Significance
Tight junctions (TJs) direct cell adhesion and paracellular transport of solutes in vertebrate epithelia. Claudins are integral components of TJs, and disruptions to their assembly cause various diseases. Clostridium perfringens disassembles TJs between gut epithelium by secreting an enterotoxin (CpE) that binds claudins via its C terminus (cCpE). Here, we report crystal structures of human claudin-9 (hCLDN-9) in complex with cCpE, the cytotoxic effects of CpE on hCLDN-9–expressing cells, and binding kinetics and affinities of hCLDN-9/enterotoxin interactions. Together, structural and functional data provide molecular bases for mutants involved in ion selectivity, deafness, hepatitis C, and CpE binding. Further, they suggest mechanisms for CpE-driven disruption of claudin assemblies, which leads to TJ dissociation and breakdown of the gastrointestinal barrier.
Keywords: claudin, tight junction, membrane protein, X-ray structure, toxin
Abstract
The human pathogenic bacterium Clostridium perfringens secretes an enterotoxin (CpE) that targets claudins through its C-terminal receptor-binding domain (cCpE). Isoform-specific binding by CpE causes dissociation of claudins and tight junctions (TJs), resulting in cytotoxicity and breakdown of the gut epithelial barrier. Here, we present crystal structures of human claudin-9 (hCLDN-9) in complex with cCpE at 3.2 and 3.3 Å. We show that hCLDN-9 is a high-affinity CpE receptor and that hCLDN-9–expressing cells undergo cell death when treated with CpE but not cCpE, which lacks its cytotoxic domain. Structures reveal cCpE-induced alterations to 2 epitopes known to enable claudin self-assembly and expose high-affinity interactions between hCLDN-9 and cCpE that explain isoform-specific recognition. These findings elucidate the molecular bases for hCLDN-9 selective ion permeability and binding by CpE, and provide mechanisms for how CpE disrupts gut homeostasis by dissociating claudins and TJs to affect epithelial adhesion and intercellular transport.
Tight junctions (TJs) are macromolecular assemblies that govern cell–cell adhesion and paracellular transport of solutes through endo- and epithelial sheets (Fig. 1A). The molecular composition of TJs is finely tuned in tissues to control movements of select compounds, imparting unique functions to individual epithelial clusters or layers. Claudins are tetra-spanning membrane proteins with 27 human isoforms and are the major structural components of TJs, forming permselective pores in paracellular space (1, 2). Epithelia achieve their vast functionalities by regulating the number and types of claudins that constitute their TJs (3). Claudins have convergent structural topologies that include an intracellular N terminus, 4 α-helical transmembrane (TM) segments, 2 extracellular segments (ECSs), and an intracellular C terminus; the latter is tethered to the cytoskeleton via accessory proteins (4). How claudins simultaneously direct epithelial cell adhesion and paracellular transport is driven by polymerization of the TM and ECS, which interact side-by-side within the same cell membrane (cis) and/or with claudins from adjacent cell membranes across intercellular space (trans) (Fig. 1A). Disruption to claudin organization leads to TJ dissociation and is a hallmark of tissue-specific diseases (5).
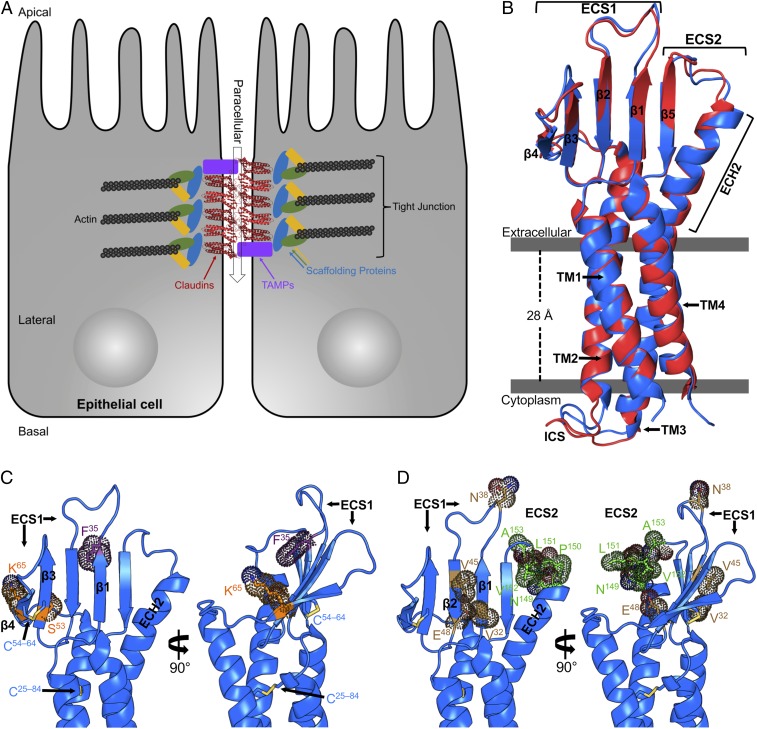
Fig. 1.
Structures of TJs and hCLDN-9. (A) Schematic diagram of epithelial TJs with claudins (red), TJ-associated MARVEL proteins (TAMPs; purple), scaffolding proteins (various), and actin (black). Paracellular transport is shown as a transparent arrow (black). (B) hCLDN-9 closed (blue) and open (red) in complex with cCpE, with cCpE omitted for clarity, viewed parallel to the membrane. Bars (gray) are approximate membrane boundaries. ICS, intracellular segment. (C) hCLDN-9 closed sites of ion selectivity (orange), the F35 site of deafness mutations (purple), and disulfide bonds (yellow) shown as sticks with dot surfaces. (D) hCLDN-9 closed HCV entry sites (brown) and the NPLVA153 motif (green) that binds CpE are shown as in C.
Claudin-9 is the highest expressing claudin in the inner ear and is essential for hearing (6, 7). In epithelial cells of the endolymphatic system, claudin-9 acts as a cation barrier and reduces Na+ and K+ permeability (7). In a mouse model for deafness, claudin-9 mutations disrupt subapical TJs, resulting in elevated K+ levels that flood ear sensory hair cells with cations, leading to cell death (7). Human claudin-9 (hCLDN-9), along with hCLDN-1 and hCLDN-6, is an entry coreceptor for hepatitis C virus (HCV) entry (8–10). In liver hepatocytes, HCV entry is facilitated by envelope protein recognition of ECS1, which contains the conserved claudin-defining W30-GLW51-C54-C64 motif (11, 12). HCV recognizes a patch of residues on ECS1 of hCLDN-9, and mutations of these amino acids impair viral entry (10).
hCLDN-9 overexpression decreases cation permeability in kidney cells, hinting at a cation barrier function (13). hCLDN-9 is not preferentially expressed in any single tissue. Based on RNA sequencing, the parathyroid gland, bone marrow, spleen, and fallopian tube contain a greater number of transcripts than the pancreas, duodenum, and small intestine (14, 15). Tissue-specific messenger RNA expression levels show mostly ubiquitous expression of hCLDN-9, apart from high amounts found in heart and skeletal muscle (16, 17). It was therefore surprising when Winkler et al. (18) reported that a 20-mer peptide from mouse CLDN-9 (mCLDN-9) could bind a toxin from Clostridium perfringens, a known gut pathogen. C. perfringens enterotoxin (CpE) is released upon ingestion, causing gastrointestinal maladies due to CpE targeting and binding receptor claudins, resulting in disordering of TJs and eventual epithelial cell death by Ca+ influx (19). Notwithstanding its involvement in these key biological processes, hCLDN-9 is a poorly described claudin family member.
To better understand the molecular bases for hCLDN-9 cation barrier function and HCV and CpE toxin recognition, we determined crystal structures of full-length wild-type hCLDN-9 in complex with the C-terminal receptor-binding domain of CpE (cCpE). Previous crystal structures of mutated and C-terminally truncated mCLDN-19 and hCLDN-4 in complex with cCpE show nonphysiological antiparallel multimeric arrangements in the crystallographic asymmetrical unit, which could influence claudin/cCpE interactions and biophysical interpretation (20, 21). For both structures presented, a single hCLDN-9/cCpE complex exists, providing an unperturbed view of the intermolecular interactions that cCpE utilizes to bind hCLDN-9. Through structure–function analyses, we identify hCLDN-9 as a high-affinity receptor for CpE and show that CpE is cytotoxic to hCLDN-9–expressing cells. Taken together, these findings reveal molecular bases for hCLDN-9 permselectivity and CpE binding, and provide a model for the mechanisms by which CpE dissociates TJs, leading to breakdown of the gut epithelial barrier.
Results and Discussion
Structures of hCLDN-9.
hCLDN-9 was expressed in and isolated from insect cells as detailed in SI Appendix, Supplementary Methods. We discovered after incubation of hCLDN-9 with cCpE using size-exclusion chromatography (SEC) that they formed a complex in vitro. Two closely related crystal forms of hCLDN-9 bound to cCpE were obtained under the same conditions. Two single-crystal datasets of each form were determined to ∼3.2 Å in space group P212121, with cell dimensions of 70.9, 111.6, and 121.7 Å (closed form) and 71.0, 114.8, and 115.6 Å (open form). The structures were determined by molecular replacement (MR) using the C terminus of CpE (22) and a homology model of hCLDN-9, each differing in conformation of cCpE. Data collection and refinement statistics are summarized in SI Appendix, Table S1.
Readily interpretable maps were obtained post-MR and allowed placement of >82% of hCLDN-9 and >91% of cCpE residues in both complexes. For each, the 2 Fo − Fc electron density is continuous, allowing interpretation of both hCLDN-9 and cCpE peptide chains without any structural breaks. Asymmetrical units for both structures contain 1 hCLDN-9/cCpE complex, with 2 crystal contacts made by hCLDN-9/cCpE and 1 via a cCpE/cCpE interaction. This crystal packing containing a single hCLDN-9/cCpE complex is unique compared with other structures of claudins bound to cCpE and allows both molecules to interact without additional steric constraints (SI Appendix, Fig. S1).
The overall structures of hCLDN-9 reveal a left-handed 4-TM helix bundle that forms from over half of the primary sequence, with TM3 maintaining helical structure through the membrane into extracellular space (Fig. 1B). The ECS of hCLDN-9 contains a β-sheet composed of 5 antiparallel strands, with 4 residing in ECS1 and 1 in ECS2 (Fig. 1B). This creates a stable epitope where both ECSs are tethered on a similar plane. ECS1 is further fortified by a conserved Cys54–Cys64 disulfide bond that covalently links β3 to β4. An extracellular helix (ECH) between β4 and TM2, ECH1, is not present in either structure of hCLDN-9. The ECH extended from TM3, which we term ECH2, splays outward compared with the TM helical portion, with Ile143 and Asp146 forming interactions with β5. Buried into the 4-helix bundle core of hCLDN-9 is a Cys25–Cys84 disulfide linking TM1 to TM2 at the apex of the extracellular leaflet (Fig. 1C). ECS1 and ECS2 form an intricately associated epitope resembling a left hand: with ECH2 as the “thumb,” followed by β5, β1, β2, and disulfide linked β3–β4 as the “little” finger (Fig. 1B). While the global folds of hCLDN-9 are similar to other claudins (SI Appendix, Fig. S2), the improved resolution allows visualization of all nonterminal residues resulting in a structure of a complete claudin.
hCLDN-9 Ion Selectivity.
hCLDN-9 is thought to function as a cation barrier and anion pore (7, 13), although residues that direct ion selectivity have not been defined. To help identify hCLDN-9 ion selectivity, we compared its sequence and structure with the well-defined anion barrier hCLDN-2 (23) and the cation barrier-forming hCLDN-4 (24) (SI Appendix, Fig. S3). Asp65 in hCLDN-2 (25) and Lys65 in hCLDN-4 (26) have the greatest influence on pore ion selectivity. To understand the structural basis for this, we created a homology model of hCLDN-2 and compared it with crystal structures of hCLDN-9 and hCLDN-4 (21). Sequence and structure alignments show hCLDN-9 Lys65 resides in close proximity to these defining residues (SI Appendix, Fig. S3). The structures reveal that Lys65 is the penultimate residue of β4, lying below Cys64 and its disulfide bond (Fig. 1C). Superposition of hCLDN-2 with hCLDN-9 and hCLDN-4 shows that residues 31, 48, 53, and 65 all lie on the same plane in β1–β4 of ECS1 and could create a single-sided half-pore with each side chain facing the claudin palm (SI Appendix, Fig. S3B). In these claudins, residue 48 has the opposite charge of residue 65. Our sequence and structural analyses between claudins with known ion selectivity and hCLDN-9 show how Lys65 could form a cation barrier and how anions could selectively permeate through its pore (SI Appendix, Fig. S3B). Because residue 65 has most influence for hCLDN-2 and hCLDN-4, we propose Lys65 governs hCLDN-9 anion selectivity, as this residue sits on top of the pore and lies on a pore-forming plane. hCLDN-9 anion selectivity imparts unique function to ear (7), eye (27), kidney (13), and liver (10) epithelia. Understanding the molecular basis of its selectivity is required for development of therapeutics that treat tissue-specific diseases.
hCLDN-9–Based Deafness.
In a mouse model of deafness, a F35L missense mutation in hCLDN-9 compromises barrier function, making inner ear epithelia leakier to cations (7). Structural analysis shows that Phe35 is a terminal residue of β1, forming backbone hydrogen bonds with β2 and hydrophobic side-chain interactions with residues in β1, β2, and β5 (Fig. 1C). In the “hand” analogy, Phe35 is interior, facing the palm. Modeling Leu35 on the structure of hCLDN-9 results in the loss of 5 to 6 hydrophobic interactions with neighboring residues, potentially explaining this mutant’s reduced cation barrier function. As the F35L mutant alters ion selectivity but lies outside the permselective pore (Fig. 1C and SI Appendix, Fig. S3), Phe35 may play a role in structural maintenance of the pore or in facilitating trans interactions. F35L could disrupt interactions with surrounding residues due to nonpolar contact losses, altering the aperture of the pore. Alternately, since Phe35 is adjacent to residues 156 and 158, which are known to direct trans interactions (28), loss of an aromatic could disrupt trans assembly and barrier function. Our hCLDN-9 structures illustrate how a mutant that causes deafness may affect inner ear epithelial barrier function.
Isoform-Specific Disulfide Bond.
hCLDN-9 contains a Cys54–Cys64 disulfide that is conserved across claudins, as well as an additional disulfide (Fig. 1C). Cys25–Cys84 joins TM1–TM2 at the extracellular interface where phospholipids head groups would lie. Claudin sequence alignments reveal that Cys25 is present in TM1 of hCLDN-3 through hCLDN-6 and hCLDN-9 (SI Appendix, Fig. S4). However, only hCLDN-9 and hCLDN-6 have a pairing Cys84 required for disulfide formation. hCLDN-9 and hCLDN-6 share 71% sequence identity, are highly expressed in the neonate proximal tubule (29), are entry cofactors for HCV (10), and bind CpE (30), so there is functional overlap between these paralogs. This isoform-specific disulfide has an as yet unassigned function and may impart these and other specific functionalities to hCLDN-9 and hCLDN-6.
hCLDN-9 and HCV Entry.
HCV replicates in liver and within peripheral blood mononuclear cells (31). hCLDN-9 is present in both and acts as a coreceptor for HCV entry (10), along with hCLDN-1 and hCLDN-6 (9). Val32, Asn38, Val45, and Glu48 in hCLDN-9 are known to mediate HCV entry, with Val45, and then Val32, having greatest influence (10). The structures reveal these 4 residues reside on 3 distinct regions of the hCLDN-9 “hand”: Val32 on β1 and Val45 on β2 lie 4 Å apart on the “knuckle” side, Asn38 lies on the loop connecting β1 to β2, and Glu48 on β2 faces the palm (Fig. 1D). Because scattering of important HCV entry side chains appeared counterintuitive, we performed sequence analysis with other claudins to explain hCLDN-9 Val32- and Val45-mediated HCV entry. Analysis reveals high conservation (52%) of a short hydrophobic side chain, particularly a Val, at position 32 (SI Appendix, Fig. S4). However, only hCLDN-1, hCLDN-6, hCLDN-9, and hCLDN-20 have short hydrophobic side chains at both positions 32 and 45. We suggest that these 2 side chains distinguish HCV-receptor claudins from nonreceptors, as Val32 and Val45 are spatially close and HCV requires both residues for entry (10). hCLDN-1 Ile32 is vital for HCV1 entry (11), but Ala45 was not tested to verify our hypothesis. Our analyses show that claudin/virus interactions essential for HCV entry are focused on a single nonconserved epitope and how this epitope may be used by HCV for isoform-selective entry.
Structural Analysis of cCpE Binding to hCLDN-9.
CpE binding to claudins in intestinal epithelia initiates a cascade of events that dissociate TJs, subsequently causing cell death by intracellular influx of Ca2+ through a cation-selective pore in claudin-bound CpE oligomers, leading to eventual breakdown of the gut TJ barrier (32). CpE is a 35-kDa toxin with an N-terminal domain that functions in oligomerization and cytotoxicity, and a C-terminal domain (cCpE) that binds receptor claudins (22, 33). cCpE binds a subset of claudins, including a peptide of CLDN-9 (18, 32). An ECS2 motif with sequence NP(V/L)(V/L)(P/A153) is required for claudins to interact with CpE (18, 34) (Fig. 1D). In hCLDN-9 structures, cCpE penetrates measurably deeper into the palm of the closed form relative to the open form due to a rigid body motion away from the palm, but interacts with the NPLVA153 motif in both.
Structures of cCpE bound to hCLDN-9 superimpose onto unbound cCpE (35) with rmsds between Cα atoms of 0.8 Å (closed) and 0.9 Å (open), and onto unbound CpE (22) with rmsds of 0.4 Å (closed) and 0.6 Å (open) (SI Appendix, Fig. S5A). cCpE bound to hCLDN-9 more closely resembles unbound CpE rather than unbound cCpE, and demonstrates that cCpE undergoes minimal conformational change upon binding claudins (SI Appendix, Fig. S5B). However, the toxin appears to “rock” in a rigid body-like manner within the hand of claudin structures (SI Appendix, Fig. S6A), as the ECS conformationally adapts to the rocking movements (SI Appendix, Fig. S6B). The physiological significance of these movements is hard to ascertain as there are no distinguishing in vivo data. The crystallographically trapped hCLDN-9 closed cCpE conformer resembles cCpE from mCLDN-19, while the open conformer appears distinct.
The overall folds of hCLDN-9 in open and closed complex with cCpE are similar (Fig. 2), but intermolecular interactions between hCLDN-9 and cCpE differ. The cCpE has polar interactions (distances 2.4 to 4.0 Å) with both ECSs in the hCLDN-9 closed and open complexes. In the closed complex, 7 residues in ECS2 make 11 hydrogen bonds with cCpE, 9 mediated by the main chain, and there is a salt bridge linking hCLDN-9 Asp146 to cCpE Arg227 (SI Appendix, Fig. S7A); in ECS1, there are 8 polar interactions that occur along the β1–β4 strand (SI Appendix, Fig. S7 B and C). In the open complex, 6 ECS2 residues make 8 hydrogen bonds with cCpE, 7 with the main chain, and 2 salt bridges form between Asp146 and Arg227 and between Glu154 and Lys257 of hCLDN-9 and cCpE (SI Appendix, Fig. S8A); in ECS1, 7 polar interactions occur on the loops connecting β-strands at the finger tips (SI Appendix, Fig. S8 B and C). In both closed and open complexes, nonpolar interactions [distances 3.0 to 4.0 Å and Protein Data Bank Protein Interfaces, Surfaces and Assemblies (PDBePISA) buried surface areas >40 Å2 (36)] are largely focused at the NPLVA153 motif on ECS2 (SI Appendix, Figs. S7A and S8A), but Phe35 and Leu156 contribute too (SI Appendix, Figs. S7C and S8C). The hCLDN-9 closed and open structures show that both ECSs support cCpE binding, although there are fundamental differences between complexes.
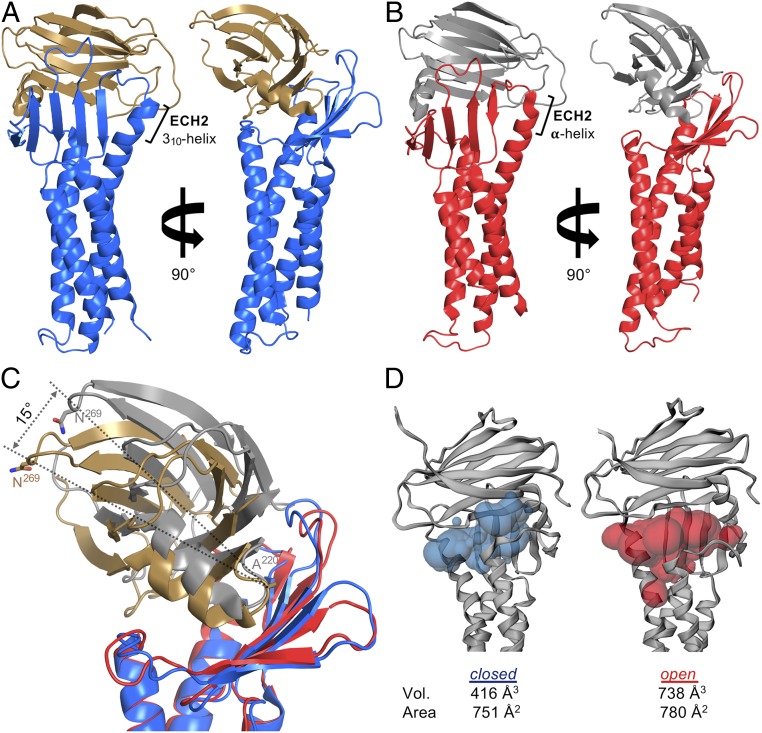
Fig. 2.
hCLDN-9 in closed and open complex with cCpE reveals different conformations of toxin. (A) hCLDN-9 (blue) and cCpE (copper) closed form. (B) hCLDN-9 (red) and cCpE (gray) open form. (C) Superposition of hCLDN-9 closed and open structures colored as in A and B. The cCpE undergoes a 15° movement from closed to open. (D) Solvent-accessible surface volumes (Vol.) and areas between hCLDN-9 and cCpE in the closed and open forms calculated by CASTp (46). Cavity imprints are shown as translucent blobs for hCLDN-9 closed (blue) and open (red).
Clear conformational movements of cCpE are observed between hCLDN-9 closed and open complexes (Fig. 2). All hCLDN-9 palm surface residues interact with cCpE in the closed complex, with Lys65, Gln75, and Gln145 forming hydrogen bonds with cCpE (Fig. 2A and SI Appendix, Fig. S7B). In the open complex, these hydrogen bonds are lost, resulting in a rigid body movement of cCpE out of the palm (Fig. 2B and SI Appendix, Fig. S8B). The cCpE undergoes a 15° rocking movement from the closed to open complex (Fig. 2C), creating a 77% larger solvent-accessible surface volume and 4% increase in solvent-accessible surface area (Fig. 2D). Comparative analysis using cavity-detecting algorithms reveals that an 11% decrease in hCLDN-9/cCpE interface area in hCLDN-9 open creates an ∼36% larger interface volume compared to the hCLDN-9 closed form complex (SI Appendix, Fig. S9). Creation of this solvent-accessible cavity results in claudin finger side chains compensating for a loss of interactions at the palm base (SI Appendix, Fig. S8B). When compared with other CLDN/cCpE structures, both cCpE-bound hCLDN-9 crystal conformers exhibit side-chain conformational heterogeneity at 2 epitopes known to enable claudin self-assembly.
CpE Is Cytotoxic to Claudin-Expressing Sf9 Cells.
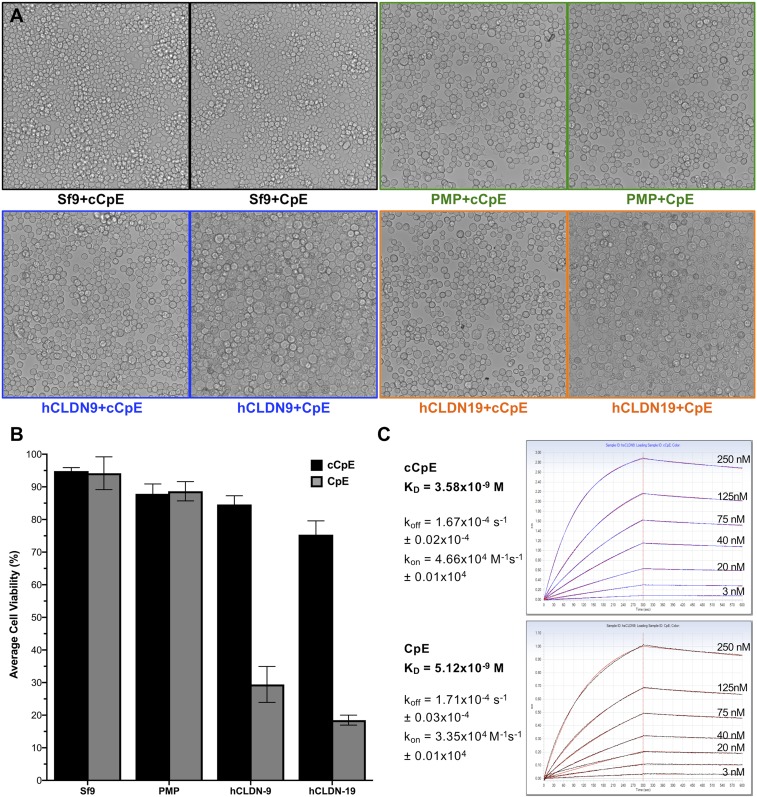
CpE is cytotoxic, and cCpE causes claudin delocalization from TJs (33, 37, 38). We previously determined that cCpE binds hCLDN-9 in vitro, so we sought to characterize cellular consequences after CpE treatment. Using baculovirus-mediated transduction of Sf9 cells that express hCLDN-9 and hCLDN-19, we show that CpE-treated cells form large swollen cells resembling toxin effects on naturally sensitive cells (Fig. 3A). As controls, transduced cells expressing nonclaudin protein (PMP) and nontransduced cells exhibit no CpE sensitivity, indicating that CpE effects are claudin-specific. Further, cells treated with cCpE, which lacks a cytotoxic-essential domain, exhibited no damage, showing that morphological changes require the N-terminal domain of CpE. We then measured cell viability of the toxin-treated Sf9 cells. CpE-treated cells expressing hCLDN-9 and hCLDN-19 exhibited 59.2% and 70.2% decreases in viability, respectively, compared with those not expressing claudins, due to cell death (Fig. 3B). Likewise, CpE-treated cells expressing hCLDN-9 and hCLDN-19 showed viability declines of 55.2% and 56.9%, respectively, compared with cells treated with cCpE, due to the presence of a cytotoxic domain. No measurable effects on viability of nontransduced or PMP-expressing cells were observed after incubation with either CpE or cCpE (Fig. 3B).
Fig. 3.
Cytotoxicity and binding affinity of toxins to hCLDN-9. (A) Morphology of baculovirus-transduced Sf9 cells treated with purified cCpE or CpE and expressing hCLDN-9 (blue), hCLDN-19 (orange), or nonclaudin protein (PMP, green). Sf9 cells with no baculovirus (control, black) were treated identically. (B) Average viability with SD of Sf9 cells from A. (C) cCpE (blue trace) and CpE (black trace) binding to hCLDN-9 shown with overlaid fits (red). koff, dissociation rate constant; kon, association rate constant.
Our cell-based cytotoxicity assay results support a model where the presence of a single TJ-forming claudin receptor is sufficient for CpE cytotoxicity because Sf9 cells lack TJs and endogenous claudins (39), but form TJ-like strands when transduced with claudin baculoviruses (40). Since a “small complex” between hCLDN-9 and CpE in 1:1 stoichiometry alone is insufficient for cytotoxicity, these proteins must interact and form a cytotoxic-inducing pore on Sf9 membranes (41). Superpositioning CpE onto cCpE in hCLDN-9 structures (SI Appendix, Fig. S10A) shows a conformational change in CpE must occur to position its pore-forming helix (residues 93 to 105) perpendicular to the membrane (SI Appendix, Fig. S10B).
Affinity of the Claudin/Enterotoxin Interaction.
As the affinity of enterotoxin to full-length hCLDN-9 was undetermined, we characterized it and its kinetics. Robust association and dissociation curves using biolayer interferometry (BLI) with immobilized toxins gave equilibrium dissociation constants (KDs) of 5.1 nM for CpE and 3.6 nM for cCpE, due, in large part, to very slow dissociation rate constants (koff) (Fig. 3C). CpE affinity for hCLDN-9 is as high as for the well-characterized toxin receptors hCLDN-3 and hCLDN-4. Shinoda et al. (42) reported a KD of 3.4 nM for cCpE binding to hCLDN-4 using surface plasmon resonance. Hence, hCLDN-9 is a high-affinity CpE receptor.
Previous indirect toxin-binding assays have yielded affinities from 8 to 800 nM for CLDN-3 and CLDN-4 and cannot explain why hCLDN-3–expressing cells display lower CpE-binding capacity compared with hCLDN-4 (20, 43), obscuring subtle affinity variations between receptor and nonreceptor claudins. While a structural basis for this remains unknown, sequence comparisons between hCLDN-9, hCLDN 4, and hCLDN-3 reveal 4 amino acid changes that may explain lower CpE affinity for hCLDN-3. Whereas high-affinity receptors hCLDN-9 and hCLDN-4 contain sequence V41Q145L151A153 in ECS1 and ECS2, hCLDN-3 contains I41R145V151P153 (SI Appendix, Fig. S11). We postulate changes L151V and A153P from hCLDN-4/9 to hCLDN-3 in ECS2 decrease CpE affinity for hCLDN-3 based on numerous interactions with cCpE (SI Appendix, Figs. S7A and S8A) in 2 polar surface cavities (34) (SI Appendix, Fig. S12). Analyzing the structural and chemical properties of hCLDN-9/cCpE interfaces in both complexes using PDBePISA shows Leu151 has the greatest sole influence on this association. In mCLDN-19, S151L increased the affinity for cCpE 20-fold, indicating that the hydrophobic length of the side chain is key for high-affinity binding (20). These structure–function analyses verify that the NPLVA153 motif distinguishes high- versus low-affinity CpE receptors and explains hCLDN-3 reduced binding ability.
Mechanism for CpE-Induced Dissociation of Claudin/TJ Assembly.
Whether CpE binding occurs before a claudin reaches TJs to prevent assembly or CpE binds high-affinity receptors within intact TJs and breaks cis and/or trans interactions has not been established. It is also undetermined whether direct disruption of cis interactions by CpE occurs or is required for TJ dissociation. Yet, structures of cCpE-bound claudins led to the inference that TJ disassembly is driven by cCpE destabilization of claudin cis and trans interactions upon binding (20, 21). Our and other cCpE-bound structures clearly show how toxin would sterically hinder claudin trans interactions in paracellular space by interacting with both ECSs (Fig. 2 and SI Appendix, Fig. S10). Authors of cCpE-complexed mCLDN-19 and hCLDN-4 argued that toxin could further disrupt cis interactions by disordering ECH1 and TM3 (20, 21) based on homology models made from unbound mCLDN-15 crystallized in lipidic mesophase, which stated a cis interaction occurred in crystallo (40). Their argument is that cCpE binding would alter Met68, Phe146, and Phe147 (Leu70, Phe147, and Tyr148 in hCLDN-9), disrupting contacts between neighboring mCLDN-15 protomers and enabling TJ disassembly. However, recent evidence by Zhao et al. (44) shows that mCLDN-15 Phe146 and Phe147, as well as Ser67, Arg79, and Glu157 (Ser69, Arg81, and Glu159 in hCLDN-9), are the interface residues that form the cis interaction epitope. This result demonstrates that the mechanism explaining how claudins and TJs are dissociated by CpE remains unclear.
To better understand cCpE-induced effects on claudin structure in the context of that new finding, we superimposed unbound mCLDN-15 and cCpE-bound hCLDN-9, hCLDN-4, and mCLDN-19. The side chains Arg81 on TM2 and Glu159 on β5 show minimal movements, while Ser69 on ECH1 and ECH2 aromatic positions varies widely (SI Appendix, Fig. S13). ECH1 in mCLDN-15 becomes unwound upon cCpE binding (SI Appendix, Fig. S13A). Concomitantly, cCpE interactions with the NPLVA153 motif alter the C terminus of ECH2 (SI Appendix, Fig. S13B), forming a 310-helix in hCLDN-9 closed (Fig. 2A). Crucial perturbations to claudin side chains and secondary structure in ECH1 and ECH2 occur together upon cCpE binding. This finding would explain why CpE binds both ECS and how it could disrupt trans and cis assembly.
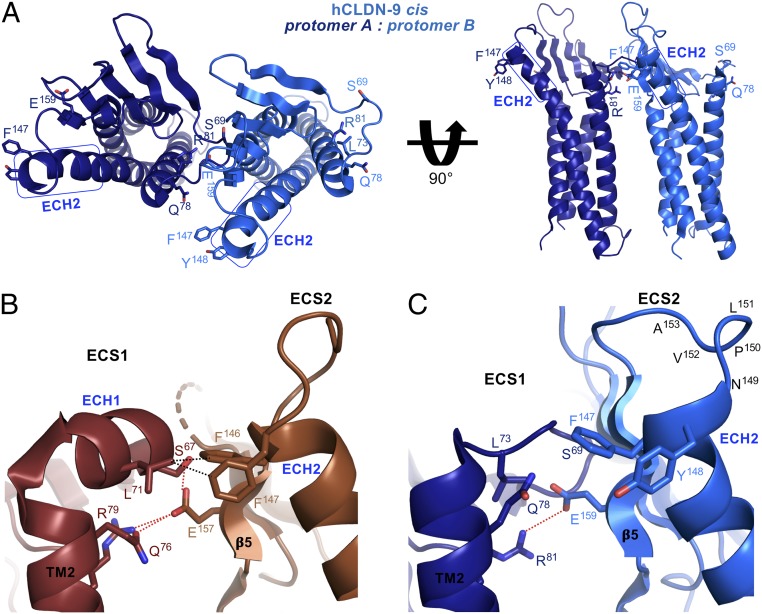
To clarify how claudin cis assembly may be disabled or broken by cCpE, we used our monomeric structures of cCpE-bound hCLDN-9 and manually docked them to each other as cis interacting homodimers (Fig. 4A and SI Appendix, Fig. S14) based on the model of unbound mCLDN-15 from Zhao et al. (44) (SI Appendix, Fig. S15A). The mCLDN-15 cis interaction epitope shows 5 bonds between protomers, polar between Glu157 and Ser67, Gln76, and Arg79, and nonpolar between Phe146/147 and Leu71 (Fig. 4B and SI Appendix, Fig. S15B). In hCLDN-9 closed bound to cCpE, only 1 bond, Glu159–Arg81, is maintained (Fig. 4C and SI Appendix, Fig. S15C). The mCLDN-15 mutant S67A with the same preserved Glu–Arg ionic bond shows 10.6-fold shorter and 7.2-fold more discontinuous TJ strands compared with wild type, emphasizing in vivo effects of cis bond proximity (44). Distances between residue Cαs are not largely increased by cCpE binding, but bonding distances between paired atoms are mostly broken compared with mCLDN-15. Our study reveals 2 conserved claudin side chains that may maintain cis interactions, Leu71/73 and Gln76/78 (SI Appendix, Figs. S4 and S16). Comparisons between homodimeric cis interacting models of unbound mCLDN-15 and cCpE-bound hCLDN-9 show cCpE-binding effects on the cis interaction epitope; explain the loss of TJ strand formation in Ser67, Glu157, Phe146, and Phe147 mutants of mCLDN-15 (44); and elucidate how claudin cis assembly could be dissociated by cCpE.
Fig. 4.
Model of the effect of cCpE on claudin cis interactions. cCpE-bound hCLDN-9 closed (blue) and unbound mCLDN-15 (SI Appendix, Fig. S15A) were manually docked based on the technique of Zhao et al. (44). (A) Full view of homodimeric cis interacting hCLDN-9. Residues purportedly involved in cis interactions are labeled and colored accordingly. ECH1 and ECH2 are highlighted with rectangles (blue). (B) Zoomed-in view of the cis interacting epitope of mCLDN-15 (brown). Polar (red) and nonpolar (black) interactions between protomers are shown as dashed lines. (C) Zoomed-in view of the cis interacting epitope of hCLDN-9 closed. One polar bond is shown as in B. Note disruptions to ECH1, ECH2, and the NPLVA153 motif between C and B.
The mCLDN-15 cis model proposes 3 cis interaction sites between claudin protomers: 1) ECH1–β5; 2) TM2–β5; and 3) ECH1–ECH2 (SI Appendix, Fig. S15B). Although 3 sites may form an ideal cis homomer, cCpE abolishes sites 1 and 3 and part of site 2 in hCLDN-9 closed (SI Appendix, Fig. S15C). Of 5 possible interatomic bonds between homomers, cCpE binding results in a loss of 4. These losses are not offset by other hCLDN-9 side chains. Comparison with hCLDN-9 open shows no interactions exist between cis sites, and the Glu–Arg ionic bond is severed. From unbound mCLDN-15 to hCLDN-9 closed, and then open, the number of bonds at the cis interaction epitope goes from 5 to 1 and then to none. Based on these structural changes between unbound and cCpE-complexed claudins, we propose mechanisms for disordering claudin cis assembly and subsequent dissociation of TJs by CpE. In one mechanism, CpE binds ECS1 and ECS2, disrupting trans interactions due to its high affinity for the NPLVA153 motif, and then positions itself deep within the claudin palm, perturbing ECH1 and ECH2, while breaking 80% of bonds at the cis interaction epitope (closed form). In another mechanism, part 1 occurs, and then CpE rocks as a rigid body from the palm toward the fingers (Movie S1), akin to a first-class lever, abolishing the cis epitope (open form) (Movie S2). Breaking trans interactions could facilitate toxin rocking by paracellular water influx into the claudin/cCpE interface during creation of a solvent-accessible cavity (Fig. 2D and SI Appendix, Fig. S9).
As both cCpE orientations observed in the closed and open structures of hCLDN-9 result in a major loss of cis interactions, we infer both could destabilize claudin/TJ assembly by stabilizing conformations of ECH1 and ECH2 that disfavor cis interactions (SI Appendix, Fig. S13). Such ECH conformations would prevent assembly if CpE binds claudins prior to reaching TJs, while a lever mechanism could disable assembly and physically break claudin self-association if CpE binds receptors within TJs. Uniquely, neither mechanism requires steric clash between adjacent cCpE molecules, as none exist (21) (SI Appendix, Fig. S14). Whether a lever mechanism is used by CpE to directly break cis interactions for TJ dissociation needs further testing. However, the process of CpE-induced cytotoxicity, which requires creation of a heterododecameric claudin/CpE pore complex, would demand local claudin cis disorganization; a lever mechanism would be effective for formation of this complex during CpE-mediated cytotoxicity (41).
Summary.
Claudins are the major structural components of TJs and simultaneously drive lateral adhesion between cells and paracellular permeation of ions between sheets, each of which is vital for epithelial tissue homeostasis. CpE binds claudins and dissociates TJs in gut epithelium, resulting in altered gastrointestinal balance. Understanding the mechanisms for claudin ion selectivity and TJ disassembly by CpE are vital first steps to remedy TJ-related diseases. Here, we present crystal structures of hCLDN-9 that inform on anion selectivity, mutations that cause deafness, epitopes coopted for HCV viral entry that may become targets for therapeutics, and how cCpE disables or breaks cis and trans claudin assembly. We show that hCLDN-9 is a high-affinity CpE receptor and that CpE is cytotoxic to hCLDN-9–expressing cells. Our findings elucidate the molecular bases for hCLDN-9 selective ion permeability and CpE isoform-specific recognition of claudins. Further, they provide mechanisms for CpE-induced dissociation of TJs, which disrupt epithelial adhesion, paracellular transport, and gastrointestinal homeostasis.
Methods
Protein Expression and Purification.
Plasmid pFastBac1 encoding hCLDN-9–His10, CpE-His10, and cCpE-His10 were expressed in insect cells. Membranes containing hCLDN-9–His10 were solubilized in 1% (wt/vol) n-decyl-β-d-maltopyranoside and purified on nickel-nitrilotriacetic acid (Ni-NTA) resin. Resin containing bound hCLDN-9 was captured, washed, and treated with thrombin. Cleaved hCLDN-9 was eluted and then used for binding studies and crystallization. For CpE-His10 and cCpE-His10, cells were lysed and centrifuged, and supernatant was incubated with Ni-NTA. Toxin purification followed hCLDN-9 apart from addition of detergent. Pure hCLDN-9 and cCpE were mixed, incubated at 4 °C, concentrated, and loaded onto an SEC column. Peak fractions were pooled, concentrated, and used for crystallization. More detailed procedures are described in SI Appendix, Supplementary Methods.
Crystallization and Structure Determination.
Crystals grew from a mother liquor containing 150 mM Na(C2H5COO), NaO2As(CH3)2, Bis-Tris propane (pH 4.5), and 25% polyethylene glycol 1500. Crystals were cryoprotected and then flash-frozen in liquid nitrogen. Diffraction data were collected at Advanced Light Source beamline 8.3.1. Datasets were processed in space group P212121, and structures were determined by MR using a homology model of hCLDN-9 (45) and cCpE (22) as search models. SI Appendix, Table S1 shows data and refinement statistics, with further details given in SI Appendix, Supplementary Methods.
Cavity Detection and Measurement.
Coordinates for hCLDN-9 in closed and open complex with cCpE were uploaded to CASTp (46) and other cavity detection programs listed in SI Appendix, Supplementary Methods. Measurements from these analyses are shown in SI Appendix, Fig. S9D.
Cytotoxicity and Binding Assays.
Baculovirus was added to adherent Sf9 cells and then placed at 27 °C. After 34 h, cCpE and CpE were added to the medium of wells expressing claudins and placed back at 27 °C. Fourteen hours postincubation with toxins, Sf9 morphology was imaged; cells were then were stained with trypan blue, and viability was measured in duplicate with an automated cell counter.
Purified CpE-His10 and cCpE-His10 were immobilized on Ni-NTA biosensors, and BLI was performed using an Octet RED384 System (FortéBIO). Kinetics experiments included a wash, toxin loading, baseline, and association and dissociation steps. During association, toxins were incubated with 0 to 250 nM free hCLDN-9. Data from triplicate trials were analyzed, modeled, and fitted. More detailed procedures are described in SI Appendix, Supplementary Methods.
Supplementary Material
Acknowledgments
We thank Alan Yu (University of Kansas Medical Center) for biology-specific guidance during initiation of this research. This research was supported by NIH Grant R01GM024485 (to R.M.S.) and initially by Postdoctoral Fellowship F32GM103277 (to A.J.V.). Beamline 8.3.1 at the Advanced Light Source is supported by the University of California Office of the President, Multicampus Research Programs and Initiatives Grant MR-15-338599 and by the Program for Breakthrough Biomedical Research, and is partially funded by the Sandler Foundation. The Advanced Light Source is a national user facility operated by the Lawrence Berkeley National Laboratory on behalf of the US Department of Energy under Contract DE-AC02-05CH11231, Office of Basic Energy Sciences, through the Integrated Diffraction Analysis Technologies program, supported by the US Department of Energy Office of Biological and Environmental Research.
Footnotes
The authors declare no conflict of interest.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID codes 6OV2 [closed form] and 6OV3 [open form]).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1908929116/-/DCSupplemental.
References
- 1.Van Itallie C. M., Anderson J. M., Claudin interactions in and out of the tight junction. Tissue Barriers 1, e25247 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Furuse M., Fujita K., Hiiragi T., Fujimoto K., Tsukita S., Claudin-1 and -2: Novel integral membrane proteins localizing at tight junctions with no sequence similarity to occludin. J. Cell Biol. 141, 1539–1550 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Günzel D., Yu A. S., Claudins and the modulation of tight junction permeability. Physiol. Rev. 93, 525–569 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lal-Nag M., Morin P. J., The claudins. Genome Biol. 10, 235 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sawada N., Tight junction-related human diseases. Pathol. Int. 63, 1–12 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kitajiri S. I., et al. , Expression patterns of claudins, tight junction adhesion molecules, in the inner ear. Hear. Res. 187, 25–34 (2004). [DOI] [PubMed] [Google Scholar]
- 7.Nakano Y., et al. , A claudin-9-based ion permeability barrier is essential for hearing. PLoS Genet. 5, e1000610 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fofana I., et al. , Functional analysis of claudin-6 and claudin-9 as entry factors for hepatitis C virus infection of human hepatocytes by using monoclonal antibodies. J. Virol. 87, 10405–10410 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Meertens L., et al. , The tight junction proteins claudin-1, -6, and -9 are entry cofactors for hepatitis C virus. J. Virol. 82, 3555–3560 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zheng A., et al. , Claudin-6 and claudin-9 function as additional coreceptors for hepatitis C virus. J. Virol. 81, 12465–12471 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Evans M. J., et al. , Claudin-1 is a hepatitis C virus co-receptor required for a late step in entry. Nature 446, 801–805 (2007). [DOI] [PubMed] [Google Scholar]
- 12.Krause G., et al. , Structure and function of claudins. Biochim. Biophys. Acta 1778, 631–645 (2008). [DOI] [PubMed] [Google Scholar]
- 13.Sas D., Hu M., Moe O. W., Baum M., Effect of claudins 6 and 9 on paracellular permeability in MDCK II cells. Am. J. Physiol. Regul. Integr. Comp. Physiol. 295, R1713–R1719 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.The Human Protein Atlas , hCLDN-9. https://www.proteinatlas.org/ENSG00000213937-CLDN9/tissue. Accessed 14 September 2018.
- 15.Uhlen M., et al. , Towards a knowledge-based Human Protein Atlas. Nat. Biotechnol. 28, 1248–1250 (2010). [DOI] [PubMed] [Google Scholar]
- 16.BioGPS , hCLDN-9. http://biogps.org/#goto=genereport&id=9080. Accessed 14 September 2018.
- 17.Wu C., et al. , BioGPS: An extensible and customizable portal for querying and organizing gene annotation resources. Genome Biol. 10, R130 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Winkler L., et al. , Molecular determinants of the interaction between Clostridium perfringens enterotoxin fragments and claudin-3. J. Biol. Chem. 284, 18863–18872 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Freedman J. C., Shrestha A., McClane B. A., Clostridium perfringens enterotoxin: Action, genetics, and translational applications. Toxins (Basel) 8, E73 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Saitoh Y., et al. , Tight junctions. Structural insight into tight junction disassembly by Clostridium perfringens enterotoxin. Science 347, 775–778 (2015). [DOI] [PubMed] [Google Scholar]
- 21.Shinoda T., et al. , Structural basis for disruption of claudin assembly in tight junctions by an enterotoxin. Sci. Rep. 6, 33632 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kitadokoro K., et al. , Crystal structure of Clostridium perfringens enterotoxin displays features of beta-pore-forming toxins. J. Biol. Chem. 286, 19549–19555 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Amasheh S., et al. , Claudin-2 expression induces cation-selective channels in tight junctions of epithelial cells. J. Cell Sci. 115, 4969–4976 (2002). [DOI] [PubMed] [Google Scholar]
- 24.Hou J., Renigunta A., Yang J., Waldegger S., Claudin-4 forms paracellular chloride channel in the kidney and requires claudin-8 for tight junction localization. Proc. Natl. Acad. Sci. U.S.A. 107, 18010–18015 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yu A. S., et al. , Molecular basis for cation selectivity in claudin-2-based paracellular pores: Identification of an electrostatic interaction site. J. Gen. Physiol. 133, 111–127 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Colegio O. R., Van Itallie C. M., McCrea H. J., Rahner C., Anderson J. M., Claudins create charge-selective channels in the paracellular pathway between epithelial cells. Am. J. Physiol. Cell Physiol. 283, C142–C147 (2002). [DOI] [PubMed] [Google Scholar]
- 27.Kratzer I., et al. , Complexity and developmental changes in the expression pattern of claudins at the blood-CSF barrier. Histochem. Cell Biol. 138, 861–879 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Piontek J., et al. , Formation of tight junction: Determinants of homophilic interaction between classic claudins. FASEB J. 22, 146–158 (2008). [DOI] [PubMed] [Google Scholar]
- 29.Abuazza G., et al. , Claudins 6, 9, and 13 are developmentally expressed renal tight junction proteins. Am. J. Physiol. Renal Physiol. 291, F1132–F1141 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lal-Nag M., Battis M., Santin A. D., Morin P. J., Claudin-6: A novel receptor for CPE-mediated cytotoxicity in ovarian cancer. Oncogenesis 1, e33 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Blackard J. T., Kemmer N., Sherman K. E., Extrahepatic replication of HCV: Insights into clinical manifestations and biological consequences. Hepatology 44, 15–22 (2006). [DOI] [PubMed] [Google Scholar]
- 32.Mitchell L. A., Koval M., Specificity of interaction between clostridium perfringens enterotoxin and claudin-family tight junction proteins. Toxins (Basel) 2, 1595–1611 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sonoda N., et al. , Clostridium perfringens enterotoxin fragment removes specific claudins from tight junction strands: Evidence for direct involvement of claudins in tight junction barrier. J. Cell Biol. 147, 195–204 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Veshnyakova A., et al. , Mechanism of Clostridium perfringens enterotoxin interaction with claudin-3/-4 protein suggests structural modifications of the toxin to target specific claudins. J. Biol. Chem. 287, 1698–1708 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Van Itallie C. M., Betts L., Smedley J. G. 3rd, McClane B. A., Anderson J. M., Structure of the claudin-binding domain of Clostridium perfringens enterotoxin. J. Biol. Chem. 283, 268–274 (2008). [DOI] [PubMed] [Google Scholar]
- 36.Krissinel E., Henrick K., Inference of macromolecular assemblies from crystalline state. J. Mol. Biol. 372, 774–797 (2007). [DOI] [PubMed] [Google Scholar]
- 37.Katahira J., Inoue N., Horiguchi Y., Matsuda M., Sugimoto N., Molecular cloning and functional characterization of the receptor for Clostridium perfringens enterotoxin. J. Cell Biol. 136, 1239–1247 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Robertson S. L., Smedley J. G. 3rd, McClane B. A., Identification of a claudin-4 residue important for mediating the host cell binding and action of Clostridium perfringens enterotoxin. Infect. Immun. 78, 505–517 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mitic L. L., Unger V. M., Anderson J. M., Expression, solubilization, and biochemical characterization of the tight junction transmembrane protein claudin-4. Protein Sci. 12, 218–227 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Suzuki H., et al. , Crystal structure of a claudin provides insight into the architecture of tight junctions. Science 344, 304–307 (2014). [DOI] [PubMed] [Google Scholar]
- 41.Smedley J. G. 3rd, Uzal F. A., McClane B. A., Identification of a prepore large-complex stage in the mechanism of action of Clostridium perfringens enterotoxin. Infect. Immun. 75, 2381–2390 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shinoda T., et al. , Cell-free methods to produce structurally intact mammalian membrane proteins. Sci. Rep. 6, 30442 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Katahira J., et al. , Clostridium perfringens enterotoxin utilizes two structurally related membrane proteins as functional receptors in vivo. J. Biol. Chem. 272, 26652–26658 (1997). [DOI] [PubMed] [Google Scholar]
- 44.Zhao J., et al. , Multiple claudin-claudin cis interfaces are required for tight junction strand formation and inherent flexibility. Commun. Biol. 1, 50 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang Y., I-TASSER server for protein 3D structure prediction. BMC Bioinformatics 9, 40 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tian W., Chen C., Lei X., Zhao J., Liang J., CASTp 3.0: Computed atlas of surface topography of proteins. Nucleic Acids Res. 46, W363–W367 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.