Significance
Inhibitory receptors specific for major histocompatibility class I (MHC-I) molecules regulate NK cell responses. Here, we describe that cluster of differentiation 8αα (CD8αα) works as a coreceptor to enhance interaction between MHC-I and one of these receptors, KIR3DL1. We further demonstrate that inhibition of NK cell activation by MHC-I and KIR3DL1 interaction is fine-tuned by CD8αα. By testing cytokine secretion ability, we found CD8αα+ NK cells are generally better educated, and thus more responsive to abnormal cells. All these results suggest that CD8αα is an important regulator of NK cell activation.
Keywords: KIR3DL1, CD8αα, coreceptor, NK cell activation
Abstract
Cluster of differentiation 8 (CD8) is a cell surface glycoprotein, which is expressed as 2 forms, αα homodimer or αβ heterodimer. Peptide-loaded major histocompatibility complex class I (pMHC-I) molecules are major ligands for both forms of CD8. CD8αβ is a coreceptor for the T cell receptor (TCR) and binds to the same cognate pMHC-I as the TCR, thus enabling or augmenting T cell responses. The function of CD8αα homodimers is largely unknown. While CD8αβ heterodimer is expressed exclusively on CD8+ T cells, the CD8αα homodimer is present in subsets of T cells and human natural killer (NK) cells. Here, we report that the CD8αα homodimer functions as a coreceptor for KIR3DL1, an inhibitory receptor of NK cells that is specific for certain MHC-I allotypes. CD8αα enhances binding of pMHC-I to KIR3DL1, increases KIR3DL1 clustering at the immunological synapse, and augments KIR3DL1-mediated inhibition of NK cell activation. Additionally, interactions between pMHC-I and CD8αα homodimers regulate KIR3DL1+ NK cell education. Together, these findings reveal another dimension to the modulation of NK cell activity.
The cluster of differentiation 8 (CD8) molecule is widely known as a marker for cytotoxic T lymphocytes (CTLs), which mediate immunity against intracellular infectious agents and cancers. CD8 is a coreceptor for the T cell receptor (TCR) and it binds to the membrane-proximal region of a peptide-loaded major histocompatibility complex class (pMHC) complex, when the TCR recognizes the membrane-distal domains of the same pMHC molecule (1). CD8 enhances binding of MHC-I to TCR and increases TCR signaling via Lck-induced CD3ζ chain tyrosine phosphorylation (1, 2). There are 2 forms of CD8, the αα homodimer and αβ heterodimer, both of which are expressed by CTLs. However, only the CD8αβ heterodimer has an important role in T cell development and functions, although the binding affinity of CD8αα/pMHC is similar to that of CD8αβ (3, 4).
Different from CD8αβ, which is exclusively expressed in CTLs, CD8αα is present on other cell types, such as γδ T cells (5), human natural killer (NK) cells (6), and murine dendritic cells (7). The function of CD8αα is largely unknown. Interestingly, CD8αα+ cells are generally more functional compared with their CD8αα− counterparts. For instance, CD8αα+ NK cells are generally more cytotoxic than their CD8αα− counterparts (6); CD8αα+ dendritic cells are more efficient at cross-presentation (8). These findings suggest CD8αα might be more than just a cell marker. On intestinal intraepithelial T lymphocytes, CD8αα binds to the nonclassical MHC class I-like thymus leukemia antigen, independently of TCR/CD3, and modifies TCR signaling (9). It has also been speculated that CD8αα functions as a TCR corepressor that negatively regulates cellular activation (10). These data suggest important functions for CD8αα.
NK cells are an important component of the innate immune response. NK cells express an array of antigen receptors that monitor cellular changes induced by cell transformation or viral infection. Changes in MHC-I expression are sensed by polymorphic receptors on NK cells. Among these receptors, the killer cell Ig-like receptors (KIRs) recognize different MHC allotypes (11). There are a lot of similarities between KIR and TCR. Both KIR and TCR bind MHC-I as a ligand. The footprint of KIR binding on the MHC-I overlaps partially with those of TCR binding on MHC-I (12). KIR and TCR also share a similar signaling mechanism (i.e., tyrosine phosphorylation by Lck) (13, 14). As CD8 is a coreceptor for TCR, we hypothesized that CD8 might also function as a coreceptor for KIRs. In this study, we analyzed the effects of CD8αα on MHC-I binding to a specific KIR, and on KIR signaling and downstream functional effects.
Results
CD8αα Enhances the Binding of Human Leukocyte Antigen-Bw4 to KIR3DL1.
Specific interactions of KIRs with their respective human leukocyte antigen-I (HLA-I; human MHC-I) ligands inhibit or enhance NK activity, which forms the basis for immune recognition of missing self or altered self. One of the best-defined allospecific pairs is KIR3DL1 and HLA-I molecules carrying the Bw4 serological motif (15), which comprises an epitope spanning residues 77 to 83 of certain HLA-B and HLA-A heavy chains. Compared with other KIRs, KIR3DL1 is quite prevalent on NK cells and is generally expressed at high levels. We found that all KIR3DL1+ donors have a subset of KIR3DL1+ NK cells that express CD8 (SI Appendix, Figs. S1 and S2). Whereas the majority of CD3+CD8+ T cells from blood are double-positive for both CD8α and CD8β (SI Appendix, Fig. S1A), CD8+ NK cells (CD3−CD56+ subset) are predominantly single-positive for CD8α (SI Appendix, Fig. S1B), as previously described (16). Immunoblotting analysis of NK cell lysates indicated expression of CD8αα in a homodimeric form (SI Appendix, Fig. S1C), consistent with previous reports (17). Based on these findings, we examined effects of CD8αα upon the interactions of HLA-Bw4 with KIR3DL1 and the resulting functional consequences for NK activity.
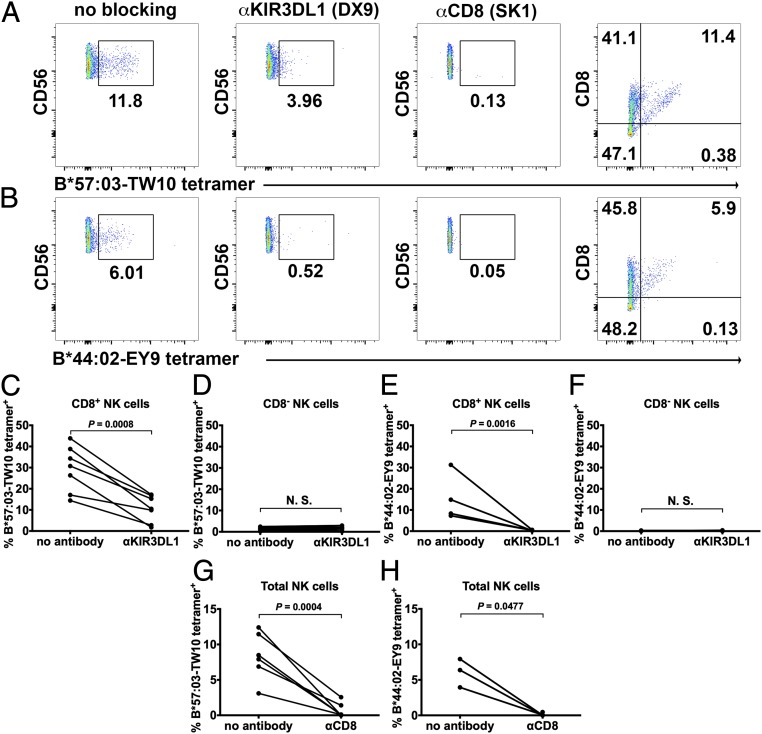
First, HLA-I tetramer staining assays were performed to determine whether CD8αα influences HLA-I binding to KIR3DL1. Peptide-bound tetramers of HLA-B*44:02 and B*57:03, 2 HLA-Bw4 allotypes, were prepared using previously described methods (18) after verification of the integrity of the protein preparations using native gels of their monomeric forms (SI Appendix, Fig. S2A). Binding to peptide enhanced HLA-B migration rates on native gels, consistent with the negative charge of the peptides loaded onto both allotypes (SI Appendix, Fig. S2A). Tetramer staining was carried out with human peripheral blood mononuclear cells (PBMCs) from multiple donors. Donors were first grouped as KIR3DL1+ or KIR3DL1− based on sequence-specific priming or by flow cytometry after staining with a KIR3DL1-specific antibody. Both KIR3DL1+ and KIR3DL1− donors were tested for staining with HLA-B*57:03-TW10 and B*44:02-EY9 tetramers. As expected, only KIR3DL1+ donor NK cells were stained by the HLA-Bw4 tetramers (SI Appendix, Fig. S2B), indicating that the staining is largely mediated by the interaction between HLA-Bw4 and KIR3DL1. This was confirmed by blocking of the tetramer signal by a KIR3DL1-specific antibody (Fig. 1 A and B, column II and Fig. 1 C and E). Intriguingly, the tetramer-binding signal is also blocked by an anti-CD8α antibody (SK1) (Fig. 1 A and B, column III and Fig. 1 G and H). The binding between CD8αα and peptide HLA-I (pHLA-I) is very weak (3, 4, 18), and CD8 alone is insufficient to support staining by pHLA-I, as KIR3DL1−CD8+ NK cells did not stain with the tetramers (SI Appendix, Fig. S2). However, there is a strong correlation between tetramer staining intensity and CD8αα expression levels on the NK cells (Fig. 1 A and B, column IV), suggesting that CD8αα enhances pHLA-I binding to KIR3DL1 in an expression level-dependent manner. Similar effects were also observed using NK cells from additional KIR3DL1+ donors (SI Appendix, Fig. S3). Under our experimental conditions, and using B*57:03-TW10 and B*44:02-EY9, we did not observe significant CD8αα-independent staining (Fig. 1 D and F; essentially no tetramer binding within the CD8− gates of KIR3DL1+ NK cells). Thus, CD8αα, expressed on NK cells, is critical for stable binding between HLA-Bw4 and KIR3DL1 (Fig. 1 D and F–H). However, under physiological conditions, pHLA-I might engage KIR3DL1 even in the absence of CD8αα, but with reduced efficiency compared with engagement in the presence of CD8αα, which is further discussed below.
Fig. 1.
CD8 augments specific HLA-B staining to KIR3DL1. NK cells (CD3−CD56+ PBMCs) were stained with B*57:03-TW10 (A) or B*44:02-EY9 (B) tetramers (column I). Tetramer staining was blocked by anti-KIR3DL1 (column II) or anti-CD8 (column III) antibodies. There is a correlation of tetramer staining with CD8 expression level on NK cells (column IV). Representative staining data from one of 7 donors for B*57:03 and 4 donors for B*44:02 are shown in A and B. Tetramer staining data with additional donors are shown in SI Appendix, Fig. S3. (C–H) Compiled tetramer staining data are shown. N.S., not significant.
CD8αα Enhances KIR3DL1 Clustering.
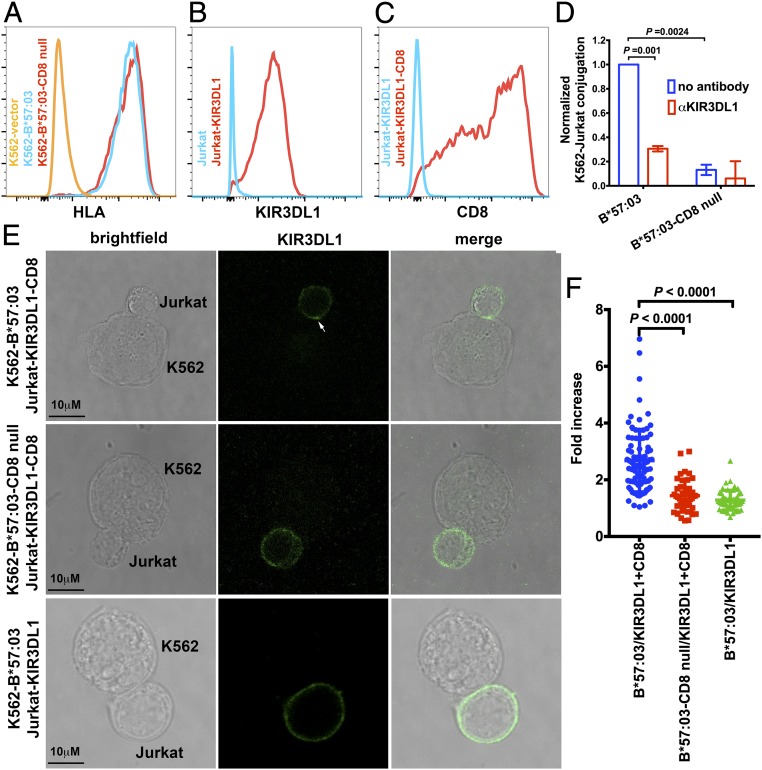
Since HLA-Bw4 tetramer staining to KIR3DL1+ NK cells is blocked by anti-CD8, the interaction with CD8 was expected to enhance the efficiency of engagement of HLA-Bw4–expressing cells to KIR3DL1-expressing cells. We performed cell–cell contact assays to verify this. We expressed HLA-B*57:03 and HLA-B*57:03 mutated at the CD8 binding residues (D227K/T228A; B*57:03-CD8null) (2) in the HLA-I–deficient erythroleukemic cell line K562. Both proteins are readily detectable on the cell surface (Fig. 2A). Jurkat cells were first infected with a retrovirus to express KIR3DL1 and then transduced to express CD8α (Fig. 2 B and C). Jurkat is a human T lymphocytic cell line that does not express HLA-Bw4 alleles (19), which prevents potential cis interactions with KIR3DL1. Cell conjugation between the K562 cells and Jurkat cells was investigated by flow cytometry. K562 cells were labeled with carboxyfluorescein succinimidyl ester (CFSE) and then incubated with CD8+KIR3DL1+ Jurkat cells. CFSE and CD8 double-positive cell populations were identified as K562-Jurkat cell conjugates, and used to quantify the effect of interactions of B*57:03-CD8 on cell adhesion. K562-B*57:03 showed stronger adhesion to CD8+KIR3DL1+ Jurkat cells as compared with K562-B*57:03-CD8null based on a flow cytometry assay (Fig. 2D and SI Appendix, Table S1), reflecting the significance of B*57:03-CD8 binding upon enhancement of cell adhesion.
Fig. 2.
CD8 facilitates cell conjugation induced by KIR3DL1–Bw4 interactions and KIR3DL1 clustering induced by HLA-Bw4. (A) Surface expression levels of B*57:03 or B*57:03-CD8null on the surface of K562 cells were assessed with flow cytometry after staining with the HLA-I–specific antibody W6/32. KIR3DL1 (B) and CD8 (C) expression on the surface of Jurkat cells was confirmed by flow cytometry. (D) Cell conjugation between Jurkat-KIR3DL1-CD8 cells and K562 cells expressing no HLA-I, B*57:03, or B*57:03-CD8null was tested by flow cytometry. K562 cells were labeled with CFSE. Cell conjugation was either blocked with anti-KIR3DL1 or medium. CFSE and CD8 double-positive cells are quantified as conjugates. Cell conjugation was normalized to that between Jurkat-KIR3DL1-CD8 cells and K562-B*57:03 cells after background correction (based on conjugates between Jurkat-KIR3DL1-CD8 cells and K562 cells). Data before normalization are shown in SI Appendix, Table S1. Conjugation between Jurkat-KIR3DL1-CD8 cells and K562-B*57:03 cells is partially blocked by anti-KIR3DL1. K562-B*57:03 cells are more efficient than K562-B*57:03-CD8null cells in forming conjugates with Jurkat-KIR3DL1-CD8 cells (n = 3 replicates). (E and F) Jurkat-KIR3DL1-CD8 cells were incubated with K562 cells expressing B*57:03 or B*57:03-CD8null and then fixed and stained with anti-KIR3DL1 before analysis by confocal microscopy. As another control, Jurkat-KIR3DL1 cells incubated with K562-B*57:03 were also assessed. The arrowhead in E indicates clustering at the interface between the Jurkat and K562 cells. The intensity of staining of the Jurkat cells at cell–cell interfaces was compared with that measured at a noncontact area. (F) Data are plotted as the fold increase in intensity above the noncontact area background. The results from 5 independent experiments for K562-B*57:03/Jurkat-KIR3DL1-CD8 and 2 independent experiments for K562-B*57:03-CD8null/Jurkat-KIR3DL1-CD8 and K562-B*57:03/Jurkat-KIR3DL1 with a total of >40 conjugates per condition are shown. Data are shown as mean ± SD. Statistical analyses were performed with paired Student’s t tests using GraphPad Prism version 7.
Similar to T cells, NK cells form an immunological synapse (IS) at their interfaces with target cells. Segregation of KIR at the IS and KIR phosphorylation within the IS are important for downstream signaling (20, 21). To further investigate the effect of CD8αα on KIR3DL1 function, we used a clustering assay to determine whether pHLA-CD8 engagement enhances KIR3DL1 clustering. CD8+KIR3DL1+ Jurkat cells were coincubated with K562 cells expressing HLA-B*57:03 or HLA-B*57:03-CD8null. There is clear KIR3DL1 clustering at the interface between Jurkat cells and K562-B*57:03 cells after incubation (Fig. 2 E and F). In contrast, K562-B*57:03-CD8null cells induced significantly reduced KIR3DL1 clustering, indicating CD8αα binding to HLA-Bw4 synergizes KIR3DL1 enrichment in the IS (Fig. 2 E and F). Additionally, K562-B*57:03 did not induce strong KIR3DL1 clustering in the absence of CD8αα (Fig. 2 E and F). We also analyzed CD8αα distribution by confocal microscopy and found that CD8αα was clustered at the interface between K562-B*57:03 cells and Jurkat-KIR3DL1-CD8 cells and colocalized with KIR3DL1 (SI Appendix, Fig. S4), consistent with a coreceptor function for CD8.
CD8αα Enhances the Inhibitory Ability of HLA-Bw4.
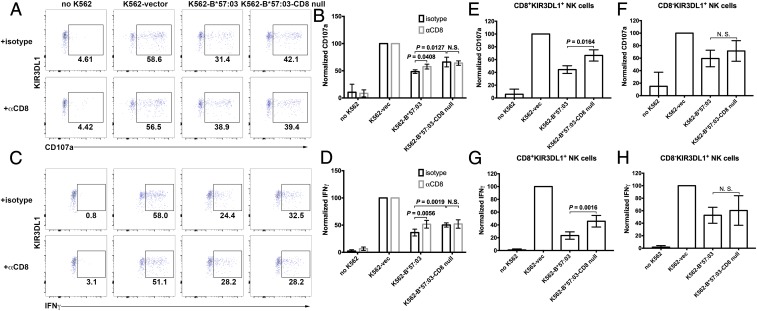
We further tested whether CD8αα can augment the inhibitory function of KIR3DL1 in the activation of primary NK cells. Donors with a high percentage of CD8+KIR3DL1+ NK cell population were chosen for this assay. PBMCs were incubated with the K562, a prototypical NK cell target due to its MHC class I deficiency (22), or K562 cells engineered to express HLA-B. KIR3DL1+CD3−CD56+ cells were gated, and expression of interferon-γ (IFN-γ) and the degranulation marker CD107a were measured by flow cytometry. This assay allowed us to determine the NK populations with the potential for cytotoxicity and cytokine secretion.
B*57:03 expression on K562 cells significantly reduced the percentage of NK cells producing CD107a (Fig. 3 A and B and SI Appendix, Fig. S5 A and C and Tables S2, S4, and S6) or IFN-γ (Fig. 3 C and D and SI Appendix, Fig. S5 B and D and Tables S3, S5, and S7). This reduction was partially rescued by blocking cell surface CD8, suggesting that CD8αα augments the inhibitory function of KIR3DL1 on primary NK cell activation. Compared with wild-type (WT) B*57:03, the B*57:03-CD8null mutant demonstrated a weakened ability to inhibit NK cell activation. Additionally, blocking surface CD8 had little effect on NK cell activation with the B*57:03-CD8null mutant, different from the WT. The data were further analyzed to compare the effects of the CD8 binding site mutation of B*57:03 on the inhibition of activation of CD8+ (Fig. 3 E and G) and CD8− (Fig. 3 F and H) NK cells. The different effects of B*57:03 and B*57:03-CD8null on inhibition of CD107a (Fig. 3 E and F) or IFN-γ (Fig. 3 G and H) production by KIR3DL1+ NK cells were only significant in CD8+ cells, excluding the possibility that the distinct behavior of WT and mutant were caused by other effects of the CD8 binding site mutations.
Fig. 3.
CD8 enhances the inhibitory ability of HLA-Bw4 toward KIR3DL1+ NK cell activation. NK cells (CD3−CD56+ PBMCs) were activated by incubation with K562 cells or K562 cells expressing exogenous B*57:03 or B*57:03-CD8null. KIR3DL1+ NK cell activation was determined by testing CD107a (A and B) and IFN-γ (C and D) induction. A and C are representative data, while B and D are compiled data (n = 4). Cell activation was normalized to the NK cell + K562-vec condition after background correction (based on untreated NK cells). vec, empty vector. Data before normalization are shown in SI Appendix, Tables S2 and S3. Anti-CD8 antibody can partially rescue B*57:03-mediated KIR3DL1+ NK cell inhibition. B*57:03-CD8null showed reduced inhibition of KIR3DL1+ NK cell activation, and anti-CD8 antibody has no significant effect on B*57:03-CD8null-induced inhibition of KIR3DL1+ NK cell activation. (E–H) B*57:03 showed stronger inhibition of NK cell activation than B*57:03-CD8null only in CD8+, but not CD8−, KIR3DL1+ NK cells. Statistical analyses were performed with paired Student’s t tests. N.S., not significant.
CD8αα Is Important in NK Cell Education.
Mechanisms behind the higher cytolytic activity of human NK cells expressing CD8αα compared with CD8–negative counterparts (6) are not elucidated. The intrinsic functional activities of NK cells are determined by a process called NK cell education or licensing. Self–MHC-I recognition by NK inhibitory receptors is known to mediate NK education and the extent of their functional activity (22, 23). Since CD8αα enhances HLA-Bw4 binding to KIR3DL1 and its inhibitory signaling, we hypothesized that CD8αα also enhances NK cell education and empowers NK cells with stronger cytolytic activity.
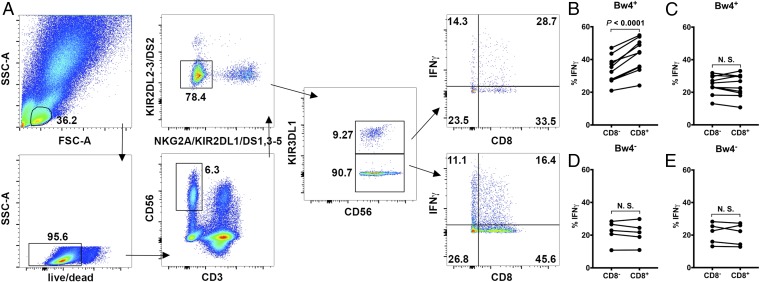
We analyzed the expression of IFNγ in primary NK cells following their coincubation with K562 cells. Besides KIR, other well-characterized NK cell-inhibitory receptors that bind classical or nonclassical HLA-I as ligands and could contribute to NK cell education include NKG2A, which recognizes HLA-E (24), and LILRB1 and LILRB2, which compete with CD8 for binding HLA-I (25), and thus should not show any CD8 dependency for NK signaling. Using established methods (22), we focused on 2 NK cell subsets to examine the influences of CD8 on NK education: KIR−NKG2A− NK cells (which do not express KIR2DL1, KIR2DL2, KIR2DL3, KIR2DS1, KIR2DS2, KIR2DS3, KIR2DS4, KIR2DS5, KIR3DL1, or NKG2A) and KIR3DL1+“others−” (22) NK cells (which express only KIR3DL1, but not KIR2DL1, KIR2DL2, KIR2DL3, KIR2DS1, KIR2DS2, KIR2DS3, KIR2DS4, KIR2DS5, or NKG2A).
Upon coincubation with K562, a larger IFNγ+ population was observed in the KIR3DL1+ NK cells than in the KIR−NKG2A− NK cells, i.e., those lacking the HLA-I–specific receptors (Fig. 4A). Since K562 cells do not express HLA-I, the low responsiveness of KIR−NKG2A− NK cells might result from their education by novel non–HLA-specific inhibitory receptors, as previously suggested (22).
Fig. 4.
CD8 modulates NK cell education by facilitating HLA-Bw4 binding to KIR3DL1. (A) Gating strategy for selective detection of KIR3DL1+ and KIR3DL1− NK cells. Live lymphocytes were first gated, from which CD3−CD56+ cells were selected. For selective gating, CD3−CD56+ NK cells negative for KIR2DL1/L2/L3/S1/S2/S3/S4/S5 and NKG2A were selected. These cells were further gated to identify KIR3DL1+ and KIR3DL1− cells. Selectively gated KIR3DL1+ and KIR3DL1− NK cells were then examined for IFN-γ expression. Functional gates were set on KIR3DL1+ or KIR3DL1− NK cells in unstimulated PBMCs. The NK cell activation rate was characterized by the fraction of activated cells among all cells within the CD8+ or CD8− gate. (B and C) In HLA-Bw4+ donors, KIR3DL1+ (i.e., KIR3DL1+others−) NK cells (B), but not KIR3DL1− (i.e., KIR−NKG2A−) NK cells (C), showed CD8-dependent activation by K562 cells (n = 8 donors). (D and E) In HLA-Bw4− donors, neither KIR3DL1+ (D) nor KIR3DL1− (E) NK cells showed CD8 dependency (n = 5 donors). Statistical analyses were performed with paired Student’s t tests. N.S., not significant.
The CD8 dependence of education of different NK cell subsets was evaluated by assessing the ratio of K562 cell responsiveness of CD8+ NK cells relative to CD8− NK cells. In donors carrying at least 1 HLA-Bw4 allele, KIR3DL1+ (i.e., KIR3DL1+others−) NK cells, but not KIR3DL1− (i.e., KIR−NKG2A−) NK cells, showed CD8-dependent activation by K562 cells (Fig. 4 B and C), indicated by higher responsiveness of CD8+ than CD8− NK cells. In contrast, in donors not carrying any HLA-Bw4 allele, neither KIR3DL1+ nor KIR3DL1− NK cells showed CD8 dependency (Fig. 4 D and E). These findings suggest that CD8 enhances the intrinsic sensitivity of KIR3DL1+ NK cells to target cells, implying that CD8 facilitates KIR3DL1+ NK cell education.
Discussion
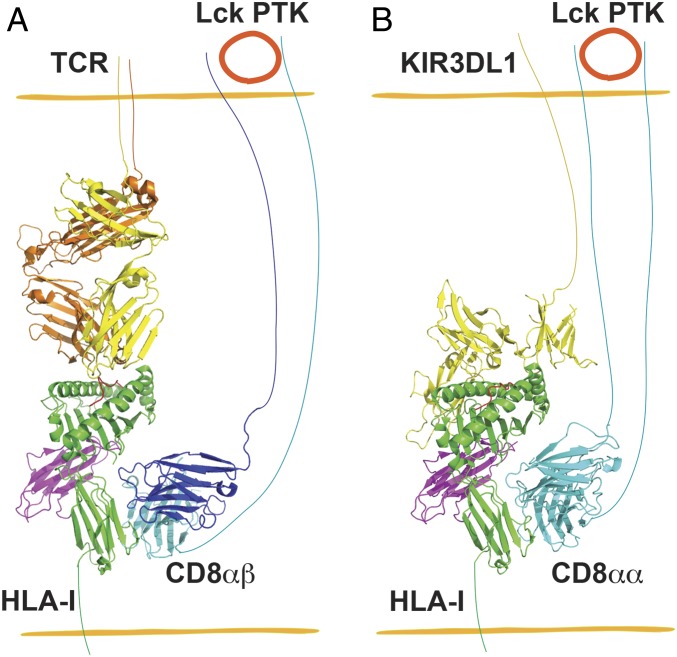
NK cells maintain immune surveillance over virally infected and transformed cells. It was shown that CD8+ NK cells are generally more cytolytic than CD8− NK cells (6). CD8+ NK cells were proposed as potential novel candidates for cancer therapy (26). High frequencies of CD8+ NK cells in chronic HIV-1 infection were also shown to correlate with delayed AIDS progression (27). However, our understanding of molecular mechanisms that regulate CD8+ NK cell function is quite limited. Previous studies showed that cross-linking CD8 on the surface of NK cells either induced (28, 29) or inhibited (6) NK cell apoptosis. In this study, we demonstrated that CD8αα functions as a coreceptor for the NK cell-inhibitory receptor KIR3DL1 and modulates NK cell activity through enhancing interactions between HLA-Bw4 and KIR3DL1. The KIR3DL1 and CD8 binding sites of HLA-I are predicted to be nonoverlapping based on previous structural studies of HLA-B*57:01–KIR3DL1 complexes (15) and MHC-I–CD8 complexes (30) (Fig. 5).
Fig. 5.
Schematic description of the parallels of CD8 coreceptor function in T cells and NK cells. (A) CD8αβ heterodimers are coreceptors for CD8+ T cell binding to target cells displaying the appropriate pHLA-I. The protein tyrosine kinase (PTK) Lck associates with the CD8αβ cytoplasmic tail, triggering enhanced T cell signaling. (B) Similar to A, CD8αα interacts with the nonpolymorphic region of HLA-I and enhances HLA-I engagement to KIR3DL1 of NK cells and the inhibitory function of KIR3DL1. In A, the TCR–HLA-A*02:01 complex structure (Protein Data Bank [PDB] ID code 5c0a) (47) was superimposed onto H2-Dd-CD8αβ (PDB ID code 3dmm) (48) by aligning Cαs of HLA-A*02:01 and H2-Dd, followed by deletion of H2-Dd. Similarly, in B, the KIR3DL1–HLA-B*57:01 complex structure (PDB ID code 3vh8) (15) was superimposed onto HLA-A*02:01-CD8αα complex (PDB ID code 1akj) (30) by aligning Cαs of HLA-B*57:01 and HLA-A*02:01, and HLA-A*02:01 was then deleted. The CD8αα and KIR3DL1 binding sites on HLA-I are predicted to be nonoverlapping and synergistic for enhancing KIR function.
NK cells share many characteristics with cytotoxic T cells, such as IFN-γ release and perforin/granzyme-mediated killing. We showed here the interaction between MHC-I and CD8 modulates NK cell activation via KIR3DL1, which adds another element of similarity between NK cells and CTL. KIR enrichment at the IS is emerging as a key mechanism for the control of NK cell activation by influencing downstream signaling (21). CD8αα strongly elevated KIR3DL1 clustering (Fig. 2), providing direct evidence that CD8αα functions as a coreceptor for KIR3DL1. Engagement of KIR3DL1 transduces negative signals through its cytoplasmic domain. It has been shown that ligation of KIR3DL1 also leads to its phosphorylation by Lck (13, 14). Previous studies have shown that the CD8α chain contains a binding site for Lck within its cytoplasmic domain (31). It is possible that ligation of CD8αα by HLA-I also results in elevated phosphorylation of KIR3DL1 by Lck, resulting in enhanced inhibitory signaling (Figs. 3–5).
To date, 14 functional KIR genes, including inhibitory and activating genes, have been characterized (32). KIR molecules share similar structure and functional mechanisms (33). Therefore, it is reasonable to hypothesize that other KIR inhibitory receptors are also CD8-dependent, especially those whose ligands are strong CD8 binders. KIR activation receptors might also engage CD8 as a coreceptor, which needs further investigation. Previous studies showed that TCRs with high binding affinity to their cognate pMHC-I are less dependent on CD8 (34). By analogy, strong interactions between pMHC-I and KIR pairs may reduce dependency on CD8, whereas CD8 might be more important in enhancing interactions between KIR and weak ligands. Additionally, CD8 binding affinity varies among HLA-I allotypes (35). Thus, we can expect variable influences of HLA-I polymorphisms on CD8 dependence.
Previous studies have revealed a remarkable degree of NK cell diversity (36). Diversity of antigen-specific receptors is a well-documented asset in adaptive immunity, which increases the likelihood of a responsive subpopulation. Although NK cells have conventionally been classified as innate, they also show features of adaptive immune cells (37). The high functional diversity of NK cells allows flexibility in pathogen-specific responses. KIR and HLA-I allelic polymorphism (38–40), KIR gene copy number (41), levels of HLA-I and KIR expression (42) at the cell surface, and binding affinity (42) between HLA-I and KIR are known to calibrate NK education. Our findings here that CD8 fine-tunes NK cell activity add another dimension to NK cell functional modulation.
Methods
Study Approval.
Blood was collected from consenting healthy donors for HLA genotyping and functional studies in accordance with a University of Michigan Institutional Review Board-approved protocol (HUM00071750).
HLA Genotyping and KIR Expression.
HLA genotyping was performed as described previously (43). The presence of KIR3DL1 was assessed by 4% agarose gel electrophoresis following PCR–sequence-specific priming (44) with primers 5′-CCCTGGTGAAATCAGGAGAGAG-3′ and 5′-TGTAGGTCCCTGCAAGGGCAA-3′.
Viruses and Cell Infections.
HLA-B*57:03 in a murine stem cell virus retroviral vector (pMSCVneo) was prepared as described previously (45). The HLA-B*57:03 mutant that is deficient for binding CD8 (HLA-B*57:03-CD8null) was made by introducing D227K/T228A mutations into HLA-B*57:03. Jurkat cells (clone E6-1), a gift from Alice Telesnitsky, University of Michigan, Ann Arbor, MI were infected with retrovirus encoding KIR3DL1 (clone ID 75056; DNASU Plasmid Repository) and then nucleofected with pCI-neo-CD8α, a gift from Lei Lu, Nanyang Technological University, Singapore (46) (Addgene plasmid 86050; Research Resource Identifier: Addgene_86050).
Tetramer Staining.
Peptide–HLA-B tetramers were prepared and verified as described previously (18). After CD8 and KIR3DL1 blocking with anti-CD8 (clone SK1; BioLegend) or anti-KIR3DL1 (clone 177407; R&D Systems), freshly prepared tetramers were added typically at 20 μg/mL, and anti–CD8-AF700 (clone HIT8a; BioLegend), anti–CD3-Pacific Blue (clone UCHT1; BioLegend) and anti–CD56-phycoerythrin-Cy7 (clone CMSSB; eBioscience) were added at concentrations indicated by the manufacturers and incubated for another 30 min at room temperature.
KIR3DL1 Clustering Assay.
The Jurkat cells expressing KIR3DL1 and CD8 were mixed with K562 cells expressing B*57:03 or B*57:03-CD8null, centrifuged briefly, and incubated for 10 min at 37 °C to allow IS formation. Cells were fixed and stained with anti–KIR3L1-allophycocyanin (clone DX9; BioLegend) or together with anti–CD8-AF594 (clone RPA-T8; BioLegend). Cells were then imaged using a Leica SP8 confocal microscope. Data were processed using Leica Imaging software and ImageJ software. The intensity of KIR3DL1 or CD8 at the interface was compared with the membrane at a noncontact area and plotted as the fold increase above background.
NK Cell Activation Assay.
NK cells were gated as the CD3−CD56+ cells within the lymphocyte forward scatter/side scatter gate. Cytolytic potential was assayed based on CD107a expression. Intracellular IFN-γ was detected with anti–IFN-γ–FITC or anti-IFN-γ–AF700. Statistical analysis was performed with paired Student’s t tests.
A detailed description of materials and methods is provided in SI Appendix, Materials and Methods.
Supplementary Material
Acknowledgments
We thank the NIH tetramer core facility staff for providing the LZ-ELBM HLA-B constructs. We also thank all blood donors and the staff at the Michigan Clinical Research Unit. We thank Brogan Yarzabek for donor sample collections and preparations for HLA genotyping. We also thank Dr. Lei Lu (Nanyang Technological University) for the pCI-neo-CD8 plasmid and Dr. David Margulies (National Institute of Allergy and Infectious Diseases, Bethesda, MD) for helpful suggestions. We thank the University of Michigan DNA Sequencing Core for sequencing analyses, and Elizabeth Smith (University of Michigan Hybridoma Core) for antibody production. This work was supported by the National Institute of Allergy and Infectious Diseases of the National Institutes of Health (Grant R01AI044115 to M.R.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1905943116/-/DCSupplemental.
References
- 1.Cole D. K., Gao G. F., CD8: Adhesion molecule, co-receptor and immuno-modulator. Cell. Mol. Immunol. 1, 81–88 (2004). [PubMed] [Google Scholar]
- 2.Purbhoo M. A., et al. , The human CD8 coreceptor effects cytotoxic T cell activation and antigen sensitivity primarily by mediating complete phosphorylation of the T cell receptor ζ chain. J. Biol. Chem. 276, 32786–32792 (2001). [DOI] [PubMed] [Google Scholar]
- 3.Garcia K. C., et al. , CD8 enhances formation of stable T-cell receptor/MHC class I molecule complexes. Nature 384, 577–581 (1996). [DOI] [PubMed] [Google Scholar]
- 4.Huang J., Edwards L. J., Evavold B. D., Zhu C., Kinetics of MHC-CD8 interaction at the T cell membrane. J. Immunol. 179, 7653–7662 (2007). [DOI] [PubMed] [Google Scholar]
- 5.Hayday A., Theodoridis E., Ramsburg E., Shires J., Intraepithelial lymphocytes: Exploring the third way in immunology. Nat. Immunol. 2, 997–1003 (2001). [DOI] [PubMed] [Google Scholar]
- 6.Addison E. G., et al. , Ligation of CD8α on human natural killer cells prevents activation-induced apoptosis and enhances cytolytic activity. Immunology 116, 354–361 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shortman K., Heath W. R., The CD8+ dendritic cell subset. Immunol. Rev. 234, 18–31 (2010). [DOI] [PubMed] [Google Scholar]
- 8.den Haan J. M., Lehar S. M., Bevan M. J., CD8(+) but not CD8(−) dendritic cells cross-prime cytotoxic T cells in vivo. J. Exp. Med. 192, 1685–1696 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Leishman A. J., et al. , T cell responses modulated through interaction between CD8alphaalpha and the nonclassical MHC class I molecule, TL. Science 294, 1936–1939 (2001). [DOI] [PubMed] [Google Scholar]
- 10.Cheroutre H., Lambolez F., Doubting the TCR coreceptor function of CD8alphaalpha. Immunity 28, 149–159 (2008). [DOI] [PubMed] [Google Scholar]
- 11.Vilches C., Parham P., KIR: Diverse, rapidly evolving receptors of innate and adaptive immunity. Annu. Rev. Immunol. 20, 217–251 (2002). [DOI] [PubMed] [Google Scholar]
- 12.Radaev S., Sun P. D., Structure and function of natural killer cell surface receptors. Annu. Rev. Biophys. Biomol. Struct. 32, 93–114 (2003). [DOI] [PubMed] [Google Scholar]
- 13.Binstadt B. A., et al. , Sequential involvement of Lck and SHP-1 with MHC-recognizing receptors on NK cells inhibits FcR-initiated tyrosine kinase activation. Immunity 5, 629–638 (1996). [DOI] [PubMed] [Google Scholar]
- 14.Marti F., et al. , LCK-phosphorylated human killer cell-inhibitory receptors recruit and activate phosphatidylinositol 3-kinase. Proc. Natl. Acad. Sci. U.S.A. 95, 11810–11815 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vivian J. P., et al. , Killer cell immunoglobulin-like receptor 3DL1-mediated recognition of human leukocyte antigen B. Nature 479, 401–405 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Terry L. A., DiSanto J. P., Small T. N., Flomenberg N., Differential expression and regulation of the human CD8 α and CD8 β chains. Tissue Antigens 35, 82–91 (1990). [DOI] [PubMed] [Google Scholar]
- 17.Hennecke S., Cosson P., Role of transmembrane domains in assembly and intracellular transport of the CD8 molecule. J. Biol. Chem. 268, 26607–26612 (1993). [PubMed] [Google Scholar]
- 18.Geng J., Altman J. D., Krishnakumar S., Raghavan M., Empty conformers of HLA-B preferentially bind CD8 and regulate CD8+ T cell function. eLife 7, e36341 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zimbwa P., et al. , Precise identification of a human immunodeficiency virus type 1 antigen processing mutant. J. Virol. 81, 2031–2038 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Davis D. M., et al. , The human natural killer cell immune synapse. Proc. Natl. Acad. Sci. U.S.A. 96, 15062–15067 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Treanor B., et al. , Microclusters of inhibitory killer immunoglobulin-like receptor signaling at natural killer cell immunological synapses. J. Cell Biol. 174, 153–161 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Anfossi N., et al. , Human NK cell education by inhibitory receptors for MHC class I. Immunity 25, 331–342 (2006). [DOI] [PubMed] [Google Scholar]
- 23.Kim S., et al. , Licensing of natural killer cells by host major histocompatibility complex class I molecules. Nature 436, 709–713 (2005). [DOI] [PubMed] [Google Scholar]
- 24.Braud V. M., et al. , HLA-E binds to natural killer cell receptors CD94/NKG2A, B and C. Nature 391, 795–799 (1998). [DOI] [PubMed] [Google Scholar]
- 25.Shiroishi M., et al. , Human inhibitory receptors Ig-like transcript 2 (ILT2) and ILT4 compete with CD8 for MHC class I binding and bind preferentially to HLA-G. Proc. Natl. Acad. Sci. U.S.A. 100, 8856–8861 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Suck G., et al. , Highly cytotoxic CD56+CD8+ NK cells as potential novel candidates for cancer cellular therapy. Clin. Cancer Res. 13 (suppl. 22), B13 (2007). [Google Scholar]
- 27.Ahmad F., et al. , High frequencies of polyfunctional CD8+ NK cells in chronic HIV-1 infection are associated with slower disease progression. J. Virol. 88, 12397–12408 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Contini P., et al. , Soluble HLA-A,-B,-C and -G molecules induce apoptosis in T and NK CD8+ cells and inhibit cytotoxic T cell activity through CD8 ligation. Eur. J. Immunol. 33, 125–134 (2003). [DOI] [PubMed] [Google Scholar]
- 29.Spaggiari G. M., et al. , Soluble HLA class I molecules induce natural killer cell apoptosis through the engagement of CD8: Evidence for a negative regulation exerted by members of the inhibitory receptor superfamily. Blood 99, 1706–1714 (2002). [DOI] [PubMed] [Google Scholar]
- 30.Gao G. F., et al. , Crystal structure of the complex between human CD8α(α) and HLA-A2. Nature 387, 630–634 (1997). [DOI] [PubMed] [Google Scholar]
- 31.Shaw A. S., et al. , Short related sequences in the cytoplasmic domains of CD4 and CD8 mediate binding to the amino-terminal domain of the p56lck tyrosine protein kinase. Mol. Cell. Biol. 10, 1853–1862 (1990). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Marsh S. G., et al. , Killer-cell immunoglobulin-like receptor (KIR) nomenclature report, 2002. Tissue Antigens 62, 79–86 (2003). [DOI] [PubMed] [Google Scholar]
- 33.Campbell K. S., Purdy A. K., Structure/function of human killer cell immunoglobulin-like receptors: Lessons from polymorphisms, evolution, crystal structures and mutations. Immunology 132, 315–325 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Laugel B., et al. , Different T cell receptor affinity thresholds and CD8 coreceptor dependence govern cytotoxic T lymphocyte activation and tetramer binding properties. J. Biol. Chem. 282, 23799–23810 (2007). [DOI] [PubMed] [Google Scholar]
- 35.Gao G. F., et al. , Classical and nonclassical class I major histocompatibility complex molecules exhibit subtle conformational differences that affect binding to CD8alphaalpha. J. Biol. Chem. 275, 15232–15238 (2000). [DOI] [PubMed] [Google Scholar]
- 36.Horowitz A., et al. , Genetic and environmental determinants of human NK cell diversity revealed by mass cytometry. Sci. Transl. Med. 5, 208ra145 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sun J. C., Beilke J. N., Lanier L. L., Adaptive immune features of natural killer cells. Nature 457, 557–561 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Saunders P. M., et al. , Killer cell immunoglobulin-like receptor 3DL1 polymorphism defines distinct hierarchies of HLA class I recognition. J. Exp. Med. 213, 791–807 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Carr W. H., Pando M. J., Parham P., KIR3DL1 polymorphisms that affect NK cell inhibition by HLA-Bw4 ligand. J. Immunol. 175, 5222–5229 (2005). [DOI] [PubMed] [Google Scholar]
- 40.Yawata M., et al. , Roles for HLA and KIR polymorphisms in natural killer cell repertoire selection and modulation of effector function. J. Exp. Med. 203, 633–645 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Béziat V., et al. , Influence of KIR gene copy number on natural killer cell education. Blood 121, 4703–4707 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Boudreau J. E., Mulrooney T. J., Le Luduec J. B., Barker E., Hsu K. C., KIR3DL1 and HLA-B density and binding calibrate NK education and response to HIV. J. Immunol. 196, 3398–3410 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yarzabek B., et al. , Variations in HLA-B cell surface expression, half-life and extracellular antigen receptivity. eLife 7, e34961 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kulkarni S., Martin M. P., Carrington M., KIR genotyping by multiplex PCR-SSP. Methods Mol. Biol. 612, 365–375 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rizvi S. M., et al. , Distinct assembly profiles of HLA-B molecules. J. Immunol. 192, 4967–4976 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Madugula V., Lu L., A ternary complex comprising transportin1, Rab8 and the ciliary targeting signal directs proteins to ciliary membranes. J. Cell Sci. 129, 3922–3934 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cole D. K., et al. , Hotspot autoimmune T cell receptor binding underlies pathogen and insulin peptide cross-reactivity. J. Clin. Invest. 126, 2191–2204 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang R., Natarajan K., Margulies D. H., Structural basis of the CD8 alpha beta/MHC class I interaction: Focused recognition orients CD8 beta to a T cell proximal position. J. Immunol. 183, 2554–2564 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.