Abstract
Frontotemporal dementia is a group of early onset dementia syndromes linked to underlying frontotemporal lobar degeneration (FTLD) pathology that can be classified based on the formation of abnormal protein aggregates involving tau and two RNA binding proteins, TDP-43 and FUS. Although elucidation of the mechanisms leading to FTLD pathology is in progress, recent advances in genetics and neuropathology indicate that a majority of FTLD cases with proteinopathy involving RNA binding proteins show highly congruent genotype–phenotype correlations. Specifically, recent studies have uncovered the unique properties of the low-complexity domains in RNA binding proteins that can facilitate liquid–liquid phase separation in the formation of membraneless organelles. Furthermore, there is compelling evidence that mutations in FTLD genes lead to dysfunction in diverse cellular pathways that converge on the endolysosomal pathway, autophagy, and neuroinflammation. Together, these results provide key mechanistic insights into the pathogenesis and potential therapeutic targets of FTLD.
Keywords: FTD, frontotemporal dementia, FTLD, frontotemporal lobar degeneration, ALS, amyotrophic lateral sclerosis, RNA binding proteins, TDP-43, FUS, low-complexity domain, hydrogels, liquid droplets
1. INTRODUCTION
Frontotemporal dementia (FTD) refers to a group of dementia syndromes with early age of onset that are linked to underlying frontotemporal lobar degeneration (FTLD) pathology (1, 2). The first description of FTD came from the Czech neurologist Arnold Pick, who described patients suffering from slowly progressive language and behavioral disorders (3, 4). Postmortem neuropathological examination of one of Pick’s patients revealed focal atrophy in the anterior temporal lobes, suggesting that dementia can be clinically and anatomically focal. Using Bielschowsky silver staining methods, Alois Alzheimer identified key microscopic features in the brain of Pick’s patient, including silver-positive, or argyrophilic, neuronal cytoplasmic inclusions in affected brain regions (5). These inclusions, now known as Pick bodies, were noted to be different from the neurofibrillary tangles seen in patients with Alzheimer’s disease (AD).
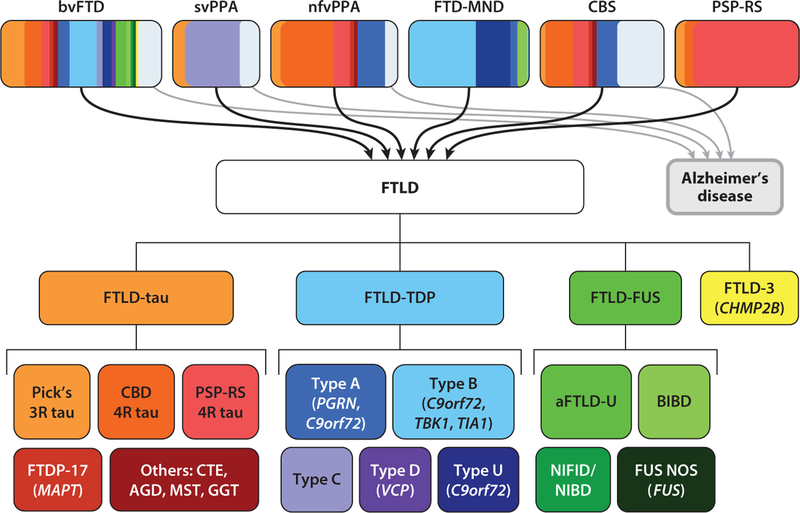
Advances in clinical assessment, neuroimaging, human genetics, and neuropathological examinations have transformed our ability to characterize neurodegenerative diseases and explore disease mechanisms. Clinically, FTD can be categorized into six major subtypes, including behavioral variant FTD (6), the semantic variant and the nonfluent/agrammatic variant of primary progressive aphasia (7), frontotemporal dementia–motor neuron disease, and two dementia–movement disorder syndromes, corticobasal syndrome and progressive supranuclear palsy–Richardson syndrome (PSP-RS; also known as Steele–Richardson–Olszewski syndrome) (Figure 1) (8, 9). Clarifying the clinical features of FTD enabled a deeper understanding of the neuroanatomy of each major syndrome. In parallel, the advent of network-sensitive neuroimaging approaches, including task-free functional magnetic resonance imaging and diffusion tensor imaging, has allowed researchers to make new maps describing the topology of human brain networks. Increasingly, these converging data streams have revealed that each FTD syndrome reflects the degeneration of a specific large-scale neural network (10, 11) and connectomic mapping can predict how degeneration will spread throughout brain networks (12, 13).
Figure 1.
Summary of the clinical syndromes, neuropathology, and genetics of FTD. The six clinical syndromes include bvFTD, svPPA, nfvPPA, FTD-MND, CBS, and PSP-RS. A small number of patients with bvFTD, svPPA, nfvPPA, or CBS have Alzheimer’s disease as the underlying pathology. Based on the proteinopathy involved, the neuropathology of FTLD can be classified as FTLD-tau, FTLD-TDP, or FTLD-FUS. Genetic mutations associated with each molecular subtype are italicized. A rare subtype of FTLD (FTLD-3) has been reported in patients with mutations in the CHMP2B gene. The designations 3R and 4R indicate, respectively, three microtubule binding repeats and four tau repeats. Abbreviations: aFTLD-U, atypical FTLD-ubiquitin; AGD, argyrophilic grain disease; BIBD, basophilic inclusion body disease; bvFTD, behavioral variant FTD; CBD, corticobasal degeneration; CBS, corticobasal syndrome; CTE, chronic traumatic encephalopathy; FTD, frontotemporal dementia; FTD-MND, FTD with motor neuron disease; FTDP-17, familial FTD with mutations on the MAPT gene on chromosome 17; FTLD, frontotemporal lobar degeneration; GGT, globular glial tauopathy; MST, multisystem tauopathy; nfvPPA, nonfluent/agrammatic variant primary progressive aphasia; NIFID/NIBD, neuronal intermediate filament inclusion disease/neurofilament inclusion body disease; NOS, not otherwise specified; PSP-RS, progressive supranuclear palsy–Richardson syndrome; svPPA, semantic variant primary progressive aphasia.
Another exciting new development in FTD research relates to the potential association between patients with specific subtypes of FTLD pathology and autoimmune disorders. This association was first noted in case–control studies that evaluated the prevalence of autoimmune diseases in control, FTLD, and AD patients. The results showed that certain FTLD patient cohorts, especially those with semantic variant primary progressive aphasia and mutations in the progranulin (GRN) and C9orf72 genes, have a higher rate of nonthyroid autoimmune diseases, such as inflammatory arthritis, cutaneous disorders, and gastrointestinal conditions (14, 15). This association is further strengthened by a recent report that analyzes data from large-scale genome-wide association studies (GWASs) provided by the International FTD-Genomics Consortium. The results identify several immune-associated genetic loci, particularly genes within the HLA loci, which increase the risk for FTD (16). Many of these FTD immune genes have been implicated in microglial and macrophage functions. Section 4 provides further discussions on the newly discovered roles of GRN and C9orf72 in microglia and the brain innate immunity system (17, 18).
The unique neural circuit vulnerability and immune dysfunctions in FTLD represent two rapidly developing and interwoven research areas that are pushing the boundaries of understanding these devastating neurodegenerative diseases. In addition, the expansion in knowledge of the genetics of FTLD has further facilitated the understanding of the disease mechanisms. Given the tremendous progress made during the past few years, we feel that this is the right time to provide a review that focuses on the congruent phenotype–genotype correlation between the neuropathology and genetic mutations in FTLD and how genetic mutations implicated in familial FTLD provide instructive information for guiding research into the molecular and cell biological bases of FTLD in general. Our discussion focuses on the unique properties of the low-complexity (LC) domain of RNA binding proteins and its contribution to liquid–liquid phase separation (LLPS) in the formation of membraneless organelles. We also discuss how mutations in FTLD genes lead to dysfunction in diverse cellular pathways that converge on RNA binding proteins, most notably FUS and TARDBP (TAR DNA binding protein 43 kDa, or TDP-43), and ultimately contribute to disease pathogenesis.
2. THE PATHOLOGY, GENETICS, AND CELL BIOLOGY OF FTLD
2.1. Proteinopathy in FTLD
The FTD syndromes are united by their association with underlying FTLD pathology, although AD pathology contributes in a small but important minority of patients whose clinical presentations mimic FTD (Figure 1). From a neuropathological perspective, FTLD can be divided into three major molecular classes: FTLD-tau, FTLD-TDP, and FTLD-FUS; each of these features distinct subtypes based on the morphology and distribution of disease-associated protein inclusions (Figure 2) (19, 20). As highlighted in Figure 2a, patients with Pick’s disease develop tau protein aggregates that predominantly contain three microtubule binding repeats (3R tau) in the Pick bodies, whereas patients with progressive supranuclear palsy or corticobasal degeneration develop tau aggregates with four repeats (4R tau) in thorny astrocytes or tufted astrocytes (Figure 2b,c). In addition, tau proteinopathy can be detected in familial FTD with mutations on the MAPT gene on chromosome 17. Other forms of tau proteinopathy have been described, including chronic traumatic encephalopathy (21), argyrophilic grain disease (22), globular glial tauopathy, and others (23).
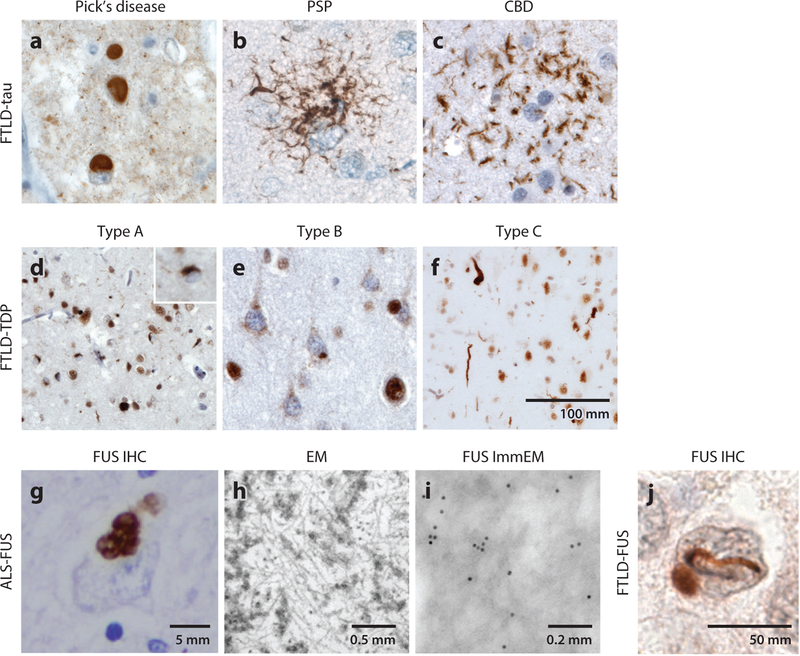
Figure 2.
Neuropathology of FTLD. Subtypes of (a–c) FTLD-tau and (d–f) FTLD-TDP are distinguished by the morphology and distribution of their characteristic lesions. (a) Pick bodies in Pick’s disease; (b) a tufted astrocyte in PSP; (c) an astrocytic plaque in CBD; (d) small compact or crescentic neuronal cytoplasmic inclusions and short, thin neuropil threads in FTLD-TDP type A; (e) diffuse granular or speckled neuronal cytoplasmic inclusions, with a relative paucity of neuropil threads, in FTLD-TDP type B; and (f) long, tortuous dystrophic neurites in FTLD-TDP type C. TDP-43 can be seen within the nucleus in neurons lacking inclusions, but it mislocalizes to the cytoplasm within inclusion-bearing neurons. (g) Immunohistochemical stain for FUS highlights the prominent FUS-positive staining in basophilic inclusions in the cytoplasm of spinal motor neurons in an ALS-FUS case. (h,i) Ultrastructural analyses of the same case as shown in panel g show that these inclusions (h) contain filamentous aggregates measuring 15–20 nm in diameter and (i) are immunoreactive for FUS antibody. (j) The most common subtype of FTLD-FUS is aFTLD-U, characterized by signature FUS immunoreactive vermiform intranuclear inclusions. Immunostains were performed using antibodies to (a) 3R tau, (b,c) phospho-tau (CP-13), (d–f) TDP-43, and (g,i,j) FUS. The scale bar in panel f is applicable to panels a–e. Abbreviations: aFTLD-U, atypical FTLD-ubiquitin; ALS, amyotrophic lateral sclerosis; CBD, corticobasal degeneration; FTLD, frontotemporal lobar degeneration; EM, electron microscopy; IHC, immunohistochemistry; ImmEM, immunogold electron microscopy; PSP, progressive supranuclear palsy.
Although FTLD-tau was the first molecular class discovered, FTLD-TDP is the most frequent class, accounting for more than half of most large series. Indeed, the discovery of TDP-43 as the other major disease protein in FTLD and as the dominant aggregating disease protein in amyotrophic lateral sclerosis (ALS) provided a major breakthrough for research into neurodegenerative diseases (24, 25). Furthermore, TDP-43 inclusions can be observed in patients with AD and as part of cognitively normal aging, suggesting that the abnormal formation of TDP-43 protein aggregates may have broader implications (26, 27). Following the identification of TDP-43 as a major disease protein, two genetic discoveries provided important insights into the pathogenesis of FTLD-TDP. The first is autosomal dominant mutations in the GRN gene, located on chromosome 17q21.31 near the MAPT locus (28, 29). The second mutation is the (GGGGCC)n hexanucleotide repeat expansion (HRE) that is located between exons 1a and 1b of the C9orf72 gene on chromosome 9p21.2 (30, 31). C9orf72 mutations are the most common cause of familial and sporadic FTLD and ALS. Importantly, patients with GRN or C9orf72 mutations almost invariably develop abnormal TDP-43 protein aggregates as the major pathological signature.
Based on the laminar distribution, morphology, and subcellular localization of TDP-43 protein aggregates in the cerebral cortex, FTLD-TDP can be classified into four major subtypes, A, B, C, and D (19). FTLD-TDP type A is characterized by numerous short, dystrophic neurites and compact, crescent-shaped or ovoid neuronal cytoplasmic inclusions, predominantly in the upper cortical layers (Figure 2d). Although rare, neuronal intranuclear inclusions can also be detected. In type B, a few to a moderate amount of fine threads or grains may be present, but the predominant inclusion type is a fine, granulofilamentous or speckled neuronal cytoplasmic inclusion found in the upper and deep cortical layers. More compact, ovoid neuronal cytoplasmic inclusions may also be seen in the same distribution (Figure 2e). FTLD-TDP type C is characterized by its predominant long, swollen dystrophic neurites that are most prominent in the upper cortical layers but also extend into the deeper layers (Figure 2f). These neurites dominate the picture, and few neuronal cytoplasmic inclusions are seen in the cerebral cortex, although compact or round neuronal cytoplasmic inclusions are identified in some subcortical structures, particularly the ventral striatum. Finally, type D is a rare form characterized by numerous short, dystrophic neurites and frequent round or lentiform neuronal intranuclear inclusions. In familial FTLD-TDP, the specific gene mutation strongly predicts the pathological subtype. FTLD with a GRN mutation is accompanied by type A pathology, whereas most patients with C9orf72 mutations develop type B or, less often, type A or a TDP-43 proteinopathy that is too sparse or atypical to classify. In addition, early reports suggested that patients with mutations in TIA1 (T cell–restricted intracellular antigen 1) develop type B (32, 33), whereas those with mutations in the TBK1 (TANK binding kinase 1) gene may develop either type A or B (Figure 1) (34–36). Finally, mutations in the VCP (valosin-containing protein) gene, which is a member of the AAA-ATPase gene superfamily, are identified in the rare patients with FTD associated with inclusion body myositis and Paget’s disease of bone. These patients universally develop type D, which is not seen in other genetic or sporadic cases (37).
Once FTLD cohorts had been sorted into patients with FTLD-tau and those with FTLD-TDP, there remained 5–10% in most large series in whom ubiquitin-positive, tau- and TDP-43-negative inclusions were seen; the most common subtype among this group was termed atypical FTLD-ubiquitin (aFTLD-U). Three years after the discovery of TDP-43 as a major insoluble protein in FTLD patients, mutations in the FUS (TLS) [fused in sarcoma (translocated in liposarcoma)] gene were shown to cause familial ALS, and FUS protein–containing inclusions were identified in these patients (Figure 2g–i) (38, 39). The link between FTLD and ALS then helped catalyze the discovery that aFTLD-U is characterized by inclusions containing FUS, the protein product of FUS (TLS) (Figure 2j) (40, 41). Further work showed that the inclusions in this and other non-tau, non-TDP-43 FTLD subtypes—including neuronal intermediate filament inclusion disease and basophilic inclusion body disease—contained FUS protein as well as the other proteins from the FET family, including EWS (Ewing sarcoma) protein and TAF15 (TATA-box binding protein associated factor 15).
Due to mutations in FUS, in ALS-FUS the characteristic neuropathological features include well-delineated single or multilobulated basophilic inclusions in the cytoplasm of spinal motor neurons, neurons in the brainstem nuclei, and cortical neurons. These inclusions are strongly immunoreactive for FUS protein (Figure 2g), but the levels of ubiquitin in these inclusions vary. Interestingly, these inclusions contain only FUS and not other FET family proteins (40). Ultrastructurally, these inclusions contain abundant filamentous protein aggregates that are immunoreactive for FUS (Figure 2h,i) (42). These filamentous FUS-positive structures, measuring 15–20 nm in diameter, are intermixed with disorganized intracellular organelles, including the endoplasmic reticulum, mitochondria, and lipid droplets, suggesting that the formation of these inclusions may interfere with the normal functions of these organelles. FTLD-FUS pathology varies by subtype, but in aFTLD-U the inclusions include characteristic serpentine or vermiform neuronal intranuclear inclusions and compact or ovoid neuronal cytoplasmic inclusions (Figure 2j). Curiously, unlike ALS-FUS, which is always the result of a FUS mutation, genetic analyses in most FTLD-FUS patients show no evidence of mutations in the FUS gene. Most patients with FTLD-FUS present with behavioral variant FTD or FTD–motor neuron disease and tend to be younger and have a more rapidly progressive clinical course, with extreme anterior striatal atrophy as well as atrophy in typical FTD-related cortical areas (43).
2.2. The Genetics of FTLD
The idea that FTD and ALS are on a disease spectrum is further supported by evidence that patients with familial FTD and ALS often share mutations in the same genes. A variety of genetic mutations have been found to cause familial FTD, including the three most common FTLD disease genes, MAPT, GRN, and the (GGGGCC)n HRE in the C9orf72 gene (44). In addition, rare FTLD disease-associated genes include CHMP2B (charged multivesicular body protein 2B), VCP, hnRNPA1 (heterogeneous nuclear ribonucleoprotein A1), hnRNPA2B1 (heterogeneous nuclear ribonucleoprotein A2/B1), SQSTM1 (sequestosome 1; p62), TBK1, OPTN (optineurin), TARDBP, FUS, TIA1, CHCHD10 (coiled-coil-helix-coiled-coil-helix domain containing 10), UBQLN2 (ubiquilin 2), and DCTN1 (dynactin 1). Finally, mutations in CSF1R (colony stimulating factor 1 receptor), TREM2 (triggering receptor expressed on myeloid cells 2), and PRKAR1B (protein kinase cAMP-dependent type I regulatory subunit beta) have been identified in FTLD patients with atypical clinical presentations. Intriguingly, several of these FTLD genes—most notably the C9orf72 HRE, TARDBP, and FUS—are also identified in patients with sporadic and familial ALS, again supporting the idea of a disease spectrum containing FTD and ALS. Unlike for AD, relatively few FTLD genetic risk factors have been identified in GWASs. To date, the most replicated FTLD risk gene is TMEM106B (transmembrane protein 106B), especially in patients with GRN or C9orf72 mutations (44). Based on the studies described in Reference 44, carriers of the T allele on a single nucleotide polymorphism (rs1990622) in TMEM106B show increased risk for FTLD, whereas the C allele appears to be protective. Other genes implicated in increasing the risk for FTLD include the HLA locus on chromosome 6p21.3, RAB38 on chromosome 6p21.3, and CTSC (cathepsin C).
2.3. Cellular Pathways Targeted by FTLD
The highly significant genotype–phenotype association between genetic mutations and TDP-43 proteinopathy (Figure 1) strongly suggests that further investigations into the functions of these FTLD genes should provide critical insights into disease pathogenesis. Here, we summarize several themes that have emerged about the key cell biological pathways that are affected by the FTLD genes. The first major theme indicates that dysfunction and protein misfolding in RNA binding proteins are prominent features of FTLD, as highlighted by pathology and genetics. Several FTLD mutations are identified in genes that encode RNA binding proteins, such as TARDBP, hnRNPA1, and hnRNPA2B1. In particular, TDP-43 is a major nuclear protein that regulates many important aspects of RNA metabolism, including RNA splicing, transcription, and nuclear– cytoplasmic transport (45–48). Similar to TDP-43, FUS also has broad and critical functions in RNA splicing, RNA transport, DNA damage repair, and the assembly of nucleoli, nuclear speckles, and stress granules (41, 48).
The second theme is the assembly and disassembly of RNA binding proteins and RNA granules. As is discussed in more detail in Section 3, a large number of recent studies provide exciting evidence that RNA binding proteins possess the unique properties of the LC domains that promote the formation of membraneless organelles. These properties can be interfered with by two major products made by the (GGGGCC)n HRE mutations in the C9orf72 gene, namely the RNA foci and dipeptide repeat (DPR) proteins (30, 31, 48).
Finally, a majority of FTLD genes—including GRN, C9orf72, VCP, OPTN, CHMP2B, and SQSTM1—are involved in vesicular trafficking in the endolysosomal pathway, autophagy and the formation of the autophagosome, and the protein quality control process. There is compelling evidence from animal models and human pathology that many of these genes regulate these cellular pathways in a gene dose–dependent manner. Given the broader roles of these pathways in diverse cell types, it is conceivable that mutations that interfere with the normal functions of these genes can trigger chain reactions with devastating consequences.
In Section 3, we discuss in depth the discovery of the LC properties of RNA binding proteins and how these unique properties offer new insights into the fundamental disease mechanisms of FTLD. In Section 4, we use the two most common FTLD mutations, of C9orf72 and GRN, to highlight how dysfunctions in the endolysosomal and autophagy pathways may facilitate the formation of TDP-43 proteinopathy. In addition, we discuss how mutations in SQSTM1, UBQLN2, OPTN, and VCP could negatively impact the autophagy pathway. Finally, we discuss the newly revealed and exciting roles of GRN and C9orf72 in neuroinflammation and how this phenotype may contribute to neurodegeneration and dysfunctions in the immune system in subsets of FTLD patients.
3. LOW-COMPLEXITY DOMAINS IN RNA BINDING PROTEINS
3.1. Low-Complexity Properties of FUS
Through serendipity, a series of studies has uncovered a unique structural and biochemical prop-erty that underscores the aggregation-prone nature of FUS (49). Using biotinylated isoxazole (b-isox)—a synthetic organic chemical that promotes neuronal fate in embryonic stem cells (50)— McKnight and colleagues (51, 52) isolated hundreds of proteins that form spontaneous precipitates with b-isox, presumably due to the increased propensity of b-isox to crystallize into orderly structures that resemble extended β strands in proteins. Mass spectrometry shows that the majority of the proteins in the precipitates with b-isox are RNA binding proteins that possess highly homologous LC domains—of about 200 amino acids in length—to provide the interaction interface with b-isox. The three proteins with the highest affinity for precipitating with b-isox are the RNA binding proteins FUS, EWSR1 (EWS RNA binding protein 1), and TAF15, which are collectively known as the FET proteins, given their close homology in amino acid sequences. A series of biochemical assays further revealed two properties in the FUS LC domain. First, when purified as a fusion protein with green fluorescent protein, maltose binding protein, or glutathione S-transferase, the FUS LC domain can self-assemble into amyloid-like polymers via homotypic interactions. This allows the FUS LC domain to undergo physical phase transition from soluble proteins into gel-like structures called hydrogels (51, 52). Second, a simple binding assay, which involves trapping these hydrogels, allows visualization of FUS and other LC domain–containing proteins under microscopy. This assay has been instrumental in characterizing the LC domain in other RNA binding proteins in the b-isox precipitates and identifying other potential binding partners for the FUS LC domain via heterotopic interactions. One salient example is the identification of the C-terminal domain (CTD) of RNA polymerase II as a binding partner for the LC domain of FET proteins (53). The CTD of mammalian RNA polymerase II—which contains 52 repeats of almost exclusively 4 amino acids, tyrosine, serine, proline, and threonine—is by definition an LC domain itself. Intriguingly, the CTD does not self-assemble into amyloid-like polymers, but it can bind to hydrogels formed by the LC domain of the FET proteins. The polymer formation between the FUS LC domain and the CTD can be regulated by cyclin-dependent kinase-mediated phosphorylation of the CTD. These results have been independently validated using a conventional protein purification scheme to identify FUS interacting partners (54, 55). In addition to the CTD of RNA polymerase II, several serine:arginine repeat domain–containing proteins implicated in pre-mRNA splicing are also identified in the b-isox precipitates. Similar to the CTD, the serine:arginine domains of these splicing factors do not self-assemble, but they can be readily detected in FUS LC domain hydrogels and then reversed upon phosphorylation by cyclin-dependent kinases.
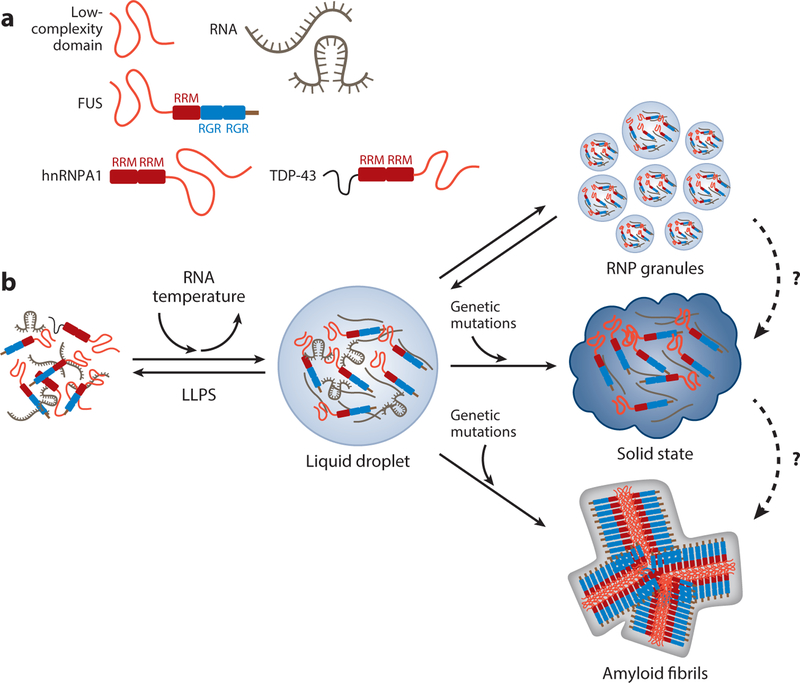
Despite the compelling evidence supporting the unique hydrogel properties of the FUS LC domain, several studies offer a different interpretation regarding its biophysical properties. In a study designed to investigate the assembly of multivalent macromolecules, Rosen and colleagues (56) discovered that when the SRC homology 3 (SH3) domain and its proline-rich motif (PRM) ligand are organized in tandem repeats, they exhibit phase separation properties to form liquid-like droplets (a process also known as LLPS). Structural analyses show that these droplets do not represent aberrant aggregate formation, but rather they are the result of a highly ordered and thermodynamically coupled molecular sol–gel transition within the droplet phase. Such behavior of SH3n and PRMn repeats can be detected in live cells and in other signaling molecules, such as the nephrin/NCK/N-WASP system (56). Following the publication of the study, a series of papers showed that the LC domains in many RNA binding proteins—such as FUS, hnRNPA1, Lsm4 (involved in P-body assembly in yeast), and stress granule protein TIA1 and its yeast homolog Pub1—are intrinsically disordered and can undergo LLPS (Figure 3) (57–60). The formation of liquid droplets from the LC domains of these proteins shows progressive maturation over time, and this is further supported by the presence of RNA. Similar to the reported heterotypic interactions of the FUS LC domain with other RNA binding proteins in hydrogels, in liquid droplets it can recruit additional LC domains from other RNA binding proteins. Most impressively, disease-related mutations in FUS and hnRNPA1 LLPS can promote solid-state or fibrillary transformation (57, 58). Together, these results provide key mechanistic insights into how the unique biophysical properties of LC domains in FUS and hnRNPA1 are pathogenic in neurodegeneration (Figure 3).
Figure 3.
Liquid–liquid phase separation (LLPS) governs the formation of membraneless organelles. (a) Schematic representing the low-complexity (LC) domain, RNA (with relaxed and stem-loop configurations), and the RNA binding proteins FUS, TDP-43, and hnRNPA1, each containing an LC domain, RNA recognition domain, and arginine-glycine-rich motifs. (b) LLPS promotes the formation of membraneless liquid droplets or hydrogels, a process which can be facilitated by the presence of RNA and low temperatures. The metastable property of liquid droplets is critical for the assembly and disassembly of many membraneless organelles, such as RNA granules, stress granules, the nucleolus, and nuclear speckles. Under pathological conditions, liquid droplets can transition into a solid-state structure or form amyloid fibrils, which are more stable and whose formation is likely irreversible.
While data about both the hydrogels and liquid droplets highlight the unique properties of the FUS LC domain to undergo reversible liquid-to-solid phase transition, there are some fundamental differences regarding the nature of protein–protein interactions between these two physical states. Structural studies of the hydrogels indicate that the interactions of the LC domains of FET proteins and the CTD form cross-β polymers, which are a key characteristic in many yeast proteins with prion-like properties (61). Data from electron microscopy and nuclear magnetic resonance spectroscopy further reveal that the FUS LC domain polymers structurally resemble other pathologically aggregated proteins, such as the Aβ40 and Aβ42 amyloid peptides seen in AD. However, unlike Aβ40, Aβ42, and α-synuclein, the FUS LC domain lacks hydrophobic amino acids that provide a hydrophobic protein interaction interface to promote the formation of pathogenic prion-like polymers. In addition, FUS LC domains can be phosphorylated by DNA-dependent protein kinase, which promotes the dissolution of FUS nuclear puncta and the translocation of FUS from the nucleus to the cytoplasm (62–64). Indeed, contrary to the cross-β polymer model, solution nuclear magnetic resonance assays combined with photobleaching show that the FUS LC domain is intrinsically disordered, both as a monomer and within the liquid droplet state (59, 60). One significant physical property of the FUS LC domain liquid droplets is that the degenerate tyrosine and glutamine residues in the repeats provide high-valency, hydrophobic, but not electrostatic, interactions that stabilize FUS LC phase separation. Such formation of liquid droplets can be further promoted by the presence of RNA, low temperatures, and low salt (59). While the exact cause or causes accounting for the differences in the biophysical properties of hydrogels and liquid droplets remain unclear, these unique properties of FUS LC domains have tremendous impacts on the biological functions of nuclear compartmentalization, RNA granule formation, and the biogenesis of intracellular organelles (65–67).
3.2. The FUS Low-Complexity Domain and RNA Granule Assembly
The discovery of the unique biophysical properties of the FUS LC domain in hydrogels or LLPS opens up multiple lines of investigation into the broader role of RNA binding proteins, such as FUS, hnRNPA1, and TDP-43, in the assembly of RNA granules and whether perturbations to this process may contribute to the pathogenesis of FTD and ALS. RNA granules are membraneless structures that control mRNA translation, localization, transport, and degradation in eukaryotes (68). The involvement of FUS in RNA granules was first implicated in a study that aimed to identify the role of conventional kinesin (KIF5) in the transportation of RNA granules along dendrites in neurons (69). Among the 42 proteins in the RNA granules fully characterized by mass spectrometry in the study, FUS shared similar elution profiles on KIF5 affinity chromatography with several RNA binding proteins, including DDX3 (DEAD-box polypeptide 3, an RNA helicase), SYNCRIP (synaptotagmin binding cytoplasmic RNA interacting protein, a multifunctional heterogeneous ribonucleoprotein), several FMRPs (fragile X mental retardation proteins), and ALY (a transcriptional coactivator and nuclear export factor). These results suggest that FUS may be involved in multiple steps in the formation of RNA granules, from nuclear–cytoplasmic transport to assembly and local protein translation. Consistent with these results, the majority of FUS proteins are present in the nuclear fractions, although a significant amount can also be detected in the cytoplasm. The localization of FUS to RNA granules seems to be disrupted by ribonuclease treatment, suggesting that the presence of FUS in these granules may depend on its binding to RNA.
If the KIF5 study provides a glimpse of the potential roles of FUS in RNA granule biology, the identification of the unique properties of the FUS LC domain opens up tremendous opportunities to interrogate its interactomes and in vivo functions under physiological conditions. One remarkable finding from the mass spectrometry analyses of the b-isox precipitates is that the majority of the proteins are involved in neuronal RNA granules, stress granules, processing bodies, or polar granules (52). Intriguingly, there is extensive overlap between the protein constituents in the KIF5 RNA granules and b-isox precipitates, supporting a critical role for the FUS LC domain in the assembly and functions of RNA granules. Indeed, FUS LC domain hydrogels have been used to isolate and characterize bound and unbound RNAs from mouse brain lysates. Deep sequencing revealed 11 mRNAs that are enriched in the RNA granules in the neuronal dendrites that are critical for dendritic growth and synaptic functions (51). These results support the idea that the FUS LC domain may regulate the assembly of RNA granules. In support of this, fluorescently tagged bacteriophage MS2 stem-loop RNAs can bind to FUS LC domain hydrogels. Furthermore, FUS has also been shown to form high-order RNA–protein complexes with other RNAs that have stem-loop structures. Several studies report similar binding of FUS to stem-loop RNA structures (70, 71) or to the secondary and tertiary structures formed by the short RNA repeats r(UUAGGG) in the G-quadruplex telomeric repeat–containing RNA (or TERRA) (72).
3.3. FUS Mutations and Proteinopathy
The above-referenced studies establish several key insights regarding the general principles employed by FUS and other RNA binding proteins in regulating the assembly of RNA granules and other membraneless structures. First, the FUS LC domain is structurally disordered and can form a hydrogel or LLPS state, which provides a versatile protein–protein or protein–RNA interaction interface that has a critical role in RNA granule assembly. These interactions are tightly regulated by phosphorylation on serine residues in the LC domain, suggesting that the FUS LC domain has an active role in the assembly and disassembly of the constituents of the RNA granules. Second, the hydrogel or LLPS properties of the LC domain in FUS and other RNA binding proteins have broader implications for the spatiotemporal control of signal transduction and the partitioning of subcellular compartments in the nucleus and cytoplasm. Finally, the dynamic progression of the FUS LC domain from LLPS to liquid droplets to the formation of spike-like fibers raises the possibility that disease-causing mutations in FUS and other RNA binding proteins, such as hnRNPA1 and TDP-43, may directly contribute to abnormal protein aggregation and cellular pathology in FTD and ALS. In support of this idea, liquid droplets that include the FUS LC domain that carries an ALS-related point mutation, FUS G165E, show a significant decrease in their ability to fuse with other liquid droplets and an increased propensity to convert from droplets to fibrous structures (58). Consistent with these results, ALS-related mutations in the CTD of FUS—including FUS R495X, FUS R522G, FUS R524S, and FUS P525L—also show reduced reversibility in liquid droplet assembly and disassembly, which correlates with an increased propensity to form protein aggregates and increased neurotoxicity when these proteins are expressed in Caenorhabditis elegans (73). Interestingly, small interfering RNA knockdown of stress granule components partially rescues the neurotoxicity, suggesting that mutant FUS protein aggregates may dominantly sequester RNP granule formation (Figure 3). Similar to FUS mutations, missense mutations in the LC domain of hnRNPA1, which are causally linked to the neurodegenerative diseases ALS and multisystem proteinopathy (74), also promote fibrillization in hnRNPA1 (57).
Taken together, these results support the model of the LC domains of RNA binding proteins providing multivalent interfaces that promote hydrogel formation or LLPS phase transition that are critical for reversible assembly and disassembly of membraneless structures, such as nuclear bodies, speckles, RNP granules, and stress granules (Figure 3). These structures critically regulate diverse cellular physiology, including the DNA damage response, RNA splicing, RNA transport, and protein synthesis. Most importantly, the discovery of the unique biophysical properties of the FUS LC domain provides critical mechanistic insights into understanding the pathological features of FUS protein aggregates in ALS-FUS and FTLD-FUS. Disease-related mutations in FUS or other pathological conditions alter the multiphase characteristics and the metastability of these RNP granules, leading to irreversible protein aggregations and devastating pathology in vulnerable neurons.
4. PATHWAYS LEADING TO FTLD-TDP
4.1. C9orf72 Mutations
The identification of HRE mutations in the C9orf72 gene in FTD patients has captured tremendous attention because of the high prevalence of these mutations in familial FTD cases. It is increasingly clear that C9orf72 mutation carriers in the same family can present with diverse clinical presentations, underscoring the complexity of the pathogenesis caused by this mutation. Here, we summarize several key features that can potentially contribute to the mechanisms of disease in patients with C9orf72 mutations.
4.1.1. RNA foci and repeat-associated non-ATG-mediated protein translation.
At the outset of the discovery of C9orf72 HRE mutations, it was reported that the sense transcripts of the (GGGGCC)n repeats can be detected in the nuclei of cortical neurons and spinal motor neurons in postmortem tissues from patients with C9orf72 mutations, but not in other ALS or FTD patients (30). Since similar repeat-associated transcripts have been reported in other noncoding repeat expansion diseases—such as myotonic dystrophy, fragile X–associated tremor/ataxia syndrome, and spinocerebellar ataxias (75)—these RNA foci garnered immediate attention as the potential pathogenic factor in the neurodegeneration associated with C9orf72 mutations. Indeed, similar RNA foci can be detected in neurons derived from induced pluripotent stem cells (iPSCs) from patients with C9orf72 mutations (76–78) and in neurons in transgenic mice that express bacterial artificial chromosome (BAC) isolated from patients with C9orf72 mutations (79–82). The foci of the antisense transcript, composed of (CCCCGG)n repeats, are also detected, indicating that bidirectional transcription occurs at the C9orf72 HRE (83).
One mechanism by which HRE RNA may aggregate into foci is by the formation of G-quadruplexes, in which four guanine bases interact around a central cation, often potassium, to form a planar structure, called a guanine tetrad, in a process known as Hoogsteen bonding, and multiple tetrads can stack to form a G-quadruplex. This is a physiologic mechanism of transcriptional and translational regulation and protection of telomere ends. (GGGGCC)n repeats form extremely stable, multimolecular G-quadruplexes that incorporate multiple strands of RNA, and they are more readily formed with increased repeat length and concentration (84). Presumably, the accumulation of these (GGGGCC)n-containing RNA foci near the nucleoli can have toxic gain-of-function properties that interact with RNA binding proteins and interfere with gene expression (76, 77). In addition to the formation of RNA foci, (GGGGCC)n repeats can also form DNA–RNA hybrids and G-quadruplexes that further promote the sequestration of RNA binding proteins (76, 85) and interfere with the machinery that regulates the nuclear–cytoplasmic transport pathway (86, 87).
Another major discovery related to C9orf72 mutations is that the (GGGGCC)n-containing RNA might be aberrantly translated into DPR proteins from both sense and antisense orientations—including poly(glycine–alanine) (GA), poly(glycine–proline) (GP), poly(glycine– arginine) (GR), poly(proline–alanine) (PA), and poly(proline–arginine) (PR)—via repeat-associated non-ATG-mediated (or RAN) translation (88, 89). As predicted, all DPR proteins can be detected in the neurons of the cerebral cortex, cerebellum, hippocampus, thalamus, and several other brain regions in patients with C9orf72 mutations, in neurons derived from C9orf72 iPSCs, and in C9orf72 BAC transgenic mouse models. These DPR proteins are reported in neurons, but not in glia, and are most frequently located in SQSTM1-positive structures immediately adjacent to the nucleus.
Several animal models have been developed to characterize the potential impacts of C9orf72 DPR proteins. In Drosophila, the expression of arginine-containing GR and PR causes robust degenerative phenotypes in the eyes, whereas the expression of GA and PA shows no effects (90). Although DPR proteins can also be detected in C9orf72 BAC transgenic mice, the neurodegenerative phenotypes in these models have been variable, perhaps due to the low expression levels (79–82). Using the adeno-associated virus (AAV2)–based approach, Petrucelli and colleagues (91) showed that intraventricular delivery of AAV2 (GGGGCC)66 produces a robust expression of RNA foci and DPR proteins—including GR, GP, and GA—in neurons in the cerebral cortex, hippocampus, cerebellum, and spinal cord. In addition, ∼7–8% of DPR-containing neurons show phosphorylated TDP-43-positive inclusions. Mice injected with AAV2 (GGGGCC)66 show evidence of neuronal loss in the cerebral cortex and exhibit behavioral phenotypes, including hyperactivity and motor coordination deficits.
4.1.2. Dipeptide repeat proteins and RNA binding proteins.
Since the discovery of DPR proteins associated with C9orf72 mutations, there have been many reports validating the presence of these proteins in postmortem brain tissue from these patients. The ongoing debates, however, are whether the distribution of DPR proteins correlates with TDP-43 proteinopathy in the same patient; why there is a paucity of DPR proteins in the spinal motor neurons of patients with C9orf72 mutations, even in those who present with ALS; and how DPR proteins could interfere with cellular functions and ultimately contribute to neurodegeneration. While it might take more time to resolve all of these issues, several recent studies provide critical insights into these questions. In a study that investigates the clinical, neuroimaging, and neuropathological features of two FTD patients with C9orf72 mutations that illuminate these issues, Seeley and colleagues (92) show that RNA foci and DPR proteins are present before symptom onset and these features can be detected before TDP-43 proteinopathy is fully developed and also in the absence of it. These results suggest that C9orf72-associated molecular changes, including the development of RNA foci and DPR proteins, may impact brain function and structure before symptom onset and also impact the formation of TDP-43 protein aggregates.
Indeed, several studies have uncovered surprising properties of DPR proteins and their potential interactions with RNA binding proteins, lending critical mechanistic insights into these interesting pathological observations in C9orf72-mutation-associated FTLD. The first evidence comes from a study showing that the DPR proteins GRn and PRn can interact with the hnRNPA1 hydrogel in manners similar to serine-arginine-rich pre-mRNA splicing factors (93). Remarkably, the DPR proteins GRn and PRn can be taken up by cells and translocated to the nucleus where they interact with nucleoli and interfere with pre-mRNA splicing, including the splicing of EAAT2 (excitatory amino acid transporter 2), which has been previously reported to be downregulated in ALS disease models. Two independent studies reveal the unique properties of GRn and PRn in interacting with the LC domains of many of the RNA binding proteins implicated in LLPS using assays that involve either hydrogels or liquid droplets. The DPR interactomes include proteins involved in the organization of the nuclear pore complex, nucleolus, nuclear speckles, and stress granules (94, 95). The presence of DPR proteins increases the propensity to form filamentous aggregates and interferes with the assembly of stress granules and protein translation. Taken together, these results support the idea that DPR proteins with GRn or PRn repeats possess toxic gain-of-function properties that can have negative impacts on several fundamental cellular functions, including the assembly of the intranuclear structure, nuclear–cytoplasmic transport machinery, and stress granule formation (Figure 4).
Figure 4.
Intracellular pathways targeted by disease pathogenesis in FTLD. The most common genetic mutation in FTLD is caused by (GGGGCC)n hexanucleotide repeat expansion in the C9orf72 gene. C9orf72 mutations are known to generate RNA foci, which reside primarily in the nucleus. In addition, C9orf72 mutations use RAN-mediated translation to generate DPRs that disrupt the nuclear pore complex and interfere with liquid droplet formation. Finally, the protein product of C9orf72 has been detected in complex with WDR41 and SMCR8 as regulating the initiation of autophagy and the formation of autophagosomes. Several other FTLD genes, including SQSTM1, UBQLN2, OPTN, and VCP, have also been implicated in autophagy and the formation of autophagosomes. GRN, another major FTLD gene, encodes progranulin, a glycoprotein that can be either secreted and endocytosed or directly transported to the endolysosomal pathway where it regulates the formation, turnover, and possibly the exophagy of lysosomes. Abbreviations: DPR, dipeptide repeat; FTLD, frontotemporal lobar degeneration; MVB, multivesicular body; RAN, repeat-associated non-ATG.
4.1.3. Cell biology of the C9orf72 protein.
The position of (GGGGCC)n repeats in the 5 regulatory region (between noncoding exons 1a and 1b) of the C9orf72 gene raises the possibility that the insertion of these repeats may affect the expression of C9orf72 mRNA and protein levels. In support of this idea, there is evidence of hypermethylation in the CpG islands 5 to the repeats and within the repeats (96–98), which correlates with the repeat size and age of onset for disease. Although it remains unclear how hypermethylation could impact C9orf72 protein levels, results from several studies indicate that C9orf72 expression is reduced in brain tissues and lymphoblasts from patients with C9orf72 mutations and that C9orf72 proteins may have important functions in regulating vesicular trafficking in the endolysosomal and autophagy pathways. For instance, bioinformatics predicts that the C9orf72 protein contains an amino acid sequence that is homologous to the DENN (differentially expressed in normal and neoplastic cells) domain, which is highly conserved among members of the Rab GEF (guanine nucleotide exchange factor) family (99). The mechanistic insights into C9orf72 proteins come from several studies, which independently characterize C9orf72 interactomes and show that C9orf72 can indeed form a stable multiprotein complex with proteins that are implicated in autophagy and lysosomal pathways, including SMCR8 (Smith–Magenis syndrome chromosome region, candidate 8), WDR41 (WD repeat domain 41), and ATG101 (autophagy related 101) (100–103). Several models have been proposed for C9orf72 functions. First, the C9orf72–WDR41–SMCR8 complex may serve as a GEF that regulates the activity of the small GTPases Rab8a and Rab39b, which are involved in membrane trafficking and the initiation of autophagy (Figure 4). Second, C9orf72 can be detected in the lysosomes where it regulates the activity of the mTOR (mechanistic target of rapamycin) complex and lysosomal morphology. Although the exact role of C9orf72 requires further investigation, it is clear that the loss of C9orf72 enhances autophagic flux and leads to prominent lysosomal defects.
4.1.4. C9orf72 loss of function and neuroinflammation.
One surprising role of C9orf72 comes from the analysis of mice that have homozygous deletion of C9orf72 (C9orf72−/−). While C9orf72−/− mice initially showed no evidence of neurodegeneration, these mutant mice developed age-dependent lymphadenopathy and splenomegaly due to macrophage infiltration and the mis-regulation of genes in the inflammation-lysosome-phagosome formation-, ribosome-, and amino acid–metabolism pathways (103–105). Consistent with these results, abnormal lysosomes are reported in C9orf72-deficient macrophages and microglia, although the functional consequences for the latter cell type remain to be determined. In at least one study, C9orf72−/− mice exhibited features of severe autoimmune dysfunction, leading to early mortality (104). Unlike in rodents, knocking down C9orf72 in zebrafish leads to axonal and motor neuron pathologies that synergize with Ataxin-2 that has intermediate polyQ repeats (101, 106). Collectively, these studies uncover the critical roles of C9orf72 proteins in regulating membrane trafficking in the autophagy and lysosomal pathways, which can profoundly impact the physiological functions of neurons and immune cells. It is conceivable that these membrane trafficking defects caused by C9orf72 deficiency may synergize with RNA foci and DPR proteins to promote pathology in patients with C9orf72 mutations.
4.2. GRN Mutations
Although they were initially isolated as growth factors that regulate tumorigenesis and angiogenesis, recently identified dominant mutations in the GRN gene of familial FTD patients herald many new directions to understanding the in vivo function of progranulin during the aging process in the brain. Many new discoveries challenge the conventional view of progranulin as a secreted growth factor. Instead, these results uncover previously unrecognized roles of progranulin in intracellular vesicular trafficking, lysosomal biogenesis, and neuroinflammation.
4.2.1. Cell biology of progranulin.
As described in the previous sections, dominant mutations in the human GRN gene constitute a major genetic underpinning of FTD (28, 29). Pathologically, GRN mutations invariably lead to aberrant TDP-43 protein aggregates that have a characteristic distribution in the superficial layers of the cerebral cortex (FTLD-TDP type A) (Figure 2) (19, 107). This tight association between GRN mutations and TDP-43 proteinopathy raises the intriguing hypothesis that progranulin (PGRN) deficiency can impact fundamental cell biological processes that contribute to the aberrant formation of TDP-43 aggregates. Consistent with this idea, several lines of evidence indicate that PGRN regulates vesicular trafficking in the endolysosomal pathway and the biogenesis of autophagosomes. First, in addition to its role as a secreted factor that promotes vasculogenesis, tumorigenesis, and neurite outgrowth, several studies indicate that secreted PGRN can interact with sortilin and undergo endocytosis from endosomes to lysosomes, where it may be cleaved or degraded to maintain a balanced level of PGRN and granulins (108–110). Another alternative, and perhaps more efficient and plausible, route for PGRN to regulate the endolysosomal pathway is that following its synthesis in the Golgi apparatus, PGRN may be directly trafficked to the endosomes and then ultimately reach the lysosomes (Figure 4) (17). In support of this model, a proteomic screen identified prosaposin (PSAP) as a key component in the PGRN interactome (111). In addition, a GWAS identified the human PSAP gene as a genetic locus that regulates PGRN plasma levels (112). Like PGRN, PSAP can undergo proteolytic cleavage to release cysteine-rich peptides called saposins, which promote sphingolipid hydrolysis and lysosomal functions (113–115). PSAP forms a heterodimer with PGRN and regulates both intracellular and secreted PGRN levels. In addition, PSAP interacts with M6PR (mannose 6-phosphate receptor) or LRP1 (low-density lipoprotein receptor–related protein 1) and facilitates the trafficking of PGRN to the lysosomes (Figure 4). The cooperative role of PGRN and PSAP in lysosomal targeting supports the notion that both have critical roles in regulating lysosomal functions.
While the exact role of PGRN in lysosomal functions remains unclear, there is strong evidence that PGRN deficiency causes severe lysosomal defects in a dose-dependent manner. Individuals with two GRN mutant alleles completely lack PGRN and develop a form of lysosomal storage disease known as neuronal ceroid lipofuscinosis (NCL, or ceroid neuronal lipofuscinosis 11) (116). These patients have no detectable PGRN in their plasma and between the ages of 20 and 30 years develop neurological deficits, such as vision loss, myoclonus seizure, and cerebellar ataxia (117, 118). Skin biopsy samples from these patients reveal lipofuscin accumulation, supporting the diagnosis of NCL (116). Remarkably, several studies show that cortical neurons and lymphoblasts from FTLD patients with GRN mutations also contain a marked increase in lysosomal storage materials (119, 120). Consistent with the human pathology, Grn knockout (Grn−/−) mice show an age-dependent accumulation of lipofuscin in neurons and glia (120–125), which upon ultrastructural analyses contains multilamellar fingerprint profiles or onionskin-like storage materials that are diagnostic for NCL (116). Remarkably, confocal scanning laser ophthalmoscopy reveals that similar lipofuscin accumulation phenotypes can be detected in the retina of Grn−/− mice and in both presymptomatic and symptomatic GRN mutation carriers (120).
Together, these results lead to several important conclusions regarding the functions of PGRN. First, PGRN has dose-dependent effects in regulating endolysosomal pathways. This is supported by the robust NCL phenotype in Grn−/− mice, homozygous GRN patients, and FTLD patients with GRN mutations, but not in Grn+/− mice (126). These results support the idea that a critical threshold of intracellular PGRN is required to maintain a homeostatic balance in the vesicular trafficking from late endosomes to lysosomes and the subsequent degradation or clearance of lysosomal structures. Second, although the exact mechanism of PGRN in the biogenesis and function of lysosomes remains to be determined, given the interaction between PGRN and PSAP, it is tempting to speculate that PGRN and its granulin peptides may cooperatively control sphingolipid hydrolysis and lysosomal functions. In support of this idea, lipidomic mass spectrometry analyses in mouse embryonic fibroblasts and lysosome-enriched organelles from liver show that the loss of PGRN results in the upregulation of polyunsaturated triacylglycerides and a concomitant reduction in diacylglycerides and phosphatidylserines (127). Finally, the NCL phenotype is a robust disease manifestation of the PGRN deficiency state. The fact that the drusen-like lipofuscin deposits can be detected in the retinas of asymptomatic GRN mutation carriers presents a highly efficient, noninvasive approach for detecting disease pathology and monitoring clinical progression.
4.2.2. Lysosomes and TDP-43 proteinopathy.
Although it remains unclear how lysosomal defects caused by PGRN deficiency lead to aberrant formation of TDP-43 protein aggregates, several lines of evidence suggest that defects in lysosomes and their affiliated autophagy pathway can contribute to the formation of TDP-43 protein aggregates. As a major site of macromolecular breakdown and recycling, lysosomes and the tightly associated autophagy machinery are critical for cellular control of proteostasis and the response to nutrient deprivation. In addition, dysfunction in autophagy and lysosomes has been implicated as a key player in the pathogenesis of neurodegenerative diseases (128). For instance, mutations in the Gaucher’s disease gene GBA, which encodes β-glucocerebrosidase, cause juvenile onset lysosomal storage disease and can increase the risk of Parkinson’s disease (129, 130). At least two NCL-associated genes, CTSD (cathepsin D) and ATP13A2, have been linked to, respectively, AD and Parkinson’s disease (131, 132). Although the exact mechanism remains to be determined by which lysosomal defects caused by PGRN deficiency can result in the aberrant formation of TDP-43 protein aggregates, several studies show that TDP-43 has an increased propensity to accumulate in the cytoplasm of cortical neurons in aged Grn−/− mice and in mice that have two copies of a common GRN R493X (1477C→T) mutation (GrnR493X) (119, 123, 133). Intriguingly, direct comparisons between Grn−/− and Ctsd−/− mouse brains showed the prominent accumulation of a number of lysosomal proteins and increased phosphorylation of TDP-43 (serine 409/410), a key signature of TDP-43 proteinopathy in FTLD (119). In addition, the loss of PGRN leads to the upregulation of several autophagy-related proteins. Since PGRN has not been directly implicated in autophagy, these results suggest that the lysosomal defects in PGRN-deficient cells may indirectly promote the activation of the autophagy pathway, which in turn interferes with the function of the autophagosome in the clearance of TDP-43 in the cytoplasm and ultimately results in the formation of TDP-43 protein aggregates (Figure 4). Alternatively, the persistent accumulation of lysosomal abnormalities in PGRN-deficient cells may impact the unconventional secretory pathway regulated by autophagy and lysosomes (134). However, it remains to be determined how these defects might have differential effects in neurons and glia.
Another lysosome-based mechanism that may potentially promote TDP-43 proteinopathy is that certain cleaved granulins may perturb lysosomal functions and thereby interfere with the degradation of TDP-43. Using the nematode C. elegans as a model system, a recent study shows that when granulins are expressed in the absence of full-length PGRN, they can increase TDP-43 levels in neurons and exacerbate its toxicity (110). These results suggest that, as with PSAP and saposins, a delicate balance of PGRN and granulins in lysosomes is crucial to regulate the biogenesis and function of these organelles, which are important in controlling proteostasis. Both the complete lack of PGRN and aberrant or excessive levels of granulins may negatively modulate lysosomal functions. Finally, perhaps in an effort to overcome abnormal lysosomal function, PGRN-deficient cells show activation of mTOR and TFEB (transcription factor EB), a master regulator of lysosomal genes and biogenesis (135–137). The persistent activation of the mTOR and TFEB pathways is likely to aggravate lysosomal defects in PGRN-deficient cells.
4.2.3. Neuroinflammation and TDP-43 proteinopathy.
While the lysosomal defects in PGRN deficiency can be detected in a wide range of cell types—from lymphoblasts to fibroblasts, neurons, and glia—the functional consequences caused by these defects vary in different cell types. Indeed, one prominent feature in Grn−/− mice is the age-dependent defects in the brain’s innate immune system. Several studies show that the loss of PGRN causes increased phagocytosis and elevated production of proinflammatory cytokines in microglia and macrophages (122, 123, 138). In young Grn−/− mice and microglia-specific Grn conditional mutant mice (Cd11b-Cre;Grnfl/fl), acute exposure to the neurotoxin MPTP (1-methyl-4-phenyl-1,2,3,6-tetrahydropyridine) induces a robust microglial response that promotes the engulfment of injured neurons (122). To investigate the role of microglial activation in the context of neurodegeneration, a recent study carried out transcriptomic analyses of several brain regions from a large cohort of aging Grn+/+, Grn+/−, and Grn−/− mice with the goal of identifying age-dependent, genotype-specific changes in gene expression (139). The results showed an age-dependent upregulation of lysosomal genes and genes related to innate immunity, which upon gene ontological and systems-level analyses revealed extensive functional interactions. Similar transcriptomic signatures of neuroinflammation in Grn−/− mice have been independently validated by several groups (125, 127, 140, 141). Collectively, these results are in agreement with the prominent expression of Grn mRNA in the microglia, and the results support the idea that PGRN is required to suppress excessive microglial activation in the aging process. Indeed, cell biological characterizations show that Grn−/− microglia exhibit increased efficiency in trafficking endocytosed materials from endosomes to lysosomes. In addition, Grn−/− microglia produce abundant complement proteins, including C1qa, C1qb, C1qc, and C3, which promote synaptic pruning activity that preferentially targets the ventral thalamus, thus leading to profound dysfunctions in the thalamocortical circuit (139). Consistent with the important role of complement-mediated synaptic pruning (142, 143), the removal of C1qa mitigates the dysfunction in the thalamocortical circuit, alleviates aberrant behavioral phenotypes, and prolongs survival in Grn−/− ;C1qa−/− mice. Similar to the profound neuroinflammation phenotype found in Grn−/− mice, postmortem examinations of the frontal cortex in FTLD patients with the GRN mutation also showed extensive microglial activation. Furthermore, cerebrospinal fluid samples from these patients showed significantly increased levels of C1qa and C3 that positively correlate with progressive cognitive decline.
The identification of profound microglial activation in Grn−/− mice and FTLD patients with GRN mutations raises a number of intriguing possibilities about how glial pathology contributes to TDP-43 proteinopathy and neurodegeneration. For example, microglial and astroglial activation are known to be tightly coupled during neuroinflammatory responses. Thus, it is possible that PGRN-deficient microglia could recruit and activate Grn−/− astrocytes to promote neuroinflammation via mechanisms similar to the lipopolysaccharide model (141). Alternatively, the loss of PGRN in Grn−/− astrocytes may lead to spontaneous aberrant activation independently of Grn−/− microglia. Regardless of the mechanism, it is likely that both Grn−/− microglia and astrocytes may have synergistic effects in promoting the neuronal stress that facilitates the accumulation of TDP-43 in intracellular organelles, such as stress granules and lysosomes, and ultimately leads to cytoplasmic aggregation of TDP-43 (Figure 4). Given the remarkably similar lysosomal defects seen in microglia and macrophages lacking PGRN or the gene product of C9orf72 (105), it is tempting to speculate that aberrant glial pathology may be a shared disease-driving factor in FTLD caused by GRN or C9orf72 mutations.
4.3. Autophagy Defects and TDP-43 Proteinopathy
There is abundant evidence that defects in the autophagy pathway play a central part in ALS-FTD pathogenesis. This is underscored by the variety of autophagy-related genes, including SQSTM1, UBQLN2, OPTN, and VCP. Autophagy is a catabolic process by which the cell degrades proteins, macromolecules, and organelles, either because they are damaged or unneeded or because their degradation produces resources that can be used for vital cellular functions during starvation (144). To ensure that only appropriate proteins are degraded, E3 ubiquitin ligases select targets with specific features, such as exposed hydrophobic domains, that indicate improper folding. The human proteome includes hundreds of distinct E3 ubiquitin ligases, which individually are somewhat specific, but together can target a broad range of undesirable molecules. E3 ubiquitin ligases transfer a molecule of ubiquitin from an E2 enzyme to a lysine residue on its specific target. As ubiquitin itself can be ubiquitinated on one of several lysine residues, ubiquitin molecules can be chained together. The polyubiquitin linkage pattern is used to direct the target protein for either proteasomal degradation (often through K48 linkages) or lysosomal degradation via autophagy (often through K63 linkages) (145).
Autophagy begins with the formation of a double-membrane vesicle called a phagophore, which elongates and ultimately wraps around and isolates the portion of the cytoplasm that contains the molecules and organelles that have been selected for degradation (Figure 4). The phagophore will mature into an autophagosome, which will fuse with a lysosome to initiate the degradation of its contents. As the phagophore forms, the soluble protein LC3-I (microtubule-associated protein 1 light chain 3) is conjugated to phosphatidylethanolamine to form LC3-II, which associates with the phagophore membrane. SQSTM1, UBQLN2, and OPTN serve as adaptors (or autophagy receptors), which bind to polyubiquitinated proteins and LC3-II, thus collecting autophagy targets within the autophagosome. VCP is involved in autophagosome maturation and fusion with lysosomes (146). Mutations in any of the genes encoding these proteins can cause ALS and FTD, suggesting that autophagy plays a central part in the pathogenesis of these diseases.
Mutations in OPTN that cause ALS-FTD include a deletion of exons 13–15 and missense, non-sense, and splicing defect–inducing intronic mutations. These mutations result in reduced mRNA and protein expression of OPTN (36). OPTN mutations result in TDP-43- and ubiquitin-positive inclusions. One ALS-causing mutation, E478G, lies in the ubiquitin binding domain, suggesting that the inability of OPTN to recruit polyubiquitinated proteins to the autophagosome gives rise to ALS (147). Consistent with this observation, wild-type OPTN colocalizes with TDP-43 aggregates in a TARDBP mutation model of ALS, whereas OPTN E478G has reduced colocalization. Furthermore, the overexpression of wild-type OPTN reduces TDP-43 aggregates in this model, and transgenic expression of mutant OPTN with an impaired ubiquitin binding domain prevents clearance of aggregates in a dominant negative fashion and reduces overall autophagic flux (148).
UBQLN2 mutant ALS-FTD exhibits striking parallels to OPTN mutant disease. UBQLN2 also can bind to TDP-43 at its CTD and enhances its clearance (149). ALS caused by UBQLN2 mutation results in inclusions positive for UBQLN2, SQSTM1, TDP-43, FUS, and OPTN. Furthermore, transgenic expression of ALS-associated mutant UBQLN2 leads to the aggregation of TDP-43 (150), and in rats, expressing mutant UBQLN2 leads to the formation of aggregates that contain both mutant and wild-type UBQLN2, suggesting a dominant negative mechanism (151).
SQSTM1 is found in toxic aggregates in a variety of neurodegenerative diseases, including AD, and Parkinson’s and Huntington’s diseases. A variety of mutations in SQSTM1 have been found to cause ALS-FTD, including mutations within several exons, introns, and the promoter. The mutations in coding regions are found in domains that interact with ubiquitin, LC3, or Keap1 (Kelch-like ECH associated protein 1) (152). The nonspecific nature of these mutations is most consistent with a loss-of-function mechanism of disease. Similarly, mutations causing ALS-FTD in VCP are also found indiscriminately throughout the gene, also supporting a loss-of-function mechanism. VCP mutations result in the accumulation of ubiquitin-positive vesicles and SQSTM1, which is consistent with a disruption of autophagy (146). Another more specific effect of VCP mutations is impairments in processing stress granules. VCP is involved in stress granule clearance by autophagy (153), and VCP knockdown in cell culture decreases stress granule formation and alters stress granule morphology and composition. However, autophagy that is impaired by other means has similar effects on stress granules, so it is possible that the effect of VCP mutations on stress granule processing occurs secondary to impaired autophagy (154).
The large number of autophagy-related genes in which mutations can cause both ALS and FTD suggests that autophagy dysfunction may be important in the pathogenesis of all etiologies of ALS-FTD. In one study, spinal cord tissue from 47 patients with ALS from a variety of etiologies, including familial and sporadic, was evaluated, and all samples had UBQLN2 in neuronal inclusions (155). A major role for autophagy in disease pathogenesis is also consistent with the ubiquity of protein aggregates found in all cases of ALS and FTD.
5. CONCLUDING REMARKS
Recent advances in human genetics, neuroimaging, and neuropathology have dramatically enhanced our ability to understand the pathogenesis of FTLD. In parallel, there are mounting biophysical, biochemical, and cell biology results supporting the model that RNA binding proteins undergo LLPS to facilitate the assembly and disassembly of many membraneless organelles. These results support the critical role of these organelles in RNA transcription, translation, transport, localization, and degradation, and they underscore the importance of maintaining homeostasis in these membraneless organelles in neurons and glia. Furthermore, many FTLD genes have critical roles in intracellular pathways, including endolysosomal trafficking, the formation of autophagosomes, nuclear–cytoplasmic transport, and protein quality control process. Perturbations to these pathways disrupt the homeostasis of membraneless organelles and lead to the progressive accumulation of proteins.
Looking ahead, there are many crucial unanswered questions that need to be addressed before we will fully understand the disease mechanisms. First, does the LLPS property of RNA binding proteins offer broader implications for the pathogenesis of FTLD? Interestingly, one recent study shows that another FTLD-related protein, tau, similar to RNA binding proteins, also is intrinsically disordered and can interact with transfer RNA to form liquid droplets (156). This exciting discovery suggests a unifying theory in which the intrinsically disordered LC domain may be a common property in human proteomes. Perturbations to the properties of these proteins can promote the initiation and progression of abnormal protein aggregate formation. Second, what are the impacts of FTLD genes in neuron–glia interactions? Microglia, astroglia, and oligodendroglia have critical functions in maintaining homeostasis in the nervous system. As discussed, there is compelling evidence that loss of function in the FTLD genes C9orf72 and GRN has profound impacts on microglial functions. It will be important to further elucidate the underlying mechanisms of microglial dysfunction and determine whether similar aberrant activation can be detected in other glial cell types. Finally, given the increased prevalence of autoimmune disorders in FTLD patients, it will be important to determine whether mutations in FTLD genes may lead to broader dysfunction in the innate and adaptive immune systems.
ACKNOWLEDGMENTS
We thank our colleagues at the Memory and Aging Center at UCSF for many stimulating discussions and the center’s patients and caregivers for their invaluable contributions to FTD and ALS research. This work has been supported by grants from the US National Institutes of Health (grants NS098516, AG057462, and AA027074 to E.J.H., and AG019724 and AG023501 to W.W.S.), the Departments of Veterans Affairs (grants BX001108 and BX002978 to E.J.H.), and the Tau Consortium and Muscular Dystrophy Association (to W.W.S.). E.J.H. and W.W.S. are investigators of the Bluefield Project to Cure Frontotemporal Dementia.
Footnotes
DISCLOSURE STATEMENT
The authors are not aware of any affiliations, memberships, funding, or financial holdings that might be perceived as affecting the objectivity of this review.
LITERATURE CITED
- 1.Miller BL. 2013. Frontotemporal Dementia New York: Oxford Univ. Press [Google Scholar]
- 2.Ratnavalli E, Brayne C, Dawson K, Hodges JR. 2002. The prevalence of frontotemporal dementia. Neurology 58:1615–21 [DOI] [PubMed] [Google Scholar]
- 3.Pick A 1892. Uber die Beziehungen der senilen Hirnatrophie zur Aphasie. Prag. Med. Wochenschr 17:165–67 [Google Scholar]
- 4.Pick A 1904. Zur Symptomatologie der linksseitigen Schläfenlappenatrophie. Monatsschrift Psychiatr. Neurol 16:378–88 [Google Scholar]
- 5.Alzheimer A 1911. Uber eigenartige Krankheitsfälle des späteren Alters. Z. Gesamte Neurol. Psychiatr 4:356–85 [Google Scholar]
- 6.Rascovsky K, Hodges JR, Knopman D, Mendez MF, Kramer JH, et al. 2011. Sensitivity of revised diagnostic criteria for the behavioural variant of frontotemporal dementia. Brain 134:2456–77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gorno-Tempini ML, Hillis AE, Weintraub S, Kertesz A, Mendez M, et al. 2011. Classification of primary progressive aphasia and its variants. Neurology 76:1006–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Boeve BF, Lang AE, Litvan I. 2003. Corticobasal degeneration and its relationship to progressive supranuclear palsy and frontotemporal dementia. Ann. Neurol 54(Suppl. 5):S15–19 [DOI] [PubMed] [Google Scholar]
- 9.Litvan I, Mangone CA, McKee A, Verny M, Parsa A, et al. 1996. Natural history of progressive supranuclear palsy (Steele–Richardson–Olszewski syndrome) and clinical predictors of survival: a clinicopathological study. J. Neurol. Neurosurg. Psychiatry 60:615–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Seeley WW. 2017. Mapping neurodegenerative disease onset and progression. Cold Spring Harb. Perspect. Biol 9:a023622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Seeley WW, Crawford RK, Zhou J, Miller BL, Greicius MD. 2009. Neurodegenerative diseases target large-scale human brain networks. Neuron 62:42–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Raj A, Kuceyeski A, Weiner M. 2012. A network diffusion model of disease progression in dementia. Neuron 73:1204–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhou J, Gennatas ED, Kramer JH, Miller BL, Seeley WW. 2012. Predicting regional neurodegeneration from the healthy brain functional connectome. Neuron 73:1216–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Miller ZA, Rankin KP, Graff-Radford NR, Takada LT, Sturm VE, et al. 2013. TDP-43 frontotemporal lobar degeneration and autoimmune disease. J. Neurol. Neurosurg. Psychiatry 84:956–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Miller ZA, Sturm VE, Camsari GB, Karydas A, Yokoyama JS, et al. 2016. Increased prevalence of autoimmune disease within C9 and FTD/MND cohorts: completing the picture. Neurol. Neuroimmunol. Neuroinflammation 3:e301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Broce I, Karch CM, Wen N, Fan CC, Wang Y, et al. 2018. Immune-related genetic enrichment in frontotemporal dementia: an analysis of genome-wide association studies. PLOS Med 15:e1002487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kao AW, McKay A, Singh PP, Brunet A, Huang EJ. 2017. Progranulin, lysosomal regulation and neurodegenerative disease. Nat. Rev. Neurosci 18:325–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lall D, Baloh RH. 2017. Microglia and C9orf72 in neuroinflammation and ALS and frontotemporal dementia. J. Clin. Investig 127:3250–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mackenzie IR, Neumann M, Baborie A, Sampathu DM, Du Plessis D, et al. 2011. A harmonized classification system for FTLD-TDP pathology. Acta Neuropathol 122:111–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Forman MS, Farmer J, Johnson JK, Clark CM, Arnold SE, et al. 2006. Frontotemporal dementia: clinicopathological correlations. Ann. Neurol 59:952–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McKee AC, Cairns NJ, Dickson DW, Folkerth RD, Keene CD, et al. 2016. The first NINDS/NIBIB consensus meeting to define neuropathological criteria for the diagnosis of chronic traumatic encephalopathy. Acta Neuropathol 131:75–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rodriguez RD, Suemoto CK, Molina M, Nascimento CF, Leite RE, et al. 2016. Argyrophilic grain disease: demographics, clinical, and neuropathological features from a large autopsy study. J. Neuropathol. Exp. Neurol 75:628–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ahmed Z, Bigio EH, Budka H, Dickson DW, Ferrer I, et al. 2013. Globular glial tauopathies (GGT): consensus recommendations. Acta Neuropathol 126:537–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Arai T, Hasegawa M, Akiyama H, Ikeda K, Nonaka T, et al. 2006. TDP-43 is a component of ubiquitin-positive tau-negative inclusions in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Biochem. Biophys. Res. Commun 351:602–11 [DOI] [PubMed] [Google Scholar]
- 25.Neumann M, Sampathu DM, Kwong LK, Truax AC, Micsenyi MC, et al. 2006. Ubiquitinated TDP-43 in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Science 314:130–33 [DOI] [PubMed] [Google Scholar]
- 26.Nag S, Yu L, Capuano AW, Wilson RS, Leurgans SE, et al. 2015. Hippocampal sclerosis and TDP-43 pathology in aging and Alzheimer disease. Ann. Neurol 77:942–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nelson PT, Trojanowski JQ, Abner EL, Al-Janabi OM, Jicha GA, et al. 2016. “New old pathologies”: AD, PART, and cerebral age-related TDP-43 with sclerosis (CARTS). J. Neuropathol. Exp. Neurol 75:482–98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Baker M, Mackenzie IR, Pickering-Brown SM, Gass J, Rademakers R, et al. 2006. Mutations in progranulin cause tau-negative frontotemporal dementia linked to chromosome 17. Nature 442:916–19 [DOI] [PubMed] [Google Scholar]
- 29.Cruts M, Gijselinck I, van der Zee J, Engelborghs S, Wils H, et al. 2006. Null mutations in progranulin cause ubiquitin-positive frontotemporal dementia linked to chromosome 17q21. Nature 442:920–24 [DOI] [PubMed] [Google Scholar]
- 30.DeJesus-Hernandez M, Mackenzie IR, Boeve BF, Boxer AL, Baker M, et al. 2011. Expanded GGGGCC hexanucleotide repeat in noncoding region of C9ORF72 causes chromosome 9p-linked FTD and ALS. Neuron 72:245–56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Renton AE, Majounie E, Waite A, Simon-Sanchez J, Rollinson S, et al. 2011. A hexanucleotide repeat expansion in C9ORF72 is the cause of chromosome 9p21-linked ALS–FTD. Neuron 72:257–68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hirsch-Reinshagen V, Pottier C, Nicholson AM, Baker M, Hsiung GR, et al. 2017. Clinical and neuropathological features of ALS/FTD with TIA1 mutations. Acta Neuropathol. Commun 5:96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mackenzie IR, Nicholson AM, Sarkar M, Messing J, Purice MD, et al. 2017. TIA1 mutations in amyotrophic lateral sclerosis and frontotemporal dementia promote phase separation and alter stress granule dynamics. Neuron 95:808–16.e9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Van Mossevelde S, van der Zee J, Gijselinck I, Engelborghs S, Sieben A, et al. 2016. Clinical features of TBK1 carriers compared with C9orf72, GRN and non-mutation carriers in a Belgian cohort. Brain 139:452–67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Koriath CA, Bocchetta M, Brotherhood E, Woollacott IO, Norsworthy P, et al. 2017. The clinical, neuroanatomical, and neuropathologic phenotype of TBK1-associated frontotemporal dementia: a longitudinal case report. Alzheimer’s Dement 6:75–81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pottier C, Bieniek KF, Finch N, van de Vorst M, Baker M, et al. 2015. Whole-genome sequencing reveals important role for TBK1 and OPTN mutations in frontotemporal lobar degeneration without motor neuron disease. Acta Neuropathol 130:77–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Guinto JB, Ritson GP, Taylor JP, Forman MS. 2007. Valosin-containing protein and the pathogenesis of frontotemporal dementia associated with inclusion body myopathy. Acta Neuropathol 114:55–61 [DOI] [PubMed] [Google Scholar]
- 38.Kwiatkowski TJ Jr., Bosco DA, Leclerc AL, Tamrazian E, Vanderburg CR, et al. 2009. Mutations in the FUS/TLS gene on chromosome 16 cause familial amyotrophic lateral sclerosis. Science 323:1205–8 [DOI] [PubMed] [Google Scholar]
- 39.Vance C, Rogelj B, Hortobagyi T, De Vos KJ, Nishimura AL, et al. 2009. Mutations in FUS, an RNA processing protein, cause familial amyotrophic lateral sclerosis type 6. Science 323:1208–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mackenzie IRA, Neumann M. 2017. Fused in sarcoma neuropathology in neurodegenerative disease. Cold Spring Harb. Perspect. Med 7:a024299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shang Y, Huang EJ. 2016. Mechanisms of FUS mutations in familial amyotrophic lateral sclerosis. Brain Res 1647:65–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Huang EJ, Zhang J, Geser F, Trojanowski JQ, Strober JB, et al. 2010. Extensive FUS-immunoreactive pathology in juvenile amyotrophic lateral sclerosis with basophilic inclusions. Brain Pathol 20:1069–76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Perry DC, Brown JA, Possin KL, Datta S, Trujillo A, et al. 2017. Clinicopathological correlations in behavioural variant frontotemporal dementia. Brain 140:3329–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pottier C, Ravenscroft TA, Sanchez-Contreras M, Rademakers R. 2016. Genetics of FTLD: overview and what else we can expect from genetic studies. J. Neurochem 138(Suppl. 1):32–53 [DOI] [PubMed] [Google Scholar]
- 45.Polymenidou M, Lagier-Tourenne C, Hutt KR, Huelga SC, Moran J, et al. 2011. Long pre-mRNA depletion and RNA missplicing contribute to neuronal vulnerability from loss of TDP-43. Nat. Neurosci 14:459–68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tollervey JR, Curk T, Rogelj B, Briese M, Cereda M, et al. 2011. Characterizing the RNA targets and position-dependent splicing regulation by TDP-43. Nat. Neurosci 14:452–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chou CC, Zhang Y, Umoh ME, Vaughan SW, Lorenzini I, et al. 2018. TDP-43 pathology disrupts nuclear pore complexes and nucleocytoplasmic transport in ALS/FTD. Nat. Neurosci 21:228–39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ling SC, Polymenidou M, Cleveland DW. 2013. Converging mechanisms in ALS and FTD: disrupted RNA and protein homeostasis. Neuron 79:416–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kato M, McKnight SL. 2017. A solid-state conceptualization of information transfer from gene to message to protein. Annu. Rev. Biochem 87:351–90 [DOI] [PubMed] [Google Scholar]
- 50.Schneider JW, Gao Z, Li S, Farooqi M, Tang TS, et al. 2008. Small-molecule activation of neuronal cell fate. Nat. Chem. Biol 4:408–10 [DOI] [PubMed] [Google Scholar]
- 51.Han TW, Kato M, Xie S, Wu LC, Mirzaei H, et al. 2012. Cell-free formation of RNA granules: bound RNAs identify features and components of cellular assemblies. Cell 149:768–79 [DOI] [PubMed] [Google Scholar]
- 52.Kato M, Han TW, Xie S, Shi K, Du X, et al. 2012. Cell-free formation of RNA granules: Low complexity sequence domains form dynamic fibers within hydrogels. Cell 149:753–67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kwon I, Kato M, Xiang S, Wu L, Theodoropoulos P, et al. 2013. Phosphorylation-regulated binding of RNA polymerase II to fibrous polymers of low-complexity domains. Cell 155:1049–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Schwartz JC, Ebmeier CC, Podell ER, Heimiller J, Taatjes DJ, Cech TR. 2012. FUS binds the CTD of RNA polymerase II and regulates its phosphorylation at Ser2. Genes Dev 26:2690–95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Schwartz JC, Wang X, Podell ER, Cech TR. 2013. RNA seeds higher-order assembly of FUS protein. Cell Rep 5:918–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Li P, Banjade S, Cheng HC, Kim S, Chen B, et al. 2012. Phase transitions in the assembly of multivalent signalling proteins. Nature 483:336–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Molliex A, Temirov J, Lee J, Coughlin M, Kanagaraj AP, et al. 2015. Phase separation by low complexity domains promotes stress granule assembly and drives pathological fibrillization. Cell 163:123–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Patel A, Lee HO, Jawerth L, Maharana S, Jahnel M, et al. 2015. A liquid-to-solid phase transition of the ALS protein FUS accelerated by disease mutation. Cell 162:1066–77 [DOI] [PubMed] [Google Scholar]
- 59.Burke KA, Janke AM, Rhine CL, Fawzi NL. 2015. Residue-by-residue view of in vitro FUS granules that bind the C-terminal domain of RNA polymerase II. Mol. Cell 60:231–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lin Y, Protter DS, Rosen MK, Parker R. 2015. Formation and maturation of phase-separated liquid droplets by RNA-binding proteins. Mol. Cell 60:208–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Alberti S, Halfmann R, King O, Kapila A, Lindquist S. 2009. A systematic survey identifies prions and illuminates sequence features of prionogenic proteins. Cell 137:146–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Murray DT, Kato M, Lin Y, Thurber KR, Hung I, et al. 2017. Structure of FUS protein fibrils and its relevance to self-assembly and phase separation of low-complexity domains. Cell 171:615–27.e16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Deng Q, Holler CJ, Taylor G, Hudson KF, Watkins W, et al. 2014. FUS is phosphorylated by DNA-PK and accumulates in the cytoplasm after DNA damage. J. Neurosci 34:7802–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Monahan Z, Ryan VH, Janke AM, Burke KA, Rhoads SN, et al. 2017. Phosphorylation of the FUS low-complexity domain disrupts phase separation, aggregation, and toxicity. EMBO J 36:2951–67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Feric M, Vaidya N, Harmon TS, Mitrea DM, Zhu L, et al. 2016. Coexisting liquid phases underlie nucleolar subcompartments. Cell 165:1686–97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Shin Y, Berry J, Pannucci N, Haataja MP, Toettcher JE, Brangwynne CP. 2017. Spatiotemporal control of intracellular phase transitions using light-activated optoDroplets. Cell 168:159–71.e14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Shin Y, Brangwynne CP. 2017. Liquid phase condensation in cell physiology and disease. Science 357:eaaf4382 [DOI] [PubMed] [Google Scholar]
- 68.Protter DS, Parker R. 2016. Principles and properties of stress granules. Trends Cell Biol 26:668–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kanai Y, Dohmae N, Hirokawa N. 2004. Kinesin transports RNA: isolation and characterization of an RNA-transporting granule. Neuron 43:513–25 [DOI] [PubMed] [Google Scholar]
- 70.Hoell JI, Larsson E, Runge S, Nusbaum JD, Duggimpudi S, et al. 2011. RNA targets of wild-type and mutant FET family proteins. Nat. Struct. Mol. Biol 18:1428–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ishigaki S, Masuda A, Fujioka Y, Iguchi Y, Katsuno M, et al. 2012. Position-dependent FUS–RNA interactions regulate alternative splicing events and transcriptions. Sci. Rep 2:529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Takahama K, Takada A, Tada S, Shimizu M, Sayama K, et al. 2013. Regulation of telomere length by G-quadruplex telomere DNA- and TERRA-binding protein TLS/FUS. Chem. Biol 20:341–50 [DOI] [PubMed] [Google Scholar]
- 73.Murakami T, Qamar S, Lin JQ, Schierle GS, Rees E, et al. 2015. ALS/FTD Mutation-induced phase transition of FUS liquid droplets and reversible hydrogels into irreversible hydrogels impairs RNP granule function. Neuron 88:678–90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kim HJ, Kim NC, Wang YD, Scarborough EA, Moore J, et al. 2013. Mutations in prion-like domains in hnRNPA2B1 and hnRNPA1 cause multisystem proteinopathy and ALS. Nature 495:467–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Todd PK, Paulson HL. 2010. RNA-mediated neurodegeneration in repeat expansion disorders. Ann. Neurol 67:291–300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Donnelly CJ, Zhang PW, Pham JT, Haeusler AR, Mistry NA, et al. 2013. RNA toxicity from the ALS/FTD C9ORF72 expansion is mitigated by antisense intervention. Neuron 80:415–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Sareen D, O’Rourke JG, Meera P, Muhammad AK, Grant S, et al. 2013. Targeting RNA foci in iPSC-derived motor neurons from ALS patients with a C9ORF72 repeat expansion. Sci. Transl. Med 5:208ra149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Almeida S, Gascon E, Tran H, Chou HJ, Gendron TF, et al. 2013. Modeling key pathological features of frontotemporal dementia with C9ORF72 repeat expansion in iPSC-derived human neurons. Acta Neuropathol 126:385–99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.O’Rourke JG, Bogdanik L, Muhammad A, Gendron TF, Kim KJ, et al. 2015. C9orf72 BAC transgenic mice display typical pathologic features of ALS/FTD. Neuron 88:892–901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Peters OM, Cabrera GT, Tran H, Gendron TF, McKeon JE, et al. 2015. Human C9ORF72 hexanucleotide expansion reproduces RNA foci and dipeptide repeat proteins but not neurodegeneration in BAC transgenic mice. Neuron 88:902–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Liu Y, Pattamatta A, Zu T, Reid T, Bardhi O, et al. 2016. C9orf72 BAC mouse model with motor deficits and neurodegenerative features of ALS/FTD. Neuron 90:521–34 [DOI] [PubMed] [Google Scholar]
- 82.Jiang J, Zhu Q, Gendron TF, Saberi S, McAlonis-Downes M, et al. 2016. Gain of toxicity from ALS/FTD-linked repeat expansions in C9ORF72 is alleviated by antisense oligonucleotides targeting GGGGCC-containing RNAs. Neuron 90:535–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Lagier-Tourenne C, Baughn M, Rigo F, Sun S, Liu P, et al. 2013. Targeted degradation of sense and antisense C9orf72 RNA foci as therapy for ALS and frontotemporal degeneration. PNAS 110:E4530–39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Reddy K, Zamiri B, Stanley SY, Macgregor RB Jr., Pearson CE. 2013. The disease-associated r(GGGGCC)n repeat from the C9orf72 gene forms tract length–dependent uni- and multimolecular RNA G-quadruplex structures. J. Biol. Chem 288:9860–66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Haeusler AR, Donnelly CJ, Periz G, Simko EA, Shaw PG, et al. 2014. C9orf72 nucleotide repeat structures initiate molecular cascades of disease. Nature 507:195–200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Freibaum BD, Lu Y, Lopez-Gonzalez R, Kim NC, Almeida S, et al. 2015. GGGGCC repeat expansion in C9orf72 compromises nucleocytoplasmic transport. Nature 525:129–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Zhang K, Donnelly CJ, Haeusler AR, Grima JC, Machamer JB, et al. 2015. The C9orf72 repeat expansion disrupts nucleocytoplasmic transport. Nature 525:56–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Ash PE, Bieniek KF, Gendron TF, Caulfield T, Lin WL, et al. 2013. Unconventional translation of C9ORF72 GGGGCC expansion generates insoluble polypeptides specific to c9FTD/ALS. Neuron 77:639–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Mori K, Weng SM, Arzberger T, May S, Rentzsch K, et al. 2013. The C9orf72 GGGGCC repeat is translated into aggregating dipeptide-repeat proteins in FTLD/ALS. Science 339:1335–38 [DOI] [PubMed] [Google Scholar]
- 90.Mizielinska S, Gronke S, Niccoli T, Ridler CE, Clayton EL, et al. 2014. C9orf72 repeat expansions cause neurodegeneration in Drosophila through arginine-rich proteins. Science 345:1192–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Chew J, Gendron TF, Prudencio M, Sasaguri H, Zhang YJ, et al. 2015. Neurodegeneration. C9ORF72 repeat expansions in mice cause TDP-43 pathology, neuronal loss, and behavioral deficits. Science 348:1151–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Vatsavayai SC, Yoon SJ, Gardner RC, Gendron TF, Vargas JN, et al. 2016. Timing and significance of pathological features in C9orf72 expansion–associated frontotemporal dementia. Brain 139:3202–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Kwon I, Xiang S, Kato M, Wu L, Theodoropoulos P, et al. 2014. Poly-dipeptides encoded by the C9orf72 repeats bind nucleoli, impede RNA biogenesis, and kill cells. Science 345:1139–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Lee KH, Zhang P, Kim HJ, Mitrea DM, Sarkar M, et al. 2016. C9orf72 dipeptide repeats impair the assembly, dynamics, and function of membrane-less organelles. Cell 167:774–88.e17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Lin Y, Mori E, Kato M, Xiang S, Wu L, et al. 2016. Toxic PR poly-dipeptides encoded by the C9orf72 repeat expansion target LC domain polymers. Cell 167:789–802.e12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Gijselinck I, Van Mossevelde S, van der Zee J, Sieben A, Engelborghs S, et al. 2016. The C9orf72 repeat size correlates with onset age of disease, DNA methylation and transcriptional downregulation of the promoter. Mol. Psychiatry 21:1112–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Xi Z, Rainero I, Rubino E, Pinessi L, Bruni AC, et al. 2014. Hypermethylation of the CpG-island near the C9orf72 G4C2–repeat expansion in FTLD patients. Hum. Mol. Genet 23:5630–37 [DOI] [PubMed] [Google Scholar]
- 98.Xi Z, Zhang M, Bruni AC, Maletta RG, Colao R, et al. 2015. The C9orf72 repeat expansion itself is methylated in ALS and FTLD patients. Acta Neuropathol 129:715–27 [DOI] [PubMed] [Google Scholar]
- 99.Levine TP, Daniels RD, Gatta AT, Wong LH, Hayes MJ. 2013. The product of C9orf72, a gene strongly implicated in neurodegeneration, is structurally related to DENN Rab-GEFs. Bioinformatics 29:499–503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Amick J, Roczniak-Ferguson A, Ferguson SM. 2016. C9orf72 binds SMCR8, localizes to lysosomes, and regulates mTORC1 signaling. Mol. Biol. Cell 27:3040–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Sellier C, Campanari ML, Julie Corbier xC, Gaucherot A, Kolb-Cheynel I, et al. 2016. Loss of C9ORF72 impairs autophagy and synergizes with polyQ Ataxin-2 to induce motor neuron dysfunction and cell death. EMBO J 35:1276–97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Yang M, Liang C, Swaminathan K, Herrlinger S, Lai F, et al. 2016. A C9ORF72/SMCR8-containing complex regulates ULK1 and plays a dual role in autophagy. Sci. Adv 2:e1601167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Sullivan PM, Zhou X, Robins AM, Paushter DH, Kim D, et al. 2016. The ALS/FTLD associated protein C9orf72 associates with SMCR8 and WDR41 to regulate the autophagy–lysosome pathway. Acta Neuropathol. Commun 4:51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Burberry A, Suzuki N, Wang JY, Moccia R, Mordes DA, et al. 2016. Loss-of-function mutations in the C9ORF72 mouse ortholog cause fatal autoimmune disease. Sci. Transl. Med 8:347ra93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.O’Rourke JG, Bogdanik L, Yanez A, Lall D, Wolf AJ, et al. 2016. C9orf72 is required for proper macrophage and microglial function in mice. Science 351:1324–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Ciura S, Lattante S, Le Ber I, Latouche M, Tostivint H, et al. 2013. Loss of function of C9orf72 causes motor deficits in a zebrafish model of amyotrophic lateral sclerosis. Ann. Neurol 74:180–87 [DOI] [PubMed] [Google Scholar]
- 107.Mackenzie IR. 2007. The neuropathology and clinical phenotype of FTD with progranulin mutations. Acta Neuropathol 114:49–54 [DOI] [PubMed] [Google Scholar]
- 108.Hu F, Padukkavidana T, Vaegter CB, Brady OA, Zheng Y, et al. 2010. Sortilin-mediated endocytosis determines levels of the frontotemporal dementia protein, progranulin. Neuron 68:654–67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Capell A, Liebscher S, Fellerer K, Brouwers N, Willem M, et al. 2011. Rescue of progranulin deficiency associated with frontotemporal lobar degeneration by alkalizing reagents and inhibition of vacuolar ATPase. J. Neurosci 31:1885–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Salazar DA, Butler VJ, Argouarch AR, Hsu TY, Mason A, et al. 2015. The progranulin cleavage products, granulins, exacerbate TDP-43 toxicity and increase TDP-43 levels. J. Neurosci 35:9315–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Zhou X, Sun L, Bastos de Oliveira F, Qi X, Brown WJ, et al. 2015. Prosaposin facilitates sortilin-independent lysosomal trafficking of progranulin. J. Cell Biol 210:991–1002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Nicholson AM, Finch NA, Almeida M, Perkerson RB, van Blitterswijk M, et al. 2016. Prosaposin is a regulator of progranulin levels and oligomerization. Nat. Commun 7:11992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Kishimoto Y, Hiraiwa M, O’Brien JS. 1992. Saposins: structure, function, distribution, and molecular genetics. J. Lipid Res 33:1255–67 [PubMed] [Google Scholar]
- 114.Tayama M, O’Brien JS, Kishimoto Y. 1992. Distribution of saposins (sphingolipid activator proteins) in tissues of lysosomal storage disease patients. J. Mol. Neurosci 3:171–75 [DOI] [PubMed] [Google Scholar]
- 115.Meyer RC, Giddens MM, Coleman BM, Hall RA. 2014. The protective role of prosaposin and its receptors in the nervous system. Brain Res 1585:1–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Cotman SL, Karaa A, Staropoli JF, Sims KB. 2013. Neuronal ceroid lipofuscinosis: impact of recent genetic advances and expansion of the clinicopathologic spectrum. Curr. Neurol. Neurosci. Rep 13:366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Almeida MR, Macario MC, Ramos L, Baldeiras I, Ribeiro MH, Santana I. 2016. Portuguese family with the co-occurrence of frontotemporal lobar degeneration and neuronal ceroid lipofuscinosis phenotypes due to progranulin gene mutation. Neurobiol. Aging 41:200.e1–5 [DOI] [PubMed] [Google Scholar]
- 118.Smith KR, Damiano J, Franceschetti S, Carpenter S, Canafoglia L, et al. 2012. Strikingly different clinicopathological phenotypes determined by progranulin-mutation dosage. Am. J. Hum. Genet 90:1102–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Gotzl JK, Mori K, Damme M, Fellerer K, Tahirovic S, et al. 2014. Common pathobiochemical hallmarks of progranulin-associated frontotemporal lobar degeneration and neuronal ceroid lipofuscinosis. Acta Neuropathol 127:845–60 [DOI] [PubMed] [Google Scholar]
- 120.Ward ME, Chen R, Huang HY, Ludwig C, Telpoukhovskaia M, et al. 2017. Individuals with progranulin haploinsufficiency exhibit features of neuronal ceroid lipofuscinosis. Sci. Transl. Med 9:eaah5642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Ahmed Z, Sheng H, Xu YF, Lin WL, Innes AE, et al. 2010. Accelerated lipofuscinosis and ubiquitination in granulin knockout mice suggest a role for progranulin in successful aging. Am. J. Pathol 177:311–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Martens LH, Zhang J, Barmada SJ, Zhou P, Kamiya S, et al. 2012. Progranulin deficiency promotes neuroinflammation and neuron loss following toxin-induced injury. J. Clin. Investig 122:3955–59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Yin F, Banerjee R, Thomas B, Zhou P, Qian L, et al. 2010. Exaggerated inflammation, impaired host defense, and neuropathology in progranulin-deficient mice. J. Exp. Med 207:117–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Yin F, Dumont M, Banerjee R, Ma Y, Li H, et al. 2010. Behavioral deficits and progressive neuropathology in progranulin-deficient mice: a mouse model of frontotemporal dementia. FASEB J 24:4639–47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Klein ZA, Takahashi H, Ma M, Stagi M, Zhou M, et al. 2017. Loss of TMEM106B ameliorates lysosomal and frontotemporal dementia-related phenotypes in progranulin-deficient mice. Neuron 95:281–96.e6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Filiano AJ, Martens LH, Young AH, Warmus BA, Zhou P, et al. 2013. Dissociation of frontotemporal dementia–related deficits and neuroinflammation in progranulin haploinsufficient mice. J. Neurosci 33:5352–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Evers BM, Rodriguez-Navas C, Tesla RJ, Prange-Kiel J, Wasser CR, et al. 2017. Lipidomic and transcriptomic basis of lysosomal dysfunction in progranulin deficiency. Cell Rep 20:2565–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Nixon RA. 2013. The role of autophagy in neurodegenerative disease. Nat. Med 19:983–97 [DOI] [PubMed] [Google Scholar]
- 129.Aharon-Peretz J, Badarny S, Rosenbaum H, Gershoni-Baruch R. 2005. Mutations in the glucocerebrosidase gene and Parkinson disease: phenotype–genotype correlation. Neurology 65:1460–61 [DOI] [PubMed] [Google Scholar]
- 130.Mitsui J, Mizuta I, Toyoda A, Ashida R, Takahashi Y, et al. 2009. Mutations for Gaucher disease confer high susceptibility to Parkinson disease. Arch. Neurol 66:571–76 [DOI] [PubMed] [Google Scholar]
- 131.Ramirez A, Heimbach A, Grundemann J, Stiller B, Hampshire D, et al. 2006. Hereditary parkinsonism with dementia is caused by mutations in ATP13A2, encoding a lysosomal type 5 P-type ATPase. Nat. Genet 38:1184–91 [DOI] [PubMed] [Google Scholar]
- 132.Schuur M, Ikram MA, van Swieten JC, Isaacs A, Vergeer-Drop JM, et al. 2011. Cathepsin D gene and the risk of Alzheimer’s disease: a population-based study and meta-analysis. Neurobiol. Aging 32:1607–14 [DOI] [PubMed] [Google Scholar]
- 133.Nguyen AD, Nguyen TA, Zhang J, Devireddy S, Zhou P, et al. 2018. Murine knockin model for progranulin-deficient frontotemporal dementia with nonsense-mediated mRNA decay. PNAS 115:E2849–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Rabouille C 2017. Pathways of unconventional protein secretion. Trends Cell Biol 27:230–40 [DOI] [PubMed] [Google Scholar]
- 135.Laplante M, Sabatini DM. 2012. mTOR signaling in growth control and disease. Cell 149:274–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Sardiello M, Palmieri M, di Ronza A, Medina DL, Valenza M, et al. 2009. A gene network regulating lysosomal biogenesis and function. Science 325:473–77 [DOI] [PubMed] [Google Scholar]
- 137.Settembre C, Di Malta C, Polito VA, Garcia Arencibia M, Vetrini F, et al. 2011. TFEB links autophagy to lysosomal biogenesis. Science 332:1429–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Kao AW, Eisenhut RJ, Martens LH, Nakamura A, Huang A, et al. 2011. A neurodegenerative disease mutation that accelerates the clearance of apoptotic cells. PNAS 108:4441–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Lui H, Zhang J, Makinson SR, Cahill MK, Kelley KW, et al. 2016. Progranulin deficiency promotes circuit-specific synaptic pruning by microglia via complement activation. Cell 165:921–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Chang MC, Srinivasan K, Friedman BA, Suto E, Modrusan Z, et al. 2017. Progranulin deficiency causes impairment of autophagy and TDP-43 accumulation. J. Exp. Med 214:2611–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Takahashi H, Klein ZA, Bhagat SM, Kaufman AC, Kostylev MA, et al. 2017. Opposing effects of progranulin deficiency on amyloid and tau pathologies via microglial TYROBP network. Acta Neuropathol 133:785–807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Stephan AH, Barres BA, Stevens B. 2012. The complement system: an unexpected role in synaptic pruning during development and disease. Annu. Rev. Neurosci 35:369–89 [DOI] [PubMed] [Google Scholar]
- 143.Stevens B, Allen NJ, Vazquez LE, Howell GR, Christopherson KS, et al. 2007. The classical complement cascade mediates CNS synapse elimination. Cell 131:1164–78 [DOI] [PubMed] [Google Scholar]
- 144.Cadwell K, Debnath J. 2018. Beyond self-eating: the control of nonautophagic functions and signaling pathways by autophagy-related proteins. J. Cell Biol 217:813–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Shaid S, Brandts CH, Serve H, Dikic I. 2013. Ubiquitination and selective autophagy. Cell Death Differ 20:21–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Tresse E, Salomons FA, Vesa J, Bott LC, Kimonis V, et al. 2010. VCP/p97 is essential for maturation of ubiquitin-containing autophagosomes and this function is impaired by mutations that cause IBMPFD. Autophagy 6:217–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Maruyama H, Morino H, Ito H, Izumi Y, Kato H, et al. 2010. Mutations of optineurin in amyotrophic lateral sclerosis. Nature 465:223–26 [DOI] [PubMed] [Google Scholar]
- 148.Shen WC, Li HY, Chen GC, Chern Y, Tu PH. 2015. Mutations in the ubiquitin-binding domain of OPTN/optineurin interfere with autophagy-mediated degradation of misfolded proteins by a dominant-negative mechanism. Autophagy 11:685–700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Cassel JA, Reitz AB. 2013. Ubiquilin-2 (UBQLN2) binds with high affinity to the C-terminal region of TDP-43 and modulates TDP-43 levels in H4 cells: characterization of inhibition by nucleic acids and 4-aminoquinolines. Biochim. Biophys. Acta 1834:964–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Osaka M, Ito D, Suzuki N. 2016. Disturbance of proteasomal and autophagic protein degradation pathways by amyotrophic lateral sclerosis–linked mutations in ubiquilin 2. Biochem. Biophys. Res. Commun 472:324–31 [DOI] [PubMed] [Google Scholar]
- 151.Wu Q, Liu M, Huang C, Liu X, Huang B, et al. 2015. Pathogenic Ubqln2 gains toxic properties to induce neuron death. Acta Neuropathol 129:417–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Rubino E, Rainero I, Chio A, Rogaeva E, Galimberti D, et al. 2012. SQSTM1 mutations in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Neurology 79:1556–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Buchan JR, Kolaitis RM, Taylor JP, Parker R. 2013. Eukaryotic stress granules are cleared by autophagy and Cdc48/VCP function. Cell 153:1461–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Seguin SJ, Morelli FF, Vinet J, Amore D, De Biasi S, et al. 2014. Inhibition of autophagy, lysosome and VCP function impairs stress granule assembly. Cell Death Differ 21:1838–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Deng HX, Chen W, Hong ST, Boycott KM, Gorrie GH, et al. 2011. Mutations in UBQLN2 cause dominant X-linked juvenile and adult-onset ALS and ALS/dementia. Nature 477:211–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Zhang X, Lin Y, Eschmann NA, Zhou H, Rauch JN, et al. 2017. RNA stores tau reversibly in complex coacervates. PLOS Biol 15:e2002183. [DOI] [PMC free article] [PubMed] [Google Scholar]