Abstract
Accounting for more than 70% of ovarian cancer cases, epithelial ovarian malignancy has a low 5-year survival rate. MicroRNAs may be targeted in the clinical treatment of the disease. In this study, we first found that miR-1251-5p was significantly upregulated in human ovarian cancer cell lines and tissues with the cancer progression and stages. Overexpression or inhibition of miR-1251-5p promoted or impeded cell proliferation and cell cycle progression. Subsequently, TBCC, one of the tubulin-binding cofactors (TBCs), was identified as a target of miR-1251-5p to be negatively associated with cell cycle and autophagy. Exogenous overexpression of TBCC inhibited the expressions of CDK4 and LC3BII, but it promoted the expressions of α/β-tubulin and p62 to suppress cell growth and autophagy, particularly under the starving condition; whereas the introduction of miR-1251-5p in TBCC-overexpressing cells rescued the suppressive effects of TBCC on cell cycle and autophagy through the inverse regulation of the above proteins. Finally, miR-1251-5p was proven to enhance xenograft tumor growth through the downregulation of TBCC but upregulation of Ki67 and LC3B in xenograft tumor tissues. Collectively, these results suggest that miR-1251-5p functions as an oncogene to suppress TBCC and α/β-tubulin expression. Thus, the miR-1251-5p/TBCC/α/β-tubulin axis may be targeted for ovarian cancer treatment.
Keywords: ovarian cancer, miR-1251-5p, TBCC, α/β-tubulin, autophagy
This study demonstrated that miR-1251-5p was upregulated in ovarian cancer (OC) cell lines and closely related to OC recurrence and stages. Overexpression of miR-1251-5p promoted cell proliferation and cell cycle progression in OC cells. TBCC was proven to be a direct target of miR-1251-5p. Upregulation or downregulation of TBCC repressed or promoted cell proliferation, cell cycle progression, and autophagy, whereas miR-1251-5p inversely regulated these effects. miR-1251-5p could also drive tumor growth in vivo. Thus, our findings suggest that the miR-1251-5p/TBCC/α/β-tubulin axis may function as an oncogenic pathway to promote cell cycle and autophagy through the dysregulation of microtubular formation and folding.
Introduction
According to the latest report in 2018, ovarian cancer (OC) ranks at the fifth highest mortality rate among all cancers in women. Epithelial OC (EOC) accounts for nearly 70% of all OC cases, and its 5-year overall survival rate is around 45%.1, 2 The general treatment strategy for OC and EOC mainly includes debulking surgery in combination with platinum-taxane chemotherapy.3 New targeted treatments such as antiangiogenic drugs, poly (adenosine diphosphate-ribose) polymerase inhibitors, and immune checkpoint inhibitors may be used for the recurrent disease,4 whereas bevacizumab may be selected to treat the patients resistant to platinum and taxane.5 Despite major advances in the clinical treatment of the disease, problems such as the difficulties of early diagnosis and recurrence are still the biggest obstacles to improving survival. Owing to the development of deep sequencing, some new potential strategies for targeted therapies have emerged. For example, a number of noncoding RNAs, including microRNAs (miRNAs), have been identified in OC, and, thus, they may be targeted in OC treatment.
miRNAs are small (20- to 23-nt) noncoding, endogenous, single-stranded RNAs regulating gene expression at the post-transcriptional level. In multiple studies, various miRNAs have been reported in OC progression. miR-141 promotes cell proliferation, tumor growth, and anoikis resistance via targeting KLF12 in EOC. miR-141/KLF12/Sp1/Survivin play an important part in enhancing anoikis resistance.6 miR-551b-3p was shown to contribute to OC cell resistance to apoptosis, survival, and proliferation and to stimulate STAT3 transcription and expression through the interaction with the promoter of STAT3.7
miR-1251-5p is located at chromosome 12q23.1, and not many reports to date have focused on the study of this miRNA. Microarray analysis of miRNAs showed that the expression of miR-1251-5p was lower in gastric cancer cells than in normal cells.8 In clear cell renal cell carcinoma, miR-1251-5p was also downregulated.9 However, we observed that miR-1251-5p was upregulated in OC, indicating that the biological function of miR-1251-5p is diversified in different cancers.
Microtubules are involved in a series of biological activities, including cell division, cell migration, and intracellular transport.10 Microtubules consist of α/β-tubulin heterodimers, and they can form correct folds with the help of cytosolic chaperonin and tubulin-binding cofactors (TBCs). Five TBCs, TBCA, TBCB, TBCC, TBCD, and TBCE, may bind to α/β-tubulin in a particular order to form a supercomplex. With the help of hydrolysis of guanosine triphosphate (GTP), the activated α/β-tubulin heterodimers are released by the supercomplex and polymerized into microtubules.11, 12 The roles of TBCs in cancer cells are unclear, but TBCs may likely be involved in more biological activities than microtubule folding.13, 14 Despite limited knowledge of this, we hypothesized that TBCs might have a role in OC progression and treatment through their involvement in the protein folding of α/β-tubulin, although no studies on this topic have been reported.
In the current study, we verified that miR-1251 was upregulated in OC cell lines. miR-1251-5p affected cell proliferation, cell cycle, and autophagy via targeting TBCC and functioned as a tumor driver in vivo.
Results
miR-1251-5p Is Upregulated in OC
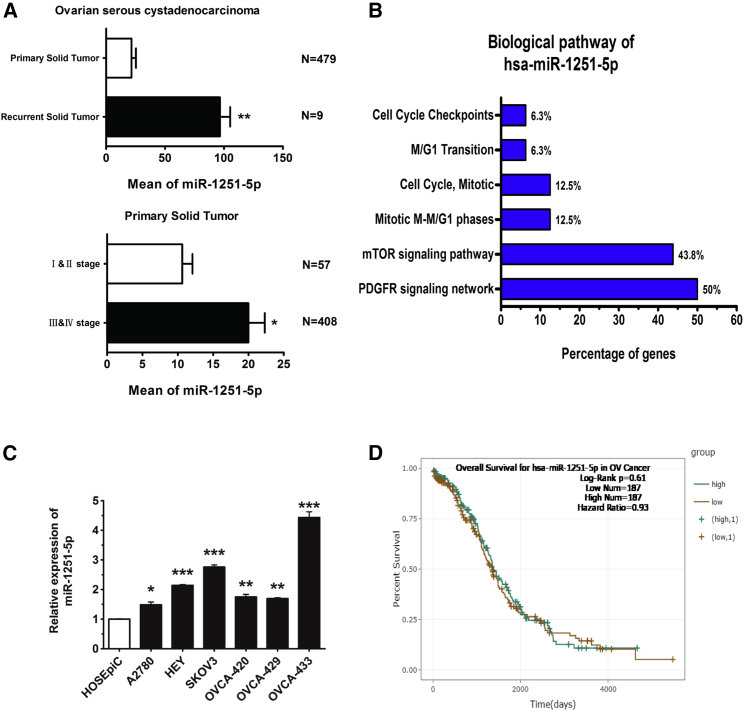
We collected 488 OC cases from the YM500v2 database (http://ngs.ym.edu.tw/ym500v2/index.php), consisting of 479 primary tumor cases and 9 recurrent tumor cases. Among the 479 primary tumor cases, 57 cases were in stages I and II, 408 cases were in stages III and IV, and 14 cases were in unclear stages. Based on analysis of these cases, we found that the RNA level of miR-1251-5p was much higher in recurrent tumors than in primary tumors. In primary tumors, the expression of miR-1251-5p was upregulated to a greater extent in stages III and IV than in stages I and II (Figure 1A). The biological pathway of miR-1251-5p might be associated with cell cycle transition, as analyzed by using FunRich 3.1 (Figure 1B). Compared with normal human ovarian epithelial cell line HOSEpiC, miR-1251-5p was upregulated in human OC cell lines (Figure 1C). However, based on analysis of 368 cases from starBase (http://starbase.sysu.edu.cn/), miR-1251-5p had a weak effect on patient overall survival, indicating that miR-1251-5p might not be an independent prognostic factor (Figure 1D). These data suggest that miR-1251-5p may promote OC progression and recurrence other than directly predicting the patient overall survival rate.
Figure 1.
miR-1251-5p Levels in OC Tissues and Cell Lines and in Association with Recurrence and Stages in OC
(A) YM500 datasets of 479 OC primary tumors and nine OC recurrent tumors were analyzed. OC primary tumors consisted of 57 cases in stages I and II, 408 cases in stages III and IV, and 14 cases without stage data. (B) The biological pathway of miR-1251-5p was analyzed using FunRich 3.1. (C) miR-1251-5p expression in human normal ovarian epithelial cell line HOSEpiC and human OC cell lines A2780, HEY, SKOV3, OVCA420, OVCA429, and OVCA433 detected by qRT-PCR. (D) Kaplan-Meier curve for starBase datasets of 368 OC patients. *p < 0.05, **p < 0.01, ***p < 0.001.
miR-1251-5p Promotes Cell Proliferation
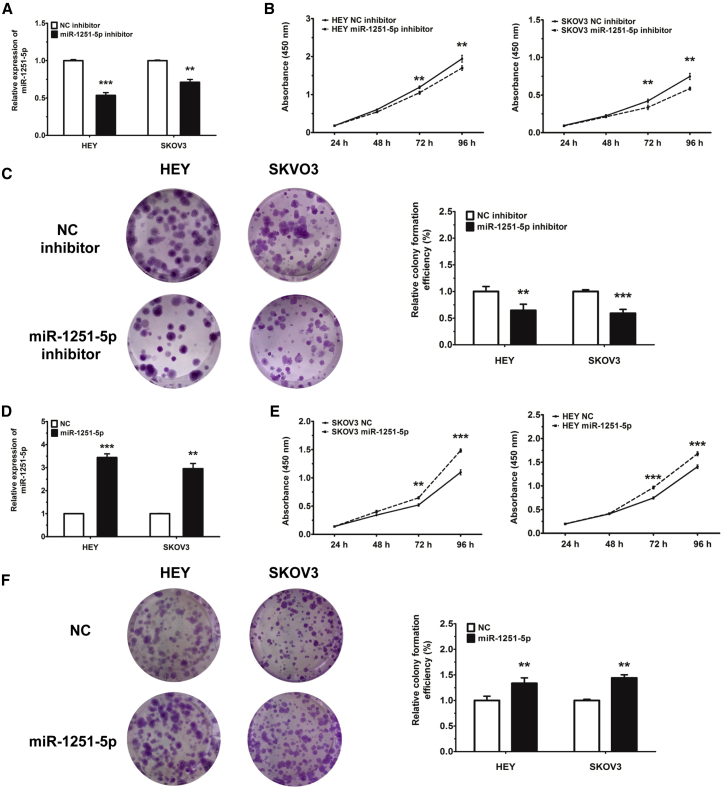
We first used an inhibitor to reduce the level of miR-1251-5p in HEY and SKOV3 cells. We found that transfection of miR-1251-5p inhibitor downregulated the RNA level of miR-1251-5p in HEY and SKOV3 cells compared with a negative control (NC) mimic (Figure 2A), which dramatically restrained cell proliferation tested by cell counting kit 8 (CCK-8) and colony formation assays (Figures 2B and 2C). Stable transfection of HEY and SKOV3 cells with miR-1251-5p (pLenti-miR-1251-5p) shown in Figures S1 and 2D also promoted more cell proliferation than that with an empty vector (pLenti-NC), as tested by CCK-8 and colony formation assays (Figures 2E and 2F).
Figure 2.
miR-1251-5p Promotes Cell Proliferation
(A) The expression of miR-1251-5p in HEY and SKOV3 cells transfected with NC or miR-1251-5p inhibitor. (B) Cell proliferation ability of HEY and SKOV3 cells transiently transfected with NC or miR-1251-5p inhibitor. (C) Colony formation of cells transfected with miR-1251-5p or NC inhibitor. Representative images and quantitative data are shown. (D) Expression of miR-1251-5p in HEY and SKOV3 cells infected with empty vector or miR-1251-5p expression vector using qRT-PCR. (E) Cell proliferation of HEY and SKOV3 cells stably transfected with empty vector or miR-1251-5p expression vector. (F) Colony formation of cells stably transfected with empty vector or miR-1251-5p expression vector. Representative images and quantitative data are shown. *p < 0.05, **p < 0.01, ***p < 0.001.
miR-1251-5p Promotes Cell Cycle Progression
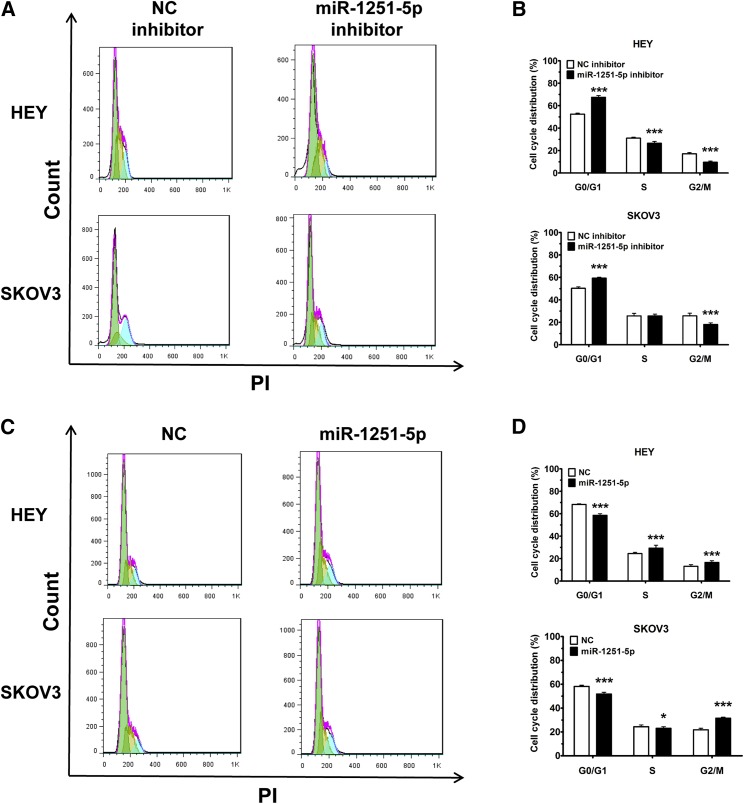
Cells transfected with miR-1251-5p inhibitor were treated with serum-free RPMI-1640 medium for 6 h for cell cycle synchronization, and then they were cultured in RPMI-1640 medium with 2% fetal bovine serum (FBS) for 48 h. The results showed that transfection with miR-1251-5p inhibitor impeded cell cycle progression through the induction of G0/G1 phase arrest (Figures 3A and 3B). Similarly, a decreased proportion of cells in G0/G1 phase was found in miR-1251-5p-expressing cells compared with NC-expressing cells (Figures 3C and 3D). These data suggest that miR-1251-5p promotes the cell cycle progression of OC cells.
Figure 3.
miR-1251-5p Drives Cell Cycle Progression
(A and B) Cell cycle distributions of HEY and SKOV3 cells transfected with NC or miR-1251-5p inhibitor. Representative images (A) and quantitative data (B) are shown. Green, yellow, and blue represent the populations of cells in G0/G1, S, and G2/M phases, respectively. (C and D) Treated cells were cultured for 48 h, harvested, fixed using ethanol, and stained with PI. Cell cycle distributions of HEY and SKOV3 cells stably transfected with empty vector or miR-1251-5p expression vector were assessed using flow cytometry. Representative images (C) and quantitative data (D) are shown. Green, yellow, and blue represent the populations of cells in G0/G1, S, and G2/M phases, respectively. *p < 0.05, **p < 0.01, ***p < 0.001.
TBCC Is a Direct Target of miR-1251-5p
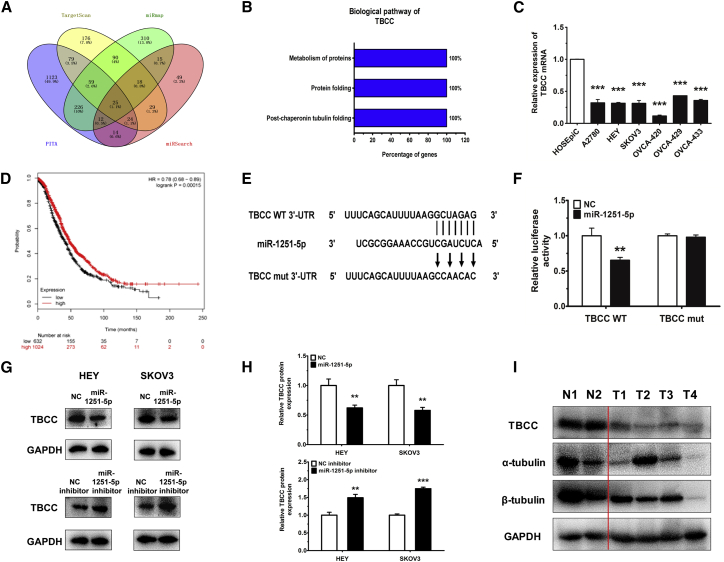
To study the mechanism by which miR-1251-5p promotes OC progression at the molecular level, we predicted a couple of potential target genes using TargetScan (http://www.targetscan.org/vert_72/), miRmap (https://mirmap.ezlab.org/), PITA (https://genie.weizmann.ac.il/pubs/mir07/mir07_data.html), and miRSearch (https://www.exiqon.com/mirsearch?tokenRefresh=true) (Figure 4A). Based on FunRich 3.1 analysis, TBCC involved in α/β-tubulin protein folding was identified as a direct target of miR-1251-5p to suppress cell proliferation and cell cycle (Figure 4B).
Figure 4.
TBCC Is a Target of miR-1251-5p
(A) TargetScan, miRmap, PITA, and miRSearch were used to predict direct target genes of miR-1251-5p. (B) The biological pathway of TBCC was analyzed using FunRich 3.1. (C) TBCC expression in human normal ovarian epithelial cell line and human OC cell lines. (D) Kaplan-Meier curve for Kaplan-Meier Plotter datasets of 1,657 OC patients. (E) Construction of pGL3 miReport plasmid, containing wild-type (WT) or mutant (mut) TBCC 3′ UTR. (F) Dual luciferase assay in HEK293T cells co-transfected with WT or mut TBCC 3′ UTR luciferase vectors and miR-1251-5p mimic or NC mimic. (G and H) Protein levels of TBCC in miR-1251-5p-overexpressing and -silencing cells. Representative images (G) and quantitative data (H) are shown. (I) The protein levels of TBCC, α-tubulin, and β-tubulin detected in two normal ovarian tissues (N1 and N2) and four ovarian cancer tissues (T1–T4). *p < 0.05, **p < 0.01, ***p < 0.001.
Further analysis showed that TBCC was downregulated in human OC cell lines (Figure 4C) and that high expression of TBCC had a longer overall survival rate in OC (http://kmplot.com/analysis/index.php?p=service&cancer=ovar) (Figure 4D). The wild-type and mutant binding sites of miR-1251-5p to TBCC are shown in Figure 4E. To confirm the inhibition of TBCC by miR-1251-5p, HEK293T cells were co-transfected with pGL3-TBCC wild-type or mutant 3′ UTR along with miR-1251-5p mimic and pRL (plasmid expressing a renilla luciferase gene) vector. We found that the relative luciferase activity was remarkably decreased in TBCC wild-type 3′ UTR-transfected cells compared to TBCC mutant 3′ UTR-transfected cells, suggesting a potential inhibitor binding activity of miR-1251-5p to TBCC 3′ UTR (Figure 4F).
The above results were further confirmed by using western blot (WB) showing that miR-1251-5p decreased the protein level of TBCC in HEY and SKOV3 cells (Figures 4G and 4H). Moreover, the expressions of TBCC, α-tubulin, and β-tubulin were downregulated in four OC tissues (T1–T4) compared with two normal ovarian tissues (N1 and N2) (Figure 4I).
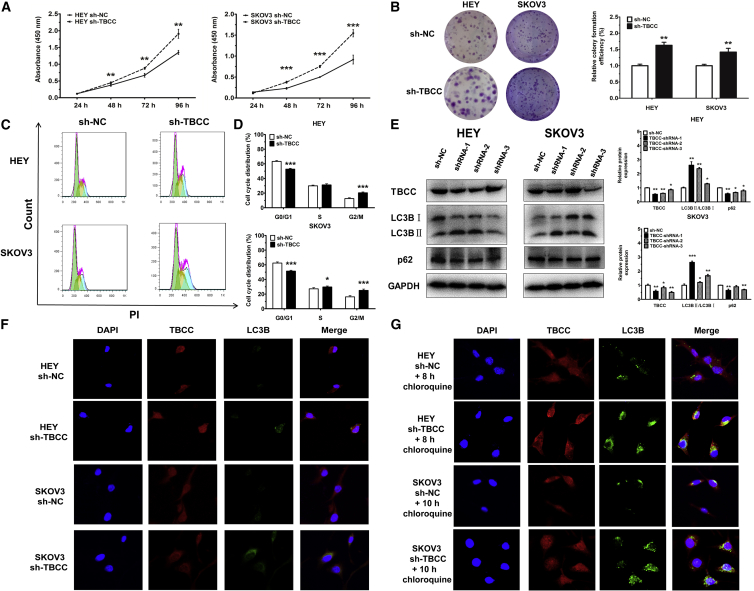
Knockdown of TBCC Induces Cell Proliferation, Cell Cycle Progression, and Autophagy
To investigate the biological function of TBCC in OC cells, TBCC was silenced by using short hairpin RNA (shRNA; sh-TBCC) as shown in Figure 5E, which resulted in the decrease of cell proliferation (Figures 5A and 5B) and a reduced cell proportion in G0/G1 phase after starvation (Figures 5C and 5D).
Figure 5.
Knockdown of TBCC Promotes Cell Proliferation and Cell Cycle Progression and Induces Autophagy
(A) Proliferation of cells with or without TBCC silencing. (B) Colony formation of cells with or without TBCC silencing. Representative images and quantitative data are shown. (C and D) Cell cycle distributions of cells with or without TBCC silencing. Representative images (C) and quantitative data (D) are shown. Green, yellow, and blue represent the populations of cells in G0/G1, S, and G2/M phases, respectively. (E) Protein levels of TBCC, LC3B, and p62 in TBCC-silencing or control cells. Representative images and quantitative data are shown. (F) Representative images of cells with or without TBCC silencing by immunofluorscent staining of DNA (DAPI, blue), TBCC (red), and LC3B (green). Autophagosome formation was monitored via confocal microscopy. (G) Autophagosome formation was monitored via confocal microscopy in TBCC-downexpressing cells treated with 25 μM chloroquine for 8 and 10 h, respectively. *p < 0.05, **p < 0.01, ***p < 0.001.
Since TBCC is associated with the protein folding of α/β-tubulin and some studies reported that α/β-tubulin might participate in autophagy,15, 16 we also tested for and found that the suppression of TBCC expression induced autophagy in OC cells (Figure 5F). Treatment of HEY and SKOV3 cells with chloroquine (25 μM, 8 and 10 h) induced more autophagosomes in TBCC-knockdown cells than in NC cells, whereas TBCC was highly stained along with LC3B in the cytoplasm of OC cells after chloroquine treatment (Figure 5G). At the protein level, an increased ratio of LC3BII:LC3BI and a low expression level of p62 were conceived after the inhibition of TBCC compared with controls (Figure 5E).
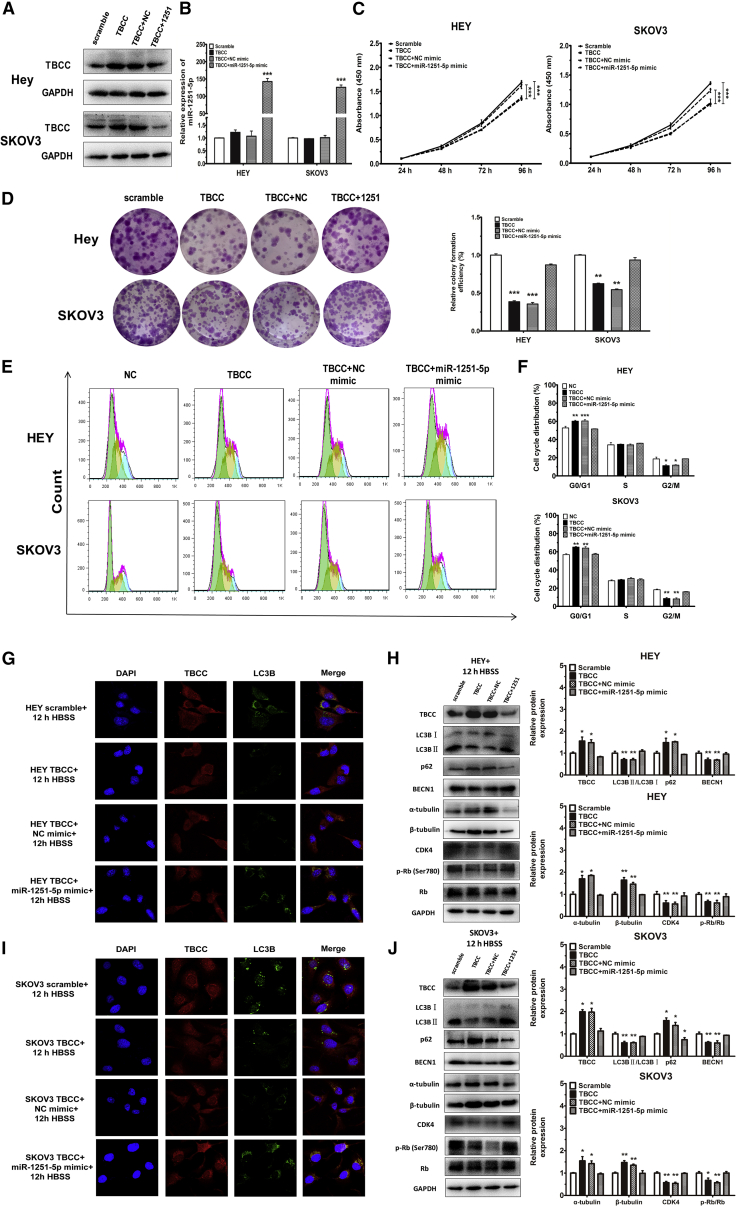
Overexpression of miR-1251-5p Rescues the Impact of TBCC on Cell Proliferation, Cell Cycle, and Autophagy
To determine whether overexpression of miR-1251-5p was able to reverse the effect of TBCC on OC cells, we first overexpressed TBCC in HEY and SKOV3 cells (Figures 6A and 6B), and we found that TBCC overexpression reduced cell proliferation and caused G0/G1 phase arrest (Figures 6C and 6D). We then transfected a miR-1251-5p mimic or NC mimic into TBCC-overexpressing cells, and we found that miR-1251-5p rescued the inhibitory effects of TBCC on cell proliferation and cell cycle (Figures 6E and 6F). Autophagosomes were highly reduced in TBCC-overexpressing cells compared with NC cells after treatment with Hank’s balanced salt solution (HBSS; Procell, Wuhan, China) for 12 h at room temperature. However, when miR-1251-5p mimic was transfected into cells, the suppressive effect of TBCC on autophagy was largely prohibited (Figures 6G and 6I).
Figure 6.
miR-1251-5p Reverses the Effects of TBCC on Cell Proliferation, Cell Cycle, and Autophagy
(A and B) The protein level of TBCC and the expression of miR-1251-5p in TBCC-overexpressing cells and those further transfected with NC or miR-1251-5p mimic. (C and D) Proliferation and colony formation. (E and F) Cell cycle distributions assessed using flow cytometry. Green, yellow, and blue represent the populations of cells in G0/G1, S, and G2/M phases, respectively. (G and I) Autophagosome formation was monitored via confocal microscope. (H and J) The protein levels of TBCC, LC3B, p62, α-tubulin, β-tubulin, CDK4, Rb, and p-Rb (Ser780). *p < 0.05, **p < 0.01, ***p < 0.001.
The protein levels of TBCC, LC3B, p62, BECN1, α-tubulin, and β-tubulin were correspondingly altered in association with autophagy. Treatment of cells overexpressing TBCC with HBSS for 12 h decreased the ratio of LC3BII:LC3BI and inhibited the expression of BECN1, but it increased the expressions of p62, α-tubulin, and β-tubulin. Moreover, the regulation of the ratio of LC3BII:LC3BI and the expressions of BECN1, p62, α-tubulin, and β-tubulin by TBCC were reversed when a miR-1251-5p mimic was transfected into TBCC-overexpressing cells. Besides, the expression of CDK4 and the phosphorylation of Rb (Ser780) were downregulated by TBCC, but the delivery of miR-1251-5p could turn down these effects (Figures 6H and 6J; Figure S2). The mRNA levels of TBCC, LC3B, p62, and BECN1 were also detected using qRT-PCR, and the results were in accordance with their protein levels (Figure S3). These findings indicate that upregulation of TBCC can reverse the biological function of miR-1251-5p in OC cells.
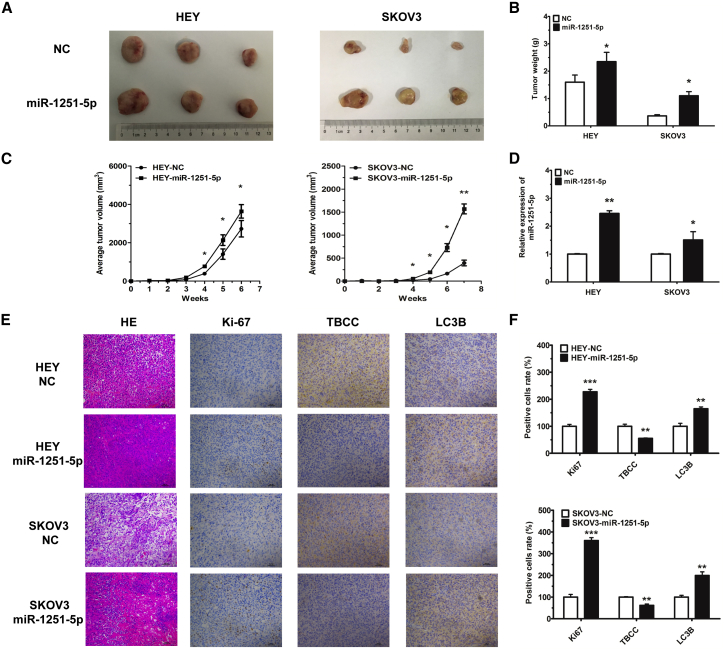
miR-1251-5p Drives In Vivo Tumor Growth
To investigate the tumorigenic role of miR-1251-5p, we generated xenograft mouse models by subcutaneous injection of HEY cells (3 × 106) or SKOV3 cells (5 × 106) expressing miR-1251-5p or NC into nude mice, and we measured tumor volumes every week. The results showed that overexpression of miR-1251-5p significantly induced tumor growth in mice within 6 or 7 weeks (Figures 7A–7C). Moreover, the expression of miR-1251-5p was upregulated, as tested by using real-time qPCR (Figure 7D). Additionally, by immunohistochemistry, we showed that TBCC was downregulated whereas Ki67 and LC3B were upregulated in miR-1251-5p-overexpressing tumor tissues (Figures 7E and 7F). These data revealed that miR-1251-5p might drive tumor growth via the suppression of TBCC and the induction of autophagy in tumor cells.
Figure 7.
miR-1251-5p Drives OC Tumor Growth In Vivo
(A and B) Images (A) and quantification of weights (B) of tumors isolated from mice after 6 or 7 weeks of subcutaneous injection. (C) Tumor growth curves with volume measured every week after injection. (D) The expression of miR-1251-5p detected in tumors by using qRT-PCR. (E and F) Tumor tissues stained with H&E solution; Ki-67, TBCC, and LC3B detected by immunohistochemistry (original magnification × 100) (E) and the number of positively stained cells with protein antibodies (F). *p < 0.05, **p < 0.01, ***p < 0.001.
Discussion
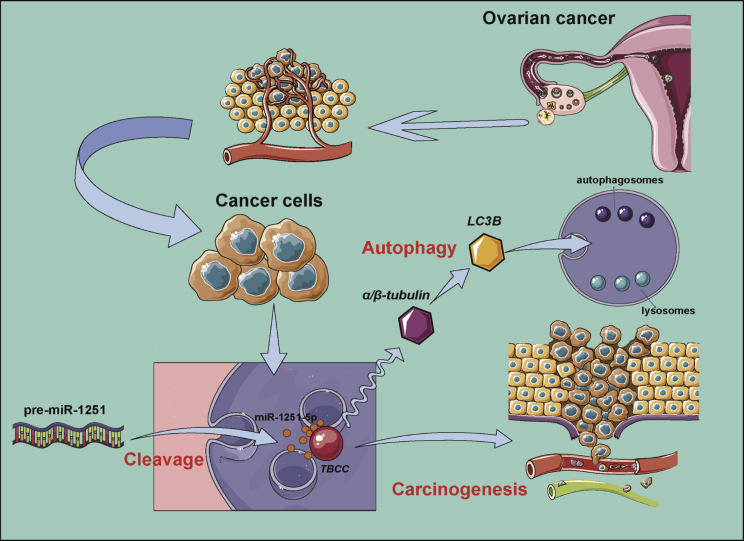
This study demonstrated that miR-1251-5p was upregulated in OC cell lines and closely related to OC recurrence and stages. Overexpression of miR-1251-5p promoted cell proliferation and cell cycle progression in OC cells. TBCC was proven to be a direct target of miR-1251-5p. Upregulation or downregulation of TBCC repressed or promoted cell proliferation, cell cycle progression, and autophagy, whereas miR-1251-5p inversely regulated these effects. Our data proved that miR-1251-5p could drive tumor growth in vivo. Thus, our findings suggest that the miR-1251-5p/TBCC/α/β-tubulin axis may function as an oncogenic pathway to promote cell cycle and autophagy through the dysregulation of microtubular formation and folding (Figure 8).
Figure 8.
Possible Mechanism of miR-1251-5p/TBCC/α/β-tubulin Axis in OC Carcinogenesis and Autophagy
The biological function of TBCC in cancer is only reported in a few studies. TBCC may function as a tumor suppressor in breast cancer to inhibit microtubule dynamicity of breast cancer cells, which thereby regulate cell cycle distribution and mitotic duration.17 TBCC overexpression can also cause a higher proportion of nonpolymerizable tubulins, leading to the enhanced in vitro and in vivo chemosensitivity to anti-microtubule agents.18 Additionally, the N-terminal domain of TBCC provides a platform for tubulin interaction. TBCC can cooperate with other TBC members in a certain order to form a supercomplex of TBCE/α-tubulin/TBCC/TBCD/β-tubulin, indicating that TBCC functions as a post-chaperonin in the folding and assembly of α/β-tubulin. Depletion of TBCC can cause mitotic failure and apoptosis in cervical cancer.19 Herein, we found that the higher expression of TBCC could lead to a higher overall survival rate. Through interaction with miR-1251-5p and α/β-tubulin, TBCC may have a potential role in OC recurrence and therapeutic efficiency, particularly in terms of OC treatment with anti-microtubule agents.
Autophagy is a complicated activity possibly playing a role in the progression of cancers. Autophagy is activated to avoid the accumulation of damaged protein and organelles, including mitochondria, which is closely associated with apoptosis in cancers.20, 21 Both autophagy and apoptosis can suppress tumorigenesis, and if one of their functions is impeded, the other becomes active to induce cancer cell death. On the other hand, autophagy is also an important factor in tumor progression. By degrading some non-essential components, autophagy can provide nutrition for cancer cell survival.15, 22, 23 As a result, autophagy can increase the resistance of cancer cells to hypoxia,24, 25 starvation,26, 27 the epithelial-mesenchymal transition,28, 29 and therapy-induced cell death,30, 31, 32 and it can enable the survival of senescent cancer cells and maintenance of cancer stem cells.33, 34 In OC cells, TXNDC17 may participate in BECN1-induced autophagy to enhance paclitaxel resistance.35 MARCH is involved in transforming growth factor β (TGF-β)1-induced autophagy, migration, and invasion in OC. Acting as a competing endogenous RNA, MARCH can regulate the expressions of SMAD2 and ATG5.36 In our study, rather than participating in the classical mammalian target of rapamycin (mTOR)-signaling pathway, the miR-1251-5p/TBCC/α/β-tubulin axis may regulate the formation and folding of microtubules through the activation other than inhibition of autophagy flux. This may be the main reason that TBCC overexpression in OC cells inhibited autophagy under starvation, whereas miR-1251-5p overexpression turned down the suppressive effect of TBCC on autophagy.
In conclusion, our results suggest that miR-1251-5p promotes ovarian carcinogenesis via directly targeting TBCC. In this regulatory mechanism, autophagy functions to promote tumorgenesis in association with miR-1251-5p and TBCC. Thus, the miR-1251-5p/TBCC/α/β-tubulin axis may play an important role in molecular therapy for cancer with anti-microtubule agents, which deserves further investigation.
Materials and Methods
Meta-analysis and Tissue Samples
OC tissue data were obtained from the YM500v2 database, and they consisted of 479 primary OC tumor cases and nine recurrent OC tumor cases. Among the 479 primary OC tumor cases, there were 57 cases in stages I and II and 408 cases in stages III and IV, whereas 14 cases had no information on stage. A total of 368 cases from starBase and 1,657 cases from the Kaplan-Meier Plotter Database were collected to analyze overall survival rates. The tissue samples were obtained from Fudan University Shanghai Cancer Center, and the ethical approval was granted by the ethics committee of Fudan University Shanghai Cancer Center.
Cell Culture
Human normal ovarian epithelial cell line HOSEpiC; human ovarian epithelial cancer cell lines A2780, HEY, SKOV3, OVCA420, OVCA429, and OVCA433; and lentiviral packaging cells HEK293T were obtained from American Type Culture Collection (ATCC, USA) or the Cell Bank, China Academy of Sciences (Shanghai, China).
HOSEpiC, A2780, HEY, SKOV3, OVCA420, OVCA429, and OVCA433 cells were cultured in RPMI-1640 medium (Corning Cellgro, Manassas, VA, USA). HEK293T was cultured in DMEM (Corning Cellgro, Manassas, VA, USA). All media were supplemented with 10% FBS (Gibco, Gaithersburg, MD, USA) and an antibiotic cocktail of 100 U/mL penicillin and 100 μg/mL streptomycin (HyClone, Logan, UT, USA). Cells were cultured at 37°C in a 5% CO2 environment.
Plasmid Construction, RNAi, and Viral Infection
To overexpress miR-1251-5p, hsa-pre-miR-1251 was inserted into a pLenti lentiviral vector. Lentiviral particles were produced from HEK293T cells co-transfected with pLenti-miR-1251 or pLenti vector by a packaging system including pSPAX2 and pMD2G vector, and then they were collected at 24, 48, and 72 h and filtered with a 0.45-μm filter lip after centrifugation at 4,000 rpm for 5 min at 4°C. HEY and SKOV3 cells were infected at 70% density for 24 h and sorted by green fluorescence via flow cytometry (Beckman Coulter, Brea, CA, USA) to generate the miR-1251-5p overexpression cell line.
To enhance or silence the expression of TBCC, human wild-type cDNA or shRNA sequences of TBCC were inserted into a pcDH-CMV-MCS-EF1-Puro vector or pLKO.1-Puro vector. The pcDH-TBCC or pcDH and pLKO.1-TBCC or pLKO.1 vectors were co-transfected into HEK293T cells using a packaging system, including pSPAX2 and pMD2G vector to generate lentiviral particles, and then they were collected at 24, 48, and 72 h and filtered with a 0.45-μm filter lip after centrifugation at 4,000 rpm for 5 min at 4°C. HEY and SKOV3 cells were infected at 70% density for 24 h and selected with puromycin (1.5–2.0 μg/mL; Sigma-Aldrich, St. Louis, MO, USA) for 7 days to establish the TBCC overexpression and knockdown cell lines. The shRNA sequences are provided in Table S1.
Cell Transfection
Cells were transfected transiently with 200 nM chemically synthesized miR-1251-5p inhibitor or NC inhibitor (Ribobio, Guangzhou, China) using Lipofectamine 2000 (Thermo Fisher Scientific, Boston, MA, USA), following the manufacturer’s instructions. RNA levels were measured using qRT-PCR at 48 h post-transfection, and the following experiments were completed within 96 h.
RNA Extraction and qRT-PCR Analysis
TRIzol reagent (Thermo Fisher Scientific, Boston, MA, USA) was used to extract total RNA, following the manufacturer’s instructions. The reverse transcription of miRNAs was performed using a PrimeScript miRNA FirstStrand cDNA Synthesis SuperMix Kit (Transgen Biotec, Beijing, China), and a cDNA library of mRNAs was constructed using PrimeScript RT Master Mix (TaKaRa Biotech, Dalian, China). The mRNA or miRNA level was quantified by qRT-PCR using a SYBR Premix Ex Taq (TaKaRa Biotech, Dalian, China) with 18S RNA and U6 small nuclear RNA (snRNA) as the endogenous controls, respectively. Data were analyzed using the relative quantification (2−ΔΔCT) method. Primers are listed in Table S1.
Cell Proliferation Assay
CCK-8 (Dojindo, Tokyo, Japan) assay was performed to assess cell proliferation. Specifically, cells were seeded at 1.5 × 103 cells/well in a 96-well plate. 8 μL CCK-8 plus 100 μL FBS-free medium was added to each well and incubated for 2 h at 37°C in a 5% CO2 environment. Finally, the 450-nm absorbance was detected with a microplate reader (Bio-Rad, Hercules, CA, USA) every 24 h for 4 days.
Colony Formation Assay
500–600 cells were plated every well in a six-well plate and cultured for 10 days at 37°C in a 5% CO2 environment. Colonies were first fixed using methanol at room temperature for 15 min, followed by staining with crystal violet (0.5% w/v) at room temperature for 15 min and counting.
Cell Cycle Analysis
Cell cycle distribution was measured using propidium iodide (PI; Sigma-Aldrich, St. Louis, MO, USA) staining. 2 × 105 cells were collected to fix in 75% ethanol overnight at −20°C. Treated cells were washed once with PBS, and they were resuspended in 250 μL RNaseA buffer (100 ng/mL; Sigma-Aldrich, St. Louis, MO, USA) at room temperature for 20 min. Then, the cells were stained with a 2× solution of PI (100 ng/mL) in the dark for 20 min and filtered using a 200-mesh filter membrane. Experiments were performed using a MoFlo XDP flow cytometer sorting system (Beckman Coulter, Brea, CA, USA).
Dual Luciferase Reporter Assay
Direct targets of miR-1251-5p were screened based on TargetScan, miRmap, PITA, and miRSearch. The 3′ UTRs of TBCC with a binding site of miR-1251-5p were sub-cloned downstream of the firefly luciferase reporter gene in pGL3 miReport vector (Promega, Madison, WI, USA), named pGL3-TBCC-3′ UTR. TBCC mutant 3′ UTRs containing a mutated binding site to miR-1251-5p were cloned into pGL3 and defined as pGL3-EGFR-3′ mUTRs.
200 ng luciferase vector pGL3-3′ UTR or pGL3-3′ mUTR, 2.5 μL miR-1251-5p mimic or NC mimic, and 15 ng pRL (Promega, Madison, WI, USA) were transiently co-transfected into HEK293T cells in 24-well plates. Cells were lysed to detect luciferase activity using an Orion II Microplate Illuminometer (Titertek-Berthold, Pforzheim, Germany) after 48 h post-transfection. The relative firefly luciferase activity was normalized to renilla luciferase activity, and the fold changes of the reporters were calculated and are shown (Figure 4).
Protein Extraction and Western Blot
Radioimmunoprecipitation assay (RIPA) lysis buffer (CWBIO, Beijing, China) was used to extract total protein that was quantified with a Protein BCA Assay Kit (Bio-Rad, Hercules, CA, USA). The protein was transferred to a polyvinylidene difluoride (PVDF) membrane (Millipore, Billerica, MA, USA) following SDS-PAGE, and then it was blocked in 5% non-fat powdered milk for 1.5 h at room temperature, followed by incubation at 4°C overnight with rabbit anti-TBCC (PA5-21710, 1:1,000; Thermo Fisher Scientific, Boston, MA, USA), anti-LC3B, anti-BECN1, anti-β-tubulin, anti-CDK4 (A7198/A7353/AC008/A0366, 1:1,000; Abclonal, Wuhan, China), anti-SQSTM1/p62, anti-Rb, anti-phospho-Rb (Ser780), anti-glyceraldehyde-3-phosphate dehydrogenase (GAPDH) (8025S/9309S/8180S/2118S, 1:1,000; Cell Signaling Technology, Boston, MA, USA), and anti-α-tubulin (sc-5286, 1:500; Santa Cruz Biotechnology, Santa Cruz, CA, USA) antibodies, respectively. After 12-h incubation, the PVDF membrane was washed with Tris-buffered saline Tween 20 (TBST) three times and incubated with a goat-anti-rabbit (7074S, 1:10,000; Cell Signaling Technology, Boston, MA, USA) or goat-anti-mouse (AS003, 1:10,000; Abclonal, Wuhan, China) secondary antibody conjugated to horseradish peroxidase (HRP) for 1.5 h at room temperature, before being washed with PBST three times repeatedly. Protein bands were determined with a chemiluminescent HRP substrate (Millipore, Billerica, MA, USA) and exposed using an E-Gel Imager (Tanon, Shanghai, China).
Immunofluorescence
Cells were washed three times by PBS and fixed in methanol for 15 min. Fixed cells were washed three times with PBS and treated with 0.5% Triton X-100 for 10 min. After treatment, cells were washed three times with PBS before being blocked in 5% BSA at 4°C overnight. After blocking, cells were washed three times with PBS and incubated in primary antibodies. For double staining, the samples were incubated in a mixture of two primary antibodies, including TBCC (PA5-21710, 1:200; Thermo Fisher Scientific, Boston, MA, USA) and LC3B (sc-376404, 1:50; Santa Cruz Biotechnology, Santa Cruz, CA, USA), at 4°C overnight. After incubation, cells were washed three times with PBS Tween 20 (PBST), incubated in a mixture of two secondary antibodies at room temperature for 1.5 h before being washed with PBST three times repeatedly, and finally mounted with DAPI Fluoromount-G (SouthernBiotech, Birmingham, AL, USA). The secondary antibodies included Alexa Fluor 594 AffiniPure goat anti-rabbit immunoglobulin G (IgG) (111-585-003, 1:200; Jackson ImmunoResearch Laboratories, West Grove, PA, USA) and Alexa Fluor 647 AffiniPure goat anti-mouse IgG (115-645-003, 1:200; Jackson ImmunoResearch Laboratories, West Grove, PA, USA). All stained cells were examined and photographed using a Zeiss LSM710 confocal fluorescence microscope (Carl Zeiss, Jena, Germany).
Tumor Xenograft Assay
The 6-week-old female nude mice were obtained from the SLRC Laboratory Animal Center (Shanghai, China) and fed in a specific-pathogen-free condition for further experiments. Animals were randomly assigned to four groups. 3 × 106 HEY cells and 5 × 106 SKOV3 cells stably transfected with pLenti-miR-1251 or pLenti (resuspended in 100 μL PBS) were inoculated subcutaneously into the right flank of each mouse. Tumor size was measured with calipers every week, and the following formula was used to calculate tumor volumes: volume = length × width2/2. Mice injected with HEY cells and their derivatives were sacrificed 6 weeks post-injection, whereas those injected with SKOV3 cells and their derivatives were sacrificed 7 weeks post-injection, and the xenograft tumors were excised and weighed. The experimental protocols were approved by the rules of the Institutional Animal Care and Use Committee of Fudan University Shanghai Cancer Center.
Immunohistochemistry Assay
After being fixed in 10% formalin, primary tumor tissues were embedded in paraffin. Samples were cut into sections at 4-μm thickness. The sections were deparaffinized and dehydrated through xylene and graded alcohols, and then they were rehydrated with demineralized water. After being placed in boiled antigen retrieval buffer for 5 min, tissue sections were incubated in 3% H2O2 solution for 25 min at room temperature in the dark. Next the sections were treated with primary antibodies against TBCC (PA5-21710, 1:200; Thermo Fisher Scientific, Boston, MA, USA), Ki67 (GB13030-2, 1:200; Servicebio, Wuhan, China), and LC3B (A7198, 1:200; Abclonal, Wuhan, China), followed by application of goat anti-rabbit HRP-conjugated antibodies, staining with a 3, 3′-diaminobenzidine reaction solution, and imaging using a digitalized microscope camera.
Statistical Analysis
SPSS version (v.)19.0 software was used for statistical analysis, and GraphPad Prism 5 software was used to make graph presentations. Flow cytometry data were analyzed using FlowJo 7.6.2, and immunofluorescence images were edited using the ZEN software. Data were expressed as the mean ± SD and analyzed using t tests in two-group comparisons and two-way ANOVA for comparisons of three groups or more, with statistical significance set as p < 0.05.
Author Contributions
Conception and Design, Y.S., Z.M., and G.Y.; Development of Methodology, Y.S., Z.M., and G.Y.; Analysis and Interpretation of Data (e.g., statistical analysis, biostatistics, and computational analysis), Y.S., X.L., and J.M.; Writing, Review, and/or Revision of the Manuscript, Y.S., Z.M., and G.Y.; Administrative, Technical, or Material Support (i.e., reporting or organizing data and constructing databases), Y.S., X.L., J.M., and X.Z.
Conflicts of Interest
The authors declare no competing interests.
Acknowledgments
This study was supported by grants from the National KEY R&D Program of China (2016YFC1303100) and the National Natural Science Foundation of China (81572553, 81772789, and 81372797 for G.Y.). The authors thank Dr. Yanli Li for comments over the course of this work.
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.ymthe.2019.06.005.
Contributor Information
Zhongliang Ma, Email: zlma@shu.edu.cn.
Gong Yang, Email: yanggong@fudan.edu.cn.
Supplemental Information
References
- 1.Siegel R.L., Miller K.D., Jemal A. Cancer statistics, 2018. CA Cancer J. Clin. 2018;68:7–30. doi: 10.3322/caac.21442. [DOI] [PubMed] [Google Scholar]
- 2.Bax H.J., Josephs D.H., Pellizzari G., Spicer J.F., Montes A., Karagiannis S.N. Therapeutic targets and new directions for antibodies developed for ovarian cancer. MAbs. 2016;8:1437–1455. doi: 10.1080/19420862.2016.1219005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Marchetti C., Muzii L., Romito A., Benedetti Panici P. First-line treatment of women with advanced ovarian cancer: focus on bevacizumab. OncoTargets Ther. 2019;12:1095–1103. doi: 10.2147/OTT.S155425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Armbruster S., Coleman R.L., Rauh-Hain J.A. Management and Treatment of Recurrent Epithelial Ovarian Cancer. Hematol. Oncol. Clin. North Am. 2018;32:965–982. doi: 10.1016/j.hoc.2018.07.005. [DOI] [PubMed] [Google Scholar]
- 5.Tomao F., Marchetti C., Romito A., Di Pinto A., Di Donato V., Capri O., Palaia I., Monti M., Muzii L., Benedetti Panici P. Overcoming platinum resistance in ovarian cancer treatment: from clinical practice to emerging chemical therapies. Expert Opin. Pharmacother. 2017;18:1443–1455. doi: 10.1080/14656566.2017.1328055. [DOI] [PubMed] [Google Scholar]
- 6.Mak C.S., Yung M.M., Hui L.M., Leung L.L., Liang R., Chen K., Liu S.S., Qin Y., Leung T.H., Lee K.F. MicroRNA-141 enhances anoikis resistance in metastatic progression of ovarian cancer through targeting KLF12/Sp1/survivin axis. Mol. Cancer. 2017;16:11. doi: 10.1186/s12943-017-0582-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chaluvally-Raghavan P., Jeong K.J., Pradeep S., Silva A.M., Yu S., Liu W., Moss T., Rodriguez-Aguayo C., Zhang D., Ram P. Direct Upregulation of STAT3 by MicroRNA-551b-3p Deregulates Growth and Metastasis of Ovarian Cancer. Cell Rep. 2016;15:1493–1504. doi: 10.1016/j.celrep.2016.04.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bibi F., Naseer M.I., Alvi S.A., Yasir M., Jiman-Fatani A.A., Sawan A., Abuzenadah A.M., Al-Qahtani M.H., Azhar E.I. microRNA analysis of gastric cancer patients from Saudi Arabian population. BMC Genomics. 2016;17(Suppl 9):751. doi: 10.1186/s12864-016-3090-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liang B., Zhao J., Wang X. A three-microRNA signature as a diagnostic and prognostic marker in clear cell renal cancer: An In Silico analysis. PLoS ONE. 2017;12:e0180660. doi: 10.1371/journal.pone.0180660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jordan M.A., Wilson L. Microtubules and actin filaments: dynamic targets for cancer chemotherapy. Curr. Opin. Cell Biol. 1998;10:123–130. doi: 10.1016/s0955-0674(98)80095-1. [DOI] [PubMed] [Google Scholar]
- 11.Nithianantham S., Le S., Seto E., Jia W., Leary J., Corbett K.D., Moore J.K., Al-Bassam J. Tubulin cofactors and Arl2 are cage-like chaperones that regulate the soluble αβ-tubulin pool for microtubule dynamics. eLife. 2015;4:e08811. doi: 10.7554/eLife.08811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tian G., Cowan N.J. Tubulin-specific chaperones: components of a molecular machine that assembles the α/β heterodimer. Methods Cell Biol. 2013;115:155–171. doi: 10.1016/B978-0-12-407757-7.00011-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang P., Ma X., Song E., Chen W., Pang H., Ni D., Gao Y., Fan Y., Ding Q., Zhang Y., Zhang X. Tubulin cofactor A functions as a novel positive regulator of ccRCC progression, invasion and metastasis. Int. J. Cancer. 2013;133:2801–2811. doi: 10.1002/ijc.28306. [DOI] [PubMed] [Google Scholar]
- 14.Pode-Shakked B., Barash H., Ziv L., Gripp K.W., Flex E., Barel O., Carvalho K.S., Scavina M., Chillemi G., Niceta M. Microcephaly, intractable seizures and developmental delay caused by biallelic variants in TBCD: further delineation of a new chaperone-mediated tubulinopathy. Clin. Genet. 2017;91:725–738. doi: 10.1111/cge.12914. [DOI] [PubMed] [Google Scholar]
- 15.Geeraert C., Ratier A., Pfisterer S.G., Perdiz D., Cantaloube I., Rouault A., Pattingre S., Proikas-Cezanne T., Codogno P., Poüs C. Starvation-induced hyperacetylation of tubulin is required for the stimulation of autophagy by nutrient deprivation. J. Biol. Chem. 2010;285:24184–24194. doi: 10.1074/jbc.M109.091553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chang W.T., Liu W., Chiu Y.H., Chen B.H., Chuang S.C., Chen Y.C., Hsu Y.T., Lu M.J., Chiou S.J., Chou C.K., Chiu C.C. A 4-Phenoxyphenol Derivative Exerts Inhibitory Effects on Human Hepatocellular Carcinoma Cells through Regulating Autophagy and Apoptosis Accompanied by Downregulating α-Tubulin Expression. Molecules. 2017;22:E854. doi: 10.3390/molecules22050854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hage-Sleiman R., Herveau S., Matera E.L., Laurier J.F., Dumontet C. Silencing of tubulin binding cofactor C modifies microtubule dynamics and cell cycle distribution and enhances sensitivity to gemcitabine in breast cancer cells. Mol. Cancer Ther. 2011;10:303–312. doi: 10.1158/1535-7163.MCT-10-0568. [DOI] [PubMed] [Google Scholar]
- 18.Hage-Sleiman R., Herveau S., Matera E.L., Laurier J.F., Dumontet C. Tubulin binding cofactor C (TBCC) suppresses tumor growth and enhances chemosensitivity in human breast cancer cells. BMC Cancer. 2010;10:135. doi: 10.1186/1471-2407-10-135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Garcia-Mayoral M.F., Castaño R., Fanarraga M.L., Zabala J.C., Rico M., Bruix M. The solution structure of the N-terminal domain of human tubulin binding cofactor C reveals a platform for tubulin interaction. PLoS ONE. 2011;6:e25912. doi: 10.1371/journal.pone.0025912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chung Y., Lee J., Jung S., Lee Y., Cho J.W., Oh Y.J. Dysregulated autophagy contributes to caspase-dependent neuronal apoptosis. Cell Death Dis. 2018;9:1189. doi: 10.1038/s41419-018-1229-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Robin M., Issa A.R., Santos C.C., Napoletano F., Petitgas C., Chatelain G., Ruby M., Walter L., Birman S., Domingos P.M. Drosophila p53 integrates the antagonism between autophagy and apoptosis in response to stress. Autophagy. 2019;15:771–784. doi: 10.1080/15548627.2018.1558001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang X.B., Wang H., Long H.Q., Li D.Y., Zheng X. LINC00641 regulates autophagy and intervertebral disc degeneration by acting as a competitive endogenous RNA of miR-153-3p under nutrition deprivation stress. J. Cell. Physiol. 2019;234:7115–7127. doi: 10.1002/jcp.27466. [DOI] [PubMed] [Google Scholar]
- 23.Poillet-Perez L., Xie X., Zhan L., Yang Y., Sharp D.W., Hu Z.S., Su X., Maganti A., Jiang C., Lu W. Autophagy maintains tumour growth through circulating arginine. Nature. 2018;563:569–573. doi: 10.1038/s41586-018-0697-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhao P., Zhang B.L., Liu K., Qin B., Li Z.H. Overexpression of miR-638 attenuated the effects of hypoxia/reoxygenation treatment on cell viability, cell apoptosis and autophagy by targeting ATG5 in the human cardiomyocytes. Eur. Rev. Med. Pharmacol. Sci. 2018;22:8462–8471. doi: 10.26355/eurrev_201812_16546. [DOI] [PubMed] [Google Scholar]
- 25.Feng Y., Kang H.H., Wong P.M., Gao M., Wang P., Jiang X. Unc-51-like kinase (ULK) complex-independent autophagy induced by hypoxia. Protein Cell. 2019;10:376–381. doi: 10.1007/s13238-018-0584-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yang Z., Sun Q., Guo J., Wang S., Song G., Liu W., Liu M., Tang H. GRSF1-mediated MIR-G-1 promotes malignant behavior and nuclear autophagy by directly upregulating TMED5 and LMNB1 in cervical cancer cells. Autophagy. 2019;15:668–685. doi: 10.1080/15548627.2018.1539590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pyo K.E., Kim C.R., Lee M., Kim J.S., Kim K.I., Baek S.H. ULK1 O-GlcNAcylation Is Crucial for Activating VPS34 via ATG14L during Autophagy Initiation. Cell Rep. 2018;25:2878–2890.e4. doi: 10.1016/j.celrep.2018.11.042. [DOI] [PubMed] [Google Scholar]
- 28.Zhang J., Chu D., Kawamura T., Tanaka K., He S. GRIM-19 repressed hypoxia-induced invasion and EMT of colorectal cancer by repressing autophagy through inactivation of STAT3/HIF-1α signaling axis. J. Cell. Physiol. 2019;234:12800–12808. doi: 10.1002/jcp.27914. [DOI] [PubMed] [Google Scholar]
- 29.Weng J., Xiao J., Mi Y., Fang X., Sun Y., Li S., Qin Z., Li X., Liu T., Zhao S. PCDHGA9 acts as a tumor suppressor to induce tumor cell apoptosis and autophagy and inhibit the EMT process in human gastric cancer. Cell Death Dis. 2018;9:27. doi: 10.1038/s41419-017-0189-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Aga T., Endo K., Tsuji A., Aga M., Moriyama-Kita M., Ueno T., Nakanishi Y., Hatano M., Kondo S., Sugimoto H. Inhibition of autophagy by chloroquine makes chemotherapy in nasopharyngeal carcinoma more efficient. Auris Nasus Larynx. 2019;46:443–450. doi: 10.1016/j.anl.2018.10.013. [DOI] [PubMed] [Google Scholar]
- 31.Wang S., Wu J., Ren J., Vlantis A.C., Li M.Y., Liu S.Y.W., Ng E.K.W., Chan A.B.W., Luo D.C., Liu Z. MicroRNA-125b Interacts with Foxp3 to Induce Autophagy in Thyroid Cancer. Mol. Ther. 2018;26:2295–2303. doi: 10.1016/j.ymthe.2018.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shao Y., Song X., Jiang W., Chen Y., Ning Z., Gu W., Jiang J. MicroRNA-621 Acts as a Tumor Radiosensitizer by Directly Targeting SETDB1 in Hepatocellular Carcinoma. Mol. Ther. 2019;27:355–364. doi: 10.1016/j.ymthe.2018.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Offei E.B., Yang X., Brand-Saberi B. The role of autophagy in morphogenesis and stem cell maintenance. Histochem. Cell Biol. 2018;150:721–732. doi: 10.1007/s00418-018-1751-0. [DOI] [PubMed] [Google Scholar]
- 34.Smith A.G., Macleod K.F. Autophagy, cancer stem cells and drug resistance. J. Pathol. 2019;247:708–718. doi: 10.1002/path.5222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang S.F., Wang X.Y., Fu Z.Q., Peng Q.H., Zhang J.Y., Ye F., Fu Y.F., Zhou C.Y., Lu W.G., Cheng X.D., Xie X. TXNDC17 promotes paclitaxel resistance via inducing autophagy in ovarian cancer. Autophagy. 2015;11:225–238. doi: 10.1080/15548627.2014.998931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hu J., Meng Y., Zhang Z., Yan Q., Jiang X., Lv Z., Hu L. MARCH5 RNA promotes autophagy, migration, and invasion of ovarian cancer cells. Autophagy. 2017;13:333–344. doi: 10.1080/15548627.2016.1256520. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.