Abstract
The advancement of precision medicine requires new methods to coordinate and deliver genetic data from heterogeneous sources to physicians and patients. The eMERGE III Network enrolled >25,000 participants from biobank and prospective cohorts of predominantly healthy individuals for clinical genetic testing to determine clinically actionable findings. The network developed protocols linking together the 11 participant collection sites and 2 clinical genetic testing laboratories. DNA capture panels targeting 109 genes were used for testing of DNA and sample collection, data generation, interpretation, reporting, delivery, and storage were each harmonized. A compliant and secure network enabled ongoing review and reconciliation of clinical interpretations, while maintaining communication and data sharing between clinicians and investigators. A total of 202 individuals had positive diagnostic findings relevant to the indication for testing and 1,294 had additional/secondary findings of medical significance deemed to be returnable, establishing data return rates for other testing endeavors. This study accomplished integration of structured genomic results into multiple electronic health record (EHR) systems, setting the stage for clinical decision support to enable genomic medicine. Further, the established processes enable different sequencing sites to harmonize technical and interpretive aspects of sequencing tests, a critical achievement toward global standardization of genomic testing. The eMERGE protocols and tools are available for widespread dissemination.
Keywords: eMERGE, electronic health record, clinical sequencing, harmonization, next generation sequencing
Introduction
The identification, interpretation, and return of actionable clinical genetic findings is an increasing focus of precision medicine. There is also growing awareness that the discovery of genes underlying human diseases is dependent upon access to samples from carefully phenotyped individuals with (and without) clinical conditions. As clinical visits provide the ideal opportunity to record patient phenotypes, with appropriate consent, the medical care of specific patient groups can drive the accumulation of clinical data and knowledge of the genetic underpinnings of disease and the penetrance of DNA risk variants. This “virtuous cycle” of data flow from the bench to the bedside and back to the bench will be a key driver of progress in genetic and genomic translation.
While conceptually straightforward, there are many challenges that must be overcome for integrating clinical and research agendas across global populations. Clinical visits are often brief, focused upon measurement related to specific symptoms and constrained by fiscal and practical concerns. On the other hand, ascertainment for research is often open ended, longitudinal, and accompanied by rigorous consent procedures. The types of data that are recorded for each purpose can be different in both depth and quality. As a result, ideal research and clinical records often diverge.
The current phase (III) of the United States National Institute of Health’s Electronic Medical Records and Genomics (eMERGE) program (see Web Resources) aims to study and improve these processes for coordinated delivery of clinical and research data, in a multi-center network, while providing actionable genetic results derived from a next-generation sequencing platform to eMERGE research participants. In previous phases, the network sampled data from large collections of volunteers (>100,000) for research and discovery purposes, as well as to establish parameters that might influence clinical data reporting. In the current phase, multiple clinical collection sites with access to predominantly healthy participants, who are willing to undergo genetic testing and to have their results returned by their physicians, were identified. The network used the opportunity to build upon experience with participant consent, to obtain clinical data from the EHR, and to return genetic testing results.1 The program addressed challenges arising from the heterogeneity of collection sites and tools used to collect patients’ and participants’ data. Points of standardization were established (Table 1) and overcame obstacles of use of different instruments and molecular reagents at different sites.
Table 1.
Items Harmonized across the Two Sequencing Centers
| Item | Challenge | Comments |
|---|---|---|
| Collection sites | sample type | agreed to blooda,b |
| sample quality | minimal quantity specifieda | |
| intake formats | standard tables supplied to sites | |
| phenotypes | not shared unless indication for testing | |
| patient ID structure | naming conventions | |
| indications for testing | selected 40 “hard coded” | |
| Assay development | gene targets | selected by consensus |
| capture strategy | agreed exons (+/− 15 bases)/SNPs; capture probes spanned min 100 bases | |
| capture reagents | two platforms supported (Nimblegen and Illumina Rapid Capture) | |
| Sanger validation | rare variants always Sanger validated; for common SNVs, stopped validation after 5 confirmations | |
| CNV validation | all CNVs by orthogonal technology | |
| Validation/proficiency | technical performance/coverage | min standards (200×; 95% coverage, etc.) |
| ongoing proficiency | interlaboratory exchange or eMERGE samples and use of standard CAP NGS PT | |
| Primary analysis | CNV calling parameters | 3+ exons |
| pharmacogenomics | report variants and inferred diplotypes | |
| Variant classification | initial harmonization | required harmonization of all medically significant differences observed 5 or more times in tested genes |
| ongoing classifications | required consensus between labs or elevation to Clinical Annotation WG for network consensus | |
| Report contenta | consensus content | 67 genes and 14 SNVs |
| site-specific genes and SNVs | see Figure 4 and Table S7 | |
| updates | variant reclassifications provided | |
| Data delivery | physician clinical reports | PDFs, consumable xml structure; GeneInsight |
| network access to interpreted variants and de-identified reports | GeneInsight de-identified case repository, DNAnexus Commons |
|
| community data sharing | dbGaP and ClinVar submissions | |
| Progress reporting | specimen progress | sequencing and reporting timelines |
| aggregate statistic reporting | rates of secondary findings; detection rates for indications |
Exceptions contributed to extended TAT
BCM-HGSC accepted saliva from some sites for a predetermined number of samples
Addressing these challenges advanced precision medical care by standardizing methods for phenotyping, sequencing, and genetic variant interpretation. Further, the harmonized flow, storage, and management of data provided a cohesive vehicle to access data to facilitate research while maintaining respect for patient privacy (e.g., HIPAA laws) and the ability to return important clinical findings to individuals.
Subjects and Methods
More details of certain methods are included in the Supplemental Subjects and Methods.
eMERGEseq Panel Overview
Panel Design and Content
A gene panel comprising a total of 109 genes and 1,551 SNV sites was developed with input from eMERGE site investigators. The design process considered potential actionability of findings and local research interests, as well as gene size. The 109 genes included 56 based upon the American College of Medical Genetics and Genomics (ACMG) actionable finding list.2 Additionally, each site nominated 6 genes relevant to their specific aims, including discovery-focused genes with varying degrees of evidence for association with clinical phenotypes in need of further study. All nominated genes apart from titin (TTN [MIM: 188840]), which was excluded due to its large size, were included in the final panel design for a total of 109 genes. Further, eMERGEseq content included several categories of single-nucleotide variants (SNVs): (1) ancestry informative markers and QC/fingerprinting loci (n = 425), (2) a suite of SNVs selected to inform HLA type (n = 272), (3) pathogenic SNVs in genes not included on the panel for which return of results was planned (n = 14), (4) pathogenic or likely pathogenic SNVs in genes not included on the panel for which return of results was not planned (n = 55; for some, penetrance is poorly understood), (5) SNVs related to site-specific discovery efforts (n = 718), and (6) pharmacogenomic variants (n = 125), selected based on potential actionability, allele frequency, and space available on the platform. A summary of all eMERGEseq content can be found in Tables 2 and 3, with additional details provided in Table S1. All sequence and SNV data are shared across the network for research, and a subset of the content, namely the clinically actionable variants associated with disease or drug response, are included in clinical reports for return to the participants.
Table 2.
List of 109 eMERGE Genes, PGx, and Actionable SNVs
| Disease Category | Gene |
|---|---|
| Cancer susceptibility and tumor diseases | APC, BLM (rs113993962), BMPR1A, BRCA1, BRCA2, , MEN1, MLH1, MSH2 (including rs193922376), MSH6, MUTYH, NF2, PALB2,PMS2, POLD1, POLE, PTEN, RB1, RET, SDHAF2, SDHB, SDHC, SDHD, SMAD4, STK11, TP53, TSC1, TSC2, VHL, WT1 |
| Cardiac diseases | ACTA2, ACTC1, , , DSC2, DSG2, DSP, GLA, KCNE1, KCNH2, KCNJ2, KCNQ1, LMNA, MYBPC3, MYH7, MYL2, MYL3, PKP2, PRKAG2, RYR2, SCN5A, TMEM43, TNNI3, TNNT2, TPM1 |
| Cholesterol and lipid disorders | , ,, APOB, , LDLR, PCSK9, , |
| Endocrine disorders | CYP21A2 (rs6467), HNF1A,HNF1B, , |
| Connective tissue disorders | COL3A1, COL5A1, FBN1, MYH11, MYLK, SMAD3, , TGFBR1, TGFBR2 |
| Neuromuscular diseases | , , CACNA1S, RYR1 |
| Inborn errors of metabolism | ACADM (rs77931234), ALDOB (rs77931234), BCKDHB (rs386834233, rs79761867), FAH (rs80338898), G6PC (rs1801175), CPT2 (rs397509431), OTC, |
| Immunological/inflammatory disorders | , , MEFV (rs28940579, rs61752717), , |
| Neurological/psychiatric disorders | , , , , , , , , , , , |
| Respiratory disorders/hypertension | , , , |
| Renal disorders | , |
| Skeletal disorders | , |
| Other | F5 (clotting disorder; rs6025), (dermatological), HFE (iron storage disorder; rs1800562), (Pitt-Hopkins syndrome), (association with many complex disorders) |
| PGx SNVs | CYP2C9 (rs1799853, rs1057910), CYP2C19 (rs12248560, rs28399504, rs41291556, rs4244285, rs4986893, rs56337013, rs72552267, rs72558186), TPMT (rs1142345, rs1800460, rs1800462, rs1800584), SLCO1B1 (rs4149056), IFNL3/IFNL4 (aka IL28B; rs12979860), VKORC1 (rs9923231), DPYD (rs67376798, rs3918290, rs55886062) |
ACMG56 genes are indicated in italics without underlining, consensus site non-PGx TOP-6 genes are underlined, non-consensus TOP-6 genes are , and actionable SNVs are indicated by their rs number.
Table 3.
Additional Information on eMERGEseq SNVs
| SNV Category | Total |
|---|---|
| Ancestry | 241 |
| Fingerprinting | 184 |
| Pharmacogenomics | 125 |
| HLA (imputed) | 272 |
| Actionable clinically significant (P/LP) | 14(see above for more details) |
| Non-actionable clinically significant (P/LP) | 55 |
| Non-actionable, not clinically significant (VUS and below) | 660 |
| TOTAL | 1,551 |
Panel Sequencing
Reagents
The gene and SNV list was used to direct construction of targeted capture platforms at two sequencing centers (SCs): The Baylor College of Medicine Human Genome Sequencing Center (BCM-HGSC), Houston TX and the Broad Institute and Partners Laboratory for Molecular Medicine, Cambridge, MA. Broad used Illumina Rapid Capture probes for this panel and the BCM-HGSC used Roche-Nimblegen methods. Each group created in-solution capture probes spanning the entire targeted regions of the eMERGEseq panel. Probes were designed to be complementary to specified exons or SNV sites with a minimum span of 100 nucleotides. Tiling was limited to exonic sequence, and analyses included ±15 intronic flanking bases (Figure S1).
Sample Preparation
Clinical sites were requested to submit 2 μg of extracted DNA within a concentration range of 30–50 ng/μL. Although DNA derived from blood was the specified sample for the program, BCM-HGSC revalidated the clinical assay and accepted saliva as a DNA source for a limited number of cases due to clinical site requirements. Once received by the sequencing center, specimens were quantified using a picogreen assay, and quality was assessed by gel. Specimens with a minimum of 600 ng of DNA that did not display high levels of degradation passed sample QC and were accepted for eMERGEseq testing.
Ethics Approval and Consent to Participate
All 11 sample collection sites consented participants under Institutional Review Board (IRB)-approved protocols and the two sequencing centers had IRB-approved protocols that deferred consent to the participating sites. Protocol numbers are as follows: Partners Healthcare (2015P000929), Baylor College of Medicine (#H-40455).
Sequencing and Primary Analysis
Samples from DNA capture using the custom capture reagents were sequenced using standard Illumina technologies. Post-sequence processing at each site utilized preferred alignment and variant calling algorithms. The variant calling pipeline at Broad incorporates Picard deduplication, BWA alignment, and GATK variant calling for SNVs and short indels.3 At the BCM-HGSC, alignment using BWA-MEM and variant calling using Atlas were instantiated within the Mercury Pipeline.4
Panel Fill-in
A common set of reference samples were initially sequenced at each SC. The chosen parameters to monitor performance were coverage of targeted sequence and percentage of the targeted bases at or above 20× coverage. Both groups sequenced cohorts of control samples and identified systematically poorly covered bases as those with less than 20× coverage in >10% of tested samples. Based on this conservative threshold, both groups went through a process of enriching with more targeting probes (“fill-in”), to boost underperforming regions, prior to final validation. The reagent performance is described in Table 4, with additional details in Table S2.
Table 4.
Assay Performance and Optimization at the Sequencing Sites
| BCM-HGSC |
Broad |
|||||
|---|---|---|---|---|---|---|
| Acceptance Criteria | Original | Low Input | Acceptance Criteria | Measured at ∼250× MTC | Measured at ∼400× MTC | |
| Assay sensitivity (SNV + indel) | 100% | 100% | ≥95% | 100% | 100% | |
| Assay sensitivity (CNV) | 97.7% | 98.3% | n/a | 100%a | n/a | |
| Assay specificity (point variant + indel) | 100% | 100% | ≥95% | 100% | 100% | |
| Assay reproducibility | ≥95% | >98% | >97% | ≥95% | 98.5% | 99.6% |
| % of >20× coverage for targeted regions | ≥99% | >99% | >99% | ≥95% | 99% | 99% |
| Depth of mean coverage | >200× | >200× | >200× | n/a | ≥250× | ≥400× |
CNV sensitivity at Broad/LMM is for events ≥3 consecutive exons.
Copy Number Variant (CNV) Calling
CNV calling at Partners/Broad was performed using VisCap, which infers copy number changes from targeted sequence data by comparing the fractional coverage of each exon in a gene to the median of these values across all samples in a given sequencing run.5 BCM-HGSC CNV calls were made via Atlas-CNV, an in-house software that combines outputs from XHMM6, 7 and the GATK DepthOfCoverage tool.6 Like VisCap, Atlas-CNV infers the presence of CNVs from normalized coverage differences to other samples in the same sequencing batch and refines these predictions with a pair of quality control metrics.8 CNV calls were confirmed by orthogonal technology: Droplet Digital PCR (Bio-Rad) at Partners/Broad and Multiplex Ligation-dependent Probe Amplification (MRC-Holland) at the BCM-HGSC. Detected CNVs were filtered based on the clinical site’s gene reporting preferences and ClinGen haplosensitivity and tri-sensitivity scores (see Gene Dosage Curations in Web Resources) and then manually reviewed. Partners/Broad required a minimum of three contiguous exons for reporting; BCM-HGSC required two.
Analytical Validation
To validate sensitivity, specificity, and reproducibility of the eMERGEseq panel, the performance of both SCs was compared using a common reference sample (NA12878). In addition, each group separately examined previously tested clinical samples, containing known pathogenic variants that were uniquely available to their laboratory. Subsequent additional validation analyses were performed to accommodate lower DNA input amounts, based on sample availability (BCM-HGSC).
Ongoing Proficiency
Ongoing proficiency testing to monitor laboratories’ continuing performance for the eMERGEseq panel involved interlaboratory exchange of previously tested eMERGE samples as part of a proficiency testing program for general sequencing platforms, with all results being concordant to date (see the Supplemental Subjects and Methods for further details).
Variant Interpretation
General Approach to Interpretation
Variant classifications from both laboratories were based on ACMG/Association of Medical Pathology (ACMG/AMP) criteria9 with ClinGen Sequence Variant Interpretation Working Group modifications as well as additional specifications for some of the eMERGEseq genes as established by ClinGen Expert Panels (see Sequence Variant Interpretation in Web Resources). Additional local data accrued from previous case studies were combined with manual literature and public data review for final decisions. Non-ACMG 56 genes underwent an in-depth clinical curation effort using the ClinGen framework for gene-disease validity assessment,10 followed by actionability assessment by the eMERGE Clinical Annotation Working Group (WG). The WG included more than 6 active MDs (including clinical geneticists) and more than 6 members with clinical laboratory genetics training, among approximately 50 members. The gene-disease pairs were presented at in-person and teleconference meetings attended by WG members of each site. The WG created the consensus list that all sites considered actionable.
Legacy Variant Interpretation
In order to harmonize prior interpretations and to assess likely ongoing differences, the BCM-HGSC and Partners LMM exchanged data from 1,047 previously interpreted variants in the 109 eMERGE genes and evaluated discrepancies (see Results).
Ongoing Harmonization
Monthly data exchanges identified any differences of interpretation of non-PGx variants intended for clinical reporting. These discrepancies were reviewed during a bi-weekly interpretation/harmonization teleconference call. Cases of unresolvable variants were presented to the eMERGE Clinical Annotation WG to attempt resolution and/or track their occurrence. All reported variants are submitted to ClinVar along with their interpretations.
Pharmacogenomics (PGx)
The SCs worked with the eMERGE PGx working group to select variants to be included on the clinical reports provided to participants, to interpret diplotypes, and to select drugs for therapeutic recommendations, guided by the Clinical Pharmacogenomics Implementation Consortium (CPIC) guidelines (Web Resources). Twenty PGx variants in seven genes were deemed to be clinically actionable and were therefore selected for return to participants. Table S3 includes details of the PGx genes and variants reported and the drugs associated. For two PGx genes, CYP3A5 (MIM: 605325) and SLCO1B1 (MIM: 604843), the gene panel included only one of three variants discussed in the CPIC guidelines. CYP3A5 was deemed not reportable, as two SNVs important for predicting phenotype for African Americans and Latinos are not included on the gene panel. SLCO1B1 was deemed reportable, as the one SNV included in the panel serves as a tag SNV for the remaining two SNVs.
The BCM-HGSC included PGx results on individual patient reports, while Partners LMM produced a batch report that accommodates one to hundreds of patients for bulk consumption and EHR integration by sites. Sample PGx reporting formats can be found in the supplemental data (Sample Clinical Reports and Table S8). The CPIC drugs that were included in the PGx report were largely the same with some minor differences (see Table S3).
Data Management
Sample Intake
Each site was provided barcoded tubes by the SC for DNA shipping. Sample identifiers and metadata were uploaded using an “eMERGE requisitioning sheet” via secure portals (see eMERGE Sample Submission Portal and Clinical Research Sequencing Platform in Web Resources). The requisitioning spreadsheet contains fields for sample information (name [optional], sex, date of birth/age, US state of residence, site-specific ID), as well as eMERGE-specific metadata including patient “disease area” (from a list defined by the network, see Table S4 details), disease status and test indication, eMERGE project ID, and barcode number on the tube. An additional option was to add phenotype terms in a free-text field, primarily based on the MonDO ontology and occasionally additional local codes largely derived from Human Phenotype Ontology (HPO) terms (Table S4). A simple .csv file structure was used by both SCs so that sites could upload all metadata at the time of sample batch shipment. For the BCM-HGSC SC, the sample accession was directly into a cloud environment, managed by DNAnexus, while for the Partners-Broad SC, a custom portal operating in the Broad’s local environment was employed for intake followed by transfer to the GeneInsight system for analysis and reporting, with all systems being HIPAA compliant. Local identifiers were then generated to track the samples as they progressed through DNA sequencing and variant calling. Orders were reviewed and approved by the SCs prior to sample shipping and accession. Upon receipt, the samples were subjected to volume and concentration quality control checks.
Data Delivery and Reporting
Each SC developed custom reporting methods (see Supplemental Subjects and Methods for examples). Partners/Broad site users have a unique, password-protected account and are able to view only orders and metadata from their own site. The Broad portal authorization procedures are customized to allow for secure transfer of sequencing output files and metadata to both Partners and DNAnexus via APIs. The BCM-HGSC sites are delivered reports from the DNAnexus environment via DNAnexus APIs. Users were provided individual logins for accessing PDF reports and structured content in a harmonized .xml format.
GeneInsight
Partners/Broad sites used the commercial tool, GeneInsight (Sunquest Information Systems), for local report management.11 This tool was configured to create a De-identified Case Repository (DCR) which contains a de-identified record of all cases and associated variants from both Partners/Broad and the BCM-HGSC supported sites.
DNAnexus Data Commons
The BCM-HGSC clinical sites were provided with two data access points in the DNAnexus infrastructure. One provides a restricted space for accessing protected health information (PHI)-containing clinical reports, while another acts as a general space for the de-identified records of each case and associated variants. Users were provided individual logins and selectively granted access to one or both access points. Data for sites that were served by the BCM-HGSC were provided both .xml and .pdf formats, at the time of reporting. De-identified, structured versions of the Partners-Broad reports are downloaded from the DCR and also stored in the DNAnexus Data Commons projects, creating a comprehensive repository of de-identified clinical reports.
Variant Updates
Two complementary mechanisms were developed to enable delivery of variant updates from the SCs to the sites as new evidence leading to a classification change becomes available. At Partners/Broad, individual participant results are stored in an eMERGE-specific instance of the GeneInsight database that is linked to Partners LMM’s GeneInsight instance enabling communication of variant updates.12 If Partners updates a variant, sites that have signed up receive proactive notification emails if a reported variant identified in one or more of their cases is updated. Hyperlinks are provided in those emails that allow sites to directly access updated information on the variant in each case, which facilitates the choice to return an updated result to a participant. In addition, Partners is generating an .xml file for each variant interpretation change alert, which sites can consume through other electronic interfaces. At the BCM-HGSC, participant results are stored in a database that is routinely queried for variants with new actionable interpretations. If such a variant is found in a previously reported sample, an amended report is issued via DNAnexus and sites are notified. Variant updates are included in the ongoing variant interpretation harmonization process described above.
eMERGE III Samples and Raw Data Storage
Results were analyzed from the eMERGE III eMERGEseq data, which consisted of 25,015 samples. These included 14,515 from Baylor and 10,500 from Partners-Broad. The associated BAM, xml, and vcf files are available on the eMERGE Commons, accessible to sites as well as outside investigators who apply for access (see eMERGE Network in Web Resources). Data are also being submitted to dbGaP for controlled public access (phs001616.v1.p1).
Results
Network Overview
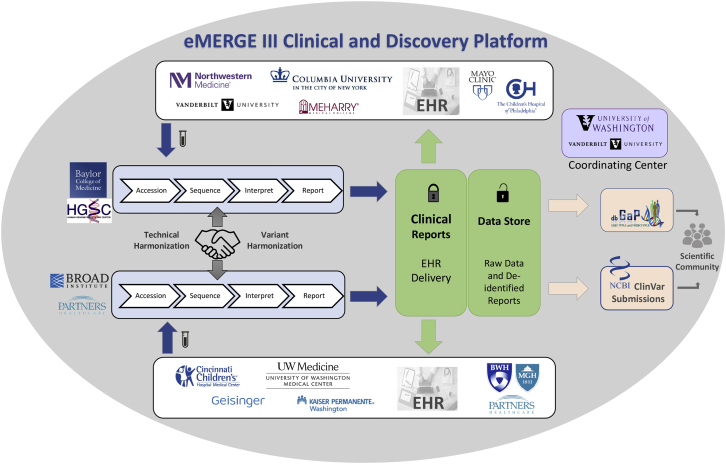
The eMERGE III network established a Clinical and Discovery Platform that consists of 11 clinical study sites, 2 DNA SCs, and a coordinating center (CC) (Figure 1). Participants were enrolled at each site, where blood was collected, and DNA was extracted locally and sent to one of two SCs for targeted sequencing. Analysis and interpretation of the DNA sequence data was performed at each SC, and the data were returned to the clinical sites for return to participants. Raw data were accrued for data mining purposes by eMERGE investigators and approved affiliates. Subsequently, raw data are released to dbGaP and interpreted variants to ClinVar.
Figure 1.
eMERGE III Network Overview
The eMERGE III network is comprised of 11 study sites, 2 sequencing centers (SCs), and a coordinating center (CC). The different components and processes involved in the data flow across both the clinical and research discovery arms of the network are highlighted in this figure and described in more detail in Network Overview.
An early decision of the program was to utilize DNA capture “panels” of approximately 500 kb, in order to generate genomic data from the eMERGE participants, as an alternative to whole-exome sequencing (WES) or whole-genome sequencing (WGS). This choice reflected a balance between available fiscal resources and a reasonable selection of content to explore return of actionable results and focused discovery efforts. It should be noted that there are other efforts within eMERGE to support discovery from research platforms, including more than 100,000 GWAS arrays and more than 5,000 exome and genome sequences generated to date. However, this effort was distinct in focusing on a CLIA platform intended for clinical return of actionable results. The use of the panel enabled testing of 109 genes and 1,551 additional sites of single-nucleotide variation in each sample. Across the network, ∼25,000 samples were assayed, ∼2,500 from each site (Table S5). The study is therefore large enough to allow robust analysis of specific phenotypes as well as to gain experience with a sufficient number of patients at each site to develop processes to support the return of actionable genetic results.
Prior population studies suggested that the genes included on the panels would reveal thousands of newly identified single-nucleotide and structural variants. A small subset of these would be expected to be pathogenic, and the program aimed to report to participants only those variants that were pathogenic or likely pathogenic according to the ACMG/AMP guidelines9 or those with actionable pharmacogenomic associations. Each site would have the option of a customized clinical reporting framework, as well as full access to all network data to guide decisions and harmonize interpretations.
This elaborate network reflects a real-world situation, where a full complement of testing, reporting, and research require coordination and harmonization of many components. First, the selection of gene targets and the rules for reporting must agree. Next, the technical aspects of DNA capture and sequencing required standardization and ongoing comparison. The DNA changes must be interpreted and reported with the same conclusions, regardless of where testing occurred. Finally, file structure standardizations and data management practices must be organized. A detailed list of components (Table 1) that require coordination and harmonization illustrates the magnitude of the challenge.
Technical Validation of Capture Panels
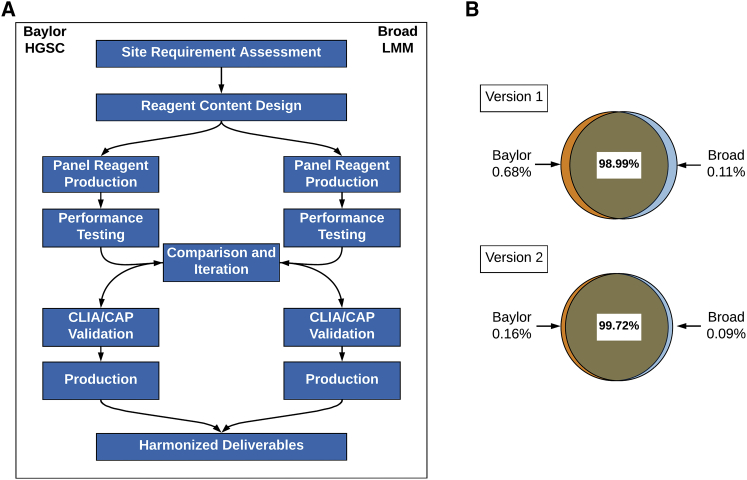
Coordination and harmonization of the DNA capture panel process at the two CAP/CLIA-certified DNA sequencing laboratories was demanding because in addition to different DNA capture reagents, the local processes of sample preparation, library construction, hybrid capture, and sequencing represented complex workflows with many variables. As an alternative to compelling each laboratory to adopt unfamiliar methods, the harmonization was achieved through phases of coordinated design, comparing initial high-level technical performance, and via ongoing monitoring of proficiency (Figure 2A). The harmonization process aimed to reduce any impact on the overall program due to the heterogeneity of capture reagents or sequencing methods between two sites and for the end users to be able to compare data from each laboratory without batch effects.
Figure 2.
eMERGEseq Panel Test Development and Validation
(A) Technical harmonization of two DNA capture panels. Coordination and harmonization of all the components of the DNA gene capture panel process at the two sequencing centers.
(B) Base coverage. Percentage of bps covered ≥20× across sequencing centers. Percent of bases in the panel targeted region covered in each version of the panel design and the extent to which these bases overlap between the genome centers is shown. Version 2 is the final version used for data generation.
Design was coordinated by first agreeing on the intended limits to reporting, e.g., number of bases adjacent to exons to be reported (see Subjects and Methods and Figure S1). Each laboratory employed slightly different criteria for the selection of the range of transcripts to be tested, reflecting a lack of harmony of public databases. Possible differences in design were resolved by selection of the union of all possible exons to be considered and validated by iterative sharing of the capture design files (“bed files”). The detailed design specifications can be found in Table S1.
Preliminary testing of the technical performance of the two capture reagents utilized both local test samples and a shared sample reference set (see Subjects and Methods). The technical performance was shared between the SCs by measuring the coverage of individual bases and other key technical metrics (Tables 4 and S2). Overall sequence coverage goals and the extent to which poorly covered regions could be tolerated were agreed upon a priori, and the technical comparison was straightforward between SCs. In general, the sequencing reagents performed well, although the presence of some uncovered bases in the first panel designs led each group to modify the initial reagents to optimize performance (Figure 2B). Throughout, the comparative performance of the two reagents informed the progress of technical development and illustrated the synergism from closely monitoring similar processes.
For final validation, both groups measured overall sensitivity and specificity on a reference sample (NA12878) as well as sensitivity to detect known pathogenic variants from previously tested clinical samples that were uniquely available to them. Groups also incorporated evaluation of variance in processing including varying coverage from ∼250× to 400× (Broad) and input amounts of 250 ng and 500 ng (Baylor). Summary results of the respective validation studies are shown in Table 4. Panel optimization results and coverage analyses can be found in Table S2. The impact of the ∼0.2% of targeted bases that were not effectively covered via the optimized panel designs was evaluated by the network for impact on clinical decision making. The majority of missing data was judged to be of little consequence although small regions of some genes (e.g., RYR1 [MIM: 180901], CACNA1B [MIM: 601012]) could not be recovered by either platform (Table S2).
Once the data production phase of the program was initiated, the ongoing performance was monitored by sharing production metrics and via the ongoing CAP/CLIA proficiency program that included exchange of samples and comparison of DNA variation data. As of this publication, mean coverage of Broad production samples is ∼420×, percent of targeted bases covered ≥20× is 99.7%, and percent of targeted bases with zero coverage is 0.17%. These metrics, collected from >7,000 production samples, closely match the performance of the validation set. Mean coverage of the BCM-HGSC production samples is ∼340×, percent of targeted bases covered ≥20× is 99.8%, and percent of targeted bases with zero coverage is 0.04%. These metrics, collected from >9,600 production samples, also closely match the performance of the validation set.
eMERGE III Cohort
The eMERGEseq cohort is comprised of 25,015 biobank or prospectively recruited participants representing 11 eMERGE sites. These were either unselected for any specific phenotype or were enriched for specific phenotypes depending on site-specific clinical and research interests. A brief summary of the nature of each site-specific sample repository, including the total number of participants per site can be found in Table S5. A more detailed description of the clinical cohorts involved in this study, including enrollment criteria, are reported elsewhere (A. Gordon et al., 2018, American Society of Human Genetics, abstract).
Genetic Ancestry
Genetic ancestry within the diverse eMERGEseq dataset was determined by using common variants throughout the eMERGEseq panel, including ancestry informative marker SNVs. Principal component analysis of genetic ancestry (Figure S2) and qualitative comparison to self-reported ancestry (Table S6) were performed as a part of various quality control analyses applied on the cohort. The self-reported race and genetically determined race appear to generally match.
Clinical Content Validation and Site-Specific Return of Results Plans
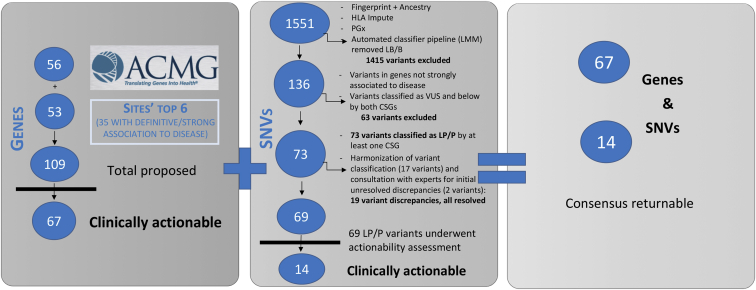
Gene selection by sites for inclusion on the eMERGEseq panel was driven by both clinical and research needs leading to a final list for panel design of 109 genes, including the “ACMG56”2 and 53 additional site selected genes. Evidence review using the ClinGen gene-disease validity framework identified 35 of the additional 53 genes as having definite or strong association to disease. These genes were considered for further actionability analyses (see Figure 3). Most of the 18 genes with lower levels of validity were included by sites to enable research on these genes, reflecting the diverse goals of the eMERGE network including discovery as well as return of results.
Figure 3.
Content Development for the eMERGEseq Panel
Left: ClinGen gene-disease validity assessment for all site top six proposed genes. Those with definite and strong association to disease were considered for further actionability analyses.
Middle: Clinical assessment for a subset of single nucleotide variants (SNVs). Those deemed P/LP were considered for actionability analyses.
Right: Final consensus list of returnable content. This included all the ACMG56 genes, in addition to 11 genes and 14 variants that were deemed actionable by the eMERGE Clinical Annotation Working Group.
A subset of the genotyping SNVs were also evaluated for possible return. This excluded 1,415 SNVs submitted for HLA analyses, fingerprinting, and ancestry typing or already designated for PGx return. Of those remaining, some had been previously classified as likely benign or benign and were thus excluded from further analyses of potential pathogenicity. The remaining 136 variants were considered for further clinical assessment. Seventy-three variants were classified as either likely pathogenic or pathogenic by at least one of the SCs. Of these, 19 had discrepant classifications between the two SCs. These were resolved by variant re-assessment and scoring on published evidence as well as combined internal evidence from both SCs. For two variants, the eMERGE Clinical Annotation WG was consulted to assist in resolving interpretation differences. A final list of 69 pathogenic/likely pathogenic (P/LP) variants was established and further considered for actionability analyses (Figure 3).
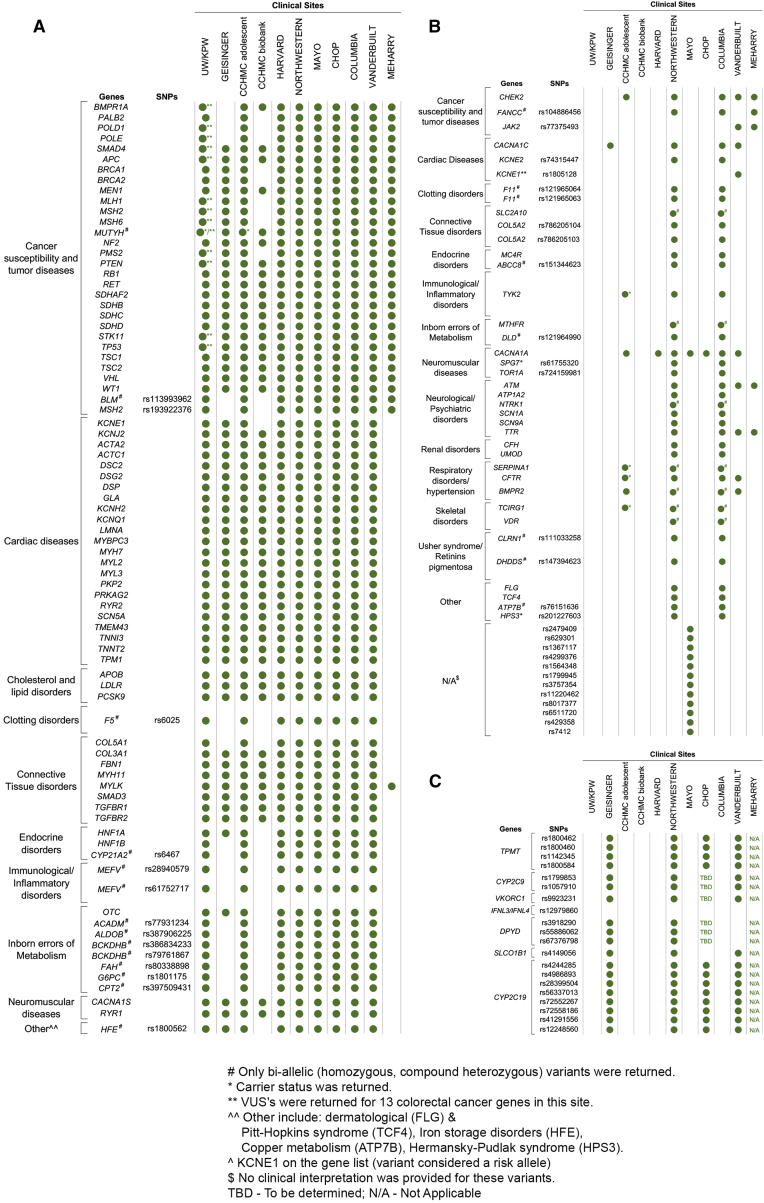
The eMERGE Clinical Annotation WG evaluated the medical actionability of the 35 non-ACMG56 genes for which we had applied ClinGen criteria and defined as having at least one strong/definitive disease association, as well as 69 P/LP pathogenic variants, based on whether there was a substantially increased risk of serious disease that could be prevented or managed differently if the risk were known. In addition to the ACMG56, 11 genes and 14 variants were deemed actionable by the eMERGE Clinical Annotation WG and placed on a consensus list of returnable content (Tables 2 and 3, Figure 3). While sites agreed that this list represented content that would generally be medically actionable in adults, some sites did not return results from all genes on the consensus list and/or chose to return additional content based on their research interests, patient populations, and IRB-approved return of results protocols (Figure 4). For example, not all sites chose to return HFE (MIM: 613609) p.Cys282Tyr homozygotes. Additionally, of the 11 sites, one that included pediatric biobank participants opted not to report variants in genes that increase risk of adult-onset diseases but are not actionable during childhood. Another site limited its actionable gene-disease pair return list to cancer-associated genes. Four other sites requested return for additional genes and SNVs that were not on the consensus list, again due to study differences. For example, a clinical site whose research included the creation and return of a polygenic risk score requested genotypes at 12 SNP sites associated with low-density lipoprotein cholesterol (LDL-C) risk be included on their report. Another site returned variants of uncertain significance in 13 colorectal cancer (CRC [MIM: 114500])-associated genes for a subset of their samples derived from a cohort of participants with CRC or polyps. A full list of the content that was returned for each site can be found in Table S7 and summarized in Figure 4.
Figure 4.
Site-Specific Reportable List of Genes/SNPs for Which Pathogenic or Likely Pathogenic Variants Will Be Returned
(A) Consensus list of returnable SNPs/genes. Inclusions are indicated with a green dot and exclusions are indicated by no dot.
(B) Site-specific list. Non-consensus genes/SNPs with site-specific inclusions indicated with a green dot
(C) Site-specific PGx list. Pre-determined SNPs for return in PGx genes. Inclusions are indicated with a green dot. UW/KPW, University of Washington/Kaiser Permanente Washington; CHOP, Children’s Hospital of Philadelphia; CCHMC, Cincinnati Children’s Hospital Medical Center
For PGx returnable content, 20 variants in 7 genes were deemed clinically actionable by the PGx working group, yet only 4 sites chose to return PGx results to participants, and for those that did, they did not return results from all genes (Figure 4). For example, none of the sites elected to return diplotypes associated with IFNL3/IFNL4. Return of PGx results was in part influenced by which sequencing center was assigned to a site, due to differences in the types of reports being issued (PGx results included on individual patient reports for BCM-HGSC versus separate batched reports with PGx results from Partners-Broad).
Data Intake and Delivery
Data intake and delivery represented challenges for the network due to the plan to test distributed, heterogeneous EHR systems and other data sources used by sites and the need to deliver updated data interpretations. All demands were required to be met while managing issues of compliance and security for PHI protection. These challenges mimicked real-world situations as these are identical needs for any health care organization opting to interact with a research enterprise or reference laboratory. The data management required the development of three main informatic components: data intake, clinical reporting, and the de-identified case repository and data commons.
Firstly, data intake and accessioning for each site was facilitated by an agreement of the specific PHI metadata to be supplied with each sample, as well as an agreement of a set of required “indications for testing” that represented the primary phenotype data that tracked each sample through the network (see Subjects and Methods).
The second is clinical reporting. Within each pipeline, the standard validated product was a PDF report that was returned to the clinical investigators (see Supplemental Data for examples of reports). Each clinical site had custom requirements for the report content that reflected local preferences for data to be returned to patients. Each SC also had different reporting requirements; for example, some sites requested negative reports, others returned only positive reports.1 Most sites also requested data in structured formats to enable direct integration onto their local EHRs (see Data S1 and S2 for examples).
The five clinical sites served by the Partners-Broad SCs received results delivered through the GeneInsight platform, which enabled storage and query of clinical reports. The six sites served by the BCM-HGSC utilized custom applications developed for report delivery. Possible difficulties in data sharing between different parts of the network were anticipated and obviated by development of an agreed .xml standard. This standard was based upon the GeneInsight system specifications and facilitated communication across all components (see Subjects and Methods and Aronson et al.13). The clinical sites therefore had two options—they could either use a stand-alone tool for report data management or alternatively the report data could be parsed into local customized systems.
For those sites using the GeneInsight platform, automated alerts were delivered immediately upon LMM variant reclassifications that affected an eMERGE report. Most alerts then led to requests for report amendments with a total of 16 amendments delivered by LMM for 7 variant reclassifications to date. In addition, ten amendments were issued by BCM after routine queries for variant updates. For PGx data, in addition to receiving results in PDF reports (either individual reports by the BCM-HGSC or batch reports by Partners-Broad), a standardized data format was also developed to deliver structured PGx data in the form of both variant level and diplotype results allowing sites to directly integrate PGx results into the EHR for clinical decision support.
Finally, the network required all deidentified data to reside together, to enable data mining for both basic research and to better inform clinical decision making with access to larger clinical datasets. There were two independent but complementary mechanisms for this. First, the GeneInsight tool maintains a record of all returned variant data from both sites in a de-identified case repository allowing an easy search interface for clinically reported variants. A second site maintained the full set of eMERGE raw data in a cloud environment, managed by DNAnexus. This “eMERGE Commons” was structured to house each DNA sequence file in the BAM format, as well as the annotations for the data in a vcf format. As clinical report delivery for the data generated in the Baylor SC also utilized the DNAnexus infrastructure, the full set of identified clinical reports and de-identified raw data were both resident in the cloud. The access permissions for the data were managed to allow only the clinical providers to access their patients’ clinical reports. The full set of raw data was available to all eMERGE investigators after PHI information had been removed.
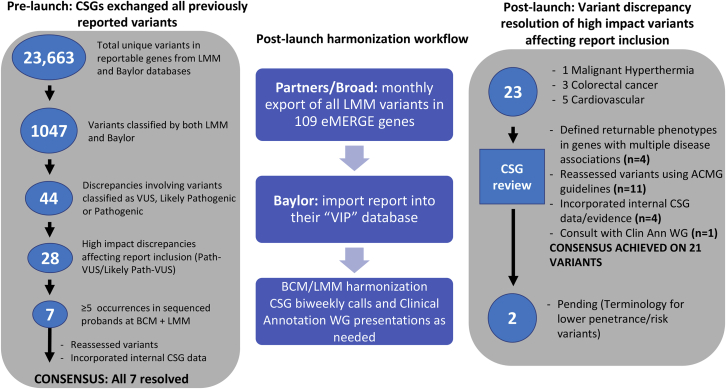
Variant Interpretation Harmonization
To ensure consistency of results being returned across the eMERGE consortium, variant interpretation was harmonized between the SCs (Figure 5). In a pre-test launch, both SCs exchanged variants in reportable genes from their respective databases, totaling 23,663 unique variants. Of those, 1,047 were previously classified by both SCs. The pre-test launch data exchange showed 90% concordance in variant classification among variants classified as VUS, likely pathogenic, and pathogenic by at least one SC. When likely pathogenic and pathogenic variants were grouped together, the concordance was 93%. When all variant classifications were considered, including benign versus likely benign, the data showed a 67.5% concordance. However, only 28, or 3% of the variants were deemed to affect reporting (VUS versus pathogenic 1.9%, VUS versus likely pathogenic 1.1%). The two SCs resolved all differences that would affect inclusion on clinical reports (i.e., P/LP versus VUS).
Figure 5.
Variant Harmonization Process Overview
Pre-launch and post-launch harmonization processes involving the exchange of variants in reportable genes between the sequencing centers and the identification, prioritization, and the resolution of discrepancies affecting report inclusion.
An ongoing process was also developed to ensure continuous harmonization of variant interpretation (Figure 5). As of May 2018, 23 initial discrepancies of interpretation of variants from five disease areas were considered, based upon potential to affect report inclusion. Most discrepancies in variant interpretation (83%) were immediately resolved when re-assessed by the SCs by using ACMG guidelines, incorporating additional laboratory-specific evidence, after defining returnable phenotypes in genes with multiple disease associations (for example malignant hyperthermia [MIM: 145600] versus myopathy [MIM: 117000] for RYR1), or defining terminology for lower penetrance/risk variants. For one variant, resolution required input from additional eMERGE investigators through the eMERGE Clinical Annotation WG.
Three variants (p.Ile1307Lys in APC [MIM: 611731], p.Met54Thr in KCNE2 [MIM: 603796], and p.Asp85Asn in KCNE1 [MIM: 176261]) were noteworthy as the interpretations were more discrepant upon initial assessment (i.e., “two-steps:” pathogenic versus likely benign), although the evidence used by both centers was identical. These represented variants that have significantly reduced penetrance, leading to difficulties applying the ACMG/AMP classification framework, which is designed primarily for highly penetrant Mendelian disorders. Nevertheless, some sites chose to return the APC variant as it imparts a 2-fold risk of CRC in Ashkenazi Jewish individuals, even though its effect in other populations in unclear. Other sites elected to return the KCNE2 variant, as it has been associated with variable presentations such as arrhythmias (MIM: 611493) and long QT syndrome (MIM: 613693).14, 15, 16 This type of classification discordance highlights the need for guidance on classification terminology for low penetrance variants for not only the eMERGE network but for the entire medical genetics community.
Aggregate Findings and Return of Results
A total of 8,437,788 variants were detected among the 25,015 case subjects that have been collected and analyzed via the eMERGEseq panel. A subset of these were excluded from further analyses due to a LB/B classification by the SCs or by an auto-classification pipeline based on allele frequency thresholds or for having a low-quality score. The remaining variants underwent a filtration process which returns (1) predicted loss-of-function variants with a minor allele frequency (MAF) < 1%, (2) variants previously classified by the SCs as likely pathogenic (LP)/pathogenic (P) regardless of MAF, and (3) ClinVar P/LP as well as HGMD “DM” variants with a MAF < 5%. This pipeline resulted in 9,653 unique variants requiring further assessment. After expert review, these were further categorized as benign (1%), likely benign (8%), VUS (69%), LP (7%), P (12%), or deemed as low penetrance risk alleles (0.5%). In addition, 205 unique copy number variants have been detected across the reviewed samples, with 141 gains and 64 losses. Of these, 30% were deemed reportable and were returned to sites. In summary, these data led to a total of 1,497 case subjects that have a LP/P variant that would require a positive report to be issued.
Results being returned to sites currently fall into three categories: (1) indication-based returnable results that include all sequence and copy number variants related to the site-provided indication for testing, (2) non indication-based consensus returnable results that include all sequence and copy number variants in genes and SNVs comprising the consensus list of returnable content (see Clinical Content Validation and Site-Specific Return of Results Plans) that are not related to the indication for testing, and thus considered secondary findings, and (3) non indication-based site-specific returnable results which include variants in additional site-requested genes that are not on the consensus list and not related to the indication for testing. Additionally, both SCs are returning results on pre-selected PGx SNVs as either an addendum to individual patient reports or in a batch report that contains up to ∼185 samples (see Subjects and Methods).
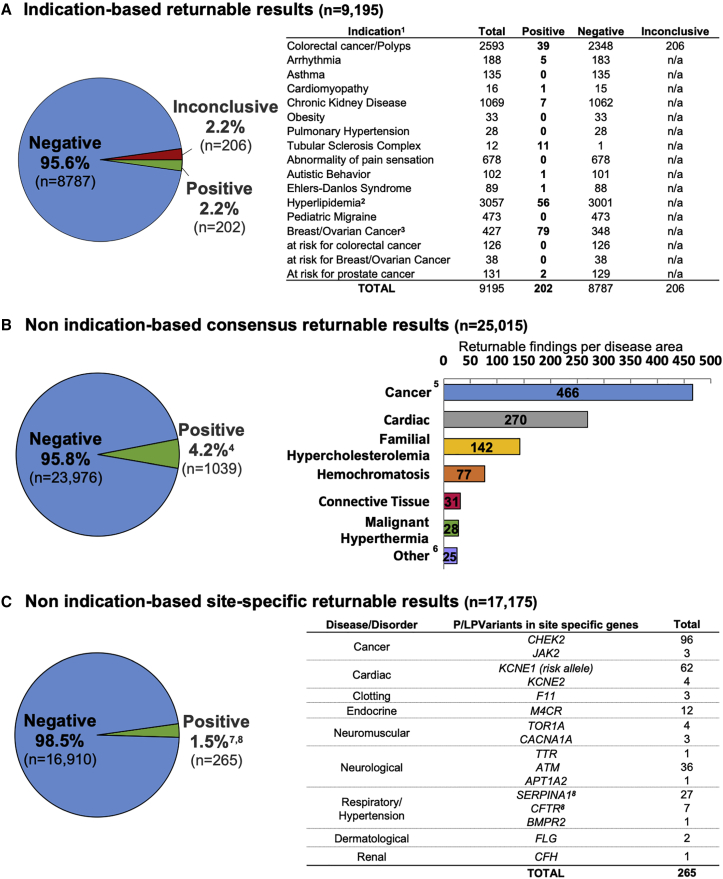
The positive rate for each category of findings is depicted in Figure 6. For all 25,015 case subjects that have been reviewed, 9,195 (37%) had an indication for testing. Of these, 202 (2.2%) had positive findings relevant to the indication for testing (Figure 6A). Moreover, of all individuals sequenced, 1,039 (4.2%) had additional/secondary findings of medical significance in genes and SNVs from the consensus list, that are being returned to participants (Figure 6B). 17,175 participants (69%) were enrolled in sites who were interested in returning pathogenic and/or likely pathogenic variants in additional genes or SNVs that were not on the consensus list. In 265 cases (1.5%), a non-indication based, site-specific returnable pathogenic or likely pathogenic variant was identified (Figure 6C). 37% of these variants were in CHEK2 (MIM: 604373), a tumor suppressor gene, and are associated with an increased risk for a variety of cancers. A full list of all positive findings returned to participants with and without indications are listed in Table S10.
Figure 6.
Aggregate Findings Returned to Sites
The positive rate for each category of returnable findings for all 25,015 participants from the eMERGE III study is shown.
(A) Indication-based returnable results. For those with an indication for testing, the different indications are depicted. 1Four positive and two inconclusive reports had an additional secondary finding; 2587 patients had colorectal cancer and hyperlipidemia;3findings from 67 consensus genes except for 2 in CHEK2.
(B) Non indication-based consensus returnable results. Secondary findings from the consensus gene list across the entire eMERGE III cohort are broken down per disease area. 414 reports had two pathogenic variants. Skewed positive rate due to one site with sample selection based on suspicious genotype (11% positive); 5colorectal cancer (40%), breast/ovarian cancer (37%), other cancers (22%); 6other: includes immunological/inflammatory disorders, inborn errors of metabolism, endocrine disorders, neurological disorders, clotting disorders, Myhre syndrome, and neuromuscular diseases.
(C) Non indication-based site-specific returnable results. For a subset of participants, the number of pathogenic and likely pathogenic variants in site-specific additional genes that are not on the consensus list are shown. 7Ten participants had a site-specific variant and an additional consensus returnable variant. Of these ten site-specific variants returned, three were relevant to the indication for testing and seven were non-indication-based findings; 814 SERPINA1 and 5 CFTR variants were reported as carrier status.
Other variants from consensus list genes and SNVs that were not related to the indication of testing were associated with cancer, cardiac disease, familial hypercholesterolemia (MIM: 143890, 144010, 603776) and hemochromatosis (MIM: 235200) (Figure 6). For indication-based assessments, detection rates were highest for breast/ovarian cancer (MIM: 114480, 604370, 600185) (39%), hyperlipidemia (28%), and CRC/polyps (19%). Some phenotypes had no disease-causing variants identified due to either the absence of genes causative for the disorders on the eMERGEseq panel or the lack of a clear monogenic disease etiology for the disorder (e.g., abnormality of pain sensation [MIM: 243000], pediatric migraine [MIM: 188840]). The rate of P/LP variants detected in participants without a clinical indication differed from site to site, ranging from 2% to 11%, depending upon the basis for participant selection, which were reflective of the underlying study designs of the individual sites. The overall positive rate for secondary findings was skewed higher for one site (Geisinger), where a subset of participants were preselected for a suspicious variant(s) previously identified in an exome study.17 On the other hand, two sites had lower rates than expected either because their cohort had an indication related to genes in the secondary findings list that led to the removal of these genes from secondary findings reporting or because the site did not choose to return all results from the consensus list. When data from Geisinger participants preselected for suspicious variants were excluded, the frequency of secondary findings was similar across sites, ranging from 1.8% to 5.1%, suggesting that the complexity of the network did not otherwise distort these results, and reflecting the success of the data and process harmonization. A further analysis of the factors that influence the rate of secondary findings return is underway (A. Gordon et al., 2018, American Society of Human Genetics, abstract).
For PGx results, reports depicting genotype and related diplotype data, including whether the reported diplotype for each gene and resulting phenotype would result in a recommendation to modify dosage, have been issued for all participants from 11 sites. The frequency of the reported diplotypes were concordant with the CPIC published frequency tables for each major race/ethnic group (see CPIC in Web Resources). One difference for diplotype interpretation was particularly illustrative of the role of harmonization. When both rs1800460 and rs1142345 are identified in TPMT (MIM: 187680), it cannot be ascertained whether these variants are in cis, resulting in a TPMT∗1/∗3A diplotype and intermediate metabolizer phenotype, or in trans, resulting in a TPMT∗3B/∗3C diplotype and a poor metabolizer phenotype. One SC emphasized the more common diplotype in their report, while the other emphasized the higher risk of the rarer diplotype under some drug regimens. With input from the sites and the eMERGE PGx working group, it was decided that the more common genotype would be reported with a warning that the rarer genotype could not be ruled out.
Across the 20 loci (7 genes) and 11 drug types, diplotype analysis prompted recommendation for potential non-standard drug dosing in at least one drug in 93% (23,232/25,015) of participants. Overall, the percentage of participants with actionable PGx results, resulting in a recommendation to potentially adjust standard drug dosing or use of an alternate drug based on their metabolizer phenotype, ranged from 2% (for DPYD [MIM: 612779] genotypes associated to response to Fluoropyrimidines) to 57% (for IFNL3 [MIM: 607402]/IFNL4 [MIM: 615090] genotypes associated to response to pegylated interferon-α (PEG-IFN-α) and Ribavarin). Site-specific PGx results across all tested genes leading to potential dosage adjustment recommendations for 11 drug types can be found in Table S9.
The majority of returned data reflected variants with relatively clear interpretations for participants, with variants that either had a large body of published evidence or were straightforward to interpret. However, understanding the actual risk to patients to develop disease in those without an indication is more challenging, with risk being dependent on what is known about the penetrance of disease for the gene and variant as well as other individual factors such as family history and environmental factors (e.g., diet, exercise, exposures, etc.). In addition, in several cases, there were more interesting and unexpected findings.
The first finding involved what appeared to be a whole chromosome gain of chromosome 12. An NGS-based CNV calling algorithm detected a gain in all exons of six eMERGEseq genes on chromosome 12 (CACNA1C [MIM: 114205], PKP2 [MIM: 602861], VDR [MIM: 601769], MYL2 [MIM: 160781], HNF1A [MIM: 142410], and POLE [MIM: 174762]), which was confirmed by ddPCR. CACNA1C, and POLE are located near the telomeric end of the chromosome 12 p and q arms, respectively, supporting a whole chromosome gain. Given that chromosome 12 trisomies are embryonic lethal, this CNV was assumed to be either of somatic origin or occurring as a mosaic variant. The former scenario is more likely as trisomy 12 is the most common somatic chromosomal aberration in chronic lymphocytic leukemia (CLL [MIM: 151400]) (see Atlas of Genetics in Web Resources) but has also been observed in other B cell lymphoproliferative disorders and is associated with a less favorable prognosis.18 Rarely, trisomy 12 has been reported as a mosaic variant in individuals with a variety of clinical phenotypes ranging from reportedly normal to multiple congenital anomalies, dysmorphic features, and developmental delay.18, 19, 20, 21, 22 Most of these were identified prenatally, with less than ten case subjects reported postnatally and even fewer detected in peripheral blood (for reviews see Chen et al.21 and Hong et al.22). Additional clinical information provided by the site indicated that this patient has a complex medical history including diabetes, heart disease, and a diagnosis of CRC at 87. While this finding is from a blood draw in early January 2016, this individual’s last complete blood count in 2010 showed no evidence of increased lymphocytes or any other abnormality suggesting a CLL diagnosis. While this type of result was not anticipated within the reporting scope for eMERGE III, upon further consultation with the site, this finding was included in the clinical report of the individual to encourage additional testing and/or management.
A second case with unexpected findings was associated with another copy number variant call. A duplication for all exons of OTC (MIM: 300461) and GLA (MIM: 300644), confirmed by ddPCR, was observed in a 40-year-old male not selected for phenotype. These genes are the only two present on the X chromosome on the eMERGEseq panel. Given that OTC and GLA are on the p and q arms, respectively, the observed duplication is most likely a single event spanning the entire X chromosome. This is most consistent with a male with Klinefelter syndrome (47,XXY). Additional clinical information provided by the site confirmed a prior diagnosis of Klinefelter syndrome that had been confirmed by chromosomal karyotyping. Although a clinical report was not issued for this individual, these findings serve to further validate the sensitivity of NGS-based copy number calling.
The third unexpected category of findings was that six individuals presented with apparently mosaic variants in genes that predispose to cancer or cardiomyopathy (TP53 [MIM: 191170], CHEK2, ATM [MIM: 607585], MYH7 [MIM: 160760]). The presence of mosaics was based upon the ascertainment of allelic variants that were present in <30% of the DNA sequence reads at the variant site. Initial observations were screened manually to eliminate false positives due to mis-mapping to pseudogene sites or other technical errors. The presence of the mosaic variants was subsequently confirmed by Sanger sequencing and clinical reporting offered to the referring sites.
Discussion
The introduction of clinical sequencing into the phase III of the eMERGE network has provided a framework for large-scale clinical translation of genomic data in healthcare, as well as for the seamless integration of research studies into clinical data management. The network integrated many research groups with diverse interests and a common mission to deliver genomic health care. To stimulate and address challenges for the delivery of genomic medicine, a large number of samples were tested and state of the art methods for interpretation and data delivery were applied.
A primary driver for the study design was cost and a focus on exploring the return of actionable genetic findings and therefore a gene-panel was chosen as a primary platform for genomic analyses. Whole-exome sequencing was considered. However, while exomes would have offered increased flexibility and saved time in design and testing, the network determined that a more focused target of ∼100 genes was needed to stay within the budget for testing all 25,015 participants and focus on a primary goal of developing experience around return of actionable results in biobank participants. In addition, sites individually contributed research data on subjects using high density genotyping arrays allowing for genome-wide association studies which are not discussed here.
Initially, predictions were made as to the major challenges that would be faced and the most likely obstacles to achieving a smooth flow of clinical results, while maintaining access to research data. However, most of the actual challenges were not anticipated. For example, the variety of different consents used to support the process sometimes stipulated requirements inconsistent with the network-wide decisions being made. As each site’s sequencing got started, these types of site-specific challenges were uncovered. Many sites altered their decisions around the reportable content and details of their reporting needs (e.g., which genes were reportable; whether negative reports were needed; whether reports should contain certain recommendations for genetic counseling, etc.). There was evolving work around how to structure pharmacogenomic results to flow into EHRs and work to ensure the accurate provision of phenotypes from the sites to the SCs. One site needed accommodation for lower DNA input. These “hiccups” led to significant delays in getting each site started with their sequencing and clinical reports. However, once a smooth workflow was developed for each site, the SCs were able to ramp up the rate of sequencing, interpretation, and reporting. For example, during the first half of the project, 9,245 cases were completed, versus 15,770 cases completed during the second half.
The work described here supports one of the major goals of the eMERGE III project, which is to study the return of actionable genetic variants to biobank participants and assess clinical outcomes. The outcomes being tracked include the ordering of any additional tests, starting new medication, and undergoing new procedures as well as overall healthcare utilization. The protocols for returning results in eMERGE III, including consent processes and the various components involved in the return of results process such as timing, mechanism of delivery, options to receive primary versus secondary findings, and the return of positive versus neutral results, have been previously described.1 For those sites that are returning negative results, most are doing so via letters to the participants. Both quantitative and qualitative studies, in the form of surveys and interviews, respectively, are being conducted by two sites to better understand how participants perceive such results, in particular the dissonance that may result when such results are received in the setting of a known family history of a disease (for example breast cancer). Data are currently being collected and analyzed and results will be reported separately. Furthermore, a follow-up study to explore variants of uncertain significance (VUSs), that were not reported but were in a “VUS leaning pathogenic” subcategory, is now beginning to allow phenotypes present within the EHR data to inform pathogenicity of these variants.
Conclusions
An important outcome of the study is the generation of real data that reflects the practicality of such a large-scale biobank study. The network has provided an accurate estimate of the frequency of returnable results within the interrogated gene set. Further, the study has established the ability for two sequencing centers to adequately harmonize both the technical and interpretive aspects of clinical sequencing tests, a critical achievement to the standardization of genomic testing. Furthermore, the eMERGE network has accomplished the integration of structured genomic results directly into multiple electronic health record systems, setting the stage for the use of clinical decision support to enable genomic medicine.
Consortia Members Contribution
Manuscript Leadership: Hana Zouk,∗ Eric Venner,∗ Donna M. Muzny, Niall J. Lennon, Heidi L. Rehm,# Richard A. Gibbs.#
Test Design, Validation, Metric Tracking, CNV detection: Niall J. Lennon,∗ Kimberly Walker,∗ Adam S. Gordon, Mark Bowser, Maegan V. Harden, Theodore Chiang, Elizabeth D. Hynes, Jianhong Hu, Matthew S. Lebo, Alyssa Macbeth, Lisa Mahanta, Eric Venner, Tsung-Jung Wu, Gail P. Jarvik, Hana Zouk, Heidi L. Rehm, Richard A. Gibbs, Birgit Funke,# Donna M. Muzny.#
Validity and Actionability Assessment for Genes and SNPs: Hana Zouk, Magalie Leduc, Emily Kudalkar, Adam S. Gordon, Clinical Annotation WG, Eric Venner, Heidi L. Rehm, Gail P. Jarvik, Birgit Funke.
Data Intake and Delivery: Lawrence J. Babb, Mullai Murugan, EHRI WG, Darren C. Ames, Maegan V. Harden, Chet Graham, Lisa Mahanta, Matthew S. Lebo, Heidi L. Rehm, Richard A. Gibbs, Samuel Aronson, Eric Venner.
Pharmacogenomics Reporting: Barbara Klanderman, Hana Zouk, Eric Venner, Elizabeth D. Hynes, Chiao-Feng Lin, Chet Graham, Lisa Mahanta, Donna M. Muzny, Steven Scherer, Matthew S. Lebo, Richard A. Gibbs, Birgit Funke, Magalie Leduc.
Variant Interpretation Harmonization and Result Interpretation: Yunyun Jiang,∗ Leora Witkowski,∗ Yaping Yang, David R. Murdock, Christine M. Eng, Matthew Varugheese, Eric Venner, Clinical Annotation WG, Birgit Funke, Richard A. Gibbs, Heidi L. Rehm, Magalie Leduc,# Hana Zouk.#
Clinical Site Representatives for Return of Results: Nancy D. Leslie (CCHMC), Melanie F. Myers (CCHMC), Cynthia A. Prows (CCHMC), Wendy Chung (Columbia), David Fasel (Columbia), Maddalena Marasa (Columbia), Hila Milo Rasouly (Columbia), Chunhua Weng (Columbia), Julia Wynn (Columbia), Melissa A. Kelly (Geisinger), Marc S. Williams (Geisinger), Gail P. Jarvik (UW), Eric B. Larson (KPW), Kathleen A. Leppig (KPW), James D. Ralston (KPW), David C. Kochan (Mayo), Iftikhar J. Kullo (Mayo), Noralane M. Lindor (Mayo), Philip Lammers (Meharry), Rajbir Singh (Meharry), Duane T. Smoot (Meharry), Christin Hoell (Northwestern), Laura J. Rasmussen-Torvik (Northwestern), Maureen E. Smith (Northwestern), Robert C. Green (Partners), Jordan W. Smoller (Partners), Josh F. Peterson (VUMC), Sara L. Van Driest (VUMC), Quinn S. Wells (VUMC), Georgia L. Wiesner (VUMC).
BCM-Human Genome Sequencing Center: Donna M. Muzny, Eric Venner, Jianhong Hu, Kimberly Walker, Sara E. Kalla, Theodore Chiang, Tsung-Jung Wu, Ritika Raj, Andrea Foster, Adithya Balasubramanian, Jesse Muniz, Shawn Denson, Gauthami Chandanavelli, Wen Liu, Harshad Mahadeshwar, Sean M. Vargas, Lan Zhang, Xiuping Liu, Qiaoyan Wang, Joy C. Jayaseelan, Darren C. Ames, Divya Kalra, Beenish Riza, Jessica De La Cruz, Liwen Wang, Viktoriya Korchina, Christie Kovar, Yunyun Jiang, Magalie Leduc, David R. Murdock, Yaping Yang, Victoria Yi, Mullai Murugan, Christine M. Eng, Richard A. Gibbs.
Partners Laboratory for Molecular Medicine/Broad Institute Sequencing Center: Hana Zouk, Maegan V. Harden, Leora Witkowski, Samuel Aronson, Lawrence J. Babb, Samantha Baxter, Mark Bowser, Wendy Brodeur, Sheila Dodge, Phil Dunlea, Christopher Friedrich, Tim DeSmet, Michael J. Dinsmore, Mark Fleharty, Chet Graham, Elizabeth D. Hynes, Emily Kudalkar, Matthew S. Lebo, Chiao-Feng Lin, Hayley Lyon, Alyssa Macbeth, Lisa Mahanta, Jim Meldrim, Thomas E. Mullen, Robert C. Onofrio, Kara Slowik, Gina Vicente, Michael W. Wilson, Betty Woolf, Stacey Gabriel, Birgit Funke#, Niall J. Lennon#, Heidi L. Rehm#.
Coordinating Center: Melissa Basford, Brittany City, David R. Crosslin, Adam S. Gordon, Kayla M. Howell, Gail P. Jarvik, Jodell E. Linder, Ian B. Stanaway, Laura A. Woods, Josh F. Peterson (PI).
eMERGE Working Group Co-Chairs: Clinical Annotation: Gail P. Jarvik and Heidi Rehm; EHR Integration: Samuel Aronson and Casey Overby-Taylor; Genomics: David R. Crosslin, Megan J. Puckelwartz and Patrick M.A. Sleiman; Outcomes: Hakon Hakonarson, Josh F. Peterson and Marc S. Williams; PGx: Cynthia A. Prows and Laura Rasmussen-Torvik; Phenotyping: George Hripcsak, Wei-Qi Wei and Chunhua Weng; Return of Results/ELSI: Ingrid Holm and Iftikhar J. Kullo.
eMERGE Principal Investigators: Rex L. Chisholm (Steering Committee Chair), Samuel E. Adunyah, David R. Crosslin, Josh C. Denny, Ali Gharavi, Richard A. Gibbs, Hakon Hakonarson, John Harley, George Hripcsak, Gail P. Jarvik, Elizabeth W. Karlson, Iftikhar J. Kullo, Eric B. Larson, Niall J. Lennon, Shawn Murphy, Josh F. Peterson, Heidi L. Rehm, Dan M. Roden, Marylyn D. Ritchie, Richard R. Sharp, Maureen E. Smith, Jordan W. Smoller, Chunhua Weng, Scott T. Weiss, Marc S. Williams.
NHGRI Staff: Rongling Li, Ken L. Wiley Jr., Jyoti Dayal, Sheethal Jose, Teri Manolio.
Additional Consortia Members
Debra Abrams, Ladia Albertson-Junkans, Berta Almoguera, Paul Appelbaum, Samuel Aronson, Sharon Aufox, Hana Bangash, Lisa Bastarache, Meckenzie Behr, Barbara Benoit, Elizabeth Bhoj, Suzette J. Bielinski, Sarah T. Bland, Carrie Blout, Erwin P. Bottinger, Harrison Brand, Murray Brilliant, Pedro Caraballo, David S. Carrell, Andrew Carroll, Lisa Castillo, Victor Castro, Kurt D. Christensen, Christopher G. Chute, Beth L. Cobb, John J. Connolly, Paul Crane, Katherine Crew, Mariza De Andrade, Ozan Dikilitas, Todd L. Edwards, Alex Fedotov, Qiping Feng, Robert Freimuth, Stephanie M. Fullerton, Vivian Gainer, M. Geoffrey Hayes, Andrew M. Glazer, Joseph T. Glessner, Jessica Goehringer, Justin H. Gundelach, Heather S. Hain, Margaret Harr, Andrea Hartzler, Scott Hebbring, Nora Henrikson, Andrew Hershey, Yoonjung Yoonie Joo, Navya Shilpa Josyula, Anne E. Justice, Brendan J. Keating, Eimear E. Kenny, Dustin Key, Krzysztof Kiryluk, Terrie Kitchner, Eric Klee, Leah Kottyan, Ming Ta (Michael) Lee, Wayne H. Liang, Todd Lingren, James G. Linneman, Cong Liu, John Lynch, Bradley Malin, Keith Marsolo, Michelle L. McGowan, Elizabeth McNally, Frank Mentch, Jonathan Mosley, Shubhabrata Mukherjee, Bahram Namjou, Yizhao Ni, Aniwaa Owusu Obeng, Thomas N. Person, Lynn Petukhova, Cassandra J. Pisieczko, Siddharth Pratap, Alanna Kulchak Rahm, Arvind Ramaprasan, Andrea Ramirez, Luke Rasmussen, Soumya Raychaudhuri, Catherine Rives, Elisabeth A. Rosenthal, Avni Santani, Dan Schaid, Stuart Scott, Aaron Scrol, Soumitra Sengupta, Ning Shang, Himanshu Sharma, Joshua C. Smith, Sunghwan Sohn, Justin Starren, Mary Stroud, Jessica Su, Kasia Tolwinski, David Veenstra, Miguel Verbitsky, Michael Wagner, Theresa Walunas, Peter S. White, Janet L. Williams, Ge Zhang.
Equal contributions indicated with symbols (∗#).
Declaration of Interests
Samuel Aronson is employed at Partners HealthCare, which receives royalties on sales of GeneInsight Software. Samuel Aronson’s team receives funding from Sunquest and has received funding from Novartis for development of SMART on FHIR apps. Andrew Carroll is employed at Google Inc. and is a former employee of DNAnexus. Paul Crane served as a consultant to Eisai on efforts unrelated to this manuscript. David Crosslin serves on a consulting board for UnitedHealth Group with precision medicine efforts, which is unrelated to this manuscript. Christine Eng is a full-time employee/faculty member of Baylor College of Medicine. Through a professional services agreement, she serves as Chief Medical Officer and Chief Quality Officer of Baylor Genetics. Richard A. Gibbs declares that Baylor College of Medicine receives payments from Baylor Genetics Laboratories, which provides services for genetic testing; Baylor College of Medicine is part owner of Codified Genomics. Robert C. Green receives personal compensation from AIA, Applied Therapeutics, Helix, Prudential, Verily, and Veritas for speaking or consulting and is co-founder of Genome Medical, Inc. Eimear E. Kenny has received speaker honorariums from Illumina and Regeneron Pharmaceuticals. Niall J. Lennon is an advisor to Genturi Inc. Elizabeth McNally serves or has served as a consultant to Invitae, Tenaya, Exonics, Pfizer, AstraZeneca, Cytokinetics, and 4D Molecular Therapeutics and founded Ikaika Therapeutics. Thomas E. Mullen is employed at and a shareholder at Quest Diagnostics. Heidi Rehm is employed at Massachusetts General Hospital, which receives royalties on sales of GeneInsight Software. Avni Santani receives royalties from Agilent Technologies and a founder of Opus Genomics. Jordan W. Smoller is an unpaid member of the Bipolar/Depression Research Community Advisory Panel of 23andMe. Eric Venner is a cofounder of Codified Genomics. Theresa Walunas completed (in 2018) legal consulting for by Pfizer Inc., Wyeth LLC, Genetics Institute, LLC, Merck KGaA, and EMD Serono, Inc. Georgia L. Wiesner is a member of the External Advisory Panel for the ClinGen Clinical Genome Resource Project. Hana Zouk is employed at Massachusetts General Hospital which receives royalties on sales of GeneInsight Software.
Acknowledgments
The eMERGE Phase III Network was initiated and funded by the National Human Genome Research Institute (NHGRI) through the following grants: U01HG8657 (Kaiser Permanente Washington Health Research Institute/University of Washington), U01HG8685 (Brigham and Women’s Hospital), U01HG8672 (Vanderbilt University Medical Center), U01HG8666 (Cincinnati Children’s Hospital Medical Center), U01HG6379 (Mayo Clinic), U01HG8679 (Geisinger Clinic), U01HG8680 (Columbia University Health Sciences), U01HG8684 (Children’s Hospital of Philadelphia), U01HG8673 (Northwestern University), MD007593 (Meharry Medical College), U01HG8701 (Vanderbilt University Medical Center serving as the Coordinating Center), U01HG8676 (Partners Healthcare/Broad Institute), and U01HG8664 (Baylor College of Medicine).
Published: August 22, 2019
Footnotes
Supplemental Data can be found online at https://doi.org/10.1016/j.ajhg.2019.07.018.
Contributor Information
The eMERGE Consortiumagibbs@bcm.eduhrehm@mgh.harvard.edu:
Hana Zouk, Eric Venner, Niall J. Lennon, Donna M. Muzny, Debra Abrams, Samuel Adunyah, Ladia Albertson-Junkans, Darren C. Ames, Paul Appelbaum, Samuel Aronson, Sharon Aufox, Lawrence J. Babb, Adithya Balasubramanian, Hana Bangash, Melissa Basford, Lisa Bastarache, Samantha Baxter, Meckenzie Behr, Barbara Benoit, Elizabeth Bhoj, Suzette J. Bielinski, Sarah T. Bland, Carrie Blout, Kenneth Borthwick, Erwin P. Bottinger, Mark Bowser, Harrison Brand, Murray Brilliant, Wendy Brodeur, Pedro Caraballo, David Carrell, Andrew Carroll, Berta Almoguera, Lisa Castillo, Victor Castro, Gauthami Chandanavelli, Theodore Chiang, Rex L. Chisholm, Kurt D. Christensen, Wendy Chung, Christopher G. Chute, Brittany City, Beth L. Cobb, John J. Connolly, Paul Crane, Katherine Crew, David Crosslin, Mariza De Andrade, Jessica De la Cruz, Shawn Denson, Josh Denny, Tim DeSmet, Ozan Dikilitas, Christopher Friedrich, Stephanie M. Fullerton, Birgit Funke, Stacey Gabriel, Vivian Gainer, Ali Gharavi, Andrew M. Glazer, Joseph T. Glessner, Jessica Goehringer, Adam S. Gordon, Chet Graham, Robert C. Green, Justin H. Gundelach, Jyoti Dayal, Heather S. Hain, Hakon Hakonarson, Maegan V. Harden, John Harley, Margaret Harr, Andrea Hartzler, M. Geoffrey Hayes, Scott Hebbring, Nora Henrikson, Andrew Hershey, Christin Hoell, Ingrid Holm, Kayla M. Howell, George Hripcsak, Jianhong Hu, Gail P. Jarvik, Joy C. Jayaseelan, Yunyun Jiang, Yoonjung Yoonie Joo, Sheethal Jose, Navya Shilpa Josyula, Anne E. Justice, Sara E. Kalla, Divya Kalra, Elizabeth Karlson, Melissa A. Kelly, Brendan J. Keating, Eimear E. Kenny, Dustin Key, Krzysztof Kiryluk, Terrie Kitchner, Barbara Klanderman, Eric Klee, David C. Kochan, Viktoriya Korchina, Leah Kottyan, Christie Kovar, Emily Kudalkar, Iftikhar J. Kullo, Philip Lammers, Eric B. Larson, Matthew S. Lebo, Magalie Leduc, Ming Ta (Michael) Lee, Kathleen A. Leppig, Nancy D. Leslie, Rongling Li, Wayne H. Liang, Chiao-Feng Lin, Jodell Linder, Noralane M. Lindor, Todd Lingren, James G. Linneman, Cong Liu, Wen Liu, Xiuping Liu, John Lynch, Hayley Lyon, Alyssa Macbeth, Harshad Mahadeshwar, Lisa Mahanta, Brad Malin, Teri Manolio, Maddalena Marasa, Keith Marsolo, Michael J. Dinsmore, Sheila Dodge, Elizabeth Duffy Hynes, Phil Dunlea, Todd L. Edwards, Christine M. Eng, David Fasel, Alex Fedotov, Qiping Feng, Mark Fleharty, Andrea Foster, Robert Freimuth, Michelle L. McGowan, Elizabeth McNally, Jim Meldrim, Frank Mentch, Jonathan Mosley, Shubhabrata Mukherjee, Thomas E. Mullen, Jesse Muniz, David R. Murdock, Shawn Murphy, Mullai Murugan, Melanie F. Myers, Bahram Namjou, Yizhao Ni, Aniwaa Owusu Obeng, Robert C. Onofrio, Casey Overby Taylor, Thomas N. Person, Josh F. Peterson, Lynn Petukhova, Cassandra J. Pisieczko, Siddharth Pratap, Cynthia A. Prows, Megan J. Puckelwartz, Alanna Kulchak Rahm, Ritika Raj, James D. Ralston, Arvind Ramaprasan, Andrea Ramirez, Luke Rasmussen, Laura Rasmussen-Torvik, Hila Milo Rasouly, Soumya Raychaudhuri, Marylyn D. Ritchie, Catherine Rives, Beenish Riza, Dan Roden, Elisabeth A. Rosenthal, Avni Santani, Dan Schaid, Steven Scherer, Stuart Scott, Aaron Scrol, Soumitra Sengupta, Ning Shang, Himanshu Sharma, Richard R. Sharp, Rajbir Singh, Patrick M.A. Sleiman, Kara Slowik, Joshua C. Smith, Maureen E. Smith, Jordan W. Smoller, Sunghwan Sohn, Ian B. Stanaway, Justin Starren, Mary Stroud, Jessica Su, Kasia Tolwinski, Sara L. Van Driest, Sean M. Vargas, Matthew Varugheese, David Veenstra, Miguel Verbitsky, Gina Vicente, Michael Wagner, Kimberly Walker, Theresa Walunas, Liwen Wang, Qiaoyan Wang, Wei-Qi Wei, Scott T. Weiss, Georgia L. Wiesner, Quinn Wells, Chunhua Weng, Peter S. White, Ken L. Wiley, Jr., Janet L. Williams, Marc S. Williams, Michael W. Wilson, Leora Witkowski, Laura Allison Woods, Betty Woolf, Tsung-Jung Wu, Julia Wynn, Yaping Yang, Victoria Yi, Ge Zhang, Lan Zhang, Heidi L. Rehm, and Richard A. Gibbs
Data and Code Availability
The datasets generated and/or analyzed during the current study will be publicly available in the dbGaP repository under phs001616.v1.p1 and pre-dbGaP submission access can also be requested on the eMERGE Network website (see Web Resources).
Web Resources
Atlas of Genetics and Cytogenetics in Oncology and Haematology, Michaux, L. (2000). +12 or trisomy 12, http://atlasgeneticsoncology.org/Anomalies/tri12ID2024.html
Clinical Research Sequencing Platform, https://portals.broadinstitute.org/portal/CRSP
CPIC Publications, https://cpicpgx.org/publications
eMERGE, https://www.genome.gov/27540473/electronic-medical-records-and-genomics-emerge-network/
eMERGE Network, https://emerge.mc.vanderbilt.edu/collaborate/
eMERGE Sample Submission Portal, https://emerge.hgsc.bcm.edu/workflow/sample-submission
Gene Dosage Curations, https://search.clinicalgenome.org/kb/gene-dosage
OMIM, https://www.omim.org/
Sequence Variant Interpretation, https://www.clinicalgenome.org/working-groups/sequence-variant-interpretation
Supplemental Data
References
- 1.Fossey R., Kochan D., Winkler E., Pacyna J.E., Olson J., Thibodeau S., Connolly J.J., Harr M., Behr M.A., Prows C.A., et al. Ethical Considerations Related to Return of Results from Genomic Medicine Projects: The eMERGE Network (Phase III) Experience. J. Pers. Med. 2018;8:2. doi: 10.3390/jpm8010002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Green R.C., Berg J.S., Grody W.W., Kalia S.S., Korf B.R., Martin C.L., McGuire A.L., Nussbaum R.L., O’Daniel J.M., Ormond K.E., et al. American College of Medical Genetics and Genomics ACMG recommendations for reporting of incidental findings in clinical exome and genome sequencing. Genet. Med. 2013;15:565–574. doi: 10.1038/gim.2013.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.DePristo M.A., Banks E., Poplin R., Garimella K.V., Maguire J.R., Hartl C., Philippakis A.A., del Angel G., Rivas M.A., Hanna M., et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat. Genet. 2011;43:491–498. doi: 10.1038/ng.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Reid J.G., Carroll A., Veeraraghavan N., Dahdouli M., Sundquist A., English A., Bainbridge M., White S., Salerno W., Buhay C., et al. Launching genomics into the cloud: deployment of Mercury, a next generation sequence analysis pipeline. BMC Bioinformatics. 2014;15:30. doi: 10.1186/1471-2105-15-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pugh T.J., Amr S.S., Bowser M.J., Gowrisankar S., Hynes E., Mahanta L.M., Rehm H.L., Funke B., Lebo M.S. VisCap: inference and visualization of germ-line copy-number variants from targeted clinical sequencing data. Genet. Med. 2016;18:712–719. doi: 10.1038/gim.2015.156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.McKenna A., Hanna M., Banks E., Sivachenko A., Cibulskis K., Kernytsky A., Garimella K., Altshuler D., Gabriel S., Daly M., DePristo M.A. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fromer M., Purcell S.M. Using XHMM Software to Detect Copy Number Variation in Whole-Exome Sequencing Data. Curr. Protoc. Hum. Genet. 2014;81:1–21. doi: 10.1002/0471142905.hg0723s81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chiang T., Liu X., Wu T.-J., Hu J., Sedlazeck F.J., White S., Schaid D., de Andrade M., Jarvik G.P., Crosslin D., et al. Atlas-CNV: a validated approach to call Single-Exon CNVs in the eMERGESeq gene panel. Genet. Med. 2018 doi: 10.1038/s41436-019-0475-4. Published online March 20, 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Richards S., Aziz N., Bale S., Bick D., Das S., Gastier-Foster J., Grody W.W., Hegde M., Lyon E., Spector E., et al. ACMG Laboratory Quality Assurance Committee Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet. Med. 2015;17:405–424. doi: 10.1038/gim.2015.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Strande N.T., Riggs E.R., Buchanan A.H., Ceyhan-Birsoy O., DiStefano M., Dwight S.S., Goldstein J., Ghosh R., Seifert B.A., Sneddon T.P., et al. Evaluating the Clinical Validity of Gene-Disease Associations: An Evidence-Based Framework Developed by the Clinical Genome Resource. Am. J. Hum. Genet. 2017;100:895–906. doi: 10.1016/j.ajhg.2017.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Aronson S.J., Clark E.H., Babb L.J., Baxter S., Farwell L.M., Funke B.H., Hernandez A.L., Joshi V.A., Lyon E., Parthum A.R., et al. The GeneInsight Suite: a platform to support laboratory and provider use of DNA-based genetic testing. Hum. Mutat. 2011;32:532–536. doi: 10.1002/humu.21470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Aronson S.J., Clark E.H., Varugheese M., Baxter S., Babb L.J., Rehm H.L. Communicating new knowledge on previously reported genetic variants. Genet. Med. 2012;14:713–719. doi: 10.1038/gim.2012.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Aronson S., Babb L., Ames D., Gibbs R.A., Venner E., Connelly J.J., Marsolo K., Weng C., Williams M.S., Hartzler A.L., et al. eMERGE Network EHRI Working Group Empowering genomic medicine by establishing critical sequencing result data flows: the eMERGE example. J. Am. Med. Inform. Assoc. 2018;25:1375–1381. doi: 10.1093/jamia/ocy051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Abbott G.W., Sesti F., Splawski I., Buck M.E., Lehmann M.H., Timothy K.W., Keating M.T., Goldstein S.A. MiRP1 forms IKr potassium channels with HERG and is associated with cardiac arrhythmia. Cell. 1999;97:175–187. doi: 10.1016/s0092-8674(00)80728-x. [DOI] [PubMed] [Google Scholar]
- 15.Kapplinger J.D., Tester D.J., Salisbury B.A., Carr J.L., Harris-Kerr C., Pollevick G.D., Wilde A.A.M., Ackerman M.J. Spectrum and prevalence of mutations from the first 2,500 consecutive unrelated patients referred for the FAMILION long QT syndrome genetic test. Heart Rhythm. 2009;6:1297–1303. doi: 10.1016/j.hrthm.2009.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nawathe P.A., Kryukova Y., Oren R.V., Milanesi R., Clancy C.E., Lu J.T., Moss A.J., Difrancesco D., Robinson R.B. An LQTS6 MiRP1 mutation suppresses pacemaker current and is associated with sinus bradycardia. J. Cardiovasc. Electrophysiol. 2013;24:1021–1027. doi: 10.1111/jce.12163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Carey D.J., Fetterolf S.N., Davis F.D., Faucett W.A., Kirchner H.L., Mirshahi U., Murray M.F., Smelser D.T., Gerhard G.S., Ledbetter D.H. The Geisinger MyCode community health initiative: an electronic health record-linked biobank for precision medicine research. Genet. Med. 2016;18:906–913. doi: 10.1038/gim.2015.187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.English C.J., Goodship J.A., Jackson A., Lowry M., Wolstenholme J. Trisomy 12 mosaicism in a 7 year old girl with dysmorphic features and normal mental development. J. Med. Genet. 1994;31:253–254. doi: 10.1136/jmg.31.3.253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hsu L.Y., Yu M.T., Neu R.L., Van Dyke D.L., Benn P.A., Bradshaw C.L., Shaffer L.G., Higgins R.R., Khodr G.S., Morton C.C., et al. Rare trisomy mosaicism diagnosed in amniocytes, involving an autosome other than chromosomes 13, 18, 20, and 21: karyotype/phenotype correlations. Prenat. Diagn. 1997;17:201–242. doi: 10.1002/(sici)1097-0223(199703)17:3<201::aid-pd56>3.0.co;2-h. [DOI] [PubMed] [Google Scholar]
- 20.DeLozier-Blanchet C.D., Roeder E., Denis-Arrue R., Blouin J.L., Low J., Fisher J., Scharnhorst D., Curry C.J. Trisomy 12 mosaicism confirmed in multiple organs from a liveborn child. Am. J. Med. Genet. 2000;95:444–449. doi: 10.1002/1096-8628(20001218)95:5<444::aid-ajmg7>3.0.co;2-x. [DOI] [PubMed] [Google Scholar]
- 21.Chen C.-P., Chang S.-D., Su J.-W., Chen Y.-T., Wang W. Prenatal diagnosis of mosaic trisomy 12 associated with congenital overgrowth. Taiwan. J. Obstet. Gynecol. 2013;52:454–456. doi: 10.1016/j.tjog.2013.06.008. [DOI] [PubMed] [Google Scholar]
- 22.Hong B., Zunich J., Openshaw A., Toydemir R.M. Clinical features of trisomy 12 mosaicism-Report and review. Am. J. Med. Genet. A. 2017;173:1681–1686. doi: 10.1002/ajmg.a.38194. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated and/or analyzed during the current study will be publicly available in the dbGaP repository under phs001616.v1.p1 and pre-dbGaP submission access can also be requested on the eMERGE Network website (see Web Resources).