Abstract
Background
Staphylococcus aureus (S. aureus) has the potential to opportunistically cause infectious diseases, including osteomyelitis, skin infections, pneumonia, and diarrhea. We previously reported that ASyycG RNA reduced the transcripts of virulent genes, and biofilm formation of S. aureus. Currently, graphene oxide (GO) nanosheets are used to efficiently deliver nucleic acids with favorable biocompatibility.
Methods
In the current study, a GO-based recombinant pDL278 ASyycG vector transformation strategy was developed. The particle size distributions and zeta-potential of the GO-PEI-based ASyycG were evaluated. The ASyycG plasmids were labeled with gene-encoding enhanced green fluorescent protein (ASyycG-eGFP). Quantitative real-time PCR assays were performed to investigate the expression of yycF/G/H and icaADB genes. Biofilm biomass and bacterial viability of S. aureus were evaluated by scanning electron microscopy and confocal laser scanning microscopy. We found that the expression of the yycG gene was inversely correlated with levels of the ASyycG transcripts and that the GO-PEI-ASyycG strain had the lowest expression of biofilm organization-associated genes.
Results
The results showed that the GO-based strategy significantly increased ASyycG transformation as a delivery system compared to the conventional competence-stimulating peptide strategy. Furthermore, GO-PEI-ASyycG suppressed bacterial biofilm aggregation and improved bactericidal effects on S. aureus after 24 h biofilm establishment.
Conclusions
Our findings demonstrated that nano-GO with antisense yycG RNA is a more effective and relatively stable strategy for the management of S. aureus infections.
Electronic supplementary material
The online version of this article (10.1186/s13018-019-1356-x) contains supplementary material, which is available to authorized users.
Keywords: Staphylococcus aureus, Antisense RNA, YycG, Graphene oxide, Biofilm
Introduction
Staphylococcus aureus (S. aureus), a gram-positive coccus, is carried by about 20–30% of healthy individuals and mostly colonizes the nasopharynx [1]. S. aureus has the potential to cause a wide range of diseases, including osteomyelitis, skin infections, pneumonia, and even life-threatening infective endocarditis associated with considerable global human morbidity and mortality [2]. However, in some cases, S. aureus is resistant to multiple types of antibiotics, which has been attributed to the abuse of antibiotics, resulting in the emergence of methicillin-resistant Staphylococcus aureus (MRSA) [3]. Presently, more than 50% of S. aureus in clinical isolates in hospitals worldwide are methicillin-resistant [4]. Therefore, the identification of novel antibacterial strategies is of the utmost importance.
Graphene oxide (GO) is a graphene sheet containing functional organic groups, such as carboxyl, hydroxyl, carbonyl, and epoxy, on its basal plane [5]. The sharp edges on the GO sheet structures physically disrupt cell membranes and cause oxidative stress reactions. Therefore, it is expected to act as a novel type of bactericidal agent with a low risk of developing resistance from pathogenic bacteria [6]. In addition, the large surface area of GO sheets makes them ideal candidates for gene delivery [7]. Although GO sheets can be used to effectively deliver single-stranded nucleic acids, the ability of GO to carry double-stranded DNA (dsDNA) is limited. Polyethyleneimine (PEI) is a well-studied cationic polymer that has been used as a common non-viral gene delivery vector when combined with GO. Compared with the PEI polymer, GO–PEI has been reported to have lower cytotoxicity and higher transfection efficiency, and thus, has high potential as a gene vector [8].
Two-component signal transduction systems (TCSs) are essential pathways for bacterial responses to environmental stimuli. Typical TCS components consist of a transmembrane histidine kinase sensor and a corresponding cytoplasmic response regulator, which can bind to specific regions to regulate the expression of targeted genes [9]. YycFG is the only essential TCS in S. aureus, contributing to its physiology, and biofilm metabolism [10]. Biofilms are microbial communities embedded within self-produced extracellular substances and are closely related to the development of infections in humans [11]. In S. aureus, polysaccharide intercellular adhesion (PIA) encoded by icaADBC is a functional factor involved in biofilm organization [12].
Antisense RNAs (asRNAs) are a type of single-strand RNA that recognize mRNA by base-pairing and inhibiting the transcription and transduction of target mRNA [13]. Antisense RNA strategy is a promising approach for novel gene-specific antisense antibiotics to cure bacterial infections [14]. However, the efficiency of transforming antisense RNA into bacterial cells is limited without a suitable carrier system [15]. Since GO-PEI complexes are highly positively charged, effective loading with DNA plasmids can be achieved. In this study, a GO-based plasmid transformation system was developed using GO-PEI complexes that were loaded with antisense yycG plasmid (GO-PEI-ASyycG). We hypothesized that the antibacterial properties of GO to S. aureus could be enhanced by loading the gene vector with antisense ASyycG plasmids. A potential role for the clinical application GO-PEI-ASyycG as a novel antibiotic agent was proposed for the management of the S. aureus infections.
Methods and materials
Preparation of GO-PEI-ASyycG and cytotoxicity evaluation
The antisense yycG sequences (ASyycG) were synthesized by Sangon Biotech (Shanghai, China). To generate a recombinant pDL278 ASyycG plasmid, the ASyycG sequences were inserted into the BamHI and EcoRI restriction sites of a pDL278 vector [16]. To synthesize the GO-PEI complexes, GO powder (XFNANO Materials Tech, Nanjing, China) was added to ddH2O to a final concentration of 0.1 mg/mL. Next, the solution was slowly mixed with branched polyethyleneimine (BPEI, 10 kDa; Sigma-Aldrich, St. Louis, MO, USA). Then, the solution was processed with 10 cycles of ultrasonication for 60 s, with 60 s rest on ice between each sonication. The obtained solution was mixed on a shaking table overnight at room temperature. To remove redundant PEI compounds, the mixtures were washed three times with ddH2O by centrifugation (12000×g, 1 min) and resuspended with ddH2O to a final concentration of 0.1 mg/mL. pDL278 ASyycG plasmid (100ng/μL) was added to the GO-PEI complexes at a volume ratio of 1:125 and the mixtures were incubated for 1 h at room temperature.
The working concentration of GO-PEI-ASyycG was determined by cytotoxicity assays. Briefly, the mouse embryonic fibroblast NIH/3T3 cell line (Sigma-Aldrich) at a density of 1000 cells/well were seeded into 96-well plates in 100 μL of Dulbecco’s modified Eagle’s medium (DMEM), supplemented with 10% fetal bovine serum (FBS), and mixed with the GO-PEI-ASyycG solutions at dilutions ranging from 100 μg/mL to 0 μg/mL. After incubation for 48 or 72 h (37 °C, 5% CO2), the plates were removed and each well was washed with phosphate buffer solution (PBS, pH = 7.4) twice. The CCK-8 cell counting kit (Dojindo Laboratories, Kumamoto, Japan) was used to test the cell viability and each well was incubated with 10 μL of CCK-8 reagent. After 2 h of culture, the OD values of each well were measured using a microplate reader (ELX800, Gene, Hong Kong, China) at 540 nm.
Particle size distribution, zeta potential, and atomic force microscopy measurements
The particle size distribution of the GO, GO-PEI, and GO-PEI-ASyycG solutions (0.1 mg/mL) was measured by dynamic light scattering (DLS) and the zeta-potential was examined by a Malvern instrument (Zetasizer Malvern Nano ZS, Instruments, Worcestershire, UK). A total of 50 μL of GO, GO-PEI, or GO-PEI-ASyycG solution was dropped onto sterile coverslips and films were prepared and air-dried in room temperature. The roughness of the films was assessed using an atomic force microscope (AFM) (SPM-9500J2, Shimadzu, Tokyo, Japan) in the contact mode. Micrographs of all films were evaluated by scanning electron microscopy (SEM; Inspect F50, FEI, Hillsboro, OR, USA) as previously described [17].
Bacterial culture and transformation
A single colony of S. aureus was selected from a tryptic soy agar (TSA) plate and cultured in tryptic soy broth (TSB) medium to the mid-exponential phase, which was determined by an OD600 value of 0.5. For the S. aureus GO group, 250 μL of mid-exponential S. aureus was incubated with 2 μL GO solution (final concentration determined by cell viability assay). In the ASyycG group, 2 μL of recombinant pDL278 ASyycG plasmid was mixed with 250 μL of mid-exponential S. aureus as in our previous studies [16]. For the GO-PEI-ASyycG strains, 250 μL of mid-exponential S. aureus was co-cultured with prepared GO-PEI-ASyycG. All S. aureus strains were cultured at 37 °C in 5% CO2 for 1 hour, then diluted into 5 mL of fresh TSB medium.
Transformation efficiency of GO-PEI-ASyycG in vitro
The ASyycG plasmids were labeled with gene encoding enhanced green fluorescent protein (ASyycG-eGFP). The sequences of ASyycG and eGFP were synthesized by Sangon Biotech (Shanghai, China) and are listed in the Additional file 1. The ASyycG-eGFP and GO-PEI-ASyycG-eGFP strains were constructed based on the transformation procedures described above. Both strains were grown in TSB medium until an OD600 value of 0.5 was reached. A total of 50 μL of bacterial suspensions was dropped onto coverslips and air-dried at room temperature for 30 min. Confocal laser scanning microscopy (CLSM) was applied to determine the expression level of eGFP. The transfection efficiency was determined by comparing the green fluorescence intensities.
Real-time polymerase chain reaction (RT-PCR) assays were conducted to assess the expression of ASyycG in all S. aureus strains. Briefly, total RNA was extracted from S. aureus suspensions from the mid-logarithmic growth phase in TSB medium using an RNA purification Kit (MasterPure, Epicentre, Madison, WI, USA) according to the manufacturer’s instructions. Total RNA was reverse transcribed using an RT Reagent Kit (PrimeScript, Takara, Kyoto, Japan). Quantitative RT-PCR assays were carried out using the primers listed in Table 1 using a LightCycler 480 system (Roche, Basel, Switzerland). The 16S rRNA gene was used as an internal control [17].
Table 1.
Sequences of primers used for qRT-PCR analysis
| Primers | Sequence 5′-3′ (forward/reverse) | Reference |
|---|---|---|
| RT-qPCR | ||
| icaA |
5′-GATTATGTAATGTGCTTGGA-3′/ 5′-ACTACTGCTGCGTTAATAAT-3′ |
This study |
| icaD |
5′-ATGGTCAAGCCCAGACAGAG-3′/ 5′-CGTGTTTTCAACATTTAATGCAA-3′ |
This study |
| icaB |
5′-CACATACCCACGATTTGCAT-3′/ 5′-TCGGAGTGACTGCTTTTTCC-3′ |
This study |
| yycF |
5′-TGGCGAAAGAAGACATCA-3′/ 5′-AACCCGTTACAAATCCTG-3′ |
This study |
| yycG |
5′-CGGGGCGTTCAAAAGACTTT-3′/ 5′-TCTGAACCTTTGAACACACGT-3′ |
This study |
| 16S rRNA |
5′-GTAGGTGGCAAGCGTTATCC-3′/ 5′-CGCACATCAGCGTCAACA-3′ |
This study |
Growth conditions of S. aureus strains
After transformation, all strains were diluted in TSB at a ratio 1:20 and incubated in 96-well plates. The bacterial growth curves were monitored by measuring the OD600nm with a microplate reader (ELX800, Gene, Hong Kong, China) every 60 min for 24 h. The proportions of live bacteria cells were estimated by confocal laser scanning microscopy (CLSM, FV1000; Olympus Corporation, Tokyo, Japan) at × 40 magnification. Live cells were stained with SYTO9 dye (LIVE/DEAD Bacterial Viability Kit reagent; BacLight, Invitrogen, Grand Island, NY, USA) and dead cells were labeled with propidium iodide (PI). Three-dimensional reconstruction was conducted and analyzed using Imaris 7.0.0 software (Imaris 7.0.0, Bitplane, Zurich, Switzerland) as previously described [18].
Evaluation of S. aureus biofilms
Crystal violet (CV) assays were applied to compare the biomass of S. aureus biofilms cultured in 24-well polystyrene plates for 24 h. As previously described, the biofilms were stained with 0.1% (w/v) crystal violet for 15 min at room temperature [17]. The dye bound on the biofilms was collected using 1 mL of de-staining solution (8:2 ethanol: acetone). Then, the solution was transferred to a new plate and the OD600nm was read by a microplate reader (ELX800, Gene, Hong Kong, China).
Sterile coverslips were immersed in 24-well plates with different S. aureus strain suspensions (OD600nm = 0.5). After 24 h of co-culturing, the planktonic suspensions were removed and the biofilms grown on the coverslips were washed three times with PBS (pH7.2). Then, the biofilms were fixed in 2.5% glutaraldehyde for 4 h at room temperature and dehydrated with serially concentrated ethanol solutions (30%, 50%, 70%, 80%, 95%, and 100%). The prepared biofilms were dried to critical-point at room temperature and coated with gold powder. Scanning electron microscopy (SEM; Inspect F50, FEI, Hillsboro, OR, USA) was used to estimate the morphologies of all S. aureus biofilms by selecting three random areas from each sample [19].
Data analyses
All data were processed using SPSS software (SPSS version 20, IBM, Armonk, NY, USA). The values are expressed as mean ± standard deviation (SD) for the indicated number of samples. The quantification cycles for describing gene expression were relatively quantified by real-time PCR using 16S as an internal control and calculated based on the ATCC29213 expression, which was set to 1.0. Bartlett’s test was employed to assess the homogeneity of data variance and the Shapiro-Wilk test was conducted to determine the normal distribution of the data. One-way analysis of variance was used to compare the data, followed by pairwise multiple comparisons. The differences were considered significant if the p value was < 0.05.
Results
Cytotoxicity and characteristics of GO-PEI-ASyycG films
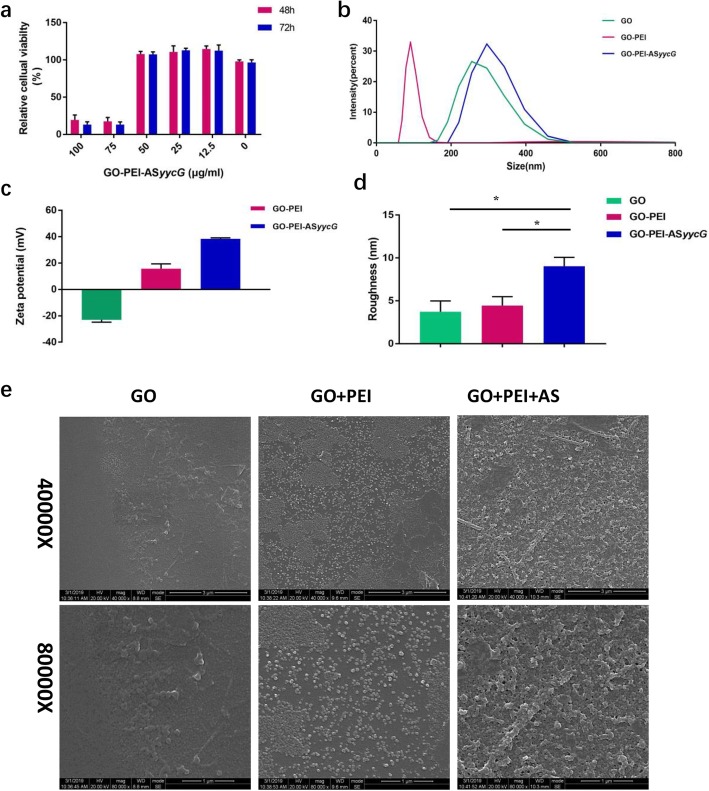
The viability of the 3T3 fibroblasts cells was significantly decreased after 48 or 72 h treatment GO-PEI-ASyycG concentrations higher than 50 μg/mL (Fig. 1a). Our results indicated that the GO-PEI-ASyycG complexes were not toxic at concentrations lower than 50 μg/mL (Fig. 1a). Dynamic light scattering measurements were applied to determine the hydrodynamic sizes of GO, GO-PEI, and GO-PEI-ASyycG in deionized water. Z-average sizes of 420 nm and 280 nm were obtained for GO and GO-PEI, which were much smaller than the average size of GO-PEI-ASyycG (490 nm) (Fig. 1b). The surface charge (zeta potential) value of GO was approximately at − 23.1 mV. After mixing with the cationic PEI polymer, the GO-PEI and GO-PEI-ASyycG complexes demonstrated positive surface charges of 15.7 mV and 38.4 mV, respectively (Fig. 1c). Using AFM, the roughness analysis of the membranes revealed that the GO-PEI-ASyycG nanosheet roughness averaged 8.9 nm, significantly higher than the GO and GO-PEI films, which averaged 3.4 nm and 4.5 nm, respectively (n = 10, p < 0.05; Fig. 1d). Using SEM, the results showed rougher and denser surface morphologies on the GO-PEI-ASyycG films compared to GO and GO-PEI (Fig. 1e).
Fig. 1.
Cytotoxicity and characterization of GO-PEI-yycG. a The cell viability were determined with CCK-8 after 48 h or 72 h of incubation with GO-PEI-ASyycG (Dulbecco’s modified Eagle’s medium supplemented with 10% fetal bovine serum was the blank control). b The particle size distributions were measured using dynamic light scattering. c The zeta potential of the GO-PEI-based ASyycG was assessed by a Malvern Zetasizer. d AFM confirmed the roughness parameters of GO, GO-PEI, and GO-PEI-ASyycG material films (n = 10,*p < 0.05). e SEM images of GO, GO-PEI, and GO-PEI-ASyycG material films
GO-PEI-ASyycG increased ASyycG transformation and significantly reduced biofilm formation-associated gene expression
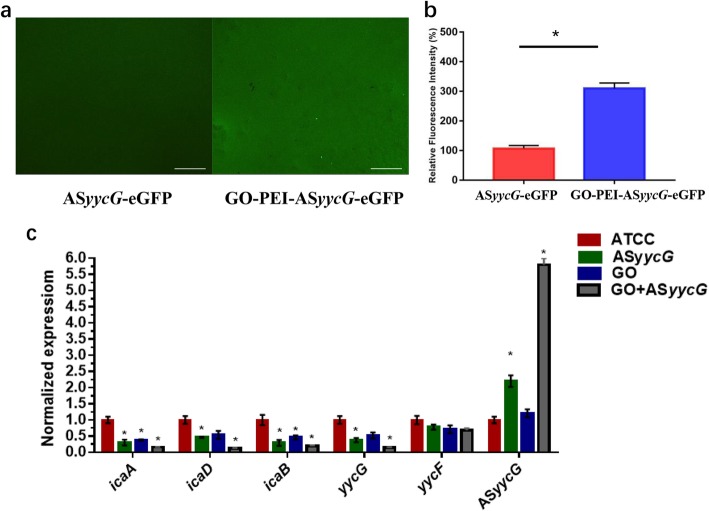
Using confocal laser scanning microscopy, higher levels of GFP-expression were observed in samples induced with GO-PEI-ASyycG compared to pure ASyycG (Fig. 2a). A 200% increase in GFP-expression transcripts in GO-PEI-ASyycG transformed strains was found, a statistically significant increase compared to the expression in ASyycG cells (n = 10, p < 0.05; Fig. 2b). Quantitative RT-PCR assays demonstrated that expression levels of ASyycG RNA in the ASyycG and GO-PEI-ASyycG strains were significantly increased 2.8-fold and 6.5-fold, respectively, compared to S. aureus ATCC29213 strains (n = 10, p < 0.05; Fig. 2c). Accordingly, the gene expression levels of yycG were significantly downregulated in the ASyycG and GO-PEI-ASyycG strains (n = 10, p < 0.05). Consequently, the expression levels of PIA synthesis-associated genes icaA/D/B/C were the lowest in the GO-PEI-ASyycG strain among all groups (n = 10, p < 0.05).
Fig. 2.
GO-PEI-ASyycG increased ASyycG transformation and inhibited virulence gene expressions. a ASyycG plasmids were labeled with gene encoding enhanced green fluorescent protein (ASyycG-eGFP) and CLSM was applied to determine the expression level of eGFP. b The transfection efficiency was determined by comparing the green fluorescent intensities (n = 10, *p < 0.05). c Quantitative RT-PCR analysis showed gene transcription in untreated S. aureus and ASyycG-, GO-, and GO-PEI-ASyycG-treated strains; S. aureus gene expression was quantified relatively using 16sR as an internal control and calculated based on untreated S. aureus ATCC29213 expression, which was set as 1.0. (n = 10, *p < 0.05)
GO-PEI-ASyycG decreased cells viability and suppressed biofilm formation
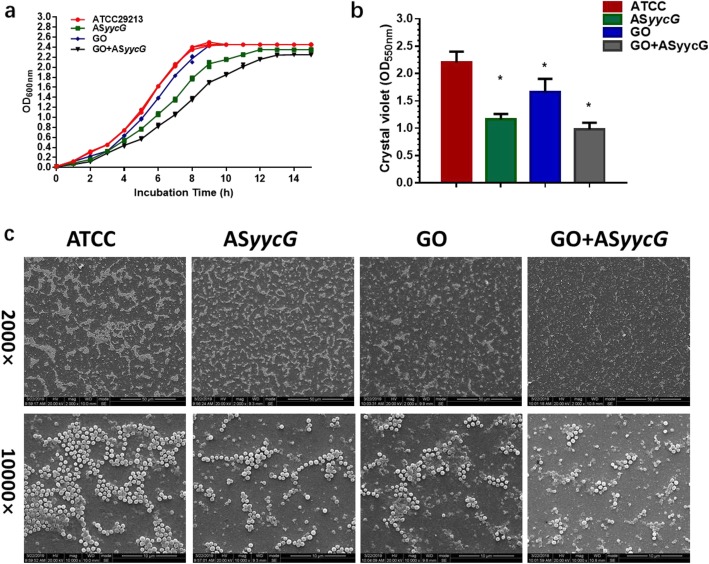
When growth was monitored in the different strains, the results showed that the time before entry into the log phase in the GO-PEI-ASyycG strains was obviously prolonged compared to that of the ASyycG strains (Fig. 3a). The most impaired formation of biofilms was observed in GO-PEI-ASyycG strains compared to GO, ASyycG, and ATCC29213 (Fig. 3b). Quantitatively, these results were confirmed by the OD values of the biofilm biomasses, which were the lowest in the GO-PEI-ASyycG strains (n = 10, p < 0.05, Fig. 3b).
Fig. 3.
GO-PEI-ASyycG suppressed S. aureus growth and biofilm formation. a S. aureus and ASyycG-, GO-, and GO-PEI-ASyycG-treated strains were cultured and growth was monitored every hour. b Biomass was quantified by crystal violet staining. Optical densities at 600 nm were measured (n = 10, *p < 0.05). c SEM of untreated S. aureus and ASyycG-, GO-, and GO-PEI-ASyycG-treated strains for 24 h biofilms. Scale bar for × 2000 magnification, 50 μm. Scale bar for × 10,000 magnification, 10 μm
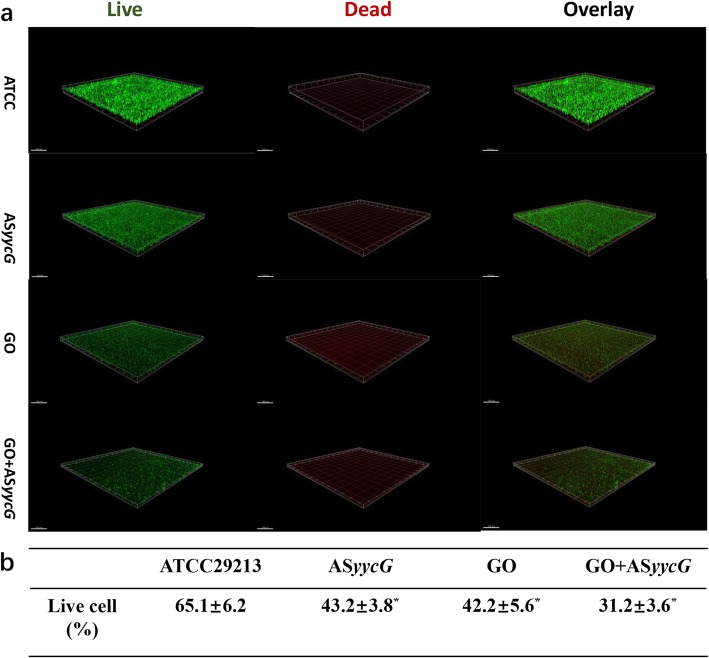
SEM observation demonstrated reduced levels of extracellular matrix components in the biofilms of the GO, ASyycG, and GO-PEI-ASyycG cells that were separated by blank areas (Fig. 3c). Exopolysaccharide-enmeshed cell clusters were greatly decreased in the GO-PEI-ASyycG strain biofilms (Fig. 3c). Using CLSM, we demonstrated that the proportion of viable bacteria was significantly decreased in the GO, ASyycG, and GO-PEI-ASyycG strains compared to the S. aureus ATCC29213 strain (Fig. 4a). The lowest percentage of live bacteria was observed in the GO-PEI-ASyycG strain, at 14.83 ± 0.5 % (n = 10, p < 0.05, Fig. 4b).
Fig. 4.
GO-PEI-ASyycG suppressed the vital cells in S. aureus biofilms. a Double labeling of the biofilms in the untreated S. aureus and ASyycG-, GO-, and GO-PEI-ASyycG-treated strains. Green, vital cells (SYTO 9); red, dead cells (PI); scale bars, 100 μm. The three-dimensional reconstruction of the biofilms was performed using Imaris 7.0.0. b Volume ratio of the vital bacterial biomass in the biofilms (n = 10, *p < 0.05)
Discussion
Our previous work indicated that antisense yycG RNA (ASyycG) could inhibit the target gene yycG in the MRSA strain. We found that ASyycG strains inhibited biofilm organization and increased antibiotic sensitivity [20]. However, one of the major obstacles to the use of antisense oligonucleotides is that, without a suitable and effective vector, the uptake by bacterial cells is limited [21]. In the current study, a GO-based recombinant pDL278 ASyycG vector transformation strategy was used to electrostatically combine the vector with cationic GO-PEI complexes. We showed that GO-PEI could efficiently deliver ASyycG plasmid into S. aureus cells with efficient transcripts of ASyycG. GO has been reported to ionically bind to cationic PEI polymers [8]. These positive surface charges could interact with the negatively charged cellular surface and promote bacterial transformation [22]. A previous study reported that 50 μg/mL of GO-PEI (or lower) did not have toxic effects on the cellular apoptosis rate [23]. In the present study, we demonstrated that the synthesized GO-PEI-ASyycG was not toxic at concentrations less than 50 μg/mL (Fig. 1a). Therefore, 50 μg/mL of GO-PEI-ASyycG was adopted as the working concentration.
The results of AFM observation indicated that the surface roughness of the GO-PEI-ASyycG nanosheets was increased compared to GO and GO-PEI material films (Fig. 1d). The surface characteristics of the GO nanosheets films were assessed using SEM, which showed a rougher and denser surface morphology in the GO-PEI-ASyycG films compared to those of GO and GO-PEI (Fig. 1e). Because the surface roughness of membrane films could influence bacterial colonization and adhesion, the increased surface characteristics of the GO-PEI-ASyycG material films probably indicated enhanced adhesive force.
To evaluate the vector transformation efficiencies, ASyycG recombinant plasmids were labeled with gene encoding enhanced green fluorescent protein. The levels of GFP-expression indicated the presence of ASyycG transcripts and revealed higher transformation efficiencies in S. aureus cells induced by GO-PEI-ASyycG compared to pure ASyycG plasmids. In particular, the quantitative RT-PCR assays showed that the fold change in ASyycG expression in the GO-PEI-ASyycG strain was roughly 3-fold higher in the ASyycG strain transformed with competence-stimulating peptide. We speculated that the GO-based strategy significantly increased ASyycG transformation as a delivery system and reduced transcripts of the yycG gene.
Furthermore, GO-PEI-ASyycG significantly suppressed bacterial growth and biofilm aggregation (Fig. 3). Using SEM observation, few randomly distributed microcolonies were identified and exopolysaccharide-enmeshed cell clusters were greatly decreased in the GO-PEI-ASyycG strain biofilms compared to the GO and ASyycG strains. After 24 h of biofilm establishment, CLSM findings revealed that GO-PEI-ASyycG greatly reduced the cellular viability (Fig. 4). These results suggested that S. aureus was markedly inhibited by GO-PEI-ASyycG, which improved the bactericidal effects of ASyycG on the S. aureus biofilms.
Biofilm-forming capacity is an essential factor in the development of S. aureus-induced infections and results in significant increases in morbidity and mortality [24]. In S. aureus, PIA is a crucial component for biofilm organization [25], which is mostly synthesized by glycosyltransferase enzymes encoded by the ica operon [12, 26]. In the current study, the GO-PEI-ASyycG strain had the lowest expression of yycF/G/H and icaA/D genes and biofilm formation, indicating that the pathogenesis of S. aureus was further decreased by the GO-PEI complexes, improving ASyycG transformation. Injectable GO-PEI-ASyycG could be useful in orthopedic applications to manage osteomyelitis lesions and reduce the use of antibiotics. Future directions will need to extend the applications of GO-PEI-ASyycG strategy as a potential way of managing the antibiotic resistance of S. aureus infections. At an appropriate concentration, GO-PEI-ASyycG could potentially improve the antibacterial properties of irrigation fluid. However, a limitation of the current study was the lack of in vivo experiments which are needed to confirm the effective concentration of this novel antibacterial agent before clinical application.
In the current study, a GO-based recombinant pDL278 ASyycG vector transformation strategy was developed. We found that the expression of the yycG gene was inversely correlated with the levels of ASyycG transcripts and that the GO-PEI-ASyycG strain had the lowest expression of biofilm organization-associated genes. The GO-based strategy significantly increased ASyycG transformation as a delivery system compared to the conventional competence-stimulating peptide strategy. Furthermore, GO-PEI-ASyycG suppressed aggregation of bacterial biofilms and improved the bactericidal effects on S. aureus after 24 h of biofilm establishment. Thus, our data demonstrated that nano-GO with antisense yycG RNA may be an effective and relatively stable strategy for the management of S. aureus infections.
Additional files
The sequences of ASyycG and eGFP were synthesized by Sangon Biotech (Shanghai, China) and were inserted into BamHI and EcoRI restriction sites of a pDL278 vector. The initial sites of eGFP were underlined and bold in red. (DOCX 14 kb)
Acknowledgements
The authors would like to give their special thanks to Huiqi Xie for her skilled technical assistance.
Abbreviations
- AFM
Atomic force microscopy
- CLSM
Confocal laser scanning microscope
- CV
Crystal violet
- EPS
Extracellular polymeric substances
- GO
Graphene oxide
- PIA
Polysaccharide intercellular adhesion
- S. aureus
Staphylococcus aureus
- SEM
Scanning electron microscopy
Authors’ contributions
Setting up the research was done by LL and ZH. Experiment section was major done by WSZ and YJL. Statistical analysis was done by WSZ and LL. Manuscript preparation was done by WSZ. Supervising was done by ZH and LL. All authors read and approved the final manuscript.
Funding
This study was supported by National Natural Science Foundation of China (no. 81800964) and Sichuan Provincial Natural Science Foundation of China (nos. 2018SZ0125, and 2019YFS0270)
Availability of data and materials
All data generated or analyzed during this study are included in this published article and its supplementary information files.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Shizhou Wu, Email: wushizhou120@163.com.
Yunjie Liu, Email: 675357414@qq.com.
Hui Zhang, Phone: +86 18980601437, Email: caesarzh@163.com.
Lei Lei, Phone: +86 18780229047, Email: leilei@scu.edu.cn.
References
- 1.van Belkum A, Verkaik NJ, de Vogel CP, Boelens HA, Verveer J, Nouwen JL, Verbrugh HA, Wertheim HF. Reclassification of Staphylococcus aureus nasal carriage types. J Infect Dis. 2009;199:1820–1826. doi: 10.1086/599119. [DOI] [PubMed] [Google Scholar]
- 2.Boada A, Pons-Vigués M, Real J, Grezner E, Bolíbar B, Llor C. Previous antibiotic exposure and antibiotic resistance of commensal Staphylococcus aureus in Spanish primary care. Eur J Gen Pract. 2018;24:125–130. doi: 10.1080/13814788.2018.1444748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Harkins CP, Pichon B, Doumith M, Parkhill J, Westh H, Tomasz A, de Lencastre H, Bentley SD, Kearns AM, Holden MTG. Methicillin-resistant Staphylococcus aureus emerged long before the introduction of methicillin into clinical practice. Genome Biol. 2017;18:130. doi: 10.1186/s13059-017-1252-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Stefani S, Chung DR, Lindsay JA, Friedrich AW, Kearns AM, Westh H, Mackenzie FM. Meticillin-resistant Staphylococcus aureus (MRSA): global epidemiology and harmonisation of typing methods. Int J Antimicrob Agents. 2012;39:273–282. doi: 10.1016/j.ijantimicag.2011.09.030. [DOI] [PubMed] [Google Scholar]
- 5.Zarafu I, Turcu I, Culiță DC, Petrescu S, Popa M, Chifiriuc MC, Limban C, Telehoiu A, Ioniță P. Antimicrobial features of organic functionalized graphene-oxide with selected amines. Materials (Basel). 2018;11:E1704. doi: 10.3390/ma11091704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liu Y, Yuan C, Cheng Y, Yao G, Xie L, Xu B. Graphene oxide affects growth and resistance to Sclerotinia sclerotiorum in Brassica napus L. J Nanosci Nanotechnol. 2018;18:8345–8351. doi: 10.1166/jnn.2018.16398. [DOI] [PubMed] [Google Scholar]
- 7.Di Santo R, Digiacomo L, Palchetti S, Palmieri V, Perini G, Pozzi D, Papi M, Caracciolo G. Microfluidic manufacturing of surface-functionalized graphene oxide nanoflakes for gene delivery. Nanoscale. 2019;11:2733–2741. doi: 10.1039/C8NR09245A. [DOI] [PubMed] [Google Scholar]
- 8.Feng L, Zhang S, Liu Z. Graphene based gene transfection. Nanoscale. 2011;3:1252–1257. doi: 10.1039/c0nr00680g. [DOI] [PubMed] [Google Scholar]
- 9.Fukuchi K, Kasahara Y, Asai K, Kobayashi K, Moriya S, Ogasawara N. The essential two-component regulatory system encoded by yycF and yycG modulates expression of the ftsAZ operon in Bacillus subtilis. Microbiology. 2000;146:1573–1583. doi: 10.1099/00221287-146-7-1573. [DOI] [PubMed] [Google Scholar]
- 10.Dubrac S, Msadek T. Tearing down the wall: peptidoglycan metabolism and the WalK/WalR (YycG/YycF) essential two-component system. Adv Exp Med Biol. 2008;631:214–228. doi: 10.1007/978-0-387-78885-2_15. [DOI] [PubMed] [Google Scholar]
- 11.Flemming HC, Wingender J, Szewzyk U, Steinberg P, Rice SA, Kjelleberg S. Biofilms: an emergent form of bacterial life. Nat Rev Microbiol. 2016;14:563–575. doi: 10.1038/nrmicro.2016.94. [DOI] [PubMed] [Google Scholar]
- 12.O’Gara JP. ica and beyond: biofilm mechanisms and regulation in Staphylococcus epidermidis and Staphylococcus aureus. FEMS Microbiol Lett. 2007;270:179–188. doi: 10.1111/j.1574-6968.2007.00688.x. [DOI] [PubMed] [Google Scholar]
- 13.Saberi F, Kamali M, Najafi A, Yazdanparast A, Moghaddam MM. Natural antisense RNAs as mRNA regulatory elements in bacteria: a review on function and applications. Cell Mol Biol Lett. 2016;21:6. doi: 10.1186/s11658-016-0007-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bai H, Xue X, Hou Z, Zhou Y, Meng J, Luo X. Antisense antibiotics: a brief review of novel target discovery and delivery. Curr Drug Discov Technol. 2010;7:76–85. doi: 10.2174/157016310793180594. [DOI] [PubMed] [Google Scholar]
- 15.Zhang Y, Ma W, Zhu Y, Shi S, Li Q, Mao C, Zhao D, Zhan Y, Shi J, Li W, Wang L, Fan C, Lin Y. Inhibiting methicillin-resistant Staphylococcus aureus by tetrahedral DNA nanostructure-enabled antisense peptide nucleic acid delivery. Nano Lett. 2018;18:5652–5659. doi: 10.1021/acs.nanolett.8b02166. [DOI] [PubMed] [Google Scholar]
- 16.Lei L, Stipp RN, Chen T, Wu SZ, Hu T, Duncan MJ. Activity of Streptococcus mutans VicR Is Modulated by Antisense RNA. J Dent Res. 2018;97:1477–1484. doi: 10.1177/0022034518781765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wu S, Huang F, Zhang H, Lei L. Staphylococcus aureus biofilm organization modulated by YycFG two-component regulatory pathway. J Orthop Surg Res. 2019;14:10. doi: 10.1186/s13018-018-1055-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wu S, Liu Y, Zhang H, Lei L. The susceptibility to calcium hydroxide modulated by the essential walR gene reveals the role for Enterococcus faecalis biofilm aggregation. J Endod. 2019;45:295–301. doi: 10.1016/j.joen.2018.11.011. [DOI] [PubMed] [Google Scholar]
- 19.Lei L, Yang Y, Mao M, Li H, Li M, Yang Y, Yin J, Hu T. Modulation of biofilm exopolysaccharides by the Streptococcus mutans vicX Gene. Front Microbiol. 2015;6:1432. doi: 10.3389/fmicb.2015.01432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wu Shizhou, Liu Yunjie, Lei Lei, Zhang Hui. Antisense yycG Regulation of Antibiotic Sensitivity of Methicillin-Resistant Staphylococcus aureus in Chronic Osteomyelitis. Surgical Infections. 2019;20(6):472–479. doi: 10.1089/sur.2019.016. [DOI] [PubMed] [Google Scholar]
- 21.Bessa LJ, Ferreira M, Gameiro P. Evaluation of membrane fluidity of multidrug-resistant isolates of Escherichia coli and Staphylococcus aureus in presence and absence of antibiotics. J Photochem Photobiol B. 2018;181:150–156. doi: 10.1016/j.jphotobiol.2018.03.002. [DOI] [PubMed] [Google Scholar]
- 22.Quijano E, Bahal R, Ricciardi A, Saltzman WM, Glazer PM. Therapeutic peptide nucleic acids: principles, limitations, and opportunities. Yale J Biol Med. 2017;90:583–598. [PMC free article] [PubMed] [Google Scholar]
- 23.Dou C, Ding N, Luo F, Hou T, Cao Z, Bai Y, Liu C, Xu J, Dong S. Graphene-based microRNA transfection blocks preosteoclast fusion to increase bone formation and vascularization. Adv Sci (Weinh). 2017;5:1700578. doi: 10.1002/advs.201700578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Moormeier DE, Bayles KW. Staphylococcus aureus biofilm: a complex developmental organism. Mol Microbiol. 2017;104:365–376. doi: 10.1111/mmi.13634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Archer NK, Mazaitis MJ, Costerton JW, Leid JG, Powers ME, Shirtliff ME. Staphylococcus aureus biofilms: properties, regulation, and roles in human disease. Virulence. 2011;2:445–459. doi: 10.4161/viru.2.5.17724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jenkins A, Diep BA, Mai TT, Vo NH, Warrener P, Suzich J, Stover CK, Sellman BR. Differential expression and roles of Staphylococcus aureus virulence determinants during colonization and disease. MBio. 2015;6:e02272–e02214. doi: 10.1128/mBio.02272-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The sequences of ASyycG and eGFP were synthesized by Sangon Biotech (Shanghai, China) and were inserted into BamHI and EcoRI restriction sites of a pDL278 vector. The initial sites of eGFP were underlined and bold in red. (DOCX 14 kb)
Data Availability Statement
All data generated or analyzed during this study are included in this published article and its supplementary information files.