear-encoded mitochondrial transcription complex is tied to key cellular signaling pathways.
Acknowledgments
This work was supported by the grants from the NIH: R01CA090465 (SBM) and R01CA141070 (SBM). In addition, this work was partially supported by funds from the Commonwealth Universal Research Enhancement Program, Pennsylvania Department of Health. We also thank Drs. Xiao-yong Zhang and Amanda Norvell for helpful discussions.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Skrtic M, Sriskanthadevan S, Jhas B, Gebbia M, Wang X, Wang Z, Hurren R, Jitkova Y, Gronda M, Maclean N, Lai CK, Eberhard Y, Bartoszko J, Spagnuolo P, Rutledge AC, Datti A, Ketela T, Moffat J, Robinson BH, Cameron JH, Wrana J, Eaves CJ, Minden MD, Wang JC, Dick JE, Humphries K, Nislow C, Giaever G, Schimmer AD, Inhibition of mitochondrial translation as a therapeutic strategy for human acute myeloid leukemia, Cancer Cell 20 (2011) 674–688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Anderson S, Bankier AT, Barrell BG, de Bruijn MH, Coulson AR, Drouin J, Eperon IC, Nierlich DP, Roe BA, Sanger F, Schreier PH, Smith AJ, Staden R, Young IG, Sequence and organization of the human mitochondrial genome, Nature 290 (1981) 457–465. [DOI] [PubMed] [Google Scholar]
- [3].Reid GA, Schatz G, Import of proteins into mitochondria. Extramitochondrial pools and post-translational import of mitochondrial protein precursors in vivo, J Biol Chem 257 (1982) 13062–13067. [PubMed] [Google Scholar]
- [4].Tiranti V, Savoia A, Forti F, D'Apolito MF, Centra M, Rocchi M, Zeviani M, Identification of the gene encoding the human mitochondrial RNA polymerase (h-mtRPOL) by cyberscreening of the Expressed Sequence Tags database, Hum Mol Genet 6 (1997) 615–625. [DOI] [PubMed] [Google Scholar]
- [5].Chang DD, Clayton DA, Precise identification of individual promoters for transcription of each strand of human mitochondrial DNA, Cell 36 (1984) 635–643. [DOI] [PubMed] [Google Scholar]
- [6].Montoya J, Christianson T, Levens D, Rabinowitz M, Attardi G, Identification of initiation sites for heavy-strand and light-strand transcription in human mitochondrial DNA, Proc Natl Acad Sci U S A 79 (1982) 7195–7199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Shutt TE, Gray MW, Bacteriophage origins of mitochondrial replication and transcription proteins, Trends Genet 22 (2006) 90–95. [DOI] [PubMed] [Google Scholar]
- [8].Cermakian N, Ikeda TM, Miramontes P, Lang BF, Gray MW, Cedergren R, On the evolution of the single-subunit RNA polymerases, J Mol Evol 45 (1997) 671–681. [DOI] [PubMed] [Google Scholar]
- [9].Falkenberg M, Gaspari M, Rantanen A, Trifunovic A, Larsson NG, Gustafsson CM, Mitochondrial transcription factors B1 and B2 activate transcription of human mtDNA, Nat Genet 31 (2002) 289–294. [DOI] [PubMed] [Google Scholar]
- [10].Daga A, Micol V, Hess D, Aebersold R, Attardi G, Molecular characterization of the transcription termination factor from human mitochondria, J Biol Chem 268 (1993) 8123–8130. [PubMed] [Google Scholar]
- [11].Parisi MA, Clayton DA, Similarity of human mitochondrial transcription factor 1 to high mobility group proteins, Science 252 (1991) 965–969. [DOI] [PubMed] [Google Scholar]
- [12].Fisher RP, Clayton DA, Purification and characterization of human mitochondrial transcription factor 1, Mol Cell Biol 8 (1988) 3496–3509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Fisher RP, Topper JN, Clayton DA, Promoter selection in human mitochondria involves binding of a transcription factor to orientation-independent upstream regulatory elements, Cell 50 (1987) 247–258. [DOI] [PubMed] [Google Scholar]
- [14].Rubio-Cosials A, Sidow JF, Jimenez-Menendez N, Fernandez-Millan P, Montoya J, Jacobs HT, Coll M, Bernado P, Sola M, Human mitochondrial transcription factor A induces a U-turn structure in the light strand promoter, Nat Struct Mol Biol 18 (2011) 1281–1289. [DOI] [PubMed] [Google Scholar]
- [15].Ngo HB, Kaiser JT, Chan DC, The mitochondrial transcription and packaging factor Tfam imposes a U-turn on mitochondrial DNA, Nat Struct Mol Biol 18 (2011) 1290–1296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].O'Farrell HC, Pulicherla N, Desai PM, Rife JP, Recognition of a complex substrate by the KsgA/Dim1 family of enzymes has been conserved throughout evolution, RNA 12 (2006) 725–733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Lodeiro MF, Uchida A, Bestwick M, Moustafa IM, Arnold JJ, Shadel GS, Cameron CE, Transcription from the second heavy-strand promoter of human mtDNA is repressed by transcription factor A in vitro, Proc Natl Acad Sci U S A 109 (2012) 6513–6518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Litonin D, Sologub M, Shi Y, Savkina M, Anikin M, Falkenberg M, Gustafsson CM, Temiakov D, Human mitochondrial transcription revisited: only TFAM and TFB2M are required for transcription of the mitochondrial genes in vitro, J Biol Chem 285 (2010) 18129–18133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Metodiev MD, Lesko N, Park CB, Camara Y, Shi Y, Wibom R, Hultenby K, Gustafsson CM, Larsson NG, Methylation of 12S rRNA is necessary for in vivo stability of the small subunit of the mammalian mitochondrial ribosome, Cell metabolism 9 (2009) 386–397. [DOI] [PubMed] [Google Scholar]
- [20].Fernandez-Silva P, Martinez-Azorin F, Micol V, Attardi G, The human mitochondrial transcription termination factor (mTERF) is a multizipper protein but binds to DNA as a monomer, with evidence pointing to intramolecular leucine zipper interactions, Embo J 16 (1997) 1066–1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Chomyn A, Martinuzzi A, Yoneda M, Daga A, Hurko O, Johns D, Lai ST, Nonaka I, Angelini C, Attardi G, MELAS mutation in mtDNA binding site for transcription termination factor causes defects in protein synthesis and in respiration but no change in levels of upstream and downstream mature transcripts, Proc Natl Acad Sci U S A 89 (1992) 4221–4225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Martinez-Azorin F, The mitochondrial ribomotor hypothesis, IUBMB Life 57 (2005) 27–30. [DOI] [PubMed] [Google Scholar]
- [23].Zhang J, Corden JL, Identification of phosphorylation sites in the repetitive carboxyl-terminal domain of the mouse RNA polymerase II largest subunit, J Biol Chem 266 (1991) 2290–2296. [PubMed] [Google Scholar]
- [24].Lee JM, Greenleaf AL, A protein kinase that phosphorylates the C-terminal repeat domain of the largest subunit of RNA polymerase II, Proc Natl Acad Sci U S A 86 (1989) 3624–3628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Attardi G, Schatz G, Biogenesis of mitochondria, Annu Rev Cell Biol 4 (1988) 289–333. [DOI] [PubMed] [Google Scholar]
- [26].Asin-Cayuela J, Gustafsson CM, Mitochondrial transcription and its regulation in mammalian cells, Trends Biochem Sci 32 (2007) 111–117. [DOI] [PubMed] [Google Scholar]
- [27].Li F, Wang Y, Zeller KI, Potter JJ, Wonsey DR, O'Donnell KA, Kim JW, Yustein JT, Lee LA, Dang CV, Myc stimulates nuclearly encoded mitochondrial genes and mitochondrial biogenesis, Mol Cell Biol 25 (2005) 6225–6234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Kim J, Lee JH, Iyer VR, Global identification of Myc target genes reveals its direct role in mitochondrial biogenesis and its E-box usage in vivo, PLoS ONE 3 (2008) e1798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Wise DR, DeBerardinis RJ, Mancuso A, Sayed N, Zhang XY, Pfeiffer HK, Nissim I, Daikhin E, Yudkoff M, McMahon SB, Thompson CB, Myc regulates a transcriptional program that stimulates mitochondrial glutaminolysis and leads to glutamine addiction, Proc Natl Acad Sci U S A 105 (2008) 18782–18787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Gao P, Tchernyshyov I, Chang TC, Lee YS, Kita K, Ochi T, Zeller KI, De Marzo AM, Van Eyk JE, Mendell JT, Dang CV, c-Myc suppression of miR-23a/b enhances mitochondrial glutaminase expression and glutamine metabolism, Nature 458 (2009) 762–765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Cotney J, Wang Z, Shadel GS, Relative abundance of the human mitochondrial transcription system and distinct roles for h-mtTFB1 and h-mtTFB2 in mitochondrial biogenesis and gene expression, Nucleic Acids Res 35 (2007) 4042–4054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Morrish F, Giedt C, Hockenbery D, c-MYC apoptotic function is mediated by NRF-1 target genes, Genes Dev 17 (2003) 240–255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Zhang H, Gao P, Fukuda R, Kumar G, Krishnamachary B, Zeller KI, Dang CV, Semenza GL, HIF-1 inhibits mitochondrial biogenesis and cellular respiration in VHL-deficient renal cell carcinoma by repression of C-MYC activity, Cancer Cell 11 (2007) 407–420. [DOI] [PubMed] [Google Scholar]
- [34].Bruni F, Polosa PL, Gadaleta MN, Cantatore P, Roberti M, Nuclear respiratory factor 2 induces the expression of many but not all human proteins acting in mitochondrial DNA transcription and replication, J Biol Chem 285 (2010) 3939–3948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Mohammadzadeh M, Halabian R, Gharehbaghian A, Amirizadeh N, Jahanian-Najafabadi A, Roushandeh AM, Roudkenar MH, Nrf-2 overexpression in mesenchymal stem cells reduces oxidative stress-induced apoptosis and cytotoxicity, Cell Stress Chaperones (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Yanaka A, Zhang S, Tauchi M, Suzuki H, Shibahara T, Matsui H, Nakahara A, Tanaka N, Yamamoto M, Role of the nrf-2 gene in protection and repair of gastric mucosa against oxidative stress, Inflammopharmacology 13 (2005) 83–90. [DOI] [PubMed] [Google Scholar]
- [37].Weber K, Bruck P, Mikes Z, Kupper JH, Klingenspor M, Wiesner RJ, Glucocorticoid hormone stimulates mitochondrial biogenesis specifically in skeletal muscle, Endocrinology 143 (2002) 177–184. [DOI] [PubMed] [Google Scholar]
- [38].Psarra AM, Sekeris CE, Glucocorticoids induce mitochondrial gene transcription in HepG2 cells: role of the mitochondrial glucocorticoid receptor, Biochim Biophys Acta 1813 (2011) 1814–1821. [DOI] [PubMed] [Google Scholar]
- [39].Demonacos C, Djordjevic-Markovic R, Tsawdaroglou N, Sekeris CE, The mitochondrion as a primary site of action of glucocorticoids: the interaction of the glucocorticoid receptor with mitochondrial DNA sequences showing partial similarity to the nuclear glucocorticoid responsive elements, J Steroid Biochem Mol Biol 55 (1995) 43–55. [DOI] [PubMed] [Google Scholar]
- [40].Psarra AM, Solakidi S, Sekeris CE, The mitochondrion as a primary site of action of steroid and thyroid hormones: presence and action of steroid and thyroid hormone receptors in mitochondria of animal cells, Molecular and cellular endocrinology 246 (2006) 21–33. [DOI] [PubMed] [Google Scholar]
- [41].Yang SH, Liu R, Perez EJ, Wen Y, Stevens SM Jr., Valencia T, Brun-Zinkernagel AM, Prokai L, Will Y, Dykens J, Koulen P, Simpkins JW, Mitochondrial localization of estrogen receptor beta, Proc Natl Acad Sci U S A 101 (2004) 4130–4135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Scheller K, Sekeris CE, Krohne G, Hock R, Hansen IA, Scheer U, Localization of glucocorticoid hormone receptors in mitochondria of human cells, Eur J Cell Biol 79 (2000) 299–307. [DOI] [PubMed] [Google Scholar]
- [43].Casas F, Rochard P, Rodier A, Cassar-Malek I, Marchal-Victorion S, Wiesner RJ, Cabello G, Wrutniak C, A variant form of the nuclear triiodothyronine receptor c-ErbAalpha1 plays a direct role in regulation of mitochondrial RNA synthesis, Mol Cell Biol 19 (1999) 7913–7924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Wiesner RJ, Kurowski TT, Zak R, Regulation by thyroid hormone of nuclear and mitochondrial genes encoding subunits of cytochrome-c oxidase in rat liver and skeletal muscle, Mol Endocrinol 6 (1992) 1458–1467. [DOI] [PubMed] [Google Scholar]
- [45].Mutvei A, Kuzela S, Nelson BD, Control of mitochondrial transcription by thyroid hormone, Eur J Biochem 180 (1989) 235–240. [DOI] [PubMed] [Google Scholar]
- [46].Solakidi S, Psarra AM, Nikolaropoulos S, Sekeris CE, Estrogen receptors alpha and beta (ERalpha and ERbeta) and androgen receptor (AR) in human sperm: localization of ERbeta and AR in mitochondria of the midpiece, Hum Reprod 20 (2005) 3481–3487. [DOI] [PubMed] [Google Scholar]
- [47].Hayden MS, Ghosh S, NF-kappaB, the first quarter-century: remarkable progress and outstanding questions, Genes Dev 26 (2012) 203–234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Cogswell PC, Kashatus DF, Keifer JA, Guttridge DC, Reuther JY, Bristow C, Roy S, Nicholson DW, Baldwin AS Jr., NF-kappa B and I kappa B alpha are found in the mitochondria. Evidence for regulation of mitochondrial gene expression by NF-kappa B, J Biol Chem 278 (2003) 2963–2968. [DOI] [PubMed] [Google Scholar]
- [49].Johnson RF, Witzel II, Perkins ND, p53-dependent regulation of mitochondrial energy production by the RelA subunit of NF-kappaB, Cancer Res 71 (2011) 5588–5597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Cammarota M, Paratcha G, Bevilaqua LR, Levi de Stein M, Lopez M, Pellegrino de Iraldi A, Izquierdo I, Medina JH, Cyclic AMP-responsive element binding protein in brain mitochondria, J Neurochem 72 (1999) 2272–2277. [DOI] [PubMed] [Google Scholar]
- [51].Lee J, Kim CH, Simon DK, Aminova LR, Andreyev AY, Kushnareva YE, Murphy AN, Lonze BE, Kim KS, Ginty DD, Ferrante RJ, Ryu H, Ratan RR, Mitochondrial cyclic AMP response element-binding protein (CREB) mediates mitochondrial gene expression and neuronal survival, J Biol Chem 280 (2005) 40398–40401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Ogita K, Okuda H, Kitano M, Fujinami Y, Ozaki K, Yoneda Y, Localization of activator protein-1 complex with DNA binding activity in mitochondria of murine brain after in vivo treatment with kainate, J Neurosci 22 (2002) 2561–2570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Heyne K, Mannebach S, Wuertz E, Knaup KX, Mahyar-Roemer M, Roemer K, Identification of a putative p53 binding sequence within the human mitochondrial genome, FEBS Lett 578 (2004) 198–202. [DOI] [PubMed] [Google Scholar]
- [54].Gaines G, Rossi C, Attardi G, Markedly different ATP requirements for rRNA synthesis and mtDNA light strand transcription versus mRNA synthesis in isolated human mitochondria, J Biol Chem 262 (1987) 1907–1915. [PubMed] [Google Scholar]
- [55].DasGupta SF, Rapoport SI, Gerschenson M, Murphy E, Fiskum G, Russell SJ, Chandrasekaran K, ATP synthesis is coupled to rat liver mitochondrial RNA synthesis, Mol Cell Biochem 221 (2001) 3–10. [DOI] [PubMed] [Google Scholar]
- [56].Amiott EA, Jaehning JA, Mitochondrial transcription is regulated via an ATP "sensing" mechanism that couples RNA abundance to respiration, Mol Cell 22 (2006) 329–338. [DOI] [PubMed] [Google Scholar]
- [57].Amiott EA, Jaehning JA, Sensitivity of the yeast mitochondrial RNA polymerase to +1 and +2 initiating nucleotides, J Biol Chem 281 (2006) 34982–34988. [DOI] [PubMed] [Google Scholar]
- [58].Narasimhan N, Attardi G, Specific requirement for ATP at an early step of in vitro transcription of human mitochondrial DNA, Proc Natl Acad Sci U S A 84 (1987) 4078–4082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Sologub M, Litonin D, Anikin M, Mustaev A, Temiakov D, TFB2 is a transient component of the catalytic site of the human mitochondrial RNA polymerase, Cell 139 (2009) 934–944. [DOI] [PMC free article] [PubMed] [Google Scholar]