Abstract
Respiratory complexes are complicated multi-subunit cofactor-containing machines that allow cells to harvest energy from the environment. Maturation of these complexes requires protein folding, cofactor insertion, and assembly of multiple subunits into a final, functional complex. Because the intermediate states in complex maturation are transitory, these processes are poorly understood. This review gives an overview of the process of maturation in respiratory complex II with a focus on recent structural studies on intermediates formed during covalent flavinylation of the catalytic subunit, SDHA. Covalent flavinylation has an evolutionary significance because variants of complex II enzymes with the covalent ligand removed by mutagenesis cannot oxidize succinate, but can still perform the reverse reaction and reduce fumarate. Since succinate oxidation is a key step of aerobic respiration, the covalent bond of complex II appears to be important for aerobic life.
Introduction
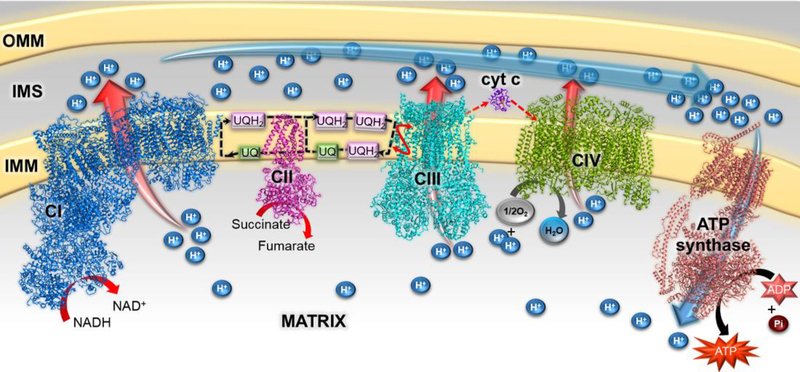
The process of cellular respiration allows cells to harvest chemical energy from the environment via redox reactions and to convert this energy into a biologically useful form. Mitochondrial aerobic respiration (also called as oxidative phosphorylation), depends on the synergetic efforts of four integral-membrane respiratory complexes of the electron transport chain (Figure 1). Often termed respiratory complex I – complex IV, these multi-subunit and multi-cofactor-containing machines use electrons as substrates or products in a series of linked reactions that physically occur in the mitochondrial matrix or within the inner mitochondrial membrane [1]. These respiratory complexes likely function within the context of a megacomplex that optimizes the transfer of intermediate electron carriers between individual complexes [2].The redox reactions catalyzed during respiration are coupled to transmembrane proton translocation, resulting in the formation of a transmembrane electrochemical gradient (Figure 1). The ATP synthase, also termed complex V, uses the energy stored in this gradient to drive ATP synthesis [3].
Figure 1. The mammalian electron transport chain.
Structural models of bovine complex I (PDB 5LNK [4]), porcine complex II (PDB 1ZOY;[5]), human complex III (PDB 5XTE [2]), bovine cytochrome c (PDB 2B4Z [6], bovine complex IV (PDB 1V54;[7]), and Saccharomyces cerevisiae ATP synthase (PDB 6B8H;[3]) are colored blue, magenta, cyan, violet, green, and light brown, respectively. The inner and outer mitochondrial membranes (IMM, OMM) are shown in yellow, and the inter membrane space (IMS) shown in grey. Ubiquinone (UQ, Coenzyme Q10)/ubiquinol (UQH2) are the oxidized and reduced forms of the membrane soluble small molecule carrier of protons and electrons between complex I, complex II and complex III. These are stylistically shown as green and pink boxes. Complexes in this figure are shown side-by-side for clarity, but may exist in a megacomplex in the mitochondria [2].
Mitochondrial respiratory complexes I, II, III and IV contain 44, 4, 11 and 14 subunits, respectively. Bacterial homologs of complexes I-IV can differ in the number of subunits, but the functional “core subunits” are conserved [1]. Distributed among these respiratory complexes are multiple types of cofactors including nicotinamides (NAD+), flavins (FMN, FAD), iron-sulfur (Fe-S) clusters, hemes, and metal ions (Mg, Cu, Zn) [2–7]. Given the complexity of these molecular machines, it is perhaps unsurprising that bacterial expression of the individual subunits commonly results in aggregation. It has been proposed that this results from either the exposure of the hydrophobic surfaces that would normally be buried within subunit interfaces [8••] or because of inappropriate cofactor insertion during heterologous expression [9••].
The maturation of individual respiratory complex subunits involves folding of each subunit and cofactor insertion. This is followed by assembly of the subunits into functional holo-respiratory complexes. These processes are facilitated by both non-specific factors and dedicated assembly factors. The non-specific factors include proteins involved in the biosynthesis of the cofactors, proteins that translocate cofactors or subunits into the mitochondria, and folding chaperones [10]. In contrast, the dedicated assembly factors are specific for each respiratory complex. These dedicated assembly factors are key for the function of the respiratory complexes, but are not a part of the final, assembled complex. Fully validated assembly factors are listed in Table 1, however it should be noted that there are hints that additional assembly factors exist for complex I, III, IV, and V, and it is not clear whether all relevant assembly factors have been discovered. A subset of these assembly factors has homologs in prokaryotes.
Table 1.
The reported assembly factors of respiratory complexes.
| Respiratory complex | Assembly Factors | Ref. |
|---|---|---|
| Complex I | NADH-dehydrogenase alpha subcomplex F assembly factors: NDUFAF1, NDUFAF2, NDHFAF3, NDUFAF4, NDUFAF5, NDUFAF6, NDUFAF7 Nucleotide-binding protein-like protein, NUBPL (Ind1) Acyl-CoA dehydrogenase 9, ACAD9 Evolutionary conserved signaling intermediate in Toll pathways, Ecsit Mitochondrial dysfunction protein A, MidA |
[51] |
| Complex II | Succinate dehydrogenase assembly factors: SDHAF1, SDHAF2, SDHAF3 and SDHAF4 | [52] |
| Complex III | Ubiquinol-Cytochrome C Reductase Complex Chaperone, BCS1 Tetratricopeptide Repeat Domain 19, TTC19 LYR Motif-Containing Protein LYRM7/MZM1L Ubiquinol-Cytochrome C Reductase Complex Assembly Factor: UQCC1 (CBP3), UQCC2 (CBP6) and UQCC3 (CBP4) |
[53] |
| Complex IV | Cytochrome oxidase interacting protein 1, Coi1 Cytochrome C Oxidase Assembly Proteins: COX14, COX16, COX23, COX Assembly Mitochondrial Protein: CMC1, CMC2 Hypoxia Inducible Domain Family Member: HIGD1A, HIGD2A |
[54] |
| ATP Synthase |
Proteins encoded by ATP genes: ATP10, ATP11, ATP12 and ATP23 | [55] |
Complex II and its assembly factors
Complex II (SDH) is the smallest and simplest respiratory complex. Mammalian SDH is nuclearly-encoded and harbors covalently-linked FAD in SDHA (flavoprotein subunit), three types of Fe-S cluster (2Fe-2S, 4Fe-4S, and 3Fe-4S) in SDHAB (Fe-S subunit) and non-covalent b-type heme between SDHC and SDHD (integral membrane subunits). Among these four polypeptide chains, the two soluble subunits, SDHA and SDHB, are conserved from bacteria to mammals [5,11]. In contrast, the two membrane-spanning subunits, SDHC and SDHD, are not well conserved. Indeed, the membrane subunits may be either one or two polypeptide chains and contain 0, 1 or 2 b-type hemes. Sequence analysis classifies the membrane subunits of complex II into five evolutionarily-distinct origins, prompting the division of complex II into subfamilies [12]. Nevertheless, the membrane-spanning subunits of complex II perform a conserved function in quinone oxidoreduction (Figure 1).
Loss of function in any subunit of human SDH via missense or nonsense mutations can manifest as hereditary tumors of neuroendocrine tissues (pheochromocytomas and paragangliomas) or neurodegenerative presentations such as those associated with Leigh’s disease [13]. Moreover, inhibition of complex II via the small molecule 3-nitropropionate forms a covalent adduct with an active site proton shuttle and results in Huntingtin Disease-like symptoms [14]. Mutations in the complex II assembly factors, SDHAF1–4, can result in complex II insufficiency and can recapitulate the clinical symptoms associated with complex II mutation, truncation, or inhibition [9••,15••,16••,17••].
Discovered around the same time [9••,15••], the complex II assembly factors SDHAF1 (Succinate dehydrogenase assembly factor 1) and SDHAF2 have different biological roles. It has been suggested that SDHAF1 promotes insertion of Fe-S clusters in SDHB and its mutation is associated with infantile leukoencephalopathy [15••]. Mutations in SDHAF1 lead to the incorporation of non-functional SDHB into SDH, which is associated with significantly reduced SDH activity [15••]. A second factor, SDHAF3, was later found to work in synergy with SDHAF1 [17••] and may protect the Fe-S clusters in unassembled SDHB from solvent prior to the formation of the SDHA-SDHB heterodimer [17••].
In contrast to the role of SDHAF1 and SDHAF3 in maturation of the SDHB subunit, SDHAF2 is reported as essential for SDHA maturation [9••,16••]. Mutation of a highly conserved RGxxE motif of SDHAF2 results in complex II insufficiency and paraganglioma [9••]. Complementary studies using yeast and bacterial homologs as model systems revealed that loss of function of the homologous proteins (termed Sdh5 in yeast and SdhE in bacteria) via deletion or mutation of the RGxxE motif results in loss of the covalent bond between FAD and the SDHA subunit (termed Sdh1 in yeast and SdhA in bacteria) [18,19]. This suggests a role of SDHAF2/Sdh5/SdhE in promoting covalent flavinylation [9••]. A second assembly factor, termed SDHAF4, also likely interacts with SDHA, and may contribute to maturation of complex II by promoting SDHA-SDHB assembly into an SDHAB subcomplex [16••,20]. SDHAF4 currently has no identifiable sequence homologs in bacteria.
Maturation of the complex II Flavoprotein Subunit, SDHA
Of the four subunits of complex II, the maturation of the flavoprotein subunit, SDHA, is the best understood. Both in unassembled subunits [21••] and in the context of assembled complex II [11], the SDHA subunit folds into two distinct domains, termed a flavin-binding domain and a capping domain [11]. The active site for succinate-fumarate interconversion is positioned at the interface between these two domains [11]. Maturation of SDHA requires the synthesis, mitochondrial import, and folding of the SDHA apoprotein, the biosynthesis of FAD, the insertion of FAD into the SDHA apoprotein, and the covalent attachment of FAD to SDHA. Of these processes, covalent flavinylation appears to depend upon each of the other steps and is therefore believed to be the terminal step of maturation of the isolated SDHA subunit prior to assembly into the complex. Monitoring covalent FAD attachment to SDHA is accordingly used as a reporter of subunit maturation.
Import of SDHA into the mitochondrion, folding, and processing
While the initial stages of SDHA maturation are poorly understood, what is known has been suggested by studies on the yeast homolog, where the complex II subunits are termed Sdh1, Sdh2, Sdh3, and Sdh4 rather than SDHA, SDHB, SDHC, and SDHD. It is not known whether the nuclearly-encoded Sdh1 is folded in the cytosol following translation, however it targets to the mitochondria via a presequence. There, if the Sdh1 subunit is folded, it must be first unfolded prior to translocation across both mitochondrial membranes, which likely depends on the translocase of the outer membrane (TOM) and the translocase of the inner membrane (TIM23) complex, both of which are general translocases [22].
Once in the mitochondrion, Sdh1 both folds and has the targeting presequence removed prior to covalent flavinylation and full maturation, although the temporal order of these two events is currently not known. As shown by gene deletion studies and immunoprecipitation, folding of SDH subunits in the matrix depends upon the activity of a membrane-spanning chaperonin, termed TCM62 in S. cerevisiae, which interacts directly with Sdh1, Sdh2, and Sdh3 [23]. Sdh1 also appears to interact with Hsp60, but deletion of Hsp60 does not impact covalent flavinylation [24]. While this initially suggested that unfolded Sdh1 could be covalently flavinylated, later structural studies on bacterial homologs reveal that a fully formed active site is almost certainly required for the flavinlation reaction to occur efficiently [21••], an observation consistent with biochemical studies showing that substrate analogs, which presumably bind to the active site, enhance flavinylation [25]. Moreover, truncated Sdh1 cannot be covalently flavinylated [26], consistent with folding as a prerequisite for covalent flavinylation. Thus, the significance of the interaction between Sdh1 and Hsp60 in maturation remains unclear.
Pulse-chase studies in S. cerevisiae also strongly suggest that proteolytic cleavage of the targeting presequence is required for covalent FAD attachment to Sdh1 [26], although the mechanistic rationale for this remains to be elucidated. Potential proteases that could cleave the presequence include the mitochondrial processing peptidase, which cleaves most presequence-containing proteins in mitochondria [27], and/or octapeptidyl aminopeptidase 1, which recognizes a conserved consensus sequence found in Sdh1 [28].
FAD Biosynthesis
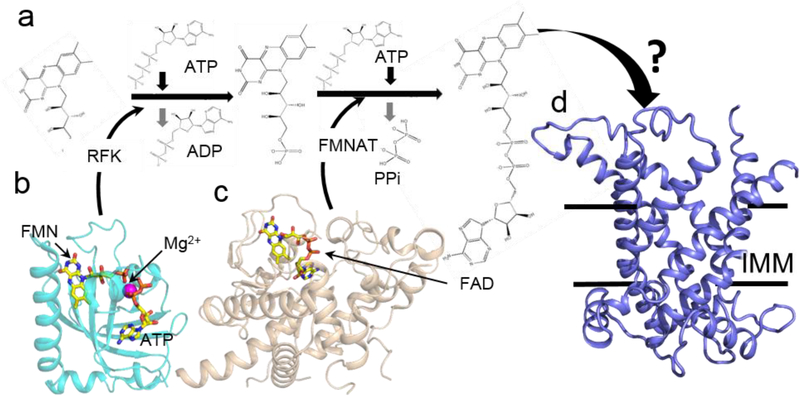
The next step in the maturation is binding of non-covalent FAD to SDHA, which may occur concomitantly with folding. This process requires FAD biosynthesis and colocalization of the FAD cofactor with the SDHA apoprotein. The isoalloxazine ring of FAD is derived from riboflavin (vitamin B2), which is converted to FAD in a two-step process (Figure 2a). First is the addition of a phosphate group to from flavin mononucleotide (FMN). This is followed by the addition of an ADP moiety to form FAD. Higher organisms accomplish this through the coupled activity of two distinct enzymes. Riboflavin kinase (RFK; Figure 2b), synthesizes FMN from riboflavin, and FMN adenylyltransferase (FMNAT; Figure 2c) converts FMN into FAD [29,30]. In contrast, prokaryotes utilize a single bifunctional enzyme, termed FAD synthetase, that possesses both catalytic modules i.e. a C-terminal RFK module and an N-terminal FMNAT module [31]. The mechanism of these enzymes in FAD biosynthesis has been reviewed elsewhere [32].
Figure 2.
Mechanism of FAD biosynthesis. a Biosynthesis of FAD is a two step process that converts riboflavin (RF) to FMN, followed by conversion to FMN to FAD. Each step is ATP dependent and involves the enzymes riboflavin kinase (RFK) and flavin mononucleotide adenylyltransferase (FMNAT) b Structure of human RFK in complex with FMN and ADP (PDB 1Q9S, [47]). c Structure of S. cerevisiae FMNAT (Fad1, PDB 2WSI [48]) with bound FAD. d A homology model of the Flx1FAD transporter constructed using mitochondrial uncoupling protein 2 (UCP2) as a template (PDB 2LCK [32]). Sequence identity and similarity between Flx1 and UCP2 is 24% and 40%, respectively. IMM is inner mitochondrial membrane.
This reaction clearly occurs in bacteria, which are often thought of as evolutionary precursors to mitochondria. Surprisingly, however, it is not currently clear for eukaryotic cells whether the enzymatic conversion of riboflavin to FAD can occur in mitochondria or whether FAD must be imported. If the latter occurs, then FAD would require two transporters, one to cross the outer membrane, and one to traverse the inner membrane. No outer membrane protein has yet been implicated in this process, although the Voltage-gated anion channel, VDAC, has been implicated in the transport of similarly-sized and charged cofactors, such as ATP, ADP, and NADH. In terms of an inner membrane transporter, early reports suggested that the flavin exchange protein, Flx1 could fill this role. Flx1 belongs to the mitochondrial anion carrier superfamily [33], with Δflx1 S. cerevisiae exhibiting a low mitochondrial FAD/FMN ratio and negligible Sdh1 flavinylation [9••,34]. However, an increasing number of subsequent reports describe Flx1 as facilitating FAD export from the mitochondrial matrix [35]. With this function, the mechanism underlying the effects of Flx1 deletion on SDH activity becomes unclear.
Covalent flavinylation of SDHA
The final step in SDHA maturation is formation of the covalent bond between FAD and SDHA. As we recently reviewed elsewhere [36] a covalent flavin can involve one of several positions on the FAD and can use a variety of protein side chains. In the case of complex II, the covalent bond is found between the C8α methylene carbon of the FAD isoalloxazine and the Nε atom of an absolutely conserved histidine of SDHA (Figure 3a). Covalent attachment of any flavin to a protein significantly increases the redox potential of the cofactor, usually by ~80 mV [36]. This increase in redox potential is required for SDHA to oxidize succinate, a key chemical transformation in both aerobic respiration and the Krebs cycle [37].
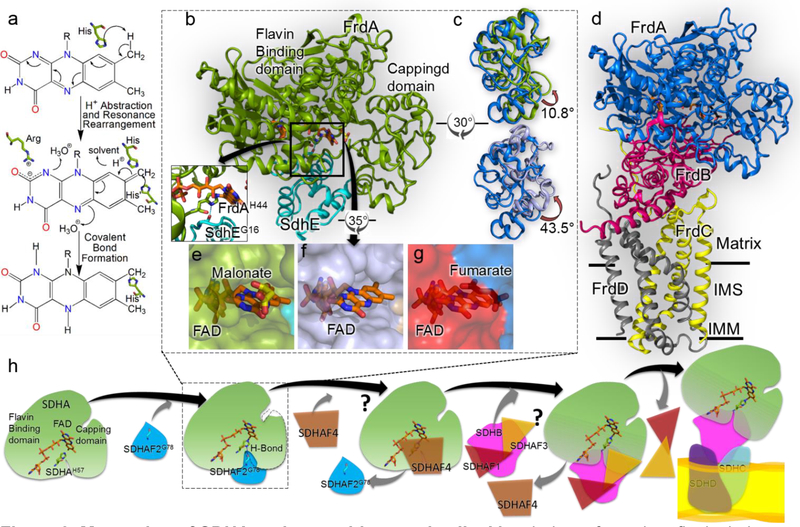
Figure 3. Maturation of SDHA and assembly complex II.
a Mecahnism of covalent flavinylation, as proposed by Walsh [49]. The first step is proton abstraction from the C8α of the isoalloxazine ring. The second step is resonance rearrangement to delocalize the resultant negative charge between N1 and C2. The third step is the attack by a histidyl ligand which results in formation of covalent bond. b The structure of the E. coli FrdA-SdhE assembly intermediate (PDB 6B58, [21••]). FrdA is colored green and SdhE is colored cyan. Inset The hydrogen bond between bacterial SdhEG16 of the RGxxE motif and FrdAH44 shows how SdhE orients the histidyl ligand. c Overlay of the capping domains of assembled FrdABCD complex (blue, PDB 3P4P, [50]) with the position observed in the FrdA-SdhE intermediate (top, green, PDB 6B58, [21••]) or SdhA-SdhE intermediate (bottom, purple, PDB 6C12 [42•]). A rotation of 10.8° and 43.5° is observed in the capping domains, respectively. d The structure of the E. coli fumarate reductase (PDB 3P4P [50]) with FrdA oriented as in Fig. 3b. IMM is inner mitochondrial membrane and IMS is inter-mitochondrial space. e, f Surface representation view of the FrdA-SdhE (PDB 6B58) and SdhA-SdhE (PDB 6C12) intermediates [21••,42•] highlights a tunnel from the solvent to the active site. g A similar surface representation of the FrdA subunit in the context of assembled complex II (PDB 3P4P, [50]) indicates that this tunnel closes following assembly. h A schematic representation of the proposed SDH assembly pathway. The details of SDHA-SDHAF2 interaction are known but structural states of interaction between assembly factors SDHAF1, SDHAF3 and SDHAF4 with SDH subunits is still unclear, and marked as “?”. Understanding how these assembly factors work is the focus of current research in the field.
Covalent flavinylation in many proteins is likely autocatalytic and is proposed to proceed via a mechanism that capitalizes on the reactive properties of the isoalloxazine ring itself (Figure 3a). Although the covalent FAD of complex II was the first covalent flavin to be discovered, characterization of the mechanisms of covalent flavinylation have predominantly relied on other model systems, for example p-cresol methylhydroxylase, monomeric sarcosine oxidase and Vanillyl-alcohol oxidase [38–40]. Based upon extrapolation of these mechanisms, it was long believed that covalent flavinylation of SDHA was similarly autocatalytic. The discovery of SDHAF2/Sdh5 in 2009 and SdhE in 2012, proteins that stimulate covalent flavinylation and are conserved in all kingdoms, was therefore a surprise [9••,19].
Initial hypotheses for the role of SDHAF2/Sdh5/SdhE in covalent flavinylation were wide-ranging. For example, using the yeast system, a proposal that Sdh5 might bind to FAD and transfer the cofactor to Sdh1 was consistent with studies showing that overexpression Sdh5 in Δflx1 S. cerevisiae, which have impaired covalent flavinylation of Sdh1, restored flavinylation up to 50% of the wild type level [9••]. One interpretation of this result is that Sdh5 increases the FAD availability for Sdh1. However, later NMR titration analysis was inconsistent with this hypothesis for function as excess of FAD or FADH2 did not induce chemical shift perturbations in yeast Sdh5, indicating that Sdh5 does not bind FAD directly [41•]. While these analyses do not preclude an indirect interaction between Sdh5 and FAD, the same structural study identified a conserved surface of Sdh5 that appeared to be a protein-protein binding interface [41•] and was later shown to be critical for activity.
A major breakthrough in understanding the role of SDHAF2/Sdh5/SdhE came from the recently reported crystal structures of bacterial homologs of the SDHA-SDHAF2 assembly intermediate (Figure 3b) [21••,42•]. In both reported structures, the Escherichia coli complex II was used as a model system. E. coli is a facultative anaerobe and contains two complex II homologs that contribute to respiration under distinct respiratory conditions. One homolog, called succinate dehydrogenase or succinate:quinone oxidoreductase (SdhABCD) participates in aerobic respiration, while a second homolog, called fumarate reductase or quinol:fumarate reductase (FrdABCD) catalyzes the final step of anaerobic respiration with fumarate as the terminal electron acceptor.
The first structure was that of E. coli FrdA-SdhE (Figure 3b) [21••]. This structure was determined in the presence of the substrate analog malonate, which is a dicarboxylate molecule that is also an enhancer of covalent flavinylation [43•]. For these studies, the FrdA-SdhE assembly intermediate was stabilized by a site-specific photoaffinity crosslinker introduced via an unnatural amino acid [21••,37,43•]. This structure also identified that the role of the highly-conserved RGxxE motif in SdhE was to stabilize the orientation of the histidyl ligand for FAD via a hydrogen-bonding interaction between the carbonyl of the glycine and the histidine Nδ (Figure 3b inset).
The second structure was that of E. coli SdhA-SdhE [42•], which was performed in the absence of a substrate analog or crosslinker. These two structures provided a satisfying explanation for how SDHAF2/Sdh5/SdhE enhances covalent flavinylation in this system. Specifically, the SdhE assembly factor bound between the two domains of the FrdA or SdhA subunit. This is the same position as the FrdB/SdhB subunit binds in fully assembled complex II (Figure 3b,d). Not only does SdhE shield this hydrophobic surface and prevent inappropriate interactions in a chaperone-like manner, but this protein-protein interaction subtly rearranges the active site architecture as compared to the FrdA/SdhA subunit bound to FrdB/SdhB in mature complex II. This has two distinct impacts. First is that the normal catalytic reaction of assembled complex II, i.e. the interconversion of succinate and fumarate, is disfavored, as validated kinetically in both the E. coli and human systems [21••,44]. The second is that the geometry of the malonate-bound active site was optimized to enhance covalent flavinylation [21••]. Or to put it another way, the assembly factor acts as a regulatory subunit of SdhA/FrdA catalytic activity, and enhances the covalent flavinylation reaction allosterically [21••].
These structures also were associated with conformational changes that have created new controversies and new open questions in the field. For example, one of the conformational changes linked to the tuning of the catalytic activity of the SdhA/FrdA subunits was an interdomain rotation between the flavin-binding domain and the capping domain as compared to the positions of these domains in the assembled complex II (Figure 3c). While both the E. coli FrdA-SdhE [21••] and SdhA-SdhE [42•] exhibited an interdomain rotation, the magnitude was profoundly different, 11° in E. coli FrdA-SdhE and 43° in E. coli SdhA-SdhE. As proposed by Maher et al. [42•], one hypothesis for this difference in magnitude is that the E. coli FrdA-SdhE was stabilized by a crosslinker, which may have impaired domain rotation. However, there are multiple competing hypotheses for the observed differences. For example, the E. coli SdhA-SdhE structure lacks a substrate analog, which is a long-known enhancer of covalent flavinylation [37,43•]. As the malonate assists in aligning the interdomain orientation, the 43° rotation observed in the uncrosslinked E. coli SdhA-SdhE could be a catalytically irrelevant state. Indeed, one would anticipate that the physiologically-relevant catalytic complex would be relatively unstable. A third hypothesis is that both structures represent relevant intermediates of the flavinylation reaction. Addressing how the inderdomain angle influences flavinylation is a topic of ongoing work.
A second new question in the field arose from the observation that binding of the SdhE assembly factor to either FrdA or SdhE was associated with the loss of electron density for two large loop regions (residues 50–58 and 103–129 of the E. coli FrdA) [21••,42•]. This likely reflects loss of a discrete position for these loops and increased flexibility. If these loops are no longer tightly associated with FrdA/SdhA, it would result in the formation of a tunnel to the active site that would be large enough to allow access of substrate (Figure 3e,f,g). The introduction of engineered disulfide bonds to lock these loops in place substantially reduced covalent flavinylation in the E. coli FrdA-SdhE system, a result that strongly suggests that these loops must become flexible during the covalent flavinylation reaction [21••]. Nevertheless, the exact role of this active site tunnel in the mechanism of covalent flavinylation is currently not known and is a direction of future research.
Formation of the SDHAB subcomplex
Following successful SDHA maturation, the folded and covalently-flavinylated SDHA subunit forms a stable complex with SDHB, which may or may not harbor all three Fe-S clusters. Neither the maturation of SDHB nor the formation of the SDHAB subcomplex is well understood at this time, and both are subjects of active investigation in the field.
SDHB maturation requires the insertion of three Fe-S clusters into two protein domains. An N-terminal plant-type ferredoxin 2Fe-2S containing domain and a, bacterial ferredoxin C-terminal 4Fe-4S and 3Fe-4S domain. Insertion of the Fe-S clusters may rely upon general Fe-S insertion mechanisms in both prokaryotes and eukaryotes [8••,45]. Eukaryotes also contain the SDHAF1 and SDHAF3 assembly factors, which have no identified sequence homologs in prokaryotes [17••]. It is notable that if the isolated SDHB subunit is anticipated to have the oxygen- and ROS-sensitive 2Fe-2S and 3Fe-4S clusters surface exposed. Thus, one possible role for the SDHAF1 and SDHAF3 assembly factors would be to shield these cluster prior to assembly of mature SDHB into the SDHAB subcomplex and the final, mature complex II.
The formation of the SDHAB subcomplex is not a simple meeting of two unassembled subunits. Indeed, SDHA and SDHB likely encounter each other in the context of an SDHA-SDHAF2 complex and an SDHB-SDHAF1-SDHAF3 complex. It is clear from crystal structures [21••,42•] that SDHAF2 physically occludes the site of SDHB interaction, and it is likely that either SDHAF1 or SDHAF3 physically occludes the site of SDHB that interacts with SDHA (Figure 3h). Therefore, these assembly factors must be displaced in order to proceed with complex II assembly. In eukaryotes, an additional assembly factor, termed SDHAF4 in human and Sdh8 in yeast, binds directly to SDHA/Sdh1 [16••] and likely facilitates this process. Indeed, studies of Arabidopsis Δsdhaf4 showed that SDHAF4 is essential for SDHB stability and assembly of SDHAB subcomplex [20]. As no sequence or functional homologs of SDHAF4 have been identified in bacteria to date, future investigations of SDHAB subcomplex formation require the development of techniques and expression systems for the eukaryotic complex II subunits, a process that is the focus of intense current efforts [21••,37,44,46].
Conclusion
While not all structural states of complex II maturation and assembly have been determined, the structures that are available provide valuable information into the role assembly factors in maturation of complex II and suggest general themes of how assembly factors contribute to the maturation of other respiratory complexes. For example, each of the four SDH-associated assembly factors potentially acts as specific chaperones for sensitive cofactors that are normally found at subunit interfaces. As exemplified by SDHAF2, assembly factors may also alter the conformation of an active site and temporarily induce distinct catalytic reactivity for isolated subunits, particularly for chemical reactions required only one time during assembly. The precise mechanisms underlying full assembly of complex II are still in the process of elucidation.
Acknowledgements
The work from the authors laboratories was supported by National Institutes of Health award GM61606. G.C. is the recipient of a Senior Research Career Scientist award 1K6B004215 from the Department of Veterans Affairs. PS supported by Postdoctoral Fellowship 19POST34450093 from the American Heart Association. The authors thank I Yamakawa, AI Kaya, NA Perry, and PK Singh for critical reading.
Footnotes
Declarations of interest
None
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References and recommended reading
Papers of particular interest, published within the period of review, have been highlighted as of special interest of outstanding interest
• of special interest
•• of outstanding interest
- 1.Rich PR, Marechal A: The mitochondrial respiratory chain. Essays Biochem 2010, 47:1–23. [DOI] [PubMed] [Google Scholar]
- 2.Guo R, Zong S, Wu M, Gu J, Yang M: Architecture of Human Mitochondrial Respiratory Megacomplex I2III2IV2. Cell 2017, 170:1247–1257 e1212. [DOI] [PubMed] [Google Scholar]
- 3.Guo H, Bueler SA, Rubinstein JL: Atomic model for the dimeric FO region of mitochondrial ATP synthase. Science 2017, 358:936–940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fiedorczuk K, Letts JA, Degliesposti G, Kaszuba K, Skehel M, Sazanov LA: Atomic structure of the entire mammalian mitochondrial complex I. Nature 2016, 538:406–410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sun F, Huo X, Zhai Y, Wang A, Xu J, Su D, Bartlam M, Rao Z: Crystal structure of mitochondrial respiratory membrane protein complex II. Cell 2005, 121:1043–1057. [DOI] [PubMed] [Google Scholar]
- 6.Mirkin N, Jaconcic J, Stojanoff V, Moreno A: High resolution X-ray crystallographic structure of bovine heart cytochrome c and its application to the design of an electron transfer biosensor. Proteins 2008, 70:83–92. [DOI] [PubMed] [Google Scholar]
- 7.Tsukihara T, Shimokata K, Katayama Y, Shimada H, Muramoto K, Aoyama H, Mochizuki M, Shinzawa-Itoh K, Yamashita E, Yao M, et al. : The low-spin heme of cytochrome c oxidase as the driving element of the proton-pumping process. Proc Natl Acad Sci U S A 2003, 100:15304–15309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Boniecki MT, Freibert SA, Muhlenhoff U, Lill R, Cygler M: Structure and functional dynamics of the mitochondrial Fe/S cluster synthesis complex. Nat Commun 2017, 8:1287.•• This report demonstrates the architecture of human “core Iron-Sulfur Cluster (ISC) complex” using crystallization supplemented by solution (SAXS) studies. This work describes how eukaryotic specific assembly factor ISD11-ACP connects Fe/S cluster synthesis to mitochondrial fatty acid synthesis.
- 9.Hao HX, Khalimonchuk O, Schraders M, Dephoure N, Bayley JP, Kunst H, Devilee P, Cremers CW, Schiffman JD, Bentz BG, et al. : SDH5, a gene required for flavination of succinate dehydrogenase, is mutated in paraganglioma. Science 2009, 325:1139–1142.•• This is the first report describing the yeast Sdh5 (SDHAF2) interaction with the catalytic subunit of complex II. This work further identifies that Sdh5 is required for Sdh1 (SDHA) flavinylation and mutations in this protein are associated with paraganglioma.
- 10.Van Vranken JG, Na U, Winge DR, Rutter J: Protein-mediated assembly of succinate dehydrogenase and its cofactors. Crit Rev Biochem Mol Biol 2015, 50:168–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Iverson TM, Luna-Chavez C, Cecchini G, Rees DC: Structure of the Escherichia coli fumarate reductase respiratory complex. Science 1999, 284:1961–1966. [DOI] [PubMed] [Google Scholar]
- 12.Hagerhall C, Hederstedt L: A structural model for the membrane-integral domain of succinate: quinone oxidoreductases. FEBS Lett 1996, 389:25–31. [DOI] [PubMed] [Google Scholar]
- 13.Iverson TM, Maklashina E, Cecchini G: Structural basis for malfunction in complex II. J Biol Chem 2012, 287:35430–35438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Huang LS, Sun G, Cobessi D, Wang AC, Shen JT, Tung EY, Anderson VE, Berry EA: 3-nitropropionic acid is a suicide inhibitor of mitochondrial respiration that, upon oxidation by complex II, forms a covalent adduct with a catalytic base arginine in the active site of the enzyme. J Biol Chem 2006, 281:5965–5972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ghezzi D, Goffrini P, Uziel G, Horvath R, Klopstock T, Lochmuller H, D’Adamo P, Gasparini P, Strom TM, Prokisch H, et al. : SDHAF1, encoding a LYR complex-II specific assembly factor, is mutated in SDH-defective infantile leukoencephalopathy. Nat Genet 2009, 41:654–656.•• This publication identified the first dedicated SDH assembly factor, SDHAF1. This work also demonstrated that loss of SDHAF1 function is linked to impaired SDHB and leads to bilateral leukoencephalopathy.
- 16.Van Vranken JG, Bricker DK, Dephoure N, Gygi SP, Cox JE, Thummel CS, Rutter J: SDHAF4 promotes mitochondrial succinate dehydrogenase activity and prevents neurodegeneration. Cell Metab 2014, 20:241–252.•• This is the first report demonstrating that SDHAF4 is a dedicated assembly factor for SDH catalytic subunit, SDHA. The data suggests that SDHAF4 facilitates SDHA-SDHB association.
- 17.Na U, Yu W, Cox J, Bricker DK, Brockmann K, Rutter J, Thummel CS, Winge DR: The LYR factors SDHAF1 and SDHAF3 mediate maturation of the iron-sulfur subunit of succinate dehydrogenase. Cell Metab 2014, 20:253–266.•• This work describes the physiological function of two assembly factors, Sdh6 (SDHAF1) and Sdh7 (SDHAF3), as key chaperones in maturation of the Fe/S cluster containing subunit Sdh2 (SDHB). This work also reveals that SDHAF3 exhibits muscular and neuronal dysfunction. Together both assembly factors promote Sdh2 maturation by binding to an Sdh1/Sdh2 intermediate, shielding it form the deleterious effects of oxidants.
- 18.McNeil MB, Fineran PC: The conserved RGxxE motif of the bacterial FAD assembly factor SdhE is required for succinate dehydrogenase flavinylation and activity. Biochemistry 2013, 52:7628–7640. [DOI] [PubMed] [Google Scholar]
- 19.McNeil MB, Clulow JS, Wilf NM, Salmond GP, Fineran PC: SdhE is a conserved protein required for flavinylation of succinate dehydrogenase in bacteria. J Biol Chem 2012, 287:18418–18428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Belt K, Van Aken O, Murcha M, Millar AH, Huang S: An Assembly Factor Promotes Assembly of Flavinated SDH1 into the Succinate Dehydrogenase Complex. Plant Physiol 2018, 177:1439–1452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sharma P, Maklashina E, Cecchini G, Iverson TM: Crystal structure of an assembly intermediate of respiratory Complex II. Nat Commun 2018, 9:274.•• First structural study demonstrating the interaction between complex II flavoprotein and its dedicated assembly factor. This structure reveals that bacterial assembly factor SdhE acts as a transient regulatory subunit that alters conformational equilibrium of flavoprotein active site towards covalent flavinylation. Structural predictions were validated biochemically.
- 22.Schmidt O, Pfanner N, Meisinger C: Mitochondrial protein import: from proteomics to functional mechanisms. Nat Rev Mol Cell Biol 2010, 11:655–667. [DOI] [PubMed] [Google Scholar]
- 23.Dibrov E, Fu S, Lemire BD: The Saccharomyces cerevisiae TCM62 gene encodes a chaperone necessary for the assembly of the mitochondrial succinate dehydrogenase (complex II). J Biol Chem 1998, 273:32042–32048. [DOI] [PubMed] [Google Scholar]
- 24.Robinson KM, Lemire BD: Covalent attachment of FAD to the yeast succinate dehydrogenase flavoprotein requires import into mitochondria, presequence removal, and folding. J Biol Chem 1996, 271:4055–4060. [DOI] [PubMed] [Google Scholar]
- 25.Brandsch R, Bichler V: Covalent cofactor binding to flavoenzymes requires specific effectors. Eur J Biochem 1989, 182:125–128. [DOI] [PubMed] [Google Scholar]
- 26.Robinson KM, Lemire BD: A requirement for matrix processing peptidase but not for mitochondrial chaperonin in the covalent attachment of FAD to the yeast succinate dehydrogenase flavoprotein. J Biol Chem 1996, 271:4061–4067. [DOI] [PubMed] [Google Scholar]
- 27.Gakh O, Cavadini P, Isaya G: Mitochondrial processing peptidases. Biochim Biophys Acta 2002, 1592:63–77. [DOI] [PubMed] [Google Scholar]
- 28.Vogtle FN, Prinz C, Kellermann J, Lottspeich F, Pfanner N, Meisinger C: Mitochondrial protein turnover: role of the precursor intermediate peptidase Oct1 in protein stabilization. Mol Biol Cell 2011, 22:2135–2143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Santos MA, Jimenez A, Revuelta JL: Molecular characterization of FMN1, the structural gene for the monofunctional flavokinase of Saccharomyces cerevisiae. J Biol Chem 2000, 275:28618–28624. [DOI] [PubMed] [Google Scholar]
- 30.Wu M, Repetto B, Glerum DM, Tzagoloff A: Cloning and characterization of FAD1, the structural gene for flavin adenine dinucleotide synthetase of Saccharomyces cerevisiae. Mol Cell Biol 1995, 15:264–271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Herguedas B, Martinez-Julvez M, Frago S, Medina M, Hermoso JA: Oligomeric state in the crystal structure of modular FAD synthetase provides insights into its sequential catalysis in prokaryotes. J Mol Biol 2010, 400:218–230. [DOI] [PubMed] [Google Scholar]
- 32.Kim HJ, Winge DR: Emerging concepts in the flavinylation of succinate dehydrogenase. Biochim Biophys Acta 2013, 1827:627–636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Robinson AJ, Kunji ER: Mitochondrial carriers in the cytoplasmic state have a common substrate binding site. Proc Natl Acad Sci U S A 2006, 103:2617–2622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tzagoloff A, Jang J, Glerum DM, Wu M: FLX1 codes for a carrier protein involved in maintaining a proper balance of flavin nucleotides in yeast mitochondria. J Biol Chem 1996, 271:7392–7397. [DOI] [PubMed] [Google Scholar]
- 35.Bafunno V, Giancaspero TA, Brizio C, Bufano D, Passarella S, Boles E, Barile M: Riboflavin uptake and FAD synthesis in Saccharomyces cerevisiae mitochondria: involvement of the Flx1p carrier in FAD export. J Biol Chem 2004, 279:95–102. [DOI] [PubMed] [Google Scholar]
- 36.Starbird CA, Maklashina E, Cecchini G, Iverson T: Flavoenzymes: Covalent versus Noncovalent In eLS. Edited by: In eLS, John Wiley & Sons, Ltd; (Ed.). 2015. doi: 10.1002/9780470015902.a0026073 [DOI] [Google Scholar]
- 37.Starbird CA, Maklashina E, Sharma P, Qualls-Histed S, Cecchini G, Iverson TM: Structural and biochemical analyses reveal insights into covalent flavinylation of the Escherichia coli Complex II homolog quinol:fumarate reductase. J Biol Chem 2017, 292:12921–12933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hassan-Abdallah A, Zhao G, Jorns MS: Covalent flavinylation of monomeric sarcosine oxidase: identification of a residue essential for holoenzyme biosynthesis. Biochemistry 2008, 47:1136–1143. [DOI] [PubMed] [Google Scholar]
- 39.Jin J, Mazon H, van den Heuvel RH, Heck AJ, Janssen DB, Fraaije MW: Covalent flavinylation of vanillyl-alcohol oxidase is an autocatalytic process. FEBS J 2008, 275:5191–5200. [DOI] [PubMed] [Google Scholar]
- 40.Mewies M, McIntire WS, Scrutton NS: Covalent attachment of flavin adenine dinucleotide (FAD) and flavin mononucleotide (FMN) to enzymes: the current state of affairs. Protein Sci 1998, 7:7–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Eletsky A, Jeong MY, Kim H, Lee HW, Xiao R, Pagliarini DJ, Prestegard JH, Winge DR, Montelione GT, Szyperski T: Solution NMR structure of yeast succinate dehydrogenase flavinylation factor Sdh5 reveals a putative Sdh1 binding site. Biochemistry 2012, 51:8475–8477.• First structure of yeast Sdh5 using NMR spectroscopy. This structure suggests the interaction interface between Sdh1-Sdh5. Complementary chemical shift perturbation measurements indicate that Sdh5 is not responsible for cofactor transport since negligible binding between Sdh5 and FAD was observed.
- 42.Maher MJ, Herath AS, Udagedara SR, Dougan DA, Truscott KN: Crystal structure of bacterial succinate:quinone oxidoreductase flavoprotein SdhA in complex with its assembly factor SdhE. Proc Natl Acad Sci U S A 2018, 115:2982–2987.• Structural study demonstrating the interaction between bacterial SdhA (SDHA) and its dedicated assembly factor SdhE. This study reveals that SdhE constrains the conformation of SdhA to facilitate covalent flavinylation, supporting previous work by Sharma et al. 2018.
- 43.Maklashina E, Rajagukguk S, Starbird CA, McDonald WH, Koganitsky A, Eisenbach M, Iverson TM, Cecchini G: Binding of the Covalent Flavin Assembly Factor to the Flavoprotein Subunit of Complex II. J Biol Chem 2016, 291:2904–2916.• The authors developed photoaffinity cross-linking method to stabilize the transient interaction between bacterial assembly factor SdhE with SdhA/FrdA (SDHA) in E. coli. This tool was used for later structural and biochemical work that assessed the function of SdhE.
- 44.Maklashina E, Rajagukguk S, Iverson TM, Cecchini G: The unassembled flavoprotein subunits of human and bacterial complex II have impaired catalytic activity and generate only minor amounts of ROS. J Biol Chem 2018, 293:7754–7765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Blauenburg B, Mielcarek A, Altegoer F, Fage CD, Linne U, Bange G, Marahiel MA: Crystal Structure of Bacillus subtilis Cysteine Desulfurase SufS and Its Dynamic Interaction with Frataxin and Scaffold Protein SufU. PLoS One 2016, 11:e0158749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zafreen L, Walker-Kopp N, Huang LS, Berry E: In-vitro, SDH5-dependent flavinylation of immobilized human respiratory complex II flavoprotein. Arch Biochem Biophys 2016, 604:47–56. [DOI] [PubMed] [Google Scholar]
- 47.Karthikeyan S, Zhou Q, Osterman AL, Zhang H: Ligand binding-induced conformational changes in riboflavin kinase: structural basis for the ordered mechanism. Biochemistry 2003, 42:12532–12538. [DOI] [PubMed] [Google Scholar]
- 48.Leulliot N, Blondeau K, Keller J, Ulryck N, Quevillon-Cheruel S, van Tilbeurgh H: Crystal structure of yeast FAD synthetase (Fad1) in complex with FAD. J Mol Biol 2010, 398:641–646. [DOI] [PubMed] [Google Scholar]
- 49.Walsh C: Flavin coenzymes: at the crossroads of biological redox chemistry. Accounts of Chemical Research 1980, 13:148–155. [Google Scholar]
- 50.Tomasiak TM, Archuleta TL, Andrell J, Luna-Chavez C, Davis TA, Sarwar M, Ham AJ, McDonald WH, Yankovskaya V,Stern HA, et al. : Geometric restraint drives on- and off-pathway catalysis by the Escherichia coli menaquinol:fumarate reductase. J Biol Chem 2011, 286:3047–3056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mimaki M, Wang X, McKenzie M, Thorburn DR, Ryan MT: Understanding mitochondrial complex I assembly in health and disease. Biochim Biophys Acta 2012, 1817:851–862. [DOI] [PubMed] [Google Scholar]
- 52.Signes A, Fernandez-Vizarra E: Assembly of mammalian oxidative phosphorylation complexes I-V and supercomplexes. Essays Biochem 2018, 62:255–270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Smith PM, Fox JL, Winge DR: Biogenesis of the cytochrome bc(1) complex and role of assembly factors. Biochim Biophys Acta 2012, 1817:276–286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Timon-Gomez A, Nyvltova E, Abriata LA, Vila AJ, Hosler J, Barrientos A: Mitochondrial cytochrome c oxidase biogenesis: Recent developments. Semin Cell Dev Biol 2018, 76:163–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Song J, Pfanner N, Becker T: Assembling the mitochondrial ATP synthase. Proc Natl Acad Sci U S A 2018, 115:2850–2852. [DOI] [PMC free article] [PubMed] [Google Scholar]