Abstract
Non-small cell lung cancer (NSCLC) is a highly malignant type of cancer with a poor 5-year survival rate. The development of prognostic biomarkers and novel drug targets are required in order to improve the survival for NSCLC patients. Signal transducer and activator of transcription (STAT) proteins are cytoplasmic transcription factors known to play key roles in many cellular biological processes. However, the roles of STAT family members in the development and progression of NSCLC have not yet been apparently determined. Our study investigated the roles of STATs in the prognosis of NSCLC using cBioPortal, Human Protein Atlas, ONCOMINE, and Kaplan-Meier Plotter databases. High mutation rate of STATs existed in both lung adenocarcinoma (ADE) patients and squamous cell carcinoma (SCC) patients. High mRNA expression of STAT2 was significantly associated with shorter overall survival (OS) in NSCLC patients, while increased STAT5 and STAT6 were associated with better OS in NSCLC patients. We further found that increased mRNA expressions of STAT2 and STAT3 predicted unfavorable overall survival (OS) while high mRNA expression of STAT5B and STAT6 related to favorable OS for lung ADE patients. However, no significant correlation was identified for lung SCC patients. In stratified survival analysis, high expression of STAT2 predicted poor prognosis in stage II NSLCC patients, surgical margins negative patients and female patients. Taken together, our results illustrated that STAT5B and STAT6 could be effective prognostic biomarkers for survivals of NSCLC patients. And STAT2 might be a promising therapeutic target for the treatment of NSCLC as well as ADE.
Keywords: STAT, non-small cell lung cancer, online database, prognosis, therapeutic target
Introduction
Lung cancer ranks first among the causes of cancer deaths in men and women worldwide [1]. Non-small cell lung cancer (NSCLC) accounts for 80-85% of all cases of lung cancer and classifies into two distinct histological subtypes: squamous cell carcinoma (SCC) and the more common adenocarcinoma (ADE) [2]. Despite advances in the early detection and therapeutic strategies, median overall survival of advanced NSCLC was marginally improved and the overall 5-year survival rate of NSCLC remains low [3]. Approximately 25% of NSCLC patients are first diagnosed with stage III NSCLC and most of these patients have unresectable disease [4]. Even in patients with early-stage and resectable disease receiving definitive chemoradiation, up to 90% of patients eventually relapse [3]. NSCLC remains a challenge to cure, thus investigations and attempts at identifying novel molecular target therapy to improve patients’ outcomes still continue.
The Signal transducer and activator of transcription (STAT) family members mediate the effects of cytokines, growth factors, several hormones and various pathogens. STAT family comprises seven sub-members (STAT1, STAT2, STAT3, STAT4, STAT5A, STAT5B and STAT6), and each family member possesses unique functions in signal transduction [5]. Inactive STATs are located in the cytosol under basal conditions [6]. Once activated, STAT proteins form dimers and/or tetramers, and STAT pathway is connected upstream with Janus kinases (JAK) family protein. They transmit signals from the cell surface to the nucleus where they bind to specific promoter regions of DNA to regulate the transcription of target genes [7,8]. Their variety of functions include proliferation, angiogenesis, inflammation, immunity and survival [9-11].
Numerous studies have reported that overexpression of STAT proteins enhances carcinogenesis and affect the prognosis of cancer patients [12]. STAT family members notably STAT3 and STAT5 play important roles in tumor progression whereas STAT1 is conversely involved in suppressing tumor growth in certain instances [13]. The functions of STAT signaling on tumor initiation and progression make the STAT proteins favorite targets for drug development and cancer therapy. STAT proteins are critically involved in tumorigenesis in various cancers, including lung cancer [14]. Previous studies have found that disruption of STAT3 signaling promotes KRAS-induced lung tumorigenesis [15] and downregulation of JAK/STAT3/SOCS3 signaling pathway by MiR-410 induced the apoptosis of lung cancer A549 cells [16]. A Recent study revealed that JAK/STAT inhibitor ruxolitinib, in combination with vesicular stomatitis virus, would enhance oncolytic virotherapy for NSCLC cancer through preventing PDL-1 expression of tumor cells in response to virotherapy [17]. These evidences suggested that targeting STATs might enhance the effect of other therapy approaches and improve outcomes for NSCLC patients. Nevertheless, the prognostic value of each STAT family member in NSCLC is not well established.
In the present study, we addressed this problem for the first time by analyzing the expression and mutations of different STAT members, and the correlation between STAT members’ mRNA expressions and OS of NSCLC patients. We further evaluate the clinical data including clinical stages, pathological grade, smoking history, gender, surgical margins status, and therapy approaches to systemically explore the roles of STATs. The comprehensive study of distinct STAT members in NSCLC will help to provide perspectives on new biomarkers for predicting the prognosis of NSCLC, and highlight the noteworthy therapeutic targets for NSCLC treatment.
Materials and methods
Oncomine
Oncomine database (www.oncomine.org) is an integrated online cancer microarray database and web-based data mining platform by which the transcriptome data of most cancers and respective normal tissues could be compared [18]. Transcriptional expression of STAT members between NSCLC tissues and normal tissues were observed by Oncomine. Cut-off of p value and fold change were as following: p value: 0.05, fold change: 1.5, gene rank: 10%, data type: mRNA. The 10th, 25th, 50th, 75th and 90th percentile data of each STAT member in both cancer and normal tissues were plotted.
cBioPortal
cBioPortal (www.cbioportal.org) is an open-access website resource for exploring and visualizing multidimensional cancer genomics dataset [19]. We selected Lung Adenocarcinoma (TCGA, Provisional) dataset and Lung Squamous Cell Carcinoma (TCGA, provisional) dataset to analyze genetic alterations of STAT family genes in NSCLC. The genomic profiles included mutations, putative copy-number alterations, and mRNA expressions (RNA Seq V2 RSEM with z-scores = ±2).
Human Protein Atlas
The Human Protein Atlas (HPA, http://www.proteinatlas.org) is a database that contains transcriptomes, proteomes and immunohistochemistry-based expression data. This database consists of Tissue Atlas, Cell Atlas and Pathology Atlas [20]. We got the Fragments Per Kilobase of exon per Million fragments mapped (FPKM) values and protein expression patterns of each STAT gene in NSCLC patients from HPA database.
Kaplan-Meier plotter
The Kaplan-Meier Plotter (www.kmplot.com) is an online database comprises gene expression information and clinical outcome parameters of liver cancer, breast cancer, ovarian cancer, lung cancer and gastric cancer [21]. The prognostic value of mRNA expression of distinct STAT members in Lung cancer was analyzed using Kaplan-Meier plotter. In Kaplan-Meier plotter, patients with lung cancer were divided into two groups by median expression (high vs. low expression) and validated by Kaplan-Meier survival curves. The Affymetrix ID of each STAT gene was valid (Supplementary Table 1). In this study, “array quality control” was selected “exclude biased arrays”.
Results
Genetic mutations and mRNA expression of STAT family members in NSCLC patients
Firstly, we analyzed the genetic alteration of individual STAT members in Lung cancers via cBioPortal database. As shown in Figure 1A and 1B, high mutation rates of STATs were observed in both lung ADE patients (36%) and lung SCC patients (28%). Among all types of mutations, mRNA deregulation was the most common alteration. We also compared the mRNA level of different STAT family members in NSCLC through HPA database. The STAT1 mRNA level was the relative highest in all the genes, whereas STAT4 expressions was very low (Figure 1C). Given that STAT4 was mainly involved in autoimmune and inflammatory disorders through modulating type 1 T helper cells differentiation [22], we assessed the STAT genes except STAT4 in the following analysis.
Figure 1.
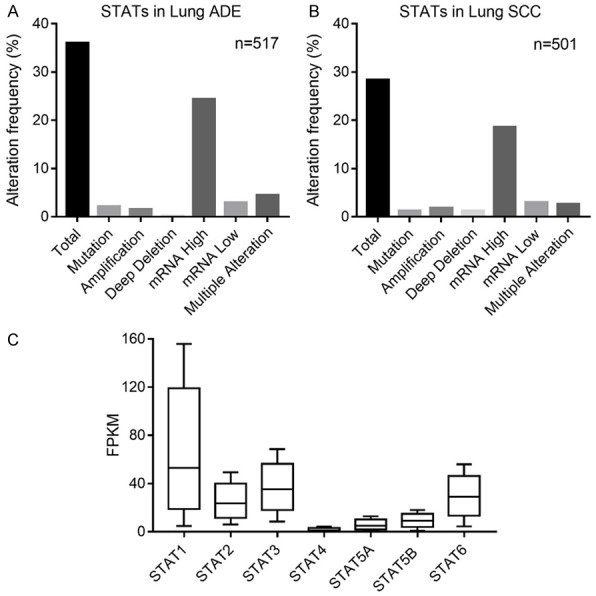
Genetic alterations of STAT family members in NSCLC. (A, B) Alteration frequency of STATs in lung ADE (A) and SCC (B) via cBioPortal. (C) The FPKM value of each STAT member in NSCLC via HPA.
Next, mRNA expressions of seven STAT family members in lung cancer were measured and compared to normal tissues by ONCOMINE database. Among all the STAT members, only higher mRNA expressions of STAT1 were found in lung cancer tissues in multiple datasets (Supplementary Table 2). In Okayama Lung dataset [23], overexpression of STAT1/2/3 was found in lung ADE tissues compared with normal tissues, while STAT5A expression in lung ADE tissues was lower and STAT5B/6 exhibited similar expression level (Figure 2). In Hou Lung database [24], mRNA expressions of STAT1/2/5B were found to be significantly up-regulated in lung SCC tissues compared to normal samples, whereas STAT3/5A/6 expression levels in lung SCC tissues were lower (Figure 3). We also assessed mRNA expression of individual STAT members in other datasets including Selamat [25], Landi [26] and Talbot [27]. STAT genes expression in normal lung tissues and different subtypes of NSCLC tissues were summarized in Table 1. The mRNA levels of STAT1/2 in lung ADE tissues were higher than in normal tissues in Selamat and Landi Lung databases. STAT1/3/6 in lung SCC tissues showed increased expression levels in Talbot database. The inconsistent conclusions from these different datasets most probably due to sample size, study design and detection methods. It is notable that STAT1/2 expressions were markedly higher in these two types of NSCLC compared with that in normal controls.
Figure 2.
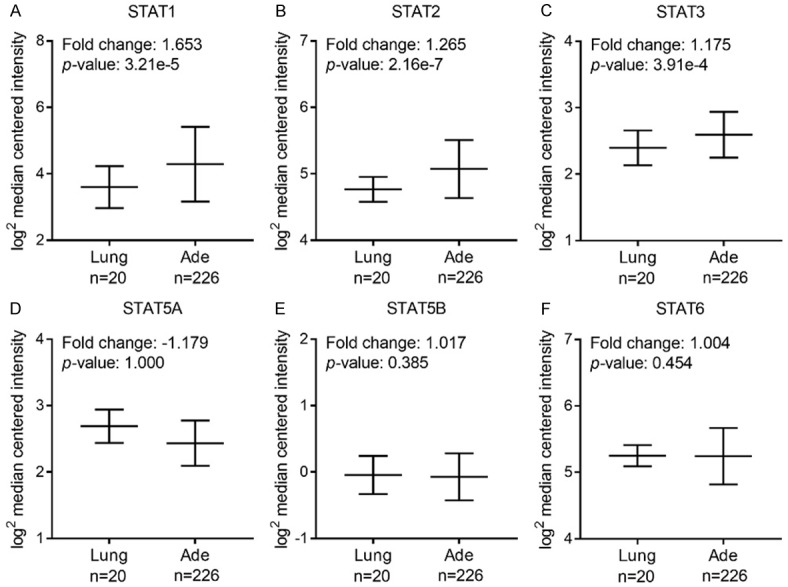
mRNA expression of distinct STAT members in lung ADE and normal tissues using Oncomine (threshold: p-value = 0.05, 1.5 fold change, top 10% gene rank).
Figure 3.
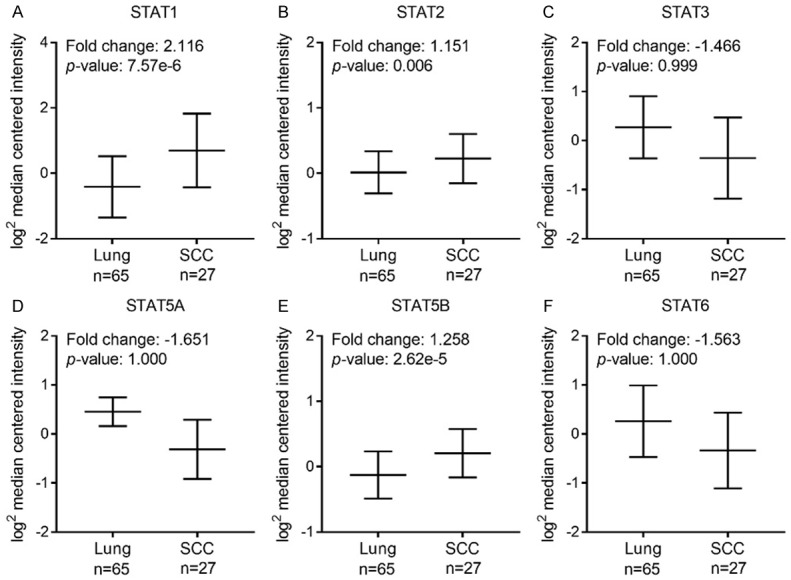
mRNA expression of distinct STAT members in lung SCC and normal tissues using Oncomine (threshold: p-value = 0.05, 1.5 fold change, top 10% gene rank).
Table 1.
Elevated STAT family genes expression in NSCLC cancer
| Gene | p-value | Fold Change | Dataset | Normal | Cancer | Total |
|---|---|---|---|---|---|---|
| Normal vs. ADE | ||||||
| STAT1 | 1.09e-15 | 2.516 | Selamat | 58 | 58 | 116 |
| 7.23e-8 | 1.756 | Landi | 49 | 58 | 107 | |
| 7.09e-9 | 1.453 | Selamat | 58 | 58 | 116 | |
| STAT2 | 0.013 | 1.069 | Landi | 49 | 58 | 107 |
| 0.049 | 1.080 | Selamat | 58 | 58 | 116 | |
| STAT3 | 0.587 | -1.010 | Landi | 49 | 58 | 107 |
| 1.000 | -1.415 | Selamat | 58 | 58 | 116 | |
| STAT5A | 1.000 | -1.258 | Landi | 49 | 58 | 107 |
| 1000 | -1.277 | Selamat | 58 | 58 | 116 | |
| STAT5B | 0.572 | -1.005 | Landi | 49 | 58 | 107 |
| 0.998 | -1.176 | Selamat | 58 | 58 | 116 | |
| STAT6 | 0.726 | -1.127 | Landi | 49 | 58 | 107 |
| Normal vs. SCC | ||||||
| STAT1 | 2.63e-10 | 2.625 | Talbot | 2 | 34 | 36 |
| STAT2 | 0.095 | 1.803 | Talbot | 2 | 34 | 36 |
| STAT3 | 8.23e-5 | 1.382 | Talbot | 2 | 34 | 36 |
| STAT5A | 0.994 | -1.112 | Talbot | 2 | 34 | 36 |
| STAT5B | 0.992 | -1.140 | Talbot | 2 | 34 | 36 |
| STAT6 | 1.20e-7 | 1.483 | Talbot | 2 | 34 | 36 |
Different subtypes of lung cancer were analyzed and p-values, fold changes, datasets and the number of clinical specimen were included.
We further explored the protein expression patterns of STATs in NSCLC by the HPA database and summaried in Supplementary Table 3. However, these data could not well explain the protein expression of STATs in NSCLC due to the small sample size.
Prognostic values of STAT family members in NSCLC patients
The prognostic values of STAT mRNA expression were evaluated in the Kaplan-Meier Plotter database and the OS curves were plotted for NSCLC patients. As shown in Figure 4, mRNA expressions of most STAT family members were significantly associated with NSCLC patients’ prognosis. Increase in STAT2 mRNA expression level (HR = 1.19, 95% CI: 1.05-1.36, P = 0.0057) was significantly associated with shorter OS for all NSCLC patients, while higher mRNA expression of STAT5A/5B/6 significantly related to favorable OS (STAT5A, HR = 0.87, 95% CI: 0.77-0.99, P = 0.033; STAT5B, HR = 0.75, 95% CI: 0.66-0.86, P = 1.4e-05; STAT6, HR = 0.69, 95% CI: 0.6-0.78, P = 7.7e-09). However, mRNA expression of STAT1/3 showed no correlation with prognosis of NSCLC patients (STAT1, HR = 0.95, 95% CI: 0.84-1.07, P = 0.4; STAT3, HR = 0.97, 95% CI: 0.86-1.1, P = 0.64). These results revealed that mRNA expressions of STAT2/5A/5B/6 were significantly associated with NSCLC patients’ prognosis and they might be exploited as useful biomarkers for predicting NSCLC patients’ survival. Most notably, STAT2 might be a potential therapeutic target for NSCLC treatment.
Figure 4.
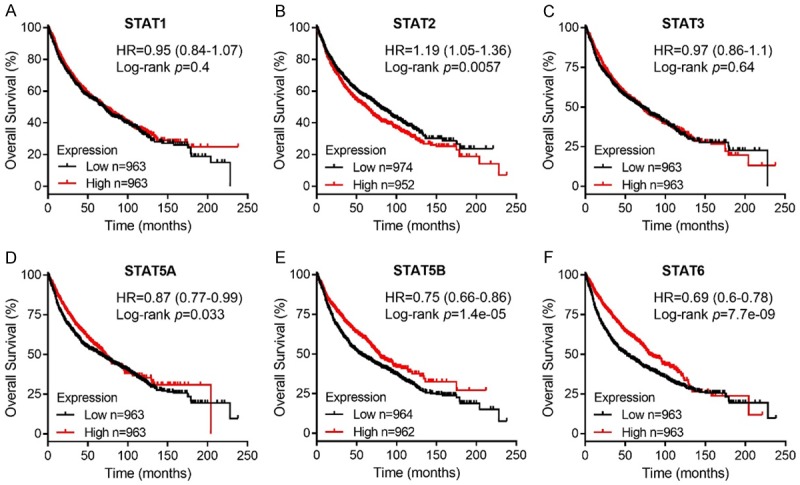
Prognostic value of STAT family members in NSCLC patients using Kaplan-Meier plotter.
We also analyzed the prognostic values of STAT genes in lung ADE patients and SCC patients. High expression of STAT2/3 mRNA were significantly associated with poor rates of OS in lung ADE patients (STAT2, HR = 1.6, 95% CI: 1.26-2.02, P = 8e-05; STAT3, HR = 1.33, 95% CI: 1.05-1.69, P = 0.017; Figure 5B and 5C), while high STAT5B/6 mRNA expression showed correlation with favorable OS in ADE patients (Figure 5E and 5F). In addition, STAT1/4/5A expression exhibited no correlation with OS for ADE patients (Figure 5). As for lung SCC patients, there was no significant association between all the STAT members and OS rates (Figure 6). These results implied that mRNA expression of STATs could significantly affect lung ADE patients’ prognosis.
Figure 5.
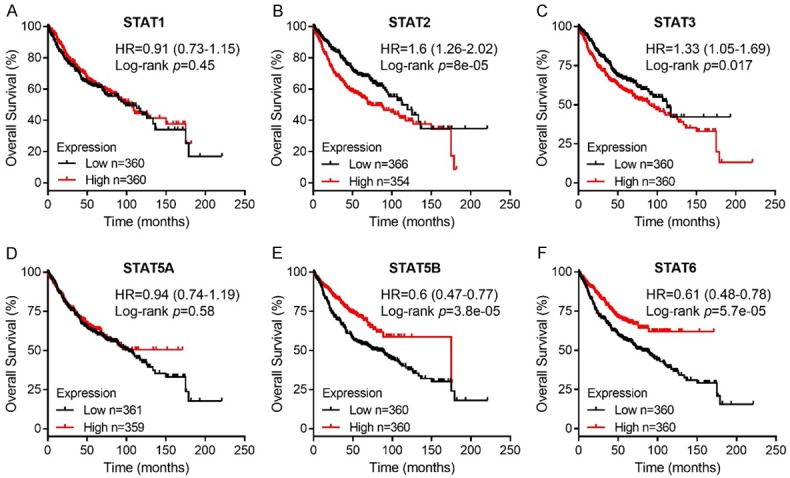
Prognostic value of mRNA expression of STAT family members in lung ADE patients using Kaplan-Meier plotter.
Figure 6.
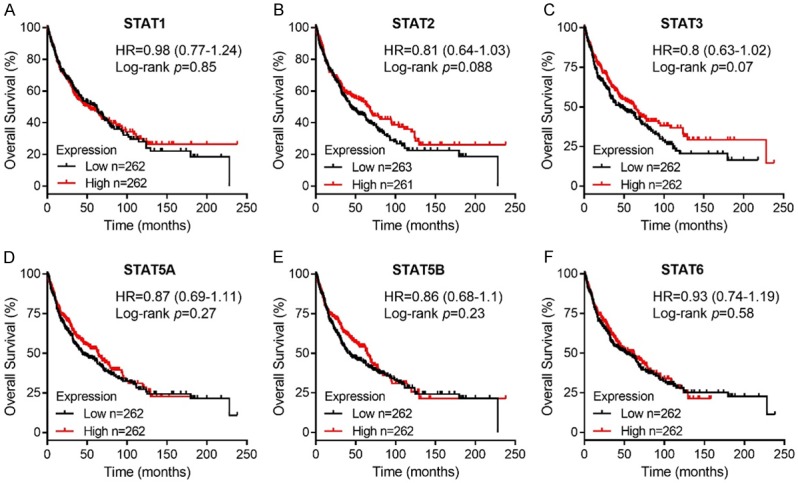
Prognostic value of mRNA expression of STAT family members in lung SCC patients using Kaplan-Meier plotter.
Prognostic values of STATs in NSCLC patients with different clinicopathological features
To assess whether the prognostic value of the mRNA expression status of individual STAT family members depend on other clinicopathological characteristics, we analyzed the influence of high vs. low STAT expression on OS for NSCLC patients with differenet clinicopathological characteristics including clinic stage, pathological grade, smoking history, gender, surgical margins status, different treatment approaches. As illustrated in Table 2, we found that overexpression of STAT2 was correlated with unfavorable OS in stage II NSCLC patients, and high STAT3 was associated with worse OS in stage I NSCLC patients. Increased expressions of STAT4/5B/6 were correlated with better OS in stage I NSCLC patients, and high STAT2 predicted better OS in stage III patients.
Table 2.
Correlation of STAT family members with tumor stages of NSCLC patients
| Gene | Stages | Low (cases) | High (cases) | HR (95% CI) | p-value |
|---|---|---|---|---|---|
| STAT1 | I | 288 | 289 | 0.83 (0.63-1.09) | 0.18 |
| II | 122 | 122 | 0.75 (0.52-1.08) | 0.12 | |
| III | 35 | 35 | 0.79 (0.46-1.36) | 0.39 | |
| STAT2 | I | 293 | 284 | 1.25 (0.96-1.64) | 0.1 |
| II | 122 | 122 | 1.64 (1.14-2.37) | 0.0078* | |
| III | 35 | 35 | 0.55 (0.32-0.96) | 0.033* | |
| STAT3 | I | 288 | 289 | 1.38 (1.05-1.81) | 0.02* |
| II | 122 | 122 | 1.45 (1-2.09) | 0.049* | |
| III | 35 | 35 | 1.57 (0.91-2.7) | 0.1 | |
| STAT5A | I | 288 | 289 | 0.76 (0.58-1) | 0.052 |
| II | 122 | 122 | 0.9 (0.63-1.3) | 0.59 | |
| III | 35 | 35 | 0.93 (0.53-1.61) | 0.79 | |
| STAT5B | I | 288 | 289 | 0.45 (0.33-0.6) | 6.5e-08** |
| II | 122 | 122 | 0.83 (0.57-1.2) | 0.32 | |
| III | 35 | 35 | 0.67 (0.39-1.16) | 0.15 | |
| STAT6 | I | 288 | 289 | 0.45 (0.33-0.6) | 3.7e-08** |
| II | 122 | 122 | 0.89 (0.62-1.29) | 0.55 | |
| III | 35 | 35 | 0.51 (0.29-0.89) | 0.016* |
Low/High (cases): low/high expression of the corresponding gene (patient number).
P < 0.05;
P < 0.01.
In Table 3 we investigated the association between STATs expression and grades in NSCLC patients. No significant association with OS was identified for any of the STAT members in patients with different pathological grades. Table 4 showed prognostic significance between STAT mRNA expression and smoking history in NSCLC patients. High mRNA expression of STAT2 was correlated with worse OS in patients with and without smoking habit. STAT3 overexpression also predicted unfavorable OS in no smoking patients. However, high STAT5B predicted worse OS in both smoking and no smoking patients. Overexpression of STAT6 was associated with favorable OS in smoking patients.
Table 3.
Correlation of STAT family members with tumor grades of NSCLC patients
| Gene | Grades | Low (cases) | High (cases) | HR (95% CI) | p-value |
|---|---|---|---|---|---|
| STAT1 | I | 100 | 101 | 0.94 (0.66-1.35) | 0.75 |
| II | 155 | 155 | 0.93 (0.68-1.28) | 0.67 | |
| III | 38 | 39 | 1.14 (0.59-2.2) | 0.7 | |
| STAT2 | I | 101 | 100 | 1.18 (0.82-1.69) | 0.37 |
| II | 156 | 154 | 1 (0.73-1.37) | 1 | |
| III | 38 | 39 | 0.78 (0.4-1.51) | 0.46 | |
| STAT3 | I | 100 | 101 | 0.96 (0.67-1.37) | 0.81 |
| II | 155 | 155 | 0.95 (0.69-1.3) | 0.76 | |
| III | 38 | 39 | 0.72 (0.37-1.39) | 0.32 | |
| STAT5A | I | 100 | 101 | 0.98 (0.69-1.4) | 0.92 |
| II | 155 | 155 | 0.86 (0.63-1.17) | 0.33 | |
| III | 38 | 39 | 0.56 (0.29-1.08) | 0.079 | |
| STAT5B | I | 100 | 101 | 1.22 (0.85-1.75) | 0.28 |
| II | 155 | 155 | 0.91 (0.66-1.24) | 0.55 | |
| III | 38 | 39 | 0.67 (0.35-1.29) | 0.23 | |
| STAT6 | I | 100 | 101 | 0.94 (0.66-1.35) | 0.76 |
| II | 155 | 155 | 0.94 (0.69-1.29) | 0.71 | |
| III | 38 | 39 | 0.67 (0.34-1.29) | 0.23 |
Low/High (cases): low/high expression of the corresponding gene (patient number). *P < 0.05, **P < 0.01.
Table 4.
Correlation of STAT family members with smoking history of NSCLC patients
| Gene | Smoking history | Low (cases) | High (cases) | HR (95% CI) | p-value |
|---|---|---|---|---|---|
| STAT1 | Never smoked | 103 | 102 | 0.73 (0.41-1.28) | 0.27 |
| Smoked | 410 | 410 | 0.92 (0.75-1.13) | 0.41 | |
| STAT2 | Never smoked | 103 | 102 | 1.88 (1.06-3.34) | 0.027* |
| Smoked | 410 | 410 | 1.27 (1.03-1.56) | 0.024* | |
| STAT3 | Never smoked | 102 | 103 | 2.17 (1.21-3.91) | 0.0079** |
| Smoked | 410 | 410 | 1.08 (0.88-1.33) | 0.48 | |
| STAT5A | Never smoked | 102 | 103 | 1.12 (0.64-1.96) | 0.68 |
| Smoked | 410 | 410 | 0.89 (0.73-1.1) | 0.28 | |
| STAT5B | Never smoked | 102 | 103 | 0.33 (0.18-0.6) | 0.00013** |
| Smoked | 410 | 410 | 0.81 (0.66-1) | 0.044* | |
| STAT6 | Never smoked | 102 | 103 | 0.87 (0.5-1.51) | 0.61 |
| Smoked | 410 | 410 | 0.77 (0.63-0.95) | 0.013* |
Low/High (cases): low/high expression of the corresponding gene (patient number).
P < 0.05;
P < 0.01.
The results in Table 5 revealed prognostic value between STAT mRNA expression and gender in NSCLC patients. STAT2 expression related to unfavorable OS for female NSCLC patients. Overexpression of STAT4/5A were significantly correlated with favorable OS in male patients. STAT5B/6 showed correlations with better OS in both female and male patients. In Table 6 we analyzed the correlation between STATs expression and surgery margins status in NSCLC patients. Overexpression of STAT2/3 were correlated with worse OS in patients with negative surgery margins, while STAT5B was associated with better OS in patients with negative surgery margins. Lastly, Tables 7 and 8 demonstrated the correlation of STATs mRNA expression with OS in NSCLC patients who received chemotherapy or radiotherapy. We found that high expression of STAT2 predicted favorable OS in patients receiving chemotherapy and increased STAT5A was associated with better OS in patients without radiotherapy.
Table 5.
Correlation of STAT family members with gender of NSCLC patients
| Gene | Gender | Low (cases) | High (cases) | HR (95% CI) | p-value |
|---|---|---|---|---|---|
| STAT1 | female | 358 | 357 | 1.21 (0.96-1.52) | 0.11 |
| male | 550 | 550 | 0.9 (0.77-1.06) | 0.2 | |
| STAT2 | female | 361 | 354 | 1.33 (1.05-1.68) | 0.016* |
| male | 557 | 543 | 1.13 (0.97-1.33) | 0.12 | |
| STAT3 | female | 358 | 357 | 1.06 (0.84-1.33) | 0.64 |
| male | 550 | 550 | 0.94 (0.8-1.1) | 0.43 | |
| STAT5A | female | 359 | 356 | 0.96 (0.76-1.21) | 0.71 |
| male | 551 | 549 | 0.76 (0.65-0.89) | 0.00086** | |
| STAT5B | female | 358 | 357 | 0.69 (0.54-0.87) | 0.0018** |
| male | 554 | 546 | 0.83 (0.71-0.97) | 0.022* | |
| STAT6 | female | 359 | 356 | 0.73 (0.58-0.93) | 0.0092** |
| male | 550 | 550 | 0.71 (0.6-0.83) | 1.9e-05** |
Low/High (cases): low/high expression of the corresponding gene (patient number).
P < 0.05;
P < 0.01.
Table 6.
Correlation of STAT family members with negative surgical margins of NSCLC patients
| Gene | Surgical margins | Low (cases) | High (cases) | HR (95% CI) | p-value |
|---|---|---|---|---|---|
| STAT1 | negative | 363 | 363 | 0.81 (0.65-1.02) | 0.071 |
| STAT2 | negative | 364 | 362 | 1.54 (1.22-1.94) | 0.00022** |
| STAT3 | negative | 364 | 362 | 1.62 (1.28-2.05) | 5.9e-05** |
| STAT5A | negative | 363 | 363 | 1.06 (0.84-1.33) | 0.63 |
| STAT5B | negative | 364 | 362 | 0.65 (0.52-0.82) | 0.00025** |
| STAT6 | negative | 363 | 363 | 0.82 (0.66-1.04) | 0.096 |
Low/High (cases): low/high expression of the corresponding gene (patient number).
P < 0.01.
Table 7.
Correlation of STAT family members with chemotherapy of NSCLC patients
| Gene | Chemotherapy | Low (cases) | High (cases) | HR (95% CI) | p-value |
|---|---|---|---|---|---|
| STAT1 | yes | 88 | 88 | 1.07 (0.71-1.61) | 0.74 |
| no | 155 | 155 | 0.9 (0.65-1.26) | 0.56 | |
| STAT2 | yes | 88 | 88 | 0.61 (0.4-0.91) | 0.015* |
| no | 158 | 152 | 1.05 (0.75-1.47) | 0.76 | |
| STAT3 | yes | 88 | 88 | 1 (0.66-1.52) | 1 |
| no | 155 | 155 | 0.78 (0.56-1.09) | 0.14 | |
| STAT5A | yes | 88 | 88 | 0.96 (0.64-1.43) | 0.82 |
| no | 155 | 155 | 0.75 (0.54-1.05) | 0.094 | |
| STAT5B | yes | 89 | 87 | 1.13 (0.75-1.69) | 0.56 |
| no | 155 | 155 | 1.06 (0.76-1.49) | 0.72 | |
| STAT6 | yes | 88 | 88 | 0.89 (0.59-1.33) | 0.56 |
| no | 155 | 155 | 0.82 (0.59-1.15) | 0.25 |
Low/High (cases): low/high expression of the corresponding gene (patient number).
P < 0.05.
Table 8.
Correlation of STAT family members with radiotherapy of NSCLC patients
| Gene | Radiotherapy | Low (cases) | High (cases) | HR (95% CI) | p-value |
|---|---|---|---|---|---|
| STAT1 | yes | 35 | 35 | 1.07 (0.63-1.82) | 0.81 |
| no | 136 | 135 | 0.94 (0.66-1.34) | 0.72 | |
| STAT2 | yes | 35 | 35 | 1.18 (0.69-2.01) | 0.54 |
| no | 136 | 135 | 0.94 (0.66-1.34) | 0.74 | |
| STAT3 | yes | 35 | 35 | 0.74 (0.43-1.28) | 0.28 |
| no | 136 | 135 | 0.8 (0.56-1.14) | 0.21 | |
| STAT5A | yes | 35 | 35 | 1.47 (0.86-2.52) | 0.16 |
| no | 136 | 135 | 0.69 (0.48-0.99) | 0.043* | |
| STAT5B | yes | 35 | 35 | 1.43 (0.84-2.45) | 0.19 |
| no | 138 | 133 | 1 (0.7-1.42) | 0.99 | |
| STAT6 | yes | 35 | 35 | 0.95 (0.56-1.63) | 0.86 |
| no | 136 | 135 | 1.02 (0.71-1.45) | 0.93 |
Low/High (cases): low/high expression of the corresponding gene (patient number).
P < 0.05.
Discussion
Being important components of signaling transduction, STAT family proteins are implicated in the development of multiple cancers, including NSCLC. However, not all STAT proteins participate in the initiation and progression of malignancy in human [13]. Despite some members of STAT have already been confirmed to play key roles in NSCLC, the prognostic value of each STAT member mRNA expression in NSCLC patients, to our knowledge, had not been clarified by any previous study thus far. Personalized treatment decisions based on the genetics of the individual tumor will be paramount to combat malignancies in the future. Therefore, understanding the prognostic values of STAT family members in NSCLC might show light in discovering new biomarkers and noteworthy therapeutic targets for NSCLC. The aim of our study was to indicate the influence of STATs expression status on the OS rate of NSCLC patients, and the association between STATs and the clinical features of NSCLC.
Previous studies demonstrated that activated STAT1 seemed to exhibit pro-apoptotic and anti-proliferative effect since STAT1-null mice had higher risk of tumor development than controls [28,29]. Modified STAT1 that was hyper-responsive to interferons (IFN) improved the antitumor response of IFNs in lung cancer cells [30]. STAT1 is unlikely to promote tumor cell growth in human and has the tumor-suppressing properties like TP53. The role of STAT1 was puzzling for the reason that STAT1 acted as a tumor promotor in other researches [31,32]. This probably attributes to cancer specific. Here, although STAT1 mRNA expression was significantly higher in NSCLC tissues than that in normal tissues, no correlation of STAT1 on OS rate of NSCLC patients was observed.
STAT2 is a well-known pivotal and specific effector of type I interferon (IFN-I) signaling which is important in tumor immunosurveillance [33,34]. A deficiency in STAT2 impaired IFN-I mediated anti-tumorigenic effects in dendritic cells thus allowing tumors to thrive. It has already reported that the growth of melanoma and colon adenocarcinoma were accelerated in STAT2-/- mice [35]. Similarly, loss of STAT2 markedly inhibited the ability of inflammatory breast cancer cells to proliferate, migrate, invade, and form 2-D colonies [36]. And STAT2 as an important molecular target has been confirmed in skin squamous cell carcinoma cells [37]. These were similar to discoveries made in our study, in which low mRNA expression of STAT2 was associated with better prognosis in NSCLC patients, apart from SCC patients. In addition, STAT2 could regulate the expression of genes involved in the differentiation and recruitment of immune effector cells to the tumor site [35]. To date, little is known about the function of STAT2 in NSCLC. Our data suggested that STAT2 might be a crucial drug target for the treatment of lung ADE. However, the converse prognostic values of STAT2 in stage II and stage III NSCLC patients possibly due to specimen’s specific and sample size. Importantly, existing therapeutic approaches plus STAT2 inhibition might effectively improve outcomes of lung ADE patients. Future mechanistic researches and clinical trials are needed to explore the clinical application of STAT2 in the treatment of NSCLC.
Among all the STAT family members, STAT3 has been the most researched and investigated member in human cancers. The aberrant activation of STAT3 occurs in many solid tumor [38-42] and hematopoietic malignancies [43,44]. STAT3 Activation was observed in nearly 50% of Lung cancers [45]. Previous studies found that STAT3 inhibitor regressed human breast and lung cancer xenografts [46], and direct inhibition of STAT3 even induced apoptosis in NSCLC cells with acquired Erlotinib resistance [47]. Moreover, STAT3 has been thought to play a tumor-promoting role in NSCLC and during acquired drug resistance [48,49]. Targeting STAT3 is currently proposed as therapeutic intervention. A number of potent small-molecule inhibitors of STAT3 have already been published [46,50-52]. In our study, STAT3 mRNA was overexpressed in lung ADE tissues compared with that in normal controls. Increased STAT3 was significantly associated with a poorer OS rate only in ADE patients but not in SCC patients. Taken together, STAT3 might be a therapeutic target for lung ADE not SCC. It should be noted that most STAT3 inhibitors reported to date have not undergone an in vivo efficacy, pharmacology or toxicity testing. There is the need to further assess the role of STAT3 in lung ADE cancer and identify suitable anti-STAT3 agents for development into clinically useful anti-ADE therapeutics.
STAT5A and STAT5B, two isoforms of STAT5, are transcribed from separate genes, but share 94% structural homology [53]. STAT5 proteins are of particular interest for their critical roles in cellular functions such as proliferation, differentiation, and survival [54]. In addition, STAT5 proteins critically regulate the maintenance of normal im-mune function and homeostasis [53], and has prominent roles in mammary development and function [55]. Similar to STAT3, constitutive activation of STAT5 is the leading causes of tumorigenesis [56]. STAT5 activation was essential for cancer progression in chronic myelogenous leukemia (CML) and myeloproliferative disease [57,58]. There are strong evidences showing that targeting STAT5 with small molecules could interfere with a substantial proportion of human tumors [59,60]. A variety of STAT5 inhibitors have been identified that induce antitumor cell effects [61-63]. Several STAT5 inhibitors for hematological malignancy have already advanced to clinical studies [13]. However, our data indicated that STAT5B was associated with an improved prognosis in all NSCLC patients combined and ADE patients alone. These results suggested that STAT5A and STAT5B might be prognostic biomarkers not therapeutic targets for NSCLC.
The function of STAT6 mainly involved in immune function, tumor immunosurveillance, lymphomagenesis and inflammatory-related tumorigenesis [56,64]. STAT6 is constitutively stimulated in a number of human cancers, such as colon and prostate cancer, mediastinal large B-cell lymphoma and Hodgkin’s lymphoma [65-67]. Thus, STAT6 is considered as a therapeutic target for various cancer types. For instance, Binnemars-Postma K et al. observed that targeting the STAT6 pathway in tumor-associated macrophages reduced tumor growth and metastatic niche formation in breast cancer [68]. Leon-Cabrera SA et al. demonstrated that lack of STAT6 attenuated inflammatory responses and cell recruitment, which in turn prevented the early development of colitis-associated colon cancer [69]. A recent study also revealed that direct inhibition of STAT6 induced prostate cancer cell apoptosis [70]. However, the function of STAT6 in NSCLC is less clear. Here, STAT6 mRNA expression was correlated with an improved prognosis in all NSCLC patients combined, as well as ADE patients alone. In detail, increased STAT6 predicted better OS in stage I patients.
All the individual STAT members were not significantly associated with pathological grades of NSCLC patients. Overexpression of STAT2 was correlated with favorable OS in stage III patients, suggesting that STAT2 inhibitors might not be suitable for advanced NSCLC patients. No STAT member was identified to be a valid prognostic factor in NSCLC patients with a smoking history. We also investigated the association between STATs expression and therapy strategies in NSCLC patients. Increased STAT2 expression exhibited correlations with better OS when patients received chemotherapy. Given that most lung cancer patients would be treated with chemotherapy, it should be paid more attention to the usage of inhibitors targeting STAT2. Conversely, patients with negative surgery margins should to consider combined with STAT2 or/and STAT3 inhibitors to improve therapeutic efficacy of NSCLC including ADE.
In conclusion, our data showed that mRNA expression of STAT1/2/3 were increased in lung ADE and STAT1/2/5B were up-regulated in lung SCC. Increased mRNA expression of STAT2 was significantly associated with shorter OS in NSCLC patients, while increased expressions of STAT4/5A/5B/6 were significantly related to favorable OS. In lung ADE patients, high STAT2/3 predicted unfavorable OS and high STAT5B/6 indicated favorable OS. A stratified analysis for all NSCLC patients combined was performed, but included patients with SCC, no correction between STAT members and OS was observed. These data suggested that STAT5B/6 could be potential biomarkers for the prognosis of lung ADE and STAT2 might be promising therapeutic targets for NSCLC treatment. Considered that all the data in our study was obtained from online databases, further studies consist of larger sample sizes are required to validate our findings and to investigate the possible underlying mechanism between distinct STATs and NSCLC. Importantly, the potential function of STAT2 in lung ADE should be verified with more evidence.
Acknowledgements
We are grateful to the contributors of data to cBioPortal, HPA, Oncomine, and Kaplan-Meier plotter. This study was supported by the National Natural Science Foundation of China (Grant #81672619).
Disclosure of conflict of interest
None.
Supporting Information
References
- 1.PDQ cancer information summaries. Bethesda (MD): 2002. Lung Cancer Screening (PDQ(R)): Health Professional Version. [Google Scholar]
- 2.Minna JD, Roth JA, Gazdar AF. Focus on lung cancer. Cancer Cell. 2002;1:49–52. doi: 10.1016/s1535-6108(02)00027-2. [DOI] [PubMed] [Google Scholar]
- 3.Tan WL, Jain A, Takano A, Newell EW, Iyer NG, Lim WT, Tan EH, Zhai W, Hillmer AM, Tam WL, Tan DSW. Novel therapeutic targets on the horizon for lung cancer. Lancet Oncol. 2016;17:e347–e362. doi: 10.1016/S1470-2045(16)30123-1. [DOI] [PubMed] [Google Scholar]
- 4.Ryan KJ, Skinner KE, Fernandes AW, Punekar RS, Pavilack M, Walker MS, VanderWalde NA. Real-world treatment patterns among patients with unresected stage III non-small-cell lung cancer. Future Oncol. 2019;15:2943–2953. doi: 10.2217/fon-2018-0939. [DOI] [PubMed] [Google Scholar]
- 5.Berg T. Signal transducers and activators of transcription as targets for small organic molecules. Chembiochem. 2008;9:2039–2044. doi: 10.1002/cbic.200800274. [DOI] [PubMed] [Google Scholar]
- 6.Richard AJ, Stephens JM. The role of JAK-STAT signaling in adipose tissue function. Biochim Biophys Acta. 2014;1842:431–439. doi: 10.1016/j.bbadis.2013.05.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Darnell JE Jr. Transcription factors as targets for cancer therapy. Nat Rev Cancer. 2002;2:740–749. doi: 10.1038/nrc906. [DOI] [PubMed] [Google Scholar]
- 8.Rawlings JS, Rosler KM, Harrison DA. The JAK/STAT signaling pathway. J Cell Sci. 2004;117:1281–1283. doi: 10.1242/jcs.00963. [DOI] [PubMed] [Google Scholar]
- 9.Miklossy G, Hilliard TS, Turkson J. Therapeutic modulators of STAT signalling for human diseases. Nat Rev Drug Discov. 2013;12:611–629. doi: 10.1038/nrd4088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pencik J, Pham HT, Schmoellerl J, Javaheri T, Schlederer M, Culig Z, Merkel O, Moriggl R, Grebien F, Kenner L. JAK-STAT signaling in cancer: from cytokines to non-coding genome. Cytokine. 2016;87:26–36. doi: 10.1016/j.cyto.2016.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.O’Shea JJ, Plenge R. JAK and STAT signaling molecules in immunoregulation and immune-mediated disease. Immunity. 2012;36:542–550. doi: 10.1016/j.immuni.2012.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Heppler LN, Frank DA. Targeting oncogenic transcription factors: therapeutic implications of endogenous STAT inhibitors. Trends Cancer. 2017;3:816–827. doi: 10.1016/j.trecan.2017.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Loh CY, Arya A, Naema AF, Wong WF, Sethi G, Looi CY. Signal transducer and activator of transcription (STATs) proteins in cancer and inflammation: functions and therapeutic implication. Front Oncol. 2019;9:48. doi: 10.3389/fonc.2019.00048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pastuszak-Lewandoska D, Domanska-Senderowska D, Kordiak J, Antczak A, Czarnecka KH, Migdalska-Sek M, Nawrot E, Kiszalkiewicz JM, Brzezianska-Lasota E. Immunoexpression analysis of selected JAK/STAT pathway molecules in patients with non-small-cell lung cancer. Pol Arch Intern Med. 2017;127:758–764. doi: 10.20452/pamw.4115. [DOI] [PubMed] [Google Scholar]
- 15.Grabner B, Schramek D, Mueller KM, Moll HP, Svinka J, Hoffmann T, Bauer E, Blaas L, Hruschka N, Zboray K, Stiedl P, Nivarthi H, Bogner E, Gruber W, Mohr T, Zwick RH, Kenner L, Poli V, Aberger F, Stoiber D, Egger G, Esterbauer H, Zuber J, Moriggl R, Eferl R, Gyorffy B, Penninger JM, Popper H, Casanova E. Disruption of STAT3 signalling promotes KRAS-induced lung tumorigenesis. Nat Commun. 2015;6:6285. doi: 10.1038/ncomms7285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li M, Zheng R, Yuan FL. MiR-410 affects the proliferation and apoptosis of lung cancer A549 cells through regulation of SOCS3/JAK-STAT signaling pathway. Eur Rev Med Pharmacol Sci. 2018;22:5987–5993. doi: 10.26355/eurrev_201809_15933. [DOI] [PubMed] [Google Scholar]
- 17.Patel MR, Dash A, Jacobson BA, Ji Y, Baumann D, Ismail K, Kratzke RA. JAK/STAT inhibition with ruxolitinib enhances oncolytic virotherapy in non-small cell lung cancer models. Cancer Gene Ther. 2019 doi: 10.1038/s41417-018-0074-6. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 18.Rhodes DR, Yu J, Shanker K, Deshpande N, Varambally R, Ghosh D, Barrette T, Pandey A, Chinnaiyan AM. ONCOMINE: a cancer microarray database and integrated data-mining platform. Neoplasia. 2004;6:1–6. doi: 10.1016/s1476-5586(04)80047-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, Jacobsen A, Byrne CJ, Heuer ML, Larsson E, Antipin Y, Reva B, Goldberg AP, Sander C, Schultz N. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2:401–404. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Roszik J, Subbiah V. Mining public databases for precision oncology. Trends Cancer. 2018;4:463–465. doi: 10.1016/j.trecan.2018.04.008. [DOI] [PubMed] [Google Scholar]
- 21.Ren J, Niu G, Wang X, Song T, Hu Z, Ke C. Overexpression of FNDC1 in gastric cancer and its prognostic significance. J Cancer. 2018;9:4586–4595. doi: 10.7150/jca.27672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.O’Shea JJ, Lahesmaa R, Vahedi G, Laurence A, Kanno Y. Genomic views of STAT function in CD4+ T helper cell differentiation. Nat Rev Immunol. 2011;11:239–250. doi: 10.1038/nri2958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Okayama H, Kohno T, Ishii Y, Shimada Y, Shiraishi K, Iwakawa R, Furuta K, Tsuta K, Shibata T, Yamamoto S, Watanabe S, Sakamoto H, Kumamoto K, Takenoshita S, Gotoh N, Mizuno H, Sarai A, Kawano S, Yamaguchi R, Miyano S, Yokota J. Identification of genes upregulated in ALK-positive and EGFR/KRAS/ALK-negative lung adenocarcinomas. Cancer Res. 2012;72:100–111. doi: 10.1158/0008-5472.CAN-11-1403. [DOI] [PubMed] [Google Scholar]
- 24.Hou J, Aerts J, den Hamer B, van Ijcken W, den Bakker M, Riegman P, van der Leest C, van der Spek P, Foekens JA, Hoogsteden HC, Grosveld F, Philipsen S. Gene expression-based classification of non-small cell lung carcinomas and survival prediction. PLoS One. 2010;5:e10312. doi: 10.1371/journal.pone.0010312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Selamat SA, Chung BS, Girard L, Zhang W, Zhang Y, Campan M, Siegmund KD, Koss MN, Hagen JA, Lam WL, Lam S, Gazdar AF, Laird-Offringa IA. Genome-scale analysis of DNA methylation in lung adenocarcinoma and integration with mRNA expression. Genome Res. 2012;22:1197–1211. doi: 10.1101/gr.132662.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Landi MT, Dracheva T, Rotunno M, Figueroa JD, Liu H, Dasgupta A, Mann FE, Fukuoka J, Hames M, Bergen AW, Murphy SE, Yang P, Pesatori AC, Consonni D, Bertazzi PA, Wacholder S, Shih JH, Caporaso NE, Jen J. Gene expression signature of cigarette smoking and its role in lung adenocarcinoma development and survival. PLoS One. 2008;3:e1651. doi: 10.1371/journal.pone.0001651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Talbot SG, Estilo C, Maghami E, Sarkaria IS, Pham DK, O-charoenrat P, Socci ND, Ngai I, Carlson D, Ghossein R, Viale A, Park BJ, Rusch VW, Singh B. Gene expression profiling allows distinction between primary and metastatic squamous cell carcinomas in the lung. Cancer Res. 2005;65:3063–3071. doi: 10.1158/0008-5472.CAN-04-1985. [DOI] [PubMed] [Google Scholar]
- 28.Bromberg JF, Horvath CM, Wen Z, Schreiber RD, Darnell JE Jr. Transcriptionally active Stat1 is required for the antiproliferative effects of both interferon alpha and interferon gamma. Proc Natl Acad Sci U S A. 1996;93:7673–7678. doi: 10.1073/pnas.93.15.7673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shankaran V, Ikeda H, Bruce AT, White JM, Swanson PE, Old LJ, Schreiber RD. IFNgamma and lymphocytes prevent primary tumour development and shape tumour immunogenicity. Nature. 2001;410:1107–1111. doi: 10.1038/35074122. [DOI] [PubMed] [Google Scholar]
- 30.Chen J, Zhao J, Chen L, Dong N, Ying Z, Cai Z, Ji D, Zhang Y, Dong L, Li Y, Jiang L, Holtzman MJ, Chen C. STAT1 modification improves therapeutic effects of interferons on lung cancer cells. J Transl Med. 2015;13:293. doi: 10.1186/s12967-015-0656-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Meissl K, Macho-Maschler S, Muller M, Strobl B. The good and the bad faces of STAT1 in solid tumours. Cytokine. 2017;89:12–20. doi: 10.1016/j.cyto.2015.11.011. [DOI] [PubMed] [Google Scholar]
- 32.O’Reilly LA, Putoczki TL, Mielke LA, Low JT, Lin A, Preaudet A, Herold MJ, Yaprianto K, Tai L, Kueh A, Pacini G, Ferrero RL, Gugasyan R, Hu Y, Christie M, Wilcox S, Grumont R, Griffin MDW, O’Connor L, Smyth GK, Ernst M, Waring P, Gerondakis S, Strasser A. Loss of NF-kappaB1 causes gastric cancer with aberrant inflammation and expression of immune checkpoint regulators in a STAT-1-dependent manner. Immunity. 2018;48:570–583. e578. doi: 10.1016/j.immuni.2018.03.003. [DOI] [PubMed] [Google Scholar]
- 33.Darnell JE Jr, Kerr IM, Stark GR. Jak-STAT pathways and transcriptional activation in response to IFNs and other extracellular signaling proteins. Science. 1994;264:1415–1421. doi: 10.1126/science.8197455. [DOI] [PubMed] [Google Scholar]
- 34.Arimoto KI, Lochte S, Stoner SA, Burkart C, Zhang Y, Miyauchi S, Wilmes S, Fan JB, Heinisch JJ, Li Z, Yan M, Pellegrini S, Colland F, Piehler J, Zhang DE. STAT2 is an essential adaptor in USP18-mediated suppression of type I interferon signaling. Nat Struct Mol Biol. 2017;24:279–289. doi: 10.1038/nsmb.3378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yue C, Xu J, Tan Estioko MD, Kotredes KP, Lopez-Otalora Y, Hilliard BA, Baker DP, Gallucci S, Gamero AM. Host STAT2/type I interferon axis controls tumor growth. Int J Cancer. 2015;136:117–126. doi: 10.1002/ijc.29004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ogony J, Choi HJ, Lui A, Cristofanilli M, Lewis-Wambi J. Interferon-induced transmembrane protein 1 (IFITM1) overexpression enhances the aggressive phenotype of SUM149 inflammatory breast cancer cells in a signal transducer and activator of transcription 2 (STAT2)-dependent manner. Breast Cancer Res. 2016;18:25. doi: 10.1186/s13058-016-0683-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li G, Yi S, Yang F, Zhou Y, Ji Q, Cai J, Mei Y. Identification of mutant genes with high-frequency, high-risk, and high-expression in lung adenocarcinoma. Thorac Cancer. 2014;5:211–218. doi: 10.1111/1759-7714.12080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yeh HH, Lai WW, Chen HH, Liu HS, Su WC. Autocrine IL-6-induced Stat3 activation contributes to the pathogenesis of lung adenocarcinoma and malignant pleural effusion. Oncogene. 2006;25:4300–4309. doi: 10.1038/sj.onc.1209464. [DOI] [PubMed] [Google Scholar]
- 39.Sriuranpong V, Park JI, Amornphimoltham P, Patel V, Nelkin BD, Gutkind JS. Epidermal growth factor receptor-independent constitutive activation of STAT3 in head and neck squamous cell carcinoma is mediated by the autocrine/paracrine stimulation of the interleukin 6/gp130 cytokine system. Cancer Res. 2003;63:2948–2956. [PubMed] [Google Scholar]
- 40.Garcia R, Bowman TL, Niu G, Yu H, Minton S, Muro-Cacho CA, Cox CE, Falcone R, Fairclough R, Parsons S, Laudano A, Gazit A, Levitzki A, Kraker A, Jove R. Constitutive activation of Stat3 by the Src and JAK tyrosine kinases participates in growth regulation of human breast carcinoma cells. Oncogene. 2001;20:2499–2513. doi: 10.1038/sj.onc.1204349. [DOI] [PubMed] [Google Scholar]
- 41.Flowers LO, Subramaniam PS, Johnson HM. A SOCS-1 peptide mimetic inhibits both constitutive and IL-6 induced activation of STAT3 in prostate cancer cells. Oncogene. 2005;24:2114–2120. doi: 10.1038/sj.onc.1208437. [DOI] [PubMed] [Google Scholar]
- 42.Wehbe H, Henson R, Meng F, Mize-Berge J, Patel T. Interleukin-6 contributes to growth in cholangiocarcinoma cells by aberrant promoter methylation and gene expression. Cancer Res. 2006;66:10517–10524. doi: 10.1158/0008-5472.CAN-06-2130. [DOI] [PubMed] [Google Scholar]
- 43.Catlett-Falcone R, Landowski TH, Oshiro MM, Turkson J, Levitzki A, Savino R, Ciliberto G, Moscinski L, Fernandez-Luna JL, Nunez G, Dalton WS, Jove R. Constitutive activation of Stat3 signaling confers resistance to apoptosis in human U266 myeloma cells. Immunity. 1999;10:105–115. doi: 10.1016/s1074-7613(00)80011-4. [DOI] [PubMed] [Google Scholar]
- 44.Grandis JR, Drenning SD, Chakraborty A, Zhou MY, Zeng Q, Pitt AS, Tweardy DJ. Requirement of Stat3 but not Stat1 activation for epidermal growth factor receptor-mediated cell growth in vitro. J Clin Invest. 1998;102:1385–1392. doi: 10.1172/JCI3785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li CJ, Li YC, Zhang DR, Pan JH. Signal transducers and activators of transcription 3 function in lung cancer. J Cancer Res Ther. 2013;9(Suppl 2):S67–73. doi: 10.4103/0973-1482.119100. [DOI] [PubMed] [Google Scholar]
- 46.Zhang X, Yue P, Page BD, Li T, Zhao W, Namanja AT, Paladino D, Zhao J, Chen Y, Gunning PT, Turkson J. Orally bioavailable small-molecule inhibitor of transcription factor Stat3 regresses human breast and lung cancer xenografts. Proc Natl Acad Sci U S A. 2012;109:9623–9628. doi: 10.1073/pnas.1121606109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lis C, Rubner S, Roatsch M, Berg A, Gilcrest T, Fu D, Nguyen E, Schmidt AM, Krautscheid H, Meiler J, Berg T. Development of Erasin: a chromone-based STAT3 inhibitor which induces apoptosis in Erlotinib-resistant lung cancer cells. Sci Rep. 2017;7:17390. doi: 10.1038/s41598-017-17600-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lee HJ, Zhuang G, Cao Y, Du P, Kim HJ, Settleman J. Drug resistance via feedback activation of Stat3 in oncogene-addicted cancer cells. Cancer Cell. 2014;26:207–221. doi: 10.1016/j.ccr.2014.05.019. [DOI] [PubMed] [Google Scholar]
- 49.Haura EB, Zheng Z, Song L, Cantor A, Bepler G. Activated epidermal growth factor receptor-Stat-3 signaling promotes tumor survival in vivo in non-small cell lung cancer. Clin Cancer Res. 2005;11:8288–8294. doi: 10.1158/1078-0432.CCR-05-0827. [DOI] [PubMed] [Google Scholar]
- 50.Mandal PK, Gao F, Lu Z, Ren Z, Ramesh R, Birtwistle JS, Kaluarachchi KK, Chen X, Bast RC Jr, Liao WS, McMurray JS. Potent and selective phosphopeptide mimetic prodrugs targeted to the Src homology 2 (SH2) domain of signal transducer and activator of transcription 3. J Med Chem. 2011;54:3549–3563. doi: 10.1021/jm2000882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mandal PK, Liao WS, McMurray JS. Synthesis of phosphatase-stable, cell-permeable peptidomimetic prodrugs that target the SH2 domain of Stat3. Org Lett. 2009;11:3394–3397. doi: 10.1021/ol9012662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Haftchenary S, Luchman HA, Jouk AO, Veloso AJ, Page BD, Cheng XR, Dawson SS, Grinshtein N, Shahani VM, Kerman K, Kaplan DR, Griffin C, Aman AM, Al-Awar R, Weiss S, Gunning PT. Potent targeting of the STAT3 protein in brain cancer stem cells: a promising route for treating glioblastoma. ACS Med Chem Lett. 2013;4:1102–1107. doi: 10.1021/ml4003138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rani A, Murphy JJ. STAT5 in cancer and immunity. J Interferon Cytokine Res. 2016;36:226–237. doi: 10.1089/jir.2015.0054. [DOI] [PubMed] [Google Scholar]
- 54.Able AA, Burrell JA, Stephens JM. STAT5-interacting proteins: a synopsis of proteins that regulate STAT5 activity. Biology (Basel) 2017;6 doi: 10.3390/biology6010020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Reichenstein M, Rauner G, Kfir S, Kisliouk T, Barash I. Luminal STAT5 mediates H2AX promoter activity in distinct population of basal mammary epithelial cells. Oncotarget. 2016;7:41781–41797. doi: 10.18632/oncotarget.9718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bowman T, Garcia R, Turkson J, Jove R. STATs in oncogenesis. Oncogene. 2000;19:2474–2488. doi: 10.1038/sj.onc.1203527. [DOI] [PubMed] [Google Scholar]
- 57.Lin TS, Mahajan S, Frank DA. STAT signaling in the pathogenesis and treatment of leukemias. Oncogene. 2000;19:2496–2504. doi: 10.1038/sj.onc.1203486. [DOI] [PubMed] [Google Scholar]
- 58.Schwaller J, Parganas E, Wang D, Cain D, Aster JC, Williams IR, Lee CK, Gerthner R, Kitamura T, Frantsve J, Anastasiadou E, Loh ML, Levy DE, Ihle JN, Gilliland DG. Stat5 is essential for the myelo- and lymphoproliferative disease induced by TEL/JAK2. Mol Cell. 2000;6:693–704. doi: 10.1016/s1097-2765(00)00067-8. [DOI] [PubMed] [Google Scholar]
- 59.Kraskouskaya D, Duodu E, Arpin CC, Gunning PT. Progress towards the development of SH2 domain inhibitors. Chem Soc Rev. 2013;42:3337–3370. doi: 10.1039/c3cs35449k. [DOI] [PubMed] [Google Scholar]
- 60.Morlacchi P, Robertson FM, Klostergaard J, McMurray JS. Targeting SH2 domains in breast cancer. Future Med Chem. 2014;6:1909–1926. doi: 10.4155/fmc.14.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Cumaraswamy AA, Lewis AM, Geletu M, Todic A, Diaz DB, Cheng XR, Brown CE, Laister RC, Muench D, Kerman K, Grimes HL, Minden MD, Gunning PT. Nanomolar-potency small molecule inhibitor of STAT5 protein. ACS Med Chem Lett. 2014;5:1202–1206. doi: 10.1021/ml500165r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Page BD, Khoury H, Laister RC, Fletcher S, Vellozo M, Manzoli A, Yue P, Turkson J, Minden MD, Gunning PT. Small molecule STAT5-SH2 domain inhibitors exhibit potent antileukemia activity. J Med Chem. 2012;55:1047–1055. doi: 10.1021/jm200720n. [DOI] [PubMed] [Google Scholar]
- 63.Elumalai N, Berg A, Rubner S, Blechschmidt L, Song C, Natarajan K, Matysik J, Berg T. Rational development of Stafib-2: a selective, nanomolar inhibitor of the transcription factor STAT5b. Sci Rep. 2017;7:819. doi: 10.1038/s41598-017-00920-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Pastuszak-Lewandoska D, Domanska-Senderowska D, Antczak A, Kordiak J, Gorski P, Czarnecka KH, Migdalska-Sek M, Nawrot E, Kiszalkiewicz JM, Brzezianska-Lasota E. The expression levels of IL-4/IL-13/STAT6 signaling pathway genes and SOCS3 could help to differentiate the histopathological subtypes of non-small cell lung carcinoma. Mol Diagn Ther. 2018;22:621–629. doi: 10.1007/s40291-018-0355-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Das S, Roth CP, Wasson LM, Vishwanatha JK. Signal transducer and activator of transcription-6 (STAT6) is a constitutively expressed survival factor in human prostate cancer. Prostate. 2007;67:1550–1564. doi: 10.1002/pros.20640. [DOI] [PubMed] [Google Scholar]
- 66.Wang CG, Ye YJ, Yuan J, Liu FF, Zhang H, Wang S. EZH2 and STAT6 expression profiles are correlated with colorectal cancer stage and prognosis. World J Gastroenterol. 2010;16:2421–2427. doi: 10.3748/wjg.v16.i19.2421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Skinnider BF, Elia AJ, Gascoyne RD, Patterson B, Trumper L, Kapp U, Mak TW. Signal transducer and activator of transcription 6 is frequently activated in Hodgkin and Reed-Sternberg cells of Hodgkin lymphoma. Blood. 2002;99:618–626. doi: 10.1182/blood.v99.2.618. [DOI] [PubMed] [Google Scholar]
- 68.Binnemars-Postma K, Bansal R, Storm G, Prakash J. Targeting the Stat6 pathway in tumor-associated macrophages reduces tumor growth and metastatic niche formation in breast cancer. FASEB J. 2018;32:969–978. doi: 10.1096/fj.201700629R. [DOI] [PubMed] [Google Scholar]
- 69.Leon-Cabrera SA, Molina-Guzman E, Delgado-Ramirez YG, Vazquez-Sandoval A, Ledesma-Soto Y, Perez-Plasencia CG, Chirino YI, Delgado-Buenrostro NL, Rodriguez-Sosa M, Vaca-Paniagua F, Avila-Moreno F, Gutierrez-Cirlos EB, Arias-Romero LE, Terrazas LI. Lack of STAT6 attenuates inflammation and drives protection against early steps of colitis-associated colon cancer. Cancer Immunol Res. 2017;5:385–396. doi: 10.1158/2326-6066.CIR-16-0168. [DOI] [PubMed] [Google Scholar]
- 70.Xu B, Lu X, Zhao Y, Liu C, Huang X, Chen S, Zhu W, Zhang L, Chen M. MicroRNA-135a induces prostate cancer cell apoptosis via inhibition of STAT6. Oncol Lett. 2019;17:1889–1895. doi: 10.3892/ol.2018.9791. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


