Abstract
Objective
To evaluate the feasibility of adjusted mitochondrial DNA quantification in human embryos as a biomarker for implantation potential.
Design
Double-blind, observational, prospective analysis of an Asian population in a single university-affiliated in vitro fertilization center. A total of 1617 embryos derived from 380 infertile couples were collected. The DNA from blastomere biopsy (n = 99) or trophectoderm biopsy (n = 1518) were analyzed with next-generation sequencing.
Results
The adjusted mtDNA quantification followed a non-normal distribution in both types of the embryos. When stratified by ploidy status, the adjusted mtDNA quantification was significantly higher in aneuploid trophectoderm than in euploid cells, but not in blastomeres. The adjusted mtDNA quantification of embryos showed significant but very weak positive correlation in total trophectoderm cells with maternal age (Spearman’s correlation, r = 0.095, p = 0.0028) but neither in blastomeres nor stratified by ploidy status. The median adjusted mtDNA quantification was also significantly higher in aneuploid blastocysts than in euploid ones while corrected with embryo morphology. Viable embryos did not contain significantly different quantities of adjusted mtDNA compared with nonviable embryos (implanted n = 103, non-implanted n = 164; median 0.00097 vs. 0.00088, p = 0.21) in 267 transferred blastocysts.
Conclusion
Quantification of adjusted mitochondria DNA in human embryos was significantly lower in euploid blastocysts than in aneuploid blastocysts. However, no statistically significant differences regarding implantation outcome were evident. To our best knowledge, this study provides the largest scale and the first correlation data between mitochondria copy number and human embryo implantation potential in Asians.
Keywords: Mitochondrial DNA copy number, Correction factors, PGT-A, IVF, Biomarker
Introduction
Despite the rapid progress of advanced technology for IVF and the improvements in embryo culture system over the past few decades, implantation still fails in a significant proportion of embryos [1]. Many factors affect embryonic viability. These include the immunologic status, morphological abnormality of the uterus, asynchrony between the endometrial receptivity and embryo stage, and, most importantly, the embryonic nature [2]. The morphology-based grading system has traditionally been the primary technique used in IVF to evaluate and select the most competent embryos for transfer. Other technologies, including genomics, proteomics, and time-lapse imaging, have also been developed to assist in selecting the best embryos [3]. In recent years, comprehensive 24-chromosome screening has been a focus for the evaluation of the ploidy status and to select and transfer the diagnosed “euploid” embryos. This approach is also termed preimplantation genetic testing for aneuploidy (PGT-A) [4]. Of all factors used to evaluate the embryo implantation potential, the chromosomal status appears to be the most pivotal. However, there are many other factors relevant to embryo viability, as evidenced by the fact that a viable pregnancy cannot be guaranteed even after transfer of a euploid embryo. Other methods for evaluation of embryo viability are required.
Mitochondria are maternally inherited organelles involved in a variety of essential cellular processes, such as apoptosis, amino acid synthesis, calcium homeostasis, and the generation of energy required for all cellular functions [5]. Mitochondrial deoxyribonucleic acid (mtDNA) is a circular plasmid composed of 16,569 base pairs of double-stranded DNA, which encode 37 genes. The multi-copy mtDNA has a copy number that varies greatly depending on the cell type. In human mature oocytes, there are an estimated 4000~150,000 copies of mtDNA, which are greater than in other somatic cells [6]. Mitochondria have long been of interest in reproductive biology. They play a central role in follicular atresia and could be the main target of the ooplasmic factors determining oocyte quality that is adversely affected by aging [7]. Mitochondria also play an important role in regulating metabolism and providing energy for embryos [8]. Previous studies had shown that decreased mitochondrial gene expression [9] or accumulation of mitochondrial DNA deletion [10] led to mitochondrial dysfunction and impaired ATP production, which affected the oocyte quality including fertilization and further embryo development. In other words, mitochondrial efficiency might have a significant impact on the viability of embryos in terms of implantation potential.
The possibility that various mitochondrial features could serve as potential biomarkers for the viability of the preimplantation embryo has been investigated [11]. Recent interest has focused on assessing the copy number of human embryo mtDNA or the relative amount of mtDNA as a potential biomarker for embryo viability. Several groups reported exciting findings of the correlation of elevated mtDNA copy number with aneuploidy and lower implantation potential in women undergoing IVF treatment [12–15]. These studies proposed the use of mtDNA copy number in the clinical setting for assisted selection of the most competent embryos. The process has been commercialized as the MitoGrade™ (Reprogenetics, USA) and MitoScore™ (Igenomix, Canada) technologies. However, later studies with different genetic testing platforms and methods of mtDNA quantification reported opposite results [16, 17].
Whether or not mtDNA quantification is an effective predictor of embryo implantation potential remains undetermined at present. Here, we present our results concerning the quantification of adjusted mtDNA and correlation with clinical outcomes to evaluate the feasibility of the adjusted mtDNA quantification in human embryos as a biomarker for implantation potential.
Materials and methods
Study design and clinical procedure
This non-selection, double-blind, prospective study was conducted in a single IVF center (Division of Infertility, Taipei Medical University Hospital) affiliated with a medical laboratory specializing in genetic testing (Genetics Generation Advancement Corporation, Taiwan). All of the embryos underwent either cleavage stage blastomere biopsy or trophectoderm biopsy from blastocysts at the request of the infertile couples for NGS-based PGT-A. The mtDNA content of the embryos was quantified using surplus whole genome amplification (WGA) DNA product. All embryos were vitrified post-biopsy and subsequently underwent embryo transfer with euploid embryos selected on the basis of morphological criteria, and without knowledge of the results of mtDNA content for both the doctors and the patients. Frozen-thawed single embryo transfer was performed in all cycles, and endometrial preparation with either nature or hormone supplementation was determined by the physicians’ clinical judgement. After the clinical outcome regarding the presence/absence of the intrauterine pregnancy (IUP) with positive fetal heart beat (FHB) at 4 weeks after embryo transfer was available, the mtDNA quantification data were collected for further bioinformatics processes and statistical analysis. The presence of IUP with positive FHB was considered as a viable embryo. The pregnancy outcomes were blinded at the time of mtDNA assessment to the clinician and the bioinformatics analyst, and decoded by the third independent individual at the end of the study.
Ethical approval
This study was reviewed and approved by the Taipei Medical University joint institutional review board (TMU-JIRB, approval number: N201707027). The research described in this paper involved analysis of surplus amplified DNA only after PGT-A had been completed. The embryos did not receive any additional interventions and the clinical treatment of the patients was not altered. Patient informed consent for analysis of surplus DNA product was obtained with the approval of the ethical committee.
Patient population and embryo sample processing
The study included 99 biopsied blastomeres and 1518 blastocysts derived from 380 couples performed between January 2016 and September 2018. The indications of receiving PGT-A included advanced maternal age, recurrent implantation failure, and known parental genetic abnormalities. The blastomeres were produced by couples with an average maternal age of 35.1 years (range 29–46 years) and the blastocysts were produced by couples with an average maternal age of 38.4 years (range 25–55 years). The standard process for PGT-A in our collaborated laboratory involves WGA with the SurePlex system (Illumina, USA), followed by comprehensive chromosomal screening via NGS (VeriSeq, Illumina) according to the manufacturers’ instructions. The results obtained were analyzed to determine the embryo ploidy status using BlueFuse Multi v4.1 software at the laboratory.
Relative quantification of mtDNA copy number and adjusted calculation
Samples from the 1617 embryos assessed for relative mtDNA level with MiSeq (Illumina) during this investigation consisted of surplus WGA material remaining after routine PGT-A. The protocol for NGS was as suggested by the manufacturer (Illumina). Briefly, MiSeq Reporter Software analysis determined the number of reads aligning to the mtDNA reference genome as per Genome Reference Consortium (GRC) h37 for each sample. Embryo ploidy was assessed with Bluefuse Multi Software (Illumina). In short, embryos displayed a median chromosomal copy number deviated from the default copy number with a possible trisomy (100% gain) or monosomy (100% loss) of autosomal chromosomes was seen as a copy number > 2 or < 2, respectively [18]. The embryos were diagnosed as euploidy when median chromosomal copy number deviation less than 50% (i.e., < 2.5 or > 1.5), mosaicism when deviation fall between 50 and 80%, and aneuploidy when deviation more than 80% in our laboratory.
After bioinformatics analysis using BlueFuse Multi Software, the resulting values were further subjected to the mathematical correction factors as previously described [16]. The accuracy of mtDNA content acquirement is the cornerstone of clinical utility. In brief, the mtDNA copy number for each sample with NGS was obtained by the number of read mapping to the mitochondrial genome divided by the number of read mapping to the nuclear genome to normalize for technical batch-to-batch variability during WGA and NGS, as well as number of cells collected during biopsy. The resulting value was then multiplied by the correction factor regarding embryo gender and ploidy status so that we could obtain the accurate nuclear genomes. The diploid human genome displayed a 1.58% difference between female and male embryos according to GRCh37 (Ensembl Release 68). Similarly, the aneuploid embryos contained more or less nuclear DNA per cell compared with euploid embryos in the condition of polysomy or nullisomy [16]. By multiplying the correction factors tailored to individual embryos’ chromosomal composition, the accurate adjusted mtDNA quantification was acquired for each sample.
Statistical analyses
Group analysis of data was conducted using the non-parametric Mann-Whitney test and Spearman’s rank-order correlation in Prism 7 (GraphPad Software, USA) and logistic regression analysis in SPSS (IBM Statistics, ver. 25). A p value < 0.05 was considered statistically significant. The results were expressed as mean ± SD if not specified.
Results
Distribution of quantified adjusted mtDNA and embryo ploidy status
Of the 99 blastomeres and 1518 trophectoderm biopsy samples, the results of PGT-A other than euploidy and aneuploidy were excluded. A total of 39 blastomeres and 988 trophectoderm biopsies were available for analysis. The euploid rates were 24.2% (24/99) in blastomeres and 39.7% (602/1518) in trophectoderm biopsies.
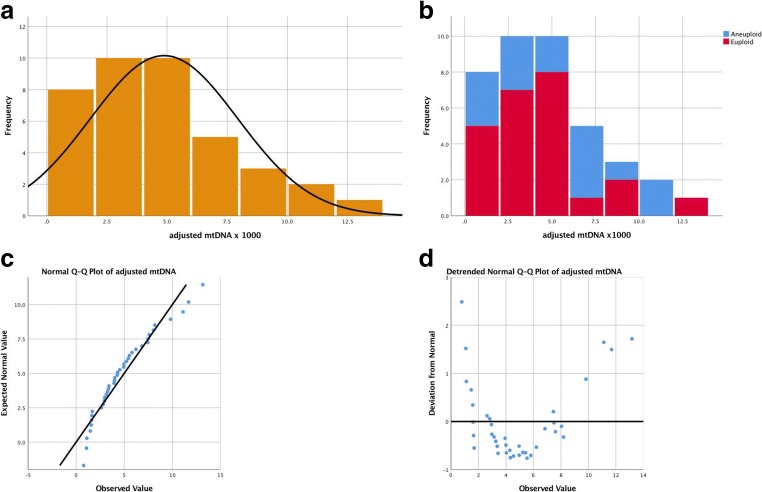
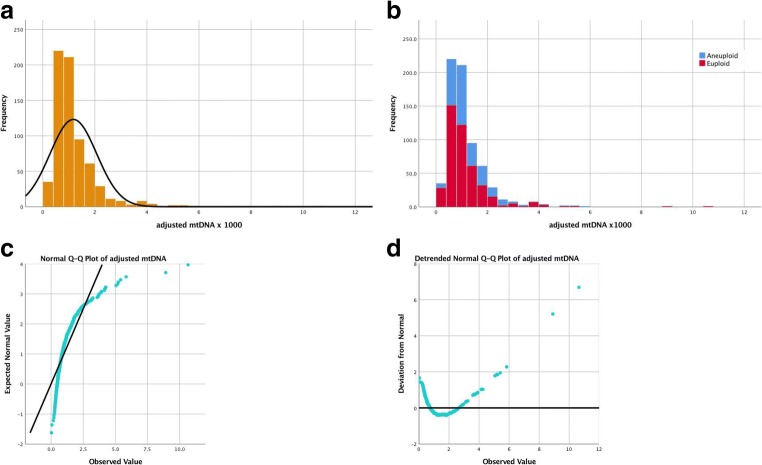
The quantified adjusted mtDNA followed a non-normal distribution in both types of embryos (Figs. 1 and 2). The descriptive analysis yielded a kurtosis of 29.12 ± 0.19 and skewness of 4.09 ± 0.09 in the trophectoderm and a kurtosis of 0.51 ± 0.94 and skewness 0.94 ± 0.38 in blastomeres. Quantile-quantile (Q-Q) plots also revealed the non-parametric distribution of the adjusted mtDNA level (Figs. 1c, d and 2c, d) in both samples. The adjusted mtDNA level was significantly higher in blastomeres than in trophectoderm cells (0.0049 ± 0.003 vs. 0.0012 ± 0.0001, p < 0.0001).
Fig. 1.
Non-normal distribution of adjusted mtDNA quantification in human blastomeres. a Total blastomeres. b Stratified by ploid status. c Normal Q-Q plot. d Detrended normal Q-Q plot. We used original value of adjusted mtDNA x 1000 for clear visualized images. mtDNA =mitochondrial DNA
Fig. 2.
Non-normal distribution of adjusted mtDNA quantification in human trophectoderm cells. a Total trophectoderm. b Stratified by ploid status. c Normal Q-Q plot. d Detrended normal Q-Q plot. We used original value of adjusted mtDNA x 1000 for clear visualized images. mtDNA =mitochondrial DNA
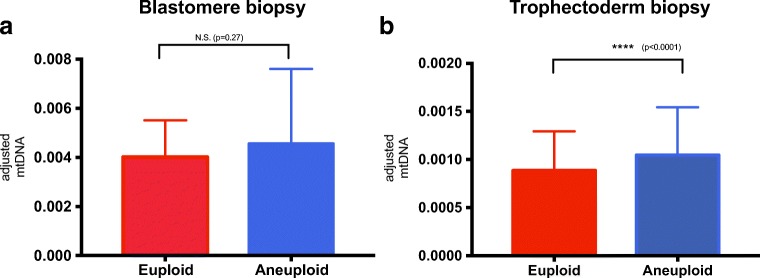
When stratified by ploidy status, the adjusted mtDNA quantification showed significantly higher values in aneuploid trophectoderm cells than in euploid cells (median 0.00089 vs. 0.001, p < 0.0001) (Fig. 3b). However, the results did not show a significant difference in blastomeres (median 0.004 vs. 0.0046, p = 0.27) (Fig. 3a).
Fig. 3.
Adjusted mtDNA quantification of embryos stratified by ploid status resulted in statistically significant difference in trophectoderm cells (b) but not in blastomeres (a). The result was calculated with the Mann-Whitney test, and the figures were expressed as median with interquartile range. N.S. not significant. ****=statistically significant. We used original value of adjusted mtDNA x 1000 for clear visualized images. mtDNA =mitochondrial DNA
Adjusted mtDNA quantification and maternal age
The maternal age was considered a continuous variable. We analyzed the correlation between adjusted mtDNA quantification and maternal age with nonparametric Spearman’s rank-order correlation. The adjusted mtDNA level showed significant but very weak positive correlation with maternal age in total trophectoderm cells (Spearman’s correlation coefficient r = 0.095, p = 0.0028, Fig. 4). However, no significant correlation was revealed when stratified by ploidy status (euploid: p = 0.2, r = 0.05; aneuploid: p = 0.16, r = 0.07). The adjusted mtDNA level also showed insignificant correlation with maternal age in blastomeres either in total or stratified by ploidy status.
Fig. 4.
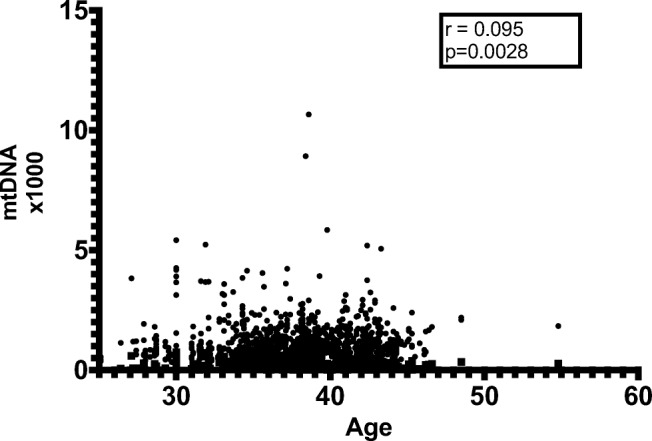
Correlation of maternal age and adjusted mtDNA quantification of embryos showed significant but very weak positive correlation in total trophectoderm cells (Spearman’s rank-order correlation). We used original value of adjusted mtDNA x 1000 for clear visualized images. mtDNA =mitochondrial DNA
Correlation among adjusted mtDNA quantification, embryo morphology, and blastocyst ploidy status
We analyzed the relationship of embryo morphology with ploidy status and adjusted mtDNA quantification in blastocyst but not in blastomeres because the sample size of blastomeres was small. The embryo morphology was determined using the Gardner grading system [19, 20] and we analyzed the correlation of degree of expansion, inner cell mass (ICM) morphology, and trophectoderm morphology (TE) with ploidy status and adjusted mtDNA quantification, respectively. Logistic regression analysis revealed that the chance of euploid status was significantly positively correlated with expansion degree (OR 1.71 in expansions 3–4 and 3.67 in expansions 5–6) and negatively correlated with ICM grading (OR 0.68 in grade B and 0.42 in grade C) and TE grading (OR 0.55 in grade B and 0.42 in grade C), respectively (Table 1). Following, the variable of adjusted mtDNA quantification was included. When stratified with embryo morphology individually, the median adjusted mtDNA was significantly higher in aneuploid blastocysts than euploid ones in the expansion 3–4 group, ICM grade B and C group, and TE grade B and C group. However, when stratified with ploidy status, the median adjusted mtDNA revealed significantly higher in ICM grade A group compared with ICM grade B and grade C (Table 1). There were no significant differences in other group comparisons (Table 1).
Table 1.
Correlation among blastocyst embryo morphology regarding degree of expansion, inner cell mass (ICM) morphology, and trophectoderm morphology (TE) and the ploidy status and adjusted mtDNA quantification
| Euploidy (N = 602) | Aneuploidy (N = 386) | Odd ratio (OR) | 95% CI | p value | Euploidy mtDNAb | Aneuploidy mtDNAb | p valuec | ||
|---|---|---|---|---|---|---|---|---|---|
| Expansion | 1–2 | 184 (30.6%) | 172 (44.6%) | 1a | 0.00096 | 0.01129 | 0.168 | ||
| 3–4 | 367 (61.0%) | 201 (52.1%) | 1.71 | 1.30–2.24 | < 0.001* | 0.00087 | 0.00099 | 0.005* | |
| 5–6 | 51 (8.5%) | 13 (3.4%) | 3.67 | 1.93–6.98 | < 0.001* | 0.00068 | 0.01089 | 0.214 | |
| p valued | 0.168 | 0.223 | |||||||
| ICM | A | 122 (20.3%) | 50 (13%) | 1a | 0.00107 | 0.00113 | 0.867 | ||
| B | 350 (58.1%) | 210 (54.4%) | 0.68 | 0.47–0.99 | 0.044* | 0.00088 | 0.01073 | < 0.001* | |
| C | 130 (21.6%) | 126 (32.6%) | 0.42 | 0.28–0.64 | < 0.001* | 0.00080 | 0.00095 | 0.018* | |
| p valued | 0.002*e | 0.309 | |||||||
| TE | A | 91 (15.1%) | 32 (8.3%) | 1a | 0.00100 | 0.01173 | 0.28 | ||
| B | 360 (59.8%) | 231 (59.8%) | 0.55 | 0.36–0.85 | 0.007 | 0.00088 | 0.00099 | 0.001* | |
| C | 151 (25.1%) | 123 (31.9%) | 0.42 | 0.27–0.69 | < 0.001* | 0.00085 | 0.00107 | 0.008* | |
| p valued | 0.064 | 0.14 |
*Statistically significant
aReference class for euploid
bExpressed as median
cp value was calculated according to morphology rank
dp value was calculated according to ploidy status
ePost hoc analysis: p value A vs. B: 0.035; A vs. C: 0.001
Adjusted mtDNA quantification and clinical outcome
The embryo transfer rate was 25% (6/24) in blastomeres and 44% (267/602) in blastocysts, respectively. We analyzed the relationship between adjusted mtDNA quantification and clinical outcome of blastocyst transfer only, since the sample size of blastomere transfer was too small. In our results, there was no significant difference in the adjusted mtDNA level between implanted (n = 103) and non-implanted (n = 164) blastocysts (median 0.00097 vs. 0.00088, p = 0.21) (Fig. 5).
Fig. 5.
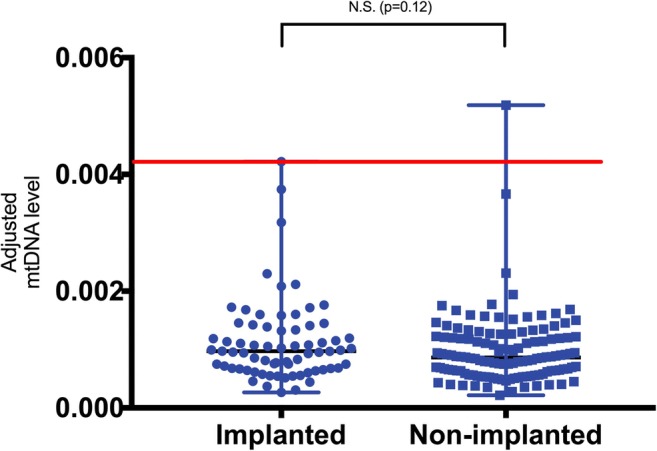
Adjusted mtDNA quantification stratified by embryo implantation status for the 267 transferred blastocysts in the current study. The red line indicated the upper limit (0.0042, set as threshold) adjusted mtDNA level of the implanted blastocysts. We used original value of adjusted mtDNA x 1000 for clear visualized images. mtDNA =mitochondrial DNA
Discussion
Following its introduction in assisted reproductive technology, PGT-A has resulted in significantly improved clinical outcomes [21, 22]. However, the routine use of PGT-A remains contentious, with its benefits and risks continuing to be debated. The most recent committee opinion published by the American Society of Reproductive Medicine [23] concluded that there is insufficient evidence to recommend the routine use of PGT-A in all infertile patients at present. Therefore, the search for identifying biomarkers for embryonic reproductive competence outside of embryo chromosomal screening is imperative.
Among the many novel investigations, the mtDNA copy number is of great interest and has been proposed to serve as an independent biomarker for embryo viability. However, there are many discrepancies in different studies with conflicting results, which may relate to numerous confounding variables. The different methodologies and discordant results from the published studies are summarized in Table 2. In the reviewed studies, four [12–15] reported that the relative quantity of mtDNA was significantly higher in old age, aneuploid, and non-implanted blastocysts. On the other hand, the use of correcting factors tailored to the embryo’s gender and chromosomal status produced no difference of the corrected mtDNA level regarding ploidy, age, and implantation potential [16]. Other authors also reported no systematic difference in mtDNA quantity between implanted and non-implanted embryos with double embryo transfer [17]. However, the mtDNA level was negatively correlated with oocyte age, and high-quality embryos were correlated with lower mtDNA level [17].
Table 2.
Comparison of different methodologies for mitochondria DNA quantification and results from published articles
| Ref. | Sample size | Settings | nDNA method | mtDNA method | Result | Comments |
|---|---|---|---|---|---|---|
| Fragouli 2015 [12] |
379 embryos D3: 39 D5: 340 |
Single center |
D3: aCGH (24Sure, Illumina) D5: 302 → aCGH 38 → NGS (MiSeq or HiSeq, Illumina) |
RT-PCR (n = 341) NGS (n = 38) (PGM, Life Technologies) |
The relative quantity of mtDNA was significantly higher in old age, aneuploid, and non-implanted blastocysts, and cleavage stage in young age patient. Overall implantation rate: 38% (16/42); “Normal” mtDNA implantation rate: 59% All euploid blastocysts with mtDNA levels above the threshold failed to implant (100%, n = 14) |
Relative mtDNA threshold: 0.003 |
| Diez-Juan 2015 [13] |
270 embryos D3: 205 D5: 65 |
Single genetics laboratory | aCGH (24Sure, BlueGnome) | RT-PCR |
Mitochondrial score (Ms): mtDNA/nDNA Day 3: < 34 (MsA): IR = 59% (n = 51) 34–52 (MsB): IR = 44% (n = 52) 52–97 (MsC): IR = 42% (n = 50) > 97 (MsD): IR = 25% (n = 52) Ms > 160 (n = 22) IR = 0 Day 5: < 18.19 (MsA): IR = 81%(n = 16) 18.19–24.15 (MsB): IR = 50% (n = 16) 24.15–50.58 (MsC): IR = 62% (n = 16) > 50.58 (MsD): IR = 18% (n = 17) > 60 (n = 7): IR = 0 |
Less is better |
| Ravichandran 2017 [14] | 1505 euploid blastocysts | Multi-center | aCGH (24Sure, Illumina) or NGS (VeriSeq, Illumina) |
qRT-PCR 16s ribosomal RNA MajArc probe sequence |
Relative mtDNA levels (normalization against nuclear DNA sequence) Overall implantation rate: 65.6% (185/282) “Normal” mtDNA implantation rate: 74.3% Negative predictive value: 100% (n = 33) * elevated mtDNA embryos: 9.2% (139/1505) |
Relative mtDNA threshold: 0.0004 for 16s ribosomal RNA; 0.00035 for the MajArc sequence |
| Victor 2017 [16] |
1396 blastocysts Euploid: 775 Aneuploid: 621 |
Single center |
NGS (MiSeq, Illumina) qPCR validation: |
NGS (n = 1396) (MiSeq, Illumina) qPCR (n = 150) |
No difference regarding ploidy, age, and implantation potential (P vs. NP: 101/140 via NGS; 49/51 via qPCR) |
With correction factors tailored to gender and chromosomal status |
| Treff 2017 [17] |
374 blastocysts DET with 69 singletons |
Single center | qPCR | qPCR |
No systematic difference in relative mtDNA quantity between implanted and non-implanted embryos. MtDNA copy number was negatively correlated with oocyte age. Embryos biopsied on day 5 were more likely to have higher quantities of mtDNA compared with biopsied on day 6. Greater blastocyst expansion scores were correlated with lower mitochondrial content. Higher quality trophoblast and ICM were similarly correlated with lower mtDNA quantity. |
Highlight the importance of patient-specific variables |
| Fragouli 2017 [15] | 199 euploid blastocysts | Single center | NGS (VeriSeq, Illumina) | qPCR |
Ongoing pregnancy rate: Normal/low levels of mtDNA: 64% (121/190) Elevated mtDNA levels: 0% (0/9) * elevated mtDNA embryos: 4.5% (9/199) |
Blinded prospective non-selection study |
| Santos 2018 [23] | 1641 blastocysts | Single center | NGS (PGM, Thermo Fisher) | NGS (PGM, Thermo Fisher) | The probability of mtDNA content above the median increased by 6% when BMI increased; lowered by 39% when P4 increased, and increased by 42% in aneuploid blastocysts. | Potential variables that affect the mtDNA content |
In our study, we analyzed both the relationship between the mtDNA copy number and maternal age, embryo ploidy status, embryo morphology, and embryo viability in both blastomeres and trophectoderm biopsy. The blastomeres contained significantly higher mtDNA quantities compared with trophectoderm cells, which was expected given that the mtDNA replication does not take place before blastocyst stage and the number of mtDNA copies per cell is progressively diluted after each embryonic cell division [8]. The euploid rate in blastomere biopsies was lower compared to trophectoderm in our result despite a lower maternal age. The possible explanation included small sample size of blastomeres and technical artefacts. Also, these patients were more prone to be indicated for PGT-A due to repeated implantation failure. In our results, we did not find significant correlation between the adjusted mtDNA contents and the maternal age, and embryo ploidy status within blastomeres. Presently, the adjusted mtDNA level was significantly higher in aneuploid blastocysts than euploid blastocyst, which was consistent with previous study proposed by Fragouli et al. [12]. However, our data echoed with other studies that there were no significant differences regarding maternal age [16] and embryo implantation potential [16, 17].
We further analyzed the correlation among the embryo morphology, ploidy status, and adjusted mtDNA quantification of blastocysts. The logistic regression analysis revealed that the chance of euploid status was significantly positively correlated with expansion degree and negatively correlated with ICM and TE grading, respectively. We shared similar opinion with previous studies [24, 25] that the morphological evaluation cannot currently be improved and applied independently to determine embryo quality and to predict implantation although there seems to be a relationship between the ploidy status and blastocyst development. Moreover, we analyzed the relationship of adjusted mtDNA quantification stratified with embryo morphology and ploidy status. The results showed that the median adjusted mtDNA quantification was significantly higher in aneuploid blastocysts than euploid ones in the expansion 3–4 group, ICM grade B and C group, and TE grade B and C group when corrected with embryo morphology independently. These results implied that the aneuploid embryos contained more mtDNA copy content per cell compared with euploid embryos in the same developmental stage. This phenomenon might be related to the intrinsic factors of the embryo (i.e., ploidy status) but not simply to slower cell division and development. The reason why this phenomenon did not show in other groups may due to bias from small sample size. When stratified with ploidy status, the median adjusted mtDNA revealed no significant differences among group comparisons except that ICM grade A group showed significant higher adjusted mtDNA compared with ICM grade B and grade C in euploid blastocysts, which might be also biased by sample size. The results further strengthened the evidence that the adjusted mtDNA quantification was not influenced by embryo morphology and development. Taking together, our results revealed that the adjusted mtDNA quantification was significantly higher in aneuploid blastocyst than in euploid ones, which was independent to the influence of embryo development and morphological status.
Interestingly, we adopted the same methodology (NGS) and mathematic correction of mtDNA level as used previously [16], but obtained different results. The possible explanation might be related to the inappropriate statistical analysis. We demonstrated that the actual distribution of the mtDNA content was a non-normal distribution in both blastomeres and trophectoderm cells. Therefore, non-parametric analysis should be used instead of the parametric analysis used previously [16]. Moreover, various confounding factors, including patient-specific variables, variations between different IVF facilities, methodology, sample preparation and storage, and technical bias, might result in the discrepant results [26–28]. Ethnicity is another concern that previous studies did not take into account. To our best knowledge, we presented the first data for the correlation between mtDNA copy number and human embryo nature in Asians.
The distribution of the quantified adjusted mtDNA revealed high kurtosis and positive asymmetry, which means a left-shift distribution with a high concentration of data that fall within a narrow range of low values and a small number of data with very high value. This is consistent with a recent study [26]. In our results, the majority of the assessed blastocysts (98.4%) displayed an adjusted mtDNA level below the threshold. Ravichandran [14] reported that 9.2% (139/1505) of assessed blastocysts contained mtDNA level above the viability threshold. However, the percentage of elevated mtDNA levels ranged from 1 to 25% among the 35 clinics enrolled in their study. Moreover, half of the clinics (17/35) reported no cases of elevated mtDNA levels. Fragouli [15] also reported a 4.5% (9/199) prevalence of blastocysts with elevated mtDNA levels. The collective prior and present findings indicate that the elevation of mtDNA copy number is not a universal phenomenon. Rather, it only exists at a small percentage with an unusually high level.
When we adopted the upper limit of the adjusted mtDNA level of implanted blastocysts of 0.0042 as the normal threshold proposed by Ravichandran [14], we found that most blastocysts in our data had a normal adjusted mtDNA level (below the threshold; 99.3%, 981/988). Among them, the proportion of euploid and aneuploid blastocysts with an adjusted mtDNA level above the threshold was 0.66% (4/602) and 0.78% (3/386), respectively. Moreover, only two euploid blastocysts in the 267 embryos transferred contained an adjusted mtDNA level above this threshold. The predictive value of adjusted mtDNA quantification in blastocysts seemed too low to be used clinically.
Although we demonstrated a significantly lower level of adjusted mtDNA level in euploid blastocysts, which was consistent with several previous studies, we failed to demonstrate a correlation of the adjusted mtDNA level with embryo implantation potential. Since the ploidy status is clearly identified in PGT-A, there is no role of the adjusted mtDNA level to predict the embryo’s ploidy status. We also demonstrated a very weak positive correlation between adjusted mtDNA level and maternal age in trophectoderm cells, but not in blastomeres and when stratified by ploidy status. Accordingly, the small percentage of elevated mtDNA copy numbers in the general population make it unsuitable for clinical use as a biomarker for embryo implantation potential, although the negative predictive value was previously reported as high as 100% [12, 14, 15]. We share the view of others that the mtDNA quantification does not have clinical utility as a biomarker of implantation [29, 30].
Among all the previous studies, only two early studies [12, 13] involved blastomeres. One study [12] reported that the mtDNA quantities in blastomeres were significantly higher in younger patients, while an opposite trend was evident in blastocysts with mtDNA levels increasing significantly with advanced maternal age. These results were based on a small sample size with 39 blastomeres and 340 trophectoderm samples. The other study [13] examined 205 blastomeres and 65 trophectoderms and failed to demonstrate the relationship between mtDNA quantities and maternal age. The authors created the MitoScore and presented the implantation rates in different MitoScore intervals. Nevertheless, there was not a completely positive correlation between the MitoScore and the implantation potential. Previous studies had suggested that each cell in the embryo reduces its mtDNA amount and remains stable during the first 3 days of embryo development [31, 32]. Significant replication of mtDNA is not thought to be initiated until after an embryo completes the first cellular differentiation into the trophectoderm and inner cell mass and becomes a blastocyst [8, 33, 34]. Considering the significant mtDNA replication that occurs after blastocyst formation, the observation concerning mtDNA quantification at the cleavage stage might represent the corresponding oocytes instead of the embryos. In other words, the quantitative analysis of mtDNA at the early preimplantation stage may only be of scientific interest, with no clinical utility.
The limitations of our study include relatively small transfer percentage and sample size. Long-term data collection is needed since surplus embryos may be transferred years later after a successful implantation. Moreover, we assessed only euploid and aneuploid embryos, since the mathematic correction did not fit the embryos with mosaicism, segmental imbalance, and other complex abnormalities. The definition and the percentage of mosaicism is another concern. In our laboratory, the median nuclear copy number deviation less than 50% is reported as euploid, which may be different from that at other facilities.
Conclusions
The quantification of adjusted mitochondria DNA in human embryos is significantly lower in euploid blastocysts than in aneuploid blastocysts, which may be related to embryo stress and developmental kinetics. However, no statistically significant differences were evident regarding the blastomere ploidy status and in embryos stratified by maternal age and implantation outcome. Based on the current evidence available, adopting the mtDNA copy number as a clinical biomarker for human embryo implantation potential is not feasible at the present time. To our best knowledge, this study provides the first data set of correlation between mitochondria copy number and human embryo implantation potential in Asian ethnics.
Acknowledgments
The authors thank all the embryologists of the reproductive center of Taipei Medical University Hospital for embryo biopsy and sample preparation. We also thank the bioinformatics support provided by the Genomics Center for Clinical and Biotechnological Applications of the National Yang-Ming University VGH Genome Research Center. Genomics Center for Clinical and Biotechnological Applications is supported by the National Core Facility for Biopharmaceuticals, Ministry of Science and Technology. We also thank the Taipei Medical University Hospital for financial support. Finally, we thank the Genetics Generation Advancement Corporation for supporting this study in various forms, including sample preparation and interpretation, technical support, and extremely helpful discussions.
Funding
This study was partially supported by research grants NSC96-2314-B-038-019 and MOST-104-2314-B-038-063-MY2 from the Ministry of Science and Technology, Taiwan, Academia Sinica (BM10501010036, BM10601010024), National Healthy Research Institute (MG-1050SP-07, MG-106-SP-07), and Taipei Medical University Hospital (108TMUH-NE-01). The funders played no role in the conduct of the study or the writing of the manuscript.
Compliance with ethical standards
This study was reviewed and approved by the Taipei Medical University joint institutional review board (TMU-JIRB, approval number: N201707027). Patient informed consent for analysis of surplus DNA product was obtained with the approval of the ethical committee.
Conflict of interest
Dr. Yi-Hui Lin serves as a medical consultant for Genetics Generation Advancement Corporation. The other authors declare that they have no conflict of interest.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Yi-Xuan Lee, Phone: 886-2-2737218, Email: xuanshiva@gmail.com.
Chii-Ruey Tzeng, Phone: +886 2-2737-2181, Email: tzengcr@tmu.edu.tw.
References
- 1.Chang J, Boulet SL, Jeng G, Flowers L, Kissin DM. Outcomes of in vitro fertilization with preimplantation genetic diagnosis: an analysis of the United States Assisted Reproductive Technology Surveillance Data, 2011–2012. Fertil Steril. 2016;105(2):394–400. doi: 10.1016/j.fertnstert.2015.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fragouli E, Alfarawati S, Spath K, Jaroudi S, Sarasa J, Enciso M, Wells D. The origin and impact of embryonic aneuploidy. Hum Genet. 2013;132(9):1001–1013. doi: 10.1007/s00439-013-1309-0. [DOI] [PubMed] [Google Scholar]
- 3.Rosenwaks Z. Introduction: biomarkers of embryo viability: the search for the “holy grail” of embryo selection. Fertil Steril. 2017;108(5):719–721. doi: 10.1016/j.fertnstert.2017.10.011. [DOI] [PubMed] [Google Scholar]
- 4.Zegers-Hochschild F, Adamson GD, Dyer S, Racowsky C, de Mouzon J, Sokol R, Rienzi L, Sunde A, Schmidt L, Cooke ID, Simpson JL, van der Poel S. The international glossary on infertility and fertility care, 2017. Fertil Steril. 2017;108(3):393–406. doi: 10.1016/j.fertnstert.2017.06.005. [DOI] [PubMed] [Google Scholar]
- 5.Chan DC. Mitochondria: dynamic organelles in disease, aging, and development. Cell. 2006;125(7):1241–1252. doi: 10.1016/j.cell.2006.06.010. [DOI] [PubMed] [Google Scholar]
- 6.Wai T, Ao A, Zhang X, Cyr D, Dufort D, Shoubridge EA. The role of mitochondrial DNA copy number in mammalian fertility. Biol Reprod. 2010;83(1):52–62. doi: 10.1095/biolreprod.109.080887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.May-Panloup P, Boucret L, Chao de la Barca JM, Desquiret-Dumas V, Ferré-L'Hotellier V, Morinière C, Descamps P, Procaccio V, Reynier P. Ovarian ageing: the role of mitochondria in oocytes and follicles. Hum Reprod Update. 2016;22(6):725–743. doi: 10.1093/humupd/dmw028. [DOI] [PubMed] [Google Scholar]
- 8.St John JC, et al. Mitochondrial DNA transmission, replication and inheritance: a journey from the gamete through the embryo and into offspring and embryonic stem cells. Hum Reprod Update. 2010;16(5):488–509. doi: 10.1093/humupd/dmq002. [DOI] [PubMed] [Google Scholar]
- 9.Hsieh R-H, Au HK, Yeh TS, Chang SJ, Cheng YF, Tzeng CR. Decreased expression of mitochondrial genes in human unfertilized oocytes and arrested embryos. Fertil Steril. 2004;81:912–918. doi: 10.1016/j.fertnstert.2003.11.013. [DOI] [PubMed] [Google Scholar]
- 10.Hsieh R-H, Tsai NM, Au HK, Chang SJ, Wei YH, Tzeng CR. Multiple rearrangements of mitochondrial DNA in unfertilized human oocytes. Fertil Steril. 2002;77(5):1012–1017. doi: 10.1016/S0015-0282(02)02994-1. [DOI] [PubMed] [Google Scholar]
- 11.Van Blerkom J. Mitochondria in human oogenesis and preimplantation embryogenesis: engines of metabolism, ionic regulation and developmental competence. Reproduction. 2004;128(3):269–280. doi: 10.1530/rep.1.00240. [DOI] [PubMed] [Google Scholar]
- 12.Fragouli E, Spath K, Alfarawati S, Kaper F, Craig A, Michel CE, Kokocinski F, Cohen J, Munne S, Wells D. Altered levels of mitochondrial DNA are associated with female age, aneuploidy, and provide an independent measure of embryonic implantation potential. PLoS Genet. 2015;11(6):e1005241. doi: 10.1371/journal.pgen.1005241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Diez-Juan A, Rubio C, Marin C, Martinez S, al-Asmar N, Riboldi M, Díaz-Gimeno P, Valbuena D, Simón C. Mitochondrial DNA content as a viability score in human euploid embryos: less is better. Fertil Steril. 2015;104(3):534–541. doi: 10.1016/j.fertnstert.2015.05.022. [DOI] [PubMed] [Google Scholar]
- 14.Ravichandran K, et al. Mitochondrial DNA quantification as a tool for embryo viability assessment: retrospective analysis of data from single euploid blastocyst transfers. Hum Reprod. 2017:1–11. [DOI] [PubMed]
- 15.Fragouli E, McCaffrey C, Ravichandran K, Spath K, Grifo JA, Munné S, Wells D. Clinical implications of mitochondrial DNA quantification on pregnancy outcomes: a blinded prospective non-selection study. Hum Reprod. 2017;32(11):2340–2347. doi: 10.1093/humrep/dex292. [DOI] [PubMed] [Google Scholar]
- 16.Victor AR, Brake AJ, Tyndall JC, Griffin DK, Zouves CG, Barnes FL, Viotti M. Accurate quantitation of mitochondrial DNA reveals uniform levels in human blastocysts irrespective of ploidy, age, or implantation potential. Fertil Steril. 2017;107(1):34–42. doi: 10.1016/j.fertnstert.2016.09.028. [DOI] [PubMed] [Google Scholar]
- 17.Treff NR, Zhan Y, Tao X, Olcha M, Han M, Rajchel J, et al. Levels of trophectoderm mitochondrial DNA do not predict the reproductive potential of sibling embryos. Hum Reprod. 2017:1–9. [DOI] [PMC free article] [PubMed]
- 18.Fiorentino F, Bono S, Biricik A, Nuccitelli A, Cotroneo E, Cottone G, Kokocinski F, Michel CE, Minasi MG, Greco E. Application of next-generation sequencing technology for comprehensive aneuploidy screening of blastocysts in clinical preimplantation genetic screening cycles. Hum Reprod. 2014;29(12):2802–2813. doi: 10.1093/humrep/deu277. [DOI] [PubMed] [Google Scholar]
- 19.Gardner DK, Lane M, Stevens J, Schlenker T, Schoolcraft WB. Blastocyst score affects implantation and pregnancy outcome: towards a single blastocyst transfer. Fertil Steril. 2000;73(6):1155–1158. doi: 10.1016/S0015-0282(00)00518-5. [DOI] [PubMed] [Google Scholar]
- 20.Nasiri N, Eftekhari-Yazdi P. An overview of the available methods for morphological scoring of pre-implantation embryos in in vitro fertilization. Cell J. 2015;16(4):392–405. doi: 10.22074/cellj.2015.486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Scott RT, et al. Blastocyst biopsy with comprehensive chromosome screening and fresh embryo transfer significantly increases in vitro fertilization implantation and delivery rates: a randomized controlled trial. Fertil Steril. 2013;100(3):697–703. doi: 10.1016/j.fertnstert.2013.04.035. [DOI] [PubMed] [Google Scholar]
- 22.Rubio C, Bellver J, Rodrigo L, Castillón G, Guillén A, Vidal C, Giles J, Ferrando M, Cabanillas S, Remohí J, Pellicer A, Simón C. In vitro fertilization with preimplantation genetic diagnosis for aneuploidies in advanced maternal age: a randomized, controlled study. Fertil Steril. 2017;107(5):1122–1129. doi: 10.1016/j.fertnstert.2017.03.011. [DOI] [PubMed] [Google Scholar]
- 23.Practice Committees of the American Society for Reproductive M et al. The use of preimplantation genetic testing for aneuploidy (PGT-A): a committee opinion. Fertil Steril. 2018;109(3):429–436. doi: 10.1016/j.fertnstert.2018.01.002. [DOI] [PubMed] [Google Scholar]
- 24.Capalbo A, Rienzi L, Cimadomo D, Maggiulli R, Elliott T, Wright G, Nagy ZP, Ubaldi FM. Correlation between standard blastocyst morphology, euploidy and implantation: an observational study in two centers involving 956 screened blastocysts. Hum Reprod. 2014;29(6):1173–1181. doi: 10.1093/humrep/deu033. [DOI] [PubMed] [Google Scholar]
- 25.Minasi MG, Colasante A, Riccio T, Ruberti A, Casciani V, Scarselli F, Spinella F, Fiorentino F, Varricchio MT, Greco E. Correlation between aneuploidy, standard morphology evaluation and morphokinetic development in 1730 biopsied blastocysts: a consecutive case series study. Hum Reprod. 2016;31(10):2245–2254. doi: 10.1093/humrep/dew183. [DOI] [PubMed] [Google Scholar]
- 26.de Los Santos MJ, et al. Variables associated with mitochondrial copy number in human blastocysts: what can we learn from trophectoderm biopsies? Fertil Steril. 2018;109(1):110–117. doi: 10.1016/j.fertnstert.2017.09.022. [DOI] [PubMed] [Google Scholar]
- 27.Wells D. Mitochondrial DNA quantity as a biomarker for blastocyst implantation potential. Fertil Steril. 2017;108(5):742–747. doi: 10.1016/j.fertnstert.2017.10.007. [DOI] [PubMed] [Google Scholar]
- 28.Humaidan P, Kristensen SG, Coetzee K. Mitochondrial DNA, a new biomarker of embryonic implantation potential: fact or fiction? Fertil Steril. 2018;109(1):61–62. doi: 10.1016/j.fertnstert.2017.10.017. [DOI] [PubMed] [Google Scholar]
- 29.Viotti M, Victor AR, Zouves CG, Barnes FL. Is mitochondrial DNA quantitation in blastocyst trophectoderm cells predictive of developmental competence and outcome in clinical IVF? J Assist Reprod Genet. 2017;34(12):1581–1585. doi: 10.1007/s10815-017-1072-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Scott RT., Jr Enhanced techniques to “power” embryonic mitochondria research. Fertil Steril. 2017;107(1):59–60. doi: 10.1016/j.fertnstert.2016.11.024. [DOI] [PubMed] [Google Scholar]
- 31.Barritt JA, K.M., Cohen J, Steuerwald N, Brenner CA, Quantification of human ooplasmic mitochondria. Reprod BioMed Online, 2002. 4: p. 243–247. [DOI] [PubMed]
- 32.Lin D. Comparison of mitochondrial DNA contents in human embryos with good or poor morphology at the 8-cell stage. Fertil Steril. 2004;81(1):73–79. doi: 10.1016/j.fertnstert.2003.05.005. [DOI] [PubMed] [Google Scholar]
- 33.Eichenlaub-Ritter U, Wieczorek M, Lüke S, Seidel T. Age related changes in mitochondrial function and new approaches to study redox regulation in mammalian oocytes in response to age or maturation conditions. Mitochondrion. 2011;11(5):783–796. doi: 10.1016/j.mito.2010.08.011. [DOI] [PubMed] [Google Scholar]
- 34.Tsai T, John JCS. The role of mitochondrial DNA copy number, variants, and haplotypes in farm animal developmental outcome. Domest Anim Endocrinol. 2016;56:S133–S146. doi: 10.1016/j.domaniend.2016.03.005. [DOI] [PubMed] [Google Scholar]