Abstract
Purpose: Lung adenocarcinoma (LUAD) of non-small cell lung cancer (NSCLC) is a highly prevalent cancer with high mortality. The gene basigin (BSG) is strongly expressed in certain tumors. This study investigated the expression level of BSG in LUAD and its role in the poor prognosis of LUAD. Methods: The mRNA expression of BSG in LUAD was from GEO, Oncomine and TCGA database. Prognostic data were provided by SurvExpress. The expression of BSG protein was detected by immunohistochemistry (IHC). Cell function experiments of viability, proliferation and apoptosis assays were performed in A549. Clustering analysis of BSG co-expressed genes was generated by Gene Ontology (GO) Enrichment, KEGG pathway and protein-protein interaction (PPI) network. Results: BSG mRNA level was significantly over-expressed in LUAD (pooled SMD = 0.564, 0.448-0.681, P<0.001). The analysis in SurvExpress revealed that high expression of BSG indicated significantly poor prognosis (pooled HR = 1.20, 1.10-1.30, P<0.0001). IHC assay also showed that BSG protein expression was significantly up-regulated in LUAD (P<0.001), and positive BSG expression was notably associated with higher pathology grade (P = 0.041) and lymphatic metastasis (P = 0.014). Moreover, BSG can enhance the viability and proliferation ability (both P<0.001) and weaken cell apoptosis (P<0.001) in A549. The most enriched GO terms in the co-expressed genes of BSG were translation related enrichment. The KEGG pathway showed that these genes were markedly involved in Ribosome pathways. Conclusion: Up-regulated BSG in LUAD is related to advanced progression and poor prognosis by influencing cell viability, proliferation and apoptosis. BSG could be a potential biomarker for the targeted therapy of LUAD.
Keywords: Lung adenocarcinoma (LUAD), basigin (BSG), proliferation, viability, apoptosis, bioinformatics analysis
Introduction
It has been well-understood that lung cancer, with high morbidity and mortality in humans, ranks as the most threatening cancer in a suburban area of China [1], and even worldwide [2]. Approximately 80%-85% of lung cancer is determined to be non-small cell lung cancer (NSCLC), which has a poor prognosis. The curative effect and prognosis of the present treatments for NSCLC, which consist of surgical resection, radiofrequency ablation, radiotherapy, platinum-based chemotherapies and targeted therapy, are unsatisfactory [3,4]. Researchers are studying the invasive and metastatic mechanism of NSCLC at various molecular levels and signal pathways [5,6]. In this study, we investigated the related molecular mechanism of lung adenocarcinoma (LUAD), which belongs to NSCLC.
Increasing evidence shows that basigin (BSG, also named TCSF, CD147, EMMPRIN) could stimulate para-cancerous fibroblast secreting matrix metalloproteinases (MMP) [7]. The increased concentration and viability of MMP can cause excessive degradation of extracellular matrix and basement membrane of blood vessels. Consequently, cancer cells can easily penetrate the basement membrane of blood vessels and extracellular matrix causing invasion and distant metastasis [8].
Large-scale studies reported that BSG was overexpressed in various human cancers, such as hepatocellular carcinoma [9], colon cancer [10] and breast cancer [11]. Moreover, the role of BSG in NSCLC [12] had been examined to regulate MMP secretion and influence tumor invasion and metastasis. Additionally, evidence showed that BSG may also induce chemoresistance in cancer cells by promoting the synthesis of hyaluronan [13].
In this study, we made a deep investigation into BSG from high-throughput data-mining, clinical immunohistochemistry verification, in vitro validation and in silico investigation to have a comprehensive understanding of BSG.
Material and methods
Data source and selection criteria
Public high-throughput RNA-sequencing and microarray data were acquired from Gene Expression Omnibus (GEO) of National Center of Biotechnology Information (NCBI) database (https://www.ncbi.nlm.nih.gov/geo/), Oncomine database (https://www.oncomine.org/resource/main.html), and The Cancer Genome Atlas (TCGA) database (https://cancergenome.nih.gov/) [14]. SurvExpress database (http://bioinformatica.mty.itesm.mx:8080/Biomatec/SurvivaX.jsp) was used to explore the survival data of BSG in lung cancer tissues, which included 23 available databases [15]. The keywords searched in GEO were (BSG OR Basigin OR CD147 OR “Extracellular Matrix Metalloproteinase Inducer” OR EMMPRIN OR “M6 activation antigen” OR “collagenase stimulatory factor protein” OR HAb18G OR “human leukocyte activation antigen M6” OR “5F7 protein” or “CD147 Antigen”). In the GEO dataset search, GEO series, which met the following criteria were selected for our study: (1) Samples contained LUAD group and noncancerous control group. (2) Samples were from clinical patients. (3) Series detected expression profiling by RT-qPCR/array/high-throughput sequencing data.
In-house validation with immunohistochemistry
In this study, a total of 127 LUAD and 30 normal lung tissues were included in a tissue microarray. All of the patient tissues were collected from the Pathology Department, First Affiliated Hospital of Guangxi Medical University, P. R. China (January 2010 to February 2014). The research was authorized by the Ethical Committee of the First Affiliated Hospital of Guangxi Medical University and the usage of the tissue study had consent from clinical doctors and patients. All the clinicopathological features were obtained to analyze the relationship to BSG expression.
Immunohistochemistry and scoring criteria
The expression of BSG protein was detected in the aforementioned LUAD patients by IHC, which was performed on formalin-fixed and paraffin-embedded tissue array sections. After being deparaffinized and boiled in sodium citrate buffer (0.01 mol/L, pH = 6.0) in microwave for ten minutes, tissue sections were applied the anti-CD147 (BSG) antibody [EPR4052] (1:250 dilution, Abcam, USA), stained by avidin-biotin-peroxidase method and counterstained with hematoxylin.
Every sample was scored by two independent pathologists (Gang Chen and Kang-lai Wei). Five visions were randomly captured and assessed by the percentage and intensity of the stained area. The positive BSG area <10%, 10-25%, 25-50%, 50-75% and 75-100% graded 0, 1, 2, 3 and 4 point, respectively. The staining intensity weakly, moderately, strongly graded 1, 2 and 3 point, respectively. If the added two scores were greater than 2, the tissue sample was regarded as BSG positive; otherwise, it was negative [16].
Cell culture and establishment of siRNA infected cell line
The human LUAD cell line A549 was cultured at Roswell Park Memorial Institute (RPMI); 1640 medium (GIBCO, Los Angeles, CA, USA), which contained 10% fetal bovine serum (FBS) and 100 U/mL penicillin-streptomycin sulfate. The culture dishes were placed in the incubator with a humidified 5% CO2 37°C environment. Lipofectamine 2000 reagent (Invitrogen, Carlsbad, CA, USA) was applied to the cell transfection. Cells were transfected according to the manufacturer’s protocol. The siRNA targeting human BSG with the sequence of 5’-GGUCAGAGCUACACAUUGA-3’ and the siRNA used as negative control were purchased from Ambion, Life Technologies Europe B.V., Ghent, Belgium.
Cell proliferation trial
The cell proliferation trial was conducted by Promega MTS kit (G3580, Promega, USA). The cell line A549 was prepared in a 96-well plate. Next, reagent was added into the 96-well plate in the quantity of 20 μl per well. Two hour later, the absorbance fluorescence values were measured at 490 nm [17].
Cell viability assay
The cell viability assay was conducted by CellTiter-Blue Cell Viability Assay kit (G8080, Promega, USA). The cell line A549 was prepared in a 96-well plate. Next, reagent was added into the 96-well plate in the quantity of 20 μl per well. Two hours later, the absorbance fluorescence values were measured at 560/590 nm.
To obtain a reliable result of cell viability, Hoechst 33342 and Propidium Iodide (PI) (Sigma, USA) double fluorescent staining assay were used to stain the cells. The stained cells were observed under microscope (200×). Normal cells showed weak red and weak blue fluorescence. Apoptotic cells showed strong blue and weak red fluorescence. Necrotic cells showed strong blue and red fluorescence [18].
Cell apoptosis experiment
The Apo-ONE Homogeneous Caspase-3/7 Assay (G7790, Promega, USA) was applied to cell apoptosis experiment. The cell line A549 was prepared in a 96-well plate. The reagent was mixed into the 96-well plate containing blank, control and samples. One hour later, the absorbance fluorescence values were measured with the excitation wavelength for detection of 499 nm, and the emission maximum at a wavelength of 521 nm.
To get a reliable result of cell apoptosis, Hoechst 33342 and PI double fluorescent staining assay was also adopted to stain the cells as above [19].
Clustering analysis of BSG co-expressed genes
To analyze the potential pathways related to BSG, co-expressed genes associated with BSG were obtained from Multi Experiment Matrix (MEM) database (http://biit.cs.ut.ee/mem/index.cgi) [20,21] and cBioPortal for Cancer Genomics (cBioPortal) database (http://www.cbioportal.org). Venny 2.1.0 (By Juan Carlos Oliveros) tool (http://bioinfogp.cnb.csic.es/tools/venny/index.html) was applied to achieve the intersection co-expression genes of the two databases. Next, the obtained BSG co-expressed genes were analyzed by DAVID Bioinformatics Resources 6.8 database (https://david.ncifcrf.gov/) for Gene Ontology (GO) clustering analysis and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis. STRING: Functional protein association networks 10.5 (https://string-db.org/) was the approach for protein-protein interaction (PPI) analysis [22].
Statistical analysis
All the original data of high-throughput expression databases were transformed by exponential log2. The mean value of BSG expression and standard deviation (SD) were calculated by SPSS 22.0 (IBM, New York, USA). Also, the standard mean difference (SMD) and 95% confidence interval (CI) were calculated by Stata 12.0 (StataCorp, College Station, TX, USA).
Z-test was performed to evaluate the significance of pooled estimates with two-sided p-values and was considered significant when P<0.05. Q-test chi-square and the Higgins I-squared were estimated to assess heterogeneity across studies [23]. I-squared values of 75%, 50% and 25% manifested high, moderate and low heterogeneity, respectively. A fixed-effects model (Mantel-Haenszel method) was used to estimate the overall pooled results, if P<0.1 in Q-test. Otherwise, a random-effect model (DerSimonian-Laird method) would be applied. Sensitivity analysis was adopted by the means of omitting one study at a time to evaluate the reliability of the pooled estimates. Publication bias was assessed by the symmetry of funnel plot visually and by Egger’s and Begg’s linear regression statistically. All statistical results were obtained by Stata 12.0 (StataCorp, College Station, TX, USA).
Results
Expression of BSG in public high-throughput datasets
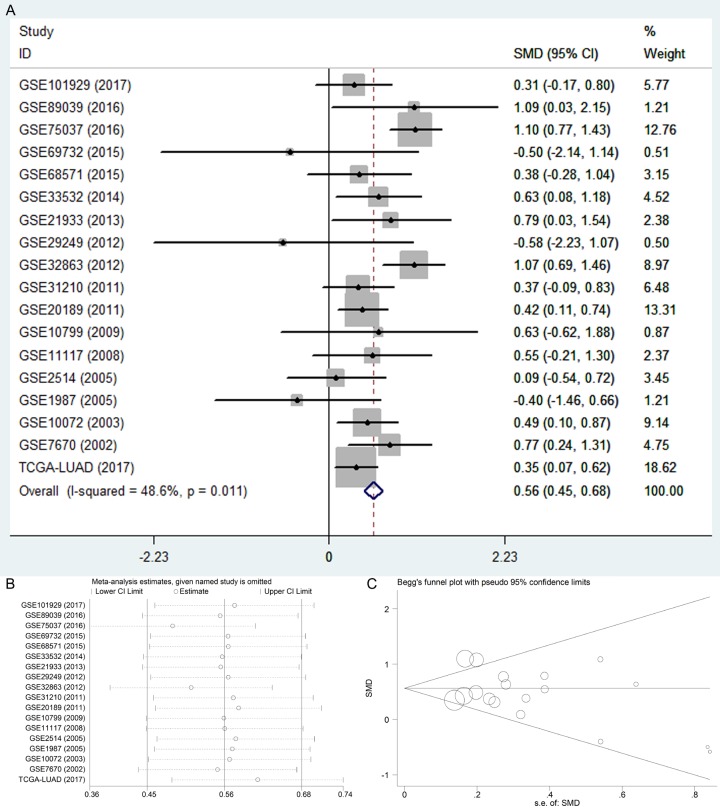
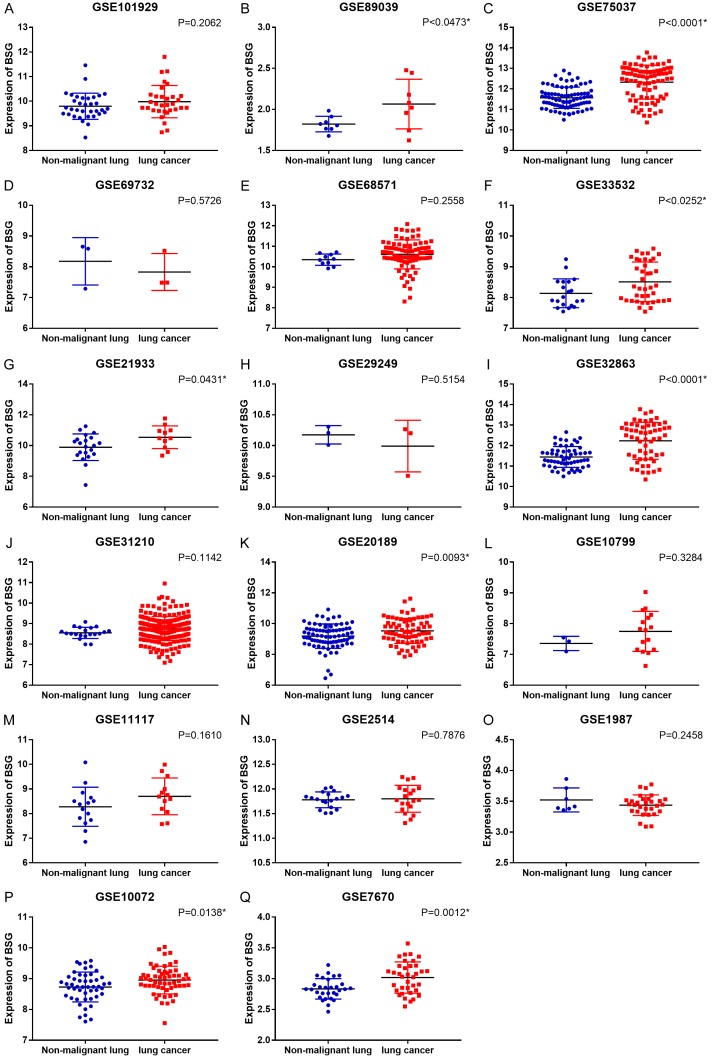
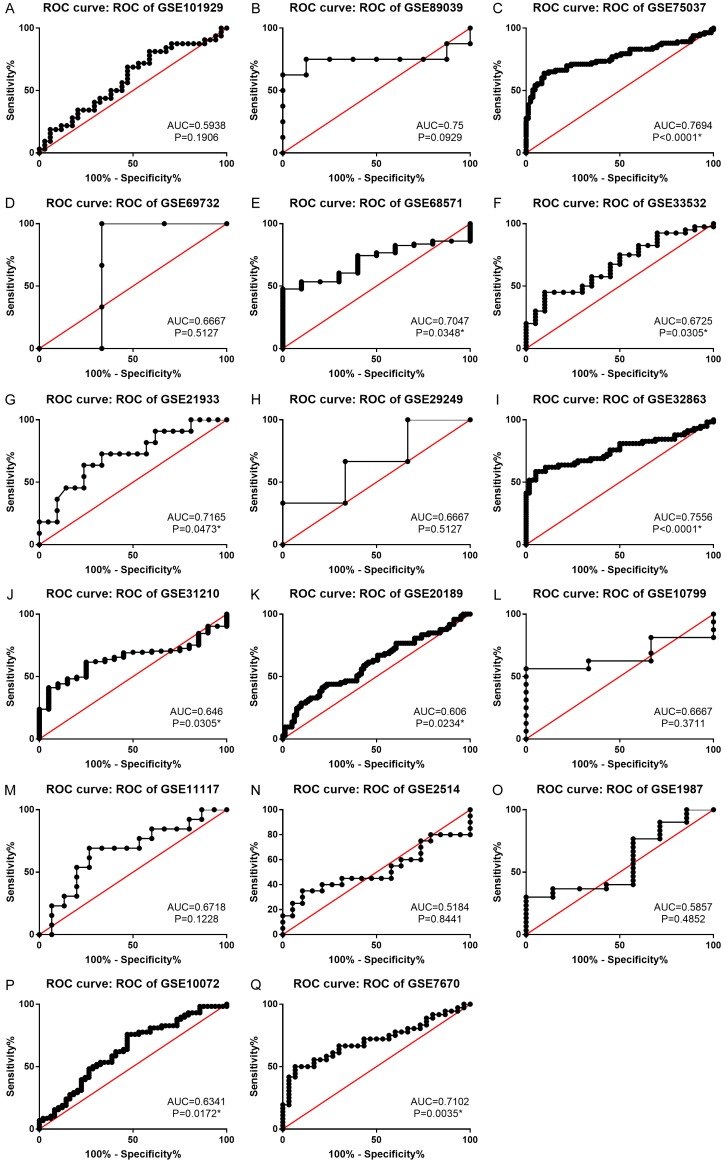
After searching from the GEO datasets, eighteen BSG expression datasets obtained from GEO or Oncomine and the LUAD data from TCGA database were included into this study (Table 1). We calculated the expression level of BSG in LUAD from the eighteen high-throughput datasets of RNA-sequencing and microarray data. The pooled SMD and 95% CI were conducted by a fix-effects model when we deleted eight datasets to make the heterogeneity I-squared <50% among the eighteen studies (I-squared = 48.6%, P = 0.011). The expression of BSG in LUAD was significantly higher than the control group (pooled SMD = 0.564, 95% CI: 0.448-0.681, P<0.001, Figure 1A). The expression of BSG in every study of GEO datasets is shown in Figure 2. The ROC curves of BSG expression in GEO are shown in Figure 3.
Table 1.
Main characteristics and results of the eligible studies of GEO, Oncomine and TCGA database
| Data source | Series | Author | Country | Platform | Source | Total sample | LUAD group | Control group | t | p-value | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
|
|
|||||||||||||
| Number | Mean | SD | Number | Mean | SD | |||||||||
| GEO | GSE101929 | Mitchell | USA | GPL570 | tissue | 66 | 32 | 9.9831 | 0.6555 | 34 | 9.7959 | 0.5319 | 1.277 | 0.206 |
| GEO | GSE89039 | Zhang | China | GPL17077 | tissue | 16 | 8 | 2.0661 | 0.30253 | 8 | 1.8223 | 0.09485 | 2.175 | 0.06 |
| GEO | GSE75037 | Girard | USA | GPL6884 | tissue | 166 | 83 | 12.328 | 0.8279 | 83 | 11.5692 | 0.51708 | 7.082 | <0.001 |
| GEO | GSE69732 | Luo | Taiwan, China | GPL14951 | blood | 6 | 3 | 7.8316 | 0.59863 | 3 | 8.177 | 0.7698 | -0.614 | 0.573 |
| GEO, Oncomine | GSE68571 | Heiskanen | USA | GPL80 | tissue | 96 | 86 | 10.6057 | 0.70797 | 10 | 10.3465 | 0.27471 | 1.143 | 0.256 |
| GEO | GSE33532 | Meister | Germany | GPL570 | tissue | 100 | 40 | 8.5114 | 0.64197 | 20 | 8.1396 | 0.46914 | 2.547 | 0.014 |
| GEO | GSE21933 | Wang | Taiwan, China | GPL6254 | tissue | 42 | 11 | 10.5396 | 0.73921 | 21 | 9.8929 | 0.86131 | 2.112 | 0.043 |
| GEO | GSE29249 | Ma | China | GPL10558 | tissue | 12 | 3 | 9.9916 | 0.41926 | 3 | 10.1748 | 0.14972 | -0.713 | 0.515 |
| GEO, Oncomine | GSE32863 | Laird-Offringa | USA | GPL6884 | tissue | 116 | 58 | 12.2309 | 0.90385 | 58 | 11.443 | 0.50775 | 5.788 | <0.001 |
| GEO, Oncomine | GSE31210 | Kohno | Japan | GPL570 | tissue | 246 | 226 | 8.761 | 0.58791 | 20 | 8.5503 | 0.27429 | 2.897 | 0.006 |
| GEO | GSE20189 | Rotunno | USA | GPL571 | blood | 162 | 73 | 9.5296 | 0.80979 | 81 | 9.1888 | 0.7948 | 2.633 | 0.009 |
| GEO | GSE10799 | Streichert | Germany | GPL570 | tissue | 19 | 16 | 7.7469 | 0.65006 | 3 | 7.3571 | 0.23229 | 1.006 | 0.328 |
| GEO | GSE11117 | Budach | Switzerland | GPL6650 | tissue | 56 | 13 | 8.7007 | 0.74643 | 15 | 8.2783 | 0.7944 | 1.443 | 0.161 |
| Oncomine | GSE2514 | Stearman | USA | GPL8300 | tissue | 39 | 20 | 11.8006 | 0.27324 | 19 | 11.781 | 0.16047 | 0.275 | 0.785 |
| GEO | GSE1987 | Dehan | Israel | GPL91 | tissue | 37 | 7 | 11.4312 | 0.70249 | 7 | 11.7023 | 0.64842 | -0.75 | 0.468 |
| GEO, Oncomine | GSE10072 | Lee | USA | GPL96 | tissue | 107 | 58 | 8.956 | 0.4501 | 49 | 8.7295 | 0.48385 | 2.505 | 0.014 |
| GEO, Oncomine | GSE7670 | Su | Taiwan, China | GPL96 | tissue | 66 | 31 | 9.8428 | 0.73624 | 27 | 9.3547 | 0.4858 | 3.014 | 0.004 |
| TCGA | TCGA-LUAD | TCGA | USA | None | tissue | 562 | 503 | 13.375 | 0.605 | 59 | 13.174 | 0.321 | 4.06 | <0.001 |
Figure 1.
The expression of BSG in LUAD from GEO, Oncomine and TCGA database. A. Forest plot. B. Sensitivity analysis plot. C. The Begg’s funnel plot of the publication bias. Abbreviations: LUAD, lung adenocarcinoma; CI, confidence interval; SMD, standard mean difference.
Figure 2.
Differential expression of BSG in cancer and non-cancerous tissues from GEO datasets. *P<0.05.
Figure 3.
ROC curve of BSG expression in LUAD. *P<0.05. Abbreviations: LUAD, lung adenocarcinoma; AUC, area under the curve; ROC, receive operator characteristic curve.
The sensitivity analysis was conducted to verify that the pooled SMD was stable (Figure 1B). The Begg’s and Egger’s tests showed the publication bias was not significant when the p-values were greater than 0.05 (Begg’s P = 0.544, Egger’s P = 0.303, Figure 1C).
Prognostic role of BSG in lung cancer
SurvExpress database showed that the expression of BSG in lung cancer patients indicated significantly poor prognosis (lung cancer vs non-cancerous tissue, HR = 1.20, 95% CI: 1.10-1.30, P<0.0001) (Figure 4A). The Begg’s funnel plot and Egger’s publication bias plot are shown in Figure 4B, 4C.
Figure 4.
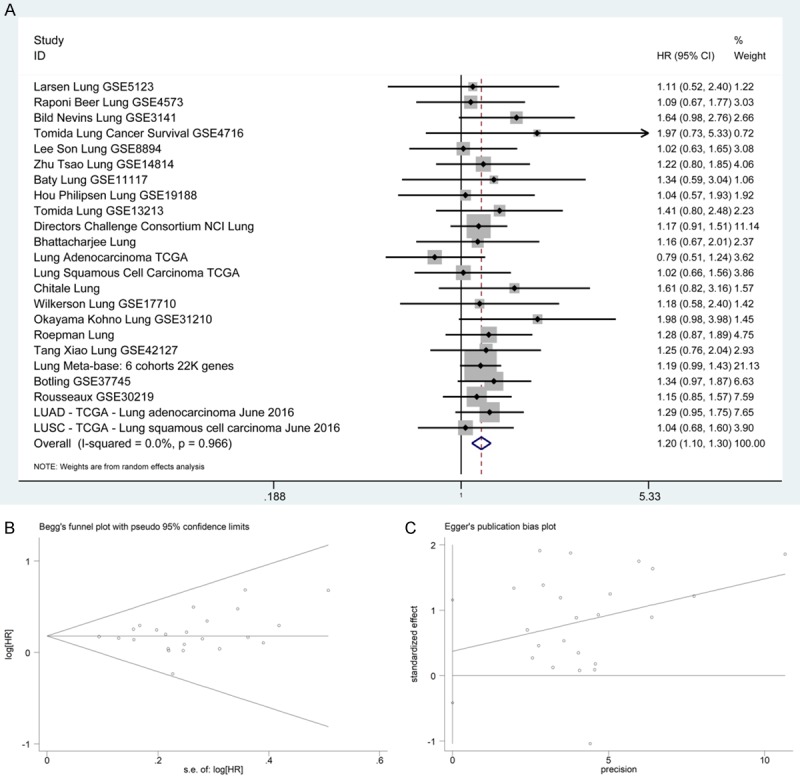
The association between BSG expression and OS in lung cancer from SurvExpress. A. Forest plot. B. Begg’s funnel plot. C. Egger’s publication bias plot. Abbreviations: OS, overall survival; HR, hazard ratio; CI, confidence interval.
Expression of BSG in patient tissues by immunohistochemistry
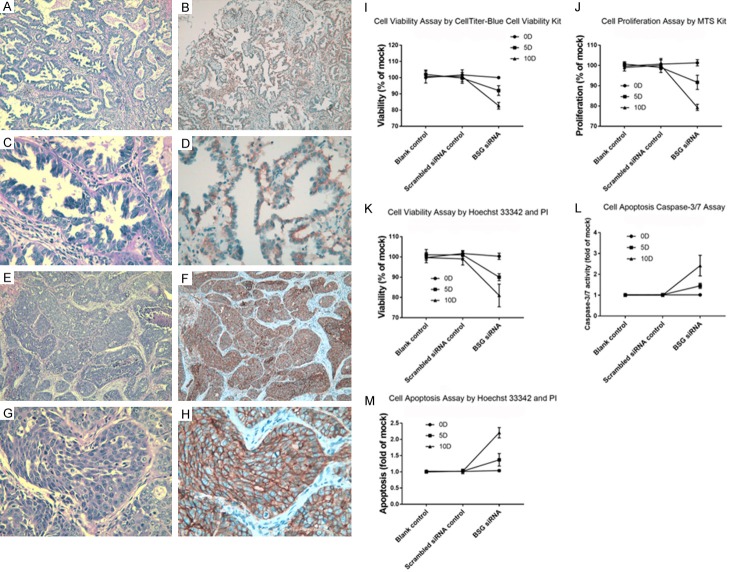
As shown in Table 2, the expression status of BSG was significantly different between LUAD and para-carcinoma tissue (P<0.001) by IHC assay. For the clinicopathological parameters, BSG expression was significantly associated with pathology grade (P = 0.041) and lymphatic metastasis (P = 0.014). The Hematoxylin and Eosin (HE) staining and IHC images of BSG-stained LUAD are shown in Figure 5A-H. The staining of BSG protein was primarily expressed in cytomembrane.
Table 2.
Relationship of BSG expression and clinicopathological parameters by immunohistochemistry
| Total number | BSG (-) | BSG (+) | Z score | P-value | |
|---|---|---|---|---|---|
| Tissue | |||||
| LUAD | 127 | 64 | 63 | -3.601 | <0.001* |
| pT | 30 | 26 | 4 | ||
| Gender | |||||
| Male | 71 | 36 | 35 | -0.629 | 0.529 |
| Female | 34 | 15 | 19 | ||
| Age | |||||
| ≤60 | 70 | 37 | 33 | -1.237 | 0.216 |
| >60 | 35 | 14 | 21 | ||
| Pathology grade | |||||
| I | 8 | 5 | 3 | 6.368a | 0.041* |
| II | 45 | 24 | 21 | ||
| III | 33 | 9 | 24 | ||
| T stage | |||||
| T1-T2 | 106 | 52 | 54 | -0.674 | 0.5 |
| T3-T4 | 21 | 12 | 9 | ||
| Distant metastasis | |||||
| No | 96 | 49 | 47 | -1.646 | 0.1 |
| Yes | 9 | 2 | 7 | ||
| Lymphatic metastasis | |||||
| No | 82 | 48 | 34 | -2.468 | 0.014* |
| Yes | 45 | 16 | 29 | ||
| Tumor size | |||||
| ≤7 cm | 105 | 51 | 54 | -0.894 | 0.371 |
| >7 cm | 22 | 13 | 9 |
P<0.05.
Figure 5.
H-E staining and IHC staining for BSG expression in LUAD and detection of the effect of BSG on LUAD cell line A549 viability, proliferation and apoptosis ability. A. HE staining of LUAD (×100); B. IHC of LUAD (×100); C. HE staining of LUAD (×400); D. IHC of LUAD (×400); E. HE staining of LUAD (×100); F. IHC of LUAD (×100); G. HE staining of LUAD (×400); H. IHC of LUAD (×400). I. Cell viability detected by CellTiter-Blue cell viability kits. J. Cell proliferation detected by MTS kit. K. Cell viability detected by Hoechst 33342 and PI double fluorescent staining assay. L. Cell apoptosis detected by Caspase-3/7 assay. M. Cell apoptosis detected by Hoechst 33342 and PI double fluorescent staining assay. Abbreviations: LUAD, lung adenocarcinoma; H-E, Hematoxylin-eosin; IHC, immunohistochemical; PI, Propidium Iodide.
Cell viability and proliferation inhibited by BSG siRNA
The LUAD cell line A549 was adopted to undertake the cell function experiments. Cell viability trial by CellTiter-Blue Cell Viability Assay kit (Figure 5I) and by Hoechst 33342 and PI double fluorescent staining assay (Figure 5K) both showed that the viability of cells decreased significantly on the tenth day after BSG siRNA transfection and the down-regulated expression of BSG (both P<0.001). In the cell proliferation assay, the result showed that when BSG siRNA was transfected, the proliferation ability of A549 was significantly weakened (P<0.001, Figure 5J).
Cell apoptosis promoted by BSG siRNA
The Apo-ONE Homogeneous Caspase-3/7 Assay (Figure 5L) and Hoechst 33342 and PI double fluorescent staining assay (Figure 5M) both presented that cell apoptosis was remarkably up-regulated after BSG transfection (P<0.001) on the tenth day.
Potential pathways related to BSG
The 450 common elements were from BSG co-expressed genes selected between the Affymetrix GeneChip Human Genoma U133 Plus 2.0 collection (containing 1794 datasets) by MEM and the mRNA Expression z-Scores (RNA Seq V2 RSEM) profile by cBioPortal. From the DAVID Bioinformatics Resources, the GO cluster and KEGG pathway are listed in Tables 3, 4. The most significant Biological process (BP) categories were translation and translational elongation. The most significant Cellular component (CC) categories were ribonucleoprotein complex, ribosome and ribosomal subunit. The most significant Molecular function (MF) categories were structural constituent of ribosome, RNA binding and structural molecule activity. KEGG pathway showed that the most significant pathways were Ribosome and Oxidative phosphorylation. Within the 450 selected genes, the hub genes were acquired from STRING. Next, we placed the top 100 hub genes into STRING again and generated the PPI network (Figure 6).
Table 3.
GO functional annotations for most significantly co-expressed genes of BSG
| GO ID | GO term | Count (%) | P-Value | Genes symbol |
|---|---|---|---|---|
| Biological process | ||||
| GO:0006412 | translation | 47 (11.41) | 1.12E-22 | RPL18, NACA, MRPL41, RPL14, MRPS12, MRPS11, RPL35, RPS6KB2, RPL36, EIF5A,etc |
| GO:0006414 | translational elongation | 26 (6.31) | 6.05E-19 | RPL18, RPL14, RPL35, RPL36, RPLP2, EEF2, RPS5, RPL28, RPL29, RPS3, etc |
| GO:0022900 | electron transport chain | 20 (4.85) | 2.20E-11 | NDUFA2, NDUFA3, UQCRC1, NDUFB10, NDUFB7, NDUFA7, NDUFA13, NDUFC1, UQCRQ, NDUFA11, etc |
| GO:0006091 | generation of precursor metabolites and energy | 30 (7.28) | 3.83E-10 | ATP5D, ALDOA, UQCRC1, NDUFB7, ATP5G2, UQCRQ, ATP6V0B, NDUFS7, CYB561D2, NDUFS5, etc |
| GO:0006119 | oxidative phosphorylation | 17 (4.13) | 1.19E-09 | ATP5D, NDUFA2, NDUFA3, NDUFB10, UQCRC1, NDUFB7, NDUFA7, ATP5G2, NDUFC1, ATP6V0B, etc |
| GO:0006396 | RNA processing | 39 (9.47) | 2.83E-09 | POLR2F, POLR2E, PRPF4B, RPL14, POLR2L, PUSL1, POLR2I, LSM7, TRMT1, TRPT1, etc |
| GO:0042775 | mitochondrial ATP synthesis coupled electron transport | 13 (3.16) | 5.89E-09 | NDUFA2, NDUFA3, NDUFB10, UQCRC1, NDUFB7, NDUFA7, NDUFC1, NDUFS7, NDUFS5, UQCR11, etc |
| GO:0042773 | ATP synthesis coupled electron transport | 13 (3.16) | 5.89E-09 | NDUFA2, NDUFA3, NDUFB10, UQCRC1, NDUFB7, NDUFA7, NDUFC1, NDUFS7, NDUFS5, UQCR11, etc |
| GO:0022904 | respiratory electron transport chain | 13 (3.16) | 2.92E-08 | NDUFA2, NDUFA3, NDUFB10, UQCRC1, NDUFB7, NDUFA7, NDUFC1, NDUFS7, NDUFS5, UQCR11, etc |
| GO:0034470 | ncRNA processing | 20 (4.85) | 1.05E-07 | RPL14, EXOSC4, PUSL1, TRPT1, TRMT1, RRP9, EXOSC1, QTRT1, NOP10, URM1, etc |
| Cellular component | ||||
| GO:0030529 | ribonucleoprotein complex | 68 (16.50) | 2.03E-30 | RPL18, SRP14, MRPS34, MRPL41, PRPF4B, RPL14, LSM7, RPLP2, RPS6KB2, RPS3, etc |
| GO:0005840 | ribosome | 44 (10.68) | 3.18E-27 | RPL18, MRPS34, MRPL41, RPL14, MRPS12, MRPS11, RPL35, RPS6KB2, RPL36, RPLP2, etc |
| GO:0033279 | ribosomal subunit | 34 (8.25) | 7.89E-25 | RPL18, MRPL41, RPL14, MRPS12, MRPS11, RPL35, RPLP2, RPL36, RPS3, MRPL12, etc |
| GO:0005739 | mitochondrion | 79 (19.17) | 1.17E-18 | ATP5D, MRPS34, TSPO, MRPL41, PTGES2, UQCRC1, PRDX5, ROMO1, COX5A, C14ORF2, etc |
| GO:0022626 | cytosolic ribosome | 23 (5.58) | 1.88E-17 | RPL18, RPL14, RPL35, RPLP2, RPL36, RPS5, RPL28, RPS3, RPL29, RPS19, etc |
| GO:0015934 | large ribosomal subunit | 20 (4.85) | 1.37E-15 | RPL18, MRPL41, RPL14, RPL35, RPLP2, RPL36, RPL28, RPL29, MRPL23, MRPL12, etc |
| GO:0044429 | mitochondrial part | 52 (12.62) | 3.61E-15 | ATP5D, TSPO, UQCRC1, MRPL41, COX5A, UQCRQ, COX5B, NDUFAF3, NDUFS7, LONP1, etc |
| GO:0044445 | cytosolic part | 27 (6.55) | 4.53E-15 | RPL18, RPL14, PIK3C2A, RPL35, RPL36, PRDX5, RPLP2, RPS5, RPL28, RPL29, etc |
| GO:0005743 | mitochondrial inner membrane | 33 (8.01) | 4.25E-12 | ATP5D, UQCRC1, NDUFB7, MTX1, ATP5G2, COX5A, UQCRQ, COX5B, NDUFAF3, NDUFS7, etc |
| GO:0019866 | organelle inner membrane | 34 (8.25) | 6.11E-12 | ATP5D, UQCRC1, NDUFB7, MTX1, ATP5G2, COX5A, UQCRQ, COX5B, NDUFAF3, NDUFS7, etc |
| Molecular function | ||||
| GO:0003735 | structural constituent of ribosome | 39 (9.47) | 1.29E-27 | RPL18, MRPL41, RPL14, MRPS12, MRPS11, RPL35, RPL36, RPLP2, RPS3, MRPL12, etc |
| GO:0003723 | RNA binding | 46 (11.17) | 3.75E-10 | RPL18, SRP14, RPL14, PUSL1, LSM7, RPL35, EIF5A, RPLP2, TRMT1, CALR, etc |
| GO:0005198 | structural molecule activity | 41 (9.95) | 3.32E-09 | RPL18, MRPL41, CLTB, RPL14, MRPS12, MRPS11, RPL35, RPL36, RPLP2, RPS3, etc |
| GO:0016651 | oxidoreductase activity, acting on NADH or NADPH | 15 (3.64) | 4.27E-09 | NDUFA2, NDUFA3, NDUFB10, NDUFB7, NDUFA7, NDUFA13, CYB5RL, NDUFC1, ECSIT, NDUFS7, etc |
| GO:0008137 | NADH dehydrogenase (ubiquinone) activity | 11 (2.67) | 2.92E-08 | NDUFS7, NDUFS5, NDUFA2, NDUFA3, NDUFB10, NDUFB7, NDUFS8, NDUFA7, NDUFA13, NDUFC1, etc |
| GO:0050136 | NADH dehydrogenase (quinone) activity | 11 (2.67) | 2.92E-08 | NDUFS7, NDUFS5, NDUFA2, NDUFA3, NDUFB10, NDUFB7, NDUFS8, NDUFA7, NDUFA13, NDUFC1, etc |
| GO:0003954 | NADH dehydrogenase activity | 11 (2.67) | 2.92E-08 | NDUFS7, NDUFS5, NDUFA2, NDUFA3, NDUFB10, NDUFB7, NDUFS8, NDUFA7, NDUFA13, NDUFC1, etc |
| GO:0015078 | hydrogen ion transmembrane transporter activity | 14 (3.40) | 1.05E-07 | ATP5D, UQCRC1, UQCR11, UQCRH, COX8A, COX6B1, COX4I1, COX6A1, ATP5G2, COX5A, etc |
| GO:0016655 | oxidoreductase activity, acting on NADH or NADPH, quinone or similar compound as acceptor | 11 (2.67) | 1.11E-07 | NDUFS7, NDUFS5, NDUFA2, NDUFA3, NDUFB10, NDUFB7, NDUFS8, NDUFA7, NDUFA13, NDUFC1, etc |
| GO:0015077 | monovalent inorganic cation transmembrane transporter activity | 14 (3.40) | 5.88E-07 | ATP5D, UQCRC1, UQCR11, UQCRH, COX8A, COX6B1, COX4I1, COX6A1, ATP5G2, COX5A, etc |
Table 4.
KEGG pathway enrichment analysis of BSG co-expressed genes
| KEGG ID | KEGG Term | Count (%) | P-Value | Genes symbol |
|---|---|---|---|---|
| hsa03010 | Ribosome | 23 (5.58) | 2.43E-15 | RPL18, RPL14, RPL35, RPLP2, RPL36, RPS5, RPL28, RPS3, RPL29, RPS19, etc |
| hsa00190 | Oxidative phosphorylation | 26 (6.31) | 2.52E-14 | ATP5D, UQCRC1, NDUFB7, ATP5G2, COX5A, COX5B, UQCRQ, ATP6V0B, NDUFS7, NDUFS5, etc |
| hsa05016 | Huntington’s disease | 29 (7.04) | 1.66E-13 | ATP5D, POLR2F, UQCRC1, CLTB, POLR2E, POLR2L, NDUFB7, AP2S1, POLR2I, ATP5G2, etc |
| hsa05012 | Parkinson’s disease | 22 (5.34) | 8.69E-11 | ATP5D, NDUFA2, NDUFA3, UQCRC1, NDUFB10, NDUFB7, NDUFA7, COX8A, COX4I1, ATP5G2, etc |
| hsa05010 | Alzheimer’s disease | 24 (5.83) | 2.40E-10 | ATP5D, NDUFA2, NDUFA3, UQCRC1, NDUFB10, NDUFB7, NDUFA7, COX8A, COX4I1, ATP5G2, etc |
| hsa04260 | Cardiac muscle contraction | 10 (2.43) | 4.48E-04 | UQCRC1, UQCR11, UQCRH, COX8A, COX6B1, COX4I1, COX6A1, COX5A, UQCRQ, COX5B |
| hsa00240 | Pyrimidine metabolism | 10 (2.43) | 0.00188043 | NME4, ITPA, NME2, POLR2F, POLR2E, NME3, POLR2L, POLD2, POLR2I, ZNRD1 |
| hsa03040 | Spliceosome | 11 (2.67) | 0.00398349 | DHX8, LSM7, PQBP1, SNRPB, SNRPA, LSM4, SF3B5, SF3A2, BUD31, NAA38, etc |
| hsa00230 | Purine metabolism | 12 (2.91) | 0.00546642 | NME4, ITPA, NME2, POLR2F, POLR2E, NME3, POLR2L, POLD2, POLR2I, ZNRD1, etc |
| hsa03020 | RNA polymerase | 5 (1.21) | 0.00877957 | POLR2F, POLR2E, POLR2L, POLR2I, ZNRD1 |
| hsa00051 | Fructose and mannose metabolism | 4 (0.97) | 0.0786479 | ALDOA, GMPPA, TSTA3, PHPT1 |
| hsa05110 | Vibrio cholerae infection | 5 (1.21) | 0.08368437 | SEC61B, ARF1, KDELR1, ATP6V0B, ATP6V1F |
| hsa03018 | RNA degradation | 5 (1.21) | 0.08802319 | EXOSC4, LSM7, LSM4, EXOSC1, NAA38 |
Figure 6.
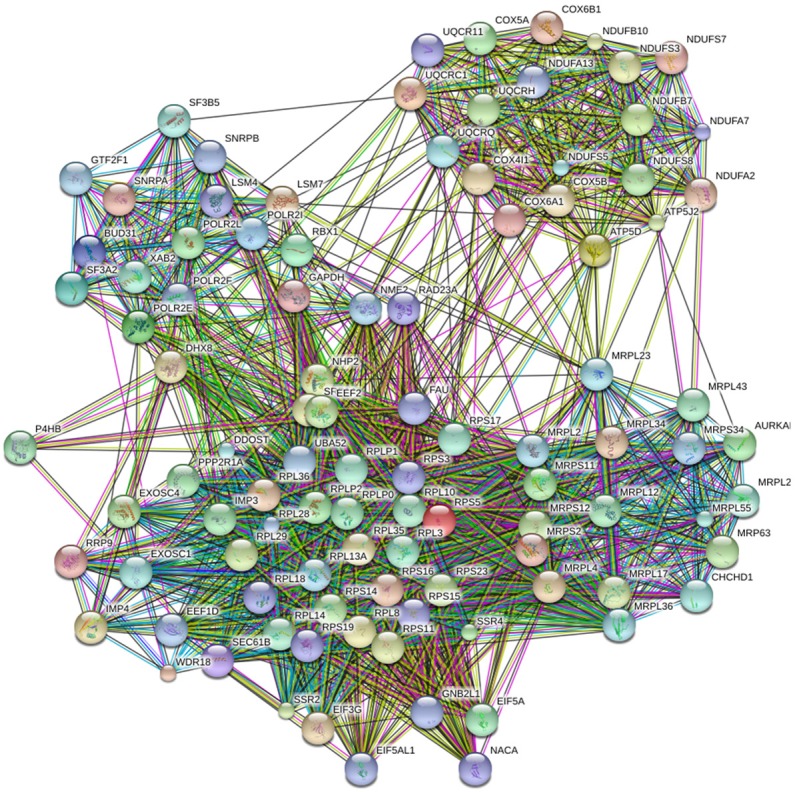
PPI network of the top 100 BSG co-expressed hub genes. Abbreviations: PPI, protein-protein interaction.
Discussion
In this study, the considerable evidence from high throughput analysis and clinical study all represented that BSG was up-regulated in LUAD tissue. BSG participated in various biological processes, especially in processes related to tumor progression.
The meta-analysis of BSG expression data from GEO showed that BSG was significantly up-regulated in LUAD. To reduce heterogeneity, we excluded studies that contained negative numbers (GSE74706, GSE66759, GSE19188, GSE11969 and GSE1037) because the array author may have processed the data by other means, which was different from making the original data log2 transformed. We thought different processing methods were not suitable together for statistics. We also excluded the studies (GSE85841, GSE18549) whose LUAD or control group SD>1, and one study (GSE68465) whose SMD (95% CI) drifted too far from the invalid line. Finally, we obtained a significant result (pooled SMD = 0.564, 95% CI: 0.448-0.681, P<0.001) with an applicable heterogeneity (I-squared = 48.6%, P = 0.011).
Within our research, BSG co-expressed genes were mainly enriched in translation related GO terms and pathways. We could deduce that BSG is an important gene which participates in various proteins translation process biological mechanism. It could be a possible reason that the function of BSG is considerable in diverse biological process.
To date, the molecular mechanism of BSG has not been fully investigated. BSG could have functions in tumor cell biological characteristics of proliferation, metastasis and apoptosis via specific signal pathways in various malignant neoplasms. Studies of BSG were searched in pulmonary disease as follows (BSG can be called CD147 or EMMPRIN in the following). It has been reported that CD147 could form a complex with CD98 heavy chain (CD98hc) on cell plasma membrane in certain cancers. Also, this complex led to the proliferation of tumor cells and poor prognosis in NSCLC patients through the PI3K/Akt pathway [24]. One research group found that EMMPRIN had a positive regulation on the canonical Wnt/β-catenin signaling pathway, which could gain the ability for tumor cell proliferation, differentiation and metastasis after altering cell-cell interactions [25]. β-catenin/canonical Wnt signaling pathway was also reported to have a function in the chronic progressively fatal disease, idiopathic pulmonary fibrosis (IPF). TGF-β1 induced this pathway to release EMMPRIN, and overexpressed EMMPRIN may be relevant to the persistent fibro-proliferative state of IPF [26]. Regarding other diseases, CD147 could be blocked by a murine antibody HAb18 to inhibit hepatocellular carcinoma invasion and migration. Inducing the integrin signaling pathway, cHAb18 also suppressed cell motility by rearranging actin cytoskeleton, which made a new tumor therapy targeting CD147 [27]. Another group recently published that elevated copper levels promoted cancer cell metastasis as an extracellular signaling role, for the reason that Cu2+-mediated self-association of CD147 was able to activate PI3K/Akt signaling pathway, leading to up-regulation of the level of matrix metalloproteinase MMP-2 and MMP-14 in HCC cells [28]. Smad4 was found to be a new interaction copartner of CD147 and could inhibit the Smad4/p21 WAF1 signal, which promotes cell proliferation [29]. Physcion 8-O-β-glucopyranoside (PG) was proved to have an effect on EMMPRIN to regulate the ROS-miR-27a/ZBTB10-Sp1 transcription factor pathway in the osteosarcoma cell to induce cell apoptosis and cause cell cycle arrest [30]. Such compelling rationales of BSG mechanism in different pathways provided tremendous benefits for clinical anticancer therapies, especially terminal cancers.
Regarding the future research prospects, the function of BSG was unclear and clinical researchers should explore the association between BSG and various cancer types in the future. Finally, to provide evidence to the clinical therapy of LUAD, considerably more research on BSG alone or with other potential relevant molecules should be conducted.
Conclusions
Since the abnormal over-expressed BSG in LUAD is correlated with advanced progression and poor prognosis and could influence the tumor cell function at the molecular level, BSG could be a potential diagnostic and prognostic biomarker, as well as a potential candidate for target therapy in patients with LUAD.
Acknowledgements
The study was supported by Fund of National Natural Science Foundation of China (NSFC 81560469), Guangxi Degree and Postgraduate Education Reform and Development Research Projects, China (JGY2019050), Medical Excellence Award Funded by the Creative Research Development Grant from the First Affiliated Hospital of Guangxi Medical University, Innovation Project of Guangxi Graduate Education (YCBZ2018038, YCBZ2019041 and YCBZ2019043), Natural Science Foundation of Guangxi, China (2015GXNSFCA139009) and Natural Science Foundation of Guangxi, China (2017GXNSFAA198016). Guangxi Medical University Training Program for Distinguished Young Scholars.
Disclosure of conflict of interest
None.
References
- 1.Chen W, Zheng R, Baade PD, Zhang S, Zeng H, Bray F, Jemal A, Yu XQ, He J. Cancer statistics in China, 2015. CA Cancer J Clin. 2016;66:115–32. doi: 10.3322/caac.21338. [DOI] [PubMed] [Google Scholar]
- 2.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2017. CA Cancer J Clin. 2017;67:7–30. doi: 10.3322/caac.21387. [DOI] [PubMed] [Google Scholar]
- 3.Bhatia S, Pereira K, Mohan P, Narayanan G, Wangpaichitr M, Savaraj N. Radiofrequency ablation in primary non-small cell lung cancer: what a radiologist needs to know. Indian J Radiol Imaging. 2016;26:81–91. doi: 10.4103/0971-3026.178347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dinh TK, Fendler W, Chalubinska-Fendler J, Acharya SS, O’Leary C, Deraska PV, D’Andrea AD, Chowdhury D, Kozono D. Circulating miR-29a and miR-150 correlate with delivered dose during thoracic radiation therapy for non-small cell lung cancer. Radiat Oncol. 2016;11:61. doi: 10.1186/s13014-016-0636-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lan D, Zhang X, He R, Tang R, Li P, He Q, Chen G. MiR-133a is downregulated in non-small cell lung cancer: a study of clinical significance. Eur J Med Res. 2015;20:50. doi: 10.1186/s40001-015-0139-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.He RQ, Li XJ, Liang L, Xie Y, Luo DZ, Ma J, Peng ZG, Hu XH, Chen G. The suppressive role of miR-542-5p in NSCLC: the evidence from clinical data and in vivo validation using a chick chorioallantoic membrane model. BMC Cancer. 2017;17:655. doi: 10.1186/s12885-017-3646-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liu HY, Gu WJ, Wang CZ, Ji XJ, Mu YM. Matrix metalloproteinase-9 and -2 and tissue inhibitor of matrix metalloproteinase-2 in invasive pituitary adenomas: a systematic review and meta-analysis of case-control trials. Medicine (Baltimore) 2016;95:e3904. doi: 10.1097/MD.0000000000003904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li Z, Ren Y, Wu QC, Lin SX, Liang YJ, Liang HZ. Macrophage migration inhibitory factor enhances neoplastic cell invasion by inducing the expression of matrix metalloproteinase 9 and interleukin-8 in nasopharyngeal carcinoma cell lines. Chin Med J (Engl) 2004;117:107–14. [PubMed] [Google Scholar]
- 9.Zhu S, Li Y, Zhang Y, Wang X, Gong L, Han X, Yao L, Lan M, Zhang W. Expression and clinical implications of HAb18G/CD147 in hepatocellular carcinoma. Hepatol Res. 2015;45:97–106. doi: 10.1111/hepr.12320. [DOI] [PubMed] [Google Scholar]
- 10.Abraham D, Zins K, Sioud M, Lucas T, Aharinejad S. Host CD147 blockade by small interfering RNAs suppresses growth of human colon cancer xenografts. Front Biosci. 2008;13:5571–9. doi: 10.2741/3100. [DOI] [PubMed] [Google Scholar]
- 11.Grass GD, Tolliver LB, Bratoeva M, Toole BP. CD147, CD44, and the epidermal growth factor receptor (EGFR) signaling pathway cooperate to regulate breast epithelial cell invasiveness. J Biol Chem. 2013;288:26089–104. doi: 10.1074/jbc.M113.497685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xu X, Liu S, Lei B, Li W, Lin N, Sheng W, Huang A, Shen H. Expression of HAb18G in non-small lung cancer and characterization of activation, migration, proliferation, and apoptosis in A549 cells following siRNA-induced downregulation of HAb18G. Mol Cell Biochem. 2013;383:1–11. doi: 10.1007/s11010-013-1722-7. [DOI] [PubMed] [Google Scholar]
- 13.Toole BP, Slomiany MG. Hyaluronan, CD44 and Emmprin: partners in cancer cell chemoresistance. Drug Resist Updat. 2008;11:110–21. doi: 10.1016/j.drup.2008.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang Y, Huang JC, Cai KT, Yu XB, Chen YR, Pan WY, He ZL, Lv J, Feng ZB, Chen G. Long non-coding RNA HOTTIP promotes hepatocellular carcinoma tumorigenesis and development: a comprehensive investigation based on bioinformatics, qRT-PCR and meta-analysis of 393 cases. Int J Oncol. 2017;51:1705–1721. doi: 10.3892/ijo.2017.4164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Aguirre-Gamboa R, Gomez-Rueda H, Martinez-Ledesma E, Martinez-Torteya A, Chacolla-Huaringa R, Rodriguez-Barrientos A, Tamez-Peña JG, Treviño V. SurvExpress: an online biomarker validation tool and database for cancer gene expression data using survival analysis. PLoS One. 2013;8:e74250. doi: 10.1371/journal.pone.0074250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gao L, Zhong JC, Huang WT, Dang YW, Kang M, Chen G. Integrative analysis of BSG expression in NPC through immunohistochemistry and public high-throughput gene expression data. Am J Transl Res. 2017;9:4574–4592. [PMC free article] [PubMed] [Google Scholar]
- 17.Zhong DN, Luo YH, Mo WJ, Zhang X, Tan Z, Zhao N, Pang SM, Chen G, Rong MH, Tang W. High expression of long non-coding HOTAIR correlated with hepatocarcinogenesis and metastasis. Mol Med Rep. 2018;17:1148–1156. doi: 10.3892/mmr.2017.7999. [DOI] [PubMed] [Google Scholar]
- 18.Li JJ, Luo J, Lu JN, Liang XN, Luo YH, Liu YR, Yang J, Ding H, Qin GH, Yang LH, Dang YW, Yang H, Chen G. Relationship between TRAF6 and deterioration of HCC: an immunohistochemical and in vitro study. Cancer Cell Int. 2016;16:76. doi: 10.1186/s12935-016-0352-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang Y, Huang S, Leng Y, Chen X, Liu T, Wang H, Wei F, Luo D, Chen G, Wei Z. Effect of DcR3-specific siRNA on cell growth suppression and apoptosis induction in glioma cells via affecting ERK and AKT. Onco Targets Ther. 2016;9:5195–202. doi: 10.2147/OTT.S108395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Adler P, Kolde R, Kull M, Tkachenko A, Peterson H, Reimand J, Vilo J. Mining for coexpression across hundreds of datasets using novel rank aggregation and visualization methods. Genome Biol. 2009;10:R139. doi: 10.1186/gb-2009-10-12-r139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kolde R, Laur S, Adler P, Vilo J. Robust rank aggregation for gene list integration and meta-analysis. Bioinformatics. 2012;28:573–80. doi: 10.1093/bioinformatics/btr709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gao L, Li SH, Tian YX, Zhu QQ, Chen G, Pang YY, Hu XH. Role of downregulated miR-133a-3p expression in bladder cancer: a bioinformatics study. Onco Targets Ther. 2017;10:3667–3683. doi: 10.2147/OTT.S137433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Higgins JP, Thompson SG, Deeks JJ, Altman DG. Measuring inconsistency in meta-analyses. BMJ. 2003;327:557–60. doi: 10.1136/bmj.327.7414.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fei F, Li X, Xu L, Li D, Zhang Z, Guo X, Yang H, Chen Z, Xing J. CD147-CD98hc complex contributes to poor prognosis of non-small cell lung cancer patients through promoting cell proliferation via the PI3K/Akt signaling pathway. Ann Surg Oncol. 2014;21:4359–68. doi: 10.1245/s10434-014-3816-1. [DOI] [PubMed] [Google Scholar]
- 25.Sidhu SS, Nawroth R, Retz M, Lemjabbar-Alaoui H, Dasari V, Basbaum C. EMMPRIN regulates the canonical Wnt/beta-catenin signaling pathway, a potential role in accelerating lung tumorigenesis. Oncogene. 2010;29:4145–56. doi: 10.1038/onc.2010.166. [DOI] [PubMed] [Google Scholar]
- 26.Hasaneen NA, Cao J, Pulkoski-Gross A, Zucker S, Foda HD. Extracellular Matrix Metalloproteinase Inducer (EMMPRIN) promotes lung fibroblast proliferation, survival and differentiation to myofibroblasts. Respir Res. 2016;17:17. doi: 10.1186/s12931-016-0334-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang Y, Yuan L, Yang XM, Wei D, Wang B, Sun XX, Feng F, Nan G, Wang Y, Chen ZN, Bian H. A chimeric antibody targeting CD147 inhibits hepatocellular carcinoma cell motility via FAK-PI3K-Akt-Girdin signaling pathway. Clin Exp Metastasis. 2015;32:39–53. doi: 10.1007/s10585-014-9689-7. [DOI] [PubMed] [Google Scholar]
- 28.Ding P, Zhang X, Jin S, Duan B, Chu P, Zhang Y, Chen ZN, Xia B, Song F. CD147 functions as the signaling receptor for extracellular divalent copper in hepatocellular carcinoma cells. Oncotarget. 2017;8:51151–51163. doi: 10.18632/oncotarget.17712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Qin H, Rasul A, Li X, Masood M, Yang G, Wang N, Wei W, He X, Watanabe N, Li J, Li X. CD147-induced cell proliferation is associated with Smad4 signal inhibition. Exp Cell Res. 2017;358:279–289. doi: 10.1016/j.yexcr.2017.07.003. [DOI] [PubMed] [Google Scholar]
- 30.Wang Z, Yang H. EMMPRIN, SP1 and microRNA-27a mediate physcion 8-O-beta-glucopyranoside-induced apoptosis in osteosarcoma cells. Am J Cancer Res. 2016;6:1331–44. [PMC free article] [PubMed] [Google Scholar] [Retracted]