Abstract
The chemokine receptor CCR5 is a drug target to prevent transmission of HIV/AIDS. We studied four analogs of the native chemokine regulated, on activation, normal T-cell-expressed, and secreted (RANTES) (CCL5) that have anti-HIV potencies of around 25 pM, which is more than four orders of magnitude higher than that of RANTES itself. It has been hypothesized that the ultrahigh potency of the analogs is due to their ability to bind populations of receptors not accessible to native chemokines. To test this hypothesis, we developed a homogeneous dual-color fluorescence cross-correlation spectroscopy assay for saturation- and competition-binding experiments. The fluorescence cross-correlation spectroscopy assay has the advantage that it does not rely on competition with radioactively labeled native chemokines used in conventional assays. We prepared site-specifically labeled fluorescent analogs using native chemical ligation of synthetic peptides, followed by bioorthogonal fluorescent labeling. We engineered a mammalian cell expression construct to provide fluorescently labeled CCR5, which was purified using a tandem immunoaffinity and size-exclusion chromatography approach to obtain monomeric fluorescent CCR5 in detergent solution. We found subnanomolar binding affinities for the two analogs 5P12-RANTES and 5P14-RANTES and about 20-fold reduced affinities for PSC-RANTES and 6P4-RANTES. Using homologous and heterologous competition experiments with unlabeled chemokine analogs, we conclude that the analogs all bind at the same binding site, whereas the native chemokines (RANTES and MIP-1α) fail to displace bound fluorescent analogs even at tens of micromolar concentrations. Our results can be rationalized with de novo structural models of the N-terminal tails of the synthetic chemokines that adopt a different binding mode as compared to the parent compound.
Significance
Dual-color fluorescence cross-correlation spectroscopy enables quantification of all the components in a binding equilibrium to give a comprehensive description of the ligand-binding reaction without the need to physically separate bound and free components. We applied fluorescence cross-correlation spectroscopy to study the G-protein-coupled receptor human chemokine receptor 5, which is the primary coreceptor for transmission of the HIV-1 virus. We show that the chemokine analog 5P12-RANTES has ultra-high-affinity binding at chemokine receptor 5 and rationalize the results in the context of recent structural studies.
Introduction
G-protein-coupled receptors (GPCRs) are cell-surface heptahelical transmembrane receptors that mediate many important physiological processes and are also involved in the transmission and virulence of several infectious diseases. For example, certain viruses can employ GPCRs during their life cycle to gain cellular entry, enhance dissemination, or evade immune detection (1). The human immunodeficiency virus 1 (HIV-1) utilizes the human chemokine receptor 5 (CCR5) as a coreceptor to infect immune cells such as dendritic cells and T cells (2). Homozygous carriers of the CCR5-Δ32 mutation are resistant to HIV-1 infection, highlighting the importance of CCR5 in transmission of the virus (3, 4, 5). HIV-1 expresses envelope glycoprotein gp120, which recognizes the cluster of differentiation 4 (CD4) receptor (6). CD4 engagement causes gp120 to undergo a conformational change that exposes its variable loop 3, which binds to CCR5. Maraviroc is the only commercial HIV-1 therapeutic agent that targets CCR5 to prevent cellular entry of R5-tropic strains, but case studies have shown the emergence of HIV-1 resistance to maraviroc (7).
The native chemokine ligands regulated, on activation, normal T-cell-expressed, and secreted (RANTES) and macrophage inflammatory protein 1α (MIP-1α) also inhibit HIV-1 entry but have very low potencies (8, 9). To increase the anti-HIV-1 potency, chemically modified RANTES analogs were developed (10, 11) that efficiently block viral transmission in macaques (12). PSC-RANTES, one of these analogs, shows picomolar anti-HIV potency, but it is also a strong CCR5 agonist, which may cause undesirable inflammatory effects (13). Several additional, fully recombinant RANTES analogs were developed using a phage display library by mutating the first nine residues (13). From this screen, we selected three analogs—5P12-RANTES (5P12), 5P14-RANTES (5P14), and 6P4-RANTES (6P4)—that inhibit HIV-1 with similar potencies as PSC-RANTES (Table S1; (13)). It is noteworthy that 5P12 is actively developed as a highly effective microbicide candidate in the prevention of human-to-human HIV-1 transmission. Like PSC, 6P4 shows strong agonist activity by calcium flux and internalization assays. In contrast, 5P14 induces receptor internalization but does not activate calcium flux signaling. Last, 5P12 binds to CCR5 but displays no functional activity on CCR5 (13). However, a subsequent study showed that 5P14 can induce Gαi signaling in cyclic adenosine monophosphate (cAMP) inhibition assays in disagreement with earlier conclusions of a lack of G-protein-linked activity (14). Lorenzen et al. investigated this discrepancy further and showed that 5P12, 5P14, 6P4, PSC, RANTES, and MIP-1α all cause Gαi activation and that PSC and 6P4 and, to a lesser extent, RANTES and MIP-1α also induce Gαq activation. Based on these data, we proposed a model in which the RANTES analogs can bind to both G-protein-uncoupled (“naked”) and precoupled CCR5, whereas the native chemokines bind only to the G-protein precoupled receptor (15). Radioligand competition-binding assays using 35S-gp120 showed that the viral glycoprotein recognizes both the “naked” receptor and the precoupled receptor fraction (16). Thus, the RANTES analogs efficiently block HIV because they can bind to both the G-protein precoupled and the “naked” CCR5 receptor fractions.
To test the hypothesis that the RANTES analogs can bind to “naked” CCR5 with high affinity, we proposed to develop a strategy by which we could measure equilibrium dissociation and competition-binding affinities to different receptor fractions. Previously, it was shown that RANTES and MIP-1α require CCR5 G-protein precoupling for high-affinity binding using a nonhydrolyzable GTP analog that locks the G-protein to the receptor. However, they could not observe the “naked” receptor fraction using 125I-MIP-1α as the tracer. Instead, 35S-gp120 was employed because it recognizes both receptor fractions. Yet, the measured chemokine-binding affinities using 35S-gp120 were different from the affinities derived using 125I-MIP-1α. Because radioligand binding measurements of GPCRs in cell-based systems or crude membrane preparations tend to yield inconsistent results because of their susceptibility to the reaction conditions, we realized that there was an unmet need for a methodology to characterize the ligand-binding affinities of the RANTES analogs and the native chemokines with CCR5 in a chemically defined environment.
Single-molecule methods can be used to determine precise receptor-ligand binding parameters of purified components in a chemically defined system. Fluorescence correlation spectroscopy (FCS) is a method that measures the diffusion of a fluorescently labeled species across an excitation volume. FCS can be used to detect ligand binding of a fluorescently labeled ligand in the presence of a receptor by observing changes in the diffusion time (τD) of the ligand. If the ligand and receptor are both labeled, then fluorescence cross-correlation spectroscopy (FCCS) can be used to measure the diffusion of the receptor-ligand complex by cross-relating the fluorescence fluctuations from the ligand with the fluorescence fluctuations from the receptor (17). Absolute concentrations of the ligand, the receptor, and the receptor-ligand complex can also be derived from FCS and FCCS measurements to determine equilibrium dissociation constants and inhibition parameters.
To establish a, to our knowledge, novel FCCS assay to measure ligand-binding parameters of four RANTES analogs with CCR5, we first expressed the CCR5-SNAP construct with a cleavable signal peptide, a SNAP-tag for fluorescent labeling, and N-terminal FLAG and C-terminal 1D4 tags for affinity purification. We then purified monomeric CCR5-SNAP labeled with Alexa-488 (CCR5-SNAP-488) to homogeneity and quantified its concentration by FCS. We prepared the fluorescent chemokines by a modular synthetic scheme that simplifies attachment of different fluorophores to larger peptide ligands. The scheme uses solid-phase peptide synthesis, native chemical fragment ligation, oxime ligation to introduce an azide handle, and strain-promoted azide-alkyne cycloaddition to attach the fluorescent label. Then, we used the materials to perform a detailed set of FCCS saturation- and competition-binding experiments with the CCR5-SNAP-488 and Alexa-647-labeled RANTES analogs. We found that 5P14 and 5P12 bind CCR5 with a very high affinity (subnanomolar). In contrast, 6P4 and PSC bind the receptor with more than an order of magnitude lower affinity. Native ligands, such as RANTES, failed to compete at 10 μM concentrations with the RANTES analogs binding to CCR5-SNAP-488. We generated homology models of 5P12, 6P4, and RANTES in complex with CCR5 based on the crystal structure of 5P7-RANTES (5P7), another RANTES analog similar to 5P12, bound to CCR5 (18). The homology models revealed that the N-termini of the RANTES analogs bind almost identically to 5P7 in the crystal structure but differently from RANTES.
Materials and Methods
Materials
Recombinant RANTES and MIP-1α were from PeproTech, (Rocky Hill, NJ). Coelenterazine 400A for BRET2 (bioluminescence resonance energy transfer) experiments was from Biotium (Hayward, CA). Forskolin and poly-D-lysine were from Sigma-Aldrich (St. Louis, MO), and the anti-CCR5 mAb (Clone 2D7) directly coupled to phycoerythrin (PE) was from R&D Systems (Minneapolis, MN), anti-CCR5 (Clone T21/8) directly coupled to PE was from eBioscience (San Diego, CA), and anti-FLAG PE was from Biolegend (San Diego, CA). All other chemicals were obtained as ACS reagent grade or equivalent from Sigma-Aldrich. The mammalian expression plasmid pcDNA3.1(+) was obtained from Invitrogen (Waltham, MA). The wild-type CCR5 (CCR5 WT) expression construct (MK101) is human CCR5 complementary DNA (cDNA) tagged with the C-terminal 1D4 epitope, TETSQVAPA in pcDNA3.1(+) (19). Dulbecco’s modified Eagle’s medium Glutamax (DMEM-Q), 1% penicillin-streptomycin, and Lipofectamine 2000 were from Life Technologies (Carlsbad, CA). Bovine serum albumin (BSA) fraction V, fatty-acid-free was from EMD Millipore (Burlington, MA). IgG-free, heat-shock-fractionated BSA solution (BSA, IgG-free, Sigma A0336) was further purified by a column of Pierce Detergent Removal Resin (Thermo Fisher Scientific, Waltham, MA) to remove colored impurities. 1D4 mAb Sepharose 2B resin was prepared by coupling purified 1D4 mAb to freshly cyanogen-bromide-activated resin (19). 96-well white microplates with clear bottom and 384-well black microplates with clear bottom plates were from Corning (Corning, NY). Nonlabeled RANTES analogs used in this study were prepared by total chemical synthesis as described previously (13). Soluble CD4 was obtained from the National Institutes of Health AIDS reagent program (catalog number 7356, lot number 130168), and monomeric BG 505 gp120 (2G12 purified) was a gift from Dr. John P. Moore (Weill Cornell Medicine, New York, NY). Benzylguanine (BG)-Alexa-488 was acquired from New England Biolabs (Ipswich, MA).
Sequence design and molecular cloning of CCR5-SNAP
We engineered the CCR5-SNAP expression construct in several iterations. First, we designed a synthetic construct encoding the human CCR5 gene with several functional tags for affinity purification and surface immobilization (TH1006, Fig. S1). TH1006 contains the following elements detailed in Table S2: 5′-adaptor-hCCR5-Linker-KpnI-OLLAS (2-14)-1D4-Stop-KpnI-OLLAS-Spacer4-EcoRV-Spacer4-OLLAS-EcoRV-AgeI-BamHI-BstEII-Strep-III-BstEIII-1D4-Stop-3′-adaptor.
The 5′-adaptor includes several restriction sites and a Kozak sequence, which correspond to a stretch of untranslated amino acids in frame with the receptor that facilitates future applications, such as the generation of N-terminal SNAP-tag fusion constructs. The 5′- and 3′-adaptors contained additional restriction sites for subcloning purposes. TH1006 was then codon-optimized using the codon usage in Homo sapiens using GeneArt (Invitrogen, Waltham, MA). We protected the restriction sites in the construct during the codon optimization. TH1006 was inserted into the pcDNA3.1(+) plasmid via double digestion with NheI and NotI enzymes.
The pairs of identical restriction sites (KpnI, EcoRV, and BstEII) were included to facilitate the removal of several modular elements in the synthetic gene. In this study, we used the KpnI sites to remove the alternative C-terminal epitope tag pair (OLLAS (2-14)-1D4-Stop) and to obtain the construct TH1007. To generate TH1007, we digested TH1006 with KpnI and purified the longer fragment, re-ligated the fragment using T4 DNA ligase, and transformed TOP10 cells with the ligated reaction product. Single colonies were screened for the correct re-ligated fragment using DNA sequencing. The TH1007 construct encodes hCCR5 followed by an eight-residue spacer (TETAST-GT), two OLLAS-tags (20) separated by a 10-residue spacer (SGGG-DI-SGGG), followed by another 10-residue spacer (DI-TGAGS-GSP), Strep-III (the double Strep-tag separated by a 12-residue spacer (GGGS)4) (21), a three-residue spacer (GSP), and the 18-residue 1D4-tag (22). The double OLLAS-tag and the Strep-III-tag are included for future applications to enable alternative affinity purification and surface immobilization schemes.
To introduce the SNAP-tag downstream CCR5 and generate TH1008 (Fig. S1), we performed a double digestion of TH1007 and the pSNAPf plasmid (New England Biolabs) with AgeI and BamHI. BamHI and AgeI flank the SNAPf sequence in the pSNAPf plasmid backbone, leading to a 570 bp fragment. The 570 bp fragment encoding the SNAPf sequence was isolated and ligated into the AgeI-BamHI double-digested TH1008 backbone using T4 DNA ligase. TOP10 cells were transformed with the ligation reaction, and single colonies were screened for the correct ligated construct. Western blot analysis of TH1008 expression in HEK293T cells showed multiple bands, suggesting that the expressed protein was partially degraded. To overcome this issue and generate a homogeneous product suitable for FCCS ligand-binding measurements, we devised a strategy to incorporate the cleavable signal peptide from the murine 5HT3A serotonin receptor and a FLAG-tag (SP-FLAG) sequence upstream TH1008.
The FLAG and 1D4 epitopes allowed us to perform a tandem affinity purification to isolate the full-length receptor from truncations. We introduced the cleavable signal peptide to ensure proper translocation of the N-terminal FLAG epitope. The signal peptide and FLAG sequences were encoded without linker between them. The SP-FLAG module was designed as part of a longer construct (TH1031) that will be used as a template to swap tags for future applications (Table S3). The construct was codon-optimized for expression in H. sapiens using GeneArt (Invitrogen). The synthetic gene of TH1031 was inserted into the pUC57 plasmid using double digestion with NheI and XmaI. To introduce the SP-FLAG sequence into the CCR5 constructs, we digested TH1031 with MlyI and purified the 165-bp blunt-ended fragment for use as a “megaprimer” using agarose gel electrophoresis. To obtain an unmethylated megaprimer, we amplified the 165 bp fragment using the Pfx Platinum polymerase (Invitrogen, Grand Island, NY) using the primers listed in Table S4. The SP-FLAG fragment was inserted upstream TH1007 (see Fig. S1) using the QuikChange Lightning mutagenesis kit (Agilent, Santa Clara, CA) with slight modifications. Briefly, 25 ng of TH1007 was added to 250 ng of purified SP-FLAG in the presence of dNTPs, QuikChange buffer, and polymerase in 25 μL total volume. The PCR reaction was cycled using the parameters as described by Agilent. XL10-Gold cells were then transformed with the PCR reaction per Agilent’s instructions. Single colonies were isolated and screened for the correct construct using DNA sequencing. The construct derived from TH1007 with the SP-FLAG upstream CCR5 is called TH1040.
To introduce SNAPf into TH1040, we performed a double digestion of TH1040 and TH1008 using HindIII and KpnI. We resolved the desired DNA fragments using agarose gel electrophoresis and purified them using DNA extraction centrifugal filter units (Millipore, Billerica, MA). The 1204 bp fragment from TH1040 was inserted into the TH1008 backbone while retaining the sequence in-frame. The sequences were ligated using T4 DNA ligase per the manufacturer’s instructions. TOP10 cells were transformed with the ligation reaction, and single colonies were isolated and screened for the correct construct using DNA sequencing. We refer to the new construct TH1030 with the SP-FLAG sequence upstream of CCR5 in TH1008 subsequently as CCR5-SNAP (see Fig. S1).
Cell culture and transfection
HEK293T cells (passage number 5 to 15; ATCC, Manassas, VA) were maintained in DMEM-Q, 1% penicillin-streptomycin, and 10% fetal bovine serum (FBS; Atlanta Biologicals, Flowery Branch, GA). Unless otherwise noted, transient transfections including high-throughput in-plate transfections were performed using Lipofectamine 2000 according to manufacturer’s instructions, with some modifications as described previously (23). Total transfected plasmid DNA was kept constant by adding empty vector pcDNA3.1(+) when necessary. The total plasmid DNA in all our experiments was 8 μg in 10-cm dishes, 2 μg in six-well plates, 100 ng in 96-well plates, and 20 ng in 384-well plates.
Flow cytometry
HEK293T cells were transfected in six-well plates with 0.75 μg CCR5 WT, 2.0 μg CCR5-SNAP, or 2 μg of empty vector pcDNA3.1(+). Cells were detached in ice-cold phosphate-buffered saline (PBS). Cells were then distributed in 96-well round-bottom plates, spun down, and resuspended in BRET buffer (PBS with 0.5 mM MgCl2 and 0.1% BSA) containing anti-CCR5 mAb (Clone 2D7), anti-CCR5 PE (Clone T21/8), or anti-FLAG PE for 45 min at 4°C. Cells were then washed three times in ice-cold PBS. Cell surface expression was quantified by flow cytometry using the Accuri C6 flow cytometer (BD Biosciences, San Jose, CA).
Adenylyl cyclase activity
HEK293T cells were co-transfected in a high-throughput in-plate manner with 12 ng RLuc3-EPAC-GFP10, a BRET2 cAMP sensor (a gift from Dr. Bouvier, Université de Montréal) and 23 ng CCR5 WT or 60 ng CCR5-SNAP or 88 ng of empty vector. The total DNA was adjusted with the empty vector pcDNA3.1(+) to 100 ng when necessary. Cells were then plated into 96-well, white microplates with clear bottoms coated with 0.1 mg/mL poly-D-lysine at a density of 100,000 cells/well. 24 h post-transfection, media was replaced with BRET buffer. Coelenterazine 400A was added at a final concentration of 5 μM, followed by a 5 min incubation at room temperature. Cells were then stimulated with ligand in the presence or absence of 5 μM of forskolin at room temperature for 5 min. Luminescence and fluorescence readings were collected using the Synergy NEO2 plate reader from Biotek (Winooski, VT) and Gen5 software. BRET2 readings between Rluc3 and GFP10 were collected by simultaneous integration of the signals detected in the 365–435 nm (Rluc3) and 505–525 nm (GFP10) windows. BRET2 ratios were calculated as described previously (24, 25). Dose-response curves were fitted using a three-parameter logistic equation (Eq. 1) in GraphPad Prism:
| (1) |
where x is the logarithm of agonist concentration, y is the response, bottom is the bottom plateau, top is the top plateau (also known as Emax), and EC50 is the effective concentration that yields 50% response.
Calcium flux assay
For each well of a 384-well plate, 20,000 HEK293T cells in 20 μL DMEM were transfected with 7.5 ng CCR5 WT or 20 ng CCR5-SNAP. Total DNA was kept constant at 20 ng by adding empty vector when necessary. Transfected HEK293T cells were plated into 384-well plates (Corning) coated with poly-D-lysine hydrobromide (Sigma-Aldrich) at 20 μL/well. 24 h post-transfection, 20 μL/well FLIPR calcium 6 dye (Molecular Devices, Sunnyvale, CA) was added to the cells and incubated for 1.5 h at 37°C with 5% CO2. The dye was dissolved in HBSS-H (Hank’s Balanced Salt Solution with 20 mM HEPES (pH 7.4)) and supplemented with 0.4% BSA. Before measurement, the plate was incubated at 37°C for an additional 30 min in a prewarmed FlexStation II 384 Plate Reader (Molecular Devices, San Jose, CA). Ligands at a 5× final concentration were diluted in HBSS-H supplemented with 0.2% BSA. Fluorescence readings were collected using the FlexStation plate reader with excitation at 485 nm, emission at 535 nm, and dichroic mirror at 525 nm. The FlexStation took measurements over a 100 s time course, with 10 μL of ligand added to the cells 20 s after the start of measurement. Relative fluorescence units (RFU) are reported as the peak magnitude signal subtracted by the basal signal in each well. Dose-response curves were fitted using the same 3-parameter logistic equation (Eq. 1) employed to fit the dose-response data from cAMP inhibition experiments.
Expression, labeling, and purification of CCR5-SNAP
Ten 100 × 20-mm polystyrene dishes were plated with HEK293T cells at 4.0 × 106 cells/dish in DMEM-Q + 10% FBS. 24 h postplating, 100 μL of Plus Reagent was mixed with 80 μg of CCR5-SNAP in 7.5 mL of DMEM-Q. In a separate vessel, 170 μL of lipofectamine reagent was mixed with 5 mL of DMEM. After 15 min, the transfection solutions were mixed and incubated for an additional 15 min. Media was removed from HEK293T cells and substituted with 2.8 mL of DMEM-Q. 1.2 mL of the transfection solution was added to each plate, and the cells were incubated for 4 h before supplementing the media with 4 mL of DMEM-Q + 20% FBS. 24 h post-transfection, media was removed from the cells, and cells were harvested in 2 mL/dish of PBS and 1 mM phenylmethylsulfonyl fluoride. Cells were pelleted in a 50 mL vessel at 1500 rpm using a Beckman GS-6R centrifuge (Beckman Coulter, Atlanta, GA) at 4°C for 5 min. The harvesting solution was removed, and the cell pellet was solubilized in 5 mL of buffer L (20 mM HEPES (pH 7.4), 0.1 M (NH4)2SO4, 1 mM CaCl2, 5 mM MgCl2, 10% glycerol, 0.1% cholesteryl hemisuccinate [CHS], 1.0% n-dodecyl-β-D-maltopyranoside [DDM], 1.0% 3-[(3-cholamidopropyl)-dimethylammonio]-1-propane sulfonate] [CHAPS]) supplemented with protease inhibitor cocktail (cOmplete EDTA-free; Sigma-Aldrich) for 2 h at 4°C. Cell lysates were then centrifuged at 55,000 rpm for 30 min, 4°C, using a TLA 100.3 rotor rpm using a Beckman TL-100 tabletop ultracentrifuge. The supernatant was added to 600 μL of 50% slurry 1D4 mAb Sepharose 2B resin and incubated overnight at 4°C. Resin was pelleted in a GS-6R for 5 min, 2000 rpm, 4°C and then transferred to an Ultrafree-MC-HV Durapore polyvinylidene difluoride (PVDF) 0.45 μm centrifugal unit (Sigma-Aldrich). CCR5-SNAP was labeled in 400 μL of buffer N (20 mM HEPES (pH 7.4), 0.1 M (NH4)2SO4, 1 mM CaCl2, 5 mM MgCl2, 10% glycerol, 0.07% CHS, 0.33% DDM, 0.33% CHAPS, 0.018% 1,2-dioleoyl-sn-glycero-3-phosphocholine [DOPC], 0.008% 1,2-dioleoyl-sn-glycero-3-phospho-L-serine [DOPS]) with 50 μM SNAP substrate and 1 mM 1,4-dithiothreitol (DTT) for 30 min at room temperature. Resin was then washed 3 × 0.5 mL in buffer N for 30 min each at 4°C. CCR5-SNAP was eluted from the 1D4 resin by incubating the sample with 1D5 peptide in buffer N (0.33 mg/mL) twice for 30 min on ice and eluting by centrifugation. 1D4 purified CCR5-SNAP was added to 100 μL of FLAG M2 resin and incubated overnight at 4°C. FLAG resin was transferred to a separate Durapore spin filter and washed three times with 0.5 mL of buffer N for 30 min each at 4°C. CCR5-SNAP was eluted by incubating the resin twice with 100 μL of buffer N and FLAG peptide (200 μg/mL) for 30 min on ice. FLAG-purified CCR5-SNAP was loaded into a Superdex 200 10/300 GL column (Sigma-Aldrich) previously equilibrated with buffer N and 0.1 mg/mL BSA (IgG-free). CCR5-SNAP was eluted over one column volume into 0.5 mL fractions. Near-infrared (NIR)-immunoblotting and FCS were employed to analyze the size-exclusion chromatography (SEC) fractions.
Immunostaining and TIRF microscopy
HEK293T cells were plated onto 35 mm glass-bottom (1.5) MatTek plates (Ashland, MA) coated with poly-D-lysine hydrobromide at 300,000 cells per dish. Cells were transfected with CCR5 WT (0.75 μg), CCR5-SNAP (2.0 μg), or pcDNA3.1(+). (2.0 μg) at a total DNA/dish ratio of 2.0 μg using Lipofectamine 2000 per manufacturer’s instructions. 24 h post-transfection, media were aspirated from the plates, and cells were washed with 1 × 2 mL of PBS supplemented with Ca2+ and Mg2+ (Ca/Mg). Cells were then permeabilized with 1 mL of cold methanol for 5 min at −20°C. Cells were then washed with 3 × 1 mL of cold PBS (Ca/Mg) before blocking overnight in 0.5% BSA in PBS (Ca/Mg) at 4°C. Blocking solution was removed, and 1D4 monoclonal antibody at a dilution of 1:2000 in 0.5% BSA-PBS (Ca/Mg) was added for 1 h at room temperature. Cells were then washed with 3 × 1 mL of PBS (Ca/Mg). Secondary antibodies conjugated to Alexa-488 were added at a final dilution of 1:500 in 0.5% BSA-PBS (Ca/Mg) for 1 h at room temperature. Cells were washed again with 3 × 1 mL of PBS (Ca/Mg), and then Fluoromount-G mounting media (SouthernBiotech, Brimingham, AL) containing DAPI was added to the cells. Cells were visualized on a Nikon TiE inverted total internal reflection fluorescence (TIRF)-FLIM microscope (Nikon, Tokyo, Japan) using an Apo TIRF 100× oil N2 objective (NA 1.49). Images were collected on an Andor NEO sCMOS camera (Andor, Belfast, UK) using 405- and 488-nm excitation with a total exposure of 150 ms per image. Images were acquired at room temperature using the following dimension order: XYCZT, which are 2048, 2048, 3, 1, and 1 pixels, respectively. Filters used were 525/50 and 450/40. Images were processed using ImageJ and Adobe Illustrator.
SDS-PAGE analysis and NIR-immunoblotting
Sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) samples were mixed with DTT at 150 mM final concentration and NuPAGE loading buffer. Samples were loaded into a NuPAGE 4–12% Bis-Tris gel in MES-SDS buffer. Electrophoresis was conducted at a constant voltage of 115 V. The gel was removed from the cassette and rinsed in water before equilibrating in Western transfer buffer (48 mM Tris, 39 mM glycine, 1.3 mM SDS, 20% MeOH (pH 9.2)). One piece of Immobilon PVDF membrane-Fl was incubated for 1 min at room temperature in 100% MeOH. The PVDF membrane and two pieces of extra thick blot papers (Bio-Rad, Hercules, CA) were rinsed in Western transfer buffer. Western transfer was performed in a semidry apparatus for 45 min with a constant voltage of 18 V. After electrophoresis, the membrane was placed in 10 mL of Odyssey blocking buffer (PBS) and incubated for 1 h at room temperature. The membrane was then placed in 10 mL of blocking buffer with anti-1D4 mouse monoclonal (1:1000), anti-FLAG rabbit polyclonal (1:1000) antibodies, and 0.2% Tween-20. The membrane was incubated overnight at 4°C. Membrane was then washed 5 × 5 min in 1× PBS-T (0.1% Tween-20). Membrane was incubated for 1 h at room temperature in 10 mL blocking buffer supplemented with 0.2% Tween-20, 0.01% SDS, goat anti-mouse IRDye680RD (1:10,000), and goat anti-rabbit IRDye800CW (1:10,000). Membrane was washed again 5 × 5 min in 1× PBS-T and then 2 × 5 min in 1× PBS buffer. Membranes were visualized using a LI-COR Odyssey SA using 100 μm resolution and intensity level 7 for both 700 and 800 nm excitations. Images were processed using Image Studio Lite Version 4.0 and ImageJ. For the line scan analysis, a rectangle of 45 × 120 pixels was drawn around the desired gel lane and set as First Lane under Analyze, Gels, in ImageJ. The command “Plot Lanes” was then selected with vertical and horizontal scale factors set to 1.0 with uncalibrated optical density. Using the magic wand, an area under the curve was selected and saved as x and y coordinates for replotting in GraphPad Prism 7.
Synthesis of fluorescently labeled chemokines
Fluorescent chemokines were prepared by total chemical synthesis. First, N-terminal and C-terminal fragments corresponding to residue numbers 1–33 and 34–68, respectively, were made by polymer-supported organic synthesis using Boc chemistry. The C-terminal fragment was common to all chemokines and consisted of RANTES (34–68). The C-terminal fragment additionally carried the K(S)G sequence, in which the serine was coupled to the ε-position of the lysine, as a C-terminal extension. After deprotection, cleavage from the resin, and purification, N-terminal fragments were coupled to the extended C-terminal fragment by native chemical ligation under previously described conditions (10). Crude products were analyzed by high-performance liquid chromatography (HPLC) and electrospray ionization mass spectrometry (ESI-MS) before protein refolding and formation of the intramolecular disulfide bridges. The refolded material was then further analyzed by HPLC on a C8 column (shorter retention time; Aeris WIDEPORE 3.6u XB-C8; Phenomenex, Torrance, CA) and ESI-MS (lower mass due to disulfide bridge formation; instrument: Bruker Esquire 3000+ ion trap mass spectrometer; Bruker, Billerica, MA).
To introduce a reactive handle for chemical derivatization of the chemokines, the serine residue on the C-terminal extension was selectively oxidized to yield a glyoxylyl residue, whose aldehyde reacts efficiently and specifically with aminooxy compounds. The oxidation was performed with 10 equivalents of NaIO4 in 1% NH4HCO3 buffer at pH 7.2 in the presence of 50 equivalents of methionine (26). The mixture was left to react in the dark for 10 min before the reaction was quenched by the addition of a 10,000-fold excess of ethylene glycol. After 10 additional minutes, the solution was acidified and the chemokines were isolated on a C18 Sep-Pak column (Waters Corporation, Milford, MA) and analyzed by ESI-MS (for PSC, expected: 8134.5, found: 8135.3 ± 0.8; 5P12: expected 8181.7, found 8181.2 ± 0.8; 6P4: expected 8118.5, found 8118.3 ± 0.4; 5P14: expected 8186.0, found 8185.8 ± 0.6; RANTES: expected 8088.3, found 8088.0 × 0.3). The material was then freeze-dried. To prevent oxidation of the N-terminal serine of RANTES during the procedure, serine was temporarily protected with a 2-(methylsulfonyl)ethyl oxycarbonyl group (27). The protection was removed by dissolving the compound in a 1:1 water/dimethylformamide mixture, cooling it to 0°C, and then adding NaOH at a final concentration of 0.5 M. After 30 s, acetic acid was added to the solution, and the material was isolated again on a C18 Sep-Pak column.
To couple Alexa-647 to the chemokines via strain-promoted azide-alkyne cycloaddition reaction, a bifunctional linker carrying on one side an aminooxyacetate (AOA) moiety and on the other side an azide was synthesized. First, desalted Boc-protected AOA dicyclohexylamine was converted into its N-hydroxysuccinimidyl (NHS) ester derivative following standard procedures (28). Briefly, 2.35 mmol of the amino acid were left to react overnight with 2.4 mmol of N-hydroxysuccinimide and 2.4 mmol of N,N′-dicyclohexylcarbodiimide in 10 mL ethyl acetate. The mixture was then filtrated, dried, resuspended in dichloromethane, and dried again. 2.23 mmol of Boc-protected AOA NHS ester were recovered. Subsequently, 1 mg of Boc-protected AOA NHS ester in 2 mL of dichloromethane was slowly added to 5 mmol of ethylenediamine (en) and left to react for 3 h. Boc-AOA-en was then isolated by HPLC on a C8 column (87 mg of product recovered). 75 mg (0.22 mmol) of this compound was further reacted for 24 h with 72 mg (0.22 mmol) of Boc-3-azidoalanine (Ala(N3)) NHS ester (made from Boc-3-azidoalanine and N-hydroxysuccinimide according to the procedure described above) in 4 mL of acetonitrile in the presence of 0.66 mmol N-methylmorpholine. The desired linker with the overall structure Boc-AOA-en-Ala(N3)-Boc was isolated by HPLC on a C8 column and its mass confirmed by ESI-MS (expected: 445.8; found: 446.3 (M+H)). Boc-AOA-en-Ala(N3)-Boc was then deprotected in trifluoroacetic acid for 10 min, dried, analyzed by ESI-MS, and resuspended in water. 20 equivalents of AOA-en-Ala(N3) were added to 150 nmol of the different oxidized chemokines at a concentration of 100–200 μM and left to react overnight in 90 mM sodium formate buffer at pH 3.0. The product of the reaction (140 nmol) was isolated on a C18 Sep-Pak column.
Dibenzocyclooctyne (DIBO)-modified Alexa Fluor 647 (DIBO-AF647, C10408; Life Technologies) was dissolved at 5 mM concentration in dimethyl sulfoxide. The chemokines derivatized with the Ala(N3) linker were labeled with DIBO-AF647 by incubating a mixture of protein (300–400 μM) with 1.8 reactive dye equivalents and letting the reaction proceed at room temperature overnight. The labeled protein was recovered on an S-2000 column (Phenomenex Biosep-SEC-S-2000) equilibrated in 50% MeOH, and its mass was confirmed by ESI-MS (PSC: expected 9523.8, found 9524.0 ± 0.5; RANTES: expected 9477.5, found 9476.8 ± 0.5; 6P4: expected 9472.6, found 9472.2 ± 0.2; 5P12: expected 9535.7, found 9535.2 ± 0.3; 5P14: expected 9593.1, found 9591.2 ± 0.3). The absence of a peak in the MS spectrum at the mass of the unlabeled protein further indicated that the labeling reaction was quantitative. Fig. S2 shows the chemical structure of the C-terminal extension on lysine for all the chemokines synthesized in this study.
Ligand-binding assays
Saturation ligand-binding assays were set up in PCR tubes by serially diluting the ligand in buffer N supplemented with 0.1 mg/mL BSA (IgG-free, detergent-free). CCR5-SNAP-A488 was then added in equal volume for a total reaction volume of 20 μL. Samples were equilibrated at room temperature for 4 h protected from ambient light. 15 μL of each sample were loaded into individual wells of a 384-well plate previously blocked with 1.0 mg/mL BSA (IgG-free, detergent-free) in water for 15 min at room temperature. To prevent sample evaporation, 5–10 μL of paraffin oil was applied to the top of each sample. Competition-binding assays were set up in a similar fashion except that the labeled chemokine was kept at constant concentration and the competitor was serially diluted. 5 μL of the labeled chemokine was mixed with 5 μL of nonlabeled chemokine, and then 10 μL of CCR5-SNAP-A488 was added for a 20 μL total reaction volume. Samples were equilibrated for ≥16 h at room temperature before imaging by FCCS. For competition with the sCD4-gp120 complex, sCD4 and gp120 were incubated for 1 h at a molar ratio of 10:1, respectively, and a final complex concentration of 20 μM. Complex was then serially diluted in buffer N before adding labeled 5P12- or 6P4-647 and CCR5-SNAP-488. Samples were then incubated for ≥16 h at room temperature before FCCS measurements.
Fluorescence correlation and cross-correlation spectroscopy measurements
Samples were loaded into #1.5 glass-bottom 96- or 384-well black plates (SensoPlate, black, 384 well reference number: 788892, 96 well plate reference number: 655892; Greiner Bio-One, Monroe, NC) and mounted on an inverted laser scanning confocal microscope LSM 780 (Zeiss, Oberkochen, Germany). Alexa-488 was excited using an argon 488-nm laser at 0.2 or 0.8% transmission, and Alexa-647 was excited using a helium-neon 633-nm laser line at 1.0% transmission. Laser excitation was focused into the sample by using a 40× C-Apochromat NA 1.2 water immersion objective. The correction collar of the objective was adjusted to 0.17 mm and room temperature. To minimize distortion of the point spread function due to glycerol in the solution, the excitation volume was focused exactly 50 μm above the glass-buffer interface, which was identified by performing a line scan using reflected light from the 488 nm laser line. For 488-nm excitation, a 488-only main beam splitter was used, and for 633 nm and dual excitation, a main beam splitter 488/561/633 was used. Emission from Alexa-488 was collected in the range of 516–596 nm using a GaAsP detector and emission from Alexa-647 in the range of 650–694 nm using a separate GaAsP detector. Pinholes for both excitations were set to 1.0 airy units and aligned along the xy plane using a solution of free dye or the sample itself. Count-rate binning time was set to 1 ms, and the correlator binning time was set to 0.2 μs. Count rates were never greater than 500 kHz, and traces showing large deviations from the average or traces with decaying or increasing fluorescence were manually removed from the analysis. Counts per minute values were between 1 and 16 kHz for all measurements to avoid optical saturation while maximizing counts above background. For single-dye measurements, 10 repetitions of 10 s each were collected and averaged, whereas for receptor-ligand binding experiments, 50 repetitions of 30 s each were collected and averaged.
Correlation traces model fitting
FCS and FCCS raw traces were fitted using the ZEN software. Correlation and cross-correlation traces were fitted using equations that modeled the diffusion, triplet, and/or blinking processes of the fluorophores and labeled proteins. For diffusion, we assumed that the fluorescent species undergo free three-dimensional (3D) translational diffusion. To model free 3D translational diffusion, we employed the following relation
| (2) |
where τ is the correlation time arising from diffusion. Equation 2 holds true in the case in which there is a single fluorescent species. The suffix i can correspond to g, r, or x, representing the three channels green, red, and cross correlation. Interchangeably, we sometimes use for clarity the labels R, L, or X to designate the green channel for the receptor labeled with Alexa-488, the red channel for the ligand labeled with Alexa-647, and the cross-correlation channel. Triplet state transitions were fitted using the following equation:
| (3) |
where Ft,i is the fraction of fluorescent species in the triplet state and τt,i is the triplet state relaxation time of the fluorophore. For Alexa-488, τt,i was set to a constant value of 4 μs, and for Alexa-647, τt,i was set to a constant value of 7 μs. Blinking state transitions were only observed when the Alexa-647 labeled chemokines were added to CCR5-SNAP-488. To model blinking state transitions, we employed the following equation,
| (4) |
where Fb,i is the fraction of fluorescent species in the blinking state and τb,i is the blinking state relaxation time of the fluorophore. For Alexa-647, Fb,i and τb,i were fitted as free parameters.
Autocorrelation and cross-correlation traces were analyzed from 2 μs to 10 s to remove afterpulsing artifacts from the detectors. For single diffusing Alexa-488 and CCR5-SNAP-488, we modeled the correlation traces using the following equation:
| (5) |
where Ag is the correlation amplitude for when Gg(τ) = 0, Dg(τ) is the diffusion correlation given by Eq. 2, Tg(τ) is the triplet correlation given by Eq. 3, and Eq. 1 is an offset. Ag, the correlation amplitude, is given by
| (6) |
For single diffusing Alexa-647 and Alexa-647-labeled chemokines, the correlation traces were modeled using the following equation:
| (7) |
where Ar is the correlation amplitude for when Gr(τ) = 0 and Br(τ) is the blinking correlation given by Eq. 4. In the absence of CCR5-SNAP-488, Alexa-647 blinking is not observed, and the correlation amplitude was fitted using Eq. 5. Ar, the correlation amplitude, is given by
| (8) |
To fit the cross-correlation traces, we employed the following equation:
| (9) |
where the cross-correlation amplitude, Ax, is given by
| (10) |
Note that the cross-correlation amplitude Ax has a unique dependence on particle numbers (Eq. 10). The amplitude increases with increasing numbers of double-labeled particles Nx, but it decreases with increasing numbers of either green- or red-labeled particles, Ng or Nr. In contrast, Eqs. 6 and 8 show that the autocorrelation amplitudes Ag and Ar both decrease with increasing Ng or Nr, respectively.
Equations 3 and 4 are normalized, which means that the correlation amplitude and the calculated number of particles (Eqs. 6, 8, and 10) are independent of fluctuations because of triplet states and blinking. For all three channels, γ is set to 0.35 and the structural parameter to 8. To estimate errors for each measurement, the total repetitions were divided into three independent sets of measurements and each set was averaged and analyzed using the equations above. From these three averages, the SD was calculated for the number of particles. In those cases in which τD,x for the complex deviated significantly from previously measured values, it was fixed to 550 μs so that the fit would converge. From the calculated number of particles, we derived concentrations using Eqs. 14, 15, and 16 below, which were used for global analysis of ligand binding.
Confocal volume determination
Solutions of Alexa-488 and Alexa-647 as cadaverine conjugates were diluted in buffer N at various concentrations from 25 to 0.8 nM. FCS measurements were acquired as described above. The number of particles derived from the fits of the correlation traces was plotted as a function of concentration (Fig. S3). The concentration of the Alexa-488 and Alexa-647 stock solutions used for the dilutions (nominally 10 μM) was determined by ultraviolet–visible (UV-Vis) spectroscopy using the extinction coefficients 73,000 M−1 cm−1 and 245,000 M−1 cm−1, respectively. The UV-Vis-derived concentration was used instead of the nominal fluorophore concentrations for calculating the concentrations of the calibration solutions. To determine the confocal volume for the cross-correlation channel, we employed a 40 bp DNA duplex dual-labeled with Alexa-488 and Alexa-647 at both ends to minimize FRET, similar to the original design from Schwille et al., who used rhodamine green and Cy5 as fluorophores (29).
The oligos were synthesized by IDT. The forward sequence for the oligonucleotide is 5′-[AminoC6Alexa488]GCCGTCTCTGACTGCTGATGACTACTATCGTATAGTGCGG[BioTEG-Q]-3′, and the sequence for the reverse oligonucleotide is 5′-[AminoC6Alexa647]CCGCACTATACGATAGTAGTCATCAGCAGTCAGAGACGGC-3′. Single-strand oligonucleotides were resuspended in 10 mM Tris (pH 8.0), 50 mM NaCl, 1 mM EDTA to a final concentration of 1 μM in 100 μL of buffer. Samples were added to a water bath at 94°C in a Dewar flask and allowed to anneal until the temperature in the water bath reached less than 40°C. Oligos were then placed at room temperature while a C4 column was equilibrated with 0.1 M ammonium acetate at pH 6.6. Oligos were loaded into the column and eluted with a 0–50% (v/v) gradient of acetonitrile in water. HPLC fractions were analyzed by UV-Vis absorbance for both Alexa-488 and Alexa-647. The peak fraction was aliquoted into 50 μL aliquots and stored at −20°C for long-term storage. The final concentration of the oligo was 110 nM. Stock solutions were employed to dilute the oligonucleotide as done for the free dyes.
We determined the confocal volumes (Vg, Vr, or Vx) using the observed number of particles (Ng, Nr, or Nx) from the linear fits of the experimentally observed number of particles as a function of concentration (Cg, Cr, or Cgr) (Eqs. 11, 12, and 13):
| (11) |
| (12) |
| (13) |
The slope of the linear fit divided by Avogadro’s number yields the confocal volumes (see Fig. S3). The following relations for the green and red channels relate the sample concentrations to the correlation amplitudes and the number of particles derived from the amplitudes:
| (14) |
| (15) |
where Cg and Cr are the concentrations of the green and red particles, respectively, and Cgr is the concentration of the double-labeled species. In the case in which there is only a single fluorescent species or no binding, the Cgr term vanishes to 0. NA is Avogadro’s number; Vg and Vr are the confocal volumes of the green and red channels, respectively; Ag and Ar are the correlation amplitudes for the green and red channels, respectively; Ng and Nr are the number of particles of the green and red particles, respectively; and γ is a correction factor to account for the fact that the confocal volume deviates from the 3D Gaussian approximation.
For the cross correlation, we employed the following relation for the concentration,
| (16) |
where Vx is the cross-correlation confocal volume, Ax is the cross-correlation amplitude, and Nx is the number of particles that are double-labeled.
The confocal volume is related to its dimensions by the following relations for the green and red channels:
| (17) |
| (18) |
Vg,eff and Vr,eff are the effective confocal volumes of the green and red channels, respectively; rg and rr are the radii of the confocal volumes along the xy plane; and Sg and Sr are the structural parameters of the green channel and red channels, respectively. The structural parameter is defined S = rz/rxy, where rz and rxy are the radii along the z and xy planes, respectively, for each channel.
Because the cross-correlation confocal volume is defined as the overlap between the green and red confocal volumes, then the equation describing the cross-correlation confocal volume is given by
| (19) |
Equation 19 is only valid for the case in which there are no chromatic aberrations in the system. The radii of the confocal volumes relate the diffusion coefficients to the observable diffusion times:
| (20) |
| (21) |
| (22) |
where τD,i is the diffusion time of species i and D is the diffusion coefficient of the same species.
Cross talk determination
Cross talk from the green channel to the red channel was determined using the protocol by Bacia and Schwille (17). Briefly, 25 nM Alexa-488 in buffer N was excited using 488-nm laser line and the count rates were recorded using a GaAsP detector in the green channel and simultaneously using a separate GaAsP detector in the red channels. Count rates from the two measurements were used to calculate the bleedthrough ratio:
| (23) |
Under our experimental conditions, the bleedthrough ratio is 0.0072. In a sample containing dual-labeled oligonucleotide or CCR5-SNAP-488 and labeled chemokine, the ratio of the measured green/red count rates was taken and then multiplied with the bleedthrough ratio calculated previously to obtain the cross-correlation amplitude relative to the green autocorrelation as expected from cross talk only.
| (24) |
We obtained a value of 0.0033, which is less than 0.4% of the observed relative cross correlation not corrected for cross talk G0,x/G0,g. Therefore, cross talk plays only a minor role and can be neglected unless otherwise noted. In cases with substantial bleedthrough, the cross-talk-corrected relative cross correlation may be obtained following Eq. 25:
| (25) |
Global fitting analysis of saturation- and competition-binding curves
For ligand-binding assays, we assumed that the chemokines recognize a single binding site on CCR5. Based on this, we then set the concentration of complexes, Cgr, equal to
| (26) |
where [RL] is the total concentration of receptor-ligand complexes. In the green channel, the calculated number of particles arise from both receptor and receptor-ligand complexes. Therefore,
| (27) |
where [Rnf] is equal to the concentration of nonfunctional receptor species, [R] is the concentration of ligand-free receptor, [RC] is the concentration of receptor-competitor complex, [RL] is the concentration of receptor-ligand complex, and [Rt] is the total concentration of receptor. In our experiments, the Alexa-488-labeled receptor R is observed in the green channel and the Alexa-647-labeled ligand L is observed in the red channel, hence AR = Ag and AL = Ar. For consistency, we use an uppercase letter to designate the cross-correlation amplitude in the experiments with AX = Ax. To define the observable fractional occupancy [RL]/[Rt], we employ the following relations derived from Eqs. 14, 16, and 26 to 27:
| (28) |
which shows that fractional occupancy can be derived from the ratio Nx/Ng. Alternatively, the fractional occupancy is can also be derived from the ratio Ax/Ar. Note that the ratio of the number of particles used the cross-correlation and green channels, whereas the ratio of the amplitudes uses the cross-correlation and red channels. Interestingly, the cross-correlation amplitude AX is directly proportional to the concentration of receptor-ligand complex [RL] and indirectly proportional to both the total ligand and the total receptor concentration, whereas the autocorrelation amplitudes AL and AR are both indirectly proportional to the total ligand and the total receptor concentrations. The ratio AX/AL eliminates the ligand concentration dependence, and it is proportional to the fractional occupancy of the receptor [RL]/[Rt].
To account for the presence of a nonfunctional receptor species, we define [Rnf] as follows:
| (29) |
where c is a constant to define the fraction of receptor that is functional. Therefore, we can state that
| (30) |
The fractional occupancy for ligand binding to the functional receptor becomes
| (31) |
and the fractional occupancy for competitor binding to the functional receptor is
| (32) |
In the red channel, we observe fluorescence from the free ligand and the receptor-ligand complex; therefore, the concentrations of each are given by
| (33) |
where [Lt] is the total concentration of ligand.
For a single binding site, the equilibrium dissociation and inhibition constants are defined as follows:
| (34) |
| (35) |
where Kd is the equilibrium dissociation constant of the ligand L and Ki is the equilibrium dissociation constant of the inhibitor C. [C] and [L] are the free concentrations of competitor and ligand, respectively. [R] is the free concentration of the receptor. [RL] is the concentration of the receptor-ligand complex RL. [RC] is the concentration of the receptor-competitor complex RC.
Using the expressions above, we derived the following equations for fractional occupancy in terms of free ligand, free competitor, Kd, and Ki:
| (36) |
| (37) |
Because the FCCS observables for concentrations are the total number of species for each channel, we developed an iterative algorithm to calculate the concentrations of free ligand and competitor by correcting their total concentrations with the concentrations of their receptor-bound complexes. With this algorithm, the concentrations of free ligand, free competitor, equilibrium dissociation constant, and equilibrium inhibition constant are fitted as free parameters on the saturation- and competition-binding isotherms. The algorithm has two parts, a nonlinear least-squares optimization and a self-consistent correction to account for ligand depletion.
The algorithm is repeated multiple times until the free ligand and competitor concentrations converge. For each iteration, we first minimize the reduced χ2 function (Eq. 38) to obtain a set of fitting parameters (Kd, Ki, and cm with m = 1…M):
| (38) |
Here, [RL]/[Rt] is derived from FCCS and FCS measurements, the suffix m is to represent each individual experiment, cm is the active receptor fraction in each experiment m, and the suffix n is to represent each data point per experiment. We set the variance to 1. The parameter o corresponds to an offset to include a small cross-talk correction, which accounts for the fact that the competition-binding isotherms do not fully reach 0 under saturating concentrations of the competitor. The value of o was fixed to 0.01. We initialize the algorithm by setting the free ligand and competitor concentrations to their respective total concentrations:
| (39) |
| (40) |
Next, we use the set of fitting parameters (Kd, Ki, and cm with m = 1…M) to solve the binding equilibrium and to calculate the concentration of ligand-receptor and competitor-receptor complexes . These allow us to re-estimate the free ligand and receptor concentrations as the difference of total and bound concentrations:
| (41) |
| (42) |
To avoid an oscillatory behavior of the algorithm, we update the free concentrations for the next iteration more gradually instead of using these new values directly in the next iteration:
| (43) |
| (44) |
where k → k + 1 is the update of the iteration counter. The parameter α can be used to tune the algorithm. We use α = 0.25 with 20 iterations. Using Eqs. 41, 42, and 44 for a self-consistent correction for ligand depletion, we minimize Eq. 38 in each iteration to yield a converged set of global fitting parameters that optimally fit the data.
Global analysis with nonlinear least-square fitting of the binding isotherms yielded parameters that describe the complete data set. To eliminate any impact of the competition experiments on the dissociation constants obtained from the saturation-binding experiments, we first performed the global analysis of the saturation-binding experiments, and then we used the obtained dissociation constants as fixed parameters in the global analysis of the competition experiments.
To facilitate visualization of data from independent experiments, we accounted for the fraction of functional receptor in each data set and plotted normalized saturation- and competition-binding isotherms as a two-dimensional function (θ, log[L]) or a 3D surface (θ, log[C], L), respectively. To determine the errors associated with each affinity, we performed a statistical bootstrapping error analysis by random data resampling with replacement. We resampled the data 100 times and calculated the means and SDs of all model parameters. In the tables, we report the model parameters determined from the original fit together with the SDs of these parameters from the bootstrap analysis. The means from the bootstrap analysis were always within the error bounds of the original solution.
Results
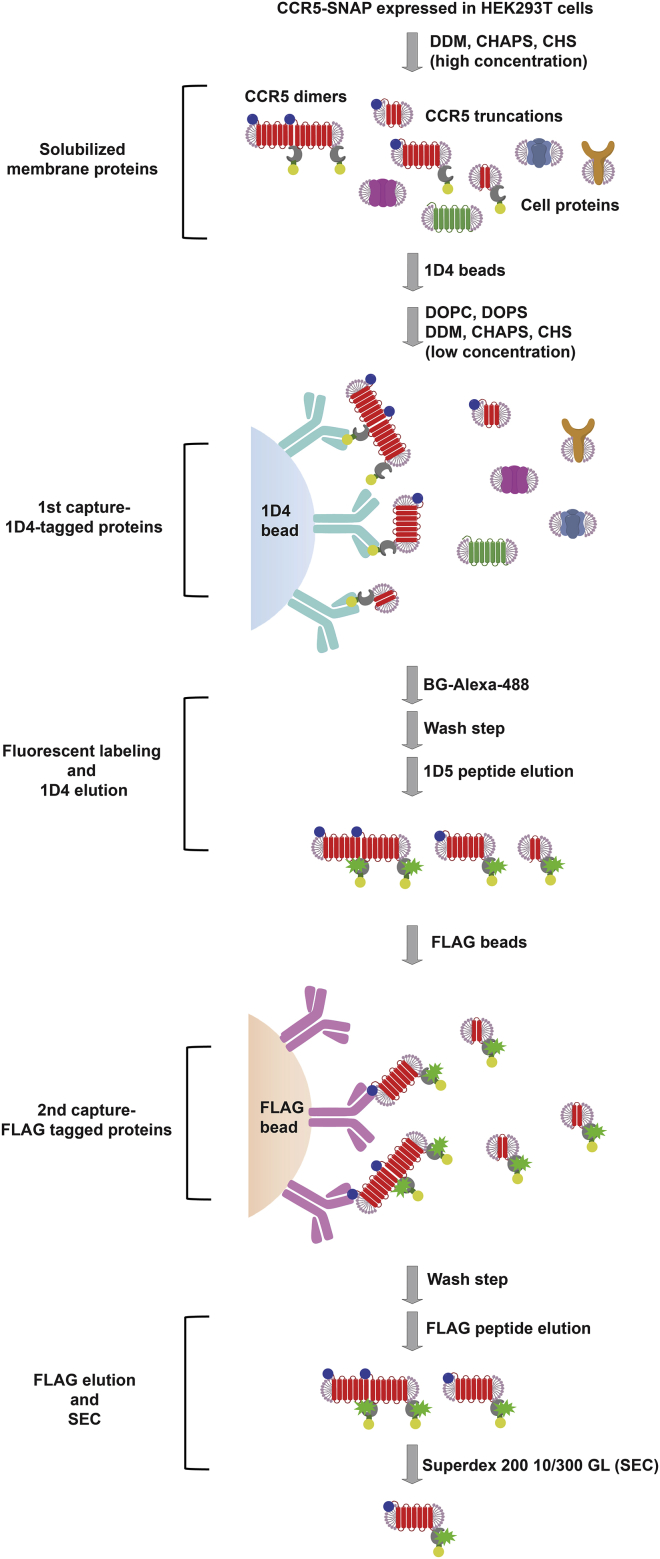
We designed a codon-optimized human CCR5 synthetic gene with a C-terminal SNAP-tag fusion to facilitate high-level expression and covalent fluorescent labeling. We positioned the SNAP-tag on the intracellular C-terminal tail of CCR5 to minimize FRET with the fluorescently labeled RANTES analog ligands that bind at the extracellular surface. Although this construct expresses at higher levels than CCR5 WT in HEK293T cells (Fig. S4), we found that the expressed receptor was subject to proteolytic degradation at the N- and C-terminal tails. To obtain homogeneous full-length receptor for quantitative fluorescence measurements, we modified the CCR5 construct with a cleavable signal peptide from the serotonin 5-HT3A receptor to enhance proper membrane insertion of the receptor with added N-terminal FLAG-tag to allow tandem affinity purification (Figs. 1 a and 2; (30)). Two OLLAS mAb epitope tags (Escherichia coli OmpF linker and mouse Langerin fusion sequence) (20) and the StrepIII-tag (21) were included for future applications, such as receptor immobilization on functionalized surfaces and alternative purification procedures (Fig. 1 a). We analyzed expression of this new construct, referred to as simply CCR5-SNAP, by dual-color NIR-immunoblot analysis and observed that the full-length receptor migrated as a 1D4/FLAG dual-stained (red and green) band with an apparent molecular mass of ∼70 kDa (Fig. 1 b).
Figure 1.
Expression, labeling, and purification of CCR5-SNAP. (a) The CCR5-SNAP construct schematic shows the receptor (blue) fused to the signal peptide (violet) for proper receptor insertion into the membrane, FLAG (cyan) and 1D4 (green) epitopes for tandem affinity purification, double OLLAS (orange) and Strep-III (red) epitopes for optional surface immobilization and purification, and SNAP-tag (yellow) for fluorescent labeling. (b) Reducing SDS-PAGE and NIR-immunoblot of cell lysate, 1D4, and FLAG elutions from the 1D4/FLAG tandem affinity purification are shown. Full-length CCR5-SNAP (∼70 kDa, yellow band) was detected using antibodies against the 1D4 (red) and FLAG (green) epitopes. (c) Representative reducing SDS-PAGE and NIR-immunoblot from one CCR5-SNAP-488 SEC purification are shown. Average values from FCS-derived concentrations for SEC fractions 20–30 are from five independent CCR5-SNAP-488 SEC purifications. Error bars are ± SEM. The purification protocol is shown schematically in Fig. 2.
Figure 2.
CCR5-SNAP purification scheme. Schematic displays the combined tandem affinity and size-exclusion purification strategy to yield full-length, monomeric CCR5-SNAP-488 from mammalian cells. HEK293T cells transiently expressing CCR5-SNAP are lysed in buffered solution containing DDM, CHAPS, and CHS (see Materials and Methods). Cell lysate, which contained cellular proteins, full-length CCR5-SNAP, and CCR5-SNAP truncations, was added to 1D4 resin. Lipids DOPC and DOPS were added to the buffered solution, and then CCR5-SNAP was labeled with the SNAP substrate BG-Alexa-488. Excess fluorophore was washed away, and the labeled receptor was eluted using excess 1D5 peptide. The 1D4 eluate was added to FLAG beads to bind full-length receptors and remove receptor truncations. FLAG resin was washed, and full-length CCR5-SNAP was eluted using excess FLAG peptide. FLAG elution was applied to a Superdex 200 10/300 GL column to purify monomeric CCR5-SNAP from receptor aggregates.
We then employed a tandem affinity purification strategy to isolate full-length CCR5-SNAP from receptor truncation products. Transiently transfected HEK293T cells expressing CCR5-SNAP were lysed with a buffer containing DDM, CHAPS, and CHS. The solubilized receptor was immobilized onto the 1D4-Sepharose immunoaffinity matrix (Fig. 2). The detergent concentration was lowered, lipids (DOPC and DOPS) were added to enhance receptor stability (19, 31, 32), and CCR5-SNAP was fluorescently labeled on resin with BG-Alexa-488, washed several times to remove excess dye, and then eluted using 1D4 nonapeptide. The first step of the purification removes cellular components and any C-terminal receptor truncations but not N-terminal truncations (Fig. 1 b). To purify full-length receptor from these truncations, we performed a second immunoaffinity purification step using anti-FLAG M2 agarose, which removed N-terminal truncations to yield the desired full-length receptor (Fig. 1 b). NIR-immunoblot analysis of 1D4/FLAG-purified CCR5-SNAP-488 shows the presence of minor bands corresponding to SDS-resistant CCR5-SNAP dimers (∼150 kDa) and oligomers (∼250 kDa) (Fig. 1 b).
Because oligomers complicate the ligand-binding analysis, we used SEC to purify receptor monomers from undesired oligomers. We determined the concentration of CCR5-SNAP-488 in the SEC fractions by FCS (Figs. 1 c and S5) because the concentrations were too low for reliable quantification by other methods, including 280-nm absorbance. The peak CCR5-SNAP fraction had an average concentration of (4.5 ± 0.6) nM. Because we observed a single peak in the FCS-derived chromatogram, we also analyzed the SEC fractions by NIR-immunoblot to evaluate the separation of monomeric CCR5-SNAP-488 from oligomers. Monomeric CCR5-SNAP-488 eluted as a single species, whereas it coeluted with receptor oligomers in earlier fractions (Figs. 1 c and S5). Monomeric full-length receptor was employed for all FCCS ligand-binding measurements.
We characterized expressed CCR5-SNAP in cell-based functional assays to determine whether the engineered tags interfere with receptor function. Because CCR5-SNAP expressed at different levels than WT 1D4-tagged CCR5 (CCR5), we optimized the gene dosage of CCR5-SNAP to obtain similar surface expression as CCR5 to accurately compare their function and pharmacology. We quantified CCR5-SNAP and CCR5 cell surface expression by flow cytometry using the anti-CCR5 antibody 2D7 conjugated to PE (Fig. S6 a). CCR5-SNAP and CCR5 expressed at similar levels when HEK293T cells were transfected with 2.0 and 0.75 μg of plasmid DNA, respectively. To validate the results, we repeated the experiment using the T21/8-PE antibody, which recognizes a different epitope than 2D7. CCR5-SNAP and CCR5 expressed at similar levels, in agreement with the results obtained with the 2D7-PE antibody (Fig. S6 a). As a further control, we quantified CCR5-SNAP expression using the FLAG PE antibody and we observed fluorescence only from CCR5-SNAP-expressing cells but not from CCR5-expressing cells (Fig. S6 a). We also performed immunofluorescence TIRF imaging on CCR5-SNAP or CCR5 expressing HEK293T cells, and we observed similar receptor expression patterns between CCR5-SNAP and CCR5 (Fig. S6 d).
We next measured the activation of Gi/o-proteins by inhibition of forskolin-stimulated cAMP formation by adenylate cyclase. We quantified the cAMP levels in HEK293T cells transfected with CCR5-SNAP and CCR5 in response to various chemokine concentrations using the RLuc3-EPAC-GFP10 BRET2 reporter (15, 23, 25). We fitted the dose-response curves for CCR5 WT and CCR5-SNAP as a function of chemokine concentrations using a three-parameter logistic equation to derive EC50 and Emax values (Fig. S6 b). Both CCR5-SNAP and CCR5 inhibited cAMP production for all the tested chemokines with similar efficacies and potencies (Table S5).
We also measured calcium mobilization after chemokine stimulus in CCR5-SNAP- and CCR5-expressing HEK293T cells using the FLIPR calcium 6 dye and generated dose-response curves (Fig. S6 c). 5P12 and 5P14 did not induce calcium flux on CCR5-SNAP- or CCR5-expressing cells in agreement with previous literature reports (13). For the remaining chemokines tested, we did not observe any meaningful differences in efficacy or potency between CCR5-SNAP and CCR5 (Table S6). We conclude from these functional control experiments that the SNAP-tag does not seem to interfere with biological function of the receptor.
In FCS and FCCS experiments, autocorrelation and cross-correlation amplitudes, denoted as A, are inversely proportional to the concentration of the fluorescent species (Materials and Methods). To derive concentrations from A, we determined the size of the confocal volumes for 488-nm excitation (0.18 ± 0.02 fL), 633-nm excitation (0.35 ± 0.03 fL), and their overlap volume (0.22 ± 0.03 fL) using serial dilutions of Alexa-488, Alexa-647, and a 40 bp Alexa-488/Alexa-647 dual-labeled oligonucleotide duplex, respectively (Fig. S3; (29, 33)).
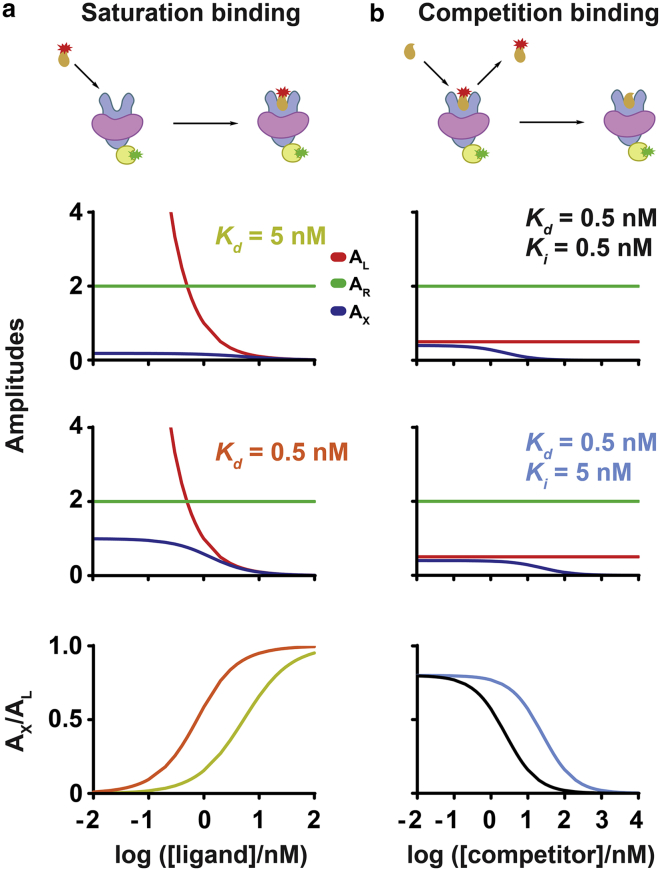
To illustrate the changes of auto- and cross-correlation functions in a ligand-binding experiment, we modeled the correlation function amplitudes as a function of titrating ligand concentration for receptor, ligand, and receptor-ligand complex at equilibrium dissociation constants of Kd = 5 nM and Kd = 0.5 nM (Fig. 3 a). AR, the receptor correlation amplitude (green curve), remains constant, whereas AL, the ligand correlation amplitude (red curve), grows hyperbolically with decreasing concentration. In contrast, AX, the receptor-ligand complex fluorescence cross-correlation amplitude (blue curve), plateaus at low ligand concentrations and asymptotically goes to zero at high ligand concentrations. Higher ligand affinities yield larger AX amplitudes as compared with lower ligand affinities because more receptor-ligand complexes are present. Assuming equal confocal volumes for all channels, the ratio AX/AL is equal to the fractional receptor occupancy, and a plot of AX/AL as a function of ligand concentration yields a saturation-binding isotherm (Fig. 3 a). We also simulated two different competition-binding cases in which ligand and receptor concentrations were kept constant while an unlabeled competitor concentration was titrated (Fig. 3 b). For these simulations, we assumed the following ligand and competitor affinities: 1) Kd = 0.5 nM and Ki = 0.5 nM and 2) Kd = 0.5 nM and Ki = 5 nM (Fig. 3 b). AR and AL remain constant while AX decreases with increasing competitor concentration. Similarly, we can derive competition-binding isotherms by plotting AX/AL as a function of competitor concentration (Fig. 3 b). Thus, we show that fractional occupancy can be derived from correlation amplitudes to derive saturation- and competition-binding isotherms.
Figure 3.
Modeling binding parameters from FCCS measurements. Fractional receptor occupancy can be derived from correlation amplitudes to derive (a) saturation- and (b) competition-binding isotherms. Cartoons show the binding interactions analyzed by FCCS. Fluorescent ligand (orange with red star) recognizes a lipid-embedded (violet) membrane receptor (blue). The receptor is fused to a functional tag (yellow) labeled with a fluorophore (green). The amplitudes are modeled as a function of titrating ligand concentration for receptor, ligand, and receptor-ligand complex. For saturation binding (a), correlation amplitude plots are shown for receptor (green, AR), ligand (red, AL), and complex (blue, AX) as a function of fluorescent ligand concentrations at which ligand dissociation constants were set to 5 nM (yellow) or 0.5 nM (orange). Ligand concentration varied from 0.01 to 100 nM, and receptor concentration was kept constant at 0.5 nM. For competition-binding analysis (b), competitor concentration varied from 0.01 to 10,000 nM, and ligand and receptor concentrations were kept constant at 2 and 0.5 nM, respectively. Competition-binding isotherms can be derived by plotting AX/AL as a function of competitor concentration. Two different competition-binding cases were simulated in which ligand and receptor concentrations were kept constant while an unlabeled competitor concentration was titrated (b). The ligand and competitor affinities were assumed to be Kd = 0.5 nM, Ki = 0.5 nM (black) or Kd = 0.5 nM, Ki = 5 nM (cyan). AR and AL remain constant while AX decreases with increasing competitor concentration.
The simulations assume that there is no cross talk from the green channel to the red channel. Cross talk from the green channel to the red channel increases AX, leading to an overestimation of the concentration of receptor-ligand complexes. Given this, we quantified the cross-talk contribution to AX using the method by Bacia and Schwille (17) and we measured negligible cross-talk contribution to AX, corresponding to an error in receptor occupancy of less than 1%.
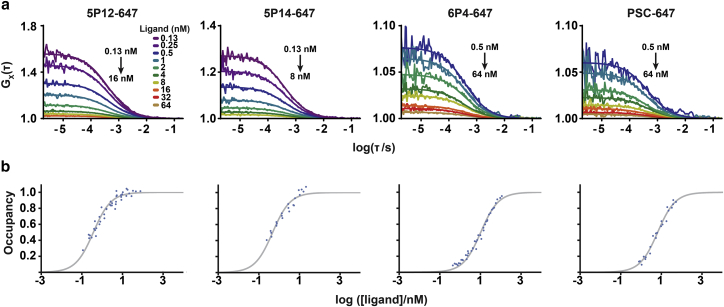
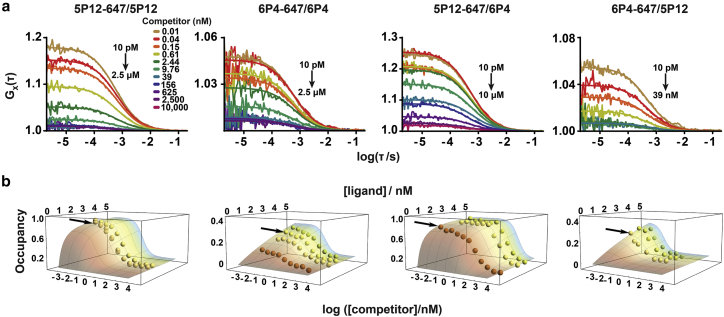
We then performed saturation-binding experiments with synthetic RANTES analogs site-specifically and covalently labeled with Alexa-647 (Fig. S2) and purified, monomeric CCR5-SNAP-488 by FCCS to determine their equilibrium dissociation constants. Representative autocorrelation and cross-correlation traces and fits are shown in Figs. 4 and S7. We also performed homologous and heterologous competition-binding FCCS experiments using Alexa-647-labeled 5P12 (5P12-647) and 6P4 (6P4-647) with unlabeled 5P12 and 6P4 (Figs. 5 and S8). Before further analyzing the data, we plotted the experimental G(0) values for the ligand, receptor, and ligand-receptor complex as a function of nominal ligand concentration for both the saturation- and competition-binding experiments, and we observed nonrandom variations in G(0) for all three components (Figs. S7 d and S8 d) that correlate with nonrandom variations in the diffusion time (Fig. S9), most likely due to changes in the point-spread function across the glass-bottom microtiter plate. A more complete discussion of variations in the diffusion time and molecular brightness is presented in the Supporting Text and Figs. S9 and S10.
Figure 4.
Saturation binding of Alexa-647-labeled RANTES analogs to CCR5-SNAP-488 by FCCS. (a) Cross-correlation traces and fits from one representative experiment of 5P12-647, 5P14-647, 6P4-647, and PSC-647 binding to CCR5-SNAP-488 at various ligand concentrations are shown. Cross-correlation traces were fitted to one translational component with 3D diffusion. (b) Saturation-binding isotherms for 5P12-647, 5P14-647, 6P4-647, and PSC-647 binding to CCR5-SNAP-488 are shown. Ligand-binding data were analyzed by global nonlinear regression of a model with a shared dissociation constant Kd and one active receptor fraction parameter for each of three to five independent experiments. The binding isotherms show the pooled receptor occupancies with respect to the active receptor fractions.
Figure 5.
Competition binding of Alexa-647-labeled 5P12 and 6P4 with nonlabeled chemokines by FCCS. (a) Cross-correlation traces and fits from one representative experiment of homologous and heterologous competition binding with 5P12-647 and 6P4-647 at various competitor and ligand concentrations are shown. Cross-correlation traces were fitted to one translational component with 3D diffusion. (b) Binding isotherms for homologous and heterologous competition results for 5P12-647 and 6P4-647 are shown. Competition-binding data were plotted as 3D surfaces to show the effect of ligand variability on the normalized occupancy. Arrows point to the representative experiments shown in (a). The concentrations of the labeled ligands were 2.37 ± 0.05, 2.37 ± 0.05, and 2.86 ± 0.06 nM in the 5P12-647/5P12 competition; 1.21 ± 0.01, 2.55 ± 0.01, 3.30 ± 0.02, and 4.02 ± 0.03 nM in the 6P4-647/6P4 competition; 1.15 ± 0.03, 2.91 ± 0.03, and 3.75 ± 0.04 nM in the 5P12-647/6P4 competition; and 2.65 ± 0.01, 3.66 ± 0.02, and 4.36 ± 0.02 nM in the 6P4-647/5P12 competition experiments, respectively. Ligand-binding fits were performed using data from at least three independent experiments.
Because the fractional occupancy derived from saturation-binding experiments was less than 100% for each of the RANTES analogs tested, we postulated that some fraction of CCR5-SNAP-488 becomes inactive during purification. We accounted for this inactive fraction by introducing the parameter, c, in the ligand-binding model. The parameter c is the active receptor fraction in each experimental data set. The active receptor fraction is a free fitting parameter independently optimized for each data set. We found on average that the active receptor fraction was 20 ± 5% (mean ± SD). This value compares well with reported specific activities of purified detergent-solubilized receptors. For example, the β2-adrenergic receptor has 30% of theoretical ligand-binding activity after His6- and FLAG-tag tandem affinity purification, and only after ligand-affinity purification may the full activity be obtained (34).
Similarly, a YFP-tagged μ-opioid receptor purified to homogeneity had only 20% of the predicted binding sites calculated from molecular mass (35). The loss of 80% of binding sites is likely the result of irreversible receptor denaturation during detergent solubilization. In a recent molecular dynamics simulation study to investigate the molecular basis of detergent denaturation of the adenosine A2A receptor, the authors found that nonionic detergents lead to receptor denaturation by decreasing intramolecular hydrophobic contacts, which leads to reduced α-helicity (36).
We performed global fitting of the binding curves using an iterative algorithm that accounts for ligand depletion by reducing the χ2function defined in the Materials and Methods. Fig. 4 b shows saturation-binding isotherms and fits for the RANTES analogs. From these fits, we calculated Kd = 0.37 ± 0.06 nM for 5P12-647 and Kd = 0.48 ± 0.08 nM for 5P14-647 (Table 1). In contrast, we derived Kd = 10.4 ± 1.2 nM for 6P4-647 and Kd = 6.6 ± 0.8 nM for PSC-647 (Table 1). Based on these results, we conclude that if 6P4- and PSC-647 are already high-affinity binders to CCR5-SNAP-488 given their nanomolar affinity, then 5P12- and 5P14-647 should be considered ultra-high-affinity binders.
Table 1.
FCCS-Derived Binding Affinities of RANTES Analogs at CCR5-SNAP-488
| Chemokine | Kd/nM |
| 5P12-647 | 0.368 ± 0.064 |
| 5P14-647 | 0.475 ± 0.082 |
| 6P4-647 | 10.4 ± 1.2 |
| PSC-647 | 6.57 ± 0.81 |
| Labeled/Unlabeled | Ki/nM |
| 5P12-647/5P12 | 0.141 ± 0.033 |
| 5P12-647/6P4 | 11.8 ± 2.0 |
| 6P4-647/6P4 | 3.49 ± 0.77 |
| 6P4-647/5P12 | 0.047 ± 0.099 |
Equilibrium dissociation (Kd) and inhibition (Ki) constants for the RANTES analogs binding to the CCR5-SNAP-488 derived from saturation and competition binding are given. Errors were derived using global analysis with nonlinear least-square fitting of the binding isotherms with bootstrapping.
The RANTES analogs are labeled with Alexa-647, which contains negatively charged sulfonate groups that might perturb the binding of the positively charged chemokines to the receptor. We therefore performed homologous competition-binding experiments with 5P12-647 and 6P4-647 with titrating concentrations of nonlabeled 5P12 and 6P4, respectively. We performed the same global fitting analysis as with the saturation-binding measurements, and we accounted for variations in the concentrations of the labeled ligand by plotting fractional occupancy as a function of both ligand and competitor. Fig. 5 b shows the 3D competition-binding surface plots for 5P12 and 6P4 homologous competition binding. We derived Ki = 0.14 ± 0.03 nM for 5P12, and Ki = 3.5 ± 0.8 nM for 6P4 (Table 1). The calculated Ki-values for 5P12 and 6P4 are approximately threefold lower as compared to the Kd-values of the labeled ligands calculated for saturation binding, indicating that Alexa-647 slightly perturbs chemokine binding to CCR5-SNAP-488, likely as a result of the electrostatic repulsion of the negatively charged fluorophore and the receptor in the negatively charged mixed micelle.
Discussion
FCS is a single-molecule-sensitive method in which fluorescence fluctuations from molecules diffusing through a subfemtoliter volume of focused excitation are analyzed (37). An autocorrelation analysis is performed on the fluorescence fluctuations to derive physical parameters such as diffusion coefficients, triplet state fractions, and concentrations. Because of the small size of the diffraction-limited volume in FCS measurements, even subnanomolar concentrations of ligand can be studied. In contrast, techniques such as surface plasmon resonance and isothermal calorimetry generally require at least micromolar concentrations of bound ligand in the detection volume to measure ligand-binding affinities, which complicates determination of nanomolar affinities. Another advantage of FCS is that binding interactions are observed in solution, overcoming immobilization artifacts present in surface plasmon resonance and in some other imaging techniques (33). However, FCS alone cannot discern binding between two species that have similar molecular weights. To separate the free and bound species in the FCS autocorrelation curve, a fluorescently labeled ligand must undergo at least an eightfold change in molecular mass upon binding to its receptor. This limitation can be surmounted if the receptor and ligand can be labeled with different fluorophores emitting at distinct wavelengths so that two autocorrelation curves can be measured simultaneously to determine a so-called cross-correlation function using FCCS. We used the FCCS approach to discover and study ultra-high-affinity interactions between CCR5 and a series of RANTES analogs of biological significance.
To enable quantitative measurements of ligand affinities for CCR5 using FCCS, we purified to near homogeneity a monomeric fluorescently labeled CCR5-SNAP-488 from receptor oligomers, aggregates, and truncation products originating from both N- and C-terminal degradation. The purified receptor was quantified in SEC fractions using FCS. Previously, Nisius et al. purified CCR5 from insect cells and observed in the size-exclusion chromatograms two distinct peaks corresponding to equal fractions of monomeric and dimeric receptor (38). In comparison, we only observed a very small number of dimers, most likely because of the mammalian cell expression system and the different detergents employed, which may alter the extent of receptor oligomerization. We demonstrated that the expressed CCR5-SNAP construct signals like WT CCR5 in cAMP inhibition and calcium mobilization assays in response to agonist ligands, showing that the SNAP-tag and additional affinity tags do not interfere with receptor function.
Using the FCCS method, we calculated the equilibrium binding affinities for the fluorescently labeled RANTES analogs at the purified functional CCR5-SNAP-488. Antoine et al. performed similar ligand-binding measurements by FCCS using several GFP receptor fusion proteins with fluorescently labeled small molecules and antibodies (39). Our approach differs in two major ways. First, we used highly purified receptor samples for ligand-binding measurements instead of GPCRs solubilized in crude cell lysates without any purification. As such, effectors that regulate ligand binding are not removed, and the measured affinities are not representative of the “naked” receptor. Also, we used the SNAP-tag technology to label receptors rather than GFP as a fluorescent label fused to the target receptor. GFP is susceptible to misfolding during biosynthesis, leading to substoichiometric receptor fluorescent labeling. Using the SNAP-tag, we achieved stoichiometric labeling of functional CCR5-SNAP by supplementing the reaction buffer with 1 mM DTT.
We also observed that the native chemokines RANTES and MIP1α could not displace 5P12-647 or 6P4-647 from CCR5-SNAP-488 (Supporting Text; Figs. S13–S16). This result parallels earlier reports that PSC-biotin binding could not be displaced with RANTES or MIP-1α but only with PSC (40). We expected that sCD4/gp120 complex should also recognize the “naked” receptor fraction, but we could not displace either 5P12-647 or 6P4-647 in our competition-binding experiments with the viral complex. In previous cell-based assays, the radiolabeled sCD4/gp120 complexes in cells could be displaced by the RANTES analogs (16), but it is unknown whether the viral complex can compete off the RANTES analogs in cells.
We demonstrated that the RANTES analogs bind with high affinity to the “naked” CCR5 receptor. In contrast, the native chemokines RANTES and MIP-1α did not bind to the CCR5 in detergent solution. We ruled out that the tags on the CCR5-SNAP construct cause the lack of binding of the native chemokines because the construct shows WT functionality in cell-based assays. One possibility is that the native chemokines require G-protein precoupling to bind with high affinity to CCR5 (15, 16). Another possibility that could explain the lack of observed native chemokine binding to CCR5 in our assay is that upon detergent solubilization, the receptor adopts a conformation that is specific only to the RANTES analogs. To discern between these different possibilities, future experiments will probe the role of G-protein precoupling on the ligand-binding interactions of the RANTES analogs and native chemokines to CCR5-SNAP in a more native lipid environment, such as nanoscale apolipoprotein bound bilayers (19, 41), because CCR5 solubilized in detergent micelles cannot efficiently bind G-protein.
We generated homology models of 5P12 and 6P4 binding to CCR5 based on the crystal structure of the 5P7-CCR5 complex to elucidate the structural determinants of their different affinities (18). Note that 5P7 and 5P12 differ by only one amino acid (13). Fig. 6 shows the lowest energy conformations for 5P12-RANTES and 6P4-RANTES bound to CCR5. 5P12-RANTES and 6P4-RANTES adopt a conformation that closely mimics the conformation observed for 5P7 in the crystal structure. In our model, we observe that the 6P4-CCR5 complex has a negatively charged residue, Asp5 in 6P4, in the binding pocket not stabilized by a counterion. We speculate that the lack of stabilizing counterion interactions with this residue may help to explain the lower affinity observed for 6P4 relative to 5P12.
Figure 6.
CCR5-ligand homology structural models based on the crystal structure of the CCR5-5P7 complex. Homology structural models of CCR5 bound to 5P12-RANTES (a), 6P4-RANTES (b), RANTES (c), and maraviroc (d) are given. The structural models are based on the crystal structures of the CCR5-5P7 complex and the CCR5-maraviroc complex. The N-terminal tail residues of the chemokines are shown as red beads on a string. The remaining part of the chemokines is shown in ribbon representation colored in green. Maraviroc is shown using a blue space-filling model. CCR5 is represented as a ribbon structure with each transmembrane segment colored differently. The figure illustrates the different orientations observed for the bound chemokine analogs relative to Glu283 in CCR5. The panels on the right show the region marked within the gray rectangle in higher magnification. Chemokine residues 1–9 are labeled, as well as residues K26, K197, and E283 in CCR5, using single-letter amino-acid abbreviations. The pyroglutamate residue 0 of the analogs is labeled as p0.
Glu283 in CCR5 is another charged residue with no stabilizing counterions in the 5P12- and 6P4-CCR5 complex models. Zheng et al. observed that Glu283 makes a hydrogen bond with Leu4 in the 5P7-CCR5 crystal structure (18). We observe a similar hydrogen-bond interaction with Glu283 and Leu4 in the 5P12-CCR5 complex model, whereas the backbone of 6P4 was removed from Glu283. Choi et al. demonstrated that Glu283 is a key residue for the antiviral activities of 5P12 and 6P4 (42). Using the CCR5 E283A mutant, Choi et al. measured a 100-fold loss in antiviral potency for 5P12 but for 6P4 a 100-fold increase in potency, supporting the hypothesis that Glu283 destabilizes 6P4 binding to CCR5. Therefore, we hypothesize that the lower affinity observed for 6P4 in our FCCS measurements relative to 5P12 may also be due to the lack of stabilizing interactions with Glu283 in CCR5.
We also modeled RANTES binding to CCR5 (Fig. 6 c), and we observed that the RANTES N-terminal tail adopts a different conformation in the ligand-binding pocket relative to that of 6P4 and 5P12. The free amino terminus of Ser1 in RANTES makes a salt bridge with Glu283 in CCR5, and we attribute the different conformations observed between RANTES and the chemokine analogs primarily to this salt bridge. Previous experimental and theoretical studies have shown that Glu283 is necessary for native chemokine binding to CCR5 (43). Moreover, the maraviroc-CCR5 crystal structure (Fig. 6 d) shows that the positively charged tropane nitrogen forms a salt bridge with Glu283 (44). Likewise, previous alanine scanning mutagenesis in CCR5 and modeling studies have shown that Glu283 is necessary for the binding of positively charged small-molecule antagonists to CCR5 (45, 46, 47).
Another difference between RANTES and 5P12/6P4 in our binding models is that the hydroxyl of Tyr3 in RANTES is close to the ε-amine of Lys197, alluding to the possibility that Tyr3 forms a salt bridge with Lys197 as a tyrosinate. Lys197 in CCR5 is buried in the binding pocket with no obvious stabilizing charges in proximity. Tyr3 is close to the hydrophobic pocket that is occupied by the 1,1-difluorocyclohexane moiety in the maraviroc-CCR5 complex and the pyroglutamate residue in the 5P12 and 6P4 complexes. In contrast to the isolated Asp5 in the CCR5-6P4 complex, Asp6 in RANTES makes a salt bridge with Lys26 of CCR5.
In conclusion, we have presented methods to prepare functional, monomeric, stoichiometrically fluorophore-conjugated receptors and fluorescent chemokine ligands that provide a foundation for single-molecule studies of ligand-GPCR interactions. We demonstrate convenient quantification of nanomolar receptor concentration by FCS, and we developed FCCS saturation- and competition-binding assays for ligand-receptor interactions with nanomolar affinities. In a chemically defined, purified system that is not affected by cellular heterogeneity, we have demonstrated high-affinity binding of high potency anti-HIV chemokines to “naked” receptor consistent with a model proposed for G-protein-biased signaling in this system.
Author Contributions
C.A.R., A.F., O.H., T.P.S., and T.H. designed the research. C.A.R., Y.A.B., M.H., E.L., H.T., M.A.K., H.F., and T.H. performed the research. J.C.P. provided new reagents. C.A.R., Y.A.B., E.L., A.F., O.H., T.P.S., and T.H. analyzed the data. C.A.R. and T.H. developed the algorithms and analytical tools. C.A.R., Y.A.B., E.L., A.F., T.P.S., and T.H. wrote the article.
Acknowledgments
We thank the Rockefeller University Bio-Imaging Resource Center for training, assistance, and discussions related to fluorescent imaging and FCCS experiments. We thank the High-Throughput and Spectroscopy Resource Center for their training and access to the LI-COR instrument for NIR-immunoblots. We thank Dr. W Vallen Graham for expression and purification of the 1D4 mAb and for technical assistance with the receptor purifications. Soluble CD4 from Progenics Pharmaceuticals, Inc. was obtained through the AIDS Reagent Program, Division of AIDS, NIAID, NIH. Monomeric BG 505 gp120 was a gift from Dr. John P. Moore (Weill Cornell Medicine, New York, NY).
E.L. was supported by the David Rockefeller Graduate program. We acknowledge grant support from the Robertson Therapeutic Development Fund, the Crowley Family Fund, and the Danica Foundation to T.H. O.H. was supported by the Swiss National Science Foundation (310030_163085). C.A.R. and M.H. were supported by the Tri-institutional Training Program in Chemical Biology and National Institute of General Medical Sciences training grant T32 GM115327. C.A.R. was supported by the National Science Foundation Graduate Research Fellowship grant DGE-1325261.
Editor: Jochen Mueller.
Footnotes
Supporting Material can be found online at https://doi.org/10.1016/j.bpj.2019.07.043.
Supporting Material
References
- 1.Sodhi A., Montaner S., Gutkind J.S. Viral hijacking of G-protein-coupled-receptor signalling networks. Nat. Rev. Mol. Cell Biol. 2004;5:998–1012. doi: 10.1038/nrm1529. [DOI] [PubMed] [Google Scholar]
- 2.Barmania F., Pepper M.S. C-C chemokine receptor type five (CCR5): an emerging target for the control of HIV infection. Appl. Transl. Genomics. 2013;2:3–16. doi: 10.1016/j.atg.2013.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dean M., Carrington M., O’Brien S.J. Genetic restriction of HIV-1 infection and progression to AIDS by a deletion allele of the CKR5 structural gene. Hemophilia growth and development study, multicenter AIDS cohort study, multicenter hemophilia cohort study, san francisco city cohort, ALIVE study. Science. 1996;273:1856–1862. doi: 10.1126/science.273.5283.1856. [DOI] [PubMed] [Google Scholar]
- 4.Huang Y., Paxton W.A., Koup R.A. The role of a mutant CCR5 allele in HIV-1 transmission and disease progression. Nat. Med. 1996;2:1240–1243. doi: 10.1038/nm1196-1240. [DOI] [PubMed] [Google Scholar]
- 5.Wei X., Nielsen R. CCR5-Δ32 is deleterious in the homozygous state in humans. Nat. Med. 2019;25:909–910. doi: 10.1038/s41591-019-0459-6. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 6.Shaik M.M., Peng H., Chen B. Structural basis of coreceptor recognition by HIV-1 envelope spike. Nature. 2019;565:318–323. doi: 10.1038/s41586-018-0804-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Roche M., Salimi H., Gorry P.R. A common mechanism of clinical HIV-1 resistance to the CCR5 antagonist maraviroc despite divergent resistance levels and lack of common gp120 resistance mutations. Retrovirology. 2013;10:43. doi: 10.1186/1742-4690-10-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cocchi F., DeVico A.L., Lusso P. Identification of RANTES, MIP-1 α, and MIP-1 β as the major HIV-suppressive factors produced by CD8+ T cells. Science. 1995;270:1811–1815. doi: 10.1126/science.270.5243.1811. [DOI] [PubMed] [Google Scholar]
- 9.Dragic T., Litwin V., Paxton W.A. HIV-1 entry into CD4+ cells is mediated by the chemokine receptor CC-CKR-5. Nature. 1996;381:667–673. doi: 10.1038/381667a0. [DOI] [PubMed] [Google Scholar]
- 10.Hartley O., Gaertner H., Offord R. Medicinal chemistry applied to a synthetic protein: development of highly potent HIV entry inhibitors. Proc. Natl. Acad. Sci. USA. 2004;101:16460–16465. doi: 10.1073/pnas.0404802101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Simmons G., Clapham P.R., Proudfoot A.E. Potent inhibition of HIV-1 infectivity in macrophages and lymphocytes by a novel CCR5 antagonist. Science. 1997;276:276–279. doi: 10.1126/science.276.5310.276. [DOI] [PubMed] [Google Scholar]
- 12.Lederman M.M., Veazey R.S., Hartley O. Prevention of vaginal SHIV transmission in rhesus macaques through inhibition of CCR5. Science. 2004;306:485–487. doi: 10.1126/science.1099288. [DOI] [PubMed] [Google Scholar]
- 13.Gaertner H., Cerini F., Hartley O. Highly potent, fully recombinant anti-HIV chemokines: reengineering a low-cost microbicide. Proc. Natl. Acad. Sci. USA. 2008;105:17706–17711. doi: 10.1073/pnas.0805098105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bönsch C., Munteanu M., Hartley O. Potent anti-HIV chemokine analogs direct post-endocytic sorting of CCR5. PLoS One. 2015;10:e0125396. doi: 10.1371/journal.pone.0125396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lorenzen E., Ceraudo E., Huber T. G protein subtype-specific signaling bias in a series of CCR5 chemokine analogs. Sci. Signal. 2018;11:eaao6152. doi: 10.1126/scisignal.aao6152. [DOI] [PubMed] [Google Scholar]
- 16.Colin P., Bénureau Y., Lagane B. HIV-1 exploits CCR5 conformational heterogeneity to escape inhibition by chemokines. Proc. Natl. Acad. Sci. USA. 2013;110:9475–9480. doi: 10.1073/pnas.1222205110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bacia K., Schwille P. Practical guidelines for dual-color fluorescence cross-correlation spectroscopy. Nat. Protoc. 2007;2:2842–2856. doi: 10.1038/nprot.2007.410. [DOI] [PubMed] [Google Scholar]
- 18.Zheng Y., Han G.W., Handel T.M. Structure of CC chemokine receptor 5 with a potent chemokine antagonist reveals mechanisms of chemokine recognition and molecular mimicry by HIV. Immunity. 2017;46:1005–1017.e5. doi: 10.1016/j.immuni.2017.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Knepp A.M., Grunbeck A., Huber T. Direct measurement of thermal stability of expressed CCR5 and stabilization by small molecule ligands. Biochemistry. 2011;50:502–511. doi: 10.1021/bi101059w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Park S.H., Cheong C., Park C.G. Generation and application of new rat monoclonal antibodies against synthetic FLAG and OLLAS tags for improved immunodetection. J. Immunol. Methods. 2008;331:27–38. doi: 10.1016/j.jim.2007.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Junttila M.R., Saarinen S., Westermarck J. Single-step Strep-tag purification for the isolation and identification of protein complexes from mammalian cells. Proteomics. 2005;5:1199–1203. doi: 10.1002/pmic.200400991. [DOI] [PubMed] [Google Scholar]
- 22.MacKenzie D., Arendt A., Molday R.S. Localization of binding sites for carboxyl terminal specific anti-rhodopsin monoclonal antibodies using synthetic peptides. Biochemistry. 1984;23:6544–6549. doi: 10.1021/bi00321a041. [DOI] [PubMed] [Google Scholar]
- 23.Berchiche Y.A., Sakmar T.P. CXC chemokine receptor 3 alternative splice variants selectively activate different signaling pathways. Mol. Pharmacol. 2016;90:483–495. doi: 10.1124/mol.116.105502. [DOI] [PubMed] [Google Scholar]
- 24.Berchiche Y.A., Gravel S., Heveker N. Different effects of the different natural CC chemokine receptor 2b ligands on beta-arrestin recruitment, Gαi signaling, and receptor internalization. Mol. Pharmacol. 2011;79:488–498. doi: 10.1124/mol.110.068486. [DOI] [PubMed] [Google Scholar]
- 25.Leduc M., Breton B., Heveker N. Functional selectivity of natural and synthetic prostaglandin EP4 receptor ligands. J. Pharmacol. Exp. Ther. 2009;331:297–307. doi: 10.1124/jpet.109.156398. [DOI] [PubMed] [Google Scholar]
- 26.Offord R.E., Gaertner H.F., Proudfoot A.E. Synthesis and evaluation of fluorescent chemokines labeled at the amino terminal. Methods Enzymol. 1997;287:348–369. doi: 10.1016/s0076-6879(97)87024-3. [DOI] [PubMed] [Google Scholar]
- 27.Gaertner H.F., Offord R.E., Rose K. Chemo-enzymic backbone engineering of proteins. Site-specific incorporation of synthetic peptides that mimic the 64-74 disulfide loop of granulocyte colony-stimulating factor. J. Biol. Chem. 1994;269:7224–7230. [PubMed] [Google Scholar]
- 28.Foillard S., Rasmussen M.O., Dumy P. 1-Ethoxyethylidene, a new group for the stepwise SPPS of aminooxyacetic acid containing peptides. J. Org. Chem. 2008;73:983–991. doi: 10.1021/jo701628k. [DOI] [PubMed] [Google Scholar]
- 29.Schwille P., Meyer-Almes F.J., Rigler R. Dual-color fluorescence cross-correlation spectroscopy for multicomponent diffusional analysis in solution. Biophys. J. 1997;72:1878–1886. doi: 10.1016/S0006-3495(97)78833-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wellerdieck C., Oles M., Hatt H. Functional expression of odorant receptors of the zebrafish Danio rerio and of the nematode C. elegans in HEK293 cells. Chem. Senses. 1997;22:467–476. doi: 10.1093/chemse/22.4.467. [DOI] [PubMed] [Google Scholar]
- 31.Navratilova I., Sodroski J., Myszka D.G. Solubilization, stabilization, and purification of chemokine receptors using biosensor technology. Anal. Biochem. 2005;339:271–281. doi: 10.1016/j.ab.2004.12.017. [DOI] [PubMed] [Google Scholar]
- 32.Tian H., Sakmar T.P., Huber T. Site-specific labeling of genetically encoded azido groups for multicolor, single-molecule fluorescence imaging of GPCRs. Methods Cell Biol. 2013;117:267–303. doi: 10.1016/B978-0-12-408143-7.00015-3. [DOI] [PubMed] [Google Scholar]
- 33.Rüttinger S., Buschmann V., Koberling F. Comparison and accuracy of methods to determine the confocal volume for quantitative fluorescence correlation spectroscopy. J. Microsc. 2008;232:343–352. doi: 10.1111/j.1365-2818.2008.02105.x. [DOI] [PubMed] [Google Scholar]
- 34.Kobilka B.K. Amino and carboxyl terminal modifications to facilitate the production and purification of a G protein-coupled receptor. Anal. Biochem. 1995;231:269–271. doi: 10.1006/abio.1995.1533. [DOI] [PubMed] [Google Scholar]
- 35.Kuszak A.J., Pitchiaya S., Sunahara R.K. Purification and functional reconstitution of monomeric mu-opioid receptors: allosteric modulation of agonist binding by Gi2. J. Biol. Chem. 2009;284:26732–26741. doi: 10.1074/jbc.M109.026922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lee S., Mao A., Vaidehi N. How do short chain nonionic detergents destabilize G-protein-coupled receptors? J. Am. Chem. Soc. 2016;138:15425–15433. doi: 10.1021/jacs.6b08742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Elson E.L. Fluorescence correlation spectroscopy: past, present, future. Biophys. J. 2011;101:2855–2870. doi: 10.1016/j.bpj.2011.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nisius L., Rogowski M., Grzesiek S. Large-scale expression and purification of the major HIV-1 coreceptor CCR5 and characterization of its interaction with RANTES. Protein Expr. Purif. 2008;61:155–162. doi: 10.1016/j.pep.2008.06.001. [DOI] [PubMed] [Google Scholar]
- 39.Antoine T., Ott D., Hannus S. Homogeneous time-resolved G protein-coupled receptor-ligand binding assay based on fluorescence cross-correlation spectroscopy. Anal. Biochem. 2016;502:24–35. doi: 10.1016/j.ab.2016.02.017. [DOI] [PubMed] [Google Scholar]
- 40.Jin J., Colin P., Brelot A. Targeting spare CC chemokine receptor 5 (CCR5) as a principle to inhibit HIV-1 entry. J. Biol. Chem. 2014;289:19042–19052. doi: 10.1074/jbc.M114.559831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Banerjee S., Huber T., Sakmar T.P. Rapid incorporation of functional rhodopsin into nanoscale apolipoprotein bound bilayer (NABB) particles. J. Mol. Biol. 2008;377:1067–1081. doi: 10.1016/j.jmb.2008.01.066. [DOI] [PubMed] [Google Scholar]
- 42.Choi W.T., Nedellec R., Mosier D.E. CCR5 mutations distinguish N-terminal modifications of RANTES (CCL5) with agonist versus antagonist activity. J. Virol. 2012;86:10218–10220. doi: 10.1128/JVI.00353-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tamamis P., Floudas C.A. Elucidating a key anti-HIV-1 and cancer-associated axis: the structure of CCL5 (Rantes) in complex with CCR5. Sci. Rep. 2014;4:5447–5456. doi: 10.1038/srep05447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tan Q., Zhu Y., Wu B. Structure of the CCR5 chemokine receptor-HIV entry inhibitor maraviroc complex. Science. 2013;341:1387–1390. doi: 10.1126/science.1241475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Maeda K., Das D., Mitsuya H. Structural and molecular interactions of CCR5 inhibitors with CCR5. J. Biol. Chem. 2006;281:12688–12698. doi: 10.1074/jbc.M512688200. [DOI] [PubMed] [Google Scholar]
- 46.Kondru R., Zhang J., Dioszegi M. Molecular interactions of CCR5 with major classes of small-molecule anti-HIV CCR5 antagonists. Mol. Pharmacol. 2008;73:789–800. doi: 10.1124/mol.107.042101. [DOI] [PubMed] [Google Scholar]
- 47.Dragic T., Trkola A., Moore J.P. A binding pocket for a small molecule inhibitor of HIV-1 entry within the transmembrane helices of CCR5. Proc. Natl. Acad. Sci. USA. 2000;97:5639–5644. doi: 10.1073/pnas.090576697. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.