Abstract
Tamoxifen is the most commonly used endocrine therapy for patients with hormone receptor (HR)-positive breast cancer. Despite its initial therapeutic efficacy, many patients eventually develop drug resistance, which remains a serious clinical challenge. To investigate roles of circular RNAs (circRNAs) in tamoxifen resistance, a tamoxifen-resistant MCF-7 cell line was established and screened for its circRNA expression profile by RNA sequencing. hsa_circ_0025202, a circRNA that was significantly downregulated, was selected for further investigation. Using a large cohort of clinical specimens, we found that hsa_circ_0025202 exhibited low expression in cancer tissues and was negatively correlated with lymphatic metastasis and histological grade. Gain- and loss-of-function assays indicated that hsa_circ_0025202 could inhibit cell proliferation, colony formation, and migration and increase cell apoptosis and sensitivity to tamoxifen. Bioinformatics and luciferase reporter assays verified that hsa_circ_0025202 could act as a miRNA sponge for miR-182-5p and further regulate the expression and activity of FOXO3a. Functional studies revealed that tumor inhibition and tamoxifen sensitization effects of hsa_circ_0025202 were achieved via the miR-182-5p/FOXO3a axis. Moreover, in vivo experiments confirmed that hsa_circ_0025202 could suppress tumor growth and enhance tamoxifen efficacy. Taken together, hsa_circ_0025202 served an anti-oncogenic role in HR-positive breast cancer, and it could be exploited as a novel marker for tamoxifen-resistant breast cancer.
Keywords: breast cancer, circRNA, hsa_circ_0025202, ceRNA, tamoxifen resistance
This report demonstrates a novel circular RNA, named hsa_circ_0025202, which is downregulated in tamoxifen-resistant breast cancer cells. hsa_circ_0025202 was identified as a tumor suppressor, and it could improve tamoxifen sensitivity through regulating the miR-182-5p/FOXO3a axis, suggesting it could be a potential therapeutic target and marker for patients with hormone receptor-positive breast cancer.
Introduction
Breast cancer (BC) is a heterogeneous cancer with the most frequent incidence and the second highest mortality in women worldwide.1 Notably, three quarters of patients with BC are hormone receptor (HR) positive.2 Previous studies have shown that estrogen signaling plays a vital role in the development and prognosis of HR-positive BC,3, 4 and adjuvant anti-hormone therapy has acted as a standard treatment for decades. Although a number of drugs have been developed, based on the reduction of estrogen level or block of ER-signaling pathway,5 tamoxifen (TAM) remains a crucial therapeutic option for patients with luminal subtype BC and the first-line therapy especially for patients who are premenopausal.6, 7 TAM, a pioneer selective estrogen receptor modulator (SERM), was reported to achieve a 50% decline in recurrence rate and 28% decline in mortality.8 The majority of HR-positive BC initially responds to TAM; however, 30%–40% of patients treated with TAM relapse within 15 years.9, 10 The tumors that develop are typically TAM resistant, and other endocrine management treatments have limited the effectiveness, leading to poor prognosis. Therefore, identifying the mechanisms of resistance and appropriate biomarkers for predicting the TAM therapy response are pressing challenges with significant clinical implications.
Circular RNAs (circRNAs), which are endogenous noncoding RNAs (ncRNAs), were recently discovered to be expressed in highly divergent eukaryotes.11, 12 As the name implies, circRNAs are characteristically different from the typical linear mRNA or ncRNA, and they have a covalently closed loop structure with neither a 5′ cap nor a 3′ poly(A) tail.13 Because of the lack of free ends and resistance to exonucleolytic degradation, circRNAs maintain high cellular stability; and, it was reported as a more abundant RNA isoform than mRNA in various cell types and organisms.14, 15, 16 With the development of RNA sequencing techniques and bioinformatics, hundreds of circRNAs have been detected, featuring developmental regulation, localization, and tissue-specific expression.17
In recent years, an increasing number of studies have indicated that circRNAs are deregulated in distinct types of human cancers,14 such as lung,18 liver,19 colorectal,20 gastric,21, 22 bladder,23 and breast24 cancers, and they are disease specific. Moreover, circRNAs play essential roles in biological processes involved in tumor progression. circRNAs also have significant roles as microRNA (miRNA) sponges, RNA-binding protein sponges, gene transcription and expression regulators, and protein-coding genes.25 Among these functions, the role of a miRNA sponge is the most common in cancer cells, especially in BC. For instance, circIRAK was reported to promote BC metastasis by acting as a sponge for miR-3607,26 and circGFRA1 was proven to decoy miR-34a and participate in triple-negative BC progression.24 Therefore, circRNAs may be important target molecules and potential biomarkers for the diagnosis and treatment of various cancers.27 However, few studies have focused on the roles of circRNAs in the regulation of endocrine-resistant BC.
In this study, we first analyzed the expression profile of circRNAs from TAM-resistant MCF-7 (MCF7/TR) and parental MCF-7 (MCF7/P) cells, and we characterized a novel circRNA, hsa_circ_0025202, which was significantly downregulated in MCF7/TR cells. Furthermore, the roles of hsa_circ_0025202 in TAM resistance and BC progression were studied.
Results
Identification of Deregulated circRNAs in TAM-Resistant BC Cells
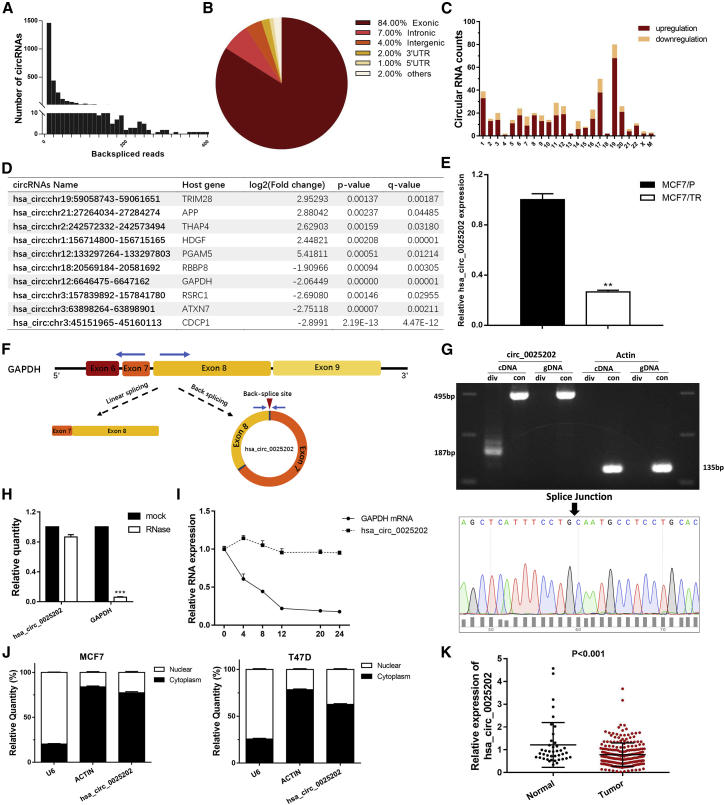
To investigate the roles of circRNAs in patients with TAM-resistant BC, we analyzed the circRNA expression profiles of MCF7/TR and MCF7/P cells using RNA sequencing (RNA-seq) analysis. We detected 36,110 distinct circRNAs, 23,357 of which contained more than one unique backspliced read (Figure 1A; Data S1). Among them, 84% of the circRNAs consisted of protein-coding exons (Figure 1B; Data S2). The length of the exonic circRNAs was mostly less than 2,000 nt. We identified 465 differentially expressed circRNAs based on log2 (fold change) > 1 (or <−1) and q-value < 0.01, 352 of which were upregulated and 113 were downregulated (Figure 1C).
Figure 1.
The Characteristics of the hsa_circ_0025202
(A) Numbers of circRNA and backspliced reads identified in MCF7/TR and MCF7/P BC cell lines. (B) The proportions of circRNAs derived from different genomic regions. (C) Numbers of differently expressed circRNAs from each chromosome. (D) Top 10 differentially expressed circRNAs between MCF7/P and MCF7/TR cell lines. (E) qRT-PCR indicating the levels of hsa_circ_0025202 expression in MCF7/P and MCF7/TR cell lines. (F) Schematic illustrates the generation of hsa_circRNA_0025202. Blue arrows represent the detection of specific primers for hsa_circ_0025202 by qRT-PCR. (G) Gel electrophoresis shows that hsa_circ_0025202 can be amplified by divergent primers using total cDNA, but not genomic DNA (gDNA). Sanger sequencing validates its splicing junction. Actin served as a control. (H) qRT-PCR analysis of hsa_circ_0025202 and GAPDH expressions in MCF7 cells after treating with RNase R. (I) qRT-PCR data for the abundance of GAPDH mRNA and hsa_circRNA_0025202 in MCF7 cells at the indicated time points. (J) qRT-PCR analysis of hsa_circ_0025202, β-actin, and U6 in the cytoplasm and nucleus in MCF7 cells. (K) qRT-PCR shows differently expressed hsa_circRNA_0025202 in 41 normal and 230 tumor tissues. Data are presented as means ± SEM of three independent experiments. **p < 0.01.
To verify these results, five upregulated and five downregulated circRNAs (Figure 1D) were chosen for further confirmation in MCF7/P and MCF7/TR cells using real-time qPCR. The results showed that the hsa_circ: chr12:6646475-6647162 was significantly downregulated in MCF7/TR cells with high abundance (Figure 1E), compared with the other 9 candidates (Figures S1A–S1H). According to the human reference genome (GRCh37/hg19) and CircBase database annotation, this circRNA is named CircBase: hsa_circ_0025202 and spliced from GAPDH. The genome length is 688 bp and the spliced length is 495 bp (Figure S1I).
Characterization of hsa_circ_0025202 in HR-Positive BC
We found that hsa_circ_0025202 is derived from regions of exons 7 and 8 of GAPDH, with short introns between each exon (Figure 1F). The specific convergent and divergent primers that specially amplified the linear and backsplicing forms of GAPDH were designed to confirm the existence of hsa_circ_0025202. Our PCR results showed that hsa_circ_0025202 could only be amplified by cDNA templates, whereas the canonical GAPDH can be obtained by both cDNA and genomic DNA (gDNA) templates (Figure 1G, top). Next, we used Sanger sequencing to directly verify the splice junction (Figure 1G, bottom). In addition, resistance to digestion by RNase R exonuclease further confirmed the circular form and loop structure of hsa_circ_0025202 (Figure 1H). Actinomycin D assay was used to investigate the stability of hsa_circ_0025202 in MCF7 cells. The half-life of this circRNA transcript exceeded 24 h, whereas that of the linear form was less than 8 h, indicating that the circRNA isoform was much more stable (Figure 1I). qRT-PCR analysis of nuclear and cytoplasmic RNAs revealed that hsa_circ_0025202 resided predominantly in the cytoplasm in MCF7 and T47D cells (Figure 1J). Taken together, our results indicate that hsa_circ_0025202 is an abundant and stable cytoplasmic circRNA derived from GAPDH.
Next, the expression level of hsa_circ_0025202 was evaluated in HR-positive BC tissues (n = 230) and adjacent normal tissues (n = 41) using qRT-PCR. hsa_circ_0025202 was significantly downregulated in BC specimens (p < 0.001) (Figure 1K). To explore the correlation between hsa_circ_0025202 and clinicopathological parameters, we categorized all patients with BC into hsa_circ_0025202 high- and low-expression groups using the median expression value as a cutoff threshold. We found the level of hsa_circ_0025202 was negatively correlated with histological grade (p = 0.005) and lymph node metastasis (p = 0.048) (Table 1). These results suggest that hsa_circ_0025202 may play an essential role in tumor progression of HR-positive BC.
Table 1.
Association between Clinicopathological Variables and hsa_circ_0025202 Expression in Patients with Hormone Receptor-Positive Breast Cancer
| Variable | Cases (n = 230) | hsa_circ_0025202 |
p Value | |
|---|---|---|---|---|
| Low | High | |||
| Age (years) | ||||
| ≤50 | 121 | 54 | 67 | 0.086 |
| >50 | 109 | 61 | 48 | |
| Menopause | ||||
| No | 119 | 54 | 65 | 0.147 |
| Yes | 111 | 61 | 50 | |
| Histologic grade | ||||
| I | 11 | 1 | 10 | 0.005 |
| II | 162 | 79 | 83 | |
| III | 57 | 35 | 22 | |
| Lymph node metastasis | ||||
| No | 119 | 52 | 67 | 0.048 |
| Yes | 111 | 63 | 48 | |
| T stage | ||||
| T1/T2 | 223 | 110 | 113 | 0.446 |
| T3/T4 | 7 | 5 | 2 | |
| N stage | ||||
| N0 | 119 | 52 | 67 | 0.139 |
| N1/N2 | 106 | 60 | 46 | |
| N3 | 5 | 3 | 2 | |
| Ki-67 score | ||||
| Low | 57 | 25 | 32 | 0.526 |
| High | 133 | 65 | 68 | |
Overexpression of hsa_circ_0025202 Can Reverse the Progressive Phenotype and TAM Resistance of MCF7/TR Cells In Vitro
To identify the phenotypic differences between the MCF7/P and MCF7/TR cells, we conducted the functional assays, and we found that, besides the stronger resistance to TAM (Figures S2A and S2B), MCF7/TR cells, compared with MCF7/P cells, presented more enhanced proliferation, colony formation, and migration ability and reduced cell apoptosis (Figures S2C–S2F). Thus, these results suggest that MCF7/TR possess a more progressive phenotype.
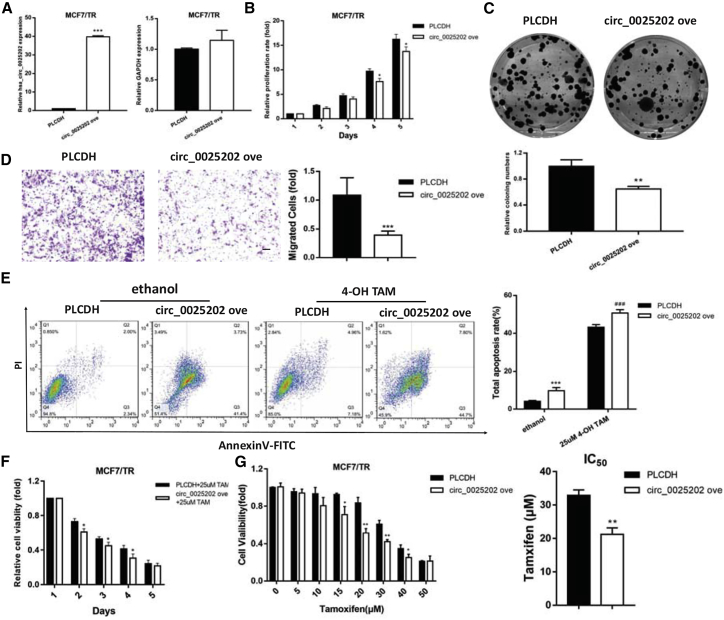
Given that hsa_circ_0025202 is downregulated in MCF7/TR cells, we built a specific vector carrying hsa_circ_0025202 to investigate its potential functional roles. qRT-PCR verified that the overexpressing vector did not influence the expression of its linear form (Figure 2A). Subsequent functional assays suggested that overexpressing hsa_circ_0025202 inhibited cell viability, colony formation ability, and migration potential (Figures 2B–2D). Since the induction of cell apoptosis is one of the major tumor cell responses to TAM treatment,28 flow cytometry was performed to analyze the effect of hsa_circ_0025202 on cell apoptosis rate, which was higher in the overexpression group (Figure 2E).
Figure 2.
Overexpression of hsa_circ_0025202 Reversed the Progressive Phenotype and TAM Resistance of MCF7/TR Cells In Vitro
(A) qRT-PCR analysis of hsa_circ_0025202 and GAPDH expressions in MCF7/TR cells transfected with PLCDH and circ_0025202 ove. (B) MTT analysis of cell proliferation in MCF7/TR cells transfected with or without circ_0025202 ove at the indicated time points. (C) The colony formation, (D) cell migration, (E) cell apoptosis, and cell viability (F) and IC50 (G) in MCF7/TR cells transfected with or without circ_0025202 ove. Data are presented as means ± SEM of three independent experiments. *p < 0.05; **p < 0.01; ***, ###p < 0.001. Scale bar, 50 μm.
To further investigate the roles of hsa_circ_0025202 in TAM resistance, MCF7/TR cells were subjected to cytotoxicity and IC50 assays under TAM treatment. Upregulation of hsa_circ_0025202 resulted in a more significant inhibition of cell growth with TAM treatment and decreased IC50 (Figures 2F and 2G). Together, these results suggest that hsa_circ_0025202 can partly reverse the progressive phenotype of MCF7/TR cells and re-sensitize MCF7/TR cells to TAM.
hsa_circ_0025202 Acts as a Tumor Suppressor and a Modulator of TAM Sensitivity in HR-Positive BC Cells
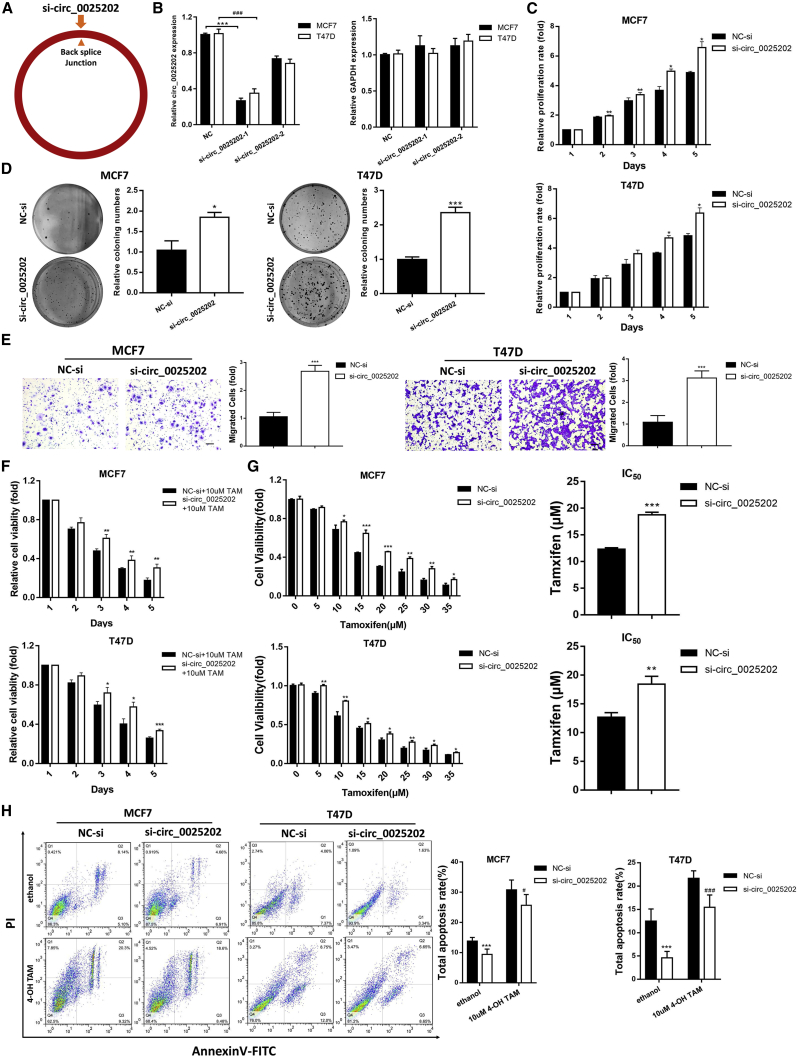
To further analyze the roles of hsa_circ_0025202 in HR-positive BC, two small interfering RNAs (siRNAs) were designed that specifically targeted the backsplice junction sequence of hsa_circ_0025202 (Figure 3A). We confirmed that can only knock down the circular transcript without prejudice to the linear form in T47D and MCF7 cells (Figure 3B). si-circ_0025202-1 was chosen for the following experiments, because of its higher knockdown efficiency. Additionally, the transfection efficiency of the hsa_circ_0025202-overexpressing vector in these two cell lines was verified by qRT-PCR (Figure S3A). The siRNA-mediated downregulation of hsa_circ_0025202 promoted cell growth, colony formation, and migration of MCF7 and T47D cells (Figures 3C–3E). Conversely, the overexpression of hsa_circ_0025202 inhibited these functions (Figures S3B–S3D).
Figure 3.
Knockdown of hsa_circ_0025202 Promotes Growth and Metastasis and Decreases TAM Sensitivity in HR-Positive BC Cells In Vitro
(A) Schematic illustrates si-circ_0025202 targeted the backsplice junction of circ_0025202. (B) The expression of hsa_circ_0025202 and GAPDH in T47D and MCF7 cells was determined with qRT-PCR transfected with the negative control of siRNA (NC-si) and si-circ_0025202. (C–H) hsa_circ_0025202 was silenced in T47D and MCF7 cells. (C) MTT assay was conducted to evaluate cell proliferative ability. (D) Colony formation is shown in T47D and MCF7 cells. (E) Transwell migration was conducted to evaluate cell migration ability (magnification, 100× ). Scale bar, 50 μm. (F) MTT assay demonstrates cell viability of T47D and MCF7 cells treated together with TAM. (G) IC50 assay shows the variation in TAM sensitivity. (H) Cells were treated with TAM (10 nM) or ethanol (control) and subjected to Annexin V-FITC and propidium iodine staining to detect apoptotic rate by flow cytometry. Data are presented as means ± SEM of at least three independent experiments. *, #p < 0.05; **p < 0.01; ***, ###p < 0.001.
Furthermore, the cytotoxicity and IC50 assays demonstrated that TAM action and cell sensitivity to TAM were significantly enhanced by hsa_circ_0025202, and its downregulation decreased TAM efficacy (Figures 3F and 3G; Figures S3E and S3F). Moreover, knockdown of hsa_circ_0025202 protected MCF7 and T47D cells from apoptosis, while overexpression induced apoptosis (Figure 3H; Figure S3G). Transfecting MCF7/TR cells with si-circ_0025202-1, similar changes in cell proliferation, migratory ability apoptosis, and TAM sensitivity were observed as in MCF7 and T47D cells (Figure S4). The representative cell proliferation assays in MCF7, MCF7/TR, and T47D cells were also confirmed by cell counting kit-8 (CCK8) assays (Figure S5). Together, these results suggest that hsa_circ_0025202 acts as a tumor suppressor and an important modulator of TAM sensitivity in HR-positive BC cells.
hsa_circ_0025202 Acts as an Efficient miRNA Sponge for miR-182-5p
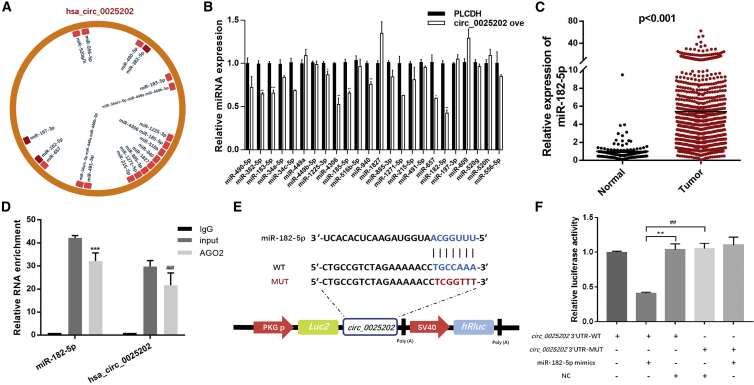
Given that many circRNAs have been shown to function as miRNA sponges, we next investigated the ability of hsa_circ_0025202 to bind miRNAs. CircInteractome and starBase version (v.)2.0 were utilized to explore the miRNA response elements (MREs) harbored by hsa_circ_0025202. Among the predicted miRNAs, we chose the 24 miRNAs most highly ranked as the candidate miRNAs (Figure 4A). Among them, miR-197-3p, miR-382-5p, and miR-182-5p were predicted by both databases.
Figure 4.
hsa_circ_0025202 Acts as an Efficient miRNA Sponge for miR-182-5p
(A) A schematic illustration demonstrating the putative binding sites of the miRNAs associated with hsa_circ_0025202. (B) qRT-PCR data show expressions of the candidate miRNAs after hsa_circ_0025202 overexpression in MCF7 cells. (C) Expression of miR-182-5p in HR-positive BC tissues and adjacent normal tissues from TCGA data. (D) qRT-PCR for the expression of hsa_circ_0025202 and miR-182-5p in MCF7 cells after pulling down of Ago2 by RIP assay. (E) A schematic of wild-type (WT) and mutant (MUT) hsa_circ_0025202 luciferase reporter vectors. (F) The luciferase activity of WT hsa_circ_0025202 3′ UTR or mutant hsa_circ_0025202 3′ UTR after transfection with miR-182-5p mimics in 293T cells. Data are presented as means ± SEM of at least three independent experiments. *p < 0.05; **, ##p < 0.01; ***, ###p < 0.001.
miR-182-5p, the most downregulated miRNA after overexpressing hsa_circ_0025202, was selected for further analysis (Figure 4B). Subsequently, we used The Cancer Genome Atlas (TCGA) sequencing data to validate the increased expression of miR-182-5p in human HR-positive BC tissues (n = 1,078), as compared to adjacent normal tissues (n = 104) (Figure 4C). We further performed RNA immunoprecipitation (RIP) assays to pull down RNA transcripts that bound to Ago2 in MCF7 cells. qRT-PCR showed that hsa_circ_0025202 and miR-182-5p can be efficiently pulled down by anti-Ago2 (Figure 4D).
Next, luciferase reporter assays were applied to determine whether miR-182-5p directly targets hsa_circ_0025202. We constructed dual-luciferase reporter vectors containing either the full length of wild-type (WT) hsa_circ_0025202 or a version where the miR-182-5p-binding site was mutated (MUT) (Figure 4E). A remarkable reduction in luciferase reporter activity was detected in 293T cells co-transfected with miR-182-5p mimics and WT, but not with the mutant vector (Figure 4F). Taken together, these experiments indicate that hsa_circ_0025202 may function as a sponge for miR-182-5p.
hsa_circ_0025202 Regulates Tumor Progression and TAM Sensitivity of BC Cells via miR-182-5p
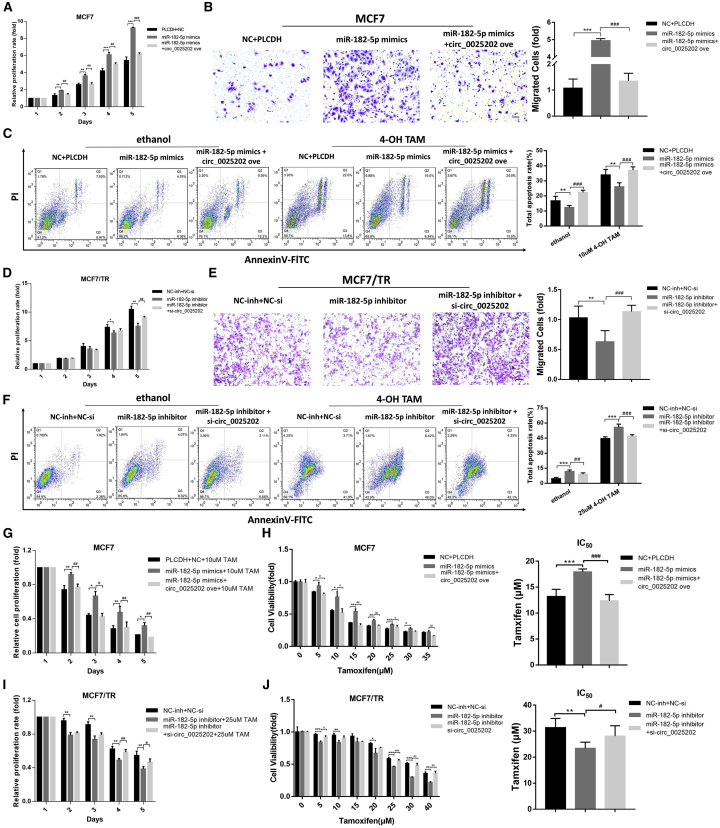
Given the apparent relationship between hsa_circ_0025202 and miR-182-5p, we next investigated the biological functions of miR-182-5p. We found that transfection of MCF7 and T47D cells with miR-182-5p mimics promoted cell proliferation and migration and reduced apoptosis. These effects were significantly reversed by hsa_circ_0025202 overexpression in MCF7 and T47D cells (Figures 5A–5C; Figures S6C–S6E). Moreover, inhibition of miR-182-5p showed the reverse of that observed when cells were transfected with miR-182-5p mimics, and these effects were weakened by si-circ_0025202 in MCF7/TR cells (Figures 5D–5F). Ectopic expression of miR-182-5p attenuated TAM cytotoxicity and increased the IC50 values in MCF7 and T47D cells, which were antagonized by hsa_circ_0025202 overexpression (Figures 5G and 5H; Figures S6F and S6G). Likewise, silencing miR-182-5p sensitized MCF7/TR cells to TAM, and knocking down hsa_circ_0025202 reversed this effect (Figures 5I and 5J). Taken together, these results demonstrate that hsa_circ_0025202 suppressed BC progression and sensitized cells to TAM via sponging miR-182-5p, thereby attenuating its oncogenic effect.
Figure 5.
hsa_circ_0025202 Sponges miR-182-5p to Regulate Tumor Progression and TAM Sensitivity of BC Cells
(A) Cell proliferation, (B) migration, and (C) apoptosis of MCF7 cells after transfection with miR-182-5p mimics combined with or without circ_0025202 ove. (D) Cell proliferation, (E) migration, and (F) apoptosis of MCF7/TR cells after transfection with miR-182-5p inhibitor combined with or without si-circ_0025202. MTT data for the cell viability (G) and IC50 (H) of MCF7 cells treated with TAM after transfection with miR-182-5p mimics combined with or without circ_0025202 ove. MTT data for the cell viability (I) and IC50 (J) of MCF7/TR cells treated with TAM after transfection with miR-182-5p inhibitor combined with or without si-circ_0025202. Data are presented as means ± SEM of at least three independent experiments. *, #p < 0.05; **, ##p < 0.01; ***, ###p < 0.001. Scale bar, 50 μm.
FOXO3a Is a Direct Target Gene of miR-182-5p and Is Regulated by hsa_circ_0025202
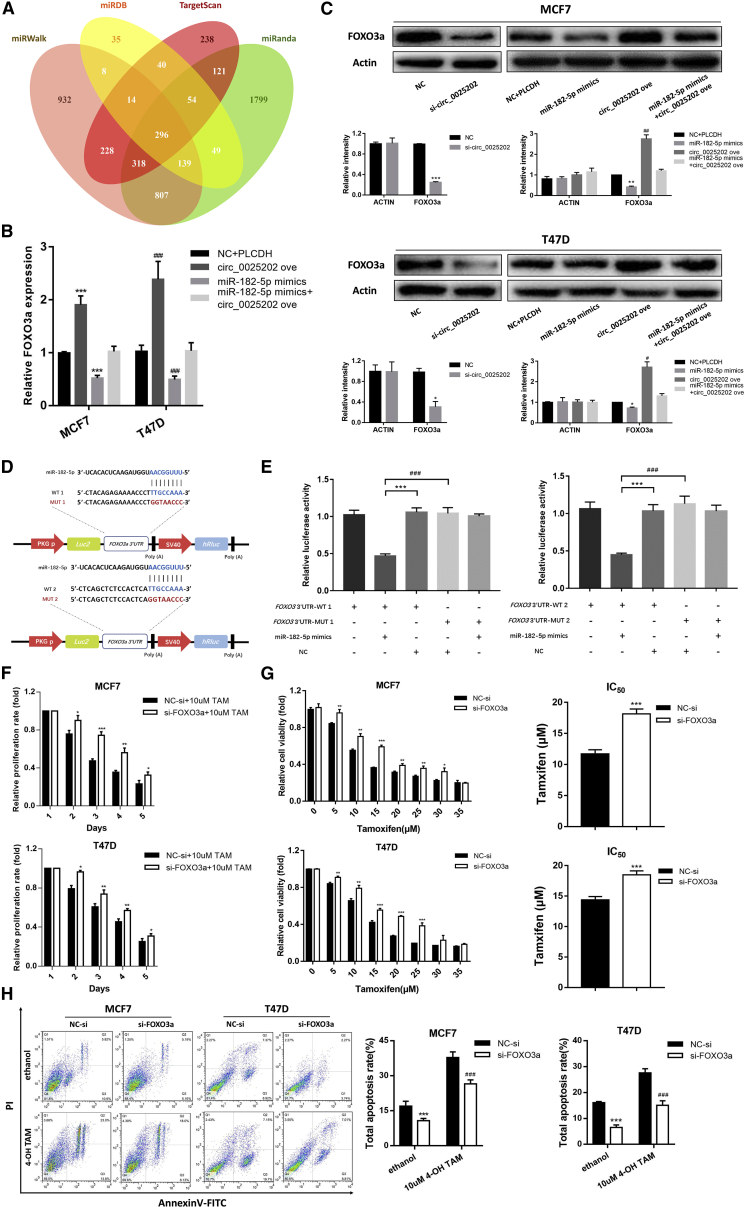
The potential targets of miR-182-5p were predicted by four miRNA target gene databases (miRWalk, miRDB, TargetScan, and miRanda), and 296 genes were listed in all four algorithms (Figure 6A). Among these genes, FOXO3a, which is a well-known tumor suppressor and is downregulated in human BC,29 was predicted with a high score. Using qRT-PCR and western blot assay, we found that the expression of FOXO3a was increased by hsa_circ_0025202 overexpression and reduced by miR-182-5p mimics. The effect of these mimics could be reversed by hsa_circ_0025202 at both the mRNA and protein levels (Figures 6B and 6C). According to MRE analysis, miR-182-5p has two potential binding sites within the 3′ UTR of FOXO3a (Figure 6D). Therefore, we inserted the WT and mutated 3′ UTR of FOXO3a containing each predicted binding site into the luciferase reporter plasmids. Luciferase activity significantly decreased after co-transfecting cells with miR-182-5p mimics and FOXO3a 3′ UTR WT1 or FOXO3a 3′ UTR WT2, while the luciferase activity of the cells that contained the mutant FOXO3a constructs was not affected by miR-182-5p (Figure 6E).
Figure 6.
FOXO3a Is a Direct Target Gene of miR-182-5p and Is Regulated by hsa_circ_0025202
(A) Venn diagram represents the mutual candidate target genes of miR-182-5p identified by miRWalk, miRDB, TargetScan, and miRanda. (B and C) FOXO3a expression assessed by qRT-PCR (B) and western blot (C), respectively, in MCF7 and T47D cells after transfection with miR-182-5p mimics and circ_0025202 ove. (C) The quantification of the bands is shown. (D) Potential binding sites of miR-182-5p with FOXO3a 3′ UTR. (E) The luciferase activity of WT FOXO3a 3′ UTR or mutant FOXO3a 3′ UTR of two potential binding sites after transfection with miR-182-5p mimics in HEK293T cell lines. (F) MTT data for the proliferation of MCF7 and T47D cell lines treated with TAM after transfection of si-FOXO3a. (G) MTT data for the cell viability and IC50 of MCF7 and T47D cell lines treated with TAM after transfection of si-FOXO3a. (H) Annexin V-FITC assay was used to assess cell apoptosis in MCF7 and T47D cells after transfection of si-FOXO3a. Data are presented as means ± SEM of at least three independent experiments. *, #p < 0.05; **, ##p < 0.01; ***, ###p < 0.001.
Previous studies have verified FOXO3a as a tumor suppressor in BC;30 however, it was unclear whether FOXO3a was involved in TAM resistance. We performed cell cytotoxicity and IC50 assays, and we found that FOXO3a knockdown induced an increase in TAM resistance and IC50 values in MCF7 and T47D cells (Figures 6F and 6G). Moreover, the TAM-mediated cell apoptosis rate was reduced following FOXO3a knockdown (Figure 6H). These biological functions were further confirmed in MCF7/TR cells (Figure S7). Consequently, these results showed that hsa_circ_0025202 functions as a competing endogenous RNA (ceRNA) for miR-182-5p and regulates the expression and activity of FOXO3a, which could further suppress tumor progression and sensitize cells to TAM in BC.
hsa_circ_0025202 Combined with TAM Treatment Exhibits an Additive Inhibitory Effect on HR-Positive BC Growth In Vivo
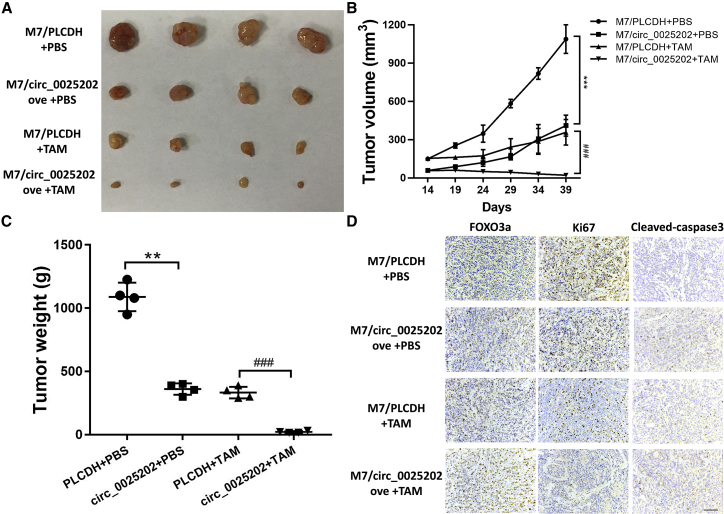
To evaluate the tumor-suppressing and TAM sensitization effects of hsa_circ_0025202 in vivo, we established MCF7 cells that stably overexpressed hsa_circ_0025202 (M7/circ_0025202 ove) and the corresponding control cells (M7/PLCDH), and the overexpression was confirmed by qRT-PCR (Figure S8). In comparison with the M7/PLCDH group, hsa_circ_0025202 overexpression significantly reduced the mean tumor volume and tumor weight (Figures 7A–7C). Moreover, TAM treatment further inhibited tumor growth as compared to the mice that received PBS. There were also significant differences in the fold change in tumor volume and tumor weight at the endpoint, which was more prominent than the fold change between the TAM group (Figures S9A and S9B). Immunohistochemistry (IHC) analysis also showed that decreased Ki67 expression, increased FOXO3a, and cleaved caspase-3 levels were observed in the M7/circ_0025202 ove group as compared with the M7/PLCDH group, as well as in the groups treated with TAM (Figure 7D). Taken together, our findings suggest that hsa_circ_0025202 can suppress tumor growth and increase TAM sensitivity in vivo.
Figure 7.
Overexpression of hsa_circ_0025202 Suppressed the Growth and TAM Resistance of BC Xenografts
(A) Images of the xenograft tumors obtain from BALB/c nude mice at the endpoint. (B) Growth curve of tumor volume. The volumes were measured every 5 days after the initial 2 weeks. (C) Weights of xenograft tumors at the endpoint. (D) FOXO3a, Ki67, and cleaved caspase-3 staining by IHC of the xenograft tumors. Scale bar, 100 μm. Data are presented as means ± SEM of at least three independent experiments. **p < 0.01; ***, ###p < 0.001.
Discussion
TAM is widely used as an adjuvant endocrine therapy for patients with HR-positive BC who are premenopausal; however, almost a third of these patients eventually developed TAM resistance.10 There are several mechanisms that underlie the acquired resistance to TAM, among which epigenetic modifications are of great importance.31 Epigenetic modifications are inheritable changes in the DNA that alter gene expression without altering the DNA sequence, including DNA methylation, histone modifications, and ncRNA. miRNAs and long ncRNAs (lncRNAs) are known to play vital roles in TAM resistance. For example, miR-375 is capable of reversing the TAM-resistant phenotype by regulating Metadherin (MTDH) expression.32 Moreover, dysregulation of miR-10b increases the resistance to TAM and attenuates TAM-mediated inhibition of migration.33 lncRNA HOTAIR contributes to TAM resistance via promoting ligand-independent estrogen receptor (ER) activities.34
Nevertheless, circRNAs are a new class of ncRNAs that have been suggested to be essential in carcinogenesis and tumor progression in BC.26, 35, 36 However, the function of circRNAs in TAM resistance remained unknown. In our study, we first identified a circRNA named hsa_circ_0025202, which was downregulated in MCF7/TR cells and HR-positive BC tumor tissues. Gain- and loss-of-function studies demonstrated that hsa_circ_0025202 could inhibit cell proliferation, colony formation, and migration and increase cell apoptosis and sensitize cells to TAM treatment. Moreover, we found that the expression of hsa_circ_0025202 was negatively correlated with histological grade and lymphatic metastasis, suggesting that it may serve as a tumor suppressor and important regulator in HR-positive BC. Furthermore, circRNAs possess not only tissue- and stage-specific expression but also a higher degree of sequence conservation and stability,37, 38 suggesting that hsa_circ_0025202 could be used as the potential biomarker of TAM sensitivity.
Previous studies have shown that some of the most common roles of circRNAs within cancer cells are to act as miRNA sponges and regulate the expression and activity of the target genes.25 The first circRNA verified as a miRNA sponge was ciRS-7, which harbors more than 70 conventional binding sites for miR-7.38 Subsequently, a string of studies have shown that, by acting as a sponge, ciRS-7 reduced the activity of miR-7 and increased the expression of its target genes, such as EGFR, RAF1, E2A, NF-κB, and HOXB13.39, 40, 41, 42 Apart from ciRS-7, many other circRNAs were also reported to act as miRNA sponges. For instance, circRNA_100290 functioned as a sponge for the miR29 family in oral cancer.43 Additionally, circHIPK3 can sponge miR-124 in hepatocellular carcinoma,44 and circLARP4 can sponge miR-424-5p in gastric cancer.22 In our study, after predicting with bioinformatics analysis and confirming with luciferase and RIP assays, we revealed that hsa_circ_0025202 can act as a miRNA sponge for miR-182-5p. Further functional studies demonstrated that ectopic expression of miR-182-5p could mimic the effects of hsa_circ_0025202 inhibition and was attenuated by hsa_circ_0025202 overexpression, suggesting that miR-182-5p was a functional target of hsa_circ_0025202 and played an oncogenic role in BC.
Since miRNAs perform their biological functions by inhibiting the translation of target mRNA or promoting the degradation of mRNA by binding to their 3′ UTRs, we used online miRNA target bioinformatics prediction databases to predict the potential targets of miR-182-5p. FOXO3a was validated to be a target gene of miR-182-5p. FOXO3a, a member of the forkhead box class O (FOXO) transcription factors, is considered a cancer suppressor in various types of cancers,45, 46, 47 including BC.29 Moreover, a growing amount of evidence has indicated that FOXO3a inhibits BC progression by suppressing cell proliferation and metastasis and increasing apoptosis.48, 49 However, the roles it might play in TAM resistance of HR-positive BC have not been studied. In this study, we conducted a series of functional experiments, and we found that knockdown of FOXO3a induces TAM resistance and diminishes the TAM-mediated apoptosis. Thus, hsa_circ_0025202 participates in tumor progression and regulates the TAM response in BC cells affecting the miR-182-5p/FOXO3a axis.
Taken together, our study demonstrated that hsa_circ_0025202 could affect BC cell proliferation, migration, and apoptosis and sensitivity to TAM. hsa_circ_0025202 acts as a potent suppressor gene for HR-positive BC, and it plays a noticeable role in the regulation of TAM sensitivity. Based on our data, we suggest that hsa_circ_0025202 could be a therapeutic target in patients with HR-positive BC, especially for those receiving TAM therapy. Moreover, our results imply that hsa_circ_0025202 could be an important and promising biomarker for TAM resistance; further clinical study with a large cohort of patients is necessary to demonstrate its clinical importance.
Materials and Methods
Analyzing the circRNA Expression Profile
Total RNA from parental MCF7 and MCF7/TR cells was extracted with TRIzol reagents (Invitrogen, Carlsbad, CA, USA), as per the manufacturer’s protocol. The rRNA was removed from approximately 2 μg total RNA from each sample by using the Epicenter Ribo-Zero rRNA Removal Kit (Illumina, USA), followed by RNase R treatment (Epicenter Technologies, Madison, WI, USA). Subsequently, strand-specific RNA-seq libraries were prepared using the NEBNext Ultra RNA Library Prep Kit from Illumina (New England Biolabs, Beverly, MA, USA), and they were subjected to deep sequencing with an Illumina HiSeq 3000 at RiboBio (Guangzhou, China).
Identification and Quantification of circRNAs
The RNA-seq FASTQ reads were first mapped to a human reference genome (GRCh37/hg19) using TopHat2.50 The unmapped reads were then used to identify circRNAs as previously described.51 Differential expression analysis of circRNAs was executed using R software package DEGseq.52 Only the circRNAs that were differently expressed with a q-value < 0.05 were chosen for further analysis. The fold change was Log2 transformed, and we used a log2 (fold change) > 1.5 (or <−1.5) and a q-value < 0.05 to sort the differently expressed circRNAs. Subsequently, to generate an overview of circRNA expression profiles between the two groups, hierarchical clustering analysis was performed.
Patient Specimens and Ethics Statement
BC tissues (n = 230) and adjacent breast tissues (n = 44) were obtained from patients who received surgical treatment at Qilu Hospital of Shandong University between 2008 and 2015. All patients received a postoperative histopathological diagnosis of invasive ductal HR-positive BC. None of these patients received preoperative chemotherapy or radiotherapy, and all tissue specimens were stored at −80°C until use. Pathological characteristics, clinicopathological classification, and staging were determined according to the American Joint Committee on Cancer Classification Criteria. Written informed consent was obtained from all patients, and our project was approved by the Ethics Committee on Scientific Research of Shandong University Qilu Hospital. Detailed clinical information is summarized in Data S3.
Cell Culture
BC cell lines MCF7 and T47D and the HEK293T cells were obtained from ATCC. MCF7 and HEK293T cells were cultured in DMEM (HyClone) with 10% fetal bovine serum (FBS, Clark Bioscience). MCF7/TR cells were established as previously described and supplemented with 10 μM TAM.53 T47D cells were maintained in RPMI-1640 medium (HyClone) with 10% FBS (Clark Bioscience) and 5 mg/mL insulin. All the cells were maintained at 37°C in a humidified atmosphere with 5% CO2.
Actinomycin D and RNase R Treatment
Transcription was inhibited by adding actinomycin D (2 mg/mL) or DMSO (Sigma-Aldrich, St. Louis, MO, USA) as a control to the culture medium. Total RNA (5 μg) was incubated with or without 3 U/μg RNase R (Epicenter Technologies) at 37°C for 30 min, and the resulting RNA was purified using an RNeasy MinElute Cleaning Kit (QIAGEN, Germany). After the treatment above, RNA was transcribed into cDNA, and the expression levels of GAPDH and hsa_circ_0025202 were determined by qRT-PCR.
Nucleic Acid Preparation and qRT-PCR
Total RNA from whole-cell lysates was isolated using TRIzol reagent (Invitrogen), genomic DNA was isolated with the TIANamp Genomic DNA Kit (TIANGEN), and cytoplasmic and nuclear RNA isolations were performed using a PARIS Kit (Invitrogen, USA), following the manufacturer’s instructions. To quantify the amounts of mRNA and circRNA, cDNA was synthesized with the PrimeScript RT Master Mix (Takara, Dalian, China) from 500 ng RNA. Mir-X miRNA First-Strand Synthesis Kit (Takara) was used to reverse transcribe total RNA into mature miRNA. The real-time PCR analyses were performed with SYBR Premix Ex TaqII (Takara) with Light Cycler 480 II Real-Time PCR System (Roche, Switzerland). Actin was used as the endogenous control for the detection of mRNA and circRNA expression levels, while U6 was used as an endogenous control for miRNA expression analysis. To determine the abundance of circRNA, the divergent primers were designed for the circular transcripts. The primers are listed in Table S1.
Plasmid Construction and Cell Transfection
To overexpress hsa_circ_0025202, the full length of 495 bp of hsa_circ_0025202 cDNA was cloned into vector pLCDH-ciR (Ribobio, Guangzhou, China). For luciferase reporter assays, the full length of hsa_circ_0025202 and 276-bp and 506-bp fragments of the FOXO3a 3′ UTR were amplified from the cDNA of MCF7 cells and cloned into vector pmiRGLO (Promega, Madison, WI, USA), respectively. All constructs were confirmed by sequencing. siRNA and miRNA mimics were synthesized by RiboBio (Guangzhou, China). The sequences that were used are shown in Table S1.
For transient transfection, MCF7 and T47D cells were transfected with Lipofectamine 2000 reagent (Invitrogen, Carlsbad, CA, USA), according to the manufacturer’s instructions. For stable cell line establishment, the lentiviral vector pLCDH-ciR carrying hsa_circ_0025202 and two assistant vectors, psPAX2 and pMD2.G, were introduced into HEK293T cells by transient transfection. After 6 h, the cell culture medium was replaced and viral supernatants were collected 48 h later. The supernatant was then collected and filtered through a 0.22-μm filter. MCF7 and T47D cells were infected at approximately 70% confluence in complete medium supplemented with 8 μg/mL polybrene (Sigma), followed by selection with puromycin at 0.5 μg/mL (Sigma). The overexpression efficiency was determined by qRT-PCR.
Cell Viability Assay and Cytotoxicity Assay
Transfected T47D and MCF7 cells were cultured in 96-well plates overnight. Subsequently, the medium was replaced with 100 μL full-growth medium solutions containing the indicated concentrations of TAM for 48 h or the indicated TAM and cultured for the indicated days. Cell proliferation was determined by a 3-(4, 5-dimethylthiazol-2-yl)-2,-5-diphenyltetrazolium bromide (MTT) assay. In brief, 20 μL MTT (5 mg/mL in PBS) was added to the medium and incubated at 37°C for 5 h. The supernatants were carefully aspirated and 100 μL DMSO was added to each well. Absorbance values at 490 nm were measured using a microplate reader (Bio-Rad).
Colony Formation Assay
For the colony formation assay, 103 MCF7 and T47D cells were seeded into 6-cm plates. After 2 weeks of incubation, the colonies were fixed with methanol for 20 min and stained with 0.1% crystal violet (Sigma) at room temperature. Cell colonies were counted and imaged.
Migration Assay
At 24 h after transfection, 1.5 × 105 MCF7 and T47D cells were suspended in 200 μL serum-free medium and seeded into the upper chambers of a transwell (8-μm pore size, Costar). 700 μL full-growth medium with 20% FBS was added to the lower well of each chamber as a chemo-attractant for MCF7 cells. With respect to the T47D cells, 700 μL medium with 20% FBS and NIH 3T3 cell-conditioned medium was used to fill the lower chamber.
The cells were incubated at 37°C in 5% CO2 for 48 h for the migration assay. After incubation, cells in the top chamber were removed with cotton swabs, and the cells that had invaded the lower surface were fixed with methanol, stained with 0.1% crystal violet, and photographed under a microscope at 100× magnification (Olympus, Japan).
Cell Apoptosis Assay
Cell apoptosis was analyzed using the Annexin V-Fluorescein Isothiocyanate (FITC)/Propidium Iodide (PI) Apoptosis Detection Kit (BD Biosciences, Franklin Lakes, NJ, USA), according to the manufacturer’s instructions. MCF7 and T47D cells were treated with 10 μM TAM or ethanol as a control for 48 h, and they were stained with FITC and PI and then analyzed by fluorescence-activated cell sorting using a FACS Calibur (BD Biosciences, Franklin Lakes, NJ, USA). The cell apoptosis data were analyzed by FlowJo 7.6.1 software (Tree Star, San Francisco, CA, USA).
Luciferase Reporter Assay
Dual fluorescein reporter gene detection was carried out using a Dual Luciferase Assay System Kit (Promega, Madison, WI, USA), according to the manufacturer’s instructions. WT and mutant hsa_circ_0025202 3′ UTRs and FOXO3a 3′ UTR were amplified and cloned into the luciferase reporter vector pmirGLO (Promega). HEK293T cells were co-transfected with luciferase plasmids and miR-182-5p mimics. After 48 h, luciferase activity was measured with the Dual-Luciferase Reporter Assay System (Promega), and firefly luciferase activity was normalized against Renilla luciferase activity.
RIP Assay
RIP assay was conducted using the Magna RIP RNA-Binding Protein Immunoprecipitation Kit (Millipore, Billerica, MA, USA), according to the manufacturer’s instructions. Antibodies for RIP assays against Ago2 and immunoglobulin G (IgG) were purchased from Millipore. Total RNA was extracted for the detection of miRNA and circRNA expressions by qRT-PCR.
Western Blotting and Immunohistochemistry Analysis
Western blotting and immunohistochemistry assays were performed as previously reported.54 The following antibodies were used: anti-FOXO3a (10849-1-AP, dilution 1:500, Proteintech), anti-actin (ab6276, dilution 1:1,000, abcam), anti-cleaved caspase-3 (9661, dilution 1:200, Cell Signaling Technology), and anti-Ki67 (27309-1-AP, dilution 1:100, Proteintech).
TAM Treatment of Xenograft Tumors
For our xenograft tumor model, we utilized 5-week-old female BALB/c nude mice (n = 16). Every mouse had a 17β-estradiol pellet (Innovative Research of America) implanted subcutaneously 1 week in advance. Before the injection, the stably transfected M7/PLCDH cells and M7/circ_0025202 ove were suspended at the concentration of 1 × 107 cells/0.2 mL PBS: Matrigel (1:3, v/v). Mice were randomly divided into two groups: eight of them were injected with M7/PLCDH cells, and the others were injected with M7/circ_0025202 ove cells. The cells were subcutaneously inoculated into the flank of each nude mouse.
Then 2 weeks after injection, the animals of each group were randomly assigned to vehicle control (PBS) or TAM (5 mg/kg) groups (n = 4 for each group). The drugs were administered by gavage once every 3 days, and tumor size was monitored at the same time. Mice were maintained until the tumor size of one group reached the endpoint (approximately 1,000 mm3). Both maximum (L) and minimum (W) lengths of the tumor were measured using a slide caliper, and the tumor volume was calculated as ½LW2.
The animal studies have been approved by the Ethics Committee on Scientific Research of Shandong University Qilu Hospital.
Statistical Analysis
Statistical analyses were performed using GraphPad Prism 7 and SPSS 20.0 (IBM, SPSS, Chicago, IL, USA). Differences between two groups of data and statistical significance were analyzed by Student’s t test or chi-square test, as appropriate. Values of p < 0.05 were considered to be statistically significant.
Author Contributions
Conceptualization, Y.S.; Methodology, Y.S., X.S., and Y. Li; Validation, B.C. and N.Z.; Investigation, Y.S., X.S., Y. Liang, D.H., H.Z., Y. Liu, T.C., and C.L.; Resources, W.Z. and L.W.; Writing – Original Draft, Y.S.; Writing – Review & Editing, Y.S., Q.Y., and B.C.; Supervision, Q.Y. and B.C.; Project Administration, Y.S.; Funding Acquisition, Q.Y. and B.C.
Conflicts of Interest
The authors declare no competing interests.
Acknowledgments
This work was supported by the National Natural Science Foundation of China (grants 81874119, 81672613, 81702602, and 31500699), the Key Research and Development Program of Shandong Province (grant 2016GSF201119), the Special Support Plan for National High-Level Talents (“Ten Thousand Talents Program”), and the China Postdoctoral Science Foundation funded project (grants 2018M630785, 2016M602146, and 2017M612292).
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.ymthe.2019.05.011.
Supplemental Information
References
- 1.Ferlay J., Soerjomataram I., Dikshit R., Eser S., Mathers C., Rebelo M., Parkin D.M., Forman D., Bray F. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int. J. Cancer. 2015;136:E359–E386. doi: 10.1002/ijc.29210. [DOI] [PubMed] [Google Scholar]
- 2.Ojo D., Wei F., Liu Y., Wang E., Zhang H., Lin X., Wong N., Bane A., Tang D. Factors Promoting Tamoxifen Resistance in Breast Cancer via Stimulating Breast Cancer Stem Cell Expansion. Curr. Med. Chem. 2015;22:2360–2374. doi: 10.2174/0929867322666150416095744. [DOI] [PubMed] [Google Scholar]
- 3.Tryfonidis K., Zardavas D., Katzenellenbogen B.S., Piccart M. Endocrine treatment in breast cancer: Cure, resistance and beyond. Cancer Treat. Rev. 2016;50:68–81. doi: 10.1016/j.ctrv.2016.08.008. [DOI] [PubMed] [Google Scholar]
- 4.Osborne C.K., Schiff R., Fuqua S.A., Shou J. Estrogen receptor: current understanding of its activation and modulation. Clin. Cancer Res. 2001;7(Suppl 12):4338s–4342s. [PubMed] [Google Scholar]
- 5.Jiang Q., Zheng S., Wang G. Development of new estrogen receptor-targeting therapeutic agents for tamoxifen-resistant breast cancer. Future Med. Chem. 2013;5:1023–1035. doi: 10.4155/fmc.13.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Arpino G., Weiss H., Lee A.V., Schiff R., De Placido S., Osborne C.K., Elledge R.M. Estrogen receptor-positive, progesterone receptor-negative breast cancer: association with growth factor receptor expression and tamoxifen resistance. J. Natl. Cancer Inst. 2005;97:1254–1261. doi: 10.1093/jnci/dji249. [DOI] [PubMed] [Google Scholar]
- 7.Rakha E.A., El-Sayed M.E., Green A.R., Paish E.C., Powe D.G., Gee J., Nicholson R.I., Lee A.H., Robertson J.F., Ellis I.O. Biologic and clinical characteristics of breast cancer with single hormone receptor positive phenotype. J. Clin. Oncol. 2007;25:4772–4778. doi: 10.1200/JCO.2007.12.2747. [DOI] [PubMed] [Google Scholar]
- 8.Osborne C.K. Tamoxifen in the treatment of breast cancer. N. Engl. J. Med. 1998;339:1609–1618. doi: 10.1056/NEJM199811263392207. [DOI] [PubMed] [Google Scholar]
- 9.Early Breast Cancer Trialists’ Collaborative Group (EBCTCG) Effects of chemotherapy and hormonal therapy for early breast cancer on recurrence and 15-year survival: an overview of the randomised trials. Lancet. 2005;365:1687–1717. doi: 10.1016/S0140-6736(05)66544-0. [DOI] [PubMed] [Google Scholar]
- 10.Musgrove E.A., Sutherland R.L. Biological determinants of endocrine resistance in breast cancer. Nat. Rev. Cancer. 2009;9:631–643. doi: 10.1038/nrc2713. [DOI] [PubMed] [Google Scholar]
- 11.Sanger H.L., Klotz G., Riesner D., Gross H.J., Kleinschmidt A.K. Viroids are single-stranded covalently closed circular RNA molecules existing as highly base-paired rod-like structures. Proc. Natl. Acad. Sci. USA. 1976;73:3852–3856. doi: 10.1073/pnas.73.11.3852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Szabo L., Salzman J. Detecting circular RNAs: bioinformatic and experimental challenges. Nat. Rev. Genet. 2016;17:679–692. doi: 10.1038/nrg.2016.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen L.L., Yang L. Regulation of circRNA biogenesis. RNA Biol. 2015;12:381–388. doi: 10.1080/15476286.2015.1020271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kristensen L.S., Hansen T.B., Venø M.T., Kjems J. Circular RNAs in cancer: opportunities and challenges in the field. Oncogene. 2018;37:555–565. doi: 10.1038/onc.2017.361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Salzman J., Chen R.E., Olsen M.N., Wang P.L., Brown P.O. Cell-type specific features of circular RNA expression. PLoS Genet. 2013;9:e1003777. doi: 10.1371/journal.pgen.1003777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Broadbent K.M., Broadbent J.C., Ribacke U., Wirth D., Rinn J.L., Sabeti P.C. Strand-specific RNA sequencing in Plasmodium falciparum malaria identifies developmentally regulated long non-coding RNA and circular RNA. BMC Genomics. 2015;16:454. doi: 10.1186/s12864-015-1603-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Salzman J. Circular RNA Expression: Its Potential Regulation and Function. Trends Genet. 2016;32:309–316. doi: 10.1016/j.tig.2016.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dai X., Zhang N., Cheng Y., Yang T., Chen Y., Liu Z., Wang Z., Yang C., Jiang Y. RNA-binding Protein Trinucleotide repeat-containing 6A Regulates the Formation of Circular RNA 0006916, with Important Functions in Lung Cancer Cells. Carcinogenesis. 2018;39:981–992. doi: 10.1093/carcin/bgy061. [DOI] [PubMed] [Google Scholar]
- 19.Han D., Li J., Wang H., Su X., Hou J., Gu Y., Qian C., Lin Y., Liu X., Huang M. Circular RNA circMTO1 acts as the sponge of microRNA-9 to suppress hepatocellular carcinoma progression. Hepatology. 2017;66:1151–1164. doi: 10.1002/hep.29270. [DOI] [PubMed] [Google Scholar]
- 20.Zeng K., Chen X., Xu M., Liu X., Hu X., Xu T., Sun H., Pan Y., He B., Wang S. CircHIPK3 promotes colorectal cancer growth and metastasis by sponging miR-7. Cell Death Dis. 2018;9:417. doi: 10.1038/s41419-018-0454-8. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 21.Chen J., Li Y., Zheng Q., Bao C., He J., Chen B., Lyu D., Zheng B., Xu Y., Long Z. Circular RNA profile identifies circPVT1 as a proliferative factor and prognostic marker in gastric cancer. Cancer Lett. 2017;388:208–219. doi: 10.1016/j.canlet.2016.12.006. [DOI] [PubMed] [Google Scholar]
- 22.Zhang J., Liu H., Hou L., Wang G., Zhang R., Huang Y., Chen X., Zhu J. Circular RNA_LARP4 inhibits cell proliferation and invasion of gastric cancer by sponging miR-424-5p and regulating LATS1 expression. Mol. Cancer. 2017;16:151. doi: 10.1186/s12943-017-0719-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhong Z., Huang M., Lv M., He Y., Duan C., Zhang L., Chen J. Circular RNA MYLK as a competing endogenous RNA promotes bladder cancer progression through modulating VEGFA/VEGFR2 signaling pathway. Cancer Lett. 2017;403:305–317. doi: 10.1016/j.canlet.2017.06.027. [DOI] [PubMed] [Google Scholar]
- 24.He R., Liu P., Xie X., Zhou Y., Liao Q., Xiong W., Li X., Li G., Zeng Z., Tang H. circGFRA1 and GFRA1 act as ceRNAs in triple negative breast cancer by regulating miR-34a. J. Exp. Clin. Cancer Res. 2017;36:145. doi: 10.1186/s13046-017-0614-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Han B., Chao J., Yao H. Circular RNA and its mechanisms in disease: From the bench to the clinic. Pharmacol. Ther. 2018;187:31–44. doi: 10.1016/j.pharmthera.2018.01.010. [DOI] [PubMed] [Google Scholar]
- 26.Wu J., Jiang Z., Chen C., Hu Q., Fu Z., Chen J., Wang Z., Wang Q., Li A., Marks J.R. CircIRAK3 sponges miR-3607 to facilitate breast cancer metastasis. Cancer Lett. 2018;430:179–192. doi: 10.1016/j.canlet.2018.05.033. [DOI] [PubMed] [Google Scholar]
- 27.Meng S., Zhou H., Feng Z., Xu Z., Tang Y., Li P., Wu M. CircRNA: functions and properties of a novel potential biomarker for cancer. Mol. Cancer. 2017;16:94. doi: 10.1186/s12943-017-0663-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cittelly D.M., Das P.M., Spoelstra N.S., Edgerton S.M., Richer J.K., Thor A.D., Jones F.E. Downregulation of miR-342 is associated with tamoxifen resistant breast tumors. Mol. Cancer. 2010;9:317. doi: 10.1186/1476-4598-9-317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Accili D., Arden K.C. FoxOs at the crossroads of cellular metabolism, differentiation, and transformation. Cell. 2004;117:421–426. doi: 10.1016/s0092-8674(04)00452-0. [DOI] [PubMed] [Google Scholar]
- 30.Zou Y., Tsai W.B., Cheng C.J., Hsu C., Chung Y.M., Li P.C., Lin S.H., Hu M.C. Forkhead box transcription factor FOXO3a suppresses estrogen-dependent breast cancer cell proliferation and tumorigenesis. Breast Cancer Res. 2008;10:R21. doi: 10.1186/bcr1872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Raha P., Thomas S., Munster P.N. Epigenetic modulation: a novel therapeutic target for overcoming hormonal therapy resistance. Epigenomics. 2011;3:451–470. doi: 10.2217/epi.11.72. [DOI] [PubMed] [Google Scholar]
- 32.Ward A., Balwierz A., Zhang J.D., Küblbeck M., Pawitan Y., Hielscher T., Wiemann S., Sahin Ö. Re-expression of microRNA-375 reverses both tamoxifen resistance and accompanying EMT-like properties in breast cancer. Oncogene. 2013;32:1173–1182. doi: 10.1038/onc.2012.128. [DOI] [PubMed] [Google Scholar]
- 33.Ahmad A., Ginnebaugh K.R., Yin S., Bollig-Fischer A., Reddy K.B., Sarkar F.H. Functional role of miR-10b in tamoxifen resistance of ER-positive breast cancer cells through down-regulation of HDAC4. BMC Cancer. 2015;15:540. doi: 10.1186/s12885-015-1561-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Xue X., Yang Y.A., Zhang A., Fong K.W., Kim J., Song B., Li S., Zhao J.C., Yu J. LncRNA HOTAIR enhances ER signaling and confers tamoxifen resistance in breast cancer. Oncogene. 2016;35:2746–2755. doi: 10.1038/onc.2015.340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chen B., Wei W., Huang X., Xie X., Kong Y., Dai D., Yang L., Wang J., Tang H., Xie X. circEPSTI1 as a Prognostic Marker and Mediator of Triple-Negative Breast Cancer Progression. Theranostics. 2018;8:4003–4015. doi: 10.7150/thno.24106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Du W.W., Yang W., Li X., Awan F.M., Yang Z., Fang L., Lyu J., Li F., Peng C., Krylov S.N. A circular RNA circ-DNMT1 enhances breast cancer progression by activating autophagy. Oncogene. 2018;37:5829–5842. doi: 10.1038/s41388-018-0369-y. [DOI] [PubMed] [Google Scholar]
- 37.Memczak S., Jens M., Elefsinioti A., Torti F., Krueger J., Rybak A., Maier L., Mackowiak S.D., Gregersen L.H., Munschauer M. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495:333–338. doi: 10.1038/nature11928. [DOI] [PubMed] [Google Scholar]
- 38.Hansen T.B., Jensen T.I., Clausen B.H., Bramsen J.B., Finsen B., Damgaard C.K., Kjems J. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495:384–388. doi: 10.1038/nature11993. [DOI] [PubMed] [Google Scholar]
- 39.Weng W., Wei Q., Toden S., Yoshida K., Nagasaka T., Fujiwara T., Cai S., Qin H., Ma Y., Goel A. Circular RNA ciRS-7-A Promising Prognostic Biomarker and a Potential Therapeutic Target in Colorectal Cancer. Clin. Cancer Res. 2017;23:3918–3928. doi: 10.1158/1078-0432.CCR-16-2541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhao Y., Alexandrov P.N., Jaber V., Lukiw W.J. Deficiency in the Ubiquitin Conjugating Enzyme UBE2A in Alzheimer’s Disease (AD) is Linked to Deficits in a Natural Circular miRNA-7 Sponge (circRNA; ciRS-7) Genes (Basel) 2016;7:7. doi: 10.3390/genes7120116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Su C., Han Y., Zhang H., Li Y., Yi L., Wang X., Zhou S., Yu D., Song X., Xiao N. CiRS-7 targeting miR-7 modulates the progression of non-small cell lung cancer in a manner dependent on NF-κB signalling. J. Cell. Mol. Med. 2018;22:3097–3107. doi: 10.1111/jcmm.13587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li R.C., Ke S., Meng F.K., Lu J., Zou X.J., He Z.G., Wang W.F., Fang M.H. CiRS-7 promotes growth and metastasis of esophageal squamous cell carcinoma via regulation of miR-7/HOXB13. Cell Death Dis. 2018;9:838. doi: 10.1038/s41419-018-0852-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chen L., Zhang S., Wu J., Cui J., Zhong L., Zeng L., Ge S. circRNA_100290 plays a role in oral cancer by functioning as a sponge of the miR-29 family. Oncogene. 2017;36:4551–4561. doi: 10.1038/onc.2017.89. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 44.Chen G., Shi Y., Liu M., Sun J. circHIPK3 regulates cell proliferation and migration by sponging miR-124 and regulating AQP3 expression in hepatocellular carcinoma. Cell Death Dis. 2018;9:175. doi: 10.1038/s41419-017-0204-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fu G., Peng C. Nodal enhances the activity of FoxO3a and its synergistic interaction with Smads to regulate cyclin G2 transcription in ovarian cancer cells. Oncogene. 2011;30:3953–3966. doi: 10.1038/onc.2011.127. [DOI] [PubMed] [Google Scholar]
- 46.Liang Z., Wang X., Xu X., Xie B., Ji A., Meng S., Li S., Zhu Y., Wu J., Hu Z. MicroRNA-608 inhibits proliferation of bladder cancer via AKT/FOXO3a signaling pathway. Mol. Cancer. 2017;16:96. doi: 10.1186/s12943-017-0664-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hong Z.Y., Lee H.J., Shin D.Y., Kim S.K., Seo M., Lee E.J. Inhibition of Akt/FOXO3a signaling by constitutively active FOXO3a suppresses growth of follicular thyroid cancer cell lines. Cancer Lett. 2012;314:34–40. doi: 10.1016/j.canlet.2011.09.010. [DOI] [PubMed] [Google Scholar]
- 48.Ananda Sadagopan S.K., Mohebali N., Looi C.Y., Hasanpourghadi M., Pandurangan A.K., Arya A., Karimian H., Mustafa M.R. Forkhead Box Transcription Factor (FOXO3a) mediates the cytotoxic effect of vernodalin in vitro and inhibits the breast tumor growth in vivo. J. Exp. Clin. Cancer Res. 2015;34:147. doi: 10.1186/s13046-015-0266-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Liu Y., Ao X., Ding W., Ponnusamy M., Wu W., Hao X., Yu W., Wang Y., Li P., Wang J. Critical role of FOXO3a in carcinogenesis. Mol. Cancer. 2018;17:104. doi: 10.1186/s12943-018-0856-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kim D., Pertea G., Trapnell C., Pimentel H., Kelley R., Salzberg S.L. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013;14:R36. doi: 10.1186/gb-2013-14-4-r36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhang X.O., Wang H.B., Zhang Y., Lu X., Chen L.L., Yang L. Complementary sequence-mediated exon circularization. Cell. 2014;159:134–147. doi: 10.1016/j.cell.2014.09.001. [DOI] [PubMed] [Google Scholar]
- 52.Wang L., Feng Z., Wang X., Wang X., Zhang X. DEGseq: an R package for identifying differentially expressed genes from RNA-seq data. Bioinformatics. 2010;26:136–138. doi: 10.1093/bioinformatics/btp612. [DOI] [PubMed] [Google Scholar]
- 53.Gao S., Li X., Ding X., Jiang L., Yang Q. Huaier extract restrains the proliferative potential of endocrine-resistant breast cancer cells through increased ATM by suppressing miR-203. Sci. Rep. 2017;7:7313. doi: 10.1038/s41598-017-07550-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Li Y., Liang Y., Sang Y., Song X., Zhang H., Liu Y., Jiang L., Yang Q. MiR-770 suppresses the chemo-resistance and metastasis of triple negative breast cancer via direct targeting of STMN1. Cell Death Dis. 2018;9:14. doi: 10.1038/s41419-017-0030-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.