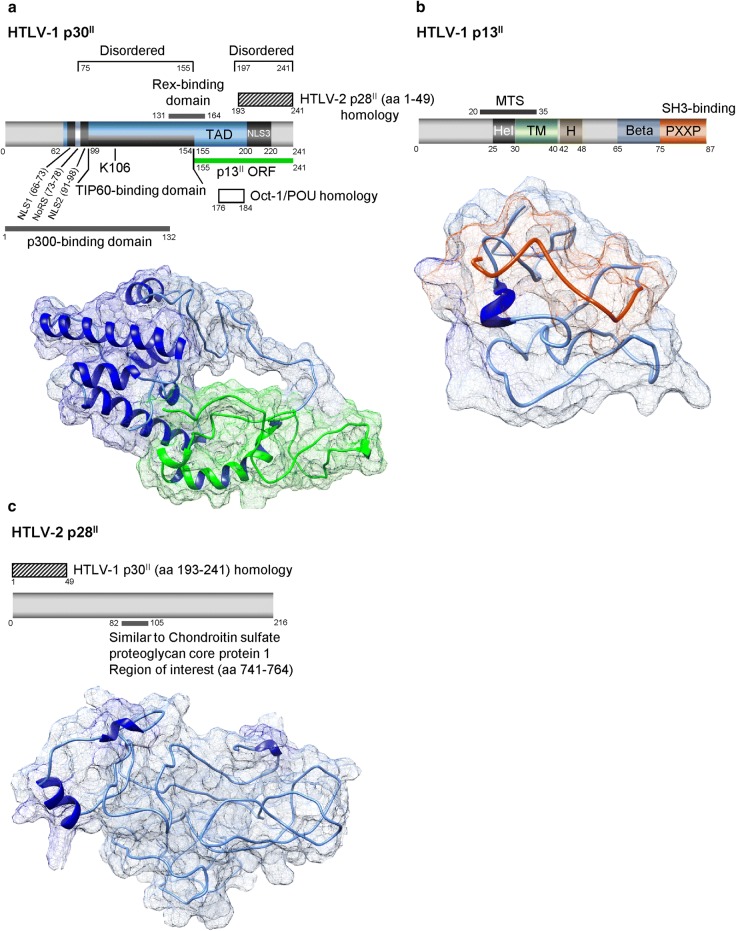
Fig. 4.
Functional domains and predicted structures of the HTLV-1 p30II, p13II, and HTLV-2 p28II proteins. The predicted structures were generated using the I-TASSER algorithm and modeled using UCSF-Chimera. a Diagram and structure of HTLV-1 p30II. TAD, transcriptional activation domain; NLS, nuclear localization signal; NoRS, nucleolar retention sequence. b The p13II aa sequence overlaps with the C-terminal region of HTLV-1 p30II (aa 155–241) and is colored green in the p30II structure (lower panel, a). The SH3-binding domain (aa 75–87) of HTLV-1 p13II which contains a PXXP motif is colored orange in the diagram and in the modeled structure (b). MTS, mitochondrial targeting sequence; Hel, helical region; TM, transmembrane domain; H, flexible hinge region; Beta, predicted beta-sheet secondary structure. c The amino-terminus of the HTLV-2 p28II protein (aa 1–49) shares 78% sequence homology with aa residues 193–241 of HTLV-1 p30II. A region of 58% similarity with a peptide sequence of the Chondroitin sulfate proteoglycan core protein 1 (aa 741–764) is also indicated