Abstract
Rothmund-Thomson syndrome (RTS) is an autosomal-recessive disorder characterized by poikiloderma, sparse hair, short stature, and skeletal anomalies. Type 2 RTS, which is defined by the presence of bi-allelic mutations in RECQL4, is characterized by increased cancer susceptibility and skeletal anomalies, whereas the genetic basis of RTS type 1, which is associated with juvenile cataracts, is unknown. We studied ten individuals, from seven families, who had RTS type 1 and identified a deep intronic splicing mutation of the ANAPC1 gene, a component of the anaphase-promoting complex/cyclosome (APC/C), in all affected individuals, either in the homozygous state or in trans with another mutation. Fibroblast studies showed that the intronic mutation causes the activation of a 95 bp pseudoexon, leading to mRNAs with premature termination codons and nonsense-mediated decay, decreased ANAPC1 protein levels, and prolongation of interphase. Interestingly, mice that were heterozygous for a knockout mutation have an increased incidence of cataracts. Our results demonstrate that deficiency in the APC/C is a cause of RTS type 1 and suggest a possible link between the APC/C and RECQL4 helicase because both proteins are involved in DNA repair and replication.
Keywords: Rothmund-Thomson syndrome, cataracts, anaphase-promoting complex, ANAPC1, RECQL4, pseudoexon, alternative splicing, poikiloderma, cryptic splice site, splicing variant
Main Text
Analysis of the clinical and molecular features of individuals with Rothmund-Thomson syndrome (RTS [MIM: 268400]), including assessing the prevalence of osteosarcoma and the mutational status of the RECQL4 gene (MIM: 603780), resulted in the definition of two distinct forms of RTS: RTS type 2, which is characterized by poikiloderma (hyper- and hypo-pigmentation, telangiectasias, and atrophy), skeletal anomalies, sparse hair, and abnormal nails; increased susceptibility to cancer, particularly osteosarcoma; and bi-allelic mutations in RECQL4; and RTS type 1, which displays the same core clinical features, including poikiloderma, sparse hair, and nail abnormalities, but is also associated with bilateral juvenile cataracts, the distinct absence of osteosarcoma, and a lack of RECQL4 mutations.1, 2 Mutations in RECQL4 have also been described in individuals with RAPADILINO (MIM: 266280) and Baller-Gerold (MIM: 268400) syndromes, both of which share clinical features, including radial anomalies, short stature, and increased risk of cancer, with RTS.3 Thus far, more than 300 individuals with RTS have been reported, and the majority of these have RECQL4 mutations and are classified as RTS type 2.1 However, the etiology of RTS type 1 remains undefined.
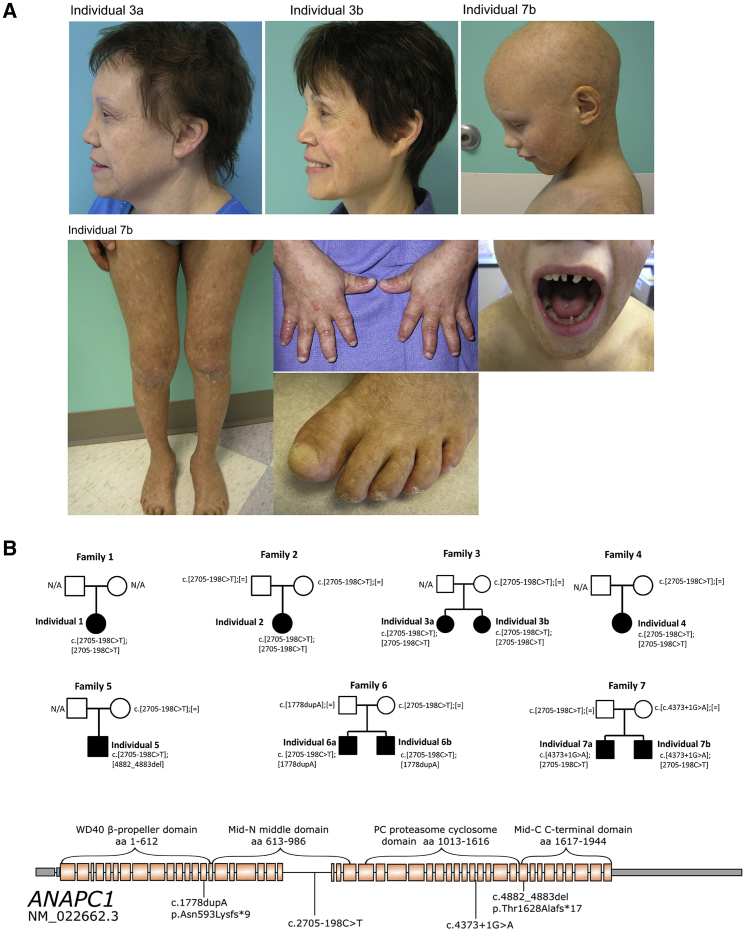
To identify the genetic cause of RTS type 1, we recruited a cohort of ten individuals, from seven families, who had a firm clinical diagnosis of RTS type 1. Informed consent was obtained from all participants, who were enrolled in a study approved by the institutional review board at Baylor College of Medicine. Families 1–3 were of Amish ancestry. All individuals presented with classical RTS type 1 features, including poikiloderma, abnormal hair and nails, bilateral juvenile cataracts, and an absence of RECQL4 mutations (see Table 1 and Figure 1A for photos and Figure 1B for pedigrees). Additional features in our cohort included undescended testes, growth retardation, and multiple skeletal and dental abnormalities (see Figure S1).
Table 1.
Clinical Characteristics of RTS Type 1-Affected Individuals with ANAPC1 Mutations
| Individuals Homozygous for the Intron 22 Mutation | Individuals Compound Heterozygous for the Intron 22 Mutation and Another Mutation | Individuals WithoutANAPC1 Mutations | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Individual | 1 | 2 | 3A | 3B | 4 | 5 | 6A | 6B | 7A | 7B | 8A | 8B | 9 | |
| Gender | f | f | f | f | f | m | m | m | m | m | m | m | f | |
| Eyes | Bilateral juvenile cataracts | + | + | + | + | + | + | + | + | + | + | − | − | − |
| Ectoderm | Poikiloderma | + | + | + | + | + | + | + | + | + | + | + | + | + |
| Sparse or absent hair, eyebrows, or eyelashes | − | + | + | + | + | + | + | + | + | + | + | + | − | |
| Abnormal teeth | − | − | − | − | + | − | + | + | + | + | − | − | − | |
| Abnormal nails | − | − | − | − | − | − | + | + | + | + | − | − | − | |
| Other systems | Endocrine and fertility | − | POF | − | POF | − | Hypogonadism | HypoT4 | HypoT4 | − | − | − | − | − |
| Undescended testes | NA | NA | NA | NA | NA | + | + | + | + | + | − | − | − | |
| Short stature | − | − | − | − | + | + | + | + | + | + | + | + | − | |
| Growth hormone therapy | − | − | − | − | − | + | + | + | + | + | − | − | − | |
| Skeletal abnormalities | + | − | − | − | − | − | + | + | + | + | − | − | − | |
| Neurological | − | − | − | − | − | DD | ID | ADHD | − | − | − | − | − | |
Abbreviations are as follows: f = female, m = male; ID = intellectual disability; DD = developmental delay; ADHD, =attention-deficit hyperactivity disorder; HypoT4 = hypothyroidism; NA = not applicable, and POF = premature ovarian failure. See Table S1 for additional details and Figure S1 for radiographs of some of the skeletal changes (sclerotic marks).
Figure 1.
ANAPC1 Mutations in Individuals with Rothmund-Thomson Syndrome Type 2
(A) Photos of three affected individuals show the alopecia and abnormal teeth, nails, and skin.
(B, top) Pedigrees of ten individuals from seven families with RTS type 1 and ANAPC1 mutations. All individuals affected have the ANAPC1 intronic splicing variant (GenBank: NM_022662.3:c.[2705−198C>T]). Individuals 1, 2, 3A, 3B, and 4 are homozygous for the ANAPC1 intronic splicing variant, whereas individuals 5, 6A, 6B, 7A, and 7B have compound-heterozygous mutations.
(B, bottom) Graphical representation of all ANAPC1 variants identified. Introns other than 22 are not drawn to scale. The domains of ANAPC1 are discussed in detail by Li et al. and Chang et al.24, 25 The WD40 domain mediates conformational changes important for stimulating the APC’s catalytic activity upon co-activator binding. The Mid-N and Mid-C domains coalesce together in the APC complex. The PC domain joins the APC platform to the tetratricopeptide repeat lobe of the APC.
Whole-exome sequencing was performed in all affected individuals. Although we found that no gene displayed bi-allelic coding variants in a significant proportion of individuals, we identified a single region of homozygosity larger than 1 Mb on chr2q13-q14.1 in the Amish individuals by using HomozygosityMapper software (hg19 position chr2:112,484,446–118,589,953).4 We then performed real-time qRT-PCR analysis in RTS type 1 and control fibroblasts for the genes in that interval. ANAPC1 (MIM: 608473) expression levels were significantly lower in RTS samples than in controls (Figures 2A and S1B–S1D). A focused search for coding and non-coding mutations of ANAPC1 in the exome data revealed a variant in intron 22 (GenBank: NM_022662.3:c.2705−198C>T) in the ANAPC1 gene, which happened to be covered in the exome. All four Amish individuals (1–3B) and one non- Amish individual (4) were homozygous for this variant. In the other families, we found compound-heterozygous ANAPC1 mutations where, in each case, the intronic mutation was in trans with any of three other mutations predicted to cause loss of function; these mutations included: GenBank: NM_022662.3:c.4882_4883del in individual 5; GenBank: NM_022662.3:c.1778dupA in individuals 6A and 6B; and GenBank: NM_022662.3:c.4373+1G>A in individuals 7A and 7B (Figures 1B and 1C). All variants were confirmed by Sanger sequencing and were found to segregate in an autosomal-recessive manner (n.b. for individual 1; parental samples were unavailable) (Figure 1A). We additionally performed concomitant exome sequencing and then screening for the intronic ANAPC1 mutation in three individuals from two families (individuals 8A and 8B are siblings) who had RECQL4-negative RTS but no juvenile cataracts, and none were found to have ANAPC1 mutations, suggesting that ANAPC1 mutations are specific to individuals with RTS type 1 with juvenile cataracts. We had complete coverage of the ANAPC1 coding sequence, and we searched the exome data for mutations in other helicases, as well as components of the anaphase-promoting complex and pathway, and no mutation was identified.
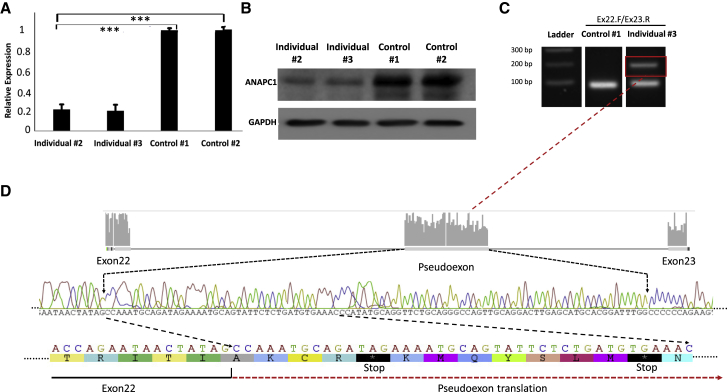
Figure 2.
Effect of the ANAPC1 Splicing Variant on Protein Levels and the Mechanism of Pseudoexon Activation in Fibroblasts Derived from Individuals 1 and 2
(A) Realtime PCR analysis demonstrating significant difference (p < 0.001) in messenger RNA from individuals 1 and 2 compared to unrelated controls.
(B) An immunoblot showing a decrease in ANAPC1 protein levels in fibroblasts from individuals 1 and 2 compared to unrelated controls.
(C) Realtime PCR with primers spanning exons 22 and 23 of ANAPC1 and subsequent agarose gel electrophoresis revealing the presence of an additional fragment in fibroblasts from affected individuals. Sanger sequencing of the additional fragment revealed the presence of a 95 bp nucleotide that is incorporated between exon 22 and 23 of ANAPC1 cDNA. Analysis of the pseudoexon revealed two stop codons leading to the nonsense-mediated decay pathway, accounting for the decrease in RTS type 1 individual transcript (A) and protein (B) levels.
There was a more complex phenotype in those individuals that had compound-heterozygous ANAPC1 mutations compared to those homozygous for the intronic splicing mutation (Table 1). All individuals with compound-heterozygous mutations in ANAPC1 had short stature and received growth hormone therapy (Table 1). This small group of individuals also had more anomalies of the teeth, nail, endocrine, genital, and skeletal systems. With regard to the skeletal anomalies, four individuals with compound-heterozygous mutations presented with one or more bone abnormalities such as osteoporosis, bone fracture, delayed bone age, short metacarpals and phalanges, dysplastic phalanges, sclerotic metaphyseal foci, and genu varum, features which are commonly associated with individuals who have type 2 RTS.5 Whether these findings will be generalizable and truly represent genotype-phenotype correlations is limited by the fact that there were only five individuals (who were all female) homozygous for the intron 22 mutation and five (who were all male) with compound-heterozygous variants.
The ANAPC1 gene encodes the APC1 protein, the largest subunit of the anaphase-promoting complex/cyclosome (APC/C). The finding of decreased ANAPC1 RNA expression was consistent with subsequent immunoblotting analysis that demonstrated decreased APC1 protein levels compared to controls (Figure 2B). To further explore a potential mechanism that could result in a reduction of ANAPC1 transcript and protein levels, we assessed the effect of this c.2705−198C>T intronic mutation on the basis of the in silico prediction that the intron 22 splicing mutation introduces a new splice donor site that could lead to the introduction of a pseudoexon with premature termination codons. To test this hypothesis, we used the protein synthesis inhibitor cycloheximide (CHX), an inhibitor of nonsense-mediated decay (NMD), to assess the potentially degraded mRNA. Indeed, CHX treatment of fibroblasts from RTS-affected individuals and control cells and then real-time PCR of RNA-derived cDNA with primers spanning the intronic splicing mutation revealed the presence of a longer fragment in the RTS type 1 samples but not in the control samples (Figure 2C). Sequencing of this band revealed a 95 nucleotide pseudoexon that is incorporated in the ANAPC1 nascent transcript (Figure 2D). As predicted, the pseudoexon introduces two premature termination codons that activate the NMD pathway, accounting for the decreased mRNA and protein levels observed in RTS type 1 cells.
The APC/C is an E3 ubiquitin ligase that targets specific proteins for degradation and associates with one of two co-activators, CDC20 and CDH1, to control cellular transition at distinct phases of the cell cycle. The APC/C also plays roles in the control of senescence, quiescence, DNA replication and repair, cell differentiation, metabolism, and neuronal function.6 In light of previous findings demonstrating a low proliferation rate and an early entry into senescence by Cdh1-deficient mouse embryonic fibroblasts7 and other studies showing that inhibition of APC-Cdh1 or APC-cdc20 prevents cells from entering the cell cycle,8 we hypothesized that RTS type 1 cells undergo defective cell cycling. Analysis by real-time cell cycle microscopy revealed a significant delay within the interphase for synchronized fibroblast cells derived from individuals with the intronic splicing mutation compared to control fibroblasts (Figure 3A). There was no difference in the time spent in mitosis (Figure S1E). Because APC1 is a major structural component of the APC/C and acts as a scaffold between the catalytic and regulatory subcomplexes,9, 10, 11 we speculate that a reduction in APC1 levels could impair the integrity and function of the APC/C. This, in turn, could prevent cells from entering the cell cycle or reduce the rate at which cell cycle substrates are degraded at the end of mitosis and at the G1-phase,12 resulting in a longer interphase in RTS type 1 cells.
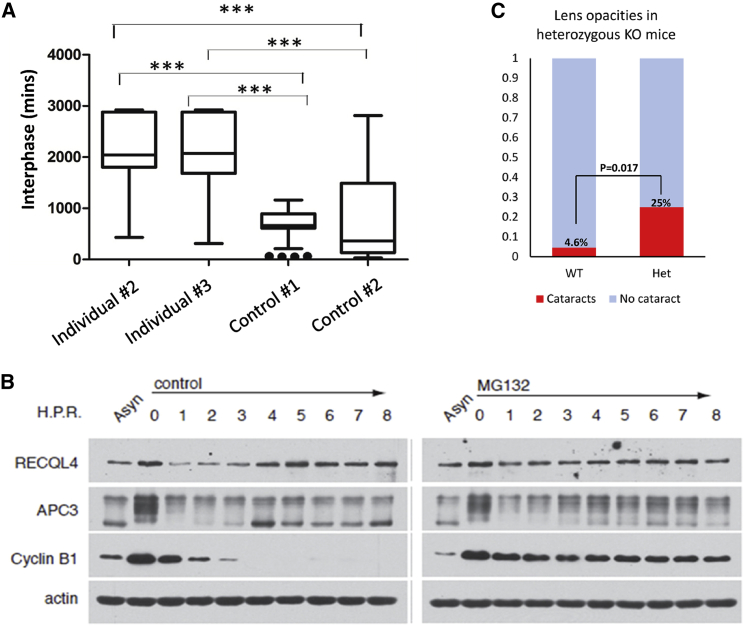
Figure 3.
Effect of ANAPC1 Deficiency and Cell Cycle Variations of RECQL4
(A) Fibroblasts derived from individuals homozygous for the ANAPC1 intronic variant spent a longer time at the interphase of the cell cycle compared to control fibroblasts (∗∗∗ = p < 0.001); the time-lapse microscopy was performed as described previously.26 The middle line represents the median, the box represents quartiles, and the whiskers represent minimum and maximum values within 1.5 times the interquartile range.
(B) Proteins from thymidine-nocodazole-synchronized-MG132-treated and non-treated HeLa cells at specific time points (0 to 8 hours) were analyzed by SDS-PAGE and immunoblot. The upward-shifted (phospho) APC3 represents a marker for mitosis. The cyclin B disappearance represents a positive control for APC/C activity, and actin is used as a loading control. RECQL4 protein levels decreased within the first 3 h of mitosis and became stable after treatment with MG132, similar to classical APC/C substrates such as cyclin B. Asyn signifies asynchronous cells.
(C) Mice heterozygous for a loss-of-function Anapc1 mutation develop cataracts more frequently than wild-type mice (detailed data: no lens opacity in 2,766 WT (wild-type) mice and 12 het (heterozygous KO) mice, lens opacity in one eye in 116 WT mice and 4 het mice, lens opacities in both eyes in 17 WT mice and no het mice).
Given the phenotypic overlap between RTS type 1 (caused by ANAPC1 mutations) and type 2 (caused by RECQL4 mutations), we hypothesize that both types of RTS might share a common pathway. Several lines of evidence suggest potential links between the APC/C and RECQL4. Defects in either the APC/C or RECQL4 lead to chromosomal instability and cell cycle delays.13, 14 The APC/C is known to degrade ctIP (CtBP-interacting protein) in a timely fashion to prevent hyper-resection of double-stranded breaks and increase genomic stability,15, 16 and RECQL4 is known to recruit ctIP to double-stranded breaks.17 Finally, in a recent review, Sommers et al. discuss the importance of helicase degradation during the cell cycle to maintain genome homeostasis.18 Our experiments show that RECQL4 levels, like the levels of most cell cycle substrates targeted by the APC/C, vary at different phases of the cell cycle in HeLa cells; the levels decrease at early mitosis and peak after the degradation of cyclin B1 (Figure 3B). A fluctuation of RECQL4 during the cell cycle has been noted by Kitao et al. in 1998,19 and RECQL4 has up to three potential D-box APC/C destruction motifs.20 Also, like the levels of most proteins targeted by the APC/C, RECQL4 levels stabilize after treatment with the proteasome inhibitor MG132 (Figure 3B).
The APC has been shown to be essential for cell proliferation and differentiation in the lens, in vitro and in zebrafish.21, 22 As part of the main project of the International Mouse Phenotyping Consortium (IMPC),23 a mouse with a knock-out mutation of Anapc1 was generated by the Knockout Mouse Phenotyping Program (KOMP2) at the Jackson Laboratory with CRISPR-Cas9 technology. The alteration results in the deletion of 221 bp, deleting exon 4 and 169 bp of a flanking intronic sequence, including the splice acceptor and donor. This alteration is predicted to cause a change of the amino acid sequence after residue 125 and early truncation four amino acids later. A homozygous mutation resulted in lethality in mice (no homozygous mice were detected at weaning). Interestingly, mice heterozygous for the mutation show an increased incidence of lens opacities when assessed at three months of age (p = 0.017; see Figure 3C for graphical representation of data from the IMPC). The IMPC phenotyping pipeline did not identify other features suggestive of RTS in these mice, but it is important to note that only one allele is mutated in the phenotyped mice.
In summary, we identified ANAPC1 mutations to be a genetic cause of RTS type 1 and demonstrated that pseudoexon activation results in decreased ANAPC1 transcript and protein levels with concomitant delay in the interphase of the cell cycle. Given the marked phenotypic similarity between RTS types 1 and 2, we further highlight a possible link between ANAPC1 and RECQL4, both of which are involved in DNA replication and repair. It is interesting to note that to date, bilateral juvenile cataracts are seen only in individuals with RTS and ANAPC1 mutations and osteosarcoma only in those with RECQL4 mutations. The cause for this association and the detailed interaction between these proteins warrant further investigation.
Declaration of Interests
The authors declare no competing interests.
Acknowledgments
We gratefully acknowledge the patients and families affected by RTS, without whom this work would not be possible. We also thank Ms. Ta-Tara Rideau for research coordination and her contribution to this manuscript. P.M.C. is supported by salary awards from the Canadian Institutes of Health Research (CIHR) and the Fonds de Recherche Québec – Santé. We thank Dr. Jacqui White from the Jackson Laboratories for help obtaining KOMP2 data. L.L.W. was supported in part by the American Society for Clinical Oncology Young Investigator Award, the Eunice Kennedy Shriver National Institute of Child Health and Human Development (HD42136), the Doris Duke Charitable Foundation Clinician Scientist Development Program, the Amschwand Sarcoma Cancer Foundation, the Carousel Young Friends of Texas Children’s Cancer Center, the Kurt Groten Family Research Scholar’s Program, and the Gillson Longenbaugh Foundation. Portions of this work were supported by the National Institutes of Health (NIH) by grants (K12 HD41648–01) to the Baylor College of Medicine Child Health Research Center, (RR000188-42) to the Baylor College of Medicine General Clinical Research Center, and (HD083092) to the Clinical Translational Core and Tissue Culture Core of the Baylor College of Medicine Intellectual and Developmental Disabilities Research Center.
Published: July 11, 2019
Footnotes
Supplemental Data can be found online at https://doi.org/10.1016/j.ajhg.2019.06.011.
Contributor Information
Lisa L. Wang, Email: llwang@bcm.edu.
Philippe M. Campeau, Email: p.campeau@umontreal.ca.
Accession Numbers
Variants have been deposited in ClinVar under submission number SUB5720241.
Web Resources
APC/C Degron Repository, http://slim.ucd.ie/apc/
HomozygosityMapper, http://www.homozygositymapper.org/
International Mouse Phenotyping Consortium, https://www.mousephenotype.org/
OMIM, https://omim.org
Supplemental Data
References
- 1.Larizza L., Roversi G., Volpi L. Rothmund-Thomson syndrome. Orphanet J. Rare Dis. 2010;5:2. doi: 10.1186/1750-1172-5-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wang L.L., Plon S.E. Rothmund-Thomson syndrome. In: Adam M.P., Ardinger H.H., Pagon R.A., Wallace S.E., Bean L.J.H., Stephens K., Amemiya A., editors. In GeneReviews. Seattle; 1993. [Google Scholar]
- 3.Siitonen H.A., Sotkasiira J., Biervliet M., Benmansour A., Capri Y., Cormier-Daire V., Crandall B., Hannula-Jouppi K., Hennekam R., Herzog D. The mutation spectrum in RECQL4 diseases. Eur. J. Hum. Genet. 2009;17:151–158. doi: 10.1038/ejhg.2008.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Seelow D., Schuelke M., Hildebrandt F., Nürnberg P. HomozygosityMapper--an interactive approach to homozygosity mapping. Nucleic Acids Res. 2009;37 doi: 10.1093/nar/gkp369. W593-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mehollin-Ray A.R., Kozinetz C.A., Schlesinger A.E., Guillerman R.P., Wang L.L. Radiographic abnormalities in Rothmund-Thomson syndrome and genotype-phenotype correlation with RECQL4 mutation status. AJR Am. J. Roentgenol. 2008;191 doi: 10.2214/AJR.07.3619. W62-6. [DOI] [PubMed] [Google Scholar]
- 6.Eguren M., Manchado E., Malumbres M. Non-mitotic functions of the Anaphase-Promoting Complex. Semin. Cell Dev. Biol. 2011;22:572–578. doi: 10.1016/j.semcdb.2011.03.010. [DOI] [PubMed] [Google Scholar]
- 7.Li M., Shin Y.H., Hou L., Huang X., Wei Z., Klann E., Zhang P. The adaptor protein of the anaphase promoting complex Cdh1 is essential in maintaining replicative lifespan and in learning and memory. Nat. Cell Biol. 2008;10:1083–1089. doi: 10.1038/ncb1768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cappell S.D., Chung M., Jaimovich A., Spencer S.L., Meyer T. Irreversible APC(Cdh1) inactivation underlies the point of no return for cell-cycle entry. Cell. 2016;166:167–180. doi: 10.1016/j.cell.2016.05.077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Thornton B.R., Ng T.M., Matyskiela M.E., Carroll C.W., Morgan D.O., Toczyski D.P. An architectural map of the anaphase-promoting complex. Genes Dev. 2006;20:449–460. doi: 10.1101/gad.1396906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schwickart M., Havlis J., Habermann B., Bogdanova A., Camasses A., Oelschlaegel T., Shevchenko A., Zachariae W. Swm1/Apc13 is an evolutionarily conserved subunit of the anaphase-promoting complex stabilizing the association of Cdc16 and Cdc27. Mol. Cell. Biol. 2004;24:3562–3576. doi: 10.1128/MCB.24.8.3562-3576.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vodermaier H.C., Gieffers C., Maurer-Stroh S., Eisenhaber F., Peters J.M. TPR subunits of the anaphase-promoting complex mediate binding to the activator protein CDH1. Curr. Biol. 2003;13:1459–1468. doi: 10.1016/s0960-9822(03)00581-5. [DOI] [PubMed] [Google Scholar]
- 12.Manchado E., Eguren M., Malumbres M. The anaphase-promoting complex/cyclosome (APC/C): Cell-cycle-dependent and -independent functions. Biochem. Soc. Trans. 2010;38:65–71. doi: 10.1042/BST0380065. [DOI] [PubMed] [Google Scholar]
- 13.Croteau D.L., Singh D.K., Hoh Ferrarelli L., Lu H., Bohr V.A. RECQL4 in genomic instability and aging. Trends Genet. 2012;28:624–631. doi: 10.1016/j.tig.2012.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Greil C., Krohs J., Schnerch D., Follo M., Felthaus J., Engelhardt M., Wäsch R. The role of APC/C(Cdh1) in replication stress and origin of genomic instability. Oncogene. 2016;35:3062–3070. doi: 10.1038/onc.2015.367. [DOI] [PubMed] [Google Scholar]
- 15.Lafranchi L., Sartori A.A. The ubiquitin ligase APC/C(Cdh1) puts the brakes on DNA-end resection. Mol. Cell. Oncol. 2015;2:e1000696. doi: 10.1080/23723556.2014.1000696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lafranchi L., de Boer H.R., de Vries E.G., Ong S.E., Sartori A.A., van Vugt M.A. APC/C(Cdh1) controls CtIP stability during the cell cycle and in response to DNA damage. EMBO J. 2014;33:2860–2879. doi: 10.15252/embj.201489017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lu H., Shamanna R.A., Keijzers G., Anand R., Rasmussen L.J., Cejka P., Croteau D.L., Bohr V.A. RECQL4 promotes DNA end resection in repair of DNA double-strand breaks. Cell Rep. 2016;16:161–173. doi: 10.1016/j.celrep.2016.05.079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sommers J.A., Suhasini A.N., Brosh R.M., Jr. Protein degradation pathways regulate the functions of helicases in the DNA damage response and maintenance of genomic stability. Biomolecules. 2015;5:590–616. doi: 10.3390/biom5020590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kitao S., Ohsugi I., Ichikawa K., Goto M., Furuichi Y., Shimamoto A. Cloning of two new human helicase genes of the RecQ family: Biological significance of multiple species in higher eukaryotes. Genomics. 1998;54:443–452. doi: 10.1006/geno.1998.5595. [DOI] [PubMed] [Google Scholar]
- 20.Davey N.E., Morgan D.O. Building a regulatory network with short linear sequence motifs: Lessons from the degrons of the anaphase-promoting complex. Mol. Cell. 2016;64:12–23. doi: 10.1016/j.molcel.2016.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wu G., Glickstein S., Liu W., Fujita T., Li W., Yang Q., Duvoisin R., Wan Y. The anaphase-promoting complex coordinates initiation of lens differentiation. Mol. Biol. Cell. 2007;18:1018–1029. doi: 10.1091/mbc.E06-09-0809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Imai F., Yoshizawa A., Fujimori-Tonou N., Kawakami K., Masai I. The ubiquitin proteasome system is required for cell proliferation of the lens epithelium and for differentiation of lens fiber cells in zebrafish. Development. 2010;137:3257–3268. doi: 10.1242/dev.053124. [DOI] [PubMed] [Google Scholar]
- 23.Dickinson M.E., Flenniken A.M., Ji X., Teboul L., Wong M.D., White J.K., Meehan T.F., Weninger W.J., Westerberg H., Adissu H., International Mouse Phenotyping Consortium; Jackson Laboratory; Infrastructure Nationale PHENOMIN, Institut Clinique de la Souris (ICS); Charles River Laboratories; MRC Harwell; Toronto Centre for Phenogenomics; Wellcome Trust Sanger Institute; RIKEN BioResource Center High-throughput discovery of novel developmental phenotypes. Nature. 2016;537:508–514. doi: 10.1038/nature19356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chang L., Zhang Z., Yang J., McLaughlin S.H., Barford D. Atomic structure of the APC/C and its mechanism of protein ubiquitination. Nature. 2015;522:450–454. doi: 10.1038/nature14471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li Q., Chang L., Aibara S., Yang J., Zhang Z., Barford D. WD40 domain of Apc1 is critical for the coactivator-induced allosteric transition that stimulates APC/C catalytic activity. Proc. Natl. Acad. Sci. USA. 2016;113:10547–10552. doi: 10.1073/pnas.1607147113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kucharski T.J., Minshall P.E., Moustafa-Kamal M., Turnell A.S., Teodoro J.G. Reciprocal regulation between 53BP1 and the anaphase-promoting complex/cyclosome is required for genomic stability during mitotic stress. Cell Rep. 2017;18:1982–1995. doi: 10.1016/j.celrep.2017.01.080. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.