Abstract
Neuroblastoma is a cancer of the developing sympathetic nervous system. It is diagnosed in 600–700 children per year in the United States and accounts for 12% of pediatric cancer deaths. Despite recent advances in our understanding of this malignancy’s complex genetic architecture, the contribution of rare germline variants remains undefined. Here, we conducted a genome-wide analysis of large (>500 kb), rare (<1%) germline copy number variants (CNVs) in two independent, multi-ethnic cohorts totaling 5,585 children with neuroblastoma and 23,505 cancer-free control children. We identified a 550-kb deletion on chromosome 16p11.2 significantly enriched in neuroblastoma cases (0.39% of cases and 0.03% of controls; p = 3.34 × 10−9). Notably, this CNV corresponds to a known microdeletion syndrome that affects approximately one in 3,000 children and confers risk for diverse developmental phenotypes including autism spectrum disorder and other neurodevelopmental disorders. The CNV had a substantial impact on neuroblastoma risk, with an odds ratio of 13.9 (95% confidence interval = 5.8–33.4). The association remained significant when we restricted our analysis to individuals of European ancestry in order to mitigate potential confounding by population stratification (0.42% of cases and 0.03% of controls; p = 4.10 × 10−8). We used whole-genome sequencing (WGS) to validate the deletion in paired germline and tumor DNA from 12 cases. Finally, WGS of four parent-child trios revealed that the deletion primarily arose de novo without maternal or paternal bias. This finding expands the clinical phenotypes associated with 16p11.2 microdeletion syndrome to include cancer, and it suggests that disruption of the 16p11.2 region may dysregulate neurodevelopmental pathways that influence both neurological phenotypes and neuroblastoma.
Keywords: neuroblastoma, genetic predisposition, pediatric cancer, 16p11.2 microdeletion, copy number variation, rare variant, chromosomal abnormalities, genome-wide association study, germline, de novo
Main Text
Current knowledge of predisposition to neuroblastoma (MIM: 256700) is incomplete. Approximately 1%–2% of cases occur in the context of familial disease with a dominant mode of inheritance and are largely explained by germline mutations in the anaplastic lymphoma kinase (ALK) gene1 (MIM: 105590) or the paired like homeobox 2B (PHOX2B) gene2, 3 (MIM: 603851). However, the vast majority of neuroblastomas arise sporadically without family history. Genome-wide association studies (GWAS) have identified common variants that confer risk for sporadic neuroblastoma at over a dozen genetic loci,4, 5, 6, 7, 8, 9, 10, 11, 12, 13 including a common copy number variant (CNV) at 1q21.1 (MIM: 613017).14 Notably, several of the genes identified through this approach have been functionally validated as neuroblastoma oncogenes or tumor suppressors that influence both tumor initiation and maintenance.15, 16, 17, 18, 19, 20 However, the common variants identified to date have only low or moderate effects on neuroblastoma risk. Higher-impact variants are expected to be rarer due to purifying selection,21 but the role of rare variants in sporadic neuroblastoma has not been extensively explored. Recent sequencing efforts have reported rare germline single-nucleotide variants (SNVs) and indels which affect cancer-associated genes in children with neuroblastoma,22, 23, 24 but power to detect true disease associations has been limited due to the lack of sufficient sample sizes available. In contrast, large CNVs can be reliably detected using high-density single-nucleotide polymorphism (SNP) arrays, and hence CNV-based studies often do not suffer the same limitations. Rare CNVs have recently been implicated in multiple complex diseases, including cancer.25, 26, 27, 28 Here, we hypothesized that rare germline CNVs contribute substantially to neuroblastoma risk with larger effect sizes than do common variants previously identified through neuroblastoma GWAS.
To identify rare CNVs associated with neuroblastoma, we analyzed a large cohort of 5,585 neuroblastoma cases and 23,505 cancer-free control children SNP-genotyped at the Children’s Hospital of Philadelphia (CHOP). All neuroblastoma specimens were obtained with informed consent at original diagnosis through Children's Oncology Group (COG) member institutions. DNA from peripheral blood lymphocytes or bone marrow was provided for genotyping. Neuroblastoma cases were not selected for tumor stage or other characteristics, so a range of clinical presentations across low-, intermediate-, and high-risk neuroblastoma were represented (Table 1). The control group included cancer-free children recruited after informed consent through the CHOP Health Care Network by the Center for Applied Genomics (CAG) and genotyped together with the neuroblastoma cases on matched genotyping platforms at the CAG. The CHOP Institutional Review Board was responsible for oversight of this study. The diverse case and control datasets included individuals of European, African, East Asian, South Asian, and Hispanic descent (Figure S1).
Table 1.
Summary of Clinical Covariates for Neuroblastoma Cases before and after Quality Control
|
- |
Discovery Cohort |
Replication Cohort |
||
|---|---|---|---|---|
| Clinical Covariate | Before Quality Control (n = 3,688) | After Quality Control (n = 3,309) | Before Quality Control (n = 2,384) | After Quality Control (n = 2,276) |
| Sex | ||||
| female | 1,668 (45.2%) | 1,496 (45.2%) | 1,149 (48.2%) | 1,093 (45.6%) |
| male | 1,967 (53.3%) | 1,777 (53.7%) | 1,232 (51.7%) | 1,180 (49.2%) |
| unavailable | 53 (1.4%) | 36 (1.1%) | 4 (0.1%) | 3 (0.1%) |
| Age at Diagnosis | ||||
| <18 months | 1,705 (46.2%) | 1,553 (46.9%) | 1,030 (43.2%) | 978 (40.8%) |
| ≥18 months | 1,930 (52.3%) | 1,720 (52.0%) | 1,351 (56.7%) | 1,295 (54.0%) |
| unavailable | 53 (1.4%) | 36 (1.1%) | 4 (0.1%) | 3 (0.1%) |
| INSSaStage | ||||
| stage 1 | 645 (17.5%) | 592 (17.9%) | 402 (16.9%) | 391 (16.3%) |
| stage 2 | 514 (13.9%) | 473 (14.3%) | 364 (15.3%) | 339 (14.1%) |
| stage 3 | 581 (15.8%) | 542 (16.4%) | 380 (15.9%) | 362 (15.1%) |
| stage 4 | 1,644 (44.6%) | 1,435 (43.4%) | 1,105 (46.4%) | 1,063 (44.4%) |
| stage 4S | 228 (6.2%) | 210 (6.3%) | 130 (5.5%) | 118 (4.9%) |
| unavailable | 76 (2.1%) | 57 (1.7%) | 4 (0.1%) | 3 (0.1%) |
| Risk | ||||
| low | 1,195 (32.4%) | 1,095 (33.1%) | 682 (28.6%) | 651 (27.2%) |
| intermediate | 753 (20.4%) | 700 (21.2%) | 567 (23.8%) | 537 (22.4%) |
| high | 1,596 (43.3%) | 1,398 (42.2%) | 1,034 (43.4%) | 995 (41.5%) |
| unavailable | 144 (3.9%) | 116 (3.5%) | 102 (4.2%) | 93 (3.9%) |
| MYCN Status | ||||
| amplified | 631 (17.1%) | 551 (16.7%) | 425 (17.8%) | 411 (17.2%) |
| non-amplified | 2,746 (74.5%) | 2,481 (75.0%) | 1,768 (74.2%) | 1,682 (70.2%) |
| unavailable | 315 (8.4%) | 277 (8.4%) | 192 (8.0%) | 183 (7.6%) |
INSS = International Neuroblastoma Staging System.
The initial dataset before quality control included 6,072 neuroblastoma cases and 24,242 cancer-free controls. We divided these samples into discovery and replication cohorts based on SNP array platform: samples genotyped on the Illumina HumanHap550 and Human610-Quad arrays were used for discovery (n = 3,688 cases and 9,229 controls) and samples genotyped more recently on the HumanOmniExpress arrays served as an independent replication cohort (n = 2,384 cases and 15,013 controls, Table S1). We restricted the analysis to high-quality SNPs (see Supplemental Material and Methods in Supplemental Data) shared across all array types within each cohort (402,743 and 506,045 SNPs for discovery and replication, respectively). CNV calls were then generated using a circular binary segmentation algorithm implemented in Nexus Copy Number (BioDiscovery) with adjustment for waves created by GC content variability (Table S2). We removed excessively noisy samples and those with contamination from circulating tumor cells and DNA (see Supplemental Material and Methods in Supplemental Data), resulting in a final discovery cohort of 3,309 cases and 8,855 controls and a replication cohort of 2,276 cases and 14,650 controls (Table S1).
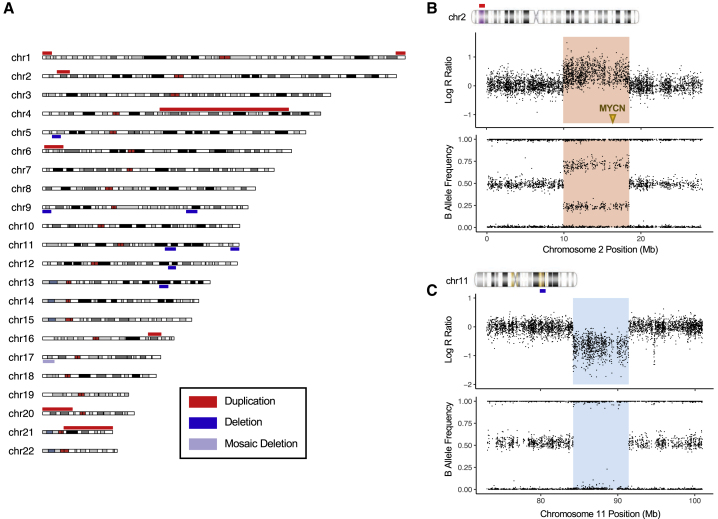
We first examined megabase-scale chromosomal abnormalities in our combined cohort of 5,585 neuroblastoma cases. Chromosomal abnormalities of this size are detectable by conventional cytogenetic analysis as well as by SNP array and comparative genomic hybridization array (array CGH) technologies. Several abnormalities have been identified in children with neuroblastoma and published as case reports and case series over the last 50 years,29 but their overall frequency has not been established. Here, we report that deletions and duplications larger than 5 Mb on autosomal chromosomes were present in 0.27% of individuals in our large unselected neuroblastoma cohort (Figure 1A and Figure S3). Notably, a 2p25.1–p24.2 duplication encompassing the oncogene MYCN (MIM: 164840) and an interstitial 11q14 deletion were observed in neuroblastoma cases but not in 23,505 cancer-free controls (Figures 1B and 1C). Additionally, a terminal deletion on 11q24.3–q25 was observed in one case and one control. MYCN amplification and 11q loss are frequent somatic alterations in neuroblastoma and are predictive of poor outcome.30, 31 Reports of germline 2p gain and 11q deletion are sparse in the literature, but several of the reported cases have presented with neuroblastoma.29, 32, 33, 34 The megabase-scale events reported here passed our quality control (see Supplemental Material and Methods in Supplemental Data) and showed allelic ratios consistent with a germline deletion or duplication. However, we cannot completely rule out the possibility of contamination from circulating tumor DNA, particularly for the 2p25.1–p24.2 gain, because matched tumor data were not available. Finally, trisomy 21 (MIM: 190685) was observed in only one out of 5,585 neuroblastoma cases in this cohort, which is low relative to the general population prevalence of approximately one out of 1,200 individuals.35 This is consistent with previous studies and suggests a decreased incidence of Down Syndrome in neuroblastoma cases.29, 36
Figure 1.
Megabase-Scale Germline Chromosomal Abnormalities Including 2p Gain and 11q Loss Are Observed in 0.27% of Neuroblastoma Cases
(A) Germline deletions and duplications larger than 5 Mb on autosomal chromosomes are plotted in a summary karyotype (karyoploteR74) showing the entire cohort of 5,585 neuroblastoma cases. We observed 16 events affecting 15 individuals (the 1q44 terminal duplication and mosaic 17p13 terminal deletion were present in the same individual).
(B) Log R ratio and B allele frequency plots for the 8.6-Mb germline duplication of 2p25.1-p24.2 (red shading) in a high-risk, MYCN-amplified case. The location of the MYCN oncogene is indicated by an arrow.
(C) Log R ratio and B allele frequency plots for the 7.2-Mb 11q14 interstitial deletion (blue shading) in an intermediate-risk, MYCN-non-amplified case.
To narrow our focus to a small number of rare, potentially high-impact CNVs for further analyses and to limit the interference of artifacts, we considered only CNVs that were longer than 500 kb and that met stringent filtering criteria (Table S2.). As expected, large CNVs were rare. We identified 4,668 high-confidence large CNVs across the entire sample set, which came to 0.160 per individual on average. Large duplications were more common than deletions (0.114 and 0.046 per individual, respectively). The overall burden of large CNVs was similar in neuroblastoma cases and cancer-free controls. We observed an average of 0.154 and 0.157 CNVs per case and control, respectively, in the discovery cohort (p = 0.70) and 0.178 and 0.161 CNVs per case and control in the replication cohort (p = 0.07). Similarly, no significant differences were observed between cases and controls when considering deletions and duplications separately.
We next carried out a regional association study to identify CNVs that were larger than 500 kb and enriched in neuroblastoma in our discovery cohort of 3,309 cases and 8,855 controls. We collapsed CNVs >500 kb in size that were recurrent in three or more cases (approximately 0.1%) into copy number variable regions (CNVRs), requiring at least 100 kb minimum overlap. Consistent with the overall rarity of large CNVs, no region was deleted or duplicated in more than 0.63% of neuroblastoma cases or 0.58% of controls in the discovery cohort. We observed 40 recurrent CNVRs: seven deletions and 33 duplications (Table S3). We tested these recurrent CNVRs for association with neuroblastoma using Fisher’s exact test, applying a Bonferroni-corrected significance threshold (p = 0.00125). With these restrictions, the only CNVR reaching genome-wide significance was a 550 kb single-copy deletion on chromosome 16p11.2 (in 0.36% of cases and 0.02% of controls; p = 8.3 × 10−6; Table 2 and Figure 2A). We validated this association in the independent replication cohort (p = 3.0 × 10−6). Meta-analysis detected no heterogeneity between the discovery and replication cohorts (p = 0.8) and yielded a combined p value of 3.34 × 10−9 and an odds ratio of 13.9 (95% confidence interval: 5.8–33.4). This represents a considerably larger impact on neuroblastoma susceptibility than was seen with common variants previously identified through GWAS, for which odds ratios range from 1.2 to 3.0.37
Table 2.
Association of 16p11.2 Microdeletion with Neuroblastoma
| Cohort | Total Cases | Cases with 16p11.2 Deletion | Total Controls | Controls with 16p11.2 Deletion | Deletion Frequency in Cases | Deletion Frequency in Controls | p Value | Odds Ratio (95% CI)a |
|---|---|---|---|---|---|---|---|---|
| All Subjects | ||||||||
| discovery | 3,309 | 12 | 8,855 | 2 | 0.36% | 0.02% | 8.28x10-6 | 16.1 (3.6–148.2) |
| replication | 2,276 | 10 | 14,650 | 5b | 0.44% | 0.03% | 2.99x10-6 | 12.9 (4.0–48.2) |
| meta-analysis | - | - | - | - | - | - | 3.34x10-9 | 13.9 (5.8–33.4) |
| European Only | ||||||||
| discovery | 2,219 | 7 | 6,236 | 2 | 0.32% | 0.03% | 1.83x10-3 | 9.9 (1.9–97.4) |
| replication | 1,340 | 8 | 12,065 | 4b | 0.60% | 0.03% | 3.35x10-6 | 18.1 (4.8–82.2) |
| meta-analysis | - | - | - | - | - | - | 4.10x10-8 | 14.5 (5.6–37.6) |
CI = Confidence Interval
One control possesses a mosaic 16p11.2 microdeletion.
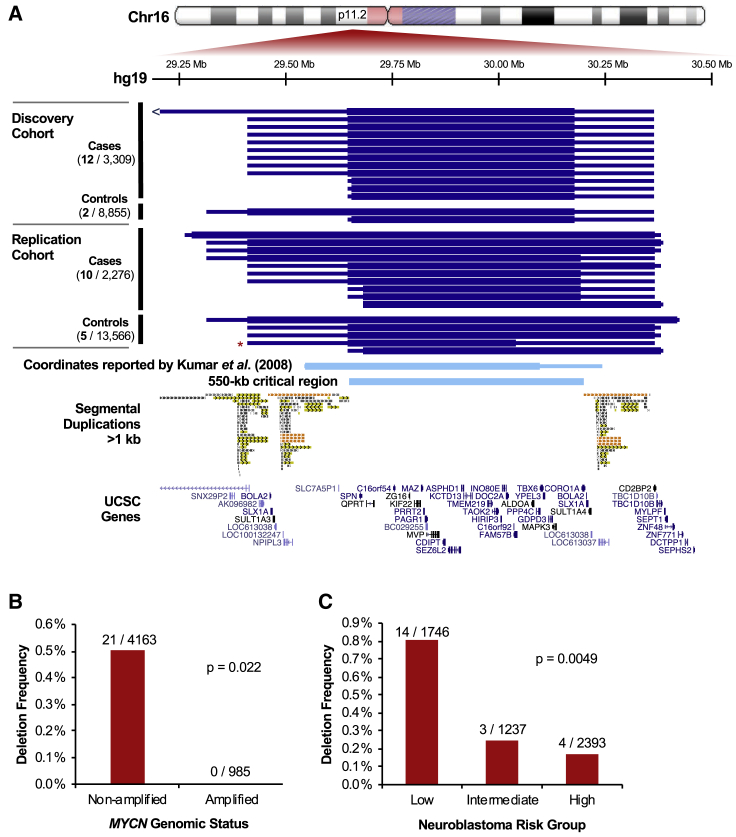
Figure 2.
16p11.2 Microdeletion Associates with Neuroblastoma and Is Enriched in MYCN Non-Amplified, Low-Risk Cases
(A) 16p11.2 microdeletions identified in neuroblastoma cases and cancer-free controls in each analysis cohort (discovery and replication) are plotted in UCSC Genome Browser.75 Minimum and maximum deletion coordinates approximated by SNP array are represented by the thick and thin blue bars, respectively. The red asterisk denotes one mosaic deletion identified in a control (the maximum end coordinate for this sample was extended to encompass a region of decreased probe intensity not called by the segmentation algorithm). The deletion coordinates reported by Kumar et al.39 are shown for reference (converted from hg18 to hg19 using UCSC liftOver76). These coordinates match the 16p11.2 proximal CNV, a 550-kb critical region flanked by segmental duplications. This unique critical region (29.65–30.20 Mb) contains 25 annotated protein-coding genes.
(B–C) 16p11.2 microdeletion was tested for association with clinical covariates in 21 cases with available annotation using Fisher’s exact test. The deletion associated significantly (p < 0.05) with MYCN amplification status (B) and Children’s Oncology Group risk classification (C).
Notably, deletion of this 550 kb region on 16p11.2 is a recognized genetic syndrome (MIM: 611913) that confers increased risk for autism spectrum disorder, developmental delay, intellectual disability, seizures, macrocephaly, early-onset obesity, increased body mass index, and birth defects.38, 39, 40, 41, 42, 43, 44, 45 Chromosome 16p is rich in segmental duplications, which serve as hotspots for non-allelic homologous recombination (NAHR) during meiosis or mitosis and give rise to reciprocal microdeletions and microduplications in several regions. Five recurrent breakpoint regions have been defined.44 The deletion we observed in neuroblastoma is referred to as the proximal (or typical) deletion and is characterized by breakpoint (BPs) in the segmental duplications near 29.6 and 30.2 Mb (BP4–BP5). The 550 kb unique segment between the segmental duplications (29.65–30.20 Mb) is considered the critical region, and this segment contains 25 annotated protein-coding genes. We did not observe any distal (BP1–BP3 or BP2–BP3) deletions in our cohort of 5,585 neuroblastoma cases, and we observed proximal and distal 16p11.2 microduplications only at frequencies consistent with their population prevalence (two and three cases, respectively).
These 16p11.2 microdeletions were observed in 0.39% of neuroblastoma cases and 0.03% of controls overall. The frequency observed in our control cohort is consistent with previous estimates of 16p11.2 deletion prevalence, which range from 0.01%–0.04% in adult populations38, 43, 45, 46 and 0.03% in a screen of French-Canadian newborns.47 We observed deletions in all ethnic groups except South Asians, likely due to the small number of South Asian individuals included in the study (Figure S2). The association of 16p11.2 microdeletions with neuroblastoma remained significant in the discovery and replication cohorts when considering European subjects only (0.42% of cases and 0.03% of controls; p = 4.10 × 10−8; odds ratio = 14.5; 95% confidence interval = 5.6–37.6; Table 2).
We next tested the 16p11.2 microdeletion for association with clinical, biological, and genetic covariates in our neuroblastoma cohort (Table S4). Strikingly, no cases with 16p11.2 microdeletion had somatic amplification of the oncogene MYCN (p = 0.022, Figure 2B), an event that is observed in approximately 20% of neuroblastoma tumors and associates with poor prognosis. The deletion was more frequent in cases classified as low- or intermediate-risk according to the COG risk stratification method (p = 0.0049, Figure 2C), even after adjusting for MYCN amplification (p = 0.046). It also trended toward association with tumor stage as described by the International Neuroblastoma Staging System (p = 0.080), and there was a slight enrichment for Stage 1 and 2 disease. There was no significant association with sex (p = 1) or age at diagnosis (p = 0.68). We compared survival rates between low-risk cases with and without the deletion and did not detect a significant difference in overall or event-free survival at current sample sizes (p = 0.54 and p = 0.23, respectively; Figure S4). Finally, we restricted our analyses to European individuals and tested 16p11.2 microdeletion for association with genotypes at eight known neuroblastoma susceptibility loci previously identified through GWAS. We did not detect any significant differences in allele frequencies after multiple testing corrections (Table S5). The enrichment of MYCN non-amplified, low-risk cases among those with 16p11.2 microdeletion may suggest that the deletion predisposes individuals to neuroblastoma through a mechanism that is more likely to give rise to low-risk disease. Some neuroblastoma-associated GWAS variants have also shown enrichment in distinct disease subsets such as high- or low-risk, MYCN amplified, or 11q-deleted cases.5, 10, 13, 48
To validate these 16p11.2 microdeletions and gain further genetic insight, we performed 30× whole-genome sequencing (WGS) on germline and tumor DNA from seven neuroblastoma cases with the microdeletion. We also utilized WGS data for five additional microdeletion-carrying neuroblastoma cases that were sequenced through the Therapeutically Applicable Research to Generate Effective Treatments (TARGET) program or the Gabriella Miller Kids First (GMKF) program. Germline deletions were confirmed in all 12 individuals, and the deletions were also detected in all 11 available matched tumors (Figure S5). As estimated by SNP array, all deletions covered the 550 kb critical region from 29.65–30.20 Mb and had breakpoints falling within the flanking segmental duplications.
We next used the WGS data for 11 tumors from 16p11.2 microdeletion cases to profile genome-wide somatic copy number changes (Figure S6). Segmental chromosomal alterations, which clinically correlate with high-risk disease,49 were observed in two individuals. Nine tumors harbored no large copy number changes or only whole-chromosome gains without segmental alterations, consistent with low-risk classification. Chromosome 16 was gained in three hyperploid low-risk tumors, but did not harbor any other large alterations. We also assessed the 16p11.2 region for focal copy number changes in these tumors (Figure S5). In most tumors, read coverage within the 16p11.2 deleted region was decreased by approximately 50% relative to the surrounding region; this result was similar to the decrease in relative coverage seen with the germline one-copy deletion. However, two tumors had slightly lower relative coverage (37%–38%), and one had higher relative coverage (65%), indicating that the region may have undergone additional somatic NAHR-mediated rearrangements in these three tumors.
We next hypothesized that damaging germline or somatic variants in the unaffected allele might exacerbate the effect of the deletion by disrupting one or more of the 25 protein-coding genes within the deleted region. We identified five rare (allele frequency < 1%), predicted-damaging (CADD score ≥ 20) germline SNVs and indels affecting these protein-coding genes among the 12 cases with germline WGS data available (Table S6), but we found no somatic SNVs or indels meeting these criteria in 11 matched tumors. We identified two germline missense variants of potential clinical relevance by using the Human Gene Mutation Database: The R386H variant in SEZ6L2 is weakly associated with autism spectrum disorder (ASD [MIM: 209850]),50 and the P216L variant in PRRT2 has been reported in Landau-Kleffner syndrome (MIM: 245570), a rare childhood neurological disorder.51 It is unclear whether these and other variants within 16p11.2 play a functional role in neuroblastoma initiation or progression, but follow-up studies on these protein-coding variants as well as noncoding and structural variants are warranted.
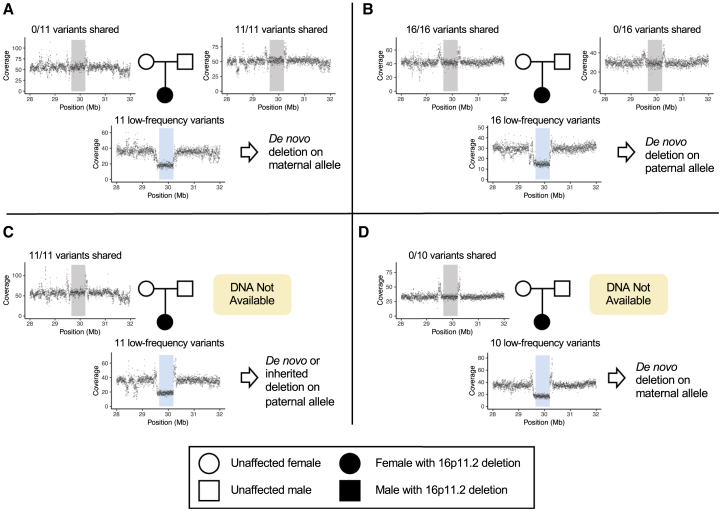
Lastly, to determine whether these 16p11.2 microdeletions arose de novo or were inherited, we considered four cases for which parental DNA had been collected through the Neuroblastoma Epidemiology in North America (NENA) study52 and WGS had been performed through the GMKF Pediatric Research Program (Figure 3 and Table S7). By comparing relative coverage and low-frequency variants within the deleted region in the children to the same data from their parents, we concluded that the deletion arose de novo in three of four children; of these, two deletions arose on the maternal allele and one on the paternal allele. The fourth deletion was either inherited from the father or arose de novo on the paternal allele. This predominantly de novo inheritance pattern was also observed for ASD-associated 16p11.2 microdeletions, where 90% of deletion cases were de novo or mosaic in the germline.53 However, in ASD, de novo 16p11.2 deletions exhibited a strong maternal bias: 89% arose on the maternal haplotype.53 In contrast, here we detected 16p11.2 deletions occurring de novo on both the maternal and paternal alleles. Maternal ages ranged from 17–32 and paternal ages from 18–42, showing no apparent trend with de novo deletion origin (Table S7).
Figure 3.
16p11.2 Microdeletion Arises de novo in Neuroblastoma without Allelic Bias
Coverage is plotted in 2-kb bins for four neuroblastoma cases with parental DNA available: two parent-child trios (A and B) and two mother-child duos (C and D). All parents show normal coverage within the 16p11.2 critical region (20.65–30.20 Mb, gray shading), whereas the children show a 50% decrease in coverage; this result confirms 16p11.2 microdeletion (blue shading). The number of low-frequency variants (<5% in 1000 Genomes) found within each child’s single remaining allele is displayed above the child’s coverage plot. The fraction of these variants the child shares with each parent is displayed above the parent’s plot.
16p11.2 microdeletion is associated with a diverse array of phenotypes including ASD, developmental delay, seizures, macrocephaly, and obesity.35, 36, 37, 38, 39, 40, 41, 42 The association of 16p11.2 microdeletion with neuroblastoma further expands the breadth of the deletion’s clinical manifestations to include cancer. Notably, this region has previously been implicated in the regulation of proliferation and growth: while 16p11.2 microdeletion associates with macrocephaly and increased body weight, the reciprocal 16p11.2 microduplication associates with microcephaly and decreased body weight.40, 43 Similar reciprocal deletion/duplication growth phenotypes are also observed with the neurodevelopment-associated CNVs at 1q21.1 (MIM: 612474, 612475)54 and 22q11.2 (MIM: 192430, 608363).55 In general, neurodevelopmental disorders are enriched for mutations in cancer-associated genes which control proliferation and differentiation.56 This supports the notion that genetic variants such as 16p11.2 microdeletion could predispose individuals to both cancer and neurodevelopmental phenotypes by perturbing these cellular functions. For 16p11.2 microdeletion, this effect may partially be attributed to dysregulation of the MAPK/ERK pathway caused by deletion of MAPK3 (MIM: 601795), encoding ERK1, and MVP (MIM: 605088). Deletion of a region syntenic to 16p11.2 causes elevated ERK activity in neural precursors in mice57, 58 but the impact on other lineages, such as those that give rise to neuroblastoma, is unknown. Aberrant MAPK signaling contributes to many cancers, including neuroblastoma, where an enrichment of mutations in the MAPK pathway has been observed at relapse.59
The association of 16p11.2 microdeletion with neuroblastoma is especially noteworthy given this cancer’s neurodevelopmental origin. Neuroblastoma is thought to arise from improper differentiation of neural crest cells of the sympathoadrenal lineage, giving rise to primary tumors in sympathetic nervous system tissues. The pleiotropic effects of 16p11.2 microdeletion on neuroblastoma and neurodevelopmental phenotypes such as ASD may be explained by dysregulation of developmental programs involved in both central and peripheral nervous system development. Most studies on 16p11.2 microdeletion syndrome have focused on central nervous system defects,60, 61, 62, 63, 64, 65, 66, 67 so additional research on the peripheral nervous system is needed. It is possible that aberrant neural crest development plays an underappreciated role in the pathology of 16p11.2 microdeletion syndrome. The neural crest has important functions in development of the brain and the adrenal and endocrine systems, facial bones, heart, and other tissues. Dysregulation of these systems could partially explain phenotypes of 16p11.2 microdeletion including language and learning impairment, macrocephaly, dysmorphic facial features, and cardiac malformation.68 This possibility is supported by a recent study which suggests that altered neural crest activity explains some of these features in the context of ASD.69 Additionally, several 16p11.2 genes were implicated in neural crest-related phenotypes in a zebrafish loss-of-function screen.70 Future research on the role of the 16p11.2 region in neural crest development could uncover novel biology and shed light on the clinical manifestations of the 16p11.2 CNV.
The phenotypes exhibited by 16p11.2 microdeletion carriers can vary dramatically, even within the same family. Some carriers have no identifiable phenotypic abnormalities, while others are severely affected with multiple deficits.68 This makes the deletion syndrome extremely challenging to diagnose and manage, and it suggests that other factors cooperate to influence pathogenesis. Both common and rare variants have been shown to modify neurodevelopmental outcomes and other phenotypes in 16p11.2 microdeletion carriers.71, 72, 73 Given that most deletion carriers do not develop clinically detectable neuroblastoma, 16p11.2 deletion alone is likely not sufficient for neuroblastoma tumorigenesis. Additional genetic, epigenetic, and environmental factors probably contributed to initiation and maintenance in individuals harboring the microdeletion. We identified five rare, predicted-damaging variants in protein-coding genes within the 16p11.2 region that could potentially function as second hits, but the significance of these variants and other genetic and epigenetic alterations requires further investigation.
Currently, complete medical histories are not available for the individuals with neuroblastoma profiled in this study. An important future direction for this work is to re-contact families of children with 16p11.2 microdeletion and define the co-occurrence of neuroblastoma with other traits typical of the microdeletion syndrome. This will help determine whether counseling or surveillance for neuroblastoma might be appropriate for children diagnosed with 16p11.2 microdeletion, or conversely, whether germline testing for children newly diagnosed with neuroblastoma should include 16p11.2 microdeletion so that the other phenotypes associated with the syndrome can be monitored.
Overall, this study identifies a rare germline CNV that substantially impacts neuroblastoma risk, highlighting the potential clinical relevance of rare variants in this often deadly pediatric cancer. This finding expands the already diverse clinical outcomes associated with 16p11.2 microdeletion syndrome to include cancer, and it raises questions regarding the role of the 16p11.2 region in neural crest development.
Data Availability
Data analyzed in this paper are available through the database of Genotypes and Phenotypes (dbGaP). SNP array data are available through accession phs000124.v3.p1. WGS data generated through TARGET are available through accession phs000218.v21.p7. WGS data generated through GMKF are available through dbGaP accession phs001436.v1.p1 and through the Kids First Data Resource Portal. Additional WGS data generated for this study have been deposited to dbGaP: phs000124.v3.p1.
Declaration of Interests
The authors declare no competing interests.
Acknowledgments
This work was supported by National Institutes of Health (NIH) awards R01CA204974, R03CA230366, R01CA132887, R01CA124709, X01HL136997, and U2CHL138346. L.E.E. was supported in part by NIH grant T32GM008216, and A.M. was supported in part by T32HG000046, also from the NIH. Neuroblastoma case DNA used for SNP array genotyping and for the WGS generated for this manuscript was collected and bio-banked through the Children’s Oncology Group (COG). The platform-matched control genotyping data were generated and supplied by the Center for Applied Genomics (CAG) at CHOP. DNA extraction from Neuroblastoma Epidemiology in North America (NENA) blood or buccal samples was carried out in the UNC BioSpecimen Processing Core. Whole-genome sequencing from Gabriella Miller Kids First (GMKF) was delivered on the CAVATICA analysis platform by the Kids First Pediatric Data Resource Center.
Published: August 29, 2019
Footnotes
Supplemental Data can be found online at https://doi.org/10.1016/j.ajhg.2019.07.020.
Accession Numbers
The dbGaP accession number for the WGS data newly generated for this study and reported in this paper is phs000124.v3.p1.
Web Resources
ANNOVAR version 2016Feb01, annovar.openbioinformatics.org/
Gabriella Miller Kids First (GMKF) Pediatric Research Program, https://commonfund.nih.gov/kidsfirst/
GATK version 4.0.2.0, https://software.broadinstitute.org/gatk/
GenomeStudio version 2.0 (Illumina), https://support.illumina.com/array/array_software/genomestudio/downloads.html
karyoploteR version 1.8.8, http://www.bioconductor.org/packages/karyoploteR
Kids First Data Resource Portal, https://kidsfirstdrc.org/
Nexus Copy Number versions 8.0 and 10.0 (BioDiscovery), https://www.biodiscovery.com/products/Nexus-Copy-Number
OMIM, http://www.omim.org/
PLINK version 1.9, https://www.cog-genomics.org/plink2
R version 3.5.2, https://www.r-project.org/
SnpEff version 4.3, http://snpeff.sourceforge.net/
Therapeutically Applicable Research to Generate Effective Treatments (TARGET) Program, https://ocg.cancer.gov/programs/target
UCSC Genome Browser, http://genome.ucsc.edu/
UCSC liftOver, https://genome.ucsc.edu/cgi-bin/hgLiftOver
VarScan2 version 2.3.9, http://varscan.sourceforge.net/
Supplemental Data
References
- 1.Mossé Y.P., Laudenslager M., Longo L., Cole K.A., Wood A., Attiyeh E.F., Laquaglia M.J., Sennett R., Lynch J.E., Perri P. Identification of ALK as a major familial neuroblastoma predisposition gene. Nature. 2008;455:930–935. doi: 10.1038/nature07261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mosse Y.P., Laudenslager M., Khazi D., Carlisle A.J., Winter C.L., Rappaport E., Maris J.M. Germline PHOX2B mutation in hereditary neuroblastoma. Am. J. Hum. Genet. 2004;75:727–730. doi: 10.1086/424530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Trochet D., Bourdeaut F., Janoueix-Lerosey I., Deville A., de Pontual L., Schleiermacher G., Coze C., Philip N., Frébourg T., Munnich A. Germline mutations of the paired-like homeobox 2B (PHOX2B) gene in neuroblastoma. Am. J. Hum. Genet. 2004;74:761–764. doi: 10.1086/383253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Maris J.M., Mosse Y.P., Bradfield J.P., Hou C., Monni S., Scott R.H., Asgharzadeh S., Attiyeh E.F., Diskin S.J., Laudenslager M. Chromosome 6p22 locus associated with clinically aggressive neuroblastoma. N. Engl. J. Med. 2008;358:2585–2593. doi: 10.1056/NEJMoa0708698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Capasso M., Devoto M., Hou C., Asgharzadeh S., Glessner J.T., Attiyeh E.F., Mosse Y.P., Kim C., Diskin S.J., Cole K.A. Common variations in BARD1 influence susceptibility to high-risk neuroblastoma. Nat. Genet. 2009;41:718–723. doi: 10.1038/ng.374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang K., Diskin S.J., Zhang H., Attiyeh E.F., Winter C., Hou C., Schnepp R.W., Diamond M., Bosse K., Mayes P.A. Integrative genomics identifies LMO1 as a neuroblastoma oncogene. Nature. 2011;469:216–220. doi: 10.1038/nature09609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Diskin S.J., Capasso M., Schnepp R.W., Cole K.A., Attiyeh E.F., Hou C., Diamond M., Carpenter E.L., Winter C., Lee H. Common variation at 6q16 within HACE1 and LIN28B influences susceptibility to neuroblastoma. Nat. Genet. 2012;44:1126–1130. doi: 10.1038/ng.2387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.McDaniel L.D., Conkrite K.L., Chang X., Capasso M., Vaksman Z., Oldridge D.A., Zachariou A., Horn M., Diamond M., Hou C. Common variants upstream of MLF1 at 3q25 and within CPZ at 4p16 associated with neuroblastoma. PLoS Genet. 2017;13:e1006787. doi: 10.1371/journal.pgen.1006787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Capasso M., Diskin S.J., Totaro F., Longo L., De Mariano M., Russo R., Cimmino F., Hakonarson H., Tonini G.P., Devoto M. Replication of GWAS-identified neuroblastoma risk loci strengthens the role of BARD1 and affirms the cumulative effect of genetic variations on disease susceptibility. Carcinogenesis. 2013;34:605–611. doi: 10.1093/carcin/bgs380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nguyen B., Diskin S.J., Capasso M., Wang K., Diamond M.A., Glessner J., Kim C., Attiyeh E.F., Mosse Y.P., Cole K. Phenotype restricted genome-wide association study using a gene-centric approach identifies three low-risk neuroblastoma susceptibility Loci. PLoS Genet. 2011;7:e1002026. doi: 10.1371/journal.pgen.1002026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Latorre V., Diskin S.J., Diamond M.A., Zhang H., Hakonarson H., Maris J.M., Devoto M. Replication of neuroblastoma SNP association at the BARD1 locus in African-Americans. Cancer Epidemiol. Biomarkers Prev. 2012;21:658–663. doi: 10.1158/1055-9965.EPI-11-0830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Capasso M., McDaniel L.D., Cimmino F., Cirino A., Formicola D., Russell M.R., Raman P., Cole K.A., Diskin S.J. The functional variant rs34330 of CDKN1B is associated with risk of neuroblastoma. J. Cell. Mol. Med. 2017;21:3224–3230. doi: 10.1111/jcmm.13226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chang X., Zhao Y., Hou C., Glessner J., McDaniel L., Diamond M.A., Thomas K., Li J., Wei Z., Liu Y. Common variants in MMP20 at 11q22.2 predispose to 11q deletion and neuroblastoma risk. Nat. Commun. 2017;8:569. doi: 10.1038/s41467-017-00408-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Diskin S.J., Hou C., Glessner J.T., Attiyeh E.F., Laudenslager M., Bosse K., Cole K., Mossé Y.P., Wood A., Lynch J.E. Copy number variation at 1q21.1 associated with neuroblastoma. Nature. 2009;459:987–991. doi: 10.1038/nature08035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Russell M.R., Penikis A., Oldridge D.A., Alvarez-Dominguez J.R., McDaniel L., Diamond M., Padovan O., Raman P., Li Y., Wei J.S. CASC15-S is a tumor suppressor lncRNA at the 6p22 neuroblastoma susceptibility locus. Cancer Res. 2015;75:3155–3166. doi: 10.1158/0008-5472.CAN-14-3613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pandey G.K., Mitra S., Subhash S., Hertwig F., Kanduri M., Mishra K., Fransson S., Ganeshram A., Mondal T., Bandaru S. The risk-associated long noncoding RNA NBAT-1 controls neuroblastoma progression by regulating cell proliferation and neuronal differentiation. Cancer Cell. 2014;26:722–737. doi: 10.1016/j.ccell.2014.09.014. [DOI] [PubMed] [Google Scholar]
- 17.Bosse K.R., Diskin S.J., Cole K.A., Wood A.C., Schnepp R.W., Norris G., Nguyen B., Jagannathan J., Laquaglia M., Winter C. Common variation at BARD1 results in the expression of an oncogenic isoform that influences neuroblastoma susceptibility and oncogenicity. Cancer Res. 2012;72:2068–2078. doi: 10.1158/0008-5472.CAN-11-3703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhu S., Zhang X., Weichert-Leahey N., Dong Z., Zhang C., Lopez G., Tao T., He S., Wood A.C., Oldridge D. LMO1 Synergizes with MYCN to Promote Neuroblastoma Initiation and Metastasis. Cancer Cell. 2017;32:310–323.e5. doi: 10.1016/j.ccell.2017.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Molenaar J.J., Domingo-Fernández R., Ebus M.E., Lindner S., Koster J., Drabek K., Mestdagh P., van Sluis P., Valentijn L.J., van Nes J. LIN28B induces neuroblastoma and enhances MYCN levels via let-7 suppression. Nat. Genet. 2012;44:1199–1206. doi: 10.1038/ng.2436. [DOI] [PubMed] [Google Scholar]
- 20.Schnepp R.W., Khurana P., Attiyeh E.F., Raman P., Chodosh S.E., Oldridge D.A., Gagliardi M.E., Conkrite K.L., Asgharzadeh S., Seeger R.C. A LIN28B-RAN-AURKA Signaling Network Promotes Neuroblastoma Tumorigenesis. Cancer Cell. 2015;28:599–609. doi: 10.1016/j.ccell.2015.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lek M., Karczewski K.J., Minikel E.V., Samocha K.E., Banks E., Fennell T., O’Donnell-Luria A.H., Ware J.S., Hill A.J., Cummings B.B., Exome Aggregation Consortium Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016;536:285–291. doi: 10.1038/nature19057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pugh T.J., Morozova O., Attiyeh E.F., Asgharzadeh S., Wei J.S., Auclair D., Carter S.L., Cibulskis K., Hanna M., Kiezun A. The genetic landscape of high-risk neuroblastoma. Nat. Genet. 2013;45:279–284. doi: 10.1038/ng.2529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Parsons D.W., Roy A., Yang Y., Wang T., Scollon S., Bergstrom K., Kerstein R.A., Gutierrez S., Petersen A.K., Bavle A. Diagnostic yield of clinical tumor and germline whole-exome sequencing for children with solid tumors. JAMA Oncol. 2016;2:616–624. doi: 10.1001/jamaoncol.2015.5699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang J., Walsh M.F., Wu G., Edmonson M.N., Gruber T.A., Easton J., Hedges D., Ma X., Zhou X., Yergeau D.A. Germline Mutations in Predisposition Genes in Pediatric Cancer. N. Engl. J. Med. 2015;373:2336–2346. doi: 10.1056/NEJMoa1508054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stefansson H., Rujescu D., Cichon S., Pietiläinen O.P.H., Ingason A., Steinberg S., Fossdal R., Sigurdsson E., Sigmundsson T., Buizer-Voskamp J.E., GROUP Large recurrent microdeletions associated with schizophrenia. Nature. 2008;455:232–236. doi: 10.1038/nature07229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pinto D., Pagnamenta A.T., Klei L., Anney R., Merico D., Regan R., Conroy J., Magalhaes T.R., Correia C., Abrahams B.S. Functional impact of global rare copy number variation in autism spectrum disorders. Nature. 2010;466:368–372. doi: 10.1038/nature09146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Park R.W., Kim T.M., Kasif S., Park P.J. Identification of rare germline copy number variations over-represented in five human cancer types. Mol. Cancer. 2015;14:1–12. doi: 10.1186/s12943-015-0292-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Marshall C.R., Howrigan D.P., Merico D., Thiruvahindrapuram B., Wu W., Greer D.S., Antaki D., Shetty A., Holmans P.A., Pinto D., Psychosis Endophenotypes International Consortium. CNV and Schizophrenia Working Groups of the Psychiatric Genomics Consortium Contribution of copy number variants to schizophrenia from a genome-wide study of 41,321 subjects. Nat. Genet. 2017;49:27–35. doi: 10.1038/ng.3725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Satgé D., Moore S.W., Stiller C.A., Niggli F.K., Pritchard-Jones K., Bown N., Bénard J., Plantaz D. Abnormal constitutional karyotypes in patients with neuroblastoma: a report of four new cases and review of 47 others in the literature. Cancer Genet. Cytogenet. 2003;147:89–98. doi: 10.1016/s0165-4608(03)00203-6. [DOI] [PubMed] [Google Scholar]
- 30.Attiyeh E.F., London W.B., Mossé Y.P., Wang Q., Winter C., Khazi D., McGrady P.W., Seeger R.C., Look A.T., Shimada H., Children’s Oncology Group Chromosome 1p and 11q deletions and outcome in neuroblastoma. N. Engl. J. Med. 2005;353:2243–2253. doi: 10.1056/NEJMoa052399. [DOI] [PubMed] [Google Scholar]
- 31.Thompson D., Vo K.T., London W.B., Fischer M., Ambros P.F., Nakagawara A., Brodeur G.M., Matthay K.K., DuBois S.G. Identification of patient subgroups with markedly disparate rates of MYCN amplification in neuroblastoma: A report from the International Neuroblastoma Risk Group project. Cancer. 2016;122:935–945. doi: 10.1002/cncr.29848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mosse Y., Greshock J., King A., Khazi D., Weber B.L., Maris J.M. Identification and high-resolution mapping of a constitutional 11q deletion in an infant with multifocal neuroblastoma. Lancet Oncol. 2003;4:769–771. doi: 10.1016/s1470-2045(03)01283-x. [DOI] [PubMed] [Google Scholar]
- 33.Passariello A., De Brasi D., Defferrari R., Genesio R., Tufano M., Mazzocco K., Capasso M., Migliorati R., Martinsson T., Siani P. Constitutional 11q14-q22 chromosome deletion syndrome in a child with neuroblastoma MYCN single copy. Eur. J. Med. Genet. 2013;56:626–634. doi: 10.1016/j.ejmg.2013.08.005. [DOI] [PubMed] [Google Scholar]
- 34.Lurie I.W. Clinical manifestations of partial trisomy 2p. Cytogenet. Genome Res. 2014;144:28–30. doi: 10.1159/000367908. [DOI] [PubMed] [Google Scholar]
- 35.Presson A.P., Partyka G., Jensen K.M., Devine O.J., Rasmussen S.A., McCabe L.L., McCabe E.R.B. Current estimate of Down Syndrome population prevalence in the United States. J. Pediatr. 2013;163:1163–1168. doi: 10.1016/j.jpeds.2013.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Satgé D., Sasco A.J., Carlsen N.L., Stiller C.A., Rubie H., Hero B., de Bernardi B., de Kraker J., Coze C., Kogner P. A lack of neuroblastoma in Down syndrome: a study from 11 European countries. Cancer Res. 1998;58:448–452. [PubMed] [Google Scholar]
- 37.Ritenour L.E., Randall M.P., Bosse K.R., Diskin S.J. Genetic susceptibility to neuroblastoma: current knowledge and future directions. Cell Tissue Res. 2018;372:287–307. doi: 10.1007/s00441-018-2820-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Weiss L.A., Shen Y., Korn J.M., Arking D.E., Miller D.T., Fossdal R., Saemundsen E., Stefansson H., Ferreira M.A.R., Green T., Autism Consortium Association between microdeletion and microduplication at 16p11.2 and autism. N. Engl. J. Med. 2008;358:667–675. doi: 10.1056/NEJMoa075974. [DOI] [PubMed] [Google Scholar]
- 39.Kumar R.A., KaraMohamed S., Sudi J., Conrad D.F., Brune C., Badner J.A., Gilliam T.C., Nowak N.J., Cook E.H., Jr., Dobyns W.B., Christian S.L. Recurrent 16p11.2 microdeletions in autism. Hum. Mol. Genet. 2008;17:628–638. doi: 10.1093/hmg/ddm376. [DOI] [PubMed] [Google Scholar]
- 40.Shinawi M., Liu P., Kang S.H.L., Shen J., Belmont J.W., Scott D.A., Probst F.J., Craigen W.J., Graham B.H., Pursley A. Recurrent reciprocal 16p11.2 rearrangements associated with global developmental delay, behavioural problems, dysmorphism, epilepsy, and abnormal head size. J. Med. Genet. 2010;47:332–341. doi: 10.1136/jmg.2009.073015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rosenfeld J.A., Coppinger J., Bejjani B.A., Girirajan S., Eichler E.E., Shaffer L.G., Ballif B.C. Speech delays and behavioral problems are the predominant features in individuals with developmental delays and 16p11.2 microdeletions and microduplications. J. Neurodev. Disord. 2010;2:26–38. doi: 10.1007/s11689-009-9037-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Walters R.G., Jacquemont S., Valsesia A., de Smith A.J., Martinet D., Andersson J., Falchi M., Chen F., Andrieux J., Lobbens S. A new highly penetrant form of obesity due to deletions on chromosome 16p11.2. Nature. 2010;463:671–675. doi: 10.1038/nature08727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jacquemont S., Reymond A., Zufferey F., Harewood L., Walters R.G., Kutalik Z., Martinet D., Shen Y., Valsesia A., Beckmann N.D. Mirror extreme BMI phenotypes associated with gene dosage at the chromosome 16p11.2 locus. Nature. 2011;478:97–102. doi: 10.1038/nature10406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zufferey F., Sherr E.H., Beckmann N.D., Hanson E., Maillard A.M., Hippolyte L., Macé A., Ferrari C., Kutalik Z., Andrieux J., Simons VIP Consortium. 16p11.2 European Consortium A 600 kb deletion syndrome at 16p11.2 leads to energy imbalance and neuropsychiatric disorders. J. Med. Genet. 2012;49:660–668. doi: 10.1136/jmedgenet-2012-101203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Macé A., Tuke M.A., Deelen P., Kristiansson K., Mattsson H., Nõukas M., Sapkota Y., Schick U., Porcu E., Rüeger S. CNV-association meta-analysis in 191,161 European adults reveals new loci associated with anthropometric traits. Nat. Commun. 2017;8:1–11. doi: 10.1038/s41467-017-00556-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Malhotra D., Sebat J. CNVs: harbingers of a rare variant revolution in psychiatric genetics. Cell. 2012;148:1223–1241. doi: 10.1016/j.cell.2012.02.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tucker T., Giroux S., Clément V., Langlois S., Friedman J.M., Rousseau F. Prevalence of selected genomic deletions and duplications in a French-Canadian population-based sample of newborns. Mol. Genet. Genomic Med. 2013;1:87–97. doi: 10.1002/mgg3.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hungate E.A., Applebaum M.A., Skol A.D., Vaksman Z., Diamond M., McDaniel L., Volchenboum S.L., Stranger B.E., Maris J.M., Diskin S.J. Evaluation of Genetic Predisposition for MYCN-Amplified Neuroblastoma. J. Natl. Cancer Inst. 2017;109:1–4. doi: 10.1093/jnci/djx093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schleiermacher G., Janoueix-Lerosey I., Ribeiro A., Klijanienko J., Couturier J., Pierron G., Mosseri V., Valent A., Auger N., Plantaz D. Accumulation of segmental alterations determines progression in neuroblastoma. J. Clin. Oncol. 2010;28:3122–3130. doi: 10.1200/JCO.2009.26.7955. [DOI] [PubMed] [Google Scholar]
- 50.Kumar R.A., Marshall C.R., Badner J.A., Babatz T.D., Mukamel Z., Aldinger K.A., Sudi J., Brune C.W., Goh G., Karamohamed S. Association and mutation analyses of 16p11.2 autism candidate genes. PLoS ONE. 2009;4:e4582. doi: 10.1371/journal.pone.0004582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Conroy J., McGettigan P.A., McCreary D., Shah N., Collins K., Parry-Fielder B., Moran M., Hanrahan D., Deonna T.W., Korff C.M. Towards the identification of a genetic basis for Landau-Kleffner syndrome. Epilepsia. 2014;55:858–865. doi: 10.1111/epi.12645. [DOI] [PubMed] [Google Scholar]
- 52.Mazul A.L., Siega-Riz A.M., Weinberg C.R., Engel S.M., Zou F., Carrier K.S., Basta P.V., Vaksman Z., Maris J.M., Diskin S.J. A family-based study of gene variants and maternal folate and choline in neuroblastoma: a report from the Children’s Oncology Group. Cancer Causes Control. 2016;27:1209–1218. doi: 10.1007/s10552-016-0799-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Duyzend M.H., Nuttle X., Coe B.P., Baker C., Nickerson D.A., Bernier R., Eichler E.E. Maternal Modifiers and Parent-of-Origin Bias of the Autism-Associated 16p11.2 CNV. Am. J. Hum. Genet. 2016;98:45–57. doi: 10.1016/j.ajhg.2015.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mefford H.C., Sharp A.J., Baker C., Itsara A., Jiang Z., Buysse K., Huang S., Maloney V.K., Crolla J.A., Baralle D. Recurrent rearrangements of chromosome 1q21.1 and variable pediatric phenotypes. N. Engl. J. Med. 2008;359:1685–1699. doi: 10.1056/NEJMoa0805384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rump P., de Leeuw N., van Essen A.J., Verschuuren-Bemelmans C.C., Veenstra-Knol H.E., Swinkels M.E., Oostdijk W., Ruivenkamp C., Reardon W., de Munnik S. Central 22q11.2 deletions. Am. J. Med. Genet. A. 2014;164A:2707–2723. doi: 10.1002/ajmg.a.36711. [DOI] [PubMed] [Google Scholar]
- 56.Ernst C. Proliferation and Differentiation Deficits are a Major Convergence Point for Neurodevelopmental Disorders. Trends Neurosci. 2016;39:290–299. doi: 10.1016/j.tins.2016.03.001. [DOI] [PubMed] [Google Scholar]
- 57.Pucilowska J., Vithayathil J., Tavares E.J., Kelly C., Karlo J.C., Landreth G.E. The 16p11.2 deletion mouse model of autism exhibits altered cortical progenitor proliferation and brain cytoarchitecture linked to the ERK MAPK pathway. J. Neurosci. 2015;35:3190–3200. doi: 10.1523/JNEUROSCI.4864-13.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Pucilowska J., Vithayathil J., Pagani M., Kelly C., Karlo J.C., Robol C., Morella I., Gozzi A., Brambilla R., Landreth G.E. Pharmacological Inhibition of ERK Signaling Rescues Pathophysiology and Behavioral Phenotype Associated with 16p11.2 Chromosomal Deletion in Mice. J. Neurosci. 2018;38:6640–6652. doi: 10.1523/JNEUROSCI.0515-17.2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Eleveld T.F., Oldridge D.A., Bernard V., Koster J., Colmet Daage L., Diskin S.J., Schild L., Bentahar N.B., Bellini A., Chicard M. Relapsed neuroblastomas show frequent RAS-MAPK pathway mutations. Nat. Genet. 2015;47:864–871. doi: 10.1038/ng.3333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Escamilla C.O., Filonova I., Walker A.K., Xuan Z.X., Holehonnur R., Espinosa F., Liu S., Thyme S.B., López-García I.A., Mendoza D.B. Kctd13 deletion reduces synaptic transmission via increased RhoA. Nature. 2017;551:227–231. doi: 10.1038/nature24470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Park S.M., Park H.R., Lee J.H. MAPK3 at the Autism-Linked Human 16p11.2 Locus Influences Precise Synaptic Target Selection at Drosophila Larval Neuromuscular Junctions. Mol. Cells. 2017;40:151–161. doi: 10.14348/molcells.2017.2307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Haslinger D., Waltes R., Yousaf A., Lindlar S., Schneider I., Lim C.K., Tsai M.M., Garvalov B.K., Acker-Palmer A., Krezdorn N. Loss of the Chr16p11.2 ASD candidate gene QPRT leads to aberrant neuronal differentiation in the SH-SY5Y neuronal cell model. Mol. Autism. 2018;9:1–17. doi: 10.1186/s13229-018-0239-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Richter M., Murtaza N., Scharrenberg R., White S.H., Johanns O., Walker S., Yuen R.K.C., Schwanke B., Bedürftig B., Henis M. Altered TAOK2 activity causes autism-related neurodevelopmental and cognitive abnormalities through RhoA signaling. Mol. Psychiatry. 2018 doi: 10.1038/s41380-018-0025-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Golzio C., Willer J., Talkowski M.E., Oh E.C., Taniguchi Y., Jacquemont S., Reymond A., Sun M., Sawa A., Gusella J.F. KCTD13 is a major driver of mirrored neuroanatomical phenotypes of the 16p11.2 copy number variant. Nature. 2012;485:363–367. doi: 10.1038/nature11091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Grissom N.M., McKee S.E., Schoch H., Bowman N., Havekes R., O’Brien W.T., Mahrt E., Siegel S., Commons K., Portfors C. Male-specific deficits in natural reward learning in a mouse model of neurodevelopmental disorders. Mol. Psychiatry. 2018;23:544–555. doi: 10.1038/mp.2017.184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lin G.N., Corominas R., Lemmens I., Yang X., Tavernier J., Hill D.E., Vidal M., Sebat J., Iakoucheva L.M. Spatiotemporal 16p11.2 protein network implicates cortical late mid-fetal brain development and KCTD13-Cul3-RhoA pathway in psychiatric diseases. Neuron. 2015;85:742–754. doi: 10.1016/j.neuron.2015.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Iyer J., Singh M.D., Jensen M., Patel P., Pizzo L., Huber E., Koerselman H., Weiner A.T., Lepanto P., Vadodaria K. Pervasive genetic interactions modulate neurodevelopmental defects of the autism-associated 16p11.2 deletion in Drosophila melanogaster. Nat. Commun. 2018;9:2548. doi: 10.1038/s41467-018-04882-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Miller D.T., Chung W., Nasir R., Shen Y., Steinman K.J., Wu B.-L., Hanson E. 16p11.2 recurrent microdeletion. In: Adam M.P., Ardinger H.H., Pagon R.A., Wallace S.E., Bean L.J.H., Stephens K., Amemiya A., editors. GeneReviews. University of Washington, Seattle; 1993. [Google Scholar]
- 69.Benítez-Burraco A., Lattanzi W., Murphy E. Language impairments in ASD resulting from a failed domestication of the human brain. Front. Neurosci. 2016;10:373. doi: 10.3389/fnins.2016.00373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Blaker-Lee A., Gupta S., McCammon J.M., De Rienzo G., Sive H. Zebrafish homologs of genes within 16p11.2, a genomic region associated with brain disorders, are active during brain development, and include two deletion dosage sensor genes. Dis. Model. Mech. 2012;5:834–851. doi: 10.1242/dmm.009944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Pizzo L., Jensen M., Polyak A., Rosenfeld J.A., Mannik K., Krishnan A., McCready E., Pichon O., Le Caignec C., Van Dijck A. Rare variants in the genetic background modulate cognitive and developmental phenotypes in individuals carrying disease-associated variants. Genet. Med. 2019;21:816–825. doi: 10.1038/s41436-018-0266-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wu N., Ming X., Xiao J., Wu Z., Chen X., Shinawi M., Shen Y., Yu G., Liu J., Xie H. TBX6 null variants and a common hypomorphic allele in congenital scoliosis. N. Engl. J. Med. 2015;372:341–350. doi: 10.1056/NEJMoa1406829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Girirajan S., Rosenfeld J.A., Cooper G.M., Antonacci F., Siswara P., Itsara A., Vives L., Walsh T., McCarthy S.E., Baker C. A recurrent 16p12.1 microdeletion supports a two-hit model for severe developmental delay. Nat. Genet. 2010;42:203–209. doi: 10.1038/ng.534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Gel B., Serra E. karyoploteR: an R/Bioconductor package to plot customizable genomes displaying arbitrary data. Bioinformatics. 2017;33:3088–3090. doi: 10.1093/bioinformatics/btx346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kent W.J., Sugnet C.W., Furey T.S., Roskin K.M., Pringle T.H., Zahler A.M., Haussler D. The human genome browser at UCSC. Genome Res. 2002;12:996–1006. doi: 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Hinrichs A.S., Karolchik D., Baertsch R., Barber G.P., Bejerano G., Clawson H., Diekhans M., Furey T.S., Harte R.A., Hsu F. The UCSC Genome Browser Database: update 2006. Nucleic Acids Res. 2006;34:D590–D598. doi: 10.1093/nar/gkj144. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data analyzed in this paper are available through the database of Genotypes and Phenotypes (dbGaP). SNP array data are available through accession phs000124.v3.p1. WGS data generated through TARGET are available through accession phs000218.v21.p7. WGS data generated through GMKF are available through dbGaP accession phs001436.v1.p1 and through the Kids First Data Resource Portal. Additional WGS data generated for this study have been deposited to dbGaP: phs000124.v3.p1.