Abstract
Rheumatoid arthritis (RA) is the most common immune-mediated arthritis. Anti-citrullinated peptide antibodies (ACPA) are highly specific to RA and assayed with the commercial CCP2 assay. Genetic drivers of RA within the MHC are different for CCP2-positive and -negative subsets of RA, particularly at HLA-DRB1. However, aspartic acid at amino acid position 9 in HLA-B (Bpos-9) increases risk to both RA subsets. Here we explore how individual serologies associated with RA drive associations within the MHC. To define MHC differences for specific ACPA serologies, we quantified a total of 19 separate ACPAs in RA-affected case subjects from four cohorts (n = 6,805). We found a cluster of tightly co-occurring antibodies (canonical serologies, containing CCP2), along with several independently expressed antibodies (non-canonical serologies). After imputing HLA variants into 6,805 case subjects and 13,467 control subjects, we tested associations between the HLA region and RA subgroups based on the presence of canonical and/or non-canonical serologies. We examined CCP2(+) and CCP2(−) RA-affected case subjects separately. In CCP2(−) RA, we observed that the association between CCP2(−) RA and Bpos-9 was derived from individuals who were positive for non-canonical serologies (omnibus_p = 9.2 × 10−17). Similarly, we observed in CCP2(+) RA that associations between subsets of CCP2(+) RA and Bpos-9 were negatively correlated with the number of positive canonical serologies (p = 0.0096). These findings suggest unique genetic characteristics underlying fine-specific ACPAs, suggesting that RA may be further subdivided beyond simply seropositive and seronegative.
Keywords: rheumatoid arthritis, MHC, major histocompatability complex, genetics, HLA, citrullinated peptides
Introduction
Genetic variation within the MHC locus harboring the HLA genes causes an altered serological response to an environmental stimulus.1, 2, 3, 4 For example, in inflammatory myositis (MIM: 160750) and type I diabetes (MIM: 222100), different HLA alleles are associated with specific antibody responses.5, 6, 7, 8 Rheumatoid arthritis (RA [MIM: 180300]) is the most common autoimmune-mediated arthritis with prevalence 0.5%–1.0% worldwide. Anti-citrullinated peptide antibodies (ACPA) are a set of highly RA-specific antibodies (Ab) and are often assayed in the clinical setting with the commercial CCP2 assay. We have shown that ACPA-positive RA has distinct MHC associations from ACPA-negative (or seronegative) RA, driven mainly by differences at position 11 (DRB1pos-11 or equivalently DRB1pos-13) in HLA-DRB1 (MIM: 142857).4, 9, 10 However, both seropositive and seronegative RA share an association to amino acid position 9 in HLA-B (MIM: 142830) (Bpos-9), driven by increase in risk associated with the presence of an aspartic acid residue at that position. However, subtle serological differences in HLA effects influencing RA risk are not yet fully recognized in RA or other diseases.
Here, we investigated whether subtle differences in ACPA serologies might be connected to different driving HLA alleles. If this is the case, it might become possible to define the precise relationship between antigen presentation and serological response in RA. In addition, we might redefine RA subsets based on ACPA serologies supported by genetic differences.
Material and Methods
Subjects
We recruited a total of 6,805 case subjects and 13,467 control subjects for this study from four cohorts in Sweden, UK, and US (see Table S1) with institutional review board approval at each institution. We obtained written informed consent from each participant. All of the cohorts are described in detail elsewhere.4, 11
Identifying Serologies of Interest
In order to determine IgG reactivities to a large set of citrullinated epitopes from proteins implicated as autoantigens in RA, we utilized a previously described multiplex chip assay.12 For each cohort we used sera from healthy individuals from that cohort that had been captured and stored in the similar way as the case subjects to set cutoffs for positive values as detailed elsewhere.13
Quantifying Ab Reactivity against Citrullinated Peptides
We quantified 18 fine-specific ACPA in RA-affected individuals using a multiplex peptide array.12 The origin of peptides and amino acid sequences are described in Table S2. We quantified the reactivity toward peptides from sera in the samples from individuals together with up to 175 healthy control subjects in each cohort.
Definition of Cut-off Levels of Serologies
For CCP2, we used commercially pre-defined levels, which corresponds to 98.4% specificity in the Swedish EIRA.14 Except for CCP2, we set cut-off levels for each serology in each cohort. We defined the cut-off levels of positivity for each serology as the 98 percentile of control levels based on the previous studies12, 14, 15 and CCP2 specificity. The serology data was then compiled into a matrix Sij, where each entry is a binary indication of the presence of fine-specific ACPA serology i in individual j.
Clustering
Prior to clustering serological profiles, we standardized them to have a mean of 0 and standard deviation of 1 and created a new matrix Pij. We applied Ward’s method for agglomerative hierarchical clustering of serologies, using the Kappa statistic to define distance metric between serological profiles. We also used the Pearson’s correlation coefficient as a secondary analysis to confirm the reliability of the clustering results. We also conducted clustering with the use of complete linkage method instead of Ward’s method.
After cutting the tree at different levels, we wanted to assess the significance of the clusters. To assess clustering of the actual data, we evaluated the fraction of the sum of squares explained by the clusters:
where WSS is the sum of squares within clusters, TSS is the total sum of squares within clusters, and rSS is the residual sum of squares representing the sum of squares between clusters. Here there are k serologies indexed with i, n individuals indexed with j, and m clusters indexed with c. represents the global mean of standardized serologies, and represents the mean of the serologies within cluster c. Higher values of this ratio indicate better clustering. We permuted the data 1,000 times by randomly reassigning positive subjects for each serology, so that the total number of individuals positive for each serology was constant. We compared the original statistics with permuted tests to assess significance of original clusters. We also conducted clustering of the subjects with the use of Ward’s method. We evaluated Gap statistics of hierarchical and k-means clustering and revealed that the two clusters best classified the subjects (Supplemental Subjects and Methods).
Principal Component Analysis
We conducted principal component analysis (PCA) of Pij to visualize the serologies and their relationship to each other and to identify outlier serologies. We also applied PCA to visualize RA-affected subjects and their relationship to each other.
Identifying Subsets of Serologies Enriched for CCP2(−) RA
We calculated the proportion of presence of fine-specific ACPAs in CCP2(−) RA in comparison of CCP2(+) RA by dividing the number of CCP2(−) RA-affected individuals expressing the Ab by the respective number among CCP2(+) RA-affected individuals. The ratios for non-canonical Ab were compared to those of canonical Ab using the one-sided Mann-Whitney rank sum test. Our alternative hypothesis was that the CCP2 ratios were larger for non-canonical Ab compared to canonical reactivities.
HLA Imputation
We had previously genotyped all samples with the Illumina Immunochip genotyping platform.16 With these data, we imputed genetic variations in the HLA region defined from 29.6 Mb to 33.27 Mb in chromosome 6 with SNP2HLA17 using a European reference panel consisting of 5,225 European individuals with European descent in the Type 1 Diabetes Genetic Consortium18 as reference. We could impute a total of 5,968 SNPs, 966 amino acid residues over 398 amino acid position in HLA-A (MIM: 142800), B, C (MIM: 142840), DRB1, DPA1 (MIM: 142880), DPB1 (MIM: 142858), DQA1 (MIM: 146880), and DQB1 (MIM: 191160) and 126 and 298 HLA-alleles in 2- and 4-digit resolution, respectively. We have previously shown that this approach obtains high accuracy.17
Association Analysis
We estimated associations between variants and susceptibility to RA subsets in logistic regression analysis. We put top 10 principal components in each cohort as covariates and cohort information as indicator variables in the model as follows:
where ORi is odds ratio for individual i, v is a variant for evaluation, and gv,i are an effect size and individual dosage of the variant v, respectively, θ represents logistic regression intercept, is an indicator variable which is 1 only if individual i is in the set j, Sj is effect size of set j, and Pi,j,k are effect sizes and individual i’s values for principal components k in set j. To assess significance of amino acid positions, we used omnibus test and compared with a baseline model and calculated the change in deviance by adding the genotype term whose details are written below. We used the same model for conditional analyses.
Genetic Risk Score for CCP2(+) RA and Ankylosing Spondylitis in CCP2(−) Subset
Since we previously showed that CCP2(−) RA potentially included subjects with atypical presentations of CCP2(+) RA and ankylosing spondylitis (AS [MIM: 106300]),4 we calculated genetic risk score for CCP2(+) RA (CCP2-GRS) and AS (AS-GRS) by the same methods as our previous study4 to associate with CCP2(−) subsets. GRS are sum of dosages and effect sizes of susceptibility markers to CCP2(+) RA and AS. GRS for CCP2(+) RA and AS were calculated separately for each individual.
Omnibus Test for Amino Acid Positions
We estimated associations between specific amino acid positions and susceptibility to subsets of RA based on positivity of fine-specific ACPA(+) in case-control analyses by using omnibus test as previously described (based on a likelihood ratio test).19 We included 10 principal components in each cohort as covariates and cohort information as indicator variables in the model as follows:
| (Equation 1) |
| (Equation 2) |
where a is the amino acid residue in the position, and ga,i are an effect size and individual dosage of the a amino acid residue, respectively, n is the number of different amino acids at the position, θ represents logistic regression intercept, is an indicator variable which is 1 only if individual i is in the set j, Sj is effect size of set j, and are effect sizes and individual i’s values for principal components k in set j. We assessed the improvement in fit for each amino acid position in comparison with null model (Equation 2) based on the deviance, which follows a distribution with n-1 degree of freedom. We also performed conditional analyses, where we added all amino acid residues from a second amino acid site, and assessed improvement in model fit over Equation 1.
We also assessed multiplicative interaction associations on the number of HLA-Bpos-9 D amino acids between number or positivity of positive canonical and that of non-canonical serologies. We used generalized linear regression framework and logarithm of the number of positive canonical and non-canonical serologies to which one was added as follows:
#binary
| (Equation 3) |
| (Equation 4) |
#quantitative
| (Equation 5) |
| (Equation 6) |
where B9D is dosage of aspartic acid at HLA-Bpos-9, , , and represent binary positivity of canonical or non-canonical, canonical and non-canonical serologies, respectively, , , and are logarithm of number of canonical or non-canonical serologies added by one, respectively, and and are effect sizes of positivity or log-transformed number of serologies and their interaction, respectively. Equations 3 and 4 were analyzed in case-control analyses and Equations 5 and 6 were in intra-case analyses.
We also assessed an association between B9D and RA status in Equation 4 by substituting RA affection status (a binary variable) for positivity of canonical or non-canonical serologies.
We performed statistical tests with the PLINK software20 or R statistical software. Stringent significant levels for the associations with RA were set as p values less than 5.0 × 10−8. In other cases, we applied Bonferroni correction.
Results
We observed that a total of 70.5% of subjects were positive for CCP2, consistent with previous studies (Table S1).21 Individual fine-specific ACPAs varied in their positivity from 8.3% (Fib β 62-81 cit 72) to 62.7% (Fib β 60-74 cit). Fib β 62-81 cit 72 corresponds to a peptide spanning from amino acid 62 to 81 from human fibrinogen beta chain with a citulline instead of an arginine at position 72. We observed that Fib β 60-74 cit was most predictive of CCP2 with an area under curve (AUC) of 0.885 and high spearman’s correlation of 0.697 (Table S3). Unsurprisingly, we recognized that serologies could have cross reactivity, and we wanted to use a statistical approach to minimize a risk of over-interpreting any single serology. Hence, we decided to cluster serologies to mitigate any technical artifacts that might be related to an individual serology.
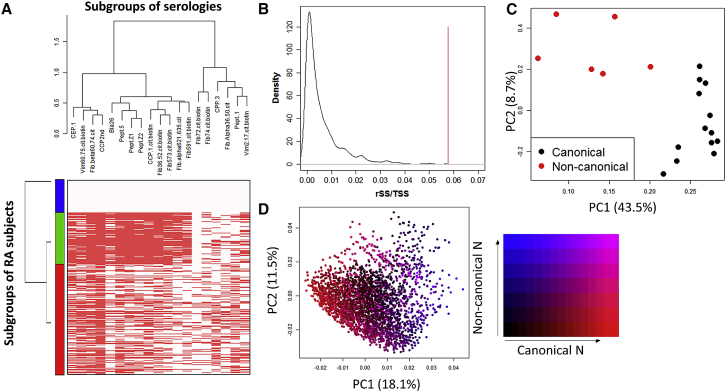
We observed two distinct clusters in the ACPAs (Figure 1A) when we clustered 19 ACPAs using kappa metric in a hierarchical clustering scheme (see Material and Methods and Figure S1). We found that two clusters classified the data best (see Material and Methods and Supplemental Subjects and Methods). We detected a cluster of tightly co-occurring Ab (median r2 within cluster: 0.53) (Figure 1A). We refer to this cluster as the canonical serologies. Separate from these are a set of Ab that are not strongly correlated to any Ab (median r2: 0.22), which we refer to as non-canonical serologies. While 4,236 individuals were positive for at least one non-canonical serologies, almost all of these individuals (96.4%) were also positive for canonical serologies. We observed the same two clusters with the alternative Pearson correlation coefficient or the alternative complete linkage method (Figure S2). Clustering each of the four cohorts separately resulted in clusters with almost identical composition (Figure S3A). We also obtained very similar results of correlation matrix of serologies across the four cohorts (Figure S3B). Taken together, the clustering results are robust and stable. To assess significance of the clusters, we permuted ACPAs to disrupt the covariance structure 1,000 times (see Material and Methods); in no instance did we observe similar variance explained by two clusters in these tests (Figure 1B). Application of PCA revealed that PC 1, comprising 43.5% of variance, divided ACPAs by their two different clusters (Figure 1C). Fib β 60-74 cit showed distinct reactivity from Fib β 62-81 cit 72 and Fib β 62-81 cit 74 although they recognize peptides with similar sequences (Figures S1 and S2 and Tables S1 and S2).
Figure 1.
Fine-Specific ACPAs Can Be Subgrouped into Two Clusters and Non-canonical Cluster ACPAs Are Enriched in CCP2(−) RA
(A) Heatmap presenting positivity of ACPAs and clusters of ACPAs is indicated. Each row of heatmap represents each individual. Red squares indicate positive ACPA. The three colored bars in the y axis indicate subgroups of RA-affected subjects.
(B) Distribution of fraction of sum square between two clusters over total sum square in the 1,000 permutation tests is indicated. A red line indicates statistic of the original two clusters.
(C) The 19 ACPAs are projected according to PC1 and 2. Black and red closed circles indicate canonical and non-canonical cluster serologies, respectively.
(D) RA-affected subjects are projected according to PC1 and 2. Samples are given colors according to positive rate of canonical and non-canonical cluster serologies.
Clustering of subjects based on serologies revealed three separate groups of individuals. These individuals can be generally described as (1) subjects negative for all serologies, (2) subjects negative for the non-canonical Fib β 62-81 cit 72 serology, or (3) subjects positive and enriched for the non-canonical serologies Fib β 62-81 cit 72 and enriched for positive Fib α 36-50 cit (Figure 1A). PCA showed that case subjects tended to be defined based on the fraction of positive serologies in canonical and non-canonical serologies (Figure 1D).
We collapsed serologies into canonical and non-canonical serologies and assessed genetic differences. We regarded case subjects as positive for a set of Ab when they were positive for at least one serology of the group. We observed that 97.6% of CCP2(+) and 42.4% of CCP2(−) subjects were positive for at least one canonical cluster serology (excluding CCP2). In contrast, 79.3% of CCP2(+) and 21.3% of CCP2(−) subjects were positive for at least one non-canonical cluster serology (Figure 1A).
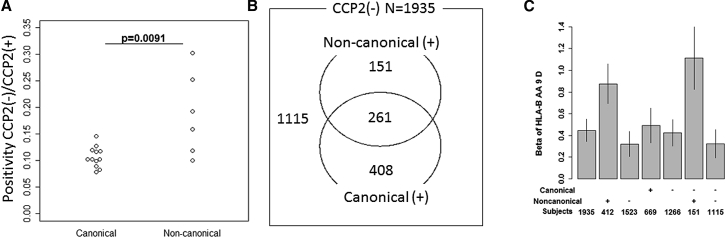
We observed that the ratio of positivity between CCP(−) and CCP(+) individuals was higher for the non-canonical clusters than canonical clusters (p = 0.0091, one-sided Mann-Whitney Rank sum test, Figure 2A, Table S4). This result matches the lower correlation we observe between non-canonical serologies and CCP2.
Figure 2.
Aspartic Acid Residue of Bpos-9, a Driver of Bpos-9 Association with RA, Is Associated with Non-canonical Cluster Serologies(+) Subset
(A) Enrichment fold of CCP2(−) RA in each ACPA is compared between the two clusters.
(B) Distribution of CCP2(−) subjects according to canonical and/or non-canonical serologies is indicated.
(C) Effect sizes of aspartic acid residue at Bpos-9 in susceptibility to subsets of CCP2(−) RA are indicated. Bar indicates 95% confidence intervals.
To investigate the role that genetics might play in driving ACPA serological phenotypes, we imputed eight HLA classical alleles (HLA-A, B, C, DRB1, DPA1, DPB1, DQA1, and DQB1) in 2- and 4-digit resolution along with amino acid residues at 966 protein sequence positions and 5,968 SNPs4 using dense SNP genotype data from the Immunochip. We applied logistic regression analysis and omnibus test across the HLA region.
We separately analyzed CCP2(+) and CCP(−) RA because of their well-documented different genetic architectures.22, 23, 24 We had previously demonstrated that HLA-B Bpos-9 is associated with RA regardless of CCP2 positivity status.4, 19 In contrast, HLA-DRB1 position 11 was associated with the two subsets in a different manner; different amino acid residues at position 11 confer strikingly different risk effects for CCP(+) and CCP(−) RA.4 For example the presence of Val residue at amino acid position 11 drives risk of CCP(+) RA, but is protective for CCP(−) RA.
We first analyzed CCP2(−) RA and found that aspartic acid residue of Bpos-9 was strongly associated with the presence of non-canonical serologies. Omnibus test using all CCP2(−) case subjects against control subjects confirmed that Bpos-9 was the strongest nominal association among amino acid positions within HLA-B (p = 5.5 × 10−15; Figure S4). This association was mainly driven by aspartic acid residue (Table S5), consistent with our previous reports. Then we analyzed subsets of CCP2(−) case subjects, subcatergorizing CCP2(−) case subjects by whether or not they were positive for canonical or non-canonical serologies and evaluated the associations of this position (Figure 2B). Since subdividing CCP2(−) case subjects led to a small dataset, we had less power to detect associations. However, focusing on Bpos-9 since this position is an established susceptibility position to RA, we found that nominal effect sizes of aspartic acid residue at Bpos-9 were different between the subsets and that the residue is associated with presence of non-canonical serologies (Figure 2C). CCP2(−) non-canonical(+) subjects constitute less than one fourth of CCP2(−) RA, conferred the association between CCP2(−) RA and Bpos-9 (omnibus_p = 9.2 × 10−17, Figures 2C and S4). This was not true for canonical serologies (Figure 2C). Canonical serologies(−) non-canonical serologies(+) subset showed the strongest effect size with significant omnibus p value at Bpos-9 in spite of the small sample size (n = 151, omnibus_p = 4.9 × 10−11, Figure 2C).
We previously showed that a subset of CCP2(−) subjects were genetically similar to CCP2(+) RA or AS.4, 14 Intra-case analysis of CCP2(−) RA revealed a significant association between CCP2-GRS and canonical cluster(+) subjects in comparison with other subjects (p ≤ 0.010, Table S6). Furthermore, subjects having multiple positive canonical cluster serologies showed higher CCP2-GRS than subjects having single positive canonical cluster serology (p = 0.013, Table S7). In contrast, we did not find significant association between non-canonical(+) RA and CCP2-GRS (p = 0.42, Table S8). These results indicate that while canonical serology positive subjects in CCP2(−) RA tends to be genetically similar to CCP2(+) RA, non-canonical serology(+) subjects are distinct from CCP2(+) RA. While we found a trend toward enrichment of AS-GRS in canonical(−) non-canonical(−) RA, it was not significant (p = 0.17, Table S9).
Next we analyzed CCP2(+) RA. Initially, we observed that Bpos-9 was not nominally associated with CCP2(+) RA. An association with Bpos-9 becomes apparent after conditioning on DRB1pos-11 (Figure S5). These results are consistent with previously published results.18
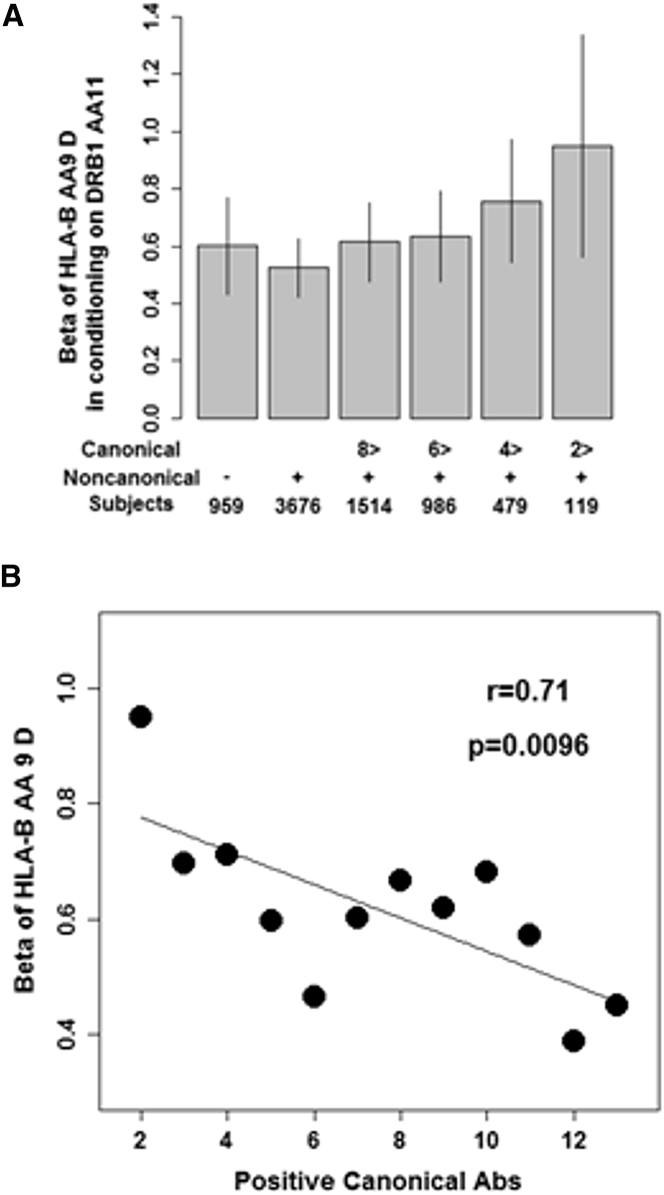
The majority of CCP2(+) RA-affected case subjects (79.3%) were positive for non-canonical serology (Figure S6). Subdivision of CCP2(+) RA based on positivity of non-canonical serology showed comparable effect sizes of aspartic acid residue of Bpos-9 after conditioning on DRB1pos-11 (Figure 3A). However, when we examined the association between Bpos-9 and susceptibility to non-canonical (+) RA according to the number of positive canonical serologies, reflecting relative contribution of purified non-canonical serology positivity, we found dose-dependent alteration of effect sizes of aspartic acid residue of Bpos-9 after conditioning on DRB1pos-11 (Figure 3A). The fewer positive canonical serologies in non-canonical (+) RA, the higher effect size for aspartic acid at Bpos-9 (p = 0.0096, Figure 3B). We observed the similar patterns after conditioning on DRB1pos-71 and DRB1pos-74 together with DRB1pos-11, the three susceptibility amino acid positions to CCP2(+) RA in the HLA-DRB1 (Figure S7). We found that attenuated effect size of valine residue at DRB1pos-11, main driver of the association of this position, on susceptibility to non-canonical cluster(+) RA according to decreased number of positive canonical serologies (Figure S8).
Figure 3.
Contribution of Bpos-9 to Susceptibility to CCP2(+) Non-canonical Cluster(+) RA
(A) Strong effect sizes of aspartic acid at Bpos-9 after conditioning on DRB1pos-11 are observed in subsets with decreased number of canonical serologies in CCP2(+) non-canonical(+) RA. Bar indicates 95% confidence intervals.
(B) Effect sizes of aspartic acid at Bpos-9 after conditioning on DRB1pos-11 are negatively correlated with number of canonical serologies when CCP2(+) non-canonical(+) RA are subdivided into bins based on number of positive canonical serologies.
These results suggest that the associations between RA and the presence of aspartic acid at Bpos-9 were driven by non-canonical serologies and not by canonical serologies. To confirm this point, we constructed a statistical model using all subjects by setting serologies of control subjects as negative and tested whether number of positive canonical or non-canonical serologies could predict dosages of aspartic acid residue at Bpos-9 (for details, see Material and Methods). As a result, case status and the number of positive non-canonical serologies showed significant positive associations, but that of positive canonical cluster serologies did not (p = 5.6 × 10−9, p = 5.1 × 10−6, and p = 0.25, respectively, Table 1). Furthermore, we found that the number of positive non-canonical serologies showed a significant negative multiplicative interactive effect with that of positive canonical serologies (p = 6.5 × 10−6, Table 1). These results indicate that positive association between non-canonical serologies and aspartic acid residue at Bpos-9 is cancelled out by canonical serologies. We also found that these association patterns were true for the analyses in only case subjects or analysis using positivity of non-canonical and canonical serologies (Table S10).
Table 1.
Non-canonical and Canonical Serologies Showed a Multiplicative Interactive Association with Aspartic Acid Residue at Bpos-9
|
Single Linear Regression |
Multiple Linear Regression |
|||||
|---|---|---|---|---|---|---|
| Items | Beta | SE | P | Beta | SE | p |
| Canonical serologies (quantitative) | 0.033 | 0.0037 | 2.0 × 10−19 | 0.0095 | 0.0082 | 0.25 |
| Non-canonical serologies (quantitative) | 0.057 | 0.0069 | 1.4 × 10−16 | 0.13 | 0.028 | 5.1 × 10−6 |
| Canonical serologies (quantitative) × Non-canonical serologies (quantitative) |
0.019 | 0.0031 | 3.3 × 10−10 | −0.059 | 0.013 | 6.5 × 10−6 |
| RA case-control status | 0.09 | 0.0073 | 7.6 × 10−35 | 0.074 | 0.013 | 5.6 × 10−9 |
SE, standard error; RA, rheumatoid arthritis
Discussion
In the current study, we showed that reactivity toward specific citrullinated peptides follow a specific pattern where a correlated cluster of reactivities is co-occurring among individuals (canonical serologies), while other reactivities are only weakly correlated to others (non-canonical serologies). These immunological reactivities have different genetic architectures. The association between RA and Bpos-9 is driven by subsets positive for non-canonical serologies. Non-canonical and canonical serologies show a multiplicative interaction with each other on the association of aspartic acid residue at Bpos-9.
This is the largest study so far to address genetic components underlying fine-specific ACPAs, and indeed to our knowledge, the largest study exploring the genetic basis of autoantibodies. We looked at ACPAs across four cohorts and observed that serological features of RA are consistent across different populations. We recognize that different thresholds might have been considered for serological positivity. We used the cut-off levels to define positivity in fine-specific Abs used in the previous studies,12, 14, 15 which insured high specificity for rheumatoid arthritis. While some of these thresholds might be overly conservative, they guarantee that positive signals represent RA-specific signals. Clustering and PCA revealed that canonical serologies are very consistent with each other, suggesting that these serologies are closely related as a consequence of epitope spreading25 or cross-reactivity. In contrast, the scattered distribution of non-canonical serologies in PCA suggest that these serologies may represent a more diverse group.
The close relationship between CCP2 and reactivity toward Fib β 60-74 cit is consistent with previous studies,14, 26 suggesting that this antibody captures a majority of CCP2(+) RA signal. Distinct clustering of Fib β 60-74 cit, Fib β 62-81 cit 72, and Fib β 62-81 cit 74, despite very similar antigen peptide amino acid sequences, indicate that slight difference in amino acid sequences of peptides strongly affect reactivity in RA. This phenomenon has been recently demonstrated from analysis of the reactivity of human monoclonal antibodies generated from single plasma cells from RA-affected individuals.27
We previously reported that Bpos-9 was the position associated with seropositive and negative RA. In addition, the associations of seropositive and negative RA were driven by Bpos-9 aspartic acid, indicating an important role of this allele position on RA pathophysiology regardless of autoantibody status. The current results showed that these associations were driven by subsets positive for non-canonical cluster serologies regardless of CCP2 status. Canonical and non-canonical serologies do largely co-occur. In fact, 96.4% of subjects positive for non-canonical serologies were also positive for canonical serologies. In our study, we found that relative abundance (ratio of the number of positive serologies) of non-canonical over canonical serologies had a specific association with Bpos9 (Table 1). The most extreme case was CCP2(−) subjects that were largely negative for canonical serologies (34.6% positive), but not necessarily negative for non-canonical serologies (21.3% positive). Consistent with this, the small subset of non-canonical(+)canonical(−)CCP2(−) subjects also demonstrated the association between CCP2(−) RA and Bpos9 (Figure 2C).
It is possible that while most of HLA-class II amino acid positions play roles in CCP2(+) RA through CD4+ T cells, Bpos-9 is involved with RA pathology regardless of CCP2 status via CD8+ T cells, which are present in RA synovium in parallel to CD4-positive T cells.28, 29 Several explanations may exist for such effects of both MHC class II and MHC class I alleles, and interactions between effects of MHC class II and class I alleles have been reported in other immune-mediated diseases, for example in multiple sclerosis.30 Thus antigen-specific CD8 cells may for example interact with antigen-presentation involved in activation of CD4 T cells providing help to antigen-specific B cells.31 Recent observation of abundant interferon gamma-producing CD8+ T cells within the inflamed synovium of RA-affected individuals supports this possibility.28 Future functional experiments will be needed to address these points.
Multiplicative interactive associations between canonical and non-canonical serologies on presence of aspartic acid residue at Bpos-9 indicate that the associations of Bpos-9 are opposite between canonical and non-canonical cluster serologies. Since, in CCP(+) individuals, most have both canonical and non-canonical cluster serologies, the association of Bpos-9 in CCP2(+) can be explained by non-canonical cluster serologies and is mainly brought by subjects with reduced number of positive canonical serologies. The negative interaction suggests that the two distinct cluster serologies were produced by exclusively different mechanisms.
HLA-DRB1∗03:01, a main driver of HLA-DRB1 associations in CCP2(−) RA32 but not in CCP2(+) RA, showed the same association patterns to Bpos-9 (data not shown). We have previously reported that HLA-DRB1∗03:01 is associated with CCP2(−) RA but is protective for CCP2(+) RA.4 HLA-DRB1∗03:01 and HLA-B∗0801, characterized by Bpos-9 aspartic acid, are located together on the 8.1 haplotype. However, they are not in complete LD (r2 = 0.68) and our previous study showed that the association of Bpos-9 aspartic acid was independent on HLA-DRB1∗03:01.4 The risk association of HLA-DRB1∗03:01 which is specific to CCP2(−) RA seems to be explained not only by Bpos-9 and non-canonical serologies, but by yet-to-be-defined components. For example, subjects with antibodies against native collagen type II at RA onset have an acute onset clinical phenotype and smoking association representing the opposite to CCP2.33 As anti-collagen II antibodies are associated with HLA-DRB1∗03, this might represent one possible explanation for the Bpos-9-independent HLA-DRB1∗03 association in CCP2(−) RA. While detailed information of clinical manifestations is not available in our subjects, it would be interesting to analyze connections between additional subphenotypes and Bpos-9 aspartic acid or DRB1∗03:01.
Our study also suggests that DRB1pos-11 effects are strongest in individual case subjects that are positive for many canonical serologies in CCP2(+) RA. This supports the existence of a common genetic architecture between CCP2(+) RA susceptibility and production of CCP2.34 This suggests a stronger involvement of DRB1pos-11 with production of canonical cluster serologies than non-canonical cluster serologies and is in agreement with our recent findings of similar shared epitope and smoking associations among ACPA peptide-positive individuals in the CCP2(−) subset as for the CCP2(+) subset.14
Previous studies of European and non-European populations have showed the association between Bpos-9 aspartic acid and seropositive RA and also seronegative RA.4, 19, 35 In this study we sought to define the specific relationship between the HLA associations and serological subtypes. While this is the largest study to our knowledge where fine-specific ACPA were quantified with the use of serum samples from subjects with RA, further replication studies are essential, including in non-European populations. These studies will require that future recruitment of RA-affected case subjects and control subjects include serum collections so that deep serotype phenotyping is possible.
Our results may have implications for other autoimmune diseases as well. Autoantibodies are associated with autoimmune thyroid diseases, myositis, systemic lupus erythematosus, autoimmune vasculitides, and other conditions. For each of these diseases, multiple antibodies have been defined. It is feasible to analyze other autoimmune diseases with autoantibodies to assess whether there are multiple serological clusters which are associated with different unique HLA genetic architectures and possible different clinical features.
Declaration of Interests
L.M.-A. is employed by Thermo Fisher Scientific. G.S. is a co-inventor of patents owned by bioMerieux (France), and licensed to Eurodiagnostica (NL), Axis-Shield and Genesis (UK) for commercialization of anti-CCP assays. As a civil service researcher, he receives a part of the royalties which are shared with University of Toulouse and Toulouse University-Hospital.
Acknowledgments
This study was supported by The Swedish Research Council, the EU/IMI project RTCure, Stockholm County Council, the insurance company AFA, the Swedish Rheumatism Association, King Gustaf V's 80-year Foundation, and the Swedish Research Council for Health, Working Life and Welfare. We thank Lena Israelsson for handling serum samples and the EIRA study group for help in individual characterization and capture of serum samples. S.R. is supported by funding from the National Institute of Arthritis, Musculoskeletal, and Skin Diseases (1R01AR063759). A.B. is supported by funding from the NIHR Manchester Biomedical Research Center.
Published: August 29, 2019; corrected online September 20, 2019
Footnotes
Supplemental Data can be found online at https://doi.org/10.1016/j.ajhg.2019.08.002.
Contributor Information
Lars Klareskog, Email: lars.klareskog@ki.se.
Soumya Raychaudhuri, Email: soumya@broadinstitute.org.
Web Resources
OMIM, https://www.omim.org/
PLINK 1.9, https://www.cog-genomics.org/plink2/
R statistical software, https://www.r-project.org/
Supplemental Data
References
- 1.Klareskog L., Stolt P., Lundberg K., Källberg H., Bengtsson C., Grunewald J., Rönnelid J., Harris H.E., Ulfgren A.K., Rantapää-Dahlqvist S. A new model for an etiology of rheumatoid arthritis: smoking may trigger HLA-DR (shared epitope)-restricted immune reactions to autoantigens modified by citrullination. Arthritis Rheum. 2006;54:38–46. doi: 10.1002/art.21575. [DOI] [PubMed] [Google Scholar]
- 2.Verpoort K.N., Cheung K., Ioan-Facsinay A., van der Helm-van Mil A.H., de Vries-Bouwstra J.K., Allaart C.F., Drijfhout J.W., de Vries R.R., Breedveld F.C., Huizinga T.W. Fine specificity of the anti-citrullinated protein antibody response is influenced by the shared epitope alleles. Arthritis Rheum. 2007;56:3949–3952. doi: 10.1002/art.23127. [DOI] [PubMed] [Google Scholar]
- 3.Kim K., Jiang X., Cui J., Lu B., Costenbader K.H., Sparks J.A., Bang S.Y., Lee H.S., Okada Y., Raychaudhuri S. Interactions between amino acid-defined major histocompatibility complex class II variants and smoking in seropositive rheumatoid arthritis. Arthritis Rheumatol. 2015;67:2611–2623. doi: 10.1002/art.39228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Han B., Diogo D., Eyre S., Kallberg H., Zhernakova A., Bowes J., Padyukov L., Okada Y., González-Gay M.A., Rantapää-Dahlqvist S. Fine mapping seronegative and seropositive rheumatoid arthritis to shared and distinct HLA alleles by adjusting for the effects of heterogeneity. Am. J. Hum. Genet. 2014;94:522–532. doi: 10.1016/j.ajhg.2014.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wedderburn L.R., McHugh N.J., Chinoy H., Cooper R.G., Salway F., Ollier W.E., McCann L.J., Varsani H., Dunphy J., North J., Davidson J.E., Juvenile Dermatomyositis Research Group (JDRG) HLA class II haplotype and autoantibody associations in children with juvenile dermatomyositis and juvenile dermatomyositis-scleroderma overlap. Rheumatology (Oxford) 2007;46:1786–1791. doi: 10.1093/rheumatology/kem265. [DOI] [PubMed] [Google Scholar]
- 6.O’Hanlon T.P., Rider L.G., Mamyrova G., Targoff I.N., Arnett F.C., Reveille J.D., Carrington M., Gao X., Oddis C.V., Morel P.A. HLA polymorphisms in African Americans with idiopathic inflammatory myopathy: allelic profiles distinguish patients with different clinical phenotypes and myositis autoantibodies. Arthritis Rheum. 2006;54:3670–3681. doi: 10.1002/art.22205. [DOI] [PubMed] [Google Scholar]
- 7.Chinoy H., Adimulam S., Marriage F., New P., Vincze M., Zilahi E., Kapitány A., Gyetvai A., Ekholm L., Novota P. Interaction of HLA-DRB1∗03 and smoking for the development of anti-Jo-1 antibodies in adult idiopathic inflammatory myopathies: a European-wide case study. Ann. Rheum. Dis. 2012;71:961–965. doi: 10.1136/annrheumdis-2011-200182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Howson J.M., Stevens H., Smyth D.J., Walker N.M., Chandler K.A., Bingley P.J., Todd J.A. Evidence that HLA class I and II associations with type 1 diabetes, autoantibodies to GAD and autoantibodies to IA-2, are distinct. Diabetes. 2011;60:2635–2644. doi: 10.2337/db11-0131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Terao C., Ohmura K., Kochi Y., Ikari K., Maruya E., Katayama M., Shimada K., Murasawa A., Honjo S., Takasugi K. A large-scale association study identified multiple HLA-DRB1 alleles associated with ACPA-negative rheumatoid arthritis in Japanese subjects. Ann. Rheum. Dis. 2011;70:2134–2139. doi: 10.1136/annrheumdis-2011-200353. [DOI] [PubMed] [Google Scholar]
- 10.Padyukov L., Seielstad M., Ong R.T., Ding B., Rönnelid J., Seddighzadeh M., Alfredsson L., Klareskog L., Epidemiological Investigation of Rheumatoid Arthritis (EIRA) study group A genome-wide association study suggests contrasting associations in ACPA-positive versus ACPA-negative rheumatoid arthritis. Ann. Rheum. Dis. 2011;70:259–265. doi: 10.1136/ard.2009.126821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Westra H.J., Martínez-Bonet M., Onengut-Gumuscu S., Lee A., Luo Y., Teslovich N., Worthington J., Martin J., Huizinga T., Klareskog L. Fine-mapping and functional studies highlight potential causal variants for rheumatoid arthritis and type 1 diabetes. Nat. Genet. 2018;50:1366–1374. doi: 10.1038/s41588-018-0216-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hansson M., Mathsson L., Schlederer T., Israelsson L., Matsson P., Nogueira L., Jakobsson P.J., Lundberg K., Malmström V., Serre G. Validation of a multiplex chip-based assay for the detection of autoantibodies against citrullinated peptides. Arthritis Res. Ther. 2012;14:R201. doi: 10.1186/ar4039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Too C.L., Murad S., Hansson M., Alm L.M., Dhaliwal J.S., Holmdahl R., Jakobsson P.J., Alfredsson L., Klareskog L., Rönnelid J., Padyukov L. Differences in the Spectrum of Anti-Citrullinated Protein Antibody Fine Specificities Between Malaysian and Swedish Patients With Rheumatoid Arthritis: Implications for Disease Pathogenesis. Arthritis Rheumatol. 2017;69:58–69. doi: 10.1002/art.39827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rönnelid J., Hansson M., Mathsson-Alm L., Cornillet M., Reed E., Jakobsson P.J., Alfredsson L., Holmdahl R., Skriner K., Serre G. Anticitrullinated protein/peptide antibody multiplexing defines an extended group of ACPA-positive rheumatoid arthritis patients with distinct genetic and environmental determinants. Ann. Rheum. Dis. 2018;77:203–211. doi: 10.1136/annrheumdis-2017-211782. [DOI] [PubMed] [Google Scholar]
- 15.Lundberg K., Bengtsson C., Kharlamova N., Reed E., Jiang X., Kallberg H., Pollak-Dorocic I., Israelsson L., Kessel C., Padyukov L. Genetic and environmental determinants for disease risk in subsets of rheumatoid arthritis defined by the anticitrullinated protein/peptide antibody fine specificity profile. Ann. Rheum. Dis. 2013;72:652–658. doi: 10.1136/annrheumdis-2012-201484. [DOI] [PubMed] [Google Scholar]
- 16.Eyre S., Bowes J., Diogo D., Lee A., Barton A., Martin P., Zhernakova A., Stahl E., Viatte S., McAllister K., Biologics in Rheumatoid Arthritis Genetics and Genomics Study Syndicate. Wellcome Trust Case Control Consortium High-density genetic mapping identifies new susceptibility loci for rheumatoid arthritis. Nat. Genet. 2012;44:1336–1340. doi: 10.1038/ng.2462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jia X., Han B., Onengut-Gumuscu S., Chen W.M., Concannon P.J., Rich S.S., Raychaudhuri S., de Bakker P.I. Imputing amino acid polymorphisms in human leukocyte antigens. PLoS ONE. 2013;8:e64683. doi: 10.1371/journal.pone.0064683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Brown W.M., Pierce J., Hilner J.E., Perdue L.H., Lohman K., Li L., Venkatesh R.B., Hunt S., Mychaleckyj J.C., Deloukas P., Type 1 Diabetes Genetics Consortium Overview of the MHC fine mapping data. Diabetes Obes. Metab. 2009;11(Suppl 1):2–7. doi: 10.1111/j.1463-1326.2008.00997.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Raychaudhuri S., Sandor C., Stahl E.A., Freudenberg J., Lee H.S., Jia X., Alfredsson L., Padyukov L., Klareskog L., Worthington J. Five amino acids in three HLA proteins explain most of the association between MHC and seropositive rheumatoid arthritis. Nat. Genet. 2012;44:291–296. doi: 10.1038/ng.1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Purcell S., Neale B., Todd-Brown K., Thomas L., Ferreira M.A., Bender D., Maller J., Sklar P., de Bakker P.I., Daly M.J., Sham P.C. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.van Venrooij W.J., Zendman A.J. Anti-CCP2 antibodies: an overview and perspective of the diagnostic abilities of this serological marker for early rheumatoid arthritis. Clin. Rev. Allergy Immunol. 2008;34:36–39. doi: 10.1007/s12016-007-8029-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Terao C., Ohmura K., Kochi Y., Ikari K., Okada Y., Shimizu M., Nishina N., Suzuki A., Myouzen K., Kawaguchi T. Anti-citrullinated peptide/protein antibody (ACPA)-negative RA shares a large proportion of susceptibility loci with ACPA-positive RA: a meta-analysis of genome-wide association study in a Japanese population. Arthritis Res. Ther. 2015;17:104. doi: 10.1186/s13075-015-0623-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Viatte S., Plant D., Bowes J., Lunt M., Eyre S., Barton A., Worthington J. Genetic markers of rheumatoid arthritis susceptibility in anti-citrullinated peptide antibody negative patients. Ann. Rheum. Dis. 2012;71:1984–1990. doi: 10.1136/annrheumdis-2011-201225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ohmura K., Terao C., Maruya E., Katayama M., Matoba K., Shimada K., Murasawa A., Honjo S., Takasugi K., Tohma S. Anti-citrullinated peptide antibody-negative RA is a genetically distinct subset: a definitive study using only bone-erosive ACPA-negative rheumatoid arthritis. Rheumatology (Oxford) 2010;49:2298–2304. doi: 10.1093/rheumatology/keq273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Elliott S.E., Kongpachith S., Lingampalli N., Adamska J.Z., Cannon B.J., Mao R., Blum L.K., Robinson W.H. Affinity Maturation Drives Epitope Spreading and Generation of Proinflammatory Anti-Citrullinated Protein Antibodies in Rheumatoid Arthritis. Arthritis Rheumatol. 2018;70:1946–1958. doi: 10.1002/art.40587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cornillet M., Sebbag M., Verrouil E., Magyar A., Babos F., Ruyssen-Witrand A., Hudecz F., Cantagrel A., Serre G., Nogueira L. The fibrin-derived citrullinated peptide beta60-74Cit(6)(0),(7)(2),(7)(4) bears the major ACPA epitope recognised by the rheumatoid arthritis-specific anticitrullinated fibrinogen autoantibodies and anti-CCP2 antibodies. Ann. Rheum. Dis. 2014;73:1246–1252. doi: 10.1136/annrheumdis-2012-202868. [DOI] [PubMed] [Google Scholar]
- 27.Steen J., Forsström B., Sahlström P., Odowd V., Israelsson L., Krishnamurthy A., Badreh S., Mathsson Alm L., Compson J., Ramsköld D. Recognition of Amino Acid Motifs, Rather Than Specific Proteins, by Human Plasma Cell-Derived Monoclonal Antibodies to Posttranslationally Modified Proteins in Rheumatoid Arthritis. Arthritis Rheumatol. 2019;71:196–209. doi: 10.1002/art.40699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang F., Wei K., Slowikowski K., Fonseka C.Y., Rao D.A., Kelly S., Goodman S.M., Tabechian D., Hughes L.B., Salomon-Escoto K. Defining Inflammatory Cell States in Rheumatoid Arthritis Joint Synovial Tissues by Integrating Single-cell Transcriptomics and Mass Cytometry. Nat. Immunol. 2018;20:928–942. doi: 10.1038/s41590-019-0378-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Klareskog L., Forsum U., Scheynius A., Kabelitz D., Wigzell H. Evidence in support of a self-perpetuating HLA-DR-dependent delayed-type cell reaction in rheumatoid arthritis. Proc. Natl. Acad. Sci. USA. 1982;79:3632–3636. doi: 10.1073/pnas.79.11.3632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Link J., Kockum I., Lorentzen A.R., Lie B.A., Celius E.G., Westerlind H., Schaffer M., Alfredsson L., Olsson T., Brynedal B. Importance of human leukocyte antigen (HLA) class I and II alleles on the risk of multiple sclerosis. PLoS ONE. 2012;7:e36779. doi: 10.1371/journal.pone.0036779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nakagawa H., Wang L., Cantor H., Kim H.J. New Insights Into the Biology of CD8 Regulatory T Cells. Adv. Immunol. 2018;140:1–20. doi: 10.1016/bs.ai.2018.09.001. [DOI] [PubMed] [Google Scholar]
- 32.Verpoort K.N., van Gaalen F.A., van der Helm-van Mil A.H., Schreuder G.M., Breedveld F.C., Huizinga T.W., de Vries R.R., Toes R.E. Association of HLA-DR3 with anti-cyclic citrullinated peptide antibody-negative rheumatoid arthritis. Arthritis Rheum. 2005;52:3058–3062. doi: 10.1002/art.21302. [DOI] [PubMed] [Google Scholar]
- 33.Manivel V.A., Mullazehi M., Padyukov L., Westerlind H., Klareskog L., Alfredsson L., Saevarsdottir S., Rönnelid J. Anticollagen type II antibodies are associated with an acute onset rheumatoid arthritis phenotype and prognosticate lower degree of inflammation during 5 years follow-up. Ann. Rheum. Dis. 2017;76:1529–1536. doi: 10.1136/annrheumdis-2016-210873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Terao C., Suzuki A., Ikari K., Kochi Y., Ohmura K., Katayama M., Nakabo S., Yamamoto N., Suzuki T., Iwamoto T. An association between amino acid position 74 of HLA-DRB1 and anti-citrullinated protein antibody levels in Japanese patients with anti-citrullinated protein antibody-positive rheumatoid arthritis. Arthritis Rheumatol. 2015;67:2038–2045. doi: 10.1002/art.39133. [DOI] [PubMed] [Google Scholar]
- 35.Okada Y., Kim K., Han B., Pillai N.E., Ong R.T., Saw W.Y., Luo M., Jiang L., Yin J., Bang S.Y. Risk for ACPA-positive rheumatoid arthritis is driven by shared HLA amino acid polymorphisms in Asian and European populations. Hum. Mol. Genet. 2014;23:6916–6926. doi: 10.1093/hmg/ddu387. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.