Abstract
The biflagellate zoospores of some phytopathogenic Phytophthora species spontaneously aggregate within minutes in suspension. We show here that Phytophthora parasitica zoospores can form aggregates in response to a K+ gradient with a particular geometric arrangement. Using time-lapse live imaging in macro- and microfluidic devices, we defined (i) spatio-temporal and concentration-scale changes in the gradient, correlated with (ii) the cell distribution and (iii) the metrics of zoospore motion (velocity, trajectory). In droplets, we found that K+-induced aggregates resulted from a single biphasic temporal sequence involving negative chemotaxis followed by bioconvection over a K+ gradient concentration scale [0–17 mM]. Each K+-sensing cell moved into a region in which potassium concentration is below the threshold range of 1–4 mM, resulting in swarming. Once a critical population density had been achieved, the zoospores formed a plume that migrated downward, with fluid advection in its wake and aggregate formation on the support surface. In the microfluidic device, the density of zoospores escaping potassium was similar to that achieved in droplets. We discuss possible sources of K+ gradients in the natural environment (zoospore population, microbiota, plant roots, soil particles), and implications for the events preceding inoculum formation on host plants.
Keywords: Phytophthora, zoospore, negative chemotaxis, bioconvection, aggregation
1. Introduction
The genus Phytophthora, which belongs to the class Oomycetes, includes some of the most destructive plant pathogens known, responsible for diseases on crops and in natural ecosystems worldwide. They grow as filamentous coenocytic hyphae producing various types of sexual and asexual propagules. In most cases, both dispersal and primary infection are mediated by airborne sporangia or waterborne zoospores [1,2]. The zoospores are uninuclear and reniform, with typical cell body dimensions of 7–10 and 5–7 µm along the anteroposterior and dorsoventral axes, respectively. They have a locomotor apparatus consisting of two flagella (10–20 µm) inserted into a longitudinal and ventral groove, enabling them to explore aqueous environments and reach potential hosts. A ‘tinsel-type’ flagellum with lateral mastigonemes along its entire length beats in front of the cell body. Behind the cell body, there is a smooth ‘whiplash-type’ flagellum [1,2].
Exogenous signals may bias zoospore motility such that the direction of movement is not random. Zoospores have sensory systems enabling them to respond to chemical gradients (chemotaxis), oxygen (aerotaxis), ionic fields (electrotaxis), gravity (negative geotaxis) and light (phototaxis). They target plant tissues by following gradients of various ions and compounds such as plant isoflavones, amino acids and sugars [3–5]. For example, Phytophthora palmivora zoospores display anodal electrotaxis in electrical fields of at least 0.5 V m−1, of similar strength to those found in the physiological fields around plant roots [6]. When a zoospore reaches a potential infection site, the cell sheds its flagella, synthesizes a primary cell wall and generates a germ tube that penetrates the host tissue [1,7–9].
Zoospores can control their motility in response to self-produced signals. They swarm even in populations composed exclusively of swimming zoospores, and they swim towards encysted spores, leading to auto-aggregation (autotaxis) [10,11] or biofilm formation on plant surfaces [12,13]. There is evidence to suggest that coordinated zoospore behaviour may regulate plant infection through interspecific or intraspecific cell-to-cell signalling [12,14]. However, it remains unclear how zoospores perceive cell density, transduce signals and control their own auto-aggregation. Mathematical and experimental data indicate that the convective zoospore movement associated with plume formation and auto-aggregation is consistent with predictions for bioconvection [15]. Bioconvection has been reported for microorganisms swimming towards the liquid–air surface and aggregating at the top layer of the suspension. This leads to a higher cell density at the surface than in the bulk of the fluid. Gravity destabilizes the dense layer of cells formed at the fluid surface, creating convective plumes. Bioconvection modelling uses cell concentration in a continuous sense assuming that the cell concentration gradient drives the instability [16–18]. Phytophthora infestans zoospores may also display sequential combinations of bioconvection followed by chemotaxis between plumes [11]. Phytophthora zoospores have been reported to produce and use molecules to monitor cell density, but no auto-attractant has been identified to date. Phytophthora parasitica zoospores produce an AI-2-like signal (but not N-acyl homoserine lactones) that could drive quorum sensing [19]. They also secrete cAMP, a putative chemoeffector, during biofilm formation [20].
The behaviour of zoospores in response to changes in ionic conditions suggests that cationic fluxes may be involved in collective motion, but their nature and precise role remain unclear. Ca2+ plays a key role in autonomous encystment, adhesion, germination and auto-aggregation [10], but does not directly trigger cooperative behaviour between zoospores, instead acting more like a secondary messenger [19,21]. K+ homeostasis influences the locomotion and encystment of zoospores. High external concentrations (5–10 mM) of potassium salts reduce the swimming speed of P. palmivora zoospores and cause them to swim in a jerky fashion [3]. Potassium sensing also triggers negative chemotaxis in the zoospores of P. palmivora [4].
We sought to characterize the signals altering the pattern of zoospore motility and initiating auto-aggregation or biofilm formation. Our findings suggest that the aggregation of P. parasitica zoospores is elicited by the perception of monocationic gradients in a particular geometric arrangement. Using macro- and microfluidic systems together with time-lapse live-cell imaging to measure the fluorescence intensity (FI) of impermeant cation probes, we defined the characteristics of the K+ gradients and zoospore motion.
2. Material and methods
2.1. Preparation of the zoospore suspension
Phytophthora parasitica (isolate 310, Phytophthora INRA collection, Sophia Antipolis) mycelium was routinely cultured on malt agar at 24°C in the dark. For zoospore production, mycelium was grown for one week in V8 liquid medium at 24°C, under continuous light. The material was then drained, macerated and incubated for a further 4 days on water agar (2%). The zoospores were released by heat shock treatment: incubation for 30 min at 4°C and then 20 min at 37°C. We added 10 ml (per Petri dish Φ100 mm) of 2 mM Mes [2-(N-morpholino) ethanesulfonic acid]-NaOH buffer pH 6.5 between incubations. Zoospore concentration was adjusted to 5 × 105 cells ml−1.
2.2. Droplet assay
Three chemotaxis set-ups were used for zoospore motion studies. The first was a droplet assay (figure 1). A controlled profile of the signal was generated by applying a [1 mM–1 M] gradient in a local oriented manner to a freshly prepared suspension of zoospores so as to achieve an essentially stable profile by diffusion. The basic operation involved the microinjection of putative chemoeffectors (1/200, V/V) at the periphery of droplets (200–500 µl) including zoospores deposited on glass slides or polydimethylsiloxane (PDMS) stamps. Various conditions (in terms of ionic composition and strength, pH, nutrient supply and cell density) were tested, and K+ sensing was identified as a primary signal for zoospore aggregation.
Figure 1.
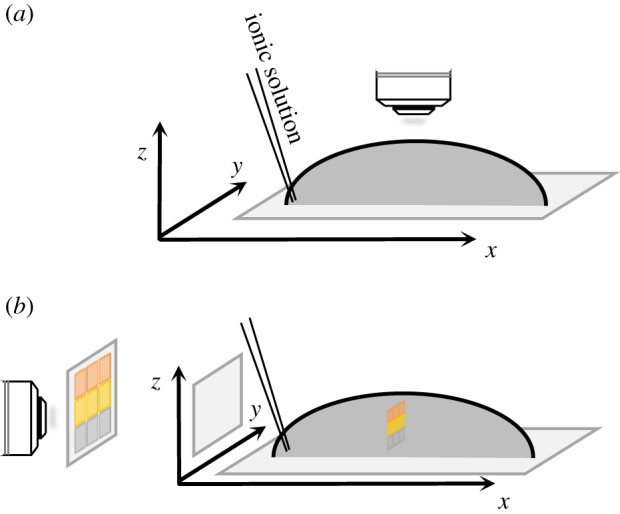
Schematic views of the droplet assay. Potassium was applied at the base of each droplet containing zoospores and at a point on the circumference. Subsequent characterization of the metrics of zoospore motion was based on micrographs generated in either the horizontal (a) or vertical (b) plane. (Online version in colour.)
2.3. Passive dispersion system for the generation of ionic gradients
A passive dispersion system was used to generate a diffusion gradient for the simultaneous measurement of extracellular potassium concentration on the one hand and tracking of zoospore distribution on the other hand. Cells were preloaded with 2 µM Asante Potassium Green-2 (APG-2 TMA+ salt; Teflabs, 3622), a potassium-specific non-permeant fluorescent dye. We placed 50 µl of cell suspension in a microchamber (µ-Slide VI—flat; Ibidi; size l: 17 mm; w: 3.8 mm; h: 400 µm), to which we added 0.5 µl of 500 mM KCl. At various time points, the content of each chamber was observed with a confocal microscope (LSM 880-Zeiss) and the Tile Scan tool. Images (1.2 mm × 1.2 mm) were generated from the point of application and along the length of the chamber. APG-2 was excited with a 488 nm argon laser and the fluorescence emitted was captured in channel mode (band pass 510–590 nm). The distance between the point of application and the highest position in the chamber allowing zoospore motion was determined with a transmission photomultiplier. The 1.2 mm × 0.4 mm area corresponding to this position was divided into three technical replicates (0.4 × 0.4) for the measurement of the mean APG-2 FI in each area. The highest ionic concentration allowing zoospore motion was calculated from the difference between the APG-2 FI measured at this position, before and after the addition of potassium, and relative to the FI values obtained with a range of discrete concentrations. Image analysis was performed with ZEN 2 software (Zeiss).
2.4. Microfluidic device
The third set-up used to capture the immediate response of zoospores at the single-cell level consisted of a PDMS microfluidic circuit in which the zoospores were subjected to a continuous flow presenting a gradient of potassium concentration. The main advantage of this approach is that it allows rapid and effective control over the conditions around the spores in the chamber.
This system is presented in §3.4. Three channels (H × W = 0.05 mm × 0.1 mm) intersect as a cross, fusing into the final channel, which is much wider (H × W = 0.2 mm × 1 mm). The spores are observed in this chemotactic chamber. Spores are injected into the middle inlet and the 100 mM KCl solution is injected into the two lateral channels. This device was produced by soft lithography techniques [22], with all the steps performed in the clean room of the Institut de Physique de Nice, Nice, France. SU-8 moulds exposed at a resolution of 50 800 dots per inch (DPI) were covered with 10 : 1 Sylgard PDMS for curation. They were unmoulded and punctured for inlets and plasma bonding on clean glass slides was then used to seal the channels. Teflon tubing was directly inserted into the PDMS holes to connect the solution reservoirs (2 ml) to the system. Liquid flows were driven with a pressure controller (Fluigent), in the 0–100 mbar range, between the three inlets and the outlet, which was at atmospheric pressure. A high-speed camera (Phantom v. 7.11) placed on a Zeiss Axiovert 200 M inverted microscope with a ×10 objective was used to film movies at a speed of 200 frames s−1.
2.5. Microscopy for image acquisition along horizontal or vertical axes
Zoospore motion was captured with several different instruments: a VHX-2000 digital microscope (Keyence); an LSM 880 inverted confocal microscope (Zeiss); an Axio Imager Z1 (Zeiss) equipped for bright-field and epifluorescence microscopy; an Axioskop (Zeiss) microscope mounted vertically but rotated by 90° with appropriate support on the back of its frame for imaging over a vertical plane. Movie acquisition was controlled by 3D profile VHX-H3M (Keyence) or ZEN (Zeiss) software, generating sequences of 10–30 s at 10–30 frames s−1.
2.6. Image analysis
The dynamics of zoospore motion were first investigated by single-particle tracking with various image-processing algorithms available as plugins from the ImageJ or Fiji software libraries. The initial processing of phase-contrast zoospore images involved TIFF format conversion, image inversion, threshold adjustment and binary conversion. Velocity_Measurement_Tool plugins were used to generate kymographs. We used MosaicSuite [23] to draw trajectories. The TrackMate plugin [24] was used to track the zoospores in droplets. Movies were analysed with the following parameters: estimated blob diameter, 16 µm and threshold, 12 µm; automatic initial thresholding; linking max distance, 50 µm; gap-closing max distance, 50 µm; gap-closing max frame, 2; spot filtering, quality above 168; track filtering process, duration of track above 1.5 s. For each remaining trajectory, we calculated instantaneous velocities between two successive points, which made it possible to calculate the mean velocity vi for each trajectory i; we calculated the mean velocity of the zoospores for a given movie as the mean vi, together with the associated standard deviation.
We obtained velocity fields with the particle image velocimetry (PIV) plugin applied to two sequential images (image size 1388 × 1040 pixels; time scale 1 s). Magnitude vector maps were generated with multiple-iteration window sizes of 128 × 128, 64 × 64 and 32 × 32 pixels.
3. Results
3.1. Potassium induces zoospore aggregation in a concentration-specific manner
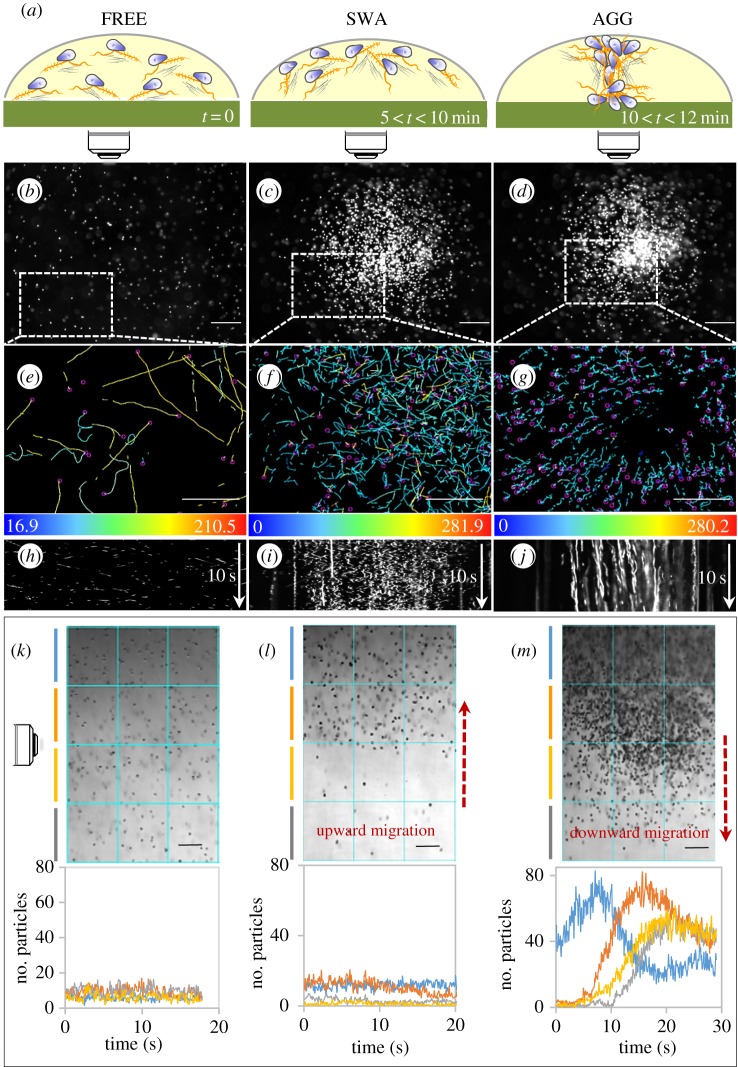
The droplet assay (figure 1) was used to evaluate specificity and to determine the optimal conditions for the detection of ion gradients by zoospores. Upright microscopes were used for observation in the horizontal plane (figure 1a). A microscope set-up was adjusted so as to position the axis of the lens horizontally for observations of displacement in the vertical plane (figure 1b). Three states were defined and reproducibly observed (figure 2a): (i) a free state (FREE) corresponding to untreated cells, uniformly distributed throughout the droplet and displaying random motion; (ii) a swarm state (SWA), corresponding to ion-treated cells, with the upward migration of zoospores to form a swarm of increasing cell density in a progressively restricted area within minutes; (iii) an aggregate state (AGG) in which the zoospores formed a plume and migrated downward to collect together on the support surface.
Figure 2.
Zoospore motion in droplets in response to K+ application. (a) Summary scheme of the distribution and displacement of the cell population in three different defined states: FREE for zoospores swimming freely and distributed randomly before gradient sensing; SWA and AGG for zoospores forming a swarm and aggregating, respectively, upon gradient sensing. The range is indicative of the time at which each sequence occurred after KCl application. (b–j) Patterns observed in the horizontal plane for zoospore distribution (b–d), swimming paths (e–g) and two-dimensional kymographs drawn for 10 s (h–j). The kymographs provide a picture of the variation of zoospore behaviour in the three states, from highly motile cells, appearing as oblique lines (h), to more static cells, represented by vertical lines (j). Swimming paths were recorded for x,y coordinates corresponding to the areas delimited by dotted rectangles in (b,c,d) for 2 s for (e,f) or 4 s for (g). The colour coding provides information about mean velocity with range-limiting values indicated at the bottom of each inset in µm s−1. On the kymograph image, the vertical axis represents time (white arrow), the horizontal axis corresponds to the intensity of the pixel along the length of the selected line: a linear region of interest of 1500 µm located at the centre of the field along the x axis. (k–m) The distribution and displacement patterns observed in the vertical plane. The upper insets show representative micrographs for the FREE, SWA and AGG states. For each state, the lower insets show plots of zoospore number per unit area versus time, at each depth identified by the colour code defined in the upper insets. The SWA state is associated with an upward migration (signified by an ascending red arrow), whereas AGG is associated with a downward migration (descending red arrow). The bidirectional and sequential movement of the cell population results in a five- to sevenfold increase in local cell density during downward migration (compare (k) with (m)). The zoospore numbers presented are means calculated from three consecutive unit areas and are based on counts of particles of 3–30 µm in diameter on converted binary images. Bar sizes: 100 µm. (Online version in colour.)
The aggregation process occurred with potassium, regardless of the associated anion (Cl−, CH3COO−, , ), over a pH range extending from 5 to 8, at a cell density of 105 to 4 × 106 zoospores ml−1, and a quorum of 5 × 103 to 2 × 104 cells. Aggregation peaked within 12–15 min when 0.5–3 µmol of potassium was applied per 100 µl of cell suspension at the starting point (electronic supplementary material, movie S1; figure 2a–d). We tested several other cations (table 1). Na+ application also induced aggregation, but larger amounts of this cation than of K+ were required. H+ also affected the swimming behaviour of zoospores but did not provoke aggregation, instead triggering a transient local gathering of zoospores (0.001–0.01 µmol) or ring formation followed by high rates of cell death for larger amounts. For subsequent analyses, the assays were performed at pH 6.5, with the application of 0.5 µmol KCl per 100 µl of a suspension of 5 × 105 cells ml−1. The objectives of this study were to explore the effects of K+ ions, which induce various behaviours of interest in the particular case of Phytophthora, and, more generally, to develop a model for the biophysics of swimming for a large number of swimming cells based on these effects. K+ has strong effects at low concentration (such as those found in natural environments), with weaker osmotic effects than Na+ ions, which would need to be used at concentrations 10 times higher. We therefore performed droplet assay experiments on various ions, but we focused exclusively on the effects of K+ ions in the milli- and microfluidic experiments.
Table 1.
Effect of the anisotropic application of cations on zoospore pattern formation. The first and second rows indicate the identity and ranges of the amount of cation (chloride salt) added to a droplet (100 µl); yes/no for the induction or non-induction of a swarm, plume or aggregate upon cation application.
| cation | amount (µmol) | swarm | plume | aggregate | volume (µl) |
|---|---|---|---|---|---|
| K+ | [0.5–3] | yes | yes | yes | 100 |
| Na+ | [5–30] | yes | yes | yes | 100 |
| H+ | [10−3–10−2] | yes | no | no | 100 |
| Ca2+ | [0.5–3] | no | no | no | 100 |
| Cu2+ | [0.5–3] | no | no | no | 100 |
| Mg2+ | [0.5–3] | no | no | no | 100 |
| Zn2+ | [0.5–3] | no | no | no | 100 |
| NH4+ | [0.5–3] | no | no | no | 100 |
3.2. Behaviour of zoospores sensing potassium
In each state, we analysed the distribution and displacement of zoospores in both the horizontal (figure 2e–j) and vertical (figure 2k–m) planes. In the FREE state, the projected velocities were 150 ± 34 µm s−1 in the horizontal plane, and 146 ± 39 µm s−1 in the vertical plane, and no preferential direction of cell trajectories was noted (figure 2b,e,h).
In the SWA state (figure 2c,f,i,l), the projected velocity in the horizontal plane was 115 ± 29 µm s−1, decreasing at the periphery of the swarm, as illustrated on the kymograph by the weak slope of the lines at the edge of the swarm for this plane (figure 2i). There was no clear preferential direction for trajectories in the horizontal plane, but all trajectories were delineated within the swarm (figure 2c,f). The velocity in the vertical plane was 78 ± 32 µm s−1, and swarm formation in this plane was correlated with a continuous upward migration (electronic supplementary material, movie S2) culminating in the accumulation of almost all cells at the air–liquid interface, with only a very small number of cells in the scanned area below (figure 2l). No such spectacular upward migration was observed for zoospores swimming freely in water (electronic supplementary material, movie S3; figure 2k).
In the AGG state and the horizontal plane (figure 2d,g,j), all zoospores migrated with centripetal trajectories (figure 2g), at very low velocity (VXY = 24 ± 11 µm s−1), towards a restricted area (1–5 × 106 µm2) in which they ceased moving linearly and formed an aggregate (electronic supplementary material, movie S1; figure 2d). The velocity in the vertical plane was 41 ± 19 µm s−1 (electronic supplementary material, movie S4; figure 2m). Once the plume had been formed, the cells abruptly began to migrate downward (plume velocity = 65 ± 23 µm s−1; electronic supplementary material, movies S4 and S5). At the end of the sequence, the zoospores were alive and motile (electronic supplementary material, movie S6), and they bore flagella (electronic supplementary material, figure S1), subsequently undergoing massive progressive encystment. Based on the number of zoospores sedimenting on the lower surface at the end of the experiment and the size of the plumes observed on microscopy, the density of zoospores in a plume has been estimated at about 40 × 103 spores µl−1.
Obviously, plume displacement included a large vertical component during downward migration (electronic supplementary material, movie S5 and figure S2). This downward migration initially involved an almost rectilinear and vertical displacement for about 10 s at a Vz of about 50 µm s−1. The trajectory then became helical, with a large decrease in Vz, probably when the cells reached the threshold K+ concentration range defined below (electronic supplementary material, figure S2C,D).
In this assay, we also analysed the effect of zoospore motion on fluid velocity, by adding fluorescent microspheres to the cell suspension. By tracking the movements of zoospores and microspheres simultaneously, we showed that fluid velocity was unaffected by zoospores swimming freely or in a swarm at the time and spatial scales studied (data not shown). By contrast, during aggregation, zoospores migrating downward from the surface of the fluid caused microsphere advection (electronic supplementary material, movie S7). This finding was confirmed by PIV analysis, providing a picture of the local streamlines of the fluid velocity fields, which closely resembled the geometry, orientation and magnitude of zoospore movement (figure 3a,b). Further confirmation was provided by the variation of local velocities calculated from zoospores and microspheres located within and outside the plume (figure 3c). Taken together, these data indicate that the downward migration of zoospores correlates with fluid advection. At the beginning, migrations cause fluid advection, but then these phenomena are linked and can be mutually supportive.
Figure 3.
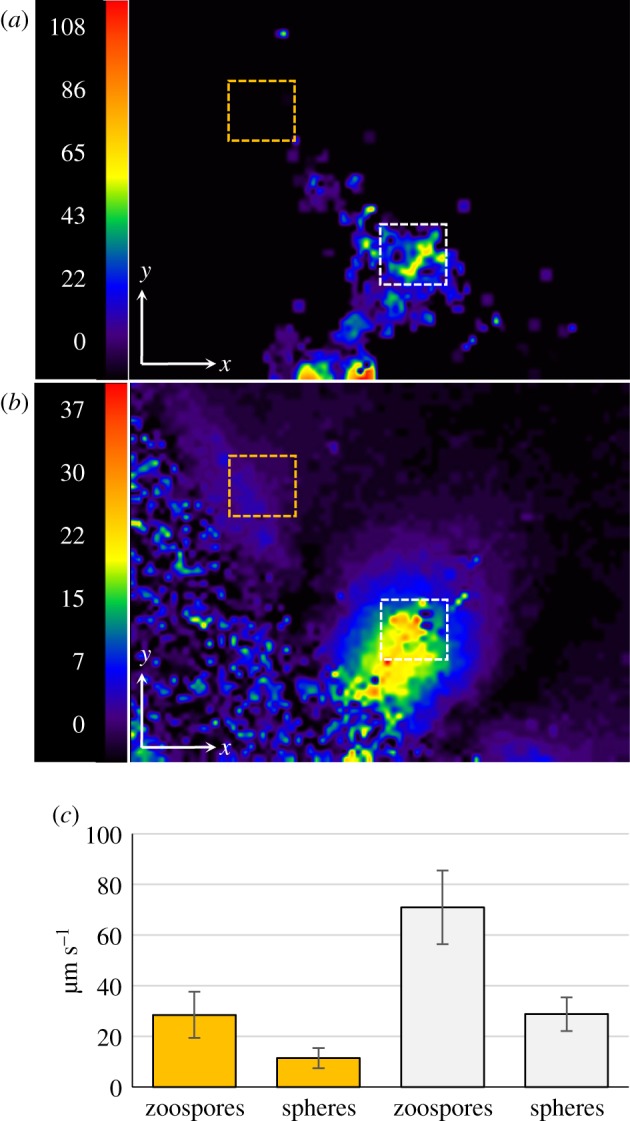
Flow field of the water and velocity field of zoospores within a plume. The flow field was analysed by adding fluorescent microspheres (Φ 0.5 µm; 1/1000, v/v) to the cell suspension. One representative instance of a zoospore swimming within a plume was visualized in the horizontal plane (electronic supplementary material, movie S7). The movement of zoospores was captured by dark-field microscopy during the first 6 s of the sequence. The movement of fluorescent microspheres was then captured for 5 s by switching to fluorescence microscopy with a rhodamine fluorescence filter set. (a) A vector magnitude plot of velocity fields, defined by the PIV of zoospore movement and generated between t = 5 s and t = 6 s. (b) The vector magnitude plot of the microsphere movement was generated between t = 7 s and t = 8 s, 1 s after the switch from dark-field to fluorescence microscopy. The colour code indicates the magnitude of velocity (μm s−1). (c) Histogram of the mean velocity of zoospores and microspheres determined from areas located outside (dotted yellow square in (a) and (b)) or within (dotted white squares in (a) and (b)) the plume. The difference between the values determined within and outside the plume was highly significant for both particles in Student's t-test (p < 0.0001). (Online version in colour.)
3.3. The distribution of potassium-sensing zoospores depends on the ionic gradient
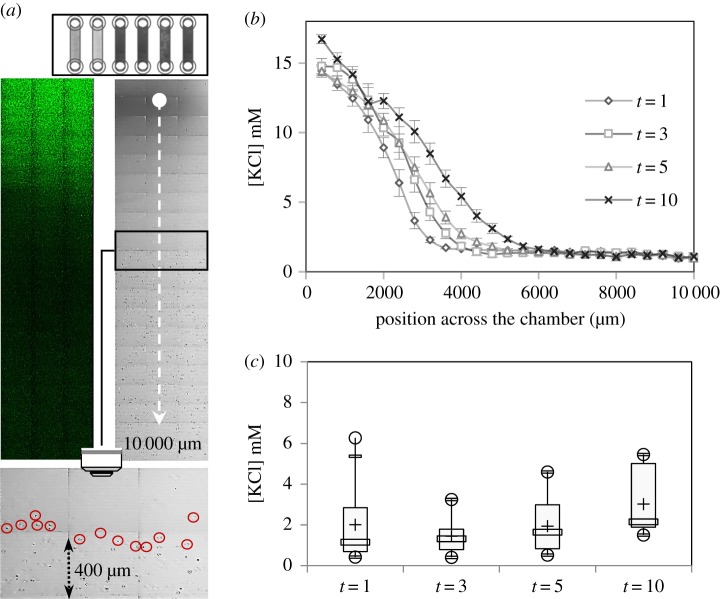
The concentration gradient was heterogeneous and difficult to control in the droplet assay. This assay was not, therefore, suitable for analyses of the relationship between zoospore distribution and the extracellular K+ concentration gradient. We investigated the behaviour of zoospores in response to various concentrations of potassium, by generating a diffusion gradient in a millifluidic device (figure 4a). This device consisted basically of a single channel, with one inlet and one outlet. The cells were loaded into the channel (H × W × L = 400 µm × 3.8 mm × 1.7 mm) in the presence of APG-2, a K+-sensing probe, before a spot application of KCl at the inlet. We followed the fluorescence dynamics of APG-2 upon ion binding and the change in zoospore distribution along the channel by confocal microscopy (figure 4a). FI was measured to determine K+ concentration, which is plotted in figure 4b. The FI profile obtained indicated that the dynamic range of K+ concentration that could be resolved in this device was [0–17 mM]. A small minority of the zoospores in the population could not swim, and these zoospores were mostly located in the immediate vicinity of the application spot. Most of the zoospores displayed no change in motion (velocity and random trajectory; data not shown), but their distribution was restricted. The cells displayed negative chemotaxis [4], with optimal conditions for swimming occurring away from the higher concentrations of potassium ions. The ‘no-swimming zone’ expanded over time as the ions diffused away from the point of application. We used the measurements of APG-2 FI obtained to determine the range of higher concentrations compatible with zoospore motion at various time points (figure 4a,c). The cell distribution depended on local K+ concentration. The difference in this concentration between the two sides of the border between the swimming and no-swimming zones lay in a narrow range (1–4 mM). This range did not change significantly over time. In the chemotaxis assay, we monitored the zoospores for a period of 30 min. We observed no change in the characteristics of zoospore motion in the swimming zone, except that the zoospores were unable to move into the areas of high potassium concentration. Thus, K+-mediated negative chemotaxis seems to have been initiated in these conditions, but was not sufficient to induce aggregation.
Figure 4.
A potassium gradient drives zoospore distribution in a passive dispersion system. (a) The millifluidic device (H × W = 400 µm × 3.8 mm) is shown at the top. The two images below show a characteristic APG-2 fluorescent signal pattern captured by confocal laser microscopy (upper left panel), and the corresponding area (12 mm2) observed by transmission with a T-PMT detector (upper right panel) in a millifluidic device. The blank point corresponds to the site of application of 1 µl of 0.5 M KCl at t = 0 and the dotted line indicates the total distance screened (10 mm) at 1, 3, 5 and 10 min. The lower panel focuses on an area (0.48 mm2) in which the fields of the device in which zoospore motion can and cannot be observed can be delimited. The location of encircled zoospores defines, at time t = 5 min, the transition between the two fields characterized by the presence/absence of cells. (b) Measurement of FI across 10 000 µm of the chamber, defining the concentration profile, which follows a nonlinear gradient. (c) Box plot of the highest ion concentrations compatible with zoospore motion. Minimum and maximum values are depicted by white dots; the box indicates the upper and lower quartiles, the mean and the median are represented by + and a white rectangle within the box, respectively, for each raw datum. The values indicated are the means of 5, 6, 5 and 4 replicates for 1, 3, 5 and 10 min, respectively.
3.4. Changes in zoospore motion in response to K+ in a microfluidic device
For the quantification of zoospore distribution and velocity following the application of a K+ gradient, zoospores were subjected to different gradients of KCl in a microfluidic system (see Material and methods). Zoospores were injected from the central inlet into a chemotaxis observation chamber, and the two side channels were perfused with KCl (figure 5a). Zoospores and KCl were first injected together, at t = 0; the flow of KCl was stopped for time T (ranging from 5 to 15 s) to achieve a partial flushing out of the KCl; the flow of zoospores was then also stopped, and zoospore movement was observed for at least 20 s. A picture of the chamber is shown in figure 5b (upper image).
Figure 5.
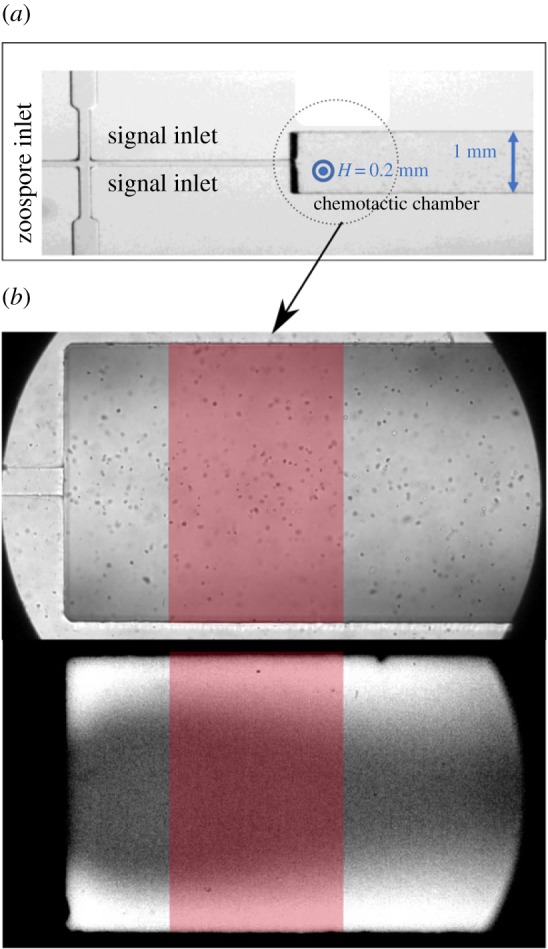
Microfluidic device. (a) Image of the device, showing the three inlets and the chemotactic chamber (H × W = 0.2 mm × 1 mm). Zoospores were injected into the central inlet and 100 mM KCl was injected into the side inlets (signal inlets). (b) Images of the observation chamber (or chemotactic chamber) in bright-field conditions (upper image), showing zoospores, and in fluorescence conditions (lower picture), for the mapping of potassium concentration in the chamber based on the fluorescence of the APG-2 probe. The red window indicates the area of the chamber taken into account in the analysis of zoospore tracking and measurement of the spatio-temporal concentration of potassium. In the lower image, the white areas in the uppermost and lowermost regions of the image indicate the areas in which the concentration of potassium is high. (Online version in colour.)
In another experiment in the same set-up but without zoospores, the steps described above were performed with KCl mixed with the potassium probe APG-2, to obtain a spatio-temporal map of potassium concentration in the chamber. Because of the geometry of the microfluidic set-up, the KCl was concentrated towards the top and bottom (on the image) of the chamber, as shown in the snapshot presented in figure 5b (lower image).
We characterized the migration process, by tracking cells from t = T to t = T + 20 s, to extract their trajectories, calculate the local density of cells and measure the velocity of individual cells. The area used for this analysis was restricted to the red window shown in figure 5b. The same area was used for the calculation of spatio-temporal potassium concentrations. Both spore density and potassium concentration were determined by the sliding window method (see §2.4.), by calculating the mean values in rectangular windows sliding along the direction of the potassium gradient (the y coordinate in our set-up).
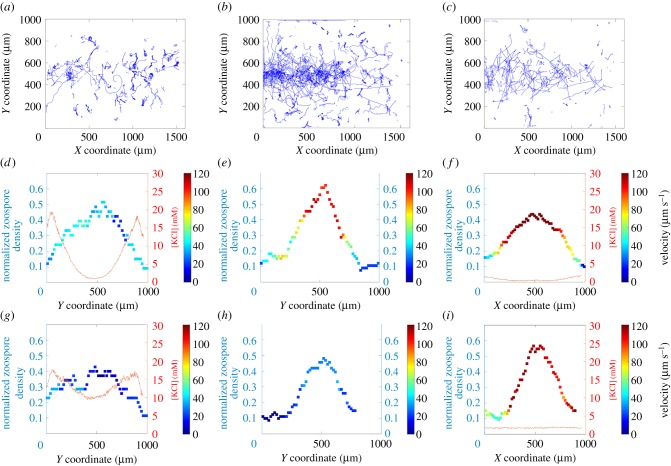
The results are shown in figure 6. After KCl was flushed out for 5 s, the vast majority of zoospores rapidly adopted circular trajectories (figure 6a; electronic supplementary material, movie S8), moving at low speed, below 40 µm s−1 (figure 6d,g). The residual concentration of potassium was therefore assumed to be higher than the threshold of 3–5 mM, resulting in the immediate ‘freezing’ of cell motion. The concentration profiles in figure 6d,g are consistent with this interpretation: after 4 s, potassium concentration exceeded 5 mM over more than half the observed area; after 16 s, potassium concentration exceeded 10 mM over the entire area.
Figure 6.
Trajectories, density profiles and the speed of the zoospores in the chamber, according to potassium concentration profile. Trajectories of the zoospores throughout the entire chamber after the flushing out of KCl for a time T equal to 5 s (a), 10 s (b) and 15 s (c, only 1/10th of the trajectories are shown for the sake of simplicity). (d–f) Local density of zoospores (squares) with a colour map for the associated velocity, and the potassium profile (red lines), the flushing out of KCl for 4 s. (g–i) Same representations after 16 s of KCl flushing. (Online version in colour.)
After KCl was flushed out for T = 10 s, some zoospores initially moved towards the central zone (figure 6b; electronic supplementary material, movie S9), as expected, to escape the high concentrations of potassium towards the top and bottom of the device. These zoospores followed linear trajectories and moved at relatively high speeds (above 100 µm s−1) after 4 s (figure 6e). Nevertheless, some cells initially present in the zones at the top and bottom of the device immediately adopted low-speed circular trajectories (figure 6b,e), suggesting that the concentration of potassium in these zones was too high for the cells to escape. After about 10 s, all the cells shifted to low-speed circular trajectories (not shown), suggesting that the potassium had diffused into the central zone and had reached the threshold concentration there too. Unfortunately, we were unable to measure the potassium concentration during this cycle. After 16 s, cells were travelling at speeds of less than 40 µm s−1 in the central zone (figure 6h), consistent with a shift to low-speed circular trajectories.
Flushing out KCl for T = 15 s resulted in behaviour similar to that observed for T = 10 s, except that the area in which the zoospores moved along linear trajectories at high speed was larger (electronic supplementary material, movie S10, figure 6c,f), and, after 16 s, the cells continued to move along linear trajectories at high speed (figure 6i). This suggests that the concentration of potassium was sufficiently low to allow the cells to move. The concentration profiles obtained after 4 and 16 s (figure 6f,i) are consistent with this interpretation, as the concentration of potassium ions remained below 3 mM. Interestingly, we found that: (i) between 4 and 16 s, the cells tended to migrate towards the central zone, despite the very low concentration of potassium; (ii) after 16 s, zoospore density peaked, whereas the concentration profile appeared to be flat. These observations suggest that motion leading to aggregation can be triggered by a cue other than potassium concentration. Further investigations are required to shed light on this behaviour. In the last 12 s, the density of zoospores swimming in a central area of the chamber (a 400 µm × 100 µm window) was estimated at 20 × 103 (s.d. = 1.103) zoospores µl−1.
4. Discussion
Phytophthora auto-aggregation has been associated with bioconvection patterns [15] or a combination of bioconvection and positive chemotaxis [11]. Potassium sensory cues regulate the behaviour of Phytophthora potassium zoospores [3,4]. In this report, we present in vitro evidence that potassium gradient sensing by P. parasitica zoospores is a primary stimulus, inducing synchronized zoospore behaviour and cell aggregation. The resulting macroscopic pattern resembles auto-aggregation. Cell behaviour changes during a single biphasic temporal sequence different from that previously described for auto-aggregation.
4.1. Negative chemotaxis and bioconvection
Negative chemotaxis in response to the sensing of potassium initially causes the cells to move into a region in which potassium concentration is below the threshold range of 1–4 mM. The arrangement of the cells results in their spatial concentration. Within a droplet, negative chemotaxis increases cell concentration through upward migration towards the upper surface of the suspensions. The simultaneous accumulation of cells in the horizontal plane suggests that positive chemotaxis may also occur. Upward migration increases local mass density in the uppermost regions of the suspension, which become denser than the regions below them. This leads to the development of overturning instability, analogous to bioconvection, resulting in the formation of descending plumes of dense cell suspension, with fluid flow advection in their wake. We can compare the measured frontal velocity (§3.2; 65 ± 23 µm s−1) with a simple model, by assuming that the plume is a single sedimenting object. We can start by comparing an individual sedimenting spore and then move up to the level of a whole plume, assimilated to a drop of a heavier liquid falling into water. The sedimentation speed of a passive zoospore is about Vss = 2 × Rs2 × g × Δρ/(9µ), where Rs is the average radius of the spore (3.2 µm), g = 9.81 m s−2 and µ = 0.001, the viscosity of water. Δρ is assumed to be 80 kg m−3, corresponding to a spore density of 1.08, a typical value for yeast cells or bacteria. We obtained a value of 1.79 µm s−1 for Vss. For plume speed, Vsp, we used the same formula as for a spherical object falling, but using R = 450 µm rather than Rs, the mass density being the mean density of the water/spore suspension (for N = 4000 spores in the plume): Δρp = Δρ × N × Rs3/R3 = 0.12 kg m−3. Vsp = 2 × R2 × g × Δρp/(9µ) = Vss × N × Rs/R, and, with Rs/R = 0.007, we obtain Vsp = 51 µm s−1. This provides us with an approximate order of magnitude close to the measured values, given that N is not precisely determined and the exact flow conditions between spores are not exactly known.
Based on the congruent results of the droplet (in the vertical and horizontal planes) and microfluidic (horizontal plane) assays, we can suggest the following scheme. If K+-treated cells are able to escape from high concentration areas, they follow a linear trajectory and head towards areas in which the potassium concentration is below the threshold of 1–4 mM. When conditions restrict the cells to areas in which the potassium concentration is beyond this threshold, two drastic changes are observed: velocity decreases sharply and the cells switch from a linear trajectory to a helical one. The K+-induced aggregation phenomenon was observed when the motion of a quorum of zoospores (5 × 103–2 × 104) explored territories delimited by the concentration field and its spatial distribution in the various set-ups used here. In both the droplet assay and the microfluidic device, the maximum density of zoospores was estimated at a few tens of thousands of zoospores per microlitre. This may constitute a threshold value for the density of P. parasitica zoospores. However, zoospores seemed to be able to remain clustered for longer at higher spatio-temporal variations of the K+ gradient in microfluidic devices, suggesting that signalling between zoospores may result in a stronger response.
4.2. Sources of K+ gradients in the natural environment
The induction of zoospore clustering by K+ in vitro demonstrates that aggregation may result from the perception of an external signal. How can such gradients be achieved in natural habitats? Such efflux mechanisms could be generated by efflux from zoospores released from other microbes present in the same biotope, rhizospheric activity and/or exchange dynamics in soil.
The self-generation of K+ by a net K+ zoospore efflux seems highly unlikely. In freshwater, the osmolality of the zoospore cytosol is always higher than that of the external environment. Throughout the course of their displacement, one of the major challenges faced by wall-less zoospores is the removal of excess cytosolic water rather than ions to maintain homeostasis. The excess water collects in the contractile vacuole complex, the osmoregulatory organelle and is discharged into the extracellular environment [7]. A calculation of the K+ diffusion potential of cells also rules out the hypothesis of K+ efflux from zoospores. If we consider high densities (106–107 ml−1) of zoospores (diameter of 10 µm), and an intracellular K+ concentration range of 100–200 mM, then the relative volume expansion rate of the cell population ranges from 0.052% to 0.52%. This would result in an extracellular K+ concentration of between 0.05 and 1 mM if the entire potassium content of the cell were released by efflux. It would not, therefore, be possible to reach the threshold concentration of 1–4 mM identified here as the stimulus eliciting a pattern reminiscent of auto-aggregation. Instead, as in most prokaryotic and eukaryotic cells [25], depolarization due to the increase in extracellular potassium concentration may lead to fluctuations of the membrane potential of zoospores, potentially modifying the beating of the flagella, cellular responses, such as osmoregulation, and/or cell-to-cell signalling.
The extent to which K+ release from the microbiota present in the same habitat as the zoospores can mediate zoospore aggregation remains to be evaluated. Potassium seems to play a key role in the displacement of bacteria, the physical composition of microbiota [25] and pathogenic processes [26]. The electrical signalling mediated by potassium ion channels regulates cell–cell dialogue within bacterial biofilms, with potassium driving the attraction of distant cells of different species [25]. The range of bacterially generated gradients effective for prokaryote attraction in vitro is of the same order of magnitude as that affecting zoospore movement in our experiments. Such gradients may, therefore, affect the distribution of P. parasitica propagules through repulsion.
In terms of rhizosphere activity and exchange dynamics in the soil, any area in which strong K+ gradients are generated will tend to repel P. parasitica zoospores. The total K+ content of soils generally ranges between 0.4 and 30 g kg−1 [27], and has two components. Soil particles, which bind about 98% of the total K+ content, may constitute a major obstacle to zoospore tracking. The film of water surrounding these particles (2% of total K+ content) has a potassium concentration in the range of 0.2–15 mM, reaching 5–10 mM in the most highly fertigated agricultural soils [27]. It is therefore reasonable to speculate that K+ exchange dynamics in the soil constitute a major parameter determining zoospore distribution and aggregation. At the scale of an infected host plant, zoospores repelled by soil particles and subject to constraints on their movement in the water film would be likely to move towards host tissues. This tendency would be enhanced by root K+ uptake, which would result in a zone of potassium depletion around the root surface [28], and by root exudates, which attract zoospores [1,29]. Thus, the physical and chemical distribution of K+ at the soil–root interface is a parameter that may contribute to the constitution of a high-density inoculum or biofilm formation on the plant surface. However, the importance of this contribution relative to other ion exchange dynamics, such as soil acidification by roots, remains to be determined. A more holistic view of the relationships between the concentrations of various ions and zoospore distribution is required to determine the influence of K+.
4.3. Concluding remarks
We show here that the sensing of a potassium gradient induces synchronized zoospore behaviour (due to negative chemotaxis) and the aggregation of P. parasitica zoospores. In all the in vitro experimental set-ups used (droplet assays, milli- and microfluidic devices), the zoospores displayed the same behaviour, with a clear, consistent response to potassium. The use of these different set-ups sheds light on particular aspects linked to the size and geometric effects of each of the set-ups used. Bioconvection patterns followed by aggregation were observed only in droplet assays. Nevertheless, in millifluidic devices, clear and quantitative responses to potassium gradients were demonstrated, and the zoospores aggregated, reaching a density of the same order of magnitude as that observed in droplets. The microfluidic devices allowed faster spatio-temporal variations of potassium concentration, making it possible to observe the clustering process, which appeared to be stronger than the response expected on the basis of quasi-static negative chemotaxis. Thus, even when the potassium concentration fell to low values compatible with zoospore movement, the zoospores remained clustered, at least transiently. Future studies should try to identify the signals released by zoospores in the presence of K+ responsible for promoting auto-aggregation behaviour. More complex microfluidics devices and a modelling approach are being developed with this aim in mind, for future studies.
Supplementary Material
Supplementary Material
Supplementary Material
Supplementary Material
Supplementary Material
Supplementary Material
Supplementary Material
Supplementary Material
Supplementary Material
Supplementary Material
Supplementary Material
Supplementary Material
Acknowledgements
We thank the Microscopy Platform-Sophia Agrobiotech Institut-INRA 1355-UNS-CNRS 7254-INRA PACA Sophia Antipolis for providing us with access to instruments and technical advice. The authors would like to thank Fernando Peruani, Emiliano Perez Ipiña (LJAD, Nice) and Laurent Counillon (LP2M, Nice) for fruitful discussions.
Data accessibility
Movies used in this work are available and uploaded as electronic supplementary material.
Authors' contributions
E.G. and X.N. designed the experiments. E.G. and C.E. carried out droplet and millifluidic analyses. X.N., C.C. and P.T. carried out microfluidic analyses. E.G., X.N., C.C. and P.T. wrote the manuscript.
Competing interests
The authors have no competing interests to declare.
Funding
This work was supported by the French government, through the UCAJEDI Investments in the Future project managed by the National Research Agency (ANR) under reference number ANR-15-IDEX-01; through the ‘Credits Scientifiques Incitatifs' of the University of Nice Sophia Antipolis and the ‘Action Recherche’ of the INRA Plant Health and Environment Division.
References
- 1.Judelson HS, Blanco FA. 2005. The spores of Phytophthora: weapons of the plant destroyer. Nat. Rev. Microbiol. 3, 47–58. ( 10.1038/nrmicro1064) [DOI] [PubMed] [Google Scholar]
- 2.Walker CA, van West P. 2007. Zoospore development in the oomycetes. Fungal Biol. Rev. 21, 10–18. ( 10.1016/j.fbr.2007.02.001) [DOI] [Google Scholar]
- 3.Appiah AA, van West P, Osborne MC, Gow NA. 2005. Potassium homeostasis influences the locomotion and encystment of zoospores of plant pathogenic oomycetes. Fungal Genet. Biol. 42, 213–223. ( 10.1016/j.fgb.2004.11.003) [DOI] [PubMed] [Google Scholar]
- 4.Cameron JN, Carlile MJ. 1980. Negative chemotaxis of zoopsores of the fungus Phytophthora palmivora. J. Gen. Microbiol. 120, 347–353. ( 10.1099/00221287-120-2-347) [DOI] [Google Scholar]
- 5.Morris PF, Ward EWB. 1992. Chemoattraction of zoospores of the soybean pathogen, Phytophthora sojae, by isoflavones. Physiol. Mol. Plant Pathol. 40, 17–22. ( 10.1016/0885-5765(92)90067-6) [DOI] [Google Scholar]
- 6.van West P, Morris BM, Reid B, Appiah AA, Osborne MC, Campbell TA, Shepherd SJ, Gow NAR. 2002. Oomycete plant pathogens use electric fields to target roots. Mol. Plant Microbe Interact. 15, 790–798. ( 10.1094/Mpmi.2002.15.8.790) [DOI] [PubMed] [Google Scholar]
- 7.Hardham AR. 2007. Cell biology of plant–oomycete interactions. Cell. Microbiol. 9, 31–39. ( 10.1111/j.1462-5822.2006.00833.x) [DOI] [PubMed] [Google Scholar]
- 8.Kebdani N, Pieuchot L, Deleury E, Panabieres F, Le Berre JY, Gourgues M. 2010. Cellular and molecular characterization of Phytophthora parasitica appressorium-mediated penetration. New Phytol. 185, 248–257. ( 10.1111/j.1469-8137.2009.03048.x) [DOI] [PubMed] [Google Scholar]
- 9.Le Berre JY, Engler G, Panabieres F. 2008. Exploration of the late stages of the tomato-Phytophthora parasitica interactions through histological analysis and generation of expressed sequence tags. New Phytol. 177, 480–492. ( 10.1111/j.1469-8137.2007.02269.x) [DOI] [PubMed] [Google Scholar]
- 10.Reid B, Morris BM, Gow NAR. 1995. Calcium-dependent, genus-specific, autoaggregation of zoospores of phytopathogenic fungi. Exp. Mycol. 19, 202–213. ( 10.1006/emyc.1995.1025) [DOI] [Google Scholar]
- 11.Savory AIM, Grenville-Briggs LJ, Wawra S, van West P, Davidson FA. 2014. Auto-aggregation in zoospores of Phytophthora infestans: the cooperative roles of bioconvection and chemotaxis. J. R. Soc. Interface 11, 20140017 ( 10.1098/rsif.2014.0017) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Larousse M, Galiana E. 2017. Microbial partnerships of pathogenic oomycetes. PLoS Pathog. 13, e1006028 ( 10.1371/journal.ppat.1006028) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Larousse M. et al. 2014. Characterization of PPMUCL1/2/3, three members of a new oomycete-specific mucin-like protein family residing in Phytophthora parasitica biofilm. Protist 165, 275–292. ( 10.1016/j.protis.2014.03.003) [DOI] [PubMed] [Google Scholar]
- 14.Kemen E. 2014. Microbe–microbe interactions determine oomycete and fungal host colonization. Curr. Opin Plant Biol. 20, 75–81. ( 10.1016/j.pbi.2014.04.005) [DOI] [PubMed] [Google Scholar]
- 15.Ochiai N, Dragiila MI, Parke JL. 2011. Pattern swimming of Phytophthora citricola zoospores: an example of microbial bioconvection. Fungal Biol. 115, 228–235. ( 10.1016/j.funbio.2010.12.006) [DOI] [PubMed] [Google Scholar]
- 16.Hill NA, Pedley TJ. 2005. Bioconvection. Fluid Dyn. Res. 37, 1–20. ( 10.1016/j.fluiddyn.2005.03.002) [DOI] [Google Scholar]
- 17.Kessler JO. 1986. Individual and collective fluid dynamics of swimming cells. J. Fluid Mech. 173, 191–205. ( 10.1017/S0022112086001131) [DOI] [Google Scholar]
- 18.Pedley TJ, Kessler JO. 1992. Hydrodynamic phenomena in suspensions of swimming micro-organisms. Annu. Rev. Fluid Mech. 24, 313–358. ( 10.1146/annurev.fl.24.010192.001525) [DOI] [Google Scholar]
- 19.Kong P, Hong C. 2010. Zoospore density-dependent behaviors of Phytophthora nicotianae are autoregulated by extracellular products. Phytopathology 100, 632–637. ( 10.1094/PHYTO-100-7-0632) [DOI] [PubMed] [Google Scholar]
- 20.Galiana E, Fourre S, Engler G. 2008. Phytophthora parasitica biofilm formation: installation and organization of microcolonies on the surface of a host plant. Environ. Microbiol. 10, 2164–2171. ( 10.1111/j.1462-2920.2008.01619.x) [DOI] [PubMed] [Google Scholar]
- 21.Zheng L, Mackrill JJ. 2016. Calcium signaling in oomycetes: an evolutionary perspective. Front. Physiol. 7, 123 ( 10.3389/fphys.2016.00123) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McDonald JC, Duffy DC, Anderson JR, Chiu DT, Wu H, Schueller OJ, Whitesides GM. 2000. Fabrication of microfluidic systems in poly(dimethylsiloxane). Electrophoresis 21, 27–40. () [DOI] [PubMed] [Google Scholar]
- 23.Sbalzarini IF, Koumoutsakos P. 2005. Feature point tracking and trajectory analysis for video imaging in cell biology. J. Struct. Biol. 151, 182–195. ( 10.1016/j.jsb.2005.06.002) [DOI] [PubMed] [Google Scholar]
- 24.Tinevez JY, Perry N, Schindelin J, Hoopes GM, Reynolds GD, Laplantine E, Bednarek SY, Shorte SL, Eliceiri KW. 2017. TrackMate: an open and extensible platform for single-particle tracking. Methods 115, 80–90. ( 10.1016/j.ymeth.2016.09.016) [DOI] [PubMed] [Google Scholar]
- 25.Humphries J, Xiong L, Liu J, Prindle A, Yuan F, Arjes HA, Tsimring L, Suel GM. 2017. Species-independent attraction to biofilms through electrical signaling. Cell 168, 200–209.e12. ( 10.1016/j.cell.2016.12.014) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yost S, Duran-Pinedo AE, Krishnan K, Frias-Lopez J. 2017. Potassium is a key signal in host-microbiome dysbiosis in periodontitis. PLoS Pathog. 13, e1006457 ( 10.1371/journal.ppat.1006457) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sparks DL. 1987. Potassium dynamics in soils. In Advances in soil science (ed. Stewart BA.), pp. 1–63. Berlin, Germany: Springer. [Google Scholar]
- 28.Maathuis FJM, Sanders D. 1996. Mechanisms of potassium absorption by higher plant roots. Physiol. Plant. 96, 158–168. ( 10.1111/j.1399-3054.1996.tb00197.x) [DOI] [Google Scholar]
- 29.Gow NAR. 2004. New angles in mycology: studies in directional growth and directional motility. Mycol. Res. 108, 5–13. ( 10.1017/S0953756203008888) [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Movies used in this work are available and uploaded as electronic supplementary material.