Abstract
Cervical cancer (CC) is the second most common cancer and the fourth leading cause of cancer-related death in women worldwide. Up to date, only a few of long noncoding RNAs (lncRNAs) have been functionally characterized. Here, we aimed to discover the functional roles of lncRNA GAS5-AS1. The GAS5-AS1 expression in CC tissues was markedly decreased when compared with that in the adjacent normal tissues. The downregulation of GAS5-AS1 was significantly correlated with the advanced FIGO stage, distant metastasis, lymphatic metastasis and poor prognosis in patients with CC. Functionally, GAS5-AS1 drastically reduced CC cell proliferation, migration and invasion in vitro, and remarkably suppressed CC tumorigenicity and metastasis in vivo. Mechanistically, it was found that GAS5-AS1 interacted with the tumor suppressor GAS5, and increased its stability by interacting with RNA demethylase ALKBH5 and decreasing GAS5 N6-methyladenosine (m6A) modification. Moreover, it was shown that m6A-mediated GAS5 RNA degradation relied on the m6A reader protein YTHDF2-dependent pathway. Our findings reveal an important mechanism of epigenetic alteration in CC carcinogenesis and metastasis.
Keywords: RNA stability, m6A modification, ALKBH5, YTHDF2
Introduction
Cervical cancer (CC) is the second most common cancer and the fourth leading cause of cancer-related death in women worldwide [1]. Even though treatments including surgery, chemotherapy, radiotherapy or targeted therapy have been used to improve clinical outcomes, the prognosis of patients with CC is still unsatisfied and the 5-year survival rate remains approximately 40% [2]. Therefore, exploring the molecular mechanisms underlying CC progression is urgently necessary for the development of the novel therapeutic targets.
Emerging evidences indicated that long non-coding RNAs (lncRNAs), a group of large transcripts (larger than 200 nucleotides in length) without protein-coding potential, play important roles in a wide variety of human diseases, including cancers. lncRNAs are involved in a variety of pathophysiological processes, such as cell proliferation, migration, invasion, apoptosis and chemotherapy resistance [3,4]. LncRNA growth arrest special 5 (GAS5), a tumor suppressor, was down-regulated in many kinds of cancers, including breast cancer, hepatocellular carcinoma (HCC), gastric cancer and CC [5]. GAS5 exerts suppressive effects on proliferation, migration, invasion, epithelial-to-mesenchymal transition (EMT) and radio resistance in cancer cells [6-8]. GAS5-AS1, the antisense RNA of GAS5, is a novel lncRNA transcript which is mapped on chromosome 1q25.1. Besides, it was downregulated in non-small cell lung cancer (NSCLC), and significantly correlated with larger tumors, higher TNM stages, and lymph node metastasis. GAS5-AS1 in NSCLC cells could inhibit EMT in NSCLC cells [9]. In addition, GAS5-AS1 was also observed to be down-regulated in HCC tissues and correlated with prognosis of HCC patients [10]. However, up to date, the functional roles of GAS5-AS1 and the regulatory mechanism between GAS5-AS1 and GAS5 remain to be elucidated.
Here, it was reported that GAS5-AS1 was significantly downregulated in CC tissues compared with that in adjacent normal tissues. Decrease of GAS5-AS1 was closely related to the FIGO stage, distant metastasis, lymphatic metastasis and poor prognosis in patients with CC. It was also revealed that GAS5-AS1 inhibited CC cell proliferation, migration and invasion by regulating GAS5 expression via interacting with RNA demethylase ALKBH5.
Materials and methods
Patients and tissue samples
A total of 66 pairs of CC tissues and adjacent normal epithelial tissues were collected from CC patients who had underwent surgical resection in The First Affiliated Hospital of Henan University of Science and Technology. No patients had undergone chemotherapy, radiotherapy or adjuvant treatment prior to surgery. The isolated tissues were immediately snap-frozen in liquid nitrogen and stored in -80°C for further analysis. Patients were clinically staged according to the International Federation of Gynecology and Obstetrics (FIGO) staging system.
Ethics approval and consent to participate
The present study was approved by the Ethics Committee of The first affiliated Hospital of Henan University of Science and Technology (approval number: 20180915). The research has been carried out in accordance with the World Medical Association Declaration of Helsinki. All patients provided written informed consent prior to their inclusion within the study.
Cell culture
Human CC cell lines (Caski, SiHa, C33A and HeLa) and the normal cervical epithelium cell line (HCvEpC) were obtained from the Chinese Academy of Science (Shanghai, China). The cells were maintained in RPMI-1640 medium (Hyclone) supplemented with 10% fetal bovine serum (FBS) (Gibco), penicillin (100 U/ml; Solarbio, Beijing, China) and streptomycin (100 μg/ml; Solarbio) solution and kept in a humidified incubator with 5% CO2 at 37°C.
Lentiviral infection
Full-length GAS5-AS1 or mutant GAS5-AS1 (mutation at the binding sites in GAS5-AS1, named GAS5-AS1-mut) were PCR-amplified and then cloned into the pLV-puro vector, and GAS5-AS1 shRNA (shGAS5-AS1, target sequence: 5’-GCTTTAGCCCAGAAACAGA-3’) was inserted into pLKO.1 vector, and empty vectors were used as negative controls (NC). All the lentiviral constructs were packaged into the HEK-293T cells and then were collected. Caski cells were infected with the GAS5-AS1-overexpressing lentiviral constructs. HeLa cells were infected with the shGAS5-AS1 lentiviral constructs into cells. After 48 hours, the stable cells were selected by using puromycin. The knockdown and overexpression efficacy was determined by qRT-PCR.
siRNA transfection
Transfection of siRNA (100 nM, GenePharma, China) was performed by using Lipofectamine RNAiMAX (Invitrogen, USA). Sequences of siRNA against specific targets were listed as follow: siALKBH5: GCGCAACAAGTACTTCTTC; siYTHDF2: CTGCCATGTCAGATTCCTA siGAS5: GTCCTAAAGAGCAAGCCTA.
RNA extraction and qRT-PCR analysis
The total RNA from tissue samples or cells was extracted using Trizol Reagent (Thermo Fisher Scientific) according to the manufacturer’s protocol. cDNA was synthesized using the PrimeScript RT reagent Kit (TaKaRa, Dalian, China), and quantitative Real-time PCR (qRT-PCR) analysis was then performed using the SYBR Premix Ex Taq™ II kit (TaKaRa) on an ABI PRISM 7500 fast Sequence Detection System. The relative expression of RNAs was calculated using the comparative Ct method. GAPDH was used as the internal control. The primer sequences for target genes were shown as follow: GAS5-AS1-Forward: TGTGCCCTTTATACCCACTTT, GAS5-AS1-Reverse: GCCCAACTAGTGATAGGCATTA; GAS5-Forward: CATTGGCACACAGGCATTAG, GAS5-Reverse: GTTACCAGGAGCAGAACCATTA; GAPDH-Forward: GGTGTGAACCATGAGAAGTATGA, GAPDH-Reverse: GAGTCCTTCCACGATACCAAAG.
Cell Counting Kit-8 assay (CCK-8)
The cells were suspended in 100 μl RPMI-1640 medium and seeded in a 96-well plate at the density of 2000 cell per well. At the different time point, the culture medium was added with 10 μL Cell Counting Kit 8 (CCK-8) reagents (Dojindo Molecular Technologies, Rockville, Japan) and then incubated for 1 hour. The cell proliferation was determined by detecting the optical density at 450 nm.
Colony formation assays
Cells were seeded at a density of 2000 cells in 6-well culture dish and cultured for 2 weeks; the colonies were fixed and stained with 1% crystal violet and counted. One clone should contain more than 50 cells.
Migration and invasion
Transwell migration was assessed using Transwell plates (8-μm pore size) without Matrigel. The upper chamber was filled with serum-free RPMI-1640 medium loading 1×105 cells and the lower chamber was filled with culture medium containing 10% FBS as chemoattractant. The invasion assay was conducted in a similar fashion with coating the filters with 30 μg of Matrigel (BD Biosciences), and 2×105 cells were added to the upper inserts. After incubation for 24 h, the cells on the upper membranes were eliminated with a cotton swab and those migrated cells were fixed by 4% paraformaldehyde and stained with 0.1% crystal violet.
RNA immunoprecipitation (RIP) and MS2-binding sequences-MS2-binding protein-based RNA immunoprecipitation assay (MS2-RIP)
RIP experiments were performed using the Magna RIP RNA-Binding Protein Immunoprecipitation kit (Millipore). Cells lysates were incubated with protein A+G magnetic beads, which were conjugated using 5 μg of ALKBH5 antibodies (Abcam), FTO (Abcam), YTHDF2 (Abcam), YTHDF3 (Abcam), YTHDC2 (Abcam) or negative control IgG (Abcam) at 4°C for overnight. After wash, the immunoprecipitated RNA was purified and then detected by qRT-PCR.
The MS2-binding sequences-MS2-binding protein-based RNA immunoprecipitation assay (MS2-RIP) was carried out according to previous study [11]. Cells were transfected with pcDNA-GAS5-AS1-MS2, pcDNA-GAS5-AS1-mut-MS2, or pcDNA-MS2 and pMS2-GFP by using lipofectamine 2000 (Invitrogen). After 48 hours, cells were used to perform RIP using an anti-GFP antibody (Abcam) and the Magna RIP RNA-Binding Protein Immunoprecipitation Kit (Millipore) according to the manufacturer’s protocol. Cells lysates were incubated with protein A+G magnetic beads, which were conjugated using 5 μg of GFP antibodies (Abcam) or negative control IgG (Abcam) at 4°C for overnight. After wash, the immunoprecipitated RNA was purified. Then the GAS5 level was analyzed by qRT-PCR.
Methylated RNA immunoprecipitation (MeRIP)
Total RNAs were first extracted from cells. Chemically fragmented RNA (~100 nucleotides) was incubated with m6A antibody for immunoprecipitation according to the standard protocol of the Magna methylated RNA immune-precipitation (MeRIP) m6A Kit (Merck Millipore). Enrichment of m6A containing RNA was then analyzed through qRT-PCR.
RNA pull-down
GAS5-AS1 or GAS5-AS1-mut were in vitro transcribed respectively from vector pSPT19- GAS5-AS1, or pSPT19-GAS5-AS1-mut, and biotin-labeled with the Biotin RNA Labeling Mix (Roche) and T7 RNA polymerase (Roche), treated with RNase-free DNase I (Roche), and purified with an RNeasy Mini Kit (Qiagen, Valencia, CA). 1 mg of whole-cell lysates from Caski and HeLa cells were incubated with 3 μg of purified biotinylated transcripts for 1 hr at 25°C, then the complexes were isolated with streptavidin agarose beads (Invitrogen). The RNA present in the pull-down material was detected by qRT-PCR analysis.
Luciferase reporter assay
The adenine residue embedded within the consensus sequence in the GAS5 was mutated (5’-RRACU-3’ to 5’-RRUCU-3’), thus abolishing m6A modification. Wild-type or mutant GAS5-fused reporter was transfected into stable HeLa and Caski cells. After 48 hours, the luciferase activity was detected by Dual-Luciferase® Reporter Assay System (Promega). The relative luciferase activity was normalized to Renilla luciferase activity.
In vivo xenografts
200 µL PBS containing 1×107 cells of stable cells were subcutaneously injected into male BALB/c athymic nude mice (6-week old, 18-20 g). Tumor volumes were measured by vernier caliper every 3 days using the following formula: volume = 1/2 × length × width2. After 38 days, the mice were killed, and the tumors were removed and weighed. A tail vein injection model was used for lung metastasis assays. Each group contained 6 mice. This animal study was approved by the Ethics Committee of The First Affiliated Hospital of Henan University of Science and Technology according to the Guidelines Followed for the Welfare of The First Affiliated Hospital of Henan University of Science and Technology.
Statistical analysis
The data were presented as mean ± standard deviation (SD). A value of P < 0.05 was determined to be statistically significant. All of the experiments were performed by triplicates. GraphPad Prism V5.0 software was used to analyze. The statistical differences among different groups were analyzed by test or one-way analysis of variance. Kaplan-Meier survival curve and log-rank test were used to determine the association between GAS5-AS1 expression and overall survival of patients.
Results
GAS5-AS1 expression is significantly decreased in human CC tissues
First, the expression pattern of GAS5-AS1 was revealed in 66 pairs of CC tissues and corresponding adjacent normal tissues. As shown in Figure 1A, GAS5-AS1 expression was significantly decreased in CC tissues than in adjacent normal tissues (Figure 1A). We then analyzed the correlation between GAS5-AS1 expression and clinical characteristics of CC patients. The 66 patients were divided into a high expression level group and a low expression level group based on the median value of GAS5-AS1 in CC tissues, and the results showed that low expression of GAS5-AS1 was significantly associated with the advanced FIGO stage, distant metastasis and lymphatic metastasis (Table 1). However, there was no association between GAS5-AS1 expression and age, tumor size and differentiation. Moreover, the Kaplan-Meier survival curve and the log-rank test were used to examine the association between GAS5-AS1 and overall survival of CC patients. Then, it was found that patients with high-level GAS5-AS1 had better overall survival than those with low-level GAS5-AS1 (median survival: 38.175 VS 68.891, Figure 1B). In addition, GAS5-AS1 expression in CC cell lines was also detected (Figure 1C), and the results revealed that GAS5-AS1 in CC cell lines was much lower than that in the normal cervical epithelium cell line (HCvEpC).
Figure 1.
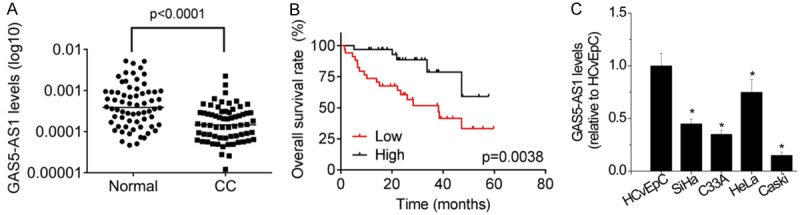
The expression of GAS5-AS1 is significantly decreased in CC tissues. A. The expression levels of GAS5-AS1 in 66 pairs of cervical cancer (CC) tissues and corresponding adjacent normal tissues were examined by qRT-PCR. B. CC samples with high GAS5-AS1 expression level or low GAS5-AS1 expression level were subjected to Kaplan-Meier analysis. The median of GAS5-AS1 expression in CC tissues was taken as cutoff. C. The relative expression of GAS5-AS1 was tested in four CC cells and the normal cervical epithelium cell HCvEpC. *P<0.05.
Table 1.
The relationship between GAS5-AS1 expression and clinical parameters of cervical cancer patients
| Clinical parameters | GAS5-AS1 expression | P value | |
|---|---|---|---|
|
| |||
| Low | High | ||
| Age (years) | |||
| <40 | 13 | 15 | 0.618 |
| >40 | 20 | 18 | |
| Tumor size | |||
| ≤4 cm | 16 | 19 | 0.459 |
| >4 cm | 17 | 14 | |
| Differentiation | |||
| Well + moderate | 15 | 20 | 0.218 |
| Poor | 18 | 13 | |
| FIGO stage | |||
| I-II | 14 | 25 | 0.006 |
| III-IV | 19 | 8 | |
| Lymph node metastasis | |||
| No | 11 | 22 | 0.007 |
| Yes | 22 | 11 | |
| Distant metastasis | |||
| No | 14 | 23 | 0.026 |
| Yes | 19 | 10 | |
GAS5-AS1 inhibits CC cell proliferation, migration and invasion in vitro
To understand the role of GAS5-AS1 in the malignant phenotypes of CC cells, both overexpression and knockdown assays were utilized. Briefly, lentiviruses for GAS5-AS1 overexpression and control lentiviruses (NC) were used to infect the Caski cells, whose GAS5-AS1 expression was relatively low (Figure 1C), whereas the specific shRNA against GAS5-AS1 (shGAS5-AS1) and the negative control shRNA (shNC) were employed in HeLa cells, whose GAS5-AS1 expression was relatively high (Figure 1C). The results of qRT-PCR showed that infection of shGAS5-AS1 lentiviruses significantly suppressed GAS5-AS1 expression in HeLa cells (Figure 2A), and infection of GAS5-AS1 overexpression lentiviruses obviously increased GAS5-AS1 expression in Caski cells (Figure 2B). The results of CCK-8 assays showed that GAS5-AS1 knockdown significantly promoted the proliferation of HeLa cells (Figure 2C), whereas the proliferation of Caski cells was dramatically suppressed by GAS5-AS1 overexpression (Figure 2D). Colony formation assays on these cells also showed the similar results (Figure 2E and 2F). Moreover, transwell migration and invasion assays were used to determine the cell metastatic behaviors. HeLa cells with GAS5-AS1 knockdown showed better migration and invasion abilities than control cells (Figure 2G). Conversely, both migration and invasion of Caski cells were repressed after GAS5-AS1 overexpression (Figure 2H).
Figure 2.
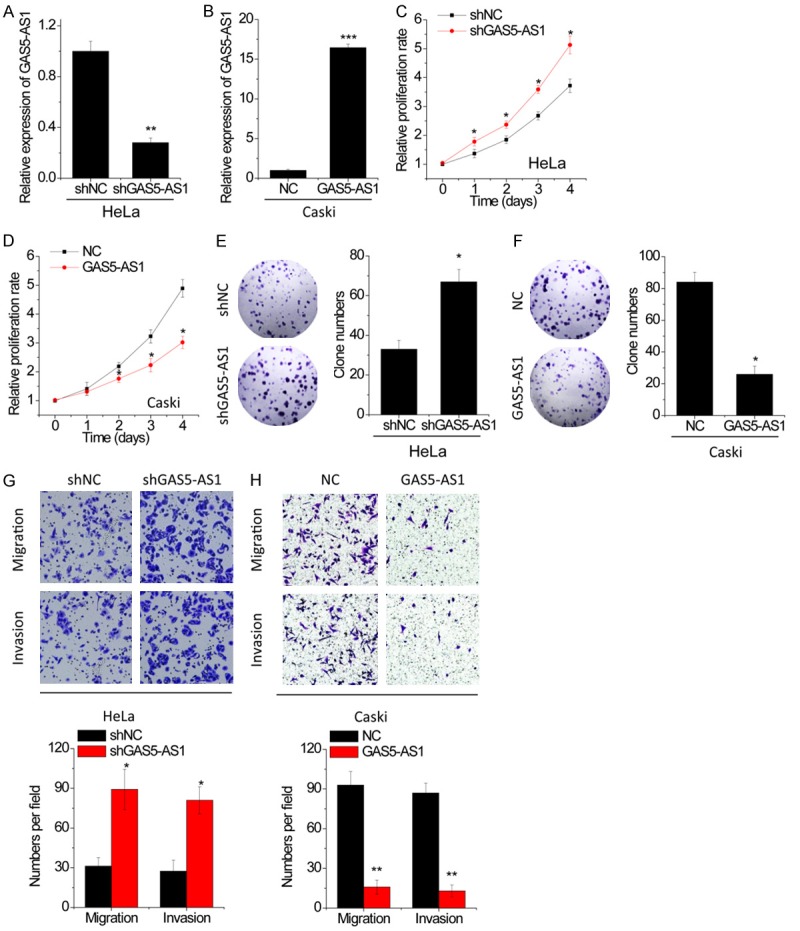
GAS5-AS1 suppresses proliferation, migration and invasion in CC cells. A. GAS5-AS1 expression was knocked down in HeLa cells, and the knockdown efficiency was validated by qRT-PCR. B. GAS5-AS1 expression was overexpressed in Caski cells, and the overexpression efficiency was validated by qRT-PCR. C. The proliferation of HeLa cells with GAS5-AS1 knockdown was detected by CCK-8 assay. D. The proliferation of Caski cells with GAS5-AS1 overexpression was detected by CCK-8 assay. E. The clone formation of HeLa cells with GAS5-AS1 knockdown. F. The clone formation of Caski cells with GAS5-AS1 overexpression. G. The migratory and invasive abilities of CC cells with GAS5-AS1 knockdown were evaluated by tranwell assay. H. The migratory and invasive abilities of CC cells with GAS5-AS1 overexpression were evaluated by tranwell assay. *P<0.05, **P<0.01, ***P<0.001.
GAS5-AS1 inhibits CC growth and metastasis in vivo
The effect of GAS5-AS1 on CC progress in vivo was then determined. The growth suppressing effect of GAS5-AS1 expression was confirmed by performing in vivo tumor growth assays. Xenograft tumors growing from HeLa cells with GAS5-AS1 knockdown had larger mean volumes and weights, and formed more rapidly, than tumors growing from control cells (Figure 3A and 3B). Conversely, tumors growing from Caski cells with GAS5-AS1 overexpression were smaller than control (Figure 3C and 3D).
Figure 3.
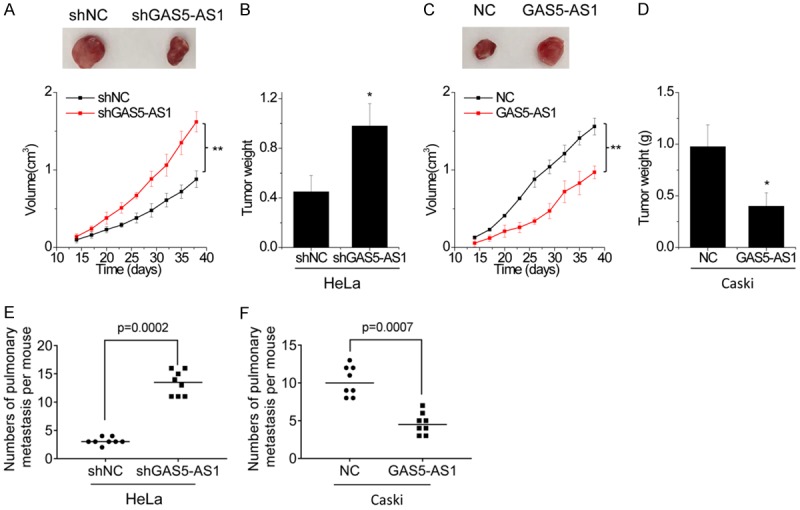
GAS5-AS1 inhibits growth and metastasis of CC cells in vivo. (A and B) Control and GAS5-AS1 knockdown HeLa cells were subcutaneously injected into nude mice. The tumor growth curve (A) and weight (B) was measured. (C and D) Control and GAS5-AS1 overexpressing Caski cells were subcutaneously injected into nude mice. The tumor growth curve (C) and weight (D) was measured. (E) Quantitative analysis was used to compare the lung metastasis nodules between control and GAS5-AS1 knockdown group. (F) Quantitative analysis was used to compare the lung metastasis nodules between control and GAS5-AS1 overexpressing group. *P<0.05, **P<0.01.
To reveal the inhibitory effect of GAS5-AS1 on CC metastasis in vivo, cells with GAS5-AS1 alteration were injected into the tail vein of mice. The lung metastases were detected after 8 weeks. The results showed that GAS5-AS1 knockdown enhanced the pulmonary metastasis of HeLa cells (Figure 3E), while the number of lung metastatic nodules formed by Caski cells was dramatically decreased by GAS5-AS1 overexpression (Figure 3F). Taken together, the data indicates that GAS5-AS1 impairs CC growth and metastasis both in vitro and in vivo.
GAS5-AS1 interacts with GAS5 and increases its stability
As the antisense RNA of GAS5, GAS5-AS1 may interact with GAS5. The result of BLAST indicated the binding sites of high complement between GAS5-AS1 and GAS5 (Figure S1). We then mutated the binding sites in GAS5-AS1 (GAS5-AS1-mut). To validate this hypothesis, the MS2-RIP with MS2-binding protein which specifically binds the RNA containing MS2-binding sequences was performed to pull down endogenous RNAs associated with GAS5-AS1. The result demonstrated that GAS5-AS1 RIP in HeLa and Caski cells is significantly enriched for GAS5 compared with the empty vector, IgG, or GAS5-AS1-mut (Figure 4A). The specific association between GAS5-AS1 and GAS5 was further confirmed by performing the RNA pull-down assay using a biotin-labeled-specific GAS5-AS1 probe (Figure 4B).
Figure 4.
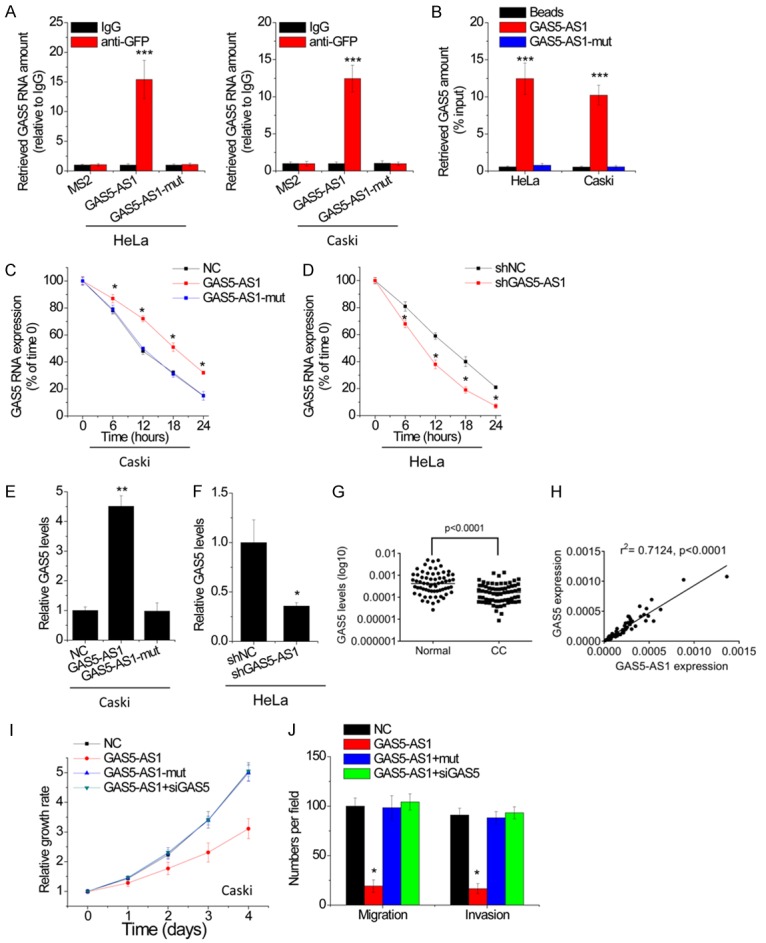
GAS5-AS1 interacts with GAS5 and increases its stability. A. The interaction between GAS5-AS1 and GAS5 was detected by MS2-RIP followed by qRT-PCR in HeLa and Caski cells. B. HeLa and Caski cell lysates were incubated with biotin-labeled GAS5-AS1; after pull-down, GAS5 was assessed by qRT-PCR. C. The stability of GAS5 over time was measured by qRT-PCR relative to time 0 after blocking new RNA synthesis with α-amanitin(50 mM) in indicated stable Caski cells and normalized to 18S rRNA (a product of RNA polymerase I that is unchanged by α-amanitin). D. The stability of GAS5 over time was measured by qRT-PCR relative to time 0 after blocking new RNA synthesis with α-amanitin(50 mM) in indicated stable HeLa cells and normalized to 18S rRNA. E. The GAS5 expression in indicated stable Caski cells was tested by qRT-PCR. F. The GAS5 expression in indicated stable HeLa cells was tested by qRT-PCR. G. The expression levels of GAS5 in 66 pairs of cervical cancer (CC) tissues and corresponding adjacent normal tissues were examined by qRT-PCR. H. The correlation between GAS5-AS1 and GAS5 level was measured in the same set of CC tissues. The expression levels were subjected to Pearson correlation analysis. I. SiRNA against GAS5 (siGAS5) was transfected into GAS5 overexpressing Caski cells. The proliferation of indicated Caski cells was tested by CCK-8 assay. J. SiRNA against GAS5 (siGAS5) was transfected into GAS5 overexpressing Caski cells. The migration and invasion of indicated Caski cells were tested by migration and invasion transwell assays. *P<0.05, **P<0.01, ***P<0.001.
Previous study reported that the lncRNA could interact with other RNAs to enhance their stability [12,13]. To test whether GAS5-AS1 could influence the stability of GAS5, CC cells with GAS5-AS1 alteration were treated with the inhibitor of RNA synthesis, α-amanitin, and the loss of GAS5 over a 24-hour period was measured. Interestingly, upregulation of GAS5-AS1, but not GAS5-AS1-mut, elongated the half-life of GAS5 in Caski cells (Figure 4C), whereas knockdown of GAS5-AS1 shortened the half-life of GAS5 in HeLa cells (Figure 4D). Moreover, GAS5 levels in different stable Caski and HeLa cells were detected, and then it was found that the overexpression of GAS5-AS1, but not GAS5-AS1-mut, significantly increased GAS5 levels in Caski cells (Figure 4E), while depletion of GAS5-AS1 significantly reduced GAS5 levels in HeLa cells (Figure 4F).
Then, the pathological correlation between GAS5-AS1 and GAS5 was determined. In this case, it was found that GAS5 expression was significantly decreased in CC tissues than in adjacent normal tissues (Figure 4G). Additionally, GAS5-AS1 was positively correlated with GAS5 expression in CC tissues (r2 = 0.7124, P<0.0001, Figure 4H). Collectively, these data demonstrates that GAS5-AS1 enhances the stability of GAS5, which depends on the binding of GAS5.
To test whether GAS5-AS1 exerted suppressive effect through GAS5, rescue experiments were conducted. Overexpression of wild-type GAS5-AS1 instead of GAS5-AS1-mut, significantly decreased cell proliferation, migration and invasion in Caski cells. Knockdown of GAS5 totally reversed these inhibitory effects (Figure 4I and 4J), indicating that GAS5-AS1 inhibits CC progression through suppression of GAS5.
GAS5-AS1 regulates the m6A modification of GAS5
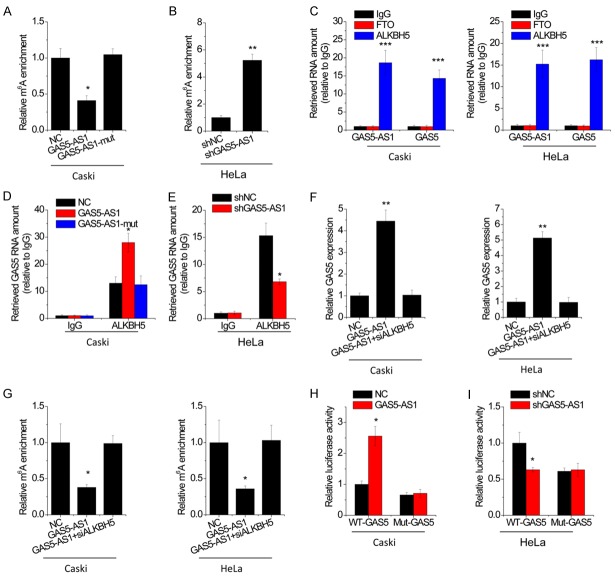
We then explored the underlying mechanism, by which GAS5-AS1 regulated GAS5 stability, and performed the nucleocytoplasmic separation followed by qRT-PCR. After that, it was found that GAS5-AS was located in both cytoplasm and the nucleus (Figure S2). A recent study showed that the antisense lncRNA could modulate the N6-Methyladenosine (m6A) modification of its target RNA [14]. The effects of RNA m6A modification on cellular processes include alterations in RNA stability, translation efficiency, subcellular localization and alternative splicing [15,16]. To validate whether GAS5-AS1 had an effect on GAS5 m6A modification, a MeRIP assay was performed. The result demonstrated that the m6A-specific antibody significantly enriched GAS5 compared with the immunoglobulin G pull-down control. Overexpression of GAS5-AS1 instead of GAS5-AS1-mut, dramatically reduced the m6A level of GAS5 in Caski cells (Figure 5A). Conversely, depletion of GAS5-AS1 significantly increased the m6A level of GAS5 in HeLa cells (Figure 5B). These results implied that m6A demethyltransferases may be involved in GAS5-AS1-mediated m6A modification. Up to now, two enzymes, namely fat mass and obesity-associated protein (FTO) and alkB homolog 5 (ALKBH5) capable of removing m6A have been identified. We performed RIP assays using anti-FTO or anti-ALKBH5 antibodies, and the results demonstrated that only ALKBH5 could interact with both GAS5-AS1 and GAS5 (Figure 5C). The interaction between GAS5 and ALKBH5 increased following GAS5-AS1 overexpression (Figure 5D), while decreased after depletion of GAS5-AS1 (Figure 5E).
Figure 5.
GAS5-AS1 decreases the m6A modification of GAS5. A. MeRIP-qPCR analysis of fragmented GAS5 RNA in Caski with GAS5-AS1 or mutant GAS5-AS1 overexpression. B. MeRIP-qPCR analysis of fragmented GAS5 RNA in HeLa with GAS5-AS1 knockdown. C. RIP assay was performed to test the interaction of FTO or ALKBH5 with GAS5-AS1 and GAS5. D. RIP assay was performed to test the interaction between ALKBH5 and GAS5 in indicated stable Caski cells. E. RIP assay was performed to test the interaction between ALKBH5 and GAS5 in HeLa cells with GAS5-AS1 knockdown. F. The GAS5 expression in GAS5-overexpressing Caski and HeLa cells transfected with siRNA against ALKBH5 (siALKBH5) was tested by qRT-PCR. G. The m6A modification of GAS5 in GAS5-overexpressing Caski and HeLa cells transfected with siALKBH5 was tested by MeRIP-qPCR. H. Wild-type or m6A consensus sequence mutant GAS5 was fused with firefly luciferase reporter. Mutation of m6A consensus sequences or overexpression of GAS5-AS1 relieved the posttranscriptional regulation of GAS5 in Caski cells. I. Wild-type or m6A consensus sequence mutant GAS5 was fused with firefly luciferase reporter. Mutation of m6A consensus sequences or knockdown of GAS5-AS1 relieved the posttranscriptional regulation of GAS5 in HeLa cells. *P<0.05, **P<0.01, ***P<0.001.
Then, by detecting, it was found that GAS5 was regulated by GAS5-AS1 in an ALKBH5-dependent manner. Transfection of the ALKBH5 siRNA abolished the upregulation of GAS5 induced by GAS5-AS1 overexpression (Figure 5F). Moreover, the decrease of the GAS5 m6A level induced by GAS5-AS1 overexpression was restored by transfection of the ALKBH5 siRNA (Figure 5G). Collectively, the data shows that GAS5-AS1 upregulates GAS5 expression through interaction with ALKBH5. m6A usually embedded within the consensus sequence 5’-RRACU-3’ (R = A or G; methylated adenosine residue) is underscored. Then, it was found that those four match the 5’-RRACU-3’ m6A consensus sequence within the GAS5 (Figure S3). Next, the adenine residue embedded within the consensus sequence in the GAS5 was mutated (5’-RRACU-3’ to 5’-RRUCU-3’), thus abolishing m6A modification. The relative luciferase activity of the wild-type or mutant GAS5-fused reporter was compared in HeLa and Caski cells. When compared with the wild-type, mutation on the m6A consensus sequences decreased the expression of the GAS5-fused reporter. The luciferase activity of the wild-type GAS5-fused reporter was also significantly augmented upon GAS5-AS1 overexpression in Caski cells (Figure 5H), while decreased after GAS5-AS1 knockdown in HeLa cells (Figure 5I). Neither downregulation nor upregulation of GAS5-AS1 showed obvious effect on the expression of the mutant GAS5-fused reporter, suggesting the modulation of GAS5 expression was under the control of GAS5-AS1-mediated m6A modification.
GAS5-AS1 upregulates GAS5 expression through an YTHDF2-dependent pathway
Previous studies demonstrated that reader protein YTHDF2, YTHDF3 and YTHDC2 could selectively bind m6A-modified mRNAs, recruited them to mRNA decaying sites and then controlled target RNA stability [17]. We then performed RIP assays using anti-YTHDF2, anti-YTHDF3 or anti-YTHDC1 antibodies, and the results showed that GAS5 could only be significantly pulled down by YTHDF2 compared with IgG. The association between GAS5 and YTHDF2 was suppressed by GAS5-AS1 overexpression in Caski cells and enhanced following GAS5-AS1 knockdown in HeLa cells (Figure 6A and 6B). Moreover, we silenced YTHDF2 expression in GAS5-AS1 knockdown HeLa cells and found that knockdown of YTHDF2 rescued the expression and stability of GAS5 decreased by GAS5-AS1 downregulation to a great extent (Figure 6C and 6D). These results indicate that GAS5-AS1 increases GAS5 expression and stability in an YTHDF2-dependent manner.
Figure 6.
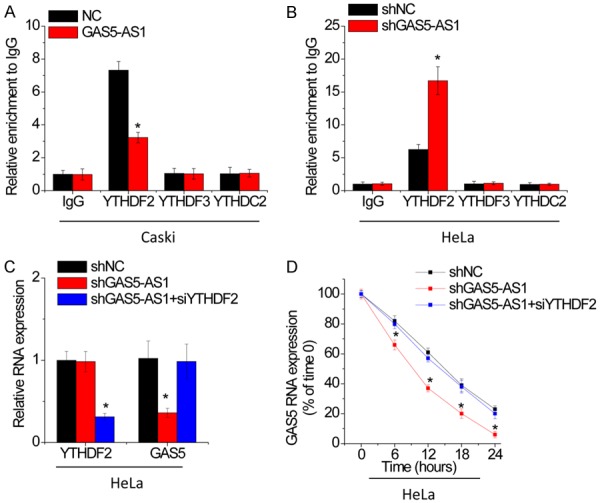
GAS5-AS1 upregulates GAS5 expression through an YTHDF2-dependent manner. A. RIP assay was performed to test the interaction of YTHDF2 or YTHDF3 or YTHDC2 with GAS5 in control and GAS5-AS1 overexpressing Caski cells. B. RIP assay was performed to test the interaction of YTHDF2 or YTHDF3 or YTHDC2 with GAS5 in control and GAS5-AS1 knockdown HeLa cells. C. The siRNA against YTHDF2 was transfected into GAS5-AS1 knockdown HeLa cells and then the GAS5 expression was detected by qRT-PCR. D. The siRNA against YTHDF2 was transfected into GAS5-AS1 knockdown HeLa cells and then the GAS5 stability was examined. *P<0.05.
Discussion
An increasing amount of evidence has reported that lncRNAs are crucial regulators of gene expression and biological processes. Here, for the first time, we demonstrated that lncRNA GAS5-AS1 was frequently downregulated in CC tissues and cell lines. Clinical data analysis showed that downregulation of GAS5-AS1 was significantly associated with the advanced FIGO stage, distant metastasis, lymphatic metastasis and shorter overall survival of CC patients. The functional roles of GAS5-AS1 in regulating malignant phenotypes of CC were then examined. Overexpression of GAS5-AS1 increased CC cell proliferation, migration and invasion, while depletion of GAS5-AS1 exerted the opposite effects. In vivo, GAS5-AS1 suppressed the growth and metastasis of CC cells.
Many lncRNAs have been reported to interact with mRNAs and inhibit their degradation. For example, lncRNA-ATB promoted organ colonization of disseminated tumor cells by binding IL-11 mRNA and increasing its stability [12]. The Sirt1 antisense lnc RNA can bind the Sirt1 3’-untranslated region, thus enhancing the stability of Sirt1 and increasing Sirt1 abundance at both the mRNA and protein levels [18]. Through antisense pairing with FGFR3 3’UTR, FGFR3-AS1 enhances FGFR3 mRNA stability and upregulates FGFR3 expression [19]. Here, it was also demonstrated that GAS5 stability was regulated by its antisense RNA, GAS5-AS1. In our present study, through RIP and RNA pull-down assays, we identified a direct interaction between GAS5-AS1 and GAS5. In addition, overexpression of GAS5-AS1 elongated the half-life of GAS5, whereas knockdown of GAS5-AS1 shortened the half-life of GAS5. In CC, decreased expression of GAS5 predicts a poor prognosis [20,21]. GAS5 acts as a tumor suppressor to inhibit proliferation, invasion, cisplatin and radio resistance through inhibiting miR-196a, miR-205, miR-21 and miR-106b [7,22,23]. Therefore, GAS5-AS1 exerts its inhibitory effect by binding with GAS5 and suppressing its degradation.
The m6A modification is the most abundant internal RNA modification and present in several classes of RNAs, such as the mRNA and lncRNAs. Internal m6A modification in RNAs is dynamic and reversible, while m6A RNA modification can be installed enzymatically by various RNA methyltransferases, including METTL3, METTL14 and WTAP. m6A in the RNA can be removed by RNA demethylases, including FTO and ALKBH5 [24,25]. ALKHB5 depletion was related to the increase of m6A, whereas its overexpression reduced m6A in mRNAs of human cell lines [26]. Alkbh5 knockout mice were viable, but showed compromised spermatogenesis due to significantly altered expression of key genes required for spermatogenic maturation [27]. ALKBH5 decreased the m6A level of FOXM1 mRNA for maintaining tumorigenicity of glioblastoma stem-like cells in an RNA-RNA interaction-dependent manner [14]. These studies suggest that altered expression of key genes, which are sensitive to the function of the m6A modulator, can give rise to a significant phenotype change. However, to date, the biological significance and the key target genes of ALKBH5 in CC remain elusive. Here, it was demonstrated that GAS5-AS1 reduced the m6A level of GAS5 through interaction with ALKBH5, and increased the stability of GAS5.
Proteins that selectively bind m6A can be defined as m6A “readers” that exert regulatory functions by selective recognition of the methylated RNA. The reader protein YTHDF2 specifically binds the m6A-containing RNA and further accelerates the decay of m6A modified transcripts evidenced by the interactions between YTHDF2 and P-bodies [28]. YTHDF2 is overexpressed in a broad spectrum of human Acute myeloid leukemia (AML) and is required for propagation in AML. YTHDF2 decreases the half-life of diverse m6A transcripts that contribute to the overall integrity of leukemic stem cell function, including the tumor necrosis factor receptor Tnfrsf2, whose upregulation in Ythdf2-deficient LSCs primes cells for apoptosis [29]. Nevertheless, YTHDF2 is capable to suppress cell proliferation, tumor growth and activation of MEK and ERK via promoting the degradation of EGFR mRNA in hepatocellular carcinoma cells [30]. So far, the relationship between YTHDF2 and CC remains unknown. Our results showed that GAS5-AS1 epigenetically upregulated GAS5 through an YTHDF2-dependent mechanism. Knockdown of YTHDF2 abolished the decrease of GAS5 mediated by GAS5-AS1 knockdown. Further investigation should be necessary to identify the m6A enrichment regions and the specific site of GAS5 for the interaction with YTHDF2.
In conclusion, it is shown that GAS5-AS1 represses CC tumorigenesis and metastasis and that tumor suppressor GAS5 is regulated by GAS5-AS1 in an ALKBH5-m6A-YTHDF2-dependent manner. These findings indicate that GAS5-AS1 is a critical molecular for CC progression and is potentially an effective therapy for CC.
Disclosure of conflict of interest
None.
Supporting Information
References
- 1.Ventriglia J, Paciolla I, Pisano C, Cecere SC, Di Napoli M, Tambaro R, Califano D, Losito S, Scognamiglio G, Setola SV, Arenare L, Pignata S, Della Pepa C. Immunotherapy in ovarian, endometrial and cervical cancer: state of the art and future perspectives. Cancer Treat Rev. 2017;59:109–116. doi: 10.1016/j.ctrv.2017.07.008. [DOI] [PubMed] [Google Scholar]
- 2.Vaccarella S, Lortet-Tieulent J, Plummer M, Franceschi S, Bray F. Worldwide trends in cervical cancer incidence: impact of screening against changes in disease risk factors. Eur J Cancer. 2013;49:3262–3273. doi: 10.1016/j.ejca.2013.04.024. [DOI] [PubMed] [Google Scholar]
- 3.D’Angelo E, Agostini M. Long non-coding RNA and extracellular matrix: the hidden players in cancer-stroma cross-talk. Noncoding RNA Res. 2018;3:174–177. doi: 10.1016/j.ncrna.2018.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lu T, Wang Y, Chen D, Liu J, Jiao W. Potential clinical application of lncRNAs in non-small cell lung cancer. Onco Targets Ther. 2018;11:8045–8052. doi: 10.2147/OTT.S178431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pickard MR, Williams GT. Molecular and cellular mechanisms of action of tumour suppressor GAS5 LncRNA. Genes (Basel) 2015;6:484–499. doi: 10.3390/genes6030484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liu B, Wu S, Ma J, Yan S, Xiao Z, Wan L, Zhang F, Shang M, Mao A. lncRNA GAS5 reverses EMT and tumor stem cell-mediated gemcitabine resistance and metastasis by targeting miR-221/SOCS3 in pancreatic cancer. Molecular therapy. Mol Ther Nucleic Acids. 2018;13:472–482. doi: 10.1016/j.omtn.2018.09.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gao J, Liu L, Li G, Cai M, Tan C, Han X, Han L. LncRNA GAS5 confers the radio sensitivity of cervical cancer cells via regulating miR-106b/IER3 axis. Int J Biol Macromol. 2018;126:994–1001. doi: 10.1016/j.ijbiomac.2018.12.176. [DOI] [PubMed] [Google Scholar]
- 8.Dong X, Kong C, Liu X, Bi J, Li Z, Li Z, Zhu Y, Zhang Z. GAS5 functions as a ceRNA to regulate hZIP1 expression by sponging miR-223 in clear cell renal cell carcinoma. Am J Cancer Res. 2018;8:1414–1426. [PMC free article] [PubMed] [Google Scholar]
- 9.Wu Y, Lyu H, Liu H, Shi X, Song Y, Liu B. Downregulation of the long noncoding RNA GAS5-AS1 contributes to tumor metastasis in non-small cell lung cancer. Sci Rep. 2016;6:31093. doi: 10.1038/srep31093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang Y, Jing W, Ma W, Liang C, Chai H, Tu J. Down-regulation of long non-coding RNA GAS5-AS1 and its prognostic and diagnostic significance in hepatocellular carcinoma. Cancer Biomark. 2018;22:227–236. doi: 10.3233/CBM-170781. [DOI] [PubMed] [Google Scholar]
- 11.Li T, Xie J, Shen C, Cheng D, Shi Y, Wu Z, Deng X, Chen H, Shen B, Peng C, Li H, Zhan Q, Zhu Z. Amplification of long noncoding RNA ZFAS1 promotes metastasis in hepatocellular carcinoma. Cancer Res. 2015;75:3181–3191. doi: 10.1158/0008-5472.CAN-14-3721. [DOI] [PubMed] [Google Scholar]
- 12.Yuan JH, Yang F, Wang F, Ma JZ, Guo YJ, Tao QF, Liu F, Pan W, Wang TT, Zhou CC, Wang SB, Wang YZ, Yang Y, Yang N, Zhou WP, Yang GS, Sun SH. A long noncoding RNA activated by TGF-beta promotes the invasion-metastasis cascade in hepatocellular carcinoma. Cancer Cell. 2014;25:666–681. doi: 10.1016/j.ccr.2014.03.010. [DOI] [PubMed] [Google Scholar]
- 13.Hu J, Qian Y, Peng L, Ma L, Qiu T, Liu Y, Li X, Chen X. Long noncoding RNA EGFR-AS1 promotes cell proliferation by increasing EGFR mRNA stability in gastric cancer. Cell Physiol Biochem. 2018;49:322–334. doi: 10.1159/000492883. [DOI] [PubMed] [Google Scholar]
- 14.Hu J, Qian Y, Peng L, Ma L, Qiu T, Liu Y, Li X, Chen X. m(6)A demethylase ALKBH5 maintains tumorigenicity of glioblastoma stem-like cells by sustaining FOXM1 expression and cell proliferation program. Cancer Cell. 2017;31:591–606. e596. doi: 10.1016/j.ccell.2017.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Batista PJ. The RNA Modification N(6)-methyladenosine and its implications in human disease. Genomics Proteomics Bioinformatics. 2017;15:154–163. doi: 10.1016/j.gpb.2017.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang S, Chai P, Jia R, Jia R. Novel insights on m(6)A RNA methylation in tumorigenesis: a double-edged sword. Mol Cancer. 2018;17:101. doi: 10.1186/s12943-018-0847-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang X, Lu Z, Gomez A, Hon GC, Yue Y, Han D, Fu Y, Parisien M, Dai Q, Jia G, Ren B, Pan T, He C. N6-methyladenosine-dependent regulation of messenger RNA stability. Nature. 2014;505:117–120. doi: 10.1038/nature12730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li B, Hu Y, Li X, Jin G, Chen X, Chen G, Chen Y, Huang S, Liao W, Liao Y, Teng Z, Bin J. Sirt1 antisense long noncoding RNA promotes cardiomyocyte proliferation by enhancing the stability of Sirt1. J Am Heart Assoc. 2018;7:e009700. doi: 10.1161/JAHA.118.009700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sun J, Wang X, Fu C, Wang X, Zou J, Hua H, Bi Z. Long noncoding RNA FGFR3-AS1 promotes osteosarcoma growth through regulating its natural antisense transcript FGFR3. Mol Biol Rep. 2016;43:427–436. doi: 10.1007/s11033-016-3975-1. [DOI] [PubMed] [Google Scholar]
- 20.Cao S, Liu W, Li F, Zhao W, Qin C. Decreased expression of lncRNA GAS5 predicts a poor prognosis in cervical cancer. Int J Clin Exp Pathol. 2014;7:6776–6783. [PMC free article] [PubMed] [Google Scholar]
- 21.Wen Q, Liu Y, Lyu H, Xu X, Wu Q, Liu N, Yin Q, Li J, Sheng X. Long noncoding RNA GAS5, which acts as a tumor suppressor via microRNA 21, regulates cisplatin resistance expression in cervical cancer. Int J Gynecol Cancer. 2017;27:1096–1108. doi: 10.1097/IGC.0000000000001028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yang W, Hong L, Xu X, Wang Q, Huang J, Jiang L. LncRNA GAS5 suppresses the tumorigenesis of cervical cancer by downregulating miR-196a and miR-205. Tumour Biol. 2017;39:1010428317711315. doi: 10.1177/1010428317711315. [DOI] [PubMed] [Google Scholar]
- 23.Yao T, Lu R, Zhang J, Fang X, Fan L, Huang C, Lin R, Lin Z. Growth arrest-specific 5 attenuates cisplatin-induced apoptosis in cervical cancer by regulating STAT3 signaling via miR-21. J Cell Physiol. 2019;234:9605–9615. doi: 10.1002/jcp.27647. [DOI] [PubMed] [Google Scholar]
- 24.Ma C, Chang M, Lv H, Zhang ZW, Zhang W, He X, Wu G, Zhao S, Zhang Y, Wang D, Teng X, Liu C, Li Q, Klungland A, Niu Y, Song S, Tong WM. RNA m(6)A methylation participates in regulation of postnatal development of the mouse cerebellum. Genome Biol. 2018;19:68. doi: 10.1186/s13059-018-1435-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lan Q, Liu PY, Haase J, Bell JL, Huttelmaier S, Liu T. The critical role of RNA m(6)A methylation in cancer. Cancer Res. 2019;79:1285–1292. doi: 10.1158/0008-5472.CAN-18-2965. [DOI] [PubMed] [Google Scholar]
- 26.Shah A, Rashid F, Awan HM, Hu S, Wang X, Chen L, Shan G. The DEAD-Box RNA Helicase DDX3 interacts with m(6)A RNA Demethylase ALKBH5. Stem Cells Int. 2017;2017:8596135. doi: 10.1155/2017/8596135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zheng G, Dahl JA, Niu Y, Fedorcsak P, Huang CM, Li CJ, Vågbø CB, Shi Y, Wang WL, Song SH, Lu Z, Bosmans RP, Dai Q, Hao YJ, Yang X, Zhao WM, Tong WM, Wang XJ, Bogdan F, Furu K, Fu Y, Jia G, Zhao X, Liu J, Krokan HE, Klungland A, Yang YG, He C. ALKBH5 is a mammalian RNA demethylase that impacts RNA metabolism and mouse fertility. Mol Cell. 2013;49:18–29. doi: 10.1016/j.molcel.2012.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xu C, Wang X, Liu K, Roundtree IA, Tempel W, Li Y, Lu Z, He C, Min J. Structural basis for selective binding of m6A RNA by the YTHDC1 YTH domain. Nat Chem Biol. 2014;10:927–929. doi: 10.1038/nchembio.1654. [DOI] [PubMed] [Google Scholar]
- 29.Paris J, Morgan M, Campos J, Spencer GJ, Shmakova A, Ivanova I, Mapperley C, Lawson H, Wotherspoon DA, Sepulveda C, Vukovic M, Allen L, Sarapuu A, Tavosanis A, Guitart AV, Villacreces A, Much C, Choe J, Azar A, van de Lagemaat LN, Vernimmen D, Nehme A, Mazurier F, Somervaille TCP, Gregory RI, O’Carroll D, Kranc KR. Targeting the RNA m(6)A reader YTHDF2 selectively compromises cancer stem cells in acute myeloid leukemia. Cell stem Cell. 2019;25:137–148. e6. doi: 10.1016/j.stem.2019.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhong L, Liao D, Zhang M, Zeng C, Li X, Zhang R, Ma H, Kang T. YTHDF2 suppresses cell proliferation and growth via destabilizing the EGFR mRNA in hepatocellular carcinoma. Cancer Lett. 2019;442:252–261. doi: 10.1016/j.canlet.2018.11.006. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.