Abstract
Understanding the relationships between glomerular endothelial cells (GECs) and glomerular mesangial cells (GMCs) is important to identify the molecular mechanisms underlying diabetic nephropathy (DN). Exosomes carried with mRNA, microRNA, and protein play important roles in cell-to-cell communication. In this study, we showed that high glucose (HG)-treated GECs secreted a higher number of exosomes enriched in circRNAs compared with normal glucose (NG)-treated GECs. Differentially expressed circRNAs (DECs) were obtained by high-throughput sequencing. Of these DECs, the expressions of 217 DECs and 484 DECs in HG-treated GEC exosomes were significantly downregulated and upregulated, respectively, compared with NG-treated GEC exosomes. The functions of the DEC target genes were involved in the PI3K/AKT and MAPK pathways. Five DECs were randomly selected for identification by quantitative real-time PCR (qRT-PCR). Two DECs (circRNF169 and circSTRN3) were further selected for functional validation. Moreover, we demonstrated that exosomes released by HG-treated GECs promoted α-smooth muscle actin (α-SMA) expression. It also inhibited proliferation and promoted epithelial-mesenchymal transition (EMT) in GMCs. In addition, cell functional studies indicated that the knockdown and over-expression of two DECs (circRNF169 and circSTRN3) effectively inhibited or promoted cell proliferation and promoted or inhibited EMT, respectively. Thus, the results of this study provide new insights into the pathogenesis of DN that involves the intercellular transfer of circRNAs from GECs to GMCs via exosomes.
Keywords: Diabetic nephropathy, exosomes, circRNAs, EMT, mesangial cells
Introduction
Diabetic nephropathy (DN) is one of the most serious chronic complications of diabetes. DN is caused by diabetic microangiopathy. At present, there are 92 million diabetic patients and 148 million pre-diabetes patients in China [1]. The number of diabetic patients is still increasing worldwide; thus, the prevalence of DN is also increasing. The main pathological feature of DN is glomerular sclerosis. In the United States, a previous study suggested that approximately 30% of the type 1 diabetes mellitus (T1DM) cases and 20% of the type 2 diabetes mellitus (T2DM) cases develop DN. Approximately 50% patients die of end-stage renal disease, which is the leading cause of death from chronic kidney disease [2]. DN causes great suffering to the patients themselves and their families and results in a heavy financial burden to the family, medical system, and society as a whole. Currently, the relevant pathogenic factors of DN include genetic background [3], abnormal glucose metabolism [4], abnormal lipid metabolism [5], microcirculatory disorders [6], cytokines [7], and inflammation [8], among others. However, the exact pathogenesis of DN has not yet been elucidated. In general, it is caused by long-term hyperglycemia in patients with certain genetic backgrounds and some relevant acquired risk factors, which could initiate cytokine networks and ultimately result in damage to kidneys and other important tissues [9].
The exosomes are vesicle-like bodies that are secreted from the cell into the extracellular space. The functions of exosomes focus on transmitting proteins, lipids, and nucleic acids. As a new type of biological marker, exosomes serve as important carriers of signal transmission between cells and participate in the development of various renal diseases [10]. In 1981, Trams et al. first proposed the concept of “exosomes” when studying the detachment of normal cells and tumor cells [11]. A few years later, Pan et al. first purified exosomes during a study of reticulocyte ripening [12]. Subsequent study have found that almost all cells secrete exosomes, which can be isolated from blood, urine, semen/prostatic fluid, amniotic fluid, and pleural effusion [13]. At present, the distinction between exosomes and other microbubble structures is mainly based on the diameter (particle diameter 100 nm-1 μm, exosome diameter 30-100 nm, apoptotic body diameter 1-4 μm). In addition, exosomes express major tissue phase endogenous molecules, such as capacitive complex (MHC) and CD63, and contain a large number of specific proteins and functional mRNAs, miRNAs, and even DNA [14]. miRNAs are a class of short non-coding ribonucleotides involved in numerous biological processes. In a variety of pathophysiological processes, cells encapsulate miRNAs in exosomes and release them into the peripheral circulation [15]. Recent studies found that 14 miRNAs (miR-320c, miR-6068, etc.), which were secreted by exosomes in the urine of DN patients, were downregulated. Downregulated miR-320c could affect the TGF-β-related signaling pathway through regulation of platelet-reactive protein-1 (TSP-1). Meanwhile, exosome-derived miRNA192 was upregulated in the urine of early DN patients with microalbuminuria [16,17]. Therefore, exosomal-derived miRNAs can truly reflect the functional status of DN kidneys.
In this study, we isolated the exosomes from glomerular endothelial cells (GEC) treated with high glucose (HG) and normal glucose (NG). The morphological size and protein markers of the exosomes were identified. Moreover, we screened the circRNAs differentially expressed between the two different groups by high-throughput sequencing. The differentially expressed circRNAs identified were further confirmed by qRT-PCR. In addition, we studied the functional roles of the screened circRNAs in glomerular mesangial cells. The results provide detailed information on the roles of exosome-derived circRNAs in DN.
Materials and methods
Cell treatment
Mouse primary kidney GEC (C57-6014G) and mouse glomerular mesangial cells (GMC) (SV40 MES 13) were obtained from Cell Biologics (Chicago, IL, USA) and China Infrastructure of Cell Line Resources (China), respectively. Mouse GECs were cultured in complete culture medium (Procell, China) and GMCs were cultured in low glucose Dulbecco’s Modified Eagle’s Medium (DMEM) with 5% (v/v) fetal bovine serum (FBS) (Invitrogen, Carlsbad, CA, USA). The cell lines were maintained in a humidified chamber at 37°C and 5% CO2. For the GEC experiments, all cells were divided into two sub-groups: GECs treated with low glucose (5 mmol/l glucose + 25 mmol/l mannitol) and GECs treated with high glucose (30 mmol/l). The ORF plasmid of mmu_circ_0001605 and mmu_circ_0000372 was obtained from Forevergen (China). The LV003 Vector was used to build a system over-expressing mmu_circ_0001605 and mmu_circ_0000372 in GMC. All lentiviral particles were generated in 293T by following a standardized protocol provided by Forevergen. Lentivirus was used to infect the GMC cells when the cell confluence reached 70%-80% in 6-well culture plates. Double-stranded siRNAs (dsRNA) targeting mmu_circ_0001605 and mmu_circ_0000372 were obtained from GenePharma (China). The sequence of the siRNAs used to knock down the circRNAs were: Si-circRNF169-1, 5’-GGCAGGAGTTTATATTCA-3’; Si-circRNF169-2, 5’-GTTCCTGGCAGGAGTTTA-3’; Si-circSTRN3-1, 5’-GAATGGGGCACGGATTGCA-3’; and Si-circSTRN3-2, 5’-GTACAGAATGGGGCACGGA-3’. The cells were seeded in 6-well plates at 5 × 105 cells per well with low glucose DMEM and 5% FBS overnight. Transfection was conducted with Lipofectamine 2000 reagent (final siRNA concentration, 100 nM) (Thermo Fisher Scientific, Waltham, MA, USA).
Exosomal extractions
GEC exosomal extractions from cell culture media were conducted with Total Exosome Isolation Reagent (Thermo Fisher Scientific), following the protocol provided by the manufacturer. Briefly, serum-free GEC culture supernatants were collected and centrifuged at 2,000 g for 30 min to remove cells and cell debris. Reagent (0.5 ml) was added to 1 ml of the supernatants. The culture media/reagent mixture was mixed well and incubated at 2°C to 8°C overnight. Then, the mixture was centrifuged at 10,000 g for 1 h, the supernatants were aspirated, and 0.1 ml of 1 × PBS was added. The isolated exosomes were kept at 2°C to 8°C for one week. NTA of the GEC exosomes was performed as previously reported [18].
GEC exosome circRNA sequencing
The MiniBEST Universal RNA Extraction Kit (Takara, Japan) was used to obtain the total RNA of the GEC exosomes. The purity and concentration of the total GEC exosomal RNA were measured with an Agilent 2100 (Agilent Technologies, USA). DNase I (Epicenter, USA) and the Epicenter Ribo-zero™ rRNA Removal Kit (Epicenter) were used to remove the contaminating DNA and ribosomal RNA, respectively. Furthermore, linear RNA was removed with RNase R digestion (Epicenter). Reverse transcription was conducted with PrimeScript™ II Strand cDNA Synthesis Kit (Takara). An ultrasonic method was used to cut the circRNAs into 140-160 bp pieces (Covaris M220, USA). The NEBNext® Ultra™ Directional RNA Library Prep Kit for Illumina® (NEB, Ipswich, MA, USA) was used following the manufacturer’s recommendations. Adaptors were ligated to the 3’adenylated ends of the DNA fragments. Suitable cDNA fragments between 150-200 bp were selected with the AMPure XP system (Beckman, USA). Then, the cDNA library was amplificated with universal PCR primers and Index (X) primer. The final library product was assessed with an Agilent Bioanalyzer 2100 system (Agilent). The Illumina HiSeq 2500 platform was used to sequence the library with 150 bp paired-end sequencing strategy.
Bioinformatics
CircRNAs were considered differentially expressed at P ≤ 0.05 and |Log2 Ratio| ≥ 1.5. GO enrichment analysis and KEGG analysis of the differentially expressed circRNAs were conducted according to genomic position and overlap with protein-coding genes using the KOBAS3.0 online database (http://kobas.cbi.pku.edu.cn/). The default parameters were strictly followed. Meanwhile, a hypergeometric test was used to calculate the major biochemical metabolic pathways.
PCR
The total RNA of the GEC exosomes and GECs was obtained by the MiniBEST Universal RNA Extraction Kit (Takara). The PrimeScript™ II Strand cDNA Synthesis Kit (Takara) was used to harvest cDNA. The primer sequences of the five selected circRNAs were: mmu_circ_0001605-F: 5’-GATGAAGCAGTGGTGCTGAA-3’, mmu_circ_0001605-R: 5’-CTCAGCTTTCTCAGGCATCC-3’; mmu_circ_0000372-F: 5’-CAGGGGATGGTACAGAATGG-3’, mmu_circ_0000372-R: 5’-CTGATTCAAAGGTGGGCATT-3’; mmu_circ_0000505-F: 5’-GTCAGCCTTCTACCCCATGA-3’; mmu_circ_0000505-R: 5’-ACACAACCTGCACATTCCAA-3’; chr3: 141922094:141940965-F: 5’-GCCCACTGCAACCAAGTTAT-3’, chr3: 141922094:141940965-R: 5’-CTACAGGCTTGAAAGCAGCA-3’; chr17: 78679313:78689706-F: 5’-GGACAGCTACGAAACGCAAG-3’, and chr17: 78679313:78689706-R: 5’-GCTGGGTAAAGGACAGCACT-3’. The primer sequences of the two circRNAs (mmu_circ_0001605 and mmu_circ_0000372) target miRNAs (miR-29c-3p and miR-93-5p) were: miR-29c-3p-F: 5’-TAGCACCATTTGAAATCG-3’, miR-29c-3p-RT: 5’-GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACTAACCG-3’; miR-93-5p-F: 5’-CAAAGTGCTGTTCGTGCA-3’, miR-93-5p-RT: 5’-GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACCTACCT-3’. Universe-R: 5’-GTGCAGGGTCCGAGGT-3’. The 10 µl q-PCR system contained 5µl 2 × SsoAdvanced™ Universal SYBR® Green Supermix (BioRad, USA), 0.5 µl primers (5pmol each), 0.5 µl cDNA template (approximately 20 ng), and 4 µl ddH2O. The thermal cycler steps: 94°C for 3 min, 40 cycles of 95°C for 15 s and 60°C for 25 seconds. GAPDH reported in a previous study was used as a reference gene [19]. The ΔΔCt method was applied to calculate circRNA expression. Four primer pairs were designed to identify the linear DNA and the corresponding circRNA (mmu_circ_0001605 and mmu_circ_0000372). The primers used were: circRNA (234 bp): mmu_circ_0001605-F: 5’-GATGAAGCAGTGGTGCTGAA-3’, mmu_circ_0001605-R: 5’-CTCAGCTTTCTCAGGCATCC-3’; liner DNA (169 bp): mmu_circ_0001605-F: 5’-CGGGAGAACTCAGTGAGGAG-3’, mmu_circ_0001605-R: 5’-TTCAGCACCACTGCTTCATC-3’. circRNA (199 bp): mmu_circ_0000372-CF: 5’-CAGGGGATGGTACAGAATGG-3’, mmu_circ_0000372-CR: 5’-CTGATTCAAAGGTGGGCATT-3’; liner DNA (178 bp): mmu_circ_0000372-LF: 5’-GCACGGATTGCATTTTTACA-3’, mmu_circ_0000372-LR: 5’-CTGATTCAAAGGTGGGCATT-3’. The PCR products were analyzed by 1.5% agarose gel electrophoresis and Sanger sequencing.
Western blots
The total exosomal protein of the different groups was extracted with pre-cooled cell lysate (volume ratio PMSF: RIPA cell lysate, 1:120). Protein loading buffer was added to the collected protein samples. The mixture was heated in a 100°C boiling water bath for 5 minutes. Subsequently, the proteins were transferred to a polyvinylidene difluoride membrane (Millipore, Bedford, MA, USA). After blocking for 1 h at room temperature, the membrane was incubated with rabbit polyclonal anti-mouse HspA8 (1:1000) and Alix (1:1500) antibody (BOSTER, USA) for 12 hours. Then, the treated proteins were incubated with the corresponding secondary antibody (1:2000 dilution) for 1 h at room temperature (BOSTER). The Odyssey Infrared Imaging System (LI-COR Biosciences, USA) was used to observe the reactions.
Enzyme-linked immunosorbent assay (ELISA)
When the GMC cell confluence reached 70%-80%, the cells were washed with 1 × PBS. Exosomes derived from the same cells, treated with low glucose or high glucose, circRNA siRNA, or circRNA over-expressing lentiviral vector were added along with medium (without FBS, low-glucose DMEM, 1% double antibody). The supernatant was incubated for 24 hours. ELISA kits were used to quantify the main components of the ECM (Col IV and FN) according to the manufacturer’s instructions (CUSABIO, USA).
Immunofluorescence
The cells were plated in 6-well chamber slides at 5 × 104 cells per well (Millicell EZ Slide, Millipore). The cells were fixed with cold 100% methanol (stored at -20°C) for 2 min, washed with 800 μl of PBS three times for five min each and blocked with 2% bovine serum albumin (BSA) for 1 hour. Each slide was incubated with 200 μl of diluted primary anti-α-SMA antibody (1:50; Abcam) and vimentin (1:200; Abcam) overnight in a refrigerator. The nuclei were stained with 4’,6-diamidino-2-phenylindole (DAPI). Each slide was washed with 500 μl of PBS three times. Subsequently, the samples were treated with 200 μl of Alexa Fluor 568 (red) or Alexa Fluor 488 (green)-labeled secondary antibody (1:1000 diluted in PBS buffer with 2% BSA) for 2 h in the dark. Images were taken under confocal microscopy. Zeiss LCM image browser software was used to produce a composite multicolor image. All images were combined with Adobe Photoshop software. In addition, the exosomes were labeled with PKH67 dye for 24 hours (Sigma, St. Louis, MO, USA). All assays were performed according to the manufacturer’s instructions. Subsequently, the GMCs were observed under a confocal laser microscope.
Cell function
An MTT kit (Beyotime Institute of Biotechnology, China) was used to study cell proliferation according to the manufacturer’s protocol. Cells (2,000) were plated in a 96-well plate. The cells were cultured in media containing 5% FBS at 37°C and 5% CO2 for 24 hours. Then, a microplate reader (Thermo Fisher Scientific) was used to measure the absorbance at 490 nm. Transwell Permeable Support was used to study cell migration (Corning Incorporated, Corning, NY, USA). A density of 1 × 106 per ml (100 µl per chamber) cells was carefully transferred to the top chamber of each transwell and the cells were allowed to migrate for 24 h at 37°C. Cells that had penetrated to the bottom side of the membrane were fixed in methanol and stained with hematoxylin.
Data analysis
SPSS V16.0 software (IBM, USA) was used to perform the statistical analyses. The mean values of the statistical results for the measured data were presented as the mean ± standard error. ANOVA was applied to calculate the differences between the groups. The differences between the two groups were calculated using Tukey’s test. P-values less than 0.05 were defined as significantly different.
Results
High glucose (HG) treatment stimulated GECs to secrete high numbers of exosomes
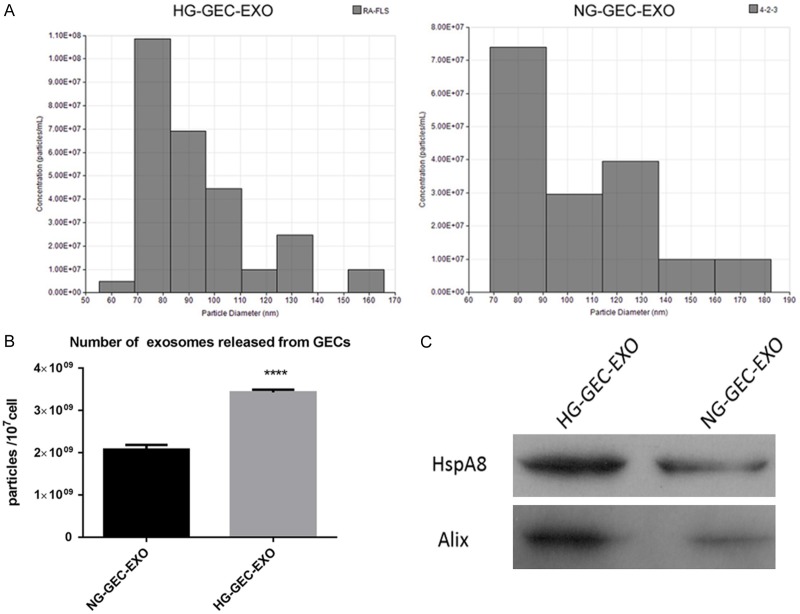
In order to investigate the effect of HG treatment on exosome release, GECs were divided into two sub-groups: HG treatment (30 mmol/l) and low glucose treatment (5 mmol/l + 25 mmol/l mannitol). Culture supernatants (1 ml) with approximately 4 × 106 GECs was used to obtain exosomes using a commercial separation kit. Nanoparticle tracer analysis (NTA) of the GEC exosomes suggested that exosomal sizes ranged from 55 nm to 180 nm (Figure 1A). No difference in GEC exosomes was observed between the two groups. And the number of exosomes was also measured by NTA. The results revealed that the number of exosomes in HG-GEC-EXO group was significantly higher than that in NG-GEC-EXO group (Figure 1B). Western blots were performed to analyze the expression of specific exosomal proteins (HspA8 and Alix). The results indicated that both exosomal biomarkers were more abundant in the HG-GEC-EXO group compared to the NG-GEC-EXO group (Figure 1C). In summary, the results in this study demonstrated that HG effectively promoted the release of exosomes from GECs.
Figure 1.
Isolation and identification of exosomes from glomerular endothelial cells treated with high glucose and low glucose. A: Exosomal size analysis of HG-GEC-EXO and NG-GEC-EXO by NTA. B: The number of exosomes analyzed in the two different groups by NTA. C: Western blot analysis of HspA8 and Alix in the exosomes of the two different groups. ****P-value < 0.001. HG-GEC-EXO: exosomes from glomerular endothelial cells treated with high glucose. NG-GEC-EXO: exosomes from glomerular endothelial cells treated with normal (low) glucose; NTA: Nanoparticle tracer analysis.
Screening and identification of differentially expressed circRNAs in exosomes
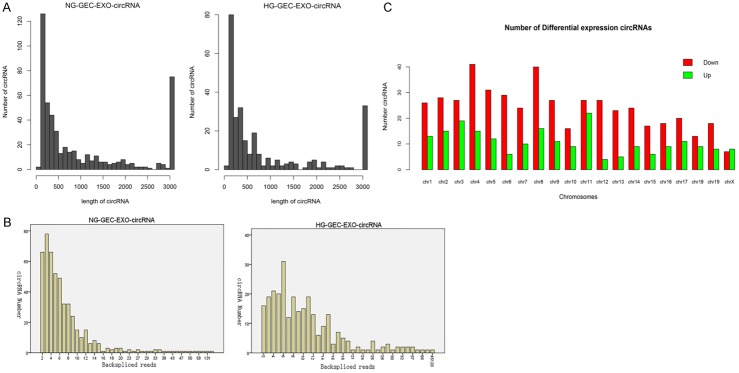
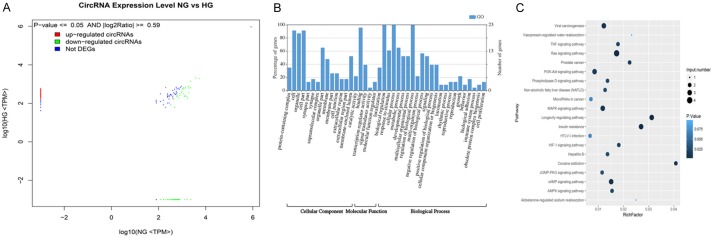
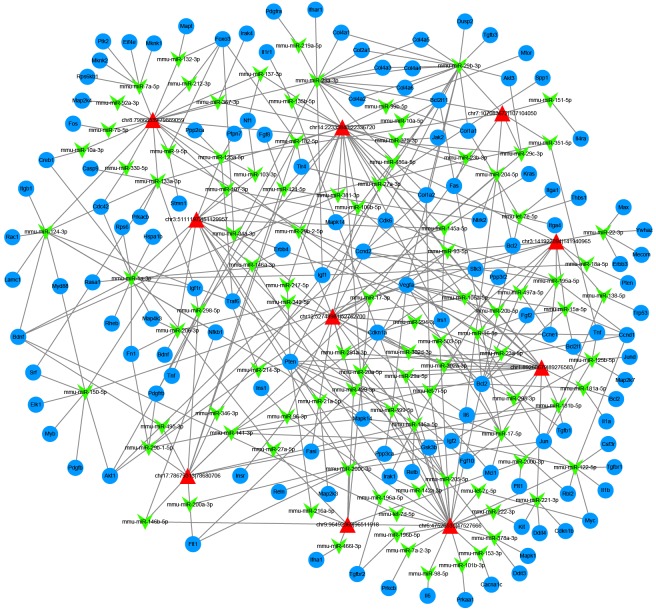
In this study, we focused on the differentially expressed circRNAs in exosomes in HG and low glucose-treated GECs. Figure 2A shows the size distribution of the sequenced exosomal circRNAs of the two groups. The main size of the two libraries was mainly concentrated in 500 bp. No differences were seen between the two groups. The copy number of back-spliced reads suggested that the exosomal circRNAs of the two groups ranged from 2 to 10 (Figure 2B). Moreover, we also analyzed the chromosomal distribution of the differentially expressed circRNAs (DECs) in the two groups. The results indicated that these DECs were located at all chromosomes. The number of upregulated DECs (217) was lower than the number of downregulated DECs (484) (Figures 2C and 3A). Furthermore, we analyzed the functions of these DECs. Gene ontology (GO) analysis revealed that gene function was mainly distributed into three sub-groups: cellular components, molecular function, and biological processes (Figure 3B). In the cellular component, cells, cell parts, and organelles were the three most enriched items. From the perspective of molecular function, localization was the most enriched item. From the perspective of biological processes, cellular processes, biological regulation, and regulation of biological processes were the most enriched items. Meanwhile, KEGG analysis suggested that items were most enriched in the PI3K/AKT and RAS signaling pathways (Figure 3C). In addition, to further examine the function of DECs, we reconstructed a network of circRNA-miRNA-mRNAs. circRNAs related to the RAS pathway were selected with the following criteria: 1) The circRNAs were sourced from exons and 2) The length of the circRNAs ranged between 300 bp and 1200 bp. Figure 4 suggested that ten circRNAs played key roles in this network. For example, chr3: 141922094:141940965 regulated mmu-miR-195a-5p, which regulated the expression of Bcl-2 mRNA. In addition, five DECs, including mmu_circ_0001605, mmu_circ_0000372, mmu_circ_0000505, chr3: 141922094:141940965, and chr17: 78679313:78689706, were randomly selected to confirm their expression levels in exosomes of different groups by qRT-PCR (Figure 5). Figure 5A shows that the expression of mmu_circ_0001605, mmu_circ_0000372, mmu_circ_0000505, and chr3: 141922094:141940965 were significantly reduced in the HG-GEC-EXO group compared to the NG-GEC-EXO group (P < 0.05). No difference was found in the expression analysis of chr17: 78679313:78689706. Meanwhile, the expression of the target miRNAs (mmu-miR-29c-3p and miR-93-5p) of mmu_circ_0001605 and mmu_circ_0000372 in exosomes were also studied (Figure 5A). The expression of mmu-miR-29c-3p and miR-93-5p were significantly higher in the HG-GEC-EXO group than in the NG-GEC-EXO group (P < 0.05). Moreover, we also examined the expression of five selected circRNAs in GECs with different treatments (Figure 5B). Results similar to the exosomal analysis results were obtained. In addition, we further identified the characteristics of mmu_circ_0001605 and mmu_circ_0000372 by PCR and Sanger sequencing (Figure 6A and 6B). The results confirmed mmu_circ_0001605 and mmu_circ_0000372 in the GEC exosomes.
Figure 2.
High-throughput sequencing analysis of circRNAs in GEC exosomes treated with low glucose and high glucose. A: Length distribution analysis of circRNAs in GEC exosomes treated with high and low glucose. The main size of the two libraries was mainly concentrated in 500 bp. B: Back-spliced read numbers of circRNAs in GEC exosomes treated with high and low glucose. The copy number of back-spliced reads ranged from 2 to 10. C: Chromosomal distribution of differentially expressed circRNAs in GEC exosomes treated with high and low glucose were located at all chromosomes. NG-GEC-EXO-circRNA: circRNA in exosomes from GECs treated with normal (low) glucose. HG-GEC-EXO-circRNA: circRNA in exosomes from GECs treated with high glucose.
Figure 3.
Differentially expressed circRNAs and their original gene functions in GEC exosomes treated with low glucose and high glucose. A: Differentially expressed circRNAs in GEC exosomes treated with low glucose and high glucose. Not DEGs means no differentially expressed circRNAs were seen between the two groups. B: GO analysis of differentially expressed circRNA original genes. All GO items were divided into three main sub-groups, cellular components, molecular functions, and biological processes. C: KEGG analysis of differentially expressed circRNA original genes. NG: circRNA in exosomes from GEC with normal glucose treatment. HG: circRNA in exosomes from GEC treated with high glucose.
Figure 4.
circRNA-miRNA-mRNA network analyses of ten circRNAs related to the RAS pathway. Red triangle indicates the selected circRNAs. Green arrow represents the circRNA-related miRNAs. Blue ellipse indicates the mRNAs regulated by circRNA-related miRNAs.
Figure 5.
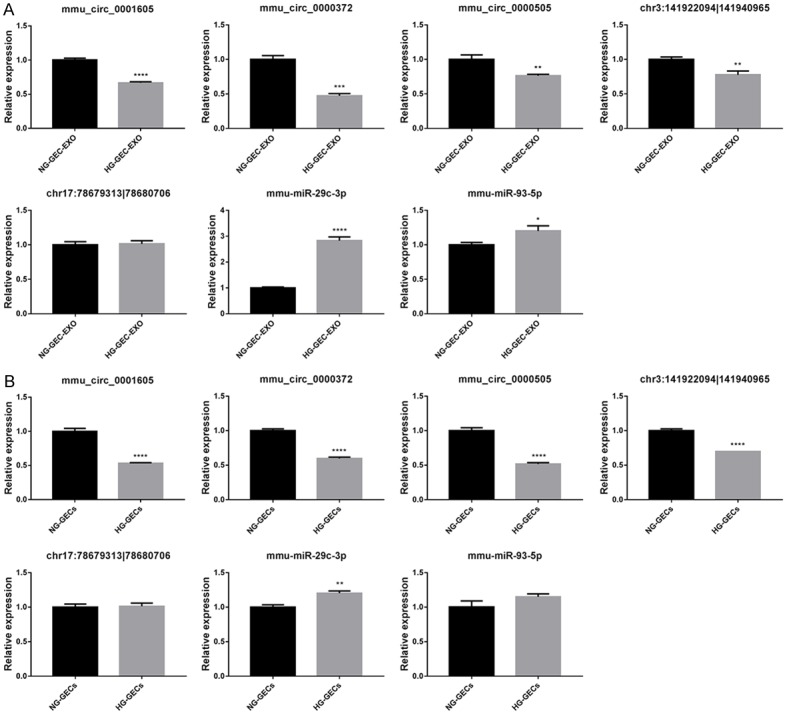
qRT-PCR analysis of five selected differentially expressed circRNAs in GEC exosomes and GECs treated with normal glucose and high glucose. A: The expressions of five selected differentially expressed circRNAs and two of their target miRNAs in GEC exosomes treated with low and high glucose. HG-GEC-EXO: exosomes from GECs treated with high glucose. NG-GEC-EXO: exosomes from GECs treated with normal glucose. B: The expression of five selected differentially expressed circRNAs and two of their target miRNA in GECs treated with low and high glucose. HG-GEC: GEC treated with high glucose. NG-GEC: GEC treated with normal glucose. *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001.
Figure 6.
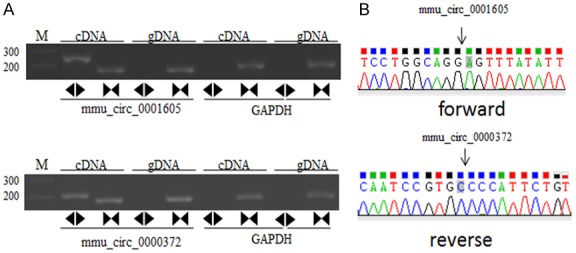
PCR and Sanger sequencing identification of mmu_circ_0001605 and mmu_circ_0000372 in GEC exosomes treated with normal glucose and high glucose. A: Divergent primers detect circular RNAs in cDNA but not genomic DNA (gDNA). GAPDH was used as an internal reference. Forward indicates the forward primer used in Sanger sequencing. Reverse indicates the reverse primer used in Sanger sequencing. B: Sanger sequencing depicts the junction of mmu_circ_0001605 and mmu_circ_0000372.
GEC exosomes in the HG-GEC-EXO group affected GMC function
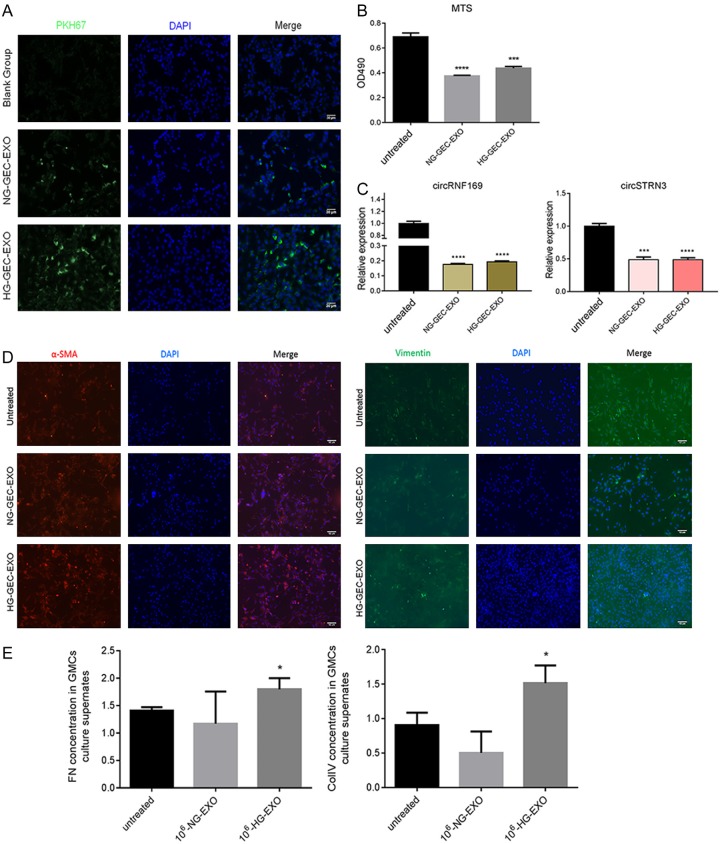
To study the function of GEC exosomes, we labeled exosomes using a PKH67 fluorescent cell linker kit. The GMCs were incubated with GEC exosomes from the blank group, the HG-GEC-EXO group, and the NG-GEC-EXO group for 24 hours. The results suggested that the PKH67-labeled exosomes were localized in the GMC cytoplasm, which indicated that GEC-derived exosomes were internalized by the GMCs. Meanwhile, endocytosis was more obvious in the HG-GEC-EXO group than in NG-GEC-EXO group (Figure 7A). We also performed a cell proliferation study in GMCs, which were treated with the same cell number of exosomes in the HG-GEC-EXO group and the NG-GEC-EXO group. The results revealed that the exosomes of both groups inhibited GMC proliferation (P < 0.0001) (Figure 7B). QRT-PCR was performed to study the expression of circRNF169 and circSTRN3 in GMCs, which were treated with the same cell number of exosomes in the HG-GEC-EXO group and the NG-GEC-EXO group. The results suggested that the expression of circRNF169 and circSTRN3 in the GMCs of both groups was significantly reduced compared with the untreated group (P < 0.001) (Figure 7C). Furthermore, we also studied the potential influence of exosomes on epithelial-mesenchymal transition (EMT). α-SMA and vimentin were selected as biomarkers of EMT. The immunofluorescence results showed that the GEC exosomes of the HG-GEC-EXO group and the NG-GEC-EXO group significantly promoted α-SMA expression in the GMCs. However, there was no difference in vimentin expression in the different groups (Figure 7D). In addition, we used enzyme-linked immunosorbent assay (ELISA) assays to study the FN and COl IV of the culture medium supernatant in the different groups. The results indicated that the GEC exosomes in the HG-GEC-EXO group effectively induced the expression of FN and COl IV in GMC supernatant (Figure 7E). Collectively, the data showed that GEC exosomes in the HG-GEC-EXO group affected GMC cell function.
Figure 7.
Effect of GEC exosomes treated with low glucose and high glucose on mesangial cells (GMC). (A) Endocytosis analysis of GMCs into GEC exosomes with different glucose treatments. GEC exosomes were labeled with PKH67. Scale bar = (B) Cell proliferation analysis of GMCs treated with different GEC exosome. (C) Expression of circRNF169 and circSTRN3 in GMCs treated with different GEC exosomes. (D) Immunofluorescence analysis of epithelial-mesenchymal transition (EMT) of GMCs treated with different GEC exosomes. (E) ELISA examination of EMT of GMCs treated with different GEC exosomes. *P < 0.05; ***P < 0.001; ****P < 0.0001. Untreated: GMC without treatment. HG-GEC-EXO: exosomes from GMCs treated with high glucose. NG-GEC-EXO: exosomes from GMCs treated with low glucose.
Knockdown and over-expression analysis of mmu_circ_0001605 and mmu_circ_0000372 in GMCs
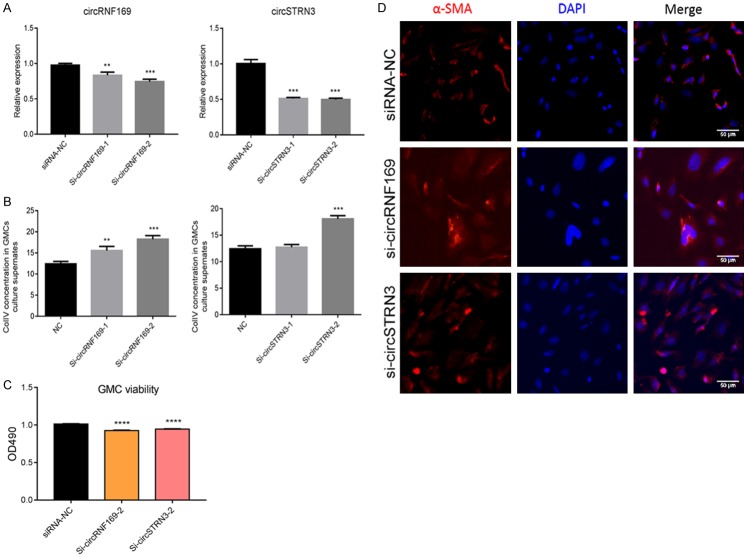
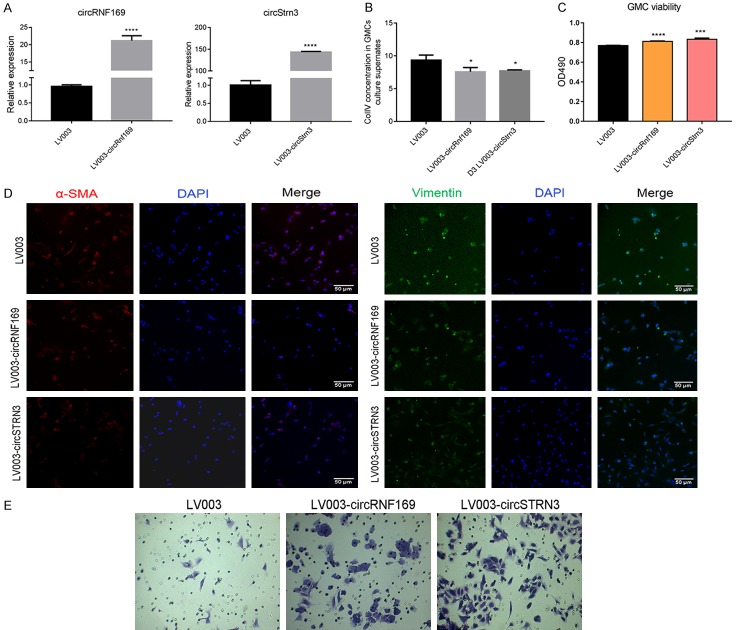
We used GMCs to further study the detailed function of mmu_circ_0001605 and mmu_circ_0000372. In the knockdown study, we designed two siRNA pairs to mmu_circ_0001605 (circRNF169) and mmu_circ_0000372 (circSTRN3) in GMCs. The transfection effect was confirmed by qRT-PCR (Figure 8A). QRT-PCR analysis revealed that Si-circRNF169-1, Si-circRNF169-2, Si-circSTRN3-1, and Si-circSTRN3-2 effectively downregulated the expression of circRNF169 and circSTRN3 (P < 0.05). Therefore, to explore the function of the two interfering circRNAs, we examined the effects of siRNA from two circRNAs on GMC cell function. The ELISA analysis indicated that Si-circRNF169-2 and Si-circSTRN3-2 effectively upregulated the expression of Col IV in GMCs supernatant (P < 0.05) (Figure 8B). Si-circRNF169-2 and Si-circSTRN3-2-treated GMCs were selected to perform the subsequent study. Cell proliferation analysis suggested that Si-circRNF169-2 and Si-circSTRN3-2 treatments effectively inhibited GMC proliferation compared to GMCs without any treatments (Figure 8C). Furthermore, immunofluorescence analysis revealed that α-SMA expression was upregulated in circRNF169 and circSTRN3 knockdown-treated GMCs (Figure 8D), which could be beneficial to EMT. In the over-expression study, the transfection effect was also confirmed by qRT-PCR (Figure 9A). QRT-PCR analysis revealed that the expression of circRNF169 and circSTRN3 were significantly upregulated (P < 0.001). Therefore, to explore the function of the two over-expressed circRNAs, we investigated how the over-expressing plasmids influenced GMC cell functions. ELISA analysis indicated that the over-expression of circRNF169 and circSTRN3 effectively downregulated the expression of Col IV in GMC supernatant (P < 0.05) (Figure 9B), while cell proliferation analysis suggested that over-expression of circRNF169 and circSTRN3 effectively promoted GMCs proliferation compared to GMCs without any treatment (Figure 9C). Furthermore, the immunofluorescence analysis revealed that α-SMA and vimentin expression were downregulated in GMCs treated with circRNF169 and circSTRN3 over-expressing plasmids (Figure 9D). In addition, we used transwell chambers to assess the in vitro migration potential of GMCs with different treatments. The results indicated that over-expression of circRNF169 and circSTRN3 effectively promoted cell migration (Figure 9E). In summary, circRNF169 and circSTRN3 were closely associated with the function of GMCs.
Figure 8.
Functional changes in GMCs after interference with mmu_circ_0001605 and mmu_circ_0000372. A: QRT-PCR confirmation of the results of knockdown treatment with mmu_circ_0001605 and mmu_circ_0000372 in GMCs. B: ELISA examination of the expression of Col IV in GMCs. C: Cell proliferation analysis of GMCs after knockdown by mmu_circ_0001605 and mmu_circ_0000372. D: Immunofluorescence analysis of α-SMA in GMCs after knockdown by mmu_circ_0001605 and mmu_circ_0000372; Scale bar = 50 µm circRNF169 refers to mmu_circ_0001605, located between chr7: 107083672-107104050. circSTRN3 refers to mmu_circ_0000372, located between chr12:52748981-52762700. *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001. siRNA-NC: GMC treated with nonsense siRNA; si-circRNF-169: mmu_circ_0001605 siRNA-treated GMCs; si-circSTRN3: mmu_circ_0000372 siRNA-treated GMCs.
Figure 9.
Functional changes in GMCs after over-expression of mmu_circ_0001605 and mmu_circ_0000372. A: QRT-PCR confirmation of the results of over-expression of mmu_circ_0001605 and mmu_circ_0000372 in GMCs. B: ELISA examination of the expression of Col IV in GMCs over-expressing mmu_circ_0001605 and mmu_circ_0000372. C: Cell proliferation analysis of GMC over-expressing mmu_circ_0001605 and mmu_circ_0000372. D: Immunofluorescence analysis of α-SMA and vimentin in GMCs over-expressing mmu_circ_0001605 and mmu_circ_0000372. Scale bar = 50 µm. E: Cell migration analysis of GMCs over-expressing mmu_circ_0001605 and mmu_circ_0000372. circRNF169 refers to mmu_circ_0001605, located between chr7: 107083672-107104050. circSTRN3 refers to mmu_circ_0000372, located between chr12: 52748981-52762700. *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001. LV003: GMCs treated with empty lentiviral vector. LV003-circRNF-169: mmu_circ_0001605 over-expressing lentiviral-treated GMCs. LV003-circSTRN3: mmu_circ_0000372 over-expressing lentiviral-treated GMCs.
Discussion
The interaction between GECs and GMCs has long been considered to be closely related to the pathogenesis of DN. However, the detailed molecular mechanism of this relationship has not been fully demonstrated. Wu et al. studied the potential role of exosomes in DN. The results indicated that high glucose-treated GECs secreted more exosomes than those exposed to NG levels. TGF-β1 mRNA was higher in high glucose-treated GEC exosomes, which promoted the proliferation of GMCs through the TGF-β1/Smds3 signaling pathway. This signaling pathway promoted the expression of α-SMA protein. Therefore, exosomes can be used as a new target for the prevention and treatment of DN [20]. Meanwhile, a previous study suggested that excessive expression of exogenous miRNA-145 in DN cells may be associated with increased expression of TGF-β [21]. Furthermore, Jiang et al. found that exosomes secreted by human urinary stem cells (USCs-Exo) reduced proteinuria, apoptosis in podocytes, and increased renal size in DN rats. The evidence mentioned above demonstrated that exosomes were released from specific membrane regions. Exosomes selectively carry cell proteins, mRNA, miRNA, DNA, and other cellular components [22]. Therefore, exosomes are the important messengers of cell communication. In contrast, there is little evidence related to exosomal circRNAs in DN. In this study, we demonstrated the DECs in exosomes from HG-treated GECs and NG-treated GECs. The HG-treated GECs produced a significantly higher number of exosomes compared to the NG-treated GECs. The functions of these DECs were closely related to the PI3K/AKT and MAPK signaling pathways. Meanwhile, exosomes extracted from HG-treated GECs affected the cell proliferation and EMT of GMCs. Functional studies of two selected DECs revealed that circRNAs played an important role in the cell proliferation and EMT of GMCs. The results of this study provide a basis for exosome-mediated GEC-GMC crosstalk.
Exosomes derived from the kidney play an important role in the progression, control, and development of renal tumors in kidney disease. Zhang et al. found that renal tubule cells secreted exosomes under hypoxic conditions. Hypoxia-inducible factor (HIF-1)-related exosomes were shown to have protective effects on renal tubular cells [23]. Meanwhile, Zhou et al. suggested that renal proximal tubular epithelial exocytosis plays an antagonistic role in epithelial cell wound healing. Activation of epidermal growth factor and its receptor (EGFR) promoted wound healing and reduced exocrine release. Meanwhile, Gildea et al. found that renal tubular epithelial cells produced more exosomes after stimulation by fenoldopam. These exosomes were transferred to the distal tubules and collecting ducts, reducing the yield of reactive oxygen species in the recipient cells [24]. In this study, we showed that the HG conditions induced GECs to release a higher number of exosomes And that circRNAs were released along with the exosomes in the communication between GECs and GMCs. Nevertheless, due to the complexity of the biomaterial in exosomes, we still cannot exclude other HG-driven proteins, mRNAs, or microRNAs in the exosomes which could potentially play a role in intercellular communication.
A previous study suggested that exosomes can be incorporated by target cells [25]. In our study, we identified that PKH67-labeled GECs exosomes were internalized by GMCs. Therefore, circRNAs were delivered from HG-treated GECs to the neighboring GMCs by exosomes, leading to the elevated expression of circRNAs in GMCs. In our study, we demonstrated that circRNAs containing exosomes were able to change the phenotype of GMCs, which has been identified in DN [26]. But when selected circRNAs were knocked down and over-expressed in GMCs, obvious cell function changes occurred, which supported the fact that DECs were functionally important in the intercellular communication of GMCs.
In conclusion, circRNAs were transferred from GECs to GMCs by means of exosomes. circRNAs carried by exosomes from HG-treated GECs activated GMCs to promote renal fibrosis. Our findings provide new insights into the pathological mechanism of DN.
Acknowledgements
Authors would like to thank the Provincial Natural Science Foundation of Guangdong (grant no. 2017A030313783) for financial support.
Disclosure of conflict of interest
None.
References
- 1.Yang W, Lu J, Weng J, Jia W, Ji L, Xiao J, Shan Z, Liu J, Tian H, Ji Q. Prevalence of diabetes among men and women in China. N Engl J Med. 2010;362:1090–101. doi: 10.1056/NEJMoa0908292. [DOI] [PubMed] [Google Scholar]
- 2.Collins AJ, Foley RN, Herzog C, Chavers B, Gilbertson D, Ishani A, Kasiske B, Liu J, Mau LW, McBean M, Murray A, St Peter W, Guo H, Gustafson S, Li Q, Li S, Li S, Peng Y, Qiu Y, Roberts T, Skeans M, Snyder J, Solid C, Wang C, Weinhandl E, Zaun D, Arko C, Chen SC, Dalleska F, Daniels F, Dunning S, Ebben J, Frazier E, Hanzlik C, Johnson R, Sheets D, Wang X, Forrest B, Constantini E, Everson S, Eggers P, Agodoa L. US renal data system 2010 annual data report. Am J Kidney Dis. 2011;57:A8, e1–526. doi: 10.1053/j.ajkd.2010.10.007. [DOI] [PubMed] [Google Scholar]
- 3.Freedman BI, Bostrom M, Daeihagh P, Bowden DW. Genetic factors in diabetic nephropathy. Clin J Am Soc Nephrol. 2007;2:1306–1316. doi: 10.2215/CJN.02560607. [DOI] [PubMed] [Google Scholar]
- 4.Stadler K, Goldberg IJ, Susztak K. The evolving understanding of the contribution of lipid metabolism to diabetic kidney disease. Curr Diab Rep. 2015;15:40. doi: 10.1007/s11892-015-0611-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stadler K, Goldberg IJ, Susztak K. The evolving understanding of the contribution of lipid metabolism to diabetic kidney disease. Curr Diab Rep. 2015;15:40. doi: 10.1007/s11892-015-0611-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.von Scholten BJ, Rosendahl A, Hasbak P, Bergholdt R, Kjaer A, Rossing P, Hansen TW. Impaired coronary microcirculation in type 2 diabetic patients is associated with elevated circulating regulatory T cells and reduced number of IL-21R(+) T cells. Cardiovasc Diabetol. 2016;15:67. doi: 10.1186/s12933-016-0378-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Donate-Correa J, Martín-Núñez E, Muros-de-Fuentes M, Mora-Fernández C, Navarro-González JF. Inflammatory cytokines in diabetic nephropathy. J Diabetes Res. 2015;2015:948417. doi: 10.1155/2015/948417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Duran-Salgado MB, Rubio-Guerra AF. Diabetic nephropathy and inflammation. World J Diabetes. 2014;5:393–8. doi: 10.4239/wjd.v5.i3.393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kanwar YS, Sun L, Xie P, Liu FY, Chen S. A glimpse of various pathogenetic mechanisms of diabetic nephropathy. Annu Rev Pathol. 2011;6:395–423. doi: 10.1146/annurev.pathol.4.110807.092150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Erdbrügger U, Le TH. Extracellular vesicles in renal diseases: more than novel biomarkers? J Am Soc Nephrol. 2016;27:12–26. doi: 10.1681/ASN.2015010074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Trams EG, Lauter CJ, Salem JN, Heine U. Exfoliation of membrane ecto-enzymes in the form of micro-vesicles. Biochim Biophys Acta. 1981;645:63–70. doi: 10.1016/0005-2736(81)90512-5. [DOI] [PubMed] [Google Scholar]
- 12.Pan BT, Johnstone RM. Fate of the transferrin receptor during maturation of sheep reticulocytes in vitro: selective externalization of the receptor. Cell. 1983;33:967–978. doi: 10.1016/0092-8674(83)90040-5. [DOI] [PubMed] [Google Scholar]
- 13.Armstrong D, Wildman DE. Extracellular vesicles and the promise of continuous liquid biopsies. J Pathol Transl Med. 2018;52:1–8. doi: 10.4132/jptm.2017.05.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kalra H, Drummen G, Mathivanan S. Focus on extracellular vesicles: introducing the next small big thing. Int J Mol Sci. 2016;17:170. doi: 10.3390/ijms17020170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang J, Li S, Li L, Li M, Guo C, Yao J, Mi S. Exosome and exosomal microRNA: trafficking, sorting, and function. Genomics Proteomics Bioinformatics. 2015;13:17–24. doi: 10.1016/j.gpb.2015.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Delić D, Eisele C, Schmid R, Baum P, Wiech F, Gerl M, Zimdahl H, Pullen SS, Urquhart R. Urinary exosomal miRNA signature in type II diabetic nephropathy patients. PLoS One. 2016;11:e0150154. doi: 10.1371/journal.pone.0150154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jia Y, Guan M, Zheng Z, Zhang Q, Tang C, Xu W, Xiao Z, Wang L, Xue Y. miRNAs in urine extracellular vesicles as predictors of early-stage diabetic nephropathy. J Diabetes Res. 2016;2016:7932765. doi: 10.1155/2016/7932765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sokolova V, Ludwig AK, Hornung S, Rotan O, Horn PA, Epple M, Giebel B. Characterisation of exosomes derived from human cells by nanoparticle tracking analysis and scanning electron microscopy. Colloids Surf B Biointerfaces. 2011;87:146–50. doi: 10.1016/j.colsurfb.2011.05.013. [DOI] [PubMed] [Google Scholar]
- 19.Yao Z, Luo J, Hu K, Lin J, Huang H, Wang Q, Zhang P, Xiong Z, He C, Huang Z. ZKSCAN1 gene and its related circular RNA (circZKSCAN1) both inhibit hepatocellular carcinoma cell growth, migration, and invasion but through different signaling pathways. Mol Oncol. 2017;11:422–437. doi: 10.1002/1878-0261.12045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wu XM, Gao YB, Cui FQ, Zhang N. Exosomes from high glucose-treated glomerular endothelial cells activate mesangial cells to promote renal fibrosis. Biol Open. 2016;5:484–91. doi: 10.1242/bio.015990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dear JW, Street JM, Bailey MA. Urinary exosomes: a reservoir for biomarker discovery and potential mediators of intrarenal signalling. Proteomics. 2013;13:1572–1580. doi: 10.1002/pmic.201200285. [DOI] [PubMed] [Google Scholar]
- 22.Rani S, O’Brien K, Kelleher FC, Corcoran C, Germano S, Radomski MW, Crown J, O’Driscoll L. Gene Expression Profiling. Springer; 2011. Isolation of exosomes for subsequent mRNA, MicroRNA, and protein profiling; pp. 181–195. [DOI] [PubMed] [Google Scholar]
- 23.Zhang W, Zhou X, Yao Q, Liu Y, Zhang H, Dong Z. HIF-1-mediated production of exosomes during hypoxia is protective in renal tubular cells. Am J Physiol Renal Physiol. 2017;313:F906–F913. doi: 10.1152/ajprenal.00178.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gildea JJ, Seaton JE, Victor KG, Reyes CM, Wang DB, Pettigrew AC, Courtner CE, Shah N, Tran HT, Van Sciver RE. Exosomal transfer from human renal proximal tubule cells to distal tubule and collecting duct cells. Clin Biochem. 2014;47:89–94. doi: 10.1016/j.clinbiochem.2014.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Skog J, Würdinger T, Van Rijn S, Meijer DH, Gainche L, Curry WT Jr, Carter BS, Krichevsky AM, Breakefield XO. Glioblastoma microvesicles transport RNA and proteins that promote tumour growth and provide diagnostic biomarkers. Nat Cell Biol. 2008;10:1470–6. doi: 10.1038/ncb1800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Makino H, Kashihara N, Sugiyama H, Kanao K, Sekikawa T, Shikata K, Nagai R, Ota Z. Phenotypic changes of the mesangium in diabetic nephropathy. J Diabetes Complications. 1995;9:282–4. doi: 10.1016/1056-8727(95)80022-7. [DOI] [PubMed] [Google Scholar]