Abstract
Mitochondria are iconic structures in biochemistry and cell biology, traditionally referred to as the powerhouse of the cell due to a central role in energy production. However, modern-day mitochondria are recognized as key players in eukaryotic cell biology and are known to regulate crucial cellular processes, including calcium signalling, cell metabolism and cell death, to name a few. In this review, we will discuss foundational knowledge in mitochondrial biology and provide snapshots of recent advances that showcase how mitochondrial function regulates other cellular responses.
Keywords: mitochondria, mitochondrial biogenesis, metabolism
1. Introduction
All modern-day eukaryotes are believed to have arisen from a primordial ancestor that engulfed an α-protobacterium with the capacity for respiration [1]. This event gave rise to modern-day mitochondria, an event that is now deeply integrated in eukaryotic cell homeostasis and survival. Mitochondria are dynamic networks capable of remodelling their morphology and activity. They provide energy and biomolecules for the cell, in addition to contributing to pathways of cell stress, immune responses, intra- and intercellular signalling, cell-cycle control and cell death. The unique biology of mitochondria underpins their influence on the cell and the ability to calibrate their structure and proteome is an efficacious means of adapting their function. As such, we will begin with a brief outline of three fundamental concepts in mitochondrial biology: (i) mitochondrial ultrastructure; (ii) mitochondrial protein import; and (iii) mitochondrial dynamics. This will inform subsequent discussion of mitochondria as key players in broad and diverse roles, including metabolism, signal transduction, immunity, cell cycle, cell differentiation, cell death and stress.
2. Mitochondrial ultrastructure, dynamics and protein import
2.1. Mitochondrial ultrastructure
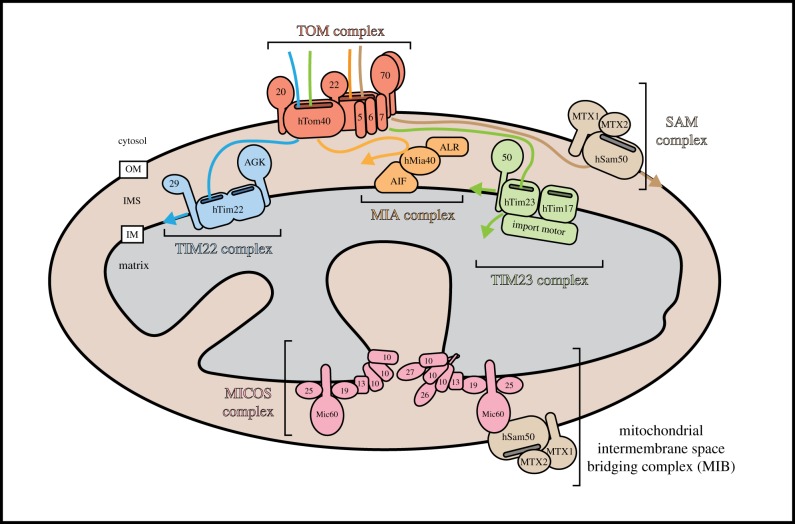
Mitochondria have a double membrane that defines four compartments: the outer membrane, the intermembrane space, the inner membrane and the matrix. The architecture of the inner membrane is malleable and typically convoluted into folded invaginations, called cristae, that dictate the spatial arrangement of proteins [2]. Remodelling cristae structure of cristae can also alter enzymatic flux between the compartments, consistent with the diverse cristae structures observed across cell types with different metabolic demands [2]. The recently described MICOS complex (mitochondrial contact site and cristae organizing system) is required to maintain cristae morphology [3] (figure 1). Loss of MICOS assembly ablates cristae junctions and manifests severe defects in energy metabolism, calcium handling and lipid trafficking [4]. However, it remains unclear how MICOS is regulated by cellular conditions to produce diverse cristae morphologies. Interestingly, disruption of MICOS alters the activity and/or abundance of mitochondrial morphology proteins [5,6]. Perturbations to organelle function have long been associated with gross morphological changes in the mitochondrial network, therefore cristae reorganization by MICOS assembly/disassembly may be an intermediary between function and dynamics. Recently identified associations between MICOS and protein import complexes point to the broad influence of MICOS on mitochondrial function [7,8].
Figure 1.
Nuclear-encoded mitochondrial proteins are imported by multi-subunit translocases. Mitochondrial proteins synthesized in the cytosol are imported into mitochondria post-translationally. The TOM complex at the outer membrane serves as a general protein entry gate. hTom40 forms the pore of the translocase, while hTom20, hTom22 and hTom70 function as receptors. hTom22 plays an additional role in the assembly of the complex. hTom5, hTom6 and hTom7, collectively called the small TOMs, regulate the dynamics and assembly of the complex. The TIM22 complex at the inner membrane mediates the import of multi-pass transmembrane proteins into the inner membrane. hTim22 forms the pore through which proteins are inserted, while AGK and hTim29 function as receptors and in complex assembly. The TIM23 complex can translocate precursor proteins into the matrix or the inner membrane. hTim23 and hTim17 form the channel pore, and hTim50 functions as a receptor for precursors. The core complex associates with an import motor that helps to translocate proteins into the matrix in an ATP-dependent manner. The MIA complex mediates the import of soluble intermembrane space proteins by catalysing the formation of disulfide bonds. hMia40 carries out the disulfide bond formation and is anchored to the inner membrane through an interaction with AIF. ALR removes electrons from hMia40 so that it can undergo further rounds of catalysis. The SAM complex of the outer membrane mediates insertion of β-barrel proteins into the outer membrane. hSam50 associates with MTX1 and MTX2. Cristae, the large invaginations of the inner mitochondrial membrane, are stabilized by a multi-subunit complex called MICOS. Mic60 is the core subunit of MICOS, which additionally contains Mic10, Mic13, Mic14, Mic19, Mic25, Mic26 and Mic27. MICOS also associates with the SAM complex at the outer membrane to form a structure known as the mitochondrial intermembrane space bridging complex (MIB).
(Recommended further reading on cristae, MICOS and ultrastructure: [2,9,10].)
2.2. Mitochondrial protein import
From their endosymbiont origins, human mitochondria have retained only 37 genes in a small circular genome known as the mitochondrial DNA (mtDNA), which encodes 13 polypeptides, 22 tRNAs and 2 rRNAs. The remaining 1000–1500 mitochondrial proteins are nuclear encoded and must be imported and sorted to the relevant mitochondrial compartment following synthesis in the cytosol. Fundamentally, mitochondrial protein import is mediated by multimeric protein complexes known as translocases, which are located at mitochondria (figure 1). Briefly, two major translocases reside in the outer membrane of mitochondria: the Translocase of the Outer Membrane (TOM) complex and the Sorting and Assembly Machinery (SAM). The TOM complex is the initial point of contact for almost all mitochondrial precursors and provides a means of entry into the organelle. Following translocation through TOM, precursor import pathways diverge based on their targeting information and ultimate location within the organelle. β-barrel proteins of the outer membrane are sorted to the SAM complex for integration into the membrane. There are two translocases embedded in the inner membrane of mitochondria: the Translocase of the Inner Membrane (TIM) 22 and 23 (TIM22 and TIM23) complexes. TIM22 mediates the insertion of non-cleavable polytopic membrane proteins into the inner membrane, while the TIM23 complex is responsible for importing precursors across the inner membrane into the matrix or in some instances can laterally release transmembrane precursors into the inner membrane. Finally, the Mitochondrial Intermembrane space Assembly (MIA) machinery mediates the import of small cysteine-rich intermembrane space proteins and couples their import to their oxidation [11]. These import pathways and machines have been predominately characterized in fungal organisms; however, in more recent years, analysis in higher eukaryotes has uncovered important physiological consequences due to perturbations in protein import. Specifically, mutations in genes encoding protein import subunits cause distinct mitochondrial diseases with phenotypes ranging from severe muscular defects to neurodegeneration and congenital growth defects [12].
(Recommended further reading on mitochondrial protein import: [13–15].)
2.3. Mitochondrial dynamics: fission, fusion and organelle contact sites
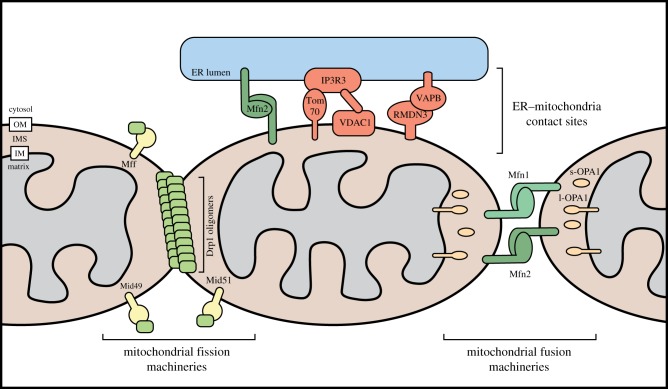
As an organellar network, mitochondria undergo fission and fusion to replicate, be recycled, and alter their bioenergetics. Fusion of the outer membrane is mediated by homotypic interactions between the GTPases Mfn1 and Mfn2 on adjacent mitochondria (figure 2) [16], but the domains involved and stepwise mechanism of fusion are still debated. Fusion of the inner membrane is controlled by Opa1, which exists as five isoforms generated by mRNA splicing and proteolytic cleavage (figure 2) [17]. It is believed that the stoichiometry of these isoforms governs Opa1 interactions with the mitochondrial-specific lipid cardiolipin and, subsequently, fusion events [18]. Mitochondrial fusion is associated with increased ATP production by oxidative phosphorylation and protects against oxidative and proteostatic stress [19]. Conversely, mitochondrial fission is concomitant with a reliance on glycolysis and precedes mitochondrial turnover. Fission is largely dependent on the dynamin-related and cytosolic protein Drp1, which oligomerizes around and constricts mitochondrial tubules (figure 2). The recruitment of Drp1 from the cytosol requires adaptor proteins on the mitochondrial outer membrane, including Mff, Mid49 and Mid51 [20,21], although human Fis1 can promote Drp1-independent mitochondrial fragmentation through inhibition of fusion proteins [22] (figure 2). While conflicting models of Drp1 recruitment have been proposed, its localization and activity are known to be regulated by numerous post-translational modifications [23]. The scission ability of Drp1 oligomers is sterically limited to tubules up to 250 nm diameter, indicating pre-constriction is required for larger mitochondria [24]. This is achieved by the endoplasmic reticulum (ER), which wraps around and constrict tubules to mark future fission sites and aid correct partitioning of mitochondrial contents [25,26].
Figure 2.
Cellular machineries mediating mitochondrial fission, fusion and formation of contact sites with the endoplasmic reticulum. Mitochondria continuously undergo fission and fusion. Fission is mediated by the GTPase Drp1, which can be recruited to the outer mitochondrial membrane by a variety of receptors, including Mff, Fis1, Mid49 and Mid51. Drp1 at the outer membrane can oligomerize into fibrils that constrict mitochondria to initiate fission. Mitochondrial fusion is initiated by tethering of mitochondria through homotypic interactions between Mfn1 and Mfn2 on opposing mitochondria. Inner membrane fusion is mediated by OPA1, which exists as long and short forms generated through proteolysis. Contact sites between the mitochondria and the endoplasmic reticulum (ER) are established and maintained through protein–protein interactions. Interactions occur between Mfn2 molecules on the ER membrane and the outer mitochondrial membrane, and between VAPB on the ER membrane and RMDN3 on the mitochondrial outer membrane. Interactions also occur between IP3R3, a calcium channel on the ER membrane, and VDAC1 and hTom70 on the mitochondrial outer membrane.
Mitochondria also engage in extensive dynamic inter-organelle contacts that coordinate functional exchanges between mitochondria and other cellular components [27]. In particular, ER–mitochondria contact sites (ERMCs) facilitate a multitude of functions including mitochondrial fission, coenzyme Q biosynthesis, lipid transfer, Ca2+ transfer, mtDNA replication and autophagy [25,26,28–31]. The ER–mitochondria encounter structure (ERMES) has been well characterized in Saccharomyces cerevisiae [32], however no human equivalent has been identified [33]. Preliminary work in humans suggests that metazoan ERMCs are tethered by interactions between hTom70 and IP3R3, VDAC1 and IP3R3, RMDN3 and VAPB, Mfn2 homodimers, Vps13a and Pdzd8 with an unknown partner (figure 2) [34–39]. Furthermore, acetylated microtubule ‘tracks’ have been proposed to maintain these contacts despite the movements and remodelling of the two organellar networks [40]. Other inter-organelle contacts have been described between mitochondria and Golgi [27,41], peroxisomes [42], lysosomes [43], lipid droplets [44] and the plasma membrane [45]. The interconnectivity of mitochondria with cellular components enables significant interplay across various pathways, examples of which we will highlight throughout this review (table 1).
Table 1.
Full names and identifiers of proteins discussed in this review.
| section | protein name |
gene | accession (NCBI; UniProt) | function(s) | |
|---|---|---|---|---|---|
| abbreviation | full name(s) | ||||
| mitochondrial dynamics | Mfn1 | Mitofusin 1 | MFN1 | 55669; Q8IWA4 | outer membrane fusion |
| Mfn2 | Mitofusin 2 | MFN2 | 9927; O95140 | outer membrane fusion; ER–mitochondria contact | |
| Opa1 | OPA1 mitochondrial dynamin-like GTPase | OPA1 | 4976; O60313 | inner membrane fusion | |
| Drp1 | dynamin-1-like protein; Drp1 | DNM1L | 10059; O00429 | mitochondrial fission | |
| Fis1 | mitochondrial fission protein 1 | FIS1 | 51024; Q9Y3D6 | mitochondrial fission | |
| Mff | mitochondrial fission factor | MFF | 56947; Q9GZY8 | mitochondrial fission | |
| Mid51 | mitochondrial dynamics protein 51 | MIEF1 | 54471; L0R8F8 | mitochondrial fission | |
| Mid49 | mitochondrial dynamics protein 49 | MIEF2 | 125170; Q96C03 | mitochondrial fission | |
| organelle contact site | hTom70 | translocase of the outer membrane 70 | TOMM70 | 9868; O94826 | protein import; ER–mitochondria contact |
| VDAC1 | voltage-dependent anion channel 1 | VDAC1 | 7416; P21796 | ER–mitochondria contact; ion permeability | |
| IP3R3 | inositol 1,4,5-trisphosphate receptor type 3 | ITPR3 | 3710; Q14573 | ER contact sites; calcium transport | |
| RMDN3 | regulator of microtubule dynamics protein 3 | RMDN3 | 55177; Q96TC7 | ER contact sites; calcium transport | |
| VAPB | VAMP associated protein B and C | VAPB | 9217; O95292 | ER contact sites | |
| Vps13a | vacuolar protein sorting 13 homolog A | VPS13A | 23230; Q96RL7 | ER contact sites; lipid transfer | |
| Pdzd8 | PDZ containing 8 | PDZD8 | 118987; Q8NEN9 | ER contact sites; calcium transport | |
| metabolism | mTOR | mechanistic target of rapamycin kinase; serine/threonine protein kinase mammalian target of rapamycin | MTOR | 2475; P42345 | metabolic regulation; cell growth |
| p53 | tumour protein 53 | TP53 | 7157; P04637 | metabolic regulation; cell survival | |
| calcium homeostasis | SLC8A3 | solute carrier family 8 member A3 | SLC8A3 | 6547; P57103 | calcium transport |
| MCU | mitochondrial calcium uniporter | MCU | 90550; Q8NE86 | calcium transport | |
| MICU1 | mitochondrial calcium uptake 1 | MICU1 | 10367; Q9BPX6 | calcium transport, regulation | |
| MICU2 | mitochondrial calcium uptake 2 | MICU2 | 221154; Q8IYU8 | calcium transport, regulation | |
| hMia40 | coiled-coil–helix–coiled-coil–helix domain containing 4; mitochondrial intermembrane space import and assembly 40 homolog | CHCHD4 | 131474; Q8N4Q1 | protein import; calcium transport regulation | |
| immune signalling | MAVS | mitochondrial antiviral signalling protein | MAVS | 57506; Q7Z434 | immune signalling |
| TRADD | TNFRSF1A associated via death domain | TRADD | 8717; Q15628 | immune signalling | |
| TRAF3 | TNF receptor-associated factor 3 | TRAF3 | 7187; Q13114 | immune signalling | |
| STING | transmembrane protein 173; stimulator of interferon genes | TMEM173 | 340061; Q86WV6 | immune signalling | |
| IRF3 | interferon regulatory factor 3 | IRF3 | 3661; Q14653 | immune signalling | |
| IRF7 | interferon regulatory factor 7 | IRF7 | 3665; Q92985 | immune signalling | |
| NLRX1 | NLR family member X1 | NLRX1 | 79671; Q86UT6 | immune signalling | |
| NLRP3 | NLR family pyrin domain containing 3 | NLRP3 | 114548; Q96P20 | immune signalling | |
| IL-1β | interleukin 1 beta | IL1B | 3553; P011584 | immune signalling | |
| cell differentiation | Ras | K-Ras proto-oncogene, GTPase | KRAS | 3845; P01116 | cell proliferation |
| Raf | Raf-1 proto-oncogene, serine/threonine kinase | RAF1 | 5894; P04049 | cell proliferation | |
| Pdk2 | pyruvate dehydrogenase kinase 2 | PDK2 | 5164; Q15119 | metabolism regulation | |
| Oct4 | POU class 5 homeobox | POU5F1 | 5460; Q01860 | stem cell differentiation | |
| Sox2 | SRY-box transcription factor 2 | SOX2 | 6657; P48431 | stem cell differentiation | |
| Nanog | Nanog homeobox | NANOG | 79923; Q9H9S0 | stem cell differentiation | |
| ZFP42 | ZFP42 zinc finger protein | ZFP42 | 132625; Q96MM3 | stem cell pluripotency | |
| cell death | Bax | BCL2 associated X, apoptosis regulator | BAX | 581; Q07812 | intrinsic apoptosis |
| Bak | BCL2 antagonist/killer1 | BAK1 | 578; Q16611 | intrinsic apoptosis | |
| Cyt c | cytochrome c, somatic | CYCS | 54205; P99999 | intrinsic apoptosis | |
| Diablo | Diablo IAP-binding mitochondrial protein | DIABLO | 56616; Q9NR28 | intrinsic apoptosis | |
| Htra2 | HtrA serine peptidase 2 | HTRA2 | 27429; O43464 | intrinsic apoptosis | |
| EndoG | endonuclease G | ENDOG | 2021; Q14249 | caspase-independent apoptosis | |
| AIF | apoptosis-inducing factor mitochondria associated 1 | AIFM1 | 9131; O95831 | caspase-independent apoptosis | |
| VDAC2 | voltage-dependent anion channel 2 | VDAC2 | 7417; P45880 | intrinsic apoptosis; ion permeability | |
| mitochondrial quality control | ATF5 | activating transcription factor 5 | ATF5 | 22809; Q9Y2D1 | mitochondrial unfolded protein response |
| ATF4 | activating transcription factor 4 | ATF4 | 468; P18848 | integrated stress response | |
| hTim17a | translocase of inner mitochondrial membrane 17A | TIMM17A | 10440; Q99595 | protein import; mitochondrial stress response | |
| MCL1 | MCL1 apoptosis regulator, BCL2 family member; myeloid cell leukaemia 1 | MCL1 | 4170; Q07820 | intrinsic apoptosis | |
| PINK1 | PTEN-induced kinase 1 | PINK1 | 65018; Q9BXM7 | mitophagy | |
| PARKIN | Parkin RBR E3 ubiquitin protein ligase | PRKN | 5071; O60260 | mitophagy | |
| PARL | presenilin-associated rhomboid like | PARL | 55486; Q9H300 | mitophagy | |
| hTom22 | translocase of outer mitochondrial membrane 22 | TOMM22 | 56993; Q9NS69 | protein import; mitophagy | |
| FUNDC1 | FUN14 domain containing 1 | FUNDC1 | 139341; Q8IVP5 | mitophagy | |
| BCL2L13 | BCL2-like 13 | BCL2L13 | 23786; Q9BXK5 | mitophagy | |
(Recommended further reading on mitochondrial dynamics: [46–48]; on organelle contacts: [49–51].)
3. Mitochondria and metabolism
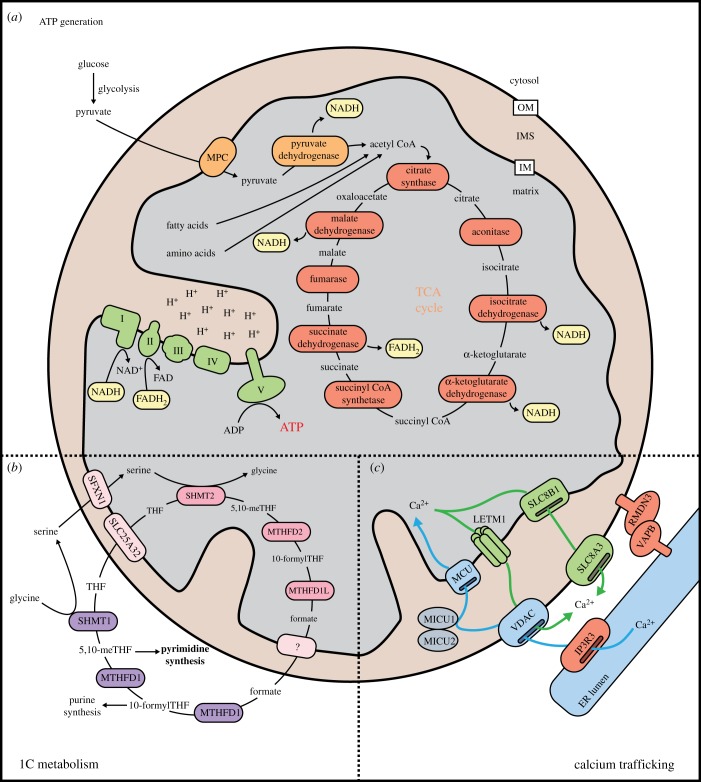
Mitochondria are well known for providing energy to the cell, predominantly by coupling the tricarboxylic acid (TCA) cycle with oxidative phosphorylation. The TCA cycle is a series of eight enzymatic reactions that occur in the matrix to harvest electrons from citrate and its catabolic intermediates (figure 3a). The typical input to the cycle is acetyl-CoA, which can be produced from glucose (via glycolysis), fatty acids (via β-oxidation) and amino acids (via deamination) (figure 3a). The electrons scavenged throughout the cycle are transferred by NADH and FADH2 to the complexes of the electron transport chain. Complexes I–IV of the electron transport chain shuttle electrons, using their energy to pump protons into the intermembrane space and establish an electrochemical gradient across the inner membrane. Complex V (ATP synthase) releases the protons back into the matrix, using the energy of the electrochemical gradient to produce ATP, the cell's energy currency (figure 3a) [52]. Although normally efficient, oxidative phosphorylation is negatively regulated by the accumulation of its toxic by-product, reactive oxygen species (ROS). If unchecked, ROS can cause damage to mitochondria, induce protein aggregation and introduce mutations in DNA [53–55]. Recent advances in cryoelectron microscopy have revealed Complexes I, III and IV can assemble to form supercomplexes thought to reduce the amount of ROS produced during electron transport, as well as enhance respiration rates [56].
Figure 3.
Mitochondria coordinate essential metabolic processes. (a) Mitochondria are best known for housing the protein machinery required for generating ATP. When oxygen is available, most cells will generate ATP through oxidative phosphorylation, where electrons harvested through catabolic reactions are used to power ATP synthase. Electrons are obtained through the TCA cycle, which occurs in the matrix and consists of eight enzymatic reactions. Acetyl-CoA is the primary input for the TCA cycle, and can be obtained through metabolism of glucose, fatty acids and amino acids. Electrons extracted during the TCA cycle are loaded onto NAD+ and FAD2+. Electrons are subsequently transferred from NADH and FADH2 onto Complexes I and II of the electron transport chain. Electrons are passed through Complexes III and IV, which transport protons into the intermembrane space. Protons are allowed to flow back into the matrix through ATP synthase (Complex V), which uses the energy of the proton gradient to convert ADP to ATP. (b) Mitochondrial one-carbon (1C) metabolism comprises a series of parallel and reversible reactions which occur in the cytosol and mitochondrial matrix. In proliferating cells, the reaction normally proceeds in a specific direction such that formate produced within mitochondria can be used for biosynthetic processes in the cytosol. Within the mitochondria, THF and serine imported from the cytosol are acted upon sequentially by SHMT2, MTHFD2 and MTHFD1 L to produce formate, which is exported back into the cytosol. Cytosolic MTHFD1 loads formate onto THF to form charged folate intermediates that can be used to synthesize purine and pyrimidine nucleotides. Mitochondrial 1C metabolism is also an important source of glycine. (c) The mitochondrial matrix functions as an important storage site for calcium ions. Mitochondrial calcium uptake often occurs at ER contact sites, where large volumes of Ca2+ can be released through IP3R3. Calcium can pass freely through the outer membrane via VDAC channels and is transported across the intermembrane space and inner membrane through the coordinated function of a MICU1/MICU2 dimer docking to MCU in the inner membrane. Calcium can exit the mitochondrial matrix through LETM1 or SLC8B1 (in exchange for H+ or Na+, respectively) and can cross the outer membrane through VDACs or NCX3.
Mitochondria also produce fatty acids, amino acids, nucleotides and haem groups for the cell through biosynthetic pathways [57–59]. One such process, one-carbon (1C) metabolism, produces glycine, methionine, nucleotides, phosphatidylcholine and 1C units (methyl-like groups) from serine catabolism through the redox chemistry of folate and its derivatives (figure 3b) [60]. These 1C units charge the universal methyl donor S-adenosylmethionine required for the methylation of proteins and chromatin [61]. There is now significant evidence of metabolic enzymes and metabolites altering gene expression as reporters of environmental conditions (nutrient availability, hypoxia, oxidative stress) or mitochondrial dysfunction. This has been shown for acetyl-CoA, TCA intermediates, ketones, lactate, fatty acids and amino acids [62–68]. Emerging studies also indicate cellular nutrient and energy sensing by mTOR kinase regulates mitochondrial biogenesis and protein synthesis [69]. Through downstream effectors of transcription and translation, mTORC1 stimulates mitochondrial biogenesis and oxidative metabolism to meet the energy demand of anabolism [70–72]. Interestingly, the tumour suppressor p53 inhibits mTOR-mediated growth and proliferation to prevent oncogenesis [73,74]. p53 activity increases electron transport chain efficacy [75], mtDNA stability [76,77] and reduced glutathione (GSH) levels [78] to limit ROS production as well as inhibiting glycolysis [79,80], which contributes to the replicative potential of tumour cells [79,81,82]. Thus, metabolism is intimately integrated with other cellular pathways, but is not the sole contribution of mitochondria to signalling mechanisms.
(Recommended further reading on metabolism: [60,83]; on metabolite signalling: [68,84]; on mTOR/p53: [85,86].)
4. Signalling
4.1. Mitochondria control calcium homeostasis
Calcium ions are common to diverse signalling pathways. The outer mitochondrial membrane is permeable to Ca2+, in part due to channel-forming VDAC proteins [87] and export via SLC8A3 [88]. The mitochondrial inner membrane calcium uniporter (MCU) complex regulates transport into the matrix (figure 3c). Permeability of the MCU complex is calibrated by two regulatory subunits, MICU1 and MICU2, that are linked by an intermolecular disulfide bond introduced by hMia40 [89,90]. The ability of mitochondria to accumulate Ca2+ up to 20-fold higher concentrations than the cytosol allows them to function as buffering systems and re-establish homeostasis following Ca2+ bursts [91,92]. Bursts of Ca2+ into the cytosol, from across the plasma membrane or intracellular stores, can initiate neurotransmitter release, muscle fibre contraction and transcriptional regulation. In neurons, mitochondrial Ca2+ buffering modulates both the propensity and duration of neurotransmitter release [93,94]. In cardiac muscle, contraction is coupled to enhanced mitochondrial ATP production via Ca2+-increased activities of TCA cycle enzymes, Complex V and the ADP/ATP transporter [95–98]; an effect maximized by local Ca2+ concentrations at ERMCs [29,99] (figure 3c). Additionally, mitochondrial Ca2+ regulation influences hormone secretion [100], tissue regeneration [101] and interferon-β signalling via the mitochondrial antiviral signalling protein, MAVS [102].
(Recommended further reading on mitochondrial Ca2+ signalling: [92,103,104].)
4.2. Roles of mitochondria in immune responses
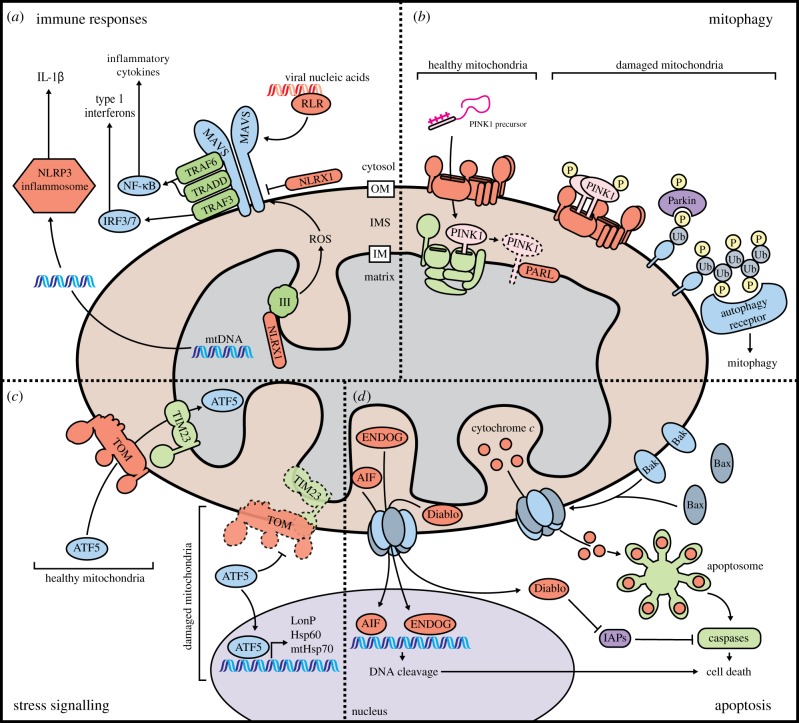
The contribution of mitochondria to immune responses is a growing area of research. Cell-autonomous immune signalling is driven by MAVS at the outer membrane, which acts as a relay point for immune signal transduction. Rig-like receptors in the cytosol undergo conformational changes upon detecting viral RNA or DNA and are recruited to MAVS, particularly at ERMCs [105]. MAVS then dimerizes to enable the binding of multiple downstream signalling adaptors including TRADD, TRAF3 and STING to activate NF-κB and IRF-3/7 transcription of interleukins and pro-inflammatory cytokines [106–108] (figure 4a). Interestingly, MAVS dimers and many of its adaptors co-immunoprecipitate with hTom70 of the TOM complex, the overexpression of which increases the signalling response [109]. MAVS signalling is also affected by ROS and negatively regulated by Nlrx1, a binding partner of Complex III and MAVS [110,111] (figure 4a). As mitochondrial protein import and oxidative metabolism can be hijacked by virulence factors [112], these interactions may make MAVS sensitive to consequences of infection. Finally, if mitochondria are compromised by infection, the increased ROS and release of mtDNA into the cytosol can activate the NLRP3 inflammasome to evoke an inflammatory response [113,114] (figure 4a).
Figure 4.
Mitochondria make crucial contributions to diverse cellular processes. (a) The mitochondrial outer membrane is the site of important signalling events during the innate immune response. Detection of viral nucleic acids by Rig-like receptors (RLRs) induces dimerization of MAVS, a protein of the mitochondrial outer membrane. Dimerized MAVS recruits signalling adaptors that initiate downstream activation of IRF3/7 and NF-κB, transcription factors that induce expression of type I interferons and pro-inflammatory cytokines. MAVS is regulated by NLRX1, a protein which downregulates MAVS when localized to the outer membrane, but activates MAVS when at the inner membrane by interacting with Complex III to induce ROS production. Release of mtDNA during infection can also activate the NLRP3 inflammasome. (b) Mitophagy is a process that allows damaged mitochondria to be identified and destroyed. Under normal conditions, PINK1 is imported into mitochondria and degraded by PARL. When mitochondria are damaged, import is impaired and PINK1 accumulates in the TOM complex at the outer membrane. Autophosphorylated and active PINK1 at the outer membrane phosphorylates monoubiquitin molecules on outer membrane proteins, recruiting and activating the E3 ubiquitin ligase Parkin. Activated Parkin synthesizes polyubiquitin chains that recruit autophagy receptors to initiate mitophagy. (c) Mitochondrial proteostatic stress is sensed through the partitioning of the transcription factor ATF5 between the mitochondria and the nucleus. Under normal conditions, ATF5 is imported into and sequestered within mitochondria. If mitochondrial protein import becomes compromised, ATF5 is trafficked into the nucleus, where it upregulates expression of genes that enhance proteostasis. (d) Mitochondria play crucial roles in the initiation of apoptosis. In response to pro-apoptotic stimuli, Bax and Bak oligomerize in the outer membrane to form pores that allow for efflux of apoptogenic proteins (Cytochrome c, Diablo, AIF and Endonuclease G) from the intermembrane space into the cytosol. Cytochrome c binds to Apaf-1 to induce formation of the apoptosome and activation of caspases. Diablo blocks inhibitors of apoptosis (IAPs) which would otherwise mitigate the effect of caspases. AIF and Endonuclease G translocate into the nucleus where they contribute to destruction of the genome.
Mitochondrial metabolism also directs rapid changes to specialized immune cells during infection. Changes in membrane potential can activate or supress M2 macrophages [115,116] and M1 macrophages shunt intermediates from the TCA cycle to generate nitrous oxide, IL-1β and the antibacterial itaconic acid [117,118]. Furthermore, the phagocytic abilities of macrophages depend on mitochondrial ROS production to destroy internalized pathogens [119]. Naive T-cells display increases in mitochondrial mass, mtDNA copy number, glycolysis, and glutamine metabolism during differentiation for rapid proliferation and to escape quiescence [120,121]. Metabolic remodelling then also decides the T-cells' mature fate [122,123], by altering cristae architecture [124] or by direct effect of metabolites on epigenetic transcription regulation [125].
(Recommended further reading on mitochondrial immune signalling: [106,118,126].)
5. Cell cycle, differentiation and death
Mitochondria are implicitly tied to cell-cycle control as providers of energy and nucleotides; however, they also coordinate checkpoints and respond to signals of proliferation. To meet the metabolic demand of mitosis, mitochondrial mass and membrane potential increase from G1/S until late mitotic stage [127]. Indeed, hyperpolarization and increased ATP production inhibit AMP kinase to allow cyclinE-mediated entry to S-phase [128]. In the late G2 stage of dividing S. cerevisiae, the cyclinB1/Cdk1 complex traffics to mitochondria to phosphorylate Complex I subunits and Tom6, stimulating oxidative metabolism both directly and indirectly via increased protein import [129,130]. During mitosis, a highly fused and reticular mitochondrial network progressively fragments to small tubular organelles that segregate in anticipation of cytokinesis [127,131]. Mitochondria can also delay cell-cycle progression to increase their biogenesis [132], because of insufficient nucleotide production [133], or because of ROS accumulation [134]. Moreover, the fusion mediator Mfn2 can sequester both Ras and Raf to inhibit proliferative signalling [135].
Stem cell differentiation also relies on mitochondria as a ‘metabolic switch’. Human embryonic stem cells are glycolytic; however, they develop mature cristae, rapidly replicate mtDNA and increase ATP production upon differentiation [136]. In haematopoietic stem cell differentiation, the downregulation of Pdk2, an inhibitor of pyruvate dehydrogenase, releases suppression of acetyl-coA production and enables oxidative phosphorylation [137]. The subsequent increase in ROS production and oxidative phosphorylation during differentiation drives upregulation of mitochondrial antioxidant proteins by the transcription factors Oct4, Sox2 and Nanog [138]. Mitochondrial fusion is believed to facilitate these metabolic changes, although the importance of specific proteins and fission/fusion balance may be cell-type specific [139–141]. This is supported by somatic cell reprogramming studies showing deletion of Mfn2 permits pluripotency as glycolysis becomes predominant over oxidative phosphorylation [142]; the same effect being achieved by the pluripotency factor ZFP42 activation of Drp1 [143].
If cellular conditions or external insults are too harsh, mitochondria can trigger multiple forms of cell death. Apoptosis, or programmed cell death, can be elicited from extrinsic signalling via the Fas, TRAIL and TNFα receptors or intrinsic insults such as DNA damage, Ca2+ overload, ROS and ER stress [144]. Mitochondria contribute to the extrinsic pathway but are the nexus of the intrinsic apoptotic pathway. In the latter pathway, cytosolic pro-apoptotic Bax oligomerizes with Bak at the outer membrane to permeabilize mitochondria and release pro-apoptotic proteins, including cytochrome c, Diablo, Htra2, Endonuclease G and AIF (figure 4d) [145]. In the cytosol, cytochrome c nucleates the formation of the apoptosome and activation of the caspases that dismantle the cell in an immunologically silent manner. Cytosolic Diablo and Htra2 block inhibitors of caspase activation, which would otherwise protect the cell from basal cytochrome c leakage [146,147]. Endonuclease G and AIF translocate to the nucleus to fragment DNA (figure 4d), AIF first requiring proteolytic cleavage of its transmembrane domain [148–150]. AIF is normally part of the intermembrane space import machinery, or MIA complex, anchoring the oxidoreductase hMia40 to the inner membrane. The outer membrane protein VDAC2 protects against apoptosis by sequestering Bak [151,152], yet new evidence suggests it may be required for Bax-mediated apoptosis [153]. Emerging research also implicates mitochondria in alternate and less-studied cell-death pathways such as ROS-induced necrosis [154], immune-activated necroptosis [155], ferroptosis [156,157] and parthanotosis [158].
(Recommended further reading on mitochondria in the cell cycle: [159,160]; on differentiation [161–163]; on cell death: [164,165].)
6. Mitochondrial quality control
The loss of mitochondrial function has profound negative effects on cellular health; therefore, multiple quality control and stress response mechanisms have evolved. The mitochondrial unfolded protein response (mtUPR) detects proteostatic stress within mitochondria [166]. Central to the mtUPR is the transcription factor ATF5. When stress causes protein import and/or electron transport chain dysfunction ATF5 accumulates in the nucleus to transcribe mitochondrial chaperones and protease genes (figure 4c) [167,168]. The Caenorhabditis elegans homologue ATFS-1 has also been shown to repress translation of the electron transport chain subunit and assembly proteins from both mitochondrial and nuclear genomes [169]. Translation of ATF5 is partly controlled by its homologue ATF4, both of which are upregulated in the integrated stress response (ISR) [170,171]. The ISR can be triggered by ER stress, amino acid starvation or degradation of hTim17A, a TIM23 complex subunit [172,173]. The ISR is characterized by phosphorylation of eIF2α, leading to global reduction of translation and selective induction of cytoprotective genes including pro-survival MCL1 and autophagy proteins. This illustrates the preference for clearance of defective organelles over controlled cell death although the response may alter with cell type or insult [174].
The selective autophagic clearance of mitochondria is termed mitophagy and is controlled by the mitochondrial serine/threonine protein kinase PINK1 and the E3 ubiquitin ligase Parkin. PINK1 is constitutively imported into healthy mitochondria through the TOM complex and laterally released into the inner membrane by TIM23 [175] before cleavage by the PARL protease (figure 4b) [176]. Depolarization of the inner membrane in defective mitochondria prevents import of PINK1, causing it to oligomerize at the outer membrane TOM complex [177], where it becomes auto-phosphorylated [178]. This triggers phospho-PINK1 phosphorylation of basal outer membrane monoubiquitin and recruits Parkin to rapidly poly-ubiquitinate outer membrane proteins for the recruitment of autophagosome factors (figure 4b) [179,180]. Recent data suggest that mitochondria can identify and initiate mitophagy of specific tubules [181], while mitophagy induced by CSNK2/CK2 phosphorylation of hTom22, FUNDC1 and BCL2L13 suggests a potential cytoplasmic influence or pathway [182–185]. Additionally, observations of transcellular mitophagy in astrocytes illustrate much is still unknown in these processes [186].
New stress responses are emerging that demonstrate the reciprocal communication between mitochondria and cytoplasm. Ablation of MIA import pathways in S. cerevisiae activates the proteasome to mitigate mitochondrial precursor accumulation in the cytosol [187]. This correlates with the mammalian, intermembrane space-specific mtUPR (mtUPRIMS) where ERRα transcriptional activity upregulates intermembrane space proteases and activates the proteasome [188,189]; the proteasome being previously shown to degrade unfolded intermembrane space proteins that retrotranslocate to the cytosol [190]. In S. cerevisiae, the proteasome is also engaged by Ubx2 to clear mitochondrial protein precursors arrested during translocation, blocking the TOM complex [191]. Reciprocally, mitochondria can degrade defective proteins to aid cytosolic proteostasis. In S. cerevisiae, cytosolic Vms1 can remove mistranslated mitochondrial precursors from stalled ribosomes and direct their import for intra-mitochondrial degradation [192] and aggregation-prone cytosolic proteins may be imported for intra-mitochondrial degradation if cytosolic Hsp70s fail [193]. Intriguing for further research are reports of lysosomal fusion of mitochondria-derived vesicles enriched for non-natively oxidized proteins [194,195] and the extracellular jettison of aggregates by neurons of C. elegans [196].
(Recommended further reading on mitochondrial quality control: [197–199]; on mitophagy: [200,201].)
7. Concluding remarks
This review illustrates the importance of mitochondria to eukaryotic cellular functions. As mitochondrial biologists we are frequently surprised by novel pathways or protein networks that involve mitochondria and/or mitochondrial proteins. Mitochondrial protein import and structural dynamics provide the means for rapid alterations in activity to facilitate biological responses to signalling molecules, nutrient availability and pathogenic insult. The temporal coordination of mitochondrial energetics and their biosynthetic capacity drives cell proliferation and differentiation. However, the highly reactive biochemistry compartmentalized in the organelle makes it capable of inducing cell death and necessitates quality control mechanisms. An understanding of this interplay between mitochondrial functions and their diverse cellular implications is therefore critical to a comprehensive holistic model of cellular homeostasis and biochemistry. The importance of this is evident in the escalating occurrence of mitochondria in post-genomic medical research [202]. Although mitochondria are undeniably hubs of cellular biochemistry, further fundamental research is required. In particular, elucidating how the mitochondrion regulates and integrates the various pathways it is associated with, in specialized cells/tissue types and in the context of health and in disease, will help uncover the true depth of influence this amazing organelle has on eukaryotic cells.
Supplementary Material
Data accessibility
This article does not contain any additional data.
Competing interests
We declare we have no competing interests.
Funding
We received no funding for this study.
References
- 1.Gray MW, Burger G, Lang BF. 1999. Mitochondrial evolution. Science 283, 1476–1481. ( 10.1126/science.283.5407.1476) [DOI] [PubMed] [Google Scholar]
- 2.Cogliati S, Enriquez JA, Scorrano L. 2016. Mitochondrial cristae: where beauty meets functionality. Trends Biochem. Sci. 41, 261–273. ( 10.1016/j.tibs.2016.01.001) [DOI] [PubMed] [Google Scholar]
- 3.Hoppins S, et al. 2011. A mitochondrial-focused genetic interaction map reveals a scaffold-like complex required for inner membrane organization in mitochondria. J. Cell Biol. 195, 323 ( 10.1083/jcb.201107053) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gödiker J, et al. 2018. QIL1-dependent assembly of MICOS complex–lethal mutation in C19ORF70 resulting in liver disease and severe neurological retardation. J. Hum. Genet. 63, 707–716. ( 10.1038/s10038-018-0442-y) [DOI] [PubMed] [Google Scholar]
- 5.Ding C, et al. 2015. Mitofilin and CHCHD6 physically interact with Sam50 to sustain cristae structure. Sci. Rep. 5, 16064 ( 10.1038/srep16064) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Darshi M, Mendiola VL, Mackey MR, Murphy AN, Koller A, Perkins GA, Ellisman MH, Taylor SS. 2011. ChChd3, an inner mitochondrial membrane protein, is essential for maintaining crista integrity and mitochondrial function. J. Biol. Chem. 286, 2918–2932. ( 10.1074/jbc.M110.171975) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Callegari S, et al. 2019. A MICOS-TIM22 association promotes carrier import into human mitochondria. J. Mol. Biol. 431, 2835–2851. ( 10.1016/j.jmb.2019.05.015) [DOI] [PubMed] [Google Scholar]
- 8.Tang J, et al. 2019. Sam50–Mic19–Mic60 axis determines mitochondrial cristae architecture by mediating mitochondrial outer and inner membrane contact. Cell Death Differ. ( 10.1038/s41418-019-0345-2) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rampelt H, Zerbes RM, van der Laan M, Pfanner N. 2017. Role of the mitochondrial contact site and cristae organizing system in membrane architecture and dynamics. Biochim. Biophys. Acta Mol. Cell Res. 1864, 737–746. ( 10.1016/j.bbamcr.2016.05.020) [DOI] [PubMed] [Google Scholar]
- 10.Kozjak-Pavlovic V. 2017. The MICOS complex of human mitochondria. Cell Tissue Res. 367, 83–93. ( 10.1007/s00441-016-2433-7) [DOI] [PubMed] [Google Scholar]
- 11.Stojanovski D, Bragoszewski P, Chacinska A. 2012. The MIA pathway: a tight bond between protein transport and oxidative folding in mitochondria. Biochim. Biophys. Acta 1823, 1142–1150. ( 10.1016/j.bbamcr.2012.04.014) [DOI] [PubMed] [Google Scholar]
- 12.Frazier AE, Thorburn DR, Compton AG. 2017. Mitochondrial energy generation disorders: genes, mechanisms and clues to pathology. J. Biol. Chem. 294, 5386–5395. ( 10.1074/jbc.R117.809194) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wiedemann N, Pfanner N. 2017. Mitochondrial machineries for protein import and assembly. Annu. Rev. Biochem. 86, 685–714. ( 10.1146/annurev-biochem-060815-014352) [DOI] [PubMed] [Google Scholar]
- 14.Pfanner N, Warscheid B, Wiedemann N. 2019. Mitochondrial proteins: from biogenesis to functional networks. Nat. Rev. Mol. Cell Biol. 20, 267–284. ( 10.1038/s41580-018-0092-0) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wasilewski M, Chojnacka K, Chacinska A. 2017. Protein trafficking at the crossroads to mitochondria. Biochim. Biophys. Acta Mol. Cell Res. 1864, 125–137. ( 10.1016/j.bbamcr.2016.10.019) [DOI] [PubMed] [Google Scholar]
- 16.Santel A, Fuller MT. 2001. Control of mitochondrial morphology by a human mitofusin. J. Cell Sci. 114, 867–874. [DOI] [PubMed] [Google Scholar]
- 17.Ehses S, et al. 2009. Regulation of OPA1 processing and mitochondrial fusion by m-AAA protease isoenzymes and OMA1. J. Cell Biol. 187, 1023–1036. ( 10.1083/jcb.200906084) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ban T, Ishihara T, Kohno H, Saita S, Ichimura A, Maenaka K, Oka T, Mihara K, Ishihara N. 2017. Molecular basis of selective mitochondrial fusion by heterotypic action between OPA1 and cardiolipin. Nat. Cell Biol. 19, 856–863. ( 10.1038/ncb3560) [DOI] [PubMed] [Google Scholar]
- 19.Mishra P, Chan DC. 2016. Metabolic regulation of mitochondrial dynamics. J. Cell Biol. 212, 379–387. ( 10.1083/jcb.201511036) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gandre-Babbe S, van der Bliek AM. 2008. The novel tail-anchored membrane protein Mff controls mitochondrial and peroxisomal fission in mammalian cells. Mol. Biol. Cell 19, 2402–2412. ( 10.1091/mbc.e07-12-1287) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Palmer CS, Osellame LD, Laine D, Koutsopoulos OS, Frazier AE, Ryan MT. 2011. MiD49 and MiD51, new components of the mitochondrial fission machinery. EMBO Rep. 12, 565–573. ( 10.1038/embor.2011.54) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yu R, Jin S-B, Lendahl U, Nistér M, Zhao J. 2019. Human Fis1 regulates mitochondrial dynamics through inhibition of the fusion machinery. EMBO J. 38, e99748 ( 10.15252/embj.201899748) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chang C-R, Blackstone C. 2010. Dynamic regulation of mitochondrial fission through modification of the dynamin-related protein Drp1. Ann. N. Y. Acad. Sci. 1201, 34–39. ( 10.1111/j.1749-6632.2010.05629.x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kamerkar SC, Kraus F, Sharpe AJ, Pucadyil TJ, Ryan MT. 2018. Dynamin-related protein 1 has membrane constricting and severing abilities sufficient for mitochondrial and peroxisomal fission. Nat. Commun. 9, 5239 ( 10.1038/s41467-018-07543-w) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Friedman JR, Lackner LL, West M, DiBenedetto JR, Nunnari J, Voeltz GK. 2011. ER tubules mark sites of mitochondrial division. Science 334, 358–362. ( 10.1126/science.1207385) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lewis SC, Uchiyama LF, Nunnari J. 2016. ER-mitochondria contacts couple mtDNA synthesis with mitochondrial division in human cells. Science 353, aaf5549 ( 10.1126/science.aaf5549) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Valm AM, et al. 2017. Applying systems-level spectral imaging and analysis to reveal the organelle interactome. Nature 546, 162–167. ( 10.1038/nature22369) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lahiri S, et al. 2014. A conserved endoplasmic reticulum membrane protein complex (EMC) facilitates phospholipid transfer from the ER to mitochondria. PLoS Biol. 12, e1001969 ( 10.1371/journal.pbio.1001969) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rizzuto R, Pinton P, Carrington W, Fay FS, Fogarty KE, Lifshitz LM, Tuft RA, Pozzan T. 1998. Close contacts with the endoplasmic reticulum as determinants of mitochondrial Ca2+ responses. Science 280, 1763–1766. ( 10.1126/science.280.5370.1763) [DOI] [PubMed] [Google Scholar]
- 30.Hamasaki M, et al. 2013. Autophagosomes form at ER–mitochondria contact sites. Nature 495, 389 ( 10.1038/nature11910) [DOI] [PubMed] [Google Scholar]
- 31.Subramanian K, et al. 2019. Coenzyme Q biosynthetic proteins assemble in a substrate-dependent manner into domains at ER-mitochondria contacts. J. Cell Biol. 218, 1353–1369. ( 10.1083/jcb.201808044) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stroud DA, et al. 2011. Composition and topology of the endoplasmic reticulum-mitochondria encounter structure. J. Mol. Biol. 413, 743–750. ( 10.1016/j.jmb.2011.09.012) [DOI] [PubMed] [Google Scholar]
- 33.Cohen S, Valm AM, Lippincott-Schwartz J. 2018. Interacting organelles. Curr. Opin. Cell Biol. 53, 84–91. ( 10.1016/j.ceb.2018.06.003) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Filadi R, et al. 2018. TOM70 sustains cell bioenergetics by promoting IP3R3-mediated ER to mitochondria Ca2+ transfer. Curr. Biol. 28, 369–382. ( 10.1016/j.cub.2017.12.047) [DOI] [PubMed] [Google Scholar]
- 35.Szabadkai G, et al. 2006. Chaperone-mediated coupling of endoplasmic reticulum and mitochondrial Ca2+ channels. J. Cell Biol. 175, 901–911. ( 10.1083/jcb.200608073) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stoica R, et al. 2014. ER–mitochondria associations are regulated by the VAPB–PTPIP51 interaction and are disrupted by ALS/FTD-associated TDP-43. Nat. Commun. 5, 3996 ( 10.1038/ncomms4996) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kumar N, et al. 2018. VPS13A and VPS13C are lipid transport proteins differentially localized at ER contact sites. J. Cell Biol. 217, 3625–3639. ( 10.1083/jcb.201807019) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hirabayashi Y, et al. 2017. ER-mitochondria tethering by PDZD8 regulates Ca2+ dynamics in mammalian neurons. Science 358, 623–630. ( 10.1126/science.aan6009) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.de Brito OM, Scorrano L. 2008. Mitofusin 2 tethers endoplasmic reticulum to mitochondria. Nature 456, 605–610. ( 10.1038/nature07534) [DOI] [PubMed] [Google Scholar]
- 40.Friedman JR, Webster BM, Mastronarde DN, Verhey KJ, Voeltz GK. 2010. ER sliding dynamics and ER–mitochondrial contacts occur on acetylated microtubules. J. Cell Biol. 190, 363–375. ( 10.1083/jcb.200911024) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dolman NJ, Gerasimenko JV, Gerasimenko OV, Voronina SG, Petersen OH, Tepikin AV. 2005. Stable Golgi-mitochondria complexes and formation of Golgi Ca2+ gradients in pancreatic acinar cells. J. Biol. Chem. 280, 15 794–15 799. ( 10.1074/jbc.M412694200) [DOI] [PubMed] [Google Scholar]
- 42.Fan J, Li X, Issop L, Culty M, Papadopoulos V. 2016. ACBD2/ECI2-mediated peroxisome-mitochondria interactions in Leydig cell steroid biosynthesis. Mol. Endocrinol. 30, 763–782. ( 10.1210/me.2016-1008) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wong YC, Ysselstein D, Krainc D. 2018. Mitochondria–lysosome contacts regulate mitochondrial fission via RAB7 GTP hydrolysis. Nature 554, 382–386. ( 10.1038/nature25486) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Benador IY, et al. 2018. Mitochondria bound to lipid droplets have unique bioenergetics, composition, and dynamics that support lipid droplet expansion. Cell Metab. 27, 869–885. ( 10.1016/j.cmet.2018.03.003) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wu M-J, et al. 2019. Epithelial-mesenchymal transition directs stem cell polarity via regulation of mitofusin. Cell Metab. 29, 993–1002. ( 10.1016/j.cmet.2018.11.004) [DOI] [PubMed] [Google Scholar]
- 46.Palmer CS, Osellame LD, Stojanovski D, Ryan MT. 2011. The regulation of mitochondrial morphology: intricate mechanisms and dynamic machinery. Cell. Signal. 23, 1534–1545. ( 10.1016/j.cellsig.2011.05.021) [DOI] [PubMed] [Google Scholar]
- 47.Tilokani L, Nagashima S, Paupe V, Prudent J. 2018. Mitochondrial dynamics: overview of molecular mechanisms. Essays Biochem. 62, 341–360. ( 10.1042/EBC20170104) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pagliuso A, Cossart P, Stavru F. 2018. The ever-growing complexity of the mitochondrial fission machinery. Cell. Mol. Life Sci. 75, 355–374. ( 10.1007/s00018-017-2603-0) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lackner LL. 2019. The expanding and unexpected functions of mitochondria contact sites. Trends Cell Biol. ( 10.1016/j.tcb.2019.02.009) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Elbaz-Alon Y. 2017. Mitochondria–organelle contact sites: the plot thickens. Biochem. Soc. Trans. 45, 477–488. ( 10.1042/BST20160130) [DOI] [PubMed] [Google Scholar]
- 51.Wu H, Carvalho P, Voeltz GK. 2018. Here, there, and everywhere: the importance of ER membrane contact sites. Science 361, eaan5835 ( 10.1126/science.aan5835) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Fillingame RH, Angevine CM, Dmitriev OY. 2003. Mechanics of coupling proton movements to c-ring rotation in ATP synthase. FEBS Lett. 555, 29–34. ( 10.1016/S0014-5793(03)01101-3) [DOI] [PubMed] [Google Scholar]
- 53.Dizdaroglu M, Jaruga P. 2012. Mechanisms of free radical-induced damage to DNA. Free Radic. Res. 46, 382–419. ( 10.3109/10715762.2011.653969) [DOI] [PubMed] [Google Scholar]
- 54.Finkel T, Holbrook NJ. 2000. Oxidants, oxidative stress and the biology of ageing. Nature 408, 239–247. ( 10.1038/35041687) [DOI] [PubMed] [Google Scholar]
- 55.Niforou K, Cheimonidou C, Trougakos IP. 2014. Molecular chaperones and proteostasis regulation during redox imbalance. Redox Biol. 2, 323–332. ( 10.1016/j.redox.2014.01.017) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Letts JA, Sazanov LA. 2017. Clarifying the supercomplex: the higher-order organization of the mitochondrial electron transport chain. Nat. Struct. Mol. Biol. 24, 800–808. ( 10.1038/nsmb.3460) [DOI] [PubMed] [Google Scholar]
- 57.Wang L. 2016. Mitochondrial purine and pyrimidine metabolism and beyond. Nucleosides Nucleotides Nucleic Acids 35, 578–594. ( 10.1080/15257770.2015.1125001) [DOI] [PubMed] [Google Scholar]
- 58.Lill R. 2009. Function and biogenesis of iron–sulphur proteins. Nature 460, 831–838. ( 10.1038/nature08301) [DOI] [PubMed] [Google Scholar]
- 59.Kastaniotis AJ, et al. 2017. Mitochondrial fatty acid synthesis, fatty acids and mitochondrial physiology. Biochim. Biophys. Acta Mol. Cell Biol. Lipids 1862, 39–48. ( 10.1016/j.bbalip.2016.08.011) [DOI] [PubMed] [Google Scholar]
- 60.Ducker GS, Rabinowitz JD. 2017. One-carbon metabolism in health and disease. Cell Metab. 25, 27–42. ( 10.1016/j.cmet.2016.08.009) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Grillo MA, Colombatto S. 2005. S-Adenosylmethionine and protein methylation. Amino Acids 28, 357–362. ( 10.1007/s00726-005-0197-6) [DOI] [PubMed] [Google Scholar]
- 62.Shimazu T, et al. 2013. Suppression of oxidative stress by beta-hydroxybutyrate, an endogenous histone deacetylase inhibitor. Science 339, 211–214. ( 10.1126/science.1227166) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Cai L, Sutter BM, Li B, Tu BP. 2011. Acetyl-CoA induces cell growth and proliferation by promoting the acetylation of histones at growth genes. Mol. Cell 42, 426–437. ( 10.1016/j.molcel.2011.05.004) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wellen KE, Hatzivassiliou G, Sachdeva UM, Bui TV, Cross JR, Thompson CB. 2009. ATP-citrate lyase links cellular metabolism to histone acetylation. Science 324, 1076–1080. ( 10.1126/science.1164097) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lee DC, et al. 2015. A lactate-induced response to hypoxia. Cell 161, 595–609. ( 10.1016/j.cell.2015.03.011) [DOI] [PubMed] [Google Scholar]
- 66.Waldecker M, Kautenburger T, Daumann H, Busch C, Schrenk D. 2008. Inhibition of histone-deacetylase activity by short-chain fatty acids and some polyphenol metabolites formed in the colon. J. Nutr. Biochem. 19, 587–593. ( 10.1016/j.jnutbio.2007.08.002) [DOI] [PubMed] [Google Scholar]
- 67.Wang S, et al. 2015. Lysosomal amino acid transporter SLC38A9 signals arginine sufficiency to mTORC1. Science 347, 188–194. ( 10.1126/science.1257132) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Frezza C. 2017. Mitochondrial metabolites: undercover signalling molecules. Interface focus 7, 20160100 ( 10.1098/rsfs.2016.0100) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Thoreen CC, Chantranupong L, Keys HR, Wang T, Gray NS, Sabatini DM. 2012. A unifying model for mTORC1-mediated regulation of mRNA translation. Nature 485, 109–113. ( 10.1038/nature11083) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Cunningham JT, Rodgers JT, Arlow DH, Vazquez F, Mootha VK, Puigserver P. 2007. mTOR controls mitochondrial oxidative function through a YY1–PGC-1α transcriptional complex. Nature 450, 736–740. ( 10.1038/nature06322) [DOI] [PubMed] [Google Scholar]
- 71.Goo CK, Lim HY, Ho QS, Too H-P, Clement M-V, Wong KP. 2012. PTEN/Akt signaling controls mitochondrial respiratory capacity through 4E-BP1. PLoS ONE 7, e45806 ( 10.1371/journal.pone.0045806) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Morita M, et al. 2013. mTORC1 controls mitochondrial activity and biogenesis through 4E-BP-dependent translational regulation. Cell Metab. 18, 698–711. ( 10.1016/j.cmet.2013.10.001) [DOI] [PubMed] [Google Scholar]
- 73.Feng Z, Zhang H, Levine AJ, Jin S. 2005. The coordinate regulation of the p53 and mTOR pathways in cells. Proc. Natl Acad. Sci. USA 102, 8204–8209. ( 10.1073/pnas.0502857102) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Horton LE, Bushell M, Barth-Baus D, Tilleray VJ, Clemens MJ, Hensold JO. 2002. p53 activation results in rapid dephosphorylation of the eIF4E-binding protein 4E-BP1, inhibition of ribosomal protein S6 kinase and inhibition of translation initiation. Oncogene 21, 5325–5334. ( 10.1038/sj.onc.1205662) [DOI] [PubMed] [Google Scholar]
- 75.Matoba S, et al. 2006. p53 regulates mitochondrial respiration. Science 312, 1650–1653. ( 10.1126/science.1126863) [DOI] [PubMed] [Google Scholar]
- 76.Kulawiec M, Ayyasamy V, Singh KK. 2009. p53 regulates mtDNA copy number and mitocheckpoint pathway. J. Carcinog. 8, 8 ( 10.4103/1477-3163.50893) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Achanta G, Sasaki R, Feng L, Carew JS, Lu W, Pelicano H, Keating MJ, Huang P. 2005. Novel role of p53 in maintaining mitochondrial genetic stability through interaction with DNA Pol gamma. EMBO J. 24, 3482–3492. ( 10.1038/sj.emboj.7600819) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Hu W, Zhang C, Wu R, Sun Y, Levine A, Feng Z. 2010. Glutaminase 2, a novel p53 target gene regulating energy metabolism and antioxidant function. Proc. Natl Acad. Sci. USA 107, 7455–7460. ( 10.1073/pnas.1001006107) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Bensaad K, Tsuruta A, Selak MA, Vidal MN, Nakano K, Bartrons R, Gottlieb E, Vousden KH. 2006. TIGAR, a p53-inducible regulator of glycolysis and apoptosis. Cell 126, 107–120. ( 10.1016/j.cell.2006.05.036) [DOI] [PubMed] [Google Scholar]
- 80.Schwartzenberg-Bar-Yoseph F, Armoni M, Karnieli E. 2004. The tumor suppressor p53 down-regulates glucose transporters GLUT1 and GLUT4 gene expression. Cancer Res. 64, 2627 ( 10.1158/0008-5472.CAN-03-0846) [DOI] [PubMed] [Google Scholar]
- 81.Elstrom RL, et al. 2004. Akt stimulates aerobic glycolysis in cancer cells. Cancer Res. 64, 3892–3899. ( 10.1158/0008-5472.CAN-03-2904) [DOI] [PubMed] [Google Scholar]
- 82.Kondoh H, et al. 2005. Glycolytic enzymes can modulate cellular life span. Cancer Res. 65, 177–185. [PubMed] [Google Scholar]
- 83.Rich PR, Maréchal A. 2010. The mitochondrial respiratory chain. Essays Biochem. 47, 1–23. ( 10.1042/bse0470001) [DOI] [PubMed] [Google Scholar]
- 84.Salminen A, Kaarniranta K, Hiltunen M, Kauppinen A. 2014. Krebs cycle dysfunction shapes epigenetic landscape of chromatin: novel insights into mitochondrial regulation of aging process. Cell. Signal. 26, 1598–1603. ( 10.1016/j.cellsig.2014.03.030) [DOI] [PubMed] [Google Scholar]
- 85.Saxton RA, Sabatini DM. 2017. mTOR signaling in growth, metabolism, and disease. Cell 168, 960–976. ( 10.1016/j.cell.2017.02.004) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Kim J, Guan K-L. 2019. mTOR as a central hub of nutrient signalling and cell growth. Nat. Cell Biol. 21, 63–71. ( 10.1038/s41556-018-0205-1) [DOI] [PubMed] [Google Scholar]
- 87.Tan W, Colombini M. 2007. VDAC closure increases calcium ion flux. Biochim. Biophys. Acta 1768, 2510–2515. ( 10.1016/j.bbamem.2007.06.002) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Scorziello A, et al. 2013. NCX3 regulates mitochondrial Ca2+ handling through the AKAP121-anchored signaling complex and prevents hypoxia-induced neuronal death. J. Cell Sci. 126, 5566–5577. ( 10.1242/jcs.129668) [DOI] [PubMed] [Google Scholar]
- 89.Patron M, et al. 2014. MICU1 and MICU2 finely tune the mitochondrial Ca2+ uniporter by exerting opposite effects on MCU activity. Mol. Cell 53, 726–737. ( 10.1016/j.molcel.2014.01.013) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Petrungaro C, Zimmermann KM, Kuttner V, Fischer M, Dengjel J, Bogeski I, Riemer J. 2015. The Ca2+-dependent release of the Mia40-induced MICU1-MICU2 dimer from MCU regulates mitochondrial Ca2+ uptake. Cell Metab. 22, 721–733. ( 10.1016/j.cmet.2015.08.019) [DOI] [PubMed] [Google Scholar]
- 91.Giorgi C, Danese A, Missiroli S, Patergnani S, Pinton P. 2018. Calcium dynamics as a machine for decoding signals. Trends Cell Biol. 28, 258–273. ( 10.1016/j.tcb.2018.01.002) [DOI] [PubMed] [Google Scholar]
- 92.Giorgi C, Marchi S, Pinton P. 2018. The machineries, regulation and cellular functions of mitochondrial calcium. Nat. Rev. Mol. Cell Biol. 19, 713–730. ( 10.1038/s41580-018-0052-8) [DOI] [PubMed] [Google Scholar]
- 93.Kang JS, Tian JH, Pan PY, Zald P, Li C, Deng C, Sheng Z-H. 2008. Docking of axonal mitochondria by syntaphilin controls their mobility and affects short-term facilitation. Cell 132, 137–148. ( 10.1016/j.cell.2007.11.024) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.David G, Barrett EF. 2003. Mitochondrial Ca2+ uptake prevents desynchronization of quantal release and minimizes depletion during repetitive stimulation of mouse motor nerve terminals. J. Physiol. 548, 425–438. ( 10.1113/jphysiol.2002.035196) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Viola HM, Arthur PG, Hool LC. 2009. Evidence for regulation of mitochondrial function by the L-type Ca2+ channel in ventricular myocytes. J. Mol. Cell. Cardiol. 46, 1016–1026. ( 10.1016/j.yjmcc.2008.12.015) [DOI] [PubMed] [Google Scholar]
- 96.McCormack JG, Halestrap AP, Denton RM. 1990. Role of calcium ions in regulation of mammalian intramitochondrial metabolism. Physiol. Rev. 70, 391–425. ( 10.1152/physrev.1990.70.2.391) [DOI] [PubMed] [Google Scholar]
- 97.Das AM, Harris DA. 1990. Control of mitochondrial ATP synthase in heart cells: inactive to active transitions caused by beating or positive inotropic agents. Cardiovasc. Res. 24, 411–417. ( 10.1093/cvr/24.5.411) [DOI] [PubMed] [Google Scholar]
- 98.Mildaziene V, Baniene R, Nauciene Z, Bakker BM, Brown GC, Westerhoff HV, Kholodenko BN. 1995. Calcium indirectly increases the control exerted by the adenine nucleotide translocator over 2-oxoglutarate oxidation in rat heart mitochondria. Arch. Biochem. Biophys. 324, 130–134. ( 10.1006/abbi.1995.9918) [DOI] [PubMed] [Google Scholar]
- 99.Paillusson S, Gomez-Suaga P, Stoica R, Little D, Gissen P, Devine MJ, Noble W, Hanger DP, Miller CCJ. 2017. α-Synuclein binds to the ER-mitochondria tethering protein VAPB to disrupt Ca2+ homeostasis and mitochondrial ATP production. Acta Neuropathol. 134, 129–149. ( 10.1007/s00401-017-1704-z) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Wiederkehr A, et al. 2011. Mitochondrial matrix calcium is an activating signal for hormone secretion. Cell Metab. 13, 601–611. ( 10.1016/j.cmet.2011.03.015) [DOI] [PubMed] [Google Scholar]
- 101.Antony AN, et al. 2016. MICU1 regulation of mitochondrial Ca2+ uptake dictates survival and tissue regeneration. Nat. Commun. 7, 10955 ( 10.1038/ncomms10955) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Cheng J, Liao Y, Zhou L, Peng S, Chen H, Yuan Z. 2016. Amplified RLR signaling activation through an interferon-stimulated gene-endoplasmic reticulum stress-mitochondrial calcium uniporter protein loop. Sci. Rep. 6, 20158 ( 10.1038/srep20158) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Rizzuto R, De Stefani D, Raffaello A, Mammucari C. 2012. Mitochondria as sensors and regulators of calcium signalling. Nat. Rev. Mol. Cell Biol. 13, 566–578. ( 10.1038/nrm3412) [DOI] [PubMed] [Google Scholar]
- 104.Bravo-Sagua R, Parra V, Lopez-Crisosto C, Diaz P, Quest AF, Lavandero S. 2017. Calcium transport and signaling in mitochondria. Compr. Physiol. 7, 623–634. ( 10.1002/cphy.c160013) [DOI] [PubMed] [Google Scholar]
- 105.Horner SM, Liu HM, Park HS, Briley J, Gale M Jr. 2011. Mitochondrial-associated endoplasmic reticulum membranes (MAM) form innate immune synapses and are targeted by hepatitis C virus. Proc. Natl Acad. Sci. USA 108, 14 590–14 595. ( 10.1073/pnas.1110133108) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.West AP, Shadel GS, Ghosh S. 2011. Mitochondria in innate immune responses. Nat. Rev. Immunol. 11, 389–402. ( 10.1038/nri2975) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Seth RB, Sun L, Ea CK, Chen ZJ. 2005. Identification and characterization of MAVS, a mitochondrial antiviral signaling protein that activates NF-κB and IRF 3. Cell 122, 669–682. ( 10.1016/j.cell.2005.08.012) [DOI] [PubMed] [Google Scholar]
- 108.Tang ED, Wang C-Y. 2009. MAVS self-association mediates antiviral innate immune signaling. J. Virol. 83, 3420–3428. ( 10.1128/JVI.02623-08) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Liu XY, Wei B, Shi HX, Shan YF, Wang C. 2010. Tom70 mediates activation of interferon regulatory factor 3 on mitochondria. Cell Res. 20, 994–1011. ( 10.1038/cr.2010.103) [DOI] [PubMed] [Google Scholar]
- 110.Tattoli I, Carneiro LA, Jehanno M, Magalhaes JG, Shu Y, Philpott DJ, Arnoult D, Girardin SE. 2008. NLRX1 is a mitochondrial NOD-like receptor that amplifies NF-κB and JNK pathways by inducing reactive oxygen species production. EMBO Rep. 9, 293–300. ( 10.1038/sj.embor.7401161) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Arnoult D, Soares F, Tattoli I, Castanier C, Philpott DJ, Girardin SE. 2009. An N-terminal addressing sequence targets NLRX1 to the mitochondrial matrix. J. Cell Sci. 122, 3161–3168. ( 10.1242/jcs.051193) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Fielden LF, Kang Y, Newton HJ, Stojanovski D. 2017. Targeting mitochondria: how intravacuolar bacterial pathogens manipulate mitochondria. Cell Tissue Res. 367, 141–154. ( 10.1007/s00441-016-2475-x) [DOI] [PubMed] [Google Scholar]
- 113.Allam R, et al. 2014. Mitochondrial apoptosis is dispensable for NLRP3 inflammasome activation but non-apoptotic caspase-8 is required for inflammasome priming. EMBO Rep. 15, 982–990. ( 10.15252/embr.201438463) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Nakahira K, et al. 2011. Autophagy proteins regulate innate immune responses by inhibiting the release of mitochondrial DNA mediated by the NALP3 inflammasome. Nat. Immunol. 12, 222–230. ( 10.1038/ni.1980) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Sanin DE, et al. 2018. Mitochondrial membrane potential regulates nuclear gene expression in macrophages exposed to prostaglandin E2. Immunity 49, 1021–1033. ( 10.1016/j.immuni.2018.10.011) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.De Simone R, Ajmone-Cat MA, Pandolfi M, Bernardo A, De Nuccio C, Minghetti L, Visentin S. et al. 2015. The mitochondrial uncoupling protein-2 is a master regulator of both M1 and M2 microglial responses. J. Neurochem. 135, 147–156. ( 10.1111/jnc.13244) [DOI] [PubMed] [Google Scholar]
- 117.Jha AK, et al. 2015. Network integration of parallel metabolic and transcriptional data reveals metabolic modules that regulate macrophage polarization. Immunity 42, 419–430. ( 10.1016/j.immuni.2015.02.005) [DOI] [PubMed] [Google Scholar]
- 118.Angajala A, Lim S, Phillips JB, Kim J-H, Yates C, You Z, Tan M. 2018. Diverse roles of mitochondria in immune responses: novel insights into immuno-metabolism. Front. Immunol. 9, 1605 ( 10.3389/fimmu.2018.01605) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Arsenijevic D, et al. 2000. Disruption of the uncoupling protein-2 gene in mice reveals a role in immunity and reactive oxygen species production. Nat. Genet. 26, 435–439. ( 10.1038/82565) [DOI] [PubMed] [Google Scholar]
- 120.van der Windt Gerritje JW, et al. 2012. Mitochondrial respiratory capacity is a critical regulator of CD8+ T cell memory development. Immunity 36, 68–78. ( 10.1016/j.immuni.2011.12.007) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Tan H, et al. 2017. Integrative proteomics and phosphoproteomics profiling reveals dynamic signaling networks and bioenergetics pathways underlying T cell activation. Immunity 46, 488–503. ( 10.1016/j.immuni.2017.02.010) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Pearce EL, et al. 2009. Enhancing CD8 T-cell memory by modulating fatty acid metabolism. Nature 460, 103–107. ( 10.1038/nature08097) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Chao T, Wang H, Ho P-C. 2017. Mitochondrial control and guidance of cellular activities of T cells. Front. Immunol. 8, 473 ( 10.3389/fimmu.2017.00473) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Buck MD, et al. 2016. Mitochondrial dynamics controls T cell fate through metabolic programming. Cell 166, 63–76. ( 10.1016/j.cell.2016.05.035) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Tyrakis PA, et al. 2016. S-2-hydroxyglutarate regulates CD8+ T-lymphocyte fate. Nature 540, 236–241. ( 10.1038/nature20165) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Mills EL, Kelly B, O'Neill LAJ. 2017. Mitochondria are the powerhouses of immunity. Nat. Immunol. 18, 488 ( 10.1038/ni.3704) [DOI] [PubMed] [Google Scholar]
- 127.Lee S, Kim S, Sun X, Lee J-H, Cho H. 2007. Cell cycle-dependent mitochondrial biogenesis and dynamics in mammalian cells. Biochem. Biophys. Res. Commun. 357, 111–117. ( 10.1016/j.bbrc.2007.03.091) [DOI] [PubMed] [Google Scholar]
- 128.Mitra K, Wunder C, Roysam B, Lin G, Lippincott-Schwartz J. 2009. A hyperfused mitochondrial state achieved at G1–S regulates cyclin E buildup and entry into S phase. Proc. Natl Acad. Sci. USA 106, 11 960–11 965. ( 10.1073/pnas.0904875106) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Gregg T, Sdao SM, Dhillon RS, Rensvold JW, Lewandowski SL, Pagliarini DJ, Denu JM, Merrins MJ. 2019. Obesity-dependent CDK1 signaling stimulates mitochondrial respiration at complex I in pancreatic beta-cells. J. Biol. Chem. 294, 4656–4666. ( 10.1074/jbc.RA118.006085) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Harbauer AB, et al. 2014. Cell cycle-dependent regulation of mitochondrial preprotein translocase. Science 346, 1109–1113. ( 10.1126/science.1261253) [DOI] [PubMed] [Google Scholar]
- 131.Martínez-Diez M, Santamaría G, Ortega ÁD, Cuezva JM. 2006. Biogenesis and dynamics of mitochondria during the cell cycle: significance of 3′UTRs. PLoS ONE 1, e107 ( 10.1371/journal.pone.0000107) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Chen Y-C, Chang M-Y, Shiau A-L, Yo Y-T, Wu C-L. 2007. Mitochondrial ribosomal protein S36 delays cell cycle progression in association with p53 modification and p21WAF1/CIP1 expression. J. Cell. Biochem. 100, 981–990. ( 10.1002/jcb.21079) [DOI] [PubMed] [Google Scholar]
- 133.Shiraki N, et al. 2014. Methionine metabolism regulates maintenance and differentiation of human pluripotent stem cells. Cell Metab. 19, 780–794. ( 10.1016/j.cmet.2014.03.017) [DOI] [PubMed] [Google Scholar]
- 134.Owusu-Ansah E, Yavari A, Mandal S, Banerjee U. 2008. Distinct mitochondrial retrograde signals control the G1-S cell cycle checkpoint. Nat. Genet. 40, 356–361. ( 10.1038/ng.2007.50) [DOI] [PubMed] [Google Scholar]
- 135.Chen K-H, Dasgupta A, Ding J, Indig FE, Ghosh P, Longo DL. 2014. Role of mitofusin 2 (Mfn2) in controlling cellular proliferation. FASEB J. 28, 382–394. ( 10.1096/fj.13-230037) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Cho YM, Kwon S, Pak YK, Seol HW, Choi YM, Park DJ, Park KS, Lee HK. 2006. Dynamic changes in mitochondrial biogenesis and antioxidant enzymes during the spontaneous differentiation of human embryonic stem cells. Biochem. Biophys. Res. Commun. 348, 1472–1478. ( 10.1016/j.bbrc.2006.08.020) [DOI] [PubMed] [Google Scholar]
- 137.Takubo K, et al. 2013. Regulation of glycolysis by Pdk functions as a metabolic checkpoint for cell cycle quiescence in hematopoietic stem cells. Cell Stem Cell 12, 49–61. ( 10.1016/j.stem.2012.10.011) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Solari C, et al. 2018. Superoxide dismutase 1 expression is modulated by the core pluripotency transcription factors Oct4, Sox2 and Nanog in embryonic stem cells. Mech. Dev. 154, 116–121. ( 10.1016/j.mod.2018.06.004) [DOI] [PubMed] [Google Scholar]
- 139.Kasahara A, Cipolat S, Chen Y, Dorn GW, Scorrano L. 2013. Mitochondrial fusion directs cardiomyocyte differentiation via calcineurin and Notch signaling. Science 342, 734–737. ( 10.1126/science.1241359) [DOI] [PubMed] [Google Scholar]
- 140.De Palma C, et al. 2010. Nitric oxide inhibition of Drp1-mediated mitochondrial fission is critical for myogenic differentiation. Cell Death Differ. 17, 1684–1696. ( 10.1038/cdd.2010.48) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Sin J, et al. 2015. Mitophagy is required for mitochondrial biogenesis and myogenic differentiation of C2C12 myoblasts. Autophagy 12, 369–380. ( 10.1080/15548627.2015.1115172) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Son MJ, et al. 2015. Mitofusins deficiency elicits mitochondrial metabolic reprogramming to pluripotency. Cell Death Differ. 22, 1957 ( 10.1038/cdd.2015.43) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Son M-Y, Choi H, Han Y-M, Sook Cho Y. 2013. Unveiling the critical role of REX1 in the regulation of human stem cell pluripotency. Stem Cells 31, 2374–2387. ( 10.1002/stem.1509) [DOI] [PubMed] [Google Scholar]
- 144.Vakifahmetoglu-Norberg H, Ouchida AT, Norberg E. 2017. The role of mitochondria in metabolism and cell death. Biochem. Biophys. Res. Commun. 482, 426–431. ( 10.1016/j.bbrc.2016.11.088) [DOI] [PubMed] [Google Scholar]
- 145.Kuwana T, Mackey MR, Perkins G, Ellisman MH, Latterich M, Schneiter R, Green DR, Newmeyer DD. 2002. Bid, Bax, and lipids cooperate to form supramolecular openings in the outer mitochondrial membrane. Cell 111, 331–342. ( 10.1016/S0092-8674(02)01036-X) [DOI] [PubMed] [Google Scholar]
- 146.Du C, Fang M, Li Y, Li L, Wang X. 2000. Smac, a mitochondrial protein that promotes cytochrome c-dependent caspase activation by eliminating IAP inhibition. Cell 102, 33–42. ( 10.1016/S0092-8674(00)00008-8) [DOI] [PubMed] [Google Scholar]
- 147.Verhagen AM, et al. 2002. HtrA2 promotes cell death through its serine protease activity and its ability to antagonize inhibitor of apoptosis proteins. J. Biol. Chem. 277, 445–454. ( 10.1074/jbc.M109891200) [DOI] [PubMed] [Google Scholar]
- 148.Sevrioukova IF. 2011. Apoptosis-inducing factor: structure, function, and redox regulation. Antioxid. Redox Signal. 14, 2545–2579. ( 10.1089/ars.2010.3445) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Polster BM, Basanez G, Etxebarria A, Hardwick JM, Nicholls DG. 2005. Calpain I induces cleavage and release of apoptosis-inducing factor from isolated mitochondria. J. Biol. Chem. 280, 6447–6454. ( 10.1074/jbc.M413269200) [DOI] [PubMed] [Google Scholar]
- 150.van Loo G, et al. 2001. Endonuclease G: a mitochondrial protein released in apoptosis and involved in caspase-independent DNA degradation. Cell Death Differ. 8, 1136 ( 10.1038/sj.cdd.4400944) [DOI] [PubMed] [Google Scholar]
- 151.Cheng EH-Y, Sheiko TV, Fisher JK, Craigen WJ, Korsmeyer SJ. 2003. VDAC2 inhibits BAK activation and mitochondrial apoptosis. Science 301, 513–517. ( 10.1126/science.1083995) [DOI] [PubMed] [Google Scholar]
- 152.Lazarou M, et al. 2010. Inhibition of Bak activation by VDAC2 is dependent on the Bak transmembrane anchor. J. Biol. Chem. 285, 36 876–36 883. ( 10.1074/jbc.M110.159301) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Chin HS, et al. 2018. VDAC2 enables BAX to mediate apoptosis and limit tumor development. Nat. Commun. 9, 4976 ( 10.1038/s41467-018-07309-4) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Angelina VV, Natalie DM, Ji K, Tsirka Stella E, Holzmann S, Moll Ute M. 2012. p53 opens the mitochondrial permeability transition pore to trigger necrosis. Cell 149, 1536–1548. ( 10.1016/j.cell.2012.05.014) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Wang Z, Jiang H, Chen S, Du F, Wang X. 2012. The mitochondrial phosphatase PGAM5 functions at the convergence point of multiple necrotic death pathways. Cell 148, 228–243. ( 10.1016/j.cell.2011.11.030) [DOI] [PubMed] [Google Scholar]
- 156.Wu C, Zhao W, Yu J, Li S, Lin L, Chen X. 2018. Induction of ferroptosis and mitochondrial dysfunction by oxidative stress in PC12 cells. Sci. Rep. 8, 574 ( 10.1038/s41598-017-18935-1) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Gao M, et al. 2019. Role of mitochondria in ferroptosis. Mol. Cell 73, 354–363. ( 10.1016/j.molcel.2018.10.042) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Fatokun AA, Dawson VL, Dawson TM. 2014. Parthanatos: mitochondrial-linked mechanisms and therapeutic opportunities. Br. J. Pharmacol. 171, 2000–2016. ( 10.1111/bph.12416) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Finkel T, Hwang PM. 2009. The Krebs cycle meets the cell cycle: mitochondria and the G1–S transition. Proc. Natl Acad. Sci. USA 106, 11 825–11 826. ( 10.1073/pnas.0906430106) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Salazar-Roa M, Malumbres M. 2017. Fueling the cell division cycle. Trends Cell Biol. 27, 69–81. ( 10.1016/j.tcb.2016.08.009) [DOI] [PubMed] [Google Scholar]
- 161.Kasahara A, Scorrano L. 2014. Mitochondria: from cell death executioners to regulators of cell differentiation. Trends Cell Biol. 24, 761–770. ( 10.1016/j.tcb.2014.08.005) [DOI] [PubMed] [Google Scholar]
- 162.Lisowski P, Kannan P, Mlody B, Prigione A. 2018. Mitochondria and the dynamic control of stem cell homeostasis. EMBO Rep. 19, e45432 ( 10.15252/embr.201745432) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Noguchi M, Kasahara A. 2018. Mitochondrial dynamics coordinate cell differentiation. Biochem. Biophys. Res. Commun. 500, 59–64. ( 10.1016/j.bbrc.2017.06.094) [DOI] [PubMed] [Google Scholar]
- 164.Tait SWG, Green DR. 2013. Mitochondrial regulation of cell death. Cold Spring Harb. Perspect. Biol. 5, a008706 ( 10.1101/cshperspect.a008706) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Bhola PD, Letai A. 2016. Mitochondria—judges and executioners of cell death sentences. Mol. Cell 61, 695–704. ( 10.1016/j.molcel.2016.02.019) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Zhao Q, Wang J, Levichkin IV, Stasinopoulos S, Ryan MT, Hoogenraad NJ. 2002. A mitochondrial specific stress response in mammalian cells. EMBO J. 21, 4411–4419. ( 10.1093/emboj/cdf445) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Fiorese CJ, Schulz AM, Lin Y-F, Rosin N, Pellegrino MW, Haynes CM. 2016. The transcription factor ATF5 mediates a mammalian mitochondrial UPR. Curr. Biol. 26, 2037–2043. ( 10.1016/j.cub.2016.06.002) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Monaco SE, Angelastro JM, Szabolcs M, Greene LA. 2007. The transcription factor ATF5 is widely expressed in carcinomas, and interference with its function selectively kills neoplastic, but not nontransformed, breast cell lines. Int. J. Cancer 120, 1883–1890. ( 10.1002/ijc.22469) [DOI] [PubMed] [Google Scholar]
- 169.Nargund AM, Fiorese CJ, Pellegrino MW, Deng P, Haynes CM. 2015. Mitochondrial and nuclear accumulation of the transcription factor ATFS-1 promotes OXPHOS recovery during the UPR(mt). Mol. Cell 58, 123–133. ( 10.1016/j.molcel.2015.02.008) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Zhou D, Palam LR, Jiang L, Narasimhan J, Staschke KA, Wek RC. 2008. Phosphorylation of eIF2 directs ATF5 translational control in response to diverse stress conditions. J. Biol. Chem. 283, 7064–7073. ( 10.1074/jbc.M708530200) [DOI] [PubMed] [Google Scholar]
- 171.Quirós PM, Prado MA, Zamboni N, D'Amico D, Williams RW, Finley D, Gygi SP, Auwerx J. 2017. Multi-omics analysis identifies ATF4 as a key regulator of the mitochondrial stress response in mammals. J. Cell Biol. 216, 2027–2045. ( 10.1083/jcb.201702058) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Harding HP, et al. 2003. An integrated stress response regulates amino acid metabolism and resistance to oxidative stress. Mol. Cell 11, 619–633. ( 10.1016/S1097-2765(03)00105-9) [DOI] [PubMed] [Google Scholar]
- 173.Rainbolt TK, Atanassova N, Genereux JC, Wiseman RL. 2013. Stress-regulated translational attenuation adapts mitochondrial protein import through Tim17A degradation. Cell Metab. 18, 908–919. ( 10.1016/j.cmet.2013.11.006) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Pakos-Zebrucka K, Koryga I, Mnich K, Ljujic M, Samali A, Gorman AM. 2016. The integrated stress response. EMBO Rep. 17, 1374–1395. ( 10.15252/embr.201642195) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Okatsu K, Kimura M, Oka T, Tanaka K, Matsuda N. 2015. Unconventional PINK1 localization to the outer membrane of depolarized mitochondria drives Parkin recruitment. J. Cell Sci. 128, 964–978. ( 10.1242/jcs.161000) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Jin SM, Lazarou M, Wang C, Kane LA, Narendra DP, Youle RJ. 2010. Mitochondrial membrane potential regulates PINK1 import and proteolytic destabilization by PARL. J. Cell Biol. 191, 933–942. ( 10.1083/jcb.201008084) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Lazarou M, Jin SM, Kane LA, Youle RJ. 2012. Role of PINK1 binding to the TOM complex and alternate intracellular membranes in recruitment and activation of the E3 ligase Parkin. Dev. Cell 22, 320–333. ( 10.1016/j.devcel.2011.12.014) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Okatsu K, et al. 2012. PINK1 autophosphorylation upon membrane potential dissipation is essential for Parkin recruitment to damaged mitochondria. Nat. Commun. 3, 1016 ( 10.1038/ncomms2016) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Sarraf SA, et al. 2013. Landscape of the PARKIN-dependent ubiquitylome in response to mitochondrial depolarization. Nature 496, 372–376. ( 10.1038/nature12043) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Ordureau A, et al. 2014. Quantitative proteomics reveal a feedforward mechanism for mitochondrial PARKIN translocation and ubiquitin chain synthesis. Mol. Cell 56, 360–375. ( 10.1016/j.molcel.2014.09.007) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Cho HM, et al. 2019. Drp1-Zip1 interaction regulates mitochondrial quality surveillance system. Mol. Cell 73, 364–376. ( 10.1016/j.molcel.2018.11.009) [DOI] [PubMed] [Google Scholar]
- 182.Kravic B, et al. 2018. In mammalian skeletal muscle, phosphorylation of TOMM22 by protein kinase CSNK2/CK2 controls mitophagy. Autophagy 14, 311–335. ( 10.1080/15548627.2017.1403716) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Chen G, et al. 2014. A regulatory signaling loop comprising the PGAM5 phosphatase and CK2 controls receptor-mediated mitophagy. Mol. Cell 54, 362–377. ( 10.1016/j.molcel.2014.02.034) [DOI] [PubMed] [Google Scholar]
- 184.Kanki T, et al. 2013. Casein kinase 2 is essential for mitophagy. EMBO Rep. 14, 788–794. ( 10.1038/embor.2013.114) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Otsu K, Murakawa T, Yamaguchi O. 2015. BCL2L13 is a mammalian homolog of the yeast mitophagy receptor Atg32. Autophagy 11, 1932–1933. ( 10.1080/15548627.2015.1084459) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Davis C-HO, et al. 2014. Transcellular degradation of axonal mitochondria. Proc. Natl Acad. Sci. USA 111, 9633–9638. ( 10.1073/pnas.1404651111) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Wrobel L, et al. 2015. Mistargeted mitochondrial proteins activate a proteostatic response in the cytosol. Nature 524, 485–488. ( 10.1038/nature14951) [DOI] [PubMed] [Google Scholar]
- 188.Papa L, Germain D. 2011. Estrogen receptor mediates a distinct mitochondrial unfolded protein response. J. Cell Sci. 124, 1396–1402. ( 10.1242/jcs.078220) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Riar AK, Burstein SR, Palomo GM, Arreguin A, Manfredi G, Germain D. 2017. Sex specific activation of the ERalpha axis of the mitochondrial UPR (UPRmt) in the G93A-SOD1 mouse model of familial ALS. Hum. Mol. Genet. 26, 1318–1327. ( 10.1093/hmg/ddx049) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Bragoszewski P, et al. 2015. Retro-translocation of mitochondrial intermembrane space proteins. Proc. Natl Acad. Sci. USA 112, 7713–7718. ( 10.1073/pnas.1504615112) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Mårtensson CU, et al. 2019. Mitochondrial protein translocation-associated degradation. Nature 569, 679–683. ( 10.1038/s41586-019-1227-y) [DOI] [PubMed] [Google Scholar]
- 192.Izawa T, Park SH, Zhao L, Hartl FU, Neupert W. 2017. Cytosolic protein Vms1 links ribosome quality control to mitochondrial and cellular homeostasis. Cell 171, 890–903. ( 10.1016/j.cell.2017.10.002) [DOI] [PubMed] [Google Scholar]
- 193.Ruan L, et al. 2017. Cytosolic proteostasis through importing of misfolded proteins into mitochondria. Nature 543, 443–446. ( 10.1038/nature21695) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Soubannier V, Rippstein P, Kaufman BA, Shoubridge EA, McBride HM. 2012. Reconstitution of mitochondria derived vesicle formation demonstrates selective enrichment of oxidized cargo. PLoS ONE 7, e52830 ( 10.1371/journal.pone.0052830) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Soubannier V, et al. 2012. A vesicular transport pathway shuttles cargo from mitochondria to lysosomes. Curr. Biol. 22, 135–141. ( 10.1016/j.cub.2011.11.057) [DOI] [PubMed] [Google Scholar]
- 196.Melentijevic I, et al. 2017. C. elegans neurons jettison protein aggregates and mitochondria under neurotoxic stress. Nature 542, 367–371. ( 10.1038/nature21362) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Shpilka T, Haynes CM. 2018. The mitochondrial UPR: mechanisms, physiological functions and implications in ageing. Nat. Rev. Mol. Cell Biol. 19, 109–120. ( 10.1038/nrm.2017.110) [DOI] [PubMed] [Google Scholar]
- 198.Samluk L, Chroscicki P, Chacinska A. 2018. Mitochondrial protein import stress and signaling. Curr. Opin. Physiol. 3, 41–48. ( 10.1016/j.cophys.2018.02.010) [DOI] [Google Scholar]
- 199.Fischer F, Hamann A, Osiewacz HD. 2012. Mitochondrial quality control: an integrated network of pathways. Trends Biochem. Sci. 37, 284–292. ( 10.1016/j.tibs.2012.02.004) [DOI] [PubMed] [Google Scholar]
- 200.Palikaras K, Lionaki E, Tavernarakis N. 2018. Mechanisms of mitophagy in cellular homeostasis, physiology and pathology. Nat. Cell Biol. 20, 1013–1022. ( 10.1038/s41556-018-0176-2) [DOI] [PubMed] [Google Scholar]
- 201.Yoo S-M, Jung Y-K. 2018. A molecular approach to mitophagy and mitochondrial dynamics. Mol. Cells 41, 18–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Picard M, Wallace DC, Burelle Y. 2016. The rise of mitochondria in medicine. Mitochondrion 30, 105–116. ( 10.1016/j.mito.2016.07.003) [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
This article does not contain any additional data.