Small proteins in plasma regulate diverse processes from metabolism to immune functions and are difficult to study, because of the large dynamic range of protein abundance. To facilitate their detection, we developed a method using protein dissociation, precipitation and size exclusion chromatography called SPEA. Herein, we demonstrate SPEA is capable of reproducible characterization of known small proteins and identification of novel factors such as C5ORF46. When applied to study humans undergoing intermittent fasting, novel changes in iron regulation were observed.
Keywords: Serum/Plasma*, Plasma or serum analysis, Hormones*, Chromatography, Insulin resistance, chemokine, hepcidin, insulin, intermittent fasting
Graphical Abstract
Highlights
Development of a method for low-abundance small protein analysis in human plasma.
Deep proteome profiling demonstrated unbiased identification of diverse factors.
Method enabled identification of a novel human plasma protein C5ORF46.
Analysis of intermittent fasting response showed changes in iron regulation.
Abstract
Unbiased and sensitive quantification of low abundance small proteins in human plasma (e.g. hormones, immune factors, metabolic regulators) remains an unmet need. These small protein factors are typically analyzed individually and using antibodies that can lack specificity. Mass spectrometry (MS)-based proteomics has the potential to address these problems, however the analysis of plasma by MS is plagued by the extremely large dynamic range of this body fluid, with protein abundances spanning at least 13 orders of magnitude. Here we describe an enrichment assay (SPEA), that greatly simplifies the plasma dynamic range problem by enriching small-proteins of 2–10 kDa, enabling the rapid, specific and sensitive quantification of >100 small-protein factors in a single untargeted LC-MS/MS acquisition. Applying this method to perform deep-proteome profiling of human plasma we identify C5ORF46 as a previously uncharacterized human plasma protein. We further demonstrate the reproducibility of our workflow for low abundance protein analysis using a stable-isotope labeled protein standard of insulin spiked into human plasma. SPEA provides the ability to study numerous important hormones in a single rapid assay, which we applied to study the intermittent fasting response and observed several unexpected changes including decreased plasma abundance of the iron homeostasis regulator hepcidin.
The specific detection and quantification of low abundance small proteins in human plasma (e.g. hormones, immune factors, metabolic regulators) plays a key role in clinical diagnosis, because of the importance of these factors in maintaining homeostasis and their functional disruption in diverse disease states. Typically, analysis of these small protein factors is performed with assays employing antibodies in either ELISAs, or in specific enrichment procedures before targeted LC-MS analysis (1, 2). Although these assays can be fast and sensitive, they are expensive and typically only quantify one protein at a time. This precludes discovery of novel factors and limits precision medicine approaches that require a global view of blood components including hormones and immune system-related factors. In addition, antibody-based assays are not always specific enough to distinguish between active protein chains, their isoforms, or degradation products (3).
Mass spectrometry-based proteomic analysis could overcome these issues, but a major problem is the large dynamic range of blood plasma protein abundance covering ∼13 orders of magnitude (4). For example, many protein hormones such as insulin exist in plasma at abundances <10 ng/ml, whereas albumin is ∼45 mg/ml. The approach taken by most groups to deeply characterize the plasma proteome and identify all these factors either employs depletion of the top 5–60 most abundant proteins using antibody-based columns before trypsin digestion, or peptide-level fractionation of tryptic peptides (e.g. high-pH reverse phase), or the sequential combination of both techniques (4–6). Although this approach is effective, it consumes a very large amount of instrument time (days to weeks) to acquire the data for each sample, which makes it difficult to apply in clinical studies.
Analysis of small-protein factors in plasma from large clinical trials is more typically performed using single-shot targeted top-down mass spectrometry, downstream of various sample preparation methods, for the primary purpose of quantifying previously known small proteins in plasma. The sample preparation methods employed in these studies include either acidic-alcohol extraction (7), basic-alcohol extraction (8), acetonitrile-based extraction (9–11), or immunoaffinity depletion (12). These extraction methods facilitate dissociation of small proteins from larger factors (13, 14) and the bulk removal of large plasma proteins by precipitation. For example, these studies have successfully quantified several commonly studied small protein hormones such as insulin (8, 15, 16), hepcidin (9, 16), and IGF-1 (7, 10, 11, 16). For quantification of analytes, these studies generally use high flow rate liquid chromatography separations with online targeted mass spectrometry detection. However, such methods do not facilitate identification of new small plasma proteins for biomarker discovery.
In contrast, single-shot untargeted mass spectrometry analysis with high-sensitivity nanoflow chromatography and high-resolution data-dependent MS acquisition offers excellent sensitivity for new protein discovery. Untargeted single-shot LC-MS/MS analysis for the identification of small plasma proteins has been described previously, but most of these studies have not identified from MS/MS spectra with standard database search approaches the active chains of a wide variety of clinically relevant small-proteins (e.g. insulin, APOC4, IGF-1, hepcidin and RANTES) (12, 16–19). The identification of peptides derived from the active chains of these small secreted proteins is key as it indicates an ability to identify new small protein factors and unexpected modifications of previously known proteins. These studies have all used depletion/precipitation extraction methods to identify small proteins in plasma, however no precipitation method is 100% efficient and small amounts of intact albumin and other large proteins likely remain in the sample, which will reduce detection sensitivity. No study has yet employed a precipitation step before size-exclusion chromatography (SEC)1 to enable the specific isolation of small proteins for subsequent unbiased mass spectrometry-based analysis.
The central aim of this study was to develop a method that was able to analyze new small protein factors in human plasma. To this end, we have developed a multi-step protocol called SPEA (small-protein enrichment assay), aimed at allowing discovery of new small proteins in plasma while also allowing quantification and identification of peptides derived from the active chains of previously known low abundance factors. Our protocol dissociates protein-protein interactions, removes large proteins >10 kDa and small degradation products <2 kDa using SEC, before trypsin digestion and high sensitivity nanoLC-MS/MS analysis. This has allowed us to explore all the components of this fraction using unbiased bottom-up mass spectrometry-based identification and quantification. Using this protocol, we have detected the presence of a new small protein factor (C5ORF46) in human plasma that may play a key role in lipid homeostasis, demonstrated the reproducibility of our method with a stable-isotope labeled protein standard, and characterized the response to intermittent fasting in a human clinical trial cohort. Together these applications demonstrate the capabilities of our workflow for the simultaneous measurement of many small proteins in human plasma, which will provide a molecular profiling tool for use in precision medicine.
EXPERIMENTAL PROCEDURES
Chemicals and Reagents
Acetonitrile (Optima grade), water (Optima grade), ammonia, formic acid and isopropanol (Optima grade) were from Thermo Fisher Scientific (Waltham, MA). Proteomics-grade trypsin (Catalogue number T6567) and all other reagents were from Sigma Aldrich (St. Louis, MO).
Plasma Small Protein Hormone Enrichment (SPEA)
Dissociation-Precipitation
Human blood plasma samples were randomized using a random number generator before processing. Plasma samples were thawed on ice and vortexed before aliquoting 50 μl into a 2 ml tube (Eppendorf, Hamburg, Germany) at room temperature that contained 450 μl of extraction buffer (0.25 m HCl, 87.5% ethanol in H2O). Samples were vortexed every 15 min and incubated for a total of 30 min at room temperature. Precipitated proteins were pelleted by centrifugation at 8400 × g for 10 min at room temperature. The supernatant was moved to new 2 ml tube and 125 μl of chloroform added and the tube vortexed. This is a key step to ensure lipid removal before SEC-based separations. To facilitate phase separation, 500 μl of water was added and the tube vortexed before centrifugation for 10 min at 7000 × g at room temperature. The clear top phase (∼850 uL) was retained (peptide supernatant) while avoiding any pellet and the yellow chloroform phase. The small-protein hormone containing supernatant was filtered using 0.45 μm Ultrafree-MC HV centrifugal filter units (Merck Millipore, Burlington, MA) before moving the filtrate into vials for uHPLC-based SEC separation.
15N-Insulin Spike-in
Heavy human insulin was purchased from Sigma (SILuProt Insulin) labeled with 15N (>97% incorporation efficiency), which was recombinantly expressed in Pichia pastoris. The entire vial was resuspended in 2% acetic acid to a final concentration of 1.722 μm and the stock aliquoted into 0.5 ml low protein binding tubes for storage at −80 °C. For generation of the standard curve, an intermediate dilution of 312.5 nm heavy insulin was made with 2% acetic acid in low protein binding tubes, which was spiked into plasma with a 10-fold dilution to make the most concentrated heavy insulin spike-in (45 μl plasma, 5 μl heavy insulin). For all other points on the standard curve 5-fold dilutions of the 312.5 nm intermediate stock were made with 2% acetic acid in low protein binding tubes, before being spiked into plasma with a 10-fold dilution as above (45 μl plasma, 5 μl heavy insulin). These 50 μl samples were then processed for dissociation-precipitation with ethanol-HCl as described above.
Denaturing Small-Protein Targeted SEC
To isolate proteins <10 kDa from larger proteins in the small-protein hormone containing supernatant a Dionex Ultimate 3000 Bio-RS uHPLC system (Thermo Fisher Scientific) was used combined with an Agilent (Santa Clara, CA) AdvanceBio SEC column with 130 Å pores, 2.7 μm particles, dimensions of 7.8 × 300 mm. The column was equilibrated with 10 column volumes of denaturing SEC running buffer (30% acetonitrile and 0.1% TFA) before sample analysis. Standards consisting of either ubiquitin (Sigma - U6253–5MG), or a HPLC peptide standard mix (Sigma - H2016–1VL) diluted in SEC running buffer were used for mass calibration. The small-protein hormone containing supernatant was stored in the auto-sampler at 4 °C before analysis and for each SEC separation, 200 μl was injected onto the column. Each SEC separation was performed for 25 min (1.5 column volumes) at a flow rate of 1 ml min−1 at a column temperature of 30 °C. The eluting proteins were monitored by UV absorbance at 215 and 280 nm. Only one fraction was collected into a low protein binding 2 ml tube or 2 ml 96-deep-well plate (Eppendorf) between 6 min to 8 min retention time. This fraction corresponded to proteins of molecular weight between <10 kDa and >2 kDa, which had a total volume of 2 ml and the fractions were stored at 4 °C before subsequent processing.
Peptide Reduction, Alkylation and Trypsin Digestion
The collected fraction was dried using a GeneVac EZ-2 (SP Industries, Warminster, PA) using the HPLC setting at 45 °C and the dried proteins were resuspended in 100 μl of 50 mm triethylammonium bicarbonate (TEAB) pH 8.5 in H2O. Disulfide bonds were reduced by addition of DTT to a final concentration of 5 mm and incubated on a thermomixer at 95 °C at 1000 rpm for 10 min. To alkylate the reduced sulfhydryl groups chloroacetamide was added to a final concentration of 20 mm and samples incubated on a thermomixer at 95 °C at 1000 rpm for 10 min. For trypsin digestion samples were cooled to room temperature and trypsin was added at a ratio of 1:20 (trypsin to protein), where each SEC fraction contained ∼ 4 μg protein and therefore 200 ng was added. The samples were incubated for 16 h at 37 °C at 500 rpm on a thermomixer to digest. To stop the digest 10% TFA in H2O was added to achieve a 1% final concentration.
Peptide Clean-Up using SDB-RPS StageTips
SDB-RPS StageTips were generated by punching double-stacked SDB-RPS discs (Sigma, Cat#66886-U) with an 18-gauge needle and mounted in 200 μl tips (Eppendorf), as shown in supplementary Methods. For clean-up using the Spin96, StageTips were inserted into a holder and placed in the top, which was then stacked onto the wash-bottom containing a polypropylene 96-well microtitre plate to collect the sample flow-through and washes. Each tip was wetted with 100 μl of 100% acetonitrile and centrifuged at 1000 × g for 1 min. Following wetting, each StageTip was equilibrated with 30% methanol/1% TFA, followed by 100 μl of 0.1% TFA in H2O, with centrifugation for each at 1000 × g for 3 min. Each StageTip was then loaded with the equivalent of ∼10 μg peptide in 1% TFA (<100 μl total volume per spin). The peptides were washed once with 100 μl of 0.2% TFA in water, which was followed by one wash with 100 μl of 99% isopropanol/1% TFA. For elution of peptides, the wash-bottom was exchanged with a bottom containing a holder supporting an unskirted PCR plate that has been trimmed to fit. To elute, 100 μl of 5% ammonium hydroxide/80% acetonitrile was added to each tip and centrifuged as above for 5 min. Samples in the PCR plate were dried using a GeneVac EZ-2 using the ammonia setting at 35 °C for 1 h 15 m. Dried peptides were resuspended in 7 μl of 5% formic acid per 200 μl SEC injection of small-protein hormone containing supernatant and stored at 4 °C until analyzed by LC-MS.
High pH Reverse Phase Chromatography
Pooled trypsin digested plasma proteins processed using SPEA (100 μg total) in 5% formic acid were subjected to high pH reverse phase chromatography on a Thermo Scientific Dionex Ultimate 3000 BioRS system with a fractionation auto-sampler, using a Waters (Milford, MA) XBridge Peptide BEH C18 column (130 Å, 3.5 μm, 4.6 mm × 250 mm, Cat No. 186003570). The column was incubated at 30 °C with a constant flow rate of 1 ml/min, with buffer A containing 2% acetonitrile (ACN) and 10 mm ammonium formate (pH 9) and buffer B containing 80% ACN and 10 mm ammonium formate (pH 9). Fractions were collected every 8.75 s from a retention time of 2 min to 16 min (96 pseudo fractions, concatenated into 16 fractions total). Peptides were separated by a linear gradient from 10% to 40% buffer B for the first 11 min and 100% buffer B for the remaining time. The fractions were collected in a 2 ml protein low-bind 96-well deep well plate (Eppendorf) across 16 wells in a concatenated pattern using tube wrapping.
LC-MS/MS Acquisition
Using a Thermo Fisher Dionex RSLCnano uHPLC, peptides in 5% (v/v) formic acid (injection volume 3 μl) were directly injected onto a 45 cm × 75 μm C18 (Dr. Maisch, Ammerbuch, Germany, 1.9 μm) fused silica analytical column with a ∼10 μm pulled tip, coupled online to a nanospray ESI source. Peptides were resolved over gradient from 5% acetonitrile to 40% acetonitrile over various gradient lengths ranging from 15 min to 140 min with a flow rate of 300 nL/min. Peptides were ionized by electrospray ionization at 2.3 kV. Tandem mass spectrometry analysis was carried out on either a Q-Exactive HF, or HFX mass spectrometer (ThermoFisher) using HCD fragmentation.
DDA
The data-dependent acquisition method used acquired MS/MS spectra of the top 10 most abundant ions at any one point during the gradient.
PRM
The parallel reaction monitoring (PRM) method used was a tier 3 analysis. This PRM method used a 15 min gradient separation and acquired targeted MS/MS spectra for the +4 charge-state tryptic peptide derived from human insulin beta-chain (FVNQHLCGSHLVEALYLVCGER) as it was the most abundant precursor, which was carbamidomethylated at both cysteine residues. Both the light (m/z = 651.3227) and heavy 15N-labeled (m/z = 659.0496) forms of this peptide were co-isolated (MSX count = 2) using the same injection time (MSX isochronous ITs = ON). The isolation window was tested across numerous runs with windows between 0.4–1.2 Th and 0.7 Th was determined as the optimal isolation window. The normalized collision energy (NCE) was tested for energies between 17 and 35, with 19 being the optimum used because of the maximal abundance of large (>800 m/z) fragment ions. Background interference was determined by comparison to blank runs. All spectra were acquired at 60,000 resolution in profile mode and a maximum injection time for MS/MS (PRM) of 100 ms.
DIA
The data-independent acquisitions were performed as described previously using variable isolation widths for different m/z ranges(20). Stepped normalized collision energy of 25 ± 10% was used for all DIA spectral acquisitions.
LC-MS/MS Data Analysis
MaxQuant Analysis
RAW data were analyzed using the quantitative proteomics software MaxQuant (21) (version 1.5.7.0). Both the PREFER plasma samples and high pH RP samples were searched together to increase identifications through the match-between-runs feature. This version of MaxQuant includes an integrated search engine, Andromeda (22). Peptide and protein level identification were both set to a false discovery rate of 1% using a target-decoy based strategy. The database supplied to the search engine for peptide identifications contained both the human UniProt database downloaded on the 30th September 2017, containing 42,325 protein sequence entries and the MaxQuant contaminants database. Mass tolerance was set to 4.5 ppm for precursor ions and MS/MS mass tolerance was 20 ppm. Enzyme specificity was set to semi-specific N-ragged trypsin (cleavage C-terminal to Lys and Arg) with a maximum of 2 missed cleavages permitted for the main search and fully specific trypsin (cleavage C-terminal to Lys and Arg) for the first search. Deamidation of Asn and Gln, oxidation of Met, pyro-Glu (with peptide N-term Gln) and protein N-terminal acetylation were set as variable modifications. Carbamidomethyl on Cys was searched as a fixed modification. Maxquant output was processed and statistical tests performed using the R software package (version 3.4.3). Processed data was plotted using Tableau (version 10.0.2).
Spectronaut Pulsar X directDIA Analysis
RAW data were analyzed using the quantitative proteomics software Spectronaut Pulsar X (version 12.0.20491.11.25225 (Jocelyn) (Biognosys, Schlieren, Switzerland)). Only the PREFER plasma samples were searched, not the high pH RP samples. The database supplied to the search engine for peptide identifications was a focused database generated in house from the fractionated plasma small-protein hormone analysis (1043 proteins, supplemental File S1). Enzyme specificity was set to semi-specific N-ragged trypsin (cleavage C-terminal to Lys and Arg) with a maximum of 2 missed cleavages permitted. Deamidation of Asn and Gln, oxidation of Met, pyro-Glu (with peptide N-term Gln) and protein N-terminal acetylation were set as variable modifications. Carbamidomethyl on Cys was searched as a fixed modification. To recalibrate for retention drift and intensity assignment for peaks the iRT profiling workflow was used (23). For peak list generation, interference (MS1 & MS2) correction was enabled, removing fragments/isotopes from potential quantitation if there was a presence of interfering signals, while keeping a minimum of three for quantitation. Spectra were de-isotoped based on RT apex distance and m/z spacing, de-multiplexing was not required. Each observed fragment ion could only be assignment to a single precursor peak list (24). The FDR was set to 1% using a target-decoy approach. Spectronaut generated a custom mass tolerance for each precursor ion. The threshold for accepting a precursor was set at a Q value <0.01 and each precursor must have >3 fragment ions. All other settings were factory default. Processed data was analyzed, and statistical tests performed using the R software package (version 3.4.3) with plot generated using Tableau (version 10.0.2).
PRM-data Analysis using Spectronaut Pulsar X
RAW data from the heavy insulin spike-in standard curve were analyzed using the quantitative proteomics software Spectronaut Pulsar X (version 12.0.20491.11.25225 (Jocelyn)). The PRM data files were analyzed using a “labeled” plasma library generated in house from the fractionated plasma small-protein hormone analysis. For each ion (either light, or heavy), the 6 most abundant fragment ions were selected for quantification and their intensity was summed. For light insulin, m/z = 651.3227 the transitions used for quantitation were: b14++ (796.886), b15++ (853.428 m/z), b16++ (934.959 m/z), y4+ (521.214 m/z), y5+ (620.282) and y6+ (733.366). For heavy insulin, m/z = 659.0496 the transitions used for quantitation were: b14++ (806.856 m/z), b15++ (863.896 m/z), b16++ (945.927 m/z), b17++ (1002.967 m/z), y4+ (528.193 m/z) and y5+ (628.258 m/z). To quantify both the 15N heavy and light forms of insulin the “labeled” library was generated using the label modifications 15N (A, C, D, E, F, G, I, L, M, P, S, T, V, Y), 15N2 (K, N, Q, W), 15N3 (H) & 15N4 (R). The workflow was set to use iRT profiling. All other settings were factory default.
Determining SPEA Peptide Recovery via Insulin ELISA
To determine the yield of insulin using the SPEA method we measured insulin using an ELISA at 2 steps in the SPEA protocol (raw plasma and the SEC fraction before digestion). The supernatant samples were dried overnight at 45 °C using a GeneVac EZ-2 on the HPLC setting, before resuspension in PBS. All samples were subjected to an insulin ELISA (Crystal Chem, Elk Grove Village, IL, Cat. No. 90095) as per manufacturer's instructions.
Hepcidin ELISA
Plasma hepcidin was measured by Human Hepcidin Quantikine ELISA Kit (R&D Systems, Minneapolis, MN; cat no. DHP250) according to the manufacturer's instructions. Briefly, 50 μl of assay diluent RD1–21 and 50 μl of either standards, or patient plasma (diluted 1:250 with calibrator diluent RD5–26), was added to assay wells and incubated for 2 h at room temperature. Following aspiration and 4 washes with wash buffer, 200 μl of human hepcidin conjugate was added to each well and incubated at room temperature for an additional 2 h at room temperature. Following 4 additional washes, 200 μl of substrate solution was added to all wells and allowed to develop over 30 min while protected from light, until color had developed in all standard wells. 50 μl of stop solution was used to cease color development and absorbance was determined at 450 nm with subtraction of a 540 nm reference on a Tecan M200 Pro plate reader (Tecan, Männedorf, Switzerland).
Intermittent Fasting Without Weight Loss Clinical Trial
The PREFER randomized controlled trial was a discovery-based, single-center study in Adelaide, South Australia and was registered with Clinicaltrials.gov (NCT01769976). The Royal Adelaide Hospital Research Ethics Committee approved the study protocol, and all participants provided written, informed consent before their inclusion. Each subject was assigned a number allowing for de-identification. A total of 88 women were enrolled in the study of which 25 were assigned to the intermittent fasting with weight maintenance group (IF100) analyzed in this manuscript, 3 participants withdrew during the diet period (2 because of time, 1 no longer wished to participate). The resulting 44 paired plasma samples were subjected to MS analysis with two samples from two different patients failing QC, leaving 40 patient samples for data analysis. Inclusion criteria were: aged 35–70 years; BMI 25–42 kg/m2; weight-stable (within 5% of their screening weight) for >6 months before study entry; no diagnosis of type 1 or type 2 diabetes; non-smoker; sedentary or lightly active (i.e. <2 moderate to high-intensity exercise sessions per week); consumed <140 g alcohol/week; no personal history of cardiovascular disease, no diagnosis of eating disorders or major psychiatric disorders (including those taking antidepressants); not pregnant or breastfeeding; and not taking medication that may affect study outcomes (e.g. phentermine, orlistat, metformin, excluding antihypertensive/lipid lowering medication). The active trial period was 10 weeks, comprised of a 2-week lead-in period, and 8 weeks of dietary intervention.
During the lead-in period, participants consumed their normal diet and maintained their weight. Following this, participants were placed on an intermittent fasting diet at 100% of calculated baseline energy requirements per week (i.e. weight maintenance). Energy requirements were calculated using an average of published equations, both of which use age, gender, height and weight variables. Because of the nature of the intervention, blinding was not possible.
Diet
On fed days, participants were provided with food equal to ∼145% of energy requirements. On fasting days, participants consumed breakfast before 8 am (∼37% of energy requirements were given at breakfast on fasting days) and were then instructed to “fast” for 24 h, until 8 am the following day. Participants were advised to fast on 3 non-consecutive week-days per week. During the fasting period participants were allowed to consume water and limited amounts of energy-free foods (e.g. “diet” drinks, chewing gum, mints), black coffee and/or tea, and were provided with 250 ml of a very low energy broth (86 kJ/250 ml, 2.0 g protein, 0.1 g fat, 3.0 g carbohydrate) for either lunch, or dinner. Participants were free-living, and foods were provided by fortnightly delivery to their home, except for fresh fruits and vegetables. Portions of fruits and vegetables were standardized, and participants allowed to self-select according to the number of serves specified in their individual menus (∼10% overall energy intake).
Blood Collection and Analysis
Blood samples were collected at 9am following a 10 h fast directly into purple K2-EDTA vacutainers (Becton Dickinson, Franklin Lakes, NJ) and placed on ice immediately after collection. Samples were centrifuged within 10 min of collection at 4 °C and the plasma frozen at −80 °C in cryotubes. Each sample was subject to <3 freeze-thaw cycles on ice.
Human Plasma for Small-protein Hormone Enrichment Assay Setup
Human plasma was obtained from 5 healthy volunteer using 4 ml purple K2-EDTA vacutainers (Becton Dickinson). Samples were immediately placed on ice and spun <15 min post-collection at 1500 × g for 15 min at 4 °C. Blood samples were frozen at −80 °C and exposed to <3 freeze-thaw cycles. The Royal Prince Alfred Hospital Research Ethics Review Committee approved the study protocol (X17–0129 and HREC/17/RPAH/183), and all participants provided written, informed consent before their inclusion.
Experimental Design and Statistical Rationale
Heavy Insulin Quantification
From previous Spin96 optimizations, we concluded three technical replicates of each sample was enough to account for experimental variation and provide statistically significant results.
PREFER Trial
The number of participants was established from past studies, which suggested n = 22 per group would allow detection of a mean difference in glucose infusion rate (GIR) of 15 μmol/kgFFM+17.7 between groups, with β = 0.8 and α = 0.05. We allowed for a 10% drop out rate, and thus recruited a total of n = 25 per group. In this manuscript we analyzed the intermittent fasting with weight maintenance group (IF100), with plasma samples collected before and after intermittent fasting. To these data we applied a Wilcox robust test to allow for proteins whose distribution for the difference between treatment groups across participants was not normally distributed. Specifically, we used Yuen's test on trimmed means for dependent (paired) samples. Fold changes comparing plasma peptide precursor abundance before and after intermittent fasting were calculated using the median. For plotting the variation in individual peptide precursor intensity measurements and clinical measures in response to IF, we calculated adjusted values that force all participants to have the same mean value for each measure. For all datasets statistical analyses were performed using R (version 3.4.3) and processed data was plotted using Tableau (version 10.0.2). Data are shown as median ± 95% confidence interval, unless otherwise stated. Significance was set at p < 0.05.
RESULTS
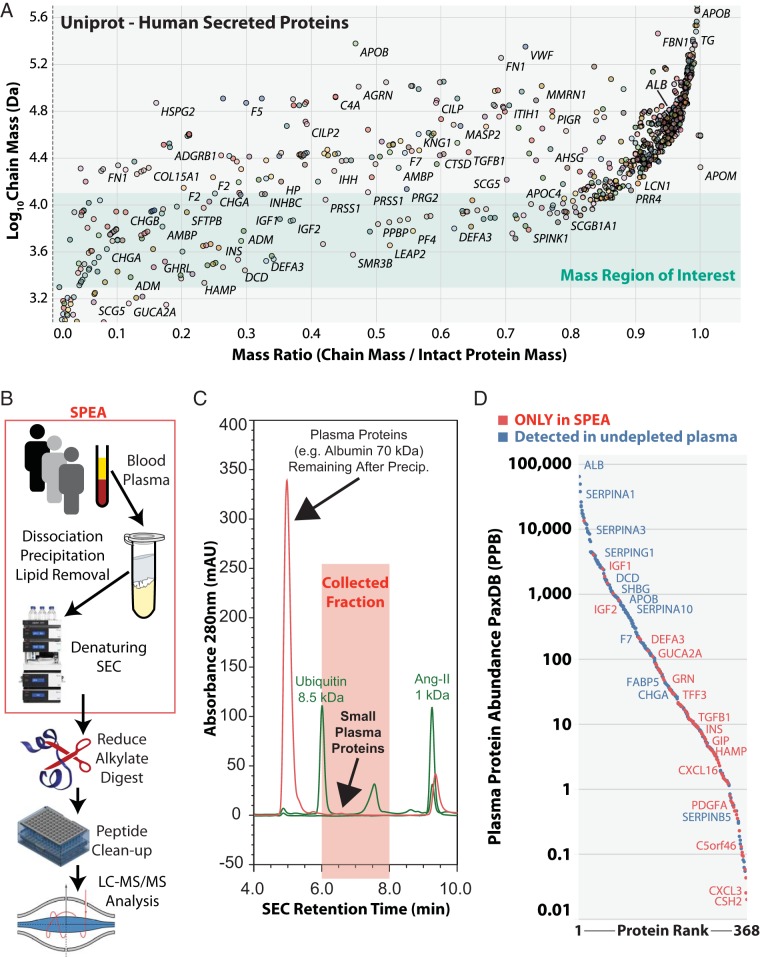
We aimed to adapt the latest developments in high-resolution size-exclusion chromatography (SEC) to solve the dynamic range problem for low abundance small-protein factors in human plasma. We analyzed the mass distribution for all protein chains derived from secreted human proteins in the Uniprot database, versus the proportion of their chain mass relative to their full-length intact protein mass (Fig. 1A). Importantly, these protein chains represent the active forms of the secreted proteins and not pro-peptides. This analysis highlighted the large number of protein chains with mass >15 kDa, many of which are among the top 200 most abundant proteins in plasma. It also showed many clinically relevant small protein chains are present between 2–10 kDa, which were significantly enriched for the gene ontology terms “regulation of signaling receptor activity” (i.e. hormones), “immune response” (e.g. chemokines, anti-microbial peptides, complement cascade), “negative regulation of proteolysis” and “response to stress” (supplemental Fig. S1). In addition, the protein chains from several apolipoprotein C family members (APOC2, APOC3, APOC4) that regulate lipid homeostasis were highly enriched in this size range, with APOC4 being known as a low abundance plasma protein (25). Therefore, we decided to target the mass region less than ∼10 kDa, as this would avoid purification of the many abundant high-mass proteins and yield these factors of interest (Fig. 1A, green zone). We also sought to avoid the purification of very low molecular weight protein degradation products below ∼2kDa. Before SEC-based separation for the isolation of small proteins in our mass region of interest, we wanted to dissociate small proteins from other protein binding partners. For example, >99% of circulating insulin-like growth factor 1 (IGF-1) is bound to insulin-like growth factor-binding proteins (IGFBPs) (26). We noted that for the analysis of total plasma IGF-1 by immunoassay, a combination of HCl and ∼80% ethanol provided an optimum for protein-protein dissociation and protein removal by precipitation (27). Interestingly, almost identical conditions were also observed by Best et al., to be optimal for purification of insulin from mammalian pancreatic extracts nearly 100 years ago (28). Lipids present in plasma samples may compromise downstream separations by SEC and reversed-phase chromatography, therefore we used a chloroform extraction step to remove lipids before downstream analysis.
Fig. 1.
Overview of the small-protein enrichment assay (SPEA). A, Analysis of the Uniprot database for human secreted proteins and their mass distribution. The scatter plot shows one point per protein chain and all chains that can be generated from each intact protein precursor (not including signal peptides or pro-peptides) are shown. Points are colored by gene name and only selected proteins have been labeled with their corresponding gene name. The green highlighted zone represents the mass range that we targeted with the SPEA method. B, Steps in the SPEA workflow to isolate small proteins from plasma and the subsequent bottom-up proteomics workflow used in this study either with, or without, high pH reversed phase fractionation. C, A representative UV absorbance chromatogram of the denaturing size exclusion chromatography (SEC) for a human plasma sample (red) and two molecular weight standards (green). The red highlighted zone between 6–8 min is collected as one fraction for subsequent digestion and LC-MS/MS analysis. D, Ranked abundance plot of plasma proteins detected in either a high-pH reversed phase fractionated proteome from SPEA (red), or a high-pH reversed phase fractionated proteome from undepleted plasma from (32) (blue). Protein abundance rank is derived from all previously detected plasma proteins from proteomics experiments in PaxDB.
We combined this dissociation-precipitation procedure before denaturing high-resolution UHPLC-based SEC analysis, which enabled 25 min molecular weight-based separations that were optimized for proteins <10 kDa, but also >2 kDa (Fig. 1B). The UV absorbance profile from these denaturing SEC separations highlighted the large amount of proteins >10 kDa remaining in the extract after the dissociation-precipitation procedure and the comparatively low levels of proteins below 10 kDa (Fig. 1C). We collected the fraction between ∼10 kDa and ∼2 kDa for all subsequent experiments. To determine the percentage recovery for a small protein factor using SPEA, we quantified the metabolic hormone insulin, which is within the mass range targeted by our SEC method, using an ELISA of both raw plasma samples and their corresponding SPEA SEC fractions. From this analysis we observed a mean insulin recovery of 44.7% (standard deviation of 4.3%, n = 3). Examining molecular weight standards run before and after plasma samples, we observed only small variations (<10% CV) in retention time, peak area, peak width and theoretical plates, during the fractionation of many plasma samples over several weeks (supplemental Fig. S2). This confirmed that our sample preparation was adequate to maintain column performance (i.e. lipid removal by chloroform extraction) and the UHPLC system was working reproducibly. This analysis also showed that the SEC resolution we achieved was very high based upon the theoretical plate values being >20,000.
For subsequent characterization of proteins in this fraction, we used the standard bottom-up workflow with trypsin digestion before LC-MS/MS. Although analysis of this fraction in the undigested state would be preferred for the quantification of certain hormones such as glucagon, which can yield many potentially overlapping protein chains (proteoforms), we wanted to achieve the best sensitivity possible for detection of potential new factors and new modifications to known proteins. Trypsin-digestion enabled us to achieve maximum sensitivity in an untargeted method, because of the much higher peak capacity in nanoflow reversed-phase chromatography and efficient fragmentation of typical tryptic peptides. The analysis of tryptic digests also greatly reduces the database search space, thereby increasing sensitivity for the same false-discovery rate cutoff, compared with “no enzyme” database searches (29, 30).
Using only 2 h single-shot LC-MS/MS data-dependent acquisitions from sample injections equivalent to 6 μl of plasma, we could identify peptides from a wide variety of small proteins including the known active chains from: APOC4, SPINK1, defensins, IGF-1, insulin, hepcidin, RANTES, and guanylin. (supplemental Table S1). The identification of peptides derived from the active chains of each protein is an important advance compared with previous methods when using untargeted single shot LC-MS/MS acquisitions. We also observed fragments of much larger proteins known to play a physiological role including the C terminus of alpha1-antitrypsin (SERPINA1), or SPAAT, which regulates neutrophil extracellular trap (NET) formation (31).
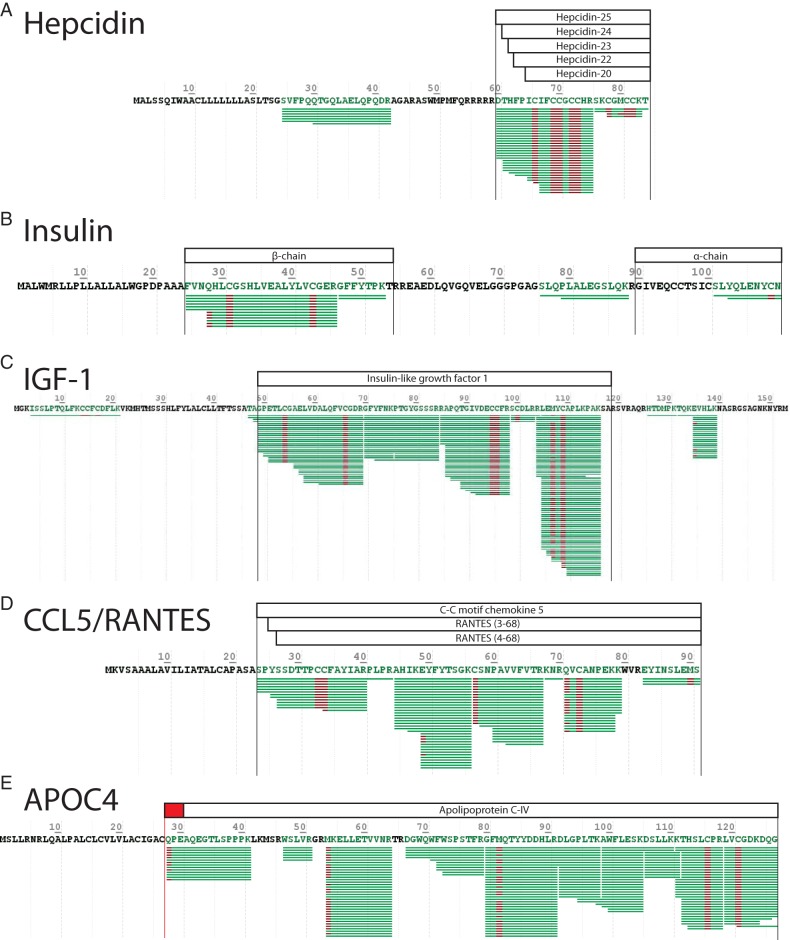
To fully characterize the components of this fraction, we performed extensive fractionation of trypsin digested peptides using offline high-pH reversed phase separation before LC-MS/MS analysis. We identified >900 proteins (supplemental Table S2) derived from >6000 peptide precursors (supplemental Table S3) (32). Importantly, these data were compared with those proteins identified previously in raw non-depleted human plasma, fractionated similarly (Fig. 1D). This analysis highlighted the depth of protein detection we were able to achieve based upon comparison with the PaxDB-based abundance estimates for >4300 human plasma proteins (33). This analysis also showed that although extensive fractionation of either sample allows detection of proteins over a wide dynamic range down to sub-ppb levels, many small proteins were only detected in our SPEA approach. Alongside the >125 known small-protein factors detected (Table I and supplemental Table S2, see supplemental Fig. S2 for annotated MS/MS spectra of selected factors), we observed peptides from uncharacterized proteins that may function as important metabolic regulators. These include the uncharacterized protein C5ORF46, which is a 7.2 kDa mammal-specific protein that we can detect for the first time in human plasma. The sequence coverage we have achieved is sufficient to observe the N-terminal degradation products of several hormones including hepcidin and insulin (Fig. 2A and 2B), and the dipeptidyl peptidase 4 catalyzed cleavage of IGF-1 and CCL5/RANTES (Fig. 2C and 2D) (34). Our data also demonstrated that the human APOC4 signal peptide cleavage site is mis-annotated in the Uniprot database, which it defined as between residues 29 and 30 (Fig. 2E). However, we consistently detected the N-terminal peptide of the secreted protein beginning at position 28, indicating signal peptide cleavage between residues 27 and 28 as suggested previously (35). To confirm this, we analyzed human APOC4 protein sequence using the SignalP predictor (http://www.cbs.dtu.dk/services/SignalP/) (36), which agreed with our data that the signal peptide cleavage site is between position 27 and 28 in the protein.
Table I. Selected small protein hormones/factors of interested detected using the SPEA method with offline high pH reversed-phase fractionation.
| Gene names | Uniprot Entry | Uniprot Protein names | Uniprot Secreted | Uniprot Propeptide | Uniprot Full Protein Mass (Da) | Uniprot Smallest Processed Chain MW (Da) | Fractionated sPHEA Intensity | Fractionated Razor + unique peptides |
|---|---|---|---|---|---|---|---|---|
| ADM | P35318 | ADM (Adrenomedullin) | + | + | 20420 | 2462 | 6.0E+08 | 3 |
| AGT | P01019 | Angiotensinogen | + | − | 53154 | 552 | 5.6E+08 | 7 |
| BDNF | P23560 | Brain-derived neurotrophic factor | + | + | 27818 | 13512 | 7.2E+07 | 1 |
| C5orf46 | Q6UWT4 | Uncharacterized protein C5orf46 | + | − | 9693 | 7216 | 6.3E+07 | 1 |
| CAMP | P49913 | Cathelicidin antimicrobial peptide | + | + | 19301 | 4493 | 4.3E+08 | 4 |
| CCL14 | Q16627 | C-C motif chemokine 14 | + | − | 10678 | 7801 | 2.8E+10 | 10 |
| CCL15 | Q16663 | C-C motif chemokine 15 | + | − | 12248 | 6928 | 3.6E+09 | 12 |
| CCL16 | O15467 | C-C motif chemokine 16 | + | − | 13600 | 11202 | 6.8E+08 | 2 |
| CCL18 | P55774 | C-C motif chemokine 18 | + | − | 9849 | 7557 | 3.4E+10 | 11 |
| CCL19 | Q99731 | C-C motif chemokine 19 | + | − | 10993 | 8800 | 1.8E+07 | 1 |
| CCL27 | Q9Y4X3 | C-C motif chemokine 27 | + | − | 12618 | 10150 | 1.1E+08 | 2 |
| CCL3 | P10147 | C-C motif chemokine 3 | + | − | 10085 | 7445 | 1.5E+07 | 1 |
| CCL5 | P13501 | C-C motif chemokine 5 | + | − | 9990 | 7504 | 8.6E+09 | 11 |
| CHGA | P10645 | Chromogranin-A | + | − | 50688 | 978 | 1.3E+09 | 9 |
| CHGB | P05060 | Secretogranin-1 | + | − | 78276 | 1342 | 5.4E+08 | 8 |
| CXCL1 | P09341 | Growth-regulated alpha protein | + | − | 11301 | 7436 | 1.3E+07 | 1 |
| CXCL12 | P48061 | Stromal cell-derived factor 1 | + | − | 10666 | 7610 | 1.4E+09 | 3 |
| CXCL3 | P19876 | C-X-C motif chemokine 3 | + | − | 11342 | 7509 | 1.1E+09 | 6 |
| CXCL5 | P42830 | C-X-C motif chemokine 5 | + | − | 11972 | 7706 | 3.5E+08 | 4 |
| CYTL1 | Q9NRR1 | Cytokine-like protein 1 | + | − | 15577 | 13371 | 4.0E+09 | 6 |
| DCD | P81605 | Dermcidin | + | + | 11284 | 2971 | 1.5E+10 | 16 |
| DEFA1 | P59665 | Neutrophil defensin 1 | + | + | 10201 | 3377 | 2.1E+10 | 5 |
| DEFA6 | Q01524 | Defensin-6 | + | + | 10975 | 3714 | 2.8E+07 | 4 |
| DEFB1 | P60022 | Beta-defensin 1 | + | + | 7420 | 3935 | 3.1E+09 | 7 |
| GAL | P22466 | Galanin peptides | + | + | 13302 | 3157 | 9.1E+08 | 4 |
| GAST | P01350 | Gastrin | + | + | 11394 | 818 | 4.2E+07 | 1 |
| GCG | P01275 | Glucagon | + | + | 20909 | 3299 | 3.2E+08 | 6 |
| GDF15 | Q99988 | Growth/differentiation factor 15 | + | + | 34140 | 12514 | 4.5E+06 | 1 |
| GHRL | Q9UBU3 | Appetite-regulating hormone | + | + | 12911 | 2548 | 7.5E+07 | 2 |
| GIP | P09681 | Gastric inhibitory polypeptide | + | + | 17108 | 4984 | 2.9E+09 | 10 |
| GRN | P28799 | Granulins | + | − | 63544 | 3149 | 4.0E+10 | 25 |
| GUCA2A | Q02747 | Guanylin | + | − | 12388 | 1463 | 7.1E+09 | 8 |
| GUCA2B | Q16661 | Guanylate cyclase activator 2B | + | + | 12069 | 1672 | 2.8E+08 | 2 |
| HAMP | P81172 | Hepcidin | + | + | 9408 | 2200 | 8.0E+09 | 8 |
| IGF1 | P05019 | Insulin-like growth factor I | + | + | 21841 | 7655 | 1.1E+11 | 21 |
| IGF2 | P01344 | Insulin-like growth factor II | + | + | 20140 | 4030 | 1.5E+11 | 27 |
| INHBB | P09529 | Inhibin beta B chain | + | + | 45122 | 12817 | 7.0E+07 | 1 |
| INHBC | P55103 | Inhibin beta C chain | + | + | 38238 | 12534 | 7.6E+07 | 2 |
| INHBE | P58166 | Inhibin beta E chain | + | + | 38561 | 12465 | 7.8E+07 | 1 |
| INS | P01308 | Insulin | + | + | 11981 | 2384 | 4.7E+08 | 2 |
| KISS1 | Q15726 | Metastasis-suppressor KiSS-1 | + | − | 14705 | 1303 | 1.7E+08 | 4 |
| KNG1 | P01042 | Kininogen-1 | + | − | 71957 | 528 | 2.1E+09 | 4 |
| MIF | P14174 | Macrophage migration inhibitory factor | + | − | 12476 | N/A | 8.8E+07 | 2 |
| MLN | P12872 | Promotilin | + | − | 12920 | 2699 | 1.7E+07 | 1 |
| MSTN | O14793 | Growth/differentiation factor 8 | + | + | 42750 | 12407 | 1.4E+07 | 2 |
| NPPA | P01160 | Natriuretic peptides A | + | + | 16708 | 3082 | 7.2E+08 | 5 |
| NPPC | P23582 | C-type natriuretic peptide | + | + | 13246 | 2200 | 1.2E+07 | 1 |
| NTS | P30990 | Neurotensin/neuromedin N | + | − | 19795 | 618 | 2.4E+08 | 6 |
| PCSK1N | Q9UHG2 | ProSAAS | + | − | 27372 | 796 | 4.5E+07 | 2 |
| PDGFA | P04085 | Platelet-derived growth factor subunit A | + | + | 24043 | 14306 | 5.4E+08 | 3 |
| PDGFB | P01127 | Platelet-derived growth factor subunit B | + | + | 27283 | 12294 | 5.4E+08 | 3 |
| PNOC | Q13519 | Prepronociceptin | + | + | 20295 | 1809 | 2.4E+08 | 2 |
| RETN | Q9HD89 | Resistin | + | − | 11419 | 9554 | 1.5E+07 | 1 |
| SCG2 | P13521 | Secretogranin-2 | + | + | 70941 | 3679 | 5.9E+08 | 5 |
| SCG5 | P05408 | Neuroendocrine protein 7B2 | + | − | 23730 | 1502 | 3.8E+06 | 1 |
| SCGB1A1 | P11684 | Uteroglobin | + | − | 9994 | 7915 | 2.4E+09 | 6 |
| SERPINA1 | P01009 | Alpha-1-antitrypsin | + | − | 46737 | 5068 | 5.5E+11 | 108 |
| SLURP1 | P55000 | Secreted Ly-6/uPAR-related protein 1 | + | − | 11186 | 8853 | 3.0E+09 | 9 |
| SPINK1 | P00995 | Serine protease inhibitor Kazal-type 1 | + | − | 8507 | 6247 | 1.4E+08 | 1 |
| SPINK5 | Q9NQ38 | Serine protease inhibitor Kazal-type 5 | + | − | 120714 | 6482 | 6.7E+09 | 25 |
| SRGN | P10124 | Serglycin | + | − | 17652 | 14712 | 3.6E+09 | 4 |
| TAC3 | Q9UHF0 | Tachykinin-3 | + | + | 13438 | 1211 | 6.4E+08 | 6 |
| TFF1 | P04155 | Trefoil factor 1 | + | − | 9150 | 6674 | 2.4E+08 | 2 |
| TFF2 | Q03403 | Trefoil factor 2 | + | − | 14284 | 11989 | 8.8E+07 | 1 |
| TFF3 | Q07654 | Trefoil factor 3 | + | − | 10181 | 6580 | 7.2E+08 | 4 |
| TGFB1 | P01137 | Transforming growth factor beta-1 | + | − | 44341 | 12795 | 4.3E+07 | 1 |
| TNFRSF12A | Q9NP84 | Tumor necrosis factor receptor superfamily member 12A | − | − | 13911 | 10925 | 7.4E+07 | 3 |
| WFDC12 | Q8WWY7 | WAP four-disulfide core domain protein 12 | + | − | 12050 | 9716 | 6.3E+07 | 2 |
| WFDC13 | Q8IUB5 | WAP four-disulfide core domain protein 13 | + | − | 10386 | 7978 | 2.5E+09 | 3 |
Fig. 2.
Characterization of SPEA-mediated small-protein sequence coverage. The sequence coverage of selected small proteins identified using SPEA with downstream high-pH reversed phase fractionation and subsequent LC-MS/MS. Each green line represents an individual peptide identified in this study with maroon areas representing a modification identified for that residue. Black boxes above each sequence represent the Uniprot-derived canonical active-chains of each protein and red boxes represent discrepancies between our observations and the Uniprot annotation. Selected proteins shown are: A, Hepcidin (HAMP); B, insulin; C, insulin-like growth factor 1 (IGF1); D, C-C motif chemokine 5 or RANTES (CCL5); and E, apolipoprotein C-IV (APOC4).
To determine if our small-protein hormone enrichment workflow provides reproducible quantification, we employed a stable isotope labeled protein standard of insulin. Using stable-isotope (15N) labeled intact human insulin that is available commercially, we established a PRM method to quantify a heavy and light insulin derived peptide in fractions from our small-protein hormone enrichment workflow (supplemental Fig. S3A). This PRM-based LC-MS/MS analysis method enabled combined quantification of the light and heavy insulin beta chain peptide fragments in the same MS/MS (PRM) spectrum (supplemental Fig. S3B). The peptide chosen for PRM analysis is from the beta-chain of insulin. For quantification, the intensities of multiple y and b fragment ions are used to generate a total intensity value for either the light, or heavy insulin. We subsequently analyzed the heavy insulin dilution series across 2 biological replicates (2 different human plasma samples) with the targeted PRM acquisition method. This demonstrated that our SPEA approach had low percentage CV values (<10%) for the heavy insulin standard down to 250 pm across the 3 technical replicates for each spike-in and we were able to detect the heavy insulin down to 50 pm with much greater variability. Given that normal plasma insulin abundance after a meal is ∼1 nm and in the fasted state is <250 pm, our limit of quantification is at the high end of the physiological range for insulin (37).
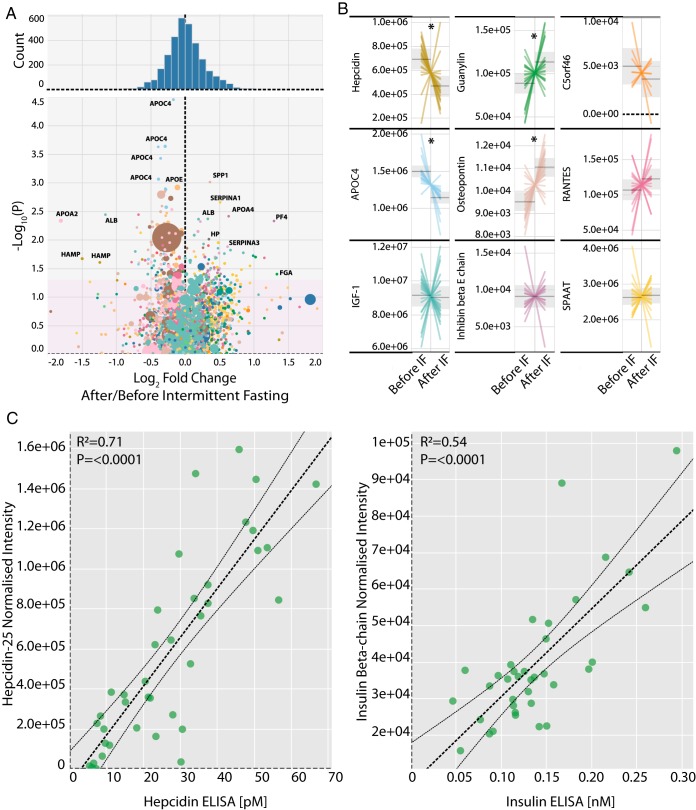
To further explore the utility of our method, we analyzed plasma samples from humans that had undergone intermittent fasting (IF), a dietary regimen that provides beneficial metabolic and lifespan phenotypes across many model organisms. This trial employed 8-weeks of IF in 22 participants with alternating feeding and fasting days, as described previously (38, 39) with plasma samples taken before and after the 8-week IF intervention. The small-protein hormones from each of the participant plasma samples were quantified by 2 h single-shot LC-MS/MS analysis using a data-independent acquisition (DIA) approach (20). This analysis quantified >3400 peptide precursors, corresponding to >2000 unique peptides across >500 proteins (supplemental Table S4). Using a paired test statistic, we detected 235 peptide precursors that were significantly changed (p < 0.05) because of the IF intervention (Fig. 3A). The protein displaying the largest fold-change was hepcidin-25, which was downregulated ∼3-fold after the IF intervention (Fig. 3B). In addition, we observed significant downregulation of the lipoprotein particle associated protein APOC4 and upregulation and osteopontin and guanylin (Fig. 3B). To validate our findings, we correlated our MS-based quantitative data with immunoassay-based analysis of the same plasma samples, which were specific for either hepcidin (ELISA), or insulin (RIA). This showed good agreement between our methods and more traditional measurement approaches (Fig. 3C).
Fig. 3.
SPEA analysis of a human intermittent fasting clinical trial. This clinical trial examined the effects of intermittent fasting (IF) in humans (n = 20), with blood plasma sampling before and after the 8-week IF intervention. A, Volcano plot for all the peptide precursors detected, with the log2 fold change plotted on the x axis versus the -log10(P) of a paired test statistic on the y axis. The pink region highlights non-significant p values (>0.05). The size of the circles represents the abundance of the precursor peptide with larger being more abundant. Precursor peptides derived from the same gene share the same color. B, Adjusted intensity plots for selected peptides of interest, where each line represents an individual patient. The gray line and surrounding ribbon represent the mean and the 95% confidence interval of the mean, respectively. Peptides that had a significant p value (p < 0.05) are marked with an asterisk. C, Correlation analysis across all patient samples of either the hepcidin-25 peptide MS intensity versus the hepcidin ELISA concentrations (left), or the insulin beta-chain peptide MS intensity versus the insulin RIA concentrations (right). The Pearson correlation and the associated Benjamini-Hochberg corrected p value are shown inside each plot. The dashed lines represent the fitted model and its 95% confidence interval.
DISCUSSION
In this study we detail the first method (SPEA) that enables rapid, specific and sensitive quantitation of >100 low abundance small protein factors in human plasma. Importantly, we identified peptides from the active chains of hormones and not just pro-peptides often characterized in previous studies. We demonstrate the utility of our method by three applications. First, we identified a previously uncharacterized plasma hormone (C5ORF46) through deep-proteome profiling. Second, we demonstrated the capacity to perform reproducible quantification using the low abundance hormone insulin, which occurs in plasma at <10 ng/ml. Finally, we applied our method to quantify changes in small proteins in human plasma during intermittent fasting and unexpectedly identified hepcidin-25 as being downregulated by the dietary intervention. The SPEA method offers the ability to study numerous important small proteins and hormones in a single rapid assay, greatly increasing feasibility while reducing cost. In addition, it provides opportunities for discovery of new biomarkers produced by a variety of tissues/conditions. We suggest this approach will be key to the future of precision medicine given the importance of these factors in metabolic homeostasis and their functional disruption in diverse disease states.
One of the key strengths in the SPEA method is our employment of hydrochloric acid to dissociate protein-protein interactions in the plasma protein precipitation step. HCl is a relatively poor protein precipitant (40) when compared with halogenated organic acids (trifluoroacetic acid or trichloroacetic acid) frequently used in proteomics studies. Halogenated organic acids typically facilitate precipitation by several mechanisms including depletion of the hydration shell around proteins to increase hydrophobic interactions and disrupting ionic interactions through pH changes. However, this is not the case for HCl treatment, which mainly mediates pH-based dissociation of ionic interactions. Likewise, ethanol is also a poor protein precipitant (although it is commonly used for albumin precipitation) when compared with acetonitrile (41). The ability to dissociate but not precipitate small protein hormones is likely the reason this reagent combination has been established for the analysis of IGF-1 (7, 27) and for insulin extraction from tissue (28). This also the likely reason why a large amount of high MW protein remained in the supernatant after the precipitation step and needs to be removed during SEC (Fig. 1B).
In this study, we chose to use trypsin digestion of the proteins in the SPEA fraction rather than undigested protein analysis by LC-MS/MS to achieve maximum sensitivity for protein identification (Fig. 1A). The chromatographic separation of tryptic peptides (average mass ∼1.5 kDa) in nanoflow reversed-phase chromatography shows much higher peak capacity than separation of intact small proteins up to ∼10 kDa in size. Likewise, tryptic peptides are fragmented (for example by HCD) for MS/MS analysis much more efficiently than small intact proteins. Another advantage of trypsin-based bottom-up proteomics is at the database search step, as it will lead to the generation of a much smaller theoretical database for peptide spectral matching to the MS/MS data, which will in turn provide for lower score thresholds and higher sensitivity for a given false-discovery rate (29, 42). Although tryptic digestion has provided an increase in sensitivity for identification, it will come at the cost of specificity for individual proteoforms (42). This is exemplified by examining the polyprotein, glucagon. Within our data, we have detected a peptide within the active peptide of glucagon (supplemental Fig. S4), but this peptide is not unique as it is also a part of the other glucagon-derived hormones, oxyntomodulin and glicentin. By digesting the peptide with trypsin, we lose the ability to properly assign the peptide to its parent protein, which may be incompatible with specific analysis of some. In many ways, this puts SPEA at the discovery end of small protein analysis from plasma, which would enable subsequent follow-up studies in larger sample cohorts using intact (top-down) analysis. For example, this would be performed using targeted methodologies on either triple-quadrupole instruments with SRM-based assays, or orbitrap instruments with PRM-based assays, downstream of robust high flow-rate LC (7, 8, 10, 11, 15). As such, SPEA exists in a semi-quantitative area that allows for much greater discovery potential with some quantitative potential that could inform further studies in a two-pronged manner.
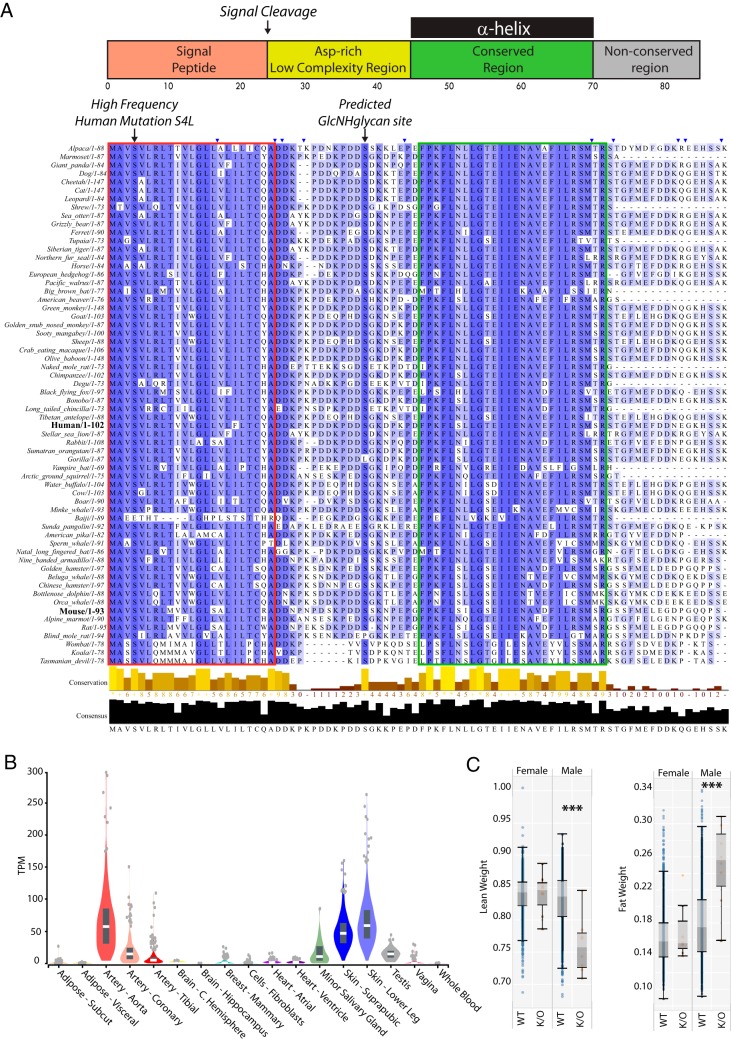
Using the SPEA method we were able to detect previously uncharacterized proteins that may act as hormones such as C5ORF46. Bioinformatic predictions have suggested that C5ORF46 is a secreted small-protein hormone (43). C5ORF46 is conserved across mammalian evolution, with two regions of the protein being highly conserved. The first is the canonical signal peptide (Fig. 4A, red box, residues 1–23) and the second is part of the secreted protein (Fig. 4A, green box, residues 45–70). This second region is also predicted to be the only ordered region in the secreted protein, which together with high sequence conservation, suggest that it is the functional element of the secreted C5ORF46 protein. This region was also predicted to form an α-helix using Phyre2 (44). In human populations, C5ORF46 has a high frequency coding SNP within the signal peptide (S4L), with a minor allele frequency of ∼0.5 across all human populations. This S4L variant has not previously been associated with a disease phenotype, but this serine is very highly conserved (Fig. 4A), and mutations within signal peptide regions have previously been demonstrated to alter the protein's secretion rate and contribute to disease (45). Future studies should determine if this occurs with this common human coding variant of C5ORF46. Prior studies have identified links between C5ORF46 and either heart disease, or heart development (46, 47). This agrees with the expression profile in human tissues and cells, where C5ORF46 is detectable at the RNA-level in major blood vessels (aorta) and the heart, but also in skin and salivary gland (Fig. 4B) (48, 49). Using a systems genetics approach the mouse homologue of C5ORF46 (named Gm94) has been demonstrated to be positively correlated with HDL particle abundance and HDL-associated proteins (APOM, APOD, PON1 and LCAT), which stimulate HDL particle function and maturation (50). Combined with the arterial and heart expression profile, we hypothesize that C5ORF46 is secreted into plasma by endothelial cells to stimulate reverse cholesterol transport by HDL. The production of C5ORF46 at key blood vessels may allow it to rapidly respond to changes in arterial health. Fortuitously, data was available from mice that were heterozygous for a null mutation in Gm94, which highlighted that these animals had significantly increased fat mass and reduced lean mass compared with wild type controls (Fig. 4C) (51). These phenotypes are very similar to that observed in a APOA1 (a core protein of HDL particles) deficient mouse model (52), which suggests C5ORF46 may play a key role in lipid homeostasis through regulation of HDL function. Our data is the first to identify C5ORF46 in human plasma, whereas the phenotypic data indicate an important role in mammalian physiology, which should be explored in future experiments including analysis of HDL particle structure/function in Gm94 null mutant animals.
Fig. 4.
Conservation and expression of novel human plasma protein C5ORF46. A, Alignment of the C5ORF46 protein sequence for a diverse cross-section of mammalian species using the tCoffee online resource (https://www.ebi.ac.uk/Tools/msa/tcoffee/) (66). Blue coloration of an amino acid residue represents conservation across species, with darker blue representing higher conservation. Blue triangles at the top of the alignment indicate hidden protein sequence information for species other than human. The red box highlights the canonical signal peptide for C5ORF46, the green box highlights a highly conserved and ordered region of C5ORF46. The prediction of the glycosaminoglycan site was performed using ELM (http://elm.eu.org/) (67) and protein secondary structure prediction was performed using Phyre2 (http://www.sbg.bio.ic.ac.uk/phyre2/) (44). B, The previously observed RNA abundance measurements of C5ORF46 in various human tissues was provided by the GTEx resource (https://gtexportal.org/home/) (49). C, Male and female adult wild-type (n>1600) and C5ORF46 mice (n = 10) were measured for changes in lean and fat weight (measurements are relative) using dual-energy X-ray absorptiometry by the International Mouse Phenotyping Consortium (IMPC, http://www.mousephenotype.org/) (68). *** Indicates p value less than 1 × 10−5.
Intermittent fasting (IF) is a dietary intervention that is of significant interest because of the beneficial effects on metabolic health. However, very few studies have examined in an unbiased way the physiological changes induced by IF. Using SPEA, we observed that hepcidin-25 was significantly downregulated by >3-fold in humans undergoing IF for 8-weeks. Hepcidin-25 is a liver-derived hormone that controls systemic iron homeostasis by inhibiting iron absorption in the small intestine and inhibiting iron release from the liver. Previous studies have demonstrated an acute increase in hepcidin mRNA and protein levels following a single fasting bout in humans and mice (53, 54). These studies offer conflicting reports on the effects of longer-term fasting on plasma hepcidin, where it has been demonstrated to either continuously increase with extended fasting, or peak after a few hours of fasting before returning to baseline. Previous studies have also shown that fasting lowers blood iron levels after 6–10 h, which is then rescued by hepcidin-25 activity (55). However, no previous study has reported a change in hepcidin abundance after IF. We propose the decrease we observe in hepcidin-25 after IF is triggered by activation of two proteases upstream of the hepcidin transcriptional network in the liver because of low blood iron levels. Production of hepcidin in the liver is dependent on coordinated signaling from hemojuvelin (HJV) and bone morphogenic protein 6 (BMP6) through canonical SMAD signal transduction pathways (56). However, decreased blood iron levels both stabilizes the serine protease matripase-2 (TMPRSS6) (57–59), and simultaneously upregulates HIF1-α that increases the abundance of the serine protease furin (60, 61). HJV is a direct target of both TMPRSS6 and furin proteolytic activity, which inhibits its downstream signaling (62, 63) and may act as a soluble decoy for BMP6 (64). This ultimately causes downregulation of hepcidin leading to greater intestinal iron uptake. The repeated fasting bouts in the EODF regime used in this study may not allow enough time for degradation of stabilized TMPRSS6, leading to a long-term decrease in plasma hepcidin abundance. One advantage to these decreased levels is that maximal iron absorption is facilitated during subsequent feeding periods. Future studies to verify this hypothesis and examine changes in iron homeostasis in human participants undergoing IF will be important to document their dietary iron intake and verify that their blood iron levels are maintained in the normal range.
The SPEA method provides many opportunities for multiplexed small protein analysis of the vast number of human plasma samples collected during clinical trials. By addressing the dynamic range problem in the analysis of plasma, we have enabled an unbiased view of many low abundance small plasma proteins in a short timescale. Clinically, our method offers the ability to study numerous important proteins in a single assay, greatly increasing feasibility while reducing cost. Our workflow also provides opportunities for discovery of new proteins and biomarkers produced by a variety of tissues/conditions. The purified small proteins could also be examined by other methods including top-down mass spectrometry, structural analysis of individual components, and different quantitative MS strategies to improve sample throughput such as isobaric tagging. It may also be possible to apply our method to analysis of small proteins from other body fluids such as cerebrospinal fluid (CSF), where neuropeptides play a key role (65). This methodology would also be applicable in other organisms where blood plasma can be easily collected including mice and rats. The small protein enrichment workflow detailed here will facilitate the quantification of global changes during many key immunological, metabolic and developmental perturbations.
Data Availability
All the RAW MS data and corresponding search outputs have been deposited to the ProteomeXchange Consortium (http://proteomecentral.proteomexchange.org) via the PRIDE partner repository with the dataset identifier PXD011797.
Supplementary Material
Acknowledgments
We thank SydneyMS for providing the instrumentation used in this study and the University of Sydney Precision Medicine Initiative for funding.
Footnotes
* This work was supported by grants from the National Health and Medical Research Council (NHMRC) APP1120475 and the clinical research study was funded by APP1023401. L.K.H. is supported by an Australian Research Council Future Fellowship FT120100027. M.L. is supported by a Cancer Institute New South Wales Future Research Leader Fellowship 15/FRL/1-06A. The authors declare that they have no conflict of interest.
 This article contains supplemental material.
This article contains supplemental material.
1 The abbreviations used are:
- SPEA
- small protein enrichment assay
- IF
- Intermittent fasting
- SEC
- size-exclusion chromatography.
REFERENCES
- 1. Stocklin R., Vu L., Vadas L., Cerini F., Kippen A. D., Offord R. E., and Rose K. (1997) A stable isotope dilution assay for the in vivo determination of insulin levels in humans by mass spectrometry. Diabetes 46, 44–50 [DOI] [PubMed] [Google Scholar]
- 2. Nedelkov D., Niederkofler E. E., Oran P. E., Peterman S., and Nelson R. W. (2018) Top-down mass spectrometric immunoassay for human insulin and its therapeutic analogs. J. Proteomics 175, 27–33 [DOI] [PubMed] [Google Scholar]
- 3. Girelli D., Nemeth E., and Swinkels D. W. (2016) Hepcidin in the diagnosis of iron disorders. Blood 127, 2809–2813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Schwenk J. M., Omenn G. S., Sun Z., Campbell D. S., Baker M. S., Overall C. M., Aebersold R., Moritz R. L., and Deutsch E. W. (2017) The Human Plasma Proteome Draft of 2017: Building on the Human Plasma PeptideAtlas from Mass Spectrometry and Complementary Assays. J. Proteome Res. 16, 4299–4310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Geyer P. E., Holdt L. M., Teupser D., and Mann M. (2017) Revisiting biomarker discovery by plasma proteomics. Mol. Syst. Biol. 13, 942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Keshishian H., Burgess M. W., Specht H., Wallace L., Clauser K. R., Gillette M. A., and Carr S. A. (2017) Quantitative, multiplexed workflow for deep analysis of human blood plasma and biomarker discovery by mass spectrometry. Nat. Protoc. 12, 1683–1701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Bystrom C., Sheng S., Zhang K., Caulfield M., Clarke N. J., and Reitz R. (2012) Clinical utility of insulin-like growth factor 1 and 2; determination by high resolution mass spectrometry. PloS One 7, e43457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Chen Z., Caulfield M. P., McPhaul M. J., Reitz R. E., Taylor S. W., and Clarke N. J. (2013) Quantitative insulin analysis using liquid chromatography-tandem mass spectrometry in a high-throughput clinical laboratory. Clin. Chem. 59, 1349–1356 [DOI] [PubMed] [Google Scholar]
- 9. Abbas I. M., Hoffmann H., Montes-Bayon M., and Weller M. G. (2018) Improved LC-MS/MS method for the quantification of hepcidin-25 in clinical samples. Anal. Bioanal. Chem. 410, 3835–3846 [DOI] [PubMed] [Google Scholar]
- 10. Barton C., Kay R. G., Gentzer W., Vitzthum F., and Pleasance S. (2010) Development of high-throughput chemical extraction techniques and quantitative HPLC-MS/MS (SRM) assays for clinically relevant plasma proteins. J. Proteome Res. 9, 333–340 [DOI] [PubMed] [Google Scholar]
- 11. Kam R. K., Ho C. S., and Chan M. H. (2016) Serum insulin-like growth factor I quantitation by mass spectrometry: insights for protein quantitation with this technology. Ejifcc 27, 318–330 [PMC free article] [PubMed] [Google Scholar]
- 12. Shen Y., Liu T., Tolic N., Petritis B. O., Zhao R., Moore R. J., Purvine S. O., Camp D. G., and Smith R. D. (2010) Strategy for degradomic-peptidomic analysis of human blood plasma. J. Proteome Res. 9, 2339–2346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Baczynskyj L., Bronson G. E., and Kubiak T. M. (1994) Application of thermally assisted electrospray ionization mass spectrometry for detection of noncovalent complexes of bovine serum albumin with growth hormone releasing factor and other biologically active peptides. Rapid Commun. Mass Spectrom. 8, 280–286 [DOI] [PubMed] [Google Scholar]
- 14. Lufrano D., Trejo S. A., Llovera R. E., Salgueiro M., Fernandez G., Martinez Damonte V., Gonzalez Flecha F. L., Raingo J., Ermacora M. R., and Perello M. (2016) Ghrelin binding to serum albumin and its biological impact. Mol. Cell. Endocrinol. 436, 130–140 [DOI] [PubMed] [Google Scholar]
- 15. Darby S. M., Miller M. L., Allen R. O., and LeBeau M. (2001) A mass spectrometric method for quantitation of intact insulin in blood samples. J. Anal. Toxicol. 25, 8–14 [DOI] [PubMed] [Google Scholar]
- 16. Kay R. G., Challis B. G., Casey R. T., Roberts G. P., Meek C. L., Reimann F., and Gribble F. M. (2018) Peptidomic analysis of endogenous plasma peptides from patients with pancreatic neuroendocrine tumours. Rapid Commun. Mass Spectrom. 32, 1414–1424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Mahboob S., Mohamedali A., Ahn S. B., Schulz-Knappe P., Nice E., and Baker M. S. (2015) Is isolation of comprehensive human plasma peptidomes an achievable quest? J. Proteomics. 127, 300–309 [DOI] [PubMed] [Google Scholar]
- 18. Parker B. L., Burchfield J. G., Clayton D., Geddes T. A., Payne R. J., Kiens B., Wojtaszewski J. F. P., Richter E. A., and James D. E. (2017) Multiplexed temporal quantification of the exercise-regulated plasma peptidome. Mol. Cell. Proteomics 16, 2055–2068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Wewer Albrechtsen N. J., Kuhre R. E., Hornburg D., Jensen C. Z., Hornum M., Dirksen C., Svane M., Gasbjerg L. S., Jorgensen N. B., Gabe M. N., Balk-Moller E., Albrechtsen R., Winther-Sorensen M., Galsgaard K. D., Meissner F., Jorsal T., Lund A., Vilsboll T., Eliasen R., Bojsen-Moller K. N., Idorn T., Deacon C. F., Knop F. K., Rosenkilde M. M., Hartmann B., Feldt-Rasmussen B., Mann M., Madsbad S., and Holst J. J. (2017) Circulating glucagon 1–61 regulates blood glucose by increasing insulin secretion and hepatic glucose production. Cell Rep. 21, 1452–1460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Bruderer R., Bernhardt O. M., Gandhi T., Miladinovic S. M., Cheng L. Y., Messner S., Ehrenberger T., Zanotelli V., Butscheid Y., Escher C., Vitek O., Rinner O., and Reiter L. (2015) Extending the limits of quantitative proteome profiling with data-independent acquisition and application to acetaminophen-treated three-dimensional liver microtissues. Mol. Cell. Proteomics 14, 1400–1410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Cox J., and Mann M. (2008) MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotech. 26, 1367–1372 [DOI] [PubMed] [Google Scholar]
- 22. Cox Jr., Neuhauser N., Michalski A., Scheltema R. A., Olsen J. V., and Mann M. (2011) Andromeda: a peptide search engine integrated into the MaxQuant environment. J. Proteome Res. 10, 1794–1805 [DOI] [PubMed] [Google Scholar]
- 23. Muntel J., Kirkpatrick J., Bruderer R., Huang T., Vitek O., Ori A., and Reiter L. (2019) Comparison of protein quantification in a complex background by DIA and TMT workflows with fixed instrument time. J. Proteome Res. 18, 1340–1351 [DOI] [PubMed] [Google Scholar]
- 24. Tsou C. C., Avtonomov D., Larsen B., Tucholska M., Choi H., Gingras A. C., and Nesvizhskii A. I. (2015) DIA-Umpire: comprehensive computational framework for data-independent acquisition proteomics. Nat. Methods 12, 258–264, 257 p following 264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Allan C. M., and Taylor J. M. (1996) Expression of a novel human apolipoprotein (apoC-IV) causes hypertriglyceridemia in transgenic mice. J. Lipid Res. 37, 1510–1518 [PubMed] [Google Scholar]
- 26. Horner J. M., Liu F., and Hintz R. L. (1978) Comparison of [125I]somatomedin A and [125I]somatomedin C radioreceptor assays for somatomedin peptide content in whole and acid-chromatographed plasma. J. Clin. Endocrinol. Metab. 47, 1287–1295 [DOI] [PubMed] [Google Scholar]
- 27. Daughaday W. H., Mariz I. K., and Blethen S. L. (1980) Inhibition of access of bound somatomedin to membrane receptor and immunobinding sites: a comparison of radioreceptor and radioimmunoassay of somatomedin in native and acid-ethanol-extracted serum. J. Clin. Endocrinol. Metab. 51, 781–788 [DOI] [PubMed] [Google Scholar]
- 28. Best C. H., and Scott D. A. (1923) The preparation of insulin. J. Biol. Chem. 57, 709–723 [Google Scholar]
- 29. Cottrell J. S. (2011) Protein identification using MS/MS data. J. Proteomics 74, 1842–1851 [DOI] [PubMed] [Google Scholar]
- 30. Eng J. K., Searle B. C., Clauser K. R., and Tabb D. L. (2011) A face in the crowd: recognizing peptides through database search. Mol. Cell. Proteomics 10, R111.009522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Yost C. C., Schwertz H., Cody M. J., Wallace J. A., Campbell R. A., Vieira-de-Abreu A., Araujo C. V., Schubert S., Harris E. S., Rowley J. W., Rondina M. T., Fulcher J. M., Koening C. L., Weyrich A. S., and Zimmerman G. A. (2016) Neonatal NET-inhibitory factor and related peptides inhibit neutrophil extracellular trap formation. J. Clin. Invest. 126, 3783–3798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Geyer P. E., Kulak N. A., Pichler G., Holdt L. M., Teupser D., and Mann M. (2016) Plasma Proteome Profiling to Assess Human Health and Disease. Cell Syst. 2, 185–195 [DOI] [PubMed] [Google Scholar]
- 33. Wang M., Weiss M., Simonovic M., Haertinger G., Schrimpf S. P., Hengartner M. O., and von Mering C. (2012) PaxDb, a database of protein abundance averages across all three domains of life. Mol. Cell. Proteomics 11, 492–500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Mulvihill E. E., and Drucker D. J. (2014) Pharmacology, physiology, and mechanisms of action of dipeptidyl peptidase-4 inhibitors. Endocr. Rev. 35, 992–1019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Kotite L., Zhang L. H., Yu Z., Burlingame A. L., and Havel R. J. (2003) Human apoC-IV: isolation, characterization, and immunochemical quantification in plasma and plasma lipoproteins. J. Lipid Res. 44, 1387–1394 [DOI] [PubMed] [Google Scholar]
- 36. Almagro Armenteros J. J., Tsirigos K. D., Sonderby C. K., Petersen T. N., Winther O., Brunak S., von Heijne G., and Nielsen H. (2019) SignalP 5.0 improves signal peptide predictions using deep neural networks. Nat. Biotechnol. 37, 420–423 [DOI] [PubMed] [Google Scholar]
- 37. Arendt J., Hampton S., English J., Kwasowski P., and Marks V. (1982) 24-hour profiles of melatonin, cortisol, insulin, C-peptide and GIP following a meal and subsequent fasting. Clin. Endocrinol. 16, 89–95 [DOI] [PubMed] [Google Scholar]
- 38. Harney D. J., Hutchison A. T., Hatchwell L., Humphrey S. J., James D. E., Hocking S., Heilbronn L. K., and Larance M. (2019) Proteomic analysis of human plasma during intermittent fasting. J. Proteome Res. 18, 2228–2240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Hutchison A. T., Liu B., Wood R. E., Vincent A. D., Thompson C. H., O'Callaghan N. J., Wittert G. A., and Heilbronn L. K. (2019) Effects of intermittent versus continuous energy intakes on insulin sensitivity and metabolic risk in women with overweight. Obesity 27, 50–58 [DOI] [PubMed] [Google Scholar]
- 40. Rajalingam D., Loftis C., Xu J. J., and Kumar T. K. (2009) Trichloroacetic acid-induced protein precipitation involves the reversible association of a stable partially structured intermediate. Protein Sci. 18, 980–993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Polson C., Sarkar P., Incledon B., Raguvaran V., and Grant R. (2003) Optimization of protein precipitation based upon effectiveness of protein removal and ionization effect in liquid chromatography-tandem mass spectrometry. J. Chromatogr. 785, 263–275 [DOI] [PubMed] [Google Scholar]
- 42. Tran J. C., Zamdborg L., Ahlf D. R., Lee J. E., Catherman A. D., Durbin K. R., Tipton J. D., Vellaichamy A., Kellie J. F., Li M., Wu C., Sweet S. M. M., Early B. P., Siuti N., LeDuc R. D., Compton P. D., Thomas P. M., and Kelleher N. L. (2011) Mapping intact protein isoforms in discovery mode using top-down proteomics. Nature 480, 254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Mirabeau O., Perlas E., Severini C., Audero E., Gascuel O., Possenti R., Birney E., Rosenthal N., and Gross C. (2007) Identification of novel peptide hormones in the human proteome by hidden Markov model screening. Genome Res. 17, 320–327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Kelley L. A., Mezulis S., Yates C. M., Wass M. N., and Sternberg M. J. (2015) The Phyre2 web portal for protein modeling, prediction and analysis. Nat. Protoc. 10, 845–858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Kiraly O., Boulling A., Witt H., Le Marechal C., Chen J. M., Rosendahl J., Battaggia C., Wartmann T., Sahin-Toth M., and Ferec C. (2007) Signal peptide variants that impair secretion of pancreatic secretory trypsin inhibitor (SPINK1) cause autosomal dominant hereditary pancreatitis. Hum. Mutat. 28, 469–476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. McKean D. M., Homsy J., Wakimoto H., Patel N., Gorham J., DePalma S. R., Ware J. S., Zaidi S., Ma W., Patel N., Lifton R. P., Chung W. K., Kim R., Shen Y., Brueckner M., Goldmuntz E., Sharp A. J., Seidman C. E., Gelb B. D., and Seidman J. G. (2016) Loss of RNA expression and allele-specific expression associated with congenital heart disease. Nat. Commun. 7, 12824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Sahara M., Santoro F., Sohlmer J., Zhou C., Witman N., Leung C. Y., Mononen M., Bylund K., Gruber P., and Chien K. R. (2019) Population and Single-Cell Analysis of Human Cardiogenesis Reveals Unique LGR5 Ventricular progenitors in embryonic outflow tract. Dev. Cell. 48, 475–490.e477 [DOI] [PubMed] [Google Scholar]
- 48. Uhlen M., Fagerberg L., Hallstrom B. M., Lindskog C., Oksvold P., Mardinoglu A., Sivertsson A., Kampf C., Sjostedt E., Asplund A., Olsson I., Edlund K., Lundberg E., Navani S., Szigyarto C. A., Odeberg J., Djureinovic D., Takanen J. O., Hober S., Alm T., Edqvist P. H., Berling H., Tegel H., Mulder J., Rockberg J., Nilsson P., Schwenk J. M., Hamsten M., von Feilitzen K., Forsberg M., Persson L., Johansson F., Zwahlen M., von Heijne G., Nielsen J., and Ponten F. (2015) Proteomics. Tissue-based map of the human proteome. Science 347, 1260419. [DOI] [PubMed] [Google Scholar]
- 49. Consortium G. (2015) Human genomics. The Genotype-Tissue Expression (GTEx) pilot analysis: multitissue gene regulation in humans. Science 348, 648–660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Pamir N., Pan C., Plubell D. L., Hutchins P. M., Tang C., Wimberger J., Irwin A., Vallim T. Q. A., Heinecke J. W., and Lusis A. J. (2019) Genetic control of the mouse HDL proteome defines HDL traits, function, and heterogeneity. J. Lipid Res. 60, 594–608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Skarnes W. C., Rosen B., West A. P., Koutsourakis M., Bushell W., Iyer V., Mujica A. O., Thomas M., Harrow J., Cox T., Jackson D., Severin J., Biggs P., Fu J., Nefedov M., de Jong P. J., Stewart A. F., and Bradley A. (2011) A conditional knockout resource for the genome-wide study of mouse gene function. Nature 474, 337–342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Lehti M., Donelan E., Abplanalp W., Al-Massadi O., Habegger K. M., Weber J., Ress C., Mansfeld J., Somvanshi S., Trivedi C., Keuper M., Ograjsek T., Striese C., Cucuruz S., Pfluger P. T., Krishna R., Gordon S. M., Silva R. A., Luquet S., Castel J., Martinez S., D'Alessio D., Davidson W. S., and Hofmann S. M. (2013) High-density lipoprotein maintains skeletal muscle function by modulating cellular respiration in mice. Circulation 128, 2364–2371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Troutt J. S., Rudling M., Persson L., Stahle L., Angelin B., Butterfield A. M., Schade A. E., Cao G., and Konrad R. J. (2012) Circulating human hepcidin-25 concentrations display a diurnal rhythm, increase with prolonged fasting, and are reduced by growth hormone administration. Clin. Chem. 58, 1225–1232 [DOI] [PubMed] [Google Scholar]
- 54. Vecchi C., Montosi G., Garuti C., Corradini E., Sabelli M., Canali S., and Pietrangelo A. (2014) Gluconeogenic signals regulate iron homeostasis via hepcidin in mice. Gastroenterology 146, 1060–1069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Nguyen L. T., Buse J. D., Baskin L., Sadrzadeh S. M. H., and Naugler C. (2017) Influence of diurnal variation and fasting on serum iron concentrations in a community-based population. Clin. Biochem. 50, 1237–1242 [DOI] [PubMed] [Google Scholar]
- 56. Wang C. Y., and Babitt J. L. (2019) Liver iron sensing and body iron homeostasis. Blood 133, 18–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Du X., She E., Gelbart T., Truksa J., Lee P., Xia Y., Khovananth K., Mudd S., Mann N., Moresco E. M., Beutler E., and Beutler B. (2008) The serine protease TMPRSS6 is required to sense iron deficiency. Science 320, 1088–1092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Folgueras A. R., de Lara F. M., Pendas A. M., Garabaya C., Rodriguez F., Astudillo A., Bernal T., Cabanillas R., Lopez-Otin C., and Velasco G. (2008) Membrane-bound serine protease matriptase-2 (Tmprss6) is an essential regulator of iron homeostasis. Blood 112, 2539–2545 [DOI] [PubMed] [Google Scholar]
- 59. Zhao N., Nizzi C. P., Anderson S. A., Wang J., Ueno A., Tsukamoto H., Eisenstein R. S., Enns C. A., and Zhang A. S. (2015) Low intracellular iron increases the stability of matriptase-2. J. Biol. Chem. 290, 4432–4446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Kuninger D., Kuns-Hashimoto R., Nili M., and Rotwein P. (2008) Pro-protein convertases control the maturation and processing of the iron-regulatory protein, RGMc/hemojuvelin. BMC biochemistry 9, 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Silvestri L., Pagani A., and Camaschella C. (2008) Furin-mediated release of soluble hemojuvelin: a new link between hypoxia and iron homeostasis. Blood 111, 924–931 [DOI] [PubMed] [Google Scholar]
- 62. Silvestri L., Pagani A., Nai A., De Domenico I., Kaplan J., and Camaschella C. (2008) The serine protease matriptase-2 (TMPRSS6) inhibits hepcidin activation by cleaving membrane hemojuvelin. Cell Metab. 8, 502–511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Zhang A. S., Anderson S. A., Wang J., Yang F., DeMaster K., Ahmed R., Nizzi C. P., Eisenstein R. S., Tsukamoto H., and Enns C. A. (2011) Suppression of hepatic hepcidin expression in response to acute iron deprivation is associated with an increase of matriptase-2 protein. Blood 117, 1687–1699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Chen W., Sun C. C., Chen S., Meynard D., Babitt J. L., and Lin H. Y. (2013) A novel validated enzyme-linked immunosorbent assay to quantify soluble hemojuvelin in mouse serum. Haematologica 98, 296–304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Secher A., Kelstrup C. D., Conde-Frieboes K. W., Pyke C., Raun K., Wulff B. S., and Olsen J. V. (2016) Analytic framework for peptidomics applied to large-scale neuropeptide identification. Nat. Commun. 7, 11436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Chojnacki S., Cowley A., Lee J., Foix A., and Lopez R. (2017) Programmatic access to bioinformatics tools from EMBL-EBI update: 2017. Nucleic Acids Res. 45, W550–W553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Gouw M., Michael S., Samano-Sanchez H., Kumar M., Zeke A., Lang B., Bely B., Chemes L. B., Davey N. E., Deng Z., Diella F., Gurth C. M., Huber A. K., Kleinsorg S., Schlegel L. S., Palopoli N., Roey K. V., Altenberg B., Remenyi A., Dinkel H., and Gibson T. J. (2018) The eukaryotic linear motif resource - 2018 update. Nucleic Acids Res. 46, D428–d434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Dickinson M. E., Flenniken A. M., Ji X., Teboul L., Wong M. D., White J. K., Meehan T. F., Weninger W. J., Westerberg H., Adissu H., Baker C. N., Bower L., Brown J. M., Caddle L. B., Chiani F., Clary D., Cleak J., Daly M. J., Denegre J. M., Doe B., Dolan M. E., Edie S. M., Fuchs H., Gailus-Durner V., Galli A., Gambadoro A., Gallegos J., Guo S., Horner N. R., Hsu C. W., Johnson S. J., Kalaga S., Keith L. C., Lanoue L., Lawson T. N., Lek M., Mark M., Marschall S., Mason J., McElwee M. L., Newbigging S., Nutter L. M., Peterson K. A., Ramirez-Solis R., Rowland D. J., Ryder E., Samocha K. E., Seavitt J. R., Selloum M., Szoke-Kovacs Z., Tamura M., Trainor A. G., Tudose I., Wakana S., Warren J., Wendling O., West D. B., Wong L., Yoshiki A., MacArthur D. G., Tocchini-Valentini G. P., Gao X., Flicek P., Bradley A., Skarnes W. C., Justice M. J., Parkinson H. E., Moore M., Wells S., Braun R. E., Svenson K. L., de Angelis M. H., Herault Y., Mohun T., Mallon A. M., Henkelman R. M., Brown S. D., Adams D. J., Lloyd K. C., McKerlie C., Beaudet A. L., Bucan M., and Murray S. A. (2016) High-throughput discovery of novel developmental phenotypes. Nature 537, 508–514 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All the RAW MS data and corresponding search outputs have been deposited to the ProteomeXchange Consortium (http://proteomecentral.proteomexchange.org) via the PRIDE partner repository with the dataset identifier PXD011797.