Abstract
This study aimed to investigate the intestinal microbiota in duodenal ulcer (DU) patients, effects of proton pump inhibitors,clarithromycin and amoxicillin, PCA) for Helicobacter pylori (H. pylori) and Bacillus subtilis and Enterococcus faecium (BSEF) on intestinal microbiota. DU patients were randomly assigned to receive either PCA (group TT) or PCA plus BSEF(group TP). The fecal microbiome was conducted using high throughput 16S rDNA gene and internal transcribed spacer sequencings. The diversity and abundance of intestinal bacteria in the DU were significantly lower than health check control (HC) group. In the TT group, the abundance and diversity of both intestinal bacteria and fungi decreased after PCA treatment, compared with those before treatment, whereas in the TP group no obvious changes were observed. In the TT group at all the time points, both the intestinal bacteria and fungi were different from those in the HC group. However, in the TP group, at 10w the bacterial flora abundance was close to that in the HC group. The results indicate that anti- H. pylori treatment induced significant decrease in the diversity of intestinal microbiota, while the combined therapy supplemented with BSEF could protect and restore the intestinal microbiota.
Subject terms: Gastroenteritis, Duodenal ulcers
Introduction
Guidelines about H. pylori eradication have stated that H. pylori infection should be eradicated in duodenal ulcer (DU) patients by the standard triple therapy1. Of the DU patients who received standard anti-H. pylori treatment, 10% to 30% develop antibiotic-associated diarrhea (AAD) and stool changes2. The reasons may relate to intestinal microbiota disorders caused by the use of anti-bacterial drugs3,4. Recent studies have reported that eradication of H. pylori caused perturbation of the gut microbiome and may indirectly affect the health of human5. However, the intestinal microbiota in DU patients is unknown. The effect on intestinal fungi caused by anti- H. pylori therapy, as well as a combined therapy with probiotic treatment in DU patients remains unclear. We hypothesized that (1) the intestinal microbiota in DU is different from that in healthy people; (2) H. pylori standard treatment disrupts intestinal microbiota; and (3) combined Bacillus subtilis and Enterococcus faecium(BSEF) treatment may improve the rate of H. pylori eradication, reduce side effects, and protect intestinal microbiota.
Methods
Study population
Healthy adults aged 18–65 years old were first screened to assess study eligibility. The exclusion criteria for the study were diabetes, hyper- or hypothyroidism, prior gastric or bariatric surgery, prior documented treatment for H. pylori, antibiotic or probiotics use within 4 weeks of enrollment, use of steroids or other immunomodulating drugs within 4 weeks of enrollment, recent vaccination and Charlson weighed comorbidity index <2. Written informed consent was obtained from qualified volunteers prior to study participation. Authors had access to the study data and reviewed and approved the final manuscript. The study protocol was reviewed and approved by the Medical Ethics Committee at the Chinese PLA General Hospital (PLAGH) (date of first registration:1/4/2013,registration number:S2013-051-01) and the Chinese Clinical Trial Registry (registration number:ChiCTR-TRC-13003228) on 19April 2013. We confirm that all methods were performed in accordance with the relevant guideline.
Each participant was asked to fill in 2 side-effect questionnaires to record the side effects according to severity: mild, moderate or severe6,7. The patients were also asked to record the improvement of symptoms and the status of ulcer healing under endoscopy before and after treatment8.
Sample collection
The active DU patients were diagnosed in the Chinese PLA General Hospital (PLAGH) from 2013 to 2016. Urea breathing test (UBT) was performed in all patients. Biopsy specimens were taken from the antrum and corpus for a rapid urease test (RUT) and histology. H. pylori was considered present if two of the three were positive. When taking a gastroscope, take 1–2 ml of gastric juice, immerse the Acilit® pH test strip into the measured gastric juice, and read the pH of the gastric juice.
After the patients met the eligibility criteria and provided informed consent, we subsequently randomized patients using concealed allocation based on a list of random numbers, which was computer-generated. Forty H. pylori infected patients with active DU were randomly divided into a triple treatment group (TT, n = 20) and a triple therapy plus probiotic treatment group (TP, n = 20). In addition, 20 normal healthy persons were included as health check controls (HC, n = 20). Healthy persons meas no digestive diseases and other systemic diseases, and have not taken any drugs in the last month. Patients in the TT group received 2 weeks of standard triple therapy (20 mg esomeprazole, 500 mg clarithromycin, 1000 mg amoxicillin). Patients in the TP group received PCA followed by 6 weeks of the probiotics Bacillus Subtilis and coated capsules BSEF 500 mg 3 times daily. Follow-up endoscopy was conducted for all patients after stopping PCA for 4 weeks to check ulcer healing and H. pylori status. The volunteers were subsequently followed up at 2, 4, and 8 weeks post-H. pylori eradication. Stool samples (group TT fecal samples, TF; group TP fecal samples, PF) were collected pre-therapy and during each visit, and were frozen immediately at −80 °C until DNA extraction. TF1, TF2 and TF3 represent fecal samples of TT group for 2, 4 and 6 weeks of anti-Hp treatment, respectively. PF1, PF2 and PF3 represent fecal samples of TP group for 2, 4 and 6 weeks of anti-Hp treatment, respectively.
Gene amplification and sequencing
The gut microbial genomic DNA was extracted from stool samples using a stool DNA extraction kit (QIAamp DNA Stool Mini Kit; cat. no. 51504). The primers of the bacterial 16S rDNA were as follows: The upstream primer was 5′-GTGCCAGCMGCCGCGGTAA-3′, and the downstream primer was 5′-GGACTACHVGGGTWTCTAAT-3′. The V4 region of stool bacterial 16S rDNA and the ITS region of fungi were amplified with a high-fidelity enzyme. The ITS1 region primer is ITS1-5F–ITS2; the ITS2 region primer is: ITS2-3F–ITS2- 4R. The library was constructed using New England Biolabs’ NEB Next® UltraTM DNA Library Prep Kit for Illumina library. Sequencing was completed using a Paired-End (PE) approach on the Illumina MiSeq high-throughput sequencing platform.
Bioinformatics analysis
Quality control of the reads was performed by the QIIME software package to pick the high-quality reads that met the requirement9,10. Sequences with similarity greater than 97% were picked up using the operation command of the QIIME quality controller and were clustered into an operational taxonomic unit (OTU)11. The Ribosomal Database Project (RDP) software package was used for the homology alignment, as well as the species and genera taxonomic identification of the longest 16S rDNA sequence fragment of OTUs12. The database referred to the Greenbank at http://greengenes.lbl.gov/cgi-bin/nph-index.cgi. The resulting UniFrac distance matrices were used to perform Principal Coordinate Analysis (PCoA) to determine the similarity between groups of samples/time-points.
Statistical methods
The raw data of the taxonomy summary results was exported to SPSS software version 20.0 (SPSS Inc., Chicago, IL) for statistical analysis. The mean abundance in percentage (%) and the 95% confidence interval (95% CI) for the phyla of stool microbiome at different time-points were calculated. Parametric paired-samples t-test was performed to compare the genera of the stool microbiome. Pearson’s Correlation Coefficient was also calculated to investigate across different time points; a two-tailed P-value of < 0.05 was considered significant.
Compliance with ethical standards
The study protocol was reviewed and approved by the Medical Ethics Committee at the Chinese PLA General Hospital (PLAGH) (date of first registration:1/4/2013,registration numberS2013-051-01) and the Chinese Clinical Trial Registry (date of first registration:19/4/2013,registration number:ChiCTR-TRC-13003228).
Results
Clinical information
There was no significant difference in gender, age, and BMI among the 3 groups (P < 0.05), suggesting the comparability of the groups (Table 1).
Table 1.
Basic information of the subjects.
| Basic information | TT group | TP group | HC group | P-value |
|---|---|---|---|---|
| Number of cases | 20 | 20 | 20 | |
| Gender | ||||
| Male | 13 | 12 | 12 | 0.9319 |
| Female | 7 | 8 | 8 | |
| Ages | 45.5 ± 8.4 | 45.2 ± 11.6 | 42.9 ± 12.3 | 0.2386 |
| BMI | 24.1 ± 2.6 | 24.4 ± 2.7 | 24.5 ± 3.0 | 0.8112 |
H. pylori eradication rate
Gastroscopy revealed that the ulcer healing rate of the TT group (90%) was lower than that of the TP group (100%), but there was no statistically significant difference (P = 0.4682).
A total of 16 cases (80%) in the TT group were successfully eradicated, and 17 cases (85%) were successfully eradicated in the TP group. No difference in the success rate of the H. pylori eradication was observed between the 2 groups (P > 0.05).
Gastric pH value
The gastric pH values of DU patients in the pre-TT and the pre-TP groups were significantly different from that in the HC group (P < 0.01), while the TT and the TP groups after treatment (post-TT and post-TP groups) showed no difference with the HC group (P = 0.274). Significant differences were observed between the pre-TT and the post-TT group (P < 0.01), as well as between the pre-TP and post-TP groups (P < 0.01) (Table 2 and Fig. 1).
Table 2.
pH value of gastric juice before and at 4w after treatment (Mean ± SD).
| Group | Number of cases | pH value Before treatment | pH value After treatment | P-value |
|---|---|---|---|---|
| TT Group | 20 | 0.720 ± 0.330 | 1.395 ± 0.613 | 0.0001* |
| TP Group | 20 | 0.720 ± 0.321 | 1.555 ± 0.610 | 0.0000* |
| HC Group | 20 | 1.265 ± 0.456 |
Figure 1.
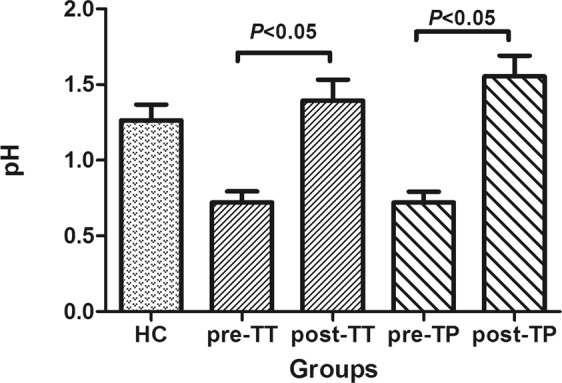
Gastric pH value before and after treatment.
Side effects
There were more side effects in the TT group than in the TP group (35% vs 20%). The most common side effects were diarrhea (35%), followed by nausea (30%), abdominal pain (25%), abnormal taste (20%), abdominal bloating (15%), anorexia (10%), and constipation (5%). These side effects mostly occurred during treatment, and were predominantly mild to moderate, whereas all the cases of the TP group were mild.
Fecal microbiome in DU patients
The average Raw PE number obtained for each bacterial sample was 65630 and fungi sample was 54549. The dilution curve was drawn by 3 methods, including the OTU numbers, the Chao1 and the Shannon 3 for mutual validation, and the results suggested a trend of reaching gentleness at 10,000 sequences.
Bacterial microbiome
OTU analysis: Statistical analysis showed that the OTU number was lower in the DU group compared with that in the HC group, either at the 97% OTU species-level (HC group 505.33 ± 133.63 vs DU group 355.07 ± 104.11 P = 0.0003) or at the 95% OTU genus level (HC group 412.17 ± 114.75 vs DU group 271.28 ± 89.73, P = 0.0001). Differences between the 2 groups were statistically significant (Fig. 2).
Figure 2.
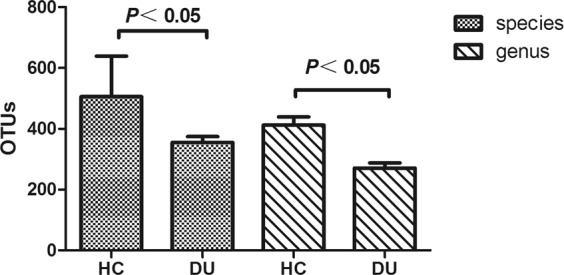
Comparison of OTUs from the 2 groups at the species and genus levels.
Annotation of species and analysis of differential bacteria: The statistical analysis revealed that the bacterial flora community was derived from 38 bacterial phyla, 110 bacterial classes, 157 bacterial orders, 230 bacterial families, and 275 bacterial genera.
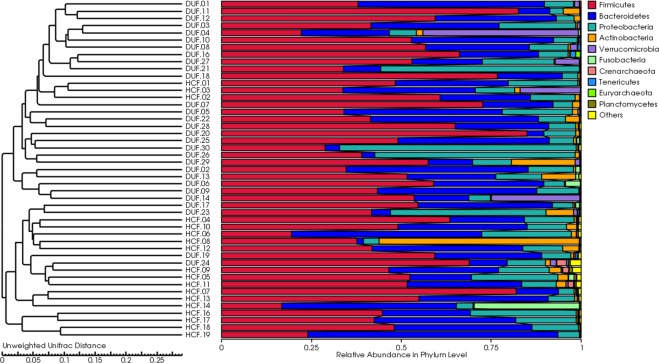
At the phylum level, the abundance distribution showed that in the HC group, the dominant phyla were Firmicutes, Bacteroidetes, and Proteobacteria. These 3 bacterial phyla were abundant in every sample with high percentages, accounting for 45.5%, 33.0%, and 13.2%, respectively, and together accounting for 91.7% of the total bacteria. In the DU group, the dominant phyla were also Firmicutes, Bacteroidetes, and Proteobacteria, accounting for 52.5%, 25.7%, and 15.7%, respectively, and together accounting for 93.9% of the total bacteria. Although the percentages of the 3 bacterial phyla have changed, the differences between the DU group and the HC group were not statistically different. Comparing the other 35 bacterial phyla between the DU group and the HC group revealed a total of 7 bacterial phyla with statistically significant differences: Actinobacteria, Gemmatimonadetes, Nitrospirae, Chlorobi, Thermi, WS3, and Caldithrix. The percentages of these 7 bacterial phyla were all lower in the DU group compared with those in the HC group.
At the genus level, the dominant genera in the HC group were Bacteroides (22.8%), Faecalibacterium (13.6%), Prevotella (5.3%), Roseburia (5.1%), Ruminococcus (4.5%), Escherichia (4.2%), Bifidobacterium (4.2%), Lachnospira (2.3%), etc.; the dominant genera in the DU group were Bacteroides (17.1%), Faecalibacterium (15.3%), Escherichia (7.8%), Ruminococcus (5.5%), Roseburia (4.7%), Prevotella (4.6%), Akkermansia (2.9%), Parabacteroides (2.4%), Bifidobacterium (2.0%), etc. There were a total of 55 differential bacteria at the genus level, 54 of which were less in the DU group than those in the HC group, except the Natronomonas genus that increased in the DU group.
Analysis of species composition: The PCoA analysis found that the intra-group distances between each sample within the HC group and the DU group were smaller than the inter-group distances, suggesting the similarity of the species compositions (Fig. 3).
Figure 3.
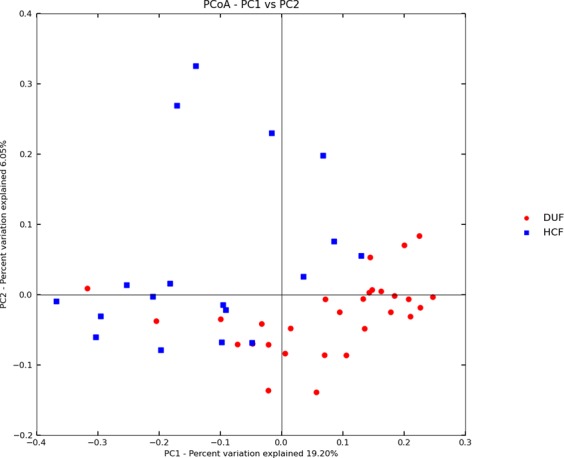
PCoA analysis (blue denotes the HC group and red denotes the DU group).
The UPGMA (Unweighted Pair Group Method with Arithmetic Mean) method was used to input the unweighted UniFrac clustering matrix, which performed cluster analysis on samples. In the cluster diagram (Fig. 4), we found that the bacterial flora structure among all the samples exhibited both clustering and various degrees of overlapping. However, only a few samples could be clustered between the DU group and the HC group, with a general trend of high similarity between samples within groups. Therefore, the DU group or the HC group was considered as a homologous group, and samples from such groups exhibited internal clustering.
Figure 4.
Unweighted UniFra distance clustering tree of samples.
Fungi microbiome
Fungi microbiome
OTU analysis: The OTU number was lower in the DU group compared with that in the HC group, either at the 97% OTU species level (HC group 60.70 ± 14.46 vs DU group 48.65 ± 10.26P = 0.0180) or at the 95% OTU genus level (HC group 51.20 ± 15.49 vs DU group 38.77 ± 9.13P = 0.0141). Differences between the 2 groups were statistically significant (Fig. 5).
Figure 5.
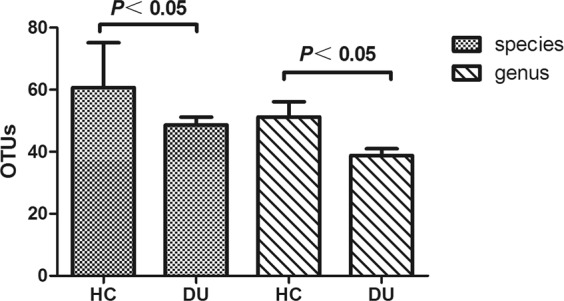
Comparison of OTUs of the 2 groups of at the species and genus level.
Annotation of species and analysis of differential fungi: According to the annotation results of species, most of the fungal communities came from 2 fungal phyla, 31 fungal classes, 53 fungal orders, 90 fungal families, and 104 fungal genera.
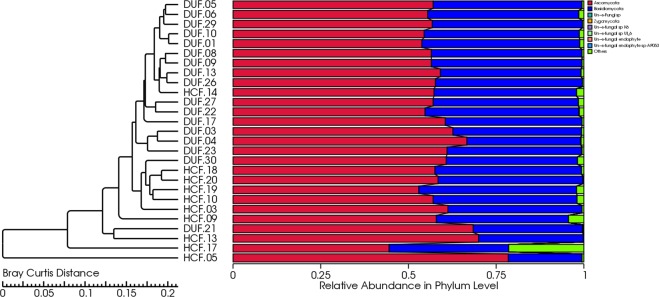
At the phylum level, the abundance distribution showed that the abundance of Ascomycota was 59.6% in the HC group and 58.8% in the DU group; the abundance of Basidiomycota was 37.0% in the HC group and 40.3% in the DU group. There was no difference in the fungal phyla between the 2 groups.
At the genus level, the abundance distribution showed that in the HC group the dominant genera with the fungal flora abundance of more than 1% were Sarcinomyces (49.2%), Malassezia (33.1%), Alternaria (5.9%), Pseudozyma (2.4%), Simplicillium (1.9%), and Aspergillus (1.5%); in the DU group, the dominant genera with the fungal flora abundance of more than 1% were Sarcinomyces (53.9%), Malassezia (35.8%), Pseudozyma (3.6%), Simplicillium (2.2%), and Aspergillus (1.0%). There were 2 differential fungi at the genus level: Pseudozyma significantly increased in DU (3.6%) compared with that in HC (2.4%) (P = 0.01499), and Candida significantly decreased in DU (0.001%) compared with that in HC (0.024%) (P = 0.02113).
Analysis of species composition: PCoA analysis was performed, and the results are shown in Fig. 6. It can be seen that the majority of the samples from the DU groups were clustered in the lower left quadrant, whereas samples from the HC group tended to locate in the upper part of the quadrant. These results suggest that the fungal flora compositions between the DU group and the HC group are not exactly the same, while the internal fungal flora structures within groups are more similar.
Figure 6.
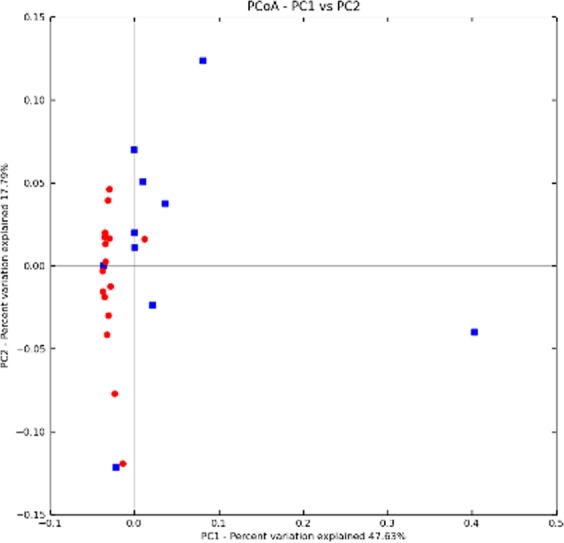
PCoA analysis (blue represents the HC group, and red represents the DU group).
Next, samples were subjected to cluster analysis (Fig. 7). In the clustering diagram, we found that the fungal flora structure among all the samples exhibited both clustering and various degrees of overlapping. However, only a few samples could be clustered between the DU group and the HC group. The general trend was that samples from the DU group or the HC group were first clustered, suggesting that either the DU group or the HC group had a high intra-group similarity.
Figure 7.
Clustering tree.
Changes of intestinal stool microbiome after anti-H. pylori treatment
Bacterial microbiome
OTU analysis: Statistical analysis of OTUs at the bacterial species level was carried out for 4 different time points in the TT group and the TP group (Table 3 and Fig. 8); 0 week (w) represents the DU group before treatment (i.e., with ulcers). Intra-group comparisons were first performed within both groups, and we found that, except in the TT group where differences were observed between DU and TF1 (P = 0.03159) and between DU and TF2 (P = 0.04526), there was no statistical difference in the rest of the pairwise comparisons. In other words, at the end of the anti-H. pylori treatment and at 6w after treatment, the intestinal bacterial flora abundance in the TT group decreased compared with that in the untreated group, whereas the TP group showed no significant changes compared with the untreated group.
Table 3.
OTUs at different time points in the two groups.
| TT Group | TP Group | |
|---|---|---|
| 0w | 347.41 ± 82.61 | 335.66 ± 85.43 |
| 2w | 302.40 ± 48.11* | 342.67 ± 68.58# |
| 6w | 310.07 ± 34.14* | 346.07 ± 63.33# |
| 10w | 316.40 ± 48.02 | 400.00 ± 141.34# |
(*denotes P < 0.05 compared with 0w, and # denotes P < 0.05 when comparing the 2 groups at the same time points).
Figure 8.
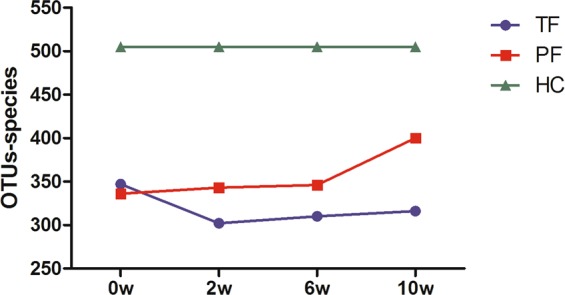
OTUs at different time points before and after treatment.
When the 2 groups were compared with the HC group before and after treatment at different time points, respectively, most of the compared groups showed statistical differences, except between the TP group and the HC group (Table 4).
Table 4.
P-values from the inter-group comparisons of the OTUs at different time points.
| Group | HC | PF1 | PF2 | PF3 |
|---|---|---|---|---|
| HC | — | 0.00068** | 0.00043** | 0.11850 |
| TF1 | 0.00000** | 0.02077* | — | — |
| TF2 | 0.00000** | — | 0.01052* | — |
| TF3 | 0.00001** | — | — | 0.03916* |
(*denotes P < 0.05, ** denotes P < 0.01).
Annotation of species and analysis of differential bacteria:
The intestinal bacterial changes between groups at the same time points. At the end of the 2w treatment, there was no significant change in intestinal bacterial flora at the phylum level between the 2 groups, whereas at the genus level, a total of 23 differential bacterial genera (P < 0.05) were discovered. After 6w of treatment, there were 28 differential bacterial genera, and at 10w, there were 11. Details of the differential bacterial genera are listed in the following table (Table 5).
The intestinal bacterial changes within groups at different time points. In the TT group, among the top ten bacterial phyla at the phylum level, pairwise comparisons within the group showed that differences were only observed in the Bacteroidetes phylum and the Tenericutes phylum: the relative abundance of the Bacteroidetes phylum increased in TF2 compared with that in DU (P = 0.03664), and the relative abundance of the Tenericutes phylum decreased in TF1 compared with that in DU (P = 0.01807). There was no statistical difference in changes of different bacterial phyla at the remaining time points (Fig. 9).
Table 5.
Differential bacterial genera between the 2 groups at different time points.
| 2w | 6w | 10w | |
|---|---|---|---|
| The bacterial genera higher in the TP group than the TT group | Actinomadura | Actinomadura | Achromobacter |
| Atopobium | Anaerofilum | Actinomyces | |
| Brevundimonas | Candidatus Nitrososphaera | Bradyrhizobium | |
| Butyrivibrio | Candidatus Solibacter | Candidatus Solibacter | |
| Coprococcus | Coprococcus | Coprococcus | |
| Coraliomargarita | Coraliomargarita | Chelativorans | |
| Corynebacterium | Dechloromonas | Cetobacterium | |
| Desulfobacca | Desulfomonile | Cupriavidus | |
| Desulfobulbus | Desulfobulbus | ||
| Eggerthella | Dok59 | ||
| Faecalibacterium | Dorea | Thiobacillus | |
| GOUTA19 | Leuconostoc | Leuconostoc | |
| Helicobacter | Luteimonas | ||
| Lewinella | Lewinella | ||
| Luteolibacter | |||
| Neisseria | |||
| Nitrosopumilus | |||
| Oscillospira | Oscillospira | ||
| Rhodobacter | Parapedobacter | ||
| Rhodoplanes | Planctomyces | ||
| Roseburia | Thauera | ||
| Roseomonas | Syntrophobacter | ||
| Rubrivivax | Syntrophomonas | ||
| Thauera | T78 | ||
| Thermomonas | |||
| Thiobacillus | Thiobacillus | ||
| The bacterial genera lower in the TP group than the TT group | Dialister | Dialister | Dialister |
| Plesiomonas |
Figure 9.
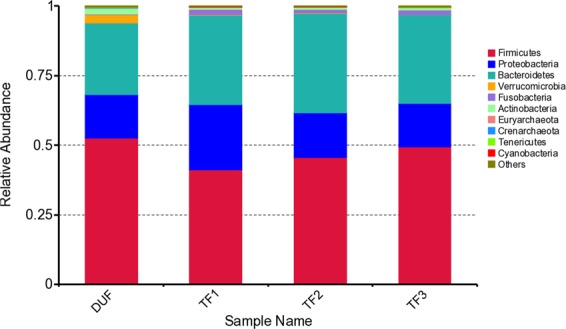
Abundance changes of bacterial phyla in the TT group before and after treatment.
In the TP group, among the top 10 bacterial phyla at phylum level, pairwise comparisons within the group showed that differences were only observed in the Firmicutes phylum, the relative abundance of which increased in PF2 compared with that in PF1 (P = 0.01356). There was no statistical difference in changes of different bacterial phyla at the remaining time points (Fig. 10).
Figure 10.
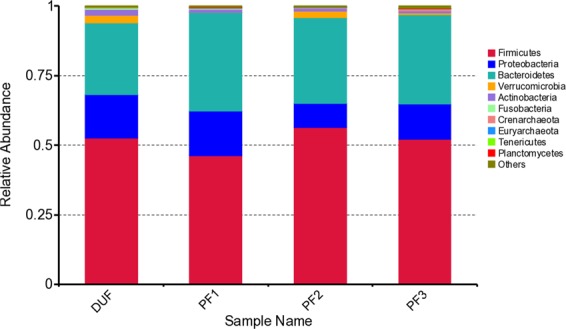
Abundance changes of bacterial phyla in the TP group before and after treatment.
At the genus level, among the 4 different time points in the TT group, pairwise comparisons within the group revealed a total of 8 significantly different (P < 0.05) bacterial genera, the bacterial flora abundance of which was more than 0.1% of the total samples.
At the genus level, among the 4 different time points in the TP group, pairwise comparisons within the group revealed a total of 5 significantly different (P < 0.05) bacterial genera, the bacterial flora abundance of which was more than 0.1% of the total samples. Such differential bacterial genera were fewer in the TP group than that in the TT group, and their abundance percentage was very low. These results suggest that the changes of bacteria at different time points in the TP group were slight, and were from only a small number of bacteria. These bacterial changes were stable, and exhibited a trend of gradual transition. In the TT group, however, the bacterial changes were larger, came from a larger number of bacteria, and presented more intense amplitudes.
Fungi microbiome
OTU analysis: The results revealed that at the 3 time points after TT treatment, the number of OTUs of the intestinal fungal flora decreased compared with that in DU, and was statistically different (P < 0.05); before and after TP treatment, however, there was no obvious change in the number of OTUs. There were differences between the 2 treatment approaches at the same time points (Table 6 and Fig. 11).
Table 6.
OTUs at different time points in the two groups.
| TT Group | TP Group | |
|---|---|---|
| 0w | 46.12 ± 9.44 | 51.00 ± 10.38 |
| 2w | 37.71 ± 8.03* | 55.62 ± 6.69# |
| 6w | 35.30 ± 5.50* | 58.14 ± 6.20# |
| 10w | 37.62 ± 6.02* | 55.25 ± 10.27# |
(*denotes P < 0.05 compared with 0w, and # denotes P < 0.05 when comparing the 2 groups at the same time points).
Figure 11.
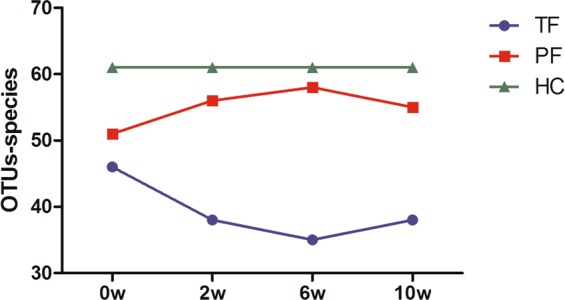
OTUs at different time points before and after treatment.
When the 2 groups were compared with the HC group before and after treatment at different time points, respectively (Table 7), it could be seen that there was no statistical difference among the 3 time points after TP treatment and the HC group, whereas there were statistically significant differences among the 3 time points in the TT group and the HC group.
Table 7.
P values from the inter-group comparisons of the OTUs at different time points.
| Group | HC | PF1 | PF2 | PF3 |
|---|---|---|---|---|
| HC | 0.28750 | 0.40350 | 0.2505 | |
| TF1 | 0.00442** | 0.00021** | ||
| TF2 | 0.00060** | 0.00043** | ||
| TF3 | 0.00318** | 0.01979* |
(*denotes P < 0.05, ** denotes P < 0.01).
Annotation of species and analysis of differential fungi:
The intestinal fungal changes between groups at the same time points. At the phylum level, the 2 treatment approaches showed no obvious difference in the intestinal fungal flora at any of the 3 time points after treatment. At the genus level, some of the fungi were significantly different (P < 0.05): at the end of 2w treatment, a total of 5 differential fungal genera were observed. Details of the differential fungal genera are listed in the following table (Table 8).
Intestinal fungal changes within groups at different time points At the phylum level, there was no difference among the 4 time points either in the TP group or in the TT group. At the genus level, among the 4 time points in the TT group, pairwise comparisons within the group revealed that only the Pseudozyma genus and the Cladosporium genus showed significant differences (P < 0.05), and the trends of the changes at the 4 time points are shown in the figure below; among the 4 time points in the TP group, pairwise comparisons within the group revealed a total of 6 differential fungal genera (P < 0.05), more than those in the TT group. Moreover, the relative abundance of the differential fungal genera was higher than that in the TT group, suggesting that the fungal changes at different time points in the TT group were slight and came from only a small number of fungi, whereas the fungal changes in the TP group were larger and came from a larger number of fungi.
Table 8.
Differential fungal genera between the two groups at different time points.
| 2w | 6w | 10w | |
|---|---|---|---|
| The fungal genera higher in the TP group than the TT group | Aspergillus | Trichoderma | Aureobasidium |
| The fungal genera lower in the TP group than the TT group | Cladosporium | Cladosporium | Cladosporium |
| Pseudozyma | Pseudozyma | Pseudozyma | |
| Simplicillium | Simplicillium | Simplicillium | |
| Sarcinomyces | Sarcinomyces | ||
| Fusarium | Fusarium | ||
| Candida |
Discussion
Eradication of H. pylori is essential for the treatment of the H. pylori-associated DU2. PCA therapy for H. pylori eradication has been in clinical use for a long time. Due to the increased rates of drug resistance and side effects, most studies have shown that the addition of probiotics reduces side effects, but whether these probiotics additives improve the eradication rate is still controversial6,13–16. Chen et al.17 found that Lactobacillus delbrueckii and Lactobacillus plantarum-18 additives had significant anti-H. pylori effects. The anti-H. pylori effects of the 2 strains mainly related to the pH value of the cells’ supernatant and their metabolites.
However, some researchers have questioned the methods that combine anti-H. pylori therapy with probiotics additives18. But the efficacy of the therapy can be increased if acid-resistant and bile salts-resistant BSEF preparations are added. Park et al.19 have shown that the therapy combining 2 live bacteria-BSEF improved eradication rates and reduced side effects. Results from our study showed that the combination therapy with BSEF did not significantly improve eradication rates but reduced the incidence of side effects.
The application of PPIs in triple therapy increased the pH value of gastric juice, resulting in a low acidic environment of the stomach. The low acidic environment weakens the bacteria killing effect of the therapy, and may change intestinal microecology20. Our study found that the pH value of the gastric juice in DU patients (0.72) was significantly lower than that in normal people (1.27). The pH value returned to normal after anti-H. pylori treatment. We have investigated, for the first time, the intestinal bacterial and fungal flora in DU, and found that the diversity and abundance of intestinal bacteria and fungi were significantly lower than those in healthy people.
The presence of human intestinal microbiota is vital for the health of the host. These microorganisms play a unique role in human nutrition, growth, metabolism, resistance to pathogens, immune regulation, and the like21–23. However, the use of antibiotics may break the steady state of the interaction between the microorganisms and the host24,25. Meanwhile, the impact of PPIs on intestinal microbiota has also been of concern26,27. PPIs weaken the role of gastric acid in eliminating exogenous bacteria, leading to easy invasion and colonization of exogenous pathogenic bacteria of the human body. A number of studies have confirmed that when receiving the combined application of PPIs and antibiotics for H. pylori infection, 10% to 30% of the patients experienced changes in defecation frequency and stool characteristics28–32. The reasons for this may relate to intestinal microbiota disturbance. This was also confirmed in our study, which has revealed that the incidence of diarrhea was as high as 35% during triple therapy.
Oh et al.28 showed that probiotic supplementation can reduce the antibiotic-induced alteration and imbalance of the intestinal microbiota composition. Despite the changes in intestinal microbiota, this study suggested that the impact of triple therapy on the intestinal microbiota was short-term about 1 month. Our study found that anti-H. pylori treatment led to an imbalance of intestinal homeostasis and the reduction of diversity, especially during the triple only treatment. After combination therapy with probiotics additives, changes of intestinal microbiota were more moderate: intestinal bacterial diversity at the 10th week was not significantly different from that in healthy people, while intestinal fungal diversity has always remained similar to that in the HC group.
Studies based on traditional microscopy have revealed a high incidence of fungal infection in patients with gastric ulcer, gastric cancer and gastric remnants33,34. So far, there is no report on sequencing results of intestinal microbiota in DU patients. Our study applied a high-throughput sequencing method, for the first time, to sequence intestinal fungal colonies in DU patients. The annotation of species found that the fungal communities were from 2 fungal phyla, 31 fungal classes, 53 fungal orders, 90 fungal families, and 104 fungal genera. In the HC group, the dominant genera were different with the DU group. The probiotic drugs used in our study contain 2 important strains of bacteria that promote the health of human intestines. In addition to the contributions from the bacteria themselves, these strains also promote the growth of other bacteria, especially thick-walled bacteria, such as Bifidobacterium. In the probiotic preparation we used, E. faecium is a gram-positive and facultative anaerobic bacterium. This bacterium reproduces rapidly, and has a strong inhibitory effect on pathogenic bacteria; B. subtilis is a gram-positive and aerobic bacterium, which consumes intestinal oxygen upon its intestinal colonization, resulting in biological oxygen deprivation. Such biological oxygen deprivation leads to a reduction in local oxygen concentration, creating an appropriate environment for the growth of normal intestinal anaerobic microbiota. This promotes the growth of Bifidobacterium, Lactobacillus, enterococci, and other beneficial bacteria, and inhibits the growth of Escherichia coli, dysentery bacili, Staphylococcus aureus, and other pathogenic bacteria, thereby restoring the balance of intestinal microbiota33. Studies have also found that B. subtilis produces a variety of digestive enzymes, further breaking down protein, fat, carbohydrates, and fibrin, etc. B. subtilis also produces lysozyme and more than 80 types of antibacterial compounds, thereby inhibiting the colonization and growth of pathogenic bacteria34. Moreover, B. subtilis inhibits the activity of all the H. pylori microbeads by secreting amicoumacin A and non-amicoumacin antibiotics. This inhibitory effect is not subject to acid restriction and is resistant to proteolytic cleavage. Based on the aforementioned mechanisms, BSEF formulations can be used against the intestinal microbiota imbalance caused by triple therapy.
Acknowledgements
This study was supported by the National High-Tech 863 Program of China (2015AA020701). The authors would like to thank this Program for its assistance and support. We would like to thank Chen Wei for her support in translation of paper.
Author Contributions
L.L.W., Z.K.W. and Y.S.Y. Study concept and design; Z.S.L. and B.Y. acquisition of samples; L.L.W., H.K. and Z.K.W. acquisition of data, analysis of data and interpretation of data; L.L.W. drafting of the manuscript; G.S., L.H.P. and Y.S.Y. critical revision of the manuscript for important intellectual content; Y.S.Y. obtained funding. All the authors contributed to manuscript revision.
Data Availability
The datasets generated and/or analysed during the current study are available from the corresponding author and first author on resonable request.
Competing Interests
The authors declare no competing interests.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Malfertheiner P, et al. Management of Helicobacter pylori infection–the Maastricht IV/ Florence Consensus Report. Gut. 2012;61:646–664. doi: 10.1136/gutjnl-2012-302084. [DOI] [PubMed] [Google Scholar]
- 2.Asaka M, et al. Follow-up survey of a large-scale multicenter, double-blind study of triple therapy with lansoprazole, amoxicillin, and clarithromycin for eradication of Helicobacter pylori in Japanese peptic ulcer patients. J. Gastroenterol. 2003;38:339–347. doi: 10.1007/s005350300061. [DOI] [PubMed] [Google Scholar]
- 3.Vesper BJ, et al. The effect of proton pump inhibitors on the human microbiota. Curr Drug Metab. 2009;10:84–89. doi: 10.2174/138920009787048392. [DOI] [PubMed] [Google Scholar]
- 4.Bartlett JG. Clinical practice. Antibiotic-associated diarrhea. N Engl J Med. 2002;346:334–339. doi: 10.1056/NEJMcp011603. [DOI] [PubMed] [Google Scholar]
- 5.Yap TW, et al. Helicobacter pylori eradication causes perturbation of the human gut microbiome in young adults. PLoS One. 2016;11:e0151893. doi: 10.1371/journal.pone.0151893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cindoruk M, Erkan G, Karakan T, Dursun A, Unal S. Efficacy and safety of Saccharomyces boulardii in the 14-day triple anti-Helicobacter pylori therapy: a prospective randomized placebo-controlled double-blind study. Helicobacter. 2007;12:309–316. doi: 10.1111/j.1523-5378.2007.00516.x. [DOI] [PubMed] [Google Scholar]
- 7.Armuzzi A, et al. The effect of oral administration of Lactobacillus GG on antibiotic-associated gastrointestinal side-effects during Helicobacter pylori eradication therapy. Aliment Pharmacol Ther. 2001;15:163–169. doi: 10.1046/j.1365-2036.2001.00923.x. [DOI] [PubMed] [Google Scholar]
- 8.De Luca L, et al. Measuring dyspepsia: a new severity index validated in Bologna. Dig Liver Dis. 2004;36:806–810. doi: 10.1016/j.dld.2004.07.010. [DOI] [PubMed] [Google Scholar]
- 9.Wang Q, Garrity GM, Tiedje JM, Cole JR. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol. 2007;73:5261–5267. doi: 10.1128/AEM.00062-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Caporaso JG, et al. Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proc Natl Acad Sci USA. 2011;108(Suppl 1):4516–4522. doi: 10.1073/pnas.1000080107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Edgar RC. UPARSE: highly accurate OTU sequences from microbial amplicon reads. Nat Methods. 2013;10:996–998. doi: 10.1038/nmeth.2604. [DOI] [PubMed] [Google Scholar]
- 12.Caporaso JG, et al. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010;7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhou L, et al. A comparative study of sequential therapy and standard riple therapy for Helicobacter pylori Infection: A Randomized Multicenter Trial. Am J Gastroenterol. 2014;109:535–541. doi: 10.1038/ajg.2014.26. [DOI] [PubMed] [Google Scholar]
- 14.Duman DG, et al. Efficacy and safety of Saccharomyces boulardii in prevention of antibiotic-associated diarrhoea due to Helicobacter pylori eradication. Eur J Gastroenterol Hepatol. 2005;17:1357–1361. doi: 10.1097/00042737-200512000-00015. [DOI] [PubMed] [Google Scholar]
- 15.Wang KY, et al. Effects of ingesting Lactobacillus- and Bifidobacterium-containing yogurt in subjects with colonized Helicobacter pylori. Am J Clin Nutr. 2004;80:737–741. doi: 10.1093/ajcn/80.3.649. [DOI] [PubMed] [Google Scholar]
- 16.Kim MN, et al. The effects of probiotics on PPI-triple therapy for Helicobacter pylori eradication. Helicobacter. 2008;13:261–268. doi: 10.1111/j.1523-5378.2008.00601.x. [DOI] [PubMed] [Google Scholar]
- 17.Chen X, et al. Antagonistic activities of lactobacilli against Helicobacter pylori growth and infection in human gastric epithelial cells. J Food Sci. 2012;77:M9–14. doi: 10.1111/j.1750-3841.2011.02498.x. [DOI] [PubMed] [Google Scholar]
- 18.Dai C, Zheng CQ, Jiang M. Efficacy of probiotics in Helicobacter pylori eradication therapy. J Clin Gastroenterol. 2013;47:814–815. doi: 10.1097/MCG.0b013e318298a32c. [DOI] [PubMed] [Google Scholar]
- 19.Park SK, et al. The effect of probiotics on Helicobacter pylori eradication. Hepatogastroenterology. 2007;54:2032–2036. [PubMed] [Google Scholar]
- 20.Guerre J, Vedel G, Gaudric M, Paul G, Cornuau J. Bacterial flora in gastric juice taken at endoscopy in 93 normal subjects. Pathol Biol (Paris) 1986;34:57–60. [PubMed] [Google Scholar]
- 21.Perez-Cobas AE, et al. Gut microbiota disturbance during antibiotic therapy: a multi-omic approach. Gut. 2013;62:1591–1601. doi: 10.1136/gutjnl-2012-303184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lynch SV, Pedersen O. The human intestinal microbiome in health and disease. N Engl J Med. 2016;375:2369–2379. doi: 10.1056/NEJMra1600266. [DOI] [PubMed] [Google Scholar]
- 23.Kahrstrom CT, Pariente N, Weiss U. Intestinal microbiota in health and disease. Nature. 2016;535:47. doi: 10.1038/535047a. [DOI] [PubMed] [Google Scholar]
- 24.Ianiro G, Tilg H, Gasbarrini A. Antibiotics as deep modulators of gut microbiota: between good and evil. Gut. 2016;16:312297. doi: 10.1136/gutjnl-2016-312297. [DOI] [PubMed] [Google Scholar]
- 25.Blaser MJ. Antibiotic use and its consequences for the normal microbiome. Science. 2016;352:544–545. doi: 10.1126/science.aad9358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jackson MA, et al. Proton pump inhibitors alter the composition of the gut microbiota. Gut. 2016;65:749–756. doi: 10.1136/gutjnl-2015-310861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Imhann F, et al. Proton pump inhibitors affect the gut microbiome. Gut. 2016;65:740–748. doi: 10.1136/gutjnl-2015-310376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Oh B, et al. The effect of probiotics on gut microbiota during the Helicobacter pylori eradication: Randomized Controlled Trial. Helicobacter. 2016;21:165–174. doi: 10.1111/hel.12270. [DOI] [PubMed] [Google Scholar]
- 29.Buhling A, Radun D, Muller WA, Malfertheiner P. Influence of anti-Helicobacter triple-therapy with metronidazole, omeprazole and clarithromycin on intestinal microflora. Aliment Pharmacol Ther. 2001;15:1445–1452. doi: 10.1046/j.1365-2036.2001.01033.x. [DOI] [PubMed] [Google Scholar]
- 30.Adamsson I, Nord CE, Lundquist P, Sjostedt S, Edlund C. Comparative effects of omeprazole, amoxycillin plus metronidazole versus omeprazole, clarithromycin plus metronidazole on the oral, gastric and intestinal microflora in Helicobacter pylori-infected patients. J. Antimicrob Chemother. 1999;44:629–640. doi: 10.1093/jac/44.5.629. [DOI] [PubMed] [Google Scholar]
- 31.Lou JG, Chen J, Huang XL, Zhao ZY. Changes in the intestinal microflora of children with Helicobacter pylori infection and after Helicobacter pylori eradication therapy. Chin Med J (Engl) 2007;120:929–931. doi: 10.1097/00029330-200705020-00014. [DOI] [PubMed] [Google Scholar]
- 32.Sheu BS, et al. Impact of supplement with Lactobacillus- and Bifidobacterium-containing yogurt on triple therapy for Helicobacter pylori eradication. Aliment Pharmacol Ther. 2002;16:1669–1675. doi: 10.1046/j.1365-2036.2002.01335.x. [DOI] [PubMed] [Google Scholar]
- 33.Tompkins TA, Hagen KE, Wallace TD, Fillion-Forte V. Safety evaluation of two bacterial strains used in Asian probiotic products. Can J Microbiol. 2008;54:391–400. doi: 10.1139/W08-022. [DOI] [PubMed] [Google Scholar]
- 34.Pinchuk IV, et al. In vitro anti-Helicobacter pylori activity of the probiotic strain Bacillus subtilis 3 is due to secretion of antibiotics. Antimicrob Agents Chemother. 2001;45:3156–3161. doi: 10.1128/AAC.45.11.3156-3161.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets generated and/or analysed during the current study are available from the corresponding author and first author on resonable request.