Significance
Nearly one-third of adults over the age of 65 have lost all their teeth. We set out to understand tooth renewal in animals that have replacement and regeneration capabilities. Using cichlid fishes and mouse models, we discovered plasticity between tooth and taste bud progenitor cell derivatives, mediated by BMP. Our results suggest that oral organs have surprising regenerative capabilities and can be manipulated to express characteristics of different tissue types.
Keywords: regeneration, teeth, taste buds, stem cells
Abstract
In Lake Malawi cichlids, each tooth is replaced in one-for-one fashion every ∼20 to 50 d, and taste buds (TBs) are continuously renewed as in mammals. These structures are colocalized in the fish mouth and throat, from the point of initiation through adulthood. Here, we found that replacement teeth (RT) share a continuous band of epithelium with adjacent TBs and that both organs coexpress stem cell factors in subsets of label-retaining cells. We used RNA-seq to characterize transcriptomes of RT germs and TB-bearing oral epithelium. Analysis revealed differential usage of developmental pathways in RT compared to TB oral epithelia, as well as a repertoire of genome paralogues expressed complimentarily in each organ. Notably, BMP ligands were expressed in RT but excluded from TBs. Morphant fishes bathed in a BMP chemical antagonist exhibited RT with abrogated shh expression in the inner dental epithelium (IDE) and ectopic expression of calb2 (a TB marker) in these very cells. In the mouse, teeth are located on the jaw margin while TBs and other oral papillae are located on the tongue. Previous study reported that tongue intermolar eminence (IE) oral papillae of Follistatin (a BMP antagonist) mouse mutants exhibited dysmorphic invagination. We used these mutants to demonstrate altered transcriptomes and ectopic expression of dental markers in tongue IE. Our results suggest that vertebrate oral epithelium retains inherent plasticity to form tooth and taste-like cell types, mediated by BMP specification of progenitor cells. These findings indicate underappreciated epithelial cell populations with promising potential in bioengineering and dental therapeutics.
Humans have evolved long life spans but often retain organs damaged during their lives. This is perhaps taken to the extreme in our dentitions. One-fifth of all humans exhibit genetic disorders affecting teeth (primary or permanent), and nearly all humans develop dental problems (e.g., cavities) with age. Thirty percent of humans worldwide over the age of 65 have none of their 32 natural teeth remaining in their mouths (World Health Organization). Whereas nonmammalian teeth are replaced de novo throughout life via various mechanisms (1), mammals have largely lost this dental regenerative ability. For example, humans replace each tooth only once, and mice never replace their teeth. Instead, mice and other rodents exhibit continuously growing incisors wherein enamel is renewed asymmetrically on the labial (outside) surface, which bears the brunt of primary mastication.
In the mouse, the base of each incisor contains a region called the cervical loop (CL), the location of an epithelial stem cell (ESC) niche that has become a powerful model for understanding stem cell (SC) biology. In the incisor CL, a histologically distinct group of mesenchymal-like epithelial cells called stellate reticulum (SR) lie sandwiched in between the inner enamel epithelium (IEE) and outer enamel epithelium (OEE). A subset of cells from within the SR serve as ESCs, differentiating into transit amplifying (TA) cells that will multiply to generate enamel-secreting ameloblasts along the IEE (2, 3). Classic work demonstrated that Sox2 marks the putative ESC niche in a cadre of mammals and reptiles (4), bony fishes (5), and sharks (1, 6). Genetic fate mapping experiments show that Sox2+ ESCs contribute to all lineages of the dental epithelium (7, 8).
Recent work in models of stem cell-driven organ renewal (e.g., tooth, intestine, hair follicle, lung) has revealed surprising plasticity and noteworthy context dependence of epithelial cellular behavior (9). In each of these systems, there are multiple stem cell types and conditions during which plasticity between types is favored. For example, in the intestinal crypt and the hair follicle, cells from differentiated organ zones can regain stem cell competence and ultimately repopulate the organ upon targeted ablation (10). Notably, epithelial cells from outside the hair follicle can migrate to the follicular stem cell niche. Once in position, these cells behave like endogenous stem cells (11). Likewise, in the mouse incisor, renewal is restored in certain circumstances by recruitment of Sox2+ cells from a Sox2− cell population (8). The degree to which such heterogeneity exists in mesenchymal stem cell populations is less well studied, but consensus is emerging. In mouse incisor mesenchyme for instance, pericytes can be reprogrammed to form odontoblasts upon injury (12) whereas neural crest-derived glia (13) and Gli1-expressing periarterial cells associated with the neurovascular bundle (14) contribute to dental pulp homeostasis. Recently, we showed that Celsr1 marks a population of quiescent cells that are mobilized to replenish CD90+ dental mesenchymal stem cells (MSCs), in specific response to incisor clipping (15). Taken together, these studies highlight the importance of the niche-signaling environment, which can impinge upon internal molecular programs to mediate reversible behavior of epithelial and mesenchymal stem cells (16) and/or mobilize populations of cells otherwise quiescent. Heterogeneity and plasticity are fundamental features of stem cell systems that must be understood as bioengineers and developmental biologists seek to manipulate cell biology for regenerative therapies.
Developmental plasticity is also apparent between organ systems. In previous work, we showed that developing teeth and taste buds in cichlid fishes share a bipotent epithelium during early patterning stages, from which dental and taste fields are specified (17). Small molecule manipulation of the Wnt, Hh, and BMP pathways, during the critical developmental window when organ fields differentiate one from the other, provides clues to the regulatory logic of cichlid tooth and taste bud copatterning. Wnt signaling couples tooth and taste bud density and acts upstream of Hh and BMP, which together mediate organ plasticity. Notably, treatment with the small molecule LDN, which inhibits BMP signaling, results in a striking phenotype wherein tooth density is reduced and taste buds infiltrate the tooth field. These results resemble studies of the integument wherein BMP-modulated transgenic K14-Noggin mice demonstrated “trans-differentiation” of sweat glands from the distal limbs into hairs (18), and both spatial and temporal modulation of BMP through development and lentivirus manipulation specified the fate of sweat glands versus hair in mice (19).
The relationship between teeth and taste bud renewal has recently been established in the shark wherein a sox2+ “odontogustatory band” (OGB) has been demonstrated to share a progenitor pool of cells associated with taste buds that migrate into the dental successional lamina (6). Similar results have now been observed during the development of the beaked dentition in pufferfish (20). Additionally, recent study demonstrates the conversion of characteristics between integument-derived epithelial appendages, including the growth of hair from teeth (21) and the transition from scales to feathers (22). In the latter case, transcriptome analyses implicated specific pathways, β-catenin and retinoic acid, in the process (23).
Given the integration of teeth and taste buds during cichlid early development (17), we were prompted to investigate the renewal and regeneration of these organs at later stages. To this end, we 1) identified label-retaining cells (LRCs) using a pulse–chase strategy and 2) examined the activity of adult stem cell markers in these adjoining structures undergoing lifelong renewal (taste buds) and replacement (teeth). We performed unbiased RNA-seq of replacement tooth and taste bud tissues and highlighted the role of BMP ligands in replacement teeth (RT) and not taste buds (TBs). Finally, in both fish teeth and on mouse tongues, we manipulated the BMP signaling environment and demonstrated striking interorgan epithelial plasticity between teeth and taste-like papillae.
Results
Expression of Putative Stem Markers Reveals a Highly Potent Oral Epithelium Connecting Teeth and Taste Buds.
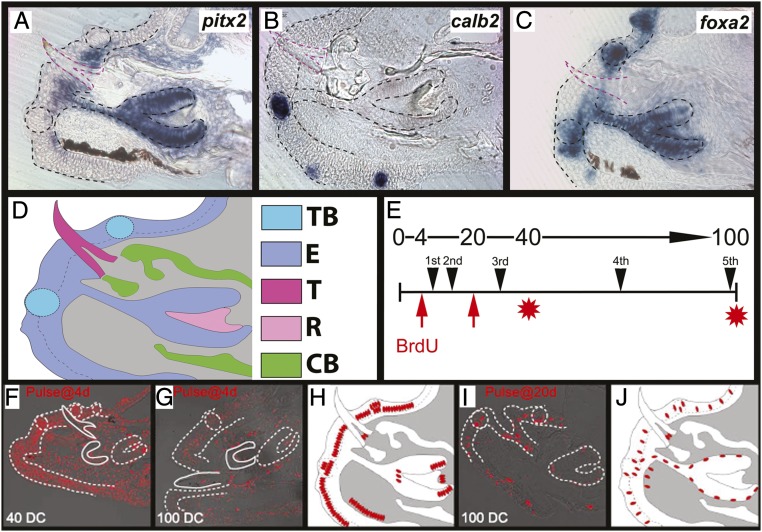
We began our study with the aim of identifying the spatial location of putative stem cell niches in both renewing TBs and successional teeth through in situ hybridization (ISH). Three distinct stages of cichlid replacement tooth maturation are known: initiation, which encompasses placode or successional dental lamina stages of odontogenesis; differentiation, which encompasses cap and bell stages; and secretion, which encompasses late bell and mineralization stages (5). We chose to focus our study on the latter because RT spend the longest time in this stage and secretion is the most consistent in histology across different aged cichlids. We conducted our ISH experiments on cichlid fry (∼30 days postfertilization [dpf]), developing their third generation of teeth. We first used pitx2, a marker of dental tissues in vertebrates, including cichlids (24) and mice (25), to identify those cells belonging to the RT germ but noted expression along the successional dental lamina, along basement membrane cells deep in the oral epithelium, both labial and lingual to the taste bud unit (Fig. 1 A and D, schematic). As we have done before in cichlids (17, 26) and has been done in trout (27), we used calretinin (calb2) to mark TBs, which more specifically was expressed in the elongated taste bud intragemmal cells and the support perigemmal cells that surround it (Fig. 1 B and D, schematic). We attempted to further characterize the taste bud unit using foxa2, a marker of the endoderm and taste buds in mice (28) and in zebrafish (29), but, to our surprise, foxa2 not only marked TBs but was strongly expressed in all RT cells and across outer oral epithelium associated with these two organs (Fig. 1C).
Fig. 1.
ISH identification of tissue in RT-TB unit. Expression of pitx2 in cichlid RT germ and oral epithelium (A), calb2 expressed in TB (B), and foxa2 across RT and TB epithelium (C) during the secretion stage of cichlid replacement tooth development. Functional tooth is outlined in magenta. Shown are vibratome sections in the sagittal plane at 15-μm thickness, imaged at 40× magnification. Labial is oriented to the bottom and oral toward the left of the page. Schematic (D) shows tissue architecture with taste bud in light blue, epithelium in purple, erupted functional tooth in magenta, replacement tooth mesenchyme in pink, and ossified tissue in green. Time scale of experimental design for pulse–chase experiments (E) with 0, 4, 20, 40, and 100 days chase (DC) on top and black arrowheads representing tooth generations below. Pulse (BrdU incorporation) was started at either 4 or 20 dpf, and killing was made at either 40 or 100 DC. Many cells labeled by IHC for BrdU after 40 DC (F) with discrete cell populations by 100 DC (G, and schema in H). When pulsed beginning at 20 dpf, again followed by a 100 DC (I), fewer cells are labeled, with discrete populations still apparent (J). Labial oriented to the bottom and oral to the left of the page. Paraffin sections in sagittal plane at 15-mm thickness, imaged at 40× magnification. Epithelium outlined in black (A–C) or white (F, G, I) with taste buds outlined in circles.
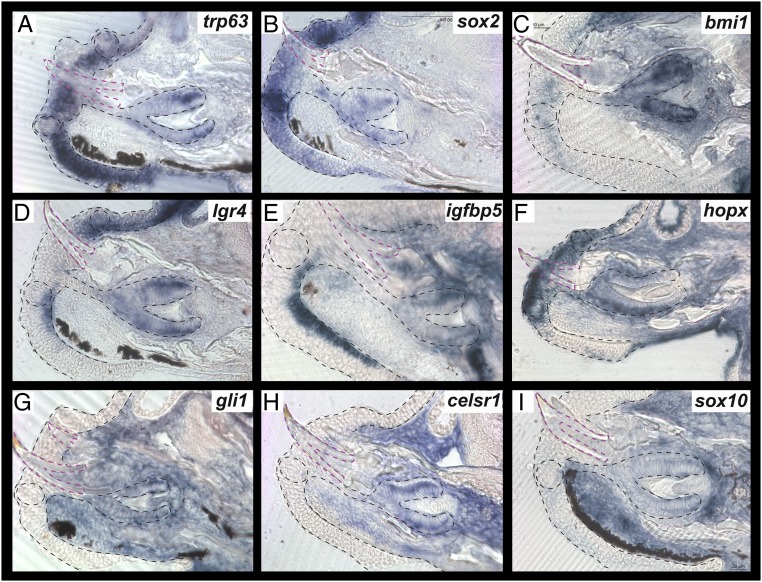
We turned our focus to markers of, or associated with, adult stem cells. trp63 is a p53 transcription factor family member and a well-known marker of proliferation and mitotic activity. Trp63-deficient mice exhibit both anodontia and a thin degenerate tongue epithelial layer (30). A hypothesized bipotent progenitor layer shared between filliform and fungiform taste papilla is marked by Trp63 in mice (31). We found expression of trp63 to mirror expression of the progenitor layer in mice (Fig. 2A), being strongly expressed in the basal cells surrounding the intragemmal TB cells continuing across the oral epithelium overlying the dental lamina and in the CLs of the RT epithelium, in a pattern complimentary to that of sox2. Sox2 is one of the most studied factors in taste bud development, stem cell biology, and, more recently, in tooth regeneration. While Sox2 is a well-known TB marker, important for both the generation of TBs as well as maintenance of TB stem cell populations (32), it has more recently been implicated in dental ESCs of both mice (7) and other vertebrates (4). It has also been demonstrated as a marker of shared progenitor cells in the shark OGB, giving rise to cells in the tooth/taste bud interface (6). We found sox2 expression in TBs, as well as RT and across epithelium associated with the two structures (Fig. 2B). Bmi1, a Polycomb group gene required for adult stem cells in a host of organs (33), is essential for incisor renewal through the repression of Ink4a/Arf and Hox genes (34) while, in TBs, it appears that a population of Bmi1+ SCs renew keratinized epithelial cells distinct from SCs that renew TB cells themselves (35). We observed expression of bmi1 distinct from that of sox2, diffusely across TBs and surrounding epithelium and in RT epithelium. The difference between sox2 and bmi1 expression is not surprising. In the mouse intestine, Bmi1 marks a population of stem cells that is relatively quiescent and activated in response to injury while another distinct population of Lgr5-positive stem cells are more active and responsible for regular renewal of the crypt unit (36), and, further, each population responds differently to tissue perturbations, such as apoptosis (37). While no homolog of Lgr5 exists in teleost fishes, lgr4 was expressed in cichlid RT and TBs more similarly to that of sox2, although more restricted to the basal layers of epithelium in and around the taste unit. This is an intriguing result, given that Lgr5+ cells replenish Sox2+ cells after genetic deletion of Sox2 in mouse incisors (8). igfbp5, shown coincident in expression to lgr5 in gecko RT dental lamina (38), indeed colabels the basal epithelial cells associated with the regenerating RT–TB unit. Meanwhile, Hopx, which has been used to label SCs in intestine (39) and hair follicles (40), is expressed in the sox2/lgr4/igfbp5+ and bmi1+ populations, as well as strongly within the dental lamina. We noted expression of bmi1, igfbp5, and hopx in mesenchyme, as well as in epithelium (Fig. 2). Markers of mesenchymal stem cells (MSCs) were indeed expressed in oral mesenchyme, but also across epithelial tissue. gli1 was expressed across dental and TB mesenchyme, as well as in dental epithelium, but absent from taste and surrounding oral epithelium (Fig. 2G). celsr1, recently described as a marker of quiescent cells in mouse dental mesenchyme (15), and sox10 were expressed in the epithelium of RT, the mesenchyme subjacent to epithelium in the TB and dental lamina, and outside of the dental papilla itself but in crypt mesenchyme near the CLs (Fig. 2 H and I). Taken together, our ISH data indicate that markers of adult ESCs and MSCs are expressed within RT, TBs and the dental successional lamina connecting the two organs.
Fig. 2.
ISH of adult proliferation and stem markers. Expression of trp63 (A), sox2 (B), bmi1 (C), lgr4 (D), igfbp5 (E), hopx (F), gli1 (G), celsr1 (H), and sox10 (I) during the secretion stage of cichlid replacement tooth development. Functional tooth is outlined in magenta. Shown are vibratome sections in the sagittal plane at 15-μm thickness, imaged at 40× magnification. Labial is oriented to the bottom and oral toward the left of the page. Epithelium outlined in black dashed lines and functional teeth in magenta, with taste buds outlined in circles.
Colabeling with Stem Markers and Nucleoside Chase Identifies Stem Cell Niches across the Oral Epithelium.
To more precisely identify SC niche environments in cichlid RT and TBs, we conducted nucleoside pulse–chase experiments, one of the primary experiments done to first identify stem cells in the mouse incisor (41). By exposing animals to the synthetic nucleosides 5-bromo-2′-deoxyuridine (BrdU) or 5-chloro-2′-deoxyuridine (CldU), it is incorporated into newly created cells, and, once removed, only those cells that are slow cycling or nondividing, a property of stem cells, will be label-retaining cells (LRCs). We bathed cichlid fry in a solution containing BrdU at pharyngula stage (4 dpf) for a period of 1 wk and then killed sequentially until LRCs were identified (Fig. 1E). Numerous cells retained the BrdU label after 40 d of chase, but, by 100 d (at least 4 cycles of replacement teeth), discrete cell populations were apparent (Fig. 1 F and G). By 100 d of chase, there was a high density of LRCs across all epithelium labial to the FT, within the TB unit and within the RT. We also detected LRCs in the mesenchyme, mostly associated in a band approximate to the epithelium and in the dental papilla. In order to compare the effects of early versus late pulse, we repeated these experiments beginning with a pulse beginning at 20 dpf, again followed by a 100-d chase period. We found that both the late pulse and long chase groups resulted in fewer LRCs (Fig. 1I), likely because many founder stem cells are formed early in development (42).
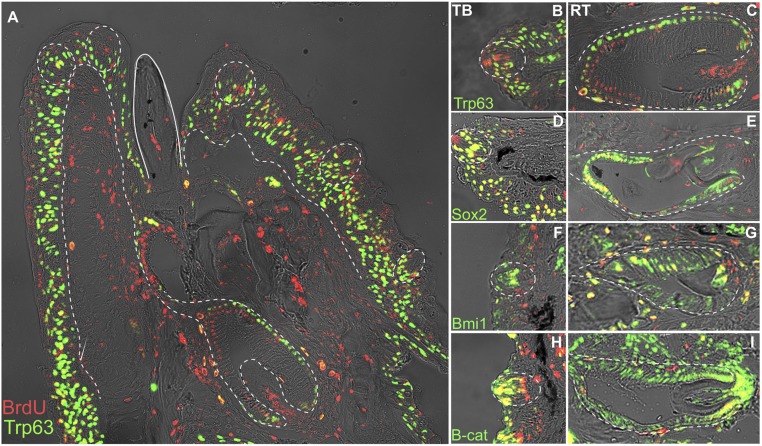
Because ISH revealed the expression of multiple adult stem markers within RT and TBs, we used double labeling of BrdU LRCs with immunohistochemistry (IHC) to better characterize putative stem niches associated with these organs. In accordance with mRNA expression, Trp63 protein, a marker of proliferation, was detected in TB support cells and coexpressed with LRCs across the basal cells of outer oral epithelium (Fig. 3 A and B). After 20-d pulse and immediate sacrifice, (SI Appendix, Fig. S1), Trp63, a marker of proliferation, and BrdU cells were overlapping at the tip of the tooth, marking proliferating cells that are likely undergoing transient amplification (TA). After 4-d pulse and subsequent chase, Trp63 was detected in outer dental epithelial (ODE) cells, analogous to mammalian OEE, and associated with the CL, while LRCs were located in a subset of cells closer to the inner dental epithelium (IDE) and the apex of the dental papilla (Fig. 3 A and C). The lack of overlap between these markers after a chase period is indicative of a distinction between putative TA cells, labeled by Trp63, and putative quiescent populations, labeled by BrdU. In contrast, Sox2 was found coexpressed with LRCs associated at the base of taste buds in perigemmal cells (Fig. 3D) and at the tips of RT (Fig. 3E). Bmi1 protein colabeled a smaller subset of LRCs; its domain in the epithelium of both organs was largely in the more superficial cells, many of which were negative for label retention (Fig. 3 F and G). Finally, β-cat, a regulator of stem cell maintenance and differentiation through Wnt signaling (43, 44), was coexpressed with epithelial and mesenchymal cells after pulse at 20 dpf and no chase period and after pulse at 4 dpf and 100-d chase. β-cat was active in LRCs not marked by Sox2, at the intragemmal tip of the taste bud and at the cervical loops and papilla of the RT (SI Appendix, Fig. S1 and Fig. 3 H and I). Taken together, we observed distinct populations of LRCs: Sox2+/LRC+ populations at the base of taste buds, in the tips of teeth, and in the CLs; β-cat+/LRC+ populations at the base of teeth and in the tips of taste buds; and distinct Trp63+ and LRC populations following a chase period. Colabeling of Sox2/β-cat/Bmi with LRCs confirmed the presence of an SC-rich epithelium, and, within it, distinct SC niches became apparent: those associated with the base of the taste bud analogous to murine TB SCs (32), those associated with the incisor CL (7, 13), and an SC niche not described in the dental literature, occupying cells at the tip of the maturing RT. This niche has been understudied likely because very little to date has been published on the stem cell populations involved in whole tooth replacement in a one-for-one replacement system.
Fig. 3.
Double label for LRC and IHC of adult proliferation and stem markers. Fluorescent labeling after BrdU pulse at 4 dpf and killing at 100 days chase with BrdU labeled in red and protein markers in green. Yellow/orange indicates coexpressing cells. Functional tooth is outlined by solid line, epithelium oral and taste epithelium by dashed lines. Trp63 (A) is a marker of proliferation; labial is oriented to the left and oral toward the top of the page. A zoom-in of taste buds (B, Trp63; D, Sox2; F, Bmi1; H, B-cat) and teeth (C, Trp63; E, Sox2; G, Bmi1; and I, B-cat) shows zones of proliferation, label retention, and stem marker expression, with labial oriented to the bottom and oral to the left of the page. Shown are paraffin sections in the sagittal plane at 15-μm thickness, imaged at 40× magnification.
RNA-Seq Reveals Unique Transcriptomes of Replacement Teeth and Taste Buds.
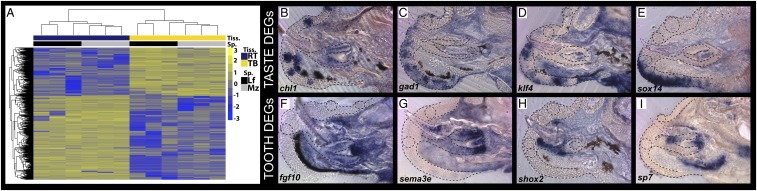
Because cichlid teeth and taste buds are copatterned during early development and because we know little about the molecular biology of vertebrate tooth replacement, we used an unbiased RNA-seq approach to characterize gene expression profiles in adult cichlid replacement teeth and taste buds. We dissected a band of epithelium, just labial to the outer row of adult cichlid functional teeth, which contained a high density of taste buds (17). We then removed the periosteum surrounding the dental bony crypts and isolated replacement tooth germs, at secretion stage, easily identified by their hypermineralized acrodin cap. We pooled taste bud-bearing epithelial tissues and replacement tooth germs for each animal, extracted RNA, prepared RNA libraries, and performed RNA-seq on the Illumina 2500 platform. High quality reads were aligned to the Malawi cichlid reference genome (on average, across all samples, over 95% of reads mapped to the reference). Fragment counts across all samples were obtained, normalized, and fit to a linear model to determine differential expression between tissue types. Genes were considered significantly differentially expressed between teeth and taste buds if they exhibited both a 2-fold expression difference or greater and an adjusted P value of <0.05. Using this criterion, we found that 3,902 genes were differentially expressed between the tissue types (Fig. 4). Of those, 2,482 were up-regulated in dental tissues while 1,420 were up-regulated in taste buds (Dataset S1). Significant tooth-biased genes included axin2, bmp2, bmb4, bmp6, bmp10, dlx4, dlx5, dlx6, edar, fgf10, msx2, pax9, pitx2, runx2, sostdc1, and wnt7b. Genes biased in taste buds included avpr2, barx2, dmbx1, egfr, osr1, six3, and sox14. For a subset of these genes, we confirmed differential expression by ISH (Fig. 4 C–I). Notably, we identified hundreds of genome paralogues expressed complementarily in RT or TBs, respectively. Paralogues included glutamate receptors, keratins, solute carriers, and zinc finger proteins (Dataset S2). Given the role of paralogous genes in evolutionary novelty (45), we were intrigued by this example of complementary paralogue expression in adjacent oral organs. Six of 15 loci implicated by GWAS in human tooth number (ajuba, bmp4, calu, cacnb2, cdon, and vcl) (46) were expressed in cichlid replacement teeth. We used GeneAnalytics (47) and ToppGene (48) to identify pathway and phenotype enrichment based on lists of differentially expressed genes (Dataset S3). Notable statistical enrichment in replacement teeth included the categories “odontogenesis,” “bone morphogenesis,” “focal adhesion,” “mesenchymal stem cell differentiation,” “Wnt signaling,” “TGF-beta signaling,” “extracellular matrix organization,” and “abnormal molar morphology.” Categories enriched in taste buds included “epidermis development,” “abnormal tongue epithelium morphology,” and “signaling by ERBB2.”
Fig. 4.
RNA-seq of RT and TB bearing epithelium. Heat map (A) of differentially expressed genes shows clustering based on sample type (x axis; with species data) and log fold change (heat; yellow to blue) for individual genes (y axis). ISH expression bias in TBs bearing epithelium chl1 (B), gad1 (C), klf4 (D), and sox14 (E), or in RT tissues fgf10 (F), sema3e (G), shox2 (H), and sp7 (I) for genes significantly differentially expressed in sequencing analysis. Shown are vibratome sections in the sagittal plane at 15-μm thickness, imaged at 40× magnification. Labial is oriented to the bottom and oral toward the left of the page. Tiss., tissue; Sp., species; DEG, deferentially expressed genes.
Manipulation of the BMP Pathway Uncovers Interorgan Plasticity between RT and TBs during Regeneration and Renewal.
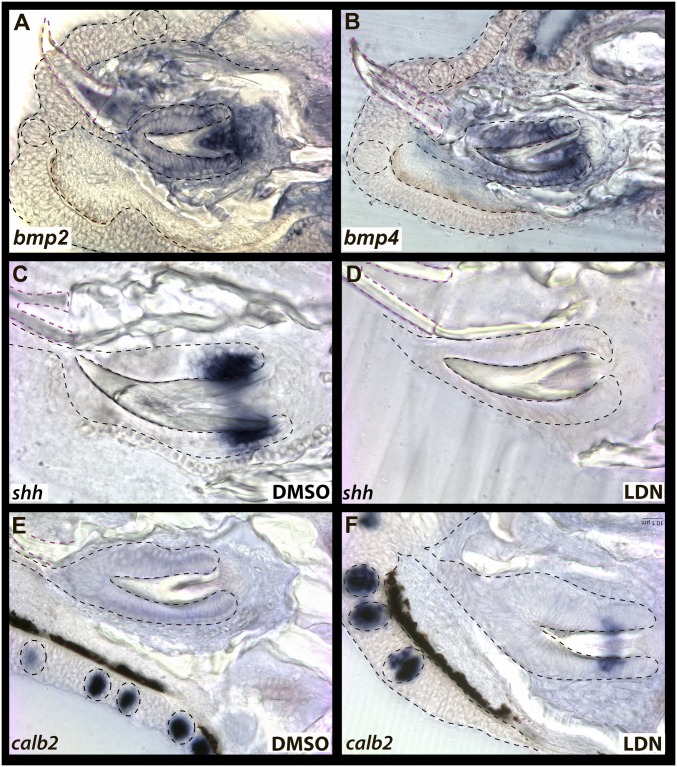
The pleiotropic effects of BMPs on multiple epithelial appendages has been well established, such as Bmpr1a control of both tooth and hair differentiation (49), and the role of BMPs in integument transdifferentiation has been demonstrated to have spatial and temporal effects in development (19). Given differential expression of BMPs, functional enrichment of this pathway in cichlid replacement teeth, and previous demonstration that BMP inhibition results in ectopic taste bud formation in dental field progenitor epithelium (17), we further explored the spatial expression of BMP ligands in cichlid replacement teeth. We observed that bmp2 and bmp4 were sharply expressed in replacement tooth epithelium and mesenchyme but excluded from all tissues in and around the taste bud (Fig. 5 A and B). In turn, we bathed cichlids in the small molecule inhibitor of BMP, LDN, for a period of 48 h and then killed to characterize treated replacement teeth at the cellular and transcript level. Shh regulates preameloblasts in the rodent incisor transitioning from stem cell-derived TACs to enamel-secreting ameloblasts (50). In control (dimethyl sulfoxide [DMSO]) animals, shh was expressed in the analogous inner dental epithelium of cichlid teeth (Fig. 5C). Upon exposure to LDN, shh expression was undetectable, and replacement teeth appeared shorter and malformed, particularly in the cervical loop regions (Fig. 5D). We then assayed expression of the taste bud marker calb2 in LDN-treated animals and observed expression of calb2 in cervical loop epithelium in regions where shh expression was abrogated (Fig. 5F). These data suggest that temporary loss of BMP signaling in cichlid replacement teeth results in dental cells adopting taste bud characteristics.
Fig. 5.
BMP expression and effect of LDN on RT SC differentiation. bmp2/4 bias to RT (A and B). Solvent control RT express shh (C) and not calb2 (E) in CL epithelium, but LDN-treated RT express calb2 (F) and not shh (D). Shown are vibratome sections in the sagittal plane at 15-μm thickness, imaged at 40× magnification. Labial is oriented to the bottom and oral toward the left of the page. Epithelium outlined in black dashed lines and functional teeth in magenta, with taste buds outlined in circles.
Tongue Papillae of Fst−/− Mice Adopt the Characteristics of Teeth.
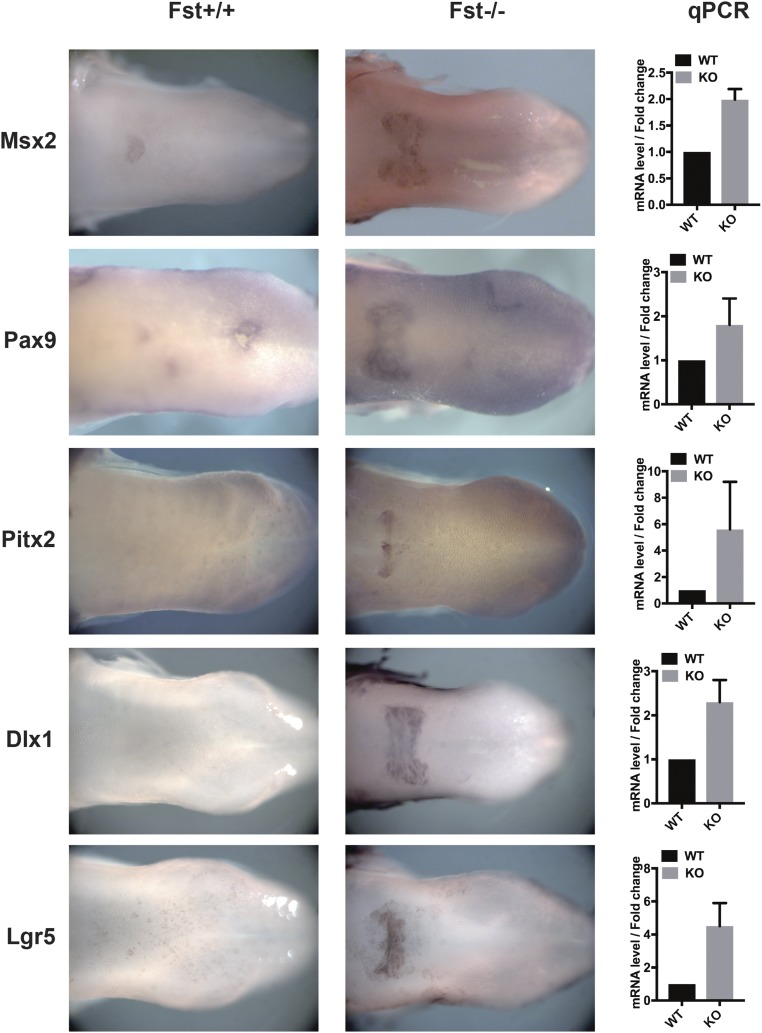
Given the protooth role of the BMP pathway in our data and elsewhere, as well as the expression of BMP antagonists (fst, sostdc1, and osr2) in embryonic taste territories in cichlids (17), we were intrigued by results from Beites et al. (51), who demonstrated that mouse tongues in Follistatin mutants (Fst−/−) exhibited dysmorphic epithelium in the posterior intermolar eminence (IE). The IE epithelium of Fst−/− mice invaginates and expresses Sox2, Shh, and Foxa2 (51), similar to cichlid teeth (ref. 17 and above), but somewhat distinct from more anterior gustatory mouse tongue papillae. Thus, we first examined the transcriptomes of dissected IE epithelium from embryonic day 17.5 (E17.5) wild-type and Fst−/− littermates. This unbiased approach identified ∼700 differentially expressed genes (Dataset S4). We hypothesized that those genes up-regulated in this context might carry a tooth-like signature. Indeed, this gene set was enriched for biological process Gene Ontology (GO) categories “bone morphogenesis,” “Wnt signaling,” and “odontogenesis” (Dataset S3). Notably, the overlap in GO biological process terms between genes up-regulated in Fst−/− IE papillae and those enriched in cichlid replacement teeth is statistically significant (Fisher’s exact test, P < 2.2−27; shown graphically in SI Appendix, Fig. S2). Next, we examined the spatial domain of tooth markers up-regulated on Fst-null tongues. Using in situ hybridization and qPCR, we confirmed that numerous dental markers are misexpressed in the IE of mouse tongues lacking Follistatin. These include Bmp7, Shh, and Foxa2 (as shown by ref. 51), as well as Gli1, Osr2, Msx1, Msx2, Pax9, Pitx2, Barx1, and Dlx1 (Fig. 6 and SI Appendix, Figs. S3 and S4). Our experiments in mouse mirror those conducted in cichlids and highlight a surprising long-term plasticity between dental and other oral organ types, mediated by BMP.
Fig. 6.
Mouse tongues lacking Follistatin misexpress dental markers in the intermolar eminence. Dorsal view of whole mount ISH for a panel of tooth markers shows misexpression of these genes in Follistatin mutants (Fst−/−). Quantification of this effect by qRT-PCR follows dissection of intermolar eminence tissue. (Magnification: 10×.)
Discussion
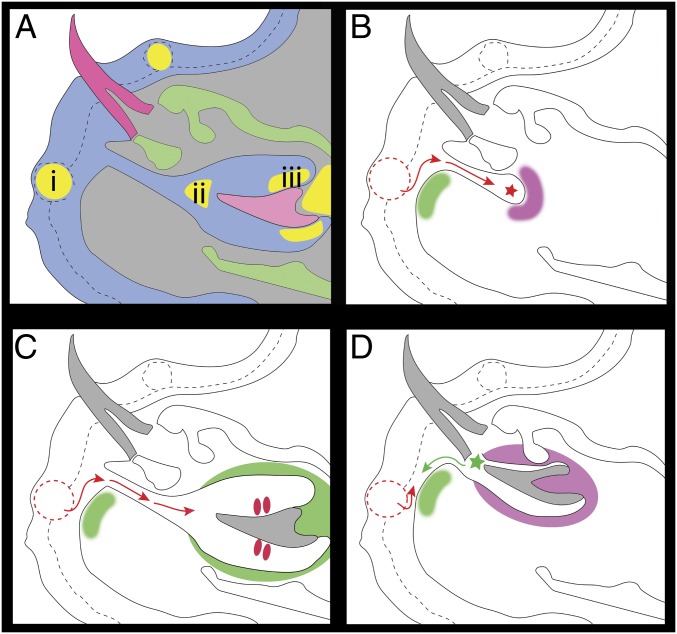
Here, we report three main discoveries. Cichlid one-for-one, cycled tooth replacement occurs in anatomical linkage with taste buds undergoing continuous renewal. These two structures, teeth and taste buds, are colocalized in the oro-pharynx of most nonmammalian vertebrates, and the two organ types share developmental precursors and deep molecular homology (17, 52). We identified 5 anatomical zones (Fig. 7), or stem cell niches, where label-retaining cells reside: 1) labial and 2) lingual cervical loops (CLs) (similar to mouse teeth), 3) at the tip of the tooth, 4) dental pulp mesenchyme, where the tooth is innervated by a neurovascular bundle (NVB), and 5) surrounding the taste bud unit. Stem cell factors Bmi1 and Sox2 colabel subsets of LRCs in the epithelium of replacement teeth and taste buds (Figs. 2 and 3). The tooth tip niche has not been observed in other systems and is particularly interesting. The tip of the replacement tooth is located in close proximity to the dental successional lamina and likely acts as a signaling center to direct tooth morphogenesis (53) (Fig. 7).
Fig. 7.
A model of stem cell niche localization and associated stages of tooth regeneration in the cichlid. Schemes are based on a sagittal section through a single functional tooth and its replacement within the lower jaw. (A) During maturation of the replacement tooth, a number of potential stem cell pockets are identified (yellow). These include regions associated with (i) taste buds in the oral epithelium, (ii) at the tip of the tooth in association with the enamel knot-like structure, and (iii) in the cervical loop-like regions of inner dental epithelium of the developing tooth. (B) As tooth regeneration initiates, an epithelial invagination is triggered by reciprocal interactions of epithelial stem/progenitor cell compartments and the underlying neural crest mesenchyme where stem cells are also thought to be active (at the onset of dental lamina invagination: green; and surrounding the proliferative progression (red arrows) of the successional lamina (red star) toward tooth regeneration: pink). (C) During the differentiation phase of successor tooth development, signals emanate from the surrounding mesenchymal papilla and dental follicle (green). Signals from the oral epithelial compartments associated with taste territories are still directing tooth development and growth (red arrows). (D) As the successor tooth matures and is ready to replace the functional tooth, signals from the epithelium and the dental follicle (pink) start to form the support tissues. As the tooth makes its way toward functionality, signals from the EK-like structure (green star) may direct the epithelial stem/progenitor niche associated with the oral taste territory (red arrow) and the underlying mesenchyme (green) to activate further tooth regeneration as the successor tooth tip (green star) moves past the site of the oral epithelial niche (green arrow). Epithelium outlined in black dashed lines, with taste buds outlined in dashed lined circles (red labial, black lingual).
As we have demonstrated previously for early tooth and taste bud development (17), these organs share syn-expression of many factors during regenerations stages, but BMP ligands appear confined to the tooth zone. Knockdown of BMP signaling results in a striking phenotype wherein dental epithelium expresses calb2, a marker of taste bud fate (Fig. 5). This effect is likely due to the loss of BMP activity in the dental zone such that cells migrating to the dental successional lamina (e.g., ref. 6) or cells already present in the replacement tooth CLs are transfated (Fig. 7). We used mice null for the BMP antagonist Follistatin (Fst) to examine predictions associated with increased BMP signaling on the tongue (51). Notably, the intermolar eminence tongue epithelium of Fst−/− mice invaginates (51), exhibits a modified tooth-like transcriptome, and expresses classic dental markers like Dlx1, Msx1, Msx2, Pax9, and Pitx2 (Fig. 6 and SI Appendix, Figs. S2–S4). The data in mouse model suggest latent plasticity among teeth and tongue papillae, even when the organs are not colocalized or linked via a successional lamina.
The last decades of research in numerous organ systems have uncovered significant intraorgan plasticity of epithelial cells in tissue regeneration (9). Examples of interorgan plasticity are perhaps even more dramatic. Such examples include the case of conditional deletion of Med1 in the mouse incisor, which switches dental to epidermal fate, such that hairs grow in the place of teeth from renewing postnatal dental epithelia (21). Similarly, Wu et al. showed how multiple pathways could each partly convert scales to feathers (22). We demonstrate a change in interorgan characteristics shown consistently in fishes and mouse. Two general concepts are central to these transformations (SI Appendix, Fig. S5) in either developing or renewing organs, as well as the more common cases of intraorgan plasticity. First is the idea of a ground state for cells (committed or multipotent) in tissues that develop from common epithelium. In the examples explored above, dental fate is layered upon a sensory (taste bud) or epidermal ground state. Manipulation of the niche signaling environment can coax precursor cells back into a ground state where differentiation into different placode derivatives can occur (SI Appendix, Fig. S5). Second is the clear spatial and temporal context dependency of the niche-signaling environment. Better understanding of the evolutionary and developmental lineages of cells that function in oral organ systems (54) may provide clues to their manipulation in regenerative medicine.
Materials and Methods
Cichlid Husbandry.
Adult Malawi cichlids were housed in recirculating aquarium systems at 28 °C (Georgia Institute of Technology) for embryo production. Species of Lake Malawi cichlids included Labeotropheus fueleborni (LF), Metriaclima zebra (MZ), Petrotilapia chitimba (PC) and were selected based on embryo availability, with a preference toward MZ, owing to their genome assemblage (55) and partial albinism morph, which permitted better imaging of histological stain. Fertilized embryos were harvested from mouth-brooding females and staged in dpf according to Nile Tilapia developmental series (56). Embryos were raised to desired stages for ISH, pulse–chase experiments, or chemical treatment and euthanized with buffered MS-222 for fixation in either 4% paraformaldehyde or 10% neutral buffered formalin.
Cichlid In Situ Hybridization.
Primers for target probe sequence were designed using the published and annotated genomes of tilapia species Oreochromis niloticus (55) and the aligned genome of Malawi cichlid M. zebra from the University of Maryland Cichlid Blast Server Tool. It has been reported that genomic sequence diversity across the Lake Malawi assemblage is 0.28%, less than reported values for laboratory strains of zebrafish (57), and riboprobes were reactive across Malawi cichlid species. Target sequences were transformed and cloned, and sequences were deposited in GenBank (26). Riboprobes were synthesized and labeled with Digoxigenin (DIG) (Roche) using the Promega System Sp6/T7. In situ hybridization was performed using previously published methods in whole mount (24) and visualized using an alkaline phosphatase (AP)-conjugated anti-digoxigenin antibody (Roche) to activate an NBT/BCIP (Roche) blue color reaction. Specimens were embedded in chick albumin and cross-fixed with 2.5% glutaraldehyde followed by being postfixed with 4% paraformaldehyde (PFA). Histological sections were cut at 18 to 20 μm using a Leica Microsystems VT1000 vibratome and then mounted with glycerine for imaging using a Leica DM2500 compound microscope with 20× to 40× objectives.
BrdU/CldU Labeling.
BrdU pulse–chase experiments were carried out to label slow-cycling cells, a property of stem cells. Specimens reared to 4 dpf were bathed in either a 2% solution of BrdU in vivo labeling reagent (00-0103; Invitrogen) or in 200 mL of fish room water at 28 °C in an Erlenmeyer flask. A similar pulse was performed on fish reared to 20 dpf using a 0.1 molar stock solution of CldU (C6891; Sigma) made with DMSO. This solution was added to 200 mL of fish water in 100-mL aliquots three times daily for a period of 1 wk. Daily, 1-mL aliquots of BrdU/CldU solution were added for a total labeling period of 1 wk to complete the “pulse” period. Embryos were rinsed 2× and then moved to fresh water at 28 °C in a recirculating aquarium system (GIT). Embryos were killed over 20-d periods up until a period of 100-d “chase.” This period was verified by immunohistochemistry as the chase time point where only discreet populations of slow-cycling cells were labeled.
Immunohistochemistry.
Embryos were killed as described and fixed in 10% NBF at room temperature at 4 °C. Embryos were then rinsed in phosphate-buffered saline (PBS) and decalcified for a period of 48 to 72 h in a mild acid (0.1 M EDTA) at room temperature before being processed through a graded series of EtOH (25%, 50%, 75%, 100%, 100%) and 2 washes in xylene. Embryos were washed in xylene for 3 h and incubated 60 °C and embedded in paraffin for sectioning on a Thermo Scientific Microm HM355S microtome at 5 µm. Slides were dried for 24 h at 42 °C and rehydrated through xylene and a graded series of EtOH for incubation in blocking solution (3% goat serum, 1% bovine serum, 0.1% Triton X-100) for 1 h at room temperature. Slides were then incubated overnight in a 1:100 dilution of anti-rabbit primary antibody (rabbit anti–β-cat [GTX26302; Genetex], rabbit anti-Sox2 [GTX124477; Genetex], rabbit anti-Trp63 [GTX124660; Genetex], and rabbit anti-Bmi1 [RA25083; Neuromics]) in conjunction with mouse IgG2a anti-BrDU (RPN202; GE Healthcare) and the provided blocking solution containing nuclease enzyme at 4 °C. Slides were then rinsed 2× 1 h in PBS and incubated in secondary antibodies at 1:400 horseradish peroxidase (HRP)-conjugated goat anti-rabbit IgG (Molecular Probes) and Alexa Fluor 568 goat anti-mouse IgG2a (Molecular Probes) in blocking solution at room temperature. Unbound secondary antibody was removed by washing 2× 1 h in PBS, and the HRP signal was amplified using a 488-tyramide chemistry signal amplification kit (Molecular Probes). Slides were again rinsed 2× 1 h and mounted with a 50:50 glycerin:Vectashield mixture for imaging using a Zeiss 710 confocal imaging system.
Chemical Treatment.
A 10-µm stock solution of LDN-193189 (LDN) (Enzo) was prepared for each chemical treatment experiment using dimethyl sulfoxide (DMSO) (MP Biomedicals). All chemical and control experiments were performed in Erlenmeyer flasks at 28 °C in an oscillating platform culture incubator (Lab-Line Max 4000; Barnstead). For changes in gene expression assayed by ISH, cichlids were raised to 20 dpf, and embryos from single broods were split into a small molecule treatment and a solvent control group. Treatments were performed at 4 µM LDN in 200 mL of fish H2O. After 48-h treatment in the small molecule dilution, fry were killed immediately and fixed in 4% PFA. ISH was then carried out to assay effects of treatment on gene expression.
Cichlid RNA Extraction and Sequencing.
Animals (∼1-y-old adult MZ and LF males) were killed and immediately dissected for RNA extraction. A ribbon ∼1 mm × 10 mm of epithelium was removed labial to the outer row of teeth from the dentary of experimental animals using a no. 12 scalpel blade. The extraosseous soft tissue was removed from the entire jaw to reduce the risk of TBs containing epithelium carryover. The bone was then shaved down using a scalpel to expose the bony crypts, and intraosseous RT were extracted with fine forceps. Extracted tissue was quickly placed in RNAlater RNA Stabilization Reagent (Qiagen). Tissues were frozen in liquid nitrogen, homogenized using a mortar and pestle, and placed in TRIzol. Following standard chloroform extraction, RNeasy mini columns (Qiagen) were utilized to purify RNA for storage at −80 °C. Total RNA was quantified using Qubit (Molecular Probes) and quality analyzed using the Agilent 2100 Bioanalyzer System for RNA library preparation. RNA input was normalized to 1 µg, and libraries were prepared using the TruSeq Stranded mRNA Sample Prep Kit (Illumina-Kit A). Libraries were again quantified, quality assessed, and normalized for sequencing on the HiSeq 2500 Illumina Sequencing System.
Cichlid Transcriptome Analyses.
Raw sequence reads from RT and TB samples were quality controlled using the NGS QC Toolkit (58). Raw reads with an average Phred quality score below 20 were filtered out. The remaining reads were further trimmed of low-quality bases at the 3′ end. Quality-controlled reads for each sample were aligned to a recently improved M. zebra reference genome (59) using TopHat v2.0.9 (60). The resulting TopHat2 output bam files were sorted and converted to sam files using samtools v0.19 (61). Sorted sam files were used as input for the HTSeq-count v0.6.1 program to obtain fragment counts for each locus (62). Fragment counts were scale-normalized across all samples using the calcNormFactors function in the edgeR package v3.6.8 (63). Relative consistency among replicates and samples was determined via the Multidimensional scaling (MDS) feature within the edgeR package in R. Scale-normalized fragment counts were converted into log2 counts per million reads mapped (cpm), with precision weights using voom, and fit to a linear model using the limma package v3.20.9 (64, 65). Pairwise contrasts were constructed between RT and TB samples. After correcting for multiple comparisons using the Benjamini–Hochberg method (66), genes were considered differentially expressed between RT and TB samples if they exhibited both a fold change ≥2 and Padj < 0.05. Data have been deposited in NCBI GEO under accession code GSE122501.
Functional Overrepresentation Analyses.
Functional enrichment of differentially expressed genes was identified using the comprehensive Gene Analytics (47) and ToppGene (48) tools.
Mouse Husbandry.
All mouse work was performed according to Home office guidelines in the United Kingdom and approved by the King’s College London animal ethics committee. Follistatin heterozygotes mice (Fst+/−) were from JAX (stock no. 002788), and C57BL/6 mice were from CRL (Charles River Laboratory). Fst+/− mutant mice were bred with C57BL/6 for mouse strain maintenance because homozygous mice die immediately after birth.
Mouse RNA-Seq.
Total RNA was extracted from intermolar eminence (IE) epithelium of E17.5 control and mutant embryos (n = 3 of each) following standard methods. In brief, tissue was homogenized with TRIzol (Invitrogen). RNA was separated with chloroform, precipitated with isopropanol, and subsequently purified with RNeasy MINI kit (74104; Qiagen). RNA quantity was measured, and quality was assessed with an Agilent Bioanalyzer (RNA 6000 nano; Agilent). One microgram of each high-quality (RIN > 7) RNA sample was sent to the Oxford Genomics Centre for sequencing. Raw FASTQ files were trimmed based on End min quality level (Phred) and aligned BAM files by BWA 0.7.12 to the mouse mm10 reference genome using Partek Flow software with default parameters. Aligned reads were then quantified to the annotation mode (mm10; Ensembl Transcripts release 83), and raw gene counts were normalized. Differential gene expression was identified by Partek Gene-specific analysis. False discovery rate (FDR) correction with default P value of <0.05 was considered statistically significant.
Mouse Whole Mount In Situ Hybridization.
Freshly dissected tongues from Follistatin homozygous (Fst−/−) and wild-type mice (Fst+/+) at E17.5 were fixed in 4% PFA overnight at 4 °C, followed by dehydration procedures through a methanol series (25%, 50%, 75%, and 100% methanol, 15 min per step). Samples were then rehydrated in PBS and bleached in 6% hydrogen peroxide at room temperature for 1 h prior to proteinase K incubation and washed with glycine in PBS and tween (PBT). Samples were refixed in 0.2% glutaraldehyde and 4% PFA in PBT before hybridization with DIG-labeled RNA probes at 70 °C overnight. After intensive posthybridization wash in solutions containing 50% formamide and RNase treatment, followed by antibody binding (anti–Digoxigenin-AP, 1:3,000; Roche) at 4 °C overnight, antibodies were stained with Alkaline Phosphate and BM-Purple (11442074001; Roche) and were used for colorimetric detection. Samples were then photographed using a Leica MZ FLIII microscope with Leica DFC300FX camera and Leica FireCam software and transmitted lateral and or/direct illumination.
Mouse Tissue Real-Time PCR.
Mouse tongues were collect at E17.5 from Fst−/− and Fst+/+ mice, and posterior intermolar eminence (IE) regions were carefully dissected in cold PBS. Dissected tissues were incubated with Dispase II (04942078001; Roche) at 37 °C for 30 min before mechanically dissociating epithelium from underneath mesenchyme tissue. Dissociated epithelial tissues were snap frozen by liquid nitrogen and stored at −80 °C prior to total RNA extraction. Total RNA was extracted from mouse tongue epithelia on IE regions using RNeasy Mini Kit (Qiagen; 74104) and purified by Ambion DNA-free DNA Removal Kit (AM1906; Invitrogen,). cDNA was then synthesized as described before (15, 67). One microgram of cDNA was used for qPCR reaction applied on a LightCycler 480 system qPCR platform (05015278001; Roche). All primers used in the experiments are listed in Dataset S3. Ct values were normalized with β-actin levels as internal controls, and data were analyzed by 2−∆∆ct methods. Error bars are presented by SDs, which were calculated from biological triplicate samples.
Supplementary Material
Acknowledgments
We thank members of the J.T.S. laboratory and P.T.S. laboratory for comments on previous drafts of this manuscript. This work was supported by National Institute of Dental and Craniofacial Research (NIDCR) Grant R01 DE019637 (to J.T.S.) and National Institutes of Health (NIH) Grant 1F30 DE023013 (to R.F.B.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, https://www.ncbi.nlm.nih.gov/geo (accession nos. GSE122501, GSE128942, and GSM3688726–GSM3688731).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1821202116/-/DCSupplemental.
References
- 1.Tucker A. S., Fraser G. J., Evolution and developmental diversity of tooth regeneration. Semin. Cell Dev. Biol. 25–26, 71–80 (2014). [DOI] [PubMed] [Google Scholar]
- 2.Wang X. P., et al. , An integrated gene regulatory network controls stem cell proliferation in teeth. PLoS Biol. 5, e159 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Harada H., Mitsuyasu T., Toyono T., Toyoshima K., Epithelial stem cells in teeth. Odontology 90, 1–6 (2002). [DOI] [PubMed] [Google Scholar]
- 4.Juuri E., et al. , Sox2 marks epithelial competence to generate teeth in mammals and reptiles. Development 140, 1424–1432 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fraser G. J., Bloomquist R. F., Streelman J. T., Common developmental pathways link tooth shape to regeneration. Dev. Biol. 377, 399–414 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Martin K. J., et al. , Sox2+ progenitors in sharks link taste development with the evolution of regenerative teeth from denticles. Proc. Natl. Acad. Sci. U S A. 113, 14769–14774 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Juuri E., et al. , Sox2+ stem cells contribute to all epithelial lineages of the tooth via Sfrp5+ progenitors. Dev. Cell 23, 317–328 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sanz-Navarro M., et al. , Plasticity within the niche ensures the maintenance of a Sox2+ stem cell population in the mouse incisor. Development 145, dev155929 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Donati G., Watt F. M., Stem cell heterogeneity and plasticity in epithelia. Cell Stem Cell 16, 465–476 (2015). [DOI] [PubMed] [Google Scholar]
- 10.Blanpain C., Fuchs E., Stem cell plasticity. Plasticity of epithelial stem cells in tissue regeneration. Science 344, 1242281 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rompolas P., Mesa K. R., Greco V., Spatial organization within a niche as a determinant of stem-cell fate. Nature 502, 513–518 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Feng J., Mantesso A., De Bari C., Nishiyama A., Sharpe P. T., Dual origin of mesenchymal stem cells contributing to organ growth and repair. Proc. Natl. Acad. Sci. U.S.A. 108, 6503–6508 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kaukua N., et al. , Glial origin of mesenchymal stem cells in a tooth model system. Nature 513, 551–554 (2014). [DOI] [PubMed] [Google Scholar]
- 14.Zhao H., et al. , Secretion of shh by a neurovascular bundle niche supports mesenchymal stem cell homeostasis in the adult mouse incisor. Cell Stem Cell 14, 160–173 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.An Z., et al. , A quiescent cell population replenishes mesenchymal stem cells to drive accelerated growth in mouse incisors. Nat. Commun. 9, 378 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nieto M. A., Epithelial plasticity: A common theme in embryonic and cancer cells. Science 342, 1234850 (2013). [DOI] [PubMed] [Google Scholar]
- 17.Bloomquist R. F., et al. , Coevolutionary patterning of teeth and taste buds. Proc. Natl. Acad. Sci. U.S.A. 112, E5954–E5962 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Plikus M., et al. , Morpho-regulation of ectodermal organs: Integument pathology and phenotypic variations in K14-Noggin engineered mice through modulation of bone morphogenic protein pathway. Am. J. Pathol. 164, 1099–1114 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lu C. P., Polak L., Keyes B. E., Fuchs E., Spatiotemporal antagonism in mesenchymal-epithelial signaling in sweat versus hair fate decision. Science 354, aah6102 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Thiery A. P., et al. , Spatially restricted dental regeneration drives pufferfish beak development. Proc. Natl. Acad. Sci. U.S.A. 114, E4425–E4434 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yoshizaki K., et al. , Ablation of coactivator Med1 switches the cell fate of dental epithelia to that generating hair. PLoS One 9, e99991 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wu P., et al. , Multiple regulatory modules are required for scale-to-feather conversion. Mol. Biol. Evol. 35, 417–430 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lai Y.-C., et al. , Transcriptome analyses of reprogrammed feather/scale chimeric explants revealed co-expressed epithelial gene networks during organ specification. BMC Genomics 19, 780 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fraser G. J., Bloomquist R. F., Streelman J. T., A periodic pattern generator for dental diversity. BMC Biol. 6, 32 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lin C. R., et al. , Pitx2 regulates lung asymmetry, cardiac positioning and pituitary and tooth morphogenesis. Nature 401, 279–282 (1999). [DOI] [PubMed] [Google Scholar]
- 26.Bloomquist R. F., Fowler T. E., Sylvester J. B., Miro R. J., Streelman J. T., A compendium of developmental gene expression in Lake Malawi cichlid fishes. BMC Dev. Biol. 17, 3 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Díaz-Regueira S. M., Lamas I., Anadón R., Calretinin immunoreactivity in taste buds and afferent fibers of the grey mullet Chelon labrosus. Brain Res. 1031, 297–301 (2005). [DOI] [PubMed] [Google Scholar]
- 28.Luo X., Okubo T., Randell S., Hogan B. L., Culture of endodermal stem/progenitor cells of the mouse tongue. In Vitro Cell. Dev. Biol. Anim. 45, 44–54 (2009). [DOI] [PubMed] [Google Scholar]
- 29.Kapsimali M., et al. , Fgf signaling controls pharyngeal taste bud formation through miR-200 and Delta-Notch activity. Development 138, 3473–3484 (2011). [DOI] [PubMed] [Google Scholar]
- 30.Mills A. A., et al. , p63 is a p53 homologue required for limb and epidermal morphogenesis. Nature 398, 708–713 (1999). [DOI] [PubMed] [Google Scholar]
- 31.Okubo T., Clark C., Hogan B. L., Cell lineage mapping of taste bud cells and keratinocytes in the mouse tongue and soft palate. Stem Cells 27, 442–450 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Okubo T., Pevny L. H., Hogan B. L., Sox2 is required for development of taste bud sensory cells. Genes Dev. 20, 2654–2659 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Molofsky A. V., et al. , Bmi-1 dependence distinguishes neural stem cell self-renewal from progenitor proliferation. Nature 425, 962–967 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Biehs B., et al. , BMI1 represses Ink4a/Arf and Hox genes to regulate stem cells in the rodent incisor. Nat. Cell Biol. 15, 846–852 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tanaka T., et al. , Identification of stem cells that maintain and regenerate lingual keratinized epithelial cells. Nat. Cell Biol. 15, 511–518 (2013). [DOI] [PubMed] [Google Scholar]
- 36.Yan K. S., et al. , The intestinal stem cell markers Bmi1 and Lgr5 identify two functionally distinct populations. Proc. Natl. Acad. Sci. U.S.A. 109, 466–471 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhu Y., Huang Y.-F., Kek C., Bulavin D. V., Apoptosis differently affects lineage tracing of Lgr5 and Bmi1 intestinal stem cell populations. Cell Stem Cell 12, 298–303 (2013). [DOI] [PubMed] [Google Scholar]
- 38.Handrigan G. R., Leung K. J., Richman J. M., Identification of putative dental epithelial stem cells in a lizard with life-long tooth replacement. Development 137, 3545–3549 (2010). [DOI] [PubMed] [Google Scholar]
- 39.Takeda N., et al. , Interconversion between intestinal stem cell populations in distinct niches. Science 334, 1420–1424 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Takeda N., et al. , Hopx expression defines a subset of multipotent hair follicle stem cells and a progenitor population primed to give rise to K6+ niche cells. Development 140, 1655–1664 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Harada H., et al. , Localization of putative stem cells in dental epithelium and their association with Notch and FGF signaling. J. Cell Biol. 147, 105–120 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nichols J., et al. , Formation of pluripotent stem cells in the mammalian embryo depends on the POU transcription factor Oct4. Cell 95, 379–391 (1998). [DOI] [PubMed] [Google Scholar]
- 43.Ring A., Kim Y.-M., Kahn M., Wnt/catenin signaling in adult stem cell physiology and disease. Stem Cell Rev. 10, 512–525 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Deschene E. R., et al. , β-Catenin activation regulates tissue growth non-cell autonomously in the hair stem cell niche. Science 343, 1353–1356 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Force A., et al. , Preservation of duplicate genes by complementary, degenerative mutations. Genetics 151, 1531–1545 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fatemifar G., et al. , Genome-wide association study of primary tooth eruption identifies pleiotropic loci associated with height and craniofacial distances. Hum. Mol. Genet. 22, 3807–3817 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ben-Ari Fuchs S., et al. , GeneAnalytics: An integrative gene set analysis tool for next generation sequencing, RNAseq and microarray data. OMICS 20, 139–151 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chen J., Xu H., Aronow B. J., Jegga A. G., Improved human disease candidate gene prioritization using mouse phenotype. BMC Bioinformatics 8, 392 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Andl T., et al. , Epithelial Bmpr1a regulates differentiation and proliferation in postnatal hair follicles and is essential for tooth development. Development 131, 2257–2268 (2004). [DOI] [PubMed] [Google Scholar]
- 50.Seidel K., et al. , Hedgehog signaling regulates the generation of ameloblast progenitors in the continuously growing mouse incisor. Development 137, 3753–3761 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Beites C. L., et al. , Follistatin modulates a BMP autoregulatory loop to control the size and patterning of sensory domains in the developing tongue. Development 136, 2187–2197 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Fraser G. J., Cerny R., Soukup V., Bronner-Fraser M., Streelman J. T., The odontode explosion: The origin of tooth-like structures in vertebrates. BioEssays 32, 808–817 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Thesleff I., Sharpe P., Signalling networks regulating dental development. Mech. Dev. 67, 111–123 (1997). [DOI] [PubMed] [Google Scholar]
- 54.Krivanek J., Adameyko I., Fried K., Heterogeneity and developmental connections between cell types inhabiting teeth. Front. Physiol. 8, 376 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Brawand D., et al. , The genomic substrate for adaptive radiation in African cichlid fish. Nature 513, 375–381 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Fujimura K., Okada N., Development of the embryo, larva and early juvenile of Nile tilapia Oreochromis niloticus (Pisces: Cichlidae). Developmental staging system. Dev. Growth Differ. 49, 301–324 (2007). [DOI] [PubMed] [Google Scholar]
- 57.Loh Y. H., et al. , Comparative analysis reveals signatures of differentiation amid genomic polymorphism in Lake Malawi cichlids. Genome Biol. 9, R113 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Patel R. K., Jain M., NGS QC Toolkit: A toolkit for quality control of next generation sequencing data. PLoS One 7, e30619 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Conte M. A., Kocher T. D., An improved genome reference for the African cichlid, Metriaclima zebra. BMC Genomics 16, 724 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kim D., et al. , TopHat2: Accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 14, R36 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Li H., et al. ; 1000 Genome Project Data Processing Subgroup , The sequence alignment/map format and SAMtools. Bioinformatics 25, 2078–2079 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Anders S., Pyl P. T., Huber W., HTSeq–A Python framework to work with high-throughput sequencing data. Bioinformatics 31, 166–169 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Robinson M. D., McCarthy D. J., Smyth G. K., edgeR: A bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Smyth G. K., Michaud J., Scott H. S., Use of within-array replicate spots for assessing differential expression in microarray experiments. Bioinformatics 21, 2067–2075 (2005). [DOI] [PubMed] [Google Scholar]
- 65.Law C. W., Chen Y., Shi W., Smyth G. K., voom: Precision weights unlock linear model analysis tools for RNA-seq read counts. Genome Biol. 15, R29 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Benjamini Y., Hochberg Y., Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. R. Stat. Soc. B 57, 289–300 (1995). [Google Scholar]
- 67.An Z., et al. , Regulation of mesenchymal stem to transit-amplifying cell transition in the continuously growing mouse incisor. Cell Rep. 23, 3102–3111 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.