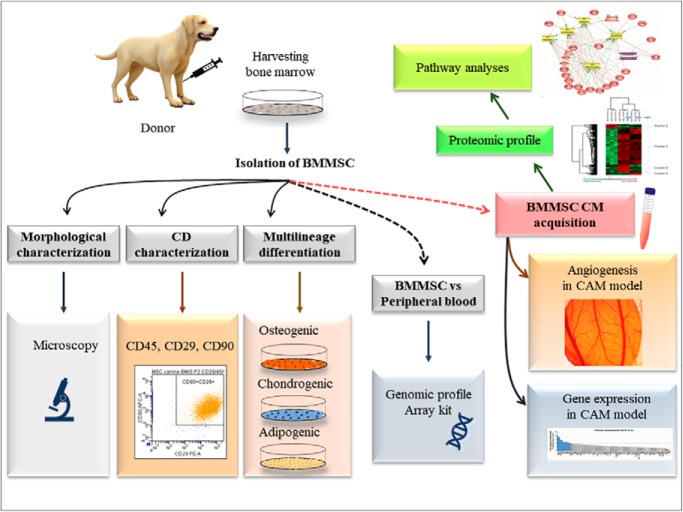
The study aim was to provide a comprehensive analyse of canine bone marrow derived mesenchymal stem cells (BMMSC) and conditioned media, isolated from healthy adult dogs of different breeds. The multipotent differentiation and specific surface markers analyses of BMMSC were supplemented with detection of gene expression, proteomic profile, and their biological function. Canine BMMSC release a variety of bioactive molecules revealing a strong paracrine component that may possess therapeutic potential in various pathologies.
Keywords: Stem Cells, Secretome, Induced Pluripotent Stem Cells, Biomarker: Prognostic, Progenitor Cells, Bone Marrow-Derived Stem Cells, Canine, Proteomic, Regenerative Medicine, Spinal Cord Injury
Graphical Abstract
Highlights
Canine bone marrow-derived mesenchymal stem (BMMSC) express genes for differentiation toward osteo-, chondro-, and tendo-genic directions.
Canine BMMSC express genes associated with angiogenic, neurotrophic, and immunomodulatory properties.
Dynamic of BMMSC conditioned medium (CM) proteome revealed transcription and translation factors, osteogenic, growth, angiogenic, and neurotrophic factors.
BMMSC CM express angiogenic activity in the chorioallantoic membrane (CAM) assay.
Abstract
Adult stem cells have become prominent candidates for treating various diseases in veterinary practice. The main goal of our study was therefore to provide a comprehensive study of canine bone marrow-derived mesenchymal stem cells (BMMSC) and conditioned media, isolated from healthy adult dogs of different breeds. Under well-defined standardized isolation protocols, the multipotent differentiation and specific surface markers of BMMSC were supplemented with their gene expression, proteomic profile, and their biological function. The presented data confirm that canine BMMSC express important genes for differentiation toward osteo-, chondro-, and tendo-genic directions, but also genes associated with angiogenic, neurotrophic, and immunomodulatory properties. Furthermore, using proteome profiling, we identify for the first time the dynamic release of various bioactive molecules, such as transcription and translation factors and osteogenic, growth, angiogenic, and neurotrophic factors from canine BMMSC conditioned medium. Importantly, the relevant genes were linked to their proteins as detected in the conditioned medium and further associated with angiogenic activity in chorioallantoic membrane (CAM) assay. In this way, we show that the canine BMMSC release a variety of bioactive molecules, revealing a strong paracrine component that may possess therapeutic potential in various pathologies. However, extensive experimental or preclinical trials testing canine sources need to be performed in order to better understand their paracrine action, which may lead to novel therapeutic strategies in veterinary medicine.
Regenerative medicine has gradually gained a significant position in current clinical veterinary practice. Stem cells have become prominent candidates for treating various diseases in companion animals (dogs and cats) where traditional treatments have proven unsuccessful (1). As part of cell therapy, various tissue-specific stem cells have a dominant strategic role, mainly because they can be obtained from different adult tissues (2). However, the diversity of tissue-specific stem cell sources suggests that these cells will also have different properties and possibly different healing effects. Currently, we know that bone marrow-derived mesenchymal stem cells (BMMSC) are suitable candidates for repairing locomotor disorders, particularly hard tissue such as bone, cartilage, and tendons (1, 3, 4). The ability of BMMSC to replace damaged bone and cartilaginous tissue is closely related to their mesodermal origin and natural multiline differentiation.
BMMSC, on the other hand, are also assigned immunomodulatory and paracrine functions because of their ability to affect the immune system and produce a wide variety of growth and proliferation factors (1, 5). These immuno-trophic properties are used to suppress strong inflammatory processes and vascularize ischemic tissue (myocardium, nerve tissue), as well as also to rejuvenate plasticity and regenerate nerve tissue (6). However, not all BMMSC-based treatments are able to evoke these beneficial properties. Mesenchymal stem cells (MSCs) isolated from bone marrow and adipose tissue are most commonly used as autologous or allogenic transplants also in veterinary medicine (7). However, MSCs have also been isolated from other tissues as well as from donors of various ages. Because there are around 400 dog breeds worldwide, the high interbreed genetic diversity may also influence the available pool of mesenchymal stem cells.
The main objective of our study was to compare BMMSC isolated from healthy adult dogs of different breeds using optimized protocols ensuring good quality of the isolated cells. Under these conditions, we analyzed mesenchymal CD markers, their multipotent potential, gene expression and proteomic profile, and their biological function. Our results confirm that canine BMMSC express not only genes for hard tissue differentiation but also genes associated with neurotrophic, angiogenic, and immunomodulatory properties. Furthermore, using proteomics, we identified for the first time the dynamics of molecules released from canine BMMSC conditioned medium and studied their pro-angiogenic effect in a CAM model. Finally, we suggest that optimized and standardized guidelines should be respected when choosing suitable donors for specific stem cell therapies. Moreover, further studies testing different canine MSC sources need to be performed in order to better understand the individual differences between healthy donors that need to be considered prior to their inclusion in treatment studies.
EXPERIMENTAL PROCEDURES
Isolation of Bone Marrow and Peripheral Blood
Bone marrow and peripheral blood were obtained from healthy adult dogs (n = 6) after informed consent of the owners was obtained. In this study, we included the following large or middle-size dog breeds, all males: Alaskan malamute (2.0 years old), Labrador (2.3 years old), American Staffordshire terrier (2.6 years old), Poodle (2.4 years old), Eurohound (2.7 years old), and Beagle (2.4 years old). The bone marrow was collected from the epiphysis of the humerus under general anesthesia using 0.2 mg/kg intravenous butorphanol (Butomidor, Richter Pharma AG, Austria) and 15 μg/kg intravenous medetomidin (Cepetor 1 mg/ml ad usum veterinarium, CP-Pharma Handelsges. mbH, Germany) as premedication and a 3 mg/kg intravenous bolus of propofol (Propofol 1% MCT/LCT Fresenius, Fresenius Kabi, Germany) as induction. Anesthesia was maintained with propofol as required. Radiological control was used to evaluate the right position of the standard Jamshidi™ Bone Marrow Biopsy Needles (16 ga). Briefly, the epiphysis of the humerus was palpated and aseptically prepared. A single stab incision was made with a No. 11 scalpel blade through the skin and subcutaneous tissue over the intended bone marrow aspiration site. The bone marrow biopsy needle was inserted into the bone with firm pressure and twisting motion. After penetration of the bone cavity, we aspirated 20 ml of bone marrow. Postprocedural analgesia was provided with single dose of meloxicam 0.2 mg/kg subcutaneous injection (Meloxidyl 5 mg/ml, Ceva Sante Animale, France).
Peripheral blood samples of venous blood were collected from each dog (3 ml) from the jugular vein, and one part (1 ml) was used for standard hematology profiling (IDEXX ProCyte Dx Hematology Analyzer, Idexx Laboratories, USA) and 2 ml as control for PCR array.
Experimental Design and Statistical Rationale
A total of six dogs were included in the study for flow cytometry (n = 6), multilineage differentiation (n = 6), proteomics, and genomic and functional studies (n = 3), all of the latter being biological replicates. The statistical analysis carried out for BMMSC cultivated at different time points (14 h, 24 h, 48 h) was based on multiple-sample ANOVA1 testing with p value = 0.01. Normalization was achieved using the Z-score. These analyses were carried out on Perseus software.
In Vitro Culture of BMMSC
From each dog we obtained 2–5 ml of bone marrow suspension, which was diluted in sterile phosphate-buffered saline solution supplemented with antibiotic (PBS; Gibco, Switzerland) and centrifuged at 500 × g for 10 min. The bone marrow cells (including erythrocytes) were counted using the trypan blue exclusion method and plated at a density of 5 × 107 cells/cm2 in alpha minimum Eagle's medium (Gibco, Switzerland) supplemented with 10% FBS), 100 units/ml penicillin, 100 mg/ml streptomycin, and 2.5 μg/ml amphotericin B (Gibco, Switzerland) in tissue culture flasks T75 and incubated at 37 °C, and 5% CO2. After 2–3 days, nonadherent cells were removed, and adherent MSCs were washed and then cultured under the above-mentioned conditions until they reached ∼80% confluence. MSCs were washed two times with Dulbecco's phosphate-buffered saline and dissociated from the adherent surface with trypsin-EDTA (0.05%, Invitrogen, 0.25% trypsin-EDTA, Thermo Fisher Scientific, USA). The trypsin was neutralized with cell culture media supplemented with 10% FBS, and the cells were centrifuged and counted using the trypan blue exclusion method, and plated at 3.6 × 103 cells/cm2 in T75 flasks. This procedure was repeated until 2–3 passages were completed. Nonadherent cells were removed after 4–5 days by means of medium change, and the remaining cells were fed twice per week. When the cultures reached 80% of confluence, the cells were passaged with 0.25% trypsin/0.53 mm EDTA (Gibco, Switzerland), centrifuged, and replated at a density of 5000 cells/cm2.
BMMSC Conditioned Media Preparation
BMMSC at passage three cultured in DMEM with low glucose and without FBS were incubated in a humidified atmosphere with 5% CO2 at 37 °C for 14 h, 24 h, and 48 h and used for BMMSC conditioned media (BMMSC CM). BMMSC phenotype was confirmed by means of flow cytometry with specific CD markers and three lineage differentiation, and afterward the samples were processed for PCR array profiling, proteomic analyses, and biological function in CAM assay.
Flow Cytometry with Canine CD Markers
Canine BMMSC were sampled to investigate the proportions of CD29 and CD90-positive and CD45-negative cells. Each suspension of cells (1 × 105/100 μl) was incubated with fluorochrome-conjugated monoclonal antibodies: anti-CD45/FITC, anti-CD29/R-phycoerythrin, anti-CD90/allophycocyanin, or isotype-matched control immunoglobulin (IgGs, 1 μg each) diluted in PBS for 45 min at room temperature and in the dark. After incubation, the cells were washed twice with 1 ml PBS (MP Biomedicals, France), followed by 5-min centrifugation at 250 × g. Finally, 100 μl PBS were added and cytometric analysis was performed on a BD FACSCanto™ flow cytometer (Becton Dickinson Biosciences, USA) equipped with a blue (488 nm) and a red (633 nm) laser and six fluorescence detectors. The percentage of cells expressing the individual CD characters was determined by means of dot plotting for the respective fluorescence. The data obtained were analyzed in the BD FACS DivaTM analysis software. For flow cytometry, the following antibodies were employed according to the supplier's recommendations: phycoerythrin anti-human CD29/IgG1 (Clone: TS2/16, human, canine, Sony Biotechnology); FITC anti-dog CD45/IgG2b (Clone: YKIX716.13, BIOPORT, CZ); allophycocyanin anti-dog CD90/IgG2b (Clone: YKIX337.217, BIOPORT, CZ); and their isotype controls: FITC dog IgG (CD29) and phycoerythrin, allophycocyanin dog IgG2b (CD45, CD90) from Biolegend.
Three-Lineage Profile of BMMSC (Osteogenic, Chondrogenic and Adipogenic Phenotypes)
The multilineage potential of canine BMMSC (passages 2–3) was investigated with commercial StemPro Differentiation Kits containing all the reagents required for inducing canine BMMSC into chondrogenic, osteogenic, and adipogenic lineages. Cultures were stimulated with the appropriate differentiation medium for 14 days according to the conditions described in the differentiation protocol for each specific lineage. Afterward, the cultures were fixed with 4% formaldehyde and stained with the following reagents: adipogenic culture with Oil Red, osteogenic culture with Alizarin Red S, and chondrogenic culture with Alcian Blue (all from Sigma-Aldrich, USA).
Analysis of Gene Expression in BMMSCs
Quantitative real-time (RT)-PCR was used to analyze gene expression in BMMSC, and PBMCs were used as comparator (“calibrator”).
First, the cell viability of samples (PBMCs and BMMSC) was determined using the Trypan blue exclusion test. Trypan blue stock solution (0.4%; 10 μl) was added to isolated cells (90 μl) and immediately loaded onto a hemocytometer. The number of blue-stained cells and the number of total cells was determined by visual inspection. Viability was found to be 90.2 ± 8.3% and 92.3 ± 4.8%, respectively.
Total RNA was extracted from the cells (PBMCs and BMMSC) using the RNeasy Mini Kit (Qiagen, Valencia, California) following the manufacturer's instructions. Contaminating genomic DNA was digested using the RNase-free DNase set (Qiagen). The RNA quality and yields were analyzed using the Nanodrop spectrophotometer. Complementary DNA was synthesized using the RT2 First Strand Kit (Qiagen): 1 μg of total RNA was used (after the genomic DNA elimination step) to prepare 20 μl of cDNA. For both cell types (PBMCs and MSCs), three independent RNA isolates were used to prepare three cDNAs.
Quantification of genes of interest in the cDNA samples was performed using the Dog Mesenchymal Stem Cells RT2 profiler PCR array (SABiosciences/Qiagen; catalogue No. PAFD-082ZA). The PCR array contained the canine homologues of human MSC array (SABiosciences/Qiagen, catalogue No. PAHS-082). The array is a 96-well optical microplate containing qPCR primer assays for 84 genes involved in maintaining pluripotency and self-renewal status in human MSCs, five housekeeping genes, and a set of controls (PCR amplification controls, reverse transcription controls, and a genomic DNA contamination control). These genes are human MSC-specific markers that distinguish adult MSCs from embryonic stem cells. The array also included differentiation markers that can be used to monitor early MSC differentiation events. RT-PCR was performed using the SYBR Green qPCR mastermix (SABiosciences/Qiagen) in Light Cycler 480 RT-PCR system (Roche Diagnostics, Rotkreuz, Switzerland). Three cDNA samples were amplified (in three PCR array plates) for both cell types (PBMCs and MSCs) under identical conditions. A quantity of 19 μl cDNA (diluted in an appropriate amount of water and mixed with qPCR mastermix) were dispensed into a 96-well plate and cycled under the following conditions: 95 °C for 10 min (initial denaturation and enzyme activation), followed by 45 cycles of 95 °C for 15 s (denaturation), 60 °C for 30 s (annealing), 72 °C for 20 s (extension), and 75 °C for 20 s (fluorescence acquiring). Amplification specificity was checked by generation of a melting curve.
Array results were analyzed using the web-based data analysis software (comparative ΔΔCt method with amplification efficiency = 2) provided by the manufacturer (SABiosciences/Qiagen, (software version 3.5, available at www.qiagen.com/sk/shop/genes-and-pathways/data-analysis-center-overview-page/). Hypoxanthine-guanine phosphoribosyltransferase 1 transcript quantity was used for normalization of target gene quantity, and the fold change in gene expression (transcript up- or down-regulation) in MSCs compared with PBMCs was calculated.
Proteomic Analyses
Protein Extraction
From each BMMSC CM (n = 3) and time course (14 h, 24 h, 48 h) 1 ml was collected, vacuum-dried, and then taken up in 100 μl of extraction buffer (4% SDS, Tris 0.1 m, pH 7.8). The samples were heated at 95 °C for 15 min and then sonicated for 15 min. Centrifugation was performed at 16,000 × g, 20 °C, for 10 min, and then the supernatant was collected. After the extraction was complete, a Bradford assay was performed to determine the protein concentration in each sample. The samples were kept at −80 °C until further experimentation.
Filter-Aided Sample Preparation Method
The samples were processed using a shotgun bottom-up proteomic approach. All samples were normalized with a final protein concentration of 1 mg/ml in 50 μl. An equivalent volume of reduction solution (dithiothreitol DTT 0.1 m) was added to each sample followed by an incubation step at 56 °C for 40 min. Then the samples were processed following the filter-aided sample preparation protocol (8, 9) using a filter with a nominal molecular weight limit of 30,000 KDa (Amicon Ultra-0.5 30K, Millipore). Briefly, each sample was mixed with 200 μl of denaturant buffer (8 m urea, Tris/HCl 0.1 m, pH 8.5) and transferred to filter-aided sample preparation filters. The samples were centrifuged at 14,000 × g, 20 °C, for 15 min. For the alkylation step, 100 μl of 0.05 m of iodoacetamide in denaturant buffer were added to each sample, followed by incubation in the dark for 20 min at room temperature. Samples were washed twice with 100 μl of denaturant buffer followed by two washes with 100 μl of buffer ammonium bicarbonate (0.05 m). After each washing step, centrifugation was performed at 14,000 × g, 20 °C, for 15 min. The proteins were digested by adding 40 μl of trypsin at 40 μg/ml in buffer ammonium bicarbonate and then incubated at 37 °C overnight. The peptides were eluted by adding 50 μl of saline solution (NaCl 0.5 m) and centrifuged at 14,000 × g, 20 °C, for 15 min. The digestion was stopped by adding 10 μl of TFA 5%. The samples were desalted using ZipTip C-18 (Millipore) and eluted with a solution of acetonitrile/0.1% TFA (7:3, v/v). The samples were dried with SpeedVac and resuspended in 20 μl of acetonitrile/0.1% formic acid (0.2:9.8, v/v) just before processing using LC-MS/MS.
LC-MS/MS
The analysis of digested proteins was performed using a nano Acquity UPLC system (Waters) coupled with a Q-Exactive Orbitrap mass spectrometer (Thermo Scientific) via a nanoelectrospray source. The samples were separated by means of online reversed-phase, using a preconcentration column (nanoAcquity Symmetry C18, 5 μm, 180 μm × 20 mm) and an analytical column (nanoAcquity BEH C18, 1.7 μm, 75 μm × 250 mm). The peptides were separated by applying a linear gradient of acetonitrile in 0.1% formic acid (5–35%) for 2 h, at a flow rate of 300 nl/min. The Q-Exactive was operated in data-dependent mode defined to analyze the 10 most intense ions of MS analysis (Top 10). The MS analysis was performed with an m/z mass range between 300 to 1600, resolution of 70,000 full width at half maximum, automatic gain control of 3e6 ions and maximum injection time of 120 ms. The MS/MS analysis was performed with an m/z mass range between 200 to 2000, automatic gain control of 5e4 ions, maximum injection time of 60 ms, and resolution set at 17,500 full width at half maximum.
Data Analysis
The proteins were identified by comparing all MS/MS data with the proteome database of Canis lupus familiaris (Uniprot, release December 2018, 25,493 entries), using the MaxQuant software version 1.5.8.3 (10, 11). The digestion parameters were defined using trypsin with two maximum missed cleavages. The oxidation of methionine and N-terminal protein acetylation were defined as variable modifications. The carbamidomethylation of cysteine was chosen as fixed modifications. The label-free quantification was done keeping the default parameters of the software. As for initial mass tolerance, 6 ppm was selected for MS mode, and 20 ppm was set for fragmentation data with regard to MS/MS tolerance. The identification parameters of the proteins and peptides were performed with a false discovery rate at 1%, and a minimum of two peptides per protein in which one was unique. The statistical analysis was done by Perseus software (version 1.6.2.1). Briefly, the label-free quantification intensity of each sample was downloaded in Perseus, and the data matrix was filtered by removing the potential contaminants (12), reverse and only identified by site. The data were then transformed using the log2(x). Before statistical analysis, three groups were defined with three replicates per group. A multiple-sample test was performed using ANOVA with a p value of 0.01, and the results were normalized by Z-score and represented as a hierarchical clustering.
Angiogenic Assays
Chorioallantoic Membrane (CAM) Assay
CAM assay was used to evaluate the angiogenic response to CM derived from BMMSC. Fertilized chicken hybrid (Gallus gallus) eggs, Ross 308 (n = 30) were purchased from a commercial farm (Parovske Haje, Nitra,SK) and delivered via courier in a temperature-controlled manner to ensure egg viability and quality. The eggs were incubated blunt end up in a forced-draft, constant-humidity incubator at 37.5 °C and 60% relative humidity until embryonic day 3 of the incubation period. At embryonic day 3, the eggs were windowed on the blunt end, and the albumen (egg white, 2 ml) and the inner shell membrane (membrana papyracea) were carefully removed. The windows were closed using insulation tape and returned to a still-draft incubator for re-incubation until the day of implantation (37.5 °C; 70% relative humidity, without rocking). On embryonic day 6, a sterilized silicone ring (d = 5 mm) was gently situated on the chorioallantoic membrane using suture tying forceps. In the control group (n = 10) 30 μl of DMEM and in the experimental group (n = 10) 30 μl of CM were injected in the space bordered by silicone ring. For visual evaluation of vascular density, we used a stereomicroscope (Leica MZ125) fitted with a Digital Single Lens Reflex (DSLR) camera (Nikon D7000, Nikon, Tokyo, Japan) for photo documentation. Photo documentation of vascular density was done at the moment of application 0 h and again after 72 h.
The number of vessels growing within the area outlined by the ring where BMMSC CM was applied was counted blinded using program FIJI ImageJ. First, we had to convert each image to 8-bit quality, and we used the AutoLocalTrashold and Otsu function, which highlights the vessels. To count the numbers of vessels and their branches in the experimental area (bordered by the silicone ring) we used the Cell Counter function, which counts every marked object (vessel or branch) automatically. Statistical analysis was carried out using Graph Pad InStat software. Data are presented as mean ± S.D. from 10 CAM. Mean values in different groups were statistically compared using one-way ANOVA and Tukey's post hoc tests. Values of p < 0.05 were considered statistically significant (*p value of < 0.05, **p value of < 0.01, ***p < 0.001).
Analysis of Angiogenesis-Related Gene Expression in the CAM Model
Quantitative RT-PCR was used to analyze gene expression of angiogenesis-related genes in CAM assay.
The CAM from the surviving chicken embryos was collected after 72 h from the control (n = 5) and experimental application (n = 9) and used for analysis of gene expression. Total RNA was extracted using the TRIreagent LS method (Sigma, Cat. T3934). CAM membranes (250 μl) were homogenized with 30 strokes in 750 μl of TRIreagent LS using a 1-ml Dounce glass homogenizer with tight pestle (Wheaton, USA). Following the cleanup, the resulting RNA pellet was dissolved in 50 μl of RNase-free water (Qiagen, Cat. 129112) and stored at −80 °C. Concentration of extracted RNA was spectrophotometrically measured with a nanophotometer (Implen, Germany), and samples were diluted to 4 μg/8 μl prior to reverse transcription. Coding DNA was synthesized using an RT2 first strand kit (Qiagen, Cat. 330401) according to the manufacturer's protocol, including the genomic DNA elimination step. Gene expression analysis was performed using the Chicken Angiogenesis RT2 Profiler PCR Array (Qiagen, Cat. 330231 PAGG-024Z). The composition of each 25 μl qPCR reaction/well was as follows: 12.5 μl RT2 SYBR Green qPCR Mastermix, 12 μl nuclease-free water, and 0.5 μl cDNA (200 ng/μl). The sealed PCR plate was mixed, centrifuged, and processed 7500 RT-PCR system (Applied Biosystems, USA). Cycling conditions were as follows: initial denaturation 10 min at 95 °C, 42 cycles of 15 s at 95 °C, and 1 min at 60 °C. To check the specificity of amplified PCR product, a melt curve analysis was performed.
Out of the 89 analyzed genes, two were not amplified (Csf3 and Egf). Fold change of gene expression in the CAM-treated sample was compared with the control membrane and calculated using the delta-delta-Cycle threshold method with Hmbs, Rpl4, and Ubc as housekeeping reference genes. Genes with fold change ≥2 were defined as differentially expressed.
RESULTS
Flow Cytometry with Canine CD Markers
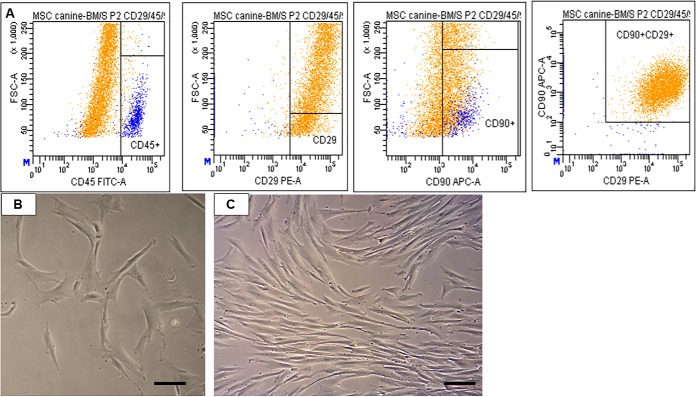
After an increasing number of passages, canine BMMSCs showed enhanced expression of mesenchymal-specific surface markers, while the hematopoietic decreased. The flow cytometry analyses revealed that cells at passages 2–3 expressed primarily CD29 (98.9% ± 0.8) and CD90 (59.1% ± 4.7), CD29+CD90+ (79% ± 2.4) (but low CD45 (2% ± 1.2), as shown by the representative flow cytometry results of canine BMMSC in Fig. 1A. The cultured cell population at passage 3, day in vitro 1 contained attached spindle-shaped and fibroblastic-like cells (Fig. 1B), which reached confluence approximately after 4–6 days of cultivation (Fig. 1C).
Fig. 1.
Representative flow cytometry analyses CD29 (98.9% ± 0.8), CD90 (59.1% ± 4.7), CD29 + CD90 (79% ± 2.4), CD45 (2% ± 1.2) for canine MSC at passage 2 (A). Morphology of cultured canine MSC at passage 3, day in vitro 1 (B) and at the stage of 80% confluency day in vitro 4 (C). Scale bars B, C = 50 μm.
Multilineage Potential
Three-lineage potential was tested with commercial StemPro® Differentiation Kits. We confirmed that canine BMMSC after 14 days of incubation in specific differentiation medium underwent a high degree of biomineralizing osteogenesis with visual staining of calcium deposits expressing Alizarin Red positivity (Fig. 2A). Furthermore, chondrocytes migrating from spherical chondrocyte-like aggregates revealed intense Alcian Blue staining, which is typical for chondrogenesis (Fig. 2B). In contrast to the high osteogenic and chondrogenic differentiation of canine MSCs, we found a low degree of adipogenesis, with limited vacuole formation and Oil Red O staining (Fig. 2C).
Fig. 2.
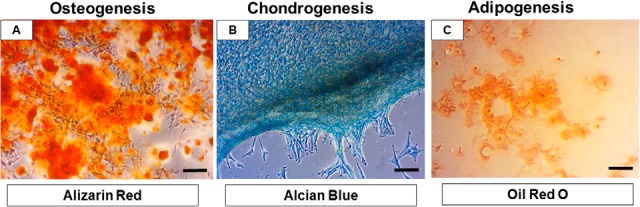
Micrographs documenting multipotent characteristics of canine MSCs, differentiated into osteocytes (A, Alizarin Red), chondrocytes (B, Alcian Blue), and adipocytes (C, Oil Red O). Scale bars A, B, C = 50 μm.
Gene Expression in Canine BMMSC
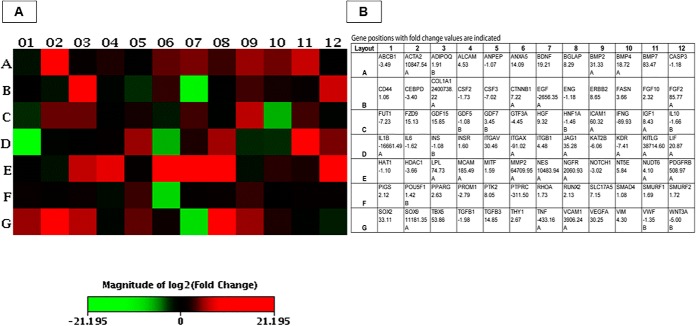
We analyzed gene expression in BMMSC and PBMC samples from the Alaskan malamute and American Staffordshire terrier, and we obtained similar results. We found different expression only in four genes: BGLAP, HGF, IGF1, and LPL were up-regulated in the Alaskan malamute but not the American Staffordshire terrier. Results obtained from the Alaskan malamute are presented in Fig. 3. Eight genes were expressed in MSCs but not in PBMCs: BMP7, ERBB2, GDF7, KITLG, SOX2, SOX9, TBX5, and COL1A1. From these genes, SOX2 is a typical stemness maker, while BMP7, ERBB2, and KITLG are MSC genes and GDF7 and SOX9 are specific chondrogenesis differentiation markers. Five genes were highly significantly (p < 0.001***) up-regulated in MSCs compared with PBMCs. Highest expression was registered for ACTA2 (10,847-fold), falling to the category of MSC differentiation markers followed by overexpression of stemness markers (Table I): ANXA5 (14-fold), FGF2 (85-fold), ITGAV (30-fold), and VEGFA (30-fold). Expression of several other genes was significantly (p < 0.01** or p < 0.05*) changed. Many genes were up-regulated: BDNF (19-fold), BGLAP (8-fold), BMP2 (31-fold), BMP4 (18-fold), FZD9 (15-fold), GDF15 (15-fold), HGF (9-fold), ICAM1 (60-fold), IGF1 (8-fold), JAG1 (35-fold), LIF (20-fold), LPL (74-fold), MCAM (185-fold), MMP2 (64,709-fold), NES (10,483-fold), NGFR (2060-fold), PDGFRB (508-fold), PTK2 (8-fold), TGFB3 (14-fold), and VCAM1 (3906-fold). In contrast, there were also genes showing down-regulation: EGF (2656-fold), IFNG (89-fold), IL1B (16,661-fold), ITGAX (91-fold), PTPRC (311-fold), and TNF (433-fold).
Fig. 3.
Summary heatmap of differentially expressed transcripts, heatmap indicates the patterns of expression for 84 selected genes in dog BMMSCs compared with PBMCs (a panel of genes analyzed in the dog mesenchymal stem cells RT2 profiler PCR array). Up-regulated genes are marked red, down-regulated genes are marked green (A). Gene names and their positions in the heatmap, and fold-change values (expression in MSCs compared with PBMCs) are indicated in the table (B).
Table I. Identification of stemness markers and specific mesenchymal and differentiation markers in canine MSCs.
| Stemness markers | Specific MSCs genes | Other MSC genes | Differentiation MSCs genes | Osteogenesis/Os Adipogenesis/AD Chondrogenesis/CH Myogenesis/MY Tendon Development/TD |
|---|---|---|---|---|
| SOX2 | ERBB2 | BMP7 | GDF7 | CH |
| FGF2 | ITGAV | KITLG | SOX9 | CH |
| LIF | FZD9 | BGLAP | TBX5 | OS |
| MCAM | HGF | ACTA2 | MY | |
| NGFR | ICAM1 | BMP2 | OS CH | |
| PDGFRB | IGF1 | BMP4 | CH TE | |
| VCAM1 | MMP2 | GDF15 | CH TE | |
| NES | JAG1 | MY | ||
| TGFB3 | LPL | AD | ||
| PTK2 | OS |
Proteomic Analyses
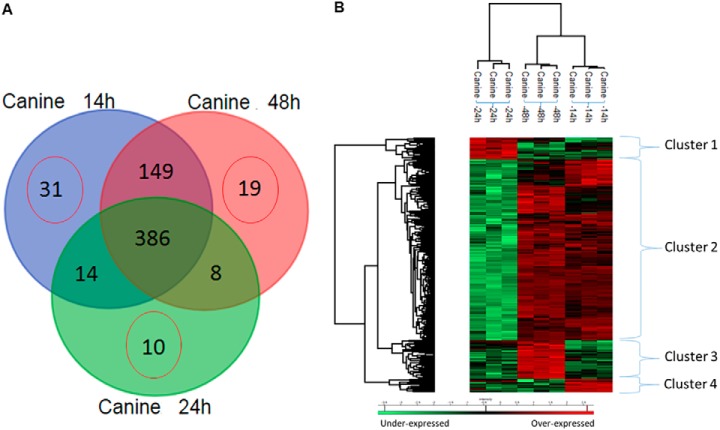
BMMSC CM collected at three different times (14 h, 24 h, and 48 h) were subjected to protein extraction followed by shotgun proteomic analyses (Supplemental Data 1. Using triplicate analyses we identified 1355 significant grouped proteins based on two peptides minimum, false discovery rate < 1% using the Canis lupus bank (507 proteins for the canine MSC isolated at 14 h, 487 proteins at 24 h, and 3 proteins at 48 h) (Supplemental Data 2–4). After statistical analysis (p value 0.01), 529 proteins were identified with 19 unique proteins at 14 h, 9 unique proteins at 24 h, and 10 unique proteins at 48 h (Supplemental Data 2–4). Three hundred and thirty five proteins were common to all three conditions, and 140 proteins were identified in common between 14 h and 48 h, 13 proteins between 14 h and 24 h, and 3 proteins between 24 h and 48 h (Fig. 4A, Supplemental Data 2–4).
Fig. 4.
Proteomic analyses. (A) Venn diagram of shotgun proteomic analyses of proteins issued from bone marrow Stem cells conditioned medium collected at 14 h, 24 h, and 48 h. Specific proteins to each condition have been highlighted as well as the ones in common between two or three conditions. (B) Shotgun proteomic analyses of proteins issued from bone marrow Stem cells conditioned medium collected at 14 h, 24 h, and 48 h. Representative heatmap of the common proteins and quantified by label free and analyses by MaxQuant with a p value = 0.01.
Proteins after 14 h Incubation
Among the 20 specific proteins identified at 14 h, 10 proteins were involved in the transcription and translation network, namely ubiquitin thiolesterase 3, constitutive photomorphogenesis 9 signalosome subunit 5, eukaryotic translation initiation factor 3 subunit F, ribosomal protein S8, S16, and S19, non-POU domain-containing octamer-binding protein (Nono), purine nucleoside phosphorylase, EAD (Asp-Glu-Ala-Asp), box helicase 42 (DDX42), and 60S acidic ribosomal protein P0. Furthermore, five proteins were involved in the immune network (myeloid marker CD109, tyrosine-protein phosphatase non-receptor type 11, matrin 3, adaptor-related protein complex 2, tripeptidyl peptidase II, and integrin alpha V).
Proteins after 24 h Incubation
In the canine MSC conditioned medium following 24-h incubation, growth factors were identified (osteomodulin, oncoprotein induced transcript 3, quiescin Q1 gene) as well as the enzymes lysyl oxidase-like, sulfhydryl oxidase 1, bis (5′-nucleosyl)-tetraphosphatase, and the interleukin 1 receptor accessory protein. At 48 h, four proteins were involved in metabolism of RNA (splicing factor 3b, serine and arginine rich splicing factor 2, eukaryotic translation initiation factor 3 subunit J, and eukaryotic translation initiation factor 4B). Furthermore, among the identified proteins, some were involved in cytoskeleton (SWAP switching B-cell complex) and aminopeptidase 1-like protein.
To better understand the modulation registered in relation to the three different times of canine MSC collection, ANOVA tests with a p value of 0.01 were performed with nonsupervised clustering of samples. Two branches were obtained separating the CM obtained at 24 h from the ones collected at 14 h and 48 h. Two subbranches separated the medium collected at 14 h from the one at 48 h. The 24 h seems as an outlier but can be explained by the nature of the proteins overexpressed in that peculiar cluster. In fact, four main clusters were highlighted (Fig. 4B). Cluster 1 corresponded to proteins overexpressed in CM obtained at 24 h, with 28 proteins identified (Supplemental Data 5). Spondin (1 2), glycans proteins (biglycan, glycan 1) and proteins involved with the cytoskeleton (collagen, type XII, microtubule-actin crosslinking factor 1, laminin, beta 2, fibronectin, tubulin alpha chain, cadherin 5) and cell growth regulator (HtrA serine peptidase 1) were detected (Supplemental Data 5). Cluster 2 was the biggest of the four, with 220 proteins, and it was common to CM collected at 14 h and 48 h (Supplemental Data 6). Five major KEGG pathways (13) were identified using String protein software, corresponding to the pathways sharing the highest e-value corresponding to the proteasome: protein processing in the endoplasmic reticulum, focal adhesion, biosynthesis of amino acid residues, and antigen processing and presentation. System biology analyses revealed that the proteins in Cluster 1 were involved in neurite outgrowth, cell growth, vascularization, and cell migration (Supplemental Fig. 1). Cluster 2 proteins were involved in ischemia and exocytosis (Supplemental Fig. 1), Clusters 3 and 4 were involved in cell differentiation, migration, and adhesion and developmental process (Supplemental Fig. 1). Reactome analysis (14) revealed that hedgehog ligand biosynthesis as well as the immune response pathways were the most predominant, with an e-value of 3.10−11. Among the proteins identified, some were of interest such as mesencephalic astrocyte-derived neurotrophic factor (MANF), which is known to selectively promote the survival of dopaminergic neurons in the ventral midbrain (15). Karyopherin-2, a member of the importin family, serves as an adaptor protein in the mediation of cargo-specific synapse-to-nucleus transport (16). Glia maturation factor (GMF) is a neurotrophic factor implicated in nervous system development, angiogenesis, and immune function (17). Osteonectin (secreted protein, acidic, cysteine-rich) is involved in embryogenesis and can modulate cytokines (18).
Cluster 3 (Supplemental Data 7), specific for CM collected at 48 h contained proteins involved in the TGF beta pathway, e.g. S-phase kinase-associated protein 1, follistatin, and latent transforming growth factor beta binding protein 1 (LTBP1) and 4. Proteins involved in the regulation of the insulin growth factor were also detected e.g. protease serine 23, follistatin-like 1, follistatin, and LTBP1. Interestingly, Dickkopf WNT signaling pathway inhibitor 3, known to be involved in embryonic development through its interactions with the Wnt signaling pathway, was found with neudesin neurotrophic factor, tissue inhibitor of metalloproteinases-2 (TIMP-2), neurogenesis and neurotophic factors (19). The sushi, nidogen, and EGF-like domains (SNED1) protein is also known as an extracellular matrixcomponent. SNED1 knockout mouse strains have revealed that SNED1 is required during neonatal survival and development, especially in the development of the skeleton and neural crest-derived craniofacial structures (20). The last Cluster 4 (Supplemental Data 8) contained osteogenic factors involved in differentiation of MSCs, such as the osteoblast-specific factor and osteonectin-2 (21). Twinfilin actin-binding protein 1 is a regulator of cytoskeletal dynamics. Serine/arginine-rich splicing factor 1 is involved in osteopontin splicing (22). Four proteins were involved in the same pathway related to translation (heterogeneous nuclear ribonucleoprotein A/B, ubiquitin-conjugating enzyme E2, phosphatidylethanolamine-binding protein 1, serine/arginine-rich splicing factor 1). Taken together, in the four clusters obtained from proteomic analysis, several growth factors linked to MSCs were detected. However, compared with transcriptomic data, only TGF beta 1 was detected.
Effects of BMMSC CM on Angiogenesis in CAM Assay
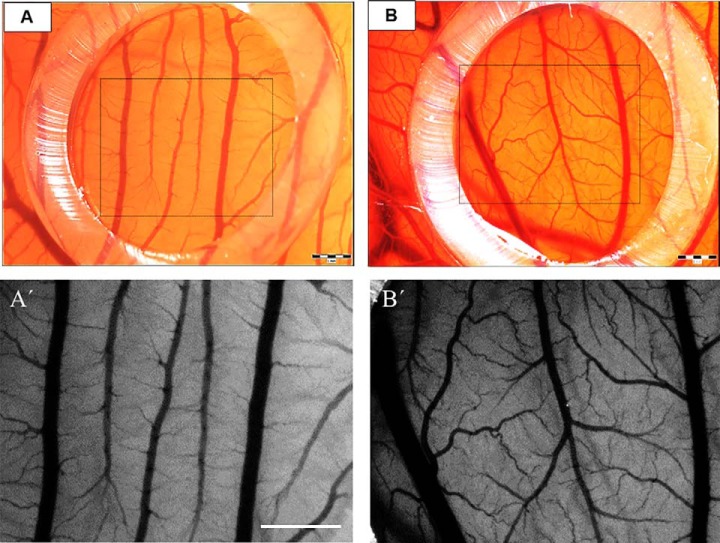
The changes in the vasculature of the CAM treated with BMMSC CM showed a positive response, with enhanced density of vessels (47.250 ± 9.7) after 72 h (Figs. 5B, 5B′, and 6). The number of vessels and bifurcations was higher compared with control (medium only, 27.625 ± 7.981) after same time (Figs. 5A, 5A′, and 6). In addition, the intervascular distance was lower, and the large vessels seemed to further enhance their diameter (Fig. 5B′). Mean values in different groups were statistically compared using one-way ANOVA and Tukey's post hoc tests, with significant differences comparing values for Control 0 h versus Experiment 72 h (*p value of < 0.05); EXP 0 h versus EXP 72 h (**p value of < 0.01) and CTR 72 h versus EXP 72 h (***p < 0.001) (Fig. 6).
Fig. 5.
The angiogenic profile in CAM assay exposed to DMEM (A, A′) and BMMSC CM (B, B′) at 72 h. Note the enhanced density of vessels in the angiogenesis zone (outlined boxed area in A, B) treated with BMMSC CM when compared with DMEM. Detail of numerous vessels and bifurcations in higher magnification (A′) compared with control (B′). Scale bars = 1 mm (A, A′, B, B′).
Fig. 6.
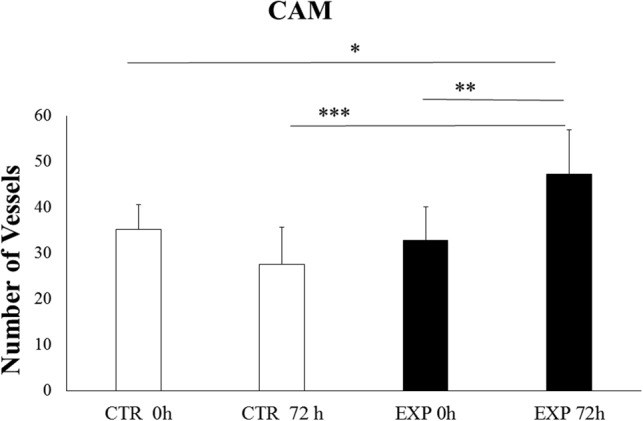
Quantification of vessels in CAM assay exposed to DMEM (A, A′) and BMMSC CM (B, B′) at 0 h and 72h. Mean values among different groups were statistically compared by one-way ANOVA and Tukey's post hock tests, with a p value = 0.05. CTR 0 h versus EXP 72 h (*p value of < 0.05); EXP 0 h versus EXP 72 h (**p value of < 0.01) and CTR 72 h versus EXP 72 h (***p < 0.001).
Angiogenesis-Related Genes Expression in CAM
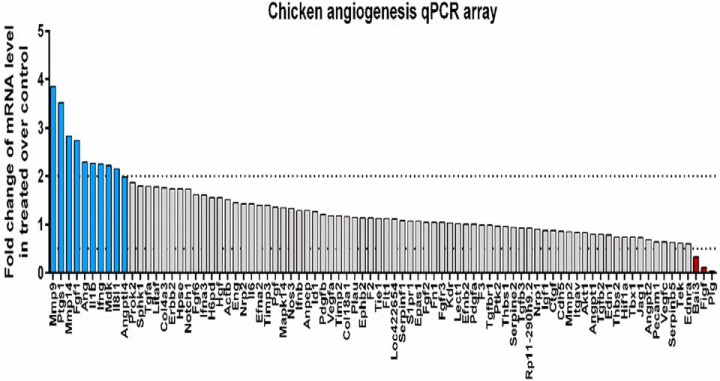
In order to define the changes after the treatment of CAM with BMMSC CM at the molecular level, we performed transcriptomic analysis targeting angiogenesis-related genes. Using a quantitative PCR array, we profiled a panel of genes involved in the development of new blood vessels, tissue remodeling, growth, inflammation, and signaling. Comparative analysis revealed 10 up-regulated and 3 down-regulated genes in the BMMSC CM-treated chicken chorioallantoic membrane when compared with control after 72 h. The group of genes with increased expression included matrix metalloproteinases: Mmp9 (3.9-fold) and Mmp14 (2.8-fold); stimulators of angiogenesis and endothelial cell outgrowth: Ptgs1 (3.5-fold), Fgf1 (2.7-fold) Ang (2.3-fold), and Mdk (2.2-fold); angiogenesis inhibitor: Angptl4 (2-fold); and modulators of inflammatory signaling: Il1b (2.3-fold), Ifng (2.3-fold), and Il8l1 (2.1-fold). The identified down-regulated genes are involved in the inhibition of angiogenesis: Bai3 (-3.1-fold) and Plg (-23-fold) and stimulation of vascular endothelial growth: Figf (-8.8-fold) (Fig. 7). In summary, transcriptomic profiling of angiogenesis-related genes following BMMSC CM treatment of CAMs revealed a predominant pro-angiogenic phenotype characterized by up-regulation of numerous angiogenic molecules (Ptgs1, Fgf1, Ang, Mdk) and simultaneous down-regulation of angiogenic inhibitors (Plg and Bai3). Experimental treatment of chicken CAM with BMMSC CM was also associated with up-regulation of genes controlling new blood vessel outgrowth (Angptl4, Mmp9, Mmp14) and modulators of inflammatory signaling (Il1b, Ifng, Il8l1) (Fig. 7). Proteomic data revealed the presence of angiogenic factors only 14 h after incubation. Among these factors MMP2 was detected, which like MMP9 is known to be involved in angiogenesis (23). The protein Myo1c, which is necessary to recruit to blood vessels together with vascular endothelial growth factor VEGFR2 (24), was also present, as well as the fibroblast-activating protein alpha, which is known to be associated with and to upregulate VEGF-A expression (25). VCAM1 and AngptL3, two factors involved directly in blood vessel formation (26, 27) were also detected. Altogether, BMMSC CM are clearly associated with the genes and proteins involved in angiogenesis.
Fig. 7.
Expression of angiogenesis-related genes. Transcriptomic profile of BM-MSC CM-treated chicken chorioallantoic membrane reveals up-regulation (blue columns) of 10 genes (Mmp9, Ptgs1, Mmp14, Fgf1, Ang, Il1b, Ifng, Mdk, Il8l1, Angptl4) and down-regulation (red columns) of three genes (Bai3, Figf, and Plg) when compared with control untreated CAM as quantified by qPCR chicken angiogenesis array (dot line represents threshold for ≥ twofold change).
DISCUSSION
Cell therapy in veterinary practice is becoming common practice, but it is often accompanied with ambiguities and contradictory results. This may be caused by the delivery of uncharacterized canine stem cell sources targeting a wider range of different diseases (1). Nowadays, we know that procedures for isolation, cultivation, freezing, and thawing of canine MSCs differ between laboratories (2), and therefore to achieve successful therapeutic efficacy of cell-based therapies, we need to know their bioactive potential under well-defined standardized protocols. Currently, MSCs isolated from fat tissue, synovia, umbilical cord, and placenta are being applied for hard tissue repair as well as for cardiovascular or neurological disorders (1). However, the conditioned medium isolated from canine MSCs is more privileged because it contains extracellular vesicles filled with a cargo of proteins, as well as important regulatory molecules (mRNA and miRNA). Although cell therapy is developing, there is still a lack of detailed studies unraveling the biological properties of MSCs derived from different sources, while cultivated under specific conditions.
In our study, therefore, under well-defined standardized protocols, the minimal characteristics of BMMSC (adherence, CD markers, and three-line differentiation) were combined with their gene expression, their proteome profile, and their biological function. We believe that it is necessary to standardize the isolation protocols ensuring the quality of both cells and their CM. In our study, we determined the basic criteria for donors, such as age, sex, breeds of dogs, health parameters, bone marrow source (humerus), and maximum time for tissue processing. Some of the most popular medium and large breeds of dogs were included. They were chosen because of their relatively long lifespan and the possibility of obtaining from them enough bone marrow and peripheral blood for further studies. First, we selected 10 dogs with matching age (between 2–3 y), weight (over 10 kg), and gender criterion (males), from which two dogs were excluded due to changes in their complete blood count from normal patterns (eosinophilia, allergies) and another two dogs because of their poor in vitro proliferation rate. Although the final number of dogs enrolled in our study was decreased to six, these dogs served as good candidates for multicharacterization of these popular breeds in order to confirm their similarities/differences.
We isolated BM from the humerus using standard biopsy sets. The operations were performed by the same surgeon, using identical guidelines for bone marrow withdrawal for each donor. The time for completion of in vitro tissue processing was a maximum of 5 h from collection.
We did not notice any side effects that might be related to the BM withdrawal procedure, such as pain, motor-sensory impairment, or inflammation. Morphologically, there were no significant differences between the cells. Initially, they exhibited spindle shape, leading into homogenous fibroblastic populations. We observed some differences in flow cytometry parameters, with some decrease in the CD90 surface marker with increasing passage number (above P3), but CD29 and CD45 markers remained stable. We assume that the canine BMMSC surface marker CD90 is more sensitive to enzymatic digestion and may be degraded after passaging (28). However, this change did not mean that the canine MSCs were losing their multipotency, which was confirmed by three-lineage characteristics. Here we used a commercial differential kit that confirmed marked osteogenic and chondrogenic differentiation. On the other hand, we were unable to induce adipogenesis, similarly as in another study (4).
After confirming minimal mesenchymal stem cell properties for canine MSCs, we performed gene expression characterization. Among the 84 genes we confirmed, only one gene specific for pluripotency (SOX2), which indicates the ability of canine BMMSC to induce self-renewal. Since no other stemness markers were overexpressed, this suggests a very low risk of developing tumorigenity (29). Furthermore, we recorded expression of 16 genes specific for canine MSCs and 9 specific differentiation genes for bone (BMP2, TBX5, PTK2) cartilage (GDF7, SOX9, BMP4, GDF15), and tendons (BMP4, GDF15). These genes directly correlate with our results, showing strong osteogenic and chondrogenic potential in vitro, thus supporting their key role in the recovery of bone, cartilage, and tendons, and they are used for treating various degenerative diseases of the locomotor system in dogs and horses (29). In addition, we also noticed overexpression of genes for several growth factors, FGF2, LIF, HGF, ICAM1, IGF1, TGFB3, and receptors (NGFR, PDGFRB) that are important for angiogenesis, the recovery of heart, and muscle (ACTA2, JAG1). MSCs are able to secrete proregenerative mediators, including chemokines, cytokines, and growth and neuroprotective factors, as well as immune-modulatory proteins affecting the adaptive and the innate cells (1, 30, 31). Thus the therapeutic delivery of MSC CM collected from expansion media has been investigated under in vitro and in vivo conditions (32–34). In line with these findings, we studied the dynamics of bioactive molecules released from canine BMMSC CM via proteomic profiling. Our data confirm that, during a two-day cultivation period, valuable molecules were released that may act through paracrine mechanisms in order to regulate regeneration processes. During 14-h cultivation, we identified proteins involved in the transcription and translation network, the immune network, and metabolism processes. Later, at 24–48 h, we found various growth factors, including neurotrophic growth factors, playing a role in neuron development and angiogenesis, as well as proteins involved in the cytoskeleton that correlate with previous studies (7).
Among the proteins identified in proteomic analysis as produced by canine MSC, some are related to neurotrophic factors (MANF, GMF, neudesin neurotrophic factor, TIMP-2, Spondin-2) and some to osteogenic factors (osteonectin-1 and 2), while others are mainly interesting for different reasons such as angiogenesis (Angioptl3, Myo1C, Vcam1, MMP2). MANF has been shown to affect stem cells directly. In fact, during embryonic development, MANF is required for neuronal migration and neurite outgrowth (35). In a rat stroke model, MANF increased the number of newborn neurons (Doublecortin X+ cells) found in the lesion area by stimulating cell migration but not NSC proliferation (36). This finding demonstrates the neuroregenerative activity of MANF, which may improve the effectiveness of endogenous repair and stem-cell-based therapies for damaged central nervous tissue. Only MANF can perform this activity because other classical neurotrophic factors such as glial cell line-derived neurotrophic factor mainly stimulate the proliferation of NSC in neurogenic zones (36). GMF is considered as a cytokine-responsive protein in erythropoietin-induced and granulocyte-colony stimulating factor-induced hematopoietic lineage development (37). It consists of two compounds: glia maturation factor-β (GMFB) and GMF-γ, while GMFB may play both protective and detrimental roles in the progression of various neuroinflammatory and neurodegenerative diseases (38). In addition, infusion of GMFB into the cavities of injured nervous tissue stimulated dendritic outgrowth and hypertrophy of specific neurons (38, 39). NENF is known to be expressed abundantly in the developing brain and spinal cord in embryos, and its neurotrophic activity may provide new insights into the development and maintenance of neurons (40). TIMP-2 is a member of the matrix metalloproteinases (MMPs) inhibitor family (41). MMP-2 for instance is inhibited by TIMP-2. MSCs are known to inhibit high levels of exogenous MMP-2 and MMP-9 through TIMP-2 and TIMP-1, respectively. Canine MSCs are revealed to be a good source of TIMP-mediated MMP inhibition, which can be highly useful for treating pathologies such as cancer. Antigen protein CD109 shows an interesting role. It is a glycosylphosphatidylinositol anchored cell-surface glycoprotein and is a member of the alpha-2-macroglobulin/C3,C4,C5 family (42). CD109 is known to be expressed at least by CD34+ bone marrow mononuclear cells and mesenchymal stem cells in humans (42, 43). Proteins related to osteonectin, osteomudulin, and spondin-2 are known to be implicated in osteoblastogenesis through the Wnt pathway (44). However, spondin-2 is also involved in midbrain dopaminergic neurogenesis and differentiation (45). Oncoprotein-induced transcript 3 protein is normally known to be produced in hepatocytes and down-regulated in hepatocellular carcinoma, but it is also present in BMMSC CM (46). SNED1 is another factor found in canine MSC CM. Recently, it has been demonstrated that SNED1 is a crucial factor during development and neonatal survival (20). We demonstrate that some of the factors released by BMMSC CM are associated with angiogenic activity in the CAM assay. BMMSC CM showed a positive response, with enhanced density of vessels, their numbers, and bifurcations. This was confirmed by transcriptomic and proteomic analyses confirming the presence of specific angiogenic factors. Thus, BMMSC CM could become a promising tool for ocular surface disorders. In fact, in cardiac ischemia repair, BMMSC CM can be used to stimulate neovascularization of infarct tissue through upregulating VEGF to improve cardiac function. Moreover, BMMSC CM could be an additional important regenerative medium for proangiogenesis, forming a provisional granulation matrix in the proliferation phase of wound healing (32, 33).
This is the first comprehensive study revealing canine BMMSC multilineage properties linked to the corresponding gene expression followed by proteomics profiling of dynamically released molecules from BMMSC CM and their pro-angiogenic effect, which may have an important correlation to their possible therapeutic function and clinical application. Before this analysis, only a few studies had attempted to identify cell surface markers and mRNA expression profiles for different MSCs (47). Importantly, here we determine the canine BMMSC properties applicable for replacement therapies in osteo-chondral disease as well as other tissues due to their capability of releasing immunomodulatory, angiogenic, and bioactive molecules via CM. By means of this comprehensive study, we demonstrate that canine BMMSC, similarly to human MSCs, under uniform characterization criteria represent an interesting source of stem cells that could be available for therapeutic and clinical application in veterinary practice.
Data Availability
Proteomic datasets including MaxQuant files and annotated MS/MS datasets were uploaded to the ProteomeXchange Consortium via the PRIDE database and then assigned the dataset identifier PXD013058.
Supplementary Material
Footnotes
* This research was supported by: APVV 15-0613 (DC), IGA UVLF 06/2018 “Influence of Regeneration Capacity of Nervous Tissue in vitro through Adult Stem Cell Products” ERANET-AxonRepair (DC), grants from Stefanick & INSERM (MS), SIRIC ONCOLille, Grant INCD a-DGOS-Inserm 6041aa (IS), VEGA 2/0146/19 (DC), and VEGA 1/0050/19 (EP). The authors declare no conflict of interest.
 This article contains supplemental material Supplemental Data 1–8 and Fig. 1.
This article contains supplemental material Supplemental Data 1–8 and Fig. 1.
1 The abbreviations used are:
- ANOVA
- analysis of variance
- BMMSC
- bone marrow-derived mesenchymal stem cells
- CD
- cluster of differentiation
- PBMCs
- peripheral blood mononuclear cells.
REFERENCES
- 1. Chow L., Johnson V., Regan D., Wheat W., Webb S., Koch P., and Dow S. (2017) Safety and immune regulatory properties of canine induced pluripotent stem cell-derived mesenchymal stem cells. Stem Cell Res. 25, 221–232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Kriston-Pál É., Czibula Á., Gyuris Z., Balka G., Seregi A., Sükösd F., Süth M., Kiss-Tóth E., Haracska L., Uher F., and Monostori É. (2017) Characterization and therapeutic application of canine adipose mesenchymal stem cells to treat elbow osteoarthritis. Can. J. Vet. Res. 81, 73–78 [PMC free article] [PubMed] [Google Scholar]
- 3. Shao J., Zhang W., and Yang T. (2015) Using mesenchymal stem cells as a therapy for bone regeneration and repairing. Biol. Res. 48, 62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bearden R. N., Huggins S. S., Cummings K. J., Smith R., Gregory C. A., and Saunders W. B. (2017) In-vitro characterization of canine multipotent stromal cells isolated from synovium, bone marrow, and adipose tissue: A donor-matched comparative study. Stem Cell Res. Ther. 8, 218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Russell K. A., Chow N. H., Dukoff D., Gibson T. W., LaMarre J., Betts D. H., and Koch T. G. (2016) Characterization and immunomodulatory effects of canine adipose tissue- and bone marrow-derived mesenchymal stromal cells. PLOS ONE 11, e0167442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Carvalho J. L., Braga V. B., Melo M. B., Campos A. C., Oliveira M. S., Gomes D. A., Ferreira A. J., Santos R. A., and Goes A. M. (2013) Priming mesenchymal stem cells boosts stem cell therapy to treat myocardial infarction. J. Cell Mol. Med. 17, 617–625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Krešić N., Šimić I., Lojkić I., and Bedeković T. (2017) Canine adipose derived mesenchymal stem cells transcriptome composition alterations: A step towards standardizing therapeutic. Stem Cells Int. 2017, 4176292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Wiśniewski J. R., Zougman A., Nagaraj N., and Mann M. (2009) Universal sample preparation method for proteome analysis. Nat. Methods 6, 359–362 [DOI] [PubMed] [Google Scholar]
- 9. Wiśniewski J. R., Ostasiewicz P., and Mann M. (2011) High recovery FASP applied to the proteomic analysis of microdissected formalin fixed paraffin embedded cancer tissues retrieves known colon cancer markers. J. Proteome Res. 10, 3040–3049 [DOI] [PubMed] [Google Scholar]
- 10. Cox J., and Mann M. (2008) MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 26, 1367–1372 [DOI] [PubMed] [Google Scholar]
- 11. Cox J., Neuhauser N., Michalski A., Scheltema R. A., Olsen J. V., and Mann M. (2011) Andromeda: A peptide search engine integrated into the MaxQuant environment. J. Proteome Res. 10, 1794–1805 [DOI] [PubMed] [Google Scholar]
- 12. Tyanova S., Temu T., Carlson A., Sinitcyn P., Mann M., and Cox J. (2015) Visualization of LC-MS/MS proteomics data in MaxQuant. Proteomics 15, 1453–1456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kanehisa M., and Goto S. (2000) KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. 28, 27–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Mi H., Huang X., Muruganujan A., Tang H., Mills C., Kang D., and Thomas P. D. (2017) PANTHER version 11: expanded annotation data from Gene Ontology and reactome pathways, and data analysis tool enhancements. Nucleic Acids Res. 45, D183–D189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Petrova P., Raibekas A., Pevsner J., Vigo N., Anafi M., Moore M. K., Peaire A. E., Shridhar V., Smith D. I., Kelly J., Durocher Y., and Commissiong J. W. (2003) MANF: A new mesencephalic, astrocyte-derived neurotrophic factor with selectivity for dopaminergic neurons. J. Mol. Neurosci. 20, 173–188 [DOI] [PubMed] [Google Scholar]
- 16. Lever M. B., Karpova A., and Kreutz M. R. (2015) An Importin Code in neuronal transport from synapse-to-nucleus? Front. Mol. Neurosci. 8, 33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kaplan R., Zaheer A., Jaye M., and Lim R. (1991) Molecular cloning and expression of biologically active human glia maturation factor-beta. J. Neurochem. 57, 483–490 [DOI] [PubMed] [Google Scholar]
- 18. Phan E., Ahluwalia A., and Tarnawski A. S. (2007) Role of SPARC—Matricellular protein in pathophysiology and tissue injury healing. Implications for gastritis and gastric ulcers. Med. Sci. Monit. 13, RA25–RA30 [PubMed] [Google Scholar]
- 19. Novais A., Silva A., Ferreira A. C., Falcão A. M., Sousa N., Palha J. A., Marques F., and Sousa J. C. (2018) Adult hippocampal neurogenesis modulation by the membrane-associated progesterone receptor family member neudesin. Front. Cell Neurosci. 12, 463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Naba A., Jan K., Barque A., Nicholas C. L., and Hynes R. O. (2018) Knockout of the gene encoding the extracellular matrix protein Sned1 results in craniofacial malformations and early neonatal lethality. bioRxiv 440081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Querques F., D'Agostino A., Cozzolino C., Cozzuto L., Lombardo B., Leggiero E., Ruosi C., and Pastore L. (2019) Identification of a novel transcription factor required for osteogenic differentiation of mesenchymal stem cells. Stem Cells Dev. 12, 463. [DOI] [PubMed] [Google Scholar]
- 22. Paavilainen V. O., Bertling E., Falck S., and Lappalainen P. (2004) Regulation of cytoskeletal dynamics by actin-monomer-binding proteins. Trends Cell Biol. 14, 386–394 [DOI] [PubMed] [Google Scholar]
- 23. Rundhaug J. E. (2005) Matrix metalloproteinases and angiogenesis. J. Cell. Mol. Med. 9, 267–285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Tiwari A., Jung J.-J., Inamdar S. M., Nihalani D., and Choudhury A. (2013) The myosin motor Myo1c is required for VEGFR2 delivery to the cell surface and for angiogenic signaling. Am. J. Physiol. Heart Circ. Physiol. 304, H687–H696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Teichgräber V., Monasterio C., Chaitanya K., Boger R., Gordon K., Dieterle T., Jäger D., and Bauer S. (2015) Specific inhibition of fibroblast activation protein (FAP)-alpha prevents tumor progression in vitro. Adv. Med. Sci. 60, 264–272 [DOI] [PubMed] [Google Scholar]
- 26. Fiedler U., and Augustin H. G. (2006) Angiopoietins: A link between angiogenesis and inflammation. Trends Immunol. 27, 552–558 [DOI] [PubMed] [Google Scholar]
- 27. Camenisch G., Pisabarro M. T., Sherman D., Kowalski J., Nagel M., Hass P., Xie M.-H., Gurney A., Bodary S., Liang X. H., Clark K., Beresini M., Ferrara N., and Gerber H.-P. (2002) ANGPTL3 stimulates endothelial cell adhesion and migration via integrin alpha vbeta 3 and induces blood vessel formation in vivo. J. Biol. Chem. 277, 17281–17290 [DOI] [PubMed] [Google Scholar]
- 28. Tsuji K., Ojima M., Otabe K., Horie M., Koga H., Sekiya I., and Muneta T. (2017) Effects of different cell-detaching methods on the viability and cell surface antigen expression of synovial mesenchymal stem cells. Cell Transplant. 26, 1089–1102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Gomes I. S., Oliveira V. C., de Pinheiro A. O., C. S. Roballo K., Araujo G. S. M. de Veronezi J. C., Martins D. S., and Ambrósio C. E. (2017) Bone marrow stem cell applied in the canine veterinary clinics. Pesquisa Veterinária Brasileira 37, 1139–1145 [Google Scholar]
- 30. Fajka-Boja R., Marton A., Tóth A., Blazsó P., Tubak V., Bálint B., Nagy I., Hegedűs Z., Vizler C., and Katona R. L. (2018) Increased insulin-like growth factor 1 production by polyploid adipose stem cells promotes growth of breast cancer cells. BMC Cancer 18, 872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Roth S. P., Schubert S., Scheibe P., Groβ C., Brehm W., and Burk J. (2018) Growth Factor-mediated tenogenic induction of multipotent mesenchymal stromal cells is altered by the microenvironment of tendon matrix. Cell Transplant. 27, 1434–1450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Asadi-Golshan R., Razban V., Mirzaei E., Rahmanian A., Khajeh S., Mostafavi-Pour Z., and Dehghani F. (2018) Sensory and motor behavior evidences supporting the usefulness of conditioned medium from dental pulp-derived stem cells in spinal cord injury in rats. Asian Spine J. 12, 785–793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Cantinieaux D., Quertainmont R., Blacher S., Rossi L., Wanet T., Noël A., Brook G., Schoenen J., and Franzen R. (2013) Conditioned medium from bone marrow-derived mesenchymal stem cells improves recovery after spinal cord injury in rats: An original strategy to avoid cell transplantation. PLoS ONE 8, e69515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Cizkova D., Cubinkova V., Smolek T., Murgoci A.-N., Danko J., Vdoviakova K., Humenik F., Cizek M., Quanico J., Fournier I., and Salzet M. (2018) Localized intrathecal delivery of mesenchymal stromal cells conditioned medium improves functional recovery in a rat model of spinal cord injury. Int. J. Mol. Sci. 19, E870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Tseng K.-Y., Danilova T., Domanskyi A., Saarma M., Lindahl M., and Airavaara M. (2017) MANF is essential for neurite extension and neuronal migration in the developing cortex. eNeuro 4, 0214–0217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Tseng K.-Y., Anttila J. E., Khodosevich K., Tuominen R. K., Lindahl M., Domanskyi A., and Airavaara M. (2018) MANF promotes differentiation and migration of neural progenitor cells with potential neural regenerative effects in stroke. Mol. Ther. 26, 238–255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Shi Y., Chen L., Liotta L. A., Wan H.-H., and Rodgers G. P. (2006) Glia maturation factor gamma (GMFG): a cytokine-responsive protein during hematopoietic lineage development and its functional genomics analysis. Genom. Prot. Bioinform. 4, 145–155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Fan J., Fong T., Chen X., Chen C., Luo P., and Xie H. (2018) Glia maturation factor-β: A potential therapeutic target in neurodegeneration and neuroinflammation. Neuropsychiatr. Dis. Treat. 14, 495–504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Lim R., and Huang L. B. (1989) Glia maturation factor-beta promotes the appearance of large neurofilament-rich neurons in injured rat brains. Brain Res. 504, 154–158 [DOI] [PubMed] [Google Scholar]
- 40. Kimura I., Yoshioka M., Konishi M., Miyake A., and Itoh N. (2005) Neudesin, a novel secreted protein with a unique primary structure and neurotrophic activity. J. Neurosci. Res. 79, 287–294 [DOI] [PubMed] [Google Scholar]
- 41. Lozito T. P., and Tuan R. S. (2011) Mesenchymal stem cells inhibit both endogenous and exogenous MMPs via secreted TIMPs. J. Cell. Physiol. 226, 385–396 [DOI] [PubMed] [Google Scholar]
- 42. Lin M., Sutherland D. R., Horsfall W., Totty N., Yeo E., Nayar R., Wu X.-F., and Schuh A. C. (2002) Cell surface antigen CD109 is a novel member of the alpha(2) macroglobulin/C3, C4, C5 family of thioester-containing proteins. Blood 99, 1683–1691 [DOI] [PubMed] [Google Scholar]
- 43. Giesert C., Marxer A., Sutherland D. R., Schuh A. C., Kanz L., and Buhring H.-J. (2003) Antibody W7C5 defines a CD109 epitope expressed on CD34+ and CD34- hematopoietic and mesenchymal stem cell subsets. Ann. N.Y. Acad. Sci. 996, 227–230 [DOI] [PubMed] [Google Scholar]
- 44. Knight M. N., Karuppaiah K., Lowe M., Mohanty S., Zondervan R. L., Bell S., Ahn J., and Hankenson K. D. (2018) R-spondin-2 is a Wnt agonist that regulates osteoblast activity and bone mass. Bone Res 6, 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Gyllborg D., Ahmed M., Toledo E. M., Theofilopoulos S., Yang S., Ffrench-Constant C., and Arenas E. (2018) The matricellular protein R-spondin 2 promotes midbrain dopaminergic neurogenesis and differentiation. Stem Cell Rep. 11, 651–664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Xu Z.-G., Du J.-J., Zhang X., Cheng Z.-H., Ma Z.-Z., Xiao H.-S., Yu L., Wang Z.-Q., Li Y.-Y., Huo K.-K., and Han Z.-G. (2003) A novel liver-specific zona pellucida domain containing protein that is expressed rarely in hepatocellular carcinoma. Hepatology 38, 735–744 [DOI] [PubMed] [Google Scholar]
- 47. de Bakker E., Van Ryssen B., De Schauwer C., and Meyer E. (2013) Canine mesenchymal stem cells: State of the art, perspectives as therapy for dogs and as a model for humans. Vet Q. 33, 225–233 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Proteomic datasets including MaxQuant files and annotated MS/MS datasets were uploaded to the ProteomeXchange Consortium via the PRIDE database and then assigned the dataset identifier PXD013058.