Significance
Colonization of the mammalian gut by trillions of commensal bacteria starts early in life and contributes to host health. However, colonization by pathogens can be the launching pad for infection. Bacterial evolution has been studied mainly under laboratory conditions or in a disease context, but we understand less the evolutionary mechanisms of commensal bacteria in a healthy host microbiome. To fill this gap, we studied real-time gut colonization by Escherichia coli in healthy mice. We observe rapid evolution of the colonizing E. coli strain by bacteriophage-mediated horizontal gene transfer, which overrides the classical evolutionary mode of mutation accumulation. By experiments and mathematical modeling, we show that bacteriophage-mediated selection can produce complex evolutionary outcomes, including the emergence of a new commensal strain.
Keywords: bacteriophage, bacterial evolution, horizontal gene transfer, mutation, gut microbiota
Abstract
Bacteria evolve by mutation accumulation in laboratory experiments, but tempo and mode of evolution in natural environments are largely unknown. Here, we study the ubiquitous natural process of host colonization by commensal bacteria. We show, by experimental evolution of Escherichia coli in the mouse intestine, that the ecology of the gut controls the pace and mode of evolution of a new invading bacterial strain. If a resident E. coli strain is present in the gut, the invading strain evolves by rapid horizontal gene transfer (HGT), which precedes and outweighs evolution by accumulation of mutations. HGT is driven by 2 bacteriophages carried by the resident strain, which cause an epidemic phage infection of the invader. These dynamics are followed by subsequent evolution by clonal interference of genetically diverse lineages of phage-carrying (lysogenic) bacteria. We show that the genes uptaken by HGT enhance the metabolism of specific gut carbon sources and provide a fitness advantage to lysogenic invader lineages. A minimal dynamical model explains the temporal pattern of phage epidemics and the complex evolutionary outcome of phage-mediated selection. We conclude that phage-driven HGT is a key eco-evolutionary driving force of gut colonization—it accelerates evolution and promotes genetic diversity of commensal bacteria.
The human gut harbors a densely populated microbial ecosystem, whose most abundant members are nonpathogenic bacteria. These commensals help in nutrient and drug metabolism and defense against pathogens, thus contributing to host health (1, 2). Intraspecies variation of commensals can also be critical for host–microbiota homeostasis (3). Yet, our understanding of the evolution of commensals during colonization of healthy hosts is lagging far behind that of pathogens. Escherichia coli is a common colonizer of the human intestine, but also a potential pathogen. Comparative genomics studies show that E. coli evolves by mutation and recombination, i.e., bacterial sex, by which genetic material is shared between organisms that are not in a parent–offspring relationship (4, 5). Recombination by horizontal gene transfer (HGT) has been recognized as a key factor in the long-term genome evolution of E. coli, but to which extent HGT generates sequence divergence compared to mutations is highly debated (6). At shorter time scales, the evolutionary speed and mode of E. coli, as well as their relative contributions to ecological adaptations during gut colonization in a noninfectious context, remain largely unknown. The mammalian gut is expected to represent a hotspot for HGT, but direct measures of within-host HGT rates are scarce, and the contribution of this mechanism to species evolution remains understudied (3). In the gut, DNA transfer between bacteria often involves temperate bacteriophages (phages) (7), which represent the vast majority of phages found in the gut ecosystem of both mice and man (8–11). Unlike lytic (virulent) phages, which destroy the cell to make new phage particles, temperate phages integrate the bacterial chromosome as prophages, a process termed lysogeny (7, 12, 13). Prophages can later excise and act as vectors for HGT. The evolution of E. coli in the gut has been studied in mouse models, which are also a classical system to study E. coli physiology (14). In previous studies, mice were continuously treated with antibiotic, which is capable of breaking colonization resistance. Under these conditions, mutation is E. coli’s dominant mode of molecular evolution, and HGT has not been reported (15–18). However, the continuous antibiotic treatment in the standard experimental model can distort tempo and mode of bacterial evolution in the gut, because it leads to a reduction in microbiota density and diversity (19). In particular, it may reduce the abundance of phage-carrying bacteria (lysogens), which are prevalent gut microbes under more natural conditions (9). Continuous antibiotic treatments can also lead to mutagenic effects and to a significant depletion of E. coli competitors.
Here, we developed a gut-colonization model that allows controlled evolution experiments in the mouse gut under close to natural conditions of gut ecology. Unlike in previous experimental studies, we perform only a short treatment with streptomycin, which maintains a complex gut microbiota. This protocol allows a new E. coli strain to colonize a virtually intact microbiota that includes resident E. coli strains, which are its closest competitors. Similar transient perturbations by antibiotics are known to occur in natural gut microbiota (20) and can produce a window of opportunity for invading strains. Importantly, our experimental protocol allows us to track the real-time evolution of the invading strain during the colonization process, which extends over hundreds of generations. We find a rapid evolution of the invader throughout this period, which is triggered and dominated by phage-mediated HGT from the resident E. coli strain. By a combination of experiments and modeling, we analyze the complex selective effects governing these evolutionary dynamics, which notably include a likely metabolic adaptive benefit of HGT.
Results
Invader and Resident E. coli Lineages Coexist during Gut Colonization.
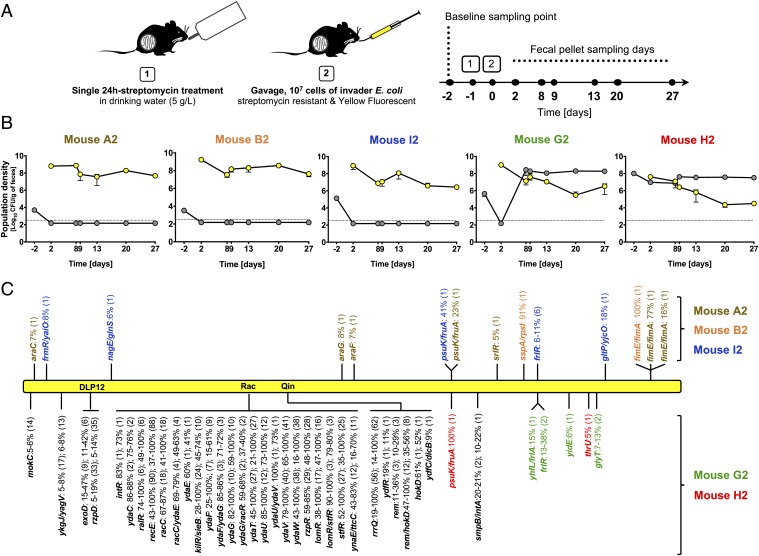
The gut-colonization model used in this study is shown in Fig. 1A. As the invader, we chose an E. coli lineage that can grow in gut ecosystems of different diversity (19) (SI Appendix, Table S1) and carries the yellow fluorescent protein (YFP) marker. This invader lineage can only colonize the gut of mice when they are treated with antibiotic (SI Appendix, Fig. S1). After a 24-h treatment with streptomycin, this strain was able to colonize the mice for 4 wk (Fig. 1B); independent experiments showed stable colonization for several months. Importantly, the transient antibiotic protocol left a higher microbiota diversity than continuous treatment (median 241 vs. 123 species); the species content showed substantial variation across 5 mice (SI Appendix, Figs. S2 and S3). Before treatment, the microbiota of all mice contained a resident E. coli lineage (SI Appendix, SI Text, Fig. S4, and Table S2) at loads between 103 and 108 CFU/g feces (Fig. 1B and SI Appendix, Table S3). This lineage is susceptible to streptomycin (SI Appendix, Fig. S5) and genetically homogeneous; randomly sampled clones of the resident belonged to phylogenetic group B1 and showed the same DNA fingerprint (n = 48 resident clones randomly isolated from different mice; SI Appendix, SI Text and Table S4).
Fig. 1.
Replacement vs. coexistence of the invader E. coli with the mouse gut resident. (A) Experimental setup for in vivo evolution. (B) Loads (log10 CFU/g of feces) of invader (yellow circles) and resident (gray circles) E. coli in mice A2, B2, I2, G2, and H2. Error bars represent 2SE, and the dotted line indicates the detection limit (330 CFU/g of feces). (C) Evolutionary pattern, analyzed by whole-genome sequencing, of large pools (>1,000 clones) of the evolved YFP invader E. coli sampled at day 27 from feces of each mouse, when compared with the genome of the ancestral invader clone. The name of each mutated gene or intergenic region (gene/gene) is indicated, along with the frequency at which the mutation was detected (shown are mutations ≥5%). For each locus, the number of distinct alleles is shown in parentheses. Schematic representation of the evolved E. coli genome (yellow bar) is shown. Mutations indicated above the yellow bar were found in the evolved E. coli clones when not coexisting with the resident E. coli: mice A2 (brown), B2 (orange), and I2 (blue). Mutations indicated below the yellow bar were found in the evolved E. coli clones when coexisting with the resident E. coli: mice G2 (green) and H2 (red). These genetic changes occurred mainly within cryptic E. coli prophage (DLP12, Rac, and Qin) sequences, with mutational parallelism shown in black.
After colonizing the mice with the invader E. coli strain, we followed the E. coli population dynamics in the gut for 1 mo (Fig. 1 A and B). We found 2 distinct colonization patterns. In 3 mice, the resident strain was lost, and the invader settled at a stable population density (Fig. 1B, 3 leftmost panels). In the other 2 mice, colonization led to a stable coexistence of both strains, with the resident being the dominant lineage in the E. coli population (Fig. 1B, 2 rightmost panels). These different outcomes of colonization most likely result from the combined effect of the initial load of the resident strain (Fig. 1B and SI Appendix, Table S3) and its susceptibility to streptomycin (SI Appendix, Fig. S5). In mice with lower initial loads of the resident [5 × 103 to 3 × 103 and 1 × 105 colony-forming units (CFU)/g feces; mice A2, B2, and I2], the 24-h antibiotic treatment presumably killed the resident, leaving the invader to evolve in the absence of that competitor. By contrast, in mice with a higher initial load (4 × 105 and 1 × 108 CFU/g feces; mice G2 and H2), coexistence of both strains occurred, and evolution of the invader took place in the presence of the resident. In mouse G2, after a strong drop in the number of resident bacteria, a rapid increase of its load was observed, and by day 8, the resident strain was dominant and sensitive to streptomycin. In mouse H2, no reduction in loads of the resident was detected, and by day 2 it was found to be streptomycin-resistant (SI Appendix, SI Text and Fig. S5). The combined load of resident and invader was maintained after the 24-h antibiotic treatment at an average of 108 CFU/g feces across all mice, suggesting that the E. coli ecological niche in this ecosystem is significantly larger than before the short streptomycin treatment (SI Appendix, SI Text, Fig. S6, and Table S3).
E. coli Coexistence Is Associated with Evolution by Phage-Mediated HGT.
The 2 patterns of colonization observed in the evolution experiment—replacement of the resident by an invader strain vs. coexistence between resident and invader strain—suggested differences in the invader’s mode of evolution (Fig. 1B). To investigate the genetic evolution taking place in the invader E. coli, we performed whole-genome sequencing of evolved invader populations (random sample of ∼1,000 YFP clones) 27 d after colonization and compared them with the ancestral invader genome (Fig. 1C). In the absence of the resident lineage (mice A2, B2, and I2), the invader evolved by successive accumulation of 2–10 mutations (Fig. 1C), similar to what has been reported under continuous antibiotic treatment (15, 21).
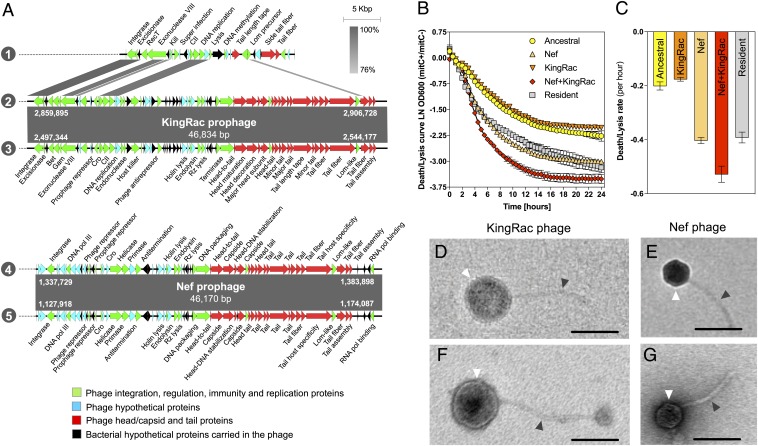
When the resident and invader lineages coexisted (mice G2 and H2), we found a much higher evolution rate in the evolved invader populations, marked by a vastly larger number of sequence changes in the same period. Specifically, numerous adjacent single-nucleotide polymorphisms (SNPs) in known lambdoid defective prophage genes (Fig. 1C and SI Appendix, Table S5) and sequence reads with no identity with the ancestor (Datasets S1 and S2) were found. This high number of adjacent SNPs in prophage genes led us to hypothesize the occurrence of phage-driven HGT events between the 2 E. coli lineages. To address this, we randomly sampled E. coli clones from mouse G2: a resident clone isolated 2 d prior to colonization and an evolved invader clone sampled on day 27 (Fig. 1A and SI Appendix, Table S1). Given the simple genetic structure of the resident E. coli lineage naturally colonizing the gut of these mice (SI Appendix, SI Text, Fig. S4, and Table S4), we hypothesized that a random resident clone should contain the DNA that had been transferred to the invader E. coli. The high frequencies of adjacent SNPs in prophage genes of the evolved invader population (SI Appendix, Table S5) suggested that a randomly sampled clone should be representative of this dominant evolutionary change. A resident and an invader clone were whole-genome sequenced with Nanopore and Illumina technology to obtain their closed genomes. The resident was found to harbor a genome of 5.2 Mbp and 2 plasmids sized 68,935 and 108,557 bp. The evolved invader clone carried no plasmids and a genome sized 4.7 Mbp, which is larger than that of its ancestor. A SNP in the frlR locus also occurred in this clone (SI Appendix, Table S6). Whole-genome comparison between ancestral, evolved, and resident clones, revealed that 2 new genomic regions were acquired by the evolved clone. These were 100% identical to DNA in the genome of the resident lineage (SI Appendix, Fig. S7). The sequences of these regions were annotated and classified as complete prophage regions (Fig. 2A and SI Appendix, Tables S7 and S8). One of the prophages, comprising a 46,834-bp sequence 100% identical to a prophage of the resident (2 and 3 in Fig. 2A and SI Appendix, Table S7), replaced the defective Rac prophage (1 in Fig. 2 and SI Appendix, Table S9) of the ancestral invader clone. Because no signs of homologous recombination were detected in the evolved lineage, the data suggest that the ancestor cryptic Rac prophage, inserted at the ttcA gene (SI Appendix, Fig. S7), was exchanged for a new Rac-like prophage acquired from the resident clone, which we named KingRac (2 and 3 in Fig. 2A). Moreover, the invader acquired an additional phage (4 and 5 in Fig. 2A) comprising 46,170 bp (SI Appendix, Table S8) inserted at the ssrA gene (SI Appendix, Fig. S7). This phage is 100% identical to another prophage of the resident, which we named Nef.
Fig. 2.
Phage-mediated HGT from resident to invader E. coli. (A) Sequence alignment of prophage DNA sequences from the ancestral invader, resident (mouse G2, day −2), and evolved invader (mouse G2, day 27) clones: 1, Rac defective prophage from ancestral; 2, KingRac prophage from resident; 3, KingRac prophage from evolved; 4, Nef prophage from resident; and 5, Nef prophage from evolved. Two new prophages were found in the genome of the evolved clone: KingRac, a Rac-related prophage, and Nef, both present and 100% identical to the resident E. coli. Arrows correspond to the coding sequences (CDS) found in the prophages. Green indicates phage integration, regulation, immunity, or replication proteins; blue indicates phage hypothetical proteins; and red indicates phage head/capsid and tail proteins, while black arrows correspond to bacterial CDS (hypothetical proteins) present in the prophage DNA sequences. Prophage length and integration positions are indicated by the numbers in white. (B) Mitomycin C-induced bacterial cell death/lysis curves. Ancestral, KingRac, Nef, Nef+KingRac, and resident E. coli clones were grown in LB medium with or without mitomycin C, an antibiotic inducing prophage excision. Ln of the ratio of OD600 values with (mitC+) and without (mitC−) mitomycin C is shown. Error bars represent 2SE. The death/lysis curve of the ancestral clone reflects direct mytomycin C bacterial killing, while for the other clones, death/lysis derives from both direct killing and lysis by prophage induction. (C) Death/lysis rate (per hour) calculated from the slope of the curves shown in B in the initial 4.5 h. Error bars represent 2SD. (D–G) Electron micrographs of the KingRac phage particle induced from the evolved KingRac clone (D), the Nef phage particle induced from the evolved Nef clone (E), the KingRac phage particle induced from the resident clone (F), and the Nef phage particle induced from the resident clone (G). White and gray arrows indicate the phage head/capsid and the tail, respectively. The ancestral clone, also induced with mitomycin C, produced no phage particles (SI Appendix, Fig. S8). (Scale bars: 100 nm.)
Acquired Prophages Have Different Life History Traits.
To characterize the process of induction of each of the prophages responsible for the HGT from the resident lineage, we performed a classical mitomycin C phage induction assay (22) on the ancestral and the evolved clone carrying Nef, an evolved clone carrying KingRac, and an evolved clone carrying both prophages (Nef+KingRac) isolated from mouse feces. Whole-genome sequencing of each evolved lysogen revealed that they only differed from the ancestor due to the acquisition of the prophages. The resident clone was also tested in the induction assay. Three distinct patterns were observed: 1) weak induction associated with the KingRac prophage; 2) medium induction for the Nef and resident clones; and 3) strong induction of the double lysogenic (Nef+KingRac) clone (Fig. 2B and SI Appendix, Table S10). We estimated clone-specific prophage induction rates from the slope of the death/lysis curves in the linear range (Fig. 2C and SI Appendix, Table S11): The induction rate of Nef was much higher than that of KingRac (slope −0.403 ± 0.01, 2SD vs. −0.176 ± 0.007, 2SD, respectively). When both prophages (Nef+KingRac) were present, the estimated induction rate was even higher (slope −0.528 ± 0.03, 2SD) (Fig. 2C). The induced lysis assay indicated that the acquired prophages can form active phage particles, and transmission electron microscopy of individual lysates, from the ancestral, resident, and evolved clones, confirmed that phage particles were produced by all clones except the ancestral (SI Appendix, Fig. S8 and Table S12). The evolved clones carrying the Nef prophage (Nef or Nef+KingRac) produced a phage particle identical to 1 of the 2 phages detected when inducing the resident E. coli clone. This phage had an average icosahedral head of 53 (±0.95, 2SE) nm and a tail length of 135 (±3.55, 2SE) nm (Fig. 2 E and G and SI Appendix, Table S12). On the other hand, the evolved clone carrying only the KingRac prophage produced a phage particle exhibiting a spherical head with 99 (±0.00, 2SE) nm and a tail length of 249 (±13.00, 2SE) nm, morphologically identical to the other phage produced by the resident clone (Fig. 2 D and F and SI Appendix, Table S12).
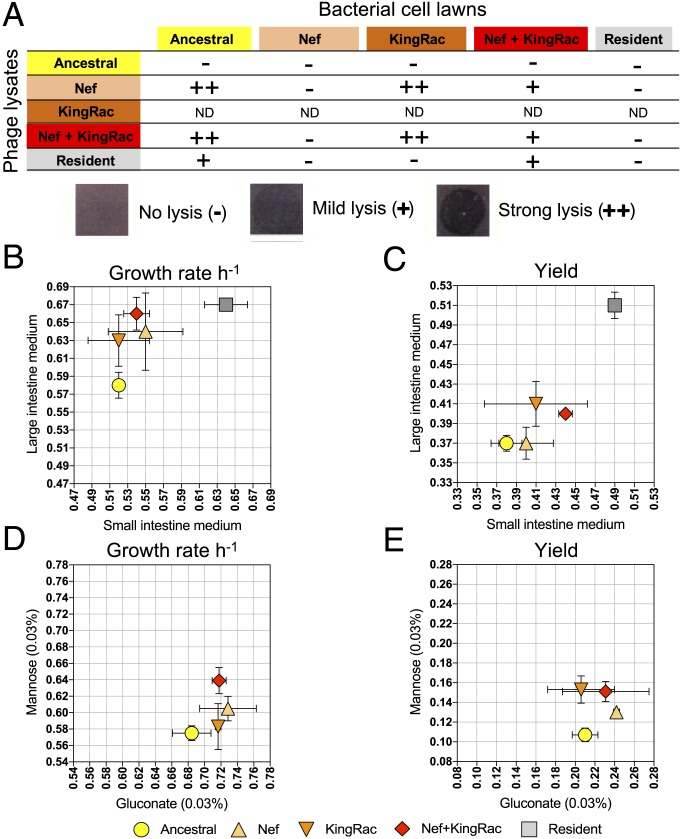
To characterize the capacity of each phage to differentially infect and lyse bacterial clones, we used a phage infection spot assay (Materials and Methods). Nef was capable of infecting and lysing the ancestral, as well as the lysogens carrying KingRac, but not lysogens carrying Nef only [the well-known phenomenon of superinfection immunity (23, 24)]. Thus, the KingRac prophage does not confer immunity to Nef phage (Fig. 3A). Interestingly, the evolved double lysogen (Nef+KingRac) was partially immune to Nef, while the resident was fully immune, suggesting that the resident genome may have other mechanisms of resistance. The observed differences in induction and infection rates (Figs. 2 B and C and 3A) are key components of our phage-mediated selection model; see Eq. 1 below.
Fig. 3.
Evolution by phage-mediated HGT is adaptive. (A) The ancestral, evolved, and resident clones were tested for phage-mediated lysis. The evolved clones include 3 different genetic backgrounds: Nef, KingRac, and Nef+KingRac prophages without any other mutations. Drops (10 µL) of phage-containing supernatant (phage lysate) were applied to growing bacterial cell lawns of each clone. The phage lysate was obtained from the clones' supernatant after mitomycin C induction (5 µg/mL). The KingRac clone is poorly induced (Fig. 2B), and the number of phage particles is expected to be small. ND, not determined. For the other clones, the exact number of phage particles is unknown, as individual phage plaques could not be scored, preventing comparison of the infection efficiency between different phage lysates. (B and C) The ancestral, evolved (Nef, KingRac, and Nef+KingRac), and resident clones were grown in intestinal medium. The maximum growth rate (±2 SE) was assessed in small and large intestinal medium (n = 3) (B), and the yield (±2 SE) after 24 h of growth in small and large intestinal medium (n = 3) was measured (C). (D and E) The ancestral and evolved (Nef, KingRac, and Nef+KingRac) clones were grown in minimal medium supplemented with mannose or gluconate (0.03%). The maximum growth rate (per hour) was assessed in mannose or gluconate medium (n = 5) (D), and the yield after 24 h of growth in mannose or gluconate medium (n = 5) was measured (E).
The Nef and KingRac phages can be classified as lambdoid by their sequence identity to the lambda phage (25, 26) (SI Appendix, Fig. S9). Thus, to understand the differences in resistance to infection between the evolved and the resident, we looked for genetic differences between their genomes at loci known to encode lambdoid phage receptors (e.g., ompC, lamB, and fhuA,) (26). Interestingly, we found 9 nonsynonymous differences in ompC and 3 in lamB genes (SI Appendix, Table S13), which are candidate loci for the increased resistance of the resident E. coli. Additionally, a phage superinfection inhibition mechanism can be invoked to explain the resistance observed for the resident lineage. This mechanism is mediated by prophage repressors that, by inhibiting incoming phage replication, turn the resident bacteria immune to phage infection (24).
Phage-Driven HGT Evolution Confers a Metabolic Growth Advantage.
The observed HGT events bring new cargo of phage and genetically linked bacterial genes (Fig. 2A). To test for selective effects of HGT (27), we compared in vitro the growth of the ancestral, evolved (Nef, KingRac, and Nef+KingRac), and resident clones in a medium derived from the mouse gut. As expected, the resident clone has higher growth rate and higher yield than the ancestral invader in media from both the small and the large intestine (2-way ANOVA; P < 0.001) (Fig. 3 B and C and SI Appendix, Table S14). Besides the resident genome being larger than that of the invader, other potential factors contributing to its increased growth could be its plasmids (28) and/or prophages (29), which have been found to enhance host bacterial fitness. Remarkably, the invader evolved Nef and Nef+KingRac clones also grew faster than their ancestor in the large intestine medium (P < 0.03), and Nef+KingRac reached higher yield in both media (P < 0.02) (Fig. 3 B and C and SI Appendix, Table S14). Additional experiments in minimal medium supplemented by mannose or gluconate, carbon sources known to be important for gut colonization (30), confirmed a net growth advantage of the evolved lysogenic clones (Fig. 3 D and E and SI Appendix, Table S15). These data indicate that the phage-driven HGT is adaptive in the mouse gut and the magnitude of the fitness advantage depends on specific nutrients.
HGT Precedes and Outweighs Adaptive Evolution by Point Mutations.
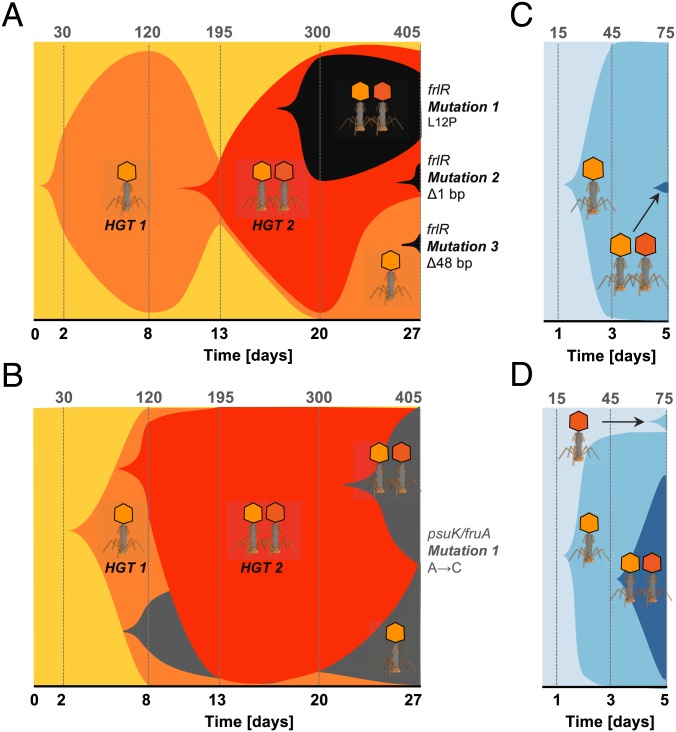
We next inferred the relative contributions and the temporal order of adaptive HGT and point mutations in the invader lineage colonizing the mouse gut. Under conditions of coexistence between invader and resident, the invader populations accumulated only a few mutations that reached a frequency ≥5% over 1 mo (2 in H2 and 6 in G2). These numbers are comparable to the mutation accumulation in the other mice (2–10 in A2, B2, and I2) (Fig. 1C). A faster dynamics of HGT relative to de novo adaptive mutations at the frlR gene and psuK/fruA intergenic region was inferred by amplicon sequencing, time-resolved target PCR, and whole-genome sequencing of pools of clones from mice G2 and H2 (Fig. 4 A and B). In both cases, the invader incurred a rapid succession of 2 phage infections driving HGT (lysogenization). The first, Nef-driven HGT event occurred just 2 d after colonization; this was followed by the integration of KingRac. We conclude that carrying Nef does not confer full immunity to infection by KingRac. Moreover, de novo adaptive mutations at the frlR locus of the invader genome spread only on the background of a Nef-driven HGT (Fig. 4A and SI Appendix, Tables S16 and S20).
Fig. 4.
Phage-mediated HGT is followed by complex clonal evolution. (A and B) Evolution experiment: Muller plots of the adaptive phage-mediated HGT and mutation dynamics in the invader E. coli population (A, mouse G2; B, mouse H2). Shaded areas are proportional to the frequency of each clone: ancestral (yellow), Nef lysogen (light orange), Nef+KingRac lysogen (dark orange), and de novo mutations frlR (1 nonsynonymous mutation, 2 deletion mutations) in mouse G2 (black) and psuK/fruA (1 intergenic mutation) in mouse H2 (gray). Phage symbols indicate the HGT events (1 refers to Nef and 2 to KingRac) for each clone. Numbers in the top row give the estimated number of generations. (C and D) Cocolonization experiment: Muller plots of phage-mediated HGT in new recipient E. coli populations in 2 mice (clones are now shaded in blue). In all cases, HGT takes place first, and de novo mutations appear on the background of high-frequency lysogenic clones.
To evaluate the reproducibility of the HGT dynamics observed in the evolution experiment (Fig. 4 A and B), we performed a cocolonization experiment in a new cohort of mice, with an evolved YFP clone and its ancestor, expressing the cyan fluorescent protein (CFP) (SI Appendix, Table S17). The evolved double lysogen carrying an adaptive mutation (Nef+KingRac+frlR) was now the phage donor and the CFP-susceptible clone should evolve via phage-mediated HGT. Indeed, the ancestral CFP clone incurred a Nef-driven HGT event within 2 d in all mice, and a KingRac-driven HGT event occurred after the Nef event (Fig. 4 C and D and SI Appendix, Table S18), bringing new adaptive traits (Fig. 3 B–E). Together, the Muller plots shown in Fig. 4 depict the speed and mode of evolutionary change at both the core and mobile genomes of the invader strain. We observed a fast and repeatable pattern of phage-driven HGT. Evolution by HGT can reach the time scale of ecological change (Fig. 1B)—it preceded and outweighed evolution by point mutations throughout the duration of the experiment.
HGT Is Followed by Complex Clonal Dynamics.
Distinct HGT events by Nef and by KingRac and subsequent mutations generated considerable genetic diversity. These events brought different clones to high frequency in rapid succession, opening windows for the establishment of de novo adaptive mutations on different lysogenic backgrounds. Together, we observed a complex pattern of clonal interference in the invader lineage (Fig. 4 A and B). Importantly, these dynamics did not eliminate the neutral genetic diversity in the invader core genome. While Nef and the linked bacterial DNA swept close to fixation, a polymorphism of CFP/YFP markers was maintained in the invader population (Fig. 4 C and D and SI Appendix, Fig. S10 and Table S17). Hence, phage-mediated HGT can lead to the spread of new genes and adaptive functions (Fig. 3 B and E), while keeping high genetic diversity in the core genome.
Phage Interactions Generate a New Mode of Selection.
The bacterial population dynamics and evolution (Figs. 1B and 4) revealed a striking pattern of increasing diversity, which was marked by coexistence of invader and resident clones at least for some hundreds of generations. This is very different from the prevalent mode of evolution in laboratory experiments, which is characterized by rapid fixation of adaptive mutations and the concurrent loss of ancestral sequence variation. The pattern of Fig. 4 is also incompatible with simple models of selection. In the initial dynamics of the invader population, the frequency of the phage-susceptible ancestral clone, x, always showed a rapid decline. This is consistent with strong negative selection; however, a model of constant selection s < 0 can be excluded by the later stabilization of susceptible frequencies (Fig. 4 A and D). A model of frequency-dependent selection, s(x), could explain the stabilization of susceptible frequencies, but would be at variance with the decline-rebound dynamics observed in Fig. 4 A and D. These data show frequency changes of opposite sign at the same frequency; that is, s cannot be a function of x only. However, the presence of phages introduces selection depending on the phage load, which provides the arguably simplest explanation of the eco-evolutionary diversity. Differences in induction and infection rates between coexisting clones, as observed in our system (Figs. 2 B and C and 3A), contribute to this selection. Specifically, the fitness of a bacterial clone i,
| [1] |
depends on the phage population density, P, and the fitness in the low-phage limit, ri − δi. Here, ri denotes the background fitness without induction or phages, δi is the fitness cost of induction, and γi is the fitness cost of infection per unit of phage density, which is proportional to the infection rate. For example, a more susceptible strain 1 competing with a more inducible strain 2(γ1 > γ2, δ2 > δ1) can have a selective advantage at low phage levels (s = f1 − f2 > 0) but a disadvantage at high levels (s < 0). In turn, the phage load at a given time depends on the population sizes of phage-producing bacteria in the recent past. This selective feedback can lead to stable coexistence of 2 bacterial clones with different infection and induction rates. Generalizing Eq. 1 to k different phages leads to a bacterial fitness of the form , which is mathematically related to fitness models used in multistrain epidemiology (31, 32). With differences in cross-immunity coefficients γij, as observed in our system (Fig. 3A), this model generates phage-dependent bacterial niches and can lead to stable coexistence of up to k bacterial clones (Materials and Methods). We now show that phage-dependent selection can explain the eco-evolutionary pattern of Figs. 1B and 4 and, in particular, facilitate successful gut colonization.
Phage Epidemics.
We describe the joint dynamics of bacterial clones and phages by a minimal eco-evolutionary model with 2 clones, referred to as the susceptible and the inducible clone, respectively, which have infection parameters γS > γI, induction parameters δ2 > δS < δI, and background fitness rS, rI. These clones compete in ecological niches with constraint c and overlap q. A fraction κ of infected susceptibles undergo lysogenic conversion. Phages are produced by infection and by induction with burst size b, and they are cleared with a constant rate λ. These processes generate a coupled dynamics of the population densities S, I, and P,
| [2] |
| [3] |
| [4] |
Details are given in Materials and Methods; related models are discussed in refs. 33 and 34.
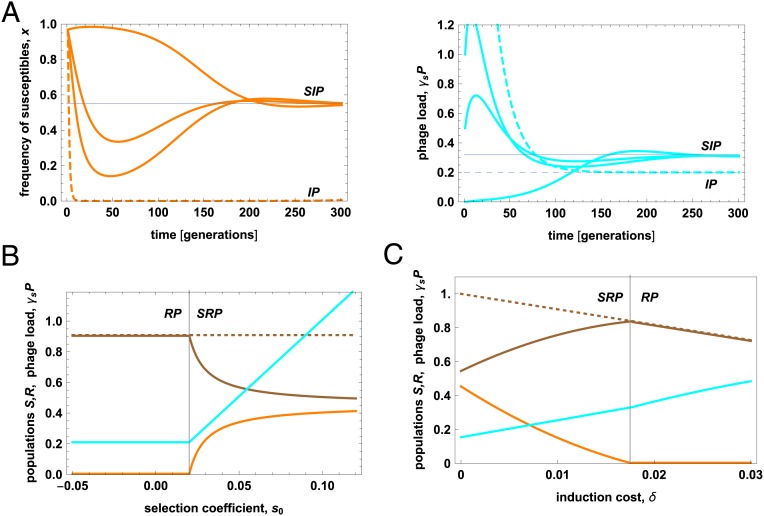
Eqs. 2–4 predict a population dynamics that critically depends on the initial phage level, P0 (Fig. 5A; see Materials and Methods for details). At low values of P0, the infection is slow, and the bacterial clones approach their equilibrium densities in a smooth way. At intermediate values of P0, the model population dynamics takes the form of an infection epidemic, with a rapid initial increase of phages causing negative selection (s < 0) and a resulting depletion of susceptibles. In the second phase, low phage production and clearance can lead to lower phage levels, positive selection (s > 0), and a rebound of susceptibles, provided the susceptible clone has a lower induction rate than its competitors. At even moderately higher values of P0, this pattern turns into a pandemic, which causes rapid loss of the susceptible clone. Both kinds of epidemic can be identified in Fig. 4. The Nef epidemic can end with a rebound of the ancestral clone (Fig. 4 A and D), and the KingRac epidemic with a rebound of the Nef clone (Fig. 4 A and B); this clone has the lowest induction rate (Fig. 2 B and C). In contrast, a Nef pandemic with rapid loss of the ancestral invader clone is observed in Fig. 4 B and C (consistently, mouse H2 had a higher initial resident population, likely producing a higher phage load). This comparison rests on the in vitro mitomycin C assay as a proxy for the innate prophage induction in the gut, which is very difficult to measure.
Fig. 5.
Phages generate epidemics and promote coexistence of bacterial clones. (A) Phage infection dynamics. The population frequency of susceptible (ancestral) bacteria in the invader population (orange) and the phage load γSP (cyan) are plotted against time. Stable equilibrium values are marked by gray lines (SIP equilibrium, solid; IP equilibrium, dashed; Materials and Methods). Time-dependent infection patterns are shown for different initial phage load: slow infection (γSP0 = 0), epidemic with initial decline of susceptibles at high phage levels and subsequent rebound at lower phage levels (γSP0 = 0.5, 1), and pandemic with rapid loss of susceptibles (γSP0 = 5, dashed). See also SI Appendix, Fig. S11. (B) The stable equilibrium state (brown, density of inducible resident strain; orange, density of susceptible invader strain; dashed brown, total bacterial population density; cyan, phage load) is plotted against the selective difference at low phage density, s0. A threshold value s0* (vertical line) marks the onset of bacterial coexistence in the presence of phages (SRP equilibrium) from a regime of displacement (RP equilibrium); Materials and Methods. (C) The same stable equilibrium is plotted against the selective cost of induction of the resident, δR. Population density and fitness of the inducible strain (brown line) reach a maximum close to the onset of coexistence, δR* (vertical line). Model parameters: γS = 0.005, b = 20 (34), δR = 0.01, κ = 0.5 (in A), background fitness rS = 0.15, 0.125 (in A and C), rI = rR = 0.11, c = 0.1, q = 1, λ = 0.05; definitions and model details are given in Materials and Methods. Population densities are shown in units of the background carrying capacity of the resident, R0 = rR∕c.
Phage-Mediated Selection Facilitates Colonization.
After the initial phage epidemics have settled down, more moderate phage-dependent fitness differences govern the subsequent fate of competing bacterial clones. The fitness model [1] predicts conditions for successful gut colonization by an invading strain. An invader clone of phage susceptibility γ can invade an immune resident clone (γR = 0) if its fitness advantage at low phage density, s0 = (r − δ) − (rR − δR), exceeds a threshold value . Notably, this includes cases where the invader has lower background fitness than the resident (r < rR). Phage-mediated colonization is consistent with the late-time pattern of the Nef clone (Fig. 4 A and B), which has (Fig. 3A). This clone has a similar induction rate as the resident (δ ≈ δR; Fig. 2 B and C) and, given its adaptive HGT fitness gain (Fig. 3 B and C), it also has a similar background fitness (r ≈ rR). Clearly, these estimates are to be taken with a grain of salt, because rates and selection coefficients may differ between hosts and from the in vitro estimates (Figs. 2 B and C and 3). The fitness balance of the Nef+KingRac clone is even more complex; it has a phage-driven fitness cost compared to the Nef clone (Figs. 2 B and C and 3A), but its fate also depends on the adaptive value of nested de novo mutations. A further factor facilitating colonization is niche diversification between invader and resident (i.e., reduction of q; Materials and Methods). This can arise from the occupation of spatial refuges by the invader (35), but also from sympatric differentiation on nutrient sources. One may speculate that the repeatedly observed mutations in the frlR gene, which affects an operon absent in the resident, contributes to niche diversification by sympatric evolution.
Phage-mediated selection is expected to promote bacterial diversity in the colonization dynamics beyond the observation period of this experiment (33, 34). In particular, the fitness model [1] predicts that the resident strain can prevent displacement, even when invader clones reach a higher background fitness, provided they retain a higher infection or induction rate. Fig. 5B shows the phage-mediated coexistence region of invader and resident as a function of their fitness difference at low phage density, s0. We note that in the absence of phages, 2 clones of different background fitness would always displace one another. Furthermore, our fitness model predicts an evolutionary optimum of the resident’s induction rate, δR, for phage-mediated defense against competing lysogenic strains. Plotting the equilibrium population sizes against δR shows that the resident population reaches a maximum at a value δ* close to the onset of coexistence (Fig. 5C; Materials and Methods). Rates close to δ* indicate the resident’s optimum investment in phage-mediated defense: They balance the cost of induction in the absence of competitors with its benefit as a defense mechanism against invaders. We note that the optimal induction rate of our model, δ* ∼ 0.01, is of the same order as induction rates observed in vivo, but higher than in vitro rates (34), which is consistent with ecological selection acting on induction in natural populations.
Discussion
The main finding of this paper is that under a minimally perturbed gut, ecosystem invasion by an external E. coli lineage can be successful in the face of competitors when it rapidly adapts via HGT followed by mutation. Our in vivo gut-colonization protocol allows us to track the evolution of E. coli when immersed in a complex ecosystem, in contrast to previous studies performed under laboratory conditions (36, 37) or under strong pressure of antibiotics or disease (15, 19, 21, 38, 39). We find a faster-paced evolutionary dynamics than in these previous studies. The evolution of the invader starts with spontaneous and rapid HGT from a resident E. coli lineage, which seeds subsequent de novo adaptive mutations in newly converted lysogenic clones (Fig. 4 A and B). Throughout the duration of the experiment, HGT dominates the genetic evolution of the invader, conferring both a phage-killing potential and an adaptive metabolic advantage to lysogenic invader lineages (Fig. 3). Interestingly, the Nef prophage was shown to induce at a higher rate (Fig. 2 B and C), which confers an increased potential to actively kill susceptible competitors (Fig. 3A), while the KingRac prophage appears to provide mainly an increased metabolic fitness to the new arising lysogens (Fig. 3). The observed metabolic advantage conferred by the Nef and KingRac prophages to the invader lineage (Fig. 3 B and C) could not be related to any specific prophage gene; its genetic basis remains an open question. But, it is tempting to speculate (29) that the prophages could alter the outer membrane proteins of the host facilitating nutrient consumption. The speedup of evolution by HGT is in accordance with previous results (40). HGT in our system turns out to be faster than observed previously in murine systems under infection (38, 39), underscoring the importance not only in disease but also in health.
By a combination of experiments and modeling, we have analyzed the complex eco-evolutionary dynamics of the invader and the resident lineage and the underlying fitness landscape. Phage-mediated selection has long been discussed in its role as a weapon in the competition between bacterial lineages (41–44). Our experiments show that strong induction of temperate phages acts as a short-term defense mechanism of the resident strain, which can rapidly kill susceptible invader bacteria by a pandemic infection (Figs. 4 B and C and 5A). However, lysogenic conversion rescues the invader in all observed cases, consistent with previous results (43). The subsequent competition between lysogenic clones is driven by a complex fitness landscape with 3 components: differences in induction rates, infection rates, and background fitness. We have shown that clones differ in all 3 fitness components (Figs. 2 B and C and 3), which explains the complex clonal dynamics found in vivo in the mouse gut (Fig. 4). In particular, phage-mediated HGT can increase the fitness of the invader; the metabolic adaptive effect of HGT established here complements previous results where HGT enhanced antibiotic resistance (45, 46). At the same time, substantial induction rates decrease the fitness of a lysogenic lineage (e.g., the resident), in tune with previous findings (34). These effects can help an external invader to stably colonize the gut of a new host (Figs. 4 and 5A). Together, our results suggest that temperate phages are a stump sword of attack that may not eradicate an invader of lower background fitness, but prevent displacement by invaders of higher background fitness (Fig. 5B). The balance between phage-mediated selective forces depends, in particular, on phage induction rates of the lysogenic (e.g., resident) lineages. These rates can change by physiology or evolution, and they are themselves subject to selection (Fig. 5C). How temperate phage-mediated selection shapes the long-term species turnover of ecosystems by colonization remains a question for future work.
Materials and Methods
Experimental Procedures and Data Analysis.
E. coli clones.
E. coli clones used in the present study are listed in SI Appendix, Table S1. E. coli clones were grown at 37 °C under aeration in brain heart infusion or Luria broth (LB). Media were supplemented with the antibiotics when specified (SI Appendix). YFP- or CFP-labeled bacterial numbers assessed as in ref. 15. For the resident E. coli, bacterial numbers were scored after plating in McConkey + 0.4% lactose plates, in which it formed red colonies due to lactose consumption. The detection limit for bacterial plating was 330 CFU/g feces.
In vivo colonization experiments.
In the gut-colonization model, mice drank water with streptomycin (5 g/L) for 24 h before a 4-h starvation period of food and water. The animals were then inoculated by gavage with 100 µL of an E. coli bacterial suspension of ∼108 CFUs. Mice A2, B2, I2, G2 and H2 were successfully colonized with the invader E. coli, while mice C2–F2 failed to be colonized. Six- to 8-wk-old C57BL/6J nonlittermate female mice were kept in individually ventilated cages under specified pathogen-free barrier conditions at the Instituto Gulbenkian de Ciência (IGC) animal facility. The gut microbiota of mice used in the experiments are natural to the animals and not the result of the introduction of a defined microbiota into germ-free animals. Fecal pellets were collected during 27 d (evolution experiment) or 5 d (cocolonization experiment) and stored in 15% glycerol at −80 °C for later analysis. This research project was ethically reviewed and approved by the Ethics Committee of the IGC (license reference A009.2010) and by the Portuguese National Entity that regulates the use of laboratory animals (Direção Geral de Alimentação e Veterinária; license reference 008958). All experiments conducted on animals followed the Portuguese (Decreto-Lei113/2013) and European (Directive 2010/63/EU) legislations concerning housing, husbandry, and animal welfare.
Bacterial growth curves, growth rate, and yield.
E. coli clones were grown in a gut-derived medium under a low-oxygen environment (culture overlaid with mineral oil) by using a Bioscreen C apparatus (Oy Growth Curves Ab Ltd.). For the gut-derived medium, we used filter-sterilized (0.22-µm syringe filters; Pall Life Sciences) mice extracts of small or large intestine (20 mL/g). Clones were also grown in minimal medium (MM9-SIGMA) with mannose or gluconate added at 0.03%. Maximum growth rate was calculated as the maximal slope of the exponential phase. Growth yield was determined by measuring the final OD600 nm after 24 h of growth.
Microbiota analysis.
DNA was extracted from feces by using the QIAamp DNA Stool Mini Kit (Qiagen), following manufacturer’s instructions with an additional mechanical disruption step (19). Sequencing of the V4 region of 16S rRNA gene was performed at the IGC Genomics Unit (SI Appendix).
Whole-genome sequencing of E. coli populations and clones.
DNA was extracted (47) with library construction and sequencing performed at the IGC Genomics Unit. For E. coli populations, DNA from a mixture of >1,000 clones was analyzed, while for clone sequencing, DNA from a single colony was used. We used Illumina and Nanopore technologies to perform whole-genome sequencing (SI Appendix).
Nef and KingRac prophage sequence annotation and representation.
Using the NCBI alignment tool (48), we identified and extracted the complete sequence of the Nef and KingRac prophages. PHASTER software (49) was used for prophage annotation and Easyfig for schematic representation (50).
Prophage induction and phage infection/lysis assay.
For prophage induction (22), we added mitomycin C (5 µg/mL) to E. coli clones growing with agitation at 37 °C in LB medium when OD600 = 0.3–0.5 (ThermoFisher Scientific Spectrophotometer). The lysis rate was calculated as the slope of the mitomycin C-induced lysis curve, from the LN OD600 between 1 and 4.5 h, noting that the data of time point zero had an anomalously large spread between replicates and that after 5 h systematic deviations from an exponential form were observed. Fig. 2C reports the mean of the estimated induction rates over 6 replicate experiments for each genetic background. Each bacterial culture was centrifuged twice (5 min, 15,871 × g, 4 °C) and the supernatant (lysate) was filtered with sterile 0.22-µm syringe filters (Pall Life sciences). The filtered lysate was added to a sucrose (33%) cushion and ultracentrifuged (45 min, 110,000 × g, 4 °C) (Beckman Optima Max XP), with the pellet (phage particles) being suspended in PBS 1× (4 °C). For the phage infection/lysis assay, we grew E. coli clones until OD600 = 0.3–0.5 and added 100 µL of each culture to 3 mL of top agar (0.7%), spreading this mix over a prewarmed (37 °C) LB agar plate. Drops (10 µL) of the lysate were then spotted onto the top agar lawn and incubated overnight at 37 °C to assess phage-induced lysis of bacteria.
Transmission electron microscopy.
Lysate samples (5 µL) were negatively stained with 2% uranyl acetate by using Formvar-carbon-coated copper grids. Images of the phage particles were acquired on a Hitachi H-7650 transmission electron microscope operating at 100 keV equipped with an XR41M midmount AMT digital camera.
Detection of phage-mediated HGT events: Lysogeny.
PCR amplification of phage-specific genes was performed in clones isolated at different time points. Using the primers listed in SI Appendix, Table S19, the Nef prophage was detected via amplification of the xerC gene, while the ydaV or asd genes identified the KingRac prophage (SI Appendix, Tables S7 and S8). PCRs were performed in a total volume of 25 μL, containing 1 μL of bacterial culture, 1× Taq polymerase buffer, 200 μM dNTPs, 0.2 μM each primer, and 1.25 U of Taq polymerase. PCR conditions were as follows: 95 °C for 3 min, followed by 35 cycles of 95 °C for 30 s, 65 °C for 30 s, and 72 °C for 30 s, finalizing with 5 min at 72 °C.
In vivo adaptive mutation frequency time series.
DNA was extracted (47) from E. coli populations (>1,000 clones) isolated at different time points, and the frlR primers described in SI Appendix, Table S19 were used to amplify a 263-bp frlR variable region. PCRs were performed in a total volume of 50 μL, containing 1 μL of bacterial culture, 1× Phusion polymerase buffer, 200 μM dNTPs, 0.5 μM each primer, and 1 U of Phusion polymerase. PCR conditions were as follows: 95 °C for 30 s, followed by 35 cycles of 98 °C for 10 s, 62 °C for 30 s, and 72 °C for 30 s, finalizing with 10 min at 72 °C. PCR amplicons were then sequenced at the IGC Genomics Unit by using Illumina MiSeq technology. Haplotype frequencies of frlR mutations along time were determined by using DADA2 software (51).
Statistical analysis.
A linear mixed-effects model was used to analyze the microbiota-driven load differences or niche size for E. coli. Two-way ANOVA with Tukey’s correction for multiple comparisons was used to analyze the bacterial growth rate and yield in the different media. The significance of groups in community structure was tested by using analysis of similarities (52). Differences in alpha diversity were evaluated by using the nonparametric Mann–Whitney test. BiodiversityR (Version 2.8-4) and vegan (Version 2.4-5) packages were used.
Eco-Evolutionary Models.
Eco-evolutionary minimal model.
We use a simple dynamical model of 2 bacterial (sub)lineages that interact with phages and differ in their response characteristics—that is, in the susceptibility to phage infection and the induction rate of phages. Similar and more detailed models of these dynamics have been discussed in refs. 33 and 34. The minimal model contains 3 populations: 1) a bacterial lineage referred to as “susceptible” has population density S, background fitness rS (in the absence of phages), and a selective cost of infection per unit of phage density γS. If this lineage is a partially susceptible lysogen, it also has a selective cost of induction, δS. 2) A lysogenic lineage referred to as “inducible” has population density I, background fitness rI, and a selective cost of induction δI > δS. This lineage may also be partially susceptible with a selective cost γI < γS. 3) Phages have population density P, are produced by infection and induction with a burst size b, and are cleared at a rate λ. The bacterial lineages compete in overlapping ecological niches with inverse carrying capacity c and overlap q. Infection of susceptibles leads to lysogenization (production of inducibles) with probability κ and to lysis (cell death) with probability (1 − κ). These processes generate the population dynamics described by Eqs. 2–4. We will use this model to describe the dominant contending lineages at a given stage of the colonization process. At the early stage, the model describes the infection and lysogenization dynamics of the invading strain. Here, S is the ancestral invader clone (i.e., δS = 0), and I is the lysogenic invader clone. Because clones belong to the same strain, we use q = 1; the carrying capacity then couples to the total population size N = S + I. At later stages, the model describes the population dynamics of lysogenic strains (i.e., κ = 0). Here, S is a partially susceptible invader clone, and I is the resident strain. The dynamics of Eqs. 2–4 define the reproductive rates FS = rS − δS − γSP − cS − cqI, FI = rI − δI − γIP − cqS − cI, FP = b(1 − κ)γS − λ, and the flux terms , , where we use the convention to integrate all population size changes proportional to the size of the same population into reproductive rates. We measure all rates in units of the inverse generation time of the inducible lineage, so that the difference in reproductive rates, s = FS − FI, becomes a standard selection coefficient. In particular, the phage load γSP measures the fitness cost of infection for the susceptible lineage; this quantity is plotted in Fig. 5 and SI Appendix, Fig. S11. The dynamical equations remain invariant under the rescaling P → P/C, b → b/C, by an arbitrary factor C > 0 . That is, if we express P in units of the burst size b, the solution does not depend on b and γS,I separately, but only on the rescaled parameters and .
The minimal eco-evolutionary model contains the key feature of phage-mediated selection between bacterial strains discussed in the main text: the fitness difference between susceptible and inducible bacteria, s = FS − FI, depends on phage density,
| [5] |
where γ ≡ γS − γI and
| [6] |
denotes the selection coefficient in the limit of low phage density. Given equal background fitness (rS = rI), the inducible lineage has a fitness advantage at high phage density, and phage selection becomes a weapon against the susceptible lineage. At low phage density, the susceptible lineage has a fitness advantage if it has a lower induction rate than the inducible lineage. These regimes are separated by a stable fixed point, which describes stable coexistence of 2 bacterial lineages at a given phage density. We will show below that, despite its simplifications, the minimal model explains the broad pattern of population sizes and frequencies observed in the gut experiments.
Model with multiple phages.
The minimal model can readily be generalized to more complex ecosystems with n bacterial lineages and k phage lineages. We assume that bacteria of type i are partially susceptible to infection by phages of type j, as given by the matrix of infection costs per unit phage density, γij(i = 1,...,n, j = 1,...,k). Such lineage-specific susceptibilities are indeed observed in our system (Fig. 3A). Given background fitness parameters ri and induction rates δi, the fitness of the bacterial lineages takes the form
| [7] |
This model bears similarities to fitness models used in multistrain epidemiology (31), previously used to describe the evolution of viral systems (32).
The multistrain model has 2 salient features. First, multiple phages diversify the weaponry of a defending resident strain, which can use different phages against invaders with different susceptibility profiles. From this point of view, one may speculate that a resident’s spectrum of inducible phages reflects its exposure to different invaders. On the other hand, multiphage systems can create complex niches of bacterial coexistence. Given k phages and a generic susceptibility matrix γij, the system of linear Eq. 7 allows coexistence of up to k bacterial lineages. Together, the multistrain model reemphasizes the double role of phage-mediated selection between bacterial lineages. A full dynamical analysis of the model is beyond the scope of this paper.
Population dynamics and phage epidemics.
We now consider the population dynamics of the minimal model under conditions allowing the equilibrium coexistence of susceptible and inducible lineages (the corresponding model parameters are discussed below). In order to describe the dynamics during the first 2 wk of gut colonization, we associate the model populations with the dominant contending lineages: the ancestral invader strain is the more susceptible lineage (S) and the lysogenic invader strain is the inducible lineage (I). This is in accordance with the observed differences in induction and infection rates between these lineages (Figs. 2 B and C and 3A). By numerical integration of Eqs. 2–4 with δS = 0 and q = 1, we obtain population trajectories (S, I, P)(t). The fraction of susceptible bacteria, x(t) = S(t)/N(t) follows a dynamical equation of the form
| [8] |
where vs(t) is the contribution of phage-dependent selection given by Eq. 5,
| [9] |
with Peq denoting the equilibrium phage level for κ = 0, and vκ is the contribution of lysogenization,
| [10] |
At high phage levels, susceptibles are negatively selected [P(t) > Peq, s < 0]; at low phage levels, selection on susceptibles becomes positive [P(t) < Peq, s > 0]. The dynamics of phages themselves introduces a memory term: The level P(t) depends on the production of phages in the recent past before t. Hence, phage-dependent selection generates a feedback loop: higher (lower) phage levels increase the fraction of inducibles (susceptibles), which, in turn, decreases (increases) the phage level. The feedback can lead to stable population equilibria of susceptible and inducible bacteria in the presence of phages. Fig. 5A shows the infection and lysogenization dynamics of this model for different values of the initial phage density P0 = P(t0). We show that the dynamical pattern depends strongly on P0. At intermediate values of P0, there is a soft phage epidemic with an initial decline of susceptibles at high phage levels, followed by a rebound at lower phage levels. At higher values of P0, the pattern changes to a phage pandemic that rapidly kills the susceptible invaders. As discussed in the main text, these 2 regimes explain the experimentally observed patterns of phage infection and lysogenization (Fig. 4).
SI Appendix, Fig. S11 shows the dependence of the population dynamics on other model parameters. In particular, variation of the fraction of lysogenization, κ, changes the value of the equilibrium population densities (and makes the analytical solution of the model more cumbersome than for κ = 0), but leaves the selective feedback loop and the pattern of the epidemic invariant (SI Appendix, Fig. S11A). The dependence on the infection cost γS and the burst size b (SI Appendix, Fig. S11B), which define the scaled parameter , shows a transition from soft epidemics to pandemics similar to Fig. 5A. We conclude that both regimes are robust features of phage-dependent selection.
Evolutionary equilibria of bacteria and phages.
In order to describe the complex outcomes of the colonization experiment, we study equilibrium states of the minimal model and their evolutionary stability or instability. Here, we associate the model populations with the dominant contending lineages in the final stage of the colonization experiment: The evolved (lysogenic) invader strain is the susceptible lineage (S) and the resident strain is the inducible lineage (I). Between these lineages, there is no lysogenization (κ = 0). The minimal model then has equilibrium states describing the coexistence of bacteria and phages, which are consistent with the equilibria derived in refs. 33 and 34. In order to display phage-mediated coexistence of lineages independently of ecological niche diversity, we evaluate these equilibria for complete niche overlap (q = 1) (the effect of niche diversity on stability will be discussed later).
SIP equilibrium.
This state describes the coexistence of a susceptible and an inducible bacterial lineage with phage-selection parameters γS > γI and δS < δI and a phage population. The total bacterial population density Neq = Seq + Ieq and the phage population density Peq can be obtained from Eqs. 2 and 3 together with the equilibrium condition s(Peq) = 0,
| [11] |
where s0 is given by Eq. 6. The frequency of susceptibles, xeq = Seq/Neq, then follows from Eq. 4,
| [12] |
These expressions are valid as long as 0 ≤ xeq ≤ 1.
SP equilibrium.
For the case of a single susceptible lineage (S) coexisting with phages, the equilibrium takes the simpler form
| [13] |
for S0 < rS/c; otherwise, P is displaced, Peq = 0 and Seq = rS/c.
RP equilibrium.
For the case of a single inducible but immune lineage (R) coexisting with phages (i.e., γR = 0), the equilibrium takes the form
| [14] |
for δR < rR.
Phage interactions facilitate colonization.
After the decay of the initial phage epidemics, the population dynamics runs closer to an evolutionary equilibrium, which is dominated by a stable resident population (Fig. 1B, G2 and H2). At this stage, the colonization is successful if a lysogenic invader clone (S) can invade the RP equilibrium (with γR = 0). This requires a positive selection coefficient s(P0) = s0 − γSP0 > 0, which amounts to the condition
| [15] |
with a selection threshold given by Eqs. 13 and 14,
| [16] |
A sufficiently high positive s0 can arise from an advantage in background fitness (rS > rR) or from a reduced induction rate induction (δS < δR). In the latter case, phages enable the invasion of a clone that would otherwise be displaced. As discussed in the main text, this condition is consistent with maintenance of the Nef clone, which has γS = 0 and δS ≈ δR (Fig. 4 B and C). Niche diversification can further facilitate colonization. A partial niche overlap between invader and resident (q < 1) lowers the selection threshold for invasion of the IP equilibrium to
| [17] |
Stable coexistence between bacterial clones, as described by the SRP equilibrium of the form [11], depends on an additional condition: R can invade the SP equilibrium, which requires
| [18] |
A stable SRP equilibrium in a single ecological niche (q = 1), which is reached under broad conditions on the model parameters, describes coexistence of strains enabled by phage interactions. In the absence of phages, S would displace R for s0 > 0 and R would displace S for s0 < 0. In Fig. 5B, we plot the crossover between a stable RP equilibrium and a stable SRP equilibrium as a function of the selection coefficient . This shows that phage-mediated selection can prevent the displacement of a resident clone by an invader with higher background fitness (rS > rR).
Selection on induction rates.
The induction rate of a lysogenic resident strain (R) determines cost and benefit of induction as a defense mechanism against susceptible competitor strains (53, 54). In Fig. 5C, we plot stable population numbers (Seq, Req, Peq) of the minimal model as a function of δR. The equilibrium population size Req(δR) can be interpreted as a fitness landscape governing selection on induction. This landscape has its maximum close to the onset of the SRP coexistence regime, which is given by
| [19] |
The fitness of an inducible lineage is reduced by the cost of induction for and by resource competition with susceptible strains for . This may imply that under recurrent exposure to invading strains and phages, the long-term evolution of commensal bacterial strains favors modest induction rates that, in turn, generate conditions for strain coexistence.
Supplementary Material
Acknowledgments
We thank the research infrastructure Congento (Project LISBOA-01-0145-FEDER-022170) and the Instituto Gulbenkian de Ciência’s Electron Microscopy, Bioinformatics, and Genomic Units for technical service. We thank M. J. Amorim, Z. Silva, and V. Cabral for expert advice, as well as the I.G. and M.L. laboratories for comments on the manuscript. This work was supported by Portuguese Science Foundation (FCT) Grant PTDC/BIA-EVF/118075/2010; Deutsche Forschungs-gemeinschaft (DFG) Grants SFB 680 and SFB 1310; and a cooperation agreement between University of Cologne and Gulbenkian Institute and Global Grant for Gut Health-623877. N.F. was supported by FCT Grant SFRH/BPD/11075/2015 and DFG Grant SFB 680; and A.S. was supported by FCT Grants SFRH/BPD/14299/2003, UID/BIM/04501/2013, and UID/BIM/04501/2019.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The data reported in this paper have been deposited in the Sequence Read Archive (SRA) database (BioProject accession no. PRJNA549246).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1906958116/-/DCSupplemental.
References
- 1.Lloyd-Price J., Abu-Ali G., Huttenhower C., The healthy human microbiome. Genome Med. 8, 51 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jandhyala S. M., et al. , Role of the normal gut microbiota. World J. Gastroenterol. 21, 8787–8803 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sousa A., Frazão N., Ramiro R. S., Gordo I., Evolution of commensal bacteria in the intestinal tract of mice. Curr. Opin. Microbiol. 38, 114–121 (2017). [DOI] [PubMed] [Google Scholar]
- 4.Narra H. P., Ochman H., Of what use is sex to bacteria? Curr. Biol. 16, R705–R710 (2006). [DOI] [PubMed] [Google Scholar]
- 5.Soucy S. M., Huang J., Gogarten J. P., Horizontal gene transfer: Building the web of life. Nat. Rev. Genet. 16, 472–482 (2015). [DOI] [PubMed] [Google Scholar]
- 6.Bobay L.-M., Traverse C. C., Ochman H., Impermanence of bacterial clones. Proc. Natl. Acad. Sci. U.S.A. 112, 8893–8900 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Colavecchio A., Cadieux B., Lo A., Goodridge L. D., Bacteriophages contribute to the spread of antibiotic resistance genes among foodborne pathogens of the Enterobacteriaceae family—A review. Front. Microbiol. 8, 1108 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mirzaei M. K., Maurice C. F., Ménage à trois in the human gut: Interactions between host, bacteria and phages. Nat. Rev. Microbiol. 15, 397–408 (2017). [DOI] [PubMed] [Google Scholar]
- 9.Kim M.-S., Bae J.-W., Lysogeny is prevalent and widely distributed in the murine gut microbiota. ISME J. 12, 1127–1141 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Reyes A., et al. , Viruses in the faecal microbiota of monozygotic twins and their mothers. Nature 466, 334–338 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lim E. S., et al. , Early life dynamics of the human gut virome and bacterial microbiome in infants. Nat. Med. 21, 1228–1234 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lwoff A., Lysogeny. Bacteriol. Rev. 17, 269–337 (1953). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Canchaya C., Fournous G., Chibani-Chennoufi S., Dillmann M.-L., Brüssow H., Phage as agents of lateral gene transfer. Curr. Opin. Microbiol. 6, 417–424 (2003). [DOI] [PubMed] [Google Scholar]
- 14.Leatham M. P., et al. , Mouse intestine selects nonmotile flhDC mutants of Escherichia coli MG1655 with increased colonizing ability and better utilization of carbon sources. Infect. Immun. 73, 8039–8049 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Barroso-Batista J., et al. , The first steps of adaptation of Escherichia coli to the gut are dominated by soft sweeps. PLoS Genet. 10, e1004182 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Barroso-Batista J., Demengeot J., Gordo I., Adaptive immunity increases the pace and predictability of evolutionary change in commensal gut bacteria. Nat. Commun. 6, 8945 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lourenço M., et al. , A mutational hotspot and strong selection contribute to the order of mutations selected for during Escherichia coli adaptation to the gut. PLoS Genet. 12, e1006420 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sousa A., et al. , Recurrent reverse evolution maintains polymorphism after strong bottlenecks in commensal gut bacteria. Mol. Biol. Evol. 34, 2879–2892 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Thompson J. A., Oliveira R. A., Djukovic A., Ubeda C., Xavier K. B., Manipulation of the quorum sensing signal AI-2 affects the antibiotic-treated gut microbiota. Cell Rep. 10, 1861–1871 (2015). [DOI] [PubMed] [Google Scholar]
- 20.Willing B. P., Russell S. L., Finlay B. B., Shifting the balance: Antibiotic effects on host-microbiota mutualism. Nat. Rev. Microbiol. 9, 233–243 (2011). [DOI] [PubMed] [Google Scholar]
- 21.Lescat M., et al. , Using long-term experimental evolution to uncover the patterns and determinants of molecular evolution of an Escherichia coli natural isolate in the streptomycin-treated mouse gut. Mol. Ecol. 26, 1802–1817 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Otsuji N., Sekiguchi M., Iijima T., Takagi Y., Induction of phage formation in the lysogenic Escherichia coli K-12 by mitomycin C. Nature 184 (suppl. 14), 1079–1080 (1959). [DOI] [PubMed] [Google Scholar]
- 23.Fuller W., Wilkens M. H. F., Brown G. L., Control of viral messenger RNA after Lambda phage infection and induction. Proc Natl Acad Sci USA 53, 378–385 (1965). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Berngruber T. W., Weissing F. J., Gandon S., Inhibition of superinfection and the evolution of viral latency. J. Virol. 84, 10200–10208 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Campbell A., Schneider S. J., Song B., Lambdoid phages as elements of bacterial genomes (integrase/phage21/Escherichia coli K-12/icd gene). Genetica 86, 259–267 (1992). [DOI] [PubMed] [Google Scholar]
- 26.Kameyama L., et al. , “Some reflections on the origin of lambdoid bacteriophages” in Bacteriophages, Kurtbke I., Ed. (InTech, London, UK, 2012). [Google Scholar]
- 27.Edlin G., Lin L., Kudrna R., λ lysogens of E. coli reproduce more rapidly than non-lysogens. Nature 255, 735–737 (1975). [DOI] [PubMed] [Google Scholar]
- 28.Lenski R. E., Simpson S. C., Nguyen T. T., Genetic analysis of a plasmid-encoded, host genotype-specific enhancement of bacterial fitness. J. Bacteriol. 176, 3140–3147 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lin L., Bitner R., Edlin G., Increased reproductive fitness of Escherichia coli lambda lysogens. J. Virol. 21, 554–559 (1977). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chang D.-E., et al. , Carbon nutrition of Escherichia coli in the mouse intestine. Proc. Natl. Acad. Sci. U.S.A. 101, 7427–7432 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gog J. R., Grenfell B. T., Dynamics and selection of many-strain pathogens. Proc. Natl. Acad. Sci. U.S.A. 99, 17209–17214 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Łuksza M., Lässig M., A predictive fitness model for influenza. Nature 507, 57–61 (2014). [DOI] [PubMed] [Google Scholar]
- 33.Levin B. R., Lenski R. E., “Coevolution in bacteria and their viruses and plasmids” in Coevolution, Futuyma D. J., Slatkin M., Eds. (Sinauer Associates Inc., Sunderland, MA, 1983), pp. 98–127. [Google Scholar]
- 34.De Paepe M., et al. , Carriage of λ latent virus is costly for its bacterial host due to frequent reactivation in monoxenic mouse intestine. PLoS Genet. 12, e1005861 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schrag S. J., Mittler J. E., Host-parasite coexistence: The role of spatial refuges in stabilizing bacteria-phage interactions. Am. Nat. 148, 348–377 (1996). [Google Scholar]
- 36.Barrick J. E., et al. , Genome evolution and adaptation in a long-term experiment with Escherichia coli. Nature 461, 1243–1247 (2009). [DOI] [PubMed] [Google Scholar]
- 37.Lenski R. E., Experimental evolution and the dynamics of adaptation and genome evolution in microbial populations. ISME J. 11, 2181–2194 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Diard M., et al. , Inflammation boosts bacteriophage transfer between Salmonella spp. Science 355, 1211–1215 (2017). [DOI] [PubMed] [Google Scholar]
- 39.Davies E. V., et al. , Temperate phages enhance pathogen fitness in chronic lung infection. ISME J. 10, 2553–2555 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Touchon M., Moura de Sousa J. A., Rocha E. P., Embracing the enemy: The diversification of microbial gene repertoires by phage-mediated horizontal gene transfer. Curr. Opin. Microbiol. 38, 66–73 (2017). [DOI] [PubMed] [Google Scholar]
- 41.Brown S. P., Le Chat L., De Paepe M., Taddei F., Ecology of microbial invasions: Amplification allows virus carriers to invade more rapidly when rare. Curr. Biol. 16, 2048–2052 (2006). [DOI] [PubMed] [Google Scholar]
- 42.Duerkop B. A., Clements C. V., Rollins D., Rodrigues J. L. M., Hooper L. V., A composite bacteriophage alters colonization by an intestinal commensal bacterium. Proc. Natl. Acad. Sci. U.S.A. 109, 17621–17626 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gama J. A., et al. , Temperate bacterial viruses as double-edged swords in bacterial warfare. PLoS One 8, e59043 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li X.-Y., et al. , Temperate phages as self-replicating weapons in bacterial competition. J. R. Soc. Interface 14, 20170563 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang X., et al. , Cryptic prophages help bacteria cope with adverse environments. Nat. Commun. 1, 147 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Modi S. R., Lee H. H., Spina C. S., Collins J. J., Antibiotic treatment expands the resistance reservoir and ecological network of the phage metagenome. Nature 499, 219–222 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wilson K., “Preparation of genomic DNA from bacteria” in Current Protocols in Molecular Biology, Ausubel F. M. et al. , Eds. (John Wiley & Sons, Inc., Hoboken, NJ, 2001), pp. 2.4.1–2.4.5. [DOI] [PubMed] [Google Scholar]
- 48.Altschul S. F., Gish W., Miller W., Myers E. W., Lipman D. J., Basic local alignment search tool. J. Mol. Biol. 215, 403–410 (1990). [DOI] [PubMed] [Google Scholar]
- 49.Arndt D., et al. , PHASTER: A better, faster version of the PHAST phage search tool. Nucleic Acids Res. 44, W16–W21 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sullivan M. J., Petty N. K., Beatson S. A., Easyfig: A genome comparison visualizer. Bioinformatics 27, 1009–1010 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Callahan B. J., et al. , DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods 13, 581–583 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Clarke K. R., Non-parametric multivariate analyses of changes in community structure. Austral Ecol. 18, 117–143 (1993). [Google Scholar]
- 53.Dedrick R. M., et al. , Prophage-mediated defence against viral attack and viral counter-defence. Nat. Microbiol. 2, 16251 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Dionisio F., Selfish and spiteful behaviour through parasites and pathogens. Evol. Ecol. Res. 9, 1199–1210 (2007). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.