Abstract
Circular RNAs (circRNAs) belong to non-coding RNAs and are known as key regulators in gene regulation. CircRNAs involve in the various biological processes of cancer. However, the functions of circRNAs in renal cell carcinoma (RCC) are still not clear. In this study, the circRNA expression profile was performed in the RCC tissues by microarray. There were 35 significantly expressed circRNAs with more than 5 folds from microarray analysis. Hsa_circ_0039569 (circ_0039569) was verified to be up-regulated in RCC and cells compared with the controls by real time RT-PCR. The assays of cellular functions showed that circ_0039569 down-regulation suppressed the proliferation and metastasis of RCC cells. The molecular mechanism of circ_0039569 in RCC cells showed that circ_0039569 promoted RCC progression by up-regulating CCL22 expression via down-regulating miR-34a-5p. Taken together, the study indicated that circ_0039569 played an important role in RCC cell survival and metastasis, which suggested that an oncogenic role of circ_0039569 in RCC progression.
Keywords: Renal cell carcinoma, hsa_circ_0039569, metastasis, cell proliferation, miR-34a-5p, CCL22
Introduction
Renal cell carcinoma (RCC) is one of the most common aggressive tumors. The morbidity and mortality of RCC are increasing in China dramatically. Although improvements are obtained in screening, diagnosis and treatment in RCC, there is a decrease in the mortality and low overall survival rates [1,2]. Various molecules like RNA, protein participate in renal cell carcinoma progression, however, the molecular mechanisms are largely unknown. These aspects demonstrate the importance of elucidating the genetic mechanisms understanding RCC and of developing novel therapeutic targets.
Recently, non-coding RNAs (ncRNAs) such as long non-coding RNA (lncRNA), microRNA (miRNAs) and circular RNAs (circRNAs) are verified to play critical roles in malignant tumor [3-6]. CircRNAs are different from linear RNA and have no protein translation capacity [6-8]. Increasing evidences indicate that circRNAs regulate RCC such as cell grow, metastasis and etc [8]. A previous report showed that circHIAT1 expression was lower in ccRCCs than adjacent normal tissues. Suppression AR expression could inhibit ccRCC cell progression via up-regulating circHIAT1 expression. The consequences of AR-suppressed circHIAT1 resulted in deregulating miR-195-5p, miR-34a-5p and miR-29c-3p expression, which increased CDC42 expression to enhance ccRCC cell migration and invasion. Finally, circHIAT1 was verified to be a metastatic inhibitor, which suppressed AR-enhanced ccRCC cell migration and invasion [9].
However, there is lack of research in the biological function of circRNAs in RCC. In present study, the circRNAs expression profiles were screened and assayed in the tissues from RCC and adjacent normal tissues with non-malignant kidney diseases using circRNA assay, and ultimately identified circ_0039569 was significantly overexpressed in RCC tissues. Further investigation was carried out to explore the role of circ_0039569 in RCC cell survival and metastasis. The molecular mechanism of oncogenic circ_0039569 was also studied. It was found that circ_0039569 functioned as an oncogene by sponging with miR-34a-5p which down-regulated CCL22 expression in RCC.
Methods
Clinical specimens
Fifty-two RCC tissues and their adjacent tissues were collected from The Second Affiliated Hospital, Harbin Medical University (Harbin, China) between Dec 2015 and Dec 2017. This study was carried out under the agreement of the Ethics Committee of the hospital. The informed consent from the patients were signed.
CircRNA microarray
The total RNA from RCC patients were collected for circular RNA array analysis. RNA extraction and microarray hybridization were performed according to the Arraystar’s standard protocols. Firstly, total RNA was treated with Rnase R to digest and remove linear RNA and then the circRNAs were enriched. Next, RNAs were amplified for cRNA and labeled with an Arraystar Super RNA Labeling Kit (Arraystar, Rockville, the USA). Finally, these labeled RNAs were hybridized using Arraystar mouse circRNA Array (V1.0, Arraystar), and scanned by the Agilent Scanner G2505C.
Cell culture
Human RCC cell lines (ACHN, A498, 786-O, 769-P and RCC4) and human normal kidney cell line (HK-2) were from the American Type Culture Collection (ATCC, Rockville, MD, USA). Cells were cultured in Dulbecco’s modified Eagle medium (DMEM, Sangon Biotech, Shanghai, China) supplemented with 10% FBS, 100 mg/ml penicillin and 100 mg/ml streptomycin (Sangon Biotech, Shanghai, China). Cells were cultured at the normal condition.
Real time RT-PCR
Total RNA from RCC tissues or cells was extracted using Trizol (Invitrogen, Carlsbad, USA). Quantitative RT-PCR was performed using the SYBR-Green PCR Master Mix kit (Takara, Dalian, China). The primers for circRNAs and miRNAs were ordered from Sangon Biotech (Shanghai, China). The relative gene expression was normalized to GAPDH or U6 snRNA and analyzed by the 2-ΔΔCt method.
Cell proliferation assay
RCC cells with circ_0039569 siRNA transfection or the controls were seeded into 96-well plates (3 × 104 in every well) and 10 μl Cell count kit-8 solution (CCK-8, Dojindo, Japan) was added to each well. Then, the cells were incubated at 37°C for two hours. The absorbance of the cells in every well at 490 nm was measured using a spectrophotometer.
Scratch wound assay
The cells with circ_0039569 shRNAs or the controls transfection were plated into a 12-well plate (2 × 105 cells in every well) and incubated to reach about 90% confluence. The monolayer was scratched using a tip, washed with serum-free medium to remove detached cells and photographed at 24 h later.
Invasion assay
The inserts in the transwell system were coated with 50 μl Matrigel (BD Biosciences, Franklin Lakes, NJ, USA). Cells (5 × 104) were suspended in 100 μl serum-free medium and then seeded on the upper floor of Transwell chambers (Corning, USA). Culture medium with 500 μl with 20% FBS were added into the lower chambers and then the cells were incubated at 37°C with 5% CO2 for 2 days. The un-invaded cells were wiped with a cotton swab, and invaded cells were fixed in methanol and stained with 0.1% crystal violet. The number was counted under a microscope.
Statistical analysis
All statistical analysis was performed by SPSS 16.0 (IBM, Chicago, IL, USA) and GraphPad Prism 6.0 (GraphPad Software, La Jolla, CA, USA). Paired t test, independent t test and one-way analysis of variance (ANOVA) were used. P < 0.05 was significant.
Results
The profile of circRNA from the tissues of RCC patients
To assess the different circRNA expression pattern of RCC, total RNA was extracted from the RCC tissues and the adjacent tissues and performed for circRNA array analysis. It was found that there were 43 significantly expressed circRNAs with more than 2 folds, including 17 downregulated and 26 up-regulated circRNAs in the heat map (Figure 1A). As Figure 1B shown, the scatter plot revealed microarray data distributions of circRNAs between RCC tissues versus the controls. All the significant circRNAs might participate in RCC progression. Some aberrant expressed circRNAs in RCC tissues were found from the circRNA microarray assay. Of the up-regulated circRNAs, six circRNAs were chosen for verifying by qRT-PCR. The data showed that six circRNAs including circ_0039569, circ_0006461, circ_0055408, circ_0049663, circ_0018064, circ_0053772 were significantly higher in the RCC tissues than the adjacent normal tissues (Figure 1C). From the down-regulated circRNAs, six circRNAs were chosen for verifying by qRT-PCR. The data showed that six circRNAs including circ_0086458, circ_0019692, circ_0037923, circ_0040865, circ_0051398, circ_0039151 were significantly lower in the RCC tissues than the adjacent normal tissues (Figure 1D). Based on the circRNA expression confirmation from real time RT-PCR, circ_0039569 was selected for further investigation.
Figure 1.
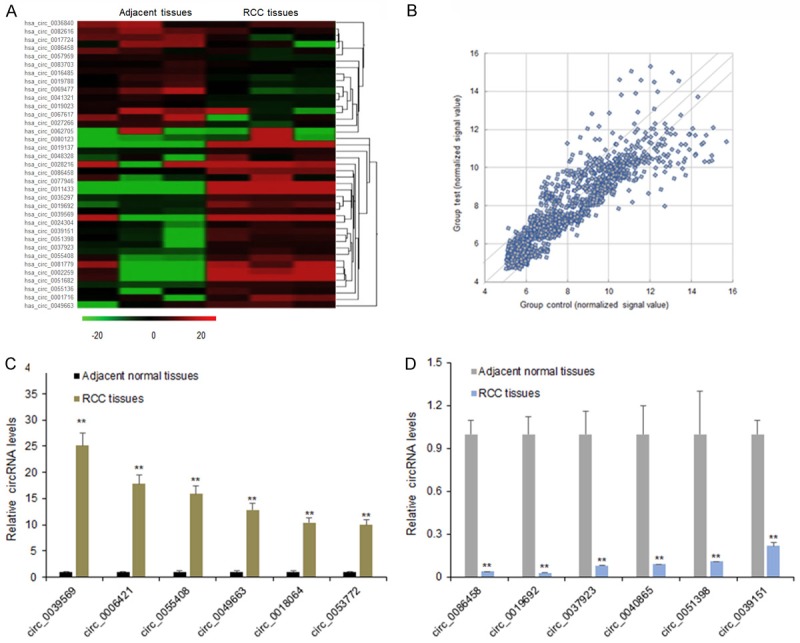
CircRNA expression profile in RCC tissues. A. Heat map showed the significantly expressed circRNAs with 5 folds were presented. In the heat map, red color: the upregulated circRNAs with high fold changes; green color: the downregulated circRNAs with high fold changes. B. Scatter plots are used to evaluate the difference in the expression of circRNAs between experiment group and control group. The values plotted on X and Y axes are the averaged normalized signal values of each group (log2 scaled). The middle green line refers to no difference between the two groups. And the flank green lines represent 2.0 fold changes. The circRNAs above the top green line and below the bottom green line indicate more than 2.0 fold changes between two groups. C. Six circRNAs including circ_0036840, circ_0082616, circ_0017724, circ_0039569, circ_0039569, and circ_0062705 were chosen for validation in the RCC tissues using qRT-PCR. D. Six circRNAs including circ_0086458, circ_0019692, circ_0037923, circ_0040865, circ_0051398, circ_0039151 were chosen for validation in the RCC tissues using qRT-PCR. Data were expressed as mean ± SD. **P < represents statistical difference.
Circ_0039569 expression was associated with metastasis in the RCC tissues
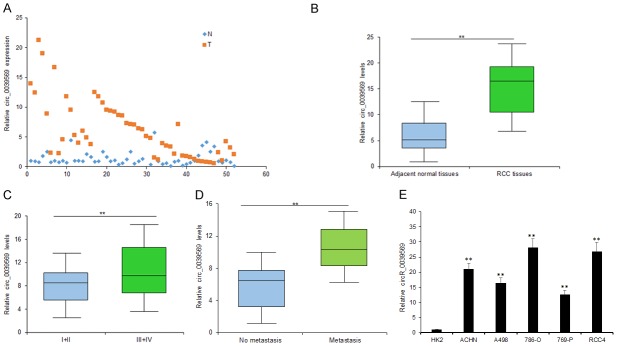
Circ_0039569 was one of the up-regulated circRNAs and its expression was further validated in the tissues of 52 RCC patients compared to the controls (Figure 2A). The average level of circ_0039569 in RCC tissues was higher than it in the compared normal tissues (Figure 2B). The RCC tissues were classified into metastatic tissues and non-metastatic tissues and the average level of circ_0039569 in metastatic tissues was higher than it in the non-metastatic tissues (Figure 2C). Circ_0039569 expression was also higher in RCC patients in advanced tumor staging than the RCC patients in early staging (Figure 2D). In RCC cells, circ_0039569 was up-regulated compared with the normal cell line (Figure 2E). So, the data suggested that circ_0039569 would be an oncogene in RCC.
Figure 2.
Circ_0039569 was up-regulated in RCC tissues and cell lines. A. Circ_0039569 was up-regulated in RCC tissues compared to the controls. Circ_0039569 expression was detected using qRT-PCR. B. Average circ_0039569 levels in RCC tissues and adjacent tissues. C. Average circ_0039569 levels in metastatic RCC tissues and non-metastatic RCC tissues. D. Circ_0039569 expression was also higher in RCC patients in advanced tumor staging than the RCC patients in early staging. E. Circ_0039569 was up-regulated in RCC cell lines. Circ_0039569 expression was detected using qRT-PCR. Data were expressed as mean ± SD. **P < represents statistical difference.
Down-regulation of circ_0039569 expression suppressed RCC cell proliferation and metastasis
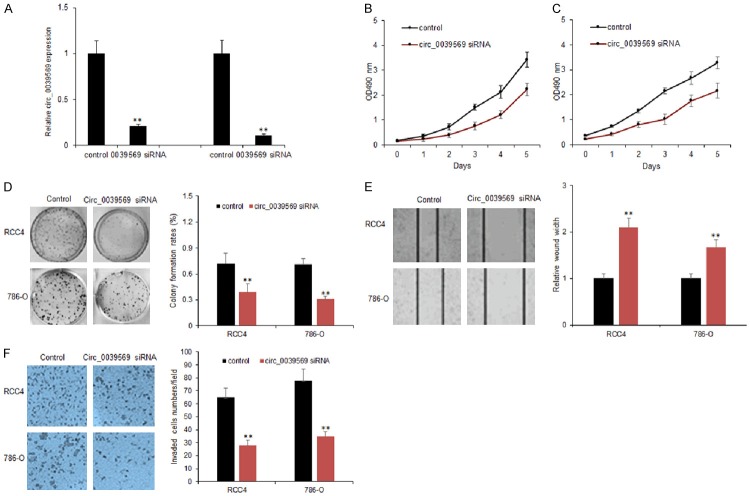
To explore the role of circ_0039569 in RCC cancer cell proliferation, RCC4 and 786-O cells were transfected with circ_0039569 siRNA using lentivirus vectors. The results indicated that circ_0039569 expression was effectively knocked down in the RCC cells (Figure 3A). Colony formation assay showed that circ_0039569 down-regulation resulted in the decreased colony numbers in the RCC cells (Figure 3B and 3C). Moreover, circ_0039569 down-regulation suppressed RCC cell survival ability (Figure 3D). In summary, results showed that the inhibition of circ_0039569 decreased the proliferation and enhanced the drug sensitivity of RCC cells. To further investigate the role of circ_0039569 on RCC cell metastasis, RCC4 and 786-O cells were transfected with circ_0039569 siRNA using lentivirus vectors. Transwell chamber with or without matrixgel treatment to analyze the migration and invasion ability of cancer cells. The data indicated that circ_0039569 down-regulation decreased the migration ability of the cancer cells (Figure 3E). Circ_0039569 down-regulation decreased the invaded cell numbers of RCC (Figure 3F).
Figure 3.
Inhibiting circ_0039569 suppressed RCC cell proliferation and metastasis. A. Circ_0039569 expression in RCC4 and 786-O cells. RCC4 and 786-O cells were transfected with circ_0039569 shRNA and circ_0039569 levels were analyzed by qRT-PCR. B, C. Down-regulation of circ_0039569 reduced RCC cell proliferation. RCC4 and 786-O cells were transfected with circ_0039569 shRNA and survival ability was analyzed by CCK8 assay. D. Down-regulation of circ_0039569 reduced RCC cell colony numbers. RCC4 and 786-O cells were transfected with circ_0039569 shRNA and survival ability was analyzed by colony formation assay. E. Down-regulation of circ_0039569 reduced migrated RCC cell numbers. RCC4 and 786-O cells were transfected with circ_0039569 shRNA and migration ability was analyzed by wound healing assay. F. Down-regulation of circ_0039569 reduced invaded RCC cell numbers. RCC4 and 786-O cells were transfected with circ_0039569 shRNA and invasion ability was analyzed by transwell chambers with matrigel treatment. Data were expressed as mean ± SD. **P < represents statistical difference.
Circ_0039569 down-regulated miR-34a-5p levels which targeted CCL2 gene in RCC cells
To further explore the underlying mechanism of circ_0039569 in RCC, Targetscan and miRanda were used to search the potential targets of circ_0039569. The analysis predicted that the miR-34a-5p was the potential sponge of circ_0039569. Luciferase reporter assay was conducted to determine the reliability. The result illustrated that upregulation of miR-34a-5p notably decreased the luciferase activity of circ_0039569-WT (wild-type), but had no effect on circ_0039569-MUT (mutant-type) (Figure 4A and 4B). Moreover, we transfected circ_0039569 shRNA to RCC4 and 786-O cells and detected the miR-34a-5p expression by qRT-PCR. We found that the RCC4 and 786-O cells with down-regulation of circ_0039569 had high expression of miR-34a-5p (Figure 4C). The result proved that circ_0039569 directly bond to miR-34a-5p in RCC cells. To know the target genes of miR-34a-5p, targetscan was applied to predict the binding site of miR-34a-5p and it was shown that CCL22 was a potential target gene. By luciferase reporter assay, the data indicated that miR-34a-5p effectively inhibited the luciferase activity of CCL22-WT (wild-type), but made no difference to CCL22-MUT (mutant-type) (Figure 4D). The qRT-PCR and western blotting result found that miR-34a-5p overexpression significantly suppressed CCL22 expression level in both transcriptional and protein levels (Figure 4E and 4F). In summary, we confirmed that CCL22 was a down-stream target gene of miR-34a-5p.
Figure 4.
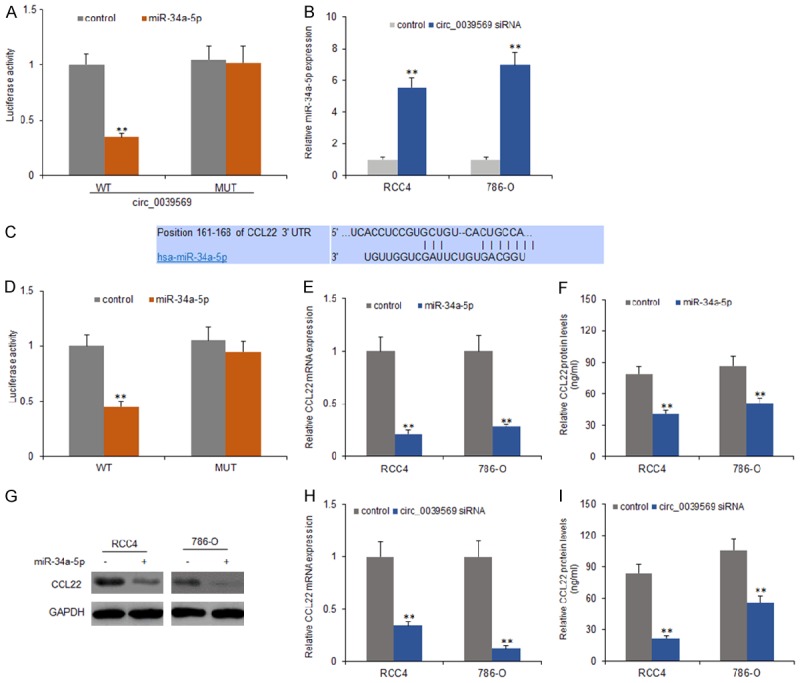
Circ_0039569 down-regulated miR-34a-5p levels by targeting CCL2 expression in RCC cells. A. MiR-34a-5p significantly inhibited luciferase activity of wild type reporter for circ_0039569, however, miR-34a-5p did not inhibit the luciferase activity of reporter vector containing the mutant binding sites of circ_0039569 in RCC4 and 786-O cells. B. MiR-34a-5p was upregulated in RCC cells that transfected by circ_0039569 siRNA. C. CCL22 was predicted as a target gene of miR-34a-5p by Targetscan. D. MiR-34a-5p significantly inhibited luciferase activity of wild type reporter for CCL22, however, miR-34a-5p did not inhibit the luciferase activity of reporter vector containing the mutant binding sites of CCL22 in both cell lines. E. RCC4 and 786-O cells were transfected with miR-34a-5p and total RNA was used for CCL22 examination by real time RT-PCR. F. CCL22 protein levels deceased in RCC4 and 786-O cells with miR-34a-5p transfection by ELISA. G. CCL22 protein levels deceased in RCC4 and 786-O cells with miR-34a-5p transfection by western blotting. H. Down-regulating circ_0039569 decreased CCL22 mRNA levels in RCC4 and 786-O cells. Total RNA was used for CCL22 examination by real time RT-PCR. I. Down-regulating circ_0039569 decreased CCL22 protein levels in RCC4 and 786-O cells. Total protein was extracted from the cells for western blotting. Data were expressed as mean ± SD. **P < represents statistical difference.
Circ_0039569 promoted RCC cell proliferation and metastasis via miR-34a-5p/CCL22
To explore the role of circ_0039569/miR-34a-5p/CCL22 axis in RCC cell proliferation and metastasis, RCC4 and 786-O cells were transfected with circ_0039569 siRNA using lentivirus vectors, or miR-34a-5p in the present of CCL12 or without CCL22. The result of MTT assay demonstrated that circ_0039569 down-regulation suppressed RCC4 and 786-O cell survival ability and also suppressed cell survival enhancement induced by CCL22 treatment with or without miR-34a-5p overexpression (Figure 5A and 5B). Transwell chamber with or without matrixgel treatment was used to analyze the migration and invasion ability of RCC cells. The result demonstrated that circ_0039569 down-regulation suppressed RCC4 and 786-O cell migration and also suppressed cell migration enhancement induced by CCL22 treatment with or without miR-34a-5p overexpression (Figure 5C). There was a similar result from invasion assay (Figure 5D).
Figure 5.
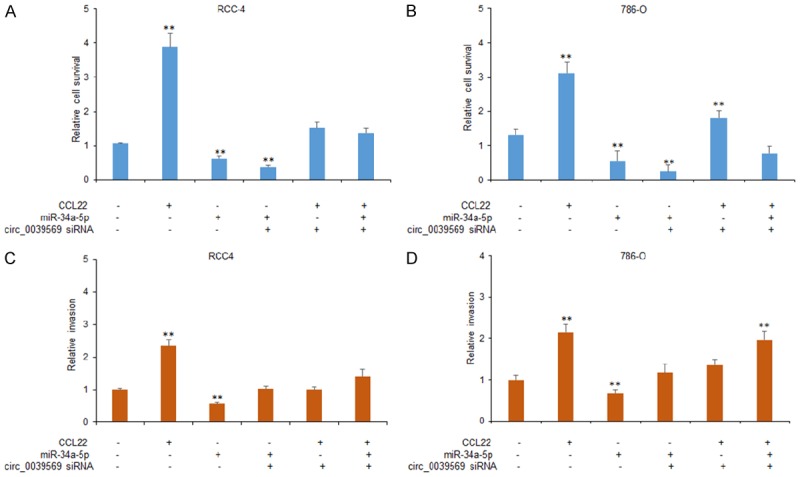
Circ_0039569 promotes RCC cell proliferation and metastasis via miR-34a-5p/CCL22. A, B. Down-regulation of circ_0039569 reduced RCC cell proliferation. RCC4 and 786-O cells were transfected with circ_0039569 siRNA or miR-34a-5p combined with or without CCL22 and the survival ability was analyzed by CCK8 assay. C. Down-regulation of circ_0039569 reduced migrated RCC cell numbers. RCC4 and 786-O cells were transfected with circ_0039569 siRNA, or miR-34a-5p combined with or without CCL22 and migration ability was analyzed by wound healing assay. D. Down-regulation of circ_0039569 reduced invaded RCC cell numbers. RCC4 and 786-O cells were transfected with circ_0039569 siRNA, or miR-34a-5p combined with or without CCL22 and invasion ability was analyzed by transwell chambers with matrigel treatment. Data were expressed as mean ± SD. **P < represents statistical difference.
Circ_0039569 was associated with miR-34a-5p, CCL22 expression in RCC tissues
To know whether circ_0039569 levels were associated with miR-34a-5p, CCL22 in RCC, the data from real time RT-PCR showed that miR-34a-5p was down-regulated in 52 primary RCC tissues in contrast to paired nontumor tissues (Figure 6A and 6B). CCL22 mRNA expression was also higher in the samples of RCC than the controls (Figure 6C and 6D). Furthermore, the relationship between circ_0039569 and miR-34a-5p expression levels was analyzed and it was shown that circ_0039569 was negatively related to miR-34a-5p expression in RCC tissues (Figure 6E). Circ_0039569 was positively related to CCL22 expression in RCC tissues (Figure 6F).
Figure 6.
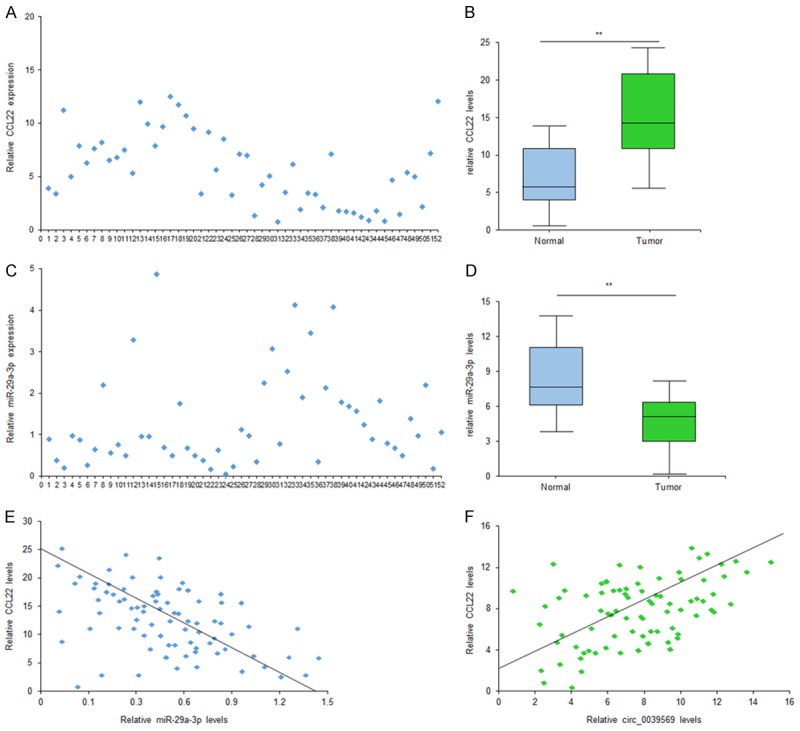
Circ_0039569 was associated with miR-34a-5p, CCL22 expression in RCC tissues. A. Circ_0039569 was up-regulated in RCC blood samples compared to the healthy persons. Circ_0039569 expression was detected using qRT-PCR. B. Average circ_0039569 levels in RCC patients with or without metastasis. C. Average circ_0039569 levels were higher in late staging RCC patients. D. Circ_0039569 was up-regulated in RCC cell lines. Circ_0039569 expression was detected using qRT-PCR. E. The relationship between circ_0039569 and CCL22 expression levels. F. The relationship between circ_0039569 and CCL22 expression levels. Data were expressed as mean ± SD. **P < represents statistical difference.
Discussion
CircRNAs have been discovered for more than twenty years and the important roles of circRNAs have been investigated in many diseases. It has been demonstrated that circRNAs regulate many molecules which involve in multiple physiological and pathological processes [4-8]. In our study, by circRNA array, we found that circ_0039569 expression was up-regulated in RCC tissues, which was also verified in RCC cell lines. Down-regulation of circ_0039569 decreased RCC cell growth and metastasis.
By high-throughput RNA sequencing and microarray analysis on the circRNA profiles screening, and many aberrantly expressed circRNAs are found [10-13]. For example, 357 circRNAs were dysregulated in the lung cancer and 5 circRNAs were identical to the microarray data using qRT-PCR [10]. Using circRNA array, the researchers found that some significant circRNAs and 12 circRNAs were confirmed in osteosarcoma such as upregulated circ_103801 and down-regulated circ_104980 in both osteosarcoma cell lines and tissues [12]. Using circRNA microarray for circRNA expression profile of RCC, circ_0004018 was found to be significantly lower in RCC compared with adjacent tissue and lower expression of circ_0004018 was correlated with AFP level, tumor diameters, differentiation, Barcelona clinic liver cancer stage and tumor-node-metastasis stage [13]. In this study, the up-regulated circ_0039569 was significantly up-regulated in 52 pairs of RCC tissue comparing to the adjacent normal tissue by circRNA array. Circ_0039569 plays important roles for in RCC occurrence and development.
CircRNAs resist the digestion of RNA enzyme due to their conservative covalently closed circular structure. So, circRNAs are stable in peripheral blood or body fluid, which determine that circRNAs are biomarkers for early diagnosis of tumor [14,15]. For instance, circ_0001649 levels were down-regulated in liver cancer, which predicted a poor prognosis of the disease [16]. CircPVT1 contributed to doxorubicin and cisplatin resistance of osteosarcoma cells by regulating ABCB1 and acted as a potential new circular RNA biomarker [17]. Circ_0014130 was verified as a circular RNA biomarker in non-small cell lung cancer [18]. In this study, circ_0039569 was significantly up-regulated in tumor tissue and cells acting as an oncogene, providing a potential biomarker for RCC cancer patients’ early detection.
C-C motif chemokine 22 (CCL22) is a protein and encoded by the CCL22 gene in human [19-23]. The protein encoded by this gene is secreted by dendritic cells and macrophages, and elicits its effects on its target cells by interacting with cell surface chemokine receptors. CCL22 gene locates in human chromosome 16 in a cluster with other chemokines called CX3CL1 and CCL17. The interactions between TAMs and prostate cancer cells could promote CCL22 expression and result in prostate cancer migration and invasion [19]. In this study, we found that CCL22 was a target gene of miR-34a-5p, which could suppress RCC cell proliferation and metastasis mediated by CCL22.
MiR-34a-5p could suppress or promote cancer progression depending on the background of cancer [24-28]. MiR-34a-5p overexpression in CAFs could inhibit the tumorigenesis of OSCC cells [24]. MiR-34a-5p suppresses colorectal cancer metastasis and predicts recurrence in patients with stage II/III colorectal cancer. MiR-34a-5p exerts oncogenic functions in human nasopharyngeal carcinoma by the regulation of long non-coding RNA XIST. LincRNA 1700020I14Rik alleviates cell proliferation and fibrosis in diabetic nephropathy via miR-34a-5p/Sirt1/HIF-1α signaling [25]. The study demonstrated that circ_0039569 down-regulated miR-34a-5p expression in RCC.
In a summary, our study reveals that the circRNAs expression profiles in RCC tissue identified the functional candidate circ_0039569 for RCC cancer tumorigenesis. The expression levels of circ_0039569 is up-regulated in RCC tissues and cells lines. In addition, circ_0039569 promotes RCC cell proliferation and metastasis via regulation miR-34a-5p/CCL22 axis. The study provides a novel insight of circRNAs for RCC cancer carcinogenesis and circ_0039569 may be a potential biomarker of RCC.
Acknowledgements
The research was supported by 2007 Heilongjiang Provincial Department of Education Overseas Scholars Research Funding Project (No. 1152hq26) and Heilongjiang Province Science and Technology Plan Project (No. LC08C16).
Disclosure of conflict of interest
None.
References
- 1.Barata PC, Rini BI. Treatment of renal cell carcinoma: current status and future directions. CA Cancer J Clin. 2017;67:507–524. doi: 10.3322/caac.21411. [DOI] [PubMed] [Google Scholar]
- 2.Joosten SC, Deckers IA, Aarts MJ, Hoeben A, van Roermund JG, Smits KM, Melotte V, van Engeland M, Tjan-Heijnen VC. Prognostic DNA methylation markers for renal cell carcinoma: a systematic review. Epigenomics. 2017;9:1243–1257. doi: 10.2217/epi-2017-0040. [DOI] [PubMed] [Google Scholar]
- 3.Rong D, Sun H, Li Z, Liu S, Dong C, Fu K, Tang W, Cao H. An emerging function of circRNA-miRNAs-mRNA axis in human diseases. Oncotarget. 2017;8:73271–73281. doi: 10.18632/oncotarget.19154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Han C, Seebacher NA, Hornicek FJ, Kan Q, Duan Z. Regulation of microRNAs function by circular RNAs in human cancer. Oncotarget. 2017;8:64622–64637. doi: 10.18632/oncotarget.19930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Han YN, Xia SQ, Zhang YY, Zheng JH, Li W. Circular RNAs: a novel type of biomarker and genetic tools in cancer. Oncotarget. 2017;8:64551–64563. doi: 10.18632/oncotarget.18350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Song X, Zeng Z, Wei H, Wang Z. Alternative splicing in cancers: from aberrant regulation to new therapeutics. Semin Cell Dev Biol. 2018;75:13–22. doi: 10.1016/j.semcdb.2017.09.018. [DOI] [PubMed] [Google Scholar]
- 7.Yang Z, Xie L, Han L, Qu X, Yang Y, Zhang Y, He Z, Wang Y, Li J. Circular RNAs: regulators of cancer-related signaling pathways and potential diagnostic biomarkers for human cancers. Theranostics. 2017;7:3106–3117. doi: 10.7150/thno.19016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zheng Y, Liu L, Shukla GC. A comprehensive review of web-based non-coding RNA resources for cancer research. Cancer Lett. 2017;407:1–8. doi: 10.1016/j.canlet.2017.08.015. [DOI] [PubMed] [Google Scholar]
- 9.Wang K, Sun Y, Tao W, Fei X, Chang C. Androgen receptor (AR) promotes clear cell renal cell carcinoma (ccRCC) migration and invasion via altering the circHIAT1/miR-195-5p/29a-3p/29c-3p/CDC42 signals. Cancer Lett. 2017;394:1–12. doi: 10.1016/j.canlet.2016.12.036. [DOI] [PubMed] [Google Scholar]
- 10.Xu T, Wu J, Han P, Zhao Z, Song X. Circular RNA expression profiles and features in human tissues: a study using RNA-seq data. BMC Genomics. 2017;18(Suppl 6):680. doi: 10.1186/s12864-017-4029-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yan N, Xu H, Zhang J, Xu L, Zhang Y, Zhang L, Xu Y, Zhang F. Circular RNA profile indicates circular RNA VRK1 is negatively related with breast cancer stem cells. Oncotarget. 2017;8:95704–95718. doi: 10.18632/oncotarget.21183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu W, Zhang J, Zou C, Xie X, Wang Y, Wang B, Zhao Z, Tu J, Wang X, Li H, Shen J, Yin J. Microarray expression profile and functional analysis of circular RNAs in osteosarcoma. Cell Physiol Biochem. 2017;43:969–985. doi: 10.1159/000481650. [DOI] [PubMed] [Google Scholar]
- 13.Fu L, Yao T, Chen Q, Mo X, Hu Y, Guo J. Screening differential circular RNA expression profiles reveals hsa_circ_0004018 is associated with hepatocellular carcinoma. Oncotarget. 2017;8:58405–58416. doi: 10.18632/oncotarget.16881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang M, Yang Y, Xu J, Bai W, Ren X, Wu H. CircRNAs as biomarkers of cancer: a meta-analysis. BMC Cancer. 2018;18:303. doi: 10.1186/s12885-018-4213-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jiang W, Zhang X, Chu Q, Lu S, Zhou L, Lu X, Liu C, Mao L, Ye C, Timko MP, Fan L, Ju H. The circular RNA profiles of colorectal tumor metastatic cells. Front Genet. 2018;9:34. doi: 10.3389/fgene.2018.00034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang X, Qiu S, Luo P, Zhou H, Jing W, Liang C, Tu J. Down-regulation of hsa_circ_0001649 in hepatocellular carcinoma predicts a poor prognosis. Cancer Biomark. 2018;22:135–142. doi: 10.3233/CBM-171109. [DOI] [PubMed] [Google Scholar]
- 17.Kun-Peng Z, Xiao-Long M, Chun-Lin Z. Overexpressed circPVT1, a potential new circular RNA biomarker, contributes to doxorubicin and cisplatin resistance of osteosarcoma cells by regulating ABCB1. Int J Biol Sci. 2018;14:321–330. doi: 10.7150/ijbs.24360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang S, Zeng X, Ding T, Guo L, Li Y, Ou S, Yuan H. Microarray profile of circular RNAs identifies hsa_circ_0014130 as a new circular RNA biomarker in non-small cell lung cancer. Sci Rep. 2018;8:2878. doi: 10.1038/s41598-018-21300-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Maolake A, Izumi K, Shigehara K, Natsagdorj A, Iwamoto H, Kadomoto S, Takezawa Y, Machioka K, Narimoto K, Namiki M, Lin WJ, Wufuer G, Mizokami A. Tumor-associated macrophages promote prostate cancer migration through activation of the CCL22-CCR4 axis. Oncotarget. 2017;8:9739–9751. doi: 10.18632/oncotarget.14185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mandal PK, Biswas S, Mandal G, Purohit S, Gupta A, Majumdar Giri A, Roy Chowdhury S, Bhattacharyya A. CCL2 conditionally determines CCL22-dependent Th2-accumulation during TGF-β-induced breast cancer progression. Immunobiology. 2018;223:151–161. doi: 10.1016/j.imbio.2017.10.031. [DOI] [PubMed] [Google Scholar]
- 21.Omland SH, Wettergren EE, Mollerup S, Asplund M, Mourier T, Hansen AJ, Gniadecki R. Cancer associated fibroblasts (CAFs) are activated in cutaneous basal cell carcinoma and in the peritumoural skin. BMC Cancer. 2017;17:675. doi: 10.1186/s12885-017-3663-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang D, Li Q, Yang Y, Hao S, Han X, Song J, Yin Y, Li X, Tanaka M, Qiu CH. Macrophage subset expressing CD169 in peritoneal cavity-regulated mucosal inflammation together with lower levels of CCL22. Inflammation. 2017;40:1191–1203. doi: 10.1007/s10753-017-0562-0. [DOI] [PubMed] [Google Scholar]
- 23.Martinenaite E, Munir Ahmad S, Hansen M, Met Ö, Westergaard MW, Larsen SK, Klausen TW, Donia M, Svane IM, Andersen MH. CCL22-specific T cells: modulating the immunosuppressive tumor microenvironment. Oncoimmunology. 2016;5:e1238541. doi: 10.1080/2162402X.2016.1238541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li YY, Tao YW, Gao S, Li P, Zheng JM, Zhang SE, Liang J, Zhang Y. Cancer-associated fibroblasts contribute to oral cancer cells proliferation and metastasis via exosome-mediated paracrine miR-34a-5p. EBioMedicine. 2018;36:209–220. doi: 10.1016/j.ebiom.2018.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gao J, Li N, Dong Y, Li S, Xu L, Li X, Li Y, Li Z, Ng SS, Sung JJ, Shen L, Yu J. miR-34a-5p suppresses colorectal cancer metastasis and predicts recurrence in patients with stage II/III colorectal cancer. Oncogene. 2015;34:4142–52. doi: 10.1038/onc.2014.348. [DOI] [PubMed] [Google Scholar]
- 26.Li A, Peng R, Sun Y, Liu H, Peng H, Zhang Z. LincRNA 1700020I14Rik alleviates cell proliferation and fibrosis in diabetic nephropathy via miR-34a-5p/Sirt1/HIF-1α signaling. Cell Death Dis. 2018;9:461. doi: 10.1038/s41419-018-0527-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Song P, Ye LF, Zhang C, Peng T, Zhou XH. Long non-coding RNA XIST exerts oncogenic functions in human nasopharyngeal carcinoma by targeting miR-34a-5p. Gene. 2016;592:8–14. doi: 10.1016/j.gene.2016.07.055. [DOI] [PubMed] [Google Scholar]
- 28.Jiang ZQ, Li MH, Qin YM, Jiang HY, Zhang X, Wu MH. Luteolin inhibits tumorigenesis and induces apoptosis of non-small cell lung cancer cells via regulation of MicroRNA-34a-5p. Int J Mol Sci. 2018;19 doi: 10.3390/ijms19020447. [DOI] [PMC free article] [PubMed] [Google Scholar]