Abstract
The kidney is one of the most complex organs composed of multiple cell types, functioning to maintain homeostasis via the filtering of metabolic wastes, balancing of blood electrolytes, and adjustment of blood pressure. Recent advances in 3D culture technologies in vitro enabled the generation of ‘organoids’ which mimic the structure and function of in vivo organs. Organoid technology has allowed for new insights into human organ development and human pathophysiology, with great potential for translational research. Increasing evidence shows that kidney organoids are a useful platform for disease modeling of genetic kidney diseases when derived from genetic patient iPSCs and/or CRISPR-mutated stem cells. Though single cell RNA-seq studies highlight the technical difficulties underlying kidney organoid generation reproducibility and variation in differentiation protocols, kidney organoids still hold great potential to understand kidney pathophysiology as applied to kidney injury and fibrosis. In this review, we summarize various studies of kidney organoids, disease modeling, genome-editing, and bioengineering, and additionally discuss the potential of and current challenges to kidney organoid research.
Keywords: pluripotent stem cell, kidney, organoid, nephron, disease modeling, regeneration
Introduction
Experimental animal models have been used extensively in the field of biomedical research to evaluate drug efficacy and toxicity, to elucidate the underlying mechanisms of varied human diseases, and to determine the role of genetic and molecular factors in developmental processes. On the other hand, these animal models often require specialized facilities, equipment and training, and are time consuming, labor intensive, low throughput and financially costly. In addition, studies suggest these models have distinct limitations in their ability to capture human pathophysiology (Mestas and Hughes, 2004). Although the human genome has many similarities to the genomes of other species, there are still key differences in gene expression patterns, DNA variations, and the relationships between genes and disease phenotypes that animal models fail to address (Goh et al., 2007; Kraja et al., 2008; Ravasi et al., 2010).
The advent of in vitro 3D culture technologies provides great potential for translational research tools. Termed ‘organoids,’ these technologies mimic in vivo organs both structurally and functionally, leading to new insights into the understanding of human organ development and human pathology. Organoid technologies have six defining features. Organoids are: (i) a collection of organ-specific cell types derived from stem cells, (ii) with complex multicellular constructs, (iii) that are self-organized, (iv) into a 3D structure, (v) in an in vitro culture system, (vi) that can recapitulate in vivo developmental programs (Lancaster and Knoblich, 2014; Morizane and Bonventre, 2017b). There are two major approaches to generate organoids: (i) generation from tissue stem cells, and (ii) generation from pluripotent stem cells (PSCs) including embryonic stem cells (ESCs) and induced pluripotent stem cells (iPSCs) (Fatehullah et al., 2016).
Organoids can be generated from tissue stem cells by mimicking the in vivo niche environment characteristic of physiological tissue self-renewal or repair processes. In 2009, Clevers and his colleagues first reported that intestinal organoids were developed from Lgr5 (leucine-rich repeat-containing G protein-coupled receptor 5)-positive stem cells by mimicking the in vivo niche environment (Sato et al., 2009). With this as a starting point, several groups reported the generation of organoids derived from mouse and/or human primary tissue stem cells including intestine (Finkbeiner et al., 2012; Schwank et al., 2013; Drost et al., 2015), colon (Sato et al., 2011; Yui et al., 2012; Matano et al., 2015; van de Wetering et al., 2015), liver (Huch et al., 2013b; Huch et al., 2015; Broutier et al., 2016), prostate (Gao et al., 2014; Karthaus et al., 2014; Drost et al., 2016), lung (Rock et al., 2009), pancreas (Huch et al., 2013a; Boj et al., 2015), breast (Williams et al., 2016), ovary (Kessler et al., 2015), stomach (Barker et al., 2010; Bartfeld et al., 2015; Bartfeld and Clevers, 2015), esophagus (Sato et al., 2011), and lingual (Hisha et al., 2013). Most of these primary tissue organoids were generated from Lgr5 positive stem cells using Wnt activators (Fatehullah et al., 2016). These organoids have not been reconstructed by dissociation and reaggregation of the primary tissue, but instead have been generated from the stem cell population residing within the tissue.
PSCs are defined by an unlimited capacity for self-renewal and the ability to differentiate into cells of the three germ layers, namely the mesoderm, ectoderm, and endoderm (Thomson et al., 1998; Takahashi et al., 2007). Wnt, fibroblast growth factor (FGF), retinoic acid (RA), and transforming growth factor (TGFβ) are the main signal pathways that induce germ layer formation and the organ primordia development (Zorn and Wells, 2009; Lippmann et al., 2015). PSCs can be transformed into particular organ tissues with a high degree of efficiency through directed differentiation, the sequential application of these growth factors and signals at specific combinations and concentrations for defined periods of time. In 2008, Sasai and his colleagues first generated self-organized layered structures resembling the cerebral cortex from human PSCs (hPSCs) by using SFEBq (serum-free floating culture of embryoid body-like aggregates with quick reaggregation) method (Eiraku et al., 2008). There are currently several published studies in which PSCs are differentiated into organoids of various organs including intestine (Spence et al., 2011; Forbester et al., 2015; Leslie et al., 2015), liver (Takebe et al., 2013), lung (Huang et al., 2014; Dye et al., 2015; Firth et al., 2015), brain (Lancaster et al., 2013; Jo et al., 2016), pancreas (Huang et al., 2015b; Hohwieler et al., 2017), stomach (McCracken et al., 2014; Huang et al., 2015a), and kidneys (Taguchi et al., 2014; Freedman et al., 2015; Morizane et al., 2015; Takasato et al., 2015; Hiratsuka et al., 2019).
Kidneys are complex organs that are responsible for many functions, including the filtering waste products and minerals from blood, maintenance of fluid and acid-base balance, and producing the erythropoietin that is essential for production of red blood cells. The functional unit of the kidney is called a nephron. There are approximately one million nephrons in one human kidney. Each nephron consists of a glomerulus, which consists of capillaries and podocytes responsible for the filtration of blood, and a multi-segmented tubule composed of a single layer of epithelial cells responsible for selective reabsorption and secretion of a large number of solutes. Loss of functional nephrons and the development of tubulointerstitial fibrosis contribute to the progression of chronic kidney disease (CKD), which affects 9–14% of the US adult population and was ranked one of the top causes of death globally (Coresh et al., 2007; Ene-Iordache et al., 2016). While adult kidneys possess an intrinsic limited capacity to self-repair following injury (Humphreys et al., 2008), the process of nephrogenesis, the formation of new nephrons, is limited to the period of embryonic development in humans (Little and McMahon, 2012). Thus, loss of nephrons is permanent and ultimately leads to end-stage kidney disease (ESKD), where patients require renal replacement therapies, such as hemodialysis, associated with high mortality and morbidity (Collins et al., 2010). In addition, there are limited human disease models for the development of treatment methods and identification of biomarkers for the vast majority of kidney diseases. Kidney organoids represent valuable platforms for the study of human pathophysiology and the discovery of novel therapeutics, providing potential sources for the generation of new nephrons in CKD/ESKD patients. Here, we summarize the current literature describing kidney organoid applications to disease modeling and regenerative medicine, and discuss the potential of and challenges to kidney organoid research.
Disease Modeling of Genetic Kidney Diseases
hiPSCs derived from patients with genetic diseases have been used as in vitro patient-specific models to study the pathophysiology of genetic diseases like Duchenne and Becker muscular dystrophy, Parkinson’s disease, Huntington’s disease, and Down syndrome/trisomy 21, for discovery of new therapeutic approaches (Dimos et al., 2008; Park et al., 2008; Ebert et al., 2009; Soldner et al., 2009). One of the advantages of using patient-derived iPSCs for disease modeling is that they would make it possible to address the symptomatic variability of human diseases with precision medicine. Moreover, hiPSCs are compatible with genome editing technologies, including clustered regularly interspaced short palindromic repeat (CRISPR)/ CRISPR associated protein 9 (Cas9) genome editing. The CRISPR/Cas9 system is a unique technology that enables the correcting of genetic variants or insertion of specific mutations at any site within the genome. CRISPR/Cas9 elements consist of two key molecules to introduce a mutation into the DNA (Cong et al., 2013; Shen et al., 2014). One is an enzyme called Cas9, which acts as a pair of molecular scissors that can cut the two strands of DNA at a specific location in the genome so that bits of DNA can then be added or removed. The other is a piece of RNA called guide RNA (gRNA), consisting of a small piece of pre-designed RNA sequence located within a longer RNA scaffold. The scaffold part binds to DNA and the pre-designed sequence guides Cas9 to the target site of the genome. CRISPR/Cas9 technology has been recently applied to disease-focused research to both i) directly introduce genetic mutations in the causative genes of hPSCs to generate disease models, which allows un-edited cells to serve as isogenic controls, and ii) correct causative genes in patient-derived iPSCs. Recent advances in genome editing using CRISPR/Cas9 provide new approaches to modeling genetic diseases using hPSCs in vitro, including Barth syndrome, Duchenne muscular dystrophy, hemophilia, β-Thalassemia, and cystic fibrosis (Schwank et al., 2013; Wang et al., 2014; Xie et al., 2014; Li et al., 2015a; Park et al., 2015; Xu et al., 2015).
There are currently many ongoing studies to model genetic kidney diseases using kidney organoid systems with patient-derived or CRISPR/Cas9-mediated hPSCs. Kidney-related studies with iPSCs and organoids have focused on polycystic kidney disease (PKD), including autosomal recessive PKD (ARPKD) and autosomal dominant PKD (ADPKD). Although ARPKD virtually always presents in childhood and is usually considered to arise from mutations in PKHD1 (Nahm et al., 2002), recent study suggested that ARPKD is not a homogeneous disorder and further implicated DZIP1L as a second gene involved in ARPKD pathogenesis (Lu et al., 2017). ADPKD, on the other hand, is a late-onset disorder, where cysts can be detected in approximately 68% of individuals aged 30 and above (Nahm et al., 2002). ADPKD is caused by heterozygous mutations in PKD1 or PKD2, which encode the transmembrane proteins polycystin-1 (PC1) and polycystin-2 (PC2) (Nahm et al., 2002). Although every cell in the kidneys of ADPKD patients has the same genetic germline mutation, only a few cells in each nephron become cystic. A hypothesis known as the two-hit model of cystogenesis was proposed to explain this phenomenon (Reeders, 1992). Per this hypothesis, a somatic second hit, a deletion or inactivating point mutation to the remaining normal PKD1 or PKD2 allele, is believed to cause kidney cyst formation in ADPKD patients through a loss of heterozygosity. For this reason, it might be difficult to find cystic phenotypes in kidney organoids derived from the cells of ADPKD patients unless one waits long periods of time. Early phenotypes of ADPKD might be potentially detected in kidney organoids or undifferentiated hiPSCs, particularly because the target genes, PKD1/PKD2, encode ciliary proteins that are expressed in many cell types (Van Adelsberg and Frank, 1995). Freedman et al. reported generation of hiPSCs from three unrelated patients with ADPKD harboring PKD1 mutations and two patients with ARPKD harboring PKHD1 mutations (Freedman et al., 2013). Western blot analysis of hiPSCs derived from ADPKD patients, ARPKD patients, and healthy controls revealed the three groups to express PC2 at similar levels. However, localization of PC2 to the primary cilium was reduced in ADPKD patient-derived hiPSCs, suggesting that normal trafficking of PKD2 to the cilium is mediated by normal PKD1 protein. This study marked the first iPSC phenotype established for a kidney disorder and might help to explain the cause of cystogenesis in ADPKD patients; nevertheless, further studies will be necessary to elucidate the mechanisms of cystogenesis due to reduced PC2 expression in the cilium.
Ameku et al. used ADPKD patient-derived hiPSCs and identified a metalloenzyme gene, matrix metalloproteinase 1 (MMP1) as a novel risk factor for intracranial aneurysms (ICA) (Ameku et al., 2016). They generated hiPSCs from seven ADPKD patients, including four with ICA, and compared gene expression profiles between the vascular endothelial cells from ADPKD-iPSCs and those from non-ADPKD subjects. They found that the expression level of MMP1, was specifically elevated in iPSC-derived vascular endothelia derived from ADPKD patients with ICAs. Furthermore, they confirmed the correlation between the serum MMP1 levels and the development of ICAs in 354 ADPKD patients, indicating that high serum MMP1 levels may be a novel risk factor for ADPKD. These results suggest that cellular disease models with ADPKD-specific iPSCs can be used to study disease mechanisms and to find novel disease-related molecules or risk factors (Ameku et al., 2016).
One caveat to note is that it might be difficult to mimic the clinical manifestations of ADPKD such as cystic expansion from tubular epithelial cells using hiPSCs derived from ADPKD patients, because ADPKD is generally thought to be caused by somatic loss of heterozygosity (the second hit hypothesis) (Reeders, 1992). Considering the long time-course of disease progression of ADPKD, alternative approaches to modeling ADPKD might need to be developed beyond the study of patient-derived hiPSCs alone, so that clinical phenotypes can be reproduced in culture systems. Freedman et al. applied the CRISPR/Cas9 genome editing system to introduce biallelic, truncating mutations in PKD1 or PKD2 in hESCs (PKD1−/− or PKD2−/− hESCs) (Freedman et al., 2015). While these PKD1−/− or PKD2−/− hESCs maintain pluripotency, large translucent cyst-like structures were formed when differentiated into kidney organoids. These cystic structures remained tethered to the underlying matrix, but moved freely in response to vibration in contrast to neighboring normal tubular structures that remained fixed in position on the surface of culture plates. Although the differentiation protocol into kidney organoids might need further refinement, cyst formation was observed in 6% of the kidney organoids derived from PKD1−/− or PKD2−/− hESCs, while isogenic control hESCs rarely formed cystic structure under same condition (Freedman et al., 2015). They hypothesized that adherent forces play a critical role in limiting tubular deformation and subsequent cyst formation. After two weeks in suspension culture, 75% of kidney organoids derived from PKD1−/− or PKD2−/− hESCs formed large, free-floating cysts, resulting in a 10-fold increase in cyst formation over adherent cultures (Cruz et al., 2017). Control organoids of identical genetic background formed cysts very rarely under under these conditions, indicating that cystogenesis remained a specific consequence of the PKD1 or PKD2 mutations. Cyclic adenosine monophosphate (cAMP), when added, induces cysts in both PKD organoids and controls. Pathway-based global gene expression analysis revealed significant enrichment of hallmark gene sets for cell cycle progression, mTOR signaling, and MYC activity in these cysts, compared to normal tubular structures (Cruz et al., 2017). These PKD1−/− or PKD2−/− hESCs can be applied to high-throughput screening compatible platforms for enhanced differentiation and phenotyping of human kidney organoids. The authors focused on candidates that might modulate interaction between cells and their surrounding microenvironment, which is important for cystogenesis in PKD organoids (Cruz et al., 2017). Screening in gene-edited kidney organoids in this system revealed an unexpected role for myosin in polycystic kidney disease. Blebbistatin, a specific inhibitor of non-muscle myosin II, induced a dose-dependent increase in cyst formation in PKD organoids. These results suggested that the polycystins may normally function to positively regulate actomyosin activation within the tubular epithelium, strengthening and tightening the tubule and preventing it from deforming into a cyst. (Czerniecki et al., 2018).
A recent study from the laboratory of Melissa Little demonstrated the utility of kidney organoids as a disease-model in the setting of prospective identification of a patient with a heritable nephronophthisis-related ciliopathy (NPHP-RC) syndrome (Forbes et al., 2018). Whole-exome sequencing of the patient and her parents identified compound-heterozygous variants in IFT140. IFT140 is a core component of the IFT-A complex, which links ciliary maintenance proteins, signaling molecules, and transmembrane receptors to the dynein motor complex in order to descend the ciliary axoneme to the basal body, a process known as retrograde intraflagellar transport (IFT). The precise downstream cellular mechanisms responsible for disease presentation remains unknown (Stepanek and Pigino, 2016). Forbes et al. generated patient-derived and isogenic gene corrected iPSCs from skin fibroblasts using simultaneous reprogramming and CRISPR genome editing of the mutation. Kidney organoids generated from both iPSC lines contained multiple nephron segments, including the contiguous immunofluorescent localization of nephrin (NPHS1, staining glomerular precursors), LTL (staining proximal tubule), cadherin 1 (CDH1, staining distal tubule), and co-immunofluorescence for CDH1 and GATA3 (collecting duct). Patient-derived organoids demonstrated primary ciliary dysmorphology, a club-shaped ciliary morphology similar to that observed in IFT140 knockout mice, whereas correction of the mutation in IFT140 rescued this disease phenotype (Forbes et al., 2018). Transcriptional profiling and differential gene expression analysis of epithelial cells isolated from these organoids showed downregulation of genes associated with apicobasal polarity, cell-cell junctions, and dynein motor assembly in the mutant cells (Forbes et al., 2018). These results suggested that this kidney organoids model would clarify the common pathogenetic mechanisms for this heterogenetic rare disease.
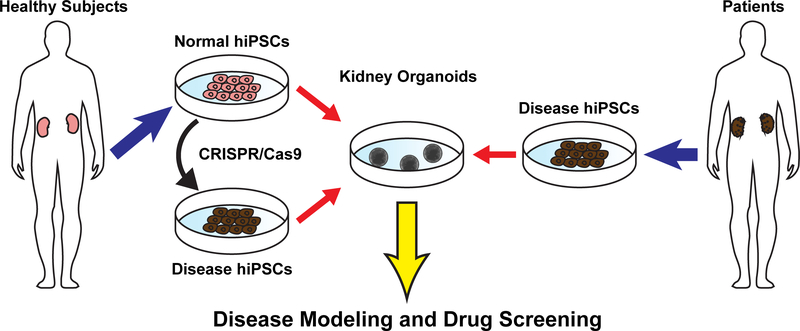
In summary, there are many attempts to model genetic kidney diseases using kidney organoids derived from hPSCs. Kidney organoids are an attractive tool to study mechanisms of genetic kidney diseases in human cells, facilitating translational research to develop candidate molecules for novel therapies of genetic kidney diseases (Fig. 1).
Fig. 1.
Disease modeling using genome editing and hPSC-derived kidney organoids.
Disease Modeling of Acute Kidney Injury and Chronic Kidney Disease
Drug nephrotoxicity is a common manifestation of the toxic effects of drugs and their metabolites, causing acute kidney injury (AKI) in hospitalized patients (Uchino et al., 2005). During drug development, 19% of failures in Phase III clinical trials are due to nephrotoxicity (Su et al., 2014). Currently there are no patient-specific models to assay nephrotoxicity in vitro. Recent reports have tested the nephrotoxicity of gentamycin or cisplatin on kidney organoids derived from hPSCs (Freedman et al., 2015; Morizane et al., 2015; Takasato et al., 2015). After kidney organoids were treated with gentamycin or cisplatin, increased expression of the DNA damage marker γH2AX and kidney injury molecule-1, clinical biomarkers of AKI (Freedman et al., 2015; Morizane et al., 2015), and/or cleaved caspase 3, a marker of apoptosis (Takasato et al., 2015), were observed in a manner that mimics in vivo kidney injury. These data support the use of kidney organoids as a novel platform for nephrotoxicity testing in human cells.
Defects in the filtration barrier of the glomerulus play a critical role in the onset of human renal diseases, which is characterized by massive proteinuria. The podocytes are cells lining the outer surface of the glomerular capillaries and maintain the filtration barrier by forming interdigitating foot processes with intervening slit diaphragms. Hale et al. sieved glomeruli from hiPSC-derived kidney organoids, which expressed podocyte-specific genes and maintained polarized protein localization expression (Hale et al., 2018). They placed organoid-derived glomeruli in a 96-well plate and exposed glomeruli to increasing concentrations of doxorubicin. The apoptosis marker caspase-3 increased at the lower doses of doxorubicin, before cell death prevailed resulting in the destruction of glomeruli (Hale et al., 2018). A single kidney organoid contains in the order of 100 glomeruli, and organoid-derived glomeruli retain marker expression in culture for 96 h. Hence, hiPSC-derived organoid glomeruli may represent an accessible approach to high content screening for podocyte toxicity.
Pericytes and peritubular fibroblasts in the kidney interstitium are generally believed to differentiate into myofibroblasts during kidney fibrosis, an important therapeutic target of CKD (Lin et al., 2008; Humphreys et al., 2010; LeBleu et al., 2013; Kramann et al., 2015). There are currently no antifibrotic drugs approved by the FDA for the treatment of CKD, which represents a major unmet medical need. Recombinant adeno-associated virus (AAV)-based gene therapy trials are currently ongoing for a range of monogenic diseases, including neuromuscular disease, hemophilia, and inherited forms of blindness (Bennett et al., 2016; George et al., 2017; Mendell et al., 2017; Bunting et al., 2018; Dunbar et al., 2018). However, protocols for transduction of kidney mesenchymal cells have not been established. Ikeda et al. evaluated the transduction profiles of various pseudotyped AAV vectors expressing either GFP or Cre recombinase reporters to kidney pericytes and fibroblasts in mouse kidneys and human kidney organoids. A synthetic AAV Anc80 were efficiently transduced into kidney mesenchymal cells in mice and human kidney organoids (Ikeda et al., 2018). These results can be the foundation for future gene therapy approaches for kidney fibrosis and CKD.
Nephron damage triggers the activation of interstitial fibrogenic cells, leading to progressive scarring and contributing to loss of kidney function. Yet, the signals that drive this process are not well understood. Our collaborative team recently identified a novel mechanism by which IL-1β, a major innate inflammatory cytokine, regulates a MYC-dependent metabolic switch using fibrotic kidney samples obtained from patients with CKD, experimental animal models of kidney fibrosis induced by unilateral ureteral obstruction (UUO), human kidney stromal cell lines, and human kidney organoids (Lemos et al., 2018). We used kidney organoids to evaluate the effects of IL-1β and MYC on kidney stromal cells in 3D environment. IL-1β induced organoid hypertrophy, proximal tubule injury confirmed by KIM-1 expression and loss of LTL, and progressive fibrotic phenotypes including thickening of the Collagen-I+ tubular basement membrane. Coincubation with MYC inhibitor significantly inhibited IL-1β-induced organoid hypertrophy, stromal cell proliferation, MYC accumulation in the nuclei of stromal cells, and upregulation of α-SMA expression (Lemos et al., 2018). These human organoid experiments highlight IL-1β and MYC as potential therapeutic targets in CKD patients.
Current Challenges in Organoid Research
A recent study from Humphreys’ laboratory performed single-cell RNA sequencing (scRNA-seq) and single-nucleus RNA-seq (snRNA-seq) to generate comprehensive molecular maps describing cell diversity in kidney organoids generated using two different protocols established by the Morizane group (Morizane and Bonventre, 2017a) and Little’s group (Takasato et al., 2016), with comparison to adult human kidney samples (Wu et al., 2018). In Wu’s study, kidney organoid-derived cell types appeared to be immature when benchmarked against fetal and adult human kidney single-cell datasets. Both proximal tubule cells and podocytes derived from these organoids expressed only a fraction of the transcription factors that are identified in the adult cell types. Comparison between the two different kidney organoid protocols showed fewer off-target cells and higher expression of nephron epithelial genes in Morizane’s protocol than Little’s, while Little’s organoids showed higher percentage of tubular cells than in Morizane’s organoids. Further, gene expression analyses of trait-relevant genes previously identified by genome-wide association studies (GWASs) suggested that kidney organoids may be limited in their ability to model kidney disease such as CKD.
However, the results might be associated with varied technical difficulties including library preparation (scRNA-seq with kidney organoids v.s. snRNA-seq in kidney biopsy samples), a higher percentage of mitochondrial genes (2.03–14.3% in organoid samples v.s. 0.18–0.33% in biopsy samples), and fewer detected gene numbers in organoids than in biopsy samples (Wu et al., 2018). Indeed, previous studies reported that high numbers of mitochondrial transcripts are indicative of unhealthy, poor-quality cells, suggesting cells with elevated mitochondrial gene expression should be excluded from data analyses (Islam et al., 2014; Ilicic et al., 2016). On the other hand, Little’s group also performed scRNA-seq in kidney organoids generated by their own differentiation protocol. They reported that directed differentiation of hPSCs to kidney organoids is robust, reproducible and transferable between stem cell lines. However, when independent differentiation experiments were performed using the same cell line but from different frozen vials, batch-to–batch variation was the greatest driver of overall variability, with primary contributions from nephron maturation and nephron patterning (Phipson et al., 2019).
In another study, despite acceptable intra-laboratory reproducibility of omics readouts in an iPSC-derived neuronal model system in vitro, neurons generated at different labs have large variation (Volpato et al., 2018). In our hands, we often experience differentiation variation due to differences in batches of reagents such as growth factors, indicating that careful optimization of the differentiation protocols is necessary to minimize batch-to-batch variation. There is currently no consensus with regard to how we should perform quality evaluation of organoid differentiation. Thus, we think that it is necessary to explore relevant parameters for the quality control of kidney organoids generated by varied differentiation protocols, especially when we use kidney organoids for disease modeling (Table 1). For example, we always evaluate differentiation efficiency at the intermediate stage of the directed differentiation as we previously reported (Morizane and Bonventre, 2017a).
Table 1.
Technical challenges and strategies for the quality control of kidney organoids
| TECHNICAL CHALLENGE | EXPERIMENTAL STRATEGY |
|---|---|
| Batch-to-Batch Variation | Evaluate the differentiation efficiency and kidney organoid characteristics |
| Plate, differentiate and assay patient or CRISPR mutant and control lines concurrently | |
| Inter-Laboratory Variation | Use same batch of regents such as growth factors, media and compounds |
| Evaluate the differentiation efficiency and kidney organoid characteristics | |
| Variability between hPSC Lines | Optimize culture and differentiation protocol by each cell line |
| Use parent cells as control (isogenic control), when using CRIPSR mutant lines | |
| Study multiple lines per patient |
Humphreys’ study also showed that off-target non-renal cell types including neural clusters were present at similar ratios (10%−20%) in all kidney organoids derived from hiPSCs and hESCs (Wu et al., 2018). To detect gene expression changes during organoid differentiation using Little’s protocol, they reconstructed kidney lineage relationships by performing pseudotemporal ordering using Monocle2. The resulting cell trajectories revealed one major branchpoint, separating loop of Henle and proximal tubular cell fates from podocyte, stromal, and neural cell fates. A second branchpoint distinguished podocyte from stromal and neural fates. Cell fates were defined by projecting marker gene expression onto the pseudotime trajectories. NTRK2, which encodes neurotrophic tyrosine kinase receptor, type 2 also known as tropomyosin receptor kinase B (TrkB), was expressed exclusively in neural clusters, and its ligand, Brain-derived neurotrophic factor (BDNF), was also strongly expressed in the podocyte-neuronstroma branch. Inhibition of BDNF pathway using K252a, NTRK2 inhibitor, reduced neuronal cell types by 90% in this study (Wu et al., 2018), though BDNF inhibition may affect nephron epithelial cells in kidney organoids. BDNF is a member of the neurotrophin family of polypeptide growth factors and binds to TrkB, which is expressed not only in neuronal cells, but also in developing glomeruli, mature tubule, collecting ducts and the juxtaglomerular apparatus in kidneys (de Girolamo et al., 2000; De Girolamo et al., 2004; Garcia-Suarez et al., 2006; Endlich et al., 2018; Tao et al., 2018). Recently, some groups reported that BDNF affects podocytes; BDNF has a TrkB-dependent trophic activity on podocyte cellular processes by microRNA-mediated increase of actin polymerization (Li et al., 2015b), and an inhibition of TrkB resulted in enhanced podocyte dedifferentiation (Tao et al., 2018). As Little’s group by themselves found fewer neuronal cells in their kidney organoids by scRNA-seq without BDNF inhibition (Phipson et al., 2019), these recent scRNA-seq studies highlight technical difficulties for kidney organoid generation in different labs or even in the same lab. Careful optimization of the differentiation protocols and reevaluation of kidney organoid characteristics would be required for reproducible organoid generation and disease modeling.
Stem Cells, Genome Editing, and Regenerative Medicine
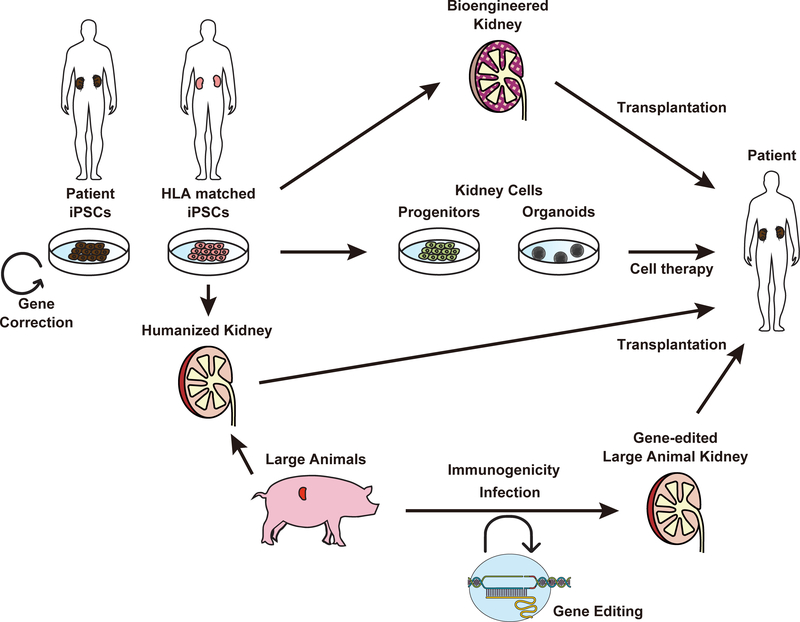
AKI is associated with higher in-hospital and long-term mortality risks (Lafrance and Miller, 2010). CKD causes varied complications such as renal anemia, mineral bone disorders, and heart diseases, and ESKD (Coresh et al., 2007; Levey et al., 2011; Jha et al., 2013). The development of stem cell therapy may provide curative care for these kidney disease patients. Currently, two major approaches are actively studied to develop curative therapy for kidney disease. One uses hPSCs-derived renal cells or tissues to regenerate nephrons, and the other uses genome editing to correct mutations in genetic kidney disease patients. In this section, we summarize current approaches for development of new curative therapy including cell therapy using hPSC-derived cells, bioengineered kidneys, and somatic cell gene editing (Fig. 2).
Fig. 2.
Approaches to develop regenerative therapy for kidney patients using hPSCs and genome editing.
The ultimate goal of cell therapy may be tissue replacement using transplanted cells, yet there are many challenges to this goal, such as the development of effective transplantation methods. Nevertheless, there are encouraging reports that suggested therapeutic potential of hPSC-derived kidney lineage cells. Toyohara et al. reported that the transplantation of hPSCs-derived OSR1+SIX2+ renal progenitors into the renal cortex ameliorated the AKI in mice (Toyohara et al., 2015). They discussed that the therapeutic effects of renal progenitors are thought to be mainly due to paracrine effects given the absence of new nephron formation from transplanted cells. The same group also reported that the renal progenitors can be enriched by flow sorting of CD9−CD140a+CD140b+CD271+ cells from differentiated hPSCs (Hoshina et al., 2018). Further, Hitomi et al. reported a method to generate erythropoietin (EPO)-producing cells from hPSCs and showed increased EPO expression and secretion in response to low oxygen conditions when subcutaneously injected into mice (Hitomi et al., 2017). These studies collectively suggest that kidney lineage cells derived from hPSCs may serve as a novel source for cell therapy against kidney diseases, yet further studies are necessary to develop better cell preparation and transplantation approaches for tissue reconstruction from hPSC-derived kidney cells.
Generation of functional kidneys as a replacement of current kidney transplantation therapy is a formidable challenge, and currently several different approaches are proposed. The approach to generate human kidneys in animals was previously suggested by studies where human kidney-like tissues were generated from human mesenchymal stem cells upon transplantation to rodent embryos (Yokoo et al., 2005). Recently, van den Berg et al. demonstrated generation of vascularized human kidney tissues derived from hPSCs upon transplantation of human kidney organoids to mouse renal capsules (van den Berg et al., 2018). Other groups transplanted human nephron progenitor cells (NPCs) derived from hPSCs to mice, which subsequently differentiated into nephron epithelial cells (Bantounas et al., 2018; Tajiri et al., 2018). These studies are encouraging towards the goal of generating functional human kidneys in animals; however, nephron structures were not thoroughly organized as functional kidneys to excrete urine. To overcome the issue of nephron organization, Yamanaka et al. showed generation of rat-mouse chimeric kidneys and proposed a combination system through which host NPCs can be eliminated by diphtheria toxin in a time- and tissue-specific manner. This elimination would allow donor NPCs to supplant host NPCs and develop into nephrons connected to host collecting ducts (Yamanaka et al., 2017). Xenogeneic kidney generation holds considerable promise as a means towards generation of the bioartificial kidney, though there are major challenges yet to be overcome. One major challenge is immunogenicity due to contamination of host animal-derived cells in kidneys. Indeed, the above studies found that kidney tissues were vascularized with host animal endothelial cells. The other major challenge is the risk of cross-species transmission of porcine endogenous retroviruses (PERVs). eGenesis recently demonstrated inactivation of all the PERVs in a porcine primary cell line and generated PERV-inactivated pigs via somatic cell nuclear transfer. Their study highlighted the value of PERV inactivation to prevent cross-species viral transmission and demonstrated the production of PERV-inactivated animals to address the safety concern in clinical xenotransplantation (Niu et al., 2017). It may become possible to use humanized kidneys or gene-edited pig kidneys as a novel source for kidney transplantation in the future.
Meanwhile, many groups have been developing protocols to generate kidney tissues from hPSCs in vitro (Taguchi et al., 2014; Morizane et al., 2015; Takasato et al., 2015; Yamaguchi et al., 2016; Morizane and Bonventre, 2017a; Przepiorski et al., 2018; Hiratsuka et al., 2019). Generated kidney organoids appeared to have different maturation states and cellular components depending on the differentiation protocols (Wu et al., 2018; Phipson et al., 2019), yet many challenges are needed for high-order kidney organogenesis. One or the common challenges in all protocols is how to generate perfusable vascular networks in order to generate vascularized glomeruli and tubules in vitro. To overcome this major challenge, our interdisciplinary team recently demonstrated that fluidic shear stress enhances vascularization and maturation of kidney organoids on 3D-printed chips (Homan et al., 2019). Of note, enhanced glomerular vascularization and morphogenesis in this system may enable production of glomerular filtrate in vitro in the near future. The next major challenges would be: further improving the chip system with organized vasculature and nephrons, overcoming anatomical problems such as incorporation of the collecting duct system and chaotic branching nephron patterns, and enhancing quality control of hPSCs and their derivatives (Merkle et al., 2015). To solve the anatomical problems of kidney organoids, some groups demonstrated that tissue recombination of induced ureteric bud and metanephric mesenchyme could facilitate higher-order architecture of kidney organoids and bioengineered branching renal tubules using PMDS scaffolds (Taguchi and Nishinakamura, 2017; Benedetti et al., 2018). Scale-up of the kidney tissue will be also a critical step towards the goal of generating functional bioengineered kidneys for the therapeutic purpose.
On the other hand, genome editing has great potential to develop curative therapy for genetic diseases (Cong et al., 2013; Ran et al., 2013a; Ran et al., 2013b). Tanigawa et al. established iPSCs from a patient with nephrotic syndrome and repaired an amino acid mutation using Cre-recombinase-mediated excision (Tanigawa et al., 2018). Genetic correction was confirmed by the expression of NPHS1 in the genetically corrected organoids, suggesting that genome editing can provide curative therapy for genetic kidney diseases. After correction of mutations in patient iPSCs, transplantation of differentiated cells or tissues may enable curative therapy. Further, somatic genome editing in vivo may also become possible to cure genetic diseases. Although studies targeting genetic kidney diseases are not yet reported, many studies demonstrated successful somatic genome editing in animals to treat genetic diseases in other organs (Nelson et al., 2016; Suzuki et al., 2016; Tabebordbar et al., 2016). Although somatic genome editing may induce serious genetic damage (Kosicki et al., 2018), further studies may enable safe and effective gene therapy for genetic kidney diseases in the future.
Conclusion
Recent advances in generating kidney organoids from hPSCs have led to new insight into understanding human kidney development and various human pathophysiology with a great potential for translational research. Kidney organoids are also usable for the disease modeling of genetic kidney diseases using patient-derived hiPSCs and CRISPR/Cas9-based gene-edited hPSCs. Furthermore, studies demonstrate the utility of kidney organoids to study disease mechanisms of AKI and CKD. Recent single cell RNA-seq studies highlight technical difficulties in reproducible organoid generation and differences of kidney organoids generated by varied differentiation protocols, indicating the need for careful quality evaluation of kidney organoids especially when used for disease modeling. Nevertheless, increasing evidence demonstrates the potential of kidney organoids for studies of kidney diseases, drug development, precision medicine, and regenerative medicine.
ACKNOWLEDGMENTS
This study was supported by the NIDDK Diabetic Complications Consortium (DiaComp, www.diacomp.org) grant (DK076169, to R.M.), NIH UG3 grant (TR002155, to R.M.), NIH UM1 grant (HG009390, to R.M.), a Brigham and Women’s Hospital Faculty Career Development Award (to R.M.) a Harvard Stem Cell Institute Seed Grant (to R.M.), AJINOMOTO Co., Inc. (to R.M.), and Toray Industries, Inc. (to R.M.).
REFERENCES
- Ameku T, Taura D, Sone M, Numata T, Nakamura M, Shiota F, Toyoda T, Matsui S, Araoka T, Yasuno T, Mae S, Kobayashi H, Kondo N, Kitaoka F, Amano N, Arai S, Ichisaka T, Matsuura N, Inoue S, Yamamoto T, Takahashi K, Asaka I, Yamada Y, Ubara Y, Muso E, Fukatsu A, Watanabe A, Sato Y, Nakahata T, Mori Y, Koizumi A, Nakao K, Yamanaka S, Osafune K. 2016. Identification of MMP1 as a novel risk factor for intracranial aneurysms in ADPKD using iPSC models. Sci Rep 6:30013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bantounas I, Ranjzad P, Tengku F, Silajdzic E, Forster D, Asselin MC, Lewis P, Lennon R, Plagge A, Wang Q, Woolf AS, Kimber SJ. 2018. Generation of Functioning Nephrons by Implanting Human Pluripotent Stem Cell-Derived Kidney Progenitors. Stem Cell Reports 10:766–779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barker N, Huch M, Kujala P, van de Wetering M, Snippert HJ, van Es JH, Sato T, Stange DE, Begthel H, van den Born M, Danenberg E, van den Brink S, Korving J, Abo A, Peters PJ, Wright N, Poulsom R, Clevers H. 2010. Lgr5(+ve) stem cells drive self-renewal in the stomach and build long-lived gastric units in vitro. Cell Stem Cell 6:25–36. [DOI] [PubMed] [Google Scholar]
- Bartfeld S, Bayram T, van de Wetering M, Huch M, Begthel H, Kujala P, Vries R, Peters PJ, Clevers H. 2015. In vitro expansion of human gastric epithelial stem cells and their responses to bacterial infection. Gastroenterology 148:126–136 e126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartfeld S, Clevers H. 2015. Organoids as Model for Infectious Diseases: Culture of Human and Murine Stomach Organoids and Microinjection of Helicobacter Pylori. J Vis Exp. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benedetti V, Brizi V, Guida P, Tomasoni S, Ciampi O, Angeli E, Valbusa U, Benigni A, Remuzzi G, Xinaris C. 2018. Engineered Kidney Tubules for Modeling Patient-Specific Diseases and Drug Discovery. EBioMedicine 33:253–268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett J, Wellman J, Marshall KA, McCague S, Ashtari M, DiStefano-Pappas J, Elci OU, Chung DC, Sun J, Wright JF, Cross DR, Aravand P, Cyckowski LL, Bennicelli JL, Mingozzi F, Auricchio A, Pierce EA, Ruggiero J, Leroy BP, Simonelli F, High KA, Maguire AM. 2016. Safety and durability of effect of contralateral-eye administration of AAV2 gene therapy in patients with childhood-onset blindness caused by RPE65 mutations: a follow-on phase 1 trial. Lancet 388:661–672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boj SF, Hwang CI, Baker LA, Chio II, Engle DD, Corbo V, Jager M, Ponz-Sarvise M, Tiriac H, Spector MS, Gracanin A, Oni T, Yu KH, van Boxtel R, Huch M, Rivera KD, Wilson JP, Feigin ME, Ohlund D, Handly-Santana A, Ardito-Abraham CM, Ludwig M, Elyada E, Alagesan B, Biffi G, Yordanov GN, Delcuze B, Creighton B, Wright K, Park Y, Morsink FH, Molenaar IQ, Borel Rinkes IH, Cuppen E, Hao Y, Jin Y, Nijman IJ, Iacobuzio-Donahue C, Leach SD, Pappin DJ, Hammell M, Klimstra DS, Basturk O, Hruban RH, Offerhaus GJ, Vries RG, Clevers H, Tuveson DA. 2015. Organoid models of human and mouse ductal pancreatic cancer. Cell 160:324–338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broutier L, Andersson-Rolf A, Hindley CJ, Boj SF, Clevers H, Koo BK, Huch M. 2016. Culture and establishment of self-renewing human and mouse adult liver and pancreas 3D organoids and their genetic manipulation. Nat Protoc 11:1724–1743. [DOI] [PubMed] [Google Scholar]
- Bunting S, Zhang L, Xie L, Bullens S, Mahimkar R, Fong S, Sandza K, Harmon D, Yates B, Handyside B, Sihn CR, Galicia N, Tsuruda L, O’Neill CA, Bagri A, Colosi P, Long S, Vehar G, Carter B. 2018. Gene Therapy with BMN 270 Results in Therapeutic Levels of FVIII in Mice and Primates and Normalization of Bleeding in Hemophilic Mice. Mol Ther 26:496–509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins AJ, Foley RN, Herzog C, Chavers BM, Gilbertson D, Ishani A, Kasiske BL, Liu J, Mau LW, McBean M, Murray A, St Peter W, Guo H, Li Q, Li S, Li S, Peng Y, Qiu Y, Roberts T, Skeans M, Snyder J, Solid C, Wang C, Weinhandl E, Zaun D, Arko C, Chen SC, Dalleska F, Daniels F, Dunning S, Ebben J, Frazier E, Hanzlik C, Johnson R, Sheets D, Wang X, Forrest B, Constantini E, Everson S, Eggers PW, Agodoa L. 2010. Excerpts from the US Renal Data System 2009 Annual Data Report. Am J Kidney Dis 55:S1–420, A426–427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, Hsu PD, Wu X, Jiang W, Marraffini LA, Zhang F. 2013. Multiplex genome engineering using CRISPR/Cas systems. Science 339:819–823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coresh J, Selvin E, Stevens LA, Manzi J, Kusek JW, Eggers P, Van Lente F, Levey AS. 2007. Prevalence of chronic kidney disease in the United States. JAMA 298:2038–2047. [DOI] [PubMed] [Google Scholar]
- Cruz NM, Song X, Czerniecki SM, Gulieva RE, Churchill AJ, Kim YK, Winston K, Tran LM, Diaz MA, Fu H, Finn LS, Pei Y, Himmelfarb J, Freedman BS. 2017. Organoid cystogenesis reveals a critical role of microenvironment in human polycystic kidney disease. Nat Mater 16:1112–1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czerniecki SM, Cruz NM, Harder JL, Menon R, Annis J, Otto EA, Gulieva RE, Islas LV, Kim YK, Tran LM, Martins TJ, Pippin JW, Fu H, Kretzler M, Shankland SJ, Himmelfarb J, Moon RT, Paragas N, Freedman BS. 2018. High-Throughput Screening Enhances Kidney Organoid Differentiation from Human Pluripotent Stem Cells and Enables Automated Multidimensional Phenotyping. Cell Stem Cell 22:929–940 e924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Girolamo P, Arcamone N, Lucini C, Castaldo L, Vega JA, Gargiulo G. 2000. The teleost kidney expresses Trk neurotrophin receptor-like proteins. Anat Embryol (Berl) 201:429–433. [DOI] [PubMed] [Google Scholar]
- De Girolamo P, Arcamone N, Lucini C, Simeoli MP, Castaldo L, Gargiulo G. 2004. TRK neurotrophin receptor-like proteins in the kidney of frog (Rana esculenta) and lizard (Podarcis sicula): an immunohistochemical study. Anat Embryol (Berl) 207:481–487. [DOI] [PubMed] [Google Scholar]
- Dimos JT, Rodolfa KT, Niakan KK, Weisenthal LM, Mitsumoto H, Chung W, Croft GF, Saphier G, Leibel R, Goland R, Wichterle H, Henderson CE, Eggan K. 2008. Induced pluripotent stem cells generated from patients with ALS can be differentiated into motor neurons. Science 321:1218–1221. [DOI] [PubMed] [Google Scholar]
- Drost J, Karthaus WR, Gao D, Driehuis E, Sawyers CL, Chen Y, Clevers H. 2016. Organoid culture systems for prostate epithelial and cancer tissue. Nat Protoc 11:347–358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drost J, van Jaarsveld RH, Ponsioen B, Zimberlin C, van Boxtel R, Buijs A, Sachs N, Overmeer RM, Offerhaus GJ, Begthel H, Korving J, van de Wetering M, Schwank G, Logtenberg M, Cuppen E, Snippert HJ, Medema JP, Kops GJ, Clevers H. 2015. Sequential cancer mutations in cultured human intestinal stem cells. Nature 521:43–47. [DOI] [PubMed] [Google Scholar]
- Dunbar CE, High KA, Joung JK, Kohn DB, Ozawa K, Sadelain M. 2018. Gene therapy comes of age. Science 359. [DOI] [PubMed] [Google Scholar]
- Dye BR, Hill DR, Ferguson MA, Tsai YH, Nagy MS, Dyal R, Wells JM, Mayhew CN, Nattiv R, Klein OD, White ES, Deutsch GH, Spence JR. 2015. In vitro generation of human pluripotent stem cell derived lung organoids. Elife 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebert AD, Yu J, Rose FF Jr., Mattis VB, Lorson CL, Thomson JA, Svendsen CN. 2009. Induced pluripotent stem cells from a spinal muscular atrophy patient. Nature 457:277–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eiraku M, Watanabe K, Matsuo-Takasaki M, Kawada M, Yonemura S, Matsumura M, Wataya T, Nishiyama A, Muguruma K, Sasai Y. 2008. Self-organized formation of polarized cortical tissues from ESCs and its active manipulation by extrinsic signals. Cell Stem Cell 3:519–532. [DOI] [PubMed] [Google Scholar]
- Endlich N, Lange T, Kuhn J, Klemm P, Kotb AM, Siegerist F, Kindt F, Lindenmeyer MT, Cohen CD, Kuss AW, Nath N, Rettig R, Lendeckel U, Zimmermann U, Amann K, Stracke S, Endlich K. 2018. BDNF: mRNA expression in urine cells of patients with chronic kidney disease and its role in kidney function. J Cell Mol Med 22:5265–5277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ene-Iordache B, Perico N, Bikbov B, Carminati S, Remuzzi A, Perna A, Islam N, Bravo RF, Aleckovic-Halilovic M, Zou H, Zhang L, Gouda Z, Tchokhonelidze I, Abraham G, Mahdavi-Mazdeh M, Gallieni M, Codreanu I, Togtokh A, Sharma SK, Koirala P, Uprety S, Ulasi I, Remuzzi G. 2016. Chronic kidney disease and cardiovascular risk in six regions of the world (ISN-KDDC): a cross-sectional study. Lancet Glob Health 4:e307–319. [DOI] [PubMed] [Google Scholar]
- Fatehullah A, Tan SH, Barker N. 2016. Organoids as an in vitro model of human development and disease. Nat Cell Biol 18:246–254. [DOI] [PubMed] [Google Scholar]
- Finkbeiner SR, Zeng XL, Utama B, Atmar RL, Shroyer NF, Estes MK. 2012. Stem cell-derived human intestinal organoids as an infection model for rotaviruses. MBio 3:e00159–00112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Firth AL, Menon T, Parker GS, Qualls SJ, Lewis BM, Ke E, Dargitz CT, Wright R, Khanna A, Gage FH, Verma IM. 2015. Functional Gene Correction for Cystic Fibrosis in Lung Epithelial Cells Generated from Patient iPSCs. Cell Rep 12:1385–1390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forbes TA, Howden SE, Lawlor K, Phipson B, Maksimovic J, Hale L, Wilson S, Quinlan C, Ho G, Holman K, Bennetts B, Crawford J, Trnka P, Oshlack A, Patel C, Mallett A, Simons C, Little MH. 2018. Patient-iPSC-Derived Kidney Organoids Show Functional Validation of a Ciliopathic Renal Phenotype and Reveal Underlying Pathogenetic Mechanisms. Am J Hum Genet 102:816–831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forbester JL, Goulding D, Vallier L, Hannan N, Hale C, Pickard D, Mukhopadhyay S, Dougan G. 2015. Interaction of Salmonella enterica Serovar Typhimurium with Intestinal Organoids Derived from Human Induced Pluripotent Stem Cells. Infect Immun 83:2926–2934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freedman BS, Brooks CR, Lam AQ, Fu H, Morizane R, Agrawal V, Saad AF, Li MK, Hughes MR, Werff RV, Peters DT, Lu J, Baccei A, Siedlecki AM, Valerius MT, Musunuru K, McNagny KM, Steinman TI, Zhou J, Lerou PH, Bonventre JV. 2015. Modelling kidney disease with CRISPR-mutant kidney organoids derived from human pluripotent epiblast spheroids. Nat Commun 6:8715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freedman BS, Lam AQ, Sundsbak JL, Iatrino R, Su X, Koon SJ, Wu M, Daheron L, Harris PC, Zhou J, Bonventre JV. 2013. Reduced ciliary polycystin-2 in induced pluripotent stem cells from polycystic kidney disease patients with PKD1 mutations. J Am Soc Nephrol 24:1571–1586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao D, Vela I, Sboner A, Iaquinta PJ, Karthaus WR, Gopalan A, Dowling C, Wanjala JN, Undvall EA, Arora VK, Wongvipat J, Kossai M, Ramazanoglu S, Barboza LP, Di W, Cao Z, Zhang QF, Sirota I, Ran L, MacDonald TY, Beltran H, Mosquera JM, Touijer KA, Scardino PT, Laudone VP, Curtis KR, Rathkopf DE, Morris MJ, Danila DC, Slovin SF, Solomon SB, Eastham JA, Chi P, Carver B, Rubin MA, Scher HI, Clevers H, Sawyers CL, Chen Y. 2014. Organoid cultures derived from patients with advanced prostate cancer. Cell 159:176–187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Suarez O, Gonzalez-Martinez T, Germana A, Monjil DF, Torrecilla JR, Laura R, Silos-Santiago I, Guate JL, Vega JA. 2006. Expression of TrkB in the murine kidney. Microsc Res Tech 69:1014–1020. [DOI] [PubMed] [Google Scholar]
- George LA, Sullivan SK, Giermasz A, Rasko JEJ, Samelson-Jones BJ, Ducore J, Cuker A, Sullivan LM, Majumdar S, Teitel J, McGuinn CE, Ragni MV, Luk AY, Hui D, Wright JF, Chen Y, Liu Y, Wachtel K, Winters A, Tiefenbacher S, Arruda VR, van der Loo JCM, Zelenaia O, Takefman D, Carr ME, Couto LB, Anguela XM, High KA. 2017. Hemophilia B Gene Therapy with a High-Specific-Activity Factor IX Variant. N Engl J Med 377:2215–2227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goh KI, Cusick ME, Valle D, Childs B, Vidal M, Barabasi AL. 2007. The human disease network. Proc Natl Acad Sci U S A 104:8685–8690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hale LJ, Howden SE, Phipson B, Lonsdale A, Er PX, Ghobrial I, Hosawi S, Wilson S, Lawlor KT, Khan S, Oshlack A, Quinlan C, Lennon R, Little MH. 2018. 3D organoid-derived human glomeruli for personalised podocyte disease modelling and drug screening. Nat Commun 9:5167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiratsuka K, Monkawa T, Akiyama T, Nakatake Y, Oda M, Goparaju SK, Kimura H, Chikazawa-Nohtomi N, Sato S, Ishiguro K, Yamaguchi S, Suzuki S, Morizane R, Ko SBH, Itoh H, Ko MSH. 2019. Induction of human pluripotent stem cells into kidney tissues by synthetic mRNAs encoding transcription factors. Sci Rep 9:913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hisha H, Tanaka T, Kanno S, Tokuyama Y, Komai Y, Ohe S, Yanai H, Omachi T, Ueno H. 2013. Establishment of a novel lingual organoid culture system: generation of organoids having mature keratinized epithelium from adult epithelial stem cells. Sci Rep 3:3224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hitomi H, Kasahara T, Katagiri N, Hoshina A, Mae SI, Kotaka M, Toyohara T, Rahman A, Nakano D, Niwa A, Saito MK, Nakahata T, Nishiyama A, Osafune K. 2017. Human pluripotent stem cell-derived erythropoietin-producing cells ameliorate renal anemia in mice. Sci Transl Med 9. [DOI] [PubMed] [Google Scholar]
- Hohwieler M, Illing A, Hermann PC, Mayer T, Stockmann M, Perkhofer L, Eiseler T, Antony JS, Muller M, Renz S, Kuo CC, Lin Q, Sendler M, Breunig M, Kleiderman SM, Lechel A, Zenker M, Leichsenring M, Rosendahl J, Zenke M, Sainz B Jr., Mayerle J, Costa IG, Seufferlein T, Kormann M, Wagner M, Liebau S, Kleger A. 2017. Human pluripotent stem cell-derived acinar/ductal organoids generate human pancreas upon orthotopic transplantation and allow disease modelling. Gut 66:473–486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Homan K, Gupta N, Kroll K, Kolesky D, Skylar-Scott M, Miyoshi T, Mau D, Valerius MT, Ferrante T, Bonventre J, Lewis J, Morizane R. in press. Flow-enhanced vascularization and maturation of kidney organoids in vitro. Nature Methods. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Homan KA, Gupta N, Kroll KT, Kolesky DB, Skylar-Scott M, Miyoshi T, Mau D, Valerius MT, Ferrante T, Bonventre JV, Lewis JA, Morizane R. 2019. Flow-enhanced vascularization and maturation of kidney organoids in vitro. Nat Methods. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoshina A, Kawamoto T, Sueta SI, Mae SI, Araoka T, Tanaka H, Sato Y, Yamagishi Y, Osafune K. 2018. Development of new method to enrich human iPSC-derived renal progenitors using cell surface markers. Sci Rep 8:6375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang JY, Sweeney EG, Sigal M, Zhang HC, Remington SJ, Cantrell MA, Kuo CJ, Guillemin K, Amieva MR. 2015a. Chemodetection and Destruction of Host Urea Allows Helicobacter pylori to Locate the Epithelium. Cell Host Microbe 18:147–156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang L, Holtzinger A, Jagan I, BeGora M, Lohse I, Ngai N, Nostro C, Wang R, Muthuswamy LB, Crawford HC, Arrowsmith C, Kalloger SE, Renouf DJ, Connor AA, Cleary S, Schaeffer DF, Roehrl M, Tsao MS, Gallinger S, Keller G, Muthuswamy SK. 2015b. Ductal pancreatic cancer modeling and drug screening using human pluripotent stem cell- and patient-derived tumor organoids. Nat Med 21:1364–1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang SX, Islam MN, O’Neill J, Hu Z, Yang YG, Chen YW, Mumau M, Green MD, Vunjak-Novakovic G, Bhattacharya J, Snoeck HW. 2014. Efficient generation of lung and airway epithelial cells from human pluripotent stem cells. Nat Biotechnol 32:84–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huch M, Bonfanti P, Boj SF, Sato T, Loomans CJ, van de Wetering M, Sojoodi M, Li VS, Schuijers J, Gracanin A, Ringnalda F, Begthel H, Hamer K, Mulder J, van Es JH, de Koning E, Vries RG, Heimberg H, Clevers H. 2013a. Unlimited in vitro expansion of adult bi-potent pancreas progenitors through the Lgr5/R-spondin axis. EMBO J 32:2708–2721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huch M, Dorrell C, Boj SF, van Es JH, Li VS, van de Wetering M, Sato T, Hamer K, Sasaki N, Finegold MJ, Haft A, Vries RG, Grompe M, Clevers H. 2013b. In vitro expansion of single Lgr5+ liver stem cells induced by Wnt-driven regeneration. Nature 494:247–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huch M, Gehart H, van Boxtel R, Hamer K, Blokzijl F, Verstegen MM, Ellis E, van Wenum M, Fuchs SA, de Ligt J, van de Wetering M, Sasaki N, Boers SJ, Kemperman H, de Jonge J, Ijzermans JN, Nieuwenhuis EE, Hoekstra R, Strom S, Vries RR, van der Laan LJ, Cuppen E, Clevers H. 2015. Long-term culture of genome-stable bipotent stem cells from adult human liver. Cell 160:299–312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humphreys BD, Lin SL, Kobayashi A, Hudson TE, Nowlin BT, Bonventre JV, Valerius MT, McMahon AP, Duffield JS. 2010. Fate tracing reveals the pericyte and not epithelial origin of myofibroblasts in kidney fibrosis. Am J Pathol 176:85–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humphreys BD, Valerius MT, Kobayashi A, Mugford JW, Soeung S, Duffield JS, McMahon AP, Bonventre JV. 2008. Intrinsic epithelial cells repair the kidney after injury. Cell Stem Cell 2:284–291. [DOI] [PubMed] [Google Scholar]
- Ikeda Y, Sun Z, Ru X, Vandenberghe LH, Humphreys BD. 2018. Efficient Gene Transfer to Kidney Mesenchymal Cells Using a Synthetic Adeno-Associated Viral Vector. J Am Soc Nephrol 29:2287–2297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ilicic T, Kim JK, Kolodziejczyk AA, Bagger FO, McCarthy DJ, Marioni JC, Teichmann SA. 2016. Classification of low quality cells from single-cell RNA-seq data. Genome Biol 17:29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Islam S, Zeisel A, Joost S, La Manno G, Zajac P, Kasper M, Lonnerberg P, Linnarsson S. 2014. Quantitative single-cell RNA-seq with unique molecular identifiers. Nat Methods 11:163–166. [DOI] [PubMed] [Google Scholar]
- Jha V, Garcia-Garcia G, Iseki K, Li Z, Naicker S, Plattner B, Saran R, Wang AY, Yang CW. 2013. Chronic kidney disease: global dimension and perspectives. Lancet 382:260–272. [DOI] [PubMed] [Google Scholar]
- Jo J, Xiao Y, Sun AX, Cukuroglu E, Tran HD, Goke J, Tan ZY, Saw TY, Tan CP, Lokman H, Lee Y, Kim D, Ko HS, Kim SO, Park JH, Cho NJ, Hyde TM, Kleinman JE, Shin JH, Weinberger DR, Tan EK, Je HS, Ng HH. 2016. Midbrain-like Organoids from Human Pluripotent Stem Cells Contain Functional Dopaminergic and Neuromelanin-Producing Neurons. Cell Stem Cell 19:248–257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karthaus WR, Iaquinta PJ, Drost J, Gracanin A, van Boxtel R, Wongvipat J, Dowling CM, Gao D, Begthel H, Sachs N, Vries RG, Cuppen E, Chen Y, Sawyers CL, Clevers HC. 2014. Identification of multipotent luminal progenitor cells in human prostate organoid cultures. Cell 159:163–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kessler M, Hoffmann K, Brinkmann V, Thieck O, Jackisch S, Toelle B, Berger H, Mollenkopf HJ, Mangler M, Sehouli J, Fotopoulou C, Meyer TF. 2015. The Notch and Wnt pathways regulate stemness and differentiation in human fallopian tube organoids. Nat Commun 6:8989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosicki M, Tomberg K, Bradley A. 2018. Repair of double-strand breaks induced by CRISPR-Cas9 leads to large deletions and complex rearrangements. Nat Biotechnol 36:765–771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraja AT, Province MA, Huang P, Jarvis JP, Rice T, Cheverud JM, Rao DC. 2008. Trends in metabolic syndrome and gene networks in human and rodent models. Endocr Metab Immune Disord Drug Targets 8:198–207. [DOI] [PubMed] [Google Scholar]
- Kramann R, Schneider RK, DiRocco DP, Machado F, Fleig S, Bondzie PA, Henderson JM, Ebert BL, Humphreys BD. 2015. Perivascular Gli1+ progenitors are key contributors to injury-induced organ fibrosis. Cell Stem Cell 16:51–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lafrance JP, Miller DR. 2010. Acute kidney injury associates with increased long-term mortality. J Am Soc Nephrol 21:345–352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lancaster MA, Knoblich JA. 2014. Organogenesis in a dish: Modeling development and disease using organoid technologies. Science 345:1247125–1247125. [DOI] [PubMed] [Google Scholar]
- Lancaster MA, Renner M, Martin CA, Wenzel D, Bicknell LS, Hurles ME, Homfray T, Penninger JM, Jackson AP, Knoblich JA. 2013. Cerebral organoids model human brain development and microcephaly. Nature 501:373–379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LeBleu VS, Taduri G, O’Connell J, Teng Y, Cooke VG, Woda C, Sugimoto H, Kalluri R. 2013. Origin and function of myofibroblasts in kidney fibrosis. Nat Med 19:1047–1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemos DR, McMurdo M, Karaca G, Wilflingseder J, Leaf IA, Gupta N, Miyoshi T, Susa K, Johnson BG, Soliman K, Wang G, Morizane R, Bonventre JV, Duffield JS. 2018. Interleukin-1beta Activates a MYC-Dependent Metabolic Switch in Kidney Stromal Cells Necessary for Progressive Tubulointerstitial Fibrosis. J Am Soc Nephrol 29:1690–1705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leslie JL, Huang S, Opp JS, Nagy MS, Kobayashi M, Young VB, Spence JR. 2015. Persistence and toxin production by Clostridium difficile within human intestinal organoids result in disruption of epithelial paracellular barrier function. Infect Immun 83:138–145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levey AS, de Jong PE, Coresh J, El Nahas M, Astor BC, Matsushita K, Gansevoort RT, Kasiske BL, Eckardt KU. 2011. The definition, classification, and prognosis of chronic kidney disease: a KDIGO Controversies Conference report. Kidney Int 80:17–28. [DOI] [PubMed] [Google Scholar]
- Li HL, Fujimoto N, Sasakawa N, Shirai S, Ohkame T, Sakuma T, Tanaka M, Amano N, Watanabe A, Sakurai H, Yamamoto T, Yamanaka S, Hotta A. 2015a. Precise correction of the dystrophin gene in duchenne muscular dystrophy patient induced pluripotent stem cells by TALEN and CRISPR-Cas9. Stem Cell Reports 4:143–154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M, Armelloni S, Zennaro C, Wei C, Corbelli A, Ikehata M, Berra S, Giardino L, Mattinzoli D, Watanabe S, Agostoni C, Edefonti A, Reiser J, Messa P, Rastaldi MP. 2015b. BDNF repairs podocyte damage by microRNA-mediated increase of actin polymerization. J Pathol 235:731–744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin SL, Kisseleva T, Brenner DA, Duffield JS. 2008. Pericytes and perivascular fibroblasts are the primary source of collagen-producing cells in obstructive fibrosis of the kidney. Am J Pathol 173:1617–1627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lippmann ES, Williams CE, Ruhl DA, Estevez-Silva MC, Chapman ER, Coon JJ, Ashton RS. 2015. Deterministic HOX patterning in human pluripotent stem cell-derived neuroectoderm. Stem Cell Reports 4:632–644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Little MH, McMahon AP. 2012. Mammalian kidney development: principles, progress, and projections. Cold Spring Harb Perspect Biol 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu H, Galeano MCR, Ott E, Kaeslin G, Kausalya PJ, Kramer C, Ortiz-Bruchle N, Hilger N, Metzis V, Hiersche M, Tay SY, Tunningley R, Vij S, Courtney AD, Whittle B, Wuhl E, Vester U, Hartleben B, Neuber S, Frank V, Little MH, Epting D, Papathanasiou P, Perkins AC, Wright GD, Hunziker W, Gee HY, Otto EA, Zerres K, Hildebrandt F, Roy S, Wicking C, Bergmann C. 2017. Mutations in DZIP1L, which encodes a ciliary-transition-zone protein, cause autosomal recessive polycystic kidney disease. Nat Genet 49:1025–1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matano M, Date S, Shimokawa M, Takano A, Fujii M, Ohta Y, Watanabe T, Kanai T, Sato T. 2015. Modeling colorectal cancer using CRISPR-Cas9-mediated engineering of human intestinal organoids. Nat Med 21:256–262. [DOI] [PubMed] [Google Scholar]
- McCracken KW, Cata EM, Crawford CM, Sinagoga KL, Schumacher M, Rockich BE, Tsai YH, Mayhew CN, Spence JR, Zavros Y, Wells JM. 2014. Modelling human development and disease in pluripotent stem-cell-derived gastric organoids. Nature 516:400–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendell JR, Al-Zaidy S, Shell R, Arnold WD, Rodino-Klapac LR, Prior TW, Lowes L, Alfano L, Berry K, Church K, Kissel JT, Nagendran S, L’Italien J, Sproule DM, Wells C, Cardenas JA, Heitzer MD, Kaspar A, Corcoran S, Braun L, Likhite S, Miranda C, Meyer K, Foust KD, Burghes AHM, Kaspar BK. 2017. Single-Dose Gene-Replacement Therapy for Spinal Muscular Atrophy. N Engl J Med 377:1713–1722. [DOI] [PubMed] [Google Scholar]
- Merkle FT, Neuhausser WM, Santos D, Valen E, Gagnon JA, Maas K, Sandoe J, Schier AF, Eggan K. 2015. Efficient CRISPR-Cas9-mediated generation of knockin human pluripotent stem cells lacking undesired mutations at the targeted locus. Cell Rep 11:875–883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mestas J, Hughes CC. 2004. Of mice and not men: differences between mouse and human immunology. J Immunol 172:2731–2738. [DOI] [PubMed] [Google Scholar]
- Morizane R, Bonventre JV. 2017a. Generation of nephron progenitor cells and kidney organoids from human pluripotent stem cells. Nat Protoc 12:195–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morizane R, Bonventre JV. 2017b. Kidney Organoids: A Translational Journey. Trends Mol Med 23:246–263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morizane R, Lam AQ, Freedman BS, Kishi S, Valerius MT, Bonventre JV. 2015. Nephron organoids derived from human pluripotent stem cells model kidney development and injury. Nat Biotechnol. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nahm AM, Henriquez DE, Ritz E. 2002. Renal cystic disease (ADPKD and ARPKD). Nephrol Dial Transplant 17:311–314. [DOI] [PubMed] [Google Scholar]
- Nelson CE, Hakim CH, Ousterout DG, Thakore PI, Moreb EA, Castellanos Rivera RM, Madhavan S, Pan X, Ran FA, Yan WX, Asokan A, Zhang F, Duan D, Gersbach CA. 2016. In vivo genome editing improves muscle function in a mouse model of Duchenne muscular dystrophy. Science 351:403–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu D, Wei HJ, Lin L, George H, Wang T, Lee IH, Zhao HY, Wang Y, Kan Y, Shrock E, Lesha E, Wang G, Luo Y, Qing Y, Jiao D, Zhao H, Zhou X, Wang S, Wei H, Guell M, Church GM, Yang L. 2017. Inactivation of porcine endogenous retrovirus in pigs using CRISPR-Cas9. Science 357:1303–1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park CY, Kim DH, Son JS, Sung JJ, Lee J, Bae S, Kim JH, Kim DW, Kim JS. 2015. Functional Correction of Large Factor VIII Gene Chromosomal Inversions in Hemophilia A Patient-Derived iPSCs Using CRISPR-Cas9. Cell Stem Cell 17:213–220. [DOI] [PubMed] [Google Scholar]
- Park IH, Arora N, Huo H, Maherali N, Ahfeldt T, Shimamura A, Lensch MW, Cowan C, Hochedlinger K, Daley GQ. 2008. Disease-specific induced pluripotent stem cells. Cell 134:877–886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phipson B, Er PX, Combes AN, Forbes TA, Howden SE, Zappia L, Yen HJ, Lawlor KT, Hale LJ, Sun J, Wolvetang E, Takasato M, Oshlack A, Little MH. 2019. Evaluation of variability in human kidney organoids. Nat Methods 16:79–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Przepiorski A, Sander V, Tran T, Hollywood JA, Sorrenson B, Shih JH, Wolvetang EJ, McMahon AP, Holm TM, Davidson AJ. 2018. A Simple Bioreactor-Based Method to Generate Kidney Organoids from Pluripotent Stem Cells. Stem Cell Reports 11:470–484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ran FA, Hsu PD, Lin CY, Gootenberg JS, Konermann S, Trevino AE, Scott DA, Inoue A, Matoba S, Zhang Y, Zhang F. 2013a. Double nicking by RNA-guided CRISPR Cas9 for enhanced genome editing specificity. Cell 154:1380–1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ran FA, Hsu PD, Wright J, Agarwala V, Scott DA, Zhang F. 2013b. Genome engineering using the CRISPR-Cas9 system. Nat Protoc 8:2281–2308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravasi T, Suzuki H, Cannistraci CV, Katayama S, Bajic VB, Tan K, Akalin A, Schmeier S, Kanamori-Katayama M, Bertin N, Carninci P, Daub CO, Forrest AR, Gough J, Grimmond S, Han JH, Hashimoto T, Hide W, Hofmann O, Kamburov A, Kaur M, Kawaji H, Kubosaki A, Lassmann T, van Nimwegen E, MacPherson CR, Ogawa C, Radovanovic A, Schwartz A, Teasdale RD, Tegner J, Lenhard B, Teichmann SA, Arakawa T, Ninomiya N, Murakami K, Tagami M, Fukuda S, Imamura K, Kai C, Ishihara R, Kitazume Y, Kawai J, Hume DA, Ideker T, Hayashizaki Y. 2010. An atlas of combinatorial transcriptional regulation in mouse and man. Cell 140:744–752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reeders ST. 1992. Multilocus polycystic disease. Nat Genet 1:235–237. [DOI] [PubMed] [Google Scholar]
- Rock JR, Onaitis MW, Rawlins EL, Lu Y, Clark CP, Xue Y, Randell SH, Hogan BL. 2009. Basal cells as stem cells of the mouse trachea and human airway epithelium. Proc Natl Acad Sci U S A 106:12771–12775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato T, Stange DE, Ferrante M, Vries RG, Van Es JH, Van den Brink S, Van Houdt WJ, Pronk A, Van Gorp J, Siersema PD, Clevers H. 2011. Long-term expansion of epithelial organoids from human colon, adenoma, adenocarcinoma, and Barrett’s epithelium. Gastroenterology 141:1762–1772. [DOI] [PubMed] [Google Scholar]
- Sato T, Vries RG, Snippert HJ, van de Wetering M, Barker N, Stange DE, van Es JH, Abo A, Kujala P, Peters PJ, Clevers H. 2009. Single Lgr5 stem cells build crypt-villus structures in vitro without a mesenchymal niche. Nature 459:262–265. [DOI] [PubMed] [Google Scholar]
- Schwank G, Koo BK, Sasselli V, Dekkers JF, Heo I, Demircan T, Sasaki N, Boymans S, Cuppen E, van der Ent CK, Nieuwenhuis EE, Beekman JM, Clevers H. 2013. Functional repair of CFTR by CRISPR/Cas9 in intestinal stem cell organoids of cystic fibrosis patients. Cell Stem Cell 13:653–658. [DOI] [PubMed] [Google Scholar]
- Shen B, Zhang W, Zhang J, Zhou J, Wang J, Chen L, Wang L, Hodgkins A, Iyer V, Huang X, Skarnes WC. 2014. Efficient genome modification by CRISPR-Cas9 nickase with minimal off-target effects. Nat Methods 11:399–402. [DOI] [PubMed] [Google Scholar]
- Soldner F, Hockemeyer D, Beard C, Gao Q, Bell GW, Cook EG, Hargus G, Blak A, Cooper O, Mitalipova M, Isacson O, Jaenisch R. 2009. Parkinson’s disease patient-derived induced pluripotent stem cells free of viral reprogramming factors. Cell 136:964–977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spence JR, Mayhew CN, Rankin SA, Kuhar MF, Vallance JE, Tolle K, Hoskins EE, Kalinichenko VV, Wells SI, Zorn AM, Shroyer NF, Wells JM. 2011. Directed differentiation of human pluripotent stem cells into intestinal tissue in vitro. Nature 470:105–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stepanek L, Pigino G. 2016. Microtubule doublets are double-track railways for intraflagellar transport trains. Science 352:721–724. [DOI] [PubMed] [Google Scholar]
- Su R, Li Y, Zink D, Loo LH. 2014. Supervised prediction of drug-induced nephrotoxicity based on interleukin-6 and −8 expression levels. BMC Bioinformatics 15 Suppl 16:S16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki K, Tsunekawa Y, Hernandez-Benitez R, Wu J, Zhu J, Kim EJ, Hatanaka F, Yamamoto M, Araoka T, Li Z, Kurita M, Hishida T, Li M, Aizawa E, Guo S, Chen S, Goebl A, Soligalla RD, Qu J, Jiang T, Fu X, Jafari M, Esteban CR, Berggren WT, Lajara J, Nunez-Delicado E, Guillen P, Campistol JM, Matsuzaki F, Liu GH, Magistretti P, Zhang K, Callaway EM, Zhang K, Belmonte JC. 2016. In vivo genome editing via CRISPR/Cas9 mediated homology-independent targeted integration. Nature 540:144–149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabebordbar M, Zhu K, Cheng JKW, Chew WL, Widrick JJ, Yan WX, Maesner C, Wu EY, Xiao R, Ran FA, Cong L, Zhang F, Vandenberghe LH, Church GM, Wagers AJ. 2016. In vivo gene editing in dystrophic mouse muscle and muscle stem cells. Science 351:407–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taguchi A, Kaku Y, Ohmori T, Sharmin S, Ogawa M, Sasaki H, Nishinakamura R. 2014. Redefining the in vivo origin of metanephric nephron progenitors enables generation of complex kidney structures from pluripotent stem cells. Cell Stem Cell 14:53–67. [DOI] [PubMed] [Google Scholar]
- Taguchi A, Nishinakamura R. 2017. Higher-Order Kidney Organogenesis from Pluripotent Stem Cells. Cell Stem Cell 21:730–746 e736. [DOI] [PubMed] [Google Scholar]
- Tajiri S, Yamanaka S, Fujimoto T, Matsumoto K, Taguchi A, Nishinakamura R, Okano HJ, Yokoo T. 2018. Regenerative potential of induced pluripotent stem cells derived from patients undergoing haemodialysis in kidney regeneration. Sci Rep 8:14919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, Tomoda K, Yamanaka S. 2007. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell 131:861–872. [DOI] [PubMed] [Google Scholar]
- Takasato M, Er PX, Chiu HS, Little MH. 2016. Generation of kidney organoids from human pluripotent stem cells. Nat Protoc 11:1681–1692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takasato M, Er PX, Chiu HS, Maier B, Baillie GJ, Ferguson C, Parton RG, Wolvetang EJ, Roost MS, Chuva de Sousa Lopes SM, Little MH. 2015. Kidney organoids from human iPS cells contain multiple lineages and model human nephrogenesis. Nature 526:564–568. [DOI] [PubMed] [Google Scholar]
- Takebe T, Sekine K, Enomura M, Koike H, Kimura M, Ogaeri T, Zhang RR, Ueno Y, Zheng YW, Koike N, Aoyama S, Adachi Y, Taniguchi H. 2013. Vascularized and functional human liver from an iPSC-derived organ bud transplant. Nature 499:481–484. [DOI] [PubMed] [Google Scholar]
- Tanigawa S, Islam M, Sharmin S, Naganuma H, Yoshimura Y, Haque F, Era T, Nakazato H, Nakanishi K, Sakuma T, Yamamoto T, Kurihara H, Taguchi A, Nishinakamura R. 2018. Organoids from Nephrotic Disease-Derived iPSCs Identify Impaired NEPHRIN Localization and Slit Diaphragm Formation in Kidney Podocytes. Stem Cell Reports 11:727–740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao YS, Piao SG, Jin YS, Jin JZ, Zheng HL, Zhao HY, Lim SW, Yang CW, Li C. 2018. Expression of brain-derived neurotrophic factor in kidneys from normal and cyclosporine-treated rats. BMC Nephrol 19:63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomson JA, Itskovitz-Eldor J, Shapiro SS, Waknitz MA, Swiergiel JJ, Marshall VS, Jones JM. 1998. Embryonic stem cell lines derived from human blastocysts. Science 282:1145–1147. [DOI] [PubMed] [Google Scholar]
- Toyohara T, Mae S, Sueta S, Inoue T, Yamagishi Y, Kawamoto T, Kasahara T, Hoshina A, Toyoda T, Tanaka H, Araoka T, Sato-Otsubo A, Takahashi K, Sato Y, Yamaji N, Ogawa S, Yamanaka S, Osafune K. 2015. Cell Therapy Using Human Induced Pluripotent Stem Cell-Derived Renal Progenitors Ameliorates Acute Kidney Injury in Mice. Stem Cells Transl Med 4:980–992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uchino S, Kellum JA, Bellomo R, Doig GS, Morimatsu H, Morgera S, Schetz M, Tan I, Bouman C, Macedo E, Gibney N, Tolwani A, Ronco C, Beginning, Ending Supportive Therapy for the Kidney I. 2005. Acute renal failure in critically ill patients: a multinational, multicenter study. JAMA 294:813–818. [DOI] [PubMed] [Google Scholar]
- Van Adelsberg JS, Frank D. 1995. The PKD1 gene produces a developmentally regulated protein in mesenchyme and vasculature. Nat Med 1:359–364. [DOI] [PubMed] [Google Scholar]
- van de Wetering M, Francies HE, Francis JM, Bounova G, Iorio F, Pronk A, van Houdt W, van Gorp J, Taylor-Weiner A, Kester L, McLaren-Douglas A, Blokker J, Jaksani S, Bartfeld S, Volckman R, van Sluis P, Li VS, Seepo S, Sekhar Pedamallu C, Cibulskis K, Carter SL, McKenna A, Lawrence MS, Lichtenstein L, Stewart C, Koster J, Versteeg R, van Oudenaarden A, Saez-Rodriguez J, Vries RG, Getz G, Wessels L, Stratton MR, McDermott U, Meyerson M, Garnett MJ, Clevers H. 2015. Prospective derivation of a living organoid biobank of colorectal cancer patients. Cell 161:933–945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Berg CW, Ritsma L, Avramut MC, Wiersma LE, van den Berg BM, Leuning DG, Lievers E, Koning M, Vanslambrouck JM, Koster AJ, Howden SE, Takasato M, Little MH, Rabelink TJ. 2018. Renal Subcapsular Transplantation of PSC-Derived Kidney Organoids Induces Neo-vasculogenesis and Significant Glomerular and Tubular Maturation In Vivo. Stem Cell Reports 10:751–765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volpato V, Smith J, Sandor C, Ried JS, Baud A, Handel A, Newey SE, Wessely F, Attar M, Whiteley E, Chintawar S, Verheyen A, Barta T, Lako M, Armstrong L, Muschet C, Artati A, Cusulin C, Christensen K, Patsch C, Sharma E, Nicod J, Brownjohn P, Stubbs V, Heywood WE, Gissen P, De Filippis R, Janssen K, Reinhardt P, Adamski J, Royaux I, Peeters PJ, Terstappen GC, Graf M, Livesey FJ, Akerman CJ, Mills K, Bowden R, Nicholson G, Webber C, Cader MZ, Lakics V. 2018. Reproducibility of Molecular Phenotypes after Long-Term Differentiation to Human iPSC-Derived Neurons: A Multi-Site Omics Study. Stem Cell Reports 11:897–911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang G, McCain ML, Yang L, He A, Pasqualini FS, Agarwal A, Yuan H, Jiang D, Zhang D, Zangi L, Geva J, Roberts AE, Ma Q, Ding J, Chen J, Wang DZ, Li K, Wang J, Wanders RJ, Kulik W, Vaz FM, Laflamme MA, Murry CE, Chien KR, Kelley RI, Church GM, Parker KK, Pu WT. 2014. Modeling the mitochondrial cardiomyopathy of Barth syndrome with induced pluripotent stem cell and heart-on-chip technologies. Nat Med 20:616–623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams KE, Lemieux GA, Hassis ME, Olshen AB, Fisher SJ, Werb Z. 2016. Quantitative proteomic analyses of mammary organoids reveals distinct signatures after exposure to environmental chemicals. Proc Natl Acad Sci U S A 113:E1343–1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H, Uchimura K, Donnelly EL, Kirita Y, Morris SA, Humphreys BD. 2018. Comparative Analysis and Refinement of Human PSC-Derived Kidney Organoid Differentiation with Single-Cell Transcriptomics. Cell Stem Cell. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie F, Ye L, Chang JC, Beyer AI, Wang J, Muench MO, Kan YW. 2014. Seamless gene correction of beta-thalassemia mutations in patient-specific iPSCs using CRISPR/Cas9 and piggyBac. Genome Res 24:1526–1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu P, Tong Y, Liu XZ, Wang TT, Cheng L, Wang BY, Lv X, Huang Y, Liu DP. 2015. Both TALENs and CRISPR/Cas9 directly target the HBB IVS2–654 (C > T) mutation in beta-thalassemia-derived iPSCs. Sci Rep 5:12065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi S, Morizane R, Homma K, Monkawa T, Suzuki S, Fujii S, Koda M, Hiratsuka K, Yamashita M, Yoshida T, Wakino S, Hayashi K, Sasaki J, Hori S, Itoh H. 2016. Generation of kidney tubular organoids from human pluripotent stem cells. Sci Rep 6:38353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamanaka S, Tajiri S, Fujimoto T, Matsumoto K, Fukunaga S, Kim BS, Okano HJ, Yokoo T. 2017. Generation of interspecies limited chimeric nephrons using a conditional nephron progenitor cell replacement system. Nat Commun 8:1719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokoo T, Ohashi T, Shen JS, Sakurai K, Miyazaki Y, Utsunomiya Y, Takahashi M, Terada Y, Eto Y, Kawamura T, Osumi N, Hosoya T. 2005. Human mesenchymal stem cells in rodent whole-embryo culture are reprogrammed to contribute to kidney tissues. Proc Natl Acad Sci U S A 102:3296–3300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yui S, Nakamura T, Sato T, Nemoto Y, Mizutani T, Zheng X, Ichinose S, Nagaishi T, Okamoto R, Tsuchiya K, Clevers H, Watanabe M. 2012. Functional engraftment of colon epithelium expanded in vitro from a single adult Lgr5(+) stem cell. Nat Med 18:618–623. [DOI] [PubMed] [Google Scholar]
- Zorn AM, Wells JM. 2009. Vertebrate endoderm development and organ formation. Annu Rev Cell Dev Biol 25:221–251. [DOI] [PMC free article] [PubMed] [Google Scholar]