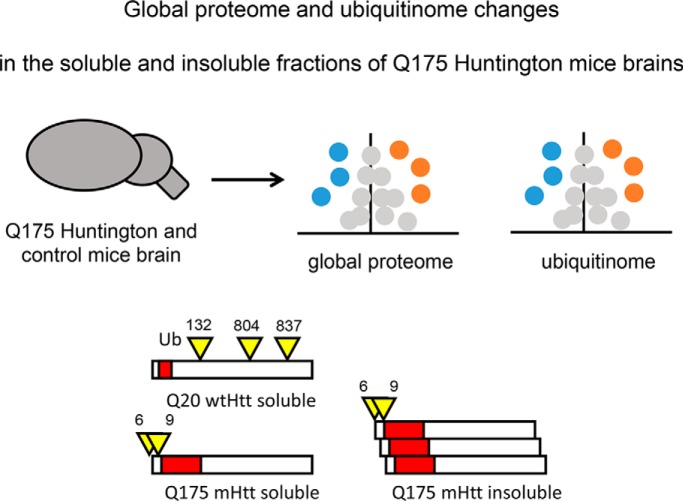
Label-free quantitative mass spectrometry techniques were applied to discover proteome and ubiquitinome changes in brain tissue of Huntington's disease mouse model Q175FDN and wild-type mice. Triton X-100 soluble and insoluble fractions were analyzed, revealing differential ubiquitination of wild-type and mutant Huntingtin in both. Cellular processes affected by the disease include vesicular transport, gene expression, translation, catabolic processes and oxidative phosphorylation. DiGly site fold changes with respect to protein fold changes were different between soluble and insoluble fractions.
Keywords: Label-free quantification, Mass Spectrometry, Neurodegenerative diseases, Post-translational modifications, Ubiquitin, Huntington
Graphical Abstract
Highlights
Quantitative changes in global proteome and ubiquitinome in Huntington's disease.
Differential ubiquitination of wild-type and mutant Htt in mice brain.
Enriched pathways include vesicle transport and mRNA processing.
Correlation between protein and diGly site fold changes.
Abstract
Huntington's disease is caused by a polyglutamine repeat expansion in the huntingtin protein which affects the function and folding of the protein, and results in intracellular protein aggregates. Here, we examined whether this mutation leads to altered ubiquitination of huntingtin and other proteins in both soluble and insoluble fractions of brain lysates of the Q175 knock-in Huntington's disease mouse model and the Q20 wild-type mouse model. Ubiquitination sites are detected by identification of Gly-Gly (diGly) remnant motifs that remain on modified lysine residues after digestion. We identified K6, K9, K132, K804, and K837 as endogenous ubiquitination sites of soluble huntingtin, with wild-type huntingtin being mainly ubiquitinated at K132, K804, and K837. Mutant huntingtin protein levels were strongly reduced in the soluble fraction whereas K6 and K9 were mainly ubiquitinated. In the insoluble fraction increased levels of huntingtin K6 and K9 diGly sites were observed for mutant huntingtin as compared with wild type. Besides huntingtin, proteins with various roles, including membrane organization, transport, mRNA processing, gene transcription, translation, catabolic processes and oxidative phosphorylation, were differently expressed or ubiquitinated in wild-type and mutant huntingtin brain tissues. Correlating protein and diGly site fold changes in the soluble fraction revealed that diGly site abundances of most of the proteins were not related to protein fold changes, indicating that these proteins were differentially ubiquitinated in the Q175 mice. In contrast, both the fold change of the protein level and diGly site level were increased for several proteins in the insoluble fraction, including ubiquitin, ubiquilin-2, sequestosome-1/p62 and myo5a. Our data sheds light on putative novel proteins involved in different cellular processes as well as their ubiquitination status in Huntington's disease, which forms the basis for further mechanistic studies to understand the role of differential ubiquitination of huntingtin and ubiquitin-regulated processes in Huntington's disease.
Huntington's disease (HD)1 is an autosomal dominant inherited neurodegenerative disorder characterized by excessive motor movements, cognitive and psychiatric deficits (1). The disease is hallmarked by intracellular aggregates and neuronal loss in various brain regions, with the striatum being mostly affected by neurodegeneration (2, 3). Patients usually live for 15–20 years after the onset of the disease (4). HD is caused by an expansion of the CAG repeat present in the exon1 domain of the huntingtin (Htt) gene. Healthy individuals exhibit between 6 to 35 CAG repeats in their Htt N termini, whereas individuals affected by HD display >39 repeats. CAG repeats are translated into polyglutamine (polyQ) repeats and mutant Htt (mHtt) proteins are thus characterized by an N-terminal polyQ-repeat expansion, as compared with the wild-type Htt (wtHtt) protein. Experimental data and the dominant inheritance pattern of HD indicate that mHtt is toxic and triggers neurodegeneration (5). Htt can be cleaved by proteases, including caspases, into shorter fragments (6, 7). Especially N-terminal fragments of the mHtt protein are aggregation-prone, resulting in proteotoxicity and protein aggregation (8).
Various studies have suggested that post-translational modifications (PTMs) of polyQ-expanded Htt modulate aggregation and toxicity (9, 10). Htt can be phosphorylated, SUMOylated, myristoylated (11) and acetylated on various residues, with phosphorylation being mostly studied. Interestingly, it has been shown that intracellular localization, cleavage, and degradation of mHtt are affected by phosphorylation of threonine 3 (12), serine 13, and serine 16, which are positioned in the N-terminal domain of Htt consisting of the first 17 amino acids (N17). Phosphorylation of these residues has also been shown to influence other post-translational modifications (13, 14). Identification of new PTMs may lead to effective therapeutic strategies, as involved enzymes may be modulated to improve the clearance of mHtt before cell toxicity.
Although differences in phosphorylation between wtHtt and mHtt have been shown by various research groups, much less is known about differences in ubiquitination between wtHtt and mHtt. Several studies have reported the effects of ubiquitin ligases and deubiquitinating enzymes (DUBs) on aggregate formation by mHtt (15–19), indicating that altering the ubiquitination of mHtt improves the turnover of the mutant protein and decreases aggregate formation (20–22). However, it is unknown whether mHtt is differentially ubiquitinated because of the polyQ expansion. Again, the N17 domain is of interest, as it contains three lysine residues that may become ubiquitinated and thereby affect clearance but also regulate modifications of other PTMs (23, 24).
To examine whether full-length (fl) mHtt is differentially ubiquitinated compared with wtHtt, we applied a mass spectrometry-based workflow in which we used K-ε-GG antibodies to purify diGly-modified peptides from both wild-type and mutant Htt mice brain samples. We used the Q175 knock-in mouse model (also called Q175F neo delete/Q175FDN) that contains a human mHtt exon1 with the expanded CAG repeat within the native mouse huntingtin gene. Both homo- and heterozygous Q175 mice show first signs of motor symptoms at the age of 3–4 months, and behavioral deficits are observed at the age of 8 months (25). The mice used for this study were 40 weeks (10 months) old representing an advanced stage of HD. Further, the Q175 mice used were homozygous, lacking wtHtt, as no discrimination between wtHtt and mHtt peptides can be made upon digestion for mass spectrometry (MS) analysis. To separate soluble Htt from aggregated Htt we used a mild lysis buffer containing Triton X-100 which does not dissolve aggregated Htt (26). Thus, insoluble fractions contain putative mHtt aggregates, among other proteins present in the lysate pellet.
Here, we show that wild-type Htt was mainly ubiquitinated at K132, K804, K837 in the soluble fraction. In contrast, the mutant Htt protein level was strongly reduced in the soluble fraction and it was ubiquitinated at K6 and K9. In the insoluble fraction both wtHtt and mHtt were ubiquitinated at K6 and K9, but label-free quantification (LFQ) revealed that mHtt ubiquitination at these sites was increased compared with wtHtt. Further, we found increased levels of ubiquitin and K48 polyubiquitin linkages in the insoluble fraction. Also, several proteins with roles in membrane organization, transport, mRNA processing, gene transcription, translation, catabolic processes and oxidative phosphorylation were differentially expressed or ubiquitinated in wild-type and mutant huntingtin brain tissues. Finally, most proteins in the soluble fraction showed stable expression levels in wtHtt and mHtt samples and thus did not affect diGly site fold changes in general. This suggests that diGly site fold changes were the result of increased ubiquitination. In the insoluble fraction several proteins showed increased protein and diGly site levels, suggesting that protein fold changes contributed to diGly site fold changes for these proteins. This was for instance the case for ubiquitin, ubiquilin-2, sequestosome-1/p62 and myosin VA (myo5a). Our data gives more insight in the ubiquitination of huntingtin and other proteins in soluble and insoluble fractions of late stage HD mice brain lysates. The findings presented here could be the basis for further mechanistic studies to understand the role of ubiquitin signaling in HD.
EXPERIMENTAL PROCEDURES
Experimental Design and Statistical Rationale
Global proteome and ubiquitinome of soluble and insoluble fractions derived from wild-type Htt mice brain (wtHtt, expressing full-length Htt with 20 polyQ repeats, control) and mutant Htt mice brain (mHtt, expressing full-length Htt with 175 polyQ repeats) were analyzed using label-free mass spectrometry. Four biological replicates of mutants and wild types were analyzed in each experiment to validate the biological reliability of measurements. For the soluble fraction, three fractions were measured for each sample. Both for global and diGly analyses, proteins and diGly sites that were identified in at least 3 out of 4 replicates both in wild-type and mutant groups were used for comparison. Student's t test was performed for comparisons between wtHtt and mHtt with 1% FDR cutoff for all datasets.
Sample Preparation for Mass Spectrometry
Snap frozen whole brains excluding cerebellum derived from 40 weeks old mice of strain HttQ20 (wild type) and strain Q175F neo delete/Q175FDN (mutant, homozygous) (25), were provided by the Jackson Lab (www.jax.org). Frozen brains were grinded in small pieces using a metal bar in a liquid nitrogen bath, and dissolved in a mild lysis buffer (50 mm Tris/HCl pH7; 150 mm NaCl; 1% Triton X-100; Roche protease inhibitor tablet (Roche Diagnostics GmbH, Mannheim, Germany); 200 μm PR-619; 1 mm EDTA; 100 mm Chloroacetamide (CAA)) using a glass douncer on ice. Lysates were centrifuged through a 70 micron filter (EASYstrainer, Greiner Bio-One B.V., Alphen a/d Rijn, The Netherlands) and DNA was sheared by sonication (Bioruptor Pico, Diagenode, Seraing, Belgium). Lysates were centrifuged for 20 min with 14,000 rpm by 4 °C to separate soluble (monomeric and oligomeric Htt) and insoluble (fibrils, aggregates and inclusion bodies) huntingtin fractions (26). Proteins of the soluble fraction were denatured by addition of 10 volumes of 8 m urea lysis buffer (8 m urea; 50 mm Tris/HCl pH8; 50 mm NaCl). The pellet or insoluble fraction was washed several times in the mild lysis buffer. Next, the Htt aggregates in the pellets were solubilized as described before (23) with some small adjustments. Briefly, aggregates were solubilized by resuspension in one pellet volume of formic acid and incubated at 37 °C for 40 min with 1000 rpm shaking and dried by speedvac O/N. Pellets were dissolved in 8 m urea-based lysis buffer. Protein quantification was performed for soluble and insoluble fractions using the Bradford assay (Serva Electrophoresis GmbH, Heidelberg, Germany). Four biological replicates of 20 mg total protein for soluble fractions and 3 mg protein for insoluble fractions were made for both wild-type and mutant samples.
Arg-C/chymotrypsin and Lys-C/trypsin Digestion of Soluble Fractions
Ten milligrams total protein of each sample was used for Arg-C/chymotrypsin digestion. Samples were diluted with 50 mm Tris-HCl pH8 to a final concentration of 2 m urea. Proteins were reduced using 5 mm 1,4-dithiothreitol (DTT) for 45 min at RT. CaAc was added to a final concentration of 1 mm. Proteins were digested for 1 h with Arg-C (Protea Biosciences, Morgantown, West Virginia; 1:200 enzyme/substrate ratio) at 37 °C and then overnight at room temperature (RT). Peptides were reduced using 5 mm DTT for 45 min at RT and subsequently alkylated using 5.5 mm chloroacetamide (CAA) for 30 min in the dark. CaCl2 was added to a final concentration of 1 mm. Samples were digested for 1 h with chymotrypsin (Pierce Chymotrypsin Protease, MS Grade, Thermo Scientific, Rockford, Illinois; 1:200 enzyme/substrate ratio) at 37 °C and then 3 h at RT. Trifluoroacetic acid (TFA) was added to 1% to stop the digestion. Digests were centrifuged at 4500 rpm for 15 min. Ten milligrams total protein of each sample was used for Lys-C/trypsin digestion. Samples were diluted with 50 mm Tris-HCl pH8 to a final concentration of 4 m urea. Proteins were reduced using 5 mm DTT for 45 min at RT and subsequently alkylated using 5.5 mm CAA for 30min in the dark. Proteins were digested for 2 h with Lys-C (Wako Pure Chemical Industries Ltd., Osaka, Japan; 1:100 enzyme/substrate ratio). Samples were diluted with 25 mm Tris-HCL pH8 to a final concentration of 1.6 m urea. CaCl2 was added to a final concentration of 1 mm. Samples were digested for 1h with trypsin (Pierce Trypsin Protease, MS-Grade, Thermo Scientific, Rockford, Illinois; 1:50 enzyme/substrate ratio) at 37 °C and then o/n at RT. TFA was added to 1% to stop the digestion. Digests were centrifuged at 4500 rpm for 15 min.
Lys-C/trypsin Digestion of Insoluble Fractions
2.75 mg of each sample was used for Lys-C/trypsin digestion. Digestion was performed as described above.
Peptide Fractionation of Soluble Fractions
Arg-C/chymotrypsin and Lys-C/trypsin digests of soluble fractions were combined. Peptide fractionation, diGly peptide enrichment and NanoLC-MS/MS analysis were performed as initially described (27), with only minor adjustments. Briefly, for peptide fractionation high pH reverse-phase (RP) chromatography was performed using polymeric PLRP-S (300Å, 50 μm, Agilent Technologies, part #PL1412–2K01). Samples were loaded onto the column and washed with 6 column volumes (CVs) of 0.1% TFA followed by 6 CVs of milliQ H2O. Peptides were then eluted in three fractions with 6 CVs of 10 mm ammonium formate solution (pH10) and 9%, 15 and 40% acetonitrile (AcN), respectively. 1/30 part of the fractions were dried to completeness by vacuum centrifugation and used for global proteome analysis. Remaining of the fractions were dried to completeness by lyophilization and used for diGly peptide enrichment.
Peptide Clean-up of Insoluble Fractions
peptide clean-up was performed with Sep-Pak C18 columns (Waters, Dublin, Ireland) according to PTMscan protocol (PTMscan, Cell Signaling Technologies, Leiden, The Netherlands).
Immunoprecipitation of diGly Peptides
Ubiquitin remnant motif (K-ε-GG) antibodies coupled to beads (PTMscan, Cell Signaling Technologies) were used according to manufacturer's advice. For the soluble fraction, each standard batch of beads was divided over the three fractions of each sample. For the insoluble fraction, 20 μl beads were used per diGly IP.
NanoLC-MS
Mass spectra were acquired on an Orbitrap Tribrid Lumos mass spectrometer (Thermo Fisher Scientific GmbH, Bremen, Germany) coupled to an EASY-nLC 1200 system (Thermo). Peptides were separated on an in-house packed 75 μm inner diameter column containing 50 cm Waters CSH130 resin (2.5 μm, 130Å, Waters, Etten-Leur, The Netherlands) with a gradient consisting of 2–30% (AcN, 0.1%FA) over 120 min at 300 nL/min. The column was kept at 50 °C in a NanoLC oven - MPI design (Sonation GmbH, Biberach, Germany). For all experiments, the instrument was operated in the data-dependent acquisition (DDA) mode using the top speed method with a cycle time of 3 s. MS1 spectra were collected at a resolution of 120,000 with an automated gain control (AGC) target of 4E5 and a max injection time of 50 ms. Precursors were filtered according to charge state (2–6z), and monoisotopic peak assignment. Previously interrogated precursors were dynamically excluded for 60 s. Peptide precursors were isolated with a quadrupole mass filter set to a width of 1.6 Th. Ion trap MS2 spectra were collected at an AGC of 7E3, a max injection time of 50 ms and HCD collision energy of 30%. Duration of all individual mass spec runs was 120 min.
Data Processing and Statistical Analysis
RAW files were analyzed using MaxQuant software suite (v1.6.5.0) (28). The data were searched against reviewed (Swiss-Prot) Mus musculus proteome from Uniprot (September 2018, contains 16981 entries) (29), along with revert decoys and standard contaminants database from MaxQuant. Methionine oxidation and N-terminal acetylation were set as variable modifications, whereas carbamidomethylation of cysteine was set as a fixed modification. In addition to these, diGly modification of Lysine was set as a variable modification for searching diGly data. Maximum number of modifications per peptide were set to 5. Default search settings for Orbitrap were used, such as a precursor tolerance of 20 ppm and 4.5 ppm for the first and main search respectively and a tolerance of 20 ppm for the fragment ions. Trypsin was selected as the search enzyme for the insoluble data; whereas Arg-C/chymotrypsin/trypsin was set as the cleavage enzyme for the soluble. Maximum number of miscleavages were set to 3. The minimal peptide length was set to 7, “second peptides” and “match between runs” were checked. The label-free quantification algorithm (LFQ) in MaxQuant was used for quantification with LFQ minimum ratio count set to 1; unique and razor peptides were selected to be used for quantification. The false discovery rate for peptide, protein, and site identifications were set to 1%. The minimum score for diGly peptides was kept at 40 (default). Statistical analyses were performed on MaxQuant outputs using the Perseus package (v1.6.2.3) (30). For global proteome data the proteinGroups files were used for statistical analyses. Proteins that were identified by less than 2 unique peptides were excluded. LFQ intensities were log2 transformed. Proteins that were identified in at least 3 (out of 4) replicates in both wtHtt and mHtt samples were selected for comparison of protein abundance. Missing values of selected proteins were then imputed from a normal distribution with width 0.3, down shift 1.8 from the general distribution. For ubiquitinome data the GlyGly (K) Sites tables were used for statistical analyses. diGly site modifications localized on C-term with a probability more than 0.75 and on any other position with a probability less than 0.75 were discarded. diGly sites that were identified in at least 3 (out of 4) experiments in both wtHtt and mHtt samples were selected for diGly site quantification. Normalization of diGly log2 transformed intensities was done manually based on average median intensities in the global evidence file. Calculated scaling factors were then applied to intensities in the GlyGly (K) sites table. In addition to these comparisons, proteins and diGly sites that were identified in all of the mHtt replicates but in none or only one of the wtHtt, and vice versa, were extracted from the tables before imputation and filtering for further exploration.
Pathway Analysis
Protein-protein association network for up and down regulated proteins was visualized using the StringApp (v1.4.1) (31) in Cytoscape (v3.7.0) (32). Cytoscape plugin BiNGO (v3.0.3) (33) was used for identification of enriched GO terms from lists of gene names of up and down regulated proteins or diGly sites (target list) versus whole annotation as a reference set. Hypergeometric statistical test was used with Benjamini & Hochberg false discovery rate correction with a significance level of 0.05. Web-based Venn diagram tool (34) from VIB/UGhent was used to plot the overlap of up- and downregulated proteins in both the soluble and insoluble fractions. Cytoscape plugin Molecular Complex Detection (MCODE) (v1.5.1) (35) was used to identify highly interconnected clusters. The following settings were used for cluster identification: Network scoring: Degree cutoff of 2. Cluster finding: haircut. Node Score cutoff: 0.2. K-core: 2. Max. Depth: 100.
SDS-PAGE and Western Blot Analysis
Antibodies used for SDS-PAGE WB analysis: α-Htt (ab109115 Abcam); α-Htt D7F7 (#5656 CST), α-Actin (SC-1616 Santa Cruz), α-Vimentin (AB1620 Sigma). SDS-PAGE and Western blotting were performed according to standard procedures, except for Htt (350 kDa). For Htt a 6% Tris/Glycine gel was used and proteins were run out of the gel until ∼80 kDa. Wet Blotting was done o/n at 45V at 4 °C. Proteins were visualized using LiCOR secondary antibodies and the Odyssey system.
RESULTS
Proteomics Workflow
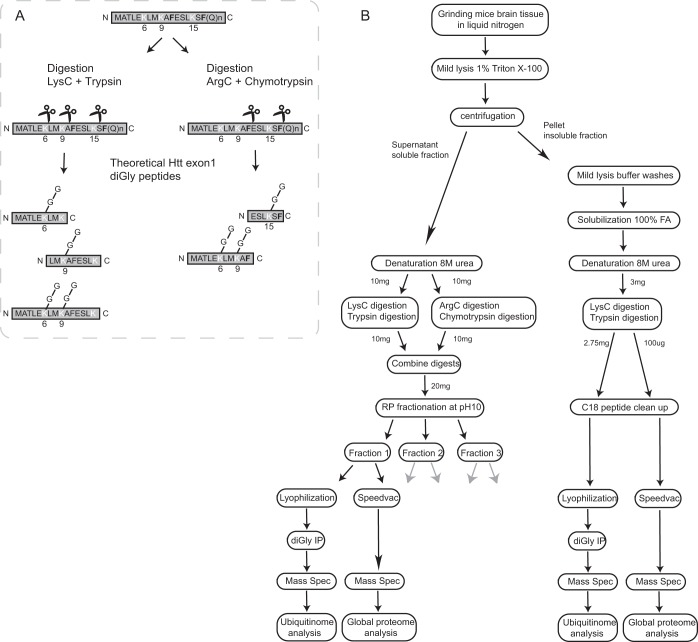
The N-terminal sequence of Htt is subjected to a variety of post-translational modifications (PTMs), such as ubiquitination (23, 24), SUMOylation (24, 36) and phosphorylation (12, 14). We aimed to identify endogenous fl Htt ubiquitination sites in mice brain lysates. We were especially interested in the putative ubiquitination sites located at the N terminus; K6, K9, and K15, as the N17 domain (consisting of the first 17 amino acids preceding the polyQ repeat) plays a crucial role in mHtt localization, aggregation and clearance. In addition to Lys-C and trypsin digestion, Arg-C and chymotrypsin digestion was used to potentially increase the number of unique N-terminal Htt diGly peptides (Fig. 1A, supplemental Fig. S1). Four biological replicates of both wild-type mice brain lysates and Q175FDN mutant Htt mice brain lysates were processed for mass spectrometric analysis. Fig. 1B shows a simplified overview of the experimental workflow for one sample. Frozen brain tissues were grinded in liquid nitrogen. To prevent resolving mHtt fibrils, aggregates and inclusion bodies, samples were lysed in a 1%Triton X-100-based mild lysis buffer (26). After sonication, a Triton X-100 soluble and insoluble fraction were obtained through centrifugation. In our workflow the soluble Htt fraction contains wtHtt- or mHtt monomers but could also contain some oligomeric states of mHtt (26). Insoluble pellet fractions were treated with formic acid to solubilize Htt aggregates. Proteins of both the soluble and insoluble fractions were denatured in an 8 m urea-based buffer. Twenty milligrams and 3 mg of total protein were digested for the soluble and insoluble fractions, respectively. Recently, we have optimized the protocol for diGly peptide detection for samples with relatively high protein amounts (27). Briefly, in this workflow digests are subjected to offline high pH reverse phase (RP) fractionation. Three fractions were collected of each sample, and of each fraction 1/30 (v/v) part was subjected to global proteome analysis, whereas the remaining part was subjected to diGly peptide enrichment. All fractions were measured individually by nanoLC-MS/MS (Fig. 1B).
Fig. 1.
Workflow of mass spectrometry sample preparation. A, Schematic representation of the used protein digestion method. A sequential digestion of Arg-C/chymotrypsin and Lys-C/trypsin was performed with the aim to obtain peptides containing all putative N-terminal Htt ubiquitination sites. The cleavage sites of trypsin and Lys-C are shown in white, whereas cleavage sites of chymotrypsin are shown in bold black. B, Simplified schematic representation of the workflow from lysate to data analysis for both the global ubiquitinome and global proteome analysis. Wild-type and mutant samples were made in quadruplicates, here the workflow is shown for just one sample. Triton X-100 soluble and insoluble fractions were obtained. Putative Htt aggregates in the insoluble fraction (pellets) were solubilized by formic acid treatment. Soluble fractions were digested as described in Fig. 1A. To reduce complexity, the soluble fractions were further fractionated by high pH RP fractionation. Insoluble fractions were digested only with Lys-C and trypsin. Further fractionation was not required based on the amount of protein in these samples. Global proteome and ubiquitin-modified proteome analyses were performed for each fraction or sample using the MaxQuant and Perseus platforms.
Quantitative Analysis of Global Proteome and Ubiquitinome Data Sets
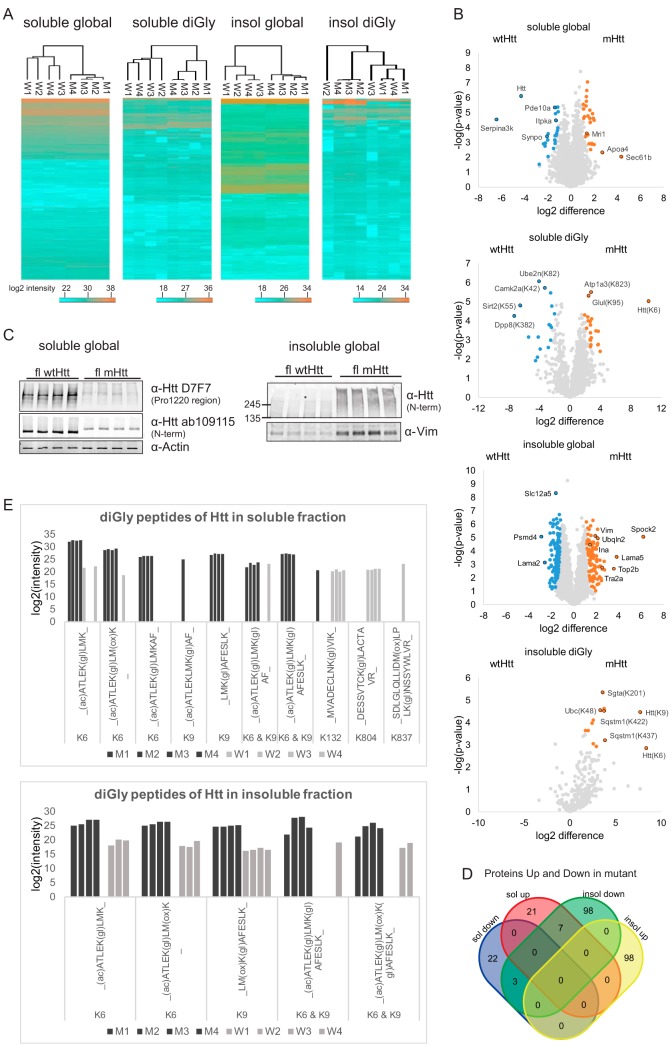
Raw files obtained for the global proteome and ubiquitinome of the soluble and insoluble fractions were analyzed using the MaxQuant and Perseus platforms. We identified 5229 protein groups in the global proteome data of the soluble fraction (supplemental Table S1), and 2590 protein groups in the global proteome data of the insoluble fraction (supplemental Table S2) (excluding contaminants and decoys). The proteins were ordered by the number of identified peptides in descending order in the protein IDs column of the proteinGroups table. The first protein, thus the protein with the highest number of identified peptides, was used from each protein group. For the diGly data we identified 5508 diGly peptides in the soluble fraction and 1146 diGly peptides in the insoluble fraction (excluding contaminants and decoys). DiGly sites were used for further analyses (supplemental Table S3 and supplemental Table S4). Hierarchical clustering (Euclidean distance) analysis was performed for all datasets and as expected we observed more variation in the ubiquitinome datasets as compared with the proteome datasets. The ubiquitinome of the insoluble fraction showed the most variation (Fig. 2A). Two-sample Student's t-tests were performed for all proteins and diGly sites that were identified in at least 3 out of 4 samples in both wild-type and mutant conditions (Fig. 2B). The relative abundances of most of the proteins and diGly sites were unaffected. Proteins and diGly sites with significant higher relative abundances in the mutant samples are shown in orange, whereas proteins and diGly sites present with significant higher relative abundances in the wild-type samples are shown in blue. For informative purposes several differentially expressed proteins and diGly sites are depicted with their gene names in the plots. Differential expression of several proteins had been associated with HD before, such as downregulation of Pde10a (37–40), synaptopodin (39), Itpka (40), and Serpina3k (40), and upregulation of Mri1 (40) in HD mice models. Also, several proteins that we found to be enriched in the insoluble fraction of the mutant Htt samples have been identified in Htt aggregates in other studies, for instance Ina, Vim, Tra2a (39), and ubqln2 (41). Further, we observed a remarkable downregulation of the Htt protein in the soluble fraction of mHtt brain lysates (Fig. 2B upper plot). Changes in protein abundances observed by mass spectrometry were confirmed by SDS-PAGE in combination with Western blotting. For instance, we observed relatively lower levels of full-length mHtt in the soluble fraction and relatively higher levels of Vimentin in the insoluble fraction by SDS-PAGE WB (Fig. 2C). Additionally, we observed the presence of both full-length mHtt as well as oligomeric species and/or fragments of the full-length protein in the insoluble fraction (Fig. 2C).
Fig. 2.
Visualization of quantitative data. A, Heatmaps (Euclidian Distance) of log2 transformed LFQ intensities of identified and quantified proteins and diGly sites for all data sets. Lowest intensities are shown in blue and highest intensities are shown in orange. B, Volcano plots showing protein or diGly site relative abundances for all data sets. Orange: proteins/diGly sites up in mutant Htt mice brain; blue: proteins/diGly sites up in wtHtt mice brain. C, SDS-PAGE and Western blot analysis of the quadruplicate wtHtt and mHtt brain lysates with antibodies against both Htt Proline 1220 region and N terminus and actin (loading control) confirm decreased Htt protein levels in mHtt mice brain in the soluble global proteome. SDS-PAGE and Western blot analysis of the quadruplicate wtHtt and mHtt brain lysates with an antibody against Vimentin confirms increased Vimentin protein levels in mHtt mice brain in the insoluble global proteome. Full-length (and probably fragments of) Htt was detected with the antibody raised against Htt N terminus only in mHtt insoluble fractions. D, Venn diagram showing number of proteins that were significantly up and down in soluble and insoluble fractions. Only the proteins that pass the fold change and p value threshold in the t test (those marked in color in the volcano plots) are shown. E, The bar plots showing the log2 intensities of the diGly peptides of the Huntingtin protein in soluble and insoluble fractions. The intensities are the raw intensities from the MaxQuant evidence file. Modified sequences that have a C-term diGly are excluded.
Although the selection of proteins identified in 3 out of 4 samples in both groups was a good basis for comparison between two groups, several putatively interesting candidates could have been omitted by using this method. In our study we focused on a late stage of HD, in which extreme differences might appear both at the protein level as well as at the level of protein ubiquitination. We therefore also investigated these extreme differences by including proteins or diGly sites identified in 4 out of 4 replicates in one group while identified exclusively in one or none of the replicates in the other group. Table I gives the amount of proteins and diGly sites in these categories, whereas supplemental Table S5 gives information about these proteins and diGly sites. The distribution of their intensities together with those that were significantly changed are shown in supplemental Fig. S2.
Table I. Number of proteins and diGly sites exclusively identified in wild-type or mutant samples. Numbers of protein and diGly sites identified exclusively (in 4 out of 4 replicates) in one group and in none or one of the replicates of the other group. Wt = wild type; Mut = mutant.
| Fraction | Proteome level | Identified exclusively in | # unique proteins or diGly sites |
|---|---|---|---|
| Soluble | global | Wt | 55 |
| Mut | 44 | ||
| diGly | Wt | 108 | |
| Mut | 233 | ||
| Insoluble | global | Wt | 39 |
| Mut | 51 | ||
| diGly | Wt | 9 | |
| Mut | 34 |
In addition to mHtt, other proteins can also become insoluble because of sequestration in mHtt aggregates. We looked whether this was the case in our analysis by plotting the overlap between significantly up and downregulated proteins in the soluble and insoluble fractions (Fig. 2D). Significant downregulation of proteins in the soluble fraction did not result in significant upregulation of those proteins in the insoluble fraction, suggesting that changes in protein relative abundances in the soluble fraction observed in our study were rather the result of changes in protein synthesis and/or degradation than in sequestration in the insoluble fraction (Fig. 2D).
We identified multiple diGly peptides for Htt. Fig. 2E shows all identified diGly peptides from Htt in the quadruplicate samples in both the soluble (upper panel) and insoluble fractions (lower panel). Intensities were log2 transformed sum of raw intensities from the MaxQuant evidence files. We identified diGly peptides with Htt K6, K9, K132, K804, and K837 in the soluble fractions while in the insoluble fraction only diGly peptides with Htt K6 and K9 were found. In the insoluble fraction, singly modified diGly peptides with Htt K6 and with Htt K9 were identified in almost all replicates and log2 intensities were clearly higher in mutant samples, suggesting that K6 and K9 were more ubiquitinated in mutant Htt insoluble fractions (Fig. 2E). Also, the Student's t test revealed that Htt K6 and K9 diGly sites were significantly upregulated in mutant Htt insoluble fractions (Fig. 2B). In contrast, in the soluble fraction diGly peptides harboring the same GlyGly sites were primarily identified in the mutant samples, whereas the Htt protein level was strongly reduced in these samples (Fig. 2B), suggesting that ubiquitination of Htt at K6 and K9 is predominantly occurring with the mutant protein in the soluble fraction of our samples (Fig. 2E). Htt could also be ubiquitinated at K6 and K9 simultaneously, as we identified two different peptides harboring both modifications (Fig. 2E). Moreover, diGly peptides for Htt K132 and K804 were primarily identified in the wild-type samples, suggesting that ubiquitination at these sites specifically occurred with the wtHtt samples under the used conditions. Finally, the Htt diGly peptide harboring K837 was only identified in 1 out of the 8 samples, indicating that ubiquitination at this site occurs almost never or only rarely in our model under the used conditions. Concluding, we observed changes in both the global proteome and ubiquitin-modified proteome of the Q175 HD mice model as compared with the wild type. Typically, the Htt protein level was remarkably decreased in the soluble fraction of the HD mice model and the mHtt protein was more ubiquitinated at K6 and K9 in both the soluble and insoluble fractions whereas K132 and K804 were more ubiquitinated in the wtHtt protein in the soluble fraction. Htt diGly peptides harboring K837 were only found in one wild-type sample.
Proteins with Roles in Membrane Organization, Translation, and Catabolic Processes Are Differentially Ubiquitinated in the Soluble Fraction of Wild-type and HD Mice Brains
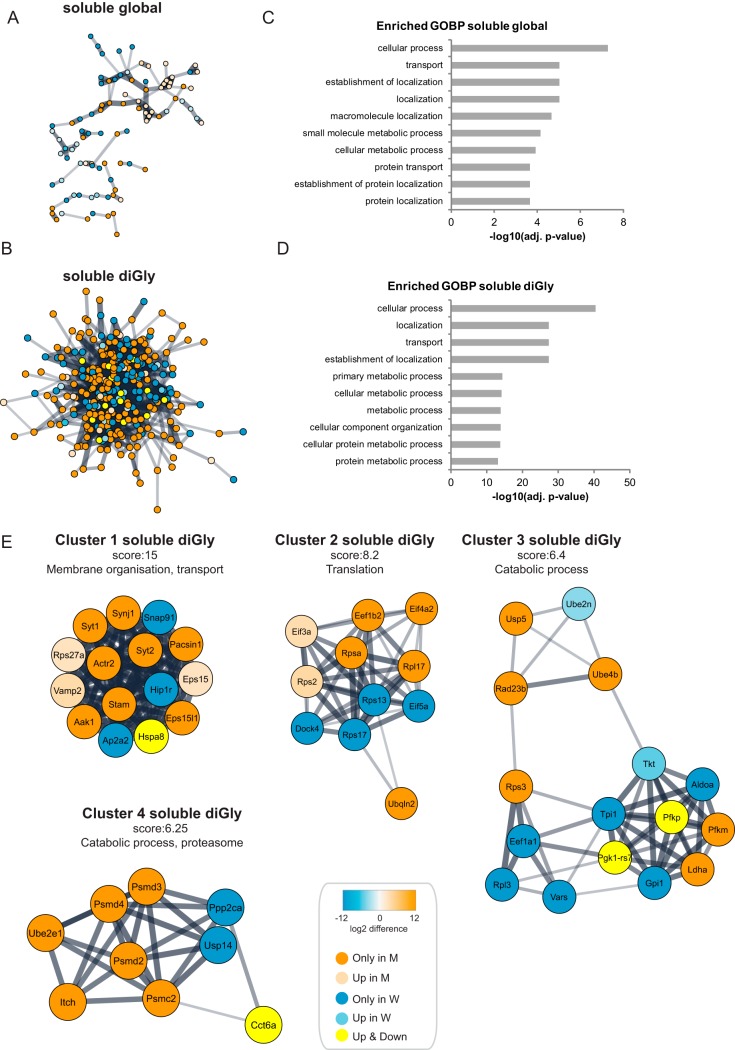
Next we performed functional analyses on differentially expressed proteins and diGly sites. Visualization of the protein-protein association network for up- and downregulated proteins and diGly sites using the stringApp (42) in Cytoscape (32) revealed that the association network for diGly sites is particularly dense as compared with the association network for proteins (Fig. 3A and 3B). Gene Ontology (GO) term analysis revealed that Biological Processes related with transport, localization and metabolic processes were enriched in the group of differentially expressed proteins and diGly sites in the soluble fraction (Fig. 3C and 3D).
Fig. 3.
Functional analysis of Huntington's disease dependent changes in the global proteome and ubiquitinome of the soluble fractions of Q175FDN mouse brain lysates. A, STRING network analysis was performed on differentially expressed proteins in the soluble fraction. Orange is used for proteins identified exclusively in the mutant Htt samples (identified in 4 mutant samples and in 0 or 1 wild-type sample), whereas blue is used for proteins identified exclusively in wild-type samples (identified in 4 wild-type samples and in 0 or 1 mutant sample). A color gradient representing the protein fold change of orange (for upregulation in mutant) and blue (for upregulation in wild type) is used for significantly differentially expressed proteins (identified in at least 3 out of 4 mutant and wild-type samples, significant proteins in volcano plots Fig. 2B). B, Same as in Fig. 3A, but for diGly sites. Proteins of diGly sites upregulated in both wild-type and mutant samples are shown in yellow. C, Gene Ontology (GO) analysis of differentially expressed proteins in the global proteome of the soluble fraction. Top ten of enriched Biological Processes is shown, enrichment probability is represented by -log10 Benjamini Hochberg adjusted p value. D, Gene Ontology (GO) analysis of differentially expressed diGly sites in the soluble fraction. Top ten of enriched Biological Processes is shown, enrichment probability is represented by -log10 Benjamini Hochberg score. E, Cytoscape plugin MCODE revealed highly interconnected clusters in the STRING network of the differentially expressed diGly sites in the soluble fraction. Clusters with highest scores are shown here. See A and B for description of node color coding.
Highly interconnected clusters were found in the protein-protein association network of up and downregulated diGly sites with the use of Cytoscape plugin MCODE (Fig. 3E). Proteins in the highest scoring clusters played roles in membrane organization and transport (cluster 1), translation (cluster 2) and catabolic processes (cluster 3 and 4).
Functional Analysis of Global Proteome and Ubiquitinome Changes in the Insoluble Fraction of Wild-type and HD Mice Brains
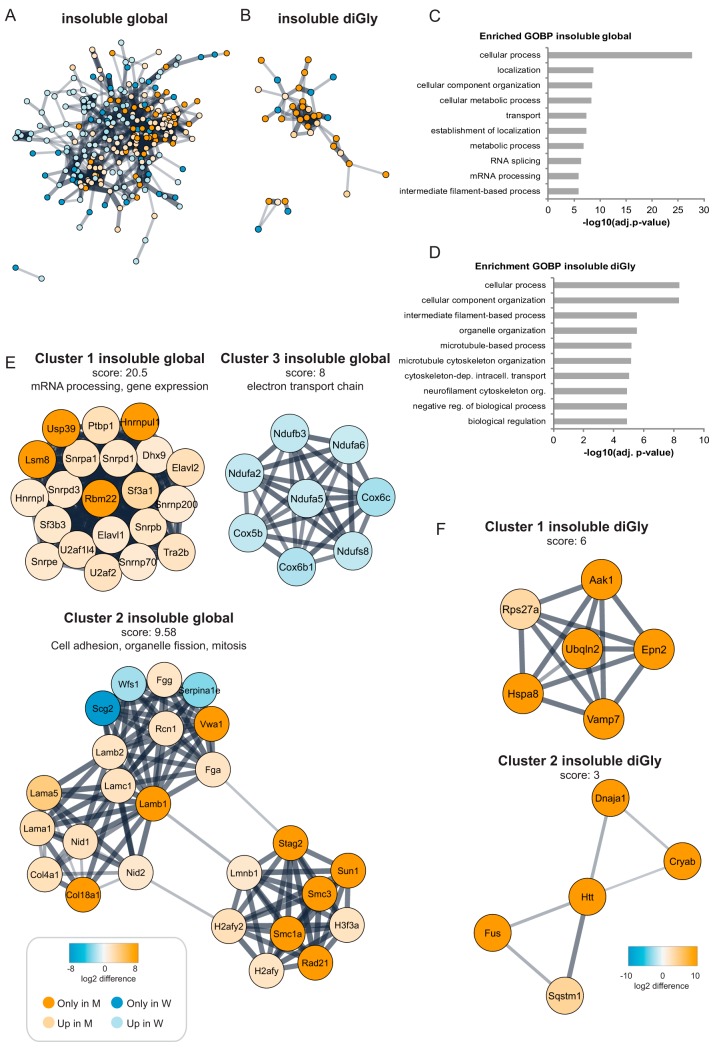
We also performed a functional network analysis of up- and downregulated proteins and diGly sites in the insoluble fractions. For the insoluble fractions we obtained a dense protein-protein association network for the proteins contributing to global proteome changes (Fig. 4A) whereas the network for up and downregulated diGly sites was relatively simple (Fig. 4B). Gene Ontology (GO) term analysis revealed that many enriched Biological Processes for the global proteome dataset of the insoluble fraction were shared with the datasets derived from the soluble fractions (transport, localization and metabolic processes). In addition, GO terms related with RNA splicing and mRNA processing were enriched in the global proteome of the insoluble fraction (Fig. 4C). GO terms enriched in the diGly data set were related to cellular organization and microtubule-based processes (Fig. 4D).
Fig. 4.
Huntington's disease dependent changes in the global proteome and ubiquitinome of the insoluble fractions of Q175FDN mouse brain lysates. A, STRING network analysis was performed on differentially expressed proteins in the insoluble fraction. Orange is used for proteins identified exclusively in the mutant Htt samples (identified in 4 mutant samples and in 0 or 1 wild-type sample), whereas blue is used for proteins identified exclusively in wild-type samples (identified in 4 wild-type samples and in 0 or 1 mutant sample). A color gradient representing the protein fold change of orange (for upregulation in mutant) and blue (for upregulation in wild type) is used for significantly differentially expressed proteins (identified in at least 3 out of 4 mutant and wild-type samples, significant proteins in volcano plots Fig. 2B). B, Same as Fig. 4A, but for diGly sites. C, Gene Ontology (GO) analysis of differentially expressed proteins in the global proteome of the insoluble fraction. Top ten of enriched Biological Processes is shown, enrichment probability is represented by -log10 Benjamini Hochberg adjusted p value. D, Gene Ontology (GO) analysis of differentially expressed diGly sites in the insoluble fraction. Top ten of enriched Biological Processes is shown, enrichment probability is represented by -log10 Benjamini Hochberg adjusted p value. E, With the use of Cytoscape plugin MCODE highly interconnected clusters were found in the STRING network of the differentially expressed proteins in the insoluble fraction. Clusters with highest scores are shown here. See A for description of node color coding. F, same as E, but for diGly sites.
Highly interconnected clusters were found in the protein-protein association network of up- and downregulated proteins in the insoluble fraction (Fig. 4E). Proteins in the highest scoring cluster (cluster 1) were all upregulated in mutant Htt samples and played roles in mRNA processing and gene expression. In contrast, cluster 3 contains only proteins that were downregulated in mutant Htt samples, and these proteins play a role in the electron transport chain in mitochondria. The second cluster contains proteins upregulated in both wild-type and mutant Htt samples and play a role in cell adhesion, organelle fission and mitosis. A few clusters were found in the association network for the proteins of which diGly sites were found in the insoluble fraction (Fig. 4F). Although their scoring was relatively low and no GO terms were found to be enriched in this selection of proteins, several of these proteins are known to be present at Htt aggregates, so their differential ubiquitination might be interesting novel information for the field.
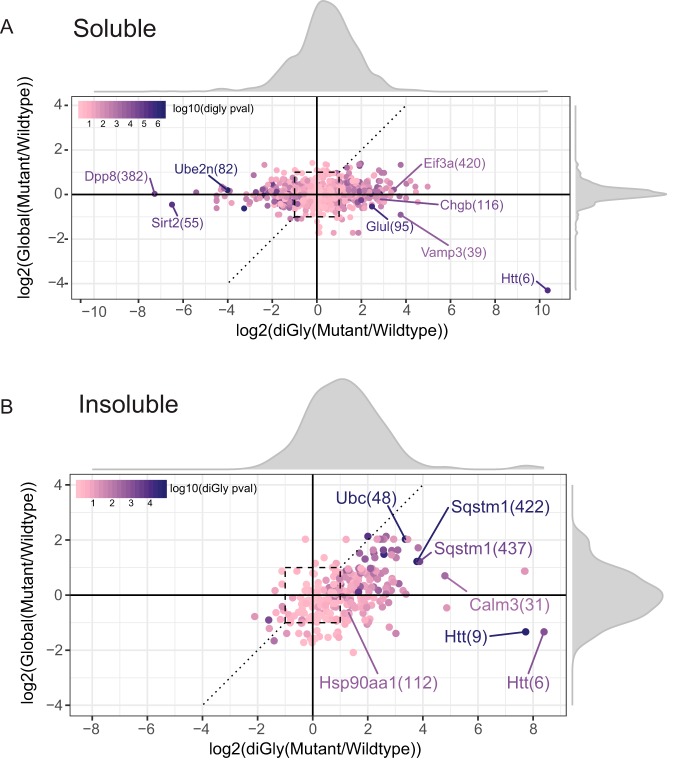
Correlation Between Protein and diGly Site Fold Changes in HD
Next, we focused on the correlation between global proteome changes and changes in the ubiquitin-modified proteome in both the soluble and insoluble fractions. Fig. 5A shows this correlation, plotted using the ggplot2 package (43) in R (44), for all proteins and diGly sites that were identified in 3 out of 4 samples in both wild-type and mutant samples of the soluble fraction. Fold changes were much larger for many diGly sites as compared with protein fold changes. Relatively increased diGly site fold changes were observed both in wild-type and mutant samples and followed a normal distribution which was only slightly skewed toward the mutant Htt samples (density plot Fig. 5A). In the soluble fraction an increase in diGly site abundance in general did not correlate with an increase in protein abundance. The relative abundance of most of the proteins was not altered in the soluble fraction. Thus, in general, changes observed in diGly site abundance were not affected by changes at the protein level for most of the proteins in the soluble fraction. Fig. 5B shows the correlation for all proteins and diGly sites that were identified in 3 out of 4 replicates in both wild-type and mutant samples of the insoluble fraction. DiGly site fold changes follow a normal distribution, which is however clearly skewed toward the mutant samples. Some diGly sites were upregulated as well as their proteins, such as the K48 diGly site of ubiquitin (Ubc K48) and both K422 and K437 diGly sites of sequestosome1 (Sqstm1/p62). There were also several diGly sites upregulated even though their proteins were not differentially up- or downregulated, such as Hsp90aa1 K112 and Calm3 K31 (Fig. 5B).
Fig. 5.
Correlation between global proteome and ubiquitinome datasets in the soluble and insoluble fraction. Log2 fold change in global protein levels versus diGly sites are shown in these figures along with the density distributions of fold changes for both soluble (A) and insoluble (B) fractions. Each diGly site identified in at least 3 out of 4 replicates both in mutant and wild type is shown as a single data point with the corresponding diGly site fold change and the fold change of the protein that the diGly peptide is coming from. p values from the diGly t test are represented using a color gradient. The dashed y = x line and the dashed square are for ease of interpretation, latter representing the area with fold changes between −1 and 1.
The correlation of diGly sites, that were only identified in one group (either wild type or mutant), to their global proteins in both the soluble and insoluble fraction are plotted in supplemental Fig. S3A and S3B, respectively. In general, the protein fold change was not altered for the majority of the diGly sites that were identified in the soluble fraction (supplemental Fig. S3A). For a few proteins the fold change was increased in wild-type samples whereas diGly sites for these proteins were only found in mutant samples. For instance, the fold change of Htt was strongly decreased in the mutant soluble fraction whereas its K9 diGly site was only found in mutant samples. In the insoluble fraction relatively more proteins showed upregulation at both the protein level and diGly site level (supplemental Fig. S3B), which is in agreement with the data in Fig. 5B. For instance, ubiquilin-2 protein levels were increased in the insoluble fraction of mHtt samples whereas the protein was also more ubiquitinated at K58 and K66 (supplemental Fig. S3B). Also, the protein level and diGly site level (K979) of Myo5a, a Htt interaction partner (45) with a role in vesicular trafficking, were both upregulated in the mHtt insoluble fraction.
Together these data show that in general protein fold changes did not affect the diGly site fold changes drastically. In the insoluble fraction more increased fold changes at the protein level were observed as compared with the soluble fraction, which is most likely the result of the relatively large differences in components of these proteomes (aggregates versus no aggregates). Increased protein fold changes were also reflected in increased diGly site fold changes in several cases.
DISCUSSION
Overlap with Other Huntington's Disease Studies
Several differentially expressed proteins in our datasets have been associated with HD before. For instance, Pde10a, a protein highly expressed in striatal medium spiny neurons (37), was found to be downregulated in affected brain regions of R6/2 mice by using a quantitative mass spectrometry approach (40). Further, Giampa et al. showed that inhibition of Pde10a alleviated neurological deficits and brain pathology in the HD mice model R6/2 (38). Pde10 expression profiling has also been suggested as a biomarker to follow early stages of HD progression (46). The actin-binding cytoskeletal protein Synaptopodin that is required for the formation of the spine apparatus was also found to be differentially expressed in R6/2 HD mice brain (39). Further, we found that Serpina3K and Inositol-trisphosphate 3-kinase A (Itpka) were downregulated in mHtt mice brain, whereas Methylthioribose-1-phosphate isomerase (Mri1) was upregulated, which is in correlation with a study of Skotte and colleagues (40). Several proteins that were enriched in the mutant Htt insoluble fractions were also identified in Htt aggregates in other studies, for instance Alpha-internexin (Ina), Vimentin (Vim), Transformer-2 protein homolog alpha (Tra2a) (39) and Ubiquilin-2 (Ubqln2) (41). Taken together, several proteins that were differentially expressed in fl Q175 mHtt brains in our study were also found to be differentially expressed in other HD models.
Comparison with transcriptome data in the striatum and cortex of the same mouse model with the same age (47) did not show any overlap with our global proteome data (Table S6), probably reflecting the difference between gene expression and protein expression.
Htt Ubiquitination Pattern in Soluble and Insoluble Fractions
In this work we identified diGly peptides for Htt K6, K9, K132, K804, and K837 in the soluble fractions, indicating that soluble Htt is ubiquitinated in mice brain samples. We observed differential ubiquitination of wild-type and mutant soluble Htt i.e. diGly peptides derived from wild-type Htt harbored primarily diGly motifs at K132 and K804, and rarely the motif was found at K837, whereas diGly peptides of soluble mHtt harbored the K6 and/or K9 diGly motifs. As proteins were digested into peptides our data does not reveal whether the full-length mHtt protein or only smaller fragments were ubiquitinated.
In the insoluble fraction we observed that mHtt was more ubiquitinated at K6 and K9 as compared with wild type. It has been established that aggregates of Htt are decorated with ubiquitin. Our lab showed previously through mutation of the three putative ubiquitination sites in mHtt exon1 that aggregated mHtt was polyubiquitinated in its N-terminal region in neuronal inclusion bodies (23). Recently, we found by a gel-based mass spectrometry approach and by SDS-PAGE WB that aggregated mHtt-exon1 was K48 polyubiquitinated at K6 in N2a cells (manuscript in preparation). Our present data shows that mHtt in the insoluble fraction is ubiquitinated at K6 and/or K9. Finding more Htt diGly sites than in the previous study might be explained by the application of a diGly peptide enrichment method which increases the depth of the analysis for diGly peptides. However, the difference might also be due to a different biological background (cell culture versus mice brain tissue, Htt exon1 versus full-length Htt). In our present study, we observed an upregulation of the K48 diGly site derived from ubiquitin in the insoluble fraction of mutant Htt samples, suggesting that mHtt aggregates are K48 polyubiquitinated, which agrees with our manuscript in preparation as well as with the study of Yau et al. They found, with the use of ubiquitin K11/K48 bispecific antibodies, that aggregated mHtt was K11 and K48 polyubiquitinated in different cell types, differentiated neurons and mice brain tissue (48).
Further, Yau et al. found that upon mHtt expression Ubiquilin-2 was K11- and K48-polyubiquitinated (48). We observed increased protein levels of Ubiquilin-2 in the insoluble fraction. Both in wild-type and mutant Htt soluble fractions Ubiquilin-2 was ubiquitinated at K58 to a similar extent. Additionally, soluble Ubiquilin-2 was ubiquitinated at K43 only in the mutant Htt samples. In the insoluble fraction Ubiquilin-2 was only ubiquitinated in the mutant samples; at K58 and K66. All these sites are in the UBL domain. Ubiquilin-2 has been shown to associate with mHtt aggregates (41) and to mediate the clearance of aggregates via the proteasome (49). The cause and role of increased ubiquitination of the UBL domain of Ubiquilin-2 in the insoluble fraction requires further investigation.
Recently two ubiquitin ligases with specificity for mHtt were reported, UBR4 and UBR5 (48). Unfortunately, our data gave no information about UBR5 because we did not identify peptides for this protein. We only identified UBR4 in the global proteome dataset of the insoluble fraction; however, no statistically significant differences were observed between wild-type and mutant samples.
Htt Protein Level Was Decreased in the Soluble Fraction of mHtt Mice as Compared with Wild Types
When examined at the age of 40 weeks, we observed a strong decrease of soluble Htt protein levels in mutant Htt mice when compared with wtHtt mice. This is in accordance with earlier work showing that Q175FDN mice display an age-dependent decline of mHtt protein levels in the cortex and striatum (25, 50). The reduction of mHtt could have been caused by reduced efficiency in transcription and/or translation of the mHtt gene and/or mRNA, respectively (25). Soluble Htt protein levels could have been additionally reduced because of sequestration in inclusion bodies. We observed significantly increased Htt K6 and K9 ubiquitination in the mHtt insoluble samples by diGly peptide enrichment. This suggests that at least a part of the Htt pool is present in the insoluble fraction. Finally, soluble Htt protein levels could have been reduced by degradation. Therefore, we tested whether soluble fl mHtt is a target for degradation in striatal cells (StHdh Q111/111). We observed a small increase in the level of soluble full-length mHtt after proteasome inhibition and after autophagy inhibition, suggesting that a part of the soluble full-length mHtt protein is targeted for degradation (supplemental Fig. S4A and S4B). It remains to be investigated whether the pool of mHtt that is targeted for degradation is the same pool as that was found to be ubiquitinated in the soluble fraction. The pool of soluble ubiquitinated Htt is however rather small (manuscript in preparation), which makes it difficult to study this in detail.
Pathway Analysis
First, we observed changes in the global proteome and ubiquitinome in proteins with a role in membrane organization and transport, translation and catabolic processes in the soluble fraction. These processes were also affected in the insoluble fraction at the protein level. Huntingtin plays different roles in cellular membrane and vesicle trafficking through interaction with microtubule-based kinesin and dynein motor proteins and with the actin-associated motor myosin VI (51). We observed that several components of the vesicle trafficking and intracellular transport network, such as huntingtin-interacting protein 1-related protein (Hip1r), AP-2 complex subunit alpha-2 (Ap2a2), Synaptotagmin-1 (Syt1) and Synaptotagmin-2 (Syt2), were differently ubiquitinated in mHtt samples (Fig. 3E). Second, we observed differential ubiquitination of proteins involved in translation in the soluble fraction and in the insoluble fraction we observed increased levels of proteins with a role in mRNA processing and gene expression. It has been shown that the Htt protein interacts with its own mRNA, RNA binding proteins and translation factors (52) and that Htt is involved in the trafficking of mRNAs (53). Third, proteins involved in catabolic processes were differentially ubiquitinated in the soluble fraction, such as several proteasome subunits and E3 ubiquitin ligase Itch. For instance, we found that USP14 was more ubiquitinated at K64 in its Ubiquitin-like (UBL) domain in wild-type samples in the soluble fraction. It was found that overexpression of USP14 reduces mHtt aggregates in cells via the ubiquitin-proteasome system (54). We found that E3 ubiquitin ligase Itch was more ubiquitinated at K407 in mutant samples of the soluble fraction. It has been shown that Itch co-localizes with polyQ-expanded Htt and Ataxin-3 aggregates and overexpression of Itch reduced the aggregation (55). Lastly, we observed significantly reduced levels of several proteins belonging to either complex I (Ndufa2, Ndufa5, Ndufa6, Ndufb3, Ndufs8) or complex IV (Cox5b, Cox6b1, Cox6c) of the oxidative phosphorylation machinery in the insoluble fraction of mutant Htt brain samples (Fig. 4E, cluster 3). These complexes are in the mitochondrial inner membrane and are required for the production of ATP. Complexes I, III and IV are responsible for the relocation of protons from the mitochondrial matrix to the intermembrane space, whereas complex V generates ATP. Deficiency of Complexes II, III and IV have been associated with HD in postmortem brains of patients (56, 57). Restoration of the activity of complex IV in HD transgenic mice was found to be neuroprotective (58). Our observations of changes in both protein levels and ubiquitination status might shed light on how these processes are affected in late stage HD disease, both at the protein level and at the level of regulation via the ubiquitin system. Knowledge about differential ubiquitination of known players and the identification of new proteins that might be involved in these processes in HD might be useful for groups who study these processes in detail.
Correlation Between Protein and diGly Site Fold Changes
We observed many changes in diGly site abundances in wild-type and mutant Htt brain samples, both in the soluble and insoluble fraction. Increased diGly site abundances could have been derived from increased levels of proteins of which the same percentage is ubiquitinated, or from increased ubiquitination of a stable amount of protein. In the soluble fraction we did not observe relative differences at the protein level for most of the proteins, which suggests that increased diGly site abundances were the result of increased protein ubiquitination for most of the cases. Comparisons like these have been made for cells treated with proteasome inhibitors. In those studies, the abundance of a small group of proteins was significantly decreased or increased, but the protein abundances of the majority of the proteins were not affected, whereas many of these proteins were significantly differentially ubiquitinated (59). In the insoluble fraction we observed a group of proteins of which both the relative abundance at the global proteome and the diGly-modified proteome were increased, suggesting that protein fold change contributed to the diGly site fold changes. Examples of proteins in this group are ubiquitin (K48 ubiquitination), Sequestosome/p62 (K422, K437), Glial fibrillary acidic protein (Gfap) (K92, K394), Neurofilament light polypeptide (nefl) (K15, K294) and Neurofilament medium polypeptide nefm (K296, K434), among others. Sequestosome-1 was ubiquitinated at K422 which is situated in the ubiquitin-associating (UBA) domain and at K437 which is flanking the UBA domain. It was shown that Htt acts as a scaffold for selective autophagy through interaction with sequestosome-1, K63-polyubiquitinated cargo proteins and LC3 (60). Glial fibrillary acidic protein is expressed in activated astrocytes and the degree of activation correlates with Huntington's disease progression (61). Neurofilament light polypeptide and Neurofilament medium polypeptide are subunits of neurofilaments and a component of the neuronal cytoskeleton. Neurofilament light protein has been proposed as a biomarker for HD as its increased levels in the cerebrospinal fluid and blood correlate with disease progression (62).
The fact that we did not observe significant differences in the relative protein levels for most ub-modified proteins in the soluble fraction does not mean that ubiquitination did not affect the protein level. The subpopulation of the ubiquitin-modified proteins is generally much smaller than the averaged protein population, as such a decreased half-life of a small ubiquitin-modified subpopulation might not be detected at the protein level (63). The use of proteasome inhibitors or autophagy inhibitors o/n or knockdown of proteasome subunits for several days might be used to accumulate the pool of ub-modified proteins targeted for degradation (59, 63, 64).
Final Remarks
Our data might reveal new players in pathways known to be affected in HD and might shed light on their regulation at the ubiquitin level. Further biochemical experiments might give mechanistic insights into these regulations, which is beyond the scope of this study. Additionally, performing global proteome and ubiquitinome studies in younger mice will be interesting as they will reveal differences between wild-type and mutant Htt mice brain in early stages of HD. Moreover, as the detection of mHtt diGly peptides in whole brain was successful, also the difference between HD affected and non-affected brain regions could be studied. Further, as in our study we could not discriminate between ubiquitination of full length Htt proteins or fragments thereof, follow-up studies could include a cut-off filter to separate proteins based on size. Understanding the role of differential ubiquitination of huntingtin as well as ubiquitin-regulated processes in Huntington's disease might lead to the identification of candidates for drug development with the aim to enhance the efficiency of degradation of toxic mHtt species via the Ubiquitin-Proteasome System.
Data Availability
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE (65) partner repository with the dataset identifier PXD010161. http://www.ebi.ac.uk/pride/archive/projects/PXD010161.
Supplementary Material
Acknowledgments
We thank Alicia Sanz Sanz for performing experiments for supplemental Fig. S4.
Footnotes
* This work was carried out on the Dutch national e-infrastructure with the support of SURF Cooperative. This work was funded by CHDI and Campagneteam Huntington.
In the discussion section we refer to a manuscript in preparation by K. Juenemann.
 This article contains supplemental Figures and Tables.
This article contains supplemental Figures and Tables.
1 The abbreviations used are:
- HD
- Huntington's disease
- CV
- column volume
- FDR
- false discovery rate
- fl
- full-length
- Htt
- huntingtin
- LFQ
- label-free quantification
- mHtt
- mutant huntingtin
- N17
- N-terminal domain of Htt protein consisting of the first 17 amino acids
- polyQ
- polyglutamine
- PTM
- post-translational modification
- Q175
- 175 polyglutamine repeats
- UBA
- ubiquitin-associated domain
- UBL
- ubiquitin-like domain
- wtHtt
- wild-type huntingtin.
REFERENCES
- 1. MacDonald M. E., Ambrose C. M., Duyao M. P., Myers R. H., Lin C., Srinidhi L., Barnes G., Taylor S. A., James M., Groot N., MacFarlane H., Jenkins B., Anderson M. A., Wexler N. S., Gusella J. F., Bates G. P., Baxendale S., Hummerich H., Kirby S., North M., Youngman S., Mott R., Zehetner G., Sedlacek Z., Poustka A., Frischauf A. M., Lehrach H., Buckler A. J., Church D., Doucette-Stamm L., O'Donovan M. C., Riba-Ramirez L., Shah M., Stanton V. P., Strobel S. A., Draths K. M., Wales J. L., Dervan P., Housman D. E., Altherr M., Shiang R., Thompson L., Fielder T., Wasmuth J. J., Tagle D., Valdes J., Elmer L., Allard M., Castilla L., Swaroop M., Blanchard K., Collins F. S., Snell R., Holloway T., Gillespie K., Datson N., Shaw D., and Harper P. S. (1993) A novel gene containing a trinucleotide repeat that is expanded and unstable on Huntington's disease chromosomes. Cell 72, 971–983 [DOI] [PubMed] [Google Scholar]
- 2. Graveland G. A., Williams R. S., and DiFiglia M. (1985) Evidence for degenerative and regenerative changes in neostriatal spiny neurons. Science 227, 770–773 [DOI] [PubMed] [Google Scholar]
- 3. Vonsattel J. P. G., and DiFiglia M. (1998) Huntington disease. J. Neuropathol. Exp. Neurol. 57, 369–384 [DOI] [PubMed] [Google Scholar]
- 4. Ross C. A., and Tabrizi S. J. (2011) Huntington's disease: From molecular pathogenesis to clinical treatment. Lancet Neurol. 10, 83–98 [DOI] [PubMed] [Google Scholar]
- 5. Imarisio S., Carmichael J., Korolchuk V., Chen C.-W., Saiki S., Rose C., Krishna G., Davies J. E., Ttofi E., Underwood B. R., and Rubinsztein D. C. (2008) Huntington's disease: from pathology and genetics to potential therapies. Biochem. J. 412, 191–209 [DOI] [PubMed] [Google Scholar]
- 6. Kim Y. J., Yi Y., Sapp E., Wang Y., Cuiffo B., Kegel K. B., Qin Z. H., Aronin N., and DiFiglia M. (2001) Caspase 3-cleaved N-terminal fragments of wild-type and mutant huntingtin are present in normal and Huntington's disease brains, associate with membranes, and undergo calpain-dependent proteolysis. Proc. Natl. Acad. Sci. U.S.A. 98, 12784–12789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Graham R. K., Deng Y., Slow E. J., Haigh B., Bissada N., Lu G., Pearson J., Shehadeh J., Bertram L., Murphy Z., Warby S. C., Doty C. N., Roy S., Wellington C. L., Leavitt B. R., Raymond L. A., Nicholson D. W., and Hayden M. R. (2006) Cleavage at the caspase-6 site is required for neuronal dysfunction and degeneration due to mutant huntingtin. Cell 125, 1179–1191 [DOI] [PubMed] [Google Scholar]
- 8. Ratovitski T., Gucek M., Jiang H., Chighladze E., Waldron E., D'Ambola J., Zhipeng H., Yideng L., Poirier M. A., Hirschhorn R. R., Graham R., Hayden M. R., Cole R. N., and Ross C. A. (2009) Mutant huntingtin N-terminal fragments of specific size mediate aggregation and toxicity in neuronal cells. J. Biol. Chem. 284, 10855–10867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Pennuto M., Palazzolo I., and Poletti A. (2009) Post-translational modifications of expanded polyglutamine proteins: Impact on neurotoxicity. Hum. Mol. Genet. 18, R40–R49 [DOI] [PubMed] [Google Scholar]
- 10. Wang Y., Lin F., and Qin Z.-H. (2010) The role of post-translational modifications of huntingtin in the pathogenesis of Huntington's disease. Neurosci. Bull. 26, 153–162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Martin D. D. O., Heit R. J., Yap M. C., Davidson M. W., Hayden M. R., and Berthiaume L. G. (2014) Identification of a post-translationally myristoylated autophagy-inducing domain released by caspase cleavage of huntingtin. Hum. Mol. Genet. 23, 3166–3179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Aiken C. T., Steffan J. S., Guerrero C. M., Khashwji H., Lukacsovich T., Simmons D., Purcell J. M., Menhaji K., Zhu Y. Z., Green K., LaFerla F., Huang L., Thompson L. M., and Marsh J. L. (2009) Phosphorylation of threonine 3: Implications for huntingtin aggregation and neurotoxicity. J. Biol. Chem. 284, 29427–29436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ehrnhoefer D. E., Sutton L., and Hayden M. R. (2011) Small changes, big impact: Posttranslational modifications and function of huntingtin in Huntington disease. Neuroscientist. 17, 475–492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Thompson L. M., Aiken C. T., Kaltenbach L. S., Agrawal N., Illes K., Khoshnan A., Martinez-Vincente M., Arrasate M., O'Rourke J. G., Khashwji H., Lukacsovich T., Zhu Y. Z., Lau A. L., Massey A., Hayden M. R., Zeitlin S. O., Finkbeiner S., Green K. N., LaFerla F. M., Bates G., Huang L., Patterson P. H., Lo D. C., Cuervo A. M., Marsh J. L., and Steffan J. S. (2009) IKK phosphorylates Huntingtin and targets it for degradation by the proteasome and lysosome. J. Cell Biol. 187, 1083–1099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Wang B., Zeng L., Merillat S. A., Fischer S., Ochaba J., Thompson L. M., Barmada S. J., Scaglione K. M., and Paulson H. L. (2018) The ubiquitin conjugating enzyme Ube2W regulates solubility of the Huntington's disease protein, huntingtin. Neurobiol. Dis. 109, 127–136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. de Pril R., Fischer D. F., Roos R. A. C., and van Leeuwen F. W. (2007) Ubiquitin-conjugating enzyme E2–25K increases aggregate formation and cell death in polyglutamine diseases. Mol. Cell Neurosci. 34, 10–19 [DOI] [PubMed] [Google Scholar]
- 17. Mishra A., Dikshit P., Purkayastha S., Sharma J., Nukina N., and Jana N. R. (2008) E6-AP promotes misfolded polyglutamine proteins for proteasomal degradation and suppresses polyglutamine protein aggregation and toxicity. J. Biol. Chem. 283, 7648–7656 [DOI] [PubMed] [Google Scholar]
- 18. Miller V. M., Nelson R. F., Gouvion C. M., Williams A., Rodriguez-Lebron E., Harper S. Q., Davidson B. L., Rebagliati M. R., and Paulson H. L. (2005) CHIP suppresses polyglutamine aggregation and toxicity in vitro and in vivo. J. Neurosci. 25, 9152–9161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Luo H., Cao L., Liang X., Du A., Peng T., and Li H. (2018) Herp promotes degradation of mutant huntingtin: involvement of the proteasome and molecular chaperones. Mol. Neurobiol. 55, 7652–7668 [DOI] [PubMed] [Google Scholar]
- 20. Yang H., Zhong X., Ballar P., Luo S., Shen Y., Rubinsztein D. C., Monteiro M. J., and Fang S. (2007) Ubiquitin ligase Hrd1 enhances the degradation and suppresses the toxicity of polyglutamine-expanded huntingtin. Exp. Cell Res. 313, 538–550 [DOI] [PubMed] [Google Scholar]
- 21. Bhat K. P., Yan S., Wang C., Li S., and Li X. (2014) Differential ubiquitination and degradation of huntingtin fragments modulated by ubiquitin-protein ligase E3A. Proc. Natl. Acad. Sci. U.S.A. 111, 5706–5711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Koyuncu S., Saez I., Lee H. J., Gutierrez-Garcia R., Pokrzywa W., Fatima A., Hoppe T., and Vilchez D. (2018) The ubiquitin ligase UBR5 suppresses proteostasis collapse in pluripotent stem cells from Huntington's disease patients. Nat. Commun. 9, 1–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Juenemann K., Wiemhoefer A., and Reits E. a. (2015) Detection of ubiquitinated huntingtin species in intracellular aggregates. Front. Mol. Neurosci. 8, 1–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Steffan J. S., Agrawal N., Pallos J., Rockabrand E., Trotman L. C., Slepko N., Illes K., Lukacsovich T., Zhu Y.-Z., Cattaneo E., Pandolfi P. P., Thompson L. M., and Marsh J. L. (2004) SUMO modification of Huntingtin and Huntington's disease pathology. Science 304, 100–104 [DOI] [PubMed] [Google Scholar]
- 25. Southwell A. L., Smith-Dijak A., Kay C., Sepers M., Villanueva E. B., Parsons M. P., Xie Y., Anderson L., Felczak B., Waltl S., Ko S., Cheung D., Cengio L. D., Slama R., Petoukhov E., Raymond L. A., and Hayden M. R. (2016) An enhanced Q175 knock-in mouse model of Huntington disease with higher mutant huntingtin levels and accelerated disease phenotypes. Hum. Mol. Genet. 25, 3654–3675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Ochaba J., Morozko E. L., O'Rourke J. G., and Thompson L. M. (2018) Fractionation for resolution of soluble and insoluble huntingtin species. J. Vis. Exp. 132, e57082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Van Der Wal L., Bezstarosti K., Sap K. A., Dekkers D. H. W., Rijkers E., Mientjes E., Elgersma Y., and Demmers J. A. A. (2018) Improvement of ubiquitylation site detection by Orbitrap mass spectrometry. J. Proteomics 172, 49–56 [DOI] [PubMed] [Google Scholar]
- 28. Tyanova S., Temu T., and Cox J. (2016) The MaxQuant computational platform for mass spectrometry – based shotgun proteomics. Nat. Protoc. 11, 2301–2319 [DOI] [PubMed] [Google Scholar]
- 29. The UniProt Consortium. (2017) UniProt: The universal protein knowledgebase. Nucleic Acids Res. 45, D158–D169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Tyanova S., Temu T., Sinitcyn P., Carlson A., Hein M. Y., Geiger T., Mann M., and Cox J. (2016) The Perseus computational platform for comprehensive analysis of (prote)omics data. Nat. Methods 13, 731–740 [DOI] [PubMed] [Google Scholar]
- 31. Doncheva N. T., Morris J. H., Gorodkin J., and Jensen L. J. (2019) Cytoscape StringApp: network analysis and visualization of proteomics data. J. Proteome Res. 18, 623–632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Shannon P., Markiel A., Owen Ozier 2, Baliga N. S., Wang J. T., Ramage D., Amin N., Schwikowski B., and Ideker T. (2003) Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2498–2504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Maere S., Heymans K., and Kuiper M. (2005) BiNGO: a Cytoscape plugin to assess overrepresentation of Gene Ontology categories in Biological Networks. Bioinformatics 21, 3448–3449 [DOI] [PubMed] [Google Scholar]
- 34. VIB-UGENT Center for Plant Systems Biology (Last accessed: 04/04/2019) Calculate and draw custom Venn diagrams. URL: http://bioinformatics.psb.ugent.be/webtools/Venn/
- 35. Bader G. D., and Hogue C. W. (2003) An automated method for finding molecular complexes in large protein interaction networks. BMC Bioinformatics 4, 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. O'Rourke J. G., Gareau J. R., Ochaba J., Song W., Rasko T., Reverter D., Lee J., Monteys A. M., Pallos J., Mee L., Vashishtha M., Apostol B. L., Nicholson T. P., Illes K., Zhu Y. Z., Dasso M., Bates G. P., Difiglia M., Davidson B., Wanker E. E., Marsh J. L., Lima C. D., Steffan J. S., and Thompson L. M. (2013) SUMO-2 and PIAS1 modulate insoluble mutant huntingtin protein accumulation. Cell Rep. 4, 362–375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Xie Z., Adamowicz W. O., Eldred W. D., Jakowski A. B., Kleiman R. J., Morton D. G., Stephenson D. T., Strick C. A., Williams R. D., and Menniti F. S. (2006) Cellular and subcellular localization of PDE10A, a striatum-enriched phosphodiesterase. Neuroscience 139, 597–607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Giampà C., Laurenti D., Anzilotti S., Bernardi G., Menniti F. S., and Fusco F. R. (2010) Inhibition of the striatal specific phosphodiesterase PDE10A ameliorates striatal and cortical pathology in R6/2 mouse model of Huntington's disease. PLoS ONE 5, 1–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Hosp F., Gutiérrez-Ángel S., Schaefer M. H., Cox J., Meissner F., Hipp M. S., Hartl F. U., Klein R., Dudanova I., and Mann M. (2017) Spatiotemporal proteomic profiling of Huntington's disease inclusions reveals widespread loss of protein function. Cell Rep. 21, 2291–2303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Skotte N. H., Andersen J. V., Santos A., Nørremølle A., Waagepetersen H. S., Nielsen M. L., Aldana B. I., Willert C. W., and Nielsen M. L. (2018) Integrative characterization of the R6/2 mouse model of Huntington's disease reveals dysfunctional astrocyte metabolism. Cell Rep. 23, 2211–2224 [DOI] [PubMed] [Google Scholar]
- 41. Rutherford N. J., Lewis J., Clippinger A. K., Thomas M. A., Adamson J., Cruz P. E., Cannon A., Xu G., Golde T. E., Shaw G., Borchelt D. R., and Giasson B. I. (2013) Unbiased screen reveals ubiquilin-1 and -highly associated with huntingtin inclusions. Brain Res. 1524, 62–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Szklarczyk D., Morris J. H., Cook H., Kuhn M., Wyder S., Simonovic M., Santos A., Doncheva N. T., Roth A., Bork P., Jensen L. J., and Von Mering C. (2017) The STRING database in 2017: Quality-controlled protein-protein association networks, made broadly accessible. Nucleic Acids Res. 45, D362–D368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Wickham H. (2009) ggplot2: Elegant Graphics for Data Analysis. Springer-Verlag New York [Google Scholar]
- 44. RCore Team. (2018) R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria: https://www.R-project.org/ [Google Scholar]
- 45. Culver B. P., Savas J. N., Park S. K., Choi J. H., Zheng S., Zeitlin S. O., Yates J. R., and Tanese N. (2012) Proteomic analysis of wild-type and mutant huntingtin-associated proteins in mouse brains identifies unique interactions and involvement in protein synthesis. J. Biol. Chem. 287, 21599–21614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Russell D. S., Barret O., Jennings D. L., Friedman J. H., Tamagnan G. D., Thomae D., Alagille D., Morley T. J., Papin C., Papapetropoulos S., Waterhouse R. N., Seibyl J. P., and Marek K. L. (2014) The phosphodiesterase 10 positron emission tomography tracer, [18F]MNI-659, as a novel biomarker for early huntington disease. JAMA Neurol. 71, 1520–1528 [DOI] [PubMed] [Google Scholar]
- 47. Langfelder P., Cantle J. P., Chatzopoulou D., Wang N., Gao F., Al-Ramahi I., Lu X. H., Ramos E. M., El-Zein K., Zhao Y., Deverasetty S., Tebbe A., Schaab C., Lavery D. J., Howland D., Kwak S., Botas J., Aaronson J. S., Rosinski J., Coppola G., Horvath S., and Yang X. W. (2016) Integrated genomics and proteomics define huntingtin CAG length-dependent networks in mice. Nat. Neurosci. 19, 623–633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Yau R. G., Doerner K., Castellanos E. R., Matsumoto M. L., Dixit V. M., Rape M., Haakonsen D. L., Werner A., Wang N., Yang X. W., and Martinez-Martin N. (2017) Assembly and function of heterotypic ubiquitin chains in cell-cycle and protein quality control. Cell 171, 1–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Hjerpe R., Bett J. S., Keuss M. J., Knebel A., Marchesi F., Kurz T., Hjerpe R., Bett J. S., Keuss M. J., Solovyova A., Mcwilliams T. G., Glickman M. H., Trost M., Knebel A., Marchesi F., and Kurz T. (2016) UBQLN2 mediates autophagy-independent protein aggregate clearance by the proteasome. Cell 166, 1–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Franich N. R., Basso M., Andre E. A., Ochaba J., Kumar A., Thein S., Fote G., Kachemov M., Lau A. L., Yeung S. Y., Osmand A., Zeitlin S. O., Ratan R. R., Thompson L. M., and Steffan J. S. (2018) Striatal mutant huntingtin protein levels decline with age in homozygous huntington's disease knock-in mouse models. J. Huntingtons. Dis. 7, 137–150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Caviston J. P., and Holzbaur E. L. F. (2009) Huntingtin as an essential integrator of intracellular vesicular trafficking. Trends Cell Biol. 19, 147–155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Culver B. P., Declercq J., Dolgalev I., Yu M. S., Ma B., Heguy A., and Tanese N. (2016) Huntington's disease protein huntingtin associates with its own mRNA. J. Huntingtons. Dis. 5, 39–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Savas J. N., §1, M‡ Deinhardt K., Culver B. P., Restituito S., Wu L., Belasco J. G., Chao M. V., and Tanese N. (2010) A role for Huntington disease protein in dendritic RNA granules. J. Biol. Chem. 285, 13142–13153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Hyrskyluoto A., Lundh S. H., Rappou E., Petersen A., Waltimo T., Lindholm D., Do H. T., Reijonen S., Bruelle C., Kivinen J., and Korhonen L. (2014) Ubiquitin-specific protease-14 reduces cellular aggregates and protects against mutant huntingtin-induced cell degeneration: involvement of the proteasome and ER stress-activated kinase IRE1. Hum. Mol. Genet. 23, 5928–5939 [DOI] [PubMed] [Google Scholar]
- 55. Chhangani D., Upadhyay A., Amanullah A., Joshi V., and Mishra A. (2014) Ubiquitin ligase ITCH recruitment suppresses the aggregation and cellular toxicity of cytoplasmic misfolded proteins. Sci. Rep. 4, 5077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Gu M., Gash M. T., Mann V. M., Javoy-Agid F., Cooper J. M., and Schapira A. H. V. (1996) Mitochondrial defect in Huntington's disease caudate nucleus. Ann. Neurol. 39, 385–389 [DOI] [PubMed] [Google Scholar]
- 57. Browne S. E., Bowling A. C., MacGarvey U., Baik M. J., Berger S. C., Muqit M. M. K., Bird E. D., and Beal M. F. (1997) Oxidative damage and metabolic dysfunction in huntington's disease: Selective vulnerability of the basal ganglia. Ann. Neurol. 41, 646–653 [DOI] [PubMed] [Google Scholar]
- 58. Bae B. II, Xu H., Igarashi S., Fujimuro M., Agrawal N., Taya Y., Hayward S. D., Moran T. H., Montell C., Ross C. A., Snyder S. H., and Sawa A. (2005) p53 mediates cellular dysfunction and behavioral abnormalities in Huntington's disease. Neuron 47, 29–41 [DOI] [PubMed] [Google Scholar]
- 59. Akimov V., Barrio-Hernandez I., Hansen S. V. F., Hallenborg P., Pedersen A. K., Bekker-Jensen D. B., Puglia M., Christensen S. D. K., Vanselow J. T., Nielsen M. M., Kratchmarova I., Kelstrup C. D., Olsen J. V., and Blagoev B. (2018) UbiSite approach for comprehensive mapping of lysine and N-terminal ubiquitination sites. Nat. Struct. Mol. Biol. 25, 631–640 [DOI] [PubMed] [Google Scholar]
- 60. Cuervo A. M., Chen D., Tito A., Rui Y.-N., Sun Y., David G., Stimming E. F., Patel B., Xu Z., Chen Z., Zhang S., and Bellen H. J. (2015) Huntingtin functions as a scaffold for selective macroautophagy. Nat. Cell Biol. 17, 262–275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Faideau M., Deglon N., Welch M., Auregan G., Cormier K., Gilmore R., Kim J., Dufour N., Guillermier M., Brouillet E., Hantraye P., Ferrante R. J., and Bonvento G. (2010) In vivo expression of polyglutamine-expanded huntingtin by mouse striatal astrocytes impairs glutamate transport: a correlation with Huntington's disease subjects. Hum. Mol. Genet. 19, 3053–3067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Byrne L. M., Rodrigues F. B., Blennow K., Durr A., Leavitt B. R., Roos R. A. C., Scahill R. I., Tabrizi S. J., Zetterberg H., Langbehn D., and Wild E. J. (2017) Neurofilament light protein in blood as a potential biomarker of neurodegeneration in Huntington's disease: a retrospective cohort analysis. Lancet Neurol. 16, 601–609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Kim W., Comb M. J., Possemato A., Rush J., Sowa M. E., Huttlin E. L., Li J., Rad R., Bennett E. J., Guo A., Harper J. W., and Gygi S. P. (2011) Systematic and quantitative assessment of the ubiquitin-modified proteome. Mol. Cell 44, 325–340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Sap K. A., Bezstarosti K., Dekkers D. H. W., Voets O., and Demmers J. A. A. (2017) Quantitative proteomics reveals extensive changes in the ubiquitinome after perturbation of the proteasome by targeted dsRNA-mediated subunit knockdown in Drosophila. J. Proteome Res. 16, 2848–2862 [DOI] [PubMed] [Google Scholar]
- 65. Perez-Riverol Y., Csordas A., Bai J., Bernal-Llinares M., Hewapathirana S., Kundu D. J., Inuganti A., Griss J., Mayer G., Eisenacher M., Pérez E., Uszkoreit J., Pfeuffer J., Sachsenberg T., Yilmaz Ş Tiwary S, Cox J., Audain E., Walzer M., Jarnuczak A. F., Ternent T., Brazma A., and Vizcaíno J. A. (2019) The PRIDE database and related tools and resources in 2019: Improving support for quantification data. Nucleic Acids Res. 47, D442–D450 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE (65) partner repository with the dataset identifier PXD010161. http://www.ebi.ac.uk/pride/archive/projects/PXD010161.