Multi-omics biomarker pipeline generated 19 proteins which were verified by a multiplex quantitative SRM assay in seminal plasma samples of 152 prostate cancer and 67 negative biopsy patients. Verification revealed a prostate-specific, secreted and androgen-regulated protein-glutamine gamma-glutamyltransferase 4 (TGM4), which detected prostate cancer on biopsies with AUC=0.66, as measured by an in-house immunoassay in seminal plasma. Perspectives of seminal plasma proteins as biomarkers of prostate cancer were reviewed.
Keywords: Prostate cancer biomarkers, serum/plasma, selected reaction monitoring, absolute quantification, assay development, machine learning, protein-glutamine gamma-glutamyltransferase 4, seminal plasma, TGM4, XGBoost
Graphical Abstract
Highlights
Seventy-six most promising proteins were qualified, and 19 proteins were verified by SRM in 219 seminal plasma samples of patients with prostate cancer and negative biopsies.
Prostate-specific, secreted and androgen-regulated protein-glutamine gamma-glutamyltransferase 4 (TGM4) was verified by SRM assay and an in-house immunoassay.
TGM4 detected prostate cancer on biopsy in seminal plasma (AUC=0.66), but not in blood serum.
Abstract
Seminal plasma, because of its proximity to prostate, is a promising fluid for biomarker discovery and noninvasive diagnostics. In this study, we investigated if seminal plasma proteins could increase diagnostic specificity of detecting primary prostate cancer and discriminate between high- and low-grade cancers. To select 147 most promising biomarker candidates, we combined proteins identified through five independent experimental or data mining approaches: tissue transcriptomics, seminal plasma proteomics, cell line secretomics, tissue specificity, and androgen regulation. A rigorous biomarker development pipeline based on selected reaction monitoring assays was designed to evaluate the most promising candidates. As a result, we qualified 76, and verified 19 proteins in seminal plasma of 67 negative biopsy and 152 prostate cancer patients. Verification revealed a prostate-specific, secreted and androgen-regulated protein-glutamine gamma-glutamyltransferase 4 (TGM4), which predicted prostate cancer on biopsy and outperformed age and serum Prostate-Specific Antigen (PSA). A machine-learning approach for data analysis provided improved multi-marker combinations for diagnosis and prognosis. In the independent verification set measured by an in-house immunoassay, TGM4 protein was upregulated 3.7-fold (p = 0.006) and revealed AUC = 0.66 for detecting prostate cancer on biopsy for patients with serum PSA ≥4 ng/ml and age ≥50. Very low levels of TGM4 (120 pg/ml) were detected in blood serum. Collectively, our study demonstrated rigorous evaluation of one of the remaining and not well-explored prostate-specific proteins within the medium-abundance proteome of seminal plasma. Performance of TGM4 warrants its further investigation within the distinct genomic subtypes and evaluation for the inclusion into emerging multi-biomarker panels.
Prostate cancer (PCa)1 is the most frequently diagnosed neoplasm and the third leading cause of cancer mortality in men. Its incidence rate has continued to increase rapidly during the past two decades, especially in men over the age of 50 years. Worldwide, close to 260,000 men die from PCa every year (1). Our best current strategy to help PCa patients is early diagnosis and administration of the most appropriate therapy, including active surveillance only (2).
The most commonly used PCa biomarker, prostate-specific antigen (PSA), is secreted by both normal prostate cells and PCa cells. There is no question that the introduction of PSA testing over the last two decades revolutionized the practice of urology. As a result of PSA screening, most men today with PCa are presented with localized disease and serum PSA values <10 ng/ml. However, the widespread use of PSA screening is not without controversy (3, 4).
Although PSA is an excellent biochemical marker, it has a number of important limitations, including lack of specificity and prognostic significance. PSA expression is prostate tissue-specific but not prostate cancer-specific. Serum PSA levels are increased in both PCa and in other non-malignant prostatic diseases, including benign prostatic hyperplasia and prostate inflammation. Because of the above limitations, clinicians currently perform on average four prostatic biopsies in order to detect one prostate cancer. PSA levels also do not predict the clinical significance or aggressiveness of PCa. Most men with PCa are destined to die of another condition before PCa becomes clinically significant (5). Lack of specificity and prognostic significance are two major limitations of PSA and constitute the major unmet needs in the current clinical diagnostics of PCa.
There have been intense efforts for the identification of novel PCa biomarkers in blood or urine. Prostatic acid phosphatase has been discovered in the 1930s (6) and for almost 50 years was used to indicate the success of hormonal therapy, whereas its clinical utility for diagnosis was limited. Apart from total PSA, which was characterized in the 1970s (7, 8) and approved in 1986 (9), current U.S. Food and Drug Administration (FDA)-approved tests include only three tests: Prostate Health Index, PCA3 and CellSearch (10). Prostate Health Index is a multivariate index assay which includes immunoassay measurements of total PSA, free PSA and [-2]proPSA in blood serum, and is intended for diagnosis of PCa in men aged ≥ 50 years with total PSA 4.0–10 ng/ml and negative digital rectal examination (11). PCA3 test measures the relative amount of a non-coding RNA PCA3 in the post-digital rectal examination urine and is indicated to aid in the decision for repeat biopsy in men aged ≥ 50 years who have had previous negative prostate biopsies (12). The CellSearch detects circulating tumor cells of epithelial origin (CD45-, EpCAM+, and cytokeratins 8, 18+, and/or 19+) in whole blood and has been only approved for monitoring patients with metastatic PCa (13). Emerging tests yet to be approved for the clinical use include 4Kscore (immunoassay measurements of kallikrein-2 and total, free and intact forms of PSA in serum) (14), STHLM3 model (6 serum proteins, 232 single nucleotide polymorphisms and clinical parameters) (15) and ConfirmMDx (hypermethylation of GSTP1, APC and RASSF1 genes in biopsies) (16). TMPRSS2-ERG fusion (17) and SPINK1 mRNA (18) measured in urine are also promising biomarkers. With diagnostic and prognostic AUCs (area under the receiver operating characteristic curve) in the range 0.66–0.70, these novel serum or urine biomarkers do not substantially outperform PSA.
Although much of the work to identify and characterize PSA was originally carried out in seminal plasma (SP) (19), there are only few comprehensive proteomic studies on identification of novel PCa biomarkers in SP (20–22) or expressed prostatic secretions (23, 24). SP has total protein concentration of 40–60 mg/ml and a dynamic range of at least nine orders of magnitude, with semenogelin-1 (20 mg/ml) and interleukin-12 (10 pg/ml) being one of the most and least abundant proteins, respectively (25). Nearly a quarter of molecular composition of SP is secreted by prostate (25), with the rest produced by seminal vesicles, epididymis, testis, and periurethral glands (26, 27). We previously completed extensive studies on the SP proteome and identified more than 3,000 proteins in SP of healthy men and patients with infertility (28, 29). Our work resulted in first-of-a-kind SP biomarkers for the differential diagnosis of male infertility (30–32). Success with male infertility biomarkers motivated us to apply a similar strategy to PCa. We previously extensively validated by targeted proteomics and immunoassays the prostate-specific kallikrein-4 as a potential PCa biomarker in SP and blood serum (20).
In this work, we hypothesized that SP could contain novel PCa biomarkers within the medium-abundance range of concentrations (0.1–100 μg/ml) of the SP proteome. Some of these proteins have never been previously studied in the context of PCa. To select the most promising biomarker candidates, we combined proteins identified through five data mining and experimental -omics approaches, such as transcriptomics, proteomics, secretomics, tissue specificity and androgen regulation. Only those proteins which were previously identified in the SP proteome were considered as candidates and were qualified and verified by mass spectrometry-based selected reaction monitoring (SRM) assays. According to the fit-for-purpose approach to biomarker measurement assays (33, 34), Tier 3 exploratory SRM assays were developed for the cost-effective qualification of dozens of candidates, followed by well-validated quantitative Tier 2 SRM assays for verification of a small number of candidates in hundreds of SP samples, followed by a high-precision Tier 1 immunoassay for orthogonal verification of a single biomarker in SP and blood serum samples. Powerful nonlinear machine-learning algorithms (35) were utilized to evaluate potential multi-marker models for PCa diagnosis and prognosis. Our study was designed to simultaneously assess biomarker candidates for the two unmet clinical needs: (1) differentiation between PCa and negative biopsies, and (2) discrimination between low- and high-grade PCa. To our knowledge, this work is one of the largest and the most comprehensive proteomic studies on SP proteins and PCa.
EXPERIMENTAL PROCEDURES
Hypothesis, Study Design and Objectives
We hypothesized that some SP proteins can emerge as novel biomarkers of primary PCa. Our study was designed to simultaneously assess biomarker candidates for the two unmet clinical needs: (1) differentiation between PCa and negative biopsies, and (2) discrimination between low- and high-grade PCa. Our objectives included selection of potential biomarker candidates, development of quantitative mass spectrometry assays, qualification of the most promising candidates, verification of candidates in a large set of SP samples by SRM assays, and verification of a top candidate by ELISA in SP and blood serum.
Study Population and Sample Collection
SP samples with relevant clinical information were obtained through the Murray Koffler Urologic Wellness Centre at Mount Sinai Hospital (REB #08-0117-E), University Health Network (#09–0830-AE) and Calgary Prostate Cancer Center (#18166). Men referred for a prostate biopsy were asked to participate in this study. None of these men had clinical signs of prostate inflammation. Semen samples were collected by masturbation into a sterile collection cup either at home or at urology clinics. Following liquefaction for 1 h at room temperature, semen samples were centrifuged at 13,000 × g for 15 min, and the supernatants were frozen at −80 °C. Blood samples were collected at the diagnostic laboratory at Mount Sinai Hospital, and blood serum was stored at −80 °C. Stability of SP and blood serum samples during long-term storage at −80 °C was not determined. However, our SRM measurements revealed 24 and 14% variability of TGM4 and KLK3 concentrations, respectively, in the SP pool stored at −20 °C for 5 weeks.
Experimental Design and Statistical Rationale for Differential Proteomics
Mass spectrometry was used to identify differentially expressed proteins in three pools of SP samples from negative biopsy, low-grade PCa and high-grade PCa patients. SP pools included: (1) low-grade PCa (GS = 6, median serum PSA 8 ng/ml [IQR 6–10 ng/ml], median age 65 y.o. [IQR 62–67 y.o.], n = 5); (2) high-grade PCa (GS = 8 or 9; median serum PSA 14 ng/ml [IQR 9–19 ng/ml], median age 66 y.o. [IQR 66–66 y.o.], n = 5); (3) no evidence of cancer (negative biopsy, median serum PSA 7 ng/ml [IQR 6–10 ng/ml], median age 63 y.o. [IQR 55–65 y.o.], n = 5). The rationale for using pooled samples was to reduce the effects of the protein biological variability between patients and to increase the likelihood of identifying consistent protein differences in clinical cohorts. According to effect size calculations, triplicate analysis of pools could identify proteins up- or downregulated at least 1.8-fold, assuming 80% power, α = 0.05, median 17% coefficient of variation (CV) for LFQ (Label-Free Quantification with MaxQuant software) values and a two-tailed MWU test (G*Power software, v3.1.7, Heinrich Heine University Dusseldorf).
Differential Proteomics
Each pool was subjected to the proteomic sample preparation and mass spectrometry analysis with three analytical replicates. We defined technical replicates as LC-SRM injections, and analytical replicates as full process replicates (independent denaturation, digestion, microextraction, and mass spectrometry analysis). Tryptic digestion (500 μg total protein per pool) was performed as previously described (30, 36). Briefly, proteins were denatured at 65 °C in the presence of 0.02% RapiGest SF (Waters, Milford, MA), reduced with 10 mm dithiothreitol (Sigma-Aldrich, Oakville, ON), alkylated with 20 mm iodoacetamide (Sigma-Aldrich) and digested overnight at 37 °C using sequencing grade modified trypsin (trypsin:total protein ratio 1:30; Promega, Madison WI). RapiGest SF was cleaved with 1% trifluoroacetic acid (Sigma-Aldrich) and removed by centrifugation. Following protein digestion, peptides were fractionated by strong-cation exchange chromatography, twenty three fractions were collected for each analytical replicate, concentrated with 10 μl OMIX C18 tips (Varian, Lake Forest, CA) and analyzed by the reverse phase liquid chromatography-tandem mass spectrometry (LTQ-Orbitrap XL, Thermo Scientific Inc., San Jose, CA), as previously described (36). A 90 min LC gradient with 5% to 10% acetonitrile for 3 min, 10% to 60% for 85 min, and 60% to 100% for 2 min was used. RAW files were generated with XCalibur software (v2.0.5; Thermo Scientific). MaxQuant software (v1.1.1.25) was used for protein identification and label-free quantification. MaxQuant executed spectral search against a concatenated International Protein Index (IPI) human protein database (v3.71) and a decoy database (86,746 entries). Parameters included: trypsin enzyme specificity, 1 missed cleavage, minimum peptide length of 7 amino acids, minimum of 1 unique peptide, top 6 MS/MS peaks per 100 Da, peptide mass tolerance of 20 ppm for precursor ion and MS/MS tolerance of 0.5 Da and fixed modification of cysteines by carbamidomethylation. Variable modifications included oxidation of methionine and acetylation of the protein at N terminus. All entries were filtered using a false positive rate of 1% both at the peptide and protein levels, and false positives were removed. MaxQuant search file proteinGroups.txt was uploaded to Perseus software (v1.4.1.3) for statistical analysis. Protein identifications annotated in the columns “Only identified by site,” “Reverse,” and “Contaminant” as well as proteins identified only in a single replicate were filtered out. Only protein entries with two or three valid non-zero values in each group were used for statistical analysis, whereas entries with single values were filtered out. LFQ intensities were log2-transformed and used to calculate means and statistical significance (t test with Benjamini-Hochberg false-discovery rate-adjusted p values) and generate volcano plots.
Development of Label-free SRM Assays for the Qualification Phase
For the cost-effective qualification of dozens of candidates, Tier 3 SRM assays were developed, as previously described (37–39). Briefly, the Peptide Atlas (www.peptideatlas.org) was used to select top 5–7 peptides (charge +2 and +3) for each of 147 candidate proteins and 12 control proteins (representing other six glands or cell types in the male urogenital system). Fully tryptic peptides with 7–20 amino acids were chosen, and peptides with methionine and N-terminal cysteine residues were avoided, if possible. A list of peptides and top 7 transitions were downloaded. All proteins were ranked according to their MaxQuant LFQ intensities and split into groups of high-, medium- and low-abundance SP proteins. Sixty survey multiplex SRM methods with 15 peptides, 7 transitions per peptide, 20 ms scan times, 8 min scheduling windows based on predicted retention times were designed in Pinpoint software (v1.4.71; Thermo Scientific), csv files were re-arranged in Microsoft Excel 2007, and SRM methods were experimentally tested in the digest of normal SP. SRM methods for high-abundance peptides were quickly developed and set aside, whereas medium- and low-abundance peptides were tested in several iterations. As a result, nearly 900 peptides and 6000 transitions were experimentally tested in the matrix of SP. Raw files were uploaded to Pinpoint, and peaks were analyzed manually. High-abundance peptides with clear peaks, high signal-to-noise intensities and multiple overlapping transitions were selected, whereas medium- and low-abundance peptides moved to the second iteration. Peptides or transitions in doubt were confirmed with our SP proteome data. In the second iteration, we designed 37 multiplexed methods (∼2500 transitions) and tested them with higher scan times (35 ms), to lower background and facilitate detection of low-abundance peptides. In the third iteration, we tested 9 methods (∼500 transitions) with 40 ms scan times. In the fourth iteration, we experimentally reconfirmed all peptides and verified, recorded or optimized the following parameters: (1) top 3 transitions; (2) retention times and scheduling intervals; (3) selectivity of transitions and possible interferences; and (4) scan times. Transitions with fragment m/z higher than precursor m/z were preferable; however, some transitions with lower m/z but high signal-to-noise ratio were also used. For proteins with multiple peptides, a single peptide with the highest SRM area was chosen. All peptides were analyzed with the Basic Local Alignment Search Tool (http://blast.ncbi.nlm.nih.gov/Blast.cgi) to ensure that peptides were unique to each UniProtKB/Swiss-Prot protein identifier. In the final iteration, peptides were scheduled in a single SRM method within 2.8-min (±1.4 min) intervals during a 60 min LC gradient (5% to 10% acetonitrile for 3 min, 10% to 60% for 55 min, and 60% to 100% for 2 min). Three most intense and reproducible transitions were monitored per each peptide. Scan times (4 to 25 ms) were further optimized for each peptide to ensure acquisition of 15–20 points per LC peak per transition.
Experimental Design and Statistical Rationale for Relative Quantification by Label-free SRM in the Qualification Phase
SP samples were randomized on a 96-well plate and included: (1) low-grade PCa (GS = 6, median serum PSA 6 ng/ml [IQR 4–8 ng/ml], median age 63 y.o. [IQR 61–67 y.o.], n = 24); (2) high-grade PCa (GS = 7, 4 + 3 and ≥ 8; median serum PSA 9 ng/ml [IQR 7–11 ng/ml], median age 67 y.o. [IQR 64–71 y.o.], n = 14); (3) negative biopsy (median serum PSA 6 ng/ml [IQR 5–7 ng/ml], median age 59 y.o. [IQR 55–62 y.o.], n = 13). According to effect size calculations, SRM analysis of 13 negative biopsy and 38 PCa samples could detect 10% change (1.1 ratio) in protein abundance among groups, assuming 80% power, α = 0.05, median 10% CV for normalized SRM intensities and a two-tailed MWU test. Ten microliters of each SP were diluted 10-fold with 50 mm ammonium bicarbonate (pH 7.8; Sigma-Aldrich), and an aliquot equivalent to 0.5 μl of SP was subjected to the proteomic sample preparation. Proteins were denatured at 60 °C with 0.1% Rapigest SF, and the disulfide bonds were reduced with 10 mm dithiothreitol. After reduction, the samples were alkylated with 20 mm iodoacetamide. Samples were then trypsin-digested overnight at 37 °C. One hundred and eighty femtomoles of heavy isotope-labeled peptide LSEPAELTDAVK (13C6, 15N2; HeavyPeptide AQUA, Thermo Scientific Inc.) of KLK3 protein were spiked into each digest and used as a quality control for C18 microextraction and data normalization. Rapigest was cleaved with 1% trifluoroacetic acid, and a 96-well plate was centrifuged at 2000 rpm for 20 min. Each digest was subjected to micro extraction with 10 μl OMIX C18 tips. Each SP sample was analyzed by SRM in technical duplicates. One to four mass spectrometry quality control samples were run every 12 injections. The LC EASY-nLC 1000 (Thermo Scientific Inc.) was coupled online to TSQ Vantage triple-quadrupole mass spectrometer (Thermo Scientific Inc.) using a nanoelectrospray ionization source. Peptides were separated on a 2 cm trap column (150 μm ID, 5 μm C18) and eluted onto a 5 cm resolving column (75 μm ID, 3 μm C18). A 60 min gradient with 5% to 10% acetonitrile for 3 min, 10% to 60% for 55 min, and 60% to 100% for 2 min was used. Peptides were scheduled within 2.8-min intervals. SRM method had the following parameters: optimized collision energy values, 0.010 m/z scan width, 4–25 ms scan times, 0.4 FWHM resolution of the first quadrupole (Q1), 0.7 FWHM resolution of the third quadrupole (Q3), 1.5 mTorr pressure of the second quadrupole, tuned S-lens values, +1 V declustering voltage. Well-to-well carryover was estimated in the range 0.05–0.2%. RAW files recorded for each sample were analyzed with Pinpoint software. Areas of all peptides were normalized to the spike-in standard heavy isotope-labeled peptide (LSEPAELTDAVK) and were used to calculate ROC AUC areas, sensitivities and specificities with GraphPad PRISM (v5.03).
Upgrade of SRM Assays for the Verification Phase
To enable verification of 19 candidates in a large set of SP samples, well-validated quantitative Tier 2 SRM assays were developed (34). Purified heavy isotope-labeled peptides (SpikeTidesTM_TQL) with trypsin-cleavable quantifying tags (serine-alanine-[3-nitro]tyrosine-glycine) and quantified amounts (1 nmol per aliquot) were obtained from JPT Peptide Technologies GmbH, Berlin, Germany. SRM transitions and TSQ Vantage parameters were optimized or corrected using the digest of synthetic peptides. Retention times and scheduling windows were optimized for a 30 min LC gradient (5% to 10% acetonitrile for 1 min, 10% to 60% for 27 min, and 60% to 100% for 2 min). Synthetic peptides were used to assess the efficiency of tryptic digestion and chemical modifications (cysteine alkylation, methionine oxidation, formation of pyroglutamate of N-terminal glutamine and deamidation of asparagines and glutamines) (40). Numerous analytical variables (technical replicate analysis, reproducibility of trypsin digestion, impact of SP total protein concentration, analytical replicate analysis, and day-to-day reproducibility) were evaluated with an upgraded and optimized SRM assay for each protein. The following steps were taken to minimize chemical modifications of internal standard and endogenous peptides (oxidation of methionines and deamidation of asparagines and glutamines) during storage and analysis: (1) supplementation of the protein digest with 0.4 m methionine (Sigma-Aldrich), (2) storage of tryptic peptides at −20 °C until use; and (3) sealing of 96-well plates with silicone rubber mats and keeping plates at 7 °C during SRM analysis.
Experimental Design and Statistical Rationale for Absolute Quantification by SRM in the Verification Phase
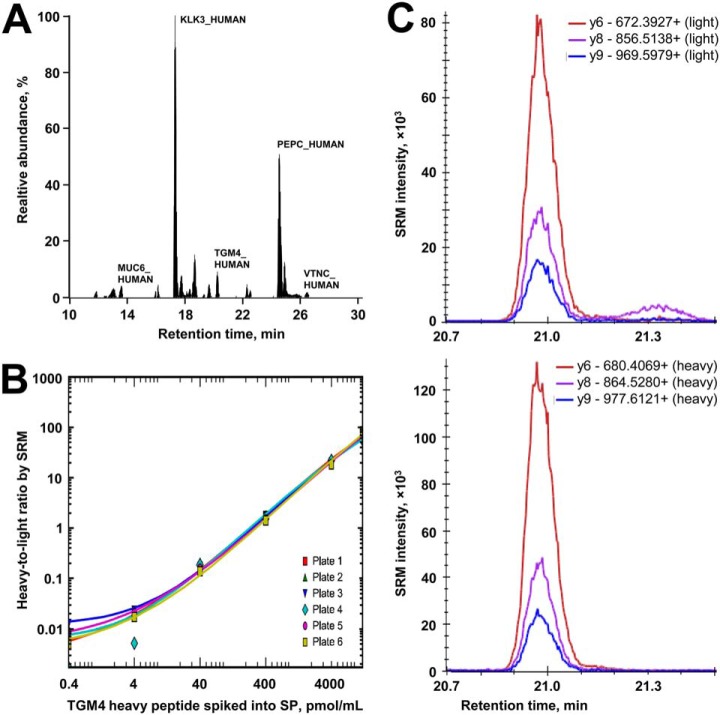
SP samples (n = 219) were randomized between six 96-well plate and included: (1) low-grade PCa (GS = 6, median serum PSA 5 ng/ml [IQR 4–8 ng/ml], median age 64 y.o. [IQR 59–68 y.o.], n = 94); (2) intermediate-grade PCa (GS = 7, 3 + 4; median serum PSA 6 ng/ml [IQR 5–7 ng/ml], median age 60 y.o. [IQR 55–64 y.o.], n = 38); (3) high-grade PCa (GS = 7, 4 + 3 and ≥ 8; median serum PSA 9 ng/ml [IQR 7–12 ng/ml], median age 66 y.o. [IQR 59–71 y.o.], n = 20); (4) negative biopsy (median serum PSA 6 ng/ml [IQR 4–7 ng/ml], median age 60 y.o. [IQR 55–65 y.o.], n = 67). According to effect size calculations, SRM analysis of 67 negative biopsy and 152 PCa samples could detect 1.1% change (1.011 ratio) in protein concentration among groups, assuming 80% power, α = 0.05, median 2.6% CV, and a two-tailed MWU test. SP samples were processes and analyzed in the same way as in the qualification phase, except the following: (1) 600 femtomoles of heavy isotope-labeled peptides (SpikeTides TM_TQL) were spiked into 5 μl of 10-fold diluted SP (∼20 μg of total protein) before proteomic sample preparation and trypsin digestion; (2) peptides were measured by TSQ Quantiva (Thermo Scientific Inc.), as previously described (20); (3) each of six plates included its own calibration curve and was generated by spiking serial dilutions of heavy internal standard peptides (0.4, 4, 40, 400, 4000 and 12000 pmol/ml) into the same SP digest. Instrumental parameters included “positive” polarity, 300°C ion transfer tube temperature, 2.0 mTorr argon pressure for the collision-induced dissociation, 10 volts source fragmentation, 0.2 and 0.7 Da FWHM resolution in the first and third quadrupoles, and 4 to 40 ms dwell times. Thirty seven peptides, 82 parent ions (including light and heavy forms as well as additional +3 forms for COR1B_HUMAN, GALT7_HUMAN and PROS_HUMAN, and an N-terminal pyroglutamate form for STEA4_HUMAN) and 250 transitions were scheduled within 2.2-min intervals during a 30 min LC gradient (5% to 10% acetonitrile for 1 min, 10% to 60% for 27 min, and 60% to 100% for 2 min). Before analysis of SP samples on each plate, eight quality control samples (10 fmoles Pierce™ BSA Protein Digest, Thermo Scientific; 10 min LC gradient) were analyzed, followed by calibration curve samples and two quality control samples. One quality control sample was analyzed after every 12 patient samples. Well-to-well carryover was found minimal (0.05–0.2%). Each SP sample was analyzed with technical duplicates. Endogenous peptide LC-SRM peak areas were normalized to the corresponding heavy peptide internal standards. Non-linear regression calibration curves were generated in the range 0.4–12,000 pmol/ml, light-to-heavy peptide ratios were fitted to corresponding calibration curves for each plate, and absolute concentrations of each peptide in each sample were calculated and averaged.
Machine-learning Analysis
Scikit-learn library for the Python programming language was used for machine learning analysis (41). Multiple algorithms including generalized linear models, support vector machines, nearest neighbors, naive Bayes, shallow and deep neural networks were tested. A nonlinear Extreme Gradient Boosting (XGBoost) algorithm (35) was selected as the most efficient approach for selection of weak features with small data sets and for generation of a single strong classifier with a set of weak classifiers. XGBoost was also relatively fast and allowed for even further speed improvements via standard parallelization strategies for multi-processor systems.
Briefly, twenty-two variables (19 candidate proteins, seminal KLK3, serum PSA and age) were used to generate all possible 1 to 5 marker combinations (35,442 combinations in total). Cross-validation was run for each combination in order to select combinations with the highest F05-scores. Compared with F1-score with the harmonic mean of precision (or positive predictive value) and recall (or sensitivity), F05-score weighted precision twice as much as recall. AUCs, sensitivities, specificities and negative predictive values were also estimated. Over-fitting was reduced using stringent 10 × 10 cross-validation which included splitting data 10 times into 10 sets in a way to keep the proportion of negative and positive samples across all sets approximately the same as in the whole dataset. The stratified 10-fold cross-validation was repeated 10 times, and the total number of train-validation runs was 100. Top combinations were verified on the whole dataset of patients to ensure that each potential marker had feature scores higher than a randomly generated feature. Finally, 100-fold bootstrapping was used to estimate mean values for performance metrics and calculate 95% confidence intervals.
Development of TGM4 ELISA
To enable orthogonal verification of TGM4 performance as a biomarker in SP and blood serum samples, a high-precision Tier 1 immunoassay was developed (34). Anti-TGM4 polyclonal mouse antibodies generated against the full length TGM4 (Met1-Lys684) were obtained from Abnova (H00007047-B01P; lot 09177 WUIZ; Walnut, CA). Antigen affinity-purified anti-TGM4 polyclonal sheep antibodies generated against the full length TGM4 (Met2-Lys684) were obtained from R&D Systems (AF5760; lot CDCX0111121; Minneapolis, MN). The full length recombinant human TGM4 Met2-Lys684 protein (R&D Systems; AAC50516; lot SIC0213111) expressed in S. frugiperda ovarian cell line Sf 21 was used during assay development. SP pool with a known concentration of endogenous TGM4 (4.65 μg/ml), as measured by SRM, was used as the assay calibrator.
To validate specificity of antibodies, we performed immunocapture-SRM analysis with recombinant human (rhTGM4) and endogenous TGM4 from SP. ELISA plates were coated with 500 ng/well of either sheep or mouse antibodies and incubated with either rhTGM4 (0, 2, 10 and 51 ng/ml) or endogenous TGM4 (0, 4, 19, and 94 ng/ml). Following that, plates were washed, proteins were digested with trypsin, and peptides were quantified with an SRM assay. As a result, endogenous TGM4 from SP was enriched equally well by both antibodies, whereas sheep antibody AF5760 could not efficiently capture rhTGM4. We then coated ELISA plates with 300 ng/well of sheep or mouse antibodies, incubated with rhTGM4 (0, 5, 20, and 100 ng/ml) or endogenous TGM4 in SP (0, 5, 20, and 100 ng/ml), and detected with biotinylated sheep or mouse antibodies and standard time-resolved fluorescence ELISA protocol (32). ELISA revealed that endogenous TGM4 from SP generated much higher signal than rhTGM4, and that the mouse-sheep format generated substantially lower background and higher signal-to-noise. Finally, mouse-sheep sandwich format with endogenous TGM4 as a calibrator was selected. Limits of blank, detection and quantification were calculated as 9, 22, and 30 pg/ml, respectively.
To measure TGM4 in SP and blood serum samples, the 96-well ELISA plates were coated with 300 ng/well of mouse antibody H00007047-B01P in 50 mm Tris-HCl buffer at pH 7.8. Plates were washed twice with the washing buffer (0.05% Tween 20 in 20 mm Tris-HCl and 150 mm NaCl at pH 7.4). Calibrator and SP samples were diluted with the assay diluent (60 g/L BSA, 25 ml/L normal mouse serum, 100 ml/L normal goat serum and 10 g/L bovine IgG in 50 mm Tris-HCl at pH 7.8). Serial dilutions of the calibrator (SP pool with 4.65 μg/ml endogenous TGM4 diluted to 0.02, 0.06, 0.18, 0.56, 1.7, 5, and 15 ng/ml, 100 μl/well) were prepared. Patient SP samples (2 μl) were diluted 400-fold with the assay diluent and added on ELISA plates (100 μl/well). Following 2 h incubation with gentle shaking, plates were washed twice with the washing buffer. Sheep polyclonal antibody AF5760 was biotinylated in-house, added to each well (25 ng in 100 μl of the assay diluent) and incubated for 1 h. Plates were washed six times, and streptavidin-conjugated alkaline phosphatase was added for 15 min with gentle shaking. After the final wash, diflunisal phosphate solution was prepared in the substrate buffer (0.1 m NaCl, 1 mm MgCl2 in 0.1 m Tris at pH 9.1), added to the plate (100 μl per well) and incubated for 10 min at room temperature with gentle shaking. Finally, the developing solution (1 m Tris-HCl, 0.4 m NaOH, 2 mm TbCl3 and 3 mm EDTA) was added and mixed for 1 min. Time-resolved fluorescence was measured with the Wallac EnVision 2103 Multilabel Reader (Perkin Elmer, Waltham, MA), as previously described (32). All SP and blood serum samples were measured in analytical duplicates. SP samples with TGM4 concentrations outside the range were re-measured with 800-, 40- and 10-fold dilutions. Blood serum samples (50 μl each) were analyzed with 2-fold dilutions.
Blood serum samples included: (1) confirmed primary PCa (all GS scores, median serum PSA 9 ng/ml [IQR 6–16 ng/ml], median age 65 y.o. [IQR 55–74 y.o.], n = 29); (2) no evidence of cancer (negative biopsy, median serum PSA 8 ng/ml [IQR 6–12 ng/ml], median age 63 y.o. [IQR 54–69 y.o.], n = 23); (3) prostate inflammation (median serum PSA 8 ng/ml [IQR 6–14 ng/ml], median age 65 y.o. [IQR 56–70 y.o.], n = 11); (4) healthy men (median age 36 y.o. [IQR 31–41 y.o.], n = 17). According to effect size calculations, ELISA analysis of 68 negative biopsy and 160 PCa samples could detect 1.3% change (1.013 ratio) in TGM4 concentration among groups, assuming 80% power, α = 0.05, median 3.0% CV, and a two-tailed MWU test. To validate TGM4 performance by ELISA in our future studies, we will need an independent set of 65 negative biopsy and 65 prostate cancer samples, assuming 80% power (α = 0.05; two-tailed MWU test).
RESULTS
Selection of Candidate Proteins
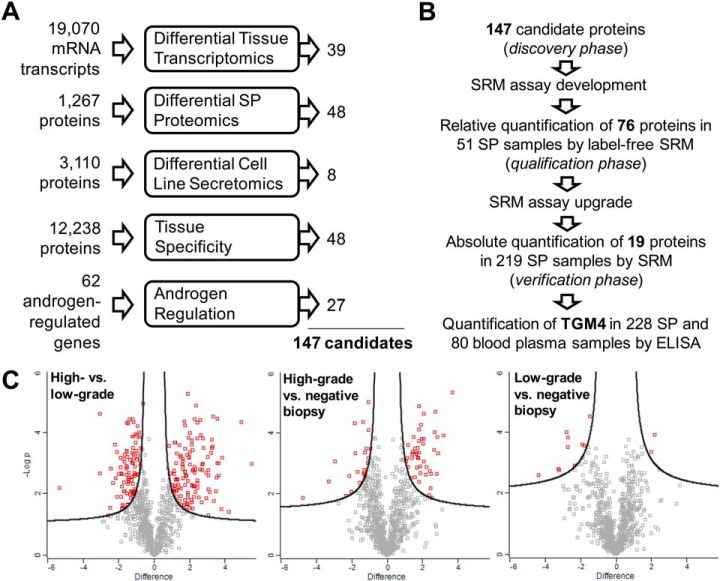
We combined proteins or transcripts identified through five independent experimental or data mining approaches, and generated a list of the most promising 147 biomarker candidates (Fig. 1A). To facilitate our diagnostic strategy, we considered as candidates only secreted and membrane-bound proteins which were previously identified in our SP proteome of more than 3,000 proteins (28, 29). We assumed that some candidates selected with large-scale -omics approaches might be false-positives because of pre-analytical, analytical, and data analysis biases. We also assumed that different -omics approaches could have different limitations (for example, discrepancy between mRNA and protein fold changes or inability to measure low-abundance proteins by mass spectrometry). As a result, our candidates were not compared across all five datasets, but were independently selected and merged into a single list. We applied more relaxed criteria for the initial selection of candidates, but then performed very stringent verification of candidates in SP by high-quality quantitative SRM assays (Fig. 1B). Our study was designed to simultaneously evaluate candidates for two clinical needs: (1) differentiation between PCa and negative biopsies, and (2) discrimination between low-grade (Gleason score (GS) = 6) and high-grade (GS≥8) PCa. Gleason score was chosen as a clinical end point for diagnosis and prognosis. We acknowledge that definition of PCa aggressiveness based on GS may not be perfect, however, the correlation between GS and the 20-year survival rate is well established (>70% for GS≤6 and <30% for GS≥8) (42).
Fig. 1.
Selection of candidate proteins. A, The most promising PCa biomarker candidates were selected with five independent experimental and data mining approaches. B, The combined 147 candidates were subjected to the SRM method development, followed by qualification and verification phases in SP and blood serum. A single peptide per protein was measured in the qualification and verification phases. C, A bottom-up proteomic approach and two-dimensional liquid chromatography followed by shotgun mass spectrometry and label-free quantification were used to identify differentially expressed proteins in pools of SP samples from patients with negative biopsy (NBx, serum PSA >4 ng/ml, n = 5), low-grade PCa (LG, GS = 6, PSA >4 ng/ml, n = 5) and high-grade PCa (GS≥8, PSA >4 ng/ml, n = 5) patients. Log2 differences and the t test with Benjamini-Hochberg false-discovery rate-adjusted p values were calculated with Perseus software, and 1% FDR was used as a cut-off to select differentially expressed proteins for each comparison.
Differential Transcriptomics
Because mRNA levels explain only some variation of protein levels (43), mRNA fold changes in tissues and protein fold changes in SP were considered as independent criteria. To identify candidate genes based on differential transcriptomics, we mined the Cancer Genomics database (www.cbioportal.org) which contained gene expression microarray data in 131 primary PCa tissues and 29 adjacent benign prostate tissues (44), as well as clinical data, such as PSA at diagnosis and GS. The following cut-off criteria were used to select candidates: (1) ≥1.5-fold increased or decreased RNA expression in PCa tissues (n = 109) versus adjacent benign prostate tissues (n = 29) and p values <0.05 (supplemental Table S1); (2) ≥1.5-fold increased or decreased expression of mRNA transcripts in PCa tissues with GS = 6 (n = 41) versus GS≥8 (n = 15) and p values <0.05 (supplemental Table S2); (3) secreted and membrane-bound proteins based on bioinformatic predictions of signal peptides or transmembrane regions (45); and (4) proteins previously identified in our SP proteome and thus amenable to quantification in SP by SRM assays. As a result, we selected 39 candidates. Interestingly, a non-coding transcript PCA3 emerged as a top candidate and differentiated between PCa and adjacent benign tissues (6-fold higher expression in PCa; supplemental Fig. S2).
Differential Proteomics
Shotgun mass spectrometry was utilized to identify differentially expressed proteins in pools of five SP samples from patients with serum PSA ≥4 ng/ml and negative biopsy, low-grade PCa (GS = 6) and high-grade PCa (GS≥8). The rationale for using pooled samples was to reduce the inter-individual variability and increase the likelihood of identifying consistent proteomic differences. A bottom-up proteomic approach and two-dimensional liquid chromatography followed by tandem mass spectrometry and label-free quantification were utilized (43). Each analytical replicate of the SP pools was fractionated by strong-cation exchange chromatography into 25 fractions, which were analyzed by reverse-phase chromatography and tandem mass spectrometry (supplemental Fig. S3). Label-free quantification with MaxQuant and Perseus software was used to prioritize candidates (Fig. 1C). The following cut-off criteria were used in Perseus to select candidates: (1) over- or under-expressed proteins (FDR≤1%, s0 = 0.22) in high-grade PCa versus negative biopsy (supplemental Table S3); (2) over- or under-expressed proteins (FDR≤1%, s0 = 0.23) in low-grade PCa versus negative biopsy (supplemental Table S4); (3) over- or under-expressed proteins (FDR≤1%, s0 = 0.27) in high- versus low-grade PCa (supplemental Table S5); (4) secreted and membrane-bound proteins based on bioinformatic predictions (45). High-abundance blood serum and testis-, seminal vesicle- and epididymis-specific proteins were excluded. As a result, 48 candidates were selected.
Differential Secretomics
Previously, we identified secretomes of a near-normal prostate epithelial cell line RWPE-1, two androgen-dependent PCa cell lines (LNCaP and VCaP) and five androgen-independent PCa cell lines (PC-3, DU-145, PPC-1, LNCaP-SF and 22Rv1) (46). Here, we hypothesized that secretomes of androgen-independent cell lines contained proteins elevated at the later stages of PCa, or in more aggressive PCa. We selected 8 most promising candidates which were identified with at least two peptides and were upregulated ≥2-fold in the androgen-independent versus androgen-dependent plus near-normal cell lines, based on spectral counting. These 8 candidates (supplemental Table S6 and supplemental Fig. S4) were secreted or membrane-bound proteins and were previously identified in the SP proteome.
Tissue Specificity
We hypothesized that aberrant changes in the level of prostate-specific proteins could indicate progressing pathological processes in the prostate. In fact, the success of PSA is mainly because of its high tissue specificity. Like PSA, leakage of other prostate-specific proteins into blood serum could indicate destruction of prostate-blood barriers because of PCa progression.
To identify proteins with exclusive or highly restricted expression in the prostate tissue, we mined the Human Protein Atlas (www.proteinatlas.org) and BioGPS (http://biogps.org) databases. Human Protein Atlas (v. 9) included 12,238 genes with immunohistochemistry-based protein expression profiles in 66 normal human tissues and cells. To identify tissue-specific proteins, we analyzed the Human Protein Atlas data and ranked proteins according to their tissue-specific expression in human tissues and cells. Proteins with high or medium immunohistochemical staining in the prostate, but not in other four glands of the male urogenital system (testis, seminal vesicles, epididymis and seminiferous tubules) were selected. We also applied a similar strategy to the BioGPS database and identified tissue-specific genes based on mRNA expression profiles in 84 normal human tissues and cells. In total, we selected 74 proteins with highly specific expression in the prostate, and 48 of these proteins were previously identified in our SP proteome (supplemental Table S7). The list of candidates included 35 secreted and membrane-bound proteins. We also hypothesized that tissue destruction because of PCa progression could result in the elevated levels of some intracellular proteins in SP and thus retained 13 prostate-specific intracellular proteins.
Androgen Regulation
Physiological role of prostate is highly dependent on androgens and on androgen receptor, which plays a pivotal role in the development and progression of PCa (47, 48). We hypothesized that androgen-regulated proteins might be differentially expressed in SP of low- versus high-grade PCa patients. To select androgen-regulated proteins, we reviewed the high-quality datasets of genes with increased expression on androgen stimulation, genes with predicted androgen-response elements, and the relevant literature (supplemental Table S8). We selected 62 androgen-regulated proteins, 27 of which were secreted or membrane-bound proteins present in our SP proteome.
Development of a Multiplex SRM Assay for the Qualification Phase
SRM is a quantitative analytical assay performed with a triple-quadrupole mass spectrometer (49). In this study, we used our previously published SRM protocols to measure putative protein biomarkers in various biological fluids (50, 51). Briefly, the Peptide Atlas database (52) was utilized to select the best proteotypic peptides for 147 candidates, as well as additional 12 tissue- and cell-specific control proteins which represented seminal vesicles, Cowper's glands, epididymis, germ cells, Sertoli cells and Leydig cells (supplemental Table S9). SRM assays in SP, however, were developed for only 82 candidate and 11 control proteins (supplemental Table S10). Such success rate of SRM assay development could be explained by the low abundance of some proteins in SP and the lack of high-quality tryptic peptides. Finally, proteins were assembled into a single multiplexed SRM assay and entered the qualification phase.
Qualification of Candidate Biomarkers
In the qualification phase, we were able to measured 76 candidate and 11 control proteins in 13 negative biopsy, 24 low-grade and 14 high-grade age-matched SP samples (6 proteins were excluded after data analysis). SRM areas for each peptide were normalized to a single spiked-in heavy isotope internal standard of KLK3 protein, and normalized areas were used to calculate concentrations and then diagnostic specificities, sensitivities and AUCs for each candidate. Proteins were ranked based on their AUCs (supplemental Table S11). Statistical analysis revealed significant upregulation of 21 proteins (p < 0.05) in all PCa versus negative biopsy groups. Control proteins, such as mucin-6 secreted by seminal vesicles, were not significant among groups, as expected. Regarding the second clinical need, statistical analysis revealed significant downregulation of 8 proteins (p < 0.05) in high- versus low-grade groups. As a result, 29 proteins were selected for the verification phase (supplemental Fig. S5).
Upgrade of the SRM Assay for the Verification Phase
To facilitate rigorous verification of top candidates and measure their absolute concentrations in SP, we utilized heavy isotope-labeled peptides with trypsin-cleavable tags. We also optimized and shortened LC gradient to 30 min, to allow for the measurement of 12 SP samples with technical duplicates per day. SRM measurements of peptide internal standards before and after trypsin digestion revealed near complete cleavage of quantifying tags. Using shotgun mass spectrometry, we discovered the following chemical modifications of our peptide internal standards following tryptic digestion: cysteine alkylation, methionine oxidation, formation of pyroglutamate of N-terminal glutamine, and deamidation of asparagines and glutamines. Using SRM, we quantified the yield of each modification: cysteine alkylation (>99% in 15 peptides), methionine oxidation (∼15% in 2 peptides), formation of pyroglutamate of N-terminal glutamine (∼50%; 1 peptide), and deamidation of asparagines and glutamines (∼20% in 7 peptides). In addition, we evaluated +2 and +3 charge states for six peptides and selected both +2 and +3 forms for three peptides. As a result, we included multiple forms of some peptides into the final SRM method (supplemental Table S12). We also fully investigated analytical and pre-analytical parameters including LC-SRM injections (technical reproducibility), trypsin digestion reproducibility, full sample preparation process (analytical reproducibility) and day-to-day reproducibility for three SP samples with different amounts of total protein (35, 67, and 102 mg/ml; supplemental Fig. S6).
Verification of Candidate Biomarkers
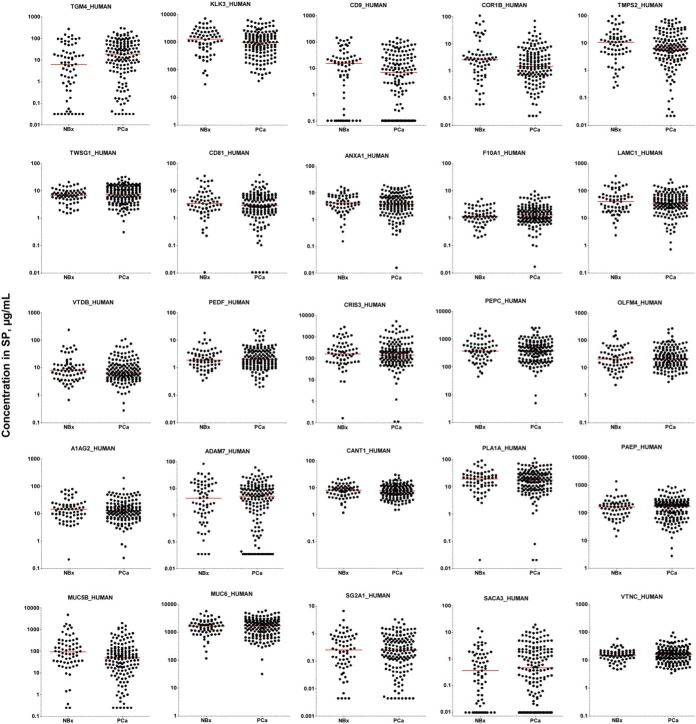
In the verification phase, top candidates were first quantified by SRM (Fig. 2A and supplemental Tables S13–S15). Digests of SP samples (n = 219) were randomized between six 96-well plates (supplemental Figs. S7) and included: 67 negative biopsy samples, 94 low-grade, 38 intermediate-grade and 20 high-grade PCa samples. SRM areas for each peptide were normalized to the corresponding spiked-in internal standards (Fig. 2C), and normalized areas and calibration curves for each protein (Fig. 2B and supplemental Figs. S8, S9) were used to calculate protein concentrations in SP (Fig. 3). As a result, TGM4 protein was found significantly upregulated (3.1-fold change, MWU p = 0.0075, AUC = 0.61) in PCa versus negative biopsy samples (Table I and supplemental Fig. S10). No individual SP proteins differentiated between high- and low-grade PCa samples, whereas serum PSA revealed significantly higher levels (10 versus 5 ng/ml, p = 0.0002). Control proteins exclusively expressed and secreted by seminal vesicles (MUC6_HUMAN), epididymis (SG2A1_HUMAN), germ cells (SACA3_HUMAN) and Leydig cells (VTNC_HUMAN) were not differentially expressed, as expected. Distribution of candidates with respect to their initial selection in the discovery phase and evaluation in qualification and verification phases is presented in supplemental Fig. S11, whereas distribution of candidates with respect to the dynamic range of SP proteome is shown in supplemental Fig. S12.
Fig. 2.
Performance of a multiplex SRM assay in the verification phase. A, Endogenous peptides and heavy peptide internal standards were multiplexed in a single SRM assay. B, Representative calibration curves used to quantify TGM4 protein in 67 negative biopsy and 152 PCa SP digests distributed between six 96-well plates. Similar curves were obtained for the rest of proteins (supplemental Fig. S8). Light-to-heavy ratios for TGM4 in each sample were plotted against the corresponding calibration curve, to derive TGM4 concentrations. C, Representative SRM transitions for the light endogenous and heavy internal standard peptides for TGM4 protein.
Fig. 3.
The most promising candidates measured in the verification phase. Using the stable-isotope dilution multiplex SRM assay, 19 candidates and 6 control proteins (KLK3, MUC6, MUC5B, SG2A1, SACA3 and VTNC) were quantified in the negative biopsy (NBx, n = 67) and PCa (n = 152) SP samples. Horizontal lines represent median values in SP.
Table I. Candidate proteins verified by Tier 2 quantitative SRM assays in 152 PCa and 67 negative biopsy SP samples. Control proteins included proteins exclusively expressed in seminal vesicles, prostate, Cowper's glands, epididymis, germ cells and Leydig cells. NBx, negative biopsy; MWU, Mann–Whitney U test; AUC, a receiver operating characteristic area under the curve.
| Gene name | Protein name | Ratio of medians PCa/NBx | p value (2-tailed MWU) | AUC | AUC 95% CI |
|---|---|---|---|---|---|
| TGM4 | TGM4_HUMAN | 3.1 | 0.0075 | 0.61 | 0.53–0.70 |
| CD9 | CD9_HUMAN | 0.5 | 0.11 | 0.59 | 0.51–0.67 |
| CORO1B | COR1B_HUMAN | 0.6 | 0.16 | 0.56 | 0.48–0.64 |
| TMPRSS2 | TMPS2_HUMAN | 0.6 | 0.19 | 0.56 | 0.47–0.64 |
| TWSG1 | TWSG1_HUMAN | 1.2 | 0.20 | 0.55 | 0.48–0.63 |
| CD81 | CD81_HUMAN | 0.9 | 0.34 | 0.54 | 0.46–0.62 |
| ST13 | F10A1_HUMAN | 1.1 | 0.32 | 0.54 | 0.46–0.62 |
| ANXA1 | ANXA1_HUMAN | 0.9 | 0.41 | 0.53 | 0.45–0.62 |
| LAMC1 | LAMC1_HUMAN | 0.9 | 0.49 | 0.53 | 0.44–0.62 |
| GC | VTDB_HUMAN | 0.8 | 0.53 | 0.53 | 0.44–0.61 |
| SERPINF1 | PEDF_HUMAN | 1.1 | 0.49 | 0.53 | 0.45–0.61 |
| CRISP3 | CRIS3_HUMAN | 0.9 | 0.64 | 0.52 | 0.44–0.60 |
| OLFM4 | OLFM4_HUMAN | 1.0 | 0.59 | 0.52 | 0.44–0.61 |
| ORM2 | A1AG2_HUMAN | 0.9 | 0.67 | 0.52 | 0.44–0.60 |
| PGC | PEPC_HUMAN | 1.0 | 0.73 | 0.51 | 0.43–0.60 |
| ADAM7 | ADAM7_HUMAN | 1.2 | 0.77 | 0.51 | 0.43–0.60 |
| PAEP | PAEP_HUMAN | 1.1 | 0.84 | 0.51 | 0.43–0.59 |
| CANT1 | CANT1_HUMAN | 1.0 | 0.95 | 0.50 | 0.42–0.59 |
| PLA1A | PLA1A_HUMAN | 0.9 | 0.98 | 0.50 | 0.42–0.58 |
| Control proteins | |||||
| KLK3 | KLK3_HUMAN | 0.8 | 0.11 | 0.57 | 0.49–0.65 |
| MUC6 | MUC6_HUMAN | 1.1 | 0.80 | 0.51 | 0.43–0.59 |
| MUC5B | MUC5B_HUMAN | 0.5 | 0.004 | 0.62 | 0.54–0.70 |
| SCGB2A1 | SG2A1_HUMAN | 1.0 | 0.93 | 0.50 | 0.42–0.59 |
| SPACA3 | SACA3_HUMAN | 1.4 | 0.41 | 0.54 | 0.46–0.61 |
| VTN | VTNC_HUMAN | 1.0 | 0.57 | 0.52 | 0.44–0.60 |
Because some clinical tests may require additional normalization to account for sample dilution (for example, levels of PCA3 transcript in urine need to be normalized by KLK3 levels), we investigated if normalization of protein concentrations by total KLK3 protein in SP would impact the performance of our markers (supplemental Fig. S13). Levels of mucin-5B, a protein exclusively expressed and secreted by the Cowper's glands (53), were significantly lower in PCa (p = 0.004), but barely significant after normalization by KLK3 (p = 0.043). Because of its exclusive expression in the Cowper's glands, mucin-5B was not considered as a candidate. Even though normalization by KLK3 slightly improved TGM4 AUC from 0.61 (p = 0.0075) to 0.63 (p = 0.0016), such improvement would not justify the measurement of an additional analyte (seminal KLK3) in the clinical lab. Thus, normalization by total KLK3 in SP was not further considered in data analysis.
Machine Learning Analysis to Identify Combinations of Markers
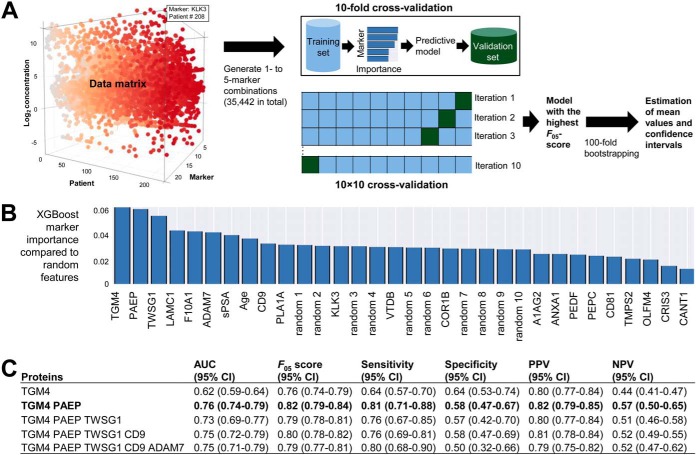
To identify combinations of SP proteins that could improve TGM4 performance to differentiate between negative biopsy and PCa, or predict high-grade PCa, we employed machine learning algorithms (Fig. 4A). A nonlinear Extreme Gradient Boosting (XGBoost) algorithm (35) was selected as the most effective algorithm for obtaining high values for AUCs, PPVs and sensitivity following 10 × 10-fold stratified cross-validation. Relative to other algorithms, XGBoost was better suitable for creating a single strong classifier with a set of weak classifiers and provided better selection of weak features with relative small data sets. Stringent cross-validation was used to reduce over-fitting. Importance of each marker, as compared with random features, was calculated (Fig. 4B), and combinations with the highest F05-scores were selected. AUCs, sensitivities, specificities, PPVs and NPVs were estimated (Fig. 4C).
Fig. 4.
Machine-learning analysis to identify multi-variable combinations of markers. A, Twenty two variables (19 candidate SP proteins, seminal KLK3, serum PSA and age) were used to generate all possible 1- to 5- marker combinations. XGBoost algorithm was applied to identify combinations with the highest F05-measure scores and calculate AUCs, sensitivities, specificities, PPVs and NPVs. Stringent 10 × 10 cross-validation was applied to reduce over-fitting. Top combinations were verified on the whole dataset of patients to ensure that each potential marker had feature scores higher than a randomly generated feature. Finally, 100-fold bootstrapping was used to estimate mean values for performance metrics and calculate 95% confidence intervals. B, XGBoost importance of individual markers to differentiate between PCa and negative biopsy, as compared with random features. C, Diagnostic performance of top combinations, with 95% confidence intervals estimated using 100-fold bootstrapping. Combination of TGM4 with PAEP protein improved AUC and sensitivity to differentiate between negative biopsy and PCa, whereas additional markers did not further increase AUCs.
Interestingly, a combination of TGM4 with PAEP protein improved AUC and sensitivity to differentiate between negative biopsy and PCa, whereas additional markers did not further increase AUCs (supplemental Table S16). This 2-protein diagnostic panel revealed AUC = 0.76 (CI95% 0.74–0.79) and was comparable to a 6-peptide diagnostic panel with AUC = 0.77 (CI95% 0.68–0.87) previously reported in the expressed prostatic secretions-urine (24). Our 3-protein SP panel (KLK3, PAEP and PEPC) revealed AUC = 0.73 (CI95% 0.66–0.80) for prediction of high-grade PCa (GS 4 + 3 and ≥8). A 4-protein SP panel in combination with serum PSA (CD9, COR1B, KLK3, TMPS2 and PSA) revealed AUC = 0.83 (CI95% 0.80–0.87) for prediction of high-grade PCa (supplemental Table S17). Such prognostic performance exceeded the performance of a previously reported 7-peptide prognostic panel in the expressed prostatic secretions-urine with AUC = 0.74 (CI95% 0.62–0.85) (24). Even though our multi-marker panels showed promise, measurements of such panels in the clinical lab may not be practical because of challenges with standardization of multi-marker assays. For the clarity of our message, we focused on a single most promising and novel protein TGM4.
Development of TGM4 ELISA
Because our SRM assay with the limit of quantification 31 ng/ml in SP was not sensitive enough to measure low levels of TGM4, we developed an in-house ELISA immunoassay. The performance of commercial polyclonal sheep and mouse anti-TGM4 antibodies was determined by immunocapture-SRM assays. ELISA with a time-resolved fluorescence detection (32) revealed that endogenous TGM4 from SP generated ∼3 times higher signal than rhTGM4, and that the mouse-sheep format generated substantially lower background (S/n = 19 at 5 ng/ml), as compared with the sheep-mouse format (S/n = 6). Following that, serial dilutions of endogenous and recombinant TGM4 were evaluated using sheep-mouse and mouse-sheep sandwich formats. A mouse-sheep format with endogenous TGM4 in SP as a calibrator provided higher signal-to-noise ratios and was chosen as the final format.
Measurement of TGM4 by ELISA in SP and Blood Serum
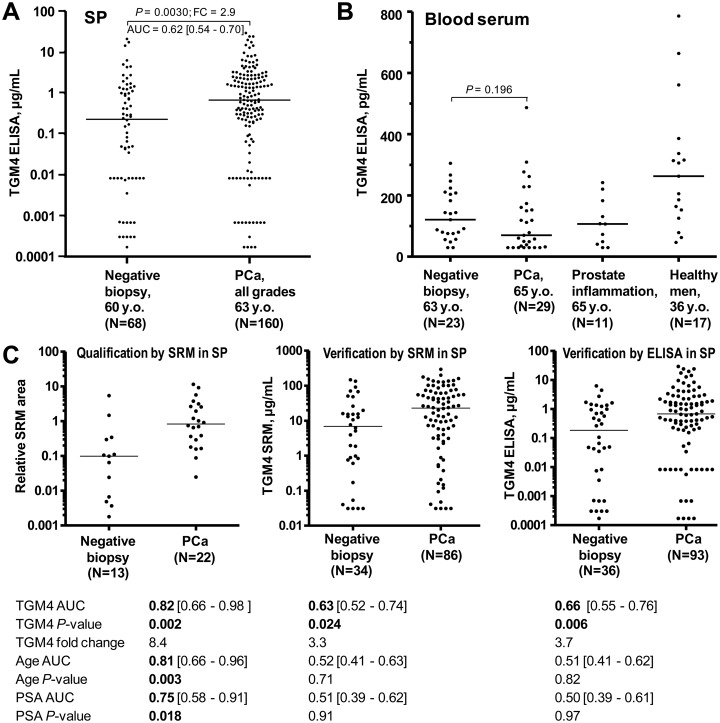
TGM4 protein was measured by ELISA in 228 SP and 80 blood serum samples (Fig. 5A, 5B and supplemental Table S18). Interestingly, diagnostic performances of TGM4 by SRM (3.1-fold change, AUC = 0.61, p = 0.0075) and ELISA (2.9-fold change, AUC = 0.62, p = 0.003) in SP were very similar. In blood serum, we were able to detect very low levels of TGM4 by ELISA (median 120 pg/ml). Unlike TGM4 in SP, TGM4 in serum could not differentiate between PCa and negative biopsy. TGM4 levels were not different because of prostate inflammation, but were elevated in serum of younger healthy men. Median TGM4 levels were ∼2,000-fold lower in blood than in SP of men with negative biopsy. To the best of our knowledge, our study could be the first report on identification and quantification of prostate-specific protein TGM4 in human blood serum.
Fig. 5.
TGM4 diagnostic performance. Performance of TGM4 protein as measured by an in-house ELISA in all 228 SP samples (A) and 80 blood serum samples (B). For comparison, age and serum PSA provided AUCs 0.60 ([95% CI 0.53–0.68]; MWU p = 0.0105) and 0.56 ([0.48–0.63]; p = 0.18) in the same cohorts of patients. C, TGM4 performance in three phases (qualification in SP by SRM, independent verification in SP by SRM, and independent verification in SP by ELISA) for the most clinically relevant patient groups (serum PSA ≥4 ng/ml and age ≥50 years old). As measured by ELISA, TGM4 provided AUC = 0.66 to predict PCa on biopsy and outperformed age and serum PSA. Concentration cut-off >1.74 μg/ml revealed 92% specificity at 31% sensitivity, and substantially increased specificity of serum PSA ≥4 ng/ml to detect PCa.
To elucidate the possible identity of TGM4 proteoforms in SP, we investigated numerous -omics aggregation databases. NextProt database (www.nextprot.org) revealed only one canonical protein-coding isoform of TGM4 (77 kDa, 684 aa) translated from the canonical transcript ENST00000296125. Another transcript ENST00000422219 undergoing a nonsense mediated decay could hypothetically encode a shorter 91 aa isoform. GTeX portal (www.gtexportal.org), however, reported no expression of that transcript in human tissues (supplemental Fig. S14). Our shotgun proteomic data identified 20 unique peptides spanning the 46–621 aa region of the canonical 684 aa isoform. TGM4 in prostate tissues was present mostly as a non-glycosylated protein (54). These data suggested that we were measuring the canonical non-glycosylated 684 aa isoform of TGM4 in SP and blood serum. We could also speculate that large 684 aa molecules of TGM4 could not easily leak through the blood-prostate barrier. Indeed, the most promising novel biomarkers of PCa currently in clinical trials are relatively small proteins: 244 aa pro-PSA, 243 aa pro-KLK2, 94 aa MSMB and 114 aa MIC1 (55). Very low levels of TGM4 in blood serum (120 pg/ml) could also originate from TGM4 expression in other tissues, like the ultra-low levels of non-prostatic PSA (10–100 pg/ml) which has been detected in female sera (56).
TGM4 Performance in Qualification and Independent Verification Sets for Patients with Serum PSA ≥4 ng/ml and Age ≥50 y.o
Finally, we investigated TGM4 performance within the most clinically relevant group of patients with serum PSA ≥4 ng/ml and age ≥50 y.o. (Fig. 5C). In all sets (qualification, independent verification by SRM and independent verification by ELISA; supplemental Table S19) TGM4 demonstrated statistical significance. As measured by ELISA (Fig. 5C), TGM4 concentration >1.74 μg/ml revealed 92% specificity at 31% sensitivity to detect PCa in the group of patients with PSA ≥4 ng/ml and age ≥50 y.o. Positive and negative predictive values were 62 and 76%, respectively. Serum PSA ≥4 ng/ml on its own demonstrated only 28% specificity at 82% sensitivity to detect PCa on biopsy for patients aged ≥50 years. Positive and negative predictive values were 33 and 78%, respectively. Thus, TGM4 > 1.74 μg/ml in SP substantially increased diagnostic specificity of serum PSA ≥4 ng/ml, which was a recognized unmet clinical need and one of the initial aims of our study.
Correlation of TGM4 Concentration with Age
We observed that TGM4 levels in blood serum were higher in samples from younger men versus older patients. To investigate the significance of such trend, we split blood serum samples measured by ELISA into groups of <40 years (n = 11), 40–49 (n = 11), 50–59 (n = 19), 60–69 (n = 21) and ≥70 (n = 18). As a result, we found a significant difference for TGM4 blood serum levels in these age groups (Kruskal-Wallis p = 0.0043). The most substantial difference (Dunn's multiple comparisons test p < 0.01) was found for groups with age <40 y.o. versus >70 y.o. (median levels 206 versus 59 pg/ml). The difference in TGM4 levels for patients in the different age groups, as measured by SRM in SP, was not significant (Kruskal-Wallis p = 0.81). However, TGM4 median levels in SP decreased 3.6-fold between age groups of 40–49 (median 36 μg/ml) and 70–79 (median 10 μg/ml). To conclude, the decrease of TGM4 levels after the age of 50 should be carefully considered in future studies and might explain the lack of consensus in previous studies which identified TGM4 as either up- or downregulated biomarker of PCa (24, 57–59).
DISCUSSION
Prostate cancer biomarkers studied up to date included genetic (60) and epigenetic markers (16), molecular markers such as RNA (12, 17, 18), proteins (11, 14, 15) and metabolites (61), and circulating tumor cells (13). These markers were studied in serum, urine, prostatic secretions, prostate tissues, and cells found in urine. The reality of PCa diagnostics, however, turned out as very challenging. Novel clinical tests only marginally improved PSA performance. Prostate Health Index (Phi), an FDA-approved blood serum test for three PSA forms, was intended for use before the initial biopsy in men with elevated PSA (>4 ng/ml), and revealed AUC = 0.68 to predict the initial biopsy outcome (62). An FDA-approved PCA3 test in urine after prostate massage was intended to predict the need for secondary biopsy, and revealed AUC = 0.70 (62). Emerging 4KScore test predicted PCa on primary biopsies with AUC = 0.69 (14). A comprehensive STHLM3 model (6 blood serum proteins, 232 risk SNPs and 5 clinical parameters) facilitated detection of GS ≥7 with the cumulative AUC = 0.76, whereas individual markers had AUCs in the range of 0.59–0.67 (15). Most of these tests re-grouped different PSA proteoforms in combination with additional clinical characteristics, whereas novel unique protein biomarkers demonstrated only marginal performance (AUC = 0.59, 0.60 and 0.59 for KLK2, MSMB and MIC1, respectively). Non-protein biomarkers, such as hypermethylation of GSTP1, APC and RASSF1 genes in tissue biopsies (ConfirmMDx) revealed AUC = 0.66 to predict GS≥7 (16).
There were only few previous studies on PCa biomarkers in SP (20–22) or expressed prostatic secretions (23, 24). Overall, SP is a highly relevant biological fluid to search for biomarkers because prostate-secreted proteins are found at much higher concentrations in SP than in serum or urine (25, 63), and might be more easily identified and quantified by mass spectrometry. No doubt that semen and SP are unconventional fluids for PCa diagnostics, and that some older patients may have difficulty in providing SP for analysis. However, discussions of our urologists with patients (50 to 75 y.o.) indicated that most of them were willing and able to provide SP for diagnostic testing, if such test would replace invasive biopsies.
We previously completed extensive studies on SP proteome, identified more than 3,000 proteins in SP of healthy men and patients with infertility (28, 29), and developed a simple 2-biomarker algorithm for the differential diagnosis of male infertility (30, 64). In this study, we designed a rigorous biomarker development pipeline with discovery, qualification and verification phases (65). Quantitative multiplex SRM assays were a foundation of our pipeline and allowed simultaneous verification of dozens of candidates in hundreds of SP samples within a realistic timeline (near 30-day continuous SRM data acquisition). We qualified 76 candidates and verified 19 candidates in SP, whereas our top candidate TGM4 protein was also evaluated in blood serum. Majority of our candidate proteins have never been previously quantified in SP or investigated in the context of PCa, and the molecular function of some proteins (olfactomedin-4, twisted gastrulation protein homolog 1, etc.) may not be well known. We demonstrated that levels of most prostate-specific proteins previously thoroughly characterized in blood (prostatic acid phosphatase, kallikreins 2 and 3, prostate-specific membrane antigen, beta-microseminoprotein, neuropeptide Y, transmembrane protease serine 2, and others) remained unchanged in SP of PCa versus negative biopsy patients. In addition, no difference was found for the levels of androgen-regulated proteins, except for TGM4. Multi-variable machine learning analysis provided a unique combination of TGM4 with a pregnancy-associated endometrial alpha-2 globulin (PAEP). Such 2-marker combination improved detection of PCa on biopsy (AUC = 0.76) and could be further investigated in detail.
Previous studies on TGM4 revealed it as a key regulator of invasiveness (66) and cell adhesion (67), and demonstrated association of TGM4 with the epithelial-mesenchymal transition and interaction between cancer and vascular endothelial cells (68). TGM4 was previously suggested as a PCa biomarker, but results were inconsistent and revealed either significant over-expression (59) or under-expression (24, 57, 58) of TGM4 in PCa versus benign disease, or slightly opposite directions based on different assays (23). TGM4 was found downregulated 1.7-fold in urinary extracellular vesicles of PCa and had AUC 0.58 to diagnose PCa on biopsy (57). Immunohistochemistry with tissue microarrays revealed under-expression of TGM4 in prostate tissues (p < 0.001) and AUC of 0.81 to detect PCa versus benign disease (57). TGM4 in the urinary extracellular vesicles also differentiated between low- and high-grade PCa with high sensitivity and specificity (p < 0.001; AUC 0.82). Mining of recent genomic and transcriptomic data sets revealed that TGM4 gene was amplified in 23% of patients with neuroendocrine PCa (69), TGM4 mRNA was significantly over-expressed (p = 0.00001) in the Cambridge cohort of 125 men with primary PCa versus matched benign tissues (70), and TGM4 mRNA was 14-fold over-expressed in primary PCa versus benign prostatic hyperplasia (71). Our data suggested that TGM4 protein levels in blood and SP might decrease with age, whereas TGM4 levels in SP were elevated in PCa versus benign disease. The complex interplay of these factors (age-dependence, androgen regulation and intra-individual variability) could explain previous inconsistency regarding the levels of TGM4 in prostate tissues and urine of PCa patients. Overall, TGM4 has all characteristics of a promising biomarker, such as exclusive prostate tissue specificity, secretion into SP and androgen regulation. Even though demonstrated performance of TGM4 as a single biomarker will unlikely result in its immediate use in the clinic, TGM4 needs to be investigated in future as a biomarker of distinct genomic subtypes, or as a protein to be included into emerging multi-marker panels.
PCa heterogeneity revealed in the recent large-scale genomic studies (72) could obstruct identification of biomarkers with high diagnostic sensitivity. However, next generation sequencing may facilitate stratification of genomic subtypes and identification of “exceptional responder” biomarkers, e.g. high diagnostic specificity-biomarkers which perform only in distinct cancer subtypes (73). For example, more than 70% of primary PCa cases are driven by elevated ETS transcription factors, either through their over-expression because of the androgen-responsive gene fusions (72) or lack of degradation in SPOP-mutated subtypes (74). A rarer and more unique subtype of PCa (8% of primary PCa cases (75)) is driven by the mutated transcription factor FOXA1, which alters selectivity of androgen receptor binding and changes the pattern of expression of androgen-regulated genes (76). In our data set (Fig. 5C, ELISA data), we had 11 patients (12%) with very high TGM4 levels >6.3 μg/ml (all patients in the negative biopsy group had TGM4 < 6.3 μg/ml). These estimates may warrant detailed investigation of TGM4 levels in SP of patients with FOXA1 mutations.
Review of our data on the abundance of SP versus blood serum proteins made us to hypothesize that sensitivity of protein assays (1 pg/ml for ultrasensitive immunoassays) could be insufficient to validate novel PCa biomarkers in blood serum. Indeed, assuming a five order concentration gradient between SP and serum for high- and medium-abundance prostate-specific proteins (KLK3, KLK2, and TGM4), and taking into account the relative abundance of top 300 SP proteins measured by LFQ in this study, it can be estimated that potential serum concentration of the unexplored low-abundance prostate-specific proteins would be much lower than 1 pg/ml, and thus be undetectable by standard immunoassays. Validation of novel biomarkers of primary PCa in blood serum may thus require development of a new generation of protein assays with fg/ml or lower analytical sensitivity.
Our study might be one of the largest and most comprehensive studies on PCa biomarkers in SP. Following our previous validation of the prostate-specific kallikrein-4 in SP and blood serum (20), we evaluated here one of the last and not well-studied prostate-specific proteins within the medium-abundance proteome of SP. However, our study was not without limitations, which included: (1) evaluation of only medium-to-high abundance protein candidates (100 ng/ml - 1 mg/ml) measurable by SRM in the unfractionated digest of SP; (2) some patients with negative biopsy might have had a missed PCa. We also recognize that the only real ground truth for PCa prognosis is a 20-year survival, whereas other clinical parameters (Gleason score, localization, staging etc.) have limitations. It is known that Gleason score correlates with PCa progression, with the 20-year survival rate >70% for GS≤6 and <30% for GS≥8 (42). In our study, Gleason score served to facilitate the easier execution of this project because the 20-year survival data was not available. Our work was primarily indented to demonstrate that our biomarker development pipeline empowered by SRM assays was suitable to search for PCa biomarkers in SP, and to demonstrate that SP could have a value as a clinical sample for PCa diagnostics. We recognize that our set of 152 PCa samples is relatively small and may not represent the true clinical heterogeneity and all distinct genomic subtypes. The only way to validate the relevance of TGM4 would be its measurement in much larger sets of prospectively collected SP samples with known genomic subtypes.
Even though extensive genomic studies on PCa did not reveal substantial correlations between genomic alterations and PCa aggressiveness (72, 77), future proteomic studies on PCa should consider distinct genomic subtypes of PCa (72). This may facilitate identification of proteomic signatures which correlate with progression of PCa and provide true biomarkers of aggressiveness within each unique genomic subtype of PCa.
Data Availability
Raw mass spectrometry shotgun data and MaxQuant output files were deposited to the ProteomeXchange Consortium via PRIDE (www.ebi.ac.uk/pride/archive/login) with the dataset identifier PXD007657. Selected annotated spectra for the single-peptide identification candidate proteins DMBT1, PVRL2, TGFB1 and TFF1 are presented in supplemental Fig. S1. Annotated spectra for single-peptide identification proteins can be viewed with MS-Viewer at the following permanent link http://msviewer.ucsf.edu/prospector/cgi-bin/mssearch.cgi?report_title=MS-Viewer&search_key=enahbvtldi&search_name=msviewer. SRM raw data and processed Pinpoint files for qualification and verification phases were deposited to the Peptide Atlas (www.peptideatlas.org/PASS/PASS00989) with the data set identifier PASS00989. Alternative link is ftp://PASS00989:BU5655ot@ftp.peptideatlas.org.
Supplementary Material
Acknowledgments
We thank Antoninus Soosaipillai for suggestions on ELISA development, Susan Lau for coordinating collection and storage of clinical samples, and Ihor Batruch for assistance with mass spectrometry.
Footnotes
* This work was supported by grants from the Canadian Institute of Health Research (#285693) to E.P.D., K.J., and A.P.D, and Prostate Cancer Canada (RS2015-01) to A.P.D.
 This article contains supplemental material.
This article contains supplemental material.
1 The abbreviations used are:
- PCa
- prostate cancer
- BH-adjusted t-test
- Benjamini-Hochberg-adjusted t-test
- CV
- coefficient of variation
- FDR
- false discovery rate
- FWHM
- full width at half maximum
- GS
- Gleason score
- ELISA
- enzyme-linked immunosorbent assay
- IQR
- interquartile range
- LFQ
- label-free quantification
- MWU
- Mann Whitney Unpaired t-test
- PSA
- prostate-specific antigen
- ROC AUC
- receiver operating characteristic area under the curve
- S/N
- signal-to-noise
- SP
- seminal plasma
- SRM
- selected reaction monitoring
- TGM4
- protein-glutamine gamma-glutamyltransferase 4
- XGBoost
- eXtreme Gradient Boosting algorithm.
REFERENCES
- 1. Jemal A., Bray F., Center M. M., Ferlay J., Ward E., and Forman D. (2011) Global cancer statistics. CA Cancer J. Clin. 61, 69–90 [DOI] [PubMed] [Google Scholar]
- 2. Wilt T. J., Brawer M. K., Jones K. M., Barry M. J., Aronson W. J., Fox S, Gingrich J. R., Wei J. T., Gilhooly P., Grob B. M., Nsouli I., Iyer P., Cartagena R., Snider G., Roehrborn C., Sharifi R., Blank W., Pandya P., Andriole G. L., Culkin D., and Wheeler T. (2012) Radical prostatectomy versus observation for localized prostate cancer. N. Engl. J. Med. 367, 203–213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Schroder F. H., Hugosson J., Roobol M. J., Tammela T. L., Ciatto S., Nelen V., Kwiatkowski M., Lujan M., Lilja H., Zappa M., Denis L. J., Recker F., Berenguer A., Määttänen L., Bangma C. H., Aus G., Villers A., Rebillard X., van der Kwast T., Blijenberg B. G., Moss S. M., de Koning H. J., Auvinen A., and ERSPC Investigators (2009) Screening and prostate-cancer mortality in a randomized European study. N. Engl. J. Med. 360, 1320–1328 [DOI] [PubMed] [Google Scholar]
- 4. McNaughton-Collins M. F., and Barry M. J. (2011) One man at a time–resolving the PSA controversy. N. Engl. J. Med. 365, 1951–1953 [DOI] [PubMed] [Google Scholar]
- 5. Konety B. R., Bird V. Y., Deorah S., and Dahmoush L. (2005) Comparison of the incidence of latent prostate cancer detected at autopsy before and after the prostate specific antigen era. J. Urol. 174, 1785–1788; discussion 1788 [DOI] [PubMed] [Google Scholar]
- 6. Gutman A. B., and Gutman E. B. (1938) An “Acid ” phosphatase occurring in the serum of patients with metastasizing carcinoma of the prostate gland. J. Clin. Invest. 17, 473–478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Wang M. C., Valenzuela L. A., Murphy G. P., and Chu T. M. (1979) Purification of a human prostate specific antigen. Invest. Urol. 17, 159–163 [PubMed] [Google Scholar]
- 8. Hara M., Koyanagi Y., Inoue T., and Fukuyama T. (1971) [Some physico-chemical characteristics of “ -seminoprotein”, an antigenic component specific for human seminal plasma. Forensic immunological study of body fluids and secretion. VII]. Nihon Hoigaku Zasshi 25, 322–324 [PubMed] [Google Scholar]
- 9. Stamey T. A., Yang N., Hay A. R., McNeal J. E., Freiha F. S., and Redwine E. (1987) Prostate-specific antigen as a serum marker for adenocarcinoma of the prostate. N. Engl. J. Med. 317, 909–916 [DOI] [PubMed] [Google Scholar]
- 10. Tanase C. P., Codrici E., Popescu I. D., Mihai S., Enciu A. M., Necula L. G., Preda A., Ismail G., and Albulescu R. (2017) Prostate cancer proteomics: Current trends and future perspectives for biomarker discovery. Oncotarget 8, 18497–18512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Lughezzani G., Lazzeri M., Haese A., McNicholas T., de la Taille A., Buffi N. M., Fossati N., Lista G., Larcher A., Abrate A., Mistretta A., Bini V., Palou Redorta J., Graefen M., and Guazzoni G. (2014) Multicenter European external validation of a prostate health index-based nomogram for predicting prostate cancer at extended biopsy. Eur. Urol. 66, 906–912 [DOI] [PubMed] [Google Scholar]
- 12. Bussemakers M. J., van Bokhoven A., Verhaegh G. W., Smit F. P., Karthaus H. F., Schalken J. A., Debruyne F. M., Ru N., and Isaacs W. B. (1999) DD3: a new prostate-specific gene, highly overexpressed in prostate cancer. Cancer Res. 59, 5975–5979 [PubMed] [Google Scholar]
- 13. Goldkorn A., Ely B., Quinn D. I., Tangen C. M., Fink L. M., Xu T., Twardowski P., Van Veldhuizen P. J., Agarwal N., Carducci M. A., Monk J. P. 3rd, Datar R. H., Garzotto M., Mack P. C., Lara P. Jr, Higano C. S., Hussain M., Thompson I. M. Jr, Cote R. J., and Vogelzang N. J. (2014) Circulating tumor cell counts are prognostic of overall survival in SWOG S0421: a phase III trial of docetaxel with or without atrasentan for metastatic castration-resistant prostate cancer. J. Clin. Oncol. 32, 1136–1142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Nordstrom T., Vickers A., Assel M., Lilja H., Grönberg H., and Eklund M. (2015) Comparison between the four-kallikrein panel and prostate health index for predicting prostate cancer. Eur. Urol. 68, 139–146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Strom P., Nordstrom T., Aly M., Egevad L., Grönberg H., and Eklund M. (2018) The stockholm-3 model for prostate cancer detection: algorithm update, biomarker contribution, and reflex test potential. Eur. Urol. 74, 204–210 [DOI] [PubMed] [Google Scholar]
- 16. Van Neste L., Partin A. W., Stewart G. D., Epstein J. I., Harrison D. J., and Van Criekinge W. (2016) Risk score predicts high-grade prostate cancer in DNA-methylation positive, histopathologically negative biopsies. Prostate 76, 1078–1087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hessels D., Smit F. P., Verhaegh G. W., Witjes J. A., Cornel E. B., and Schalken J. A. (2007) Detection of TMPRSS2-ERG fusion transcripts and prostate cancer antigen 3 in urinary sediments may improve diagnosis of prostate cancer. Clin. Cancer Res. 13, 5103–5108 [DOI] [PubMed] [Google Scholar]
- 18. Tomlins S. A., Rhodes D. R., Yu J., Varambally S., Mehra R., Perner S., Demichelis F., Helgeson B. E., Laxman B., Morris D. S., Cao Q., Cao X., Andrén O., Fall K., Johnson L., Wei J. T., Shah R. B., Al-Ahmadie H., Eastham J. A., Eggener S. E., Fine S. W., Hotakainen K., Stenman U. H., Tsodikov A., Gerald W. L., Lilja H., Reuter V. E., Kantoff P. W., Scardino P. T., Rubin M. A., Bjartell A. S., and Chinnaiyan A. M. (2008) The role of SPINK1 in ETS rearrangement-negative prostate cancers. Cancer Cell 13, 519–528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Rao A. R., Motiwala H. G., and Karim O. M. (2008) The discovery of prostate-specific antigen. BJU Int. 101, 5–10 [DOI] [PubMed] [Google Scholar]
- 20. Karakosta T. D., Soosaipillai A., Diamandis E. P., Batruch I., and Drabovich A. P. (2016) Quantification of human kallikrein-related peptidases in biological fluids by multiplatform targeted mass spectrometry assays. Mol. Cell. Proteomics 15, 2863–2876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Neuhaus J., Schiffer E., von Wilcke P., Bauer H. W., Leung H., Siwy J., Ulrici W., Paasch U., Horn L. C., and Stolzenburg J. U. (2013) Seminal plasma as a source of prostate cancer peptide biomarker candidates for detection of indolent and advanced disease. PLoS ONE 8, e67514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Flores-Morales A., and Iglesias-Gato D. (2017) Quantitative mass spectrometry-based proteomic profiling for precision medicine in prostate cancer. Front. Oncol. 7, 267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kim Y., Ignatchenko V., Yao C. Q., Kalatskaya I., Nyalwidhe J. O., Lance R. S., Gramolini A. O., Troyer D. A., Stein L. D., Boutros P. C., Medin J. A., Semmes O. J., Drake R. R., and Kislinger T. (2012) Identification of differentially expressed proteins in direct expressed prostatic secretions of men with organ-confined versus extracapsular prostate cancer. Mol. Cell. Proteomics 11, 1870–1884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kim Y., Jeon J., Mejia S., Yao C. Q., Ignatchenko V., Nyalwidhe J. O., Gramolini A. O., Lance R. S., Troyer D. A., Drake R. R., Boutros P. C., Semmes O. J., and Kislinger T. (2016) Targeted proteomics identifies liquid-biopsy signatures for extracapsular prostate cancer. Nat. Commun. 7, 11906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Drabovich A. P., Saraon P., Jarvi K., and Diamandis E. P. (2014) Seminal plasma as a diagnostic fluid for male reproductive system disorders. Nat. Rev. Urol. 11, 278–288 [DOI] [PubMed] [Google Scholar]
- 26. Robert M., and Gagnon C. (1999) Semenogelin I: a coagulum forming, multifunctional seminal vesicle protein. Cell Mol. Life Sci. 55, 944–960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Mann T., and Lutwak-Mann C. (1981) Male reproductive function and semen., Springer-Verlag, Berlin [Google Scholar]
- 28. Batruch I., Lecker I., Kagedan D., Smith C. R., Mullen B. J., Grober E., Lo K. C., Diamandis E. P., and Jarvi K. A. (2011) Proteomic analysis of seminal plasma from normal volunteers and post-vasectomy patients identifies over 2000 proteins and candidate biomarkers of the urogenital system. J. Proteome Res. 10, 941–953 [DOI] [PubMed] [Google Scholar]
- 29. Batruch I., Smith C. R., Mullen B. J., Grober E., Lo K. C., Diamandis E. P., and Jarvi K. A. (2012) Analysis of seminal plasma from patients with non-obstructive azoospermia and identification of candidate biomarkers of male infertility. J. Proteome Res. 11, 1503–1511 [DOI] [PubMed] [Google Scholar]
- 30. Drabovich A. P., Dimitromanolakis A., Saraon P., Soosaipillai A., Batruch I., Mullen B., Jarvi K., and Diamandis E. P. (2013) Differential diagnosis of azoospermia with proteomic biomarkers ECM1 and TEX101 quantified in seminal plasma. Sci. Transl. Med. 5, 212ra160. [DOI] [PubMed] [Google Scholar]
- 31. Drabovich A. P., Jarvi K., and Diamandis E. P. (2011) Verification of male infertility biomarkers in seminal plasma by multiplex selected reaction monitoring assay. Mol. Cell. Proteomics 10, M110.004127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Korbakis D., Schiza C., Brinc D., Soosaipillai A., Karakosta T. D., Légaré C., Sullivan R., Mullen B., Jarvi K., Diamandis E. P., and Drabovich A. P. (2017) Preclinical evaluation of a TEX101 protein ELISA test for the differential diagnosis of male infertility. BMC Med 15, 60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Lee J. W., Devanarayan V., Barrett Y. C., Weiner R., Allinson J., Fountain S., Keller S., Weinryb I., Green M., Duan L., Rogers J. A., Millham R., O'Brien P. J., Sailstad J., Khan M., Ray C., and Wagner J. A. (2006) Fit-for-purpose method development and validation for successful biomarker measurement. Pharm. Res. 23, 312–328 [DOI] [PubMed] [Google Scholar]
- 34. Carr S. A., Abbatiello S. E., Ackermann B. L., Borchers C., Domon B., Deutsch E. W., Grant R. P., Hoofnagle A. N., Hüttenhain R., Koomen J. M., Liebler D. C., Liu T., MacLean B., Mani D. R., Mansfield E., Neubert H., Paulovich A. G., Reiter L., Vitek O., Aebersold R., Anderson L., Bethem R., Blonder J., Boja E., Botelho J., Boyne M., Bradshaw R. A., Burlingame A. L., Chan D., Keshishian H., Kuhn E., Kinsinger C., Lee J. S., Lee S. W., Moritz R., Oses-Prieto J., Rifai N., Ritchie J., Rodriguez H., Srinivas P. R., Townsend R. R., Van Eyk J., Whiteley G., Wiita A., and Weintraub S. (2014) Targeted peptide measurements in biology and medicine: best practices for mass spectrometry-based assay development using a fit-for-purpose approach. Mol. Cell. Proteomics 13, 907–917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Chen T., and Guestrin C. (2016) XGBoost: A Scalable Tree Boosting System. arXiv 1603.02754, https://arxiv.org/abs/1603.02754
- 36. Drabovich A. P., Pavlou M. P., Schiza C., and Diamandis E. P. (2016) Dynamics of protein expression reveals primary targets and secondary messengers of estrogen receptor alpha signaling in MCF-7 breast cancer cells. Mol. Cell. Proteomics 15, 2093–2107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Drabovich A. P., Pavlou M. P., Dimitromanolakis A., and Diamandis E. P. (2012) Quantitative analysis of energy metabolic pathways in MCF-7 breast cancer cells by selected reaction monitoring assay. Mol. Cell. Proteomics 11, 422–434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Cho C. K., Drabovich A. P., Karagiannis G. S., Martínez-Morillo E., Dason S., Dimitromanolakis A., and Diamandis E. P. (2013) Quantitative proteomic analysis of amniocytes reveals potentially dysregulated molecular networks in Down syndrome. Clin. Proteomics 10, 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Schiza C., Korbakis D., Jarvi K., Diamandis E. P., Drabovich A. P. (2019) Identification of TEX101-associated proteins through proteomic measurement of human spermatozoa homozygous for the missense variant rs35033974. Mol. Cell. Proteomics 18, 338–351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Martinez-Morillo E., Nielsen H. M., Batruch I., Drabovich A. P., Begcevic I., Lopez M. F., Minthon L., Bu G., Mattsson N., Portelius E., Hansson O., and Diamandis E. P. (2014) Assessment of peptide chemical modifications on the development of an accurate and precise multiplex selected reaction monitoring assay for apolipoprotein e isoforms. J. Proteome Res. 13, 1077–1087 [DOI] [PubMed] [Google Scholar]
- 41. Pedregosa F., Varoquaux G., Gramfort A., Michel V., Thirion B., Grisel O., Blondel M., Prettenhofer P., Weiss R., Dubourg V., Vanderplas J., Passos A. Cournapeau D., Brucher M., Perrot M., and Duchesnay E. (2011) Scikit-learn: Machine Learning in Python. J. Mach. Learn. Res. 12, 2825–2830 [Google Scholar]
- 42. Albertsen P. C., Hanley J. A., and Fine J. (2005) 20-year outcomes following conservative management of clinically localized prostate cancer. JAMA 293, 2095–2101 [DOI] [PubMed] [Google Scholar]
- 43. Schwanhausser B., Busse D., Li N., Dittmar G., Schuchhardt J., Wolf J., Chen W., and Selbach M. (2011) Global quantification of mammalian gene expression control. Nature 473, 337–342 [DOI] [PubMed] [Google Scholar]
- 44. Taylor B. S., Schultz N., Hieronymus H., Gopalan A., Xiao Y., Carver B. S., Arora V. K., Kaushik P., Cerami E., Reva B., Antipin Y., Mitsiades N., Landers T., Dolgalev I., Major J. E., Wilson M., Socci N. D., Lash A. E., Heguy A., Eastham J. A., Scher H. I., Reuter V. E., Scardino P. T., Sander C., Sawyers C. L., and Gerald W. L. (2010) Integrative genomic profiling of human prostate cancer. Cancer Cell 18, 11–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Uhlen M., Fagerberg L., Hallstrom B. M., Lindskog C., Oksvold P., Mardinoglu A., Sivertsson Å., Kampf C., Sjöstedt E., Asplund A., Olsson I., Edlund K., Lundberg E., Navani S., Szigyarto C. A., Odeberg J., Djureinovic D., Takanen J. O., Hober S., Alm T., Edqvist P. H., Berling H., Tegel H., Mulder J., Rockberg J., Nilsson P., Schwenk J. M., Hamsten M., von Feilitzen K., Forsberg M., Persson L., Johansson F., Zwahlen M., von Heijne G., Nielsen J., and Pontén F. (2015) Proteomics. Tissue-based map of the human proteome. Science 347, 1260419. [DOI] [PubMed] [Google Scholar]
- 46. Saraon P., Musrap N., Cretu D., Karagiannis G. S, Batruch I., Smith C., Drabovich A. P., Trudel D., van der Kwast T., Morrissey C., Jarvi K. A., and Diamandis E. P. (2012) Proteomic profiling of androgen-independent prostate cancer cell lines reveals a role for protein S during the development of high grade and castration-resistant prostate cancer. J. Biol. Chem. 287, 34019–34031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Saraon P., Drabovich A. P., Jarvi K. A., and Diamandis E. P. (2014) Mechanisms of androgen-independent prostate cancer. EJIFCC 25, 42–54 [PMC free article] [PubMed] [Google Scholar]
- 48. Heinlein C. A., and Chang C. (2004) Androgen receptor in prostate cancer. Endocr. Rev. 25, 276–308 [DOI] [PubMed] [Google Scholar]
- 49. Lange V., Picotti P., Domon B., and Aebersold R. (2008) Selected reaction monitoring for quantitative proteomics: a tutorial. Mol. Syst. Biol. 4, 222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Drabovich A. P., and Diamandis E. P. (2010) Combinatorial peptide libraries facilitate development of multiple reaction monitoring assays for low-abundance proteins. J. Proteome Res. 9, 1236–1245 [DOI] [PubMed] [Google Scholar]
- 51. Martinez-Morillo E., Cho C. K., Drabovich A. P., Shaw J. L., Soosaipillai A., and Diamandis E. P. (2012) Development of a multiplex selected reaction monitoring assay for quantification of biochemical markers of down syndrome in amniotic fluid samples. J Proteome Res 11, 3880–3887 [DOI] [PubMed] [Google Scholar]
- 52. Kusebauch U., Campbell D. S., Deutsch E. W., Chu C. S., Spicer D. A., Brusniak M. Y., Slagel J., Sun Z., Stevens J., Grimes B., Shteynberg D., Hoopmann M. R., Blattmann P., Ratushny A. V., Rinner O., Picotti P., Carapito C., Huang C. Y., Kapousouz M., Lam H., Tran T., Demir E., Aitchison J. D., Sander C., Hood L., Aebersold R., and Moritz R. L. (2016) Human SRMAtlas: a resource of targeted assays to quantify the complete human proteome. Cell 166, 766–778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Piludu M., Hand A. R., Cossu M., and Piras M. (2009) Immunocytochemical localization of MG1 mucin in human bulbourethral glands. J Anat 214, 179–182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Cho S. Y., Jeon J. H., Kim C. W., Shin D. M., Jang G. Y., Jeong E. M., Lee S. E., Song K. Y., and Kim I. G. (2010) Monoclonal antibodies to human transglutaminase 4. Hybridoma 29, 263–267 [DOI] [PubMed] [Google Scholar]
- 55. Gronberg H., Adolfsson J., Aly M., Nordström T., Wiklund P., Brandberg Y., Thompson J., Wiklund F., Lindberg J., Clements M., Egevad L., and Eklund M. (2015) Prostate cancer screening in men aged 50–69 years (STHLM3): a prospective population-based diagnostic study. Lancet Oncol. 16, 1667–1676 [DOI] [PubMed] [Google Scholar]
- 56. Filella X., Molina R., Alcover J., Carretero P., and Ballesta A. M. (1996) Detection of nonprostatic PSA in serum and nonserum samples from women. Int. J. Cancer 68, 424–427 [DOI] [PubMed] [Google Scholar]
- 57. Sequeiros T., Rigau M., Chiva C., Montes M., Garcia-Grau I., Garcia M., Diaz S., Celma A., Bijnsdorp I., Campos A., Di Mauro P., Borrós S., Reventós J., Doll A., Paciucci R., Pegtel M., de Torres I., Sabidó E., Morote J., Olivan M. (2017) Targeted proteomics in urinary extracellular vesicles identifies biomarkers for diagnosis and prognosis of prostate cancer. Oncotarget 8, 4960–4976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Cho S. Y., Choi K., Jeon J. H., Kim C. W., Shin D. M., Lee J. B., Lee S. E., Kim C. S., Park J. S., Jeong E. M., Jang G. Y., Song K. Y., and Kim I. G. (2010) Differential alternative splicing of human transglutaminase 4 in benign prostate hyperplasia and prostate cancer. Exp. Mol. Med. 42, 310–318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Cao Z., Wang Y., Liu Z. Y., Zhang Z. S., Ren S. C., Yu Y. W., Qiao M., Zhai B. B., and Sun Y. H. (2013) Overexpression of transglutaminase 4 and prostate cancer progression: a potential predictor of less favourable outcomes. Asian J. Androl. 15, 742–746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Ahn J., Berndt S. I., Wacholder S., Kraft P., Kibel A. S., Yeager M., Albanes D., Giovannucci E., Stampfer M. J., Virtamo J., Thun M. J., Feigelson H. S., Cancel-Tassin G., Cussenot O., Thomas G., Hunter D. J., Fraumeni J. F. Jr, Hoover R. N., Chanock S. J., and and Hayes R. B. (2008) Variation in KLK genes, prostate-specific antigen and risk of prostate cancer. Nat. Genet. 40, 1032–1034; author reply 1035–1036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Wu C. L., Jordan K. W., Ratai E. M., Sheng J., Adkins C. B., Defeo E. M., Jenkins B. G., Ying L., McDougal W. S., and Cheng L. L. (2010) Metabolomic imaging for human prostate cancer detection. Sci. Transl. Med. 2, 16ra18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Stephan C., Jung K., Semjonow A., Schulze-Forster K., Cammann H., Hu X., Meyer H. A., Bögemann M., Miller K., and Friedersdorff F. (2013) Comparative assessment of urinary prostate cancer antigen 3 and TMPRSS2:ERG gene fusion with the serum [-2]proprostate-specific antigen-based prostate health index for detection of prostate cancer. Clin. Chem. 59, 280–288 [DOI] [PubMed] [Google Scholar]
- 63. Bieniek J. M., Drabovich A. P., and Lo K. C. (2016) Seminal biomarkers for the evaluation of male infertility. Asian J. Androl. 18, 426–433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Korbakis D., Brinc D., Schiza C., Soosaipillai A., Jarvi K., Drabovich A. P., and Diamandis E. P. (2015) Immunocapture-selected reaction monitoring screening facilitates the development of ELISA for the measurement of native TEX101 in biological fluids. Mol. Cell. Proteomics 14, 1517–1526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Drabovich A. P., Martinez-Morillo E., and Diamandis E. P. (2015) Toward an integrated pipeline for protein biomarker development. Biochim Biophys Acta 1854, 677–686 [DOI] [PubMed] [Google Scholar]
- 66. Davies G., Ablin R. J., Mason M. D., and Jiang W. G. (2007) Expression of the prostate transglutaminase (TGase-4) in prostate cancer cells and its impact on the invasiveness of prostate cancer. J. Exp. Ther. Oncol. 6, 257–264 [PubMed] [Google Scholar]
- 67. Jiang W. G., Ye L., Sanders A. J., Ruge F., Kynaston H. G., Ablin R. J., Mason M. D. (2013) Prostate transglutaminase (TGase-4, TGaseP) enhances the adhesion of prostate cancer cells to extracellular matrix, the potential role of TGase-core domain. J. Transl. Med. 11, 269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Jiang W. G., Ablin R. J., Kynaston H. G., Mason M. D. (2009) The prostate transglutaminase (TGase-4, TGaseP) regulates the interaction of prostate cancer and vascular endothelial cells, a potential role for the ROCK pathway. Microvasc. Res. 77, 150–157 [DOI] [PubMed] [Google Scholar]
- 69. Beltran H., Prandi D., Mosquera J. M., Benelli M., Puca L., Cyrta J., Marotz C., Giannopoulou E., Chakravarthi B. V., Varambally S., Tomlins S. A., Nanus D. M., Tagawa S. T., Van Allen E. M., Elemento O., Sboner A., Garraway L. A., Rubin M. A., and Demichelis F. (2016) Divergent clonal evolution of castration-resistant neuroendocrine prostate cancer. Nat. Med. 22, 298–305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Dunning M. J., Vowler S. L., Lalonde E., Ross-Adams H., Boutros P., Mills I. G., Lynch A. G., and Lamb A. D. (2017) Mining human prostate cancer datasets: The “camcAPP” Shiny App. EBioMedicine 17, 5–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Bhowal A., Majumder S., Ghosh S., Basu S., Sen D., Roychowdhury S., Sengupta S., and Chatterji U. (2017) Pathway-based expression profiling of benign prostatic hyperplasia and prostate cancer delineates an immunophilin molecule associated with cancer progression. Sci Rep 7, 9763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Cancer Genome Atlas Research, N. (2015) The molecular taxonomy of primary prostate cancer. Cell 163, 1011–1025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Saner F. A. M., Herschtal A., Nelson B. H., deFazio A., Goode E. L., Ramus S. J., Pandey A., Beach J. A., Fereday S., Berchuck A., Lheureux S., Pearce C. L., Pharoah P. D., Pike M. C., Garsed D. W., and Bowtell D. D. L. (2019) Going to extremes: determinants of extraordinary response and survival in patients with cancer. Nat. Rev. Cancer 19, 339–348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Gan W., Dai X., Lunardi A., Li Z., Inuzuka H., Liu P., Varmeh S., Zhang J., Cheng L, Sun Y., Asara J. M., Beck A. H., Huang J., Pandolfi P. P., and Wei W. (2015) SPOP promotes ubiquitination and degradation of the ERG oncoprotein to suppress prostate cancer progression. Mol. Cell 59, 917–930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Armenia J., Wankowicz S. A. M., Liu D., Gao J., Kundra R., Reznik E., Chatila W. K., Chakravarty D., Han G.C., Coleman I., Montgomery B., Pritchard C., Morrissey C., Barbieri C. E., Beltran H., Sboner A., Zafeiriou Z., Miranda S., Bielski C. M., Penson A. V., Tolonen C., Huang F. W., Robinson D., Wu Y. M., Lonigro R., Garraway L. A., Demichelis F., Kantoff P. W., Taplin M. E., Abida W., Taylor B. S., Scher H. I., Nelson P. S., de Bono J. S., Rubin M. A., Sawyers C. L., Chinnaiyan A.M., PCF/SU2C International Prostate Cancer Dream Team, Schultz N., and Van Allen E. M. (2018) The long tail of oncogenic drivers in prostate cancer. Nat. Genet. 50, 645–651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Robinson J. L., Holmes K. A., and Carroll J. S. (2013) FOXA1 mutations in hormone-dependent cancers. Front Oncol 3, 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Fraser M., Sabelnykova V. Y., Yamaguchi T. N., Heisler L. E., Livingstone J., Huang V., Shiah Y. J., Yousif F., Lin X., Masella A. P., Fox N. S., Xie M., Prokopec S. D., Berlin A., Lalonde E., Ahmed M., Trudel D., Luo X., Beck T. A., Meng A., Zhang J., D'Costa A., Denroche R. E., Kong H., Espiritu S. M., Chua M. L., Wong A., Chong T., Sam M., Johns J., Timms L., Buchner N. B., Orain M., Picard V., Hovington H., Murison A., Kron K., Harding N. J., P'ng C., Houlahan K. E., Chu K. C., Lo B., Nguyen F., Li C. H., Sun R. X., de Borja R., Cooper C. I., Hopkins J. F., Govind S. K., Fung C., Waggott D., Green J., Haider S., Chan-Seng-Yue M. A., Jung E., Wang Z., Bergeron A., Dal Pra A., Lacombe L., Collins C. C., Sahinalp C., Lupien M., Fleshner N. E., He H. H., Fradet Y., Tetu B., van der Kwast T., McPherson J. D., Bristow R. G., and Boutros P. C. (2017) Genomic hallmarks of localized, non-indolent prostate cancer. Nature 541, 359–364 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Raw mass spectrometry shotgun data and MaxQuant output files were deposited to the ProteomeXchange Consortium via PRIDE (www.ebi.ac.uk/pride/archive/login) with the dataset identifier PXD007657. Selected annotated spectra for the single-peptide identification candidate proteins DMBT1, PVRL2, TGFB1 and TFF1 are presented in supplemental Fig. S1. Annotated spectra for single-peptide identification proteins can be viewed with MS-Viewer at the following permanent link http://msviewer.ucsf.edu/prospector/cgi-bin/mssearch.cgi?report_title=MS-Viewer&search_key=enahbvtldi&search_name=msviewer. SRM raw data and processed Pinpoint files for qualification and verification phases were deposited to the Peptide Atlas (www.peptideatlas.org/PASS/PASS00989) with the data set identifier PASS00989. Alternative link is ftp://PASS00989:BU5655ot@ftp.peptideatlas.org.