By applying dimethyl labeling-based quantitative proteomic analysis, global alterations both at the proteome and phosphoproteome levels were determined in the Twist-induced EMT model system of HMLE cells. The epithelial and mesenchymal state specific signaling networks and the kinase-substrate interaction network of EMT were reconstructed using advanced modeling techniques. Many novel mRNA and/or protein level modulators of EMT were identified including DNAJB4 and CD81, which were validated as novel metastasis inducers in breast cancer.
Keywords: Phosphoproteome, Metastasis, Breast cancer, Kinases, Quantification, Mouse models, Networks, Cancer biomarker(s), 'omic' data integration, CD81, DNAJB4, EMT, Twist
Graphical Abstract
Highlights
314 proteins and 871 phosphopeptides were differentially regulated during EMT.
∼65% of regulated proteins do not change at the transcriptome level during EMT.
Epithelial and mesenchymal specific kinases and their substrates were identified.
DNAJB4 and CD81 were identified as novel metastasis inducers in breast cancer.
Abstract
Epithelial-mesenchymal transition (EMT) is driven by complex signaling events that induce dramatic biochemical and morphological changes whereby epithelial cells are converted into cancer cells. However, the underlying molecular mechanisms remain elusive. Here, we used mass spectrometry based quantitative proteomics approach to systematically analyze the post-translational biochemical changes that drive differentiation of human mammary epithelial (HMLE) cells into mesenchymal. We identified 314 proteins out of more than 6,000 unique proteins and 871 phosphopeptides out of more than 7,000 unique phosphopeptides as differentially regulated. We found that phosphoproteome is more unstable and prone to changes during EMT compared with the proteome and multiple alterations at proteome level are not thoroughly represented by transcriptional data highlighting the necessity of proteome level analysis. We discovered cell state specific signaling pathways, such as Hippo, sphingolipid signaling, and unfolded protein response (UPR) by modeling the networks of regulated proteins and potential kinase-substrate groups. We identified two novel factors for EMT whose expression increased on EMT induction: DnaJ heat shock protein family (Hsp40) member B4 (DNAJB4) and cluster of differentiation 81 (CD81). Suppression of DNAJB4 or CD81 in mesenchymal breast cancer cells resulted in decreased cell migration in vitro and led to reduced primary tumor growth, extravasation, and lung metastasis in vivo. Overall, we performed the global proteomic and phosphoproteomic analyses of EMT, identified and validated new mRNA and/or protein level modulators of EMT. This work also provides a unique platform and resource for future studies focusing on metastasis and drug resistance.
Carcinoma cells use epithelial-mesenchymal transition (EMT)1, a normally latent embryonic program, to facilitate the initiation and progression of metastasis. EMT can be induced via cell intrinsic and extrinsic factors based on the communication between epithelial cells and the nearby stromal cells and plays a critical role in metastasis of cancer cells (1–4). In 1982, Greenburg and Hay first reported that epithelial cells can be converted into mesenchymal cells (5). Subsequently, numerous studies described the molecular processes during EMT and suggested alternative mechanistic models that lead to EMT (6, 7). EMT does not only trigger metastatic and invasive capabilities but can also trigger cancer stem cell properties (5, 6, 8) and drug resistance (9–12).
Throughout EMT, epithelial cells lose cell-cell contacts and cell polarity (13). Several signaling pathways that facilitate the activation of EMT have been discovered (6, 14, 15) and includes TGF-β, Wnt proteins, platelet-derived growth factors, Interleukin-6 and various receptor tyrosine kinases. All of these are involved in the activation of EMT inducing transcription factors (EMT-TF). These include Snail, Slug, Zeb1, Zeb2, and Twist (13). Ectopic expression of Twist in epithelial cells leads to the loss of adherens junction protein E-cadherin and expression of mesenchymal markers such as N-cadherin and Vimentin, suggesting the induction of EMT (3, 4, 16). Gene expression profiling of immortalized human mammary epithelial (HMLE) cells that underwent EMT by expressing Goosecoid, Snail, Twist, or TGF-β, or by knocking down the expression of E-cadherin identified a “EMT core signature” that consists of 159 downregulated genes and 87 upregulated genes (4). Although transcriptomic data represents gene expression during EMT process, it may not thoroughly predict the regulation at the protein level; i.e. the mRNA levels may not be perfectly correlated with the translated proteins because of the post-transcriptional modifications and miRNA-mediated regulation (17–19).
As a comprehensive understanding of EMT requires combination of both transcriptomic and proteomic analyses, we systematically compared biochemical changes during EMT of human mammary epithelial cells that was induced by Twist and we performed detailed comparison of previous transcriptome profile of the same EMT model system with our proteome data. Our analysis revealed 314 differentially regulated proteins during EMT which was followed by the “omic” data integration to reconstruct EMT specific networks. In parallel, to profile the changes in signaling pathways during EMT, we systematically monitored phosphoproteome changes and reconstructed the network of the interactions between kinases and phosphoproteins. Finally, we validated several novel biochemical changes identified in this study by in vitro and in vivo experiments. Our data provide a framework for understanding the global regulation of EMT at phospho-/protein level and reveal a comprehensive data set to explore main molecular pathways that drive EMT.
EXPERIMENTAL PROCEDURES
Cell Culture
Human mammary epithelial (HMLE) cell line (20) was kindly provided by Dr. Robert A. Weinberg (Whitehead Institute for Biomedical Research, Boston, MA) and cultured in MEGM Bullet kit (CC-3151 & CC-4136, Lonza) in 5% CO2 at 37 °C. 4-Hydroxytamoxifen (4-OHT) treatment at a final concentration of 20 nm was performed for 16 days to induce EMT in HMLE-Twist-ER cells. MCF7 and MDA-MB-231 cells were cultured in Dulbecco's Modified Eagle's Medium (DMEM, Lonza) supplemented with 10% fetal bovine serum (Sigma), 1% penicillin/streptomycin (Sigma) and 1% MEM nonessential amino acids (Gibco) in 5% CO2 at 37 °C.
Western Blotting
Approximately 3.2 × 106 cells were lysed in 1× PBS buffer containing 0.1% TritonX-100, cOmplete EDTA-free protease inhibitor mixture (Roche) and PhosSTOP phosphatase inhibitor mixture (Roche). Inhibitor mixture tablets were used as 1 tablet for 10 ml of lysis buffer. Cells were disrupted by pulling the cell suspension through a thin needle. Cell debris was removed by centrifugation at 14,000 rpm for 5 min at 4 °C. Protein concentration was measured using BCA protein assay kit (23227, Pierce). Protein samples were prepared in 2X Laemmli sample buffer containing 4% (w/v) SDS, 20% glycerol, 120 mm Tris-Cl (pH 6.8), 0.02% (w/v) bromphenol blue and 100 mm dithiothreitol (DTT) as protein sample to 2× Laemmli sample buffer ratio of 1:1 (v/v). Samples were heated at 95 °C for 5 min, cooled before loading onto SDS-PAGE gel. 10–12% SDS-PAGE gels were used for the separation of proteins. After electrophoresis of samples at 120 V for 100 min, proteins were transferred to nitrocellulose membranes (BA85, Whatman Protran) by the Trans-Blot Turbo transfer system (Bio-Rad). After 45 min blocking with 4% milk in 0.1% Tween20 containing 1× TBS, the membrane was incubated with primary antibodies: 1:1000 E-cadherin (24E10) (3195, Cell Signaling), 1:1000 Vimentin (D21H3) (5741, Cell Signaling), 1:1000 Snail (C15D3) (3879, Cell Signaling), 1:1000 β-actin (AC15) (ab6276, Abcam), 1:100 DNAJB4 (R-07) (sc-100711, Santa Cruz) and 1:100 CD81 (B-11) (sc166029, Santa Cruz) overnight at 4 °C or 3 h at room temperature. The membrane was rinsed with 0.1% Tween20 containing 1× TBS Buffer for 10 min three times and incubated with secondary antibodies: 1:2000 anti-mouse IgG, HRP-linked antibody (7076S, Cell Signaling) or 1:2000 anti-rabbit IgG, HRP-linked antibody (7074S, Cell Signaling) at room temperature for 1.5 h. Finally, membranes were rinsed with 0.1% Tween20 containing 1× TBS Buffer for 10 min three times and proteins were visualized with the ECL Western blotting Substrate system (32106, Pierce).
Experimental Design and Statistical Rationale
Trypsin digested lysates of HMLE and HMLE-Twist cells were dimethyl labeled as light and heavy, respectively and mixed in equal amounts. Mixed lysates were separated into 40 fractions via SCX. Biological replicate 2 (BR2) was prepared by label swapping to remove labeling bias and separated into 40 fractions via SCX as in BR1. From each biological replicate, 2 technical replicates were prepared and analyzed in MS. Phosphoproteome quantification of the samples was designed similarly, 2 biological replicates with 2 technical replicates each, except the fractionation step. To enrich phosphopeptides, each SCX fraction was enriched using either in-house packed TiO2 (5 μm, Sachtleben, CAS no. 13463–67-7) or Ti4+-IMAC micro columns in which the beads were loaded onto GELoader tips using a C8 plug. For Ti4+-IMAC columns, IMAC material was prepared and used as previously described (21). The enriched samples were combined into 10–12 fractions in both biological replicates with two technical replicates for each. Ratios of proteins and peptides quantified in the replicates were combined by taking the median.
To determine the regulated proteins between epithelial and mesenchymal states, we defined epithelial, mesenchymal specific and unchanged protein ratio distributions using known epithelial and mesenchymal markers. Epithelial and mesenchymal specific protein marker groups (supplemental Table S1) were used to determine which proteins fall into the corresponding groups. By fitting normal distributions to E/M ratios of these three groups, epithelial, mesenchymal and unchanged, the likelihood of each protein belonging to the respective groups was calculated. Using the likelihoods, the relative likelihood of each protein was calculated, Prelative = Punchanged/(Pchanged-epithelial or Pchanged-mesenchymal), for epithelial or mesenchymal group memberships. Proteins with Prelative <0.05 were chosen as significantly changed proteins. Unmodified peptides of the unchanged proteins were chosen as unchanged peptides. Using the median normalized empirical distribution of the unchanged peptide ratio, we identified the respective lower 0.025 and 0.975 quantiles. These correspond to a lower and upper cutoff for the peptide ratios of −1.33404 and 1.555532, respectively. Aiming to identify those peptide ratios differing from the ones observed in the unchanged peptide ratios, phosphopeptide ratios below the lower cutoff value of the unchanged peptide distribution were identified as mesenchymal specific phosphorylations and phosphopeptide ratios above the upper cutoff of the unchanged distribution were identified as epithelial specific phosphorylations.
In-solution Digestion and Stable Isotope Dimethyl Labeling
In-solution digestion was performed as described previously (22). Briefly, cell lysate was incubated for 4 h using Lys-C (enzyme:protein ratio of 1:100) in 4 m urea, followed by overnight trypsin (enzyme:protein ratio of 1:100) incubation in 1 m urea at 37 °C. The digestion was quenched by acidification and desalted using Sep-Pak C18 cartridges. On-column dimethyl labeling was performed as described in Boersema et al. (23). Labeling reagents consisted of 4% (v/v) of formaldehyde (CH2O) (37% (v/v), Sigma, cat. no. 252549) and 0.6 m of sodium cyanoborohydride (NaBH3CN) (Fluka, cat. no. 71435) for light and 4% (v/v) of formaldehyde (13CD2O) (20%, 99% 13C, 98% D, Isotec, cat. no. 596388) and 0.6 m of sodium cyanoborodeuteride (NaBD3CN) (96% D, Isotec, cat. no. 190020) for heavy in 50 mm NaH2PO4 (Fisher) and 50 mm Na2HPO4 (Fisher). The labeled peptides were combined in equal ratios based on their average peptide intensities. The epithelial sample was labeled with the light dimethyl label and the mesenchymal sample with the heavy label. We swapped the labels for the second biological replicate.
Strong Cation Exchange Chromatography and Phosphopeptide Enrichment
Labeled peptide mixes were fractionated using a PolySULFOETHYL A 200 × 2.1 mm (PolyLC Inc.) column using an Agilent 1100 HPLC system (Agilent Technologies, Germany) into 40 fractions. Then, pooled fractions were phospho-enriched using either Ti4+-IMAC or Titanium dioxide (TiO2) (Sachtleben, Germany) beads packaged into micro-columns as described previously (24, 25). Briefly, the samples resuspended in loading buffer (80% ACN, 6% TFA) were loaded onto micro-columns pre-equilibrated with loading buffer. Ti4+-IMAC columns were sequentially washed with washing buffer 1 (50% ACN, 0.5% TFA containing 200 mm NaCl) and washing buffer 2 (50% ACN/0.1% TFA) whereas TiO2 columns were sequentially washed with loading buffer and washing buffer 2 (50% ACN/0.1% TFA). The bound peptides were eluted into a new tube (containing 35 μl of 10% formic acid) with 20 μl of 10% ammonia by centrifugation at 100 × g for 20 min. The collected eluate was further acidified by the addition of 3 μl of 100% formic acid before nano-LC-MS/MS analysis. The peptides were at very low pH (<2) and ready for LC-MS/MS analysis. 20–30% of samples were loaded into system.
Mass Spectrometric Analysis
The peptides were subjected to a reversed phase nanoLC-MS/MS (EASY-nLC, Thermo) connected to a Q Exactive quadrupole Orbitrap mass spectrometer (Thermo Fisher Scientific, Bremen) for a 120 min analysis run. The peptides were directly loaded onto an in-house packed 100 μm i.d. × 17 cm C18 column (Reprosil-Gold C18, 5 μm, 200Å, Dr. Maisch) and separated at 300 nL/min with 120 min linear gradients, starting from 5 to 30% acetonitrile in 0.1% formic acid. Mass spectrometric analysis and spectra acquisition were performed with the following settings: Orbitrap analysis: Polarity at positive mode; resolution: 35,000; mass range: 350–1500 m/z; automatic gain control (AGC) target: 3e6; maximum injection time: 250 ms. Up to 10 of the most intense ions per cycle were fragmented and analyzed in the Orbitrap. MS2 analysis consisted of collision-induced dissociation (Higher-energy collisional dissociation (HCD)) (Resolution: 17,500; AGC: 5e5; maximum injection time: 120 ms; normalized collision energy (NCE): 25; charge exclusion: unassigned, 1, 7, 8, >8; dynamic exclusion: 20 s). The isolation window for MS/MS was 1.5 m/z.
Data Processing and Analysis
All raw data files were processed and quantified with Proteome Discoverer (version 1.4, Thermo Scientific). Peak lists containing HCD fragmentation were generated with Proteome Discoverer (PD) with a signal-to-noise threshold of 1.0. Produced data was searched against a Swissprot database version 2014_08 with taxonomy Homo sapiens (20,193 sequences) using the Mascot software (version 2.5.1, Matrix Science, London, UK). Settings for PD search were as following: The enzyme was specified as Trypsin and the fragment ion type was specified as electrospray ionization quadrupole time-of-flight. A mass tolerance of ±20 ppm for precursor masses and ±0.05 Da for fragment ions was used; two missed cleavages were allowed; and cysteine carbamidomethylation as a fixed modification. Light and heavy dimethylation of peptide N termini and lysine residues; methionine oxidation; phosphorylation on serine, threonine and tyrosine were set as variable modifications. Each peptide spectral match (PSM) (Mascot peptide score >25) of a phosphorylated peptide was isolated from the data in PD. The identification of the phosphopeptide site localization was performed by phosphoRS algorithm 3.0 (26), implemented in PD. A threshold of at least 0.70 probability was used for the phospho-residue localization. The 2plex dimethyl-based quantitation method was chosen in PD, with a mass precision requirement of 2 ppm for consecutive precursor mass measurements. The filter in PD was set to select only the peptides with “high confidence.” We applied 0.2 min of retention time tolerance for isotope pattern multiplets and allowed spectra with a maximum of 1 missing channel to be quantified with the integrated Percolator-based filter using a false discovery rate of 1%.
Motif Analysis of Phosphopeptides
Phosphorylation sites with phosphorylation probabilities higher than 0.75 were chosen. ±7 amino acid sequence windows of the phosphorylation sites were used to extract motifs for epithelial and mesenchymal groups. Motif-x (27) implemented in rmotifx package (28) was used to identify enriched motifs in epithelial and mesenchymal specific phosphorylations. The significance threshold was set to 0.00018 (search length is 15 amino acids) in motif-x to make p value of each motif match to be at least 0.05 by the Bonferroni method. The motif occurrence limit is set to 5 for epithelial S phosphorylations (n = 292) and mesenchymal S phosphorylations (n = 415). The motif occurrence score is set to 2 for epithelial T phosphorylations (n = 27) and mesenchymal T phosphorylations (n = 50). The background was set to Homo sapiens proteins from Swissprot database version 2014_08 and the enriched motifs were converted to sequence logos via Weblogo (29). NetworKIN 3.0 (30) was used to generate kinase-substrate networks. We only used 4,060 phosphosites identified with more than 50% phosphorylation probabilities from epithelial and mesenchymal specific phosphosites and predictions scoring ≥5. Predicted kinase-substrate interaction networks were visualized using Cytoscape 3.4.0 (31). Cellular component annotations for each protein were fetched via the BioServices Python package. Kinase prediction of epithelial and mesenchymal specific phosphosites were performed via NetworKIN 3.0 (30). Predictions with scores 5 and above were selected and visualized in the network.
Network/Pathway Analysis
The forest module of the Omics Integrator (32) was used to construct epithelial and mesenchymal specific protein-protein interaction networks as explained in Karayel et al. (33). The iRefWeb database (34) protein-protein interaction network was used as the global proteome interactome. Significantly regulated proteins in epithelial and mesenchymal lists were used as the experimental hits (terminal proteins). Terminals are connected to each other via intermediate hidden proteins (Steiner proteins) (32) that are not identified in the experiments.
Statistical Analysis of Cell Line and Patient Data Sets
For mRNA expression analysis of genes in different breast cell lines, the GSE40059 data set (35) from NCBI GEO database was used. Epithelial and mesenchymal cell lines were assigned based on the Blick et al. study (36). SK-BR-3 and MDA-MB-468 cell lines were assigned as neither epithelial nor mesenchymal because of low expression of E-cadherin and the absence of mesenchymal markers, fibronectin and vimentin (36–38). Mean mRNA expression values of two groups were statistically analyzed by two-tailed Student's t test. TCGA mRNA data and EMT score values were downloaded from the cBioPortal for Cancer Genomics (39, 40) and TCPA database (41), respectively. Correlation analysis of mRNA z-scores with EMT scores was performed and r values were obtained for each gene. For metastasis-free survival analysis of patients, GSE2603 microarray data (42) for primary breast cancer patients (n = 82) was used. Patients were separated into two groups at the median expression. Kaplan-Meier analyses were used to determine metastasis-free survival and statistical differences were determined by Log-rank (Mantel-Cox) test.
Knockdown Experiments
shRNA sequences against targets were designed using RNAi Codex database (43). pSMP-Luc plasmid (Addgene plasmid # 36394) was used for the expression of shRNAs as described previously (44). Each shRNA sequence was cloned into pSMP-Luc plasmid after the removal of Firefly luciferase targeting shRNA sequence. DNAJB4 and CD81 targeting sequences are CTGCAACTACTTGTTATGCATA and CACACGTCGCCTTCAACTGTAA, respectively. All cloning and packaging steps of viral plasmids were carried out as described previously (44). MCF-7 and MDA-MB-231 cells were used for the virus-mediated shRNA knockdown of epithelial and mesenchymal targets, respectively.
Cell Viability and In Vitro Migration Assays
To determine cell viability, MDA-MB-231 cells were incubated with Cell Titer Glo reagent and analyzed with a luminometer according to the manufacturer's protocol (Cell Titer Glo Luminescent Cell Viability Assay kit, G7571, Promega). Real-time cell migration assays were performed on xCELLigence RTCA-DP (ACEA Biosciences Inc., CA) according to manufacturer's protocol. RTCA (Real Time Cell Analyzer) is a system that quantifies the number of cells that are migrated toward a stimulus (in this case, serum). In this assay, cells were seeded in low serum (1%) media in small inserts coated with gold particles at their bottom. The inserts were placed in chambers containing high serum media (10%), and cells were expected to migrate toward the lower chamber with a pace that positively correlates with their invasiveness. As the cells move toward the high serum media, the movement causes a change in the current passing through the gold particles which is then quantified and represented as the migration cell index. For wound healing scratch assay, 0.5 × 106 cells per well were seeded in 6-well plates and grown to subconfluence. Proliferation of cells were blocked using serum-free medium and the wound area was measured at the beginning and after 24 h following scratch formation using ImageJ software (Wayne Rasband, NIH). Three independent experiments were performed for each condition. Images were taken with 4× magnification using Nikon Eclipse inverted microscope (Nikon, Japan). The statistical analysis of results was performed using two-tailed Student's t test.
Flow Cytometry Analysis
Cells were stained with PE-conjugated anti-CD81 antibody from BD Biosciences (JS-81) (gift of Dr. Batu Erman) as described previously (45). Samples were analyzed using SONY LE-SH800 flow cytometer. All flow data were analyzed using FlowJo_V10 software.
Animal Experiments
All animal experiments were approved by the animal ethics committee of Bilkent University. Six- to eight-week-old female athymic nu/nu mice were subcutaneously injected without incision, with 1 × 106 cells into mammary fat pad (MFP). Primary tumor growth was monitored by measuring the tumor volume twice a week with a caliper after tumors became palpable. Tumor volumes were calculated as length × width2/2. Spontaneous metastasis was evaluated with a luciferase assay. Because of heterogeneous distribution of metastasis in organs, three different parts were randomly collected from each organ to make a tissue pool and weighed for normalization. Both lung and liver tissues were ground in cold PBS by tissue homogenizer and treated with cell culture lysis 5× reagent (E153A, Promega). 200 μl lysis buffer was added to 50 mg weight. After 15 min incubation, 100 μl of luciferase substrate (Promega) was added to 50 μl of the lysed sample, and luminescence was measured with a luminometer. For tail vein metastasis assay, 1.5 × 106 cells were injected into tail-vein of each 6–8-week-old female athymic nu/nu mice, with 3–5 mice per group. Lung metastasis was monitored by bioluminescence imaging (BLI). Anesthetized mice were intraperitoneally injected with 150 mg/kg d-luciferin (Perkin Elmer). Bioluminescence images were acquired with Lumina III In Vivo Imaging System (Perkin Elmer). Analysis was performed with live imaging software by measuring total photon flux. For Bouin's fixation, harvested lungs were cleaned with PBS and placed into Bouin's solution on a shaker for overnight. After fixation, they were kept in 70% alcohol.
RESULTS
Proteomic Profiling Identifies Differentially Regulated Proteins During EMT
To identify differentially expressed or phosphorylated proteins during EMT in epithelial HMLE and its mesenchymal derivative HMLE-Twist cells, we took the advantage of quantitative proteomics and phosphoproteomics approaches. Overall study design and experimental workflow is presented in supplemental Fig. S1. HMLE cells exhibiting characteristics of epithelial cells were systematically compared with HMLE cells that stably express Twist (HMLE-Twist) using dimethyl labeling (Fig. 1A). Twist ectopic expression has been known to induce epithelial cells to undergo EMT, thereby promoting mesenchymal cell characteristics (16). We validated these results by showing the protein level downregulation of epithelial markers, such as E-cadherin, and protein level upregulation of mesenchymal markers, such as N-cadherin, Vimentin, and Snail on Twist overexpression (supplemental Fig. S2).
Fig. 1.
Identification of epithelial and mesenchymal specific proteins. A, Experimental workflow as explained in the text. B, Box plots show the distribution of raw Epithelial/Mesenchymal protein ratios in the logarithmic (ln) scale for biological replicates 1 and 2 (BR1 and BR2) and corresponding technical replicates 1 and 2 (TR1 and TR2). C, Box plots show the normalized distribution of Epithelial/Mesenchymal protein ratios for BR1 and BR2 and corresponding TR1 and TR2 in the logarithmic (ln) scale. D, The correlation of the proteins between two biological replicates (Pearson correlation, r = 0.64). E, Distribution of all Epithelial/Mesenchymal protein ratios. Epithelial and mesenchymal specific proteins are labeled in green and yellow, respectively. The markers used to determine epithelial and mesenchymal groups are labeled in the histogram. F, Comparison of transcriptome versus proteome data. Epithelial/Mesenchymal ratios are compared at the mRNA (4) and protein levels (gray: no change, green: only mRNA level change, blue: only protein level change, red: both mRNA and protein levels change).
To perform a systematic quantitative profiling of EMT, the equal amount of cell lysates from HMLE and HMLE-Twist cells were digested using Trypsin. The resulting peptides were differentially labeled with light and heavy isotopes using stable isotope dimethyl labeling (23) with the labeling efficiency of ∼99%. The labels were swapped to eliminate any labeling bias in biological repeats. Two technical replicates were performed for each biological replicate. After combining differentially labeled peptides in equal amounts, the pooled peptides were separated by column-based SCX chromatography into 40 fractions, and each fraction was analyzed by LC-MS/MS in two replicates. All raw MS data were processed using the Proteome Discoverer software to identify and quantify peptides in two conditions (supplemental Fig. S1). As a result, 4,829 and 5,505 unique proteins (supplemental Table S1), as well as 29,986 and 35,591 unique peptides (supplemental Table S2A) were identified in biological replicate 1 (BR1) and biological replicate 2 (BR2), respectively with <1% FDR (supplemental Table S3). The logarithmic (ln) distributions of the raw and normalized Epithelial/Mesenchymal (E/M) ratios of the peptides identified in the technical replicates 1 (TR1) and 2 (TR2) for both BR1 and BR2 were plotted in Fig. 1B, 1C, respectively. The correlation of protein ratios identified in both biological replicates with an r = 0.64 (p value <0.005, n = 4,253) indicates the reproducibility of our workflow (Fig. 1D).
Based on our statistical analysis (see Experimental Procedures section), 168 proteins are significantly upregulated in epithelial cells whereas 146 proteins are significantly upregulated in mesenchymal ones (Fig. 1E and supplemental Table S1). As expected, many known epithelial markers, such as Desmoplakin, Occludin, and E-cadherin were significantly upregulated in epithelial cells (Fig. 1E, green) and many known mesenchymal markers, such as Twist1–2, Vimentin, and N-cadherin were significantly upregulated in mesenchymal cells (Fig. 1E, yellow). We compared our proteomics data to previously published microarray data obtained from immortalized HMLE cells underwent EMT by expressing Twist (4). Although proteomic data is significantly correlated with the transcriptomic data (r = 0.53, p value <0.005), alterations at proteome level are not thoroughly represented with transcriptional data in EMT (supplemental Table S1). Proteins only significantly regulated in proteome data (Fig. 1F, blue) comprise 67% of all regulated proteins (Fig. 1F, blue, green and red).
Phosphoproteome Is Unstable and Prone to Changes During EMT
To identify protein phosphorylation events of EMT, phosphopeptide enrichment was performed in all SCX fractions using TiO2 or Ti4+-IMAC columns (24, 25) (Fig. 1A). 7,735 and 5,685 phosphopeptides were identified with 1% FDR in BR1 and BR2, respectively (supplemental Tables S2C and S2D). Fig. 2A shows the distributions of all unmodified and phosphorylated peptides for BR1 and BR2 with E/M ratios displayed on the logarithmic (ln) scale. We normalized replicates by their median and plotted E/M ratios of merged (BR1+BR2) peptides for unmodified and phosphorylated peptides (Fig. 2B). Notably, there are more alterations in E/M ratios of phosphopeptides (std = 1.18) than the unmodified ones (std = 0.87) (Levene's test p value <0.005). Having more alterations in the phosphopeptides compared with the unmodified ones suggests a pronounced regulation at the phosphorylation status of proteins between epithelial and mesenchymal states. Combination of ratios from two replicates resulted in 4,338 phosphopeptides with at least one phosphorylated residue. Our statistical analysis (see Experimental Procedures section) revealed 357 phosphopeptides as epithelial and 514 as mesenchymal specific (supplemental Fig. S1). The correlation of phosphopeptides in both biological replicates with an r = 0.7 (p value <0.005, n = 4,338) indicates the high reproducibility (Fig. 2C). For both epithelial and mesenchymal specific phosphosites, we searched for dominant consensus motifs (supplemental Fig. S1). In epithelial cells, SP and TP motifs dominate and in mesenchymal cells, R/KxxSxD and RxxT are the enriched motifs (Fig. 2D). Next, we used NetworKIN 3.0 (30) in KinomeXplorer (46) to further predict potential kinase-substrate relationships of EMT based on known kinase motifs and protein-protein interactions in the STRINGv10 database (47) (supplemental Fig. S1). Accordingly, a kinase-substrate network was reconstructed that covers 73 kinase groups and their potential epithelial and mesenchymal specific substrates (Fig. 2E). Notably, more kinase activity and phosphorylation events take place in mesenchymal cells than in epithelial cells. ROCK, Aurora, CAMKII, and MAP kinases are among the predicted kinases for mesenchymal specific phosphorylations (Fig. 2E).
Fig. 2.
Epithelial and mesenchymal specific phosphorylation analysis. A, Distribution of raw ln(Epithelial/Mesenchymal) unmodified peptide and phosphopeptide ratios. Biological replicates 1 and 2 (BR1 and BR2) are shown. B, Distribution of normalized and combined ln(Epithelial/Mesenchymal) unmodified peptide and phosphopeptide ratios. Red dashed line illustrates the significance threshold ratio. C, The correlation of the phosphopeptides between two biological replicates (Pearson correlation, r = 0.7). D, The consensus motifs of epithelial and mesenchymal phosphorylation sites. Consensus was extracted using Motif-x. E, The predicted kinase-substrate network reveals distinct regulations of epithelial and mesenchymal states.
Reconstruction of Epithelial and Mesenchymal Specific Protein Networks Reveals Functional Subnetworks
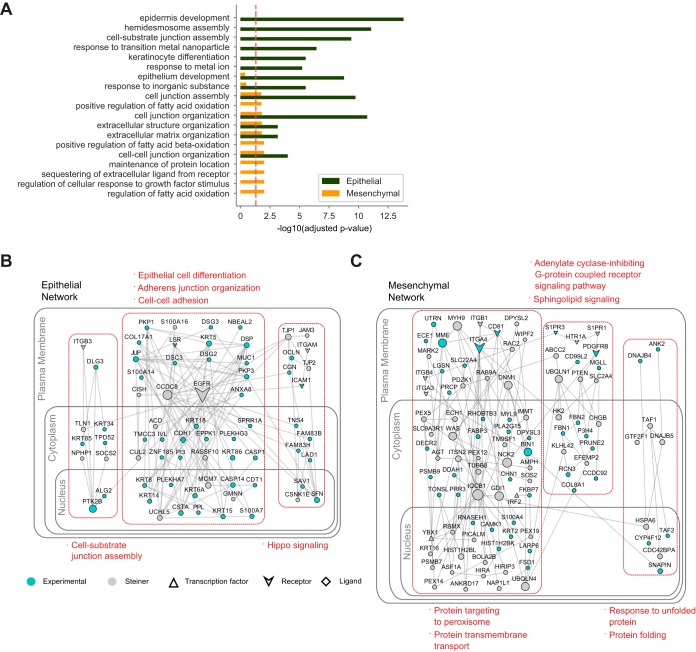
GO enrichment analysis (48) of epithelial and mesenchymal specific proteins revealed distinct annotations in epithelial and mesenchymal cells. Based on the top 10 scoring GO biological process (BP) terms (Fig. 3A), cell-cell adhesion and cell junction assembly proteins are the most significant terms in epithelial state whereas regulation of fatty acid metabolism and cellular response to growth factor stimulus are uniquely significant in mesenchymal cells (Fig. 3A).
Fig. 3.
The interaction networks of epithelial and mesenchymal states reveal distinct functional subgroups. A, Comparison of GO Biological Process (BP) enrichments in epithelial and mesenchymal specific states. Top 10 scoring GO BP terms for epithelial and mesenchymal specific proteins are illustrated. B–C, The interaction network of a subset of epithelial (B) and mesenchymal (C) specific proteins are shown with Steiner proteins added to the network. Highly interconnected subnetworks are highlighted in red rectangles and enriched GO BP terms are listed (For the networks of all identified epithelial and mesenchymal specific proteins, see supplemental Figs. S3 and S4).
Next, we sought to identify signaling pathways that represent epithelial and mesenchymal states (supplemental Figs. S3 and S4). For this purpose, we took the advantage of Omics Integrator software which optimally reconstructs a network by considering the significance of the proteomic hits and the reliability of protein-protein interactions (32). Beyond the list of proteins, it searches for the either direct interactions of the given proteins or the interactions through intermediate proteins. For epithelial state, cell-cell adhesion, cell junction assembly, and epithelial cell differentiation pathways are among the significantly enriched biological proteins in the reconstructed network (Fig. 3B). EGFR, one of the central nodes, interacts with JUP, Col17A, and numerous Keratins. Another significant pathway is Hippo signaling. Multiple components of Hippo signaling pathway SAV1, TJP1, TJP2, PARD6B, PARD6G, PRKCI, PRKCZ, LLGL2, and SFN (14–3-3 sigma, which is also known as epithelial cell marker protein) (49) are part of the epithelial specific network, which indicates their epithelial cell specific roles. Additionally, our network analysis revealed exocytosis, cell cycle, and regulation of proteolysis as notable cellular functions of epithelial specific proteins. Remarkably, seven members of S100 protein family, which is one of the largest calcium binding protein subfamily, are in multiple subnetworks of epithelial specific proteins including epithelial cell differentiation (Fig. 3B).
Multiple signaling pathways are identified in the mesenchymal specific protein network including sphingolipid signaling, regulation of antigen receptor-mediated signaling, transforming growth factor beta receptor signaling, and regulation of TOR signaling (Fig. 3C). Our network identified G-coupled receptors S1PR1, S1PR3, and HTR1A that are implicated in sphingolipid signaling as mesenchymal specific. Activation of those may regulate EMT. Interestingly, multiple mesenchymal specific proteins are linked to fatty acid metabolic processes such as TWIST1, DECR2, CPT1A, CYP4F12, FABP3, IRS2, LPIN3, PLA2G15, and MGLL. This suggests a link between fatty acid metabolism and induction of EMT (Fig. 3A, 3C). Another remarkable mesenchymal specific subnetwork is protein folding-response to unfolded protein (Fig. 3C). Within this subnetwork, two human heat shock proteins 40 family members; DNAJB4 and DNAJB5 are identified (Fig. 3C).
Novel Epithelial and Mesenchymal Specific Proteins Affect the Migration of Breast Cancer Cells
Next, we further studied a subclass of novel epithelial and mesenchymal specific proteins using real-time cell migration analysis (RTCA) (supplemental Fig. S5A). This subclass of proteins was determined compared with the transcriptome data obtained from Taube et al. study (4). We selected the proteins that are either regulated at proteome level or at both transcriptome and proteome levels. In addition to transcriptome data, we analyzed the mRNA expression levels of selected proteins in epithelial and mesenchymal breast cancer cell lines using the study by Luo et al. (35) (supplemental Fig. S5A). For the RTCA assay, we performed retrovirus-mediated shRNA knockdown of selected epithelial specific proteins in MCF7 cells and selected mesenchymal specific proteins in MDA-MB-231 invasive breast cancer cells. E-cadherin (CDH1) and N-cadherin (CDH2) depletions were used as positive controls (supplemental Fig. S5B, S5C). When epithelial specific proteins CASP14, PI3, TLR2, and TFAP2A were depleted, we observed that the migration of MCF7 cells increased (supplemental Fig. S5B). On the other hand, depletion of mesenchymal specific proteins BIN1, PDGFRB, CADM3, ANPEP, DNAJB4, and CD81 decreased the migration of MDA-MB-231 cells (supplemental Fig. S5C, Figs. 4F and 5F, respectively). We focused on two proteins, DNAJB4 and CD81, for the reasons explained below, to address their effects on EMT both in vitro and in vivo.
Fig. 4.
Increased DNAJB4 protein expression in mesenchymal state increases migration of breast cancer cells. A, mRNA expression of DNAJB4 in GSE40059 data set based on the classification of different breast cell lines as epithelial and mesenchymal. B, Western blot analysis showing the increased protein expression of DNAJB4 in HMLE-Twist cells. C, Induction of EMT with tamoxifen-inducible Twist (Twist-ER) in HMLE cells. Cells were induced to undergo EMT for 16 days of 20 nm of 4-OHT treatment and analyzed for DNAJB4 and CD81 protein expressions. D, Western blot analysis of shRNA mediated knockdown of DNAJB4 protein expression in MDA-MB-231 cells. E, Cell viability analysis of MDA-MB-231 cells stably expressing the control and DNAJB4 shRNAs. F, Real time migration analysis of MDA-MB-231 cell line with DNAJB4 knockdown compared with control cells using real time cell analyzer. shDNAJB4 cells generated a lower signal suggesting that they migrate slower than shCtrl cells. G, Wound healing scratch assay of MDA-MB-231 cells with DNAJB4 knockdown for 24 h and the quantification of percent reduction of wound area. H, Correlation analysis of the mRNA expression of DNAJB4 with the EMT score in breast cancer patients from TCGA data set. I, Metastasis-free survival of breast cancer patients in GSE2603 data set based on mRNA expression of DNAJB4.
Fig. 5.
Increased CD81 protein expression in mesenchymal state increases migration of breast cancer cells. A, mRNA expression of CD81 in GSE40059 data set based on the classification of different breast cell lines as epithelial and mesenchymal. B, Western blot analysis showing the increased protein expression of CD81 in HMLE-Twist cells. C, Cell surface expression analysis of CD81 on epithelial and mesenchymal cells by flow cytometry. D, Western blot analysis of shRNA mediated knockdown of CD81 protein expression in MDA-MB-231 cells. E, Cell viability analysis of MDA-MB-231 cells stably expressing the control and CD81 shRNAs. F, Real time migration analysis of MDA-MB-231 cell line with CD81 knockdown compared with control cells using real time cell analyzer. shCD81 cells generated a lower signal suggesting that they migrate slower than shCtrl cells. G, Wound healing scratch assay of MDA-MB-231 cells with CD81 knockdown for 24 h and the quantification of percent reduction of wound area.
DNAJB4 Knockdown Decreases Migration of Mesenchymal Breast Cancer Cells and Its mRNA Level Predicts Metastasis-free Survival
DNAJB4 is regulated at both transcriptome and proteome level during EMT and shows increased transcription in mesenchymal breast cancer cell lines (supplemental Fig. S5A and Fig. 4A). In agreement with our quantitative proteomics data, Western blot analysis showed dramatically increased DNAJB4 protein expression in HMLE-Twist cells compared with HMLE cells (Fig. 4B). To analyze temporal expression of DNAJB4 during EMT, we used EMT inducible HMLE-Twist-ER cells (50). The cells were treated with 20 nm 4-OHT for 16 days to induce EMT and collected at different days to analyze their expression pattern by Western blotting. We found that DNAJB4 expression increases gradually during EMT induction (Fig. 4C).
To examine the roles of DNAJB4 expression on mesenchymal cells, we performed shRNA-mediated knockdown of DNAJB4 in MDA-MB-231 invasive breast cancer cells (Fig. 4D). After the suppression of DNAJB4, we observed no change in cell viability (Fig. 4E), but a significant inhibition of migration in mesenchymal cells by real-time migration assay (Fig. 4F). These results were further validated by a wound healing assay (Fig. 4G). Moreover, the correlation analysis of DNAJB4 with EMT scores revealed that DNAJB4 mRNA expression levels positively correlate with the EMT scores of breast cancer patients (Fig. 4H). Similarly, the metastasis-free survival analysis of patients revealed that high level of DNAJB4 mRNA expression significantly associates with worse metastasis-free survival of breast cancer patients (Fig. 4I). In addition to DNAJB4, the mRNA expression level of DNAJB5, which was also identified as a member of mesenchymal specific network (supplemental Fig. S4 and Fig. 3C), is higher in mesenchymal breast cancer cell lines (supplemental Fig. S6A) and positively correlates with EMT scores of breast cancer patients (supplemental Fig. S6B).
CD81 Knockdown Decreases Migration of Mesenchymal Breast Cancer Cells
Another prominent hit in our migration assay was a member of tetraspanins family, CD81 (supplemental Fig. S5A). Although most of the members of tetraspanins were present in the epithelial specific network (supplemental Fig. S3), CD81 is in the mesenchymal specific protein network (supplemental Fig. S4 and Fig. 3C). CD81 is regulated only at proteome level, and its transcription level does not significantly change during EMT and in breast cancer cell lines with epithelial or mesenchymal phenotype (supplemental Fig. S5A and Fig. 5A). Our Western blot analysis confirmed that CD81 protein expression dramatically increases in HMLE-Twist cells as expected from our quantitative proteomics data (Fig. 5B). Like DNAJB4, CD81 expression also increased gradually in a time dependent manner in inducible HMLE-Twist-ER cells as EMT progresses by 4-OHT treatment (Fig. 4C). As a potential cell surface marker, we tested whether CD81 expression increases on the mesenchymal cell surface using unpermeabilized HMLE and HMLE-Twist cells in flow cytometry. Our analyses revealed that surface CD81 expression increased 79% in HMLE-Twist cells (Fig. 5C).
Next, we performed shRNA-mediated knockdown of CD81 in MDA-MB-231 invasive breast cancer cells. As a result, CD81 was effectively depleted (Fig. 5D) and viability of cells was not affected (Fig. 5E). In agreement with the real-time migration assay (Fig. 5F), the in vitro wound healing assay also revealed the impairment of migration ability of MDA-MB-231 invasive breast cancer cells upon CD81 knockdown (Fig. 5G).
Individual Knockdown of DNAJB4 and CD81 Results in Decreased Primary Tumor Growth and Metastasis
We then examined the roles of DNAJB4 and CD81 in primary tumor growth and metastasis in vivo. We performed retrovirus-mediated shRNA knockdown of DNAJB4 and CD81 in MDA-MB-231-Luc2-GFP cells. To observe the effects of DNAJB4 and CD81 knockdown on the primary tumor growth, we developed primary tumor xenografts with control and knockdown cells in nude mice. Knockdown of CD81 and DNAJB4 significantly impaired the tumor growth (Fig. 6A, 6B, 6C). To test the effect of two candidates on the spontaneous metastasis, tumor growth has been monitored until tumors in each group reached to 1000 mm3 (supplemental Fig. S7). One month after surgical removal of primary tumors, an ex vivo luciferase assay was performed. Knockdown of CD81 and DNAJB4 significantly reduced both lung and liver metastases (Fig. 6D).
Fig. 6.
Suppression of DNAJB4 and CD81 leads to decreased primary tumor growth, spontaneous metastasis and extravasation/colonization. A, Primary tumor progression in xenografts generated by mammary fat pad injection of MDA-MB-231-Luc2-GFP control (n = 3), shCD81 (n = 4) and shDNAJB4 (n = 4) into nude mice. B, Bioluminescence images of nude mice from Fig. panel A. C, Quantification of the luminescence signals from mice in Fig. panel A, given as total flux. D, Luciferase signal coming from lungs and livers of mice in Fig. panel B as quantified by luciferase assay (shCtrl n = 3, shCD81 n = 4, and shDNAJB4 n = 3). E, Bioluminescence images of nude mice intravenously injected with MDA-MB-231-Luc2-GFP control, shDNAJB4 and shCD81 cells. F, Quantification of the luminescence signals from mice in Fig. panel E, given as total flux. G, Representative images of lungs collected from the groups given in Fig. panel E. Mice were sacrificed at sixth week and lungs were fixed in Bouin's Solution. Metastatic nodules on lung surface appear white after overnight fixation in Bouin's solution. Mice injected with shCtrl cells have more nodules on the lung surface compared with shDNAJB4 and shCD81 cells injected mice. H, Representative images of lungs stained with Hematoxylin-Eosin. T: for tumor, L: for lung.
Next, we tested whether DNAJB4 and CD81 play a role in extravasation/colonization of MDA-MB-231 invasive breast cancer cells in lungs by tail-vein metastasis assay. After 6 weeks from intravenous injection of DNAJB4 and CD81 depleted cells into nude mice, we compared their lung metastasis with control cells (Fig. 6E). Mouse groups injected with either CD81 or DNAJB4 knockdown cells showed a low level of lung metastasis compared with control (Fig. 6F). Similarly, we observed less nodules in lungs in the knockdown group mice compared with control mice after fixation with Bouin's solution (Fig. 6G). Hematoxylin and eosin (H&E) staining confirmed the differences in lung metastases in control, DNAJB4 knockdown and CD81 knockdown groups (Fig. 6H). These results demonstrate that both DNAJB4 and CD81 contribute to the mesenchymal characteristics of cells undergoing EMT and have roles in primary tumor growth and metastasis of breast cancer cells in vivo.
DISCUSSION
The transition from epithelial to mesenchymal state involves a major reorganization of multiple cellular components triggered by EMT-TF. This study used quantitative proteomics to identify biochemical changes after activation of EMT-TF. Previously, Taube et al. obtained the transcriptome profiles of HMLE cells in epithelial and mesenchymal states as “EMT core signature” (4). By using HMLE-Twist cells, we profiled the biochemical changes at the protein and post-translational level. Strikingly, around 65% of significantly regulated proteins did not exhibit any detectable regulation at the mRNA level (supplemental Table S1). Similarly, a previous study analyzed the cell surface protein changes during EMT using HMLE-Twist cells by performing membrane-associated protein fractionation followed by mass spectrometry analysis (14). Based on the commonly identified proteins, our whole proteome profile largely correlated with the published cell surface proteome (76% for mesenchymal and 83% for epithelial) (supplemental Tables S1 and S4). Interestingly, when significantly changed proteins were normalized to all identified ones, Lu et al. reported more than 2-fold significantly differentially expressed proteins compared with our whole proteome data. One likely explanation for this difference is the contribution of cellular spatial organization during EMT. Altogether, these findings suggest that besides the gene expression level regulation, EMT is regulated at multiple post-transcriptional levels including protein subcellular localization and post-translational modification.
To reveal phosphorylation-driven signaling pathways during EMT, we performed a quantitative phosphoproteomic analysis. Our analysis reveals that there are more alterations in phosphoproteome than proteome. Having more modulation at the phosphorylation level suggest the importance of phosphorylation-based signal transduction pathways during EMT. Many kinases are predicted to phosphorylate either epithelial or mesenchymal specific phosphopeptides. This may suggest that phosphorylation events in EMT are important for cells to preserve their state as epithelial or mesenchymal and transition between two states must be regulated by altered phosphorylation landscape of the cells. Overall, our data points out more global phosphorylation in mesenchymal cells (514) than epithelial cells (357) suggesting an important role of kinases during EMT. We identified 73 kinase groups and their potential epithelial and mesenchymal specific substrates. Some of these roles in EMT are previously reported: CDK5 is required for TGF-β1-induced EMT (51). CK2A2 expression levels are found to be upregulated in human colorectal cancer and downregulated through miR-1228, decreasing EMT capability (52, 53). Exosomes from transformed epithelial cells that contain PAK2 are reported to induce metastatic features in endothelial cells (54). Interestingly, GSK3β's decreased activity is found to be promoting invasive abilities of epithelial cancers via Snail and Slug activation (55–57). We also identified kinases such as PAK2, ROCK2, and NEK1 are auto-phosphorylated in a mesenchymal specific manner. These auto-phosphorylated kinases might be members of the feedback mechanisms in EMT.
Besides the kinase-substrate network, reconstructing epithelial and mesenchymal specific protein-protein interaction networks revealed key signaling pathways of cells undergoing EMT including Hippo, TGF-β, mTOR, PDGF, and cAMP. In addition to expected cellular machineries that are remodeled during EMT, such as cell adhesion, cell-substrate junction assembly etc., our analysis identified understudied ones, such as lipid metabolism and sphingolipid signaling (58). We found that expression of Sphingosine-1-phosphate receptors 1 and 3 increases in mesenchymal cells. In addition, the expression of fatty acid metabolic process related proteins DECR2, CPT1A, CYP4F12, FABP3, IRS2, LPIN3, PLA2G15, and MGLL were increased in mesenchymal cells. Deciphering the molecular mechanism behind the overexpression of these proteins will provide further insights into the biology of EMT.
One of the interesting signaling pathways enriched for the mesenchymal state is unfolded protein response. DNAJB4 and DNAJB5, members of the human heat shock proteins 40 family, were identified in this subnetwork. Although previous studies have shown DNAJB4 as tumor-suppressor protein (59–61), our proteomic data and patient data set analyses describe DNAJB4 as a metastasis inducer in breast cancer cells. When we compare the mRNA expression levels of DNAJB4 in epithelial and mesenchymal breast cancer cells (35), we found that DNAJB4 mRNA expression levels are significantly increased in mesenchymal cells. Our EMT induction experiment shows that DNAJB4 protein level gradually increases as EMT progresses. Importantly, knockdown of DNAJB4 in MDA-MB-231 cells leads to a decrease in migration, impairs lung metastasis of invasive breast cancer cells, and decreases primary tumor growth rate. In parallel with this, metastasis-free survival analysis of primary breast cancer patients reveals that higher levels of DNAJB4 significantly associates with a worse metastasis-free survival. Recent research demonstrated that inducing EMT causes hyper-activation of unfolded protein response (UPR) signaling pathways (62–65). It has been shown that cells in their mesenchymal state have increased synthesis and secretion of large quantities of extracellular matrix (ECM) proteins which then triggers ER stress and activation of unfolded protein response (63). It is plausible as a chaperone protein, DNAJB4 may be induced to counteract this stress induction on EMT and its expression gradually increases because of the activation of unfolded protein response during EMT. Deciphering the molecular mechanism of DNAJB4 overexpression during EMT would bring new perspectives into the relationship between EMT and unfolded protein response.
One motivation for this study was to find biomarkers for EMT. Our network analysis identified multiple cell surface proteins as epithelial and mesenchymal specific. Some of them may serve as potential biomarkers, such as TSPAN1 and FXYD3 as epithelial and CD10 (MME), CD90 (THY1) (66), Fibulin-4 (EFEMP2) and CD81 as mesenchymal markers. Interestingly, the CD81 mRNA level does not significantly alter during EMT (4) and between epithelial and mesenchymal breast cell lines. But we identified CD81 as one of the significantly upregulated proteins during the EMT process. CD81 protein levels increase gradually as cells undergo EMT. In parallel, CD81 level increased 79% on the surface of HMLE-Twist cells. Functional assays supported its potential of being a marker for EMT process. On CD81 knockdown, we observed a decrease in the migration of mesenchymal breast cancer cells as reported in Zhang et al. (67). Moreover, in vivo experiments revealed that CD81 knockdown in MDA-MB-231 cells decreased primary tumor growth rate and impaired lung metastasis. Its strong mesenchymal characteristics suggest that CD81 may also have a value as mesenchymal stem cell marker that may target a small subpopulation of cancer cells similar to CD44 (50).
In summary, our analysis combines quantitative proteomic and phosphoproteomic approaches to provide large scale biochemical changes as HMLE cells undergo EMT and reveals key signaling pathways during this process. Considering the key involvement of EMT in both metastasis and drug resistance, detailed characterization of novel EMT regulators may open new avenues for targeting metastatic tumors and overcoming drug resistance.
Data Availability
The data sets produced in this study are available in the following databases: Protein quantification MS data: PRIDE PXD012001 http://www.ebi.ac.uk/pride/archive/projects/PXD012001, Annotated spectra: MS-Viewer search key is 9vpxyp6xgq, http://msviewer.ucsf.edu/prospector/cgi-bin/mssearch.cgi?report_title=MS-Viewer&search_key=9vpxyp6xgq&search_name=msviewer.
Supplementary Material
Acknowledgments
We gratefully acknowledge use of the services and facilities of the Koç University Research Center for Translational Medicine (KUTTAM), funded by the Republic of Turkey Ministry of Development. The content is solely the responsibility of the authors and does not necessarily represent the official views of the Ministry of Development. We thank Dr. Adema Ribic for the critical reading of the manuscript.
Footnotes
* N.O. was funded by TÜBİTAK 1001 Grant (114Z267), Science Academy (Turkey) Young Scientist Award (BAGEP) and EMBO Installation Grant. Z.C.U.K. and E.S. were funded by TÜBİTAK-BİDEB 2211/E Scholarship Program. The work in T.T.Ö.'s laboratory was supported by an EMBO Installation Grant. The work in Ö.Ş.'s laboratory was, in part, supported by EMBO Installation Grant 2791 and National Institutes of Health Grant P20 GM 109091COBRE. Ö.S. was funded by Susan G. Komen Interdisciplinary Graduate Training to Eliminate Cancer Disparities (IGniTE-CD) Scholarship GTDR17500160. N.T. acknowledges Science Academy (Turkey) for Young Scientist Award (BAGEP) and UNESCO-L'Oreal National for Women in Science Fellowship.
 This article contains supplemental material.
This article contains supplemental material.
1 The abbreviations used are:
- EMT
- epithelial-mesenchymal transition
- UPR
- unfolded protein response
- EMT-TF
- EMT inducing transcription factors
- HMLE
- human mammary epithelial cells
- IMAC
- immobilized metal ion affinity chromatography
- ACN
- acetonitrile
- HCD
- Higher-energy collisional dissociation
- NCE
- normalized collision energy
- PSM
- peptide spectral match
- GEO
- Gene Expression Omnibus database
- GSE
- Genomic Spatial Event database
- TCPA
- The Cancer Proteome Atlas
- TCGA
- The Cancer Genome Atlas
- RTCA
- real time cell analysis
- DNAJB4
- DnaJ heat shock protein family (Hsp40) member B4
- CD81
- cluster of differentiation 81.
REFERENCES
- 1. Fidler I. J. (2003) The pathogenesis of cancer metastasis: the 'seed and soil' hypothesis revisited. Nat. Rev. Cancer 3, 453–458 [DOI] [PubMed] [Google Scholar]
- 2. Hanahan D., and Weinberg R. A. (2011) Hallmarks of cancer: the next generation. Cell 144, 646–674 [DOI] [PubMed] [Google Scholar]
- 3. Onder T. T., Gupta P. B., Mani S. A., Yang J., Lander E. S., and Weinberg R. A. (2008) Loss of E-cadherin promotes metastasis via multiple downstream transcriptional pathways. Cancer Res. 68, 3645–3654 [DOI] [PubMed] [Google Scholar]
- 4. Taube J. H., Herschkowitz J. I., Komurov K., Zhou A. Y., Gupta S., Yang J., Hartwell K., Onder T. T., Gupta P. B., Evans K. W., Hollier B. G., Ram P. T., Lander E. S., Rosen J. M., Weinberg R. A., and Mani S. A. (2010) Core epithelial-to-mesenchymal transition interactome gene-expression signature is associated with claudin-low and metaplastic breast cancer subtypes. Proc. Natl. Acad. Sci. U.S.A. 107, 15449–15454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Greenburg G., and Hay E. D. (1982) Epithelia suspended in collagen gels can lose polarity and express characteristics of migrating mesenchymal cells. J. Cell Biol. 95, 333–339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Kalluri R., and Weinberg R. A. (2009) The basics of epithelial-mesenchymal transition. J. Clin. Investigation 119, 1420–1428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Lamouille S., Xu J., and Derynck R. (2014) Molecular mechanisms of epithelial-mesenchymal transition. Nat. Rev. Mol. Cell Biol. 15, 178–196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Brabletz T., Kalluri R., Nieto M. A., and Weinberg R. A. (2018) EMT in cancer. Nat. Rev. Cancer 18, 128. [DOI] [PubMed] [Google Scholar]
- 9. Du B., and Shim J. S. (2016) Targeting epithelial-mesenchymal transition (EMT) to overcome drug resistance in cancer. Molecules 21, pii: E965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Sarkar F. H., Li Y., Wang Z., and Kong D. (2009) Pancreatic cancer stem cells and EMT in drug resistance and metastasis. Minerva Chirurgica 64, 489–500 [PMC free article] [PubMed] [Google Scholar]
- 11. Singh A., and Settleman J. (2010) EMT, cancer stem cells and drug resistance: an emerging axis of evil in the war on cancer. Oncogene 29, 4741–4751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Wang Z., Li Y., Ahmad A., Azmi A. S., Kong D., Banerjee S., and Sarkar F. H. (2010) Targeting miRNAs involved in cancer stem cell and EMT regulation: An emerging concept in overcoming drug resistance. Drug Resistance Updates 13, 109–118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Tam W. L., and Weinberg R. A. (2013) The epigenetics of epithelial-mesenchymal plasticity in cancer. Nat. Med. 19, 1438–1449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Lu H., Clauser K. R., Tam W. L., Frose J., Ye X., Eaton E. N., Reinhardt F., Donnenberg V. S., Bhargava R., Carr S. A., and Weinberg R. A. (2014) A breast cancer stem cell niche supported by juxtacrine signalling from monocytes and macrophages. Nat. Cell Biol. 16, 1105–1117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Tam W. L., Lu H., Buikhuisen J., Soh B. S., Lim E., Reinhardt F., Wu Z. J., Krall J. A., Bierie B., Guo W., Chen X., Liu X. S., Brown M., Lim B., and Weinberg R. A. (2013) Protein kinase C alpha is a central signaling node and therapeutic target for breast cancer stem cells. Cancer Cell 24, 347–364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Yang J., Mani S. A., Donaher J. L., Ramaswamy S., Itzykson R. A., Come C., Savagner P., Gitelman I., Richardson A., and Weinberg R. A. (2004) Twist, a master regulator of morphogenesis, plays an essential role in tumor metastasis. Cell 117, 927–939 [DOI] [PubMed] [Google Scholar]
- 17. Filipowicz W., Bhattacharyya S. N., and Sonenberg N. (2008) Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight? Nat. Rev. Gen. 9, 102–114 [DOI] [PubMed] [Google Scholar]
- 18. Filipowicz W., Jaskiewicz L., Kolb F. A., and Pillai R. S. (2005) Post-transcriptional gene silencing by siRNAs and miRNAs. Current Opin. Struct. Biol. 15, 331–341 [DOI] [PubMed] [Google Scholar]
- 19. Selbach M., Schwanhäusser B., Thierfelder N., Fang Z., Khanin R., and Rajewsky N. (2008) Widespread changes in protein synthesis induced by microRNAs. Nature 455, 58. [DOI] [PubMed] [Google Scholar]
- 20. Elenbaas B., Spirio L., Koerner F., Fleming M. D., Zimonjic D. B., Donaher J. L., Popescu N. C., Hahn W. C., and Weinberg R. A. (2001) Human breast cancer cells generated by oncogenic transformation of primary mammary epithelial cells. Genes Develop. 15, 50–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Zhou H., Ye M., Dong J., Corradini E., Cristobal A., Heck A. J., Zou H., and Mohammed S. (2013) Robust phosphoproteome enrichment using monodisperse microsphere-based immobilized titanium (IV) ion affinity chromatography. Nat. Protocols 8, 461–480 [DOI] [PubMed] [Google Scholar]
- 22. Polat A. N., Karayel O., Giese S. H., Harmanda B., Sanal E., Hu C. K., Renard B. Y., and Ozlu N. (2015) Phosphoproteomic analysis of aurora kinase inhibition in monopolar cytokinesis. J. Proteome Res. 14, 4087–4098 [DOI] [PubMed] [Google Scholar]
- 23. Boersema P. J., Raijmakers R., Lemeer S., Mohammed S., and Heck A. J. (2009) Multiplex peptide stable isotope dimethyl labeling for quantitative proteomics. Nat. Protocols 4, 484–494 [DOI] [PubMed] [Google Scholar]
- 24. de Graaf E. L., Giansanti P., Altelaar A. F., and Heck A. J. (2014) Single-step enrichment by Ti4+-IMAC and label-free quantitation enables in-depth monitoring of phosphorylation dynamics with high reproducibility and temporal resolution. Mol. Cell. Proteomics 13, 2426–2434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Thingholm T. E., Jorgensen T. J., Jensen O. N., and Larsen M. R. (2006) Highly selective enrichment of phosphorylated peptides using titanium dioxide. Nat. Protocols 1, 1929–1935 [DOI] [PubMed] [Google Scholar]
- 26. Taus T., Kocher T., Pichler P., Paschke C., Schmidt A., Henrich C., and Mechtler K. (2011) Universal and confident phosphorylation site localization using phosphoRS. J. Proteome Res. 10, 5354–5362 [DOI] [PubMed] [Google Scholar]
- 27. Chou M. F., and Schwartz D. (2011) Biological sequence motif discovery using motif-x. Current protocols in bioinformatics Chapter 13, Unit 13.15–24 John Wiley & Sons, Inc., Hoboken, NJ: [DOI] [PubMed] [Google Scholar]
- 28. Wagih O., Sugiyama N., Ishihama Y., and Beltrao P. (2016) Uncovering Phosphorylation-Based Specificities through Functional Interaction Networks. Mol. Cell. Proteomics 15, 236–245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Crooks G. E., Hon G., Chandonia J. M., and Brenner S. E. (2004) WebLogo: a sequence logo generator. Genome Res. 14, 1188–1190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Linding R., Jensen L. J., Pasculescu A., Olhovsky M., Colwill K., Bork P., Yaffe M. B., and Pawson T. (2008) NetworKIN: a resource for exploring cellular phosphorylation networks. Nucleic Acids Res. 36, D695–D699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Shannon P., Markiel A., Ozier O., Baliga N. S., Wang J. T., Ramage D., Amin N., Schwikowski B., and Ideker T. (2003) Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 13, 2498–2504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Tuncbag N., Gosline S. J. C., Kedaigle A., Soltis A. R., Gitter A., and Fraenkel E. (2016) Network-based interpretation of diverse high-throughput datasets through the omics integrator software package. PLOS Computational Biol. 12, e1004879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Karayel Ö., Şanal E, Giese S. H., Üretmen Kagıalı Z. C., Polat A. N., Hu C.-K., Renard B. Y., Tuncbag N., and Özlü N. (2018) Comparative phosphoproteomic analysis reveals signaling networks regulating monopolar and bipolar cytokinesis. Sci. Reports 8, 2269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Turner B., Razick S., Turinsky A. L., Vlasblom J., Crowdy E. K., Cho E., Morrison K., Donaldson I. M., and Wodak S. J. (2010) iRefWeb: interactive analysis of consolidated protein interaction data and their supporting evidence. Database 2010, baq023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Luo D., Wilson J. M., Harvel N., Liu J., Pei L., Huang S., Hawthorn L., and Shi H. (2013) A systematic evaluation of miRNA:mRNA interactions involved in the migration and invasion of breast cancer cells. J. Translational Med. 11, 57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Blick T., Widodo E., Hugo H., Waltham M., Lenburg M. E., Neve R. M., and Thompson E. W. (2008) Epithelial mesenchymal transition traits in human breast cancer cell lines. Clin. Exp. Metastasis 25, 629–642 [DOI] [PubMed] [Google Scholar]
- 37. Lombaerts M., van Wezel T., Philippo K., Dierssen J. W., Zimmerman R. M., Oosting J., van Eijk R., Eilers P. H., van de Water B., Cornelisse C. J., and Cleton-Jansen A. M. (2006) E-cadherin transcriptional downregulation by promoter methylation but not mutation is related to epithelial-to-mesenchymal transition in breast cancer cell lines. Br. J. Cancer 94, 661–671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Sommers C. L., Byers S. W., Thompson E. W., Torri J. A., and Gelmann E. P. (1994) Differentiation state and invasiveness of human breast cancer cell lines. Breast Cancer Res. Treatment 31, 325–335 [DOI] [PubMed] [Google Scholar]
- 39. Cerami E., Gao J., Dogrusoz U., Gross B. E., Sumer S. O., Aksoy B. A., Jacobsen A., Byrne C. J., Heuer M. L., Larsson E., Antipin Y., Reva B., Goldberg A. P., Sander C., and Schultz N. (2012) The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discovery 2, 401–404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Gao J., Aksoy B. A., Dogrusoz U., Dresdner G., Gross B., Sumer S. O., Sun Y., Jacobsen A., Sinha R., Larsson E., Cerami E., Sander C., and Schultz N. (2013) Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci. Signaling 6, pl1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Li J., Lu Y., Akbani R., Ju Z., Roebuck P. L., Liu W., Yang J.-Y., Broom B. M., Verhaak R. G. W., Kane D. W., Wakefield C., Weinstein J. N., Mills G. B., and Liang H. (2013) TCPA: a resource for cancer functional proteomics data. Nat. Methods 10, 1046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Minn A. J., Gupta G. P., Siegel P. M., Bos P. D., Shu W., Giri D. D., Viale A., Olshen A. B., Gerald W. L., and Massague J. (2005) Genes that mediate breast cancer metastasis to lung. Nature 436, 518–524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Olson A., Sheth N., Lee J. S., Hannon G., and Sachidanandam R. (2006) RNAi Codex: a portal/database for short-hairpin RNA (shRNA) gene-silencing constructs. Nucleic Acids Res. 34, D153–D157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Onder T. T., Kara N., Cherry A., Sinha A. U., Zhu N., Bernt K. M., Cahan P., Mancarci B. O., Unternaehrer J., Gupta P. B., Lander E. S., Armstrong S. A., and Daley G. Q. (2012) Chromatin-modifying enzymes as modulators of reprogramming. Nature 483, 598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Cevik S. I., Keskin N., Belkaya S., Ozlu M. I., Deniz E., Tazebay U. H., and Erman B. (2012) CD81 interacts with the T cell receptor to suppress signaling. PloS One 7, e50396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Horn H., Schoof E. M., Kim J., Robin X., Miller M. L., Diella F., Palma A., Cesareni G., Jensen L. J., and Linding R. (2014) KinomeXplorer: an integrated platform for kinome biology studies. Nat. Methods 11, 603–604 [DOI] [PubMed] [Google Scholar]
- 47. Szklarczyk D., Franceschini A., Wyder S., Forslund K., Heller D., Huerta-Cepas J., Simonovic M., Roth A., Santos A., Tsafou K. P., Kuhn M., Bork P., Jensen L. J., and von Mering C. (2015) STRING v10: protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res. 43, D447–D452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Ashburner M., Ball C. A., Blake J. A., Botstein D., Butler H., Cherry J. M., Davis A. P., Dolinski K., Dwight S. S., Eppig J. T., Harris M. A., Hill D. P., Issel-Tarver L., Kasarskis A., Lewis S., Matese J. C., Richardson J. E., Ringwald M., Rubin G. M., and Sherlock G. (2000) Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat. Genetics 25, 25–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Zeng Q., and Hong W. (2008) The emerging role of the hippo pathway in cell contact inhibition, organ size control, and cancer development in mammals. Cancer Cell 13, 188–192 [DOI] [PubMed] [Google Scholar]
- 50. Mani S. A., Guo W., Liao M. J., Eaton E. N., Ayyanan A., Zhou A. Y., Brooks M., Reinhard F., Zhang C. C., Shipitsin M., Campbell L. L., Polyak K., Brisken C., Yang J., and Weinberg R. A. (2008) The epithelial-mesenchymal transition generates cells with properties of stem cells. Cell 133, 704–715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Liang Q., Li L., Zhang J., Lei Y., Wang L., Liu D. X., Feng J., Hou P., Yao R., Zhang Y., Huang B., and Lu J. (2013) CDK5 is essential for TGF-beta1-induced epithelial-mesenchymal transition and breast cancer progression. Sci. Rep. 3, 2932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Jia L., Wu J., Zhang L., Chen J., Zhong D., Xu S., Xie C., and Cai J. (2013) Restoration of miR-1228* expression suppresses epithelial-mesenchymal transition in gastric cancer. PloS One 8, e58637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Lin K. Y., Tai C., Hsu J. C., Li C. F., Fang C. L., Lai H. C., Hseu Y. C., Lin Y. F., and Uen Y. H. (2011) Overexpression of nuclear protein kinase CK2 alpha catalytic subunit (CK2alpha) as a poor prognosticator in human colorectal cancer. PloS One 6, e17193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Gopal S. K., Greening D. W., Hanssen E. G., Zhu H. J., Simpson R. J., and Mathias R. A. (2016) Oncogenic epithelial cell-derived exosomes containing Rac1 and PAK2 induce angiogenesis in recipient endothelial cells. Oncotarget 7, 19709–19722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Kao S. H., Wang W. L., Chen C. Y., Chang Y. L., Wu Y. Y., Wang Y. T., Wang S. P., Nesvizhskii A. I., Chen Y. J., Hong T. M., and Yang P. C. (2014) GSK3beta controls epithelial-mesenchymal transition and tumor metastasis by CHIP-mediated degradation of Slug. Oncogene 33, 3172–3182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Liu C.-W., Li C.-H., Peng Y.-J., Cheng Y.-W., Chen H.-W., Liao P.-L., Kang J.-J., and Yeng M.-H. (2014) Snail regulates Nanog status during the epithelial-mesenchymal transition via the Smad1/Akt/GSK3β signaling pathway in non-small-cell lung cancer. Oncotarget 5, 3880–3894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Wang H., Wang H.-S., Zhou B.-H., Li C.-L., Zhang F., Wang X.-F., Zhang G., Bu X.-Z., Cai S.-H., and Du J. (2013) Epithelial-mesenchymal transition (EMT) induced by TNF-α requires AKT/GSK-3β-mediated stabilization of snail in colorectal cancer. PloS One 8, e56664–e56664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Levade T., Andrieu-Abadie N., Micheau O., Legembre P., and Ségui B. (2015) Sphingolipids modulate the epithelial–mesenchymal transition in cancer. Cell Death Discovery 1, 15001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Chen H. W., Lee J. Y., Huang J. Y., Wang C. C., Chen W. J., Su S. F., Huang C. W., Ho C. C., Chen J. J., Tsai M. F., Yu S. L., and Yang P. C. (2008) Curcumin inhibits lung cancer cell invasion and metastasis through the tumor suppressor HLJ1. Cancer Res. 68, 7428–7438 [DOI] [PubMed] [Google Scholar]
- 60. Tsai M. F., Wang C. C., Chang G. C., Chen C. Y., Chen H. Y., Cheng C. L., Yang Y. P., Wu C. Y., Shih F. Y., Liu C. C., Lin H. P., Jou Y. S., Lin S. C., Lin C. W., Chen W. J., Chan W. K., Chen J. J., and Yang P. C. (2006) A new tumor suppressor DnaJ-like heat shock protein, HLJ1, and survival of patients with non-small-cell lung carcinoma. J. Natl. Cancer Inst. 98, 825–838 [DOI] [PubMed] [Google Scholar]
- 61. Wang C. C., Tsai M. F., Dai T. H., Hong T. M., Chan W. K., Chen J. J., and Yang P. C. (2007) Synergistic activation of the tumor suppressor, HLJ1, by the transcription factors YY1 and activator protein 1. Cancer Res. 67, 4816–4826 [DOI] [PubMed] [Google Scholar]
- 62. Feng Y., Jin D. X., and Gupta P. B. (2016) Abstract 2677: Unfolded protein response is required for EMT-driven metastasis by inducing CREB3L1. Cancer Res. 76, 2677 [Google Scholar]
- 63. Feng Y. X., Sokol E. S., Del Vecchio C. A., Sanduja S., Claessen J. H., Proia T. A., Jin D. X., Reinhardt F., Ploegh H. L., Wang Q., and Gupta P. B. (2014) Epithelial-to-mesenchymal transition activates PERK-eIF2alpha and sensitizes cells to endoplasmic reticulum stress. Cancer Discovery 4, 702–715 [DOI] [PubMed] [Google Scholar]
- 64. Shen X., Xue Y., Si Y., Wang Q., Wang Z., Yuan J., and Zhang X. (2015) The unfolded protein response potentiates epithelial-to-mesenchymal transition (EMT) of gastric cancer cells under severe hypoxic conditions. Med. Oncol. 32, 447. [DOI] [PubMed] [Google Scholar]
- 65. Zeindl-Eberhart E., Brandl L., Liebmann S., Ormanns S., Scheel S. K., Brabletz T., Kirchner T., and Jung A. (2014) Epithelial-mesenchymal transition induces endoplasmic-reticulum-stress response in human colorectal tumor cells. PloS One 9, e87386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Battula V. L., Evans K. W., Hollier B. G., Shi Y., Marini F. C., Ayyanan A., Wang R. Y., Brisken C., Guerra R., Andreeff M., and Mani S. A. (2010) Epithelial-mesenchymal transition-derived cells exhibit multilineage differentiation potential similar to mesenchymal stem cells. Stem Cells 28, 1435–1445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Zhang N., Zuo L., Zheng H., Li G., and Hu X. (2018) Increased expression of CD81 in breast cancer tissue is associated with reduced patient prognosis and increased cell migration and proliferation in MDA-MB-231 and MDA-MB-435S human breast cancer cell lines in vitro. Med. Sci. Monitor 24, 5739–5747 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data sets produced in this study are available in the following databases: Protein quantification MS data: PRIDE PXD012001 http://www.ebi.ac.uk/pride/archive/projects/PXD012001, Annotated spectra: MS-Viewer search key is 9vpxyp6xgq, http://msviewer.ucsf.edu/prospector/cgi-bin/mssearch.cgi?report_title=MS-Viewer&search_key=9vpxyp6xgq&search_name=msviewer.