Abstract
Acute myeloid leukemia (AML) cells often co-opt normal hematopoietic stem cell (HSC) programs to drive neoplastic proliferation, and HSC-related gene expression signatures have been identified as biomarkers for poor prognosis in AML patients. We sought to identify new regulators of HSCs and AML cells from previously published HSC and leukemia stem cell (LSC) gene expression signatures. We identified PRKCH (Protein Kinase C eta) as a gene that is highly expressed in both mouse and human HSCs, as well as in LSCs from independent cohorts of AML patients. Prkch deletion in mice resulted in impaired HSC function. PRKCH was most highly expressed in undifferentiated (FAB M0) subtype AML, and high expression correlated with TP53 and RUNX1 mutations, high risk cytogenetic features, and poor overall survival. Prkch deletion in a Flt3-ITD/Runx1 mutant mouse AML model did not extend survival. Thus, PRKCH is necessary for normal HSC function, its expression predicts poor survival in AML patients, but it is not required for AML to develop.
Keywords: PRKCH, Protein Kinase C eta, hematopoietic stem cell, acute myeloid leukemia
Introduction
Hematopoietic stem cells (HSCs) can divide extensively and give rise to undifferentiated daughter HSCs.1 This self-renewal process allows HSCs to maintain their numbers throughout life and to regenerate the hematopoietic system after transplantation or injury. Acute myeloid leukemia (AML) cells often ectopically activate HSC-associated genes to drive neoplastic proliferation.2–4 Furthermore, many AMLs are thought to follow a cancer stem cell model meaning that self-renewal capacity is restricted to subpopulations of “leukemia stem cells” (LSCs) that give rise to non-self-renewing daughter cells.2, 5 This hierarchy loosely resembles normal hematopoiesis, and it contributes to the heterogeneity of individual leukemias.3 Because HSCs and LSCs both have the capacity to divide extensively without differentiating, genes that are highly expressed in both HSCs and LSCs may encode potential drug targets.
Several HSC and LSC gene expression signatures have been described in the literature, and these signatures have prognostic significance in AML patients.2, 6, 7 Many genes within these sets have well-characterized functions in HSCs and/or AML cells.2, 7 Others have not been tested for a function in HSCs or AML cells, though they are potentially important. We therefore sought to identify genes within a published HSC/LSC gene signature, by Eppert et al. (Ref. 2), that have an unappreciated role in maintaining HSC function and that might be of prognostic or therapeutic significance in AML patients. Our approach utilized both gene expression and functional assays to identify candidate genes (Fig. 1A). Based on this approach, we have identified PRKCH as a novel regulator of HSC function and as a marker of poor prognosis in AML patients.
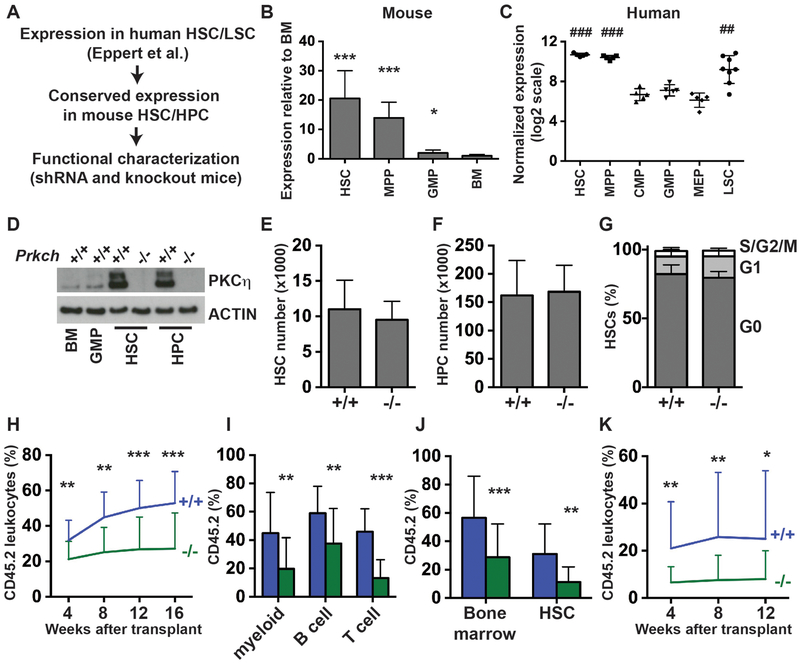
Figure 1. Prkch is highly expressed in HSCs and promotes HSC function.
(A) Overview of screen to identify novel regulators of HSC function and myeloid leukemogenesis based on a HSC/LSC gene set from Eppert et al. (Ref. 2). (B) Prkch expression in mouse HSCs, HPCs and GMPs relative to unfractionated bone marrow as determined by qRT-PCR (n=4). (C) PRKCH expression in human HSCs, MPPs, myeloid progenitors and LSCs in a microarray analysis described by Seita et al. (Ref. 8). (D) PKCη protein expression in mouse HSCs, HPCs, GMPs and bone marrow. (E, F) HSC and HPC numbers in two hind limbs (tibia + femur) from 8–10 week old Prkch+/+ or Prkch−/− mice (n=5–6). (G) Cell cycle distribution of HSCs isolated from 8–10 week old Prkch+/+ or Prkch−/− mice (n=4–5). (H) Long term repopulating assays showing CD45.2 peripheral leukocyte chimerism in recipients of Prkch+/+ or Prkch−/− bone marrow cells (n=19–20 recipients, 4 independent experiments). (I) Peripheral myeloid (CD11b+Gr1+), B-cell (B220+) and T-cell (CD3+) donor chimerism at 16 weeks after transplantation (n=19–20). (G) CD45.2 chimerism of unfractionated bone marrow cells and HSCs at 16 weeks after the primary transplant (n=17–19). (K) CD45.2 peripheral leukocyte chimerism in secondary recipients of Prkch+/+ or Prkch−/− bone marrow cells (n=19–20). For all panels, error bars indicate standard deviations. For panels B and E-K, p-values were calculated by the two-tailed Student’s t-test; *p<0.05, **p<0.01, ***p<0.001. For panel C, p-values were calculated by one-way ANOVA with the Holm-Sidak post-hoc test for multiple comparisons; ### p<0.0001 for human HSCs and MPPs relative to CMP, GMP and MEP, ## p<0.01 for LSCs relative to CMP, GMP and MEP.
Materials and Methods
A complete description of materials and methods is included within the supplementary materials.
Results and Discussion
Since important HSC regulators are likely to have conserved expression patterns in mouse and human HSCs, we tested whether genes from the Eppert et al. HSC/LSC gene set are more highly expressed in mouse HSCs (CD150+CD48−Lineage−c-kit+Sca1+) as compared to non-self-renewing granulocyte-monocyte progenitors (GMPs; Lineage−c-kit+Sca1−CD127−CD34+CD16/32+) and unfractionated bone marrow cells. We focused on 10 genes that have not been previously tested for a role in regulating HSCs or AML cells. As positive controls, we also analyzed expression of four genes − Mecom, Evi1, Soc2 and Hlf – that have been previously shown to be highly expressed in mouse HSCs. Of the 14 genes tested, all but one showed significantly higher expression in HSCs as compared to unfractionated bone marrow (Supplementary Table S1). Thus, genes within the Eppert et al. HSC/LSC gene set have well-conserved expression in mouse HSCs.
We chose to further characterize the function of Prkch using loss-of-function mice because of its high level of expression in mouse HSCs and lineage restricted hematopoietic progenitors (HPCs; CD48+Lineage−c-kit+Sca1+) relative to more committed myeloid progenitors (Fig. 1B), its high expression in human HSCs, multipotent progenitors (MPPs) and LSCs in a study separate from Eppert et al. (Fig. 1C),6, 8 and based on a preliminary screen that showed progressive depletion of Prkch shRNA-expressing 32D cells (Supplemental Fig. S1). Prkch encodes Protein Kinase C-eta (PKCη), a serine-threonine kinase that is necessary for T-cell activation and for regulatory T-cell function.9, 10 PKCη protein was highly expressed in HSCs and HPCs relative to GMPs and unfractionated bone marrow (Fig. 1D), consistent with transcript levels in these cells. Deletion of exon 2 of the mouse Prkch gene resulted in a complete loss of PKCη protein expression in the HSC and HPC cell populations (Fig. 1D). In an unperturbed state, homozygous Prkch deletion had no effect on HSC numbers, HPC numbers or HSC proliferation (Fig. 1E–G). Furthermore, peripheral blood counts were not affected by Prkch deletion (Supplemental Fig. S2A–C). Thus, Prkch is not necessary for HSC maintenance or hematopoiesis under normal homeostatic conditions.
We performed competitive transplantation assays to test whether Prkch regulates HSC function. We transplanted 300,000 wild type or Prkch−/− adult bone marrow cells (CD45.2+) along with 300,000 wild type competitor cells (CD45.1+) into lethally irradiated CD45.1 mice (N=19–20 recipients per genotype from 4 independent experiments). Recipients of Prkch−/− bone marrow cells had significantly lower levels of CD45.2 chimerism over a 16 week monitoring period as compared to recipients of wild type bone marrow (Fig. 1H). Myeloid, B-cell and T-cell lineages were all affected (Fig. 1I). Evaluation of recipient bone marrow at 16 weeks after the transplant showed a significant reduction in Prkch−/− HSC chimerism (Fig. 1J). Secondary transplants showed reduced but persistent chimerism from Prkch−/− cells (Fig. 1K). Altogether, the data show that Prkch deletion impairs HSC function, but it does not completely eliminate long-term HSC self-renewal capacity.
We next tested whether PRKCH expression is prognostic for poor survival in human AML patients, as might be expected given its expression in HSCs, HPCs and LSCs. We evaluated PRKCH transcript levels in 179 AML specimens from The Cancer Genome Atlas (TCGA) that had gene expression, mutation profiling and survival data available (Supplementary Table S2).11 PRKCH expression was significantly elevated in patients with French-American-British (FAB) subtype M0 AML as compared to other subtypes (Fig. 2A), consistent with the finding that PRKCH is highly expressed in immature hematopoietic progenitors. Patients with higher than median PRKCH expression had poorer overall survival than patients with below-median expression (Fig. 2B). To test whether this simply reflected worse survival of FAB M0 patients, we repeated the survival analysis exclusively for patients with FAB M1/M2 subtype disease. In this restricted patient population, high PRKCH expression still predicted poor overall survival (Fig. 2C). Of note, PRKCH expression correlated more strongly with poor prognosis than that other HSC-related genes that have well-established prognostic significance, including BAALC and MECOM (Supplemental Fig. S3).12, 13
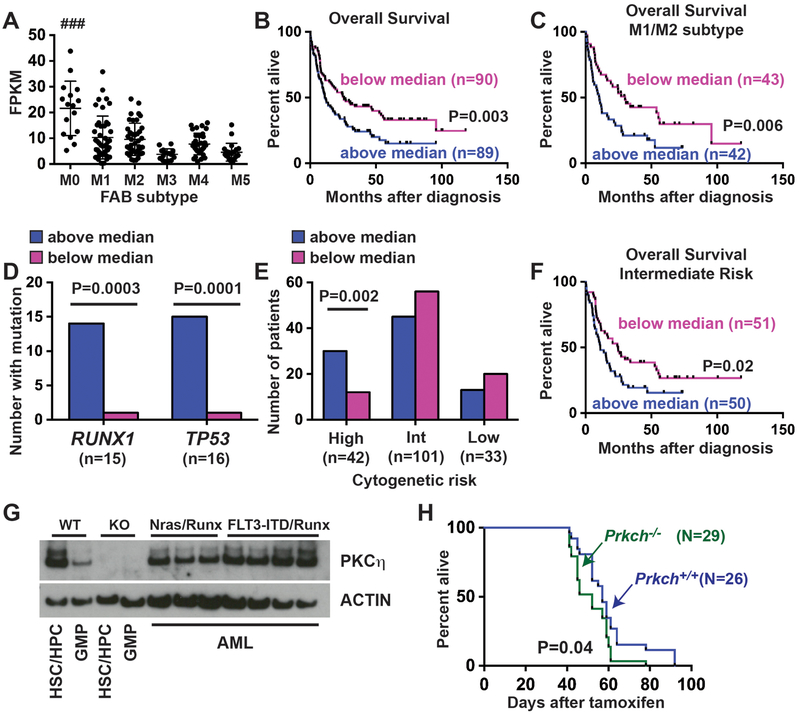
Figure 2. PRKCH expression predicts poor prognosis in AML, but it is not necessary for leukemogenesis.
(A) PRKCH expression in FAB subtypes M0 through M5 in the TCGA (Ref. 10). FPKM values were compared among all six subtypes by one-way ANOVA with the Holm-Sidak post-hoc test for multiple comparisons. ### p<0.0001 for AML M0 relative to all other subtypes. (B) Kaplan-Meier survival curves showing overall survival of TCGA patients with PRKCH expression above or below the median. (C) Kaplan-Meier curve showing overall survival of TCGA patients with M1 or M2 subtype AML. (D) RUNX1 and TP53 mutations are enriched in TCGA patients with above-median PRKCH expression (pink) as compared to patients with below-median expression (blue). P-values are shown in the panel and were calculated by the Fisher’s exact test. (E) Patients with high risk cytogenetic features are enriched within the above-median PRKCH expression group, as calculated by the Fisher’s exact test. (F) Kaplan-Meier curve showing overall survival of TCGA patients with above- and below-median PRKCH expression and intermediate risk cytogenetics. (G) Western blot showing PKCη expression in mouse AML with cooperating NrasG12D/Runx1 or Flt3ITD/Runx1 mutations. (H) Kaplan-Meier survival curves of mice transplanted with Ubc-CreER; Flt3ITD; Runx1f/f; Prkch+/+ or Ubc-CreER; Flt3ITD; Runx1f/f; Prkch−/− bone marrow cells. Survival is shown as days after Cre-ER-mediated Runx1 deletion. For all survival curves, p-values are shown in the panels and were calculated by the log-rank test.
Our findings suggest that PRKCH expression may correlate with high-risk mutations in AML. We evaluated the mutation profiles of TCGA patients with either above- or below-median PRKCH expression. Patients with RUNX1 and TP53 mutations were highly enriched within the PRKCH above-median subset (Relative Risks of 14 and 15, respectively; confidence intervals of 1.9–105 and 2.0–113, respectively; Fig. 2D). These are both high-risk mutations,14–16 and they account for ~35% of the patients with above-median PRKCH expression. High PRKCH expression was also associated with high-risk cytogenetic features such as chromosome 5 or 7q deletions (Relative Risk of 2.5, 1.4–4.6 95% Confidence Interval; Fig. 2E). However, when we restricted the survival analysis to patients with intermediate risk cytogenetic profiles, above-median PRKCH expression still predicted poor overall survival (Fig. 2F).
The association of high PRKCH expression with RUNX1 and TP53 mutations, and its association with poor overall survival, raised the question of whether PRKCH contributes to myeloid leukemogenesis. We generated Runx1-deficient leukemias in mice with cooperating NrasG12D or Flt3-Internal Tandem Duplication (Flt3ITD) mutations.17, 18 These mutations have been found in found in approximately 10% and 16–20% of patients with Runx1 mutant AML, respectively.15, 16 To generate AML we transplanted Ubc-CreER; NrasG12D; Runx1f/f and Ubc-CreER; Flt3ITD; Runx1f/f bone marrow cells into lethally irradiated mice and administered tamoxifen 6 weeks after the transplants to delete Runx1. Recipient mice developed AML between 1 and 3 months after tamoxifen treatment, and all tested AML specimens expressed PKCη (Fig. 2G) To test whether Prkch is necessary for these leukemias to form, we generated and transplanted bone marrow cells from age-matched Ubc-CreER; Flt3ITD; Runx1f/f; Prkch+/+ control and Ubc-CreER; Flt3ITD; Runx1f/f; Prkch−/− mice. Surprisingly, Prkch deletion led to a small but significant acceleration of death due to AML in this model rather than impeding disease progression (Fig. 2H). There were no differences in the morphologies or surface marker phenotypes of Prkch+/+ and Prkch−/− AML specimens (Supplementary Fig. S4). Limiting dilution assays did not reveal consistent Prkch-dependent differences in LSC frequencies (Supplementary Table S4). Western blot analyses did not reveal reproducible, Prkch-dependent differences in STAT5, MAPK, STAT3 or PI3K signal transduction (Supplementary Fig. S5A), and we did not identify consistent differences in gene expression between Prkch+/+ and Prkch−/− leukemias by RNA-sequencing (data not shown). Compensatory changes in expression of Protein Kinase C family members were not observed (Supplementary Fig. S5B, C).
Altogether, our data show that PRKCH is highly expressed in HSCs, and it is necessary for optimal HSC function. High PRKCH expression is associated with high risk mutation profiles in AML patients, but it is not required for AML formation, at least not on a Flt3ITD;Runx1Δ/Δ genetic background. High PRKCH expression may therefore reflect a more primitive cell of origin for AML bearing RUNX1 and TP53 mutations, independent of a requisite function for PKCη in these leukemias.
Supplementary Material
Acknowledgements
We thank David Spencer (Washington University) for providing annotated TCGA expression and mutation data. J.A.M. received support from the St. Baldrick’s Foundation, Hyundai Hope on Wheels, the Children’s Discovery Institute of Washington University and St. Louis Children’s Hospital and the National Heart Lung and Blood Institute (R01HL136504). J.A.M. was a Scholar of the Child Health Research Center at Washington University School of Medicine (K12-HD076224).
References
- 1.He S, Nakada D, Morrison SJ. Mechanisms of stem cell self-renewal. Annu Rev Cell Dev Biol. 2009;25:377–406. [DOI] [PubMed] [Google Scholar]
- 2.Eppert K, Takenaka K, Lechman ER, Waldron L, Nilsson B, van Galen P, et al. Stem cell gene expression programs influence clinical outcome in human leukemia. Nat Med. 2011;17(9):1086–93. [DOI] [PubMed] [Google Scholar]
- 3.Magee JA, Piskounova E, Morrison SJ. Cancer stem cells: impact, heterogeneity, and uncertainty. Cancer Cell. 2012;21(3):283–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Reya T, Morrison SJ, Clarke MF, Weissman IL. Stem cells, cancer, and cancer stem cells. Nature. 2001;414(6859):105–11. [DOI] [PubMed] [Google Scholar]
- 5.Bonnet D, Dick JE. Human acute myeloid leukemia is organized as a hierarchy that originates from a primitive hematopoietic cell. Nat Med. 1997;3(7):730–7. [DOI] [PubMed] [Google Scholar]
- 6.Gentles AJ, Plevritis SK, Majeti R, Alizadeh AA. Association of a leukemic stem cell gene expression signature with clinical outcomes in acute myeloid leukemia. JAMA. 2010;304(24):2706–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ng SW, Mitchell A, Kennedy JA, Chen WC, McLeod J, Ibrahimova N, et al. A 17-gene stemness score for rapid determination of risk in acute leukaemia. Nature. 2016;540(7633):433–7. [DOI] [PubMed] [Google Scholar]
- 8.Seita J, Sahoo D, Rossi DJ, Bhattacharya D, Serwold T, Inlay MA, et al. Gene Expression Commons: an open platform for absolute gene expression profiling. PLoS One. 2012;7(7):e40321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kong KF, Fu G, Zhang Y, Yokosuka T, Casas J, Canonigo-Balancio AJ, et al. Protein kinase C-eta controls CTLA-4-mediated regulatory T cell function. Nat Immunol. 2014;15(5):465–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fu G, Hu J, Niederberger-Magnenat N, Rybakin V, Casas J, Yachi PP, et al. Protein kinase C eta is required for T cell activation and homeostatic proliferation. Sci Signal. 2011;4(202):ra84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cancer Genome Atlas Research Network. Genomic and epigenomic landscapes of adult de novo acute myeloid leukemia. N Engl J Med. 2013;368(22):2059–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Barjesteh van Waalwijk van Doorn-Khosrovani S, Erpelinck C, van Putten WL, Valk PJ, van der Poel-van de Luytgaarde S, Hack R, et al. High EVI1 expression predicts poor survival in acute myeloid leukemia: a study of 319 de novo AML patients. Blood. 2003;101(3):837–45. [DOI] [PubMed] [Google Scholar]
- 13.Baldus CD, Tanner SM, Ruppert AS, Whitman SP, Archer KJ, Marcucci G, et al. BAALC expression predicts clinical outcome of de novo acute myeloid leukemia patients with normal cytogenetics: a Cancer and Leukemia Group B Study. Blood. 2003;102(5):1613–8. [DOI] [PubMed] [Google Scholar]
- 14.Wong TN, Ramsingh G, Young AL, Miller CA, Touma W, Welch JS, et al. Role of TP53 mutations in the origin and evolution of therapy-related acute myeloid leukaemia. Nature. 2015;518(7540):552–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gaidzik VI, Teleanu V, Papaemmanuil E, Weber D, Paschka P, Hahn J, et al. RUNX1 mutations in acute myeloid leukemia are associated with distinct clinicopathologic and genetic features. Leukemia. 2016;30(11):2282. [DOI] [PubMed] [Google Scholar]
- 16.Schnittger S, Dicker F, Kern W, Wendland N, Sundermann J, Alpermann T, et al. RUNX1 mutations are frequent in de novo AML with noncomplex karyotype and confer an unfavorable prognosis. Blood. 2011;117(8):2348–57. [DOI] [PubMed] [Google Scholar]
- 17.Porter SN, Cluster AS, Yang W, Busken KA, Patel RM, Ryoo J, et al. Fetal and neonatal hematopoietic progenitors are functionally and transcriptionally resistant to Flt3-ITD mutations. eLife. 2016;5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mead AJ, Kharazi S, Atkinson D, Macaulay I, Pecquet C, Loughran S, et al. FLT3-ITDs instruct a myeloid differentiation and transformation bias in lymphomyeloid multipotent progenitors. Cell reports. 2013;3(6):1766–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.