Significance
Targeting CDK4/6 shows great promise in the treatment of many solid cancers, including melanoma. This study has uncovered a mechanism of action of CDK4/6 inhibitors in regulating the MDM4 oncogene and the tumor suppressor, p53. In melanoma, palbociclib activates p53 via modulating PRMT5-dependent alternate MDM4 pre-mRNA splicing, which results in a robust decrease in MDM4 protein expression. Loss of palbociclib regulation of the PRMT5–MDM4 axis leads to drug resistance. Dual inhibition of CDK4/6 and PRMT5 potently suppresses melanoma tumor growth and is well tolerated in vivo. Our findings not only have immediate implications for the advancement of CDK4/6 inhibition for treating melanoma, but also more generally offer insights into CDK4/6 mechanism of action.
Keywords: acquired resistance, CDK4, PRMT5, MDM4, p53
Abstract
Cyclin-dependent kinase 4/6 (CDK4/6) inhibitors are an established treatment in estrogen receptor-positive breast cancer and are currently in clinical development in melanoma, a tumor that exhibits high rates of CDK4 activation. We analyzed melanoma cells with acquired resistance to the CDK4/6 inhibitor palbociclib and demonstrate that the activity of PRMT5, a protein arginine methyltransferase and indirect target of CDK4, is essential for CDK4/6 inhibitor sensitivity. By indirectly suppressing PRMT5 activity, palbociclib alters the pre-mRNA splicing of MDM4, a negative regulator of p53, leading to decreased MDM4 protein expression and subsequent p53 activation. In turn, p53 induces p21, leading to inhibition of CDK2, the main kinase substituting for CDK4/6 and a key driver of resistance to palbociclib. Loss of the ability of palbociclib to regulate the PRMT5–MDM4 axis leads to resistance. Importantly, combining palbociclib with the PRMT5 inhibitor GSK3326595 enhances the efficacy of palbociclib in treating naive and resistant models and also delays the emergence of resistance. Our studies have uncovered a mechanism of action of CDK4/6 inhibitors in regulating the MDM4 oncogene and the tumor suppressor, p53. Furthermore, we have established that palbociclib inhibition of the PRMT5–MDM4 axis is essential for robust melanoma cell sensitivity and provide preclinical evidence that coinhibition of CDK4/6 and PRMT5 is an effective and well-tolerated therapeutic strategy. Overall, our data provide a strong rationale for further investigation of novel combinations of CDK4/6 and PRMT5 inhibitors, not only in melanoma but other tumor types, including breast, pancreatic, and esophageal carcinoma.
Hyperactivation of CDK4/6–cyclin D1 complexes, central regulators of the G1-S transition of the cell cycle, is common in many cancers. In melanoma, hyperactivation of this pathway is observed in ∼90% of cases and is caused by several mechanisms, including loss of p16INK4A, a specific inhibitor of the CDK4/6–cyclin D1 complex (1, 2), activating mutations in CDK4 (3, 4), or increased expression of cyclin D1 (2). CDK4/6 promotes cell cycle progression mainly by phosphorylation and inhibition of the tumor suppressor RB1 and also by phosphorylation of other downstream targets, including FOXM1, SMAD3, c-MYC, and MEP50 (5).
Targeting CDK4/6 activity has long been considered a promising approach for cancer treatment. Recently, three relatively specific CDK4/6 inhibitors—palbociclib (PD0332991), abemaciclib (LY2835219), and ribociclib (LEE011)—have emerged as agents with both proven anticancer activity and tolerability (6). These inhibitors, in combination with endocrine therapy, have received FDA approval for the treatment of hormone receptor (HR)-positive, HER2-negative advanced breast cancer (7–9). There are at least 180 completed or ongoing clinical trials testing CDK4/6 inhibitors across a broad portfolio of cancers (10, 11). Preclinical studies in melanoma have shown that dual targeting of CDK4/6 and mutant-BRAF or MEK leads to robust and sustained tumor regression in BRAF-mutant and NRAS-mutant melanoma (12–16). Based on these preclinical studies, there are several clinical trials in melanoma assessing the efficacy of CDK4/6 inhibitors in combination with MAPK/ERK pathway inhibitors (10, 17). Although CDK4/6 inhibitors as combinational therapy are demonstrating clinical benefit, monotherapy is rarely effective. Therefore, developing novel combination strategies with CDK4/6 inhibitors is fundamental for the success of future development in other tumor types.
Sensitivity to CDK4/6 inhibitors is dependent on the presence of activating events of this pathway whereas primary resistance to CDK4/6 inhibitors predominantly results from the inactivation of the CDK4/6 downstream target, RB1 (18–20). In many cancers, one of the prominent mechanisms of acquired resistance to CDK4/6 inhibition is hyperactivation of the CDK2–cyclin E complex, which substitutes for CDK4/6 in phosphorylating RB1 and maintaining cell cycle progression. This can be due to elevated levels of CDK2, amplification of CCNE1 and/or loss of the CDK2 inhibitors p21 or p27 (19, 21). Other reported mechanisms of acquired resistance include loss of functional RB1 (22, 23), up-regulation of type D cyclins (24), or amplification of CDK4 or CDK6 (25, 26). Although the mechanisms of acquired resistance to CDK4/6 inhibitors in breast cancer and hematological malignancies have been reported, the mechanisms of resistance in melanoma have not been elucidated.
Herein, we have identified suppression of protein arginine methyltransferase 5 (PRMT5) activity by CDK4/6 inhibitors as being a key component in the efficacy of these drugs. PRMT5 is an epigenetic modifier that regulates gene expression through methylating arginine residues on Histones 2A, 3, and 4 (27, 28). In addition, via methylating nonhistone proteins, PRMT5 regulates many other cellular processes, including cell signaling, ribosome biogenesis, RNA transport, and pre-mRNA splicing, all of which impact on a multitude of cellular outcomes (29–31). PRMT5-mediated regulation of the spliceosome machinery, through the methylation of several spliceosomal Sm proteins (32, 33), is considered one of its most significant oncogenic roles (34), and studies have shown that MDM4 is a particularly important target of this process (35, 36). MDM4 plays a critical role as a key oncogene in melanoma and other cancers, mainly through its role in inactivating the p53 pathway (37–39). PRMT5 activity is regulated via multiple mechanisms and through a number of binding coactivators. MEP50 is one of the key coactivators of PRMT5 and is necessary for its enzymatic activity (31, 40, 41). Hyperactivated CDK4/Cyclin D has been shown to modulate PRMT5/MEP50 complex methyltransferase activation via phosphorylating MEP50 (42). In CDK4/6 inhibitor-sensitive cells, palbociclib decreased PRMT5 activity, which resulted in alterations in MDM4 pre-mRNA splicing and reduced expression of MDM4 protein. In drug-resistant cells, palbociclib failed to decrease PRMT5 activity and also MDM4 expression, and these cells exhibited heightened dependence on both PRMT5 and MDM4. Our findings have not only uncovered a link between CDK4 activity and expression of the oncogene MDM4 but also elucidate a mechanism of acquired resistance to CDK4/6 inhibition in melanoma. Furthermore, the data provide a promising combination strategy that can enhance the efficacy of CDK4/6 inhibitors and delay the emergence of resistance.
Results
Resistance to Palbociclib Is Associated with Increased Sensitivity to PRMT5 Inhibition.
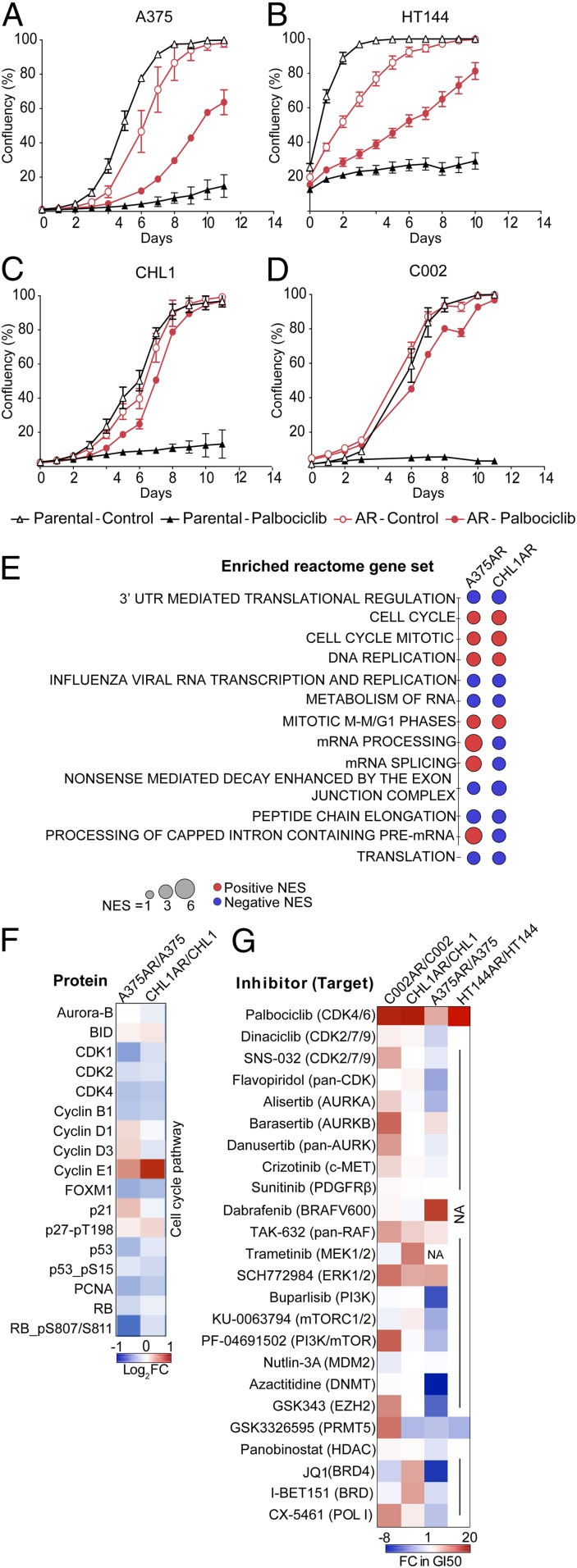
A panel of melanoma cell lines from various genomic subtypes were treated with the CDK4/6 inhibitor palbociclib (SI Appendix, Table S1). After 3 to 9 mo of treatment, cells acquired resistance (AR) and were able to proliferate in high concentrations of palbociclib (2 to 4 µM), which profoundly inhibited proliferation in the parental drug-sensitive cells (Fig. 1 A–D). All palbociclib-resistant cell lines retained the ability to proliferate with or without the drug, indicating that resistance to palbociclib was not associated with a dependency on the drug for survival, a phenomenon sometimes observed after developing acquired resistance to targeted therapies (43). Two patterns of resistance to palbociclib were observed: Partially resistant A375AR and HT144AR cells continued to proliferate while on palbociclib treatment, but at a slower rate than untreated control cells, whereas, in completely resistant CHL1AR and C002AR cells, palbociclib had no effect on cell proliferation.
Fig. 1.
Resistance to palbociclib is associated with increased sensitivity to PRMT5 inhibition. (A–D) Proliferation curves of the parental and acquired-resistant cell lines treated with 2 μΜ palbociclib or vehicle. Graphs are representative of three independent experiments. Error bars represent SEM for 3 technical replicates. (E) Dot plot of commonly enriched Reactome gene sets using RNA-seq on A375AR and CHL1AR cells compared to their parental counterparts. Normalized enrichment score (NES) ≥ 2.00 or ≤ −2.00, false discovery rate (FDR) ≤ 0.01. (F) Heat map showing RPPA analysis of changes in cell cycle targets in A375AR and CHL1AR compared to their parental counterparts. (G) Heat map showing the changes in GI50 (sensitivity) to a panel of targeted therapies in resistant cell lines compared to the parental counterparts (red represents reduced sensitivity in palbociclib-resistant cells, and blue represents increased sensitivity in palbociclib-resistant cells).
In order to identify mechanisms of acquired resistance to palbociclib, multiple high throughput approaches were employed. Transcriptome and reverse–phase protein array (RPPA) analysis of the resistant and parental cells revealed that acquired resistance to palbociclib was associated with changes in the expression of various genes and proteins involved in different cellular pathways and processes (SI Appendix, Fig. S1 A–E). Gene set enrichment analysis (GSEA) on RNA-sequencing data revealed enrichment of gene sets involved in cell cycle regulation and RNA processing and splicing in the resistant cells (Fig. 1E and Datasets S1–S4). RPPA analysis demonstrated few changes in protein expression between the resistant and sensitive cells. The most obvious change was an increase in cyclin E1, an activator of CDK2, which was consistent with the increase in cyclin E1 mRNA expression (Fig. 1F and SI Appendix, Table S2). Lastly, to identify if palbociclib resistance is associated with increased dependence on certain signaling pathways or particular drug targets, we performed a drug screen using a focused panel of clinically and preclinically advanced targeted agents (Fig. 1G). The drug screen revealed that acquisition of resistance to palbociclib was accompanied by changes in the sensitivity to other targeted therapies. All palbociclib-resistant cell lines exhibited cross-resistance to multiple inhibitors of the MAPK pathway, including TAK-632, a pan-RAF inhibitor, and SCH772984, an ERK1/2 inhibitor. Additionally, the palbociclib-resistant BRAF mutant cell line A375AR became less sensitive to dabrafenib, a selective BRAF V600 inhibitor. These data are consistent with the CDK4/6–cyclin D complex being an important downstream effector of MAPK/ERK signaling (44).
Three out of four resistant cell lines showed increased sensitivity to the PRMT5 inhibitor GSK3326595 (Fig. 1G and SI Appendix, Fig. S2 A–D). PRMT5 is an oncoprotein that regulates a range of cellular processes (45); however, recent reports suggest that the role of PRMT5 in regulating pre-mRNA alternative splicing is a dominant underlying mechanism of its oncogenicity (34). The increased sensitivity to PRMT5 inhibition and the enrichment of RNA splicing and processing gene sets in the resistant cells suggest that PRMT5 may play an important role in mediating resistance to palbociclib. Previous studies have shown that increased sensitivity to PRMT5 depletion was found to correlate with deletion of the MTAP gene (34, 46–48), a gene positioned close to CDKN2A and thus often codeleted. In cell lines where RNA sequencing was performed (A375 and CHL1), MTAP expression was not lost, and its levels were not changed in the palbociclib-resistant cells compared to the parental cells (SI Appendix, Fig. S2E), indicating that loss of MTAP was not the mechanism for the increased sensitivity to PRMT5 inhibition.
Inhibition of PRMT5 Sensitizes Melanoma Cells to Palbociclib.
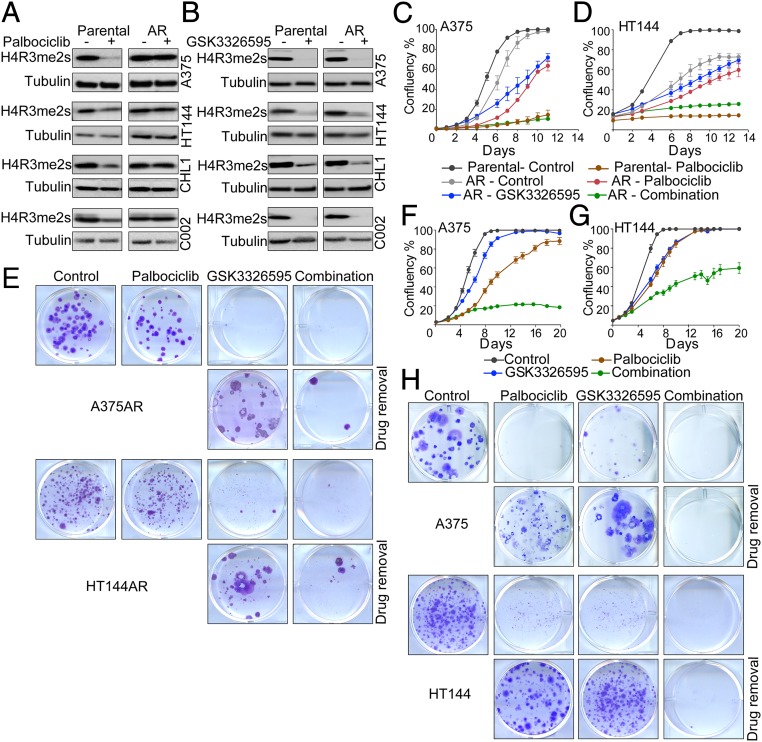
To assess if the change in sensitivity to PRMT5 inhibitors in the resistant cells was associated with alteration in PRMT5 activity, we assessed levels of PRMT5, its coactivator MEP50, and levels of H4R3 symmetric dimethylation (H4R3me2s), a marker of PRMT5 activity. PRMT5 and MEP50 protein levels were mostly unchanged between the parental and resistant cells, with or without treatment (SI Appendix, Fig. S2F). Interestingly, palbociclib reduced levels of H4R3me2s in the parental drug-naive cell lines but not in the resistant cells (Fig. 2A), indicating that CDK4 can regulate PRMT5 methyltransferase activity in melanoma cells. In contrast, in the resistant cells, the link between CDK4 and PRMT5 is disrupted, and PRMT5 retains its activity even after inhibition of CDK4/6. The inability of palbociclib to decrease H4R3me2s and the associated increase in sensitivity to PRMT5 inhibitors in the acquired resistance cells suggested that PRMT5 played an important role in mediating the response to palbociclib.
Fig. 2.
Inhibition of PRMT5 sensitizes melanoma cells to palbociclib. (A) Western blot on melanoma cell lysates after 6-d treatment with 2 μΜ palbociclib. (B) Western blot on melanoma cell lysates after 6-d treatment with 500 nM GSK3326595 for all cells except HT144, which were treated with 250 nM. (C and D) Proliferation curves of the parental and acquired resistance (AR) cells treated with 2 μM palbociclib, 250 nM (HT144), or 500 nM (A375) GSK3326595, or combination of both drugs. Graphs are representative of three independent experiments. Error bars represent SEM for 3 technical replicates. (E) Images of colony-forming assays of A375AR and HT1441AR cells treated as in C and D for 14 d, and after drug removal for 14 d. Representative of 2 biological replicates with 3 technical replicates each. (F and G) Proliferation curves of the parental A375 and HT144 cells treated with 1 μM (A375) or 250 nM (HT144) palbociclib, 500 nM (A375) or 250 nM (HT144) GSK3326595, or combination of both drugs. Graphs are representative of two independent experiments. Error bars represent SEM for 3 technical replicates. (H) Images of colony-forming assays of A375 treated as in F and G, and after drug removal for 14 d. Representative of 2 biological replicates with 3 technical replicates each.
Therefore, we tested if inhibition of PRMT5 would restore sensitivity to palbociclib in the resistant cells. We used a highly selective PRMT5 inhibitor, GSK3326595, which inhibits PRMT5 activity by competitively binding to the substrate peptide pocket (36). GSK3326595 is currently in phase 1/2 clinical trials in solid tumors, non-Hodgkin’s lymphoma, and acute myeloid leukemia (NCT03614728, NCT02783300). GSK3326595 potently inhibited the symmetric methylation of H4R3 in all cell lines (Fig. 2B). Combining GSK3326595 with palbociclib in the A375 and HT144 palbociclib-resistant cells decreased cell proliferation to a similar extent to what was observed in the parental cell line treated with palbociclib alone (Fig. 2 C and D). The responses to single agents and their combination were also confirmed by nuclei count and colony-forming assays, which showed that the combination potently suppressed cell proliferation and the ability to form colonies even after drug removal (SI Appendix, Fig. S2 G and H and Fig. 2E). These data suggest that inhibition of PRMT5 was capable of restoring sensitivity to palbociclib and indicate that the inability of palbociclib to decrease the activity of PRMT5 was conferring resistance to the drug.
Next, we tested if inhibition of PRMT5 enhances the efficacy of palbociclib in the drug-sensitive cells. Combining GSK3326595 with a submaximal concentration of palbociclib resulted in more profound growth inhibition than the single agents in the A375 and HT144 cell lines (Fig. 2 F and G and SI Appendix, Fig. S2 I and J). Furthermore, colony-forming assays also showed that the single agents and their combination reduced colony formation compared to control. However, following the removal of treatment, colonies emerged from cells treated with the single agents but not from cells treated with the combination, providing evidence to the effectiveness of this combination in the parental cells as well as the resistant cells (Fig. 2H). Collectively, these findings show that inhibition of PRMT5 is critical for a durable and robust response to CDK4/6 inhibitors, and, in addition, PRMT5 inhibition enhances the efficacy of palbociclib in both palbociclib sensitive and resistant melanoma cells. Assessing cell death showed that the combination of palbociclib and GSK3326595 did not induce further cell death compared to the single agents (SI Appendix, Fig. S2K), suggesting that the robust response observed is due to a more permanent cell cycle arrest rather than cell death.
PRMT5 Inhibition Sensitizes Melanoma Cells to Palbociclib by Suppressing CDK2 Activity.
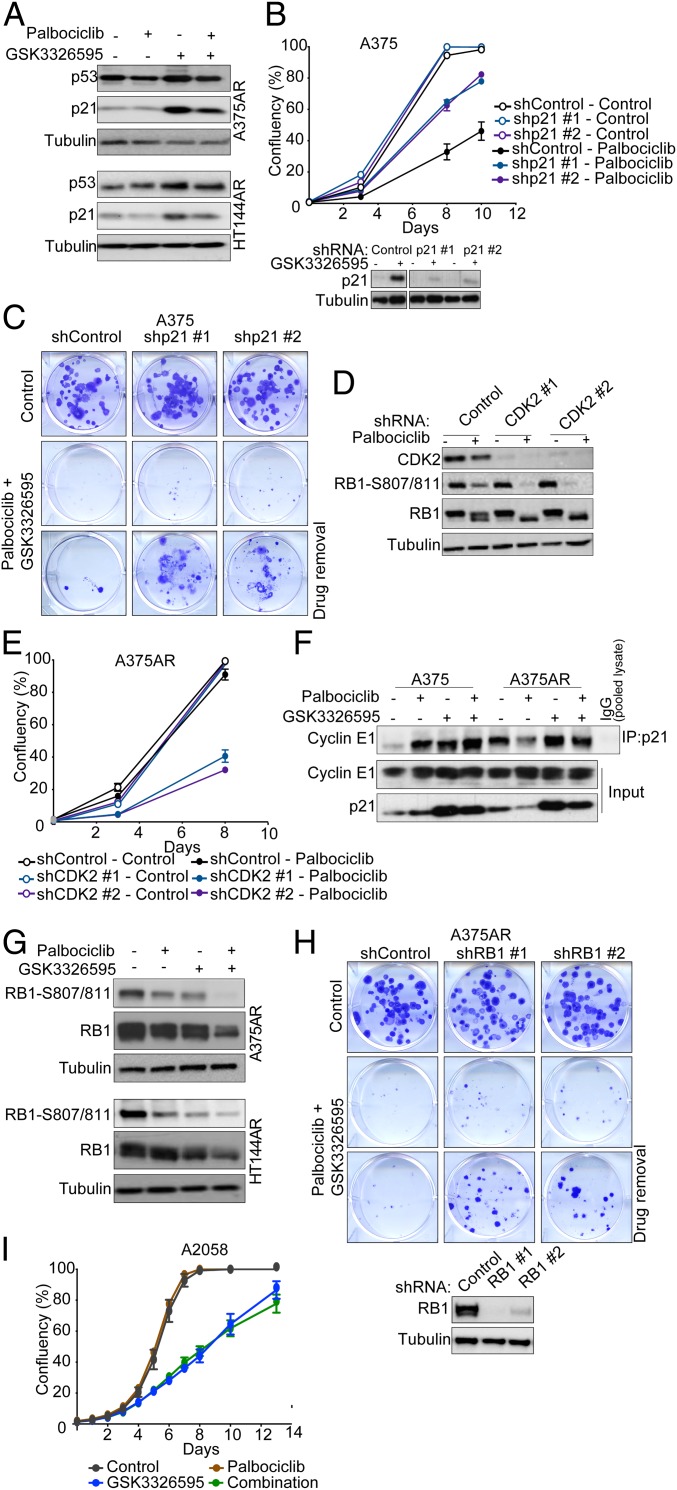
To further understand the effects of palbociclib and GSK3326595 on cell signaling, we assessed protein expression by RPPA in the A375 and A375AR cells after treatment with palbociclib, GSK3326595, or their combination. To identify potential target proteins that palbociclib alters via inhibition of PRMT5, we compiled a list of 35 proteins that were commonly changed in the A375 parental (drug-sensitive) cells in response to both single agent palbociclib and GSK3326595 (SI Appendix, Fig. S3A). This list was then used to identify palbociclib-induced changes that were diminished in the resistant cells and restored by the combination with GSK3326595 (SI Appendix, Fig. S3B); the cell cycle inhibitor p21 was identified as one of the potential targets (SI Appendix, Fig. S3C). The palbociclib-induced increase in p21 protein expression in parental cells was lost in resistant cells but induced when cells were treated with GSK3326595 alone or in combination with palbociclib. Induction of p21 by GSK3326595 in the resistant cells was confirmed by Western blot analysis in the A375AR, and an increase was also seen in the HT144AR cell lines (Fig. 3A). Notably, accumulation of p53 protein, one of the major regulators of p21, was also observed in both cell lines.
Fig. 3.
PRMT5 inhibition sensitizes melanoma cells to palbociclib by suppressing CDK2 activity. (A) Western blots on cell lysates after 6-d treatment with 2 μΜ palbociclib, 500 nM (A375AR), or 250 nM (HT144AR) GSK3326595 or combination of both. (B) Proliferation curves of A375 cells expressing shp21 or shControl treated with 2 μM palbociclib. Graphs are representative of two independent experiments. Error bars represent SEM for 3 technical replicates. Shown are Western blots confirming p21 knockdown. GSK3326595 (500 nM) was used to induce p21 expression to validate the knockdown. (C) Images of colony-forming assays of A375 cells expressing shp21 or shControl treated with a combination of 1 μM palbociclib and 500 nM GSK3326595 for 14 d, and after drug removal for 14 d. Representative of 2 biological replicates with 3 technical replicates each. (D) Western blots on lysates from A375AR cells expressing shCDK2 or shControl after 24-h treatment with 2 μΜ palbociclib. (E) Proliferation curves of A375AR cells expressing shCDK2 or shControl treated with 2 μM palbociclib. Graphs are representative of 2 independent experiments. Error bars represent SEM for 3 technical replicates. (F) Western blots on total cell lysate or after immunoprecipitation of p21 complexes from A375 and A375AR after 6-d treatment with 2 μΜ palbociclib, 500 nM GSK3326595, or combination of both. (G) Western blot on melanoma cell lysates after 6-d treatment with 2 μΜ palbociclib, 250 nM (HT144AR), or 500 nM (A375AR) GSK3326595, or combination of both drugs. (H) Images of colony-forming assays of A375AR cells expressing shRB1 or shControl treated with a combination of 2 μM palbociclib and 500 nM GSK3326595 for 14 d. Representative of 2 biological replicates with 3 technical replicates each. Western blots confirm RB1 knockdown. (I) Proliferation curves of the A2058 cells treated with 2 μM palbociclib, 500 nM GSK3326595, or combination of both drugs. Error bars represent SEM for 3 technical replicates.
Loss of p21 has been identified as one of the mechanisms of acute resistance to CDK4/6 inhibition in breast cancer (49). Consistent with this, we found that reducing p21 levels by short hairpin RNA (shRNA)-mediated gene silencing conferred a degree of resistance to palbociclib (Fig. 3B). Importantly, silencing p21 expression reversed the long-term inhibitory effect of the combination of palbociclib and GSK3326595, as shown by colony-forming ability while on drug and after drug removal (Fig. 3C). p21 is an important cell cycle regulator that suppresses the activity of the CDK2–cyclin E1 complex; therefore, loss of p21, in addition to up-regulation of cyclin E1, could result in further activation of the CDK2–cyclin E1 complex in the resistant cells. A characteristic of the acquired resistant melanoma cells was maintained RB1 phosphorylation after treatment with palbociclib (SI Appendix, Fig. S3D). Therefore, we investigated the role of CDK2 in substituting for CDK4/6 and phosphorylating RB1 in the resistant cells. In resistant cells, shRNA-mediated gene silencing of CDK2 did not decrease phosphorylation of RB1; however, palbociclib treatment of these cells led to robust inhibition of phosphorylation (Fig. 3D). Furthermore, pharmacological inhibition of CDK2 using SNS-032, a CDK2 inhibitor that also targets CDK7 and -9, sensitized the resistant cells to palbociclib (SI Appendix, Fig. S3 E and F). Similar results were seen by depleting CDK2 levels (Fig. 3E), validating its specific role in mediating CDK4/6 inhibitor resistance. These data demonstrate that palbociclib was capable of inhibiting CDK4/6 and that activation of CDK2 and the subsequent phosphorylation of RB1 play an important role in mediating the resistance to CDK4/6 inhibition.
To confirm the role of p21 in inhibiting CDK2, p21 complexes were immunoprecipitated, and the association with cyclin E was assessed (Fig. 3F). In the parental cells, there was an abundance of p21 and cyclin E1 complexes after all treatments whereas, in the resistant cells, palbociclib reduced these complexes. Inhibition of PRMT5 resulted in an increase in p21 and cyclin E1 complexes, thus restoring the response to palbociclib in the resistant cells. In line with this, coinhibition of PRMT5 and CDK4/6 resulted in strong inhibition of RB1 phosphorylation in both A375AR and HT144AR cells (Fig. 3G), recapitulating silencing of CDK2 and providing further evidence that induction of p21 by GSK3326595 led to inactivation of CDK2. To validate the importance of signaling through RB1 in mediating the response to CDK4/6 and PRMT5 inhibition, RB1 levels were reduced by shRNA-mediated gene silencing. In resistant cells, reduction of RB1 resulted in more cells bypassing the inhibitory effect of the combination and the emergence of more colonies (Fig. 3H). As expected, reducing levels of RB1 in the parental cells resulted in reduced sensitivity to palbociclib alone and also increased the ability of cells to form colonies after treatment with the combination, especially after drug removal (SI Appendix, Fig. S3 G and H). It is also worth noting that the coinhibition of CDK4/6 and PRMT5 in inherently RB1-null cells had no additional advantage over the single agents (Fig. 3I). This further highlights that reactivation of RB1 is important for the effectiveness of this combination.
Activation of the p53 Pathway Is Critical for the Response to Palbociclib and GSK3326595.
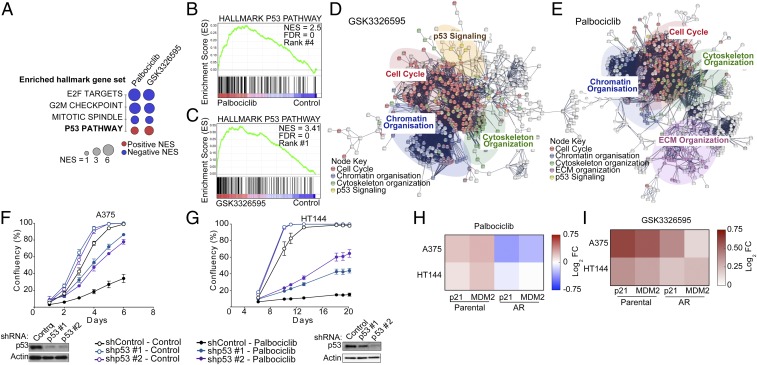
Global gene expression analysis of parental A375 cells revealed a significant enrichment of gene sets involved in the p53 pathway after treatment with GSK3326595 and, importantly, after treatment with palbociclib (Fig. 4 A–C and SI Appendix, Fig. S4 A and B). Similarly, network analysis also showed significant enrichment of the p53 network after treatment with these inhibitors (Fig. 4 D and E). Additionally, gene sets and networks involved in E2F signaling and cell cycle were also enriched in response to the drugs, which is consistent with inactivation of RB1 in response to palbociclib and GSK3326595.
Fig. 4.
Activation of the p53 pathway is critical for the response to Palbociclib and GSK3326595. (A) Dot plot of commonly enriched Hallmark gene sets in A375 cells treated with either 1 μΜ palbociclib or 500 nM GSK3326595 for 72 h compared to control. Normalized enrichment score (NES) ≥ 3.00 or ≤ −3.00, FDR P ≤ 0.01. (B and C) GSEA of genes differentially expressed in the A375 cells after treatment as in A. (D and E) Network analysis on genes that changed expression after treatment as in A. (F and G) Proliferation curves of cells expressing shTP53 (p53) or shControl treated with 2 μM palbociclib. Graphs are representative of 2 independent experiments. Error bars represent SEM for 3 technical replicates. Western blots confirm p53 knockdown. (H and I) Heat maps showing the fold change in gene expression in A375 and HT144 cells after 6-d treatment with 2 μΜ palbociclib or 500 nM GSK3326595, respectively; 3 biological replicates, each with 3 technical replicates.
Activation of p53 signaling in response to PRMT5 depletion or inhibition has been previously described (35, 36); however, CDK4/6 inhibition was shown to diminish p53 activation in a panel of sarcoma cell lines (50). Therefore, we further investigated the role of p53 signaling in palbociclib sensitivity. Reducing p53 levels conferred partial resistance to palbociclib in the A375 and HT144 cells (Fig. 4 F and G). Importantly, this reduction conferred resistance to the long-term response to the combination of palbociclib and GSK3326595 (SI Appendix, Fig. S4C), which is consistent with our observations with p21 depletion. Furthermore, palbociclib induced p21and MDM2 gene expression in the parental but not resistant cells (Fig. 4H); both genes are transcriptionally regulated by p53. This provides further evidence of the importance of activation of p53 for the response to palbociclib in melanoma. GSK3326595, on the other hand, induced p21and MDM2 gene expression in both parental and resistant cells (Fig. 4I), suggesting that one of the mechanisms by which GSK3326595 sensitizes the cell to palbociclib is by further activating the p53 pathway.
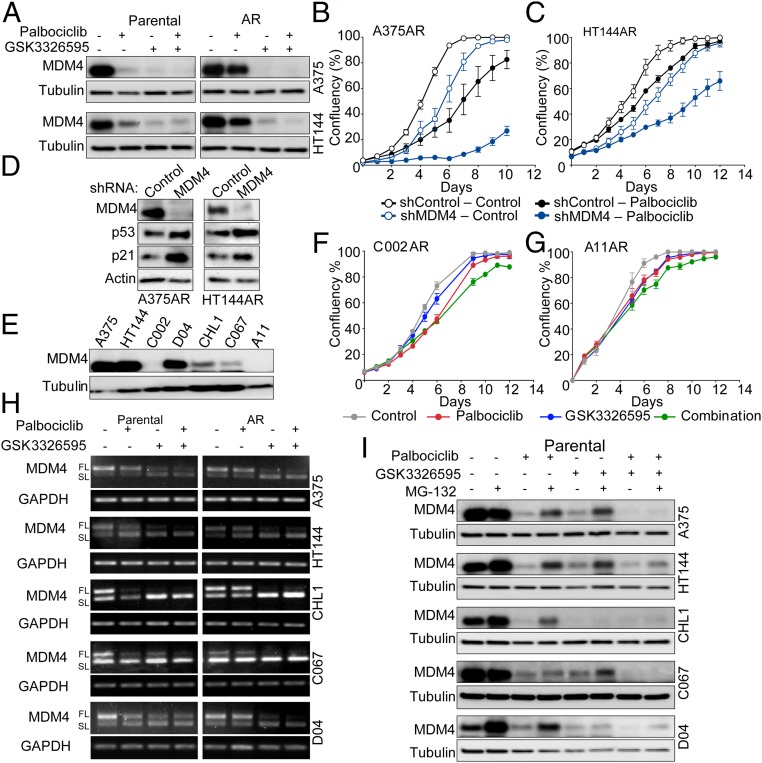
CDK4/6 and PRMT5 Inhibition Suppresses MDM4 Expression by Altering Its mRNA Splicing.
In order to elucidate the exact mechanism by which palbociclib and GSK3326595 activate the p53 pathway, we investigated upstream regulators of this pathway. Interestingly, we found that, in parental cells, inhibition of CDK4/6 by palbociclib resulted in a potent reduction in levels of MDM4 (Fig. 5A), a protein involved in modulating p53 transcriptional activity and stability in cooperation with MDM2 (37, 38). In contrast, in the resistant cells, MDM4 levels were only slightly reduced after treatment with palbociclib. PRMT5 inhibition resulted in robust reduction in MDM4 levels in both parental and resistant cell lines. To validate the importance of MDM4 as a downstream target of CDK4/6 and in mediating the response to palbociclib, we reduced its expression in the resistant cells using an inducible shRNA (51, 52). Reducing expression of MDM4 increased sensitivity to palbociclib in A375AR and HT144AR cells (Fig. 5 B and C) and induced expression of p53 and p21 (Fig. 5D). These data demonstrate that suppression of MDM4 is critical for the response to CDK4/6 inhibition. Intriguingly, we observed that, in palbociclib-resistant C002AR and A11AR cell lines, which were derived from the C002 and A11 cell lines that do not express MDM4 (Fig. 5E), the palbociclib–GSK3326595 combination did not decrease cell proliferation (Fig. 5 F and G). This further supports the role of MDM4 in the response to the drug combination.
Fig. 5.
CDK4 and PRMT5 inhibition suppresses MDM4 expression by altering its mRNA splicing . (A) Western blots on cell lysates after 6-d treatment with 2 μΜ palbociclib, 500 nM (A375) or 250 nM (HT144) GSK3326595, or combination of both. (B and C) Proliferation curves of acquired-resistant cells expressing shMDM4 or shControl with or without treatment with 2 μM palbociclib grown in media containing 10 ng/mL doxycycline. Graphs are representative of 3 independent experiments. Error bars represent SEM for 3 technical replicates. (D) Western blots on lysates from resistant cells expressing shMDM4 or shControl grown for 48 h in media containing 10 ng/mL doxycycline. (E) Western blots on melanoma cell line lysates. (F and G) Proliferation curves of C002AR and A11AR cells treated with 2 μM palbociclib, 500 nM GSK3326595, or combination of both drugs. Graphs are representative of 3 independent experiments. Error bars represent SEM for 3 technical replicates. (H) PCR products showing the expression patterns of MDM4-FL and MDM4-SL mRNA in melanoma cell lines treated with 2 μΜ palbociclib, 500 nM (A375, CHL1, C067, D04) or 250 nM (HT144) GSK3326595, or combination of both drugs for 6 d. (I) Western blots on melanoma cell lines lysates treated as in H with or without treatment with 1 μΜ MG-132 added 16 h prior to experiment end point.
This report demonstrates that CDK4/6 inhibitors potently suppress MDM4 levels. Therefore, we further investigated how palbociclib alters MDM4 expression. Given our data strongly indicate that a major part of response to palbociclib is mediated by its ability to inhibit PRMT5 activity, we hypothesized that CDK4/6 regulates MDM4 via PRMT5 activity. Previous studies indicate that PRMT5 regulates MDM4 protein expression by altering pre-mRNA splicing (35). The alternative splicing of MDM4 is based on the inclusion or the skipping of exon number 6, which results in the production of either a translatable full-length (FL) or unstable short length (SL) mRNA, respectively (53). We evaluated the alternative splicing of MDM4 pre-mRNA in 5 matched parental (sensitive) and resistant melanoma cell lines and observed a reduction in the FL form of MDM4 mRNA after treatment with GSK3326595 in all cell lines. In contrast, palbociclib resulted in a reduction in the FL transcripts in the parental but not the resistant cells (Fig. 5H). These data further support the hypothesis that palbociclib-induced suppression of PRMT5 is a key mediator of sensitivity to this drug. We also evaluated the contribution of protein degradation to the changes in MDM4 levels and observed that inhibition of proteasome activity by MG-132 did not restore MDM4 protein levels after treatment with palbociclib, GSK3326595, or their combination (Fig. 5I). This provided further evidence that pre-mRNA alternative splicing is the major mechanism by which these drugs regulate MDM4 protein levels.
In addition, and as mentioned previously, GSEA of RNA-seq data comparing sensitive to resistant cells revealed that the top commonly enriched gene sets in the resistant cells were involved in RNA splicing and processing (Fig. 1E). This suggests that global pre-mRNA splicing is a process that is rewired upon acquiring resistance to palbociclib and is potentially contributing to mediating the resistant phenotype.
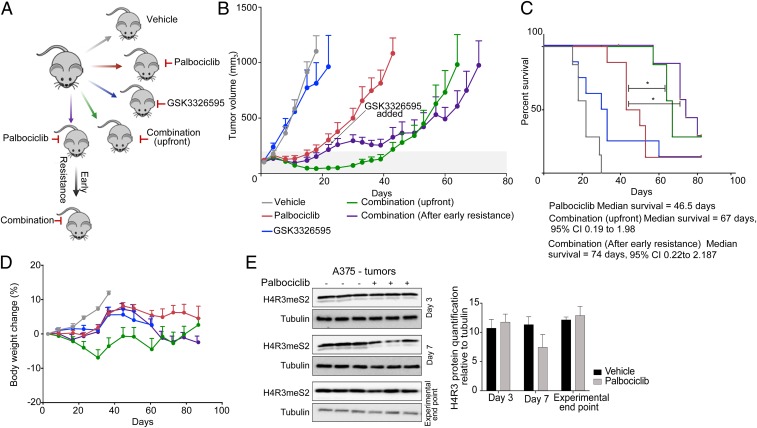
PRMT5 Inhibition Delays the Emergence of CDK4/6 Inhibitor Resistance In Vivo.
We next assessed the impact of GSK3326595 on the response to palbociclib in vivo using the A375 (drug-naive) tumor xenograft model. A375 tumor-bearing mice were treated with vehicle, palbociclib, GSK3326595, or combination palbociclib–GSK3326595. To evaluate the efficacy of GSK3326595 on tumor growth following the emergence of early resistance to palbociclib, a fifth group of tumor-bearing mice were administered palbociclib. These mice were treated with palbociclib plus GSK3326595 when tumors progressed on single agent palbociclib treatment (Fig. 6A). Compared to palbociclib alone, the combination showed robust reduction in tumor volume when given up-front or when given after development of resistance (Fig. 6B). These results demonstrate that PRMT5 inhibition delays the development of palbociclib resistance and is also effective following the emergence of resistance. The combination of palbociclib and GSK3326595, either as an up-front or sequential strategy, resulted in extended median survival compared to palbociclib alone (Fig. 6C) and was a well-tolerated combination strategy (Fig. 6D).
Fig. 6.
PRMT5 inhibition delays the emergence of CDK4/6 inhibitor resistance in vivo. (A) Schematic of in vivo drug efficacy study. (B) Tumor growth of A375 xenograft after treatment with vehicle, palbociclib, GSK3326595, or a combination of palbociclib and GSK3326595. Error bars represent SD of 6 to 8 tumors per group. (C) Kaplan–Meier curve of data in B shows survival advantage. *P < 0.05 by Log-rank (Mantel–Cox) test. Survival was measured as time for tumors to reach experimental end point volume of 1,200 mm3. (D) Weight chart body mass expressed as a percentage relative to day 0. Error bars represent SD of 6 to 8 tumors per group. (E) Western blots on A375 tumor lysates from mice treated with palbociclib and quantification of H4R3meS2 relative to tubulin. Error bars represent SD of n = 3.
To confirm the ability of palbociclib to suppress PRMT5 activity in vivo, levels of H4R3me2s were analyzed at different time points posttreatment. Consistent with our in vitro data, levels of H4R3me2s were reduced following 7 d of palbociclib treatment whereas, at experimental end point, when the tumors had progressed on palbociclib treatment, there was no change in H4R3me2s levels (Fig. 6E).
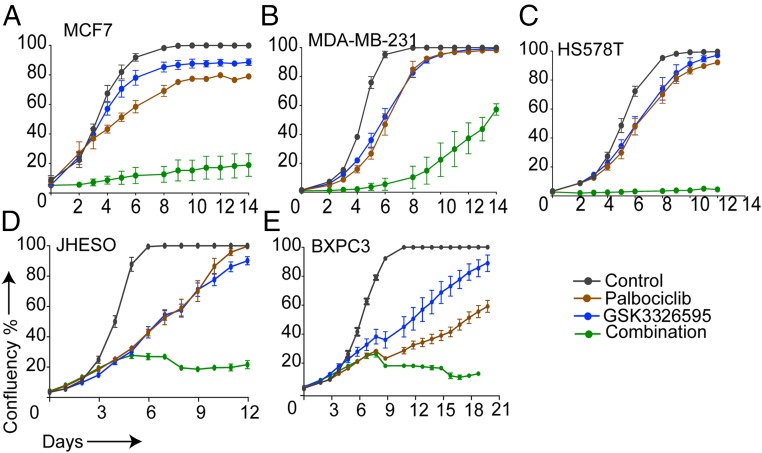
Coinhibition of CDK4/6 and PRMT5 Is Effective across Several Tumor Types.
Finally, we assessed the effectiveness of combining palbociclib and GSK3326595 across different cancer models that exhibit activated CDK4/6 pathway and express high levels of MDM4. This combination resulted in a more robust growth inhibition in breast (Fig. 7 A–C), esophageal (Fig. 7D), and pancreatic (Fig. 7E) cancer cell lines compared to the single agents. This observation provides evidence that PRMT5 inhibitors can be potentially utilized across various cancers to enhance the efficacy of CDK4/6 inhibitors.
Fig. 7.
Coinhibition of CDK4/6 and PRMT5 is effective across several tumor types. Proliferation curves of MCF7 (A), MDA-MB-231 (B), HS578T (C) breast cancer cells, JHESO (D) esophageal cancer cells, and BXPC3 (E) pancreatic cancer cells treated with 1 μM (2 μM for HS578T) palbociclib, 500 nM GSK3326595, or combination of both drugs. Graphs are representative of 2 independent experiments. Error bars represent SEM for 3 technical replicates.
Discussion
Identifying the mechanisms of response and resistance to targeted therapies is vital to develop rational combination strategies to improve antitumor efficacy. Based on the success of CDK4/6 inhibitors in combination with hormone therapy in estrogen receptor-positive breast cancer (6), there are numerous clinical trials in which CDK4/6 inhibitors are being used in combination in many other cancer types (54). Moreover, preclinical studies have demonstrated that CDK4/6 inhibitors can augment the efficacy of combination BRAF-MEK inhibitors (13, 15, 16, 55); this combination is now entering clinical development in melanoma patients.
In this study, we demonstrate that suppression of the PRMT5–MDM4 axis is key to a prolonged and potent response to CDK4/6 inhibition in melanoma and loss of control of this axis leads to resistance. CDK4/6 inhibition suppresses MDM4 protein expression by reducing PRMT5 activity. The combination of CDK4/6 and PRMT5 inhibitors potently suppressed cell proliferation and tumor growth. This combination therefore provides a promising therapeutic strategy for melanoma, in both the drug naive and resistant settings. Furthermore, our studies suggest that resistance to palbociclib is accompanied by changes in pre-mRNA splicing. CDK4/6 via regulating PRMT5 activity can regulate pre-mRNA splicing, and, through this mechanism, it regulates MDM4 expression and the p53 pathway, an important component of the response mechanism to palbociclib in p53-wild type melanoma cells.
Aggarwal et al. (42) demonstrated that only hyperactivated CDK4–cyclin D1 complex can regulate PRMT5 activity by increasing the ability of CDK4 to phosphorylate MEP50, the main coactivator of PRMT5. While, in these studies, hyperactivation was due to mutant Cyclin-D1, the melanoma cells used in our studies all exhibit hyperactivation of the CDK4/6 pathway due to either loss or mutation in CDKN2A (SI Appendix, Table S1). This suggests that only in cells that have hyperactivation of CDK4/6 will CDK4/6 inhibitors regulate PRMT5 and in turn activate p53. This could explain the inability of CDK4/6 inhibitors to activate p53 in a recent study in sarcoma cell lines (50) where CDK4/6 is not hyperactive.
Here, we showed that, in p53-wild type melanoma cells, MDM4 suppression and subsequent p53 activation were required for a robust response to CDK4/6 inhibition. Inhibition of PRMT5 by GSK3326595 resulted in profound reduction in MDM4 levels and was effective at inhibiting cell proliferation in combination with palbociclib. In contrast, in palbociclib-resistant cells that do not express MDM4 protein, GSK3326595 was unable to reduce cell proliferation or restore sensitivity to palbociclib, suggesting that MDM4 expression could be utilized as a biomarker for sensitivity to coinhibition of CDK4/6 and PRMT5. Furthermore, this observation also indicates that, in cells that lack MDM4, mechanisms independent of MDM4 can mediate acquired resistance to palbociclib.
Known mechanisms of resistance to CDK4/6 inhibitors include loss of functional RB1 by genomic deletion, mutation, or by maintenance of its phosphorylation, which is predominantly due to activation of CDK2-cyclin E1 (19, 21, 22). Recent data from the PALOMA-3 phase III clinical trial revealed that overexpression of CCNE1 was associated with poor response to palbociclib in estrogen receptor-positive metastatic breast cancer (56). In our studies, we treated p53-wild type palbociclib-resistant cell lines with a combination palbociclib-PRMT5 inhibitor that decreased RB1 phosphorylation and suppressed cell proliferation, recapitulating the response to the combination of palbociclib and CDK2 inhibitor. These data indicated that PRMT5 led to decreased CDK2 activity, potentially via inducing the CDK2 inhibitor p21. While the mechanism of action of PRMT5 inhibition may extend beyond an MDM4-p53–mediated induction of p21 and inactivation of CDK2, we believe that this is one of the key mechanisms enhancing the response to palbociclib.
In conclusion, our findings highlight the important role of PRMT5 as a downstream mediator of therapeutic efficacy of CDK4/6 inhibitors. We have also uncovered a link between CDK4/6 activity and MDM4 expression and illustrated that PRMT5-mediated regulation of alternative splicing of MDM4 pre-mRNA is a key mechanism in the response to CDK4/6 inhibitors. Given that CDK4, PRMT5, CDK2, and MDM4 are prominent oncogenes in melanoma and other cancers, the combination of CDK4/6 and PRMT5 inhibitors provides a promising therapeutic approach that can be applied not only in melanoma of various genomic subtype, but potentially in other cancers that exhibit activation of these pathways.
Materials and Methods
Cell Culture.
A375, HT144, CHL1, MCF7, MDA-MB-231, HS578T, and HEK293T cell lines were obtained from American Type Culture Collection. C002, D04, A11, and C067 were obtained from the Australasian Biospecimen Network Oncology group at QIMR, Australia. All cell lines except CHL1 and HEK293T cell lines were grown in RPMI-1640 Media (Gibco). CHL1 and HEK293T cell lines were grown in DMEM (Gibco). Routinely, media were supplemented with 10% (vol/vol) fetal bovine serum (FBS) (Gibco), 2 mM GlutaMAX-I l-alanyl-l-glutamine dipeptide (Gibco), 100 µg/mL penicillin/streptomycin (Gibco), and buffered with 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES). Cell lines were regularly tested for mycoplasma, and the identities of cell lines were confirmed by short tandem repeat (STR) analysis.
For the generation of acquired resistance melanoma cell lines, the cells were grown at increasing concentration of palbociclib. An initial concentration of 100 nM palbociclib was applied to the culture medium, and, when the cells were capable of proliferating in the presence of the drug, they were passaged and grown in double the previous palbociclib concentration. Cells were maintained in a final concentration of 2 to 4 µM palbociclib. To account for the changes associated with long-term culture, the parental cells were cultured under identical conditions and maintained in culture for the same period as the resistant cells, but in the absence of palbociclib. Before any experiment, cells were maintained in drug-free medium for 24 to 72 h.
Pharmacological Inhibitors.
All inhibitors were dissolved in dimethyl sulfoxide (DMSO) (Sigma-Aldrich). Inhibitors used were purchased from SellckChem except Palbociclib used in vivo and GSK3326595 were purchased from SYNthesis Med Chem.
Proliferation Assays.
Cell proliferation was measured by recording the confluency over time using the IncuCyte Zoom automated microscopy System (Essen BioScience). An algorithm that identifies the cell border was applied to calculate the confluency as a percentage of the vessel surface area.
Colony Formation Assay.
Cells were trypsinized, counted, and plated onto 6-well plates. Drug-containing media were added 6 to 8 h after plating. After 14 d, colonies were fixed and stained in a solution of 1% crystal violet in 35% methanol for 1 h at room temperature and washed in tap water. For drug removal assays, cells were grown in the drug for 14 d and then maintained in drug-free media for another 14 d before being fixed and stained.
High Throughput Drug Screening and Nuclei Counting.
Cells were plated in black-walled microplates (Corning). At 16 to 24 h after plating, the medium was replaced with drug-containing medium. A 10 μΜ starting concentration was used for all drugs except trametinib (1 μΜ), with 9 serial dilutions of each drug prepared at 1-in-3 ratios. Each dilution was used in four replicates. All of the plates contained media-only and solvent-only controls.
Nuclei were counted 6 d after treatment as a measure of the drug effect on cell growth. The cells were fixed using 4% paraformaldehyde (ProSciTech) in phosphate-buffered saline (PBS) for 10 min and then stained with DAPI staining solution (1 μg/mL DAPI, 50 mM Tris, pH 7.5, 0.2% Triton X-100) for 20 min. Nuclei were imaged using a Cellomics ArrayScan automated microscope (Cellomics, Life Technologies).
Cell Death Analysis.
Following treatment, tissue culture media was collected and combined with the cells after harvesting by scraping off the tissue culture plates. Cells were pelleted and then suspended in 200 µM of 1 µg/mL propidium iodide (PI) in PBS for 15 min at room temperature. Cells were analyzed using Canto II flow cytometers (BD Biosciences), and the data were processed in FlowLogic 7.2.1. PI-positive cells were counted as dead, as the cell death process is accompanied by disruption to the cellular membrane, which allows PI to penetrate and bind to nuclear DNA.
Western Blotting.
Whole-cell protein extracts were prepared by lysis in 2% sodium dodecyl sulfate (SDS) buffer (0.5 mM ethylenediaminetetraacetic acid [EDTA], 20 mM HEPES). The lysates were homogenized by passing through a 26-gauge needle, and then samples were denatured at 95 °C for 10 min. Protein quantification was performed using a detergent-compatible (DC) protein assay (Bio-RAD) according to the manufacturer’s instructions.
Equal amounts of total protein were suspended in 5× SDS sample buffer (313 mM Tris⋅HCl, pH 6.8, 50% [vol/vol] glycerol, 10% [vol/vol] β-mercaptoethanol, 10% [wt/vol] SDS, 0.05% [wt/vol] bromophenol blue) and were resolved by SDS-polyacrylamide gel electrophoresis (PAGE) with running buffer containing 25 mM Tris, 190 mM glycine, 0.1% (wt/vol) SDS using in-house or precast (Bio-Rad) gels. Protein was transferred onto polyvinylidene difluoride (PVDF) membranes (Millipore) in Tris-glycine transfer buffer (50 mM Tris, 40 mM glycine, 0.375% [wt/vol] SDS, 20% [vol/vol] methanol) using the Trans-Blot Turbo semidry transfer system (Bio-Rad) for 30 to 45 min. Immunoblots were blocked in 5% skim milk in Tris-buffered saline (TBS) containing 0.05% Tween 20 and then incubated with the appropriate primary antibody overnight at 4 °C, followed by the corresponding horseradish peroxidase (HRP)-conjugated secondary antibody for 1 h at room temperature. The signal was detected using enhanced chemiluminescent (ECL) Western blotting substrate (Amersham GE Healthcare). Actin or tubulin antibodies were used as controls for equal protein loading. Antibodies are listed in SI Appendix, Table S3.
Coimmunoprecipitation.
To prepare cell lysates for coimmunoprecipitation, cells were washed twice with ice-cold PBS and lysed on ice in 1% Nonidet P-40 nondenaturing buffer (20 mM Tris⋅HCl, pH 8.0, 137 mM NaCl, 10% glycerol, 2 mM EDTA) with freshly added Complete, EDTA-free Protease Inhibitor Mixture (ROCHE). Cell lysates were gently transferred into precooled microfuge tubes, constant agitation of the tubes was maintained for 30 min at 4 °C, and then lysates were cleared from cell debris by centrifugation at 12,000 rounds per minute (RPM) for 20 min at 4 °C. After protein quantification, equal amounts of 500 µg of protein lysate were incubated with 1.5 µg of the p21 antibody (SI Appendix, Table S3) at 4 °C overnight, followed by antibody–protein complex capture with Protein G Sepharose fast flow beads (Sigma-Aldrich) for 3 to 4 h at 4 °C with constant agitation. After extensive washing in Nonidet P-40 lysis buffer, complexes were eluted in 40 μL of 2× sample buffer without dithiothreitol (DTT) (65.8 mM Tris⋅HCl, pH 6.8, 26.3% [wt/vol] glycerol, 2% [wt/vol] SDS, 0.01% [wt/vol] bromophenol blue) and heated at 65 °C for 20 min then at 95 °C for 10 min; the beads were then pelleted, the supernatant was transferred to a new tube, and DTT was added at 100 mM. The eluted samples were boiled for 5 min and analyzed by immunoblotting.
RNA Isolation.
Following treatment, total RNA was isolated using the RNeasy Mini kit (QIAGEN) following the manufacturer’s instructions. RNA concentration and purity were determined by a NanoDrop ND1000 spectrophotometer (Thermo Scientific).
Real-Time PCR.
Complementary DNA (cDNA) was generated from 1 µg of total RNA using the High Capacity cDNA Reverse Transcription Kit (Life Technologies) following the manufacturer’s instructions. A volume of cDNA equivalent to 70 ng of total RNA was used for qPCR using Fast SYBR Green Master Mix (Applied Biosystems) and 1 µg of gene-specific primers. The reactions were performed using an Applied Biosystems StepOnePlus Real-Time PCR System (Applied Biosystems). The data were normalized to the NONO housekeeping gene. Changes in genes of interest expression were calculated as a fold increase or decrease relative to the vehicle samples. Relative expression was determined using the comparative ΔΔCt method. Primer sequences are listed in SI Appendix, Table S4.
Alternative Splicing Analysis by Conventional PCR.
A volume of cDNA equivalent to 30 ng of total RNA was used and 1 µg of total MDM4-specific primers. The reactions were performed using an S1000 Thermal cycler (Bio-RAD) using the following thermocycling conditions: 95 °C for 2 min, followed by 27 cycles of 95 °C for 45 s, 58 °C for 30 s and then 72 °C for 40 s, and finally followed by 72 °C for 5 min; products were visualized on a 2% agarose gel. Primer sequences are listed in SI Appendix, Table S4.
RNA Sequencing and Data Analysis.
RNA quality was confirmed using Agilent Bioanalyzer 2100 (Agilent Technologies), all samples had an RNA integrity number (RIN) higher than 9. Approximately 1 µg of RNA was used for library preparation according to standard protocols (TruSeq RNA; Illumina). Briefly, poly-A mRNA was purified using poly-T magnetic beads, fragmented using divalent cations under elevated temperature, and reverse transcribed to cDNA with random primers. Indexed adaptors were then ligated, and the library was amplified. Six indexed samples were pooled in a single lane of a HiSeq2500 flowcell (Illumina) to generate ∼30 million paired-end 50-bp reads per sample (resistant vs. parental samples), and a NextSeq500 (Illumina) to generate ∼30 million paired-end 150-bp reads per sample (palbociclib/GSK3326595 vs. control). RNA sequencing procedures were performed by the Molecular Genomics core facility at Peter MacCallum Cancer Centre.
The number of reads per gene was determined using HTSeq v0.5 (57) (resistant vs. parental samples) or featureCounts v1.6.2 (58) (palbociclib/GSK3326595 vs. control). The sequence was aligned against hg19v67 (resistant vs. parental samples) or hg38 (palbociclib/GSK3326595 vs. control) human reference genome. Limma-Voom (59) was used for differential expression analysis. The web-based platform Galaxy was used to perform the analysis. Differentially expressed genes were filtered for fold change ≥1.5 and adjusted P values (Padj) of <0.05. Overlaps were determined using venny. GSEA was performed using the Preranked tool within the GSEA 3.0 software (Broad Institute) (60, 61), and genes were ranked using the value of fold change (FC) and run against C2reactome and C5 (gene ontology [GO]) gene sets. Network analysis was performed using STRING 9.0 and filtered for high confidence interactions of 0.9. Enrichment analysis for GO and pathways (KEGG and biocarta) was mapped onto the network.
Viral Transfection and Transduction.
Stable gene silencing was performed using a lentiviral-mediated system. Gene-specific shRNA or scrambled constructs in pGIPZ or FH1t vectors were transfected into HEK293T cells with a pMDL/pRRE, pRSV-REV, and pCMV-VSV-G viral packaging system using PEI. Viral supernatant was collected 24 h later, filtered (0.45 μm), combined with 10 µg/mL protamine sulfate, and applied to target cells. Seventy-two hours posttransduction, the cells were selected with 1 μg/mL puromycin for 7 d. Recombinant DNA sequences are listed in SI Appendix, Table S5.
Reverse-Phase Protein Array.
The cells were washed twice with PBS and lysed in 1% Triton X-100 lysis buffer (50 mM Hepes, pH 7.4, 150 mM NaCl, 1.5 mM MgCl2, 1 mM ethylene glycol-bis(β-aminoethyl ether)-N,N,N′,N′-tetraacetic acid (EGTA), 100 mM NaF, 10 mM Na pyrophosphate, 1 mM Na3VO4, 10% glycerol) containing freshly added protease and phosphatase inhibitors: cOmplete, EDTA-free Protease Inhibitor Mixture, and PhosSTOP, respectively (Roche Applied Science). Following protein quantification, samples of 1 to 1.5 μg/μL total protein concentration were prepared in 4× SDS sample buffer (40% glycerol, 8% SDS, 0.25 M Tris⋅HCL, pH 6.8) containing freshly added 2-mercaptoethanol at 1:10 of the volume. The subsequent experimental work was performed at the MD Anderson Functional Proteomics RPPA Facility.
Xenograft Study.
Animal experiments were approved by the Peter MacCallum Animal Experimentation Ethics Committee (Protocol E569). Mice were purchased from the Garvan Institute. Tumors were implanted by s.c. injection of 3.8 × 106 A375 cells in 50 μL of 50% Matrigel, into the right flank of BALB/c nude mice (female, 6 to 7 wk old). At day 19 after injection, when the average tumor volumes reached 200 mm3, mice were randomized into 5 groups of 8 mice each. Drug treatments were delivered daily via oral gavage. Two groups of mice were administered palbociclib only (90 mg/kg in 50 mM sodium lactate, pH 4). The other three groups were either dosed with GSK3326595 (100 mg/kg in Kolliphor/PEG400), vehicle (sodium lactate with Kolliphor/PEG400), or a combination of palbociclib and GSK3326595. On day 19 after randomization, the dose of both palbociclib and GSK3326595 was reduced by 10% due to risk of weight loss. In order to evaluate the efficacy of the combination in overcoming resistance arising in vivo, GSK3326595 was added to one of the palbociclib-treated groups when the tumor volume reached twice that of the initial tumor volume when the mice were randomized. Treatment was discontinued, and mice were euthanized when the tumor size reached 1,200 mm3, a weight loss of more than 20% was recorded, or any signs of distress were observed.
Statistics.
Results obtained are presented as mean ± SEM or ± SD. Statistical significance tests are indicated in figures: *P < 0.05.
Data Sharing.
The data discussed in this publication have been deposited in NCBI's Gene Expression Omnibus database and are accessible through GEO series accession number GSE133670 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE133670) (62).
Supplementary Material
Acknowledgments
We thank the staff of the following Peter MacCallum Cancer Centre core facilities: the Victorian Centre for Functional Genomics; the Molecular Genomics facility; and the Flow Cytometry facility. We thank the MD Anderson RPPA core facility for performing the RPPA screening. We thank Rachael Walker and Susan Jackson for assisting in the in vivo study. We thank Dr. Maria Doyle (Research Computing) for bioinformatic assistance. We thank Associate Professor Marco Herold (Walter and Eliza Hall Institute) for providing the MDM4 silencing vector; Dr. Gareth Gregory and Prof. Ricky Johnston for providing CDK2 shRNA constructs; and Prof. Shereen Loi, Dr. Joyce Teo, Prof. Wayne Phillips, and Dr. Nicholas Clemmens for providing breast, pancreatic, and esophageal cancer cell lines. We thank Dr. Twishi Gulati for editorial assistance. We acknowledge Cancer Therapeutics CRC for partly funding the in vivo studies. This work was supported by the Peter MacCallum Cancer Foundation; and by Cancer Council Victoria Grant 1108149 and National Health and Medical Research Council of Australia Grant 1042986 (to K.E.S. and G.A.M.). S.A. was supported by doctoral scholarships from the University of Melbourne and Cancer Therapeutics CRC.
Footnotes
Conflict of interest statement: G.A.M and K.E.S. received research support from Celgene and Pfizer. M.A.D. is a member of the advisory boards for Novartis, GSK, BMS, Roche/Genentech, Sanofi-Aventis, and Vaccinex; a principal investigator of grants from GSK, Roche/Genentech, Sanofi-Aventis, AstraZeneca, Merck, and Oncothyreon; and a consultant to Nanostring.
This article is a PNAS Direct Submission.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, https://www.ncbi.nlm.nih.gov/geo (accession no. GSE133670).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1901323116/-/DCSupplemental.
References
- 1.Fountain J. W., et al. , Homozygous deletions within human chromosome band 9p21 in melanoma. Proc. Natl. Acad. Sci. U.S.A. 89, 10557–10561 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Curtin J. A., et al. , Distinct sets of genetic alterations in melanoma. N. Engl. J. Med. 353, 2135–2147 (2005). [DOI] [PubMed] [Google Scholar]
- 3.Zuo L., et al. , Germline mutations in the p16INK4a binding domain of CDK4 in familial melanoma. Nat. Genet. 12, 97–99 (1996). [DOI] [PubMed] [Google Scholar]
- 4.Sotillo R., et al. , Invasive melanoma in Cdk4-targeted mice. Proc. Natl. Acad. Sci. U.S.A. 98, 13312–13317 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sheppard K. E., McArthur G. A., The cell-cycle regulator CDK4: An emerging therapeutic target in melanoma. Clin. Cancer Res. 19, 5320–5328 (2013). [DOI] [PubMed] [Google Scholar]
- 6.O’Leary B., Finn R. S., Turner N. C., Treating cancer with selective CDK4/6 inhibitors. Nat. Rev. Clin. Oncol. 13, 417–430 (2016). [DOI] [PubMed] [Google Scholar]
- 7.Finn R. S., et al. , The cyclin-dependent kinase 4/6 inhibitor palbociclib in combination with letrozole versus letrozole alone as first-line treatment of oestrogen receptor-positive, HER2-negative, advanced breast cancer (PALOMA-1/TRIO-18): A randomised phase 2 study. Lancet Oncol. 16, 25–35 (2015). [DOI] [PubMed] [Google Scholar]
- 8.Turner N. C., Huang Bartlett C., Cristofanilli M., Palbociclib in hormone-receptor-positive advanced breast cancer. N. Engl. J. Med. 373, 1672–1673 (2015). [DOI] [PubMed] [Google Scholar]
- 9.Cristofanilli M., et al. , Fulvestrant plus palbociclib versus fulvestrant plus placebo for treatment of hormone-receptor-positive, HER2-negative metastatic breast cancer that progressed on previous endocrine therapy (PALOMA-3): Final analysis of the multicentre, double-blind, phase 3 randomised controlled trial. Lancet Oncol. 17, 425–439 (2016). [DOI] [PubMed] [Google Scholar]
- 10.Sherr C. J., Beach D., Shapiro G. I., Targeting CDK4 and CDK6: From discovery to therapy. Cancer Discov. 6, 353–367 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.US National Library of Medicine, CDK4/6 inhibitor. ClinicalTrials.gov. Available at https://clinicaltrials.gov/ct2/results?cond=&term=CDK4%2F6+inhibitor&cntry=&state=&city=&dist=. Accessed 29 February 2000.
- 12.Yoshida A., Lee E. K., Diehl J. A., Induction of therapeutic senescence in vemurafenib-resistant melanoma by extended inhibition of CDK4/6. Cancer Res. 76, 2990–3002 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yadav V., et al. , Co-targeting BRAF and cyclin dependent kinases 4/6 for BRAF mutant cancers. Pharmacol. Ther. 149, 139–149 (2015). [DOI] [PubMed] [Google Scholar]
- 14.Mahgoub T., et al. , Kinase inhibitor screening identifies CDK4 as a potential therapeutic target for melanoma. Int. J. Oncol. 47, 900–908 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Martin C. A., et al. , Palbociclib synergizes with BRAF and MEK inhibitors in treatment naïve melanoma but not after the development of BRAF inhibitor resistance. Int. J. Cancer 142, 2139–2152 (2018). [DOI] [PubMed] [Google Scholar]
- 16.Kwong L. N., et al. , Oncogenic NRAS signaling differentially regulates survival and proliferation in melanoma. Nat. Med. 18, 1503–1510 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shapiro G. I., et al. , Abstract CT046: Phase I dose escalation study of the CDK4/6 inhibitor palbociclib in combination with the MEK inhibitor PD-0325901 in patients with RAS mutant solid tumors. Cancer Res. 77, CT046 (2017). [Google Scholar]
- 18.Michaud K., et al. , Pharmacologic inhibition of cyclin-dependent kinases 4 and 6 arrests the growth of glioblastoma multiforme intracranial xenografts. Cancer Res. 70, 3228–3238 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dean J. L., et al. , Therapeutic response to CDK4/6 inhibition in breast cancer defined by ex vivo analyses of human tumors. Cell Cycle 11, 2756–2761 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Young R. J., et al. , Loss of CDKN2A expression is a frequent event in primary invasive melanoma and correlates with sensitivity to the CDK4/6 inhibitor PD0332991 in melanoma cell lines. Pigment Cell Melanoma Res. 27, 590–600 (2014). [DOI] [PubMed] [Google Scholar]
- 21.Herrera-Abreu M. T., et al. , Early adaptation and acquired resistance to CDK4/6 inhibition in estrogen receptor-positive breast cancer. Cancer Res. 76, 2301–2313 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.O’Leary B., et al. , The genetic landscape and clonal evolution of breast cancer resistance to palbociclib plus fulvestrant in the PALOMA-3 trial. Cancer Discov. 8, 1390–1403 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Condorelli R., et al. , Polyclonal RB1 mutations and acquired resistance to CDK 4/6 inhibitors in patients with metastatic breast cancer. Ann. Oncol. 29, 640–645 (2018). [DOI] [PubMed] [Google Scholar]
- 24.Zhang Y. X., et al. , Antiproliferative effects of CDK4/6 inhibition in CDK4-amplified human liposarcoma in vitro and in vivo. Mol. Cancer Ther. 13, 2184–2193 (2014). [DOI] [PubMed] [Google Scholar]
- 25.Olanich M. E., et al. , CDK4 amplification reduces sensitivity to CDK4/6 inhibition in fusion-positive rhabdomyosarcoma. Clin. Cancer Res. 21, 4947–4959 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yang C., et al. , Acquired CDK6 amplification promotes breast cancer resistance to CDK4/6 inhibitors and loss of ER signaling and dependence. Oncogene 36, 2255–2264 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jenuwein T., Allis C. D., Translating the histone code. Science 293, 1074–1080 (2001). [DOI] [PubMed] [Google Scholar]
- 28.Kouzarides T., Chromatin modifications and their function. Cell 128, 693–705 (2007). [DOI] [PubMed] [Google Scholar]
- 29.McBride A. E., Silver P. A., State of the arg: Protein methylation at arginine comes of age. Cell 106, 5–8 (2001). [DOI] [PubMed] [Google Scholar]
- 30.Karkhanis V., Hu Y. J., Baiocchi R. A., Imbalzano A. N., Sif S., Versatility of PRMT5-induced methylation in growth control and development. Trends Biochem. Sci. 36, 633–641 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Blanc R. S., Richard S., Arginine methylation: The coming of age. Mol. Cell 65, 8–24 (2017). [DOI] [PubMed] [Google Scholar]
- 32.Friesen W. J., et al. , The methylosome, a 20S complex containing JBP1 and pICln, produces dimethylarginine-modified Sm proteins. Mol. Cell. Biol. 21, 8289–8300 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Friesen W. J., et al. , A novel WD repeat protein component of the methylosome binds Sm proteins. J. Biol. Chem. 277, 8243–8247 (2002). [DOI] [PubMed] [Google Scholar]
- 34.Braun C. J., et al. , Coordinated splicing of regulatory detained introns within oncogenic transcripts creates an exploitable vulnerability in malignant glioma. Cancer Cell 32, 411–426.e11 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bezzi M., et al. , Regulation of constitutive and alternative splicing by PRMT5 reveals a role for Mdm4 pre-mRNA in sensing defects in the spliceosomal machinery. Genes Dev. 27, 1903–1916 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gerhart S. V., et al. , Activation of the p53-MDM4 regulatory axis defines the anti-tumour response to PRMT5 inhibition through its role in regulating cellular splicing. Sci. Rep. 8, 9711 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Marine J. C., et al. , Keeping p53 in check: Essential and synergistic functions of Mdm2 and Mdm4. Cell Death Differ. 13, 927–934 (2006). [DOI] [PubMed] [Google Scholar]
- 38.Wang X., Jiang X., Mdm2 and MdmX partner to regulate p53. FEBS Lett. 586, 1390–1396 (2012). [DOI] [PubMed] [Google Scholar]
- 39.Gembarska A., et al. , MDM4 is a key therapeutic target in cutaneous melanoma. Nat. Med. 18, 1239–1247 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Antonysamy S., et al. , Crystal structure of the human PRMT5:MEP50 complex. Proc. Natl. Acad. Sci. U.S.A. 109, 17960–17965 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ho M. C., et al. , Structure of the arginine methyltransferase PRMT5-MEP50 reveals a mechanism for substrate specificity. PLoS One 8, e57008 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Aggarwal P., et al. , Nuclear cyclin D1/CDK4 kinase regulates CUL4 expression and triggers neoplastic growth via activation of the PRMT5 methyltransferase. Cancer Cell 18, 329–340 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kong X., et al. , Cancer drug addiction is relayed by an ERK2-dependent phenotype switch. Nature 550, 270–274 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Filmus J., et al. , Induction of cyclin D1 overexpression by activated ras. Oncogene 9, 3627–3633 (1994). [PubMed] [Google Scholar]
- 45.Stopa N., Krebs J. E., Shechter D., The PRMT5 arginine methyltransferase: Many roles in development, cancer and beyond. Cell. Mol. Life Sci. 72, 2041–2059 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kryukov G. V., et al. , MTAP deletion confers enhanced dependency on the PRMT5 arginine methyltransferase in cancer cells. Science 351, 1214–1218 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Marjon K., et al. , MTAP deletions in cancer create vulnerability to targeting of the MAT2A/PRMT5/RIOK1 axis. Cell Rep. 15, 574–587 (2016). [DOI] [PubMed] [Google Scholar]
- 48.Mavrakis K. J., et al. , Disordered methionine metabolism in MTAP/CDKN2A-deleted cancers leads to dependence on PRMT5. Science 351, 1208–1213 (2016). [DOI] [PubMed] [Google Scholar]
- 49.Dean J. L., Thangavel C., McClendon A. K., Reed C. A., Knudsen E. S., Therapeutic CDK4/6 inhibition in breast cancer: Key mechanisms of response and failure. Oncogene 29, 4018–4032 (2010). [DOI] [PubMed] [Google Scholar]
- 50.Sriraman A., Dickmanns A., Najafova Z., Johnsen S. A., Dobbelstein M., CDK4 inhibition diminishes p53 activation by MDM2 antagonists. Cell Death Dis. 9, 918 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Haupt S., et al. , Targeting Mdmx to treat breast cancers with wild-type p53. Cell Death Dis. 6, e1821 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Herold M. J., van den Brandt J., Seibler J., Reichardt H. M., Inducible and reversible gene silencing by stable integration of an shRNA-encoding lentivirus in transgenic rats. Proc. Natl. Acad. Sci. U.S.A. 105, 18507–18512 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bardot B., Toledo F., Targeting MDM4 Splicing in Cancers. Genes (Basel) 8, E82 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Klein M. E., Kovatcheva M., Davis L. E., Tap W. D., Koff A., CDK4/6 inhibitors: The mechanism of action may not be as simple as once thought. Cancer Cell 34, 9–20 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Teh J. L., et al. , An in vivo reporter to quantitatively and temporally analyze the effects of CDK4/6 inhibitor-based therapies in melanoma. Cancer Res. 76, 5455–5466 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Turner N. C., et al. , Cyclin E1 expression and palbociclib efficacy in previously treated hormone receptor-positive metastatic breast cancer. J. Clin. Oncol. 37, 1169–1178 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Anders S., Pyl P. T., Huber W., HTSeq–A Python framework to work with high-throughput sequencing data. Bioinformatics 31, 166–169 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Liao Y., Smyth G. K., Shi W., FeatureCounts: An efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30, 923–930 (2014). [DOI] [PubMed] [Google Scholar]
- 59.Law C. W., Chen Y., Shi W., Smyth G. K., Voom: Precision weights unlock linear model analysis tools for RNA-seq read counts. Genome Biol. 15, R29 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Mootha V. K., et al. , PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat. Genet. 34, 267–273 (2003). [DOI] [PubMed] [Google Scholar]
- 61.Subramanian A., et al. , Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. U.S.A. 102, 15545–15550 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.AbuHammad S., et al. , Regulation of PRMT5-MDM4 axis is critical in the response to CDK4/6 inhibitors in melanoma. Gene Expression Omnibus. https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE133670. Deposited 1 July 2019.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.