Significance
Proliferation and differentiation are inversely correlated processes in many biological systems. Pu.1 is an ETS family transcription factor that promotes proliferation of erythroid progenitors and blocks their terminal differentiation. Downregulation of Pu.1 in erythroid progenitors triggers their terminal differentiation. The current work investigates the DNA sequences and transcription factors that regulate Pu.1 expression in erythroid progenitors. The results show that Runx1, acting through an upstream regulatory element, controls Pu.1 transcription in erythroid progenitors. Moreover, genome-wide analyses show that Runx1 and Pu.1 play pivotal roles in shaping the chromatin landscape during early erythropoiesis. The importance of Runx1 and Pu.1 in erythroid progenitors is further demonstrated by finding that ectopic expression of either factor in erythroid progenitors leads to their immortalization.
Keywords: gene regulation, erythropoiesis, transcription, Pu.1, Runx1
Abstract
Pu.1 is an ETS family transcription factor (TF) that plays critical roles in erythroid progenitors by promoting proliferation and blocking terminal differentiation. However, the mechanisms controlling expression and down-regulation of Pu.1 during early erythropoiesis have not been defined. In this study, we identify the actions of Runx1 and Pu.1 itself at the Pu.1 gene Upstream Regulatory Element (URE) as major regulators of Pu.1 expression in Burst-Forming Unit erythrocytes (BFUe). During early erythropoiesis, Runx1 and Pu.1 levels decline, and chromatin accessibility at the URE is lost. Ectopic expression of Runx1 or Pu.1, both of which bind the URE, prevents Pu.1 down-regulation and blocks terminal erythroid differentiation, resulting in extensive ex vivo proliferation and immortalization of erythroid progenitors. Ectopic expression of Runx1 in BFUe lacking a URE fails to block terminal erythroid differentiation. Thus, Runx1, acting at the URE, and Pu.1 itself directly regulate Pu.1 levels in erythroid cells, and loss of both factors is critical for Pu.1 down-regulation during terminal differentiation. The molecular mechanism of URE inactivation in erythroid cells through loss of TF binding represents a distinct pattern of Pu.1 regulation from those described in other hematopoietic cell types such as T cells which down-regulate Pu.1 through active repression. The importance of down-regulation of Runx1 and Pu.1 in erythropoiesis is further supported by genome-wide analyses showing that their DNA-binding motifs are highly overrepresented in regions that lose chromatin accessibility during early erythroid development.
Although cells express many transcription factors (TFs), the actions of a few TFs are critical for establishing and maintaining cellular identities. Two such factors in hematopoietic cells are Pu.1 and Gata-1. Pu.1 is an ETS family TF that is required for the generation and differentiation of myeloid cells, B cells, and T cells (1). Gata-1 is a Zn-finger TF that is required for the development of erythrocytes and the production of normal platelets (2, 3). Pu.1 and Gata-1 physically interact and repress each other’s transcriptional activation and lineage specification (4–6). In erythroid progenitors, Pu.1 controls an extensive network of genes involved in cell growth and survival (7). Indeed, mice with low Pu.1 levels have reduced numbers of erythroid progenitors (7), and Pu.1 has been shown to be critical for the proliferation of erythroid progenitors ex vivo (8). In addition, Pu.1 represses a core erythroid transcriptional network controlled by Gata-1, Klf1, and Tal1 (9). Consequently, down-regulation of Pu.1 is a key step in de-repression of this network, allowing erythroid progenitors to undergo terminal differentiation (4, 10). Despite these critical roles for Pu.1 in erythroid progenitors, the mechanisms controlling its expression in these cells are unknown.
Regulation of Pu.1 expression has been extensively studied in nonerythroid hematopoietic cells. A key enhancer of Pu.1 expression in myeloid cells and B cells is the so-called upstream regulatory element (URE) (11–18), located 14 kb upstream of the Pu.1 transcription start site (TSS). Deletion of the URE leads to a large reduction of Pu.1 in myeloid cells, and mice lacking the URE develop acute myeloid leukemia (14). Conversely, in the T cell lineage in which Pu.1 down-regulation is required for maturation of thymocyte progenitors (19), the URE is necessary for repression of Pu.1 (20), and mice lacking the URE exhibit disrupted T cell development, demonstrating key cell-type–specific differences in URE activity. In addition to the URE, a number of additional cell-type–specific regulatory elements have been discovered between the URE and the Pu.1 TSS such as additional enhancers in myeloid cells (13, 21) and a silencer in T cells (21).
A number of TFs have been identified that bind the Pu.1 URE in a cell-type–specific manner in B cells (13, 16), T cells (21), and myeloid cells (13, 15). In addition, certain factors, including Runx1, Satb1, and Pu.1 itself have been shown to be necessary for URE activity in multiple cell types (12, 16, 17, 21–24), but the TFs regulating Pu.1 expression in erythroid cells are not known.
In the current work, we investigate the cis-regulatory DNA sequences and transacting factors that govern Pu.1 expression in erythroid progenitors. Our results show that Runx1 and Pu.1 itself are major regulators of Pu.1 expression in erythroid progenitors. Down-regulation of Runx1 with erythroid differentiation coincides with dramatic, progressive remodeling of the URE and down-regulation of Pu.1, leading to a cascade that irreversibly results in terminal erythroid differentiation. Ectopic expression of Runx1 in BFUe prevents both Pu.1 down-regulation and terminal erythroid differentiation, in a URE-dependent fashion. We took advantage of these findings to determine whether Runx1 could be used to immortalize erythroid progenitors. We found that ectopic expression of Runx1 leads to continuous proliferation of erythroid progenitors for at least 4 mo. Thus, the proliferation versus terminal differentiation decision in erythroid progenitors is governed by Runx1-mediated transregulation of Pu.1.
Materials and Methods
ATAC Sequencing.
Chromatin accessibility was assayed using ATAC-seq as previously described (25) (SRA archive PRJNA491493).
Mice and Primary Cell Isolation.
Primary mouse hematopoietic cells were isolated from E14.5 murine fetal livers by FACS using markers to identify each population (SI Appendix, Tables S1 and S2).
Pu.1 TF Correlation Analysis in scRNAseq.
Cell fate was assigned using a clustering-based approach from the authors’ original analysis (Spring plots) (26, 27). Pu.1 Pearson correlation analysis was performed for all cells from multipotent progenitors (MPP) to BFUe. Enrichment or depletion of TF expression in Pu.1-expressing cells was determined by calculating the residual of the linear model for the frequency of TF expression in Pu.1+ cells versus the frequency of TF expression in Pu.1– cells.
Statistical Analysis.
Data were analyzed and plotted in GraphPad Prism. Samples were compared using the unpaired Student’s t test, and P < 0.05 was considered significant unless stated otherwise. Extended materials and methods are provided in SI Appendix.
Results
Chromatin Accessibility of the Pu.1 Locus Changes during Erythropoiesis.
To study Pu.1 regulation during erythropoiesis, we isolated Kit+Sca+Lin– hematopoietic progenitors (KSL), common myeloid progenitors (CMP), Burst-Forming Unit erythrocytes (BFUe), and late Colony-Forming Unit erythrocytes (CFUe) from murine E14.5 fetal liver using a fluorescence-activated cell sorting (FACS) protocol (28) that minimizes contamination of BFUe with KSL, CMP, and granulocyte-monocyte progenitors (GMP). The purified BFUe population forms a mixture of large immature BFUe colonies and smaller late BFUe/early multi-CFUe colonies in methylcellulose. Pu.1 mRNA levels are dramatically and progressively down-regulated as cells differentiate from CMP to BFUe to late CFUe (SI Appendix, Fig. S1A). Consistent with previous reports (7, 8, 10), the frequency of BFUe is greatly reduced in fetal livers from embryos lacking a functional copy of Pu.1 (SI Appendix, Fig. S1B) and Pu.1 null BFUe proliferate ex vivo much less than control BFUe (SI Appendix, Fig. S1C). Conversely, we found that ectopic expression of Pu.1 blocked BFUe differentiation (SI Appendix, Fig. S1D). These results support the view that Pu.1 is expressed in BFUe, required for their proliferation, and must be down-regulated to allow them to differentiate. In contrast, the frequency of late CFUe was not affected by loss of Pu.1 (SI Appendix, Fig. S1B), consistent with a previous report indicating that the number of CFUe is not affected by Pu.1 loss at E13.5, although it is affected at earlier and later stages (8).
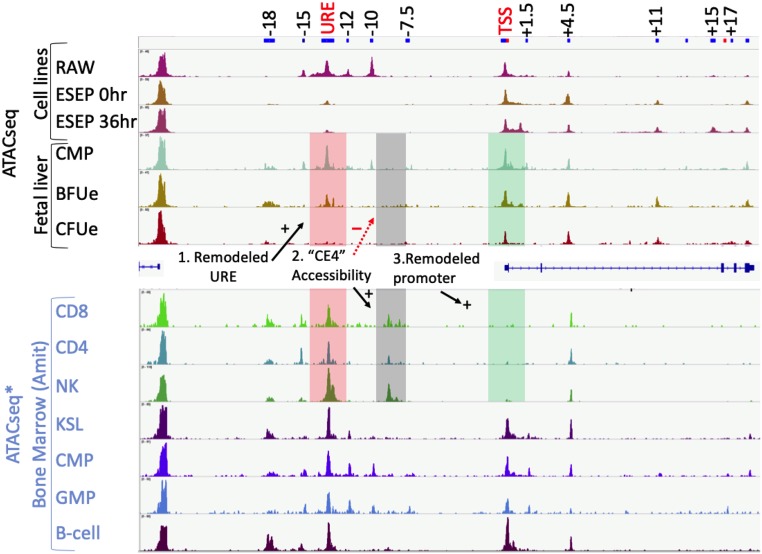
To study changes in chromatin accessibility at the Pu.1 locus during erythroid commitment, we performed ATACseq in fetal liver CMP, BFUe, and late CFUe as well as a macrophage cell line (RAW 264.7) and embryonic stem cell-derived erythroid progenitors (ESEPs) (29) that can be triggered to differentiate, whereupon Pu.1 mRNA and protein levels decline rapidly (SI Appendix, Fig. S2 A–E). Both CMP and RAW 264.7 have high levels of Pu.1 mRNA and multiple chromatin-accessible regions upstream of the Pu.1 TSS, including regions of accessibility near −15, −12, and −10 kb and a broad region of accessibility near −14 kb corresponding to the URE (Fig. 1). Accessibility of the URE, −15-, −12-, and −10-kb regions is markedly reduced in BFUe and ESEP cells. Chromatin accessibility at the URE is completely lost upon differentiation of BFUe to late CFUe (Fig. 1). These results indicate that loss of Pu.1 during erythropoiesis is associated with reduced chromatin accessibility in several regions upstream of the Pu.1 gene, including at the strong Pu.1 enhancer, the URE. There is also a small reduction in chromatin accessibility at the Pu.1 promoter in late CFUe compared to BFUe, but the extent of reduction is much less than that observed at the URE, suggesting that changes in factor binding affecting Pu.1 transcription in these cells occur within distal cis-regulatory regions rather than at the Pu.1 promoter. Interestingly, we also detected chromatin accessibility in regions within the Pu.1 gene body, near +1.5, +4.5, +11 kb downstream of the Pu.1 TSS. These regions exhibit increased accessibility in BFUe compared to CMP (Fig. 1).
Fig. 1.
Chromatin accessibility of the Pu.1 locus changes during erythropoiesis. (Top) ATACseq was performed in CMP, BFUe and late CFUe, RAW 264.7, and ESEP before and after terminal erythroid differentiation. The differences in chromatin accessibility changes at the URE (−14 kb), the “CE4” silencer element, and the Pu.1 TSS between CMP/BFUe/late CFUe versus CD4/CD8/NK cells are highlighted. (Bottom) ATACseq data from ref. 30 in multiple bone marrow populations.
Although the observed URE accessibility in purified BFUe could be due to small numbers of contaminating MPP, CMP, or GMP, it is unlikely because the total nonerythroid cell contamination level is estimated to be only 2.5% (28), which could not produce the level of accessibility we observed. Furthermore, several erythroid cell lines exhibit URE accessibility including MEL (ENCODE: ENCDO073AAA), G1E (ENCBS324ENC), and K562 (ENCSR000EPC), clearly indicating that URE accessibility is maintained in fully committed erythroid cells.
We also compared our chromatin accessibility data with ATACseq data in 7 hematopoietic cell types from bone marrow including CD8+ and CD4+ T cells (30). In erythroid cells, chromatin accessibility at the URE gradually decreases, and the Pu.1 promoter remains accessible, whereas in T cells the URE and other upstream regulatory elements are accessible, and the Pu.1 promoter is inaccessible (Fig. 1). Furthermore, erythroid cells lack a hypersensitive site “CE4” at −9 kb, which has been shown to act as a strong silencer of a Pu.1 reporter in T cells (21).
Analysis of ChIP-seq data from immature (Ter119–) and differentiating (Ter119+) fetal liver cells (31) showed that both the Pu.1 promoter and the URE are occupied by nucleosomes containing H3K9ac and H3K4me3 in Ter119– cells, whereas these post-translational modifications (PTMs) are greatly diminished in differentiating Ter119+ erythroid cells (SI Appendix, Fig. S3A). H3K9ac and H3K4me3 PTMs are associated with active promoters and enhancers, whereas chromatin without these modifications is most often observed within inactive or repressed chromatin. Together with the ATACseq data, this analysis indicates that the URE is accessible and bears a chromatin signature associated with active enhancers in immature erythroid progenitors but not in more mature erythroid cells. In contrast, the several regions within the Pu.1 gene body that become more accessible during erythroid differentiation do not contain PTMs typical of active enhancers (SI Appendix, Fig. S3A), nor do they exhibit repressive chromatin marks such as H3K27me3 (32). Thus, unlike T cell progenitors that maintain chromatin accessibility at the Pu.1 URE as they differentiate, during erythropoiesis, chromatin accessibility and PTMs associated with active chromatin are gradually lost from the URE, suggesting loss of its enhancer function.
To determine if Pu.1 is transcriptionally down-regulated during erythroid differentiation, we measured the levels of the Pu.1 unspliced primary transcript by qRT-PCR at 19 positions within the Pu.1 gene during ESEP differentiation (SI Appendix, Fig. S3 B and C). The level of the primary transcript at each intronic position declined markedly during differentiation. This uniform decline in transcript level at each position, like that obtained after treatment with the transcription initiation inhibitor 5,6-dichloro-1-beta-d-ribofuranosylbenzimidazole (DRB), suggests that transcriptional pausing is unlikely to contribute to the decline in the Pu.1 mRNA level. Thus, the decline in Pu.1 mRNA during terminal erythroid differentiation is most likely due to decreased transcription initiation rather than an interruption in transcriptional elongation or at later posttranscriptional steps.
URE Is Important for Production of BFUe In Vivo, Their Proliferation Ex Vivo, and Pu.1 Expression.
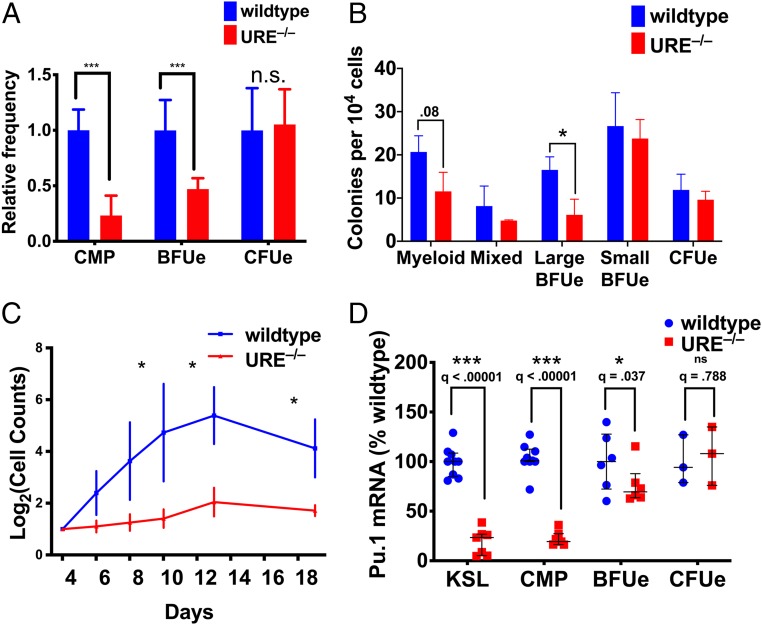
The URE exhibits a gradual loss of chromatin accessibility from CMP to BFUe to late CFUe, correlating with the decline in Pu.1 expression (Fig. 1). To determine how the URE affects BFUe cellular function and Pu.1 mRNA levels, we used a mouse strain lacking the URE (14). We observed a significant decrease in the frequency of CMP and BFUe but not late CFUe in fetal livers of URE−/− mice (Fig. 2A). Similarly, fetal liver cells from URE−/− mice produced significantly fewer large BFUe colonies in methylcellulose, whereas smaller late BFUe colonies and CFUe colonies were not reduced (Fig. 2B). BFUe from URE−/− mice also produce 10-fold fewer cells than control BFUe during ex vivo proliferation (Fig. 2C). The Pu.1 mRNA level is also reduced in URE−/− BFUe, although it is not as severely affected as in URE−/− CMP (Fig. 2D). Thus, despite the reduction in chromatin accessibility during the transition from CMP to BFUe, the URE remains a critical regulatory element for in vivo production and ex vivo proliferation of BFUe.
Fig. 2.
The URE is necessary for the normal generation of BFUe in vivo and BFUe proliferation ex vivo. (A) The frequency of each cell type was measured by FACS in E14.5 fetal livers of wild-type and URE−/− embryos. (B) CFUe colonies were grown in methylcellulose supplemented with Epo and counted at day 3. All other colonies were grown in M3234 supplemented with Epo, IL-3, IL-6, Dex, and SCF and counted after 9 d (n = 3, wild type; n = 2, URE−/−). (C) BFUe from E14.5 fetal livers of wild-type and URE−/− embryos were cultured in “proliferation medium” and counted at the indicated times (n = 3). (D) KSL, CMP, BFUe, and CFUe from E14.5 fetal livers of wild-type and URE−/− embryos; Pu.1 mRNA levels were measured by RT-qPCR (n = 6). Data were analyzed by t test using a false discovery rate of 10%. ns, not statistically significant; * q ≤ 0.05; *** q < 0.001.
We also used CRISPR–Cas9-mediated DNA deletion in ESEP to determine the effect on Pu.1 expression of removing several of the regions (at +1.5, +4.5, +15, and +17 kb) within the Pu.1 gene body that become accessible during erythroid differentiation (SI Appendix, Figs. S4 and S5A). None of the deletions altered Pu.1 mRNA levels under proliferating or differentiating conditions (SI Appendix, Fig. S5 B and C), and cells harboring individual deleted regions differentiated normally (SI Appendix, Fig. S5D). Thus, even though chromatin accessibility of these regions seems to be specific for erythroid cells, and a subset of them become more accessible during terminal differentiation, they are dispensable for normal Pu.1 regulation in ESEP.
Forced Expression of Runx1 and Pu.1 Prevents Pu.1 Down-Regulation and Blocks Terminal Erythroid Differentiation.
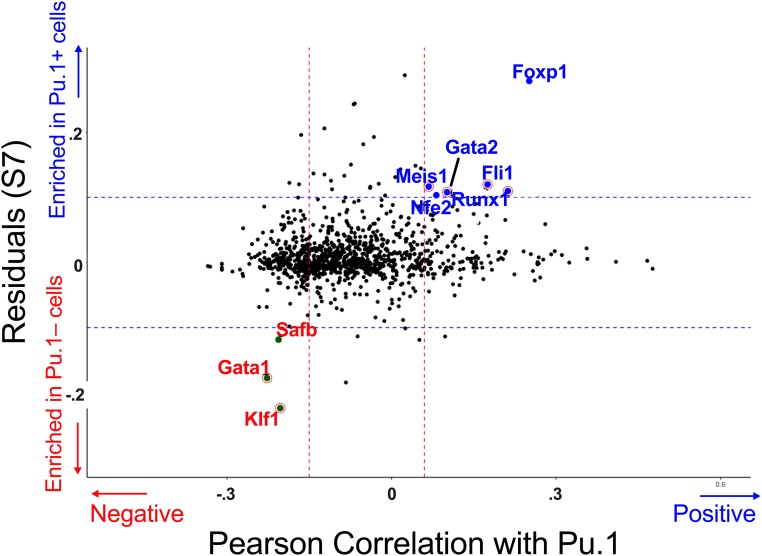
Changes in enhancer activity are likely accompanied by a loss of key effectors or a gain of repressive factors that interfere with enhancer activity. We used single-cell RNAseq data (scRNAseq) from Multipotent Progenitors (MPP) through late CFUe (26) to identify candidate regulators of Pu.1 by correlating gene expression of TFs with Pu.1 expression level. scRNAseq data were separated into 100 successive bins based on stage of erythroid differentiation using the original “Spring” clustering algorithm (26) (SI Appendix, Fig. S6A). Bins containing MPP, BFUe, and late CFUe were defined by lineage commitment and mRNA expression of characteristic FACS markers (SI Appendix, Fig. S6B). We identified expression of Runx1, Fli1, Gata-2, Meis1, Foxp1, Pu.1, and Nfe2 as positively correlated and Gata-1, Klf1, and SAFB as negatively correlated with Pu.1 mRNA level (Fig. 3 and SI Appendix, Fig. S7).
Fig. 3.
Correlation analysis of TF mRNA with Pu.1 mRNA in fetal liver scRNAseq data. scRNAseq data in Kit+ fetal liver cells from ref. 26 and 27. TFs were subset from all other genes using GO annotations for “DNA-binding TF” (GO:0003700). x axis: For each TF the Pearson correlation of its mRNA level to that of Pu.1 mRNA was determined in scRNAseq data (26) across all cells from MPP to BFUe excluding cells biased toward nonerythroid lineages (Materials and Methods). y axis: The residual for each TF mRNA was plotted from the linear regression of TF mRNA frequency in Pu.1-expressing cells versus TF mRNA frequency in Pu.1-nonexpressing cells, e.g., the residual of SI Appendix, Fig. S7.
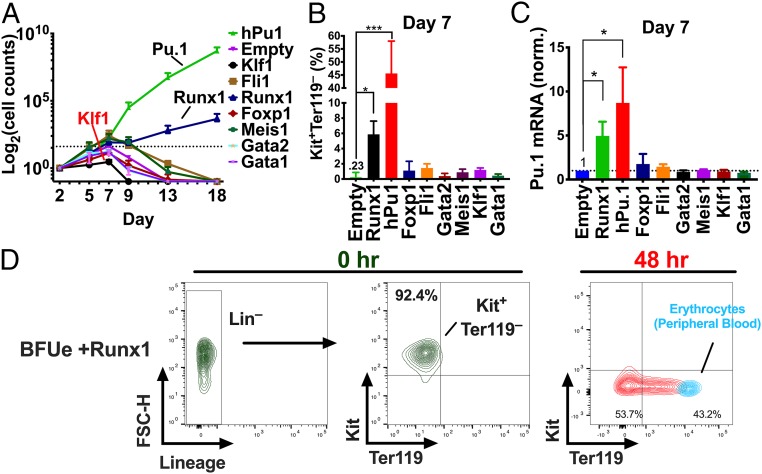
To determine whether any of these candidate regulators of Pu.1 can phenocopy ectopic expression of Pu.1, we expressed them individually in BFUe with recombinant lentiviruses and monitored proliferation for 18 d. Most cultures reached their maximum cell densities by 7 d, except those expressing human Pu.1 (hPu.1) or Runx1, which continued to proliferate for the entire length of the experiment (Fig. 4A). Moreover, in contrast to BFUe expressing the other candidates or the empty vector control, which down-regulated Kit protein and Pu.1 mRNA due to spontaneous differentiation, BFUe expressing hPu.1 or Runx1 exhibited increased levels of Kit protein (Fig. 4B) and mouse Pu.1 mRNA (Fig. 4C). Interestingly, ectopic expression of Klf1, Gata-1, Gata-2, and Foxp1 caused a reduction in the maximum number of cells produced at 7 d of culture (SI Appendix, Fig. 8A). The effect of these factors on proliferation is unlikely to be mediated through an effect on Pu.1, however, as BFUe expressing these TFs did not have reduced levels of Pu.1 expression compared with cells infected with an empty virus (Fig. 4C). Thus, of the 10 factors tested, only ectopic expression of Runx1 and Pu.1 altered the endogenous levels of Pu.1 mRNA and prevented terminal erythroid differentiation.
Fig. 4.
Ectopic expression of Runx1 and Pu.1 in BFUe prevents Pu.1 down-regulation and blocks terminal erythroid differentiation. BFUe were isolated from E14.5 fetal livers and infected with recombinant lentiviruses encoding the indicated TF. After 48 h of incubation in “proliferation medium,” GFP+ cells were collected by FACS and further incubated in the same medium. (A–C) Empty, Pu.1, and Runx1, n = 3; others, n = 2). (A) Cell counts were performed using FACS analysis at the indicated times. (B) The percentage of Kit+, Ter119- cells in each lentivirus-infected culture after 7 d of cell culture was determined by FACS. (C) After 7 d of cell culture, 10,000 viable cells were isolated by FACS and spiked with ∼100,000 Drosophila S2 cells, and Pu.1 mRNA levels were measured by RT-qPCR relative to drosophila actin. (D) BFUe ectopically expressing Runx1 were proliferated ex vivo for 4 mo in “proliferation medium.” Kit, Ter119, and expression of lineage markers (Gr1, CD11b, CD3e, B220) were measured before and after differentiation in media containing Epo, Mifepristone, and Insulin. Representative FACS plots are shown (n = 3). As a control, murine peripheral blood was stained for Kit and Ter119 and is shown in blue. *P ≤ 0.05; ***P < 0.001.
To determine whether ectopic expression of Runx1 and Pu.1 is sufficient to immortalize BFUe, we monitored cultures expressing each factor for several months. Both types of cultures continued to proliferate for 4 mo, producing Kit+Ter119– cells (Fig. 4D and SI Appendix, Fig. S8B). Nevertheless, both types of cells retained the ability to terminally differentiate in 48 to 72 h when transferred to differentiation media as measured by FACS analysis for Ter119 and Kit expression (Fig. 4D and SI Appendix, Fig. S8B) and benzidine staining for hemoglobin (SI Appendix, Fig. S8C). These cells also lost expression of GFP during differentiation, consistent with the global decrease in transcription that is characteristic of terminal erythroid differentiation (SI Appendix, Fig. S8D). The ability of these immortalized cells to terminally differentiate when placed in differentiation medium was surprising because Pu.1 readily blocks erythroid differentiation in short-term cultures (SI Appendix, Fig. S1D and ref. 10). This suggests the intriguing possibility that long-term proliferation of erythroid progenitors requires the collaboration of Pu.1 with the glucocorticoid receptor or another target of the cytokines which promote proliferation.
To determine if the immortalized cell lines that were initiated in BFUe maintained the ability to form BFUe colonies, we performed colony assays in methylcellulose. Both types of cell lines produced a mixture of large and small erythroid colonies after 8 to 9 d, suggesting that the proliferating cells are a mixture of early BFUe and late BFUe/early CFUe (SI Appendix, Fig. S8 E and F). Importantly, no nonerythroid or mixed colonies were observed, indicating that these cells are committed to the erythroid lineage.
Forced Expression of Runx1 Is Unable to Block Terminal Erythroid Differentiation in Mice Lacking the Pu.1 URE.
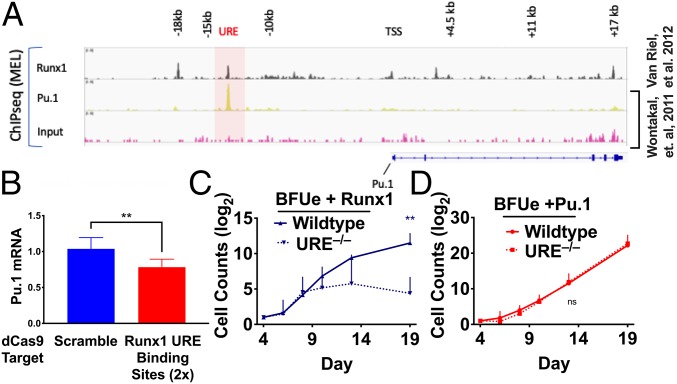
Consistent with the scRNAseq data, qRT-PCR analysis shows that the levels of both Runx1 and Pu.1 mRNAs decline during erythropoiesis (SI Appendix, Fig. S9). Forced expression of either factor in BFUe prevents down-regulation of endogenous Pu.1 mRNA (Fig. 4C). Runx1 also directly binds the Pu.1 URE in an erythroid cell line (Fig. 5A), similar to reports in several nonerythroid hematopoietic cell types (17, 33). To determine if the effect of Runx1 on Pu.1 expression is mediated through the URE in erythroid cells, we interfered with Runx1 binding to the URE in the human erythroid cell line K562 by targeting a catalytically inactive Cas9 (dCas9) to the Runx1 DNA-binding sites. Targeting of dCas9 to the Runx1-binding sites led to decreased Pu.1 expression compared with cells transfected with nontargeting guide RNA (gRNA) (Fig. 5B). To determine if the ability of Runx1 to block terminal erythroid differentiation is mediated through the URE, we tested the effect of ectopic expression of Runx1 in BFUe isolated from mice lacking the URE. Whereas wild-type BFUe ectopically expressing Runx1 begin to expand around day 6 after FACS sorting, ectopic expression of Runx1 in BFUe lacking the URE failed to promote continuous expansion, producing many fewer cells at all culture times after 10 d (Fig. 5C). As expected, loss of the URE had no effect on BFUe ectopically expressing Pu.1 (Fig. 5D). The increase we observe in proliferation of URE−/− BFUe ectopically expressing Runx1 without the URE suggests that Runx1 is able to modestly increase proliferation through activities independent of the URE, some of which we identify in the next section through gene set enrichment. Nevertheless, these results indicate that URE binding by Runx1 is required to maintain Pu.1 expression and block terminal erythroid differentiation.
Fig. 5.
Runx1 directly regulates Pu.1 and terminal differentiation through the URE. (A) ChIPseq data for Runx1 in MEL cells from ref. 39. ChIPseq data for Pu.1 in MEL cells were obtained from our previous study (7). (B) K562 cells were electroporated with plasmid vectors encoding dCas9–Cherry and either BFP and a scrambled gRNA or BFP and a gRNA targeting Runx1-binding motifs within the URE. After 48 h, 10,000 BFP+Cherry+ cells were isolated by FACS, and Pu.1 mRNA levels were measured by RT-qPCR. (C and D) BFUe were isolated from wild-type and URE−/− mice. The cells were infected with recombinant lentiviruses expressing GFP Runx1 or hPu.1 or a control (empty) vector. After 72 h of culture, GFP+ cells were isolated by FACS, and cells were further incubated in “proliferation medium.” Cell counts were performed at the indicated times by FACS analysis. Statistically significant; **P < 0.01.
Runx1 and Pu.1 Are Major Contributors to the Chromatin Landscape of Early Erythroid Cells.
To further support the importance of Runx1 and Pu.1 in erythropoiesis, we conducted an unbiased genome-wide analysis of the chromatin changes that occur during early erythroid development. We identified ∼40,000 regions that lose accessibility and ∼15,000 regions that become accessible during commitment to erythroid differentiation (CMP to BFUe) and during terminal erythroid differentiation (BFUe to late CFUe; SI Appendix, Fig. S10).
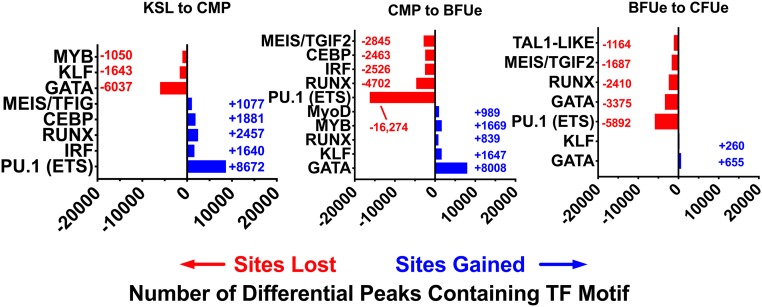
To determine the TFs that could contribute to these changes in chromatin structure, we used HOMER (34) to search each of the dynamic regions of chromatin for enrichment of TF motifs (34). The ETS motif, to which Pu.1 binds, and the Runx motif were the most overrepresented DNA-binding sequences within chromatin sites that become inaccessible during the 2 transitions (Fig. 6). On the other hand, sites that changed from closed to open were dominated by DNA-binding sequences bound by Gata-1 and Klf1. Thus, the majority of changes in chromatin structure during erythropoiesis reflect a loss of sites containing Pu.1 and Runx motifs, consistent with the down-regulation of these TFs (SI Appendix, Fig. S9), and a gain of sites containing Gata-1 and Klf1 motifs.
Fig. 6.
Loss of chromatin-accessible regions containing the Runx and Pu.1 ETS motif is the dominant chromatin change during erythropoiesis. Motif analysis was performed as in Materials and Methods for differential peaks that increase or decrease from KSL to late CFUe.
Because of the relatively large number of sites containing the ETS and Runx motifs that change between CMP and late CFUe (Fig. 6), we asked whether Runx1 and Pu.1 target an overlapping set of genes and cis-regulatory elements in immature erythroid cells. By overlapping ChIPseq data in murine erythroleukemia cells (MEL) for Runx1 and Pu.1 with nearby target genes we determined that Pu.1 and Runx1 share 40% of target genes (SI Appendix, Fig. S11A). In addition, of the 7,027 Pu.1 ChIPseq-binding sites in MEL cells, 20% of the sites occur within a 1-kb interval of Runx1-binding sites (SI Appendix, Fig. S11B). Gene Ontology and KEGG pathway enrichment show that the shared set of Pu.1 and Runx1 target genes are enriched for erythroid-specific genes including Tal1, as well as pathways associated with proliferation, such as PI3K, and cell death (SI Appendix, Fig. S11C). These observations indicate that Runx1 and Pu.1 target many of the same genes and cis-regulatory elements in erythroid progenitors and that these genes promote proliferation and negatively regulate cell death.
Discussion
The results reported here identify the action of Runx1 at the Pu.1 URE as a major regulator of Pu.1 expression during the commitment to the erythroid lineage. As cells progress from CMP to BFUe, Runx1 RNA decreases, and chromatin accessibility of the Pu.1 URE begins to decline, leading to progressive inactivation of the URE, Pu.1 down-regulation, and terminal differentiation. Ectopic expression of Runx1 in wild-type BFUe, but not in BFUe lacking the Pu.1 URE, blocks terminal differentiation, indicating that Runx1 acts directly through the Pu.1 URE. Thus, whereas the importance of Runx1 in other hematopoietic lineages is well established (35–37), the results reported here demonstrate that Runx1 also plays a crucial regulatory role in definitive erythropoiesis. Moreover, our ATACseq data show that the combination of Runx and the Pu.1 (ETS) DNA-binding motifs account for ∼70% of enriched motifs within chromatin sites that are lost during the transition from CMP to late CFUe. These results highlight the pivotal role that both Runx1 and Pu.1 play in shaping the chromatin landscape during early erythropoiesis.
The detailed mechanisms uncovered here for regulating Pu.1 gene expression in erythroid cells exhibit both similarities and differences with the mechanisms used in other hematopoietic cells. Like CMP, macrophage, and B cells (11–17), deletion of the URE in BFUe results in decreased Pu.1 expression as well as a marked decrease in the frequency of fetal liver BFUe. On the other hand, despite the importance of the URE in BFUe, the level of Pu.1 gene expression in BFUe is much lower than in these other cell types. Explanations for this difference may reside in the composition of enhancing factor complexes at the URE in these other cells versus BFUe, their level of occupancy as reflected in lower accessibility of the URE in BFUe, or in the enhancing effects of complexes occupying the −15, −12, and −10 kb regions in the other cells, compared with BFUe in which these regions appear to be unoccupied. The importance of the URE for regulation of Pu.1 in erythroid cells is also highlighted by the ATACseq analyses showing that among 9 primary hematopoietic cell types and 2 cell lines, terminally differentiating erythroid cells are the only ones that lose accessibility at the URE. Another study using an erythroid cell line suggested that a change in the occupancy of Gata factors, from Gata-2 to Gata-1, at the Pu.1 promoter is responsible for down-regulation of Pu.1 during terminal differentiation (38). However, we were unable to detect any change in Pu.1 mRNA levels after ectopic expression of either Gata factor in BFUe.
A particularly striking contrast in the mechanisms for regulating Pu.1 expression can be found in a comparison of erythroid cells and immature T cells that also down-regulate Pu.1 gene expression (19). Whereas BFUe use a passive mechanism to turn off Pu.1 transcription, namely, loss of Runx1 and Pu.1 itself, immature T cells use an active repression mechanism dependent upon the URE (20) and a T cell-specific repressor element (21), designated “CE4,” lying 9 kb upstream of the Pu.1 TSS. This element has been shown to be necessary for repression of the Pu.1 locus in T cell reporter assays. It also binds Runx1 and several other TFs in T cells. Interestingly, “CE4” exhibits chromatin accessibility only in T cells and NK cells (Fig. 1). Presumably, “CE4” does not exhibit chromatin accessibility in other hematopoietic cells because they lack T cell-specific factors necessary for protein complex binding in this region.
The importance of Runx1 for Pu.1 regulation in erythroid progenitors is further highlighted by our findings that ectopic expression of either factor in BFUe leads to immortalization of erythroid progenitors. This function of Runx1 depends on the Pu.1 URE. While Pu.1 was sufficient to block terminal erythroid differentiation in short-term cultures lacking SCF, Dex, and IGF1, we found that long-term proliferation required stimulatory cytokines, perhaps indicating a complementary function of one or more of these factors with Pu.1. This dependence also provided a convenient method to trigger terminal erythroid differentiation by simply transferring these cells into differentiation medium lacking these cytokines. These results suggest that manipulating Runx1 or Pu.1 expression in primary human erythroid cells might lead to unlimited expansion of erythroid progenitors that can be triggered at will to differentiate into human erythrocytes for blood transfusions and other therapeutic applications.
Supplementary Material
Acknowledgments
We thank Drs. Merav Socolovsky and Allon Klein for sharing their scRNAseq data with us well in advance of publication. We also thank Drs. Britta Will and Teresa Bowman for their expert advice and critical feedback on the manuscript; and members of the A.I.S. Laboratory and Cary Weiss, Daqian Sun, Matt Gamble, Charles Query, and Alyssa Casill for scientific discussion and reagents. We also thank the Einstein Flow Cytometry core facility (Grant P30CA013330).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The sequences reported in this paper have been deposited in the National Center for Biotechnology Information (NCBI) data repository under BioProject PRJNA491493 and can be found at https://www.ncbi.nlm.nih.gov/.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1901122116/-/DCSupplemental.
References
- 1.McKercher S. R., et al. , Targeted disruption of the PU.1 gene results in multiple hematopoietic abnormalities. EMBO J. 15, 5647–5658 (1996). [PMC free article] [PubMed] [Google Scholar]
- 2.Pevny L., et al. , Erythroid differentiation in chimaeric mice blocked by a targeted mutation in the gene for transcription factor GATA-1. Nature 349, 257–260 (1991). [DOI] [PubMed] [Google Scholar]
- 3.Orkin S. H., Shivdasani R. A., Fujiwara Y., McDevitt M. A., Transcription factor GATA-1 in megakaryocyte development. Stem Cells 16 (suppl. 2), 79–83 (1998). [DOI] [PubMed] [Google Scholar]
- 4.Rekhtman N., Radparvar F., Evans T., Skoultchi A. I., Direct interaction of hematopoietic transcription factors PU.1 and GATA-1: Functional antagonism in erythroid cells. Genes Dev. 13, 1398–1411 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nerlov C., Querfurth E., Kulessa H., Graf T., GATA-1 interacts with the myeloid PU.1 transcription factor and represses PU.1-dependent transcription. Blood 95, 2543–2551 (2000). [PubMed] [Google Scholar]
- 6.Zhang P., et al. , Negative cross-talk between hematopoietic regulators: GATA proteins repress PU.1. Proc. Natl. Acad. Sci. U.S.A. 96, 8705–8710 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wontakal S. N., et al. , A large gene network in immature erythroid cells is controlled by the myeloid and B cell transcriptional regulator PU.1. PLoS Genet. 7, e1001392 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Back J., Dierich A., Bronn C., Kastner P., Chan S., PU.1 determines the self-renewal capacity of erythroid progenitor cells. Blood 103, 3615–3623 (2004). [DOI] [PubMed] [Google Scholar]
- 9.Wontakal S. N., et al. , A core erythroid transcriptional network is repressed by a master regulator of myelo-lymphoid differentiation. Proc. Natl. Acad. Sci. U.S.A. 109, 3832–3837 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pop R., et al. , A key commitment step in erythropoiesis is synchronized with the cell cycle clock through mutual inhibition between PU.1 and S-phase progression. PLoS Biol. 8, e1000484 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li Y., et al. , Regulation of the PU.1 gene by distal elements. Blood 98, 2958–2965 (2001). [DOI] [PubMed] [Google Scholar]
- 12.Okuno Y., et al. , Potential autoregulation of transcription factor PU.1 by an upstream regulatory element. Mol. Cell. Biol. 25, 2832–2845 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Leddin M., et al. , Two distinct auto-regulatory loops operate at the PU.1 locus in B cells and myeloid cells. Blood 117, 2827–2838 (2011). Correction in: Blood117, 5783 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rosenbauer F., et al. , Acute myeloid leukemia induced by graded reduction of a lineage-specific transcription factor, PU.1. Nat. Genet. 36, 624–630 (2004). [DOI] [PubMed] [Google Scholar]
- 15.Hoogenkamp M., et al. , The Pu.1 locus is differentially regulated at the level of chromatin structure and noncoding transcription by alternate mechanisms at distinct developmental stages of hematopoiesis. Mol. Cell. Biol. 27, 7425–7438 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Steidl U., et al. , A distal single nucleotide polymorphism alters long-range regulation of the PU.1 gene in acute myeloid leukemia. J. Clin. Invest. 117, 2611–2620 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huang G., et al. , PU.1 is a major downstream target of AML1 (RUNX1) in adult mouse hematopoiesis. Nat. Genet. 40, 51–60 (2008). [DOI] [PubMed] [Google Scholar]
- 18.Staber P. B., et al. , Sustained PU.1 levels balance cell-cycle regulators to prevent exhaustion of adult hematopoietic stem cells. Mol. Cell 49, 934–946 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Anderson M. K., Weiss A. H., Hernandez-Hoyos G., Dionne C. J., Rothenberg E. V., Constitutive expression of PU.1 in fetal hematopoietic progenitors blocks T cell development at the pro-T cell stage. Immunity 16, 285–296 (2002). [DOI] [PubMed] [Google Scholar]
- 20.Rosenbauer F., et al. , Lymphoid cell growth and transformation are suppressed by a key regulatory element of the gene encoding PU.1. Nat. Genet. 38, 27–37 (2006). [DOI] [PubMed] [Google Scholar]
- 21.Zarnegar M. A., Chen J., Rothenberg E. V., Cell-type-specific activation and repression of PU.1 by a complex of discrete, functionally specialized cis-regulatory elements. Mol. Cell. Biol. 30, 4922–4939 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen H., et al. , PU.1 (Spi-1) autoregulates its expression in myeloid cells. Oncogene 11, 1549–1560 (1995). [PubMed] [Google Scholar]
- 23.Leddin M., et al. , Two distinct auto-regulatory loops operate at the PU.1 locus in B cells and myeloid cells. Blood 117, 2827–2838 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Spooner C. J., Cheng J. X., Pujadas E., Laslo P., Singh H., A recurrent network involving the transcription factors PU.1 and Gfi1 orchestrates innate and adaptive immune cell fates. Immunity 31, 576–586 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Buenrostro J. D., Wu B., Chang H. Y., Greenleaf W. J., ATAC-seq: A method for assaying chromatin accessibility genome-wide. Curr. Protoc. Mol. Biol. 109, 21.29.1–21.29.9 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tusi B. K., et al. , Population snapshots predict early haematopoietic and erythroid hierarchies. Nature 555, 54–60 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Weinreb C., Wolock S., Tusi B. K., Socolovsky M., Klein A. M., Fundamental limits on dynamic inference from single-cell snapshots. Proc. Natl. Acad. Sci. U.S.A. 115, E2467–E2476 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Flygare J., Rayon Estrada V., Shin C., Gupta S., Lodish H. F., HIF1alpha synergizes with glucocorticoids to promote BFU-E progenitor self-renewal. Blood 117, 3435–3444 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Carotta S., et al. , Directed differentiation and mass cultivation of pure erythroid progenitors from mouse embryonic stem cells. Blood 104, 1873–1880 (2004). [DOI] [PubMed] [Google Scholar]
- 30.Lara-Astiaso D., et al. , Immunogenetics. Chromatin state dynamics during blood formation. Science 345, 943–949 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wong P., et al. , Gene induction and repression during terminal erythropoiesis are mediated by distinct epigenetic changes. Blood 118, e128–e138 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wu W., et al. , Dynamic shifts in occupancy by TAL1 are guided by GATA factors and drive large-scale reprogramming of gene expression during hematopoiesis. Genome Res. 24, 1945–1962 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schütte J., et al. , An experimentally validated network of nine haematopoietic transcription factors reveals mechanisms of cell state stability. eLife 5, e11469 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Heinz S., et al. , Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell 38, 576–589 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lam K., Zhang D. E., RUNX1 and RUNX1-ETO: Roles in hematopoiesis and leukemogenesis. Front. Biosci. 17, 1120–1139 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kuvardina O. N., et al. , RUNX1 represses the erythroid gene expression program during megakaryocytic differentiation. Blood 125, 3570–3579 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yokomizo T., et al. , Runx1 is involved in primitive erythropoiesis in the mouse. Blood 111, 4075–4080 (2008). [DOI] [PubMed] [Google Scholar]
- 38.Chou S. T., et al. , Graded repression of PU.1/Sfpi1 gene transcription by GATA factors regulates hematopoietic cell fate. Blood 114, 983–994 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.van Riel B., et al. , A novel complex, RUNX1-MYEF2, represses hematopoietic genes in erythroid cells. Mol. Cell. Biol. 32, 3814–3822 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.