Significance
The insect-disseminated bacterium Candidatus Liberibacter asiaticus causes the destructive, incurable “citrus greening disease,” which is widespread in Asia, Africa, and the Americas, resulting in economic losses in the billions of dollars. One approach to treating infected citrus trees is application of antimicrobial compounds. However, use of broad-spectrum antibiotics in commercial citrus orchards has significant disadvantages, such as emergence of resistance and inhibition of beneficial bacteria. We designed a synthetic system for high-throughput screening of compound libraries in a closely related, culturable, and nonpathogenic bacterium, allowing us to identify small molecule inhibitors of Ca. L. asiaticus transcription activators.
Keywords: Liberibacter, Sinorhizobium, citrus, Huanglongbing (HLB), transcription regulation
Abstract
Citrus greening disease, also known as huanglongbing (HLB), is the most devastating disease of Citrus worldwide. This incurable disease is caused primarily by the bacterium Candidatus Liberibacter asiaticus and spread by feeding of the Asian Citrus Psyllid, Diaphorina citri. Ca. L. asiaticus cannot be cultured; its growth is restricted to citrus phloem and the psyllid insect. Management of infected trees includes use of broad-spectrum antibiotics, which have disadvantages. Recent work has sought to identify small molecules that inhibit Ca. L. asiaticus transcription regulators, based on a premise that at least some regulators control expression of genes necessary for virulence. We describe a synthetic, high-throughput screening system to identify compounds that inhibit activity of Ca. L. asiaticus transcription activators LdtR, RpoH, and VisNR. Our system uses the closely related model bacterium, Sinorhizobium meliloti, as a heterologous host for expression of a Ca. L. asiaticus transcription activator, the activity of which is detected through expression of an enhanced green fluorescent protein (EGFP) gene fused to a target promoter. We used this system to screen more than 120,000 compounds for compounds that inhibited regulator activity, but not growth. Our screen identified several dozen compounds that inhibit regulator activity in our assay. This work shows that, in addition to providing a means of characterizing Ca. L. asiaticus regulators, an S. meliloti host can be used for preliminary identification of candidate inhibitory molecules.
Citrus greening disease, also called huanglongbing (HLB), is catastrophic for world citrus industries (1, 2). The infecting agents are 3 bacterial Candidatus Liberibacter species, particularly Candidatus Liberibacter asiaticus (CLas) (3). CLas is spread between trees by the Asian citrus psyllid (ACP) (Diaphorina citri), a phloem-feeding insect that inoculates Citrus plants with bacteria from its salivary glands as it feeds on leaves (4).
CLas appears to cause disease by disrupting function of phloem, the essential vascular tissue that transports sugars and other nutrients from leaves (3). Early symptoms of HLB include yellowing of leaves (2), followed by dieback of both the canopy and fibrous roots (1). The few fruits that develop are misshapen, green, and bitter (2). There is no cure for HLB, and infection is terminal for the host tree. In the United States, active HLB disease was first discovered in Florida in 2005, where economic losses thus far have exceeded $4.5 billion. It has since spread to 2 other major citrus-growing states, Texas and California (1).
Since there is no effective treatment for infected trees, nor resistant commercial citrus varieties, HLB is managed mainly by controlling spread of the ACP vector and by replacing infected trees with uninfected nursery stock (1). Other measures that may ease HLB damage to citrus include maintaining optimal growth conditions, stimulating plant growth and defenses, thermotherapy of infected trees, biological control, and treatment with antimicrobials (1, 5). Regarding antimicrobials, streptomycin and oxytetracycline are permitted for foliar application in Florida under an emergency exemption (references cited in ref. 6), and there is much interest in identifying additional compounds that inhibit CLas infection and growth (1, 6, 7).
CLas is a reduced-genome, α-proteobacterium (8, 9) that cannot be cultured, precluding use of direct screens for antimicrobial discovery. The only known commensal Liberibacter, Liberibacter crescens, can be cultured and is being developed as a model system to study Liberibacter physiology and genetics, including response to antimicrobial treatments, but still lacks the tools of better studied α-proteobacteria (10–18). CLas is closely related to the beneficial nitrogen-fixing plant symbiont Sinorhizobium meliloti (Sme), which has been used as a heterologous host to express specific CLas genes (14). Construction of a flexible synthetic-model system using highly tractable Sme could allow in vivo screening for discovery of new treatments.
The vast majority of commercial antimicrobials target essential bacterial functions. However, these antimicrobials present a significant downside: By targeting essential cellular processes, they exert selective pressure that allows resistant bacterial populations to emerge. The rise of drug-resistant pathogens has resulted in increased interest in narrow-spectrum and targeted approaches against microbial pathogens, such as those focusing on specific signaling and virulence pathways (19, 20).
One approach in targeting virulence pathways is to identify inhibitors of the actual proteins that are directly responsible for the disease symptoms (19). This is not feasible for many pathogens because pathogenesis mechanisms are poorly defined: This is the case with CLas. Inhibiting regulatory proteins such as transcription factors is an alternative strategy that proved successful in other bacteria (20–25). The small number of predicted regulators encoded by CLas (8, 9) makes it feasible to systematically screen for small molecule inhibitors in a high-throughput manner. We therefore designed an in vivo synthetic system to screen for inhibitors of CLas transcription activators, using the closely related model bacterium, S. meliloti, as a heterologous host. Our approach identified candidate compounds that inhibited activity of CLas transcription activators and complements recent in vitro inhibitor screens (11, 14, 26, 27).
Results
CLas Encodes up to 19 Transcription Regulators.
Our bioinformatic analyses predict that the CLas genome encodes 19 transcription regulators, including 2 sigma factors: RpoD (housekeeping sigma factor) and RpoH (likely heat shock/stress response sigma factor) (Table 1 and SI Appendix, Table S1). All but one appear to have orthologs in the related, beneficial nitrogen fixing symbiont, Sme. Work in Sme and L. crescens (13) suggests that 6 CLas regulators are essential for viability: RpoD, RpoH, CtrA, DivK, GcrA, and CpdR1. The last 4 of these may play critical roles in CLas cell cycle regulation (28). Some of the 19 predicted CLas regulators may not modulate gene expression: The sole CLas LexA-like regulator (GenBank accession no. ACT56917) appears to lack a helix-turn-helix DNA binding domain; CLas lacks a σ54-type sigma factor to act in concert with its putative TacA-like enhancer-binding protein (ACT57389).
Table 1.
CLas transcription regulators chosen for study
| CLas regulator, GenBank accession no.* | S. meliloti (Sme) ortholog(s)† | Percent identity between CLas and Sme proteins | Regulator type | Putative function of regulator in Sme (ref.) | CLas fold change expression for plant vs. psyllid‡ |
| ACT57084 | RpoH1, RpoH2 | 72, 41 | Sigma factor | Stress response, symbiosis (30–34) | 2.6 |
| ACT57167 | VisN | 50 | LuxR | Motility; forms heterodimer with VisR (35) | 6.7 |
| ACT57166 | VisR | 49 | LuxR | Motility; forms heterodimer with VisN (35) | 4.3 |
| ACT56824 | LdtR (SMc01768) | 70 | MarR | Osmotic stress tolerance, peptidoglycan remodeling (14) | 37.1 |
| ACT56755 | LsrB (SMc01225) | 58 | LysR | LPS biosynthesis, symbiosis (42, 54) | 4.7 |
| ACT56897 | PhrR1 (SMc01110), PhrR2 (SMb21117) | 59, 48 | HTH-XRE | Quorum sensing (43) | Not reported |
| ACT57366 | CtrA | 75 | Response regulator | Cell cycle control; essential gene (40) | 3.3 |
Accession numbers are for regulators in the CLas Psy62 genome, assembled from a psyllid metagenome (8). SI Appendix, Table S1 lists other putative CLas transcription regulators not chosen for this study.
If the gene name has not been annotated in GenBank, the S. meliloti 1021 unique locus tag is given in parentheses.
Data published in Yan et al. (29). The fold change provided here was calculated from their reported log2 ratio values. Not reported means the gene was listed in Yan et al’s. supplementary table 1 as “selected for qRT-PCR analysis,” but a log2 ratio was not reported.
In deciding which CLas regulators to study, we considered 3 main factors: characteristics of the Sme ortholog(s); representation of multiple regulator families, since the 12 different regulator types present in CLas are expected to be inhibited differently; and CLas expression pattern, because regulators involved in citrus virulence may show increased in planta gene expression. For expression pattern, we consulted an RT-qPCR study that compared expression of 381 CLas genes amplified from infected sweet orange host plants versus CLas-harboring psyllid insects (29). Based on these factors, we chose the following 6 regulators, all predicted to function as transcription activators, for transcriptome analysis and high-throughput inhibitor screening: RpoH, VisNR, LdtR, LsrB, PhrR, and CtrA (Table 1).
CLas Regulators Can Be Expressed Efficiently in S. meliloti.
Our high-throughput screening approach for identifying inhibitory compounds required that the CLas regulator was expressed well in its heterologous Sme host. We optimized for plasmid copy number, exogenous promoter used for expression, ribosome binding site (RBS), and codon usage (SI Appendix, Materials and Methods). To decrease background transcription levels of target genes whose expression may be activated by both the Sme and the CLas regulator, we introduced regulator expression plasmids into strains deleted for the Sme orthologous regulator(s), except for ctrA (SI Appendix, Table S2).
We performed phenotypic assays to determine if the CLas regulator could compensate for defects caused by deletion of the Sme orthologous regulators (SI Appendix, Materials and Methods). We observed qualitative growth of all of the strains, and in certain strains we also assessed heat stress, swimming motility, cell morphology, and cell envelope integrity. To identify genes whose expression increased with ectopic expression of the CLas regulator, we performed Affymetrix GeneChip analysis on Sme strains deleted for the orthologous Sme regulator(s). These carried either a plasmid that encoded the CLas regulator or the empty vector and were induced with Isopropyl β-d-1-thiogalactopyranoside (IPTG) to provide strong expression of the heterologous regulator. Phenotypic and transcriptome results for each of these 6 strains are detailed in the sections below.
RpoH.
Sme RpoH1 and RpoH2 alternative sigma factors mediate response to various stressors, including heat, acid, hydrogen peroxide, stationary phase growth, and envelope disrupting agents (30–33). CLas RpoH is most similar to Sme RpoH1 (72% identity), which is required for an effective nitrogen-fixing symbiosis (32, 34). CLas RpoH and Sme RpoH1 both complemented a Sme ΔrpoH1rpoH2 strain for growth at restrictive temperature.
To identify transcripts whose abundance increased in Sme ΔrpoH1rpoH2 when CLas RpoH was ectopically expressed, we performed Affymetrix GeneChip analysis, with the same strain expressing Sme rpoH1 as a positive control, and the empty vector (pSRKGm) as a point of reference. Eight genes showed increased expression ≥2-fold with both CLas RpoH and Sme RpoH (Dataset S1): All were previously identified as RpoH1-dependent in other studies (30, 31).
VisNR.
VisN and VisR function as a heterodimer to positively regulate chemotaxis, flagellar, and motility genes (35) in Sme and negatively regulate the flp3 pilus gene in CLas (36). The CLas genome contains genes predicted for flagellar biogenesis and motility (8), but it is unknown if CLas forms functional flagella. The Sme ΔvisNR deletion strain was nonmotile on soft agar plates, and expression of CLas visNR restored WT motility. Transcriptomic comparisons of Sme ΔvisNR expressing CLas visNR versus the empty vector revealed that, as expected, CLas visNR stimulates expression of Sme motility and chemotaxis genes (Dataset S1). The most strongly expressed gene was rem (11-fold), which encodes a response regulator that acts downstream of Sme VisNR to activate motility gene expression during exponential growth (37).
LdtR.
Sme LdtR was postulated to play a role in response to hyperosmotic stress, perhaps by activating expression of the adjacent gene, ldtP, which encodes a putative l,d-transpeptidase (14, 38). Work in L. crescens implies that LdtR is a master regulator controlling diverse functions, including motility and cell wall biogenesis (39). Sme ΔldtR had a swimming motility defect (∼30% of WT motility), which was not suppressed by ectopically expressing CLas LdtR. Expressing CLas LdtR in WT Sme also reduced motility, implying that both too much and too little Sme LdtR is deleterious for motility. Ectopically expressing either CLas or Sme LdtR in either WT or ΔldtR strains resulted in poor growth on Luria Broth (LB) medium; because of this, and because previous work indicated a role for LdtR in cell wall remodeling (14, 38, 39), we examined cellular morphology of WT strains expressing either CLas or Sme LdtR (Materials and Methods). Compared to WT carrying the empty vector, cells expressing either CLas or Sme LdtR were elongated, branched, and had bulges (SI Appendix, Fig. S1). We compared the transcriptome of Sme ΔldtR expressing CLas LdtR versus the empty vector strain; only 8 genes showed ≥1.5-fold increase in expression (Dataset S1). Five of these may be controlled by the master cell cycle regulator, CtrA (40), including tacA, which had the highest increase in expression (∼10-fold), and whose Caulobacter crescentus ortholog was shown to be a global cell cycle regulator involved in polar development (41).
LsrB.
Sme LsrB activates expression of oxidative stress-related genes and an operon involved in lipopolysaccharide biosynthesis (lrp3-lpsCDE) (42), which likely explains why the ΔlsrB mutant grew poorly and was >1,000-fold more sensitive than WT to the envelope-disrupting detergent, deoxycholate (DOC). Expression of CLas lsrB only partially suppressed the DOC-growth defect of ΔlsrB; this strain was ∼10- to 100-fold more sensitive to DOC than was WT. Comparison of transcriptome profiles for Sme ΔlsrB expressing CLas LsrB compared to the control identified only 3 genes with expression increased ≥1.2-fold, while genes in the lrp3-lpsCDE operon failed to show increased expression (Dataset S1).
PhrR.
Sme has 2 orthologs of CLas PhrR (Table 1). PhrR1 may play a role in quorum sensing (QS) because the ExpR QS regulator represses phrR1 expression by binding upstream of phrR1, in an acyl homoserine lactone-dependent manner (43). In closely related S. medicae, phrR1 expression increases in response to stresses such as low pH, ethanol, zinc, copper, and H2O2 (44). Both ΔphrR1 and ΔphrR1phrR2 mutants grew more slowly than WT on LB plates. During RNA purification, the ΔphrR1phrR2 mutant was more resistant to lysozyme lysis, suggesting an alteration of its lipopolysaccharide or other envelope component (SI Appendix, Materials and Methods). We identified no genes whose expression increased ≥1.5-fold in Sme ΔphrR1phrR2 expressing CLas PhrR (Dataset S1).
CtrA.
CtrA is essential for viability in Sme and most other α-proteobacteria (40). When we expressed Sme ctrA from a plasmid, we could delete the genome copy of Sme ctrA but not when CLas ctrA was similarly expressed. Therefore, we used a WT Sme host to compare transcriptome profiles for cells expressing CLas ctrA or Sme ctrA vs. empty vector. Genes whose expression increased with CLas ctrA (Dataset S1) included the minCDE operon, encoding proteins that inhibit septum formation at appropriate times during the cell cycle (40), and ldtR. This latter result is intriguing given that expressing CLas ldtR appears to decrease expression of CtrA-dependent genes (see above). Sme CtrA directly represses minCDE expression during the cell cycle (40); perhaps increased expression of minCDE, caused by ectopic expression of CLas ctrA, contributes to the growth defects observed in these strains.
High-Throughput Screening Identified Compounds That May Inhibit CLas Transcription Regulators.
By identifying genes whose expression increased when IPTG induced expression of each of the 6 CLas regulators, we defined promoters targeted by each of them. Candidate promoters were fused to an enhanced green fluorescent protein (EGFP) reporter gene for high throughput compound screening. We designed the screening strains to have low basal fluorescence and to fluoresce strongly upon IPTG-induced expression of the CLas regulator. Our easy-to-use custom expression cassette has features to optimize signal over noise (SI Appendix, Fig. S2). After cloning this synthetic EGFP cassette into each of the 6 regulator gene-containing plasmids, we cloned each of 11 promoters (chosen as described in Materials and Methods) into the appropriate plasmid, then introduced each plasmid into the appropriate Sme deletion strain (SI Appendix, Table S2).
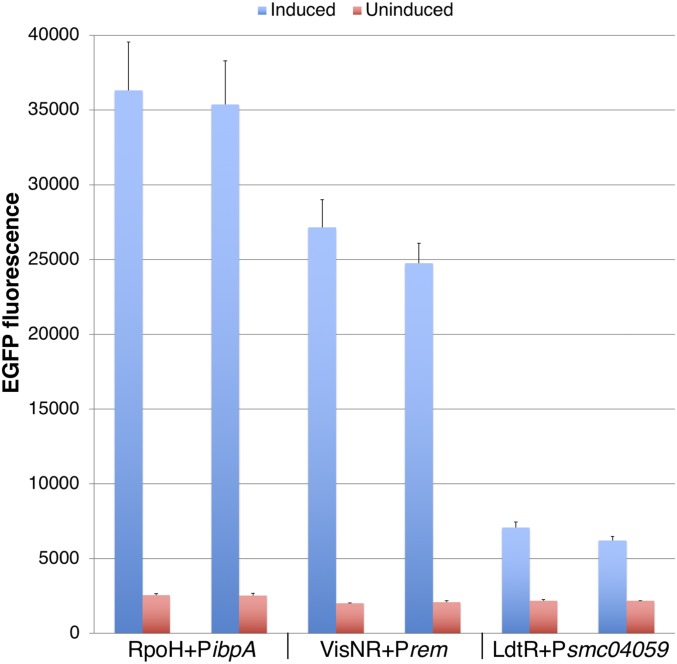
Based on qualitative fluorescence assays, we selected 3 regulator-promoter constructs for further testing at the Stanford High-Throughput Bioscience Center (HTBC): LdtR (Psmc04059), RpoH (PibpA), and VisNR (Prem). Strains with these constructs performed well in pilot experiments: fluorescence in IPTG-induced cells was high compared to the basal level (Fig. 1). High-throughput screening of 10 libraries (>120,000 compounds) was performed for each of these strains (Materials and Methods and SI Appendix, Fig. S3 and Table S3).
Fig. 1.
EGFP fluorescence for IPTG-induced and uninduced high-throughput screening strains. Each S. meliloti strain was tested in duplicate 384-well plates for EGFP fluorescence. Each column indicates the average of raw signal values for IPTG-induced (n = 352) and uninduced (n = 16) wells. Error bars indicate SD. The CLas transcription regulator and S. meliloti promoter of each strain are indicated below the x axis. Mean EGFP fluorescence for M9 sucrose medium negative controls was 293 (n = 96). Strains: ΔrpoH1rpoH2 CLas-RpoH PibpA (MB231 pMB949); ΔvisNR CLas-VisNR Prem (MB1102 pMB956); ΔldtR CLas-LdtR Psmc04059 (MB1101 pMB958).
The Known Bioactive Collection was screened first to evaluate the performance of our screening assay; most of these libraries were screened at 7 different concentrations (SI Appendix, Fig. S3 and Table S3). Screening at multiple concentrations allowed us to plot % fluorescence and absorbance inhibition versus compound concentration, and the presence of known antibacterial compounds in these libraries ensured that we would detect at least some inhibitory compounds. Over 130 compounds inhibited growth by at least 50% in all 3 testing strains (Dataset S2). As expected, these inhibitory compounds, most of which have known antibacterial activity, also inhibited EGFP fluorescence, validating the assay. However, our goal was to identify compounds that decreased function of a CLas regulator without severely inhibiting growth to avoid compounds that could be broadly toxic in a natural environment and that would increase selective pressure for resistant bacteria. The desirable compounds should show high inhibition of EGFP fluorescence but low or no inhibition of growth as measured by absorbance. We arbitrarily set cutoffs for inhibition of EGFP fluorescence ≥30%, and <50% for growth inhibition, with a difference between EGFP fluorescence and growth inhibition of ≥30%. We also eliminated from consideration compounds that inhibited EGFP fluorescence of all 3 strains, because such compounds may generally inhibit GFP activity or quench fluorescence. Dataset S2 lists 69 compounds from the Bioactive Collection meeting the above criteria. The majority of these compounds specifically inhibited EGFP fluorescence in the LdtR strain (n = 45), while 13 compounds were specific for the VisNR strain and 4 for the RpoH strain. Seven compounds inhibited 2 strains. The small number of compounds inhibiting the RpoH strain may reflect a general lack of susceptibility of RpoH sigma factors to inhibition by small molecules; however, fluorescence and absorbance measurements were more variable for the RpoH plates than for the others. This variability could have led to a high proportion of false negative compounds.
Because the 113,809 compounds from the ChemDiv, ChemBridge, and Specs libraries in the HTBC’s “Diverse Collection” were initially screened at a single concentration (SI Appendix, Fig. S3 and Table S3), we devised a ranking system for rescreening (SI Appendix, Tables S4 and S5) to ensure that efforts were evenly distributed among the regulators. The system prioritized compounds with greater differences between EGFP fluorescence and growth inhibition, and that specifically inhibited only 1 of the 3 regulators. In all, 629 compounds (0.55%) were rescreened in duplicate at 8 different concentrations. Rescreening identified 61 compounds that met the same criteria, described above for the Bioactive Collection, with strain specificities as follows: LdtR, 37; VisNR, 19; RpoH, 2; and inhibiting both RpoH and VisNR, 3 (Dataset S3). Although the candidate inhibitory compounds have diverse structures, we noticed regulator-specific patterns: 7 compounds inhibiting LdtR EGFP fluorescence possess 1,3-thiazole groups, and 9 compounds inhibiting VisNR EGFP possess sulfone groups.
To confirm the accuracy of the high-throughput screening results, we purchased 10 compounds (Fig. 2 and SI Appendix, Fig. S4) and retested them using a slightly different procedure than that of the original high-throughput screen. To gain information about specificity of inhibition, we also tested these 10 purchased compounds on each of the 3 Sme deletion strains carrying the corresponding Sme regulator (LdtR, RpoH1, VisNR) on a plasmid. Results are given in SI Appendix, Table S6, and for CLas regulators, the original screening results are provided for comparison.
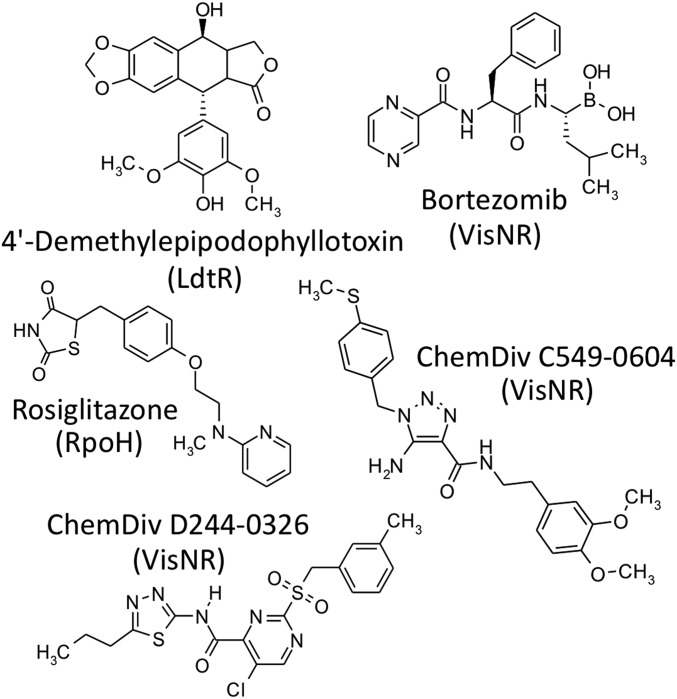
Fig. 2.
Five inhibitory compounds whose effects on the CLas and S. meliloti regulators (LdtR, RpoH/RpoH1, and VisNR) were confirmed by purchasing compounds and retesting (SI Appendix, Table S6). Regulators identified via high-throughput screening as the putative targets of the inhibitory compounds are shown below the compound name (Bioactive Collection compounds) or supplier/catalog number (Diverse Collection compounds) in parentheses.
Retesting results varied by compound and could be sorted into 3 groups by behavior (SI Appendix, Table S6). 1) One compound (ChemDiv 8013-5939) did not affect EGFP fluorescence or growth. 2) Four compounds affected growth (and therefore EGFP fluorescence) but showed no specificity for a single regulator (ChemBridge 5109513, Fisetin, Orbifloxacin, and Oxybenzone). 3) Results for 5 compounds mostly replicated our high-throughput screening data (4-Demethylepipodophyllotoxin, ChemDiv C549-0604, Bortezomib, ChemDiv D244-0326, and Rosiglitazone maleate). The latter 3 compounds inhibited EGFP fluorescence in VisNR and one or both RpoH strains. The overall pattern of results is consistent with inhibitors that affect expression, not EGFP function per se.
The most promising results were obtained with ChemDiv C549-0604, which consistently and strongly inhibited EGFP fluorescence in the CLas VisNR strain (IC50 = 0.7 μM; SI Appendix, Table S6). Since both CLas and Sme VisNR rescued a nonmotile S. meliloti ΔvisNR strain, we tested whether ChemDiv C549-0604, D244-0326, and bortezomib decreased CLas and Sme VisNR-mediated motility. ChemDiv C549-0604 decreased motility of ΔvisNR CLas pVisNR by 28%, but motility of ΔvisNR Sme pVisNR was not affected (SI Appendix, Fig. S5). Neither ChemDiv D244-0326, nor bortezomib, affected motility. In summary, these results support the validity of our high-throughput screening methods, while highlighting the importance of multiple assays to retest candidate positive results.
Discussion
We carried out a high-throughput screen to identify compounds that inhibit activity of CLas transcription activators, without substantially inhibiting bacterial growth. Because CLas cannot be cultured, use of a closely related, genetically tractable, heterologous Sme host bacterium was the key feature of our in vivo screen design. Of 6 initial CLas regulators examined (CtrA, LdtR, LsrB, PhrR, RpoH, VisNR), 3 were chosen for high-throughput screening (LdtR, RpoH, VisNR), yielding candidate inhibitory (i.e., “lead”) compounds. This work demonstrates the practicality of using the heterologous Sme host to study CLas regulator function. While CLas LdtR, RpoH, and VisNR seem to function similarly to their orthologous Sme proteins, CLas CtrA, LsrB, and PhrR may have distinct functions.
Both in vitro and in vivo assays have been used to screen for inhibitors of transcription regulators. In vitro approaches include using purified proteins to screen for compounds that differentially affect temperature-dependent protein unfolding (e.g., differential scanning fluorimetry [DSF]) (45) and cell-free DNA binding assays (21). DSF combined with DNA-binding assays successfully identified small molecule inhibitors of CLas regulators, LdtR and PrbP (11, 14, 26, 27). While in vitro approaches are valuable, there are downsides to their use: In vitro screening requires sufficient amounts of purified, active target protein. In the case of DSF, it is unclear whether the degree of thermal denaturation is an accurate surrogate for in vivo protein inhibition. In vivo whole-cell, high-throughput approaches have succeeded in targeting transcription regulators (24, 25, 46–49), but these typically require laboratory culture of the bacterial species and at least partial replication of its natural environment.
We compared our results from screening the CLas LdtR Sme strain to data from the CLas LdtR DSF study (14). Our screen included 6 compounds that Pagliai et al. identified as affecting LdtR temperature-dependent protein unfolding, DNA binding, and/or Sme growth (Benzbromarone, Diethylstilbestrol, Hexestrol, Oxantel pamoate, Phloretin, Resveratrol). Of these 6 compounds, hexestrol inhibited both EGFP fluorescence and growth of all 3 S. meliloti strains (IC50 values ranging from 7 to 20 μM). Diethylstilbestrol inhibited EGFP fluorescence of all 3 strains in just 1 of 2 different libraries we screened (IC50 values = 13–18 μM). The remaining 4 compounds reported by Pagliai et al. were inactive in our screen, perhaps because we screened at lower concentrations (SI Appendix, Table S3) than were previously found to affect DNA binding and growth (50–250 μM) (14). The compound that most specifically inhibited EGFP fluorescence of our CLas LdtR strain (4-Demethylepipodophyllotoxin) was not among the 1,312 compounds screened previously (14). A thorough comparison of in vitro vs. in vivo screening methods will require studies of additional regulators, but this preliminary comparison suggests the 2 methods are complementary.
While use of a heterologous host has an obvious advantage in probing transcription of an unculturable bacterial species, our study confirms some downsides to this approach. For example, if CLas regulator function is distinct from that of its Sme ortholog, then transcriptome analysis with Affymetrix Sme GeneChips may fail to identify Sme target promoters sufficiently activated for use in a high-throughput screen that relies on EGFP fluorescence, as was the case for CtrA, LsrB, and PhrR. For such nonhomologous regulators, one could forego transcriptome analysis and instead screen a library of short, random CLas DNAs cloned into the regulator-EGFP vectors and look for robust EGFP fluorescence. Promoters that are activated only when the regulator is induced could then be further assessed for suitability in a high-throughput screen. Another option would be use of a more closely related host such and L. crescens, which is being developed as a model system, although L. crescens grows more slowly than Sme and has complex nutritional requirements (10–17).
Another downside of our screen design is that minimizing contribution of native Sme regulators to target promoter activity fluorescence necessitated deletion of the orthologous Sme regulator(s). If one of these regulators is critical for Sme growth/viability and the CLas regulator cannot complement its function, then the bacteria may have growth or other defects that complicate high-throughput screening. For example, Sme ΔphrR exhibited slow growth that was not suppressed by expressing CLas PhrR. A more severe example is that of CLas CtrA, whose expression not only failed to rescue viability of a Sme ΔctrA strain, but was itself deleterious when expressed in WT Sme on LB growth medium. Although these issues precluded efforts using our design to screen for inhibitors of CLas CtrA, they suggest that simply screening for growth of WT Sme expressing CLas CtrA could identify compounds that inhibit activity of CLas CtrA, but not Sme CtrA.
Overall, our work demonstrates that it is feasible to identify CLas lead compounds using an in vivo fluorescence-based screen in a Sme host. This screen identified dozens of potential inhibitors of CLas transcription activators. Many of the “Known Bioactive” library compounds that inhibit EGFP fluorescence are unsuitable for treatment of citrus greening disease because of their toxicity, mutagenicity, or cost; however, results obtained with these compounds suggest that our screening system could be extended to assay nontoxic chemical fragments and related compounds for efficacy in combatting citrus greening. Screening the “Diverse Collection” library identified lead compounds, the most promising of which (ChemDiv C549-0604) appears to specifically inhibit activity of CLas VisNR.
Research on the CLas-psyllid-Citrus disease triad is hampered by many technical challenges, such as inability to culture the CLas bacterial pathogen, variable insect behavior and ecology, and a large, perennial host plant that takes years to mature (50, 51). Because a single breakthrough discovery is unlikely to win the fight against citrus greening disease, progress depends on continued broad efforts by researchers across many disciplines. Microbiologists have focused on: understanding how CLas is transmitted and survives in its hosts; characterizing multiple CLas genomes; identifying CLas virulence functions and secreted effector proteins; discerning barriers to CLas laboratory cultivation; developing the culturable L. crescens model system; defining Citrus and psyllid microbiomes; and seeking means to control CLas with antagonistic bacteria, bacteriophages, and antimicrobials (50). The ultimate goal in the war on citrus greening is CLas-resistant commercial Citrus varieties, a difficult and likely long-range goal for numerous reasons, including lack of knowledge on how CLas interacts with Citrus at a molecular level. As long as barriers to CLas laboratory cultivation exist, such molecular mechanisms will be extremely difficult to dissect; thus, research using model heterologous systems such as L. crescens, S. meliloti, and other related α-proteobacteria will continue to be an important stopgap measure. Researchers studying CLas should continue to exploit the extensive knowledge available for α-proteobacteria, and we encourage researchers studying model α-proteobacteria to participate in combatting this modern agricultural plague.
Materials and Methods
Additional information is provided in SI Appendix, Materials and Methods.
Strains and Plasmids.
Standard techniques were used for cloning, PCR amplification, strain constructions, and phenotypic assays (SI Appendix, Materials and Methods). All Sme strains used in this study are derived from CL150 (SI Appendix, Table S2). Oligonucleotide primers used in this study are listed in SI Appendix, Table S7. Regulator genes (Table 1) were cloned into the medium copy, IPTG-inducible vector, pSRKGm (52). We fully optimized the coding sequence of each CLas Psy62 (8) regulator for expression in Sme (GenBank accession nos.: MK359043–MK359048). Unmarked deletions of Sme ctrA, ldtR, lsrB, phrR1, phrR2, and the visN-visR operon were constructed.
Affymetrix GeneChip Analysis.
For transcriptome analysis of Sme, we used a custom dual genome Affymetrix Symbiosis Chip (53). Each optimized CLas regulator was ectopically expressed by inducing with 0.5 mM IPTG. Controls for comparison were Sme deletion strains carrying the empty pSRKGm plasmid (or in the case of CtrA, CL150 carrying pSRKGm). Three biological replicates were analyzed for each strain as described in SI Appendix. The Affymetrix GeneChip data have been deposited under Superseries accession no. GSE124984 in the Gene Expression Omnibus database.
High-Throughput Screening of Compound Libraries.
We designed a modular EGFP expression cassette, optimized for expression in Sme (NCBI accession no. MK387175), and constructed pSRKGm-derived plasmids containing the EGFP cassette with each CLas regulator gene, and candidate promoter sequences upstream of EGFP (SI Appendix, Table S2). Based on Affymetrix Gene Chip results for Sme strains ectopically expressing CLas regulators, candidate target promoters were amplified and cloned upstream of the EGFP cassette (SI Appendix, Table S7).
Screening of strains carrying a CLas regulator and a target promoter-EGFP fusion was performed at the HTBC (http://med.stanford.edu/htbc.html). Three strains were used for screening: MB1101 pMB958 (LdtR); RFF231 pMB949 (RpoH); and MB1102 pMB956 (VisNR). Detailed screening procedures are described in SI Appendix, Materials and Methods.
We screened a total of 10 compound libraries (SI Appendix, Table S3) using the workflow is shown in SI Appendix, Fig. S3. Data were analyzed using MDL Assay Explorer software. A description of libraries and compound concentrations screened are given in SI Appendix, Tables S3–S5.
Testing Purchased Compounds.
To evaluate selected results from the high-throughput screen, we purchased 10 compounds that showed inhibitory activity for 1 or more of the 3 CLas regulators: 6 of these were from the Known Bioactive Collection and 4 from the Diverse Collection (Fig. 2 and SI Appendix, Fig. S4). The 10 purchased compounds were also tested on each of the 3 Sme deletion strains ectopically expressing the corresponding Sme regulator (LdtR, RpoH, or VisNR) (SI Appendix, Table S2).
Supplementary Material
Acknowledgments
We are grateful to Jason Wu for technical assistance with high-throughput compound screening. We thank our laboratory members as well as James Chen, Peter Kim, and Mark Smith for helpful discussions. We appreciate Aaron Duthoy’s technical assistance with Affymetrix GeneChip experiments and Jung-Gun Kim’s help with microscopy. We thank Stephen Lynch for performing confirmatory 1D-NMR on compounds purchased from ChemDiv and ChemBridge, and Aaron Duthoy, Robert Fisher, and Reed Goodwin for critically reading the manuscript. This work was funded by Citrus Advanced Technology Program (Project 805) of the Citrus Research and Development Foundation Inc., and we thank Tom Turpen, Audrey Nowicki, and Brandi Goller of the CRDF for their administrative support and guidance. Additional funding was provided by NIH Grant R01 GM093628 and a contribution from S.R.L.
Footnotes
The authors declare no conflict of interest.
Data deposition: The sequences reported in this paper have been deposited in GenBank, https://www.ncbi.nlm.nih.gov/genbank (accession nos. MK359043–MK359048 and MK387175). Affymetrix GeneChip data have been deposited in the Gene Expression Omnibus (GEO) database, https://www.ncbi.nlm.nih.gov/geo (Superseries accession no. GSE124984).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1905149116/-/DCSupplemental.
References
- 1.Blaustein R. A., Lorca G. L., Teplitski M., Challenges for managing Candidatus Liberibacter spp. (huanglongbing disease pathogen): Current control measures and future directions. Phytopathology 108, 424–435 (2018). [DOI] [PubMed] [Google Scholar]
- 2.Bové J. M., Huanglongbing: A destructive newly emerging, century-old disease of citrus. J. Plant Pathol. 88, 7–37 (2006). [Google Scholar]
- 3.Wang N., et al. , The Candidatus Liberibacter-host interface: Insights into pathogenesis mechanisms and disease control. Annu. Rev. Phytopathol. 55, 451–482 (2017). [DOI] [PubMed] [Google Scholar]
- 4.Grafton-Cardwell E. E., Stelinski L. L., Stansly P. A., Biology and management of Asian citrus psyllid, vector of the huanglongbing pathogens. Annu. Rev. Entomol. 58, 413–432 (2013). [DOI] [PubMed] [Google Scholar]
- 5.Munir S., et al. , Huanglongbing control: Perhaps the end of the beginning. Microb. Ecol. 76, 192–204 (2018). [DOI] [PubMed] [Google Scholar]
- 6.Yang C., et al. , Antimicrobial compounds effective against Candidatus Liberibacter asiaticus discovered via graft-based assay in citrus. Sci. Rep. 8, 17288 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Coyle J. F., Lorca G. L., Gonzalez C. F., Understanding the physiology of Liberibacter asiaticus: An overview of the demonstrated molecular mechanisms. J. Mol. Microbiol. Biotechnol. 28, 116–127 (2018). [DOI] [PubMed] [Google Scholar]
- 8.Duan Y., et al. , Complete genome sequence of citrus huanglongbing bacterium, ‘Candidatus Liberibacter asiaticus’ obtained through metagenomics. Mol. Plant Microbe Interact. 22, 1011–1020 (2009). [DOI] [PubMed] [Google Scholar]
- 9.Tyler H. L., Roesch L. F., Gowda S., Dawson W. O., Triplett E. W., Confirmation of the sequence of ‘Candidatus Liberibacter asiaticus’ and assessment of microbial diversity in Huanglongbing-infected citrus phloem using a metagenomic approach. Mol. Plant Microbe Interact. 22, 1624–1634 (2009). [DOI] [PubMed] [Google Scholar]
- 10.Fagen J. R., et al. , Comparative genomics of cultured and uncultured strains suggests genes essential for free-living growth of Liberibacter. PLoS One 9, e84469 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gardner C. L., et al. , Drug repurposing: Tolfenamic acid inactivates PrbP, a transcriptional accessory protein in Liberibacter asiaticus. Front. Microbiol. 7, 1630 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jain M., Munoz-Bodnar A., Gabriel D. W., Concomitant loss of the glyoxalase system and glycolysis makes the uncultured pathogen “Candidatus Liberibacter asiaticus” an energy scavenger. Appl. Environ. Microbiol. 83, e01670-17 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lai K. K., Davis-Richardson A. G., Dias R., Triplett E. W., Identification of the genes required for the culture of Liberibacter crescens, the closest cultured relative of the Liberibacter plant pathogens. Front. Microbiol. 7, 547 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pagliai F. A., et al. , The transcriptional activator LdtR from ‘Candidatus Liberibacter asiaticus’ mediates osmotic stress tolerance. PLoS Pathog. 10, e1004101 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jain M., et al. , Liberibacter crescens is a cultured surrogate for functional genomics of uncultured pathogenic ‘Candidatus Liberibacter’ spp. and is naturally competent for transformation. Phytopathology 10.1094/phyto-04-19-0129-r (2019). [DOI] [PubMed] [Google Scholar]
- 16.Merfa M. V., et al. , Progress and obstacles in culturing ‘Candidatus Liberibacter asiaticus’, the bacterium associated with huanglongbing. Phytopathology 109, 1092–1101 (2019). [DOI] [PubMed] [Google Scholar]
- 17.Naranjo E., et al. , Liberibacter crescens biofilm formation in vitro: Establishment of a model system for pathogenic ‘Candidatus Liberibacter spp.’ Sci. Rep. 9, 5150 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cruz-Munoz M., et al. , Development of chemically defined media reveals citrate as preferred carbon source for Liberibacter growth. Front. Microbiol. 9, 668 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Johnson B. K., Abramovitch R. B., Small molecules that sabotage bacterial virulence. Trends Pharmacol. Sci. 38, 339–362 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Munguia J., Nizet V., Pharmacological targeting of the host-pathogen interaction: Alternatives to classical antibiotics to combat drug-resistant superbugs. Trends Pharmacol. Sci. 38, 473–488 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Garrity-Ryan L. K., et al. , Small molecule inhibitors of LcrF, a Yersinia pseudotuberculosis transcription factor, attenuate virulence and limit infection in a murine pneumonia model. Infect. Immun. 78, 4683–4690 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gotoh Y., et al. , Two-component signal transduction as potential drug targets in pathogenic bacteria. Curr. Opin. Microbiol. 13, 232–239 (2010). [DOI] [PubMed] [Google Scholar]
- 23.Mandal R. S., et al. , Ribavirin suppresses bacterial virulence by targeting LysR-type transcriptional regulators. Sci. Rep. 6, 39454 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ng W. L., Perez L., Cong J., Semmelhack M. F., Bassler B. L., Broad spectrum pro-quorum-sensing molecules as inhibitors of virulence in vibrios. PLoS Pathog. 8, e1002767 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.El-Mowafi S. A., et al. , Identification of inhibitors of a bacterial sigma factor using a new high-throughput screening assay. Antimicrob. Agents Chemother. 59, 193–205 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pagliai F. A., Gonzalez C. F., Lorca G. L., Identification of a ligand binding pocket in LdtR from Liberibacter asiaticus. Front. Microbiol. 6, 1314 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pan L., Gardner C. L., Pagliai F. A., Gonzalez C. F., Lorca G. L., Identification of the tolfenamic acid binding pocket in PrbP from Liberibacter asiaticus. Front. Microbiol. 8, 1591 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Collier J., Cell cycle control in Alphaproteobacteria. Curr. Opin. Microbiol. 30, 107–113 (2016). [DOI] [PubMed] [Google Scholar]
- 29.Yan Q., et al. , Global gene expression changes in Candidatus Liberibacter asiaticus during the transmission in distinct hosts between plant and insect. Mol. Plant Pathol. 14, 391–404 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Barnett M. J., Bittner A. N., Toman C. J., Oke V., Long S. R., Dual RpoH sigma factors and transcriptional plasticity in a symbiotic bacterium. J. Bacteriol. 194, 4983–4994 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.de Lucena D. K., Pühler A., Weidner S., The role of sigma factor RpoH1 in the pH stress response of Sinorhizobium meliloti. BMC Microbiol. 10, 265 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mitsui H., Sato T., Sato Y., Ito N., Minamisawa K., Sinorhizobium meliloti RpoH1 is required for effective nitrogen-fixing symbiosis with alfalfa. Mol. Genet. Genomics 271, 416–425 (2004). [DOI] [PubMed] [Google Scholar]
- 33.Lehman A. P., Long S. R., OxyR-dependent transcription response of Sinorhizobium meliloti to oxidative stress. J. Bacteriol. 200, e00622-17 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bittner A. N., Oke V., Multiple groESL operons are not key targets of RpoH1 and RpoH2 in Sinorhizobium meliloti. J. Bacteriol. 188, 3507–3515 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sourjik V., Muschler P., Scharf B., Schmitt R., VisN and VisR are global regulators of chemotaxis, flagellar, and motility genes in Sinorhizobium (Rhizobium) meliloti. J. Bacteriol. 182, 782–788 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Andrade M. O., Wang N., The Tad pilus apparatus of Candidatus Liberibacter asiaticus and its regulation by VisNR. Mol. Plant Microbe Interact. 10.1094/mpmi-02-19-0052-r (2019). [DOI] [PubMed] [Google Scholar]
- 37.Rotter C., Mühlbacher S., Salamon D., Schmitt R., Scharf B., Rem, a new transcriptional activator of motility and chemotaxis in Sinorhizobium meliloti. J. Bacteriol. 188, 6932–6942 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Coyle J. F., Pagliai F. A., Zhang D., Lorca G. L., Gonzalez C. F., Purification and partial characterization of LdtP, a cell envelope modifying enzyme in Liberibacter asiaticus. BMC Microbiol. 18, 201 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pagliai F. A., Coyle J. F., Kapoor S., Gonzalez C. F., Lorca G. L., LdtR is a master regulator of gene expression in Liberibacter asiaticus. Microb. Biotechnol. 10, 896–909 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pini F., et al. , Cell cycle control by the master regulator CtrA in Sinorhizobium meliloti. PLoS Genet. 11, e1005232 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Janakiraman B., Mignolet J., Narayanan S., Viollier P. H., Radhakrishnan S. K., In-phase oscillation of global regulons is orchestrated by a pole-specific organizer. Proc. Natl. Acad. Sci. U.S.A. 113, 12550–12555 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tang G., Wang Y., Luo L., Transcriptional regulator LsrB of Sinorhizobium meliloti positively regulates the expression of genes involved in lipopolysaccharide biosynthesis. Appl. Environ. Microbiol. 80, 5265–5273 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Charoenpanich P., Meyer S., Becker A., McIntosh M., Temporal expression program of quorum sensing-based transcription regulation in Sinorhizobium meliloti. J. Bacteriol. 195, 3224–3236 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Reeve W. G., Tiwari R. P., Wong C. M., Dilworth M. J., Glenn A. R., The transcriptional regulator gene phrR in Sinorhizobium meliloti WSM419 is regulated by low pH and other stresses. Microbiology 144, 3335–3342 (1998). [DOI] [PubMed] [Google Scholar]
- 45.Niesen F. H., Berglund H., Vedadi M., The use of differential scanning fluorimetry to detect ligand interactions that promote protein stability. Nat. Protoc. 2, 2212–2221 (2007). [DOI] [PubMed] [Google Scholar]
- 46.Gotoh Y., et al. , Novel antibacterial compounds specifically targeting the essential WalR response regulator. J. Antibiot. (Tokyo) 63, 127–134 (2010). [DOI] [PubMed] [Google Scholar]
- 47.Skredenske J. M., et al. , Identification of a small-molecule inhibitor of bacterial AraC family activators. J. Biomol. Screen. 18, 588–598 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hurt J. K., McQuade T. J., Emanuele A., Larsen M. J., Garcia G. A., High-throughput screening of the virulence regulator VirF: A novel antibacterial target for shigellosis. J. Biomol. Screen. 15, 379–387 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kim B. S., et al. , QStatin, a selective inhibitor of quorum sensing in Vibrio species. MBio 9, e02262-17 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.National Academies of Sciences, Engineering, and Medicine , A Review of the Citrus Greening Research and Development Efforts Supported by the Citrus Research and Development Foundation: Fighting a Ravaging Disease (The National Academies Press, Washington, DC, 2018). [Google Scholar]
- 51.National Research Council , Strategic Planning for the Florida Citrus Industry: Addressing Citrus Greening Disease (The National Academies Press, Washington, DC, 2010). [Google Scholar]
- 52.Khan S. R., Gaines J., Roop R. M. 2nd, Farrand S. K., Broad-host-range expression vectors with tightly regulated promoters and their use to examine the influence of TraR and TraM expression on Ti plasmid quorum sensing. Appl. Environ. Microbiol. 74, 5053–5062 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Barnett M. J., Toman C. J., Fisher R. F., Long S. R., A dual-genome Symbiosis Chip for coordinate study of signal exchange and development in a prokaryote-host interaction. Proc. Natl. Acad. Sci. U.S.A. 101, 16636–16641 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Luo L., et al. , Two new Sinorhizobium meliloti LysR-type transcriptional regulators required for nodulation. J. Bacteriol. 187, 4562–4572 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.