Significance
Multidrug efflux pumps are highly promiscuous determinants of antimicrobial resistance in bacterial pathogens. Since efflux pumps evolved long before the widespread use of antimicrobials, drug transport is likely to be a side reaction in many pumps, fortuitously beneficial to bacteria in hospitals. The AceI efflux protein from Acinetobacter baumannii is the prototype for the proteobacterial antimicrobial compound efflux (PACE) family. AceI was only known to transport the synthetic biocide chlorhexidine, which was incongruous with its ancient origin. Here we demonstrate that short-chain diamines are the physiological substrates of AceI and other PACE members, and that transport is energized by an electrochemical gradient of protons. These observations are important, because diamines play vital roles in bacterial physiology and virulence and have significant commercial uses.
Keywords: drug resistance, polyamines, transport, efflux, PACE family
Abstract
Acinetobacter baumannii has rapidly emerged as a major cause of gram-negative hospital infections worldwide. A. baumannii encodes for the transport protein AceI, which confers resistance to chlorhexidine, a widely used antiseptic. AceI is also the prototype for the recently discovered proteobacterial antimicrobial compound efflux (PACE) family of transport proteins that confer resistance to a range of antibiotics and antiseptics in many gram-negative bacteria, including pathogens. The gene encoding AceI is conserved in the core genome of A. baumannii, suggesting that it has an important primordial function. This is incongruous with the sole characterized substrate of AceI, chlorhexidine, an entirely synthetic biocide produced only during the last century. Here we investigated a potential primordial function of AceI and other members of the PACE family in the transport of naturally occurring polyamines. Polyamines are abundant in living cells, where they have physiologically important functions and play multifaceted roles in bacterial infection. Gene expression studies revealed that the aceI gene is induced in A. baumannii by the short-chain diamines cadaverine and putrescine. Membrane transport experiments conducted in whole cells of A. baumannii and Escherichia coli and also in proteoliposomes showed that AceI mediates the efflux of these short-chain diamines when energized by an electrochemical gradient. Assays conducted using 8 additional diverse PACE family proteins identified 3 that also catalyze cadaverine transport. Taken together, these results demonstrate that short-chain diamines are common substrates for the PACE family of transport proteins, adding to their broad significance as a novel family of efflux pumps.
Multidrug efflux pumps are encoded in all bacterial genomes sequenced to date (1, 2). These proteins are most widely studied for their functions in drug resistance; however, multidrug efflux pumps participate in many additional processes, such as cell adherence, invasion, biofilm formation, virulence, natural product secretion, and resistance to host encoded factors (3). This diverse array of functions reflects the promiscuous substrate recognition profiles of multidrug efflux pumps, which can extend well beyond antibiotics and biocides. Indeed, for many multidrug efflux pumps, the natural physiological substrates are likely to be endogenously produced small molecules, and the recognition of antimicrobial compounds is a fortuitous consequence of flexible substrate recognition pockets that can accommodate a diverse array of chemical structures.
A major goal of this study was to elucidate the physiological function of the AceI efflux pump, the prototype for the most recently discovered family of multidrug efflux pumps, and also of its homologs known as the proteobacterial antimicrobial compound efflux (PACE) family (4, 5). AceI is encoded by a gene that is highly conserved in the core genome of Acinetobacter baumannii. Prior to this study, the sole recognized substrate for the AceI efflux pump was the biocide chlorhexidine. A bisbiguanide compound consisting of terminal proguanil groups separated by a 1,6-diaminohexane moiety, chlorhexidine is of massive importance in pathogen control. It is listed as an “essential medicine” by the World Health Organization and is used globally in a broad range of antiseptic and disinfectant preparations. Despite its current importance, chlorhexidine was first synthesized only in the last century and so would not have been present in the environment across the evolution of the AceI pump and could not have imposed selective pressure for aceI gene maintenance during this time.
Some naturally occurring polyamines share structural similarities with chlorhexidine, since their molecules are composed of aliphatic carbon chains with interspersed and/or terminal amino groups that are typically charged at physiological pH (6). Therefore, polyamines were viewed as candidates for potential physiological substrates of AceI and other PACE pumps. Polyamines such as cadaverine, putrescine, spermidine, and spermine are common in prokaryotic and eukaryotic cells, where they may exist at high concentrations (millimolar) and have varied vital functions in protein and nucleic acid stability, metabolism, nitrogen storage, acid tolerance, cell-to-cell communication, motility, transcriptional regulation, and protein expression (7, 8). These biologically abundant polyamines also function in host immune responses and bacterial virulence (8).
A number of different transport systems that promote the uptake of polyamines into bacteria have been characterized (9). These uptake systems may help support the high cellular requirements for polyamines and facilitate catabolism of exogenous polyamines (9). Despite their high abundance and broad physiological significance, polyamines can inhibit cell growth when they are in excess, requiring cells to have detoxification mechanisms. Efflux pumps that transport polyamines out of the cell have also been identified (10, 11). Active polyamine efflux systems could help maintain cellular concentrations of polyamines at subtoxic levels and also may be required to export polyamines involved in cellular communication or bacterial motility. Bacterial multidrug efflux pumps from the small multidrug resistance (SMR; e.g., MdtIJ in Escherichia coli) family and the major facilitator superfamily (MFS; e.g., Blt from Bacillus subtilis) have previously been shown to mediate polyamine efflux (10, 11). In this study, we examined polyamines as potential physiological substrates of the prototypical PACE family member AceI from Acinetobacter baumannii (4) and some of its homologs.
Active efflux pumps need to capture the energy required to move their primary substrate against its concentration gradient out of the cell. In previously characterized families of drug efflux proteins, the energy is provided by a secondary substrate such as ATP, which provides chemical energy during hydrolysis to ADP, or a transmembrane electrochemical gradient of monovalent cations (typically H+ or Na+, and occasionally K+) that are exchanged for primary substrates in antiport reactions (12). AceI and other PACE family proteins do not include nucleotide-binding domains that are typically associated with ATP-driven pumps and are not encoded in proximity to, or coordinately regulated with, genes encoding these domains in bacterial genomes. Therefore, it was likely that PACE proteins energized the transport of their primary substrates using an electrochemical gradient, which we now show is the proton-motive force. In addition, we determine the specificities of induction and transport for polyamines using both intact cells and liposomes containing isolated AceI.
Results and Discussion
Expression of the aceI Efflux Pump Gene in A. baumannii Is Induced by Polyamines.
We used qRT-PCR analyses to determine whether the addition of exogenous polyamines would elicit a transcriptional response in aceI gene expression in A. baumannii. The cells were treated with 4 polyamines, including the short-chain diamines cadaverine and putrescine, the triamine spermidine, and the tetraamine spermine. These polyamines were chosen for analysis since they represent the major groups of polyamines found in most organisms (8). We found that both of the diamines caused strong induction of aceI expression, approximately 20-fold higher than that in untreated cells (Fig. 1A). In contrast, aceI expression was only moderately induced by spermidine (5.2-fold induction) and weakly induced by spermine (2.8-fold induction). The initial discovery of the AceI pump was based on up-regulation of aceI by chlorhexidine (4). In those investigations, chlorhexidine caused an approximately 10-fold increase in aceI expression in A. baumannii ATCC 17978 (4).
Fig. 1.
(A) Induction of aceI gene expression by polyamines. Polyamines were added to the A. baumannii AB5075-UW medium when the cells were in midexponential growth phase (SI Appendix, Methods). The bars represent the change in aceI gene expression compared with an untreated control after 30 min of growth in the presence of the polyamines. Error bars show the SDs of at least 2 biological and 4 technical replicates. (B) Accumulation of [14C]-cadaverine into A. baumannii wild-type (AB5075-UW) or an aceI inactivated mutant (ΔaceI) (13). The cells were exposed to a low concentration of cadaverine for 30 min to allow aceI expression, then washed and incubated with 50 μM [14C]-cadaverine using 1% succinate provided as an energy source. Each sample included 100 μL of cells at OD600 = 1.0 (SI Appendix, Methods). (C) Accumulation of [14C]-cadaverine into E. coli BL21 cells overexpressing the wild-type AceI protein (AceI), the inactive E15Q AceI mutant protein (E15Q), or no additional protein (negative). In these E. coli cells, expression of aceI or its E15Q variant was induced by IPTG, and the harvested, washed cells were incubated with 50 μM [14C]-cadaverine with 1% glucose provided as an energy source. Each sample included 100 μL of cells at OD600 = 1.0 (SI Appendix, Methods). Error bars show the SD of at least three independent replicate experiments.
aceI Is Required for A. baumannii to Tolerate Exogenous Diamines.
The induction of aceI by the addition of cadaverine or putrescine, and to a lesser extent by spermidine and spermine, suggested that these compounds may be substrates of the AceI efflux pump. Since these polyamines have some level of toxicity toward many species of bacteria, we tested this possibility by examining the requirement of the aceI gene for polyamine tolerance in A. baumannii. Minimum inhibitory concentration analyses were conducted to test tolerance to putrescine, cadaverine, spermidine, and spermine in wild-type A. baumannii AB5075-UW and in an isogenic aceI-inactivated mutant strain (13). The parental strain tolerated high concentrations of all 4 polyamines; the minimum inhibitory concentrations were 40 μg/mL for putrescine, cadaverine, and spermidine and 10 μg/mL for spermine (SI Appendix, Fig. S1). Inactivation of aceI in AB5075-UW resulted in at least 8-fold reductions in tolerance to cadaverine and putrescine but no change in tolerance to spermidine or spermine (SI Appendix, Fig. S1). This suggests that AceI may recognize diamines as substrates, but that it does not recognize triamines or tetraamines or does so only weakly. The minimum inhibitory concentration of chlorhexidine for A. baumannii is more than 1,000-fold lower than that of cadaverine or putrescine, but inactivation of aceI in A. baumannii causes only a 2-fold reduction in chlorhexidine tolerance (14). The high fold reductions in tolerance after aceI inactivation observed for cadaverine and putrescine suggest that efflux of short-chain diamines may be a primary role of AceI.
Expression of the AceI Protein in A. baumannii or E. coli Reduces the Accumulation of Cadaverine.
To test whether the reduced tolerance to diamines in the aceI mutant A. baumannii strain is related directly to the efflux of these compounds, we examined the accumulation of [14C]-labeled cadaverine into mutant cells compared with the parental strain. The parental AB5075-UW strain accumulated minimal amounts of [14C]-cadaverine when added to the medium (Fig. 1B). In contrast, the aceI inactivated mutant readily accumulated [14C]-cadaverine, suggesting that normal levels of AceI induced by cadaverine in A. baumannii prevent high accumulation of the diamine into the cells, consistent with cadaverine efflux activity.
Laboratory E. coli strains are excellent hosts for studies examining the function of PACE family proteins, because they do not carry genes encoding endogenous PACE family proteins that could interfere with the activity of heterologously expressed proteins. Indeed, PACE family genes have been found in <0.2% of sequenced E. coli genomes and in no laboratory strains (4, 15). To confirm that AceI isolated from A. baumannii does mediate the efflux of short-chain diamines, we examined the level of [14C]-cadaverine accumulation in E. coli cells expressing AceI compared with cells expressing an inactive AceI mutant harboring an E15Q substitution (AceI-E15Q) and control cells carrying the empty vector expression plasmid (Fig. 1C). Importantly, immunoblot analyses demonstrated that both AceI and AceI-E15Q were expressed at similar levels in the induced cells (SI Appendix, Fig. S2). [14C]-Cadaverine was readily accumulated in both the empty vector control cells and the cells expressing the AceI-E15Q mutant protein. In contrast, the concentration of [14C]-cadaverine in E. coli cells expressing the parental AceI protein did not significantly increase over the course of the experiment (Fig. 1C), suggesting that AceI can transport cadaverine out of the cell at a rate equal to or higher than its rate of accumulation. The AceI-E15Q protein was previously shown to be incapable of mediating chlorhexidine resistance or transport (4). Since cells expressing this mutant accumulated [14C]-cadaverine to a similar level as the negative control (Fig. 1C), it appears that an acidic residue at position 15 is necessary for all transport activity of AceI.
AceI Protein Promotes the Transport of [14C]-Cadaverine in a Reconstituted System.
Previous attempts to obtain measurements of AceI-mediated transport of chlorhexidine in a reconstituted system were not successful (4). At that time, chlorhexidine was the sole recognized substrate for the protein, and chlorhexidine is very poorly suited for experiments using proteoliposomes. Specifically, chlorhexidine is poorly soluble, adsorbs nonspecifically to most filter membranes and to biological membranes, and is a membrane-active biocide, so even low concentrations tend to disrupt naked proteoliposomes. The identification of cadaverine and putrescine as potential substrates of the AceI transport system in this study presented a new opportunity to examine the activities of AceI in a reconstituted membrane bilayer system.
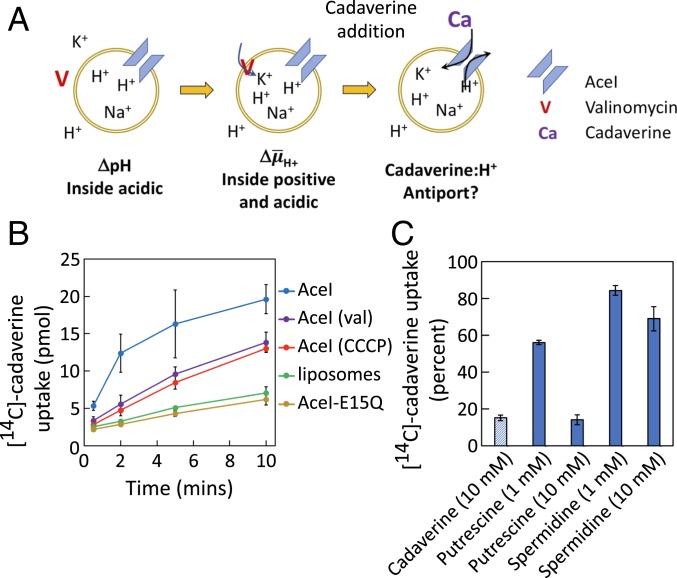
Both the wild-type AceI protein and the inactive AceI-E15Q mutant protein were purified and reconstituted into preformed liposomes composed of E. coli polar lipids (SI Appendix, Methods). For comparison, empty liposomes were also generated using the same approach. An experimental system was established to generate an electrochemical gradient across the (proteo)liposome membranes, consisting of both a chemical proton gradient (ΔpH; inside acidic) and an electrical potential (Δψ; inside positive) (Fig. 2A). The lumen of the (proteo)liposomes contained Na+ at pH 7.0. The (proteo)liposomes were diluted into a buffer containing isosmolar K+ and a low concentration of the potassium ionophore valinomycin at pH 8.0. The difference in pH of the buffers generated ΔpH, and the valinomycin promoted the downhill movement of K+ into the (proteo)liposomes, generating Δψ (Fig. 2A) (16, 17). The polarity of the pH and charge differential (inside positive and acidic) across the membrane could energize the uptake of externally applied substrates in exchange for a cation, such as a proton, by any active antiport system (16)—in this case AceI (Fig. 2A).
Fig. 2.
Uptake of [14C]-cadaverineinto (proteo)liposomes. (A) Schematic representation of the strategy used for generating a proton electrochemical gradient (ΔH+) across the proteoliposome membrane. (Proteo)liposomes were formed in Na+-containing buffer at pH 7.0, then diluted into K+ buffer at pH 8.0 containing the K+ ionophore valinomycin. (B) Liposomes and proteoliposomes containing AceI or AceI-E15Q were formed (SI Appendix, Methods) and treated as shown in A. The results show [14C]-cadaverine uptake per fraction. Uptake of [14C]-cadaverine was faster in energized AceI proteoliposomes (blue line) compared with empty liposomes (green line) or proteoliposomes containing the inactive AceI-E15Q protein (tan line). [14C]-cadaverine uptake into AceI proteoliposomes without valinomycin or with the addition of CCCP occurred at an intermediate rate (purple and red lines, respectively). (C) Inhibition of AceI-mediated [14C]-cadaverine transport by alternative AceI substrates. Transport experiments were performed as for the blue trace in B, but in the presence of 1 or 10 mM unlabeled cadaverine, putrescine, or spermidine. The values shown are the amount of [14C]-cadaverine accumulated after 10 min as a percentage of the amount accumulated with no inhibitor. Error bars show the SDs of at least 3 independent replicate experiments.
There was some uptake of cadaverine into liposomes without incorporated protein (Fig. 2B), which was probably the result of uncharged cadaverine diffusing across the membrane and accumulating in the lumen by protonation. The proteoliposomes containing wild-type AceI and energized by an electrical and pH gradient positive and acidic inside accumulated [14C]-cadaverine (Fig. 2B) much more rapidly. After subtraction of the small amount of accumulation in liposomes, the initial rate of cadaverine uptake between the first and second assay time points (30 and 120 s) was 16.7 ± 5.9 nmol/mg protein/min (SI Appendix, Fig. S3). In contrast, the rate of [14C]-cadaverine uptake into proteoliposomes containing the inactive AceI-E15Q mutant was indistinguishable from the rate of uptake into liposomes without any incorporated protein, calculated as −0.2 ± 0.7 nmol/mg protein/min between the first and second time points (Fig. 2B and SI Appendix, Fig. S3). AceI proteoliposomes that were not treated with valinomycin, and thus had ΔpH (inside acidic) but no Δψ, accumulated [14C]-cadaverine at a much lower rate of 3.9 ± 3.7 nmol/mg protein/min between the first and second assay time points (Fig. 2B and SI Appendix, Fig. S3 and Table S1). AceI proteoliposomes were also diluted into the same buffer without valinomycin, but with the protonophore CCCP that discharges both electrical and pH gradients. In this case, the proteoliposomes still took up [14C]-cadaverine, but at a much slower rate of 3.1 ± 2.0 nmol/mg protein/min between the first and second assay time points (Fig. 2B and SI Appendix, Fig. S3 and Table S1), an approximate 80% reduction in activity from the energized rate.
Based on these results and those reported below, energized AceI-mediated cadaverine transport appears to be driven by proton exchange. The intermediate levels of [14C]-cadaverine accumulation seen in the absence of valinomycin or in the presence of CCCP, which were above those seen in the liposome-negative control, may be driven by the unenergized downhill movement of [14C]-cadaverine into the proteoliposomes facilitated by AceI, i.e., a relatively low level of “uniport” (facilitated diffusion) activity, which might even reflect a variable stoichiometry of cadaverine:H+, as is seen in other secondary transporters (18). Also, low-level transport may be driven by an alternative coupling ion, such as Na+, which is present in the lumen of the proteoliposomes.
Overall, the rapid nonlinear accumulation of [14C]-cadaverine into the reconstituted proteoliposomes fully energized by both Δψ and ΔpH, markedly above the levels seen in the control and uncoupled experiments, demonstrates that AceI can promote the active transport of cadaverine in exchange for a cation.
[14C]-Cadaverine Uptake into AceI Proteoliposomes Is Inhibited by Putrescine.
With a reconstituted AceI transport assay system in place for [14C]-cadaverine, it was possible to assess whether AceI can recognize alternative substrates by examining the competitive inhibition of [14C]-cadaverine transport. Since putrescine and spermidine induced measurable increases in expression of the aceI gene (Fig. 1A), we tested whether unlabeled putrescine or spermidine reduced [14C]-cadaverine uptake into AceI proteoliposomes at 50-fold (1 mM) and 500-fold (10 mM) molar excess over [14C]-cadaverine. Putrescine caused significant inhibition of [14C]-cadaverine uptake into AceI proteoliposomes, with 43.9% inhibition of uptake by 1 mM putrescine and 85.9% inhibition by 10 mM putrescine, equivalent to the addition of excess unlabeled cadaverine (Fig. 2C). In contrast, spermidine caused only a marginal level of inhibition of [14C]-cadaverine uptake; 1 mM spermidine caused 15.7% inhibition, and 10 mM spermidine caused 31.0% inhibition (Fig. 2C). These results suggest that putrescine can be transported by AceI at an equivalent rate to cadaverine, but that spermidine is poorly recognized by AceI, consistent with the results of the polyamine tolerance experiments (SI Appendix, Fig. S1), in which inactivation of aceI in A. baumannii caused a reduction in tolerance to both putrescine and cadaverine, but not to spermidine. Therefore, of the polyamines tested, AceI appears to recognize specifically the short-chain diamines cadaverine and putrescine as substrates.
AceI-Mediated Cadaverine and Putrescine Transport into Proteoliposomes Occurs in Parallel with Internal pH Changes.
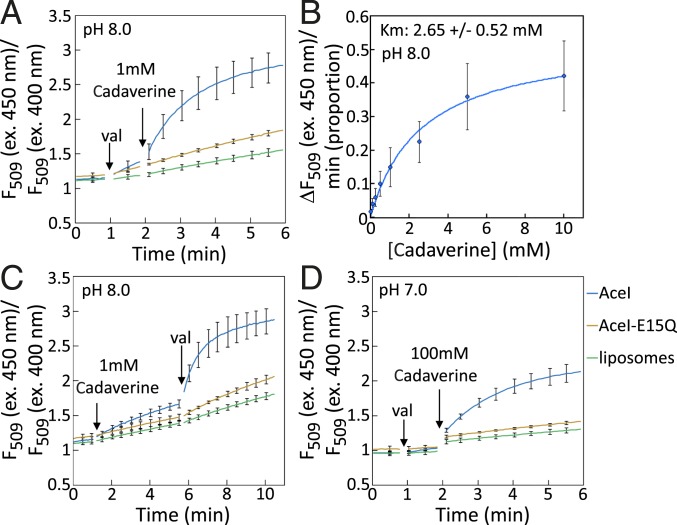
The foregoing results provided good evidence supporting AceI as a secondary active transport protein for cadaverine and putrescine; that is, the reconstituted transport experiments showed that generation of an inside positive electrical potential significantly increased the rate of uptake of externally applied [14C]-cadaverine into AceI proteoliposomes (Fig. 2). This suggested that AceI may catalyze a diamine:cation antiport reaction. To help confirm whether the cation was a proton, we examined pH changes inside the lumen of AceI proteoliposomes during transport of diamine achieved by using a membrane-impermeable pH-sensitive fluorescent dye, 8-hydroxypyrene-1,3,6-trisulfonic acid (pyranine; SI Appendix, Methods).
Experimental conditions were established to generate a proton electrochemical gradient (inside acidic and positive) as described above using [14C]-cadaverine for assaying AceI transport activity in the reconstituted system. Proteoliposomes containing AceI or the AceI-E15Q mutant and empty control liposomes were formed in Na+ buffer at pH 7.0. To establish a pH gradient in the assay, the (proteo)liposomes were diluted into buffer at higher pH (pH 8.0). An electrical gradient, Δψ, was generated by including isosmolar K+ in the external assay buffer and subsequently adding 5 nM valinomycin to catalyze the downhill movement of K+ into the lumen of the proteoliposomes (Fig. 3A). The addition of valinomycin caused a slow increase in the F509 (ex. 450 nm)/F509 (ex. 400 nm) fluorescence ratio of the AceI proteoliposomes and empty liposomes, consistent with a gradual increase in the luminal pH, likely due to the leakage of protons down their electrical gradient out of the (proteo)liposomes (Fig. 3A). The addition of 1 mM cadaverine to the energized AceI proteoliposomes resulted in a far more rapid alkalization of the proteoliposome lumen, marked by a rapid increase in the F509 (ex. 450 nm)/F509 (ex. 400 nm) fluorescence ratio of pyranine (Fig. 3A). In contrast, there was no significant change in the internal pH in empty liposomes or proteoliposomes containing AceI-E15Q after the addition of cadaverine (Fig. 3A). This result suggested that AceI catalyzes the exchange of protons for cadaverine, that is, the luminal pH increases as protons are exported in exchange for cadaverine.
Fig. 3.
Cadaverine-induced pH changes inside proteoliposomes containing AceI. Proteoliposomes were formed in buffer containing the pH-sensitive fluorescent dye pyranine (SI Appendix, Methods). The excitation maximum of pyranine shifted from 400 nm to 450 nm with increasing pH, whereas the emission maximum was stable at ∼509 nm. A, C, and D show the 450:400 nm fluorescence (509 nm) excitation ratios of pyranine in the lumen as follows: control liposomes without added protein (green), proteoliposomes containing the inactive AceI-E15Q mutant protein (tan), and proteoliposomes containing wild-type AceI (blue). An increase in this ratio is indicative of an increase in internal pH. All (proteo)liposomes were formed in Na+ buffer at pH 7.0 (SI Appendix, Methods). (A) (Proteo)liposomes were diluted into K+ buffer at pH 8.0, 5 nM valinomycin (val) was added at the first arrow, and 1 mM cadaverine was added at the second arrow. (B) AceI proteoliposomes were diluted into K+ buffer at pH 8.0 containing 5 nM valinomycin (val) and the indicated concentrations of [14C]-cadaverine. The initial rates of fluorescence change are plotted relative to the concentration of cadaverine, corrected for the small level of fluorescence change observed in liposomes containing no protein. From a Michaelis–Menten curve fitted to the data points, an apparent Km of 2.65 ± 0.52 mM was derived. (C) (Proteo)liposomes were diluted into K+ buffer at pH 8.0, 1 mM cadaverine was added at the first arrow, and 5 nM valinomycin was added at the second arrow. (D) (Proteo)liposomes were diluted into K+ buffer at pH 7.0, 5 nM valinomycin was added at the first arrow, and 100 mM cadaverine was added at the second arrow. The experiments were performed using at least 3 independent batches of (proteo)liposomes, and error bars show SDs.
The level of fluorescence change induced by different concentrations of cadaverine (0 to 10 mM) in this assay system was examined to determine whether the rate of pH change could be saturated. The results indicated that the rate of fluorescence change was saturable (Fig. 3B), consistent with the operation of a protein-catalyzed active transport activity specific for cadaverine. A Michaelis–Menten curve fit suggested an apparent Km value of 2.65 ± 0.52 mM (Fig. 3B).
We next determined the effect of excluding valinomycin from the assay until after the addition of cadaverine to determine whether ΔpH alone could promote AceI-mediated cadaverine transport. Proteoliposomes containing AceI or the AceI-E15Q mutant and empty control liposomes were formed in buffer containing Na+ at pH 7.0, then diluted into isosmolar K+ buffer at pH 8.0. Valinomycin was initially omitted, and 1 mM cadaverine was added; this induced a pH change in the lumen of AceI proteoliposomes that was above the background level seen in the liposome control or in proteoliposomes containing AceI-E15Q (Fig. 3C). However, the fluorescence change was well below that seen when cadaverine was added to AceI proteoliposomes after the generation of an electrical potential by valinomycin (Fig. 3A). The subsequent addition of valinomycin to the AceI proteoliposomes, which generated an inside positive electrical potential, again resulted in a very rapid increase in internal pH, well above that seen in the liposome control or in proteoliposomes containing AceI-E15Q (Fig. 3C). Similar to the reconstituted transport experiments using [14C]-cadaverine, these results suggest that an electrical gradient, Δψ, can promote rapid AceI-mediated cadaverine transport.
To examine the effect of Δψ in the absence of ΔpH on AceI activity in this system, proteoliposomes containing AceI or the AceI-E15Q mutant and empty control liposomes were formed in buffer containing Na+ at pH 7.0 and then diluted into isosmolar K+ buffer at pH 7.0. Valinomycin was added to generate an electrical potential, followed by cadaverine. In this system, 1 mM cadaverine did not induce a significant change in pH in the interior of AceI proteoliposomes or in control liposomes that could be reliably detected using the pyranine indicator. However, increasing the cadaverine concentration to 100 mM resulted in a large pH change inside the AceI proteoliposomes that was similar in amplitude to that seen after the addition of 1 mM cadaverine at pH 8.0 with valinomycin energization (Fig. 3 A and D). No significant change in pH was observed in the control liposomes or AceI-E15Q proteoliposomes after the addition of 100 mM cadaverine under these conditions, showing that the effect was mediated by AceI (Fig. 3D) and not by the change in osmolarity across the liposome membrane.
We also examined the effects of adding putrescine and spermidine to AceI proteoliposomes in this experimental system. The addition of 100 mM putrescine induced a significant pH change in AceI proteoliposomes, but the addition of spermidine did not (SI Appendix, Fig. S4). However, we did observe some precipitation of spermidine or buffer components on its addition. These results are consistent with our observations that expression of aceI promoted putrescine tolerance, but not spermidine tolerance, in A. baumannii (SI Appendix, Fig. S1), and that putrescine, but not spermidine, was able to significantly inhibit AceI-mediated [14C]-cadaverine transport into proteoliposomes (Fig. 2C). These results provide additional support for the idea that AceI can transport putrescine and cadaverine well, but spermidine poorly at best.
The finding that a higher concentration of cadaverine was needed to induce a strong pH change in the Δψ assay system at pH 7.0 may be related to the chemical properties of cadaverine and the nature of electrochemically driven transport reactions. Cadaverine has 2 amines, with pKa values of 10.25 and 9.13 (6). Consequently, the majority of the cadaverine added at neutral pH will be protonated and thus charged at both amines. Transport reactions driven by an electrical gradient alone must be electrogenic themselves (17). Therefore, if AceI mediates the exchange of cadaverine for protons driven by the electrical potential, the number of protons exchanged must increase with the charge state of cadaverine, that is, 1 or more protons exchanged for the neutral form of cadaverine, 2 or more protons exchanged for a singly charged form, and 3 or more protons exchanged for cadaverine charged at both amines. This may favor the transport of neutrally charged cadaverine. Since cadaverine has two ionizable groups with high pKa values, at pH 7.0 the concentration of neutral compound would be ∼100-fold lower than that at pH 8.0, since for each amine group there will be an ∼10-fold increase in the nonprotonated form for each 1 pH unit increase. This may be why a 100-fold increase in cadaverine concentration was required at pH 7.0 (Fig. 3D) to promote a pH change similar to that seen at pH 8.0 (Fig. 3A). If the transport of fully neutral or monocationic forms of cadaverine is favored by AceI under these experimental conditions, the alkalization of the proteoliposome lumen may be at least partly related to the proton-accepting potential of the deprotonated amines; that is, protons may be accepted by the cadaverine accumulating within the proteoliposome, thereby increasing the internal pH. Therefore, it is not possible to conclude with complete certainty that protons, rather than alternative cations such as Na+ ions, are exchanged for substrates by AceI.
pH Changes Induced in AceI Proteoliposomes by Chlorhexidine.
Chlorhexidine was the first substrate identified for AceI and is one of the most important biocides used in healthcare worldwide. As stated above, chlorhexidine is a membrane-active biocide, and our previous attempts to examine AceI-mediated transport in a reconstituted system were unsuccessful, due to nonspecific adsorption to filters and biological membranes and the potential lysis of proteoliposomes by chlorhexidine. The nature of the assays using pyranine-containing proteoliposomes alleviate the problems of adsorption to filters and biological membranes, since they do not require the proteoliposomes to be filtered or the total substrate associated with the proteoliposomes to be measured. Therefore, considering our success in developing this assay system to demonstrate AceI-mediated transport of cadaverine and putrescine, we attempted assays using chlorhexidine. An experimental system was used to generate a pH gradient; (proteo)liposomes were formed in K+ buffer at pH 7.0 and diluted into K+ buffer at pH 8.0. The addition of 100 μM chlorhexidine to AceI proteoliposomes caused alkalization of the AceI proteoliposome lumen, observed as an increase in pyranine fluorescence (450 nm excitation/509 nm emission; SI Appendix, Fig. S5). The addition of the same concentration of chlorhexidine to empty control liposomes caused a lower level of alkalization. Note that the addition of an equivalent amount of DMSO, the solvent for chlorhexidine, to AceI proteoliposomes did not cause a change in the internal pH (SI Appendix, Fig. S5). These observations are consistent with AceI-mediated chlorhexidine:H+ exchange but must be viewed with caution, since the chlorhexidine may have damaged the naked (proteo)liposomes.
Expression of Several Other PACE Family Proteins in E. coli Reduces Cadaverine Accumulation.
The foregoing results focused only on the AceI transport protein as the prototypical representative of the PACE family. To examine whether other members of the PACE family can catalyze the transport of cadaverine, we examined the level of [14C]-cadaverine accumulation in E. coli cells expressing 8 additional PACE family proteins: Fbal_3166 from Ferrimonas balearica, PFL_4558 from Pseudomonas protegens, 655492601 (GenBank protein ID) from Tepidiphilus margaritifer, A1S_1503 from A. baumannii, Mlut_15630 from Micrococcus luteus, PSPTO_3587 from Pseudomonas syringae pv. tomato, STY3166 from Salmonella enterica subsp. enterica serovar Typhi, and VP1155 from Vibrio parahaemolyticus (SI Appendix, Fig. S6). Each of these proteins was produced at readily detectable levels in E. coli BL21 cells on isopropyl β-d-1-thiogalactopyranoside (IPTG) induction (SI Appendix, Fig. S7). Of these additional proteins examined, 3 prevented the accumulation of [14C]-cadaverine into E. coli, similar to AceI (SI Appendix, Fig. S6). These results indicate that cadaverine, and possibly other short-chain diamines, are the likely physiological substrates of many PACE family proteins.
Conclusions
The PACE family is the most recently discovered family of multidrug efflux proteins (5). The genes encoding PACE proteins are conserved in a range of opportunistic gram-negative pathogens and may contribute to serious outbreaks of drug-resistant infections in hospitals. Owing to the recency of their identification, their physiological substrates and mechanisms of transport energization were unknown. In this study, we have made several major advances in understanding the function of the prototypical PACE family protein, AceI from A. baumannii. First, polyamines, specifically the short-chain primary diamines cadaverine and putrescine, are strong inducers of aceI in A. baumannii (Fig. 1A). Second, cadaverine and putrescine are substrates of AceI, whereas the longer polyamine spermidine is only a weak substrate of AceI, even though it induces aceI expression, albeit at a lower level than the equivalent concentrations of cadaverine and putrescine (Fig. 1A). Third, transport of cadaverine is effected by AceI protein reconstituted into liposomes. Finally, the transmembrane electrical gradient of protons is the primary source of energy for AceI-mediated cadaverine transport, and the pH gradient may make a contribution as well.
Polyamines are produced in all cells and have a raft of functions in cellular regulation, maintaining stability of nucleic acids and proteins, motility, and cell-to-cell signaling (3). Furthermore, polyamines have multifaceted roles in bacterial virulence and in host immune responses (8). Many of these effects are likely mediated by transport proteins, such as AceI, that catalyze the efflux or uptake of polyamines. Indeed, activating or deactivating the expression of polyamine transport proteins in various bacteria has been found to attenuate virulence (8, 19). Therefore, compounds that modulate polyamine transport systems may have potential uses as virulence-attenuating drugs. The design of such drugs will now be facilitated by the knowledge that the physiological substrates are short-chain polyamines. In addition to functions in human health, diamines, including cadaverine and putrescine examined in this study, have a range of industrial uses, especially as precursors of polymers related to nylon (20). The discovery that AceI is a secondary transport system for these compounds adds the potential for developing new biological platforms for their large-scale biotechnological production based around AceI-mediated efflux, which would provide much needed “green” alternatives to the petroleum-based precursors currently produced (20).
Methods
Detailed descriptions of all experimental procedures are provided in SI Appendix, Methods. In brief, for qRT-PCR analyses of aceI gene expression, we followed methods described previously (4, 21). For polyamine tolerance tests, we used strains from the Manoil laboratory collection (13) and a broth microdilution method (22). [14C]-cadaverine dihydrochloride was obtained from American Radiolabeled Chemicals. Whole-cell [14C]-cadaverine accumulation assays were performed following routine methods (4). The reconstitution method used for AceI and AceI-E15Q proteins was developed from the method described by Aires and Nikaido (23). The approaches for generating proton electrochemical gradients across the proteoliposome membranes are described in Results and Discussion, and in detail in SI Appendix, Methods.
Supplementary Material
Acknowledgments
We thank Professor Lars Jeuken for helpful discussions about the measurement of pyranine fluorescence in reconstituted transport experiments. P.J.F.H. thanks John H. Henderson, Katherine F. Henderson, Mary E. Henderson, and Helen F. Long for their dedicated support. This project was supported by Project Grants from the National Health and Medical Research Council of Australia (GNT1060895 and GNT1120298, to I.T.P., K.A.H., and P.J.F.H.), an Australian Research Council Future Fellowship (FT180100123, to K.A.H.), a Marie Skłodowska-Curie Research Fellowship from the European Commission (706499, to K.A.H. and P.J.F.H.), and an Emeritus Fellowship from the Leverhulme Trust (EM-2014-045, to P.J.F.H.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1901591116/-/DCSupplemental.
References
- 1.Ren Q., Paulsen I. T., Large-scale comparative genomic analyses of cytoplasmic membrane transport systems in prokaryotes. J. Mol. Microbiol. Biotechnol. 12, 165–179 (2007). [DOI] [PubMed] [Google Scholar]
- 2.Elbourne L. D., Tetu S. G., Hassan K. A., Paulsen I. T., TransportDB 2.0: A database for exploring membrane transporters in sequenced genomes from all domains of life. Nucleic Acids Res. 45, D320–D324 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Piddock L. J., Multidrug-resistance efflux pumps—Not just for resistance. Nat. Rev. Microbiol. 4, 629–636 (2006). [DOI] [PubMed] [Google Scholar]
- 4.Hassan K. A., et al. , Transcriptomic and biochemical analyses identify a family of chlorhexidine efflux proteins. Proc. Natl. Acad. Sci. U.S.A. 110, 20254–20259 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hassan K. A., Liu Q., Henderson P. J. F., Paulsen I. T., Homologs of the Acinetobacter baumannii AceI transporter represent a new family of bacterial multidrug efflux systems. MBio 6, e01982-14 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Blagbrough I. S., Metwally A. A., Geall A. J., Measurement of polyamine pKa values. Methods Mol. Biol. 720, 493–503 (2011). [DOI] [PubMed] [Google Scholar]
- 7.Igarashi K., Kashiwagi K., Modulation of cellular function by polyamines. Int. J. Biochem. Cell Biol. 42, 39–51 (2010). [DOI] [PubMed] [Google Scholar]
- 8.Shah P., Swiatlo E., A multifaceted role for polyamines in bacterial pathogens. Mol. Microbiol. 68, 4–16 (2008). [DOI] [PubMed] [Google Scholar]
- 9.Igarashi K., Kashiwagi K., Polyamine transport in Escherichia coli. Amino Acids 10, 83–97 (1996). [DOI] [PubMed] [Google Scholar]
- 10.Higashi K., et al. , Identification of a spermidine excretion protein complex (MdtJI) in Escherichia coli. J. Bacteriol. 190, 872–878 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Woolridge D. P., et al. , Efflux of the natural polyamine spermidine facilitated by the Bacillus subtilis multidrug transporter Blt. J. Biol. Chem. 272, 8864–8866 (1997). [DOI] [PubMed] [Google Scholar]
- 12.Paulsen I. T., Brown M. H., Skurray R. A., Proton-dependent multidrug efflux systems. Microbiol. Rev. 60, 575–608 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gallagher L. A., et al. , Resources for genetic and genomic analysis of emerging pathogen Acinetobacter baumannii. J. Bacteriol. 197, 2027–2035 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tucker A. T., et al. , Defining gene-phenotype relationships in Acinetobacter baumannii through one-step chromosomal gene inactivation. MBio 5, e01313-14 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hassan K. A., et al. , An ace up their sleeve: A transcriptomic approach exposes the AceI efflux protein of Acinetobacter baumannii and reveals the drug efflux potential hidden in many microbial pathogens. Front. Microbiol. 6, 333 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Henderson P. J. F., McGivan J. D., Chappell J. B., The action of certain antibiotics on mitochondrial, erythrocyte and artificial phospholipid membranes. The role of induced proton permeability. Biochem. J. 111, 521–535 (1969). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Henderson P. J. F., Ion transport by energy-conserving biological membranes. Annu. Rev. Microbiol. 25, 393–428 (1971). [DOI] [PubMed] [Google Scholar]
- 18.Bazzone A., Zabadne A. J., Salisowski A., Madej M. G., Fendler K., A loose relationship: Incomplete H+/sugar coupling in the MFS sugar transporter GlcP. Biophys. J. 113, 2736–2749 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rai A. N., et al. , Polyamine transporter in Streptococcus pneumoniae is essential for evading early innate immune responses in pneumococcal pneumonia. Sci. Rep. 6, 26964 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ma W. C., et al. , Advances in cadaverine bacterial production and its Applications. Engineering 3, 308–317 (2017). [Google Scholar]
- 21.Brzoska A. J., Hassan K. A., “Quantitative PCR for detection of mRNA and gDNA in environmental isolates”, Methods in Molecular Biology Environmental Microbiology, Paulsen I. T., Holmes A., Eds. (Humana Press, ed. 2, 2014), pp. 25–42. [DOI] [PubMed] [Google Scholar]
- 22.Wiegand I., Hilpert K., Hancock R. E., Agar and broth dilution methods to determine the minimal inhibitory concentration (MIC) of antimicrobial substances. Nat. Protoc. 3, 163–175 (2008). [DOI] [PubMed] [Google Scholar]
- 23.Aires J. R., Nikaido H., Aminoglycosides are captured from both periplasm and cytoplasm by the AcrD multidrug efflux transporter of Escherichia coli. J. Bacteriol. 187, 1923–1929 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.