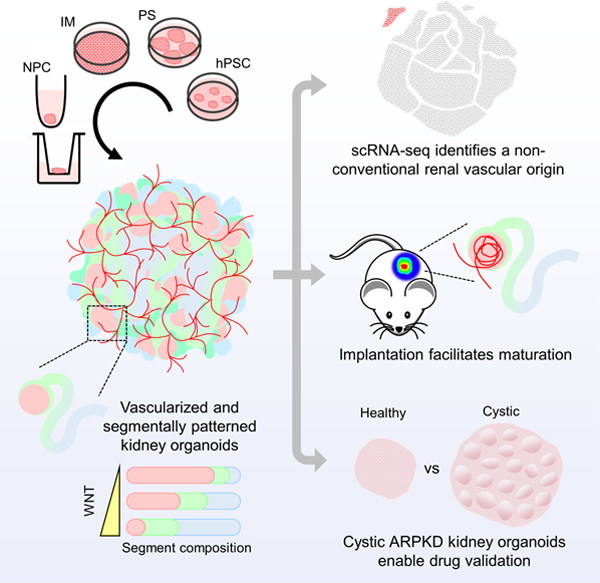
SUMMARY
Human pluripotent stem cell-derived kidney organoids recapitulate developmental processes and tissue architecture, but intrinsic limitations, such as a lack of vasculature and functionality, have greatly hampered their application. Here we establish a versatile protocol for generating vascularized three-dimensional kidney organoids. We employ dynamic modulation of WNT signaling to control the relative proportion of proximal versus distal nephron segments, thereby producing a correlative level of VEGFA to define a resident vascular network. Single-cell RNA-sequencing identifies a subset of nephron progenitor cells as a potential source of renal vasculature. These kidney organoids undergo further structural and functional maturation upon implantation. Using this kidney organoid platform, we establish an in vitro model of autosomal recessive polycystic kidney disease, the cystic phenotype of which can be effectively prevented by gene correction or drug treatment. Our studies provide new avenues for studying human kidney development, modeling disease pathogenesis, and performing patient-specific drug validation.
Graphical Abstract
In Brief
Human PSC-derived organoids represent an amenable platform for understanding human development and diseases, despite numerous limitations. Xia and colleagues establish a versatile platform for generating vascularized and patterned kidney organoids. Using this platform, they have identified a non-conventional origin of renal vasculature, as well as recapitulated ARPKD cystogenesis in vitro.
INTRODUCTION
The kidneys play critical roles in maintaining physiological homeostasis by excreting waste products and regulating the composition of body fluids. A nephron is the working unit of kidney and is patterned along its proximal-distal axis. Advances in generating complex kidney tissues from human pluripotent stem cells (hPSCs) have broadened our ability to study human kidney development and diseases (Freedman et al., 2015, Morizane et al., 2015, Takasato et al., 2015, Taguchi and Nishinakamura, 2017, Xia et al., 2013, Takasato et al., 2014, Taguchi et al., 2014). In these studies, hPSCs were differentiated into primitive streak cells, intermediate mesoderm, and ultimately into nephron progenitors through step-wise modulation of WNT, FGF, and TGFβ signaling (Freedman et al., 2015, Morizane et al., 2015, Takasato et al., 2015). Subsequently, in response to FGF and/or WNT signaling stimulation, nephron progenitors self-assembled to establish multi-compartmentalized kidney organoids that are comprised of various cellular components, including segmentally patterned epithelium, interstitium, and endothelium (Barasch et al., 1999, Li et al., 2016, Morizane et al., 2015, Takasato et al., 2015). However, current protocols cannot reliably coordinate the formation of different renal components, including both segmentally patterned nephron structures and vasculature. Furthermore, these kidney organoids did not demonstrate satisfactory intra-organoid communication between different cellular components, thus requiring additional signaling factors to support a resident vascular network (Homan et al., 2019, Czerniecki et al., 2018). Genetically engineered hPSCs have been developed to model autosomal dominant polycystic kidney disease (ADPKD) cystogenesis, but patient-specific induced PSCs (iPSCs)-derived kidney organoids have not yet been utilized for studying polycystic kidney disease (PKD) (Cruz et al., 2017, Czerniecki et al., 2018, Freedman et al., 2015). A patient-derived kidney organoid model of PKD would have great potential to transform the existing therapeutic approaches.
Here we have developed a highly robust methodology for differentiating hPSCs into three-dimensional (3D) kidney organoids comprised of segmentally patterned nephron structures and a de novo vascular network. Single-cell transcriptomics analysis was employed to demonstrate that a subpopulation of nephron progenitor cells (NPCs) contributed to the resident vasculature. Furthermore, these endothelial cells have already established a gene regulatory network that is responsible for defining endothelial sub-types. These kidney organoids are capable of coordinating the relative proportion of proximal versus distal segments based on WNT signaling. Subsequently, glomerular podocytes produce a correlative level of VEGFA to proportionally define a resident vascular network. These kidney organoids, upon implantation into a host mouse, went on to develop glomerular capillary tufts and were able to perform preliminary filtration and reabsorption, in a manner similar to wild-type mouse kidneys. Using this platform, we successfully differentiated autosomal recessive polycystic kidney disease (ARPKD) patient-derived iPSCs into 3D kidney organoids. These ARPKD iPSC-derived kidney organoids displayed drastic cystogenesis upon the upregulation of intracellular cAMP, compared to those derived from gene-corrected ARPKD iPSCs, thus enabling successful drug testing in vitro. The efficiency with which these kidney organoids can be generated, together with their high levels of versatility and functionality, provide unprecedented opportunities for studying human kidney development and diseases, as well as for performing personalized drug screening.
RESULTS
Differentiation of hPSCs into 3D Kidney Organoids with a de novo Vascular Network
We generated 3D kidney organoids from hPSCs through step-wise exposure to defined differentiation conditions. First, we treated hPSCs with 10 μM CHIR99021 (defining CHIR) for 4 days to induce primitive streak cells (T+MIXL1+) with high efficiency (Figure 1A and 1B), as previously described (Czerniecki et al., 2018, Freedman et al., 2015, Morizane et al., 2015, Takasato et al., 2015). To further differentiate primitive streak cells into intermediate mesoderm, we tested a number of culture conditions, with the goal of inducing the optimal levels of BMP signals, as BMPs specify intermediate mesoderm in a dose-dependent manner (James and Schultheiss, 2005). We found that 3 days of factor-free cell culture most effectively drove primitive streak cells toward nephrogenic intermediate mesoderm (HOXD11+WT1+) (Figure 1A, 1B, and S1A), while producing neglectable endoderm, or paraxial and lateral plate mesoderm (Figure S1A). This is in contrast to a previous report showing that 3 days of ActivinA treatment is required to induce intermediate mesoderm (Morizane et al., 2015). We then exposed the nephrogenic intermediate mesoderm to 3 μM CHIR (priming CHIR) in the presence of FGF9 (50 ng/ml), leading to the generation of SIX2+SALL1+ NPCs (Figure 1A and 1B). These cells self-assembled into clusters that morphologically resembled pre-tubular aggregates (PTAs) (Figure 1A, 1B, S1B, and S1C). These transient, PTA-like structures not only expressed NPC markers (SIX2 and SALL1) but also acquired LHX1 and PAX8 expression, indicating the initiation of nephogenesis (Figure 1B). Meanwhile, a small population of differentiating cells began to express vascular progenitor marker KDR (Figure 1B). Interestingly, it is not until NPHS1+ glomerulus-like structures appeared in the differentiation culture that these KDR+ cells acquired CD31 expression, indicating vascular maturation (Figure 1B).
Figure 1. Differentiation of hPSCs into Vascularized 3D Kidney Organoids.
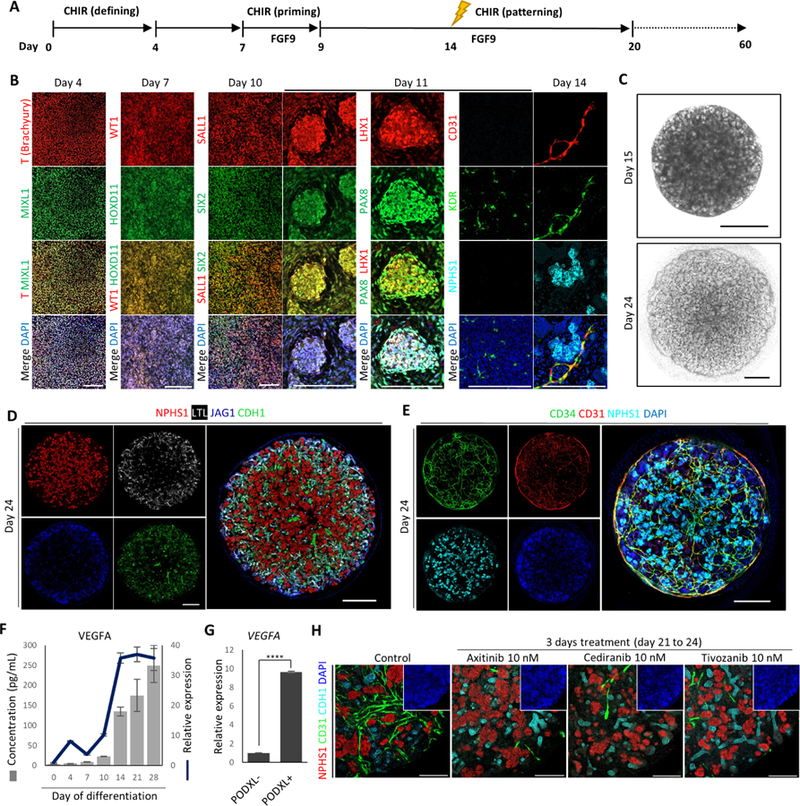
(A) Schematic of differentiation protocol.
(B) Immunofluorescence analysis for markers of primitive streak (T, MIXL1), intermediate mesoderm (WT1, HOXD11), nephron progenitor (SALL1, SIX2), pre-tubular aggregate (LHX1, PAX8), podocyte (NPHS1), vascular progenitor (KDR), and endothelial cell (CD31) during differentiation. Scale bars, 200 μm.
(C) Representative bright-field images of 3D kidney organoids (upper panel: Day 15 kidney organoid in liquid culture; lower panel: Day 24 kidney organoid in liquid-air interface culture.). Scale bars, 200 μm.
(D and E) Whole-mount immunofluorescence analysis of 3D kidney organoids (Day 24).
(F)Time course analysis of VEGFA gene expression (line) and VEGFA protein secretion (bars) during differentiation. Data were represented as mean ± SEM (n = 2 independent experiments, with 3 technical replicates).
(G) Comparison of VEGFA gene expression levels in PODXL− and PODXL+ cells of kidney organoids (Day 24). Data were represented as mean ± SEM (n = 2 independent experiments with 3 technical replicates). Statistical analysis was performed using unpaired Student’s t-test, **** p < 0.0001.
(H) Whole-mount immunofluorescence analysis of 3D kidney organoids (Day 24) treated with VEGFR inhibitors for 3 days. Scale bars, 200 μm.
After one day of 1 μM CHIR (patterning CHIR) pulse on day 14, visually distinguishable epithelial structures began to emerge, and gradually developed into highly complex tubule structures (Figure 1C and S1B). By day 24, each 3D kidney organoid became densely packed with multiple nephron components, including podocytes (NPHS1+), proximal tubules (LTL+), medial tubules (JAG1+), and distal tubules (CDH1+) (Figure 1D). Alongside these nephron structures, CD31+CD34+ endothelial cells formed a vascular network that spread throughout the entire kidney organoid (Figure 1E). To evaluate the identity of this presumptive endothelial population, we introduced human umbilical vein endothelial cells (HUVECs) into 3D kidney organoids (Figure S1D, left panel). These exogenous HUVECs formed a vascular network that seamlessly integrated with the resident vascular network, without disrupting the epithelial components (Figure S1D, right panels).
VEGFA signaling is essential for the maturation and maintenance of renal vasculature (Eremina et al., 2003). Since our protocol does not require exogenous VEGFA, the differentiating cells may autonomously secrete VEGFA to support the resident vasculature. In accordance with this hypothesis, levels of both VEGFA mRNA and secreted VEGFA protein were increased concurrently at the early stage of differentiation, and maintained at high levels from day 14 onward (Figure 1F). VEGFA is highly expressed by podocytes, and dysregulation of VEGFA expression disrupts glomerular vascular endothelium phenotypes (Eremina et al., 2003). We therefore used antibodies against PODOCALYXIN (PODXL) to label and sort podocytes from kidney organoids (Figure S1E and S1F). Compared with PODXL-cells, PODXL+ cells displayed more than 10 folds enrichment in VEGFA expression (Figure 1G). When VEGFA signaling was disrupted using 3 different VEGF receptor (VEGFR) inhibitors, the established vascular network was severely compromised, whereas nephron structures were not affected (Figure 1H and S1G). These observations demonstrate that our protocol robustly differentiates hPSCs into vascularized 3D kidney organoids. Furthermore, differentiation and maturation of the resident vascular network is highly dependent upon autonomous VEGFA production by podocytes.
Single-Cell Transcriptomics Reveal a Non-conventional Origin of Renal Vasculature
The origin of renal vasculature has not been fully elucidated, though it is widely accepted that both intra- and extra-renal vascular progenitors contribute to renal vasculature formation (Munro et al., 2017, Hu et al., 2016, Mugford et al., 2008, Risau, 1998, Abrahamson et al., 1998). On day 11, we observed a small number of NPCs co-expressing vascular progenitor marker KDR, but not mature endothelial marker CD31 (Figure S2A). Hence, we performed fluorescence-activated cell sorting (FACS) analysis to obtain a quantitative view of the kinetic expression of NPC and vascular endothelial markers. From day 11 to 13, NPCs (SIX1+ or SALL1+) constituted approximately 85%–90% of the total cell population (Figure S2B and Table S1). On day 14, the percentage of NPCs decreased to 70%, presumably due to active nephrogenesis (Figure S2B and Table S1). From day 11 to 14, we observed an increase in the percentage of NPCs (gated with SIX1 or SALL1) that co-expressed KDR (Figure S2B and Table S1). Meanwhile, more NPC+KDR+ cells acquired CD31 expression (Figure S2B and Table S1). These results suggest that a subset of NPCs display a vascular progenitor-like property, followed by differentiation into more mature endothelial cells.
We next performed single-cell RNA sequencing (scRNA-seq) to gain more insights into early nephrogenesis, which is a crucial stage for the emergence of multiple renal lineages, including endothelial cells. Using 10x Genomics Chromium Single Cell Gene Expression system, we sequenced mRNA from 15,560; 6,337; 40,609 cells harvested from day 10, 12, 14 of differentiation, respectively (Table S2). After rigorous filtering (Figure S2C), unsupervised clustering of the entire pooled dataset of 62,506 cells revealed a total of 11 transcriptionally distinct clusters, as visualized in t-distributed stochastic neighbor embedding (t-SNE) plot (Figure 2A). Based on lineage-specific marker expression defined in published datasets (Lindstrom et al., 2018a, Lindstrom et al., 2018b, Menon et al., 2018, Park et al., 2018, Phipson et al., 2019, Wang et al., 2018, Wu et al., 2018), we annotated and refined the 11 clusters into 6 broad classes of cells: early NPC (cluster 0), NPC (cluster 2), interstitial (cluster 3, 8), nascent nephron (cluster 1, 4), proliferating cells (cluster 5, 6, 7, 9), and endothelial (cluster 10) (Figure 2B and Figure S2D). Violin plots showed the expression of representative marker genes across different clusters (Figure 2B). During this stage, although most of the cells displayed a common feature of being highly proliferative yet fully defined, cluster 10 was clearly separated according to its highly distinctive endothelial gene signature (Figure S2E). Notably, while VEGFA receptors KDR and FLT1 were highly expressed in cluster 10; VEGFA itself was significantly enriched in clusters 1 and 4, which also expressed maturing podocyte marker OLFM3 (Figure S2F).
Figure 2. Single-Cell Transcriptomics Analysis of Early Nephrogenesis.
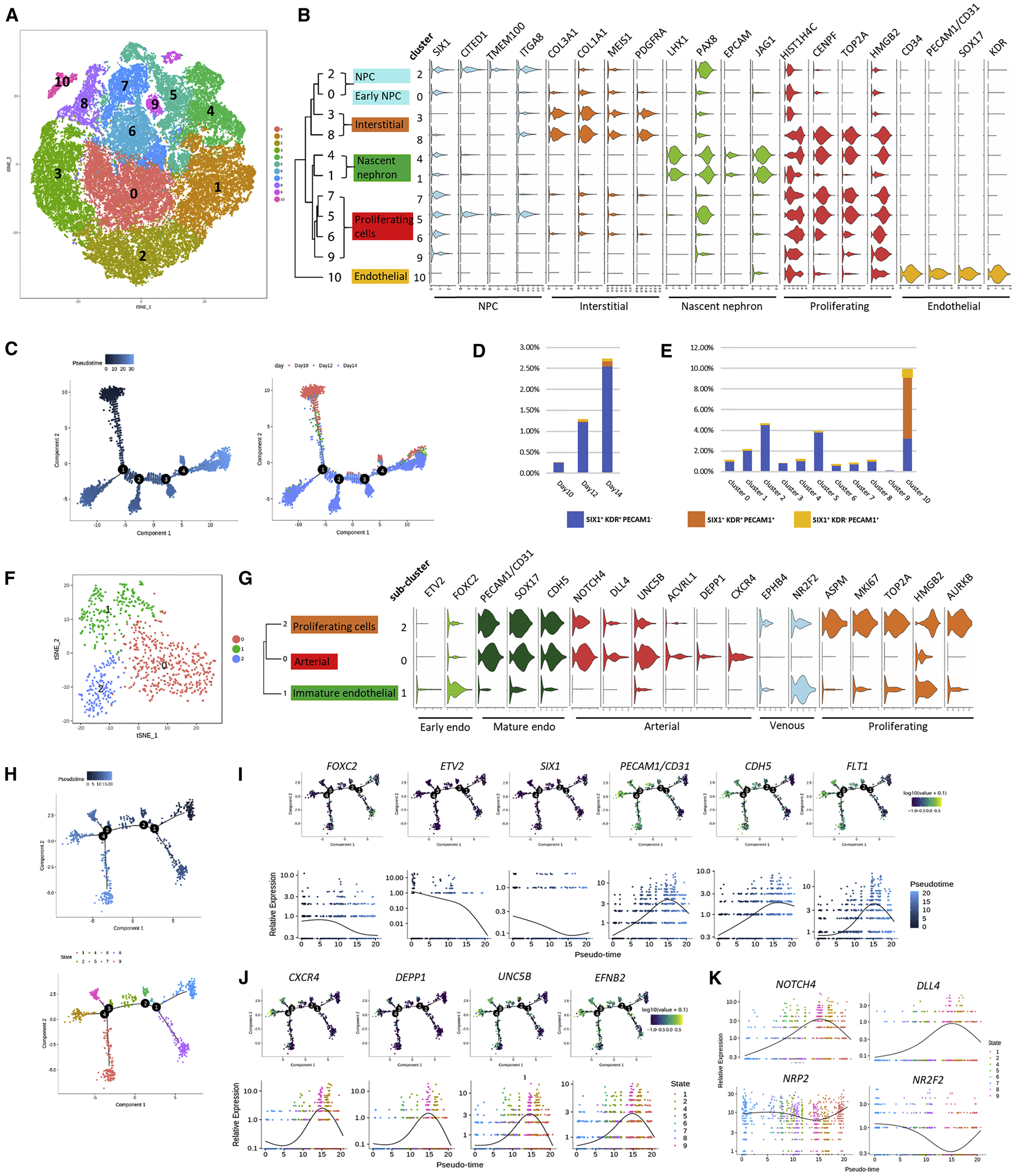
(A) tSNE plot displaying 62,506 cells from day 10, 12, and 14 of differentiation. Unsupervised clustering identified 11 clusters that are marked by different colors.
(B) Violin plots showing expression of representative marker genes.
(C) Ordering of the entire pooled scRNA-seq expression data according to pseudotime position.
(D and E) Dynamic distribution of cells that express different combinations of SIX1, KDR, and PECAM1/CD31, per differentiation time point (D), or per cluster (E).
(F) tSNE plot of cells within the endothelial sub-cluster from (A). Unsupervised clustering identified 3 sub-clusters that are marked by different colors.
(G) Violin plots showing expression of representative marker genes within the endothelial sub-cluster.
(H) Lineage trajectory of the endothelial sub-cluster, by pseudotime (upper panel), or by cell state (lower panel).
(I) Expression levels of early hemangioblast markers (FOXC2, ETV2, SIX1) and more mature endothelial markers (PECAM1/CD31, CDH5, FLT1) along endothelial lineage trajectory.
(J) Expression levels of arterial markers along endothelial lineage trajectory.
(K) Divergent trend of arterial (NOTCH4, DLL4) and venous (NRP2, NR2F2) marker expression along endothelial lineage trajectory.
Next, we reconstructed lineage relationship by performing unsupervised lineage trajectory analysis using Monocle 2. The pseudotimes were well-matched with differentiation time points, in spite of moderate stochasticity that is often seen in in vitro differentiation (Figure 2C). We also investigated differentially expressed genes (DEGs) that correspond with the bifurcation at each branchpoint, and performed gene ontology (GO) analysis to identify activated biological pathways. Along pseudotime, all branchpoints were enriched with GO terms associated with kidney development and morphogenesis, as well as vasculature development (Figure S2G and Table S3). These results indicate that within hPSC-derived kidney organoids, multiple lineages may emerge from a common origin at the onset of nephrogenesis.
To further investigate the lineage relationship between NPCs and vascular endothelial cells, we pulled out cells that expressed different combinations of markers representing NPC (SIX1), vascular progenitor (KDR), and maturing endothelial cells (PECAM1/CD31) from the entire pooled dataset. From day 10 to 14, we observed an increase in the percentage of SIX1+KDR+PECAM1-cells, the putative vascular progenitors (Figure 2D and Table S4). In the meantime, percentage of SIX1+KDR+PECAM1+ and SIX1+KDR−PECAM1+ maturing endothelial cells also increased (Figure 2D and Table S4). The NPC cluster (cluster 2) possessed both the highest number and proportion of SIX1+KDR+PECAM1−cells (Figure 2E and Table S5), whereas the endothelial cluster (cluster 10) housed the majority of maturing endothelial cells (Figure 2E and Table S5). Most importantly, we identified a subpopulation of NPCs that co-expressed CD31 within the second trimester human fetal kidneys, corroborating the lineage relationship observed during kidney organoid differentiation (Figure S2H).
Sub-clustering of the Endothelial Cluster Indicates the Emergence of Endothelial Sub-types
We proceeded to perform sub-clustering of the endothelial cluster to reveal additional substructures. After rigorous quality control (Figure S2I), unsupervised analysis revealed 3 endothelial sub-clusters (Figure 2F and Table S6) that were further annotated based on lineage-specific marker expression (Figure 2G) (De Val and Black, 2009). Compared with sub-clusters 0 and 2, sub-cluster 1 expressed high levels of early hemangioblast markers ETV2 and FOXC2, but low levels of more mature endothelial markers PECAM1/CD31, SOX17, and CDH5, suggesting an immature endothelial status (Figure 2G). Multiple arterial markers, such as NOTCH4, DLL4, and CXCR4, were all highly expressed in sub-cluster 0, indicating an arterial identity (Figure 2G). In comparison, venous markers NR2F2 and EPHB4 were completely absent in the arterial sub-cluster 0, but enriched in sub-clusters 1 and 2 (Figure 2G), both of which may have the potential to acquire a venous identity.
We also performed unsupervised lineage trajectory analysis to reconstruct the endothelial lineage relationship (Figure 2H). In agreement with vasculature development, expression levels of early hemangioblast markers FOXC2 and ETV2 displayed a time-dependent decrease along pseudotime (Figure 2I). Notably, SIX1 expression displayed a similar downregulating trend with FOXC2 and ETV2 (Figure 2I). In comparison, expression levels of more mature endothelial markers PECAM1/CD31, CDH5, and FLT1 gradually increased along pseudotime (Figure 2I). We also conducted GO analysis based on DEGs identified at each branchpoint. It is worth noting that only two biological processes were identified at branchpoint 2, and both were highly correlated with vascular development (Figure S2J and Table S7). Furthermore, multiple biological pathways at branchpoint 3 and 4 were closely associated with blood vessel formation, morphogenesis, and patterning (Figure S2J and Table S7). Within the endothelial lineage trajectory, we identified 9 distinct cell states (Figure 2H, lower panel). Examination of representative marker genes indicated that cell state 2 and 9 may represent arterial endothelial sub-types (Figure 2J). Along the endothelial lineage trajectory, we also observed a divergent trend of arterial and venous marker expression (Figure 2K). Collectively, we provide the first evidence that, in the absence of exogenous vascular growth factors (Czerniecki et al., 2018) or sheer stress (Homan et al., 2019), the vascular network within hPSC-derived kidney organoids has already established the underpinning program for specification into endothelial sub-types.
Coordinating Differentiation of Multiple Cellular Components in Kidney Organoids by Modulating WNT Signaling
During development, accurate patterning of different nephron segments along the proximal-distal axis is crucial for kidney function later in life. Over the course of differentiation, hPSCs gradually adopted a transcriptomic profile indicating acquisition of nephron segmentation (Figure S3A). In day 24 organoids, nephron-like structures have already established a distinctive proximal-distal pattern, with glomeruli (WT1+NPHS1+) located at the most proximal end, followed by proximal (LTL+LRP2+CDH6+AQP1+), medial (JAG1+), and distal-connecting tubules (CDH1+AQP2+) (Figure 3A and S3B). MEIS1, PDGFRβ, and low levels of WT1 were expressed in stromal cells (Figure S3C), whereas FOXD1 expression was mainly detected in podocytes as recently described (Lindstrom et al., 2018b, Brunskill et al., 2011). We observed a small amount of tubule segments that expressed CK8+CALB1+ (Figure 3A, white arrow) or CDH1+AQP2+ (Figure S3B). These may indicate a distal-connecting tubule identity (Horster, 2000, Kortenoeven et al., 2013), as we did not identify morphologically distinctive ureteric bud (UB)-like structures. Furthermore, although scRNA-seq revealed GO terms associated with UB development during day 10–14 of differentiation (Table S3), bulk RNA-sequencing (RNA-seq) only detected marginal induction of AQP2 and GATA3 in day 24 organoids (Figure S3D), indicating a culture condition favoring NPC-derived lineages. These kidney organoids could be maintained in liquid-air interface culture for more than 2 months without losing cellular compartmentalization (Figure S3E).
Figure 3. Modulating the Proportion of Proximal-versus-Distal Segments and the Vascular Network within 3D Kidney Organoids.
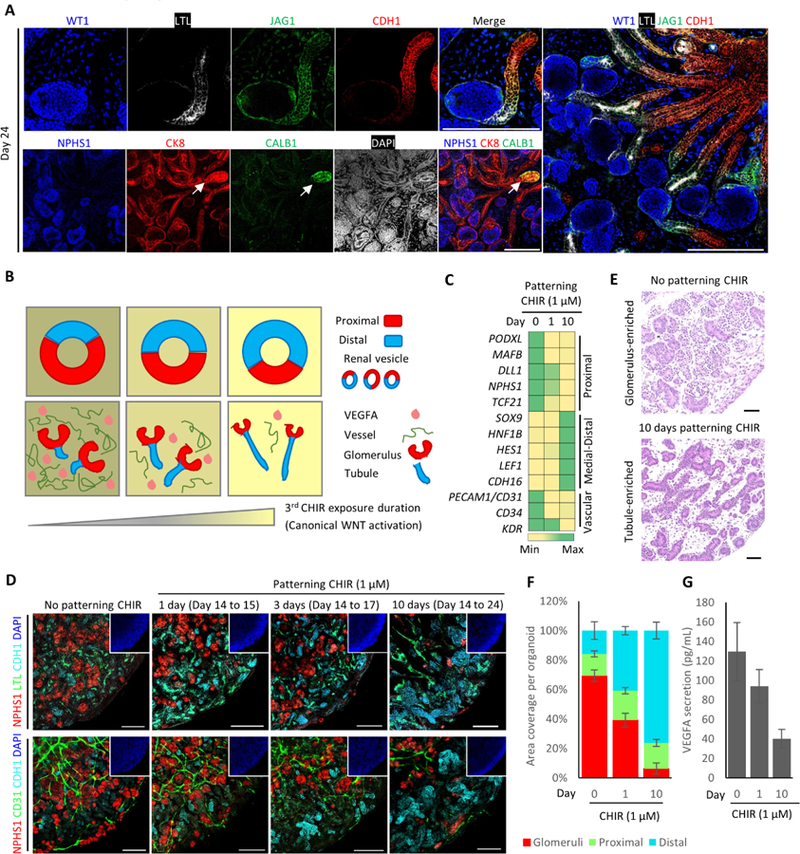
(A) Whole-mount immunofluorescence analysis of nephron segment-specific markers in 3D kidney organoids (Day 24). Scale bars, 200 μm.
(B) Proposed model for in vitro kidney organoid patterning (modified from Lindstrom et al., 2015 and GUDMAP).
(C) Gene expression profiling of 3D kidney organoids (Day 24) with different exposure lengths to patterning CHIR. Color shading correlates with the relative fold change in gene expression levels. Data were presented as mean ± SEM (n = 2 independent experiments with 3 technical replicates).
(D) Whole-mount immunofluorescence analysis of 3D kidney organoids (Day 24) with different exposure lengths to patterning CHIR. Scale bars, 200 μm.
(E) Representative H&E staining images of 3D kidney organoids (Day 24) in the absence or presence of patterning CHIR. Scale bars, 50 μm.
(F) Quantification for the ratio of multiple nephron segments in 3D kidney organoids (Day 24) with different exposure lengths to patterning CHIR. Data were presented as mean ± SEM (n = 3 independent experiments).
(G) VEGFA protein secretion by 3D kidney organoids (Day 24) with different exposure lengths to patterning CHIR. Data were presented as mean ± SEM (n = 3 independent experiments).
Preferential enrichment of specific nephron segments would greatly facilitate the study of segment-specific phenotypes, such as tubule cyst formation. Multiple signaling pathways are implicated in patterning the proximal-distal axis of nephron, such as WNT, BMP, and NOTCH (Munro et al., 2017, Mills et al., 2017, Lindstrom et al., 2015). During kidney development, renal vesicles have already acquired features that indicate the establishment of proximal-distal polarity relative to the adjacent UB (Lindstrom et al., 2018e). Thus, we asked whether modulation of WNT signaling in nascent renal vesicles could alter the relative ratio of proximal versus distal segments within kidney organoids (Figure 3B, upper panels) (Lindstrom et al., 2015, Schneider et al., 2015). To do this, we treated day 14 kidney organoids that were largely comprised of renal vesicles (Figure S3F), with 1 μM patterning CHIR for various durations (0–10 days). Gene expression profiling indicated a positive correlation between the length of exposure to patterning CHIR and the expression levels of medial-distal markers (Figure 3C). Segment-specific marker staining further validated that high levels of WNT signal favored distal segment formation (Figure 3D, upper panels) and larger organoids (Figure S3G). Following 10 days of patterning CHIR treatment, kidney organoids were almost fully comprised of tubules, whereas an absence of patterning CHIR led to an exceptionally high glomerulus-to-tubule ratio (Figure 3E and 3F).
In accordance with this observation, kidney organoids with a high glomerulus-to-tubule ratio possessed a richer vascular network than those with a high tubule-to-glomerulus ratio, due to the correlative VEGFA production by podocytes (Figure 3B–3D, lower panels). On the contrary, kidney organoids exhibited a smaller vascular network when exposed to patterning CHIR for a longer time (Figure 3B–3D, lower panels), in correlation with a reduction in VEGFA secretion (Figure 3G). To rule out the possibility that higher levels of WNT signal directly interrupted the established vasculature, we performed an additional CHIR treatment at a later differentiation time point, when all the cellular components were already defined within kidney organoids. After 20 days of differentiation, neither nephron structure nor vascular network was affected by an additional CHIR treatment (Figure S3H). In summary, by modulating WNT signaling, we successfully adjust the relative proportion of proximal versus distal segments within hPSC-derived kidney organoids. Simultaneously, the communication between glomerular podocytes and endothelial cells via VEGFA signaling patterns a correlative vascular network.
Functional Validation of hPSC-Derived Kidney Organoids
Next, we employed an in vitro dextran uptake assay to examine whether these kidney organoids exhibited physiologically relevant features (Figure 4A). After 4 hours of dextran pulse, hPSC-derived kidney organoids uptook 10 kDa and 70 kDa dextrans, but excluded 2,000 kDa dextran (Figure 4B, left panels). Likewise, 24 hours later, both 10 kDa and 70 kDa dextrans, but not 2,000 kDa dextran, were retained within LTL+ proximal tubule epithelial cells (Figure S4A and 4B, right panels). This observation was further verified by a similar dextran uptake assay conducted using kidney aggregates derived from E12.5 mouse embryonic kidneys (Figure S4B) (Unbekandt and Davies, 2010, Xia et al., 2014).
Figure 4. In Vitro Functional Validation of 3D Kidney Organoids.
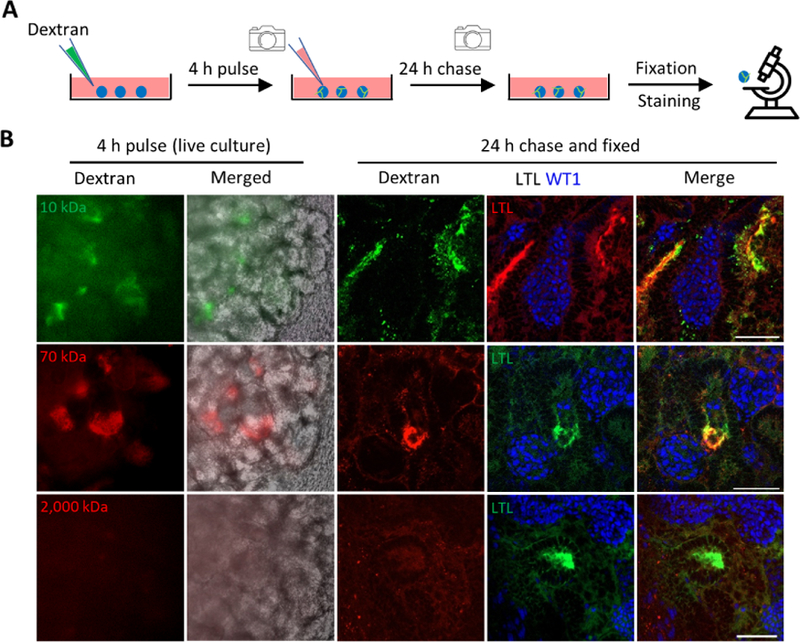
(A) Schematic of in vitro dextran uptake assay.
(B) Left panels: live images of kidney organoids incubated with fluorescence-labeled dextran of various molecule weights. Right panels: whole-mount immunofluorescence analysis of kidney organoids following dextran update assay. Scale bars, 50 μm.
It is crucial for renal microvessels to associate with respective nephrons in a timely manner to ensure the establishment of glomerular filtration apparatus. We noticed that despite the plethora of endothelial cells within these kidney organoids, most glomeruli were avascular (Figure S4C). This was possibly due to a lack of hemodynamics (Homan et al., 2019). Therefore, we implanted 4-week old kidney organoids beneath the renal capsule of immunocompromised mice (NSG) (Figure S5A). We observed a time-dependent increase in both the implant size and the number of perfused vessels that were visible on the surface of implants (Figure S5A). Upon implantation, glomeruli gradually acquired a much more mature architecture. Within implants, the slit diaphragm protein Nephrin adopted a puncta-like membrane distribution, different from the ubiquitous membrane distribution seen in in vitro organoids (Figure 5A). We also identified a PDGFRβ+ mesangial population within glomeruli after implantation (Figure 5A). Furthermore, the implanted kidney organoids established a discernible Bowman’s capsule space in between podocytes and parietal epithelial cells (Figure 5B and S5B). More importantly, the implanted kidney organoids developed morphologically distinct glomerular capillary tufts that were perfused by red blood cells (Figure 5B and S5C). A human-specific CD31 antibody revealed that human endothelial cells contributed to glomerular capillary tufts (Figure 5C). At ultrastructural level, podocytes developed highly elaborate cellular processes (Figure S5D), while proximal tubules displayed very well-developed apical microvilli and cilia (Figure S5E). Adjacent epithelial cells were perfectly sealed by tight junctions (Figure S5E). We also observed fenestrated endothelium (Figure S5F) and glomerular basement-like membrane (Figure S5F), both of which contributed to the establishment of the putative glomerular filtration barrier (Figure S5F).
Figure 5. Structural and Functional Validation of Kidney Organoid Implants.
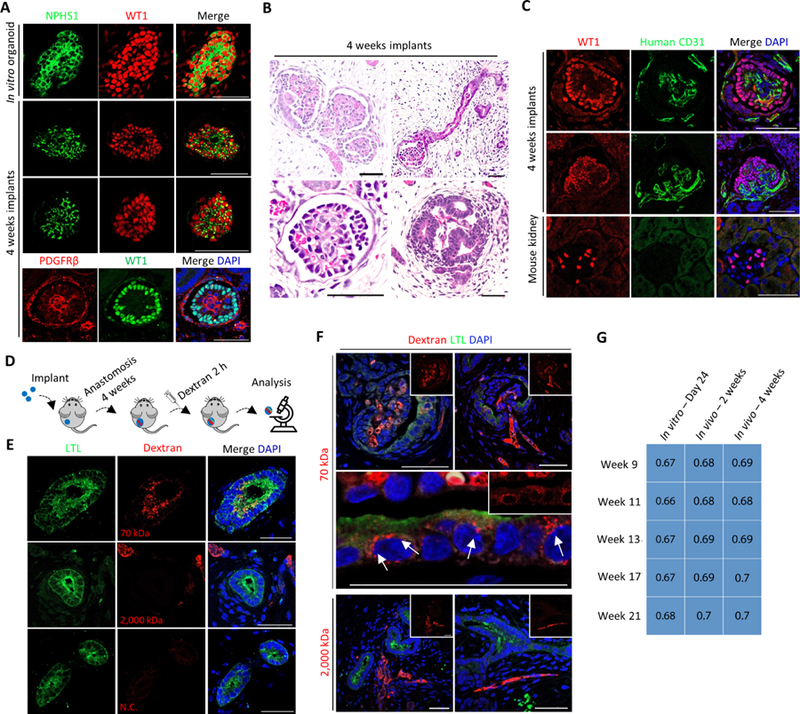
(A) Immunofluorescence analysis of in vitro kidney organoids and 4 weeks kidney organoid implants. Scale bars, 50 μm.
(B) Representative H&E staining images of kidney organoid implants (4 weeks). Scale bars, 50 μm.
(C) Immunofluorescence analysis of kidney organoid implants (upper panels) and adult mouse kidney (lower panels). Scale bars, 50 μm.
(D) Schematic of in vivo dextran uptake assay.
(E and F) Immunofluorescence analysis of kidney organoid implants, the host mice of which were systemically injected with fluorescence-labeled dextran of various molecule weights, or with PBS. N.C. denotes PBS negative control. White arrow indicates putative endosome. Scale bars, 50 μm.
(G) Transcriptome correlation plot of in vitro – Day 24, in vivo – 2 weeks, and in vivo – 4 weeks kidney organoids with human fetal kidneys at week 9, 11, 13, 17 and 21. Correlation coefficient indicated within squares. Significantly correlated features (p ≤ 0.05) were indicated by blue shading.
To further evaluate the functional maturation status of kidney organoid implants, we systemically injected dextran of different molecular weights into NSG mice, 4 weeks after they received an implant (Figure 5D). Animals were sacrificed 2 hours post injection. 70 kDa dextran was detected within glomerular capillary tufts and proximal tubules (Figure 5E and 5F, upper panels), strongly suggesting that the implanted kidney organoids could perform preliminary filtration and reabsorption. The reabsorbed 70 kDa dextran formed small puncta that resembled endosomes inside LTL+ proximal tubule epithelial cells (Figure 5F, middle panel, white arrows). In contrast, host mice injected with 2,000 kDa dextran or with phosphate buffered saline (PBS) did not exhibit co-localization of LTL and dextran within implants (Figure 5E, middle and bottom panels). Nevertheless, 2,000 kDa dextran was detected within vessel lumen, indicating successful anastomosis but a lack of filtration and reabsorption (Figure 5F, lower panels). These results suggest a size-selective filtration and reabsorption instead of capillary leaking, as wild-type mouse kidneys handled dextran of different molecular weights in a similar manner (Figure S5G and S5H). Furthermore, kidney organoid implants accumulated putative filtrate within some distal tubules, reminiscent of those observed in wild-type mouse kidneys (Figure S5I, black arrows) (Sharmin et al., 2016). Transcriptomic profiling showed that implantation increased the correlation between hPSC-derived kidney organoids and human fetal kidneys from the first and second trimester of pregnancy (Figure 5G) (Lindstrom et al., 2018c, Lindstrom et al., 2018d, Lindstrom et al., 2018e). These results point out that implantation greatly facilitated structural and functional maturation of hPSC-derived kidney organoids.
ARPKD iPSC-Derived Kidney Organoids Recapitulate Cystogenesis
To explore the translational application of our differentiation platform, we generated iPSCs from ARPKD patient-derived fibroblasts using an integration-free reprogramming approach (Okita et al., 2011). Sanger sequencing revealed that this patient harbors compound heterozygous mutations within exon 5 and exon 65 of PKHD1 locus (Figure S6A). Because ARPKD is a recessive disorder, we used the CRISPR-Cas9 gene-editing tool to correct c.11630delT mutation within exon 65, and generated isogenic iPSC lines (Figure S6B and S6C). Both ARPKD iPSCs and corrected-ARPKD iPSCs expressed pluripotency markers and presented tri-lineage differentiation potential (Figure S6D and S6E). When subjected to kidney organoid differentiation, both types of iPSCs displayed similar kinetics and efficiency (Figure S7A), and gave rise to 3D kidney organoids comprised of nephron structures that were correctly patterned along the proximal-distal axis (Figure 6A and S7B). We observed a low incidence of spontaneous cyst formation in ARPKD kidney organoids following extended culture, as previously described (Figure S7C) (Freedman et al., 2015). This may be because environmental stress is generally required, in combination with genetic aberration, for cyst formation (Takakura et al., 2009, Wang and Dong, 2016). We therefore challenged kidney organoids with either forskolin or 8-Br-cAMP to increase intracellular cAMP levels, a cellular hallmark of PKD (Harris and Torres, 2014, Torres and Harris, 2014). ARPKD kidney organoids exhibited drastic cystogenesis upon the upregulation of intracellular cAMP (Figure 6B and S7D), in a dose-dependent manner (Figure S7E). In contrast, corrected-ARPKD kidney organoids showed marginal cyst formation, resembling that of wild-type hPSC-derived kidney organoids (Figure 6B).
Figure 6. ARPKD Kidney Organoids Recapitulate Cystogenesis.
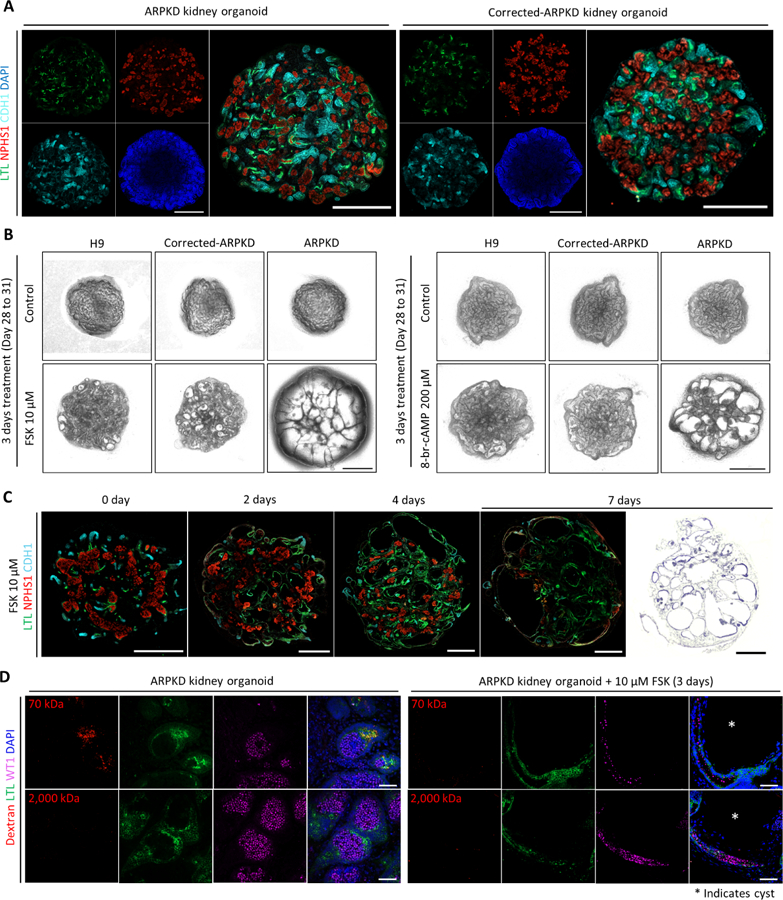
(A) Whole-mount immunofluorescence analysis of 3D kidney organoids (Day 24) derived from ARPKD iPSCs and corrected-ARPKD iPSCs. Scale bars, 500 μm.
(B) Representative bright-field images of 3D kidney organoids derived from H9 embryonic stem cells (ESCs), corrected-ARPKD iPSCs, and ARPKD iPSCs, in the absence or presence of forskolin (FSK)/8-br-cAMP for 3 days (Day 28 to 31). Scale bars, 500 μm.
(C) Left panels: time course whole-mount immunofluorescence analysis of ARPKD kidney organoids treated with 10 μM FSK. Right most panel: representative H&E staining image of ARPKD kidney organoid treated with 10 μM FSK for 7 days (Day 28 to 35). Scale bars, 500 μm.
(D) Immunofluorescence analysis of ARPKD kidney organoids in the absence or presence of 10 μM FSK for 3 days (Day 28 to 31) followed by in vitro dextran uptake assays. White asterisk indicates cyst. Scale bars, 50 μm.
During cystogenesis, ARPKD kidney organoids showed a time-dependent enlargement of tubule lumen, first proximally and later extended to distal regions (Figure 6C, left panels). This is well-aligned with a proximal tubule origin of fetal renal cysts during the first and second trimester of pregnancy (Gunay-Aygun et al., 2006, Nakanishi et al., 2000, Woollard et al., 2007). Furthermore, there was a gradual reduction in the expression of segment-specific markers as cystogenesis progressed (Figure 6C, left panels). After one week of forskolin treatment, tubule cysts covered more than 80% of the entire kidney organoid, with distorted glomeruli squeezed in between cysts (Figure 6C, right most panel). We conducted in vitro dextran uptake assay to evaluate the effect of cysts on kidney organoid function. Non-cystic ARPKD kidney organoids uptook dextran in a size-selective manner (Figure 6D, left panels), comparable with wild-type hPSC-derived kidney organoids (Figure 4B). Upon cyst formation, LTL+ proximal tubule epithelial cells completely lost the capability to uptake dextran, regardless of the molecular weight (Figure 6D, right panels), possibly due to the loss of LRP2 expression in the cyst lining proximal tubular epithelial cells (Figure S7F). Collectively, these findings validate the robustness of our differentiation protocol, allowing us to extend these efforts to a wider range of genetic kidney diseases and to tailor patient-specific therapeutics.
Testing the Effects of Chemical Compounds on ARPKD Kidney Organoid Cystogenesis
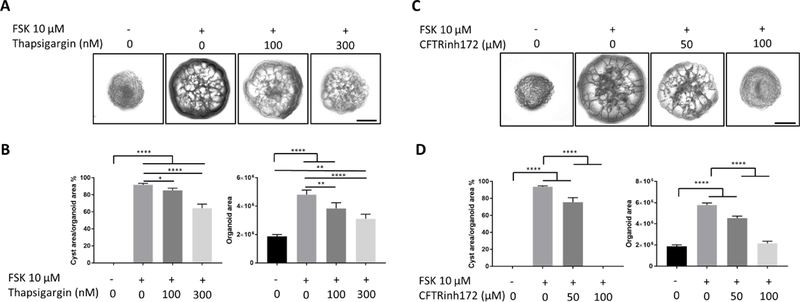
Compared to mouse genetic models or patient biopsy-derived primary culture, it is certainly appealing to use patient iPSC-derived kidney organoids for drug screening. To provide a proof-of-concept, we evaluated the effects of two chemical compounds on forskolin-induced cystogenesis in ARPKD kidney organoids. Thapsigargin, which releases intracellular Ca2+, inhibited forskolin-induced cystogenesis in a dose-dependent manner, likely by remediating defective Ca2+ homeostasis typically seen in PKD (Figure 7A and 7B) (Torres and Harris, 2006, Harris and Torres, 2009). Additionally, an inhibitor of cystic fibrosis transmembrane conductance regulator (CFTR), a chloride channel that facilitates trans-epithelial fluid secretion (Davidow et al., 1996), also effectively blocked ARPKD kidney organoid cyst formation in a dose-dependent manner, as previously described using both human primary PKD cell culture and a genetic mouse model (Figure 7C and 7D) (Hanaoka et al., 1996, Yang et al., 2008). These data strongly demonstrate that our kidney organoid platform may provide a physiological and species-relevant preclinical model for the evaluation of candidate drugs prior to clinical trials.
Figure 7. Chemical Compounds Inhibit ARPKD Kidney Organoid Cystogenesis.
(A)Representative bright-field images of ARPKD kidney organoids treated with various concentrations of Thapsigargin in the absence or presence of 10 μM FSK for 3 days (Day 28 to 31). Scale bars, 500 μm.
(B) Statistical analysis of the effect of Thapsigargin on FSK-induced ARPKD kidney organoid cystogenesis. Data were presented as mean ± SEM (n = 3 independent experiments). Statistical analysis was performed using one-way ANOVA, * p < 0.05,** p < 0.01, **** p < 0.0001.
(C) Representative bright-field images of ARPKD kidney organoids treated with various concentrations of CFTRinh172 in the absence or presence of 10 μM FSK for 3 days (Day 28 to 31). Scale bars, 500 μm.
(D) Statistical analysis of the effect of CFTRinh172 on FSK-induced ARPKD kidney organoid cystogenesis. Data were presented as mean ± SEM (n = 3 independent experiments). Statistical analysis was performed using one-way ANOVA, **** p < 0.0001.
DISCUSSION
Kidney organoids hold unprecedented advantages for understanding human kidney development and physiology, as well as for translational medicine. However, intrinsic limitations within current kidney organoids, such as variability in composition and structural organization, lack of vasculature and functionality, grievously limit their application. Hence, we have developed a highly robust protocol for efficiently differentiating hPSCs into segmentally patterned 3D kidney organoids that are associated with a de novo vascular network (Figure 1A–1E). Through precise modulation of WNT signaling, our protocol effectively controls the relative proportion of proximal versus distal segments within kidney organoids (Figure 3B–3F), a major advance over previous methods that used γ-secretase inhibitor DAPT to selectively inhibit proximal tubule formation (Cheng et al., 2007, Morizane et al., 2015). The establishment of a specific glomerulus-to-tubule ratio further determines a correlative level of podocyte-produced VEGFA, which coordinates the volume of the resident vascular network (Figure 3B–3G). Although the existence of vascular endothelial cells within hPSC-derived kidney organoids has been described, the low abundance and dependence on an exogenous source of VEGFA preclude them from presenting the physiologically relevant communication with the epithelial population (Takasato et al., 2015, Czerniecki et al., 2018). Therefore, our kidney organoid culture system may provide a reliable platform for investigating renal diseases that are associated with deterioration of renal microvessels, such as diabetic nephropathy.
Kidney organoids have been utilized to investigate various aspects of human embryonic kidney development. Several recent studies synergized scRNA-seq with organoid culture to provide a deeper understanding of the cellular components within hPSC-derived kidney organoids, as well as to recapitulate lineage relationship during kidney development (Phipson et al., 2019, Wu et al., 2018, Czerniecki et al., 2018). The potency and consistency with which vasculature can be generated within our kidney organoids provide the possibility to study the developmental origin of renal vasculature, which has yet been fully resolved (Munro et al., 2017, Hu et al., 2016, Mugford et al., 2008, Risau, 1998, Abrahamson et al., 1998). The McMahon group reported that E7.5-E8.5 OSR1+ mouse intermediate mesodermal cells contribute at least partially to renal vasculature, suggesting an intra-renal origin (Mugford et al., 2008), whereas the Davies group demonstrated that mouse kidney vasculature was formed primarily via angiogenesis (Munro et al., 2017). We have employed scRNA-seq to scrutinize the early stage of nephrogenesis, during when multiple renal lineages emerge, including endothelium. Unsupervised analysis provides the first evidence that a subpopulation of NPCs may serve as a non-conventional source of renal vasculature (Figure 2), which was further supported by the revelation of SIX1+CD31+ (SALL1+CD31+) cells in the second trimester human fetal kidneys (Figure S2H). Reconstruction of the endothelial lineage trajectory strongly indicates that, along pseudotime, the resident vascular endothelial cells acquires stage-specific lineage properties reminiscent of endothelial development (Figure 2G–2I), including the preliminary specification into endothelial sub-types (Figure 2J and 2K). Our study provides a proof-of-concept that synergism of single-cell sequencing and organoid biology can complement genetic animal models in identifying novel lineage relationship during human embryonic development.
Previous studies demonstrated that hPSC-derived kidney organoids can uptake dextran via proximal tubule epithelium in vitro (Freedman et al., 2015, Morizane et al., 2015, Takasato et al., 2015, Czerniecki et al., 2018). The advantage of the kidney organoids described here is that they can uptake dextran in a size-selective manner both in vitro and in vivo (Figure 4 and 5). This physiological feature could be harnessed as a functional readout in modeling tubular kidney diseases. For example, cystic kidney organoids have lost the capability to uptake dextran in a size-selective manner (Figure 6D). Previous studies have successfully implanted hPSC-derived kidney organoids or nephron progenitors, but in these cases the origin of glomerular capillary tufts was not clearly described (Sharmin et al., 2016, Bantounas et al., 2018, van den Berg et al., 2018), and there was a lack of functional validation. A recent report showed that chick embryo chorioallantoic membrane is highly amenable for vascularizing kidney organoid implants, although a functional evaluation was also missing (Garreta et al., 2019). In vivo implantation facilitates both structural and functional maturation of these kidney organoids, as exemplified by the formation of glomerular capillary tufts of a human origin (Figure 5C), the establishment of the putative glomerular filtration barrier (Figure S5F), and the size-selective dextran handling (Figure 5D–5F). Nonetheless, these kidney organoid implants exhibit an overall similarity to human fetal kidneys from the first and second trimester of pregnancy (Figure 5G) (Takasato et al., 2015, Lindstrom et al., 2018c, Lindstrom et al., 2018d, Lindstrom et al., 2018e). It is likely that additional approaches, such as chromatin remodeling and epigenetic reprogramming, may be needed to advance the developmental stage of hPSC-derived kidney organoids.
One of the ultimate goals for generating patient iPSC-derived kidney organoids is to model disease pathogenesis and to perform personalized drug validation. We have successfully recapitulated renal cyst formation using kidney organoids derived from ARPKD iPSCs (Figure 6B). These cystic kidney organoids exhibit defects typically seen in PKD patients (Figure 6C and 6D) (Birn and Christensen, 2006). More importantly, these cystic ARPKD kidney organoids represent an amenable platform to test drugs in a patient-specific manner (Figure 7). Earlier studies by the Freedman group used PKD1−/−and PKD2−/−hPSCs, instead of PKD patient-derived iPSCs, to model PKD cystogenesis, possibly owing to inconsistent differentiation outcomes (Freedman et al., 2015, Cruz et al., 2017, Czerniecki et al., 2018). Due to the tight correlation between patient genetic background and drug responsiveness, generating high quality kidney organoids from patient iPSCs will enable drug validation in a patient-specific manner. Combined with the recently developed high throughput methods (Czerniecki et al., 2018, Przepiorski et al., 2018), our kidney organoid differentiation platform holds great promise for realizing large-scale drug screening to improve patient outcomes.
STAR METHODS
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Yun Xia (yunxia@ntu.edu.sg).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Cell lines
H9 (WA09) female and H1 (WA01) male human embryonic stem cells (WiCell) or ARPKD iPSCs (parental ARPKD patient fibroblasts (GM10287) obtained from Coriell Institute for Medical Research) were cultured in TeSR on growth-factor-reduced Matrigel-coated plates. For routine maintenance, 70–80% confluent hPSCs were treated with Dispase (Invitrogen) for 5 mins, then dissociated into small clumps by mechanical force, and passaged onto Matrigel-coated plates at a ratio of 1:5–1:10.
To generate iPSCs from ARPKD patient, one million human fibroblasts (Coriell GM10287) were electroporated with pCXLE-hOCT3/4-shp53-F (or pCXLE-hOCT3/4), pCXLE-hSK, and pCXLE-hUL using a Nucleofector kit (Lonza), as described previously (Okita et al., 2011). Six days after nucleofection, fibroblasts were reseeded onto mitotically inactivated mouse embryonic fibroblasts (MEFs). The medium was switched to hPSC medium on the following day. The iPSC colonies were allowed to grow until they were ready to be mechanically picked and transferred onto MEFs. Upon successful expansion of the iPSC lines on MEFs, they were then cultured on Matrigel (BD Biosciences) in TeSR.
Human samples
Human fetal kidney samples from donors (17 weeks, male; 21 weeks, female) were obtained from Kandang Kerbau Women’s and Children’s Hospital (KKH) with written consent from donors. SingHealth and National Health Care Group Research Ethics Committees Singapore specifically approved this study (CIRB Ref: 2012/064/F).
Mouse strains
NOD.Cg-Prkdcscid.Il2rgtm1Wjl/SzJ (NSG) mice and CD-1 mice were purchased from Jackson Laboratory. Mice were housed in a specific pathogen-free animal facility in plastic cages on 12 h/12 h light/dark cycle and fed with standard chow diet. 8–10 weeks old NSG mice were used for implantation assay, followed by in vivo dextran assay (IACUC #A0346). 8–12 weeks old CD-1 mice were used for in vivo dextran assay (IACUC #A0346). 8–10 weeks old CD-1 mice were bred for E12.5 embryonic kidneys for generating mouse embryonic kidney aggregates (IACUC #A0289). All animal experiments were performed in accordance with institutional guidelines and approved by the Institutional Animal Care and Use Committee (IACUC) of Nanyang Technological University Singapore.
METHODS DETAILS
Induction of kidney organoids from human pluripotent stem cells (hPSCs)
hPSCs were dissociated into single cells with Accutase (STEMCELL Technologies), and plated onto Matrigel (BD Biosciences)-coated plates at a density of ~15,000 to ~24,000 cells/cm2. The plated cells were cultured in TeSR supplemented with 10 µM Y-27632 (Sigma) for 24 hours, followed by another 48–72 hours culture in TeSR to reach a confluence of 50–60%.
To induce primitive streak, cells were cultured in basal differentiation medium (Advanced RPMI 1640, 1x L-GlutaMAX, 1 × NEAA) (Life Technologies) supplemented with 10 µM CHIR99021 (Sigma) for 4 days with daily media change. Next, to induce nephrogenic intermediate mesoderm, cells were cultured in basal differentiation medium, without any growth factor for 3 days with daily media change. Subsequently, to induce nephron progenitors, cells were cultured in basal differentiation medium supplemented with 50 ng/mL FGF9 (Peprotech) and 3 µM CHIR99021 with daily media change for 2 days. From day 10 of differentiation onward, cells were cultured in basal differentiation medium supplemented with 50 ng/mL FGF9 until day 20. On day 14 of differentiation, the organoid can be cultured in differentiation medium supplemented with 1 µM CHIR99021 for 1 to 10 days (patterning CHIR) to modulate the proportion of proximal-versus-distal segments. From day 20 of differentiation onward, basal differentiation medium was used for organoid culture.
Generation and patterning of three-dimensional (3D) kidney organoids
To generate 3D kidney organoids, day 10–12 differentiating cells were dissociated into single cells by-Accutase. 2.5–5 × 104 cells (per well) were aggregated in basal differentiation medium supplemented with 50 ng/mL FGF9 and 10 µM Y-27632 using round bottom ultra-low attachment 96-well plates (Corning). 24 hours later, Y-27632 was removed. To generate kidney organoids with a high glomerulus-to-tubule ratio, no patterning CHIR was added. To generate kidney organoids with a high tubule-to-glomerulus ratio, 10 days patterning CHIR was applied from day 14 to day 24 of differentiation. 3D kidney organoids were transferred onto the upper chamber of Transwell (Corning) for liquid-air interface culture on day 16–18 of differentiation (5–7 days after aggregation), with daily media change at the bottom chamber.
Taking H9 ESCs as an example, we seeded ~21,000 cells per cm2, and started differentiation when the cells reached 50–60% confluence. On day 10–12 of differentiation, we dissociated the differentiation culture into single cells, and re-aggregated into cell clusters with a gradient of cell numbers (5,000–75,000 cells per organoid). One million starting human ESCs could give rise to approximately 3,500 kidney organoids that were aggregated with 35,000 cells per organoid.
Single-cell RNA-seq library preparation
Day 10, Day 12 and Day 14 kidney organoids were dissociated into single cells with Accutase, followed by passing through a 40 μm filter thrice to remove cell clumps. Dissociated cells were subsequently stained with trypan blue and confirmed to have at least 80% viability. A minimum of 7000 dissociated cells were loaded per well for single-cell RNA-seq library preparation with the Chromium Single Cell 3’ Gene Expression Solution according to the manufacturer’s recommendations. Libraries generated were sequenced on Illumina HiSeq 4000 platform. We captured a total of 15,834, 6,554 and 41,534 single cells from Day10, Day12 and Day14 dissociated cells loading across 2, 1 and 6 capture wells, respectively. 73,000, 56,300 and 55,856 raw mean reads were obtained per cell from the captured Day10, Day12 and Day14 single cells.
Generation of gene corrected ARPKD iPSCs
To construct sgRNA expression vectors, 19 bp target sequence were sub-cloned into pCAGmCherry-gRNA (Addgene 87110). The CRISPR/Cas9 target sequences (19 bp target and 3 bp PAM sequence (underlined)) used in this study are shown as following: PKDH1 exon 65 (ATGGCTGGCTCTGAGCTGTCGG). Cas9 expression plasmid (pCas9_GFP) was obtained from Addgene (44719).
The donor oligo for gene correction (5’-TGCTTCTTTTAAGCCAACAGCACACCAGACAGCTCAGAGCCAGCCATGAGGCCACAGAGG-3’) was purchased from Valuegene (San Diego). To generate gene-corrected iPSCs, 1.5 × 107 feeder-free cultured iPSCs were dissociated by TrypLE (Invitrogen), and resuspended in 1 ml of medium with 10 μM Y-27632. Cas9 expression plasmid (pCas9_GFP, 15 μg), sgRNA expression plasmid (15 μg) and the donor oligo (30 μg) were co-transfected by electroporation using the BioRad Gene Pulser II (a single 320 V, 200 μF pulse at room temperature) with 0.4 cm gap cuvette. Cells were plated at high density on 4 of 6-well plate coated with Matrigel. Two days after electroporation, GFP-/mCherry-double positive cells were isolated by FACS (BD FACS Aria II), and seeded on MEF-feeder plate with CDF12 medium including Y-27632. Next day, we changed the medium to CDF12 without Y-27632. After 2–3 weeks in culture, each single colony were picked into a 96-well culture plate and expanded for identification. To isolate gene-corrected clones, exon 65 of PKDH1 was PCR-amplified with PKHD1-ex65F (AAAGAAGCTGAAATTTTGCATTGGTATGGA) and PKHD1-ex65R (ATTTGGCTTTGTGTAATTTTATATTTTAGAGAAGCTCA) using PrimeSTAR GXL DNA Polymerase (Takara). Amplicons were sequenced with an ABI 3730 sequencer (Applied Biosystems).
Chemical treatment of hPSC-derived kidney organoids
VEGFR inhibition assay.
Kidney organoids cultured on liquid-air interface were changed to basal differentiation medium supplemented with 10 nM each of FDA-approved VEGFR inhibitors (Axitinib/ Cediranib/ Tivozanib) for 1 to 3 days. After treatment, kidney organoids were fixed, and subjected to whole-mount immunofluorescence staining followed by confocal microscopy imaging.
Cyst induction and inhibition.
ARPKD iPSC and corrected-ARPKD iPSC-derived 3D kidney organoids were cultured on liquid-air interface until day 28 of differentiation. To initiate cytogenesis, basal culture medium was supplemented with either forskolin (FSK, 0.1 to 50 µM) or 8-Br-cAMP (200 μM) for 2 to 7 days. Inhibition of cystogenesis was performed by co-treatment of 10 μM forskolin with either Thapsigargin (100 nM to 300 nM) or CFTRinh172 (50 µM to 100 µM) for 3 days. Medium was only added into the bottom chamber of Transwells.
Kidney organoid implantation
hPSC-derived 3D kidney organoids (8–12 organoids per kidney) were implanted beneath the renal capsule of NSG mice. 2–4 weeks later, mice were sacrificed, and implants were isolated for further analysis.
Dextran uptake assay
In vitro dextran uptake assay. Kidney organoids were incubated with 100 µg/mL of fluorescence-labeled dextran for 4 hours (pulse). 4 hours later, kidney organoids were switched to fresh medium and live cell culture images were captured using a wide-field fluorescence microscope (pulse). Kidney organoids were then cultured in the absence of dextran for another 24 hours (chase), before being fixed for immunofluorescence analysis. In vivo dextran uptake assay. 200 µL of fluorescence-labeled dextran (Thermo Fisher Scientific) (5 mg/ml, 1 mg/animal) were administered into mice through tail vein injection. The injected mice were sacrificed 2 hours later and kidneys were collected for further analysis. Phosphate buffered saline injected mice were used as negative control (N.C.).
Transmission and Scanning Electron Microscopy
Implanted organoids were recovered two weeks after implantation and fixed in 2% glutaraldehyde and 1% paraformaldehyde in 0.1 M phosphate buffer for 15 mins. The implants were dissected into small cubes (~2 mm × 2 mm), fixed in fresh fixative solution overnight at 4 °C. After fixation, samples were washed 5 × 5 mins in 0.1 M phosphate buffer and stored at 4 °C.
For TEM, samples were stained with 2% osmium tetroxide solution containing 2% potassium ferrocyanide for 2 h. Subsequently, samples were dehydrated in increasing concentrations of ethanol (25%, 50%, 75%, 95%, 100%) and acetone (100%) before infiltration with increasing concentrations of acetone : epon resin mixture (1:1 for 1 h, 1:6 for overnight; absolute resin for 1 h) (Polysciences Inc, PA, USA). Absolute resin was replaced every 1 h for 3 times before curing at 60 °C for 48 h. Blocks were sectioned using Leica EM UC7 ultramicrotome (Leica Microsystems, Wetzlar, DE) at 500 μm for semi-thin sections and 130 μm for ultra-thin sections. Sections were contrasted using 4 % (w/v) uranyl acetate solution for 5 mins, 5 × washes in distilled water, followed by Reynold’s lead citrate solution for 5 mins and 5 × washes in distilled water. Images were collected using either JEOL JEM1010 or JEOL JEM1200.
For SEM, samples were fixed with 1% osmium tetroxide solution for 2 h. Subsequently, samples were dehydrated in increasing concentrations of alcohol (25%, 50%, 75%, 95%, 100%) before drying in a critical point dryer. Samples were mounted on the stub using carbon tape before sputter coated with gold. Images were collected using FEI Quanta 650 FEG.
Histology
Implanted kidney organoids were fixed overnight in 4% paraformaldehyde (PFA) at 4°C. In vitro cell cultures and organoids were fixed for 30 mins in 4% PFA at room temperature. For frozen sections, samples were subjected to sucrose gradient treatment overnight prior to being embedded in Tissue-Tek O.C.T. Compound (Sakura Finetek). Cryosections (8 µm) were generated using a cryostat (Leica). For paraffin embedded sections, tissues were subjected to overnight processing followed by being embedded in paraffin (Leica). Tissue sections (2–4 µm) were generated using a microtome (Leica). Hematoxylin and Eosin (H&E) staining was performed according to the manufacturer’s instruction. For immunostaining, sections were de-paraffinized and subjected to antigen retrieval procedure in citrate buffer (pH 6.0, 95°C) for 45 mins before staining (see detailed staining protocol below).
Immunostaining and confocal imaging
Fixed cell cultures or 3D kidney organoids were permeabilized using phosphate buffered saline (PBS) supplemented with 0.5% Triton X-100 for 15 mins at room temperature. Blocking was performed for 1 hour at room temperature using a blocking buffer (PBS supplemented with 2% FBS, 2% BSA and 0.1% Triton X-100). Subsequently, cells were incubated with primary antibodies overnight at 4 °C with agitation. Cells were then washed three times with PBST (PBS supplemented with 0.1% Tween-20) and followed by 2 hours incubation with secondary antibodies at room temperature. Cells were then washed three times with PBST before mounting with Fluoromount-G (Southern Biotech). Confocal image was captured using a Zeiss LSM800 laser-scanning microscope (Carl Zeiss) equipped with 20x, 40x or 63x oil-immersion objectives.
RNA purification and quantitative RT-PCR (qRT-PCR)
Total RNA was isolated from cells using Trizol (Invitrogen). RNA was purified using Quick-RNA kit (Zymo Research) with DNase treatment. 500 ng total RNA of each sample was reverse transcribed using iScript cDNA synthesis kit (Bio-Rad). Quantitative real-time PCR was carried out using QuantStudio 6 (Applied Biosystems) and SYBR Green Supermix (Bio-Rad). Relative mRNA expression levels were quantified by the ΔΔCt method and normalized to GAPDH or 18s rRNA gene expression. All of the sample analyses were carried out in triplicates. In the event that the gene expression level is undetectable, a Ct value of 40 was assigned. Detailed primer information is provided in Mendeley data.
Embryonic body formation assay
hPSCs were treated with Dispase for 5 minutes, mechanically dissociated into small clumps, and transferred into low attachment 6-well plates in differentiation medium (DMEM/F12, 15% KnockOut™ serum replacement, 1xGlutaMAX, 1xNEAA, and 55 nM β-mercaptoethanol), with medium change every other day. After 21 days of culture, cells were collected for further analysis.
Fluorescence-activated cell sorting (FACS)
Cell culture was dissociated into single cells using Accutase and blocked for 30 mins in blocking buffer (PBS supplemented with 2% FBS and 2% BSA). For cell surface protein, primary antibody incubation was performed on ice for 30 mins. Cells were washed once with washing buffer (PBS supplemented with 1% FBS and 1% BSA) and incubated with secondary antibody for 30 mins on ice. Cells were then washed with ice cold PBS before analysis or sorting. For intracellular staining, cells were fixed using 4% PFA for 15 min at room temperature, followed by permeabilization (PBS supplemented with 0.2% Triton X-100) for 15 mins at room temperature prior to blocking and staining. FACS analysis was performed on the LSRFortessa X-20 and FACS sorting was performed on the FacsAria Fusion.
Introduction of HUVEC cells into 3D kidney organoids
We labelled HUVEC cells with pLV-eGFP (Addgene 36083) using the 3rd generation lentivirus packaging system. To generate virus, HEK293XT cells were transfected with pLV-eGFP and the associated packaging virus plasmid. Virus were collected 48 hours after first media change and filtered through a 0.45 μm filter. HUVEC cells were infected with freshly collected virus and the successful labelling was confirmed by visualization under fluorescence microscope.
Generation of mouse kidney aggregates from E12.5 embryonic mouse kidneys
Mouse kidney aggregates were generated as described with minor modification (Xia et al., 2014). E12.5 mouse embryonic kidneys were obtained from BALB/cJInv mice and treated with 0.25% (w/v) trypsin for 15 mins at 37 °C. After neutralizing trypsin using kidney culture medium (KCM, MEM supplemented with 10% FBS, 10 μM Y-27632 and 1:1,000 normocin) for 10 mins, mechanical force was applied to fully dissociate the kidney rudiments into single cells for aggregation. 40,000 cells were aggregated in KCM overnight in liquid culture using low attachment 96-well plates. After 24 hours aggregation, Y-27632 was removed from the culture medium, and aggregates were transferred onto liquid-air interface culture.
Enzyme-linked immunosorbent assay (ELISA)
Cell culture medium was collected 24 hours after the previous medium change, and centrifuged at 200 × g for 5 mins to remove cell debris. Human VEGFA levels were analyzed with human VEGF DuoSet ELISA (R&D systems) and DuoSet ELISA Ancillary Reagent Kit 2 (R&D systems) according to the manufacturer’s instructions. All sample analyses were carried out in triplicates.
In vivo imaging system (IVIS) assay
We injected 0.2 mg per gram body weight luciferin (Perkin Elmer) intraperitoneally into mice 5 mins prior to IVIS assay. Mice were anesthetized using isoflurane and placed in a prone position in the imaging chamber of the IVIS machine (Perkin Elmer Lumina XRMS). The injected mice were imaged over a 10 mins time period.
QUANTIFICATION AND STATISTICAL ANALYSIS
RNA-seq data processing
Sequenced reads were aligned to the Hg19 human genome (GRCh37) with STAR version 2.5.2a (Dobin et al., 2013). Cuffnorm version 2.2.1 (Trapnell et al., 2014, Trapnell et al., 2013) was used to obtain quartile normalize fragments per kilobase per million reads (FPKM) expression matrix of genes across all samples. Heatmap of expression of nephron stage markers and segment-specific markers was generated with R package pheatmap version 1.0.10 (https://cran.rproject.org/web/packages/pheatmap/index.html). Cuffdiff2 version 2.2.1 (Trapnell et al., 2013, Trapnell et al., 2010) was used to identify differential genes between samples. Significant differentially expressed genes (DEGs) which have false discovery rate (FDR) ≤ 0.05, |fold change| ≥ 1.5, and FPKM ≥ 1 in one sample group were retained for subsequent analysis. Gene ontology (GO) clustering enrichment analysis was carried out on DEGs using the Functional Annotation tool in DAVID 6.7 (Huang da et al., 2009a, Huang da et al., 2009b) under medium classification stringency for all default annotation categories except protein domains. Clustered annotation groups were considered significant when group enrichment score was ≥ 1.3, with higher scores indicative of more significant annotated GO terms.
Single-cell RNA-seq data processing
Cellranger analysis pipeline (version 2.2.0) was used to align sequenced reads to GRCh38 and aggregate output across all samples. Quality control and filtering was conducted with Seurat (version 2.3.0) (Butler et al., 2018) to retain cells with mitochondrial gene content < 5%, total genes expressed between 200 and 7000, as well as number of detected unique molecules < 50,000. 15,560 Day10 single cells, 6,337 Day12 single cells and 40,609 Day14 single cells remained following quality control and filtering. Gene expression measurements for retained cells were log-transformed, normalized by total expression per cell, and scaled to 10,000 molecules per cell. We regressed out non-biological interesting variation in gene expression that is driven by processing batch, number of detected molecules, and mitochondrial gene content. Highly variable genes across the single cells were then identified and principal components (PCs) analysis was conducted. 8 PCs were selected as input for t-Distributed Stochastic Neighbor Embedding (tSNE) for Day10, Day12, Day14 cells. We conducted unsupervised lineage trajectory analysis on a downsampled set of Day10, Day12, Day14 cells (6,251 cells, 10% of all cells) with Monocle2 (version 2.10.0) (Qiu et al., 2017, Trapnell et al., 2014). In brief, genes with mean expression value ≥ 0.05 were used to order cells along the trajectory. Significant differentially expressed genes (DEGs, q-value ≤ 1e-4) at branch points along the trajectory were identified using branched expression analysis modeling (BEAM) function of Monocle2. Significant branch specific DEGs were visualized in a branched heatmap to observe changes in both cell fates concurrently. Genes with similar lineage-dependent expression patterns were identified from the hierarchical clustering of branch specific DEGs and classified into up- or down-regulated branch specific DEGs. GO analysis was conducted on the identified up- or down-regulated branch specific DEGs to elucidate modulated biological processes at each branch point. tSNE and unsupervised lineage trajectory analysis were also conducted on the endothelial subcluster identified in Day10, Day12, Day14 (cluster 10) using the above-described method with the variation of 7 PCs selection as input for tSNE.
Correlation expression profile with human fetal kidney transcriptome
We conducted correlation analysis of our in vivo - Day 24 kidney organoids, in vivo – 2 weeks kidney organoid implants and in vivo – 4 weeks kidney organoid implants with human fetal kidney expression data from GSE100859 (Lindstrom et al., 2018b) that consisted of human kidney datasets at 9.5 week (3 replicates), 11 week (1 replicate), 13 week (2 replicates), 17 week (2 replicates) and 21 week (1 replicate). Pearson’s correlation was calculated with the cor R function and plotted as a square matrix with R package corrplot version 0.84 (Friendly, 2002, Murdoch and Chow, 1996) with significant correlated features (p ≤ 0.05) indicated by blue shaded squares.
Image quantification and statistical analysis
Organoids were stained with LTL (proximal tubule), CDH1 (distal tubule) and Nephrin (glomerulus). Whole mount confocal images were acquired and used to visualize different segments using different color channels. Images were processed and quantified using ImageJ (NIH). The scale of images was first standardized and converted to 8-bits to adjust the threshold of pixel intensity. The percentage of specific segments were calculated against the sum of three segments.
Supplementary Material
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| AQP1 (Aquaporin-1) | Santa Cruz | Cat. #SC-25287; RRID:AB_626694 |
| AQP2 (Aquaporin-2) | Santa Cruz | Cat. #SC-515770 |
| CALB1 (Calbidin-1) | Sigma | Cat. #C9848; RRID:AB_476894 |
| CDH1 (E-Cadherin) | BD Biosciences | Cat. #610182; RRID:AB_397581 |
| CDH6 (K-Cadherin) | Gift from Professor Gregory Dressler, University of Michigan | N/A |
| CD31 (PECAM1) | Abcam | Cat. #ab76533; RRID:AB_1523298 |
| CD31-PE (PECAM1) | BD Biosciences | Cat. #BD 555446; RRID:AB_395839 |
| FOXD1 | Abcam | Cat. #ab129324; RRID:AB_11150609 |
| CK8 (Cytokeratin-8) | Developmental Studies Hybridoma Bank | Cat. #TROMA-I-S |
| HOXD11 | Proteintech | Cat. #18734–1-AP |
| JAG1 (Jagged-1) | R&D Systems | Cat. #AF1277; RRID:AB_416699 |
| KDR (VEGFR2/CD309) | Biolegend | Cat. #359916; RRID:AB_2565928 |
| MEIS1 | Active Motif | Cat. #A25/39795; RRID:AB_2750570 |
| LHX1 | Developmental Studies Hybridoma Bank | Cat. #4F2-C; RRID:AB_531784 |
| LMN1 (Laminin) | Developmental Studies Hybridoma Bank | Cat. #2E8; RRID:AB_528343 |
| LRP2 (Megalin) | R&D Systems | Cat. #NB110–96417 |
| Lotus Tetragonolobus Lectin (LTL), Fluorescein labelled | Vector Laboratories | Cat. #FL-1321; RRID:AB_2336559 |
| MIXL1 | Proteintech | Cat. #22772–1-AP |
| NPHS1 (Nephrin) | Progen | Cat. #GP-N2; RRID:AB_1542487 |
| OCT4 | Santa Cruz | Cat. #sc-9081; RRID:AB_2167703 |
| PAX2 | Biolegend | Cat. #PRB-276P; RRID:AB_291611 |
| PAX8 | Proteintech | Cat. #10336–1-AP; RRID:AB_2236705 |
| 1PDGFRβ | R&D Systems | Cat. #AF385; RRID:AB_355339 |
| PODXL (Podocalyxin) | R&D Systems | Cat. #AF1658; RRID:AB_354920 |
| SALL1 | R&D Systems | Cat. #PP-K9814–00; RRID:AB_2183228 |
| SIX1 | Cell Signalling Technology | Cat. #D4A8K; RRID:AB_2753209 |
| SIX2 | Proteintech | Cat. #11562–1-AP; RRID:AB_2189084 |
| SOX2 | Millipore | Cat. #AB5603; RRID:AB_2286686 |
| T (Brachyury) | Developmental Studies Hybridoma Bank | Cat. #PCRP-T-1A2; RRID:AB_2722364 |
| WT1 | Santa-Cruz Biotechnology | Cat. #SC-7385; RRID: AB_628448 |
| WT1 | Abcam | Cat. #ab89901; RRID:AB_2273027 |
| Chicken anti-Rabbit IgG, Alexa Fluor 647 | Thermo Fisher Scientific | Cat. #A21443; RRID:AB_2535861 |
| Donkey anti-Rabbit IgG, Alexa Fluor 488 | Thermo Fisher Scientific | Cat. #A21206; RRID:AB_2535792 |
| Goat anti-Rabbit IgG, Alexa Fluor 405 | Thermo Fisher Scientific | Cat. #A31556; RRID:AB_221605 |
| Goat anti-Rabbit IgG, Alexa Fluor 594 | Thermo Fisher Scientific | Cat. #R37117; RRID:AB_2556545 |
| Donkey anti-Mouse IgG, Alexa Fluor 488 | Thermo Fisher Scientific | Cat. #A21202; RRID:AB_141607 |
| Donkey anti-Mouse IgG, Alexa Fluor 594 | Thermo Fisher Scientific | Cat. #A21203; RRID:AB_141633 |
| Donkey anti-Mouse IgG, Alexa Fluor 647 | Thermo Fisher Scientific | Cat. #A31571; RRID:AB_162542 |
| Goat anti-Mouse IgG, Alexa Fluor 633 | Thermo Fisher Scientific | Cat. #A21053; RRID:AB_1500753 |
| Goat anti-Mouse IgG, Alexa Fluor 405 | Thermo Fisher Scientific | Cat. #A31553; RRID:AB_221604 |
| Chicken anti-Goat IgG, Alexa Fluor 647 | Thermo Fisher Scientific | Cat. #A21469; RRID:AB_10374877 |
| Chicken anti-Goat IgG, Alexa Fluor 488 | Thermo Fisher Scientific | Cat. #A21467; RRID:AB_141893 |
| Donkey anti-Goat IgG, Alexa Fluor 594 | Thermo Fisher Scientific | Cat. #A11058; RRID:AB_142540 |
| Donkey anti-Goat IgG, Alexa Fluor 647 | Thermo Fisher Scientific | Cat. #A21447; RRID:AB_141844 |
| Goat anti-Guinea Pig IgG, Alexa Fluor 488 | Thermo Fisher Scientific | Cat. #A11073; RRID:AB_142018 |
| Goat anti-Guinea Pig IgG, Alexa Fluor 594 | Thermo Fisher Scientific | Cat. #A11076; RRID:AB_141930 |
| Chicken anti-Rat IgG, Alexa Fluor 647 | Thermo Fisher Scientific | Cat. #A21472; RRID:AB_1500700 |
| Donkey anti-Rat IgG, Alexa Fluor 594 | Thermo Fisher Scientific | Cat. #A21209; RRID:AB_2535795 |
| Biological Samples | ||
| Human fetal kidney samples | Human fetal kidney samples from donors 16–23 weeks of age were obtained from Kandang Kerbau Women’s and Children’s Hospital (KKH) with written consent from donors. SingHealth and National Health Care Group Research Ethics Committees Singapore specifically approved this study (CIRB Ref: 2012/064/F) | N/A |
| Chemicals, Peptides, and Recombinant Proteins | ||
| 8-Br-cAMP | Sigma-Aldrich | Cat. #B7880 |
| Activin A | Stemgent | Cat. #03–0001 |
| Axitinib | MedChemExpress (MCE) | Cat. #HY-10065 |
| Cediranib | MedChemExpress (MCE) | Cat. #HY-10205 |
| CFTR inhibitor 172 | MedChemExpress (MCE) | Cat. #HY-16671 |
| CHIR99021 | Sigma-Aldrich | Cat. #SML1046 |
| DAPI (6-diamidino-2-phenylindole dihydrochloride) | Sigma-Aldrich | Cat. #D9542 |
| Dextran, Alexa Fluor™ 647; 10,000 MW, Anionic, Fixable | Thermo Fisher Scientific | Cat. #D22914 |
| Dextran, Lucifer Yellow; 10,000 MW, Anionic, Lysine Fixable | Thermo Fisher Scientific | Cat. #D1825 |
| Dextran, Tetramethylrhodamine; 70,000 MW, Lysine Fixable | Thermo Fisher Scientific | Cat. #D1818 |
| Dextran, Tetramethylrhodamine; 2,000,000 MW, Lysine Fixable | Thermo Fisher Scientific | Cat. #D7139 |
| Forskolin | STEMCELL Technologies Inc. | Cat. #72114 |
| Growth Factor Reduced Matrigel | Corning | Cat. #354230 |
| Luciferin | Perkin Elmer | Cat. #122799 |
| Recombinant human Activin A | Stemgent, Inc. | Cat. #03–0001 |
| Recombinant human fibroblast growth factor 2 (FGF2) | Joint Protein Central | Cat. #L16F02001 |
| Recombinant human fibroblast growth factor 9 (FGF9) | PeproTech | Cat. #100–23 |
| Recombinant human Noggin | Joint Protein Central | Cat. #AA28–232 |
| Recombinant human Transforming Growth Factor-β (TGF- β) | Sigma-Aldrich | Cat. #H8541 |
| Thapsigargin | Santa Cruz | Cat. #SC-24017 |
| Tivozanib | MedChemExpress | Cat. #HY-10977 |
| Y-27632 dihydrochloride | Sigma-Aldrich | Cat. #72308 |
| Critical Commercial Assays | ||
| Chromium Single Cell A Chip Kit | 10x Genomics | Cat. #PN-1000009 |
| Chromium i7 Multiplex Kit | 10x Genomics | Cat. #PN-120262 |
| Click-iT® Edu Imaging Kit | Thermo Fisher Scientific | Cat. #C10339 |
| Direct-zol™ RNA Miniprep | Zymo Research | Cat. #R2050 |
| DuoSet ELISA Ancillary Reagent Kit 2 | R&D Systems | Cat. #DY008 |
| High Sensitivity DNA Kit | Agilent | Cat. #5067–4626 |
| Human VEGF DuoSet ELISA | R&D Systems | Cat. #DY293B |
| iScript™ cDNA Synthesis Kit | Bio-Rad | Cat. #1708841 |
| iQ™ SYBR® Green Supermix | Bio-Rad | Cat. #1725125 |
| PrimeSTAR GXL DNA Polymerase | Takara Bio Inc. | Cat. #R050B |
| TRIzol™ Reagent | Invitrogen | Cat. #15596018 |
| Chromium Single Cell 3’ Library & Gel Bead Kit v2 | 10x Genomics | Cat. #PN-120237 |
| Deposited Data | ||
| Raw/ analyzed bulk and single-cell RNA-sequencing data | This manuscript | GEO: GSE132026 |
| Mendeley Data | This manuscript | http://dx.doi.org/10.17632/mxpdwxgjy5.2 |
| Experimental Models: Cell Lines | ||
| Human embryonic stem cell H1 | WiCell | Cat. #WA01; RRID:CVCL_9771 |
| Human embryonic stem cell line H1 expressing Firefly Luciferase (H1-Luciferase) | From Professor Juan Carlos Izpisua Belmonte, Salk Institute | Cat. #WA01; RRID:CVCL_9771 |
| Human embryonic stem cell H9 | WiCell | Cat. #WA09; RRID:CVCL_9773 |
| ARPKD patient-derived and gene corrected iPSCs | From Professor Juan Carlos Izpisua Belmonte, Salk Institute | Cat. GM10287 (Coriell) (parental ARPKD patient fibroblasts); RRID:CVCL_V446 |
| HUVEC (human umbilical vein endothelial cells) | Lonza | Cat. #C2519A; RRID:CVCL_2959 |
| Experimental Models: Mouse strains | ||
| NOD.Cg-Prkdcscid Il2rgtm1Wjl/SzJ (NSG) | InVivos, Singapore | RRID:IMSR_JAX:005557 |
| BALB/cJInv (CD-1) | InVivos, Singapore | RRID:IMSR_JAX:000651 |
| Oligonucleotides | ||
| List of qRT-PCR Primers used in this study, see Mendeley data | Integrated DNA Technologies | http://dx.doi.org/10.17632/mxpdwxgjy5.2 |
| pCXLE-hOCT3/4-shp53-F | Addgene | Cat. #27077; RRID:Addgene_27077 |
| pCXLE-hOCT3/4 | Addgene | Cat. #27076; RRID:Addgene_27076 |
| pCXLE-hSK | Addgene | Cat. #27078; RRID:Addgene_27078 |
| pCXLE-hUL | Addgene | Cat. #27080; RRID:Addgene_27080 |
| pCAGmCherry-gRNA | Addgene | Cat. #87110; RRID:Addgene_87110 |
| pCas9_GFP | Addgene | Cat. #44719; RRID:Addgene_44719 |
| PKHD1 exon 65 donor oligo for gene correction | Valuegene (San Diego) | TGCTTCTTTTAAGCCAACAGCACACCAGACAGCTCAGAGCCAGCCATGAGGCCACAGAGG |
| PKHD1-ex65F | Integrated DNA Technologies | AAAGAAGCTGAAATTTTGCATTGGTATGGA |
| PKHD1-ex65R | Integrated DNA Technologies | ATTTGGCTTTGTGTAATTTTATATTTTAGAGAAGCTCA |
| Software and Algorithms | ||
| CellRanger v2.2.0 | 10x Genomics | https://support.10xgenomics.com/single-cell-gene-expression/software/downloads/latest |
| Corrplot v0.84 | (Friendly, 2002; Murdoch and Chow, 1996) | http://cran.r-project.org/package=corrplot. |
| Cuffdiff2 v2.2.1 | (Trapnell et al., 2013; Trapnell et al., 2010) | http://cole-trapnell-lab.github.io/cufflinks/cuffdiff/ ;RRID:SCR_001647 |
| Cuffnorm v2.2.1 | (Trapnell et al., 2013; Trapnell et al., 2010) | http://cole-trapnell-lab.github.io/cufflinks/cuffnorm/ |
| FlowJo | Tree Star | http://flowjo.com/solutions/flowjo; RRID:SCR_013673 |
| GraphPad Prism 7 | GraphPad | https://www.graphpad.com/ ; SCR_015807 |
| ImageJ | National Institutes of Health | http://imagej.nih.gov/ij/; RRID:SCR_003070 |
| Monocle2 v 2.10.0 | (Trapnell et al., 2014) | http://cole-trapnell-lab.github.io/monocle-release/; RRID:SCR_016339 |
| pheatmap v1.0.10 | CRAN | https://cran.r-project.org/web/packages/pheatmap/index.html; RRID:SCR_016418 |
| R 3.5.1 | R project | https://www.r-project.org; RRID:SCR_001905 |
| RSEM v1.3.0 | (Li & Dewey, 2011) | https://github.com/deweylab/RSEM; RRID:SCR_013027 |
| Seurat v2.2.0 | (Butler, Hoffman, Smibert, Papalexi, & Satija, 2018) | http://satijalab.org/seurat/; RRID:SCR_007322 |
| STAR v2.5.2a | (Dobin et al., 2013) | https://github.com/alexdobin/STAR; RRID:SCR_015899 |
| ZEN Lite | Carl Zeiss | https://www.zeiss.com/microscopy/int/products/microscope-software/zen-lite.html; RRID:SCR_013672 |
| Other | ||
| Accutase® | STEMCELL Technologies Inc. | Cat. #07920 |
| Advanced RPMI 1640 | Thermo Fisher Scientific | Cat. #12633020 |
| Clearene Solvent | Leica Biosystems | Cat. #3803600 |
| DEPEX Mounting Media | Electron Microscopy Sciences | Cat. #OT10B |
| Dispase | Invitrogen | Cat. #17105–041 |
| DMEM/F-12 | Thermo Fisher Scientific | Cat. #12400024 |
| EGM-2 (Endothelial Cell Growth Media) | Lonza | Cat. #38210090 |
| Eosin | Leica Biosystems | Cat. #HT1103128 |
| Fetal Bovine Serum (FBS) | Thermo Fisher Scientific | Cat. #10270106 |
| Glutaraldehyde | Electron Microscopy Services | Cat. #16020 |
| Fluoromount-G® Mounting Medium | SouthernBiotech | Cat. #0100–01 |
| GlutaMAX Supplement | Thermo Fisher Scientific | Cat. #35050–061 |
| Lead nitrate | Nacalai Tesque | Cat. #20230–12 |
| MEM | Thermo Fisher Scientific | Cat. #31985070 |
| Non-Essential Amino Acids | Thermo Fisher Scientific | Cat. #11140–050 |
| Normocin | Invivogen | Cat. #ant-nr-1 |
| Osmium tetroxide | Electron Microscopy Services | Cat. #19152 |
| Phosphate buffer pH 7.0 | Electron Microscopy Services | Cat. #19340–70 |
| Poly/Bed 812 – BDMA embedding kit | Polysciences Inc. | Cat. #21844–1 |
| Potassium ferrocyanide | Electron Microscopy Services | Cat. #20150 |
| SPRIselect Reagent Kit | Beckman Coulter | Cat. #B23318 |
| Surgipath Paraplast® | Leica Biosystems | Cat. #39601006 |
| Tissue-Tek® O.C.T. Compound, Sakura® Finetek | VWR | Cat. #4583 |
| Uranyl acetate | Electron Microscopy Services | Cat. #2240 |
Highlights.
Modulation of WNT generates vascularized and segmentally patterned kidney organoids
Single-cell analysis identifies a non-conventional origin for renal vasculature
Structural and functional maturation of kidney organoids occurs upon implantation
Cystic kidney organoids from ARPKD iPSCs enable personalized drug validation
ACKNOWLEDGEMENTS
We thank Drs. D. Luo, I. Liu, N. Montserrat, and D. Virshup for insightful suggestions; Chyze W. Ang, M. Schwarz, P. Schwarz, and N. Zheng for administrative help. We thank Drs. L. de Onate and H.K. Liao for technical help. We thank technical support from Next Generation Sequencing Platform and Single-Cell Omics Centre, Genome Institute of Singapore; Electron Microscopy Unit, Yong Loo Lin School of Medicine, National University of Singapore; and Advanced Bio Imaging facility of Singhealth/Duke-NUS. We thank technical assistance from 10X Genomics and NovogeneAIT Genomics Singapore Pte Ltd. Work in the laboratory of Y. X. was supported by Nanyang Assistant Professorship Grant, Lee Kong Chian School of Medicine, Nanyang Technological University Singapore; Singapore Ministry of Education grant (2017-T1–001-086); Singapore National Medical Research Council (NMRC/OFIRG/0076/2018 and NMRC/OFLCG/001/2017); NTU internal grant (JNT-14/2016-CG). J.H.L was supported by a HealthTech NTU scholarship. Work in the laboratory of J.N.F. was supported by Singapore National Research Foundation Fellowship (NRF-NRFF2016–03). Work in the laboratory of J.C.I.B was supported by The G. Harold and Leila Y. Mathers Charitable Foundation; The NIH (5 R21 AG055938); The Leona M. and Harry B. Helmsley Charitable Trust and Universidad Católica San Antonio de Murcia. K.S. was supported by Takeda Science Foundation and Mochida Memorial Foundation. Work in the laboratory of M.F. was supported by a Start Up Grant of Nanyang Technological University Singapore (M4081915); NTU-Harvard SusNano (M4082370.030). C.C.K. is supported by Singapore National Research Foundation Investigatorship (NRF-NRFI2018–01). Q.C. is supported by Singapore National Research Foundation Fellowship (NRF-NRFF2017–03).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
DECLARATION OF INTERESTS
The authors declare no competing interests.
SUPPLEMENTAL INFORMATION
Supplemental information includes seven figures, seven tables and supplemental methods, which can be found with this article online.
DATA AND SOFTWARE AVAILABILITY
The accession number for the RNA sequencing data reported in this paper is NCBI GEO: GSE132026. Additional figures can be assessed at Mendeley Data: http://dx.doi.org/10.17632/mxpdwxgjy5.2.
REFERENCES
- ABRAHAMSON DR, ROBERT B, HYINK DP, ST JOHN PL & DANIEL TO 1998. Origins and formation of microvasculature in the developing kidney. Kidney Int Suppl, 67, S7–11. [DOI] [PubMed] [Google Scholar]
- BANTOUNAS I, RANJZAD P, TENGKU F, SILAJDZIC E, FORSTER D, ASSELIN MC, LEWIS P, LENNON R, PLAGGE A, WANG Q, WOOLF AS & KIMBER SJ 2018. Generation of Functioning Nephrons by Implanting Human Pluripotent Stem Cell-Derived Kidney Progenitors. Stem Cell Reports, 10, 766–779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BARASCH J, YANG J, WARE CB, TAGA T, YOSHIDA K, ERDJUMENT-BROMAGE H, TEMPST P, PARRAVICINI E, MALACH S, ARANOFF T & OLIVER JA 1999. Mesenchymal to epithelial conversion in rat metanephros is induced by LIF. Cell, 99, 377–86. [DOI] [PubMed] [Google Scholar]
- BIRN H & CHRISTENSEN EI 2006. Renal albumin absorption in physiology and pathology. Kidney Int, 69, 440–9. [DOI] [PubMed] [Google Scholar]
- BRUNSKILL EW, GEORGAS K, RUMBALLE B, LITTLE MH & POTTER SS 2011. Defining the molecular character of the developing and adult kidney podocyte. PLoS One, 6, e24640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BUTLER A, HOFFMAN P, SMIBERT P, PAPALEXI E & SATIJA R 2018. Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat Biotechnol, 36, 411–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CHENG HT, KIM M, VALERIUS MT, SURENDRAN K, SCHUSTER-GOSSLER K, GOSSLER A, MCMAHON AP & KOPAN R 2007. Notch2, but not Notch1, is required for proximal fate acquisition in the mammalian nephron. Development, 134, 801–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CRUZ NM, SONG X, CZERNIECKI SM, GULIEVA RE, CHURCHILL AJ, KIM YK, WINSTON K, TRAN LM, DIAZ MA, FU H, FINN LS, PEI Y, HIMMELFARB J & FREEDMAN BS 2017. Organoid cystogenesis reveals a critical role of microenvironment in human polycystic kidney disease. Nat Mater, 16, 1112–1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CZERNIECKI SM, CRUZ NM, HARDER JL, MENON R, ANNIS J, OTTO EA, GULIEVA RE, ISLAS LV, KIM YK, TRAN LM, MARTINS TJ, PIPPIN JW, FU H, KRETZLER M, SHANKLAND SJ, HIMMELFARB J, MOON RT, PARAGAS N & FREEDMAN BS 2018. High-Throughput Screening Enhances Kidney Organoid Differentiation from Human Pluripotent Stem Cells and Enables Automated Multidimensional Phenotyping. Cell Stem Cell, 22, 929–940 e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DAVIDOW CJ, MASER RL, ROME LA, CALVET JP & GRANTHAM JJ 1996. The cystic fibrosis transmembrane conductance regulator mediates transepithelial fluid secretion by human autosomal dominant polycystic kidney disease epithelium in vitro. Kidney Int, 50, 208–18. [DOI] [PubMed] [Google Scholar]
- DE VAL S & BLACK BL 2009. Transcriptional control of endothelial cell development. Dev Cell, 16, 180–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DOBIN A, DAVIS CA, SCHLESINGER F, DRENKOW J, ZALESKI C, JHA S, BATUT P, CHAISSON M & GINGERAS TR 2013. STAR: ultrafast universal RNA-seq aligner. Bioinformatics, 29, 15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- EREMINA V, SOOD M, HAIGH J, NAGY A, LAJOIE G, FERRARA N, GERBER HP, KIKKAWA Y, MINER JH & QUAGGIN SE 2003. Glomerular-specific alterations of VEGF-A expression lead to distinct congenital and acquired renal diseases. J Clin Invest, 111, 707–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- FREEDMAN BS, BROOKS CR, LAM AQ, FU H, MORIZANE R, AGRAWAL V, SAAD AF, LI MK, HUGHES MR, WERFF RV, PETERS DT, LU J, BACCEI A, SIEDLECKI AM, VALERIUS MT, MUSUNURU K, MCNAGNY KM, STEINMAN TI, ZHOU J, LEROU PH & BONVENTRE JV 2015. Modelling kidney disease with CRISPR-mutant kidney organoids derived from human pluripotent epiblast spheroids. Nat Commun, 6, 8715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- FRIENDLY M 2002. Corrgrams: Exploratory displays for correlation matrices. The American Statistician, 56, 316–324. [Google Scholar]
- GARRETA E, PRADO P, TARANTINO C, ORIA R, FANLO L, MARTI E, ZALVIDEA D, TREPAT X, ROCA-CUSACHS P, GAVALDA-NAVARRO A, COZZUTO L, CAMPISTOL JM, IZPISUA BELMONTE JC, HURTADO DEL POZO C & MONTSERRAT N 2019. Fine tuning the extracellular environment accelerates the derivation of kidney organoids from human pluripotent stem cells. Nat Mater, 18, 397–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GUNAY-AYGUN M, AVNER ED, BACALLAO RL, CHOYKE PL, FLYNN JT, GERMINO GG, GUAY-WOODFORD L, HARRIS P, HELLER T, INGELFINGER J, KASKEL F, KLETA R, LARUSSO NF, MOHAN P, PAZOUR GJ, SHNEIDER BL, TORRES VE, WILSON P, ZAK C, ZHOU J & GAHL WA 2006. Autosomal recessive polycystic kidney disease and congenital hepatic fibrosis: summary statement of a first National Institutes of Health/Office of Rare Diseases conference. J Pediatr, 149, 159–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HANAOKA K, DEVUYST O, SCHWIEBERT EM, WILSON PD & GUGGINO WB 1996. A role for CFTR in human autosomal dominant polycystic kidney disease. Am J Physiol, 270, C389–99. [DOI] [PubMed] [Google Scholar]
- HARRIS PC & TORRES VE 2009. Polycystic kidney disease. Annu Rev Med, 60, 321–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HARRIS PC & TORRES VE 2014. Genetic mechanisms and signaling pathways in autosomal dominant polycystic kidney disease. J Clin Invest, 124, 2315–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HOMAN KA, GUPTA N, KROLL KT, KOLESKY DB, SKYLAR-SCOTT M, MIYOSHI T, MAU D, VALERIUS MT, FERRANTE T, BONVENTRE JV, LEWIS JA & MORIZANE R 2019. Flow-enhanced vascularization and maturation of kidney organoids in vitro. Nat Methods, 16, 255–262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HORSTER M 2000. Embryonic epithelial membrane transporters. Am J Physiol Renal Physiol, 279, F982–96. [DOI] [PubMed] [Google Scholar]
- HU Y, LI M, GOTHERT JR, GOMEZ RA & SEQUEIRA-LOPEZ ML 2016. Hemovascular Progenitors in the Kidney Require Sphingosine-1-Phosphate Receptor 1 for Vascular Development. J Am Soc Nephrol, 27, 1984–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HUANG DA W, SHERMAN BT & LEMPICKI RA 2009a. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res, 37, 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HUANG DA W, SHERMAN BT & LEMPICKI RA 2009b. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc, 4, 44–57. [DOI] [PubMed] [Google Scholar]
- JAMES RG & SCHULTHEISS TM 2005. Bmp signaling promotes intermediate mesoderm gene expression in a dose-dependent, cell-autonomous and translation-dependent manner. Dev Biol, 288, 113–25. [DOI] [PubMed] [Google Scholar]
- KORTENOEVEN ML, PEDERSEN NB, MILLER RL, ROJEK A & FENTON RA 2013. Genetic ablation of aquaporin-2 in the mouse connecting tubules results in defective renal water handling. J Physiol, 591, 2205–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LI Z, ARAOKA T, WU J, LIAO HK, LI M, LAZO M, ZHOU B, SUI Y, WU MZ, TAMURA I, XIA Y, BEYRET E, MATSUSAKA T, PASTAN I, RODRIGUEZ ESTEBAN C, GUILLEN I, GUILLEN P, CAMPISTOL JM & IZPISUA BELMONTE JC 2016. 3D Culture Supports Long-Term Expansion of Mouse and Human Nephrogenic Progenitors. Cell Stem Cell, 19, 516–529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LINDSTROM NO, DE SENA BRANDINE G, TRAN T, RANSICK A, SUH G, GUO J, KIM AD, PARVEZ RK, RUFFINS SW, RUTLEDGE EA, THORNTON ME, GRUBBS B, MCMAHON JA, SMITH AD & MCMAHON AP 2018a. Progressive Recruitment of Mesenchymal Progenitors Reveals a Time-Dependent Process of Cell Fate Acquisition in Mouse and Human Nephrogenesis. Dev Cell, 45, 651–660 e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LINDSTROM NO, GUO J, KIM AD, TRAN T, GUO Q, DE SENA BRANDINE G, RANSICK A, PARVEZ RK, THORNTON ME, BASKIN L, GRUBBS B, MCMAHON JA, SMITH AD & MCMAHON AP 2018b. Conserved and Divergent Features of Mesenchymal Progenitor Cell Types within the Cortical Nephrogenic Niche of the Human and Mouse Kidney. J Am Soc Nephrol, 29, 806–824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LINDSTROM NO, GUO J, KIM AD, TRAN T, GUO Q, DE SENA BRANDINE G, RANSICK A, PARVEZ RK, THORNTON ME, BASKING L, GRUBBS B, MCMAHON JA, SMITH AD & MCMAHON AP 2018c. Conserved and Divergent Features of Mesenchymal Progenitor Cell Types within the Cortical Nephrogenic Niche of the Human and Mouse Kidney. J Am Soc Nephrol, 29, 806–824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LINDSTROM NO, LAWRENCE ML, BURN SF, JOHANSSON JA, BAKKER ER, RIDGWAY RA, CHANG CH, KAROLAK MJ, OXBURGH L, HEADON DJ, SANSOM OJ, SMITS R, DAVIES JA & HOHENSTEIN P 2015. Integrated beta-catenin, BMP, PTEN, and Notch signalling patterns the nephron. Elife, 3, e04000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LINDSTROM NO, MCMAHON JA, GUO J, TRAN T, GUO Q, RUTLEDGE E, PARVEZ RK, SARIBEKYAN G, SCHULER RE, LIAO C, KIM AD, ABDELHALIM A, RUFFINS SW, THORNTON ME, BASKING L, GRUBBS B, KESSELMAN C & MCMAHON AP 2018d. Conserved and Divergent Features of Human and Mouse Kidney Organogenesis. J Am Soc Nephrol, 29, 785–805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LINDSTROM NO, TRAN T, GUO J, RUTLEDGE E, PARVEZ RK, THORNTON ME, GRUBBS B, MCMAHON JA & MCMAHON AP 2018e. Conserved and Divergent Molecular and Anatomic Features of Human and Mouse Nephron Patterning. J Am Soc Nephrol, 29, 825–840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MENON R, OTTO EA, KOKORUDA A, ZHOU J, ZHANG Z, YOON E, CHEN YC, TROYANSKAYA O, SPENCE JR, KRETZLER M & CEBRIAN C 2018. Single-cell analysis of progenitor cell dynamics and lineage specification in the human fetal kidney. Development, 145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MILLS CG, LAWRENCE ML, MUNRO DAD, ELHENDAWI M, MULLINS JJ & DAVIES JA 2017. Asymmetric BMP4 signalling improves the realism of kidney organoids. Sci Rep, 7, 14824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MORIZANE R, LAM AQ, FREEDMAN BS, KISHI S, VALERIUS MT & BONVENTRE JV 2015. Nephron organoids derived from human pluripotent stem cells model kidney development and injury. Nat Biotechnol, 33, 1193–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MUGFORD JW, SIPILA P, MCMAHON JA & MCMAHON AP 2008. Osr1 expression demarcates a multi-potent population of intermediate mesoderm that undergoes progressive restriction to an Osr1-dependent nephron progenitor compartment within the mammalian kidney. Dev Biol, 324, 88–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MUNRO DAD, HOHENSTEIN P & DAVIES JA 2017. Cycles of vascular plexus formation within the nephrogenic zone of the developing mouse kidney. Sci Rep, 7, 3273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MURDOCH D & CHOW E 1996. A graphical display of large correlation matrices. The American Statistician, 50, 178–180. [Google Scholar]
- NAKANISHI K, SWEENEY WE JR., ZERRES K, GUAY-WOODFORD LM & AVNER ED 2000. Proximal tubular cysts in fetal human autosomal recessive polycystic kidney disease. J Am Soc Nephrol, 11, 760–3. [DOI] [PubMed] [Google Scholar]
- OKITA K, MATSUMURA Y, SATO Y, OKADA A, MORIZANE A, OKAMOTO S, HONG H, NAKAGAWA M, TANABE K, TEZUKA K, SHIBATA T, KUNISADA T, TAKAHASHI M, TAKAHASHI J, SAJI H & YAMANAKA S 2011. A more efficient method to generate integration-free human iPS cells. Nat Methods, 8, 409–12. [DOI] [PubMed] [Google Scholar]
- PARK J, SHRESTHA R, QIU C, KONDO A, HUANG S, WERTH M, LI M, BARASCH J & SUSZTAK K 2018. Single-cell transcriptomics of the mouse kidney reveals potential cellular targets of kidney disease. Science, 360, 758–763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- PHIPSON B, ER PX, COMBES AN, FORBES TA, HOWDEN SE, ZAPPIA L, YEN HJ, LAWLOR KT, HALE LJ, SUN J, WOLVETANG E, TAKASATO M, OSHLACK A & LITTLE MH 2019. Evaluation of variability in human kidney organoids. Nat Methods, 16, 79–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- PRZEPIORSKI A, SANDER V, TRAN T, HOLLYWOOD JA, SORRENSON B, SHIH JH, WOLVETANG EJ, MCMAHON AP, HOLM TM & DAVIDSON AJ 2018. A Simple Bioreactor-Based Method to Generate Kidney Organoids from Pluripotent Stem Cells. Stem Cell Reports [DOI] [PMC free article] [PubMed]
- QIU X, MAO Q, TANG Y, WANG L, CHAWLA R, PLINER HA & TRAPNELL C 2017. Reversed graph embedding resolves complex single-cell trajectories. Nat Methods, 14, 979–982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- RISAU W 1998. Development and differentiation of endothelium. Kidney Int Suppl, 67, S3–6. [DOI] [PubMed] [Google Scholar]
- SCHNEIDER J, ARRAF AA, GRINSTEIN M, YELIN R & SCHULTHEISS TM 2015. Wnt signaling orients the proximal-distal axis of chick kidney nephrons. Development, 142, 2686–95. [DOI] [PubMed] [Google Scholar]
- SHARMIN S, TAGUCHI A, KAKU Y, YOSHIMURA Y, OHMORI T, SAKUMA T, MUKOYAMA M, YAMAMOTO T, KURIHARA H & NISHINAKAMURA R 2016. Human Induced Pluripotent Stem Cell-Derived Podocytes Mature into Vascularized Glomeruli upon Experimental Transplantation. J Am Soc Nephrol, 27, 1778–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- TAGUCHI A, KAKU Y, OHMORI T, SHARMIN S, OGAWA M, SASAKI H & NISHINAKAMURA R 2014. Redefining the in vivo origin of metanephric nephron progenitors enables generation of complex kidney structures from pluripotent stem cells. Cell Stem Cell, 14, 53–67. [DOI] [PubMed] [Google Scholar]
- TAGUCHI A & NISHINAKAMURA R 2017. Higher-Order Kidney Organogenesis from Pluripotent Stem Cells. Cell Stem Cell, 21, 730–746 e6. [DOI] [PubMed] [Google Scholar]
- TAKAKURA A, CONTRINO L, ZHOU X, BONVENTRE JV, SUN Y, HUMPHREYS BD & ZHOU J 2009. Renal injury is a third hit promoting rapid development of adult polycystic kidney disease. Hum Mol Genet, 18, 2523–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- TAKASATO M, ER PX, BECROFT M, VANSLAMBROUCK JM, STANLEY EG, ELEFANTY AG & LITTLE MH 2014. Directing human embryonic stem cell differentiation towards a renal lineage generates a self-organizing kidney. Nat Cell Biol, 16, 118–26. [DOI] [PubMed] [Google Scholar]
- TAKASATO M, ER PX, CHIU HS, MAIER B, BAILLIE GJ, FERGUSON C, PARTON RG, WOLVETANG EJ, ROOST MS, CHUVA DE SOUSA LOPES SM & LITTLE MH 2015. Kidney organoids from human iPS cells contain multiple lineages and model human nephrogenesis. Nature, 526, 564–8. [DOI] [PubMed] [Google Scholar]
- TORRES VE & HARRIS PC 2006. Mechanisms of Disease: autosomal dominant and recessive polycystic kidney diseases. Nat Clin Pract Nephrol, 2, 40–55; quiz 55. [DOI] [PubMed] [Google Scholar]
- TORRES VE & HARRIS PC 2014. Strategies targeting cAMP signaling in the treatment of polycystic kidney disease. J Am Soc Nephrol, 25, 18–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- TRAPNELL C, CACCHIARELLI D, GRIMSBY J, POKHAREL P, LI S, MORSE M, LENNON NJ, LIVAK KJ, MIKKELSEN TS & RINN JL 2014. The dynamics and regulators of cell fate decisions are revealed by pseudotemporal ordering of single cells. Nat Biotechnol, 32, 381–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- TRAPNELL C, HENDRICKSON DG, SAUVAGEAU M, GOFF L, RINN JL & PACHTER L 2013. Differential analysis of gene regulation at transcript resolution with RNA-seq. Nat Biotechnol, 31, 46–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- TRAPNELL C, WILLIAMS BA, PERTEA G, MORTAZAVI A, KWAN G, VAN BAREN MJ, SALZBERG SL, WOLD BJ & PACHTER L 2010. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nature biotechnology, 28, 511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- UNBEKANDT M & DAVIES JA 2010. Dissociation of embryonic kidneys followed by reaggregation allows the formation of renal tissues. Kidney Int, 77, 407–16. [DOI] [PubMed] [Google Scholar]
- VAN DEN BERG CW, RITSMA L, AVRAMUT MC, WIERSMA LE, VAN DEN BERG BM, LEUNING DG, LIEVERS E, KONING M, VANSLAMBROUCK JM, KOSTER AJ, HOWDEN SE, TAKASATO M, LITTLE MH & RABELINK TJ 2018. Renal Subcapsular Transplantation of PSC-Derived Kidney Organoids Induces Neo-vasculogenesis and Significant Glomerular and Tubular Maturation In Vivo. Stem Cell Reports, 10, 751–765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WANG P, CHEN Y, YONG J, CUI Y, WANG R, WEN L, QIAO J & TANG F 2018. Dissecting the Global Dynamic Molecular Profiles of Human Fetal Kidney Development by Single-Cell RNA Sequencing. Cell Rep, 24, 3554–3567 e3. [DOI] [PubMed] [Google Scholar]
- WANG S & DONG Z 2016. Environmental hit on a genetic basis in polycystic kidney disease. Am J Physiol Renal Physiol, 311, F1358–F1359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WOOLLARD JR, PUNYASHTITI R, RICHARDSON S, MASYUK TV, WHELAN S, HUANG BQ, LAGER DJ, VANDEURSEN J, TORRES VE, GATTONE VH, LARUSSO NF, HARRIS PC & WARD CJ 2007. A mouse model of autosomal recessive polycystic kidney disease with biliary duct and proximal tubule dilatation. Kidney Int, 72, 328–36. [DOI] [PubMed] [Google Scholar]
- WU H, UCHIMURA K, DONNELLY EL, KIRITA Y, MORRIS SA & HUMPHREYS BD 2018. Comparative Analysis and Refinement of Human PSC-Derived Kidney Organoid Differentiation with Single-Cell Transcriptomics. Cell Stem Cell, 23, 869–881 e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- XIA Y, NIVET E, SANCHO-MARTINEZ I, GALLEGOS T, SUZUKI K, OKAMURA D, WU MZ, DUBOVA I, ESTEBAN CR, MONTSERRAT N, CAMPISTOL JM & IZPISUA BELMONTE JC 2013. Directed differentiation of human pluripotent cells to ureteric bud kidney progenitor-like cells. Nat Cell Biol, 15, 1507–15. [DOI] [PubMed] [Google Scholar]
- XIA Y, SANCHO-MARTINEZ I, NIVET E, RODRIGUEZ ESTEBAN C, CAMPISTOL JM & IZPISUA BELMONTE JC 2014. The generation of kidney organoids by differentiation of human pluripotent cells to ureteric bud progenitor-like cells. Nat Protoc, 9, 2693–704. [DOI] [PubMed] [Google Scholar]
- YANG B, SONAWANE ND, ZHAO D, SOMLO S & VERKMAN AS 2008. Small-molecule CFTR inhibitors slow cyst growth in polycystic kidney disease. J Am Soc Nephrol, 19, 1300–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.