Abstract
The microbiome plays a key role in the biology, ecology and evolution of arthropod vectors of human pathogens. Vector-bacterial interactions could alter disease transmission dynamics through modulating pathogen replication and/or vector fitness. Nonetheless, our understanding of the factors shaping the bacterial community in arthropod vectors is incomplete. Using large-scale 16S amplicon sequencing, we examine how habitat disturbance structures the bacterial assemblages of field-collected whole-body hematophagous arthropods that vector human pathogens including mosquitoes (Culicidae), sand flies (Psychodidae), biting midges (Ceratopogonidae) and hard ticks (Ixodidae). We found that all comparisons of the bacterial community among species yielded statistically significant differences, but a difference was not observed between adults and nymphs of the hard tick, Haemaphysalis juxtakochi. While Culicoides species had the most distinct bacterial community among dipterans, tick species were composed of entirely different bacterial OTU’s. We observed differences in the proportions of some bacterial types between pristine and disturbed habitats for Coquillettidia mosquitoes, Culex mosquitoes, and Lutzomyia sand flies, but their associations differed within and among arthropod assemblages. In contrast, habitat quality was a poor predictor of differences in bacterial classes for Culicoides biting midges and hard tick species. In general, similarities in the bacterial communities among hematophagous arthropods could be explained by their phylogenetic relatedness, although intraspecific variation seems influenced by habitat disturbance.
Introduction
Bacterial communities are important components of hematophagous arthropods (e.g., blood feeders) vectoring disease-causing pathogens to humans and wildlife, and they are likely to play a key role in vector ecology, evolution and transmission capacity [1–4]. Several important human and animal diseases result from bacterial infection transmitted through the bite of arthropod vectors [5,6]. Bacteria also interact with the arthropod host to reduce or increase the transmission of pathogens or indirectly alter disease dynamics through the modification of nutrition [7], development, reproduction or the immune response of arthropod vectors [8,9]. Our understanding of the factors shaping the organization of bacterial communities in hematophagous arthropods vectoring human diseases is still limited. Studies regarding the microbiome of disease vectors have attempted to describe the structure and bacterial composition of specific taxonomic groups of arthropods, and to understand how it varies according to particular ecological or physiological factors, with the most comprehensive studies focused on mosquitoes [10] and ticks [2]. Although some studies have considered the impact of habitat or environment type on arthropod microbiota in mosquitoes [11–16], ticks [17–20] and biting midges [21], none to date have investigated the role of habitat disturbance in shaping bacterial assemblages among co-distributed hematophagous arthropods.
Mosquitoes (Diptera: Culicidae), sand flies (Diptera: Psychodidae), biting midges (Diptera: Ceratopogonidae) and hard ticks (Acari: Ixodidae) are collectively responsible for numerous medically important diseases worldwide, including arthropod-borne viruses (e.g., arboviruses) transmitted to humans (Dengue–DENV, chikungunya—CHIKV, Zika—ZIKV, Yellow Fever–YFV, West Nile–WNV, Mayaro and Oropuche) and to agriculturally important livestock (Vesicular Stomatitis–VSV, Blue-Tongue–BTV, Epizootic Hemorrhagic Disease–EHDV and African Horse Sickness–AHSV) or to both (Venezuelan Equine Encephalitis–VEEV, Eastern Equine Encephalitis–EEEV and Rift Valley Fever) [22,23]. In addition, some species in these arthropod assemblages are involved in the transmission of parasites such as filarial nematodes (Mansonella—filariasis) [24], protozoan (Leishmania—Leishmaniasis) [25] and bacteria (Rickettsia—Lyme disease and babesiosis) [26].
The ability of hematophagous arthropods to carry and transmit pathogens biologically is given by their population dynamics and feeding behaviour in relation to that of their vertebrate host, plus their immune responses to infection [9,27]. Some bacterial commensals impact the capacity of arthropods as biological vectors, through diminishing pathogen replication and dissemination in the host tissues or by reducing vector fitness and lifespan [4,9,28,29]. Studies from members of the Culicidae demonstrate the importance of the microbiome in modulating disease transmission. For example, Chromobacterium, Proteus and Paenibacillus bacteria can inhibit DENV replication in mosquitoes while the resident bacteria are required for its establishment [28]. Furthermore, the intracellular bacterium Wolbachia is known to adversely influence the transmission of DENV, CHIKV, ZIKV, YFV and WNV [30–35]. Alternatively, some bacteria are associated with an increase in disease transmission by their arthropod vectors. For example, members of Enterobacteriae are correlated with higher Plasmodium infection rates in Anopheles mosquitoes, while Serratia odorifera can increase the replication of both DENV and CHIKV in the midgut of Aedes aegypti [28,36]. Although, studies have endeavored to characterize the core microbiome of members of Psychodidae sand flies, Ceratopogonidae biting midges and Ixodidae hard ticks, it is still generally unknown how similar or different their microbiomes are, and whether some bacteria may influence disease transmission dynamics in these arthropod assemblages [18,19,21,37–42]. Nonetheless, some studies have revealed that resident bacteria are essential for the development of Leishmania parasite in Psychodidae through antibiotic treatment [37,43].
Metagenomic studies of disease vectors in the Order Diptera have revealed that different genera including those with a distinct ecology generally share a core microbiome, but often exhibit differences in bacterial composition and structure that distinguish a species [1,13,38,44–46]. Conversely, tick species may exhibit a distinct taxonomic structure in their microbiome, because they are associated with specific vertebrate hosts throughout their entire lifetime, including during the immature stages [47]. Core microbiota of Diptera are largely acquired from the environment during the immature stages, many of which persist until the adult stage [13,48–50]. Bacteria are also acquired during adult blood feeding, therefore the microbiome of arthropod vectors is likely impacted by both developmental stage and gender [1,19,49,51]. The core microbiota of ticks is either maternally-inherited, acquired from blood feeding on hosts or through the colonization of environmental microorganisms from vertebrate skin or the soil on physical contact [2,52].
Hematophagous arthropods can exhibit intra-specific variation in their bacterial associates between geographic locations, explained by differences in the quality of larval habitats or host preferences at sampling sites for both Diptera [11,37,46,53] and ticks [17,18]. Hence, it has been proposed that larval habitat conditions and geographic location are important factors shaping the bacterial community of some adult hematophagous arthropods. Conversely, some mosquitoes [44] and ticks [54] do not exhibit intra-specific variation in the bacterial community across geographical locations or habitats. This finding supports a more specific and long term association between some blood-feeding arthropods and their bacterial associates, which is likely mediated by the immune system of the host, rather than by their external environment [13,54].
Our goal herein is to test for variation in the diversity of bacteria among four epidemiologically discrete groups of hematophagous arthropods, and to identify the factors shaping this variation. Specifically, we address the following questions: (1) How do patterns of bacterial diversity and composition differ among the microbiomes of mosquitoes, biting midges, sand flies and hard ticks?, and (2) Does habitat disturbance influence the organization of bacterial communities within these arthropod assemblages? We posit that blood-feeding arthropod species in the Order Diptera will harbor comparable bacterial organizations, since they are more closely phylogenetically related, while hard ticks within the Order Ixodida are considered as an outgroup. We also postulate that intra-specific bacterial diversity and taxa composition will change owing to variation in habitat quality, but changes are only expected within Culicidae mosquitoes, Psychodidae sand flies and Ceratopogonidae biting midges. This is anticipated because host–tick interactions in obligated ectoparasites such as hard ticks are more likely to shape their microbiome than habitat disturbance. Although hard ticks can acquire surface microbiota from their environment, our study largely targets intracellular and gut bacteria colonized through vertical transmission or ingestation. We use a metabarcoding approach to compare inter-and intra- group bacterial communities among these arthropod assemblages, and also in relation to changes in habitat quality. If habitat disturbance is a significant predictor of bacterial assemblages, this could have ramifications for disease transmission through variation of the vector microbiome and correlated vectorial capacity.
Materials and methods
Arthropod collection and sample preparation
Permission was obtained from MiAmbiente under permit identification ID 8-447-900-PAN. The study was conducted in the lowland tropical rainforest ecosystem of central Panama, a region formerly known as the Panama Canal Zone. Adult specimens of mosquitoes, sand flies, biting midges and hard ticks were gathered from three forested areas that varied in their levels of anthropogenic disturbance and original habitat quality. These included a pristine site, Barro Colorado Island (BCI), which is comprised of old-growth forest with low levels of disturbance (e.g., >65% forest cover). In addition, two disturbed forest sites, Achiote (ACH) and Las Pavas (PVAS), encompass patches of secondary-growth forest subject to intermediate and high levels of disturbance (e.g., >35% and <65% forest cover) respectively [55,56]. Dipterans were collected using six Center for Disease Control (CDC) miniature light traps (John W. Hock Company, Gainesville, Florida), operating overnight in the understory (1.5 m height) and six in the canopy (> 25 m height), alternating each night. Each trap was situated along a transect and spaced at least 300 meters apart from each other to avoid pseudoreplication as in Loaiza et al. [55,57]. They were baited with 0.5 pounds of dry ice to attract blood-seeking dipterans. Adult specimens were retrieved from the traps at sunrise and taken to the laboratory in a portable freezer container holding dry ice. Individuals were sorted and identified using a chill table and taxonomic keys [58–62].
Ixodid ticks were collected with two methods at BCI and PVAS: the standard tick-dragging technique [63], and a pair of home-made cloth-pants, fabricated with white rustic fabric. Two human collectors traversed linear transects of up to 200 meters through the vegetation using either method. Adult specimens were removed from the cloth with entomological forceps, while immature stages (e.g., larvae and nymphs) were detached using transparent adhesive tape. Individuals were placed in separate cryo-vials, and subsequently transported to the laboratory. Taxonomic characters were used to identify ticks to the species level [64,65]. The samples were washed with 70% ethanol to remove surface contamination before storage in 95% ethanol. Details on the number of samples processed from each site and for each species are provided in S1 Table.
DNA extraction, 16S rRNA gene library and sequencing
Each arthropod species was processed using the following laboratory procedures independently. Each sample was rinsed in 70% ethanol before they were pooled. DNA was isolated from pools of adult female dipterans and both adults and immature ticks using a BioSprint 96 robot and associated BioSprint® 96 DNA Blood kit (Qiagen, Gaithersburg, MD, USA). Each pool was crushed individually in tissue lysis buffer using a high-speed shaking TissueLyser II and ceramic beads; the supernatant was placed in a well of a 96-well plate and followed by DNA isolation protocol from the manufacturer. DNA pools were made by combining 2 μl of DNA extract from 20 to 35 individuals of sand flies and biting midges, plus up to 5 individuals per pool of mosquitoes and ticks. Pooled DNA was used as a template to amplify the V4 region of the 16S rRNA locus using a two-step PCR protocol. The first PCR was composed of 5 μl of 2X Maxima HotStart PCR Master Mix (Thermo), 0.2 μl of each primer (which included an Illumina sequencing primer on the 5’ end (10 mM)), and 1 μl of pooled DNA. Then 1 μl of the resulting PCR product was used to add on unique barcodes and Illumina sequencing adaptors in a second PCR of six cycles. The PCR cycling conditions had an initial denaturation step of 3 min at 94° C proceeding 25 cycles of 94°C for 45 sec, 50°C for 60 sec, and 72°C for 90 sec, followed by 10 min at 72°C extension. Resulting reactions were cleaned using PCR Normalization plates (Charm Biotech, San Diego, CA, USA) and samples pooled into a library which we concentrated using Kapa magnetic beads. The DNA concentration of each library was verified with the Qubit HS assay (Invitrogen, Waltham, MA, USA) and quality checked with a Bioanalyzer dsDNA High Sensitivity assay before sequencing on an Illumina MiSeq in a 2x250 paired end run. In the Culicidae family (mosquitoes), 40 pools of adult Culex including 20 pools of each Culex coronator and Culex declarator plus 20 pools of Coquillettidia venezuelensis were sequenced. Within the Ceratopogonidae (biting midges) and Psychodidae (sand flies), 94 pools of adult Culicoides including 34 pools of Culicoides batesi, 30 of Culicoides foxi, and 30 of Culicoides heliconiae, plus 75 pools of adult Lutzomyia including 30 pools of Lutzomyia panamensis, 23 of Lutzomyia gomezi and 22 of Lutzomyia trapidoi were sequenced and analyzed. Sequences within the hard tick family Ixodidae were obtained from 37 pools in total, including 6 pools of adults and 12 pools of nymphs of Haemaphysalis juxtakochi, 12 pools of adult Amblyomma tapirellum and 7 pools of adult of Amblyomma oblongoguttatum (S1 Table).
Analysis of 16S metadata
Analysis of sequence reads was performed using the Quantitative Insights Into Microbial Ecology (QIIME) software package versions 1.9.1 and 2.0. The DADA2 data quality filtering pipeline implemented in QIIME 2.0 was used to trim sequences with base quality scores lower than 20. Operational taxonomic units (OTU’s) were assigned with a Naive Bayes classifier trained on the Greengenes 99% sequence similarity database v13.8 with sequences bound by the 515F and 806R primer pair [66]. Low abundance OTU’s (0.005%) were filtered from the resulting relative abundance table to reduce bias by sequencing error.
The feature table was rarefied to a sequencing depth of 7 000 reads before alpha and beta diversity values were calculated. The statistical test PERMANOVA was applied to the resulting UNIFRAC distance matrixes to test for significant differences between the beta diversity of metadata groups. Principle coordinates analysis (PCoA) plots were generated from unweighted UNIFRAC distance matrixes. In addition, taxonomic summary plots of the relative abundance of bacteria were generated to depict the bacterial orders with an overall proportion of > 0.1% in at least one species. Indicator species analysis was applied to identify the OTU’s unique to each species group.
Results
In total, 11 435 639 sequence reads of the bacterial 16S gene were captured from 265 sample pools, encompassing 4 916 individuals from four different hematophagous arthropod families, six genera and 12 species. After quality filtering and rarefaction to a depth of 7 000 reads, 10 838 632 sequences remained from 229 sample pools with an average of 40 900 sequences per pool (SE ± 1,209) and a total of 1 404 OTU’s composed of 13 phyla, 30 classes, 55 orders, 106 families and 137 genera. Rarefaction curves revealed that the majority of bacterial diversity for all the species of arthropods was captured with subsampling of 7 000 sequences per sample pool (S1 Fig).
Bacterial diversity and composition in mosquitoes, biting midges, sand flies and hard ticks
Among dipterans, members of the genera Culex, Coquillettidia, Culicoides and Lutzomyia had comparable proportions of bacterial OTU’s, bacterial diversity and community evenness index. In contrast, two tick species in the genus Amblyomma (i.e., Amblyomma tapirellum and Amblyomma oblongoguttatum) had higher number of OTU’s, and bacterial diversity, and the least even community composition. A third tick species, Haemaphysalis juxtakochi, had the highest overall bacterial phylogenetic diversity, although it had a lower number of OTU’s per tick pool and values of Shannon’s diversity compared to Amblyomma species (Table 1 and S1 Fig).
Table 1. Average measures of bacterial alpha diversity for 12 species of blood-feeding arthropods at a rarefaction depth of 7 000 16S sequences.
| Taxonomy | Species | Observed OTU's | Shannon's diversity | Faith's phylogenetic diversity | Evenness | |
|---|---|---|---|---|---|---|
| Acari:Ixodidae | H. juxtakochi | 43.12 | 3.51 | 14.12 | 0.66 | |
| Adults | 55.71 | 3.39 | 11.65 | 0.69 | ||
| Nymphs | 34.3 | 7.12 | 11.28 | 0.94 | ||
| A. tapirellum | 194.82 | 7.15 | 11.43 | 0.95 | ||
| A. oblongoguttatum | 144.75 | 5.64 | 11.5 | 0.81 | ||
| Diptera:Culicidae | Coq. venezuelensis | 53.35 | 3.67 | 5.83 | 0.65 | |
| Cux. coronator | 49.7 | 3.64 | 5.91 | 0.65 | ||
| Cux. declarator | 49.2 | 3.29 | 6.39 | 0.59 | ||
| Diptera:Ceratopogonidae | C. batesi | 59.38 | 4.03 | 7.13 | 0.69 | |
| C. foxi | 61.73 | 3.93 | 7.18 | 0.67 | ||
| C. heliconiae | 56.15 | 3.63 | 6.81 | 0.63 | ||
| Diptera:Psychodidae | Lu. gomezi | 55.04 | 2.99 | 7.1 | 0.52 | |
| Lu. panamensis | 62.97 | 3.59 | 6.85 | 0.61 | ||
| Lu. trapidoi | 43.59 | 2.86 | 5.76 | 0.54 | ||
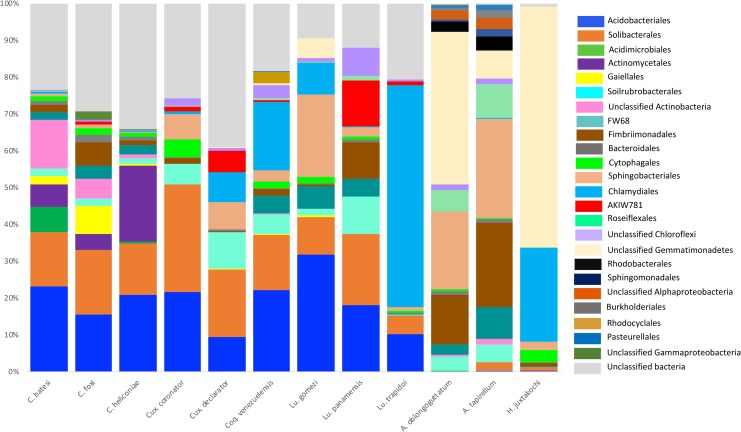
All arthropod species were dominated by the phylum Proteobacteria with proportions ranging from 48 to 72%. Other major bacteria phyla that were shared among all arthropod species included Firmicutes, Bacteriodetes and Actinobacteria. Bacterial Orders and families were generally shared among arthropod genera in the Order Diptera, although they also exhibited notable differences in their relative proportions, which are visualized to the level of Order in Fig 1 and summarised to the genus level in S2 Table. Within the bacterial phyla shared between Culex and Coquillettidia mosquitoes, Culicoides biting midges, and Lutzomyia sand flies, the major classes consisted of Gammaproteobacteria, Betaproteobacteria, Alphaproteobacteria, Bacilli, and Actinobacteria.
Fig 1. Relative abundances of bacterial orders above 0.1% summarized for each blood-feeding arthropod species.
Culicoides species share OTU’s with the other genera of dipterans, but PCA and taxonomic analysis revealed that they have a more distinct bacterial community than Lutzomyia, Culex and Coquillettidia together with unique bacterial types including a disease-causing agent in the genus Arcobacter (proteobacterial class Epsilonproteobacteria, Order Campylobacterales) [67], and Candidatus cardinium (phylum of Bacteriodetes, class Cytophagia), which is known to alter arthropod reproduction [68]. Moreover, Culicoides batesi, Culicoides foxi and Lutzomyia trapidoi had unique OTU’s in the phyla Chlamydiae.
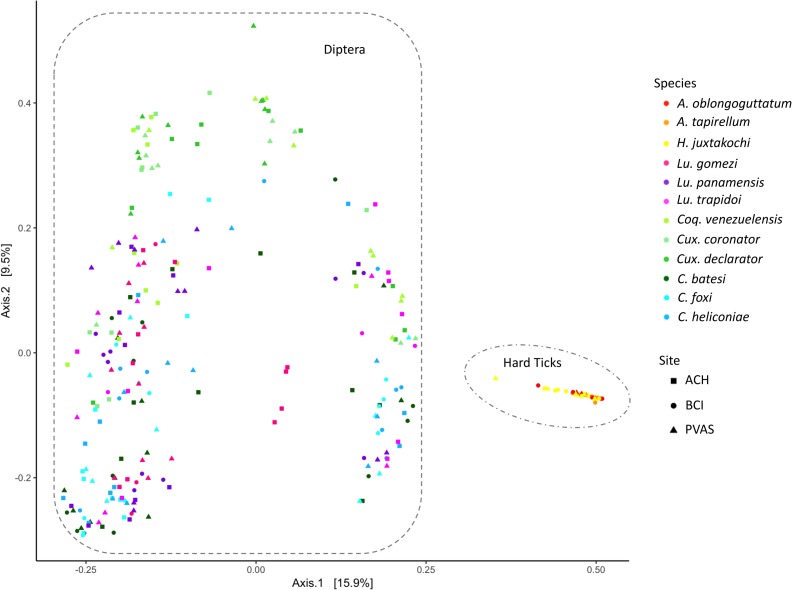
The bacterial phyla and classes of all tick species were composed of entirely different OTU’s than the other arthropod assemblages, hence they were the most distinct in terms of bacterial composition (Fig 2). Ticks in the genus Amblyomma had bacterial phyla that were not found in any other arthropod genus, including Chloroflexi, Acidobacteria, Gemmatimonadetes, Armatimonadetes and TM7. Likewise, Amblyomma ticks had a number of classes unique to this genus, including the Protobacterium Deltaproteobacteria, Saprospirae, Cytophagia within the phylum of Bacteriodetes, and the Actinobacteria Thermoleophilia and Acidimicrobiia. A. tapirellum had the largest proportion (14.7%) of OTU’s unique to its species (Fig 3).
Fig 2. PCoA ordination analysis based on UNIFRAC distances with 16S gene sequence variation of the bacterial communities from six blood-feeding arthropod genera.
Fig 3.
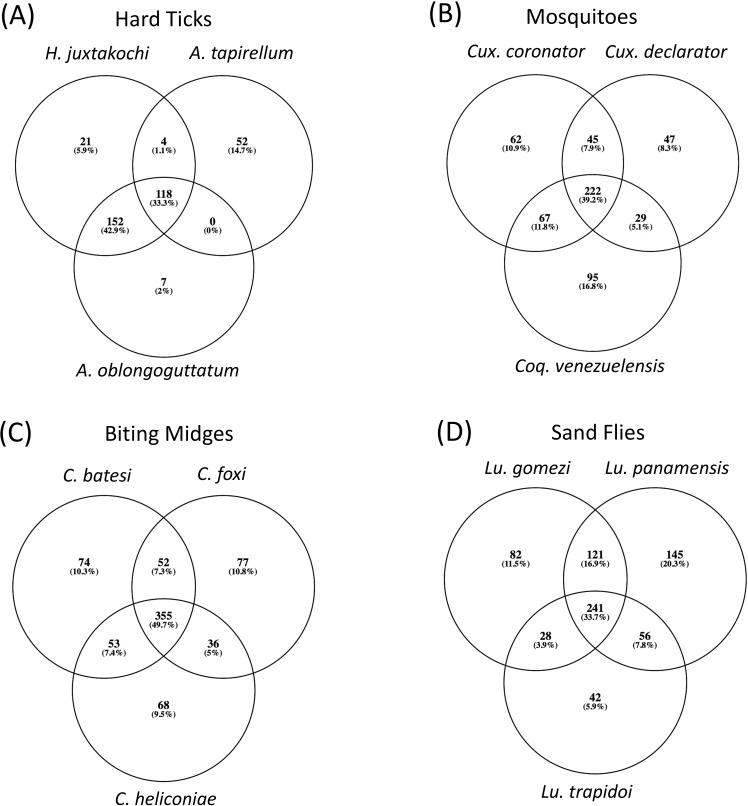
Venn diagram of shared and unique bacterial OTU’s among (a) three different species of Ixodidae; (b) two species of Culex (Culicidae) and one species of Coquillettidia (Culicidae); (c) three species of Culicoides (Ceratopogonidae); (d) three species of Lutzmyia (Psychodidae).
All comparisons of the bacterial community among the different genera and species of adult hematophagous arthropods through PERMANOVA tests yielded statistically significant differences (Table 2). Additionally, there were no statistically significant differences between the adults and nymphs of H. juxtakochi based on UNIFRAC distances of bacterial OTU’s (PERMANOVA, pseudo-F = 1.38, P = 0.247), although they share only 80 OTU’s (25%). Variation in the number of OTU’s shared among the different arthropod species are visualized in Fig 3. Arthropods within the same genus shared between ~33 to 50% of OTU’s while a smaller proportion were unique to each species (between 2 to 20%). The taxonomy of indicator OTU’s for each arthropod species in this study identified as significant and with an indicator value over 0.25 are provided in S3 Table.
Table 2. Results of PERMANOVA test for the comparison of bacterial OTU’s among pools of six different genera and 10 different species (with within genera comparisons) of blood-feeding arthropods based on unweighted UNIFRAC distances.
| Genera comparisons | No. of individuals within pools |
No. of sample pools | pseudo-F | p-value | q-value | |
|---|---|---|---|---|---|---|
| Amblyomma | Coquilettidia | 195 | 39 | 77.076 | 0.001 | 0.001 |
| Amblyomma | Culex | 295 | 59 | 95.687 | 0.001 | 0.001 |
| Amblyomma | Culicoides | 2669 | 113 | 126.282 | 0.001 | 0.001 |
| Amblyomma | Haemaphysalis | 180 | 36 | 82.764 | 0.001 | 0.001 |
| Amblyomma | Lutzomyia | 2137 | 94 | 94.831 | 0.001 | 0.001 |
| Coquilettidia | Culex | 300 | 60 | 4.267 | 0.002 | 0.002 |
| Coquilettidia | Culicoides | 2674 | 114 | 17.305 | 0.001 | 0.001 |
| Coquilettidia | Haemaphysalis | 185 | 37 | 69.818 | 0.001 | 0.001 |
| Coquilettidia | Lutzomyia | 2142 | 95 | 10.184 | 0.001 | 0.001 |
| Culex | Culicoides | 2774 | 134 | 39.82 | 0.001 | 0.001 |
| Culex | Haemaphysalis | 285 | 57 | 86.494 | 0.001 | 0.001 |
| Culex | Lutzomyia | 2242 | 115 | 23.658 | 0.001 | 0.001 |
| Culicoides | Haemaphysalis | 2574 | 111 | 113.82 | 0.001 | 0.001 |
| Culicoides | Lutzomyia | 4616 | 169 | 25.43 | 0.001 | 0.001 |
| Haemaphysalis | Lutzomyia | 2127 | 92 | 85.509 | 0.001 | 0.001 |
| Species comparisons | ||||||
| A. oblongoguttatum | A. tapirellum | 85 | 19 | 10.489 | 0.001 | 0.001 |
| Cux. coronator | Cux. declarator | 200 | 40 | 2.099 | 0.025 | 0.026 |
| C. batesi | C. foxi | 1825 | 64 | 1.499 | 0.102 | 0.102 |
| C. batesi | C. heliconiae | 1715 | 64 | 2.159 | 0.012 | 0.013 |
| C. foxi | C. heliconiae | 1608 | 60 | 1.611 | 0.064 | 0.065 |
| Lu. gomezi | Lu. panamensis | 1419 | 53 | 9.881 | 0.001 | 0.001 |
| Lu. gomezi | Lu. trapidoi | 1253 | 45 | 8.12 | 0.001 | 0.001 |
| Lu. panamensis | Lu. trapidoi | 1412 | 52 | 7.3 | 0.001 | 0.001 |
| Life Stage comparisons | ||||||
| Haemaphysalis adults | Haemaphysalis nymphs | 85 | 17 | 1.38 | 0.247 | 0.247 |
Effect of habitat disturbance on the organization of bacterial communities
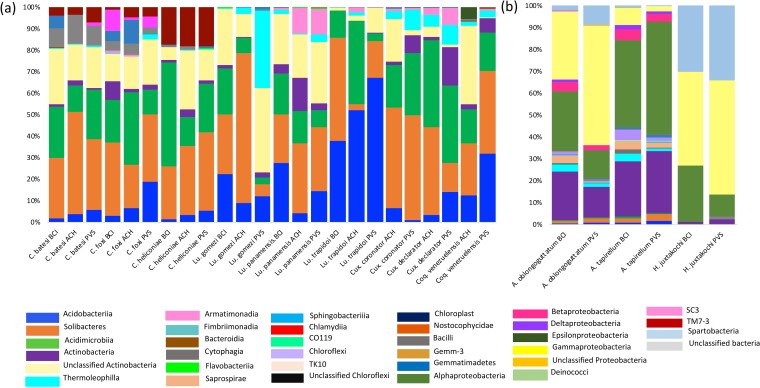
Intra-specific variation in the bacterial community was observed between sampling areas depicting different degrees of habitat disturbance for Coquillettidia, one Culex species, and all but one comparison of Lutzomyia, while another comparison between Culex coronator was close to significant (Table 3). Although bacterial diversity was comparable across pristine and disturbed habitats for most groups overall (S4 Table), we observed differences in the proportions of a number of bacterial types between pristine and disturbed habitats, although their associations differed within and among arthropod genera and species (Fig 4). For example, there was a high proportion of Cyanobacteria in both Coquillettidia and Lutzomyia from the disturbed sites at ACH and PVAS as well as an increased proportion of Chlamydiae for both Culex and Lutzomyia from the most disturbed site at PVAS. Similarly, there was an increased proportion of Betaproteobacteria, Order Burkholderias and the Flavobacteriia, family Blattabacteriaceae in pools of Culex from PVAS. Proportions of Actinobacteria, Bacteriodetes, Flavobacteria and Bacteroidia increased in Luztomyia from disturbed sites, whereas the proportion of Deltaproteobacteria increased from the pristine site BCI. Moreover, a number of bacterial classes including Nostococidae, Deltaproteobacteria, Deincoccci, Cytophagia and Chloroplast were found in Coquillettidia from the intermediately disturbed site at ACH, but not in the most disturbed site at PVAS. In contrast, within the three species of ticks, there was no difference in the proportion of bacterial classes between sampling areas or sampling method (Table 3, S4 Table and Fig 4). Similarly, no strong differences were detected in the bacterial classes of Culicoides among sampling areas or between vertical strata (i.e., forest understory or canopy) (Table 3 and S5 Table).
Table 3. Results of PERMANOVA test for the comparison of bacterial communities in pools of twelve different blood-feeding arthropod species among sampling areas based on unweighted UNIFRAC distances and 999 permutations.
Significant results are highlighted in bold.
| Taxonomy | Species | Site comparison | No. of sample pools | pseudo-F | p-value | q-value | |
|---|---|---|---|---|---|---|---|
| Diptera:Culicidae | Coq. venezuelensis | ACH | PVS | 20 | 6.920 | 0.001 | 0.001 |
| Cux. coronator | ACH | PVS | 20 | 2.042 | 0.055 | 0.055 | |
| Cux. declarator | ACH | PVS | 20 | 3.061 | 0.001 | 0.001 | |
| Diptera:Ceratopogonidae | C. batesi | ACH | BCI | 23 | 1.797 | 0.092 | 0.100 |
| ACH | PVS | 22 | 2.229 | 0.027 | 0.081 | ||
| BCI | PVS | 23 | 1.529 | 0.100 | 0.100 | ||
| C. foxi | ACH | BCI | 20 | 1.627 | 0.087 | 0.131 | |
| ACH | PVS | 20 | 1.363 | 0.196 | 0.196 | ||
| BCI | PVS | 20 | 1.882 | 0.041 | 0.123 | ||
| C. heliconiae | ACH | BCI | 15 | 1.120 | 0.356 | 0.534 | |
| ACH | PVS | 15 | 2.104 | 0.041 | 0.123 | ||
| BCI | PVS | 10 | 0.868 | 0.630 | 0.630 | ||
| Diptera:Psychodidae | Lu. gomezi | ACH | BCI | 13 | 5.341 | 0.005 | 0.006 |
| ACH | PVS | 20 | 2.709 | 0.002 | 0.006 | ||
| BCI | PVS | 13 | 3.311 | 0.006 | 0.006 | ||
| Lu. panamensis | ACH | BCI | 20 | 3.033 | 0.002 | 0.005 | |
| ACH | PVS | 20 | 1.581 | 0.070 | 0.070 | ||
| BCI | PVS | 20 | 2.645 | 0.003 | 0.005 | ||
| Lu. trapidoi | ACH | BCI | 13 | 2.936 | 0.004 | 0.005 | |
| ACH | PVS | 19 | 3.107 | 0.002 | 0.005 | ||
| BCI | PVS | 12 | 3.458 | 0.005 | 0.005 | ||
| Acari:Ixodidae | H. juxtakochi | BCI | PVAS | 17 | 0.254 | 0.918 | 0.918 |
| A. tapirellum | BCI | PVAS | 12 | 1.584 | 0.157 | 0.157 | |
| A. oblongoguttatum | BCI | PVAS | 7 | 2.006 | 0.133 | 0.133 | |
Fig 4.
Relative abundances of bacterial classes summarized for (A) dipteran species and (B) hard ticks gathered from BCI (i.e., Pristine), ACH (i.e., intermediately disturbed) and PVAS (i.e., highly disturbed).
Discussion
Habitat disturbance resulting from land use change can alter arthropod-borne disease transmission dynamics by modifying the habitat characteristics, community composition, behaviour, and patterns of dispersal and distribution of vectors or hosts [55,69,70]. Furthermore, habitat disruption can also modify the bacterial composition of natural environments, such as in the case of soil microbiota [71]. Yet, to our knowledge, no study has looked at the influence of habitat disturbance on the microbiome of human disease vectors, especially those that develop and interact with bacteria in the water, leaf litter, and soil or are acquired through animal host feeding in ecologically altered areas.
We tackled this issue by assessing bacterial communities associated with blood-feeding arthropods across sites with different degrees of habitat disturbance in the lowland tropical rainforest of central Panama. Specifically, we applied a 16S gene bacterial metagenomic approach to evaluate whether variation in the microbiome is associated with taxonomic relatedness, habitat disturbance or a combination of both. We focused on adults of Culicidae mosquitoes (i.e., Culex and Coquillettidia), Psychodidae sand flies (i.e., Lutzomyia) and Ceratopogonidae biting midges (i.e, Culicoides), which share ecological similarities in their development and adult life stages. Both Culex and Coquillettidia mosquitoes develop in aquatic sites associated with the roots of floating plants, while members of Culicoides develop in damp soil, water and organic matter [13,72–74]. All species of Lutzomyia develop in the soil within dark and humid places such as burrows and crevices associated with abundant leaf-litter or decomposing organic matter [75]. The males of Culex, Coquillettidia, Culicoides and Lutzomyia feed on nectar while the females take blood from a wide range of bird and mammal hosts. In addition, we sampled both nymphs and adult Ixodidae (i.e., Amblyomma, Ixodes, Haemaphysalis), which are distinct in their ecology compared to dipterans. Both the nymphs and adults of hard ticks adhere to and feed on vertebrate hosts throughout their lifetime [76]. Although they spend time off their host to molt through the different life stages and “quest” for a new host, they do not depend on these environments for feeding.
Our results are generally similar to those obtained in previous studies, where arthropod vectors species were dominated by Proteobacteria, including Gammaproteobacteria, Betaproteobacteria, Alphaproteobacteria, and to a lesser extent by Firmicutes, commonly Bacilli and Actinobacteria [13,18,44–46,77,78], These groups included bacterial genera previously described for Culex [13,44,46,77], Culicoides [21,38], Lutzomyia [37,79], Haemaphysalis [78,80] and Amblyomma [42,81].
We found that mosquitoes, biting midges and sand flies share a large proportion of their bacteria but statistical analysis also revealed significant differences in the OTU composition of each genera and species. It should be noted that variability at the 16S rRNA region, primer affinity and composition of the bacterial database will influence the resolution of the between-species comparisons based on OTU’s [82]. However, this finding suggests that these arthropods might encounter distinct bacterial types associated with differences in their habitat use or diet. Also, the colonization success of these bacterial types could differ among the arthropod hosts. We found that all tick species shared some bacterial OTU’s, but that this association did not extend to the dipteran assemblages. This is likely to reflect both their degree of taxonomic relatedness, since phylogenetically related species tend to share similar functional microbiomes [83], but also their distinct ecology. For instance, while all dipteran genera undergo larval development in either aquatic sites or organic soil before blood feeding as adults, hard ticks are largely associated with their host throughout their lifetime. Ticks undergo a series of molting events after each blood meal, which could be obtained from a series of animal hosts, from which they are expected to acquire much of their microbiome [80], while some symbiotic bacteria are also maternally inherited [2]. In contrast, dipteran genera also acquire bacteria through blood feeding, but their microbial community maintained through to adulthood is largely acquired during larval feeding and contact with the physical environment [13,48,49].
We observed significant differences in the bacterial community among areas with different degrees of habitat disturbance for two ecologically similar mosquito species within Culex and Coquillettidia, and three Lutzomyia sand fly species. These differences could be related to changes in the mammal or bird communities that served as feeding choices for adult arthropods as a result of habitat disruption. Alternatively, intra-specific differences could also result from changes to the pool of environmental bacteria, which might be associated with habitat disturbance. In support of these assumptions, we observed differences in a number of environmentally associated bacteria between primary forest, secondary forest and agricultural land, although changes in specific bacterial types generally vary among the different arthropod assemblages. For instance, the Cyanobacteria nostococidae, which has previously been associated with aquatic environments inhabited by mosquito larvae [13], was present in both Culex and Lutzomyia collected from secondary forest and disturbed habitats, but not from pristine forest sites. In addition, it was more common for Culex and Lutzomyia to be associated with Chlamydia in secondary forest and disturbed pastureland than in pristine forest, suggesting either differences in the mammal host reservoir or increased infection of mammals associated with changes in habitat quality.
We did not observe a significant difference in the bacterial community for any Culicoides species as a function of habitat disturbance. A potential explanation for this outcome is that Culicoides species either share a narrow ecological niche or because their optimal breeding habitats are not impacted by habitat disturbance. Cuilicoides regularly develop in areas with a high degree of organic matter known to modulate bacterial diversity [84], and are sensitive to temperature and humidity [85]. Nonetheless, the bacterial community of Culicoides in their preferred breeding sites has thus far been poorly characterized. Characterization of the differences in microhabitat features in Culicoides between land use types is required to confirm whether their breeding habitats and associated microbiota remain stable despite habitat disturbance. Furthermore, the host preferences of Culicoides, including the species in the current study are poorly classified and generally unknown within natural habitats, but some studies showed that most Culicoides species are opportunistic feeders, while others specialize on birds or mammals [86,87]. Another explanation for the lack of differences in the bacterial community of Culicoides between sites could be a stricter association of bacteria with the insect host than for other dipterans. That we did not see significant intra-specific differences in the bacterial community among tick species across areas with different habitat quality is not surprising given their specialized ecology [88].
We identified OTUs of several disease-causing bacteria as well as bacteria thought to alter life history characteristics and/or viral replication in all the arthropod genera, although these could not be identified to species. For example, we amplified Coxiella, whose members cause Q fever from all three tick species, Ehrlichia which causes ehrlichiosis infection from A. tapirellum and Rickettsia from A. oblongoguttatum and H. juxtakochi, which causes a variety of bacterial infections in humans and animals [89]. In addition, Rickettsia was also identified from Lu. trapidoi while Bartonella was detected from Lu. panamensis and Lu. gomezi plus all three species of Culicoides.
Rickettsia rickettsii, known to cause Rocky Mountain spotted fever in Panama has been previously isolated from Amblyomma mixtum, Dermacentor nitens and Haemaphysalis leporispalustris. In addition, two other Rickettsia species have been isolated from ticks in Panama including Rickettsia bellii from Amblyomma rotundatum and Rickettsia amblyommii from A. mixtum [90]. Although identification of the Rickettsia OTU’s were not to species level in this study, to our knowledge, this is first record of Rickettsia isolated from A. oblongoguttatum and H. juxtakochi in Central America as well as from Lutzomyia spp. However, agents causing bartonellosis have not yet been described from Culicoides biting midges. The ability of Culicoides to vector Bartonella requires further confirmation, but its presence in all three species is suggestive of a likely transmission role in Panama.
Congruently, we found several genera of bacteria with the potential to impact vector pathogen transmission. For instance, the genus Paenibacillus, which can inhibit DENV replication in Aedes mosquitoes was present in all Culicoides species as well as in Lu. panamensis [28]. Similarly, Serratia which can increase DENV and CHIKV in Ae. aegypti mosquitoes was present in all species of biting midges, mosquitoes and sand flies [28]. The family Enterobacteriae, which has been known to increase Plasmodium parasite infection in Anopheles mosquitoes was present in all, but A. oblongoguttatum [36]. Moreover, the bacteria Wolbachia, which impacts on vectors of arboviruses, Plasmodium infection and life history traits such as reproductive fitness and adult lifespan [91–94] was found from all Diptera.
Conclusion
Habitat disturbance has been shown to increase the likelihood of disease outbreaks of zoonotic (e.g., animal origin) infections through modifying the vector or host communities, or impacting their life history characteristics. However, the epidemiological role of bacteria associated with blood-feeding arthropods in relation to habitat disturbance is still poorly understood. Here, we observed that variation in the bacterial communities across a diverse array of hematophagous arthropods is likely to be explained by host phylogenetic relatedness, while intraspecific changes in community composition and prevalence are influenced by habitat quality. We found that the proportions of known disease-causing agents in infected arthropod species were comparable across sampling areas with different levels of habitat disturbance. However, further work is needed to determine whether the changes to the bacterial community with habitat disruption could influence disease transmission to humans. We argue further that changes in the microbiome of disease vectors should be considered when assessing the impact of habitat disturbance on disease transmission risk and emergence.
Supporting information
(TIF)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
Acknowledgments
We are grateful to Panama’s Ministry of Environment (Mi Ambiente) for supporting scientific collecting of insects in Panama.
Data Availability
Metagenomic sequence data is available under the BioProject PRJNA552605 in GenBank (https://www.ncbi.nlm.nih.gov/genbank/).
Funding Statement
This work was financed in part by the Secretariat for Science, Technology and Innovation of Panama (SENACYT), through the research grant IDDS15-047 and the National System of Investigation (SNI), supported research activities by JRL. Research activity by KLB was supported by the Smithsonian Institution Fellowship Program, George Burch Fellowship, The Edward M. and Jeanne C. Kashian Family Foundation Inc., and Nicholas Logothetis of Chartwell Consulting. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Minard G, Mavingui P, Moro CV. Diversity and function of bacterial microbiota in the mosquito holobiont. Parasit Vectors. 2013;6: 146 10.1186/1756-3305-6-146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bonnet SI, Binetruy F, Hernández-Jarguín AM, Duron O. The tick microbiome: Why non-pathogenic microorganisms matter in tick biology and pathogen transmission. Front Cell Infect Microbiol. 2017;7: 236 10.3389/fcimb.2017.00236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Telleria EL, Martins-da-Silva A, Tempone AJ, Traub-Csekö YM. Leishmania, microbiota and sand fly immunity. Parasitology. 2018;145: 1336–1353. 10.1017/S0031182018001014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cirimotich CM, Ramirez JL, Dimopoulos G. Native microbiota shape insect vector competence for human pathogens. Cell Host Microbe. 2011;10: 307–310. 10.1016/j.chom.2011.09.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gubler DJ. Resurgent vector-borne diseases as a global health problem. Emerg Infect Dis. 1998;4: 442–450. 10.3201/eid0403.980326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cantas L, Suer K. Review: The important bacterial zoonoses in “One Health” concept. Frontiers in Public Health. 2014;2: 144 10.3389/fpubh.2014.00144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Weger-Lucarelli J, Auerswald H, Vignuzzi M, Dussart P, Karlsson EA. Taking a bite out of nutrition and arbovirus infection. PLoS Negl Trop Dis. 2018;12: e0006247 10.1371/journal.pntd.0006247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Engel P, Moran NA. The gut microbiota of insects–diversity in structure and function. FEMS Microbiol Rev. 2013;37: 699–735. 10.1111/1574-6976.12025 [DOI] [PubMed] [Google Scholar]
- 9.Baxter RHG, Contet A, Krueger K. arthropod innate immune systems and vector-borne diseases. Biochemistry. 2017;56: 907–918. 10.1021/acs.biochem.6b00870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Guégan M, Zouache K, Démichel C, Minard G, Tran Van V, Potier P, et al. The mosquito holobiont: fresh insight into mosquito-microbiota interactions. Microbiome. 2018;6: 49 10.1186/s40168-018-0435-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Coon KL, Brown MR, Strand MR. Mosquitoes host communities of bacteria that are essential for development but vary greatly between local habitats. Mol Ecol. 2016;25: 5806–5826. 10.1111/mec.13877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Buck M, Nilsson LKJ, Brunius C, Dabiré RK, Hopkins R, Terenius O. Bacterial associations reveal spatial population dynamics in Anopheles gambiae mosquitoes. Sci Rep. 2016;6: 22806 10.1038/srep22806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Duguma D, Rugman-Jones P, Kaufman MG, Hall MW, Neufeld JD, Stouthamer R, et al. bacterial communities associated with Culex mosquito larvae and two emergent aquatic plants of bioremediation importance. PLoS One. 2013;8: e72522 10.1371/journal.pone.0072522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Thongsripong P, Chandler JA, Green AB, Kittayapong P, Wilcox BA, Kapan DD, et al. Mosquito vector-associated microbiota: Metabarcoding bacteria and eukaryotic symbionts across habitat types in Thailand endemic for dengue and other arthropod-borne diseases. Ecol Evol. 2017;8: 1352–1368. 10.1002/ece3.3676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yun J-H, Roh SW, Whon TW, Jung M-J, Kim M-S, Park D-S, et al. Insect gut bacterial diversity determined by environmental habitat, diet, developmental stage, and phylogeny of host. Appl Environ Microbiol. 2014;80: 5254 LP– 5264. 10.1128/AEM.01226-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tchioffo MT, Boissière A, Abate L, Nsango SE, Bayibéki AN, Awono-Ambéné PH, et al. Dynamics of bacterial community composition in the malaria mosquito’s epithelia. Front Microbiol. 2015;6: 1500 10.3389/fmicb.2015.01500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.van Overbeek L, Gassner F, van der Plas CL, Kastelein P, Nunes–da Rocha U, Takken W. Diversity of Ixodes ricinus tick-associated bacterial communities from different forests. FEMS Microbiol Ecol. 2008;66: 72–84. 10.1111/j.1574-6941.2008.00468.x [DOI] [PubMed] [Google Scholar]
- 18.Carpi G, Cagnacci F, Wittekindt NE, Zhao F, Qi J, Tomsho LP, et al. Metagenomic profile of the bacterial communities associated with Ixodes ricinus ticks. PLoS One. 2011;6: e25604 10.1371/journal.pone.0025604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Menchaca AC, Visi DK, Strey OF, Teel PD, Kalinowski K, Allen MS, et al. Preliminary assessment of microbiome changes following blood-feeding and survivorship in the Amblyomma americanum nymph-to-adult transition using semiconductor sequencing. PLoS One. 2013;8: e67129 10.1371/journal.pone.0067129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Thapa S, Zhang Y, Allen MS. Effects of temperature on bacterial microbiome composition in Ixodes scapularis ticks. Microbiologyopen. 2018;8: e00719–e00719. 10.1002/mbo3.719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Díaz-Sánchez S, Hernández-Jarguín A, Torina A, Fernández de Mera IG, Estrada-Peña A, Villar M, et al. Biotic and abiotic factors shape the microbiota of wild-caught populations of the arbovirus vector Culicoides imicola. Insect Mol Biol. 2018;27: 847–861. 10.1111/imb.12526 [DOI] [PubMed] [Google Scholar]
- 22.Weaver SC, Charlier C, Vasilakis N, Lecuit M. Zika, Chikungunya, and other emerging vector-borne viral diseases. Annu Rev Med. 2018;69: 395–408. 10.1146/annurev-med-050715-105122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sick F, Beer M, Kampen H, Wernike K. Culicoides biting midges-underestimated vectors for arboviruses of public health and veterinary importance. Viruses. 2019;11: 376 10.3390/v11040376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mediannikov O, Ranque S. Mansonellosis, the most neglected human filariasis. New microbes new Infect. Elsevier; 2018;26: S19–S22. 10.1016/j.nmni.2018.08.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Courtenay O, Peters NC, Rogers ME, Bern C. Combining epidemiology with basic biology of sand flies, parasites, and hosts to inform leishmaniasis transmission dynamics and control. PLOS Pathog. 2017;13: e1006571 10.1371/journal.ppat.1006571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wikel SK. Ticks and tick-borne infections: Complex ecology, agents, and host interactions. Vet Sci. 2018;5: 60 10.3390/vetsci5020060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Altizer S, Dobson A, Hosseini P, Hudson P, Pascual M, Rohani P. Seasonality and the dynamics of infectious diseases. Ecol Lett. 2006;9: 467–484. 10.1111/j.1461-0248.2005.00879.x [DOI] [PubMed] [Google Scholar]
- 28.Hegde S, Rasgon JL, Hughes GL. The microbiome modulates arbovirus transmission in mosquitoes. Curr Opin Virol. 2015;15: 97–102. 10.1016/j.coviro.2015.08.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Weiss B, Aksoy S. Microbiome influences on insect host vector competence. Trends Parasitol. 2011;27: 514–522. 10.1016/j.pt.2011.05.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dodson BL, Hughes GL, Paul O, Matacchiero AC, Kramer LD, Rasgon JL. Wolbachia enhances West Nile Virus (WNV) infection in the mosquito Culex tarsalis. PLoS Negl Trop Dis. 2014;8: e2965 10.1371/journal.pntd.0002965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Glaser RL, Meola MA. The native Wolbachia endosymbionts of Drosophila melanogaster and Culex quinquefasciatus increase host resistance to West Nile virus infection. PLoS One. 2010;5 10.1371/journal.pone.0011977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Aliota MT, Walker EC, Uribe Yepes A, Dario Velez I, Christensen BM, Osorio JE. The wMel strain of Wolbachia reduces transmission of Chikungunya Virus in Aedes aegypti. PLoS Negl Trop Dis. 2016;10: e0004677 10.1371/journal.pntd.0004677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Walker T, Johnson PH, Moreira LA, Iturbe-Ormaetxe I, Frentiu FD, McMeniman CJ, et al. The wMel Wolbachia strain blocks dengue and invades caged Aedes aegypti populations. Nature. 2011;476: 450 10.1038/nature10355 [DOI] [PubMed] [Google Scholar]
- 34.van den Hurk AF, Hall-Mendelin S, Pyke AT, Frentiu FD, McElroy K, Day A, et al. Impact of Wolbachia on infection with Chikungunya and Yellow Fever Viruses in the mosquito vector Aedes aegypti. PLoS Negl Trop Dis. 2012;6: e1892 10.1371/journal.pntd.0001892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Novakova E, Woodhams DC, Rodríguez-Ruano SM, Brucker RM, Leff JW, Maharaj A, et al. Mosquito microbiome dynamics, a background for prevalence and seasonality of West Nile Virus. Frontiers in Microbiology. 2017;8: 526 10.3389/fmicb.2017.00526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Boissière A, Tchioffo MT, Bachar D, Abate L, Marie A, Nsango SE, et al. Midgut microbiota of the malaria mosquito vector Anopheles gambiae and interactions with Plasmodium falciparum infection. PLoS Pathog. 2012;8 10.1371/journal.ppat.1002742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kelly PH, Bahr SM, Serafim TD, Ajami NJ, Petrosino JF, Meneses C, et al. The gut microbiome of the vector Lutzomyia longipalpis is essential for survival of Leishmania infantum. MBio. 2017;8: e01121–16. 10.1128/mBio.01121-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Campbell CL, Mummey DL, Schmidtmann ET, Wilson WC. Culture-independent analysis of midgut microbiota in the arbovirus vector Culicoides sonorensis (Diptera: Ceratopogonidae). J Med Entomol. 2004;41 10.1603/0022-2585-41.3.340 [DOI] [PubMed] [Google Scholar]
- 39.Zhang X-C, Yang Z-N, Lu B, Ma X-F, Zhang C-X, Xu H-J. The composition and transmission of microbiome in hard tick, Ixodes persulcatus, during blood meal. Ticks Tick Borne Dis. 2014;5: 864–870. 10.1016/j.ttbdis.2014.07.009 [DOI] [PubMed] [Google Scholar]
- 40.Lalzar I, Harrus S, Mumcuoglu KY, Gottlieb Y. Composition and seasonal variation of Rhipicephalus turanicus and Rhipicephalus sanguineus bacterial communities. Appl Environ Microbiol. 2012;78: 4110–4116. 10.1128/AEM.00323-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gall CA, Reif KE, Scoles GA, Mason KL, Mousel M, Noh SM, et al. The bacterial microbiome of Dermacentor andersoni ticks influences pathogen susceptibility. Isme J. 2016;10: 1846 10.1038/ismej.2015.266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Williams-Newkirk AJ, Rowe LA, Mixson-Hayden TR, Dasch GA. Characterization of the bacterial communities of life stages of free living lone star ticks (Amblyomma americanum). PLoS One. 2014;9: e102130 10.1371/journal.pone.0102130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Louradour I, Monteiro CC, Inbar E, Ghosh K, Merkhofer R, Lawyer P, et al. The midgut microbiota plays an essential role in sand fly vector competence for Leishmania major. Cell Microbiol. 2017;19: e12755 10.1111/cmi.12755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Muturi EJ, Ramirez JL, Rooney AP, Kim C-H. Comparative analysis of gut microbiota of mosquito communities in central Illinois. PLoS Negl Trop Dis. 2017;11: e0005377 10.1371/journal.pntd.0005377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bennett KL, Gómez-Martínez C, Chin Y, Saltonstall K, McMillan WO, Rovira JR, et al. Dynamics and diversity of bacteria associated with the biological competitors and disease vectors Aedes aegypti and Aedes albopictus. Sci Rep. 21;9:12160 10.1038/s41598-019-48414-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Osei-Poku J, Mbogo CM, Palmer WJ, Jiggins FM. Deep sequencing reveals extensive variation in the gut microbiota of wild mosquitoes from Kenya. Mol Ecol. 2012;21 10.1111/j.1365-294X.2012.05759.x [DOI] [PubMed] [Google Scholar]
- 47.Kurilshikov A, Livanova NN, Fomenko N V, Tupikin AE, Rar VA, Kabilov MR, et al. Comparative metagenomic profiling of symbiotic bacterial communities associated with Ixodes persulcatus, Ixodes pavlovskyiand and Dermacentor reticulatus ticks. PLoS One. 2015;10: e0131413 10.1371/journal.pone.0131413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gimonneau G, Tchioffo MT, Abate L, Boissière A, Awono-Ambéné PH, Nsango SE, et al. Composition of Anopheles coluzzii and Anopheles gambiae microbiota from larval to adult stages. Infect Genet Evol. 2014;28: 715–724. 10.1016/j.meegid.2014.09.029 [DOI] [PubMed] [Google Scholar]
- 49.Coon KL, Vogel KJ, Brown MR, Strand MR. Mosquitoes rely on their gut microbiota for development. Mol Ecol. 2014;23: 2727–2739. 10.1111/mec.12771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dickson LB, Jiolle D, Minard G, Moltini-Conclois I, Volant S, Ghozlane A, et al. Carryover effects of larval exposure to different environmental bacteria drive adult trait variation in a mosquito vector. Sci Adv. 2017;3: e1700585 10.1126/sciadv.1700585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Monteiro CC, Villegas LEM, Campolina TB, Pires ACMA, Miranda JC, Pimenta PFP, et al. Bacterial diversity of the American sand fly Lutzomyia intermedia using high-throughput metagenomic sequencing. Parasit Vectors. 2016;9: 480 10.1186/s13071-016-1767-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Narasimhan S, Fikrig E. Tick microbiome: the force within. Trends Parasitol. 2015;31: 315–323. 10.1016/j.pt.2015.03.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zouache K, Raharimalala FN, Raquin V, Tran-Van V, Raveloson LHR, Ravelonandro P, et al. Bacterial diversity of field-caught mosquitoes, Aedes albopictus and Aedes aegypti, from different geographic regions of Madagascar. FEMS Microbiol Ecol. 2011;75 10.1111/j.1574-6941.2010.01012.x [DOI] [PubMed] [Google Scholar]
- 54.Hawlena H, Rynkiewicz E, Toh E, Alfred A, Durden LA, Hastriter MW, et al. The arthropod, but not the vertebrate host or its environment, dictates bacterial community composition of fleas and ticks. ISME J. 2013;7: 221–223. 10.1038/ismej.2012.71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Loaiza JR, Dutari LC, Rovira JR, Sanjur OI, Laporta GZ, Pecor J, et al. Disturbance and mosquito diversity in the lowland tropical rainforest of central Panama. Sci Rep. 2017;7: 7248 10.1038/s41598-017-07476-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Eastwood G, Loaiza JR, Pongsiri MJ, Sanjur OI, Pecor JE, Auguste AJ, et al. Enzootic arbovirus surveillance in forest habitat and phylogenetic characterization of novel isolates of Gamboa Virus in Panama. Am J Trop Med Hyg. 2016;94: 786–793. 10.4269/ajtmh.15-0445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Loaiza JR, Dutari LC, Rovira JR, Sanjur OI, Laporta GZ, Pecor J, et al. Disturbance and mosquito diversity in the lowland tropical rainforest of central Panama. Sci Rep. 2017;7: 7248 10.1038/s41598-017-07476-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Pecor JE, Mallampalli VL, Harbach RE, Peyton EL. Catalog and Illustrated Review of the Subgenus Melanoconion of Culex (Diptera: Culicidae). Contrib Am Entomol Inst 27: 1–228. [Google Scholar]
- 59.Wilkerson RC, Peyton EL. Standardized nomenclature for the costal wing spots of the genus Anopheles and other spotted-wing mosquitoes (Diptera: Culicidae). J Med Entomol. 1990;27: 207–224. [Google Scholar]
- 60.Sallum MA, Forattini OP. Revision of the Spissipes section of Culex (Melanoconion)(Diptera: Culicidae). J Am Mosq Control Assoc. 1996;12: 517–600. [PubMed] [Google Scholar]
- 61.Chaniotis BN, Correa MA. comparative flying and biting activity of Panamanian Phlebotomine sandflies in a mature forest and adjacent open space. J Med Entomol. 1974;11: 115–116. 10.1093/jmedent/11.1.115 [DOI] [PubMed] [Google Scholar]
- 62.Young DG, Duran MA. Guide to the identification and geographic distribution of Lutzomyia sand flies in Mexico, the West Indies, Central and South America (Diptera: Psychodidae). Mem Am Entomol Inst. 1994. 54:881. [Google Scholar]
- 63.Gray JS. A carbon dioxide trap for prolonged sampling of Ixodes ricinus L. populations. Exp Appl Acarol. 1985;1: 35–44. [DOI] [PubMed] [Google Scholar]
- 64.Fairchild GB, Kohls GM, Tipton VJ. The ticks of Panama (Acarina: Ixodoidea). Ectoparasites of Panama. Field Museum Nat Hist, Chicago 1966. 167–219. [Google Scholar]
- 65.Mesa EO. Las garrapatas de la República de Colombia. Rev Fac Nac Agron. 1942;5: 57–103. [Google Scholar]
- 66.Caporaso JG, Lauber CL, Walters WA, Berg-Lyons D, Lozupone CA, Turnbaugh PJ, et al. Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proc Natl Acad Sci U S A. 2011;108: 4516–4522. 10.1073/pnas.1000080107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Collado L, Figueras MJ. Taxonomy, epidemiology, and clinical relevance of the genus Arcobacter. Clin Microbiol Rev. 2011;24: 174–192. 10.1128/CMR.00034-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Chigira A, Miura K. Detection of ‘Candidatus cardinium’ bacteria from the haploid host Brevipalpus californicus (Acari: Tenuipalpidae) and effect on the host. Exp Appl Acarol. 2005;37: 107–116. 10.1007/s10493-005-0592-4 [DOI] [PubMed] [Google Scholar]
- 69.Gottdenker NL, Streicker DG, Faust CL, Carroll CR. anthropogenic land use change and infectious diseases: A review of the evidence. Ecohealth. 2014;11: 619–632. 10.1007/s10393-014-0941-z [DOI] [PubMed] [Google Scholar]
- 70.Keesing F, Belden LK, Daszak P, Dobson A, Harvell CD, Holt RD, et al. Impacts of biodiversity on the emergence and transmission of infectious diseases. Nature. 2010;468: 647 10.1038/nature09575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Rampelotto PH, de Siqueira Ferreira A, Barboza ADM, Roesch LFW. Changes in diversity, abundance, and structure of soil bacterial communities in Brazilian savanna under different land use systems. Microb Ecol. 2013;66: 593–607. 10.1007/s00248-013-0235-y [DOI] [PubMed] [Google Scholar]
- 72.Serandour J, Willison J, Thuiller W, Ravanel P, Lemperiere G, Raveton M. Environmental drivers for Coquillettidia mosquito habitat selection: A method to highlight key field factors. Hydrobiologia. 2010. 10.1007/s10750-010-0372-y [DOI] [Google Scholar]
- 73.Reisen WK. The contrasting bionomics of Culex mosquitoes in western North America. J Am Mosq Control Assoc. 2012;28: 82–91. 10.2987/8756-971X-28.4.82 [DOI] [PubMed] [Google Scholar]
- 74.Carpenter S, Groschup MH, Garros C, Felippe-Bauer ML, Purse B V. Culicoides biting midges, arboviruses and public health in Europe. Antiviral Res. 2013;100: 102–113. 10.1016/j.antiviral.2013.07.020 [DOI] [PubMed] [Google Scholar]
- 75.Vivero RJ, Torres-Gutierrez C, Bejarano EE, Peña HC, Estrada LG, Florez F, et al. Study on natural breeding sites of sand flies (Diptera: Phlebotominae) in areas of Leishmania transmission in Colombia. Parasit Vectors. 2015;8: 116 10.1186/s13071-015-0711-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Mccoy KD, Léger E, Dietrich M. Host specialization in ticks and transmission of tick-borne diseases: A review. Front Cell Infect Microbiol. 2013;3: 57 10.3389/fcimb.2013.00057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Sant’anna MR, Darby AC, Brazil RP, Montoya-Lerma J, Dillon VM, Bates PA, et al. Investigation of the bacterial communities associated with females of Lutzomyia sand fly species from South America. PLoS One. 2012;7 10.1371/journal.pone.0042531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Khoo J-J, Chen F, Kho KL, Ahmad Shanizza AI, Lim F-S, Tan K-K, et al. Bacterial community in Haemaphysalis ticks of domesticated animals from the Orang Asli communities in Malaysia. Ticks Tick Borne Dis. 2016;7: 929–937. 10.1016/j.ttbdis.2016.04.013 [DOI] [PubMed] [Google Scholar]
- 79.Pires ACAM Villegas LEM, Campolina TB Orfanó AS, Pimenta PFP Secundino NFC. Bacterial diversity of wild-caught Lutzomyia longipalpis (a vector of zoonotic visceral leishmaniasis in Brazil) under distinct physiological conditions by metagenomics analysis. Parasit Vectors. 2017;10: 627 10.1186/s13071-017-2593-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Qiu Y, Nakao R, Ohnuma A, Kawamori F, Sugimoto C. Microbial population analysis of the salivary glands of ticks; A possible strategy for the surveillance of bacterial pathogens. PLoS One. 2014;9: e103961 10.1371/journal.pone.0103961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Budachetri K, Browning RE, Adamson SW, Dowd SE, Chao C-C, Ching W-M, et al. An insight into the microbiome of the Amblyomma maculatum (Acari: Ixodidae). J Med Entomol. 2014;51: 119–129. 10.1603/me12223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Poretsky R, Rodriguez-R LM, Luo C, Tsementzi D, Konstantinidis KT. Strengths and limitations of 16S rRNA gene amplicon sequencing in revealing temporal microbial community dynamics. PLoS One. 2014;9: e93827 10.1371/journal.pone.0093827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Degli Esposti M, Martinez Romero E. The functional microbiome of arthropods. PLoS One. 2017;12: e0176573–e0176573. 10.1371/journal.pone.0176573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Neumann D, Heuer A, Hemkemeyer M, Martens R, Tebbe CC. Importance of soil organic matter for the diversity of microorganisms involved in the degradation of organic pollutants. ISME J. 2014;8: 1289–1300. 10.1038/ismej.2013.233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Parras MA, Rosa JR, Szelag EA, Salomón OD. Identification of the natural breeding sites of sandflies (Diptera: Psychodidae: Phlebotominae), potential vectors of leishmaniasis, in the province of Chaco, Argentina. Memórias do Instituto Oswaldo Cruz. 2012. 107: 550–552. [DOI] [PubMed] [Google Scholar]
- 86.Santiago-Alarcon D, Havelka P, Schaefer HM, Segelbacher G. bloodmeal analysis reveals avian Plasmodium infections and broad host preferences of Culicoides (Diptera: Ceratopogonidae) vectors. PLoS One. 2012;7: e31098 10.1371/journal.pone.0031098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Lassen SB, Nielsen SA, Kristensen M. Identity and diversity of blood meal hosts of biting midges (Diptera: Ceratopogonidae: Culicoides Latreille) in Denmark. Parasit Vectors. 2012;5: 143 10.1186/1756-3305-5-143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Esser HJ, Herre EA, Blüthgen N, Loaiza JR, Bermúdez SE, Jansen PA. Host specificity in a diverse Neotropical tick community: an assessment using quantitative network analysis and host phylogeny. Parasit Vectors. 2016;9: 372 10.1186/s13071-016-1655-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Parola P, Raoult D. Ticks and tickborne bacterial diseases in humans: An emerging infectious threat. Clin Infect Dis. 2001;32: 897–928. 10.1086/319347 [DOI] [PubMed] [Google Scholar]
- 90.Lopes MG, May Junior J, Foster RJ, Harmsen BJ, Sanchez E, Martins TF, et al. Ticks and rickettsiae from wildlife in Belize, Central America. Parasit Vectors. 2016;9: 62 10.1186/s13071-016-1348-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Hedges LM, Brownlie JC, O’Neill SL, Johnson KN. Wolbachia and virus protection in insects. Science. 2008;322 10.1126/science.1162418 [DOI] [PubMed] [Google Scholar]
- 92.Kitrayapong P, Baimai V, O’Neill SL. Field prevalence of Wolbachia in the mosquito vector Aedes albopictus. Am J Trop Med Hyg. 2002;66: 108–111. 10.4269/ajtmh.2002.66.108 [DOI] [PubMed] [Google Scholar]
- 93.Almeida F de, Moura AS, Cardoso AF, Winter CE, Bijovsky AT, Suesdek L. Effects of Wolbachia on fitness of Culex quinquefasciatus (Diptera; Culicidae). Infect Genet Evol. 2011;11: 2138–2143. 10.1016/j.meegid.2011.08.022 [DOI] [PubMed] [Google Scholar]
- 94.Iturbe-Ormaetxe I, Walker T, O’ Neill SL. Wolbachia and the biological control of mosquito-borne disease. EMBO Rep. 2011;12 10.1038/embor.2011.84 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(TIF)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
Data Availability Statement
Metagenomic sequence data is available under the BioProject PRJNA552605 in GenBank (https://www.ncbi.nlm.nih.gov/genbank/).