Abstract
Hereditary pulmonary alveolar proteinosis (PAP) is a genetic lung disease characterized by surfactant accumulation and respiratory failure arising from disruption of GM-CSF signaling. While mutations in either CSF2RA or CSF2RB (encoding GM-CSF receptor α or β chains, respectively) can cause PAP, α chain mutations are responsible in most patients. Pulmonary macrophage transplantation (PMT) is a promising new cell therapy in development; however, no studies have evaluated this approach for hereditary PAP (hPAP) caused by Csf2ra mutations. Here, we report on the preclinical safety, tolerability, and efficacy of lentiviral-vector (LV)-mediated Csf2ra expression in macrophages and PMT of gene-corrected macrophages (gene-PMT therapy) in Csf2ra gene-ablated (Csf2ra−/−) mice. Gene-PMT therapy resulted in a stable transgene integration and correction of GM-CSF signaling and functions in Csf2ra−/− macrophages in vitro and in vivo and resulted in engraftment and long-term persistence of gene-corrected macrophages in alveoli; restoration of pulmonary surfactant homeostasis; correction of PAP-specific cytologic, histologic, and biomarker abnormalities; and reduced inflammation associated with disease progression in untreated mice. No adverse consequences of gene-PMT therapy in Csf2ra−/− mice were observed. Results demonstrate that gene-PMT therapy of hPAP in Csf2ra−/− mice was highly efficacious, durable, safe, and well tolerated.
Keywords: hereditary PAP, GM-CSF signaling, Csf2ra−/− mice, PMT, gene-therapy, surfactant homeostasis, macrophage engraftment, lentiviral vector
Arumugam et al. evaluated gene-pulmonary macrophage transplantation (PMT) therapy of hereditary pulmonary alveolar proteinosis (hPAP) in Csf2ra gene-ablated mice. Results from this non-clinical study found the approach safe, well tolerated, and efficacious, thereby supporting the translation and testing of gene-PMT therapy in human patients with hPAP caused by CSF2RA mutations.
Introduction
Hereditary pulmonary alveolar proteinosis (hPAP) is a disorder of pulmonary surfactant homeostasis caused by mutations in either CSF2RA or CSF2RB encoding the α and β subunits of granulocyte-macrophage colony-stimulating factor (GM-CSF) receptors present on the plasma membrane of alveolar macrophages and other cells.1, 2, 3, 4, 5 Surfactant normally comprises a layer of phospholipids, cholesterol, and protein at the air-liquid-tissue interface, which must be sufficient to reduce surface tension and prevent alveolar collapse and be thin enough to permit gas diffusion across the alveolar wall.6 Homeostasis is maintained by balanced secretion of surfactant from type II alveolar epithelial cells and clearance in approximately equal proportions by these cells and alveolar macrophages.7, 8 Pulmonary GM-CSF stimulates the survival, proliferation, and differentiation of alveolar macrophages and is critical for regulating numerous macrophage functions, including removal of excess surfactant from the alveolar surface.9 Binding to the GM-CSF receptor α subunit activates Janus kinase 2 (JAK2) associated with the β subunit and initiates signaling via multiple pathways, including signal transducer and activator of transcription 5 (STAT5).10, 11 Normally, signaling along the GM-CSF → PU.1 → PPARγ axis stimulates expression of ABCG1,12, 13, 14, 15, 16 which alveolar macrophages require for efflux of surfactant-derived cholesterol to high-density lipoproteins.13, 17 In hPAP, disruption of GM-CSF signaling reduces efflux of cholesterol from alveolar macrophages, which esterify and store it in cytoplasmic droplets, resulting in the formation of macrophage “foam” cells with reduced surfactant clearance capacity.12 This reduces alveolar-macrophage-mediated surfactant clearance, resulting in progressive accumulation of surfactant sediment within alveoli; displacement of alveolar air; and, ultimately, hypoxemic respiratory failure.12 While recessive and compound heterozygous mutations in either the α or the β subunit of the GM-CSF receptor cause PAP with similar disease manifestations and natural history, the majority of patients reported to date have mutations in the α subunit.1, 2, 3, 4, 5, 18 In mice, disruption of GM-CSF signaling by ablation of the genes encoding either Csf2ra or Csf2rb causes hereditary PAP (hPAP) identical to the human disease with respect to clinical, physiological, cellular, biochemical, and regulatory abnormalities.19, 20
hPAP is currently treated by whole lung lavage, an invasive procedure performed under general anesthesia in which one lung is mechanically ventilated while the other is repeatedly filled with saline and drained while percussing the chest to emulsify and remove accumulated surfactant.18, 21 This treatment results in incomplete removal of surfactant sediment but does not halt the underlying accumulation process and must be repeated, in some patients, as frequently as every 1–2 months. In Csf2rb gene-ablated mice, a validated model of hPAP, lethal irradiation followed by administration of gene-corrected hematopoietic stem progenitor cells (HSPCs) by bone marrow transplantation resulted in correction of the lung disease.22 However, bone marrow transplantation in a child with hPAP caused by CSF2RA mutations was unsuccessful, due to death from overwhelming infection prior to successful engraftment, likely due to the myeloablative conditioning regimen and immunosuppression required for bone marrow transplantation.2 Recently, we reported a novel cell therapy approach—pulmonary macrophage transplantation (PMT)—which does not require myeloablative conditioning or immunosuppression that is based on direct intrapulmonary instillation of macrophages.19 However, no reports have evaluated the feasibility of correction of hPAP caused by GM-CSF receptor α chain dysfunction in human or mouse.
In the present study, we continue preparing for a human trial of gene-PMT therapy of hPAP caused by CSF2RA mutations by conducting a preclinical study evaluating the safety and efficacy of lentiviral-vector-mediated Csf2ra gene correction and PMT therapy in a validated Csf2ra−/− mouse model of hPAP. Results show that gene-PMT therapy of hPAP in Csf2ra−/− mice is well tolerated, safe, highly efficacious, and durable.
Results
Construction and In Vitro Evaluation of a Preclinical Vector to Correct GM-CSF Receptor Function in Macrophages: mCsf2ra-LV
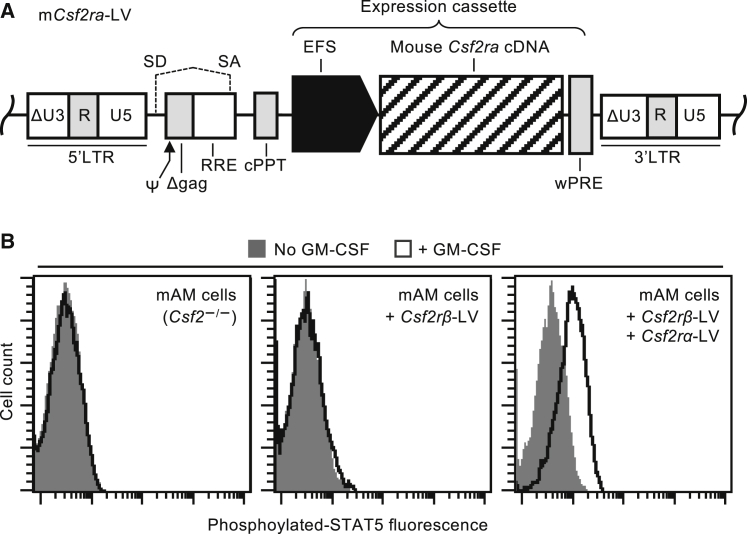
We constructed a lentiviral vector (LV) expressing murine GM-CSF receptor α encoded by the coding sequence (CDS) of Csf2ra (mCsf2ra-LV) to conduct preclinical studies of gene-PMT therapy in Csf2ra−/− mice, a validated model of hPAP caused by CSF2RA mutations.20 The design of mCsf2ra-LV (Figure 1A) is identical to our clinical grade vector expressing human GM-CSF receptor α (pRRL.cPPT.EFS.hCSF2RAcoopwpre)23 planned for use in a future human gene therapy clinical trial and was constructed by replacing the human CSF2RA cDNA with the murine Csf2ra cDNA. This was necessary because, while human and murine GM-CSF regulate their respective myeloid cell targets with remarkable similarity, they are neither functionally interchangeable nor immunologically cross-reactive.24, 25
Figure 1.
Genetic Correction of GM-CSF Receptor Signaling in Murine Macrophages
(A) Schematic of the third-generation, self-inactivating (SIN) lentiviral vector expressing the murine Csf2ra cDNA from the short elongation factor 1 promoter (EFS) (mCsf2ra-LV). See Materials and Methods for details. (B) STAT5 phosphorylation assay. Restoration of GM-CSF signaling in mAM cells by lentiviral-vector-mediated expression of both Csf2ra and Csf2rb demonstrated by flow-cytometry-based measurement of GM-CSF-stimulated STAT5 phosphorylation.
To initially evaluate mCsf2ra-LV in vitro, we used mouse alveolar macrophage (mAM) cells26 (which don’t express GM-CSF receptor α or β chains) to create a cell line that expressed Csf2rb, but not Csf2ra, by transduction with a vector expressing Csf2rb19 using routine methods.27 The resulting mAM-GM-Rβ+ cell line was used in GM-CSF receptor complementation studies to evaluate mCsf2ra-LV by measuring the phosphorylation of STAT5, a downstream target of GM-CSF signaling. GM-CSF-stimulated STAT5 phosphorylation was readily detectable in mCsf2ra-LV-transduced mAM-GM-Rβ+ cells but not in mAM or non-transduced mAM-GM-Rβ+ cells (Figure 1B). These results demonstrate that transduction by mCsf2ra-LV can restore GM-CSF receptor function in cells expressing only the GM-CSF receptor β.
Preparation and Characterization of Gene-Corrected Macrophages before PMT
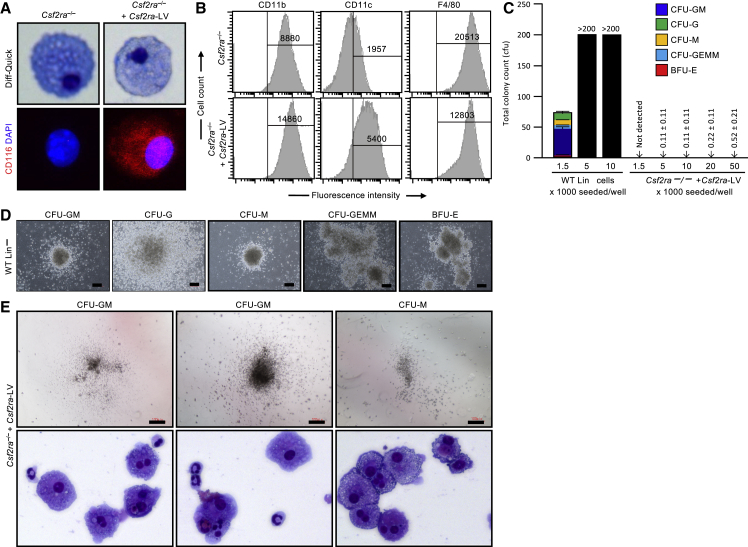
Our clinical strategy for gene-PMT therapy of human hPAP will utilize autologous, gene-corrected HSPC-derived macrophages. Therefore, in this preclinical study, murine HSPCs (lineage-negative [Lin−] Sca1+ c-Kit+ [LSK] cells) from Csf2ra−/− mice27 were transduced with mCsf2ra-LV as described earlier, expanded and differentiated into macrophages (referred to as gene-corrected macrophages hereinafter) using established methods,19 and the gene-corrected macrophages were characterized in vitro. Csf2ra−/− mouse LSK-cell-derived macrophages had the morphologic appearance of macrophages that was not affected by mCsf2ra-LV transduction (Figure 2A, top), and CD116 was detected on gene-corrected macrophages but not on non-transduced cells (Figure 2A, bottom). Phenotype marker analysis showed that gene-corrected macrophages had increased expression of markers typical of mature macrophages (CD11b, F4/80, and CD11c) (Figure 2B).
Figure 2.
Phenotypic Characterization of Csf2ra Gene-Corrected Macrophages before PMT
(A) Photomicrographs of BMDMs before and after Csf2ra cDNA transfer showing macrophage morphology (Diff-Quick stain; phase contrast) and vector-derived GM-CSF receptor α expression (CD116 immunostaining, DAPI counterstain; immunofluorescence). Magnification, 63×. (B) Phenotypic analysis. Flow-cytometric analysis of cell-surface markers on BMDMs before or after mCsf2ra-LV transduction (Csf2ra−/− or Csf2ra−/− + Csf2ra-LV, respectively) and culture in medium containing GM-CSF (n = 3). Numbers indicate the mean fluorescence intensity of cells in the indicated gate. (C) Clonogenic analysis. Hematopoietic progenitor colony count of CFU-GM, CFU-G, CFU-M, CFU-GEMM, and BFU-E is plotted for WT Lin− cells and day-14 Csf2ra gene-corrected macrophages (n = 3). Scale: 100 pixels. (D) Representative photomicrographs of typical hematopoietic progenitor colonies formed from wild-type lineage-negative cells (1,500 cells per well of a 6-well SmartDish) Scale: 100 pixels. (E) Representative photomicrographs of the colonies (top) formed from Csf2ra gene-corrected macrophages (50,000 cells per well) and colony morphology assessed by cytospin (Diff-Quick stain; phase contrast) Magnification, 40×. The colony-forming units were either CFU-GM or CFU-M.
Compared to GM-CSF signaling-deficient macrophages, GM-CSF receptor gene correction confers a profound survival advantage, which manifests as soon as transgene expression begins and complements the GM-CSF receptor defect to restore GM-CSF responsiveness.19 While the early presence of GM-CSF in the culture medium during transduction improves the overall production of gene-corrected macrophages, it obfuscates determination of initial transduction efficiency, since the newly transduced cells immediately begin to outcompete the non-transduced cells. Thus, the initial LSK cell transduction rate is difficult to measure accurately by use of vector copy number (VCN) or clonogenic colony-forming units (CFU-Cs). Notwithstanding, because such methods can provide an estimate of transduction rate, we measured VCN data for the gene-corrected cell pool used for PMT in this study and found it to be 2.40 ± 0.46 (mean ± SD; n = 6).
Analysis of hematopoietic progenitor CFUs in gene-corrected macrophages showed a complete absence of burst-forming unit-erythroid (BFU-E), CFU-G, and multipotential progenitor cell (CFU-G, erythrocyte, macrophage, and megakaryocyte [CFU-GEMM]) colonies (Figure 2C). We did observe a very low rate of granulocyte and/or macrophage progenitor cell colony formation (CFU-granulocyte and macrophage [CFU-GM] and CFU-macrophage [CFU-M] colonies) (Figures 2C and 2E). These colonies were smaller (Figure 2E, top) compared to the typical colonies derived from the hematopoietic progenitor cells (Lin− cells; Figure 2D). The frequency of CFUs calculated based on Poisson statistics using L-Calc software (Stem Cell Technologies, Vancouver, BC, Canada) was 1.0E−05 ± 5.3E−06 (n = 3). Morphologic characterization of the colonies formed from gene-corrected macrophages confirmed that they were CFU-GM and CFU-M colonies (Figure 2E, bottom). These results show that LV-mediated Csf2ra gene-corrected macrophages derived from Csf2ra−/− HSPCs are mature macrophages that are not contaminated with transduced stem cells, precluding the probability of transduced stem cells continuing to differentiate in the lung after PMT.
Functional Characterization of Csf2ra Gene-Corrected Macrophages before PMT
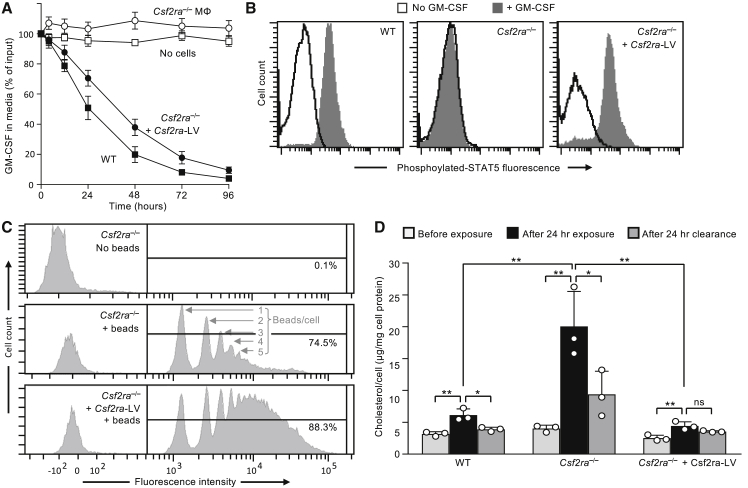
Gene-corrected macrophages rapidly cleared GM-CSF from the culture media with kinetics similar to that of wild-type (WT) LSK-cell-derived macrophages, whereas non-transduced Csf2ra−/− LSK-cell-derived macrophages did not (Figure 3A). Gene-corrected macrophages were capable of GM-CSF-stimulated phosphorylation of STAT5, similar to WT LSK-derived macrophages, while non-transduced Csf2ra−/− macrophages were not (Figure 3B). Gene-corrected macrophages had increased phagocytosis of opsonized, fluorescent microspheres, compared to non-transduced macrophages (Figure 3C). Finally, we evaluated the functional effects of restoring GM-CSF signaling in Csf2ra−/− macrophages by measuring the surfactant clearance capacity, which measures the GM-CSF-stimulated capacity to prevent cholesterol accumulation following surfactant exposure by promoting cholesterol excretion.12 Non-transduced Csf2ra−/− macrophages accumulated more cholesterol following surfactant exposure than did WT macrophages, but cholesterol clearance was normalized in gene-corrected macrophages (Figure 3D). These results show that mCsf2ra-LV transduction of Csf2ra−/− macrophages restored GM-CSF receptor α expression and GM-CSF receptor-dependent functions, including binding and clearance of GM-CSF, GM-CSF-stimulated STAT5 phosphorylation, phagocytosis, and cholesterol clearance.
Figure 3.
Functional Characterization of Csf2ra Gene-Corrected Macrophages before PMT
(A) GM-CSF clearance assay. Vector-mediated Csf2ra expression in Csf2ra−/− BMDMs restores clearance of GM-CSF from culture medium. Groups include BMDMs from wild-type mice (WT), Csf2ra−/− mice before or after mCsf2ra-LV transduction (Csf2ra−/− or Csf2ra−/− + mCsf2ra-LV, respectively), and medium without cells (No cells). Data indicate mean ± SD of 3 independent LSK-derived macrophage preparations and transductions per condition at each time point. (B) STAT5 phosphorylation assay showing that Csf2ra-LV transduction restores GM-CSF signaling in Csf2ra−/− BMDMs. (C) Phagocytosis assay. Flow-cytometry-based quantification of phagocytic internalization of opsonized latex beads. Histogram peaks showing internalization of 1, 2, 3, etc., beads per cell (indicated) by Csf2ra−/− BMDMs before and after mCsf2ra-LV transduction and culture in medium containing GM-CSF. The percentage of cells containing internalized latex beads is indicated for each condition (n = 3). (D) Surfactant clearance assay (n = 3). The ability of BMDMs (non-transduced WT or Csr2ra−/−, or mCsf2ra-LV-transduced Csf2ra−/−) to clear surfactant was evaluated by measuring surfactant-derived cholesterol content before or immediately after a 24-h “pulse” exposure to surfactant, or after pulse exposure, washing to remove extracellular surfactant, and culture for 24 h to permit clearance of surfactant-derived cholesterol (indicated). *p < 0.05; **p < 0.01; ns, not significant.
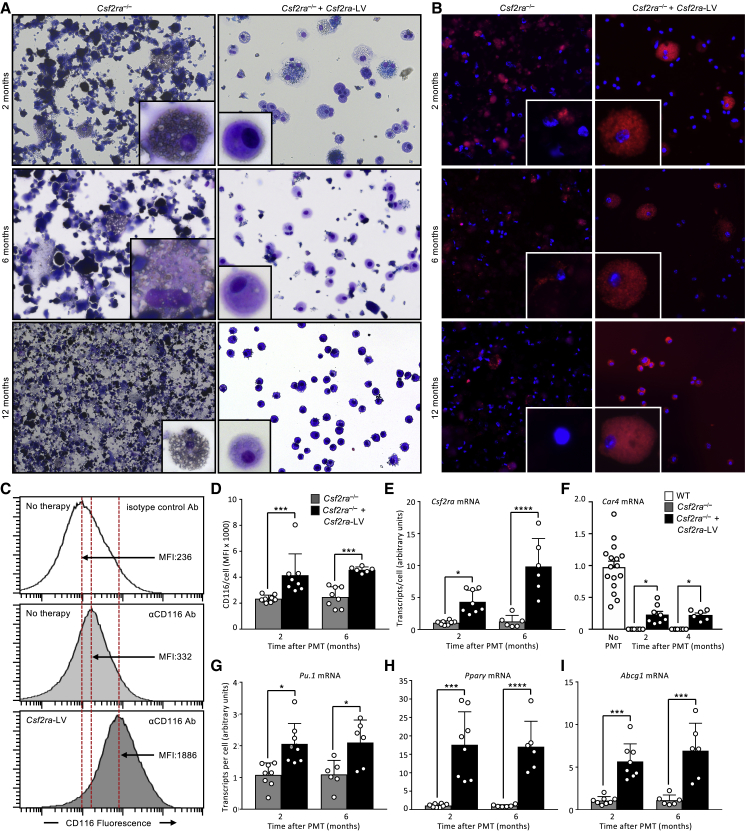
Engraftment Efficiency and Characterization of Alveolar Macrophages after PMT in Csf2ra−/− Mice
The effects of gene-PMT therapy on the alveolar macrophage population were determined after administering gene-corrected macrophages directly into the lungs of Csf2ra−/− mice (2 million cells per mouse) by recovering bronchoalveolar lavage (BAL) at subsequent times and evaluating engraftment, VCN, cytology, macrophage morphology, expression of genes normally expressed in alveolar macrophages, and alveolar macrophage biomarkers of PAP (reduced levels of Pu.1, Pparg, and Abcg1 mRNA transcripts). The percentage of donor-derived, gene-corrected macrophages recovered from the BAL cells were 31% ± 3.7%, 30.1% ± 2.11%, and 40.1% ± 7.11% at 2, 6, and 12 months after PMT. The VCN in BAL cells was 1.43 ± 0.69, 1.82 ± 0.20, and 1.69 ± 0.32 at 2, 6, and 12 months after PMT. Cytologic evaluation of BAL revealed gradual clearing of the PAP-related abnormal surfactant sediment and cellular debris over time in treated mice and progressive worsening in untreated mice (Figure 4A). Alveolar macrophages acquired a normal macrophage-like morphology in treated mice but remained large and foamy in untreated mice (Figure 4A, insets). CD116 was readily detectible on alveolar macrophages from gene-PMT-treated mice but not from non-treated mice (Figure 4B). Consistent with these results, the mean fluorescence intensity (MFI) of BAL cells after CD116 immunostaining (Figures 4C and 4D), and cellular Csf2ra mRNA levels (Figure 4E) were increased in gene-PMT-treated compared to untreated mice. The Csf2ra RNA levels in Csf2ra−/− mouse BAL cells at 2 and 6 months were 0.17 ± 0.06 and 0.6 ± 0.09 of the normal levels, respectively. Expression of mRNA for the unique alveolar macrophage marker Car4 was readily detected in BAL cells from WT mice and treated mice but was undetectable in cells from non-treated mice (Figure 4F). Further, cellular mRNA transcript levels for Pu.1, Pparg, and Abcg1 were increased in alveolar macrophages from treated mice compared to non-treated mice (Figures 4G–4I). Results from these data show that PMT of gene-corrected cells in Csf2ra−/− recipient mice resulted in stable, long-term engraftment and that alveolar macrophages have a normalized morphological appearance, express the Csf2ra transgene, and have increased expression of GM-CSF signaling axis components. Based on these results, we anticipate that PMT of LV-mediated CSF2RA gene-corrected human macrophages will engraft, persist, and have an efficacious and durable clinical benefit on the lung disease in children with hPAP.
Figure 4.
PMT of Csf2ra-LV Transduced BMDMs Restores Alveolar Macrophage Functions
Csf2ra−/− mice received PMT of non-transduced or mCsf2ra-LV-transduced Csf2ra−/− BMDMs and were evaluated at several subsequent times. (A and B) Photomicrographs of primary alveolar macrophages 2 months after PMT of the indicated BMDMs showing macrophage morphology (Diff-Quick stain; phase contrast) (A) and vector-derived GM-CSF receptor α expression (CD116 immunostain, DAPI counterstain; immunofluorescence) (B). Magnification, 20×. Inset, 40×. (C) CD116 expression on alveolar macrophages. Representative histogram of flow cytometry analysis stained with indicated antibody. (D) MFI of CD116 immunostaining of primary alveolar macrophages recovered at the indicated times after PMT. (E–I) Measurement of mRNA transcript levels by real-time qPCR for Csf2ra (E), Car4 (F), Pu.1 (G), Pparg (H), and Abcg1 (I) in primary alveolar macrophages at the indicated times after PMT. In (D)–(I), symbols represent data from one mouse. Error bars indicate mean ± SD for n = 5–8 mice per group (shown) evaluated at each time. *p < 0.05; ***p < 0.001; ****p < 0.001.
Efficacy of Gene-PMT Therapy of hPAP in Csf2ra−/− Mice
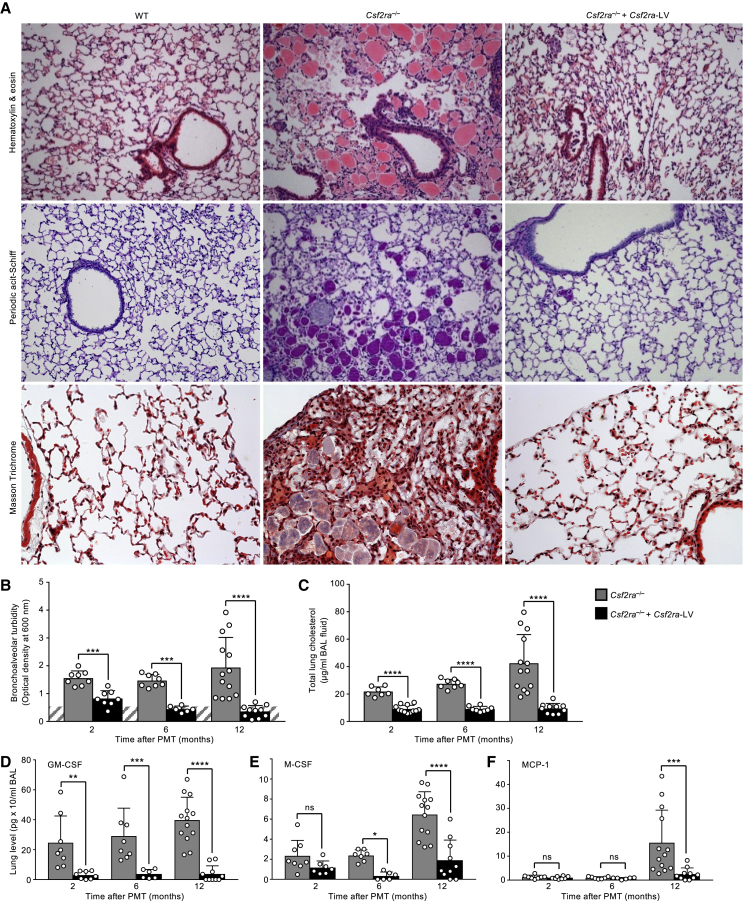
The effects of gene-PMT therapy on the severity of hPAP lung disease were determined by administering gene-corrected macrophages to Csf2ra−/− mice as described earlier and evaluating lung histopathology and the turbidity, cholesterol content, and levels of cytokine biomarkers of PAP in BAL fluid recovered at subsequent times after PMT. One year after PMT, the abnormal lung histopathology observed in age-matched, non-treated Csf2ra−/− mice was completely resolved in gene-PMT-treated mice, and the microscopic appearance of the lung parenchyma was similar to that of age-matched, non-treated WT mice (Figure 5A). We did not observe any evidence of pulmonary fibrosis in PMT-treated or non-treated Csf2ra−/− mice (Figure 5A, bottom). BAL turbidity (which reflects the extent of surfactant accumulation globally across the entire lung surface) declined over time and was within the normal range by 6 months after PMT in gene-PMT-treated mice and increased progressively in untreated mice (Figure 5B). Similarly, the cholesterol content of BAL was markedly reduced in gene-PMT-treated mice and increased progressively in untreated mice (Figure 5C). The levels of GM-CSF and M-CSF in BAL fluid also increased progressively over time in untreated mice and were significantly reduced at all times in gene-PMT-treated mice (Figures 5D and 5E). Finally, the level of MCP-1 in BAL fluid was increased in untreated mice and markedly reduced in gene-PMT-treated mice 1 year after PMT (Figure 4F). Results from multiple outcome measures demonstrate that PMT of gene-corrected macrophages is highly efficacious as therapy of hPAP in Csf2ra−/− mice.
Figure 5.
Pulmonary Instillation of Csf2ra-LV Transduced BMDMs without Myeloablation Is Efficacious as Therapy of Hereditary PAP in Csf2ra−/− Mice
Csf2ra−/− mice received one intrapulmonary instillation of mCsf2ra-LV-transduced Csf2ra−/− macrophages and were evaluated after 2, 6, or 12 months in parallel with age-matched, non-transduced WT or Csf2ra−/− mice as indicated. (A) Representative photomicrographs showing lung histology 1 year after PMT and staining with H&E (top), periodic acid-Schiff staining (middle), or Masson’s trichrome staining (bottom). Magnification, 10×, Inset, 40×. (B) BAL fluid turbidity in PMT-treated Csf2ra−/− mice and age-matched, non-treated WT or Csf2ra−/− mice. (C) Total lung surfactant-derived cholesterol recovered BAL. (D–F) Total lung levels of GM-CSF (D), M-CSF (E), and MCP-1 (F) recovered by BAL at 2, 6, and 12 months after one PMT. *p < 0.05; **p < 0.01; ***p < 0.001; ****p < 0.001; ns, not significant.
Proliferation of Transplanted Macrophages in Response to Pulmonary GM-CSF
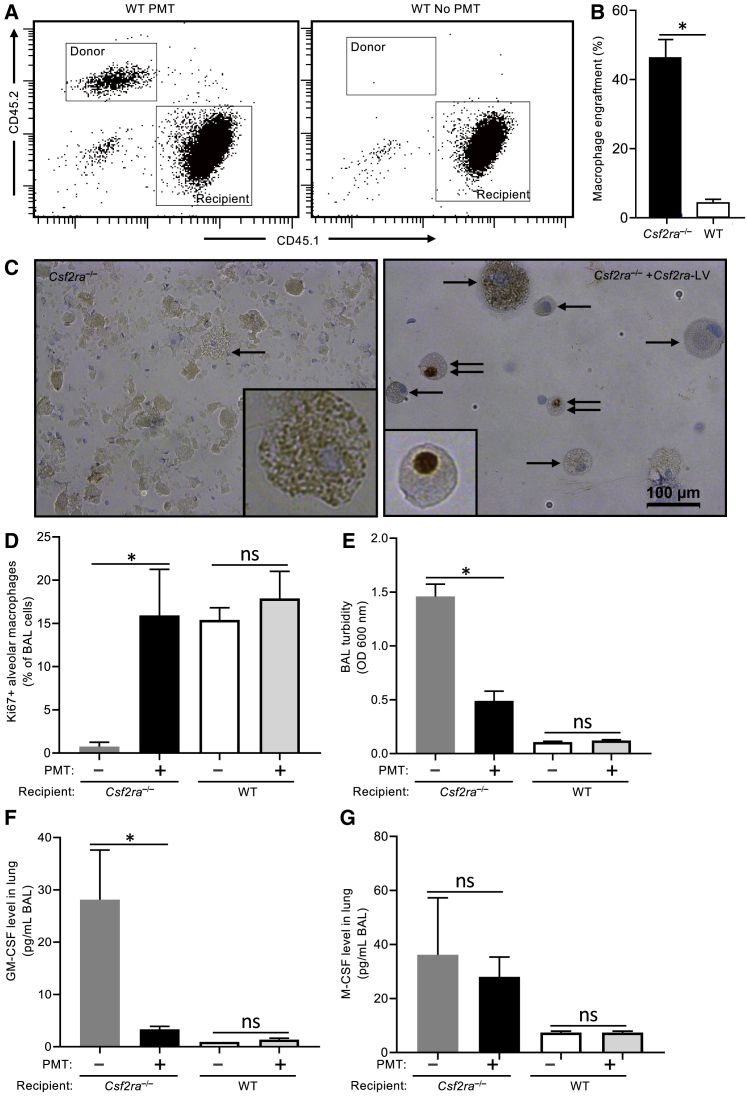
The effects of pulmonary GM-CSF on the local proliferation of transplanted macrophages was determined by transplanting Csf2ra−/− mouse LSK-cell-derived, Csf2ra-LV gene-corrected (i.e., CD116+) macrophages (CD45.2+) (2 × 106 cells per mouse) into either Csf2ra−/− (CD45.2+) or WT (CD45.1+) recipient mice. Age-matched, untreated WT and Csf2ra−/− mice served as controls. One month after PMT, mice were evaluated for the presence of donor-cell-derived CD116+ alveolar macrophages, Ki67+ alveolar macrophages, BAL turbidity (an excellent measure of global PAP disease severity19), and levels of pulmonary GM-CSF and M-CSF (biomarkers of PAP severity). Donor cell engraftment was measured by flow cytometry using antibodies recognizing donor (CD45.2) or recipient (CD45.1) cells (Figure 6A), and quantification was done by determining the percentage of cells that were CD116+ (immunofluorescence). Donor macrophage engraftment was 46.4% ± 5.1% (mean ± SEM) in Csf2ra−/− and 4.4% ± 0.97% in WT recipients (Figure 6B). The percentage of Ki67+ alveolar macrophages in PMT-treated mice was significantly higher compared to that in untreated Csf2ra−/− mice (15.74% ± 2.80% versus 0.75% ± 0.48% respectively; Figures 6C and 6D), consistent with proliferation of these cells in response to the increased pulmonary GM-CSF (28.2 ± 9.5 pg/mL versus 3.33 ± 0.58 pg/mL, respectively; Figure 6F) and the presence of functional GM-CSF receptors in the gene-corrected donor macrophages. In contrast, PMT-treated and untreated WT mice had similar levels of Ki67+ immunostaining, consistent with the presence of functional GM-CSF receptors on normal endogenous macrophages. The pulmonary GM-CSF levels in PMT recipients were also reduced significantly 1 month after PMT, consistent with clearance by the transplanted, GM-CSF receptor-positive donor macrophages (Figure 6F). In contrast, 1 month after PMT, pulmonary M-CSF levels had not yet declined significantly (Figure 6G). These results demonstrate that PMT-derived Csf2ra gene-corrected macrophages proliferate in response to high levels of pulmonary GM-CSF.
Figure 6.
Proliferation of Csf2ra Gene-Corrected Macrophages in Response to Pulmonary GM-CSF
WT (CD45.1) and Csf2ra−/− mice received one intrapulmonary instillation of mCsf2ra-LV-transduced Csf2ra−/− macrophages (CD45.2) and were evaluated at 1 month in parallel with age-matched, non-transplanted WT or Csf2ra−/− mice as indicated. (A) Representative flow plots showing donor macrophage (CD45.2) engraftment in WT recipients (CD45.1) in BAL cells. (B) Quantitation of donor macrophage engraftment in Csf2ra−/− (assessed by number of CD116+ cells by immunofluorescence) and WT (assessed by CD45.2 donor/CD45.1 recipient chimerism by flow cytometry) recipients. (C) Ki67 (proliferation marker, DAB counterstain) staining in alveolar macrophages. Single black arrows indicate Ki67− cells (blue nuclear stain), Double black arrows indicate Ki67+ cells (crisp, dark brown nuclear stain) Scale bars, 100 μm (D) Quantification of Ki67+ alveolar macrophages in Csf2ra−/− and WT recipients with or without PMT. (E) BAL fluid turbidity. (F) Total lung levels of GM-CSF. (G) Total lung levels of M-CSF. *p < 0.05; ns, not significant.
Safety of PMT Therapy of hPAP
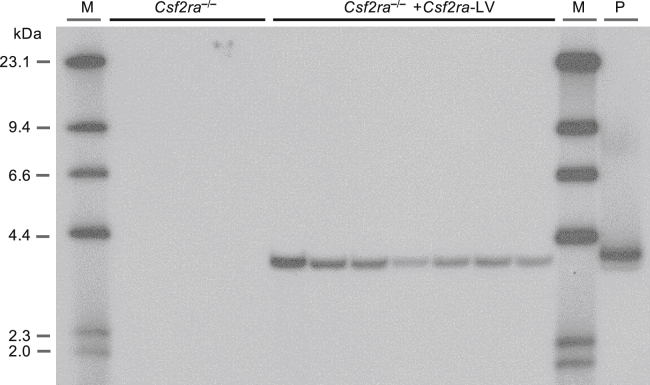
Vector-related safety was evaluated by determining the stability of the integrated provirus. This was accomplished using standard protocols23 to transduce Csf2ra−/− LSK cells with mCsf2ra-LV followed by culture in vitro to permit expansion and differentiation into gene-corrected macrophages and structural analysis of the integrated vector backbone in genomic DNA using Southern blot analysis. LSK cells from Csf2ra−/− donors (n = 7) were isolated, and transductions were performed using three different lots of mCsf2ra-LV. The transduced LSK cells were differentiated and expanded for 14 days to generate macrophages. Southern blot analysis of genomic DNA resulted in detection of a single vector-specific, 3.9-kb Eco-R1 fragment in transduced cells and no detection of mCsf2ra-LV-containing bands in non-transduced cells (Figure 7). These results demonstrate the absence of large deletions and/or splicing and/or recombination events during proviral integration, thus confirming the presence of a stable provirus.
Figure 7.
Stable Integration of mCsf2ra-LV in BMDMs
Csf2ra−/− donor-derived LSK cells (n = 7) were transduced with mCsf2ra−/−-LV at different MOIs, cultured for 21 days, and then evaluated by Southern blotting analysis. A single band of 3.9 kb was observed in Csf2ra LV gene-corrected macrophages.
Overall safety and tolerability of gene-PMT therapy was evaluated by monitoring the behavior of treated and non-treated age-matched Csf2ra−/− mice daily over the 1-year safety monitoring period, which did not identify any abnormal behaviors or differences between the two groups. Further, the body weights of gene-PMT-treated and non-treated age-matched Csf2ra−/− mice were similar at 2, 6, and 12 months after treatment (27.3 ± 1.74 g per mouse [n = 7] versus 25.6 ± 0.94 g per mouse [n = 12], p = 0.3478; 28.7 ± 1.60 g per mouse [n = 8] versus 26.9 ± 1.00 g per mouse [n = 6], p = 0.3821; and 31.4 ± 1.24 g per mouse [n = 13] versus 31.3 ± 1.64 g per mouse [n = 10], p = 0.9857, respectively).
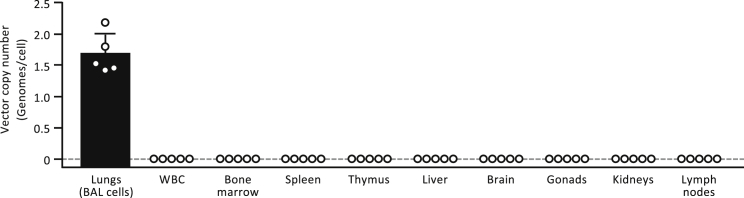
PMT-related safety was evaluated by determining the organ distribution of gene-corrected cells. This was accomplished by administering gene-corrected macrophages to Csf2ra−/− mice as described earlier, followed by quantification of mCsf2ra-LV-specific DNA (i.e., VCN) in organ-specific genomic DNA by real-time PCR analysis. One year after PMT, vector-specific DNA was readily detected in lung cells recovered by BAL in which the (mean ± SD) VCN of mCsf2ra-LV was 1.69 ± 0.32 genomes per cell (Figure 8). In contrast, vector-specific DNA was not detected in the circulation (white blood cells), other hematopoietic organs (bone marrow, spleen), organs involved in extramedullary hematopoiesis (kidney, liver, lymph nodes), or non-hematopoietic organs (brain, thymus, gonads) (Figure 8). These results show that gene-corrected macrophages remain confined to the lungs after PMT in Csf2ra−/− mice.
Figure 8.
Localization of Transplanted Macrophages after Intrapulmonary Administration
Csf2ra−/− mice received PMT of mCsf2ra-LV-transduced Csf2ra−/− BMDMs, and the indicated tissues were collected 1 year later and evaluated for the presence of transplanted macrophages by measuring the amount of mCsf2ra-LV vector DNA by qPCR using vector-specific primers. Results are expressed as the number of vector genomes per cell as described in Materials and Methods. Each circle represents results for a separate PMT-treated mouse. Error bars represent the mean ± SD vector copy number for five mice per group.
The effects of gene-PMT therapy on hematologic indices in gene-PMT-treated and non-treated age-matched Csf2ra−/− mice were evaluated 2, 6, and 12 months after PMT. No differences were observed in red blood cell indices (red blood cell count, hemoglobin, hematocrit) between groups of treated and non-treated mice at any time (Table 1). Red blood cell volume (mean corpuscular volume [MCV]), hemoglobin (mean cell hemoglobin [MCH]), and hemoglobin concentration (MCHC) were also not different between treated and non-treated mice at 2, 6, and 12 months (p > 0.1069 for all comparisons; n = 7–13 mice per group at each time point; data not shown). The total white blood count was similar in treated and non-treated mice at 2 months but increased progressively in non-treated mice thereafter and was significantly increased at 1 year after PMT in non-treated compared to gene-PMT-treated mice (Table 1). Similarly, the absolute counts of neutrophils, lymphocytes, and monocytes followed a similar pattern and were similar in treated and non-treated mice at 2 and 6 months but significantly increased in non-treated mice 12 months after PMT (Table 1). In contrast, no differences between treated and non-treated mice were observed in absolute counts of eosinophils, basophils, or platelets at any times (Table 1).
Table 1.
Effects of Intrapulmonary Instillation of mCsf2ra-LV-Transduced Macrophages on Hematological Indices in Csf2ra−/− Mice
| Hematologic Parameter | Normal Range | Time after PMT (Months) | Untreated Median [IQR] (n) | PMT Median [IQR] (n) | p Value |
|---|---|---|---|---|---|
| RBC count, × 106/μL | 8.32–11.2 | 2 | 8.02 [7.70–8.76] (7) | 8.05 [7.73–8.51] (12) | 0.8208 |
| 6 | 7.78 [7.053–8.26] (8) | 7.87 [7.44–8.35] (6) | 0.7546 | ||
| 12 | 8.48 [7.89–8.94] (13) | 8.29 [7.64–8.42] (10) | 0.3670 | ||
| Hemoglobin, g/dL | 14.8–16.3 | 2 | 13.4 [13.0–15.2] (7) | 13.9 [13.6–14.2] (12) | 0.7260 |
| 6 | 11.7 [10.2–12.2] (8) | 11.6 [10.5–11.9] (6) | 0.9201 | ||
| 12 | 12.1 [11.7–13.5] (13) | 12.35 [12.0–13.0] (10) | 0.8180 | ||
| Hematocrit, % | 40.9–54.6 | 2 | 38.3 [36.9–43.8] (7) | 39.9 [38.35–41.68] (12) | 0.7575 |
| 6 | 37.7 [33.3–49.78] (8) | 38.2 [36.4–41.0] (6) | 0.6620 | ||
| 12 | 40.4 [37.0–42.1] (13) | 38.5 [37.6–39.8] (10) | 0.2500 | ||
| WBC, × 103/μL | 1.18–14.7 | 2 | 3.00 [2.44–4.12] (7) | 3.69 [2.67–4.79] (12) | 0.6061 |
| 6 | 2.97 [2.52–3.44] (8) | 3.19 [2.55–3.55] (6) | 0.6600 | ||
| 12 | 4.68 [2.84–5.70] (13) | 1.78 [1.28–1.78] (10) | <0.0001 | ||
| Neutrophils, × 103/μL | 0.08–2.27 | 2 | 0.15 [0.07–0.26] (7) | 0.20 [0.06–0.26] (12) | 0.4192 |
| 6 | 0.19 [0.12–0.27] (8) | 0.25 [0.21–0.46] (6) | 0.0636 | ||
| 12 | 0.87 [0.62–2.07] (13) | 0.34 [0.14–0.58] (10) | 0.0015 | ||
| Lymphocytes, × 103/μL | 0.74–12.6 | 2 | 2.34 [1.98–3.4] (7) | 3.03 [2.17–3.88] (12) | 0.4824 |
| 6 | 2.29 [2.01–2.74] (8) | 1.96 [1.82–2.44] (6) | 0.1419 | ||
| 12 | 2.31 [1.62–3.54] (13) | 1.27 [0.85–1.66] (10) | 0.0011 | ||
| Monocytes, × 103/μL | 0.0–0.19 | 2 | 0.36 [0.24–0.47] (7) | 0.38 [0.25–0.48] (12) | 0.7263 |
| 6 | 0.34 [0.26–0.46] (8) | 0.33 [0.28–0.59] (6) | 0.9757 | ||
| 12 | 0.44 [0.37–0.90] (13) | 0.18 [0.3–0.26] (10) | 0.0003 | ||
| Eosinophils, × 103/μL | 0.01–0.48 | 2 | 0.01 [0.01–0.02] (7) | 0.02 [0.00–0.04] (12) | <0.9025 |
| 6 | 0.03 [0.01–0.05] (8) | 0.01 [0.01–0.17] (6) | <0.8415 | ||
| 12 | 0.08 [0.02–0.18] (13) | 0.05 [0.18–0.08] (10) | 0.2480 | ||
| Basophils, × 103/μL | 0.0–0.06 | 2 | 0.00 [0.00–0.01] (7) | 0.00 [0.00–0.01] (12) | 0.8808 |
| 6 | 0.00 [0.00–0.01] (8) | 0.00 [0.00–0.02] (6) | >0.9999 | ||
| 12 | 0.03 [0.01–0.55] (13) | 0.02 [0.01–0.03] (10) | 0.2457 | ||
| Platelets, × 103/μL | 749–2,828 | 2 | 615 [554–822] (7) | 695 [598–729] (12) | 0.4824 |
| 6 | 687 [613–857] (8) | 1059 [531–1,059] (6) | 0.9497 | ||
| 12 | 943 [747–986] (13) | 841 [686–929] (10) | 0.3758 |
Csf2ra−/− mice received intrapulmonary instillation of mCsf2ra-LV transduced macrophages (2 × 106 cells per mouse) (PMT) once, and at 2, 6, and 12 months later, blood was obtained from PMT-treated and age-matched, untreated Csf2ra−/− mice. Data are presented as medians (interquartile range [IQR]), and between-group comparisons were done using non-parametric methods (Mann-Whitney rank-sum test) for consistency, since results for some groups were either not normally distributed or of unequal variance. Normal ranges for C57BL6 mice spanning a broad range of ages from a large cohort of mice including young and old mice through 1 year are provided from the Jackson Laboratory Mouse Phenome Database (n = 93). RBC, red blood cell, WBC, white blood cell. p values < 0.05 were considered significant.
The effects of gene-PMT therapy on molecular mediators of inflammation in the lungs were measured in treated and non-treated age-matched Csf2ra−/− mice at 2, 6, and 12 months after PMT as described earlier. Interleukin (IL)-6, IL-1β, and tumor necrosis factor α (TNF-α) were absent or present in trivial amounts in BAL fluid from treated and nontreated mice at 2 and 6 months, and IL-6 and TNF-α were increased in non-treated compared to treated mice at 12 months (Table 2).
Table 2.
Effects of Intrapulmonary Instillation of mCsf2ra-LV-Transduced Macrophages on Lung Proinflammatory Cytokine Levels in Csf2ra−/− Mice
| Cytokine in BAL Fluid | Time after PMT (Months) | Untreated Median [IQR] (n) | PMT Median [IQR] (n) | p Value |
|---|---|---|---|---|
| IL-6, pg/mL | 2 | 0.00 [0.00–16.0] (8) | 0.00 [0.00–0.00] (8) | 0.4667 |
| 6 | 0.00 [0.00–0.00] (8) | 0.00 [0.00–0.00] (6) | >0.9999 | |
| 12 | 17.8 [6.70–68.5] (13) | 5.28 [1.35–19.9] (10) | 0.0436 | |
| IL-1β, pg/mL | 2 | 0.00 [0.00–8.12] (8) | 0.00 [0.00–2.67] (8) | 0.8825 |
| 6 | 0.00 [0.00–0.00] (8) | 0.00 [0.00–0.00] (6) | >0.9999 | |
| 12 | 1.05 [0.00–10.7] (13) | 7.39 [0.00–22.5] (10) | 0.4308 | |
| TNF-α, pg/mL | 2 | 3.30 [1.11–5.02] (8) | 3.04 [0.04–4.45] (8) | 0.5717 |
| 6 | 0.00 [0.00–0.00] (8) | 0.00 [0.00–0.00] (6) | >0.9999 | |
| 12 | 8.26 [5.28–16.3] (13) | 4.65 [4.22–7.33] (10) | 0.0223 |
Csf2ra−/− mice received intrapulmonary instillation of mCsf2ra-LV-transduced macrophages (2 × 106 cells per mouse) (PMT) once and at 2, 6, and 12 months later. BAL fluid was obtained from PMT-treated and age-matched, untreated Csf2ra−/− mice and evaluated as described in Materials and Methods. Data are presented as medians (interquartile range (IQR), and between-groups comparisons were done using non-parametric methods (Mann-Whitney rank-sum test) for consistency since results for some groups were either not normally distributed or of unequal variance. p values < 0.05 were considered significant.
Together, these results demonstrate that gene-PMT therapy of hPAP was safe and well tolerated in Csf2ra−/− mice and prevented the increase in cellular and molecular inflammation that occurs over time in non-treated Csf2ra−/− mice.
Discussion
In this report, we demonstrate that LV vector-mediated Csf2ra cDNA expression in Csf2ra−/− macrophages complemented the GM-CSF receptor α chain dysfunction and restored GM-CSF-dependent functions, including binding and internalization of GM-CSF, phosphorylation of STAT5, expression of macrophage markers, phagocytic uptake, and surfactant clearance. PMT of gene-corrected macrophages in Csf2ra−/− mice resulted in long-term engraftment of gene-corrected macrophage in the lungs and demonstrated that gene-PMT therapy of hPAP in Csf2ra−/− mice was highly efficacious, durable, safe and well tolerated.
The extraordinary efficacy of gene-PMT therapy of hPAP observed is supported by multiple findings, including restoration of GM-CSF signaling and functions in Csf2ra−/− macrophages in vitro and in vivo; pulmonary engraftment and long-term persistence of gene-corrected macrophages; and correction of PAP-specific cytologic, histologic, and biomarker abnormalities, indicating that gene-PMT therapy restored pulmonary surfactant homeostasis. Long-term engraftment is shown by the presence of vector-derived Csf2ra mRNA and protein expression in alveolar macrophages recovered 1 year after PMT of gene-corrected macrophages. This result is also supported by similar results seen after PMT of WT macrophages in Csf2rb−/− mice19 and is consistent with reports indicating that alveolar macrophages self-renew independently of extrapulmonary myelopoiesis and/or extrapulmonary recruitment28, 29 under control by pulmonary GM-CSF via reciprocal feedback loop regulating population size.19 The re-establishment of a functional alveolar macrophage population is supported by the presence of morphologically normal alveolar macrophages expressing Csf2ra, GM-CSF-dependent target genes (Pu.1, Pparg, and Abcg1), and an alveolar macrophage-specific marker Car4.30 The progressive increase in Csf2ra mRNA level in BAL cells after PMT indicates gradual replacement of endogenous alveolar macrophage population by gene-corrected macrophages, as was observed after PMT of WT macrophages in Csf2rb−/− mice, and is consistent with the survival advantage demonstrated for GM-CSF-responsive macrophages in vitro and in vivo.19 The magnitude and durability of the treatment effect is demonstrated by complete correction of PAP-related lung histopathology and normalization of BAL turbidity (an excellent global measure of pulmonary surfactant accumulation in PAP19) 1 year after a single PMT of gene-corrected macrophages. Study limitations pertaining to therapeutic efficacy include that the results did not establish a minimum effective dose, maximum tolerated dose, and significant dose-response relationship or exclude a possible contribution to efficacy due to expansion of a progenitor cell population. Other long-term studies are underway to address these limitations and will be reported at a later date.
The progressive parallel increase in numbers of blood neutrophils, lymphocytes, and monocytes and pulmonary IL-6, IL-1β, and TNF-α in Csf2ra−/− mice is likely a manifestation of an innate immune deficiency caused by the loss of GM-CSF stimulation of myeloid cells accompanied by a secondary systemic inflammatory process as a result of progressive worsening of lung disease over time. The absence of these abnormalities in treated mice suggests that gene-PMT therapy may improve innate immune lung host defense. GM-CSF is well-known to stimulate multiple host defense functions in human and murine alveolar macrophages and circulating neutrophils, including cell adhesion, phagocytosis, production of reactive oxygen species, and microbial killing.31, 32 In humans, disruption of GM-CSF signaling by neutralizing auto-antibodies is associated with increased infection-related morbidity and mortality from opportunistic and common microbial pathogens.31 In mice, disruption of GM-CSF signaling also increases the susceptibility to a broad range of microbial pathogens (reviewed elsewhere33) and increases in infection-related mortality.34 Further studies are needed to define this aspect of the potential therapeutic benefit of gene-PMT therapy of hPAP.
Our clinical strategy incorporates several features intended to reduce risk and improve safety, including use of a third-generation LV expressing the transgene from a weak physiological promoter, transplantation of mature macrophages rather than stem cells, and no use of myeloablative conditioning and immunosuppression. The mCsf2ra-LV-mediated integration of the transgene into murine HSPCs remained stable throughout in vitro macrophage differentiation. The observation that gene-corrected macrophages remained confined to the lungs after PMT reduces concern about potential risks from germline transmission of recombinant vector-derived DNA. Importantly, gene-PMT therapy did not result in parenchymal lung damage, systemic inflammation, germline transmission of vector DNA, or behavioral abnormalities. Safety-related study limitations include that integration site analysis was not performed after gene-PMT therapy. Further, the present study was focused on long-term safety, not the potential for acute toxicity.
Formal mouse toxicology studies are now underway to address the safety of gene correction and PMT and will be reported elsewhere. Briefly, these include a single ascending dose (SAD) study with evaluation at 24 h and 14 days to assess the potential for acute pulmonary responses such as bronchospasm, inflammation, and pneumonia and anaphylaxis. Also ongoing is a repeat ascending dose (RAD) study directly mimicking the planned clinical trial design with evaluations at 6 months to assess the potential for adverse clinical outcomes, gene-transfer vector-related safety, the bio-distribution of gene-corrected cells, and anti-GM-CSF receptor antibody responses.
In conclusion, results of this non-clinical study in a validated animal model demonstrate the safety, tolerability, and efficacy of gene-PMT therapy of hPAP and support the feasibility of testing this approach in humans with hPAP caused by CSF2RA mutations.
Materials and Methods
The procedures and assays utilized in this study have been described previously19 and are summarized briefly in the following text.
Mice
The generation of the Csf2ra−/− mice has been described previously.20 Briefly, Csf2ra−/− mice were created in C57BL/6J background (express the Ptprcb [CD45.2 or Ly5.2] allele) using standard CRISPR/Cas9 genome editing technology35 in the Transgenic Animal and Genome Editing Core (TAGE) at CCHMC. Single-strand guide RNA (sgRNA) targeting sequences were designed to recognize exon 2 (positions 1160–1179 bp) and exon 3 (positions 1322–1341) of the mouse Csf2ra gene and resulted in a deletion of 220 bp spanning DNA encoding most of the signal peptide and part of the mature protein encoded in exons 2–3. These mice do not express a functional Csf2ra protein. Congenic strain B6.SJL-PtprcaPepcb/BoyJ (stock no. 002014/B6 Cd45.1) mice were used as PMT recipients in WT transplant experiments. All animals used in this study were bred and experiments were conducted in the Cincinnati Children’s Hospital Research Foundation Vivarium using standard institutional animal care protocols and appropriate institutional animal care and use committee (IACUC) approvals.
Cell Lines
The cell line created to complement the Csf2ra gene defect was derived from an alveolar macrophage cell line, mAM, which was previously isolated from Csf2-deficient mice as described.26 Briefly, mAM cells were created without transformation by culturing Csf2-deficient mAMs in DMEM supplemented with mouse L929 cell-conditioned medium. While mAM cells do not express GM-CSF receptors and, therefore, do not undergo GM-CSF-stimulated differentiation, they spontaneously secrete M-CSF, which supports autologous cell survival and proliferation, making them useful for creating cell lines to test either Csf2ra or Csf2rb gene complementation, as we have previously reported.23
LVs, LSK-Cell Transduction, and Differentiation and Expression of Transduced Macrophages
The CDS (1,167 bp) for mouse Csf2ra (NCBI: NM_009970.2) was synthesized by GenScript (Piscataway, NJ, USA) with BspE1 and SalI restriction endonuclease sites included at the 5′ and 3′ ends, respectively, for cloning purposes. This CDS was inserted into a third-generation, self-inactivating LV (pRRL.cPPT.EFS.hCSF2RAcoopwpre)23 construct expressing a codon-optimized CSF2RA CDS driven by an internal elongation factor 1α (short; EFS) promoter, by digesting the plasmid using the restriction enzymes SgrA1 and SalI (New England Biolabs [NEB], Ipswich, MA, USA). This digestion step removes the hCSF2RA cDNA from the vector backbone. Using T4 DNA ligase (NEB) the BspE1 and SalI digestion product was ligated to the vector backbone in place of the hCSF2RA CDS. The cloned mouse Csf2ra LV vector exactly resembles the human clinical vector, except for the mouse Csf2ra CDS replacing the human CSF2RA CDS. This mouse vector permits evaluation of the safety and efficacy of the Csf2ra transgene in Csf2ra−/− mouse model. The sequence of the cloned vector plasmid (EFS.mCsf2ra-LV) was verified by DNA sequencing (DNA Sequencing and Genotyping Core, CCHMC). LV production was done by transient transfection of 293T cells in a 100-mm cell-culture dish using the calcium chloride and 2× hepes buffered saline (HBS) precipitation method, with 8 μg LV plasmid, 10 μg codon-optimized short form of Gag/Pol, and 3 μg vesicular stomatitis virus glycoprotein (VSVg). The supernatant containing the lentivirus was harvested 36 and 48 h post-transfection and was concentrated to 800- to 1,000-fold by ultracentrifugation for 1.5 h at 25,000 rpm and 4°C. To obtain high-titer lentivirus purification, the ultracentrifugation step was carried out on a 20% sucrose gradient (Viral Vector Production Facility, CCHMC). Viral titers were determined in a mAM cell line26 derived from GM-CSF-deficient mice, after transduction with serially diluted virus and measuring the percentage of mAM cells expressing CD116+ cells by flow cytometer (BD Biosciences, San Jose, CA, USA).
STAT5 Phosphorylation Assay
Restoration of GM-CSF receptor function after LV transduction in Csf2ra−/− LSK-derived macrophages was tested using a GM-CSF-stimulated STAT5 phosphorylation assay following staining using anti-phospho STAT5 antibody (47/Stat5(pY694); BD Biosciences) by flow cytometry.19 The increase in the MFI of phosphorylated STAT5 after GM-CSF stimulation is a measure of GM-CSF receptor signaling activity.
Bone Marrow-Derived Macrophages (BMDMs)
The LSK cell isolation, transduction, expansion, and differentiation were performed using culture conditions optimized to generate macrophages as reported previously.19
CFU Assay
Murine bone marrow Lin− cells or LSK-derived Csf2ra gene-corrected macrophages were evaluated for the presence of hematopoietic progenitor cells using Methocult methylcellulose-based medium. The cells (1,500–50,000) were seeded in complete Methocult GF M3434 (STEMCELL Technologies, Vancouver, BC, Canada) that contains recombinant mouse stem cell factor (rm SCF), rm IL-3, recombinant human (rh) IL-6, rh insulin, and human transferrin (iron saturated) to detect formation of erythroid progenitor cells (BFU-E), granulocyte and/or macrophage progenitor cells (CFU-GM, CFU-G, and CFU-M), and multipotential progenitor cells (CFU-GEMM) and were scored for CFUs following 12 days of incubation. The colonies were plated on a SmartDish (6-well) plate (STEMCELL Technologies), and Stemgrid-6 (STEMCELL Technologies) was used during the colony-counting procedure. The colonies were scored using a Zeiss microscope-Axiovert 25 (Ziess, Jena, Germany). The colonies from the day-14 Csf2ra gene-corrected macrophages were plucked, and cytospins were done to assess the cell morphology.
GM-CSF Clearance Assay
Receptor-mediated GM-CSF clearance was evaluated as described previously.1 Briefly, WT, Csf2ra−/−, and Csf2ra gene-corrected macrophages were seeded at a concentration of 3–4 × 105 cells per milliliter in 24-well plates. At time 0, GM-CSF (1 ng/mL) was added to the culture dishes. The culture medium was collected at subsequent time points (4, 12, 24, 48, 72, and 96 h) and analyzed for the remaining GM-CSF levels over time by ELISA (R&D Systems). The levels of GM-CSF remaining at different time points are plotted as a percentage in comparison to the initial GM-CSF concentration input.
Flow Cytometry
BAL cells were stained with F4/80 and mouse CD116. LSK-derived macrophages were stained to detect surface expression of F4/80 (BM8), CD11c (HL3), and CD11b (M1/70). The antibodies were purchased from eBioscience (Waltham, MA, USA). The macrophage phenotype was assessed using the BD FACSCanto I (Becton Dickinson), and the results were analyzed using BD FACSDiva software. The donor (Csf2ra-LV, CD45.2) cells were distinguished from the BoyJ (CD45.1) recipient cells in the BAL by differential expression of alleles of the CD45 pan-leukocyte surface marker CD45.1/CD5.2. BAL cells were stained with F4/80 monoclonal antibody, clone BM8, APC; anti-mouse CD45.1 Pacific Blue, clone A20 (BioLegend, San Diego, CA, USA); and CD45.2 anti-mouse FITC (fluorescein isothiocyanate), clone 104 (BD PharMingen, San Jose, CA, USA) according to manufacturer’s instructions and acquired with a BD FACS Canto II instrument. Donor engraftment in BAL was analyzed by first gating on F4/80+ alveolar macrophages followed by assessing CD45.2 (donor) chimerism in WT (CD45.1) PMT recipients using BD FACSDiva software.
Phagocytosis Assay
The phagocytic capacity of Csf2ra−/− and gene-corrected macrophages was assessed by exposing the cells to opsonized, fluorescent Nile red beads as previously described12 and measuring the relative bead uptake using a flow cytometer (BD FACSCanto I).
Surfactant Clearance Assay
The surfactant clearance assay was performed as previously described.36 Briefly, 3–4 × 105 macrophages (non-transduced WT or Csf2ra–/–, or mCSF2ra-LV-transduced Csf2ra–/– BMDMs) were exposed to PAP patient surfactant for 24 h to allow uptake of surfactant. After 24 h, the macrophages were washed with prewarmed PBS to remove the loaded surfactant and incubated for another 24 h to assess the surfactant clearance capacity. Cellular lipids were extracted from the macrophages by adding 100% isopropanol to the wells. The cholesterol levels were measured using the Amplex Red Cholesterol Assay Kit (Thermo Fisher Scientific) and were normalized to the cellular protein content (Pierce BCA Protein Assay Kit, Thermo Fisher Scientific) at baseline (no surfactant exposure), 24-h surfactant exposure (surfactant load), and 24 h post-surfactant exposure (surfactant clearance).
BAL Collection and Turbidity Measurement
Epithelial lining fluid and non-adherent cells were collected from the experimental mice by BAL, as described previously.1 Briefly, 1 mL PBS containing 0.5 mM EDTA was instilled into the lungs of the mice and aspirated back 4–5 times to collect the BAL cells. This step was repeated an additional 4 times. The BAL aliquots were pooled and gently mixed to ensure a homogeneous suspension. The BAL turbidity measurements were performed by diluting 250 μL BAL with 750 μL PBS; optical density was measured at a wavelength of 600 nm with a DU 640B spectrophotometer (Beckman Coulter), and the results obtained were multiplied by the dilution factor.
PMT
Age-matched Csf2ra−/− mice of both sexes were enrolled and were randomly assigned to the experimental groups. The PMT procedure was performed as described previously using a dose of 2 million Csf2ra gene-corrected macrophages.19 The mice were monitored until recovery after the PMT procedure and were returned to cages for routine care and handling. The mice were monitored daily for behavioral, morbidity, and mortality assessments.
RT-PCR
Total RNA was extracted from the BAL cells using TRIzol reagent (Life Technologies, Carlsbad, CA, USA) according to the manufacturer’s instructions. cDNA synthesis was performed using SuperScript III First-Strand Synthesis System (Thermo Fisher Scientific). 100 ng cDNA was used for real-time qPCR runs to measure the relative expression of target genes Pu.1 (Mm00488142_m1), Ppar-γ (Mm01184322_m1), Abcg1 (Mm00437390_m1), Car4 (Mm00483021), and Csf2ra (Mm00438331_g1). The target gene expression was normalized to the 18S RNA (4310893E) expression. The primer probe sets were purchased from Thermo Fisher Scientific. The real-time qPCR runs were performed on an ABI 7300 Real-Time PCR System (Thermo Fisher Scientific). Data are expressed as fold change of the mean relative to gene expression of Csf2ra−/− mice.
Lung Histology
Lung inflation and fixing were performed using standard methods as described previously.1 Briefly, fixed lungs were processed for histology sections, and the slides were stained with H&E, Periodic acid-Schiff (PAS), or Masson Trichrome.
Immunostaining for GM-CSF Receptor Alpha (CD116)
Immunostaining for CD116 was performed by fixing the BAL cells sedimented onto cytospin slides for 10 min in 4% paraformaldehyde solution. The slides were washed thrice with PBS and blocked for 1 h in PBS containing 0.25% Triton X-100 (Sigma, St. Louis, MO, USA) and 5% normal goat serum (PBST). After blocking, the slides were incubated with anti-mouse GM-CSF receptor α-PE at a dilution of 1:50 in PBST. The slides were incubated in dark conditions at room temperature for 4 h or at 4°C overnight. The slides were then washed 3 times with PBS by incubating for 10 min between subsequent washes. Counterstaining was done with DAPI (Vector Laboratories, Burlington, CA, USA) before visualization in a Zeiss Axioplan 2 microscope (Zeiss). Photographs were taken at 10×, 20×, and 40× magnifications using AxioVision software. The quantification of CD116-stained BAL cells were determined by counting CD116+ and DAPI+ cells in five or more random 20× fields for every mouse analyzed. The percentage of CD116+ cells from transplanted mice were estimated by dividing the number of CD116+ cells by the total number of DAPI+ cells in all fields and multiplying the resulting number by 100.
In Vivo Evaluation of Proliferation of Transplanted Macrophages
Immunostaining for Ki67+ cells was performed on the BAL cells sedimented onto cytospin slides by the CCHMC Pathology Core Facility using anti-Ki67 antibody-Clone 30-9 (Roche Diagnostics, Basel, Switzerland). The slides were counterstained with species-specific secondary antibodies and detected with the OptiView DAB Kit (Roche). Photographs were taken at 20× and 40× magnification using a Leica DM2700 M bright-field microscope (Leica Microsystems, Buffalo, Grove, IL, USA). The quantification of Ki67+ alveolar macrophages was performed by counting the number of Ki67+ (brown nuclear staining) AMs and the total number of alveolar macrophages in five or more random 20× fields for every mouse. The percentage of CD116+ cells from transplanted mice was estimated by dividing the number of Ki67+ cells by the total number of AM in all fields and multiplying the resulting number by 100.
BAL Cholesterol
The total cholesterol levels in the BAL were measured using the Amplex Red Cholesterol Assay Kit (Thermo Fisher Scientific) as per the manufacturer’s protocol.12
ELISA
The concentrations of GM-CSF, M-CSF, and MCP-1 (PAP biomarkers), and inflammatory cytokines IL-6, IL-1β, and TNF-α were measured in the BAL by ELISA using the mouse quantikine kits from R&D Systems as described previously.1
Southern Blotting Analysis
Genomic DNA was isolated from Csf2ra−/− LSK-derived macrophages and LV-mediated Csf2ra gene-corrected macrophages (obtained from seven independent transductions) using the QIAGEN DNA lysis and protein precipitation method and recovered by precipitation with alcohol and dissolved in hydration buffer. Briefly, 10 μg genomic DNA from the macrophages were digested with AflII (Thermo Fisher Scientific). Day-14 macrophages from non-transduced Csf2ra−/− LSK cells served as negative controls. EFS.mCsf2ra LV plasmid DNA (50 pg) served as a positive control. The woodchuck hepatitis virus posttranscriptional regulatory element (wPRE) sequence-specific probe was used for the detection of labeled DNA. The digested DNA was resolved on a Tris base, acetic acid, and EDTA (TAE) agarose using procedures described previously.23
Hematological Analysis
Blood was collected from superior vena cava, and 25 μL blood was used to perform a complete blood count using a fully automated Hemavet.850 (Drew Scientific).
Systemic and Organ-Specific Localization of PMT-Derived Cells
Genomic DNA was extracted from BAL cells; white blood cells; and single-cell suspensions of liver, spleen, gonads, lymph nodes, bone marrow, brain, kidneys, and thymus using standard procedures (described in Southern Blotting Analysis, earlier in the Materials and Methods section). VCN was assessed in these tissues to detect the presence of Csf2ra gene-corrected macrophages after PMT procedure. qPCR was performed using primers that detect the R-U5 region of integrated provirus in the Applied Biosystems ABI7900HT Fast Real-Time PCR System (Thermo Fisher Scientific). The Ct values were normalized to the levels of mouse ApoB gene as described in detail previously.1
Statistical Analysis
Numeric data were evaluated for normality and variance using the Shapiro-Wilk and Levene median tests, respectively, and presented as mean ± SD (parametric data) or median and interquartile range (nonparametric data). Statistical comparisons were made with the Student’s t test, one-way ANOVA, or Kruskal-Wallis rank-sum test as appropriate; post hoc pairwise multiple comparison procedures were done using the Student-Newman-Keuls or Dunn’s method as appropriate. p values ≤ 0.05 were considered to indicate statistical significance. All studies used male and female mice by randomly assigning mice housed in the same cage to separate experimental groups but without formal randomization or blinding. Analyses were done using Prism software (v7.0d). All experiments were repeated at least twice, with similar results.
Author Contributions
P.A., T.S., and B.C.T. designed research. P.A., T.S., K.S., A.S., M.W., Y.M., J.M., D.B., C.C., and B.C. performed research. P.A., T.S., K.S., M.W., Y.M., J.M., B.C., and B.C.T. analyzed data. P.A., C.M., and B.C.T. wrote the paper. All authors reviewed, revised, and approved the final draft of the paper.
Conflicts of Interest
The authors declare no competing interests.
Acknowledgments
This work was supported by grants from the NIH (R01 HL085453 and R01 HL118342) and the Deutsche Forschungsgesellschaft (DFG; Cluster of Excellence REBIRTH to N.L. and T.M. and LA3680/2-1), by the Cincinnati Children's Hospital Medical Center (CCHMC) trustee grant (to P.A.), and by the Translational Pulmonary Science Center. Flow-cytometric data were acquired within the Research Flow Cytometry Core in the Division of Rheumatology, CCHMC. N.L. and T.M. were supported by the German Ministry of Education and Research (01EK1602A; iMACnet).
References
- 1.Suzuki T., Sakagami T., Rubin B.K., Nogee L.M., Wood R.E., Zimmerman S.L., Smolarek T., Dishop M.K., Wert S.E., Whitsett J.A. Familial pulmonary alveolar proteinosis caused by mutations in CSF2RA. J. Exp. Med. 2008;205:2703–2710. doi: 10.1084/jem.20080990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Martinez-Moczygemba M., Doan M.L., Elidemir O., Fan L.L., Cheung S.W., Lei J.T., Moore J.P., Tavana G., Lewis L.R., Zhu Y. Pulmonary alveolar proteinosis caused by deletion of the GM-CSFRalpha gene in the X chromosome pseudoautosomal region 1. J. Exp. Med. 2008;205:2711–2716. doi: 10.1084/jem.20080759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Suzuki T., Maranda B., Sakagami T., Catellier P., Couture C.Y., Carey B.C., Chalk C., Trapnell B.C. Hereditary pulmonary alveolar proteinosis caused by recessive CSF2RB mutations. Eur. Respir. J. 2011;37:201–204. doi: 10.1183/09031936.00090610. [DOI] [PubMed] [Google Scholar]
- 4.Tanaka T., Motoi N., Tsuchihashi Y., Tazawa R., Kaneko C., Nei T., Yamamoto T., Hayashi T., Tagawa T., Nagayasu T. Adult-onset hereditary pulmonary alveolar proteinosis caused by a single-base deletion in CSF2RB. J. Med. Genet. 2011;48:205–209. doi: 10.1136/jmg.2010.082586. [DOI] [PubMed] [Google Scholar]
- 5.Hildebrandt J., Yalcin E., Bresser H.G., Cinel G., Gappa M., Haghighi A., Kiper N., Khalilzadeh S., Reiter K., Sayer J. Characterization of CSF2RA mutation related juvenile pulmonary alveolar proteinosis. Orphanet J. Rare Dis. 2014;9:171. doi: 10.1186/s13023-014-0171-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Whitsett J.A., Wert S.E., Weaver T.E. Alveolar surfactant homeostasis and the pathogenesis of pulmonary disease. Annu. Rev. Med. 2010;61:105–119. doi: 10.1146/annurev.med.60.041807.123500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hawgood S., Poulain F.R. The pulmonary collectins and surfactant metabolism. Annu. Rev. Physiol. 2001;63:495–519. doi: 10.1146/annurev.physiol.63.1.495. [DOI] [PubMed] [Google Scholar]
- 8.Ikegami M., Ueda T., Hull W., Whitsett J.A., Mulligan R.C., Dranoff G., Jobe A.H. Surfactant metabolism in transgenic mice after granulocyte macrophage-colony stimulating factor ablation. Am. J. Physiol. 1996;270:L650–L658. doi: 10.1152/ajplung.1996.270.4.L650. [DOI] [PubMed] [Google Scholar]
- 9.Trapnell B.C., Whitsett J.A. Gm-CSF regulates pulmonary surfactant homeostasis and alveolar macrophage-mediated innate host defense. Annu. Rev. Physiol. 2002;64:775–802. doi: 10.1146/annurev.physiol.64.090601.113847. [DOI] [PubMed] [Google Scholar]
- 10.Hansen G., Hercus T.R., McClure B.J., Stomski F.C., Dottore M., Powell J., Ramshaw H., Woodcock J.M., Xu Y., Guthridge M. The structure of the GM-CSF receptor complex reveals a distinct mode of cytokine receptor activation. Cell. 2008;134:496–507. doi: 10.1016/j.cell.2008.05.053. [DOI] [PubMed] [Google Scholar]
- 11.Hercus T.R., Thomas D., Guthridge M.A., Ekert P.G., King-Scott J., Parker M.W., Lopez A.F. The granulocyte-macrophage colony-stimulating factor receptor: linking its structure to cell signaling and its role in disease. Blood. 2009;114:1289–1298. doi: 10.1182/blood-2008-12-164004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sallese A., Suzuki T., McCarthy C., Bridges J., Filuta A., Arumugam P., Shima K., Ma Y., Wessendarp M., Black D. Targeting cholesterol homeostasis in lung diseases. Sci. Rep. 2017;7:10211. doi: 10.1038/s41598-017-10879-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McCarthy C., Lee E., Bridges J.P., Sallese A., Suzuki T., Woods J.C., Bartholmai B.J., Wang T., Chalk C., Carey B.C. Statin as a novel pharmacotherapy of pulmonary alveolar proteinosis. Nat. Commun. 2018;9:3127. doi: 10.1038/s41467-018-05491-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Baker A.D., Malur A., Barna B.P., Kavuru M.S., Malur A.G., Thomassen M.J. PPARgamma regulates the expression of cholesterol metabolism genes in alveolar macrophages. Biochem. Biophys. Res. Commun. 2010;393:682–687. doi: 10.1016/j.bbrc.2010.02.056. [DOI] [PubMed] [Google Scholar]
- 15.Baker A.D., Malur A., Barna B.P., Ghosh S., Kavuru M.S., Malur A.G., Thomassen M.J. Targeted PPARgamma deficiency in alveolar macrophages disrupts surfactant catabolism. J. Lipid Res. 2010;51:1325–1331. doi: 10.1194/jlr.M001651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Thomassen M.J., Barna B.P., Malur A.G., Bonfield T.L., Farver C.F., Malur A., Dalrymple H., Kavuru M.S., Febbraio M. ABCG1 is deficient in alveolar macrophages of GM-CSF knockout mice and patients with pulmonary alveolar proteinosis. J. Lipid Res. 2007;48:2762–2768. doi: 10.1194/jlr.P700022-JLR200. [DOI] [PubMed] [Google Scholar]
- 17.de Aguiar Vallim T.Q., Lee E., Merriott D.J., Goulbourne C.N., Cheng J., Cheng A., Gonen A., Allen R.M., Palladino E.N.D., Ford D.A. ABCG1 regulates pulmonary surfactant metabolism in mice and men. J. Lipid Res. 2017;58:941–954. doi: 10.1194/jlr.M075101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Suzuki T., Sakagami T., Young L.R., Carey B.C., Wood R.E., Luisetti M., Wert S.E., Rubin B.K., Kevill K., Chalk C. Hereditary pulmonary alveolar proteinosis: pathogenesis, presentation, diagnosis, and therapy. Am. J. Respir. Crit. Care Med. 2010;182:1292–1304. doi: 10.1164/rccm.201002-0271OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Suzuki T., Arumugam P., Sakagami T., Lachmann N., Chalk C., Sallese A., Abe S., Trapnell C., Carey B., Moritz T. Pulmonary macrophage transplantation therapy. Nature. 2014;514:450–454. doi: 10.1038/nature13807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Suzuki T., Shima K., Arumugam P., Ma Y., Black D., Chalk C., Carey B., Trapnell B.C. Development and validation of Csf2ra gene-deficient mice as a clinically relevant model of children with hereditary pulmonary alveolar proteinosis. Am. J. Respir. Crit. Care Med. 2017;195:A4837. [Google Scholar]
- 21.Beccaria M., Luisetti M., Rodi G., Corsico A., Zoia M.C., Colato S., Pochetti P., Braschi A., Pozzi E., Cerveri I. Long-term durable benefit after whole lung lavage in pulmonary alveolar proteinosis. Eur. Respir. J. 2004;23:526–531. doi: 10.1183/09031936.04.00102704. [DOI] [PubMed] [Google Scholar]
- 22.Kleff V., Sorg U.R., Bury C., Suzuki T., Rattmann I., Jerabek-Willemsen M., Poremba C., Flasshove M., Opalka B., Trapnell B. Gene therapy of beta(c)-deficient pulmonary alveolar proteinosis (beta(c)-PAP): studies in a murine in vivo model. Mol. Ther. 2008;16:757–764. doi: 10.1038/mt.2008.7. [DOI] [PubMed] [Google Scholar]
- 23.Hetzel M., Suzuki T., Hashtchin A.R., Arumugam P., Carey B., Schwabbauer M., Kuhn A., Meyer J., Schambach A., Van Der Loo J. Function and safety of lentivirus-mediated gene transfer for CSF2RA-deficiency. Hum. Gene Ther. Methods. 2017;28:318–329. doi: 10.1089/hgtb.2017.092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Manz M.G. Human-hemato-lymphoid-system mice: opportunities and challenges. Immunity. 2007;26:537–541. doi: 10.1016/j.immuni.2007.05.001. [DOI] [PubMed] [Google Scholar]
- 25.Willinger T., Rongvaux A., Takizawa H., Yancopoulos G.D., Valenzuela D.M., Murphy A.J., Auerbach W., Eynon E.E., Stevens S., Manz M.G., Flavell R.A. Human IL-3/GM-CSF knock-in mice support human alveolar macrophage development and human immune responses in the lung. Proc. Natl. Acad. Sci. USA. 2011;108:2390–2395. doi: 10.1073/pnas.1019682108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shibata Y., Berclaz P.Y., Chroneos Z.C., Yoshida M., Whitsett J.A., Trapnell B.C. GM-CSF regulates alveolar macrophage differentiation and innate immunity in the lung through PU.1. Immunity. 2001;15:557–567. doi: 10.1016/s1074-7613(01)00218-7. [DOI] [PubMed] [Google Scholar]
- 27.Arumugam P.I., Urbinati F., Velu C.S., Higashimoto T., Grimes H.L., Malik P. The 3′ region of the chicken hypersensitive site-4 insulator has properties similar to its core and is required for full insulator activity. PLoS ONE. 2009;4:e6995. doi: 10.1371/journal.pone.0006995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hashimoto D., Chow A., Noizat C., Teo P., Beasley M.B., Leboeuf M., Becker C.D., See P., Price J., Lucas D. Tissue-resident macrophages self-maintain locally throughout adult life with minimal contribution from circulating monocytes. Immunity. 2013;38:792–804. doi: 10.1016/j.immuni.2013.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yona S., Kim K.W., Wolf Y., Mildner A., Varol D., Breker M., Strauss-Ayali D., Viukov S., Guilliams M., Misharin A. Fate mapping reveals origins and dynamics of monocytes and tissue macrophages under homeostasis. Immunity. 2013;38:79–91. doi: 10.1016/j.immuni.2012.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lavin Y., Winter D., Blecher-Gonen R., David E., Keren-Shaul H., Merad M., Jung S., Amit I. Tissue-resident macrophage enhancer landscapes are shaped by the local microenvironment. Cell. 2014;159:1312–1326. doi: 10.1016/j.cell.2014.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Uchida K., Beck D.C., Yamamoto T., Berclaz P.Y., Abe S., Staudt M.K., Carey B.C., Filippi M.D., Wert S.E., Denson L.A. GM-CSF autoantibodies and neutrophil dysfunction in pulmonary alveolar proteinosis. N. Engl. J. Med. 2007;356:567–579. doi: 10.1056/NEJMoa062505. [DOI] [PubMed] [Google Scholar]
- 32.Fleischmann J., Golde D.W., Weisbart R.H., Gasson J.C. Granulocyte-macrophage colony-stimulating factor enhances phagocytosis of bacteria by human neutrophils. Blood. 1986;68:708–711. [PubMed] [Google Scholar]
- 33.Seymour J.F. Extra-pulmonary aspects of acquired pulmonary alveolar proteinosis as predicted by granulocyte-macrophage colony-stimulating factor-deficient mice. Respirology. 2006;11(Suppl):S16–S22. doi: 10.1111/j.1440-1843.2006.00801.x. [DOI] [PubMed] [Google Scholar]
- 34.Seymour J.F., Lieschke G.J., Grail D., Quilici C., Hodgson G., Dunn A.R. Mice lacking both granulocyte colony-stimulating factor (CSF) and granulocyte-macrophage CSF have impaired reproductive capacity, perturbed neonatal granulopoiesis, lung disease, amyloidosis, and reduced long-term survival. Blood. 1997;90:3037–3049. [PubMed] [Google Scholar]
- 35.Yuan C.L., Hu Y.C. A transgenic core facility’s experience in genome editing revolution. Adv. Exp. Med. Biol. 2017;1016:75–90. doi: 10.1007/978-3-319-63904-8_4. [DOI] [PubMed] [Google Scholar]
- 36.Suzuki T., Mayhew C., Sallese A., Chalk C., Carey B.C., Malik P., Wood R.E., Trapnell B.C. Use of induced pluripotent stem cells to recapitulate pulmonary alveolar proteinosis pathogenesis. Am. J. Respir. Crit. Care Med. 2014;189:183–193. doi: 10.1164/rccm.201306-1039OC. [DOI] [PMC free article] [PubMed] [Google Scholar]