SUMMARY
Fate-changing transcription factors (TFs) scan chromatin to initiate new genetic programs during cell differentiation and reprogramming. Yet the protein structure domains that allow TFs to target nucleosomal DNA remain unexplored. We screened diverse TFs for binding to nucleosomes containing motif-enriched sequences targeted by pioneer factors in vivo. FOXA1, OCT4, ASCL1/E12α, PU1, CEBPα, and ZELDA display a range of nucleosome binding affinities that correlate with their cell reprogramming potential. We further screened 593 full-length human TFs on protein microarrays against different nucleosome sequences, followed by confirmation in solution, to distinguish among factors that bound nucleosomes, such as the neuronal AP-2α/β/γ, versus factors that only bound free DNA. Structural comparisons of DNA binding domains revealed that efficient nucleosome binders use short anchoring α-helices to bind DNA, whereas weak nucleosome binders use unstructured regions and/or β-sheets. Thus, specific modes of DNA interaction allow nucleosome scanning that confers pioneer activity to transcription factors.
Keywords: nucleosome binding, Ascl1, Pu.1, FoxA, protein microarray, pioneer transcription factor, TFAP2A, NHLH2, RBPJ, Elk1
Graphical Abstract
eTOC blurb
Nucleosomes modulate transcription factor interactions with DNA. We show that pioneer factors recognize DNA via a short scissor-like or a helix-turn-helix module compatible with nucleosome binding, while non-pioneer TFs interact with DNA via an extended anchoring α-helix or loops. We reveal commonalities and structural limitations that associate with nucleosome binding by transcription factors.
INTRODUCTION
Diverse genomic studies have established that a subset of fate-changing transcription factors (TFs) can target nucleosomal sequences in chromatin, and hence act as pioneer TFs (Heinz et al., 2010; Iwafuchi-Doi and Zaret, 2014; Li et al., 2018; Wapinski et al., 2013). However, the principles that predict whether a given TF may target sites on nucleosomal DNA are not clear. DNA sequences containing clusters of TF binding sites at active enhancers and promoters that were considered to be “nucleosome-free regions” have been shown to harbor histones variants H3.3 and H2A.Z (de Dieuleveult et al., 2016; Jin et al., 2009), histone modifications such as H3K4me1/2, H3K27Ac (Calo and Wysocka, 2013), and exposed or “fragile” nucleosomes (Iwafuchi-Doi et al., 2016; Mieczkowski et al., 2016). Still, the presence of a nucleosome can be refractory to TF binding and transcriptional activity (Blomquist et al., 1996; Taylor et al., 1991). In this study, we investigate the intrinsic nucleosome-binding properties of diverse TFs to identify features that enable nucleosome binding.
We focused on TFs that drive cell differentiation and reprogramming. Analysis of the pioneer factor FOXA, a winged helix factor required for liver development (Lee et al., 2005), revealed that FOXA1 can bind its target sequence on nucleosomes and induce local nucleosomal accessibility in vitro (Cirillo et al., 2002; Cirillo and Zaret, 1999a). In vivo, FOXA1 can displace linker histone to promote local chromatin opening and allow cooperative binding of other TFs (Iwafuchi-Doi et al., 2016). Similar to FOXA, the hematopoietic factors PU1 and CEBPα target compacted chromatin and induce chromatin accessibility, while CEBPα binding to nucleosome-enriched regions is more dependent on PU.1 and EBF1 (Heinz et al., 2010; van Oevelen et al., 2015). EBF1 has also been shown to bind compacted chromatin and induce lineage-specific chromatin accessibility (Boller et al., 2016; Li et al., 2018).
Genomic assessment of the ectopic OCT4, SOX2, KLF4 (OSK) targeted sites and nucleosome occupancy by MNase-seq on pre-existing chromatin showed that OSK target partial motifs on nucleosomal DNA that mirrors their relative nucleosome affinity in vitro. By contrast, c-MYC bind nucleosomes poorly and localizes at its nucleosomal targets via cooperative interaction with other factors (Soufi et al., 2015a). Thus, TFs show a differential preference for nucleosomes and a hierarchy by which they enable cell fate changes. Yet the structural basis for nucleosome motif targeting, apart from the inherent adaptability of the DNA binding domains (DBDs) (Soufi et al., 2015a), remains unclear.
While there are over 2,000 predicted human TFs (Messina et al., 2004; Vaquerizas et al., 2009), only a fraction has been tested for nucleosome binding (Haswell and O’Shea, 1999; Hayes, 1992; Liu and Kraus, 2017; Perlmann, 1988). Recently, an assessment of TF DBDs interacting with nucleosomes, based on SELEX enrichment of bound populations in vitro, revealed diverse binding behaviors as revealed by the positions of motifs at different targeted positions across the nucleosome (Zhu et al., 2018). However, the study did not provide a direct quantitative assessment of free DNA vs. nucleosome binding and left open the question of how protein structure relates to nucleosome binding. Transcription factors can exhibit slow on-rates to nucleosomes, allowing time for thermal motions to elicit motif exposure, which can be compensated by slow off-rates, resulting in nucleosome binding in the nanomolar range of dissociation constant (kD) as seen for free DNA (Cirillo and Zaret, 1999b; Donovan et al., 2019)
Protein microarrays are a powerful tool for high-throughput assessment of intermolecular interactions (Chen et al., 2008; Hu et al., 2009). However, interactions determined by solid-state methods may not truly reflect interactions under physiological conditions (Sun et al., 2013) and sequence-specific binding needs to be confirmed in solution. To gain a broader view of how TFs target nucleosomes, we used a high- throughput protein microarray that contained 593 unique full-length human TFs, after filtering for technical accessibility, followed by validation in-solution of the most significant interactors. Using in vitro reconstituted nucleosomes with three different endogenous sequences enriched in TF motifs, we find that TFs bind with a range of nucleosome binding affinities that positively correlate with TF reprogramming potential. Notably, we find that strong nucleosome binding is associated with an anchoring α-helix that interacts with no more than half of the DNA’s circumference, leaving the opposite side of DNA free to engage in histone interactions. These findings improve our ability to predict transcription factors that can directly scan and bind nucleosomes and thus act as pioneer factors.
RESULTS
Recombinant Nucleosomes from Endogenous Nucleosomal Sequences Targeted by Pioneer TFs
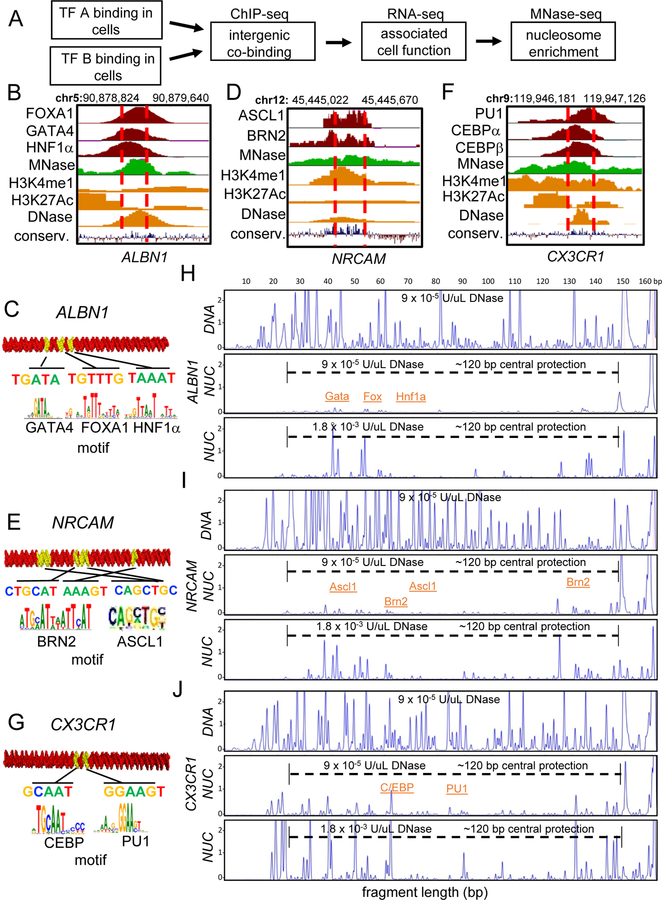
To establish conditions for a large scale study of TFs that can directly interact with nucleosomes, we selected seven TFs (Table 1) suggested to have pioneering activity on the basis of nucleosomal or closed chromatin targeting in vivo, during fibroblast reprogramming to hepatocytes, neurons, or macrophages (Heinz et al., 2010; van Oevelen et al., 2015; Wapinski et al., 2013). Pioneer TF FOXA1 can drive the conversion of fibroblasts to hepatocytes and, during liver development, directly interacts with nucleosome particle N1 within the ALB enhancer to stimulate expression (Chaya et al., 2001; McPherson et al., 1993; Sekiya and Suzuki, 2011). Therefore, FOXA1 was used as a positive control for nucleosome binding on ALBN1 in vitro, assembled into 160 bp nucleosomes containing motifs for FOXA1 and other liver TFs (Figure 1B, 1C).
Table 1 |.
Recombinant TF Dissociation Constants
| Apparent KD (nM) | FOXA1 | GATA4 | HNF1A | ASCL1/E12α | E12α | BRN2 | PU1 | CEBPα | ZLD |
|---|---|---|---|---|---|---|---|---|---|
| Total canonical | 1.3 | 2.4 | 2.7 | 3.1 | ND | 1.2 | 4.5 | ND | – |
| Specific canonical | 0.9 | 2.1 | 2.5 | 3.0 | 31.4 | 4.9 | 3.0 | 7.5 | – |
| Total DNA | 0.3 | 0.5 | 1.1 | 0.7 | – | 0.7 | 0.6 | 4.6 | 0.4 |
| Specific DNA | 0.4 | 0.5 | 1.7 | 0.9 | – | 2.3 | 0.7 | 5.3 | 0.6 |
| Total nuc | 2.5 | 1.6 | 3.5 | 1.6 | – | 1.4 | 4.9 | 13 | 1.3 |
| Specific nuc | 3.0 | 3.8 | 4.7 | 2.6 | – | 2.0 | 5.6 | 18 | 1.8 |
Apparent dissociation constants (KD) were derived from quantified EMSA bands to represent the relative affinities of TFs to their canonical sites or respective free DNA and nucleosomes (nuc). Apparent KDs were derived from two separate binding curves, each representing two experimental replicates, and fitted to the experimental data within R2 values of 0.8–0.99, expressed in nM units. Apparent KDs were quantified from the fractional decrement of free DNA or nuc, designated as “total” binding, or from the first bound-DNA/nuc complexes, representing “specific” binding. ND, not determined. “-” not measured (Figure S2C, S3A–D).
Figure 1. Endogenous TF-Nucleosomal Targets Assemble into Stable Nucleosomes In Vitro.
(A) Schematic diagram showing genomic data processing for the identification of TFs nucleosomal targets.
(B-G) ChIP-seq profile for reprogramming TFs at identified nucleosomal targets and 3D representation of the DNA sequences used for nucleosome assembly containing TFs canonical motifs indicated (yellow). (B) FOXA1, GATA4 and HNF1α ChIP-seq (red) in liver and MNase-seq profile (green) in fibroblasts across the ALBN1 enhancer within the displayed genomic location. (C) 3D representation of the 160 bp- ALBN1-DNA. (D) ASCL1 and BRN2 ChIP-seq (red) at 48 hr induction in fibroblast and MNase-seq profile in fibroblasts near the NRCAM gene. (E) 162 bp NRCAM-DNA. (F) PU1, CEBPα, and CEBPβ ChIP-seq in macrophages and MNase-seq profile in fibroblasts near the CX3CR1 gene. (G) 162 bp- CX3CR1-DNA.
(H-J) DNase-I footprinting showing the protection of (H) ALBN1-DNA, (I) NRCAM-DNA, (J) CX3CR1-DNA before and after nucleosome reconstitution in vitro. Electropherograms generated by digesting 5’−6 FAM end-labeled free DNA (top panel) and nucleosomes with low DNaseI (middle panel) and high DNaseI (bottom panel). Concentrations of DNase-I indicated. Dashed lines indicate central histone octamer protection within nucleosomes.
ASCL1 and BRN2 drive reprogramming from fibroblasts to neurons (Vierbuchen et al., 2010), with ASCL1 targeting closed fibroblast chromatin (Wapinski et al., 2013). By integration of genomic datasets (Figure 1A) we identified an ASCL1 and BRN2 nucleosomal target sequence at the neuron-glia-CAM-related cell adhesion molecule (NRCAM) locus (Figure 1D). The NRCAM gene plays a role in neurite outgrowth and schizophrenia development (Honer et al., 1997; Vawter et al., 1999). The NRCAM site was selected by consideration of its high nucleosomal MNase-seq signal enrichment in fibroblasts before Ascl1 and Brn2 expression (Figure 1D, S1A), ASCL1 and BRN2 targeting to the site by ChIP-seq, the presence of respective DNA binding motifs (Figure 1D, 1E), its enrichment for enhancer associated marks H3K4me1, H3K27Ac, DNase- hypersensitivity (Figure 1D), and gene expression associating with neuronal function by gene ontology (Figure S1A, B).
Genomic and chromatin state analysis by MNase-seq of PU1, CEBPα, and CEBPβ targeting during B cell to macrophage reprogramming suggests that hematopoietic TFs PU1 and CEBPα target nucleosome-enriched chromatin (Barozzi et al., 2014; Heinz et al., 2010; van Oevelen et al., 2015), yet the pre-bound chromatin state was not evaluated. Comparison of fibroblast nucleosome occupancy by MNase- seq signals and TFs ChIP-seq in macrophages (Figure S1C–D) revealed CX3 chemokine receptor 1 (CX3CR1) locus as a candidate nucleosomal targeted site (Figure 1F). CX3CR1-DNA shows enrichment for H3K4me1, H3K27Ac and DNase- hypersensitivity in macrophages (Figure 1F) and it contains the respective DNA binding motifs (Figure 1G). DNA fragments of 160 bp of ALBN1-DNA, and 162 bp of NRCAM- DNA and CX3CR1-DNA were PCR amplified, Cy5-end labeled (Figure S1E), and prepared as free DNA and nucleosomal templates by urea-salt step dialysis (Figure S1F).
At a low concentration of DNaseI, ALBN1, NRCAM, and CX3CR1 nucleosomes exhibited nearly complete resistance to cleavage, compared to free DNA (Figure 1H–J). Using 20 fold more DNase-I on nucleosomes elicited a markedly different cleavage pattern than seen on free DNA. Each of the nucleosomes revealed several interspersed ~10 bp cleavage repeat patterns, indicative of different rotational frames that expose diverse DNA binding motifs within 120 bp central core of the nucleosome populations (Figure 1H–J, bottom panels). Our method of curating endogenous nucleosomal sites enriched for pioneer factor binding in vivo appears robust for discovering DNA sequences that make stable nucleosomes in vitro.
ASCL1 Heterodimerization with E12α Enhances DNA Binding and Specificity
To establish conditions for a large-scale TF-nucleosome screen, we purified full-length FOXA1, GATA4, HNF1α, ASCL1, BRN2, PU1, and CEBPα from E. coli, validated their identity by predicted size and immunoblotting (Figure 2A, S2A), and measured free DNA binding activity. Apparent dissociation constants were determined by electromobility shift assays (EMSAs) with short Cy5-DNA probes (Figure S2B–D) and quantified by two methods; total dissociation constant (total KD), determined by decrement of free probe, and specific dissociation constant (specific KD), determined by the appearance of specific TF-complexes (Soufi et al., 2015a). As expected, all TFs but ASCL1 displayed nanomolar dissociation constants for probes containing canonical DNA binding motifs (Figure S2C, Table 1), but not for probes lacking binding motifs (Figure S2D). We concluded that most of the purified full-length TFs were highly active and bound DNA specifically.
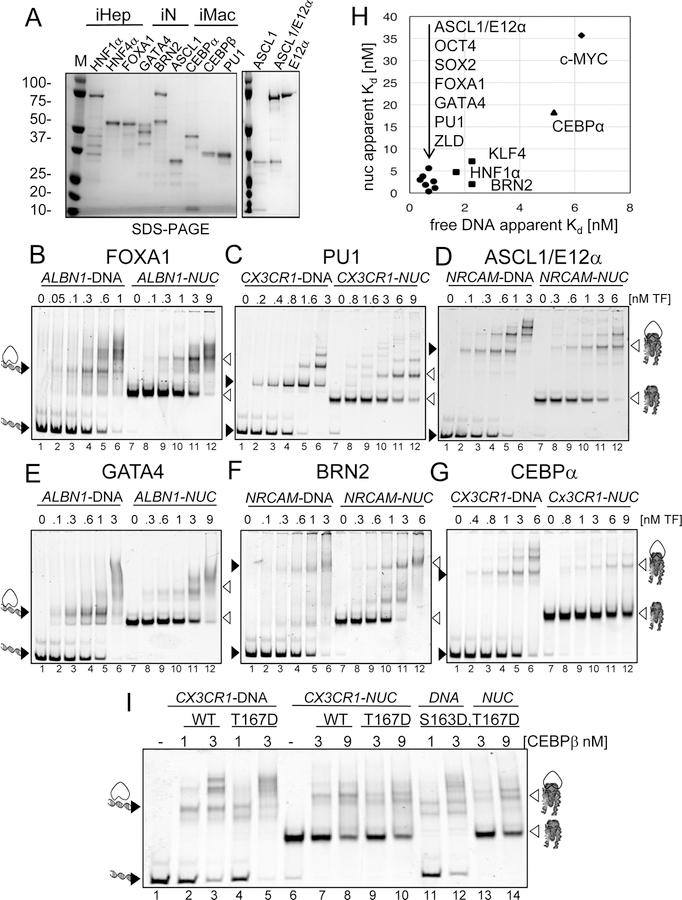
Figure 2. Reprogramming TFs bind Nucleosomes with Nanomolar Affinity.
(A) Recombinant purified full-length TFs analyzed by SDS-PAGE and Coomassie staining. The factors are grouped by reprogramming to iHEP (induced hepatocytes), iN (induced neurons), and iMAC (induced macrophages). Recombinant single purification of ASCL1, E12α, and co-purification of ASCL1/E12α (right panel).
(B-G) Representative EMSA showing the affinity of increasing amounts of TFs (B) FOXA1, (C) PU1, (D) ASCL1/E12α, (E) GATA4, (F) BRN2 and (G) CEBPα to Cy5-labelled DNA (lanes 1–6) and nucleosome (lanes 7–12). Black arrowheads indicate TF- DNA complexes. White arrowheads indicate TF-nucleosome complexes.
(H) 2D plot of TFs dissociation constants for DNA (x-axis) and nucleosomes (y-axis).
(I) Representative EMSA showing the affinity of CEBPβ WT, mutants T167D and S163D,T167D to CX3CR1-DNA and nucleosomes.
ASCL1 homodimer DNA binding was not detectable within the nM range, agreeing with previous studies estimating a high dissociation constant (KD) 140 µM (Figure S2E–F) (Meierhan et al., 1995). Basic helix-loop-helix (bHLH) TFs heterodimerize with other bHLHs (Longo et al., 2008; Powell and Jarman, 2008) and interaction network analysis revealed the ubiquitously expressed E12α (Tcf3) as an ASCL1 interacting partner (Figure S2G) (Henke et al., 2009). Therefore, we co-purified ASCL1/E12α heterodimers (Figure 2A), which formed a DNA-bound complex that migrated faster than E12α homodimers (Figure S2E, lanes 6, 11) and with a markedly increased DNA binding affinity of ~3.1 nM (Figure S2E,F, Table 1). We considered ASCL1/E12α heterodimers suitable for further analysis.
Reprogramming TFs Interact with Nucleosomal Substrates with a Nanomolar Range of Affinities
TFs bound to their respective 160 bp free DNAs of ALBN1-DNA, NRCAM-DNA, CX3CR1 within the low nM range (Figure 2B–G, lanes 1–6). As expected, the TFs show a higher affinity for the longer 160 bp DNA, compared to shorter DNA probes (Table 1) due to increased non-specific DNA interactions with longer DNA templates. Thus, at low concentrations of TFs (0.1–1 nM), each of the factors bound to the 160 bp DNA fragments in a single shifted band, while at higher concentrations we detected additional shifted complexes with distinct dissociation constants (Figure 2B–G,S3A–B). The use of an EMSA assay allowed us to make these distinctions, which could correspond to redundant “periodic” TF binding to nucleosomes detected by SELEX (Zhu et al., 2018).
We then assessed binding of the TFs to the same sequences on nucleosomes. FOXA1, GATA4, ASCL1/E12α, BRN2, HNF1α, and PU1 showed high affinity for nucleosomes, with apparent KDs of 3.0 nM, 3.8 nM, 2.6 nM, 2.0 nM, 4.7 nM, 5.6 nM, respectively (Figure 2B–G lanes 7–12, Table 1). Comparison of TFs by their K DNA and K Nuc, including prior data on iPS reprogramming factors OCT4, SOX2, KLF4, and c- MYC (Soufi et al., 2015a), showed that nucleosome binding factors segregate among each other by a slight difference in DNA affinity, with strong reprogrammers showing a slightly higher DNA affinity (lower KD) compared to BRN2, KLF4, and HNF1α which are suggested to have supporting roles in reprogramming (Figure 2H) (Chanda et al., 2014; Raposo et al., 2015; Wapinski et al., 2013). CEBPα displayed the lowest nucleosome affinity of the nucleosome-binding TFs, with a 5-fold higher KD of 18 nM compared to the other TFs (Figure 2G lanes 7–12, Table 1). In general, TF affinity for DNA was slightly higher than that for nucleosomes, demonstrating that the presence of histone octamer can attenuate, but not inhibit, DNA binding of the reprogramming pioneer TFs.
The isoforms CEBPα and CEBPβ target the same DNA motif and share a high degree of structural homology (Jakobsen et al., 2013; Nerlov, 2007). Posttranslational modifications within the DBD of CEBPs can negatively regulate DNA binding, with the few exceptions of positive DNA binding regulation by sequential phosphorylation of CEBPβ at residues T167 and S163, corresponding to S184 and T188 in CEBPβ LAP* isoforms (Li et al., 2007; Piwien-Pilipuk et al., 2002; Tang et al., 2005). We generated phospho-mimic variants of CEBPβ (T167D) and (S163D, T167D) of the LAP isoform (Figure S3E). CEBPβ T167D showed a weak increase of free DNA binding (Figure 2I, lanes 3,5) and no discernable effect on nucleosome binding (Figure 2I, lanes 8,10). CEBPβ S163D, T167D showed no DNA binding or nucleosome binding enhancement (Figure 2I, lanes 3,12,8,14). Our results show that engagement of histones with DNA limit CEBPα/β binding to DNA and that post-translational modifications of CEBPβ at T167 or S163, T167 may not enhance nucleosome binding.
Nucleosomes Enhance Reprogramming TF Binding Specificity
TFs engage DNA through specific and non-specific DNA interactions (Biggin, 2011). To measure the contribution of sequence specificity on TF binding to DNA vs. nucleosomes, we carried out EMSA in the presence of increasing amounts of short specific and non-specific double stranded DNA (dsDNA) validated as competitors (Figure S2C, D). An 80X molar excess of non-labeled short dsDNA containing specific binding sites minimally displaced free DNA-TF complexes for FOXA1, ASCL1/E12α, PU1, and C/EBPα (Figure S3F, lanes 2,5), similar to competition with non-specific probes (Figure S3F, lane 2,8); while BRN2 exhibited specific competition. Strikingly, competition experiments on nucleosomes, with a low 20X molar excess of specific competitor, completely displaced all TF-nucleosome complexes (Figure 3A, B lanes 2,3). By contrast, an 80X excess of non-specific competitor failed to displace them (Figure 3A, B lanes 2,8). Moreover, specific ASCL1/E12α binding to nucleosomes occurs without histone octamer dissociation, as determined by EMSA followed by Western blotting with antibodies for core histones H2B and H3 (Figure 3C). It appears that TFs generally engage in specific interactions with nucleosomes due to decreased DNA availability by presence of histone octamers, while free DNA is more accessible, allowing the TFs to engage in more non-specific interactions.
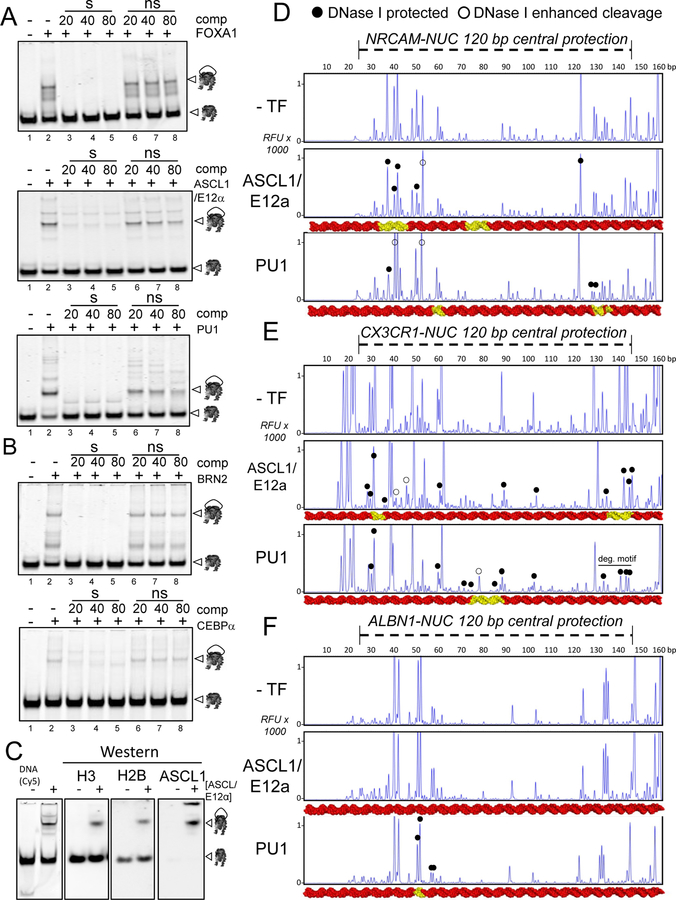
Figure 3. TFs Bind Nucleosomes with Specificity.
(A-B) Representative EMSA of competition assays showing the affinity of recombinant (A) FOXA1, ASCL1/E12α and PU1 to ALBN1-NUC, NRCAM-NUC, and CX3CR1-NUC in the presence of 20-, 40- and 80-fold molar excess of specific competitor ( “s” lanes) or non-specific competitor (“ns” lanes) or absence of competitor (“-” lanes). (B) Same as (A) for BRN2 and CEBPα.
(C) Representative WEMSA showing the binding of ASCL1/E12α to NRCAM-NUC. ASCL1/E12α :Nuc complex from EMSA were transferred onto a PVDF membrane (WEMSA) and blotted for H3, H2B, ASCL1 as indicated (the three panels on the right). White arrow heads indicate the observed TF-nucleosome complexes.
(D-F) DNase-I footprinting electropherograms of 5’−6 FAM-labeled (D) NRCAM-NUC, (E) CX3CR1-NUC, and (F) ALBN1-NUC in absence (top) or presence of ASCL1/E12α (middle) or PU1 (bottom) end-labeled free DNA (top strand). 3D DNA representation (red) with each TF motif (yellow)..Filled circles, protections; open circles, enhancements. “deg. motif” = degenerate motif for PU1.
DNase footprinting reveals that ASCL1/E12α blocks cleavages on two of its motifs on each of the NRCAM and CX3CR1 nucleosomes (Figure 3D, E, middle panels, filled circles). One of the ASCL1 motifs on the NRCAM nucleosome is in the nucleosome center and was DNase-resistant, precluding the ability to assess further protection by ASCL1/E12α. By contrast, the ALBN1 nucleosome lacks an ASCL motif and ASCL1/E12α gave no evidence of protections (Figure 3F, middle panel). PU1 exhibited protections on one of its two motifs on the NRCAM nucleosomes and protections on the single motifs of each of the CX3CR1 and ALBN1 nucleosomes (Figure 3D–F, lower panels, filled circles). ASCL1/E12α and PU1 also show protections and enhanced DNase cleavages (Figure 3D–F, closed and open circles) at additional sites, compared to the nucleosomes alone, suggesting that the TFs stabilize a preferred rotational frame that exposes the target motif (Shim et al., 1998) or induce local conformational changes in the nucleosomal DNA. The data indicate that ASCL1/E12α and PU1 bind their motifs in multiple nucleosome sequences contexts.
Zygotic Genome Activator Zelda Bind Nucleosomes Like Human Pioneer Factors
We compared the ability of the Drosophila TF Zelda (ZLD) to bind nucleosomes like the human TFs. ZLD reprograms the zygotic genome for transcriptional activation during the maternal-to-zygotic transition (Foo et al., 2014; Liang et al., 2008; Schulz et al., 2015; Sun et al., 2015) and binds in vitro assembled nucleosomes from the bottleneck locus (McDaniel, 2019). Recombinant full-length ZLD binds a short dsDNA containing a canonical ZLD-binding motif (CAGGTAG), but not to dsDNA with mutated motif, in agreement with previous studies (Figure 4A) (Hamm et al., 2015).
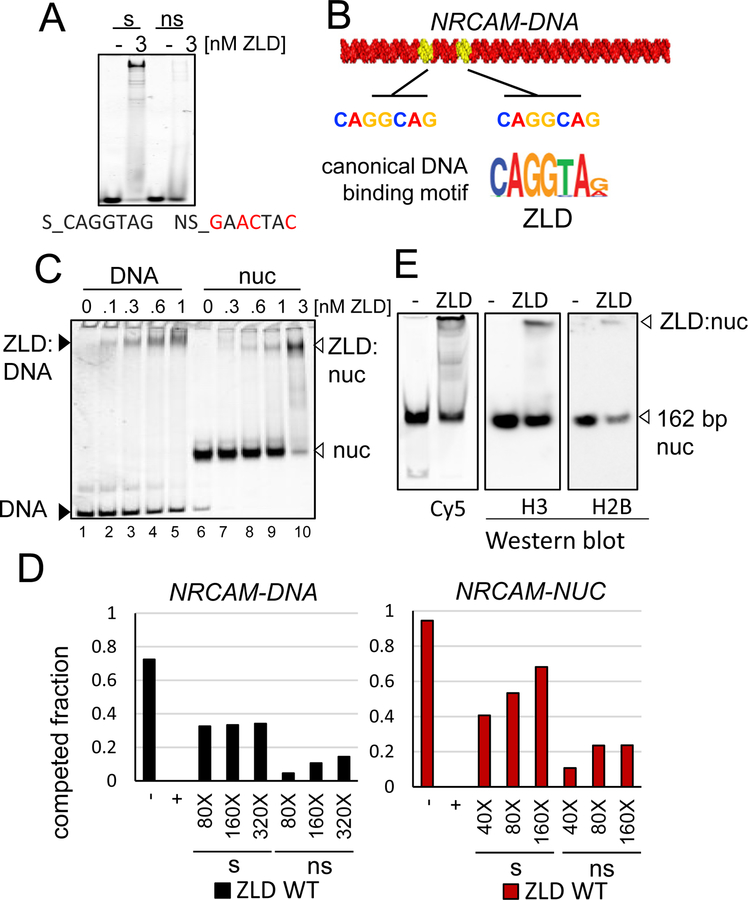
Figure 4. ZnF TF Zelda Bind to Nucleosomes.
(A) Representative EMSA of ZLD showing affinity to short dsDNA probes a containing canonical ZLD-binding motif ( “s” lanes) or mutated motif ( “ns” lanes).
(B) Graphical representation of ZLD motifs (yellow) identified on NRCAM-DNA sequence.
(C) Representative EMSA showing the affinity of increasing amounts of recombinant ZLD to Cy5- NRCAM-DNA (lanes 1–5) and nucleosome (lanes 6–10).
(D) Quantification competed fraction of ZLD:DNA (left panel) or ZLD:nucleosome (right panel) complexes by addition of molar excess of specific competitor ( “s” lanes), non- specific competitor (“ns” lanes) or absence of competitor (“-” lanes). Molar excess listed.
(E) Representative WEMSA of ZLD :Nuc complex from EMSA transferred onto a PVDF membrane (WEMSA) and blotted for H3, H2B as indicated (the two panels on the right). White arrow heads indicate the observed TF-nucleosome complexes.
We identified fortuitous ZLD binding motifs (CAGGCAG) in the NRCAM-DNA sequence (Figure 4B). ZLD robustly bound both NRCAM-DNA and NRCAM-NUC (Figure 4C), without histone octamer displacement (Figure 4E) with a KDNuc of 1.8 nM (Figure 2H), comparable to strong reprogramming TFs. Similar to the human reprogramming TFs we tested, EMSA competition experiments identified non-specific binding of ZLD to free DNA (Figure 4D left panel). However, sequence-independent features also contributed to the binding of ZLD to nucleosomes, as demonstrated by the requirement for 160X unlabeled specific competitor to compete the ZLD-NUC complex as compared to 20X competitor for the previously tested human TF-nucleosome complexes (Figure 4D, right panel). Thus, ZLD can directly engage nucleosomes, consistent with chromatin targeting seen in vivo, and supports a comparable role of nucleosome binding for reprogramming factors in multiple organisms.
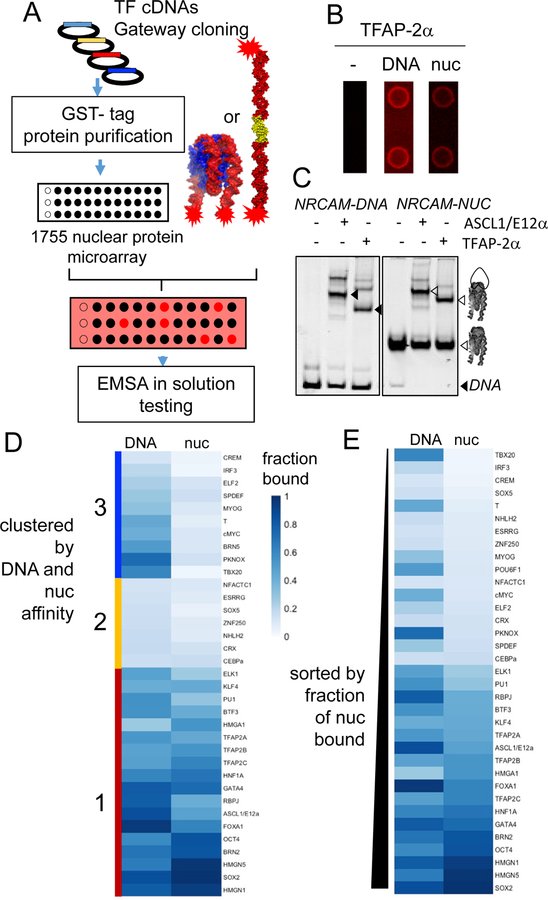
Protein Array Screen for Nucleosome Binding TFs Identified TFAP-2 and HMG Transcription Factors as Strong Nucleosome Binders
Having detailed the nucleosome binding of a subset of human-reprogramming factors and a well-studied insect TF, we were interested in more generally identifying TFs with the fundamental feature of nucleosome binding. To date only a small subset of the estimated 2,000 TFs in the human genome has been tested for direct nucleosome binding (Kummerfeld and Teichmann, 2006; Messina et al., 2004; Vaquerizas et al., 2009). We therefore screened human full-length TFs, purified from yeast and printed on microarrays in duplicates, for binding to Cy5-labeled ALBN1, NRCAM, CX3CR1 as free DNA and nucleosomes (Figure 5A) (Hu et al., 2013). The three different templates have a fortuitous occurrence of 116, 121, and 112 TF binding motifs, respectively, as determined by TRANSFAC database, enabling us to assess many TFs binding to these natural nucleosome sequences (Table S1) (Matys et al., 2006). Moreover, the occurrence of multiple rotational positions on the nucleosomes provide a myriad of accessible motifs (Figure 1H–J). Cy5 fluorescence of probed protein microarrays, incubated with Cy5-labeled ALBN1, NRCAM, CX3CR1 DNAs or nucleosomes, was measured and positive protein-nucleic acid signals were processed (Figure S4A) by protein auto-fluorescence subtraction (Figure S4B), minimal protein amount per spot cutoff (Figure S4C,D), background normalization (Figure S4E), and determination of positive versus negative signal thresholds (Figure S4F). Protein microarrays contained 1,755 nuclear proteins spotted, which after the cutoffs resulted in 1,592 proteins. Finally, out of 1,755 proteins, 593 TFs showed a fluorescent intensity after probing for free DNA and nucleosomes that passed the positive/negative signal threshold; we considered these proteins to be experimentally accessible (Table S2).
Figure 5. Systematic Assessment of Nucleosome Binding of Human TFs with Protein Microarrays.
(A) Graphical scheme used to identify nucleosome interacting human TF using DNA and nucleosome probes binding to protein microarrays.
(B) Cy5 fluorescence of TAP2-α on protein microarray printed spot in duplicate in absence (“-”) or hybridized with NRCAM-DNA or NRCAM-NUC.
(C) Representative EMSA comparing the affinity of ASCL1/E12α with TFAP-2α to NRCAM-DNA or NRCAM-NUC. Black arrow heads indicate TF-DNA complexes. White arrow heads indicate the TF-nucleosome complexes.
(D-E). Heatmap representations of TFs bound fractions to DNA (left) and nucleosome (right) by quantification from EMSA. (D) Clustered heatmap showing strong DNA and nucleosome binders (red-cluster 1), low DNA binders and (yellow-cluster 2), high DNA binders with low nucleosome affinity (blue-cluster 3) (E) Heatmap sorted on TF nucleosome bound fraction. TFs concentrations used on Figure S6A.
The protein microarray screen provisionally identified 326 TFs that interact with both DNA and nucleosomes, 104 that interact with DNA-only, and 163 that interact with nucleosomes-only. Protein microarray screening with Cy5 NRCAM free DNA and nucleosomes identified TFAP2α as a possible nucleosome binding TF (Figure 5B). AP2 TFs are crucial for neuronal crest development and regulate the fragile X mental retardation-1 gene (Schorle et al., 1996; Zhang et al., 1996). Validation of TFAP2α- nucleosome interaction with an in-solution EMSA showed that TFAP2α binds DNA and nucleosomes comparable to ASCL1/E12α (Figure 5C).
To further validate interactions detected from the screen, we purified 81 more of the proteins. Surprisingly, 57 of the proteins exhibited no free DNA or nucleosome binding by EMSA, including 9 transcription factors that scored as “nucleosome only binders” on the arrays. Twenty-four full-length human TFs resulted in detectable DNA binding by EMSA on one of the ALBN1, NRCAM, or CX3CR1 free DNA and nucleosome sequences that contained motifs for each respective TF located within the central domain of the nucleosomes (Figure S5). TF-nucleosome binding was tested at 3X, 6X, or 9X nM higher concentration of TF compared to TF-DNA concentrations (Figure S6A). Quantification of the free DNA and nucleosome bound fraction by EMSA revealed that 11 of the 24 TFs bound to DNA and nucleosomes, 13 to DNA only, and none to nucleosomes only (Figure 5D,S6B–E). Thus, the nucleosome-only interactors by protein microarrays could not be confirmed in solution. Furthermore, the DNA only TFs, IRF3 and CREM1, lacked nucleosome binding in multiple nucleosome sequences contexts harboring their target motifs (Figure S6F–G). The IRF3 DBD is a modified version of the helix-turn-helix (HTH) motif that includes a four-stranded antiparallel β- sheet (De Ioannes et al., 2011). Our results showed that lack of nucleosomes binding is not sequence context dependent.
Clustering of TFs by bound fraction, quantified from EMSAs, identified TFs as strong DNA and nucleosome binders (cluster 1), low DNA binders (cluster 2), and high DNA binders with low nucleosome affinity (cluster 3) (Figure 5D). Cluster 1 TFs with high nucleosome binding affinity also display high DNA affinity (Figure 5D). This included TFAP-2 isoforms TFAP-2β and TFAP-2γ which also showed strong nucleosome binding similar to reprogramming TFs, as quantified by EMSA (Figure 5D). In addition, we also observed that in solution, members of the high mobility group superclass HMGA1, HMGN1, HMGN5, and SOX5 show high affinity to nucleosomes (Figure S6D, lane 10, S6E, lanes 8, 12, and 16). Thus, we were able to identify the TFAP-2 and HMG families of TFs as nucleosome binding factors.
In contrast, sorting TFs by the nucleosome bound fraction revealed a more heterogeneous DNA binding preference, with IRF3, T-Box factors TBX20 and Brachyury (T), and the TALE-MEINOX TF PKNOX displaying high DNA binding but undetectable nucleosome binding, regardless of a higher 9X fold TF concentration on nucleosomes compared to DNA (Figure 5E, S6B) and the presence of DNA binding motifs (Figure S5). We conclude that high DNA affinity does not necessarily translate to nucleosome binding. Thus, nucleosome binding TFs are also strong DNA binders and yet nucleosome binding is not an obligate feature of TFs with high DNA affinity.
Common Structural Features Associated with Pioneer TFs
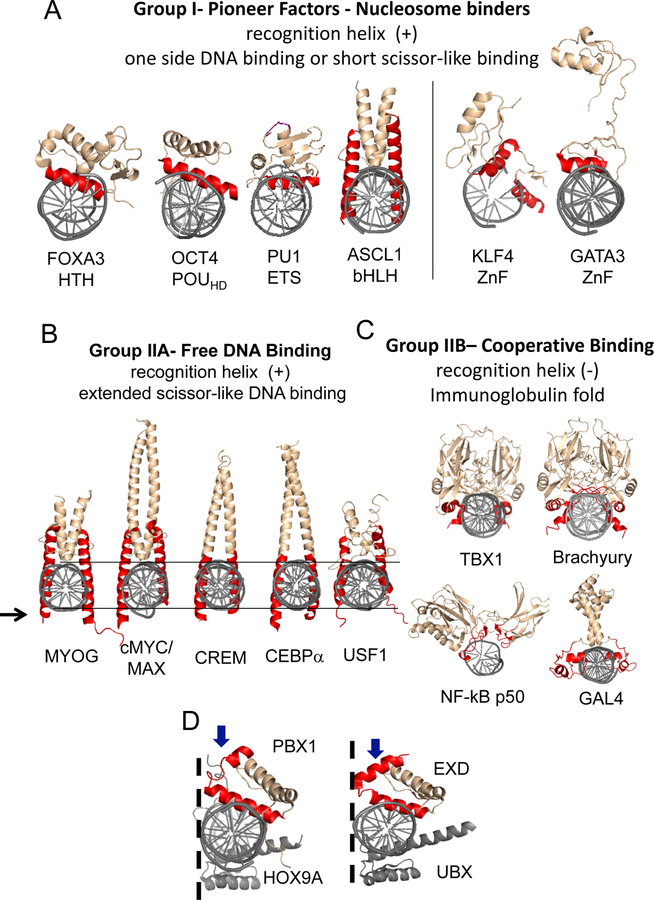
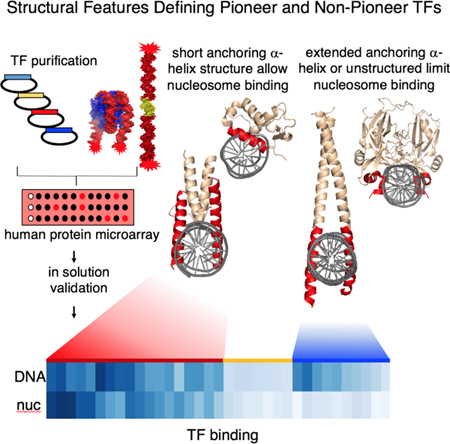
To identify structural commonalities among strong relative to weak nucleosome binders, we compared the 3D structures of the above tested transcription factors’ DBDs. TFs were classified into strong nucleosome binders (group I) and weak nucleosome binders (group II). Group I TFs, validated in solution, exclusively include structural DBD superclasses characterized by α-helical folds (Wingender et al., 2015), including basic helix-loop-helix (bHLH), helix-turn-helix (HTH), homeodomains (HD), and zinc fingers (ZnF) (Figure 6A). ZLD is a C2H2-ZnF TF that could fit within group I, even though its structure is not known. Two main types of DNA anchoring modes are observed; a scissor-like binding mode by dimers of bHLH TFs, and the HTH module characteristic of HMG TFs such as SOXs, homeodomain TFs OCT4, BRN2, and ETS TFs PU1. Although structurally different, we observed that strong nucleosome binders anchor DNA through a short recognition α-helix (Figure 6A). TALE-PBC class TFs such as PBX adopt a DNA binding arrangement with a kink in the recognition helix, resulting in a truncated recognition helix similar to group I short recognition α-helix (Figure 6D). This is in agreement with in vivo data suggesting PBX1 has pioneer activity (Berkes et al., 2004). Therefore, structural comparison of PBX1 with group I strong nucleosome binders predicts strong nucleosome binding potential.
Figure 6. Strong Nucleosome Binding TFs Recognize DNA with Short Recognition α-Helixes.
(A) Group I pioneer TFs DBDs crystal structures of FOXA3 (pdb 1VTN) (Clark, 1993), OCT4 (pdb 3L1P) (Esch et al., 2013), PU1 (pdb 1PUE) (Kodandapani et al., 1996), KLF4 (pdb 2WBS) (Schuetz et al., 2011) and, GATA3 (pdb 4HC9) (Chen et al., 2012). ASCL1 (SMR P50553).
(B) Group IIA TFs with scissor-like DBDs crystal structures and extended recognition α- helixes of cMYC/MAX (pdb 1NKP) (Nair and Burley, 2003), CEBPα (pdb 1NWQ) (Miller et al., 2003), USF (pdb 1AN4) (Ferre-D’Amare et al., 1994), MYOG (SMR P15173), and CREM (SMR Q03060).
(C) Group IIB TFs with immunoglobulin-like fold DBDs crystal structures of TBX1 (pdb 4A04) (El Omari et al., 2012), Brachyury (pdb 1XBR) (Muller and Herrmann, 1997), NF- kB p50 subunit (pdb 1SVC) (Muller et al., 1995) and, GAL4 (pbd 3COQ) (Hong et al., 2008).
(D) TALE-PBC PBX1 (pdb 1PUF) (LaRonde-LeBlanc and Wolberger, 2003) and UBX (pbd 4UUS) (Foos et al., 2015) crystal structures showing scissor-like binding in dimer form with HOX TFs showing kink in recognition α-helixes (blue arrow).
Group II TFs include weak nucleosome binders, which we further subdivided into groups IIA and IIB. Similar to group I TFs, group IIA TFs MYOG, CREM, and previously tested weak nucleosome binder USF1 (Adams and Workman, 1995) display a scissor- like arrangement, but contain an extended recognition α-helix that protrudes past the diameter of the DNA helix (Figure 6B). This suggest that a scissor-like recognition mode with an extended anchoring α-helix results in weak nucleosome binding (Soufi et al., 2015a) and would include bZIP TFs such as cMYC, MYOG, and CEBP.
Group IIB weak nucleosome binders TFs Brachyury and TBX20, homologous to the TBX1 DBD, lack a recognition α-helix and instead use short helical twist or unstructured regions (Figure 6C). These factors are members of the immunoglobulin superclass characterized by a β-sheet core structure, hypervariable loops, and DNA recognition through unstructured regions (Bork et al., 1994; Wingender et al., 2015). Interestingly, group IIB TFs have been previously shown to engage in a multitude of cooperative interactions with homeodomain TFs binding partners resembling group I strong nucleosome binders, such as PITX2 and NKX2 (Naiche and Kelly, 2005), and thus appear more dependent on direct nucleosome binders for their genetic activity.
DISCUSSION
It has been well established that the presence of nucleosomes can impair DNA binding of many transcription factors and DNA repair enzymes (Schild-Poulter et al., 1996; Taylor, 1991). We have addressed how lineage-specific TFs engage nucleosomes to initiate regulatory events in silent chromatin during development and cell programming. We previously showed that the pioneer TFs FOXA and OSK are able to target nucleosomes in vivo and comparably interact with nucleosomes in vitro (Cirillo et al., 1998; Iwafuchi-Doi et al., 2016; Soufi et al., 2015a). Here we show that TFs that drive reprogramming toward hepatic, neuronal, macrophage lineages and the maternal-to- zygotic transition can directly interact with nucleosomes. Additionally, we identify TFs with previously unreported nucleosome binding ability. Given that nucleosome binding is the defining characteristic of pioneer factors, our study defines the DNA binding domain characteristics that endow pioneer activity.
Using nucleosomes assembled with endogenous sequences, we show that ASCL1 heterodimerization with E12α markedly increases ASCL1 DNA and nucleosome binding affinity (Figure S2E,F). E12α is ubiquitously expressed in fibroblasts, suggesting that, during neuronal reprogramming, ASCL1 heterodimerizes with ubiquitously expressed fibroblast TFs to target neuron specific regulatory elements in nucleosome enriched chromatin. This agrees with previous findings, where tethering of ASCL1 to E12α results in increased transcriptional activity (Henke et al., 2009). Additionally, we find that BRN2 also directly interacts with nucleosomes (Figure 2F). This is in agreement with in vivo chromatin targeting of these factors during neuronal reprogramming where ASCL1 and BRN2 engage chromatin (Wapinski et al., 2013).
PU1 is a strong nucleosome binder, while CEBPα shows a weaker nucleosome affinity (Figure 2H). During B cell reprogramming, knockdown of PU1 results in a decreased CEBPα binding to pre-existing and de novo enhancers, suggesting that in the absence of PU1, CEBPα targeting to closed chromatin might be driven by cooperative interactions (van Oevelen et al., 2015). Comparison among reprogramming TFs show a range of DNA and nucleosome binding affinities that correlate with TF reprogramming potential and chromatin targeting strength, suggesting that the intrinsic nucleosome binding preferences of these TFs contributes to the hierarchy driving cellular reprogramming.
By utilizing protein microarrays as a discovery tool, we identified the TFAP-2 family as a family of pioneer TFs. By contrast, TBX20 and Brachyury of the T-BOX family lack detectable nucleosome binding, even while possessing high DNA affinity to sequences containing specific motifs. Nucleosome-only binders, suggested by protein microarrays, could not be validated in solution, probably due to aberrant protein folding or unnatural domain exposure in a solid-state environment. We note that our nucleosome binding results with full-length proteins agree partially but not entirely with Zhu et. al., who mostly studied DBDs in a SELEX assay. Regardless, both studies find that high DNA affinity does not necessarily result in nucleosome binding, which presents a different model than where TFs would engage nucleosomal sites simply by displacing the octamer.
Interestingly, sequence alignment of HMGN proteins and SOX2, SOX5, and SOX9 revealed a high degree of conservation between the SOX group B homology domain, with unknown function (Weina, 2014), and the C-term of the HMGN nucleosome binding domain (NBD) (Figure S6H). C-term truncation of the HMGN1 NBD greatly impairs chromatin binding (Ueda et al., 2008), suggesting that SOX homology group B, outside of the SOX DBD, is required for the nucleosome binding and chromatin targeting ability of SOX TFs.
Pioneer TFs contain diverse DBD structures (Figure 6A), suggesting diverse modes of DNA targeting in chromatin. For example, FOXA has been suggested to compete with linker histone, due to its resemblance with H1 DBD (Clark, 1993; Iwafuchi-Doi et al., 2016), while OCT4 has been suggested to initially interact with chromatin using either of its POU or homeodomain DNA binding modules (Soufi et al., 2015a). We find that DNA recognition via a short scissor-like module or a HTH module is compatible with nucleosome binding (Figure 6A). Recent studies in agreement with our findings showed that Reb1, a yeast HTH factor, displays slower dissociation rates from nucleosomes, compared to free DNA, without histone octamer eviction (Donovan et al., 2019). More recently additional HTH TFs, such as Isl1, have been proposed play a pioneering role in cardiomyocyte reprogramming (Gao et al., 2019).
In contrast, we predict that TFs of the immunoglobulin superclass, lacking an α- helix recognition helix, such as T-Box factors, would need cooperative interactions for nucleosome binding (Figure 6C). In agreement with our findings, the TBOX factor Tpit binding to closed target sites in vivo is restricted and mainly driven by the pioneer TF PAX7 (Mayran et al., 2018). Furthermore, TP53, an immunoglobulin fold TF, was shown to bind nucleosomes at their edges, at histone-free DNA (Yu and Buck, 2019). Similarly, BZLF1, a bZIP TF similar to group IIB, showed minimal nucleosome binding at high TF concentrations (Schaeffner et al., 2019).
Our study reveals that nucleosome binding positively correlates with reprogramming potential and suggests a hierarchical model where pioneer TFs work as trailblazers for epigenome regulators and modifiers during reprogramming. Furthermore, we reveal commonalities and structural length limitations among the DNA binding modes of pioneer TFs, compared to non-pioneer TFs. Our study provides insight into how diverse groups of TFs engage in nucleosomal interactions via common structural features. Further studies of the domains of pioneer factors that modulate nucleosomal interactions will unveil how other factors are enabled to bind closed, silent chromatin and initiate cell fate changes.
STAR★METHODS
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for resources should be directed to the Lead Contact, Kenneth S. Zaret (Zaret@pennmedicine.upenn.edu).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Escherichia coli bacterial strain Rosetta (DE3)pLysS Competent Cells were used for recombinant protein expression.
METHODS DETAILS
Genomic Data Analysis
The ASCL1 and BRN2 ChIP-seq were obtained from GEO GSE43916 (Vierbuchen et al., 2013); PU1, CEBPα and CEBPβ from GSE21512 (Heinz et al., 2010); FOXA1 and HNF1α from E-MTAB-941 (Faure et al., 2012); and GATA4 from GSE49132 (Zheng et al., 2013). ChIP-seq and MNase-seq data GSE40910 (Teif et al., 2012) were aligned to build version NCBI37/mm9 of the mouse genome. The MNase-seq reads were extended to 150 bp to cover one nucleosome.
Protein Expression and Purification
The bacterial expression plasmids: pET-28b-FoxA1, pET-28b-Gata4, pET-28b-Hnf1a, pET-28b-Hnf4a, pET-28b-Ascl1, pET-28b-Tcf3, pRSFDUET1-Ascl1+Tcf3, pET-28d- Brn2, pET-28b-Spi1, pET-28b-Cebpa, pET-28b-Cebpb encode the mouse FOXA1, GATA4, HNF1α, HNF4α, ASCL1, E12α, ASCL1+E12α, BRN2, PU1, CEBPα, and CEBPβ proteins respectively, fused to an N-terminal 6X histidine tag. BRN2 was expressed fused to a N-terminal 6X histidine tag followed by a GST-tag. pRSFDUET1 contained 6X his tagged Ascl1 in MCS1 between restriction sites BamH1 and HindIII and untagged E12α (tcf3 gene) in MCS2 between restriction sites NdeI and XhoI. Plasmid containing TFs cDNA were obtained as follow; FUW-TetO-Gata4 (Addgene plasmid #41084) (Buganim et al., 2012), pCMW-SPORT6-Hnf1a from Dharmacon mammalian gene collection (MMM1013–202761012), pGCDNsam-Hnf4a (Addgene plasmid # 33002) (Sekiya and Suzuki, 2011), Tet-O-FUW-Ascl1 (Addgene plasmid # 27150) (Vierbuchen et al., 2010), pBABE-E12-cTAP (Addgene plasmid # 20916)(Yang et al., 2009), pET28d-mBrn2 was a gift from Marius Wernig, LZRS PU.1 WT (Addgene plasmid #34835) (Anderson et al., 2002), CEBPalpha NGFR (Addgene plasmid #44627) (Del Real and Rothenberg, 2013), pcDNA 3.1 (−)mouse C/EBP beta (LAP) was a gift from Peter Johnson (Addgene plasmid # 12557). TF cDNA sequences were generated by PCR from the respective constructs, introducing restriction sites; NdeI and HindII for FoxA1, Hnf1a, Hnf4a; NdeI and XhoI for Gata4, Spi1, Cebpa, Cebpb, Ascl1; XbaI and XhoI for Brn2 for insertion into their respective plasmid. pET-28b-Cebpb-T167D and pET-28b-Cebpb-S163D,T167D plasmids were generated from pET-28b-Cebpb plasmid with Quick change II XL Site-Directed mutagenesis Kit (Agilent).
The histidine-tagged proteins were expressed in E. Coli Rosetta (DE3) pLysS (Novagen # 70956–3). Transformed cells were grown at 37 °C to a density of 0.5–0.7 at an absorbance of 600 nm and protein expression was induced with; 1 mM IPTG at 37 °C for 4 hr with a 30 min delay addition of 20 mg/m L rifampicin for FOXA1; 1 mM IPTG at 16 °C for 16 hr for ASCL1, BRN2 and PU1; 1 mM IP TG at 37 °C for 4 hr for HNF1 α, HNF4α, CEBPβ; 2 mM IPTG at 37 °C for 4 hr GATA4 and CEBP α. ZLD was purified as previously described (Harrison et al., 2010; McDaniel, 2019)
The proteins were purified over Hi-trap HP nickel-charged columns (GE healthcare #17–5248-01) or with Ni-NTA resin under denaturing conditions (20 mM Tris- HCl pH 8.0, 0.5M NaCl, 6M Urea) with 5 mM imidazole and 20–300 mM single step imidazole changes follow by 4M and 2M urea step dialysis. The recombinant human full length histones H2A, H2B, H3, and H4 were expressed and purified as described previously (Tanaka et al., 2004). Histone expression plasmids were a gift from Shelley Berger. Protein concentrations were calculated by quantifying the intensity of each of the protein bands running at the expected sizes in SDS-PAGE fitted to a BSA standard curve. To reduce error, the band intensities were quantified at various concentrations.
Cy5 DNA Labeling of Short Canonical DNA-Binding Sites
The DNA oligonucleotides used as binding sites are shown in Table S3. The expected binding sites are highlighted in red. The short oligonucleotide fragments were labeled with Cy5-dCTP (GE Healthcare Life Sciences) by end-repair with Klenow fragment DNA polymeraase (3ÅL→5ÅL exo-) (NEB) as follows. Cy5 5’-end-labelled double stranded probes were generated by annealing complementary single strand DNA probes to give rise to a two nucleotide 3’ overhang (with the last annealed nucleotide being a G) using the following reaction: 1 nanomoles of each ssDNA strand (10 µl of 100 µM) were mixed in 50 µl final volume annealing buffer (20 mM Tris-HCl pH7.5, 50 mM NaCl, 0.1 mM DTT, 1 mM EDTA). The reaction was incubated at 70 ºC for 10 min, and slowly cooled at room temperature overnight. Cy5-DNA labeling reactions were carried out at final concentrations of 1.26 µM dsDNA, 4 µM Cy5-dCTP and 0.5 U/uL Klenow fragment in the presence of excess 4 mM dATP, 4 mM dTTP, and 4 mM dGTP in NEB buffer 2 (50 mM NaCl, 10 mM Tris-HCl, 10 mM MgCl2, 1 mM DTT). The reaction was incubated at 37 °C for 1 hr. After Cy5 labeling, the probe was purified using Illustra MicroSpin G-25 columns (GE Healthcare Life Sciences).
Nucleosome Preparation
The 160 bp ALBN1-DNA fragment containing BamH1 flanking sequences corresponds to the genomic location: mm9 chr5:90879148–90879300
GATCCTGTCTCCTGCTCTGTCAGCAGGGCACTGTACTTGCTGATACCAGGGAATGT TTGTTCTTAAATACCATCATTCCGGACGTGTTTGCCTTGGCCAGTTTTCCATGTACA TGCAGAAAGAAGTTTGGACTGATCAATACAGTCCTCTGCCTTGGATC
The 162 bp NRCAM-DNA fragment containing BamH1 flanking sequences corresponds to the genomic location: mm9 chr12:45445284–45445450
GATCCATTACTTCTGAAACAGATGACTCCCAGCAGCTGCTGCCTGTGGCCCACAG GGCTTCCTGCCCTGCATGACAGCTGCACATCACATCCTGTGGTCATACTACTTCAG CCGCTTCTACGGCCAGATACAAAAGTGGGTGGGGAACATAGGCAAGGGATC
The 162 bp CX3CR1-DNA containing BamH1 flanking sequences fragment corresponds to the genomic location: mm9 chr9:119946611–119946762
GATCCGCAGGGCCTCTCGGCTGCTGATCTTCAGCTGGTTGCTGAGAGTTGCAGCA TTGCTGAGTCTTAGCAATGGATACTTCCCGATTCCCCTCACAAAAATAGGTCAGTCT GTCTGGCTAGTTCTGTACTTGCAGACACAGGGCATGTGGGGTTCCGGATC
The DNA sequences were created by PCR of genomic mouse DNA with EcoRI- BamHI and BamH1-XbaI restriction sites into pUC19 plasmid to generate pUC19-EB- NRCAM-BX, pUC19-EB-ALBN1-BX, and pUC19-EB-CX3CR1-BX. Plasmid were amplified in E.coli, purified, and digested with BamHI to release fragments. DNA sequences were purified by agarose gel electroelution, phenol:chloroform extraction, and ethanol precipitation followed by Cy5 enzymatic labeling as described above and purified with QIAquick PCR purification kit (Qiagen 28106)
The nucleosomes were reconstituted by dialysis as described previously (Tanaka et al., 2004). Briefly, 10 µg of Cy5 end labelled DNA sequences were mixed with purified and refolded H2A/H2B dimers and H3/H4 tetramers at a 1:1 DNA:histone- octamer molar ratio in 10 mM Tris-HCl pH 8.0, 4 M Urea, 2 M NaCl, 0.1 mg/ml BSA. The nucleosomes were assembled by salt-urea step dialysis against buffers containing 1.5, 1, 0.8, and then 0.6 M NaCl with 10 mM Tris–HCl pH 8.0, 5 M Urea, 1 mM EDTA, 10 mM 2-β mercaptoethanol for 2–4 hr at 4 °C. The nucleosome s were then dialyzed against a no urea buffer containing 0.6 M NaCl and 10 mM Tris–HCl pH 8.0, 1 mM EDTA, 1 mM 2-β mercaptoethanol and then the same buffer containing 0.1 M NaCl for 6 hrs at 4 °C. The reconstituted nucleosomes were the n heat shifted by incubating at 42 °C for 2 hr, and further purified with a 10–30% gly cerol gradient in 50 mM Tris pH 7.5, 1 mM EDTA and 0.03 mg/mL BSA at 35,000 rpm for 18 hr at 4 °C. Gradients were fractionated and nucleosome-containing factions were pooled and concentrated.
DNA and Nucleosome Binding Reactions
The end-labelled oligonucleotides containing specific or non-specific sites (see above for sequences), free DNA sequence and nucleosomes were incubated with recombinant proteins in DNA-binding buffer (10 mM Tris-HCl pH7.5, 1 mM MgCl2, 10 µM ZnCl2, 1 mM DTT, 50 mM KCl, 3 mg/ml BSA, 5% Glycerol) at room temperature for 30 min. Free and bound DNA were separated on 4% non-denaturing polyacrylamide gels run in 0.5X Tris–borate–EDTA. Gels were visualized using with an Amersham Typhoon RGB Biomolecular Imager using Cy5 fluorescence setting (excitation at 633 nm and emission filter 670 BP 30) and a high sensitivity setting. The apparent dissociation constant (Kd) were calculated as previously described (Soufi et al., 2015a). For competition assays excessive amounts (from 20 to 80 fold) of non-labelled probes containing specific and non-specific sites were added to the binding reaction and incubated for an additional 30 min at room temperature to reach equilibrium. The binding reactions were loaded on the 4% EMSA gels as described above. EMSA gels were run at 90 volts at room temperature.
Western Blotting After EMSA (WEMSA)
EMSAs were carried out as described above with 10-fold more protein and nucleosomes run on a 1.5 mm thick mini-gel cassette (life technologies # NC2015) containing 5% non-denaturing polyacrylamide gels run in 0.5X Tris–borate–EDTA. To avoid Cy5 fluorescence saturation, 90% of the nucleosomes used in binding reactions was not labelled. The gel was then visualized using Cy5 fluorescence as described above. Western blot was done as described previously (Soufi et al., 2015a). Primary antibody incubations with anti-mouse ASCL1 antibody (1:1000; Abcam # ab74065), anti-human H3 (0.5 µg/ml; abcam # ab1791), and anti-human H2B (0.8 µg/ml; abcam # ab1790) were performed for 2 hr at room temperature. The secondary antibody incubations with goat anti-rabbit IgG-HRP (1:5000 dilution; Santa Cruz # sc-2004) and donkey anti- goat IgGHRP (1:2000; Santa Cruz # sc-2020) were performed for 1 hr at room temperature. Blots were visualized by using SuperSignal West Pico chemiluminescent substrate (Thermo-Scientific # 34080) in Fujifilm LAS-4000 imaging system. The membranes were stripped by incubating with Restore Western-Blot Plus Stripping Buffer (Thermo- Scientifc # 46430) for 30 min at RT and re-blocked after blotting with each antibody. The same membrane was serially blotted and stripped with all antibodies shown.
DNase Footprinting on Free DNA and Nucleosomes
DNase footprinting reactions were carried out as previously described (Soufi et al., 2015a). In brief, 6-FAM end labeled free DNA or nucleosomes (50 ng) were incubated in the presence or absence or the purified ASCL1/E12α (60 nM) and PU1 (100 nM) for 45 min at room temperature in a total volume of 50 µl in DNA-binding buffer (10 mM Tris-HCl pH 7.5, 1 mM MgCl2, 10 µM ZnCl2, 1 mM DTT, 50 mM KCl, 3 mg/ml BSA, 5% glycerol). The binding reactions were then supplemented with an additional 50 µl of 10 mM MgCl2 and 5 mM CaCl2 at room temperature for 1 min (total 100 µl). Binding reactions were treated with DNase-I (Worthington) by addition of 3 µl of DNase-I yielding a final concentration of 9×10−5 U/ µl for free DNA or 1.8×10−3 U/ µl for nucleosomes for 1 min at room temperature. The reactions were stopped by adding 90 µl of 200 mM NaCl, 30 mM EDTA,1%SDS buffer immediately followed by phenol extraction. One tenth of reaction volume (20 µl) of 3 M NaOAc (pH 5.2) was added to the reaction before the DNA fragments were extracted with saturated phenol. The DNA fragments were further purified using MinElute PCR purification kit (QIAGEN) and eluted in 10 µl of EB buffer. The digested DNA fragments were separated by capillary electrophoresis. Briefly, the DNA fragments (5 µl) were added to 4.9 ml HiDi formamide (Applied Biosciences) and 0.1 ml GeneScan-500 LIZ size standards (Applied Biosciences). After denaturing at 95 °C for 10 min, the samples were run on an ABI 96-capillary 3730XL Sequencer, using G5 dye setting, running a genotyping module with an increased injection time of 30 s and injection voltage of 3 kV. The generated electropherograms were analyzed using the Gene mapper V4.1 (Applied Biosciences) and Peak Scanner module (Thermo Fisher Scientific Cloud).
Protein Microarrays
Full length human proteins were purified as GST fusion proteins from yeast using a high-throughput protein purification protocol as described previously (Zhu et al., 2001). Purified human proteins predicted to bind DNA were arrayed in a 384-well format and printed on FAST slides (Whatman, Germany) in duplicate, as described previously (Hu et al., 2009). To estimate protein amount printed per spot, prepared chips were incubated with anti-GST antibody conjugated with Cy5 binding targeting the printed proteins GST-tags. The protein microarrays were probed without (- control) or with Cy5- labeled ALBN1, NRCAM, CX3CR1 DNA or NUC using a similar protocol described previously (Hu et al., 2009). In brief, a protein chip was blocked for 3 hr at 4 °C with 3% BSA in 25 mM HEPES pH 8.0, 50 mM KGlu, 8 mM MgCl2, 3 mM DTT, 10% glycerol, 0.1% triton x-100 buffer and then incubated with Cy5-labeled ALBN1, NRCAM, CX3CR1 DNA or NUC at a final concentration of 20 nM in 80 µL of binding buffer (10 mM Tris-HCl pH 8.0, 1 mM MgCl2, 10 µM ZnCl2, 1 mM DTT, 50 mM KCl, 3 mg/ml BSA, 5% Glycerol) overnight at 4 °C. The chip was washed once with TBST (0.1% triton x-100) for 5 min at 4 °C, rinsed with water and spun dry. The slides were finally scanned with a GenePix 4000B scanner (Molecular Devices, Sunnyvale, CA) and the binding signals were acquired by aligning an array list over the chip, foreground and background signals for each protein spot were then extracted by the using the GenePix Pro 7 software, and fluorescence minus background (F-B) signals were obtained for each spots.
3D Structural Model Assemblies
MYOG, CREM, and ASCL1/E12α structural models (SMR P15173, SMR Q03060, SMR P50553) were constructed based on SWISS-MODEL sequence alignment (Bienert et al., 2017) with MYOD (pdb 1MDY) showing 74.2% DBD sequence identity, CREB1 (pdb 1DH3) showing 76.3% DBD sequence identity with CREM1, and NEUROD1 of the NEUROD1/E12α (pdb 2QL2) showing 42.86% DBD sequence identity with ASCL1. Structural models assemblies were aligned to published crystal structures of MYOD, CREB1, and NEUROD1/ E12α using Pymol.
QUANTIFICATION AND STATISTICAL ANALYSIS
EMSA Image Processing
For image processing, we used ImageJ version 2.0.0-rc/1.51f. Before displaying, the images were corrected for brightness and contrast; a linear range was maintained.
Dissociation Constants Analysis
Apparent KDs were derived from two separate binding curves, each representing two experimental replicates as described by (Soufi et al., 2015a). Experimental data was fitted to non-linear regressions “One site – Specific binding”, “One site – Total” or Specific binding with Hill slope” with a Bmax less than 1 constrain within R2 values of 0.8–0.99, expressed in nM units using GraphPad Prism 7 software.
Protein Microarray Analysis
To quantify the signal intensity for each spot, the signal intensities were calculated as the fluorescent median intensity minus its local background median intensity (F-B) followed by averaging of printed duplicate spots F-B (Table S2)(Hu et al., 2009). F-B signals were further processed to minimize artifacts (Figure S4A). Auto-fluorescent edging (Figure S4B) was corrected by subtraction of the F-B signal of microarrays incubated with binding buffer (F-B)-ctrl from the F-B signal of microarrays incubated with Cy5-labeled samples (F-B)+Cy5. Heterogeneity in printed protein amounts per spot (Figure S4C) was considered by estimating relative protein levels per spot amount using chips incubated with anti-GST antibody conjugated to Cy5 (F-B)GST, a (F-B)GST < 170 signal cutoff was assigned (Figure S4D). The cutoff removed proteins spots with below threshold levels amounts to yield significant binding events from our analysis. To eliminate artifacts resulting from uneven washing and drying of the chips after incubation with Cy5 probes (Figure S4E), we performed a within-chip normalization by the median normalization method. Known chromatin modifiers were removed from our analysis. Finally, to identify proteins that bind to DNA or nucleosomes, an intensity cutoff value of (F-B)+Cy5 equal to 80 was assigned (Figure S4F), where spots producing a signal greater than the cuttoff were identified as “positive hits”.
Heatmap Representation of DNA and Nucleosome Binding by Transcription Factors
Heatmaps in Figure 5D, E were generated by first quantifying the TFs total bound fraction from EMSAs as described in the methods section “DNA and Nucleosome Binding Reactions” and in (Soufi et al., 2015b). Total bound fraction for each TF was measured for their respective free DNA and nucleosome template shown in Figure S5, at the concentrations shown in Figure S6A. K-means clustering of the TF total bound fraction for free DNA and nucleosomes was done with R studio software and the pheatmap package. We defined a K means clustering function with an optimal number of 6 cluster centers (k); no Z score normalization was applied to the data. Cluster number was further simplified by manually regrouping of highly similar clusters resulting in three clusters.
Supplementary Material
Each of the ALBN1-DNA, NRCAM-DNA and CX3CR1-DNA sequences were scanned with PROMO (Messeguer et al., 2002) for identification of putative TFs binding sites (TFBS) defined by TRANSFAC databased and used as weight matrices for TFBS prediction.
Unprocessed and processed protein microarray data containing the 1,755 proteins name, identifier lists and their respective (F-B)+Cy5 signals for in each experiment, 1,592 proteins after data processing and 593 positive hits TFs in at least one experiment.
Oligonucleotide DNA sequences used as dsDNA containing TFs binding sites in EMSA competition experiments as specific or non-specific competitors.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| anti-mouse ASCL1 | Abcam | ab74065 |
| anti-human H2B | Abcam | ab1790 |
| anti-human H3 | Abcam | ab1791 |
| Cy5-dCTP | GE Healthcare Life Sciences | PA55021 |
| Klenow Fragment (3ÅL→5ÅL exo-) | New England Biolabs | M0212S |
| Bacterial and Virus Strains | ||
| 6HIS_pRSF_TEV | Gifft from Ronen Marmorstein lab | 6HIS_pRSF_TEV |
| FUW-TetO-Gata4 | Buganim et al., 2012 | Addgene 41084 |
| pCMW-SPORT6-Hnf1 | Dharmacon mammalian gene collection | MMM1013–202761012 |
| Tet-O-FUW-Ascl1 | Vierbuchen et al., 2010 | Addgene 27150 |
| pBABE-E12-cTAP | Yang et al., 2009 | Addgene 20916 |
| pET-28b-FoxA1 | This paper | pET-28b-FoxA1 |
| pET-28b-Gata4 | This paper | pET-28b-Gata4 |
| pET-28b-Hnf1a | This paper | pET-28b-Hnf1a |
| pET-28b-Ascl1 | This paper | pET-28b-Ascl1 |
| pET-28b-Tcf3 | This paper | pET-28b-Tcf3 |
| pRSFDUET1-Ascl1+Tcf3 | This paper | pRSFDUET1-Ascl1+Tcf3 |
| pET-28d-Brn2 | Gift from Marius Wernig | pET-28d-Brn2 |
| pET-28b-Spi1 | This paper | pET-28b-Spi1 |
| pET-28b-Cebpa | This paper | pET-28b-Cebpa |
| pET-28b-Cebpb | This paper | pET-28b-Cebpb |
| pET-28b-Cebpb-T167D | This paper | pET-28b-Cebpb-T167D |
| pET-28b-Cebpb- S163D,T167D | This paper | pET-28b-Cebpb- S163D,T167D |
| Biological Samples | ||
| Chemicals, Peptides, and Recombinant Proteins | ||
| His6x-FOXA1 | This paper | His6x-FOXA1 |
| His6x-GATA4 | This paper | His6x-GATA4 |
| His6x-HNF1α | This paper | His6x-HNF1α |
| His6x-ASCL1 | This paper | His6x-ASCL1 |
| His6x-E12α | This paper | His6x-E12α |
| His6x-ASCL1/E12α | This paper | His6x-ASCL1/E12α, |
| GST-His6x-BRN2 | This paper | GST-His6x-BRN2 |
| His6x-PU1 | This paper | His6x-PU1 |
| His6x-CEBPα | This paper | His6x-CEBPα |
| His6x-CEBPβ WT | This paper | His6x-CEBPβ WT |
| His6x-CEBPβ T167D | This paper | His6x-CEBPβ T167D |
| His6x-CEBPβ S163D,T167D | This paper | His6x-CEBPβ S163D,T167D |
| GST-MYOG | Hu et al., 2013 | GST-MYOG |
| GST-TBX20 | Hu et al., 2013 | GST-TBX20 |
| GST-NHLH2 | Hu et al., 2013 | GST-NHLH2 |
| GST-CREM1 | Hu et al., 2013 | GST-CREM1 |
| GST-IRF3 | Hu et al., 2013 | GST-IRF3 |
| GST-RBPJ | Hu et al., 2013 | GST-RBPJ |
| GST-HMGN1 | Hu et al., 2013 | GST-HMGN1 |
| GST-HMGN5 | Hu et al., 2013 | GST-HMGN5 |
| GST-HMGA1 | Hu et al., 2013 | GST-HMGA1 |
| GST-SOX5 | Hu et al., 2013 | GST-SOX5 |
| Critical Commercial Assays | ||
| MinElute PCR purification kit | QIAGEN | 28004 |
| Quick change II XL Site-Directed mutagenesis Kit | Agilent | 200521 |
| Deposited Data | ||
| ASCL1 and BRN2 ChIP-seq | Vierbuchen et al., 2013 | GEO GSE43916 |
| PU1, CEBPα and CEBPβ ChIP-seq | Heinz et al., 2010 | GSE21512 |
| FOXA1 and HNF1α ChIP-seq | Faure et al., 2012 | E-MTAB-941 |
| GATA4 ChIP-seq | Zheng et al., 2013 | GSE49132 |
| Fibroblast MNase-seq | Teif et al., 2012 | GSE40910 |
| FOXA3-DBD:DNA co-crystal structure | Clark et al., 1993 | RCSB PDB 1VTN |
| OCT4-DBD:DNA co-crystal structure | Esch et al., 2013 | RCSB PDB 3L1P |
| PU1-DBD:DNA co-crystal structure | Kodandapani et al., 1996 | RCSB PDB 1PUE |
| KLF4-DBD:DNA co-crystal structure | Schuetz et al., 2011 | RCSB PDB 2WBS |
| GATA3-DBD:DNA co-crystal structure | Chen et al., 2012 | RCSB PDB 4HC9 |
| ASCL1 | SWISS-MODEL | SMR P50553 |
| cMYC/MAX-DBD:DNA co-crystal structure | Nair and Burley, 2003 | RCSB PDB 1NKP |
| CEBPα-DBD:DNA co-crystal structure | Miller et al., 2003 | RCSB PDB 1NWQ |
| USF-DBD:DNA co-crystal structure | Ferre-D’Amare et al., 1994 | RCSB PDB 1AN4 |
| MYOG | SWISS-MODEL | SMR P15173 |
| CREM | SWISS-MODEL | SMR Q03060 |
| TBX1-DBD:DNA co-crystal structure | El Omari et al., 2012 | RCSB PDB 4A04 |
| Brachyury-DBD:DNA co-crystal structure | Muller and Herrmann, 1997 | RCSB PDB 1XBR |
| NF-kB p50-DBD:DNA co-crystal structure | Muller et al., 1995 | RCSB PDB 1SVC |
| GAL4-DBD:DNA co-crystal structure | Hong et al., 2008 | RCSB PDB 3COQ |
| PBX1-DBD:DNA co-crystal structure | LaRonde-LeBlanc and Wolberger, 2003 | RCSB PDB 1PUF |
| UBX-DBD:DNA co-crystal structure | Foos et al., 2015 | RCSB PDB 4UUS |
| Raw gel images | Mendeley | https://doi.org/10.17632/5dnznn9kwt.1 |
| Experimental Models: Cell Lines | ||
| Rosetta (DE3)pLysS Competent Cells | Novagen | Novagen # 70956–3 |
| Experimental Models: Organisms/Strains | ||
| Oligonucleotides | ||
| Table S3 | ||
| Recombinant DNA | ||
| GATCCTGTCTCCTGCTCTGTCAGCAGGGCACTGTACTTGCTGATACCAGGGAATGTTTGTTCTTAAATACCATCATTCCGGACGTGTTTGCCTTGGCCAGTTTTCCATGTACATGCAGAAAGAAGTTTGGACTGATCAATACAGTCCTCTGCCTTGGATC | This paper | 160 bp ALBN1-DNA |
| GATCCATTACTTCTGAAACAGATGACTCCCAGCAGCTGCTGCCTGTGGCCCACAGGGCTTCCTGCCCTGCATGACAGCTGCACATCACATCCTGTGGTCATACTACTTCAGCCGCTTCTACGGCCAGATACAAAAGTGGGTGGGGAACATAGGCAAGGGATC | This paper | 162 bp NRCAM-DNA |
| GATCCGCAGGGCCTCTCGGCTGCTGATCTTCAGCTGGTTGCTGAGAGTTGCAGCATTGCTGAGTCTTAGCAATGGATACTTCCCGATTCCCCTCACAAAAATAGGTCAGTCTGTCTGGCTAGTTCTGTACTTGCAGACACAGGGCATGTGGGGTTCCGGATC | This paper | 162 bp CX3CR1-DNA |
| Software and Algorithms | ||
| Gene Mapper v4.1 | Thermo Fisher | Cat no. 4440915 |
| Peak Scanner module | Thermo Fisher Scientific Cloud | https://www.thermofisher.com/us/en/home/life-science/sequencing/fragment-analysis/fragment-analysis-fundamentals/fragment-analysis-software-data-analysis.html |
| Pymol | Schrodinger | https://pymol.org/2/ |
| PRISM 7 | GraphPad | https://www.graphpad.com/scientific-software/prism/ |
| PROMO | Messeguer et al., 2002 | http://alggen.lsi.upc.es/cgi-bin/promo_v3/promo/promoinit.cgi?dirDB=TF_8.3 |
| GenePix Pro 7 | Molecular Devices, LLC. | http://mdc.custhelp.com/app/products/detail/p/129/~/version-7 |
| R studio | https://www.rstudio.com/ | |
| Fiji is Just ImageJ (Fiji) version 2.0.0-rc/1.51f | https://imagej.net/ | |
| SWISS-MODEL | Bienert et al., 2017 | https://swissmodel.expasy.org/ |
| Other | ||
| Human TF Protein microarrays | Hu et al., 2013 | Human TF Protein microarrays |
HIGHLIGHTS.
Diverse full-length transcription factors were tested for nucleosome binding
Strong reprogramming factors exhibit nanomolar nucleosome affinit
Various new nucleosome binding factors were identified
Pioneer TFs have a short DNA anchoring α-helix that allows nucleosome binding
ACKNOWLEDGMENTS
We thank Dr. Dario Nicetto and members of the Zaret lab for critical discussions and comments on the manuscript and Drs. Moritz Mall and Marius Wernig for sharing findings and discussions on neuronal TFs. The work was supported by NRSF F31 GM112417–02 to M.F.G and NIH GM36477 to K.Z. Work in the Harrison lab was supported by NIH-R01GM11694 and a Vallee Scholar Award.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
DECLARATION OF INTERESTS
The authors declare no competing interests.
DATA AND SOFTWARE AVAILABILITY
All the original unprocessed gel images in this manuscript have been deposited with Mendeley and can be accessed with https://doi.org/10.17632/5dnznn9kwt.1.
REFERENCES
- Adams CC, and Workman JL (1995). Binding of disparate transcriptional activators to nucleosomal DNA is inherently cooperative. Mol Cell Biol 15, 1405–1421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson MK, Weiss AH, Hernandez-Hoyos G, Dionne CJ, and Rothenberg EV (2002). Constitutive expression of PU.1 in fetal hematopoietic progenitors blocks T cell development at the pro-T cell stage. Immunity 16, 285–296. [DOI] [PubMed] [Google Scholar]
- Barozzi I, Simonatto M, Bonifacio S, Yang L, Rohs R, Ghisletti S, and Natoli G (2014). Coregulation of transcription factor binding and nucleosome occupancy through DNA features of mammalian enhancers. Mol Cell 54, 844–857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berkes CA, Bergstrom DA, Penn BH, Seaver KJ, Knoepfler PS, and Tapscott SJ (2004). Pbx marks genes for activation by MyoD indicating a role for a homeodomain protein in establishing myogenic potential. Mol Cell 14, 465–477. [DOI] [PubMed] [Google Scholar]
- Bienert S, Waterhouse A, de Beer TA, Tauriello G, Studer G, Bordoli L, and Schwede T (2017). The SWISS-MODEL Repository-new features and functionality. Nucleic Acids Res 45, D313–D319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biggin MD (2011). Animal transcription networks as highly connected, quantitative continua. Dev Cell 21, 611–626. [DOI] [PubMed] [Google Scholar]
- Blomquist P, Li Q, and Wrange O (1996). The affinity of nuclear factor 1 for its DNA site is drastically reduced by nucleosome organization irrespective of its rotational or translational position. J Biol Chem 271, 153–159. [DOI] [PubMed] [Google Scholar]
- Boller S, Ramamoorthy S, Akbas D, Nechanitzky R, Burger L, Murr R, Schubeler D, and Grosschedl R (2016). Pioneering Activity of the C-Terminal Domain of EBF1 Shapes the Chromatin Landscape for B Cell Programming. Immunity 44, 527–541. [DOI] [PubMed] [Google Scholar]
- Bork P, Holm L, and Sander C (1994). The immunoglobulin fold. Structural classification, sequence patterns and common core. J Mol Biol 242, 309–320. [DOI] [PubMed] [Google Scholar]
- Buganim Y, Itskovich E, Hu YC, Cheng AW, Ganz K, Sarkar S, Fu D, Welstead GG, Page DC, and Jaenisch R (2012). Direct reprogramming of fibroblasts into embryonic Sertoli- like cells by defined factors. Cell Stem Cell 11, 373–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calo E, and Wysocka J (2013). Modification of enhancer chromatin: what, how, and why? Mol Cell 49, 825–837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chanda S, Ang CE, Davila J, Pak C, Mall M, Lee QY, Ahlenius H, Jung SW, Sudhof TC, and Wernig M (2014). Generation of induced neuronal cells by the single reprogramming factor ASCL1. Stem Cell Reports 3, 282–296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaya D, Hayamizu T, Bustin M, and Zaret KS (2001). Transcription factor FoxA (HNF3) on a nucleosome at an enhancer complex in liver chromatin. J Biol Chem 276, 44385–44389. [DOI] [PubMed] [Google Scholar]
- Chen CS, Korobkova E, Chen H, Zhu J, Jian X, Tao SC, He C, and Zhu H (2008). A proteome chip approach reveals new DNA damage recognition activities in Escherichia coli . Nat Methods 5, 69–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Bates DL, Dey R, Chen PH, Machado AC, Laird-Offringa IA, Rohs R, and Chen L (2012). DNA binding by GATA transcription factor suggests mechanisms of DNA looping and long-range gene regulation. Cell Rep 2, 1197–1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cirillo LA, Lin FR, Cuesta I, Friedman D, Jarnik M, and Zaret KS (2002). Opening of compacted chromatin by early developmental transcription factors HNF3 (FoxA) and GATA-4. Mol Cell 9, 279–289. [DOI] [PubMed] [Google Scholar]
- Cirillo LA, McPherson CE, Bossard P, Stevens K, Cherian S, Shim EY, Clark KL, Burley SK, and Zaret KS (1998). Binding of the winged-helix transcription factor HNF3 to a linker histone site on the nucleosome. EMBO J 17, 244–254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cirillo LA, and Zaret KS (1999a). An early developmental transcription factor complex that is more stable on nucleosome core particles than on free DNA. Mol Cell 4, 961–969. [DOI] [PubMed] [Google Scholar]
- Cirillo LA, and Zaret KS (1999b). An early developmental transcription factor complex that is more stable on nucleosome core particles than on free DNA. Mol. Cell 4, 961–969. [DOI] [PubMed] [Google Scholar]
- Clark KL, Halay ED, Lai E, Burley SK (1993). Co-crystal Structure of the HNF-3 /fork head DNA-recognition otif resembles histoner H5. Nature 365, 412–420. [DOI] [PubMed] [Google Scholar]
- de Dieuleveult M, Yen K, Hmitou I, Depaux A, Boussouar F, Bou Dargham D, Jounier S, Humbertclaude H, Ribierre F, Baulard C, et al. (2016). Genome-wide nucleosome specificity and function of chromatin remodellers in ES cells. Nature 530, 113–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Ioannes P, Escalante CR, and Aggarwal AK (2011). Structures of apo IRF-3 and IRF-7 DNA binding domains: effect of loop L1 on DNA binding. Nucleic Acids Res 39, 7300–7307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Del Real MM, and Rothenberg EV (2013). Architecture of a lymphomyeloid developmental switch controlled by PU.1, Notch and Gata3. Development 140, 1207–1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donovan BT, Chen H, Jipa C, Bai L, and Poirier MG (2019). Dissociation rate compensation mechanism for budding yeast pioneer transcription factors. Elife 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El Omari K, De Mesmaeker J, Karia D, Ginn H, Bhattacharya S, and Mancini EJ (2012). Structure of the DNA-bound T-box domain of human TBX1, a transcription factor associated with the DiGeorge syndrome. Proteins 80, 655–660. [DOI] [PubMed] [Google Scholar]
- Esch D, Vahokoski J, Groves MR, Pogenberg V, Cojocaru V, Vom Bruch H, Han D, Drexler HC, Arauzo-Bravo MJ, Ng CK, et al. (2013). A unique Oct4 interface is crucial for reprogramming to pluripotency. Nat Cell Biol 15, 295–301. [DOI] [PubMed] [Google Scholar]
- Faure AJ, Schmidt D, Watt S, Schwalie PC, Wilson MD, Xu H, Ramsay RG, Odom DT, and Flicek P (2012). Cohesin regulates tissue-specific expression by stabilizing highly occupied cis-regulatory modules. Genome Res 22, 2163–2175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferre-D’Amare AR, Pognonec P, Roeder RG, and Burley SK (1994). Structure and function of the b/HLH/Z domain of USF. EMBO J 13, 180–189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foo SM, Sun Y, Lim B, Ziukaite R, O’Brien K, Nien CY, Kirov N, Shvartsman SY, and Rushlow CA (2014). Zelda potentiates morphogen activity by increasing chromatin accessibility. Curr Biol 24, 1341–1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foos N, Maurel-Zaffran C, Mate MJ, Vincentelli R, Hainaut M, Berenger H, Pradel J, Saurin AJ, Ortiz-Lombardia M, and Graba Y (2015). A flexible extension of the Drosophila ultrabithorax homeodomain defines a novel Hox/PBC interaction mode. Structure 23, 270–279. [DOI] [PubMed] [Google Scholar]
- Gao R, Liang X, Cheedipudi S, Cordero J, Jiang X, Zhang Q, Caputo L, Gunther S, Kuenne C, Ren Y, et al. (2019). Pioneering function of Isl1 in the epigenetic control of cardiomyocyte cell fate. Cell Res [DOI] [PMC free article] [PubMed]
- Hamm DC, Bondra ER, and Harrison MM (2015). Transcriptional activation is a conserved feature of the early embryonic factor Zelda that requires a cluster of four zinc fingers for DNA binding and a low-complexity activation domain. J Biol Chem 290, 3508–3518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison MM, Botchan MR, and Cline TW (2010). Grainyhead and Zelda compete for binding to the promoters of the earliest-expressed Drosophila genes. Dev Biol 345, 248–255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haswell ES, and O’Shea EK (1999). An in vitro system recapitulates chromatin remodeling at the PHO5 promoter. Mol Cell Biol 19, 2817–2827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayes JJ, WOLFF A (1992). HistonesH2A/H2B inhibittheinteractionoftranscription factor IIIA with theXenopus borealissomatic 5S RNA gene in a nucleosome. Proc. Nat. Acad. Sci 89, 1229–1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinz S, Benner C, Spann N, Bertolino E, Lin YC, Laslo P, Cheng JX, Murre C, Singh H, and Glass CK (2010). Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell 38, 576–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henke RM, Meredith DM, Borromeo MD, Savage TK, and Johnson JE (2009). Ascl1 and Neurog2 form novel complexes and regulate Delta-like3 (Dll3) expression in the neural tube. Dev Biol 328, 529–540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honer WG, Falkai P, Young C, Wang T, Xie J, Bonner J, Hu L, Boulianne GL, Luo Z, and Trimble WS (1997). Cingulate cortex synaptic terminal proteins and neural cell adhesion molecule in schizophrenia. Neuroscience 78, 99–110. [DOI] [PubMed] [Google Scholar]
- Hong M, Fitzgerald MX, Harper S, Luo C, Speicher DW, and Marmorstein R (2008). Structural basis for dimerization in DNA recognition by Gal4. Structure 16, 1019–1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu S, Wan J, Su Y, Song Q, Zeng Y, Nguyen HN, Shin J, Cox E, Rho HS, Woodard C, et al. (2013). DNA methylation presents distinct binding sites for human transcription factors. Elife 2, e00726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu S, Xie Z, Onishi A, Yu X, Jiang L, Lin J, Rho H.-s., Woodard C, Wang H, Jeong J-S, et al. (2009). Profiling the Human Protein-DNA Interactome Reveals ERK2 as a Transcriptional Repressor of Interferon Signaling. Cell 139, 610–622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwafuchi-Doi M, Donahue G, Kakumanu A, Watts JA, Mahony S, Pugh BF, Lee D, Kaestner KH, and Zaret KS (2016). The Pioneer Transcription Factor FoxA Maintains an Accessible Nucleosome Configuration at Enhancers for Tissue-Specific Gene Activation. Mol Cell 62, 79–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwafuchi-Doi M, and Zaret KS (2014). Pioneer transcription factors in cell reprogramming. Genes Dev 28, 2679–2692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakobsen JS, Waage J, Rapin N, Bisgaard HC, Larsen FS, and Porse BT (2013). Temporal mapping of CEBPA and CEBPB binding during liver regeneration reveals dynamic occupancy and specific regulatory codes for homeostatic and cell cycle gene batteries. Genome Res 23, 592–603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin C, Zang C, Wei G, Cui K, Peng W, Zhao K, and Felsenfeld G (2009). H3.3/H2A.Z double variant-containing nucleosomes mark ‘nucleosome-free regions’ of active promoters and other regulatory regions. Nat Genet 41, 941–945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kodandapani R, Pio F, Ni CZ, Piccialli G, Klemsz M, McKercher S, Maki RA, and Ely KR (1996). A new pattern for helix-turn-helix recognition revealed by the PU.1 ETS-domain-DNA complex. Nature 380, 456–460. [DOI] [PubMed] [Google Scholar]
- Kummerfeld SK, and Teichmann SA (2006). DBD: a transcription factor prediction database. Nucleic Acids Res 34, D74–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaRonde-LeBlanc NA, and Wolberger C (2003). Structure of HoxA9 and Pbx1 bound to DNA: Hox hexapeptide and DNA recognition anterior to posterior. Genes Dev 17, 2060–2072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee CS, Friedman JR, Fulmer JT, and Kaestner KH (2005). The initiation of liver development is dependent on Foxa transcription factors. Nature 435, 944–947. [DOI] [PubMed] [Google Scholar]
- Li R, Cauchy P, Ramamoorthy S, Boller S, Chavez L, and Grosschedl R (2018). Dynamic EBF1 occupancy directs sequential epigenetic and transcriptional events in B-cell programming. Genes Dev 32, 96–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Kim JW, Gronborg M, Urlaub H, Lane MD, and Tang QQ (2007). Role of cdk2 in the sequential phosphorylation/activation of C/EBPbeta during adipocyte differentiation. Proc Natl Acad Sci U S A 104, 11597–11602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang HL, Nien CY, Liu HY, Metzstein MM, Kirov N, and Rushlow C (2008). The zinc-finger protein Zelda is a key activator of the early zygotic genome in Drosophila. Nature 456, 400–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z, and Kraus WL (2017). Catalytic-Independent Functions of PARP-1 Determine Sox2 Pioneer Activity at Intractable Genomic Loci. Mol Cell 65, 589–603 e589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longo A, Guanga GP, and Rose R (2008). Crystal Structure of E47-NeuroD1/Beta2 bHLH Domain-DNA Complex: Heterodimer Selectivity and DNA Recognition. Biochemistry 47, 218–229. [DOI] [PubMed] [Google Scholar]
- Matys V, Kel-Margoulis OV, Fricke E, Liebich I, Land S, Barre-Dirrie A, Reuter I, Chekmenev D, Krull M, Hornischer K, et al. (2006). TRANSFAC and its module TRANSCompel: transcriptional gene regulation in eukaryotes. Nucleic Acids Res 34, D108–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayran A, Khetchoumian K, Hariri F, Pastinen T, Gauthier Y, Balsalobre A, and Drouin J (2018). Pioneer factor Pax7 deploys a stable enhancer repertoire for specification of cell fate. Nat Genet 50, 259–269. [DOI] [PubMed] [Google Scholar]
- McDaniel SL, Gibson TL, Schulz KN, Fernandez Garcia M, Nevil M, Jain SU, Lewis PW, Zaret KS, Harrison MM (2019). Continued activity of the pioneer factor Zelda is required to drive zygotic genome activation. Mol Cell In Press. [DOI] [PMC free article] [PubMed]
- McPherson CE, Shim EY, Friedman DS, and Zaret KS (1993). An active tissue-specific enhancer and bound transcription factors existing in a precisely positioned nucleosomal array. Cell 75, 387–398. [DOI] [PubMed] [Google Scholar]
- Meierhan D, el-Ariss C, Neuenschwander M, Sieber M, Stackhouse JF, and Allemann RK (1995). DNA binding specificity of the basic-helix-loop-helix protein MASH-1. Biochemistry 34, 11026–11036. [DOI] [PubMed] [Google Scholar]
- Messeguer X, Escudero R, Farre D, Nuñez O, Martinez J, and Alba MM (2002). PROMO: detection of known transcription regulatory elements using species-tailored searches. Bioinformatics 18, 333–334. [DOI] [PubMed] [Google Scholar]
- Messina D, Glasscock J, Gish W, and Lovett M (2004). An ORFeome-based Analysis of Human Transcription Factor Genes and the Construction of a Microarray to Interrogate Their Expression. Genome Res 14, 2041–2047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mieczkowski J, Cook A, Bowman SK, Mueller B, Alver BH, Kundu S, Deaton AM, Urban JA, Larschan E, Park PJ, et al. (2016). MNase titration reveals differences between nucleosome occupancy and chromatin accessibility. Nat Commun 7, 11485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller M, Shuman JD, Sebastian T, Dauter Z, and Johnson PF (2003). Structural basis for DNA recognition by the basic region leucine zipper transcription factor CCAAT/enhancer-binding protein alpha. J Biol Chem 278, 15178–15184. [DOI] [PubMed] [Google Scholar]
- Muller CW, and Herrmann BG (1997). Crystallographic structure of the T domain-DNA complex of the Brachyury transcription factor. Nature 389, 884–888. [DOI] [PubMed] [Google Scholar]
- Muller CW, Rey FA, Sodeoka M, Verdine GL, and Harrison SC (1995). Structure of the NF-kappa B p50 homodimer bound to DNA. Nature 373, 311–317. [DOI] [PubMed] [Google Scholar]
- Naiche LA, Harrelson Z,, and Kelly RG, Papaioannou VE, (2005). T-Box Genes in Vertebrate Development. Annu. Rev. Genet 39, 219–239. [DOI] [PubMed] [Google Scholar]
- Nair SK, and Burley SK (2003). X-ray structures of Myc-Max and Mad-Max recognizing DNA. Molecular bases of regulation by proto-oncogenic transcription factors. Cell 112, 193–205. [DOI] [PubMed] [Google Scholar]
- Nerlov C (2007). The C/EBP family of transcription factors: a paradigm for interaction between gene expression and proliferation control. Trends Cell Biol 17, 318–324. [DOI] [PubMed] [Google Scholar]
- Perlmann T, Wrange O, (1988). Specificglucocorticoidreceptor bindingto DNA reconstitutedina nucleosome. The EMBO Journal 7, 3073–3079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piwien-Pilipuk G, MacDougald O, and Schwartz J (2002). Dual regulation of phosphorylation and dephosphorylation of C/EBPbeta modulate its transcriptional activation and DNA binding in response to growth hormone. J Biol Chem 277, 44557–44565. [DOI] [PubMed] [Google Scholar]
- Powell LM, and Jarman AP (2008). Context dependence of proneural bHLH proteins. Curr Opin Genet Dev 18, 411–417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raposo AA, Vasconcelos FF, Drechsel D, Marie C, Johnston C, Dolle D, Bithell A, Gillotin S, van den Berg DL, Ettwiller L, et al. (2015). Ascl1 Coordinately Regulates Gene Expression and the Chromatin Landscape during Neurogenesis. Cell Rep 10, 1544–1556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaeffner M, Mrozek-Gorska P, Buschle A, Woellmer A, Tagawa T, Cernilogar FM, Schotta G, Krietenstein N, Lieleg C, Korber P, et al. (2019). BZLF1 interacts with chromatin remodelers promoting escape from latent infections with EBV. Life Sci Alliance 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schild-Poulter C, Sassone-Corsi P, Granger-Schnarr M, and Schnarr M (1996). Nucleosome assembly on the human c-fos promoter interferes with transcription factor binding. Nucleic Acids Res 24, 4751–4758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schorle H, Meier P, Buchert M, Jaenisch R, and Mitchell PJ (1996). Transcription factor AP-2 essential for cranial closure and craniofacial development. Nature 381, 235–238. [DOI] [PubMed] [Google Scholar]
- Schuetz A, Nana D, Rose C, Zocher G, Milanovic M, Koenigsmann J, Blasig R, Heinemann U, and Carstanjen D (2011). The structure of the Klf4 DNA-binding domain links to self-renewal and macrophage differentiation. Cell Mol Life Sci 68, 3121–3131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulz KN, Bondra ER, Moshe A, Villalta JE, Lieb JD, Kaplan T, McKay DJ, and Harrison MM (2015). Zelda is differentially required for chromatin accessibility, transcription factor binding, and gene expression in the early Drosophila embryo. Genome Res 25, 1715–1726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekiya S, and Suzuki A (2011). Direct conversion of mouse fibroblasts to hepatocyte-like cells by defined factors. Nature 475, 390–393. [DOI] [PubMed] [Google Scholar]
- Shim EY, Woodcock C, and Zaret KS (1998). Nucleosome positioning by the winged-helix transcription factor HNF3. Genes Dev 12, 5–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soufi A, Garcia MF, Jaroszewicz A, Osman N, Pellegrini M, and Zaret KS (2015a). Pioneer transcription factors target partial DNA motifs on nucleosomes to initiate reprogramming. Cell 161, 555–568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soufi A, Garcia MF, Jaroszewicz A, Osman N, Pellegrini M, and Zaret KS (2015b). Pioneer transcription factors target partial DNA motifs on nucleosomes to initiate reprogramming. Cell 161, 555–568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun H, Chen GY, and Yao SQ (2013). Recent advances in microarray technologies for proteomics. Chem Biol 20, 685–699. [DOI] [PubMed] [Google Scholar]
- Sun Y, Nien CY, Chen K, Liu HY, Johnston J, Zeitlinger J, and Rushlow C (2015). Zelda overcomes the high intrinsic nucleosome barrier at enhancers during Drosophila zygotic genome activation. Genome Res 25, 1703–1714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka Y, Tawaramoto-Sasanuma M, Kawaguchi S, Ohta T, Yoda K, Kurumizaka H, and Yokoyama S (2004). Expression and purification of recombinant human histones. Methods 33, 3–11. [DOI] [PubMed] [Google Scholar]
- Tang QQ, Gronborg M, Huang H, Kim JW, Otto TC, Pandey A, and Lane MD (2005). Sequential phosphorylation of CCAAT enhancer-binding protein beta by MAPK and glycogen synthase kinase 3beta is required for adipogenesis. Proc Natl Acad Sci U S A 102, 9766–9771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor I, Workman J, Schuetz TJ, Kingston RE, (1991). Facilitated binding of GAL4 and heat shock factor to nucleosomal templates: differential function of DNA-binding domains. Genes & Development 5, 1285–1298. [DOI] [PubMed] [Google Scholar]
- Taylor IC, Workman JL, Schuetz TJ, and Kingston RE (1991). Facilitated binding of GAL4 and heat shock factor to nucleosomal templates: differential function of DNA-binding domains. Genes Dev 5, 1285–1298. [DOI] [PubMed] [Google Scholar]
- Teif VB, Vainshtein Y, Caudron-Herger M, Mallm JP, Marth C, Hofer T, and Rippe K (2012). Genome-wide nucleosome positioning during embryonic stem cell development. Nat Struct Mol Biol 19, 1185–1192. [DOI] [PubMed] [Google Scholar]
- Ueda T, Catez F, Gerlitz G, and Bustin M (2008). Delineation of the protein module that anchors HMGN proteins to nucleosomes in the chromatin of living cells. Mol Cell Biol 28, 2872–2883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Oevelen C, Collombet S, Vicent G, Hoogenkamp M, Lepoivre C, Badeaux A, Bussmann L, Sardina JL, Thieffry D, Beato M, et al. (2015). C/EBPalpha Activates Pre-existing and De Novo Macrophage Enhancers during Induced Pre-B Cell Transdifferentiation and Myelopoiesis. Stem Cell Reports 5, 232–247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaquerizas JM, Kummerfeld SK, Teichmann SA, and Luscombe NM (2009). A census of human transcription factors: function, expression and evolution. Nature Reviews Genetics 10, 252–263. [DOI] [PubMed] [Google Scholar]
- Vawter MP, Howard AL, Hyde TM, Kleinman JE, and Freed WJ (1999). Alterations of hippocampal secreted N-CAM in bipolar disorder and synaptophysin in schizophrenia. Mol Psychiatry 4, 467–475. [DOI] [PubMed] [Google Scholar]
- Vierbuchen T, Ostermeier A, Pang ZP, Kokubu Y, Sudhof TC, and Wernig M (2010). Direct conversion of fibroblasts to functional neurons by defined factors. Nature 463, 1035–1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wapinski OL, Vierbuchen T, Qu K, Lee QY, Chanda S, Fuentes DR, Giresi PG, Ng YH, Marro S, Neff NF, et al. (2013). Hierarchical mechanisms for direct reprogramming of fibroblasts to neurons. Cell 155, 621–635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weina K, Utikal J (2014). SOX2 and cancer: current research and its implications in the clinic. Clinical and Translational Medicine 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wingender E, Schoeps T, Haubrock M, and Donitz J (2015). TFClass: a classification of human transcription factors and their rodent orthologs. Nucleic Acids Res 43, D97–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z, MacQuarrie KL, Analau E, Tyler AE, Dilworth FJ, Cao Y, Diede SJ, and Tapscott SJ (2009). MyoD and E-protein heterodimers switch rhabdomyosarcoma cells from an arrested myoblast phase to a differentiated state. Genes Dev 23, 694–707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X, and Buck MJ (2019). Defining TP53 pioneering capabilities with competitive nucleosome binding assays. Genome Res 29, 107–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Hagopian-Donaldson S, Serbedzija G, Elsemore J, Plehn-Dujowich D, McMahon AP, Flavell RA, and Williams T (1996). Neural tube, skeletal and body wall defects in mice lacking transcription factor AP-2. Nature 381, 238–241. [DOI] [PubMed] [Google Scholar]
- Zheng R, Rebolledo-Jaramillo B, Zong Y, Wang L, Russo P, Hancock W, Stanger BZ, Hardison RC, and Blobel GA (2013). Function of GATA factors in the adult mouse liver. PLoS One 8, e83723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu F, Farnung L, Kaasinen E, Sahu B, Yin Y, Wei B, Dodonova SO, Nitta KR, Morgunova E, Taipale M, et al. (2018). The interaction landscape between transcription factors and the nucleosome. Nature 562, 76–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Each of the ALBN1-DNA, NRCAM-DNA and CX3CR1-DNA sequences were scanned with PROMO (Messeguer et al., 2002) for identification of putative TFs binding sites (TFBS) defined by TRANSFAC databased and used as weight matrices for TFBS prediction.
Unprocessed and processed protein microarray data containing the 1,755 proteins name, identifier lists and their respective (F-B)+Cy5 signals for in each experiment, 1,592 proteins after data processing and 593 positive hits TFs in at least one experiment.
Oligonucleotide DNA sequences used as dsDNA containing TFs binding sites in EMSA competition experiments as specific or non-specific competitors.