Significance
Delineation of intratumor heterogeneity (ITH) has been a subject of growing interest for defining and tracking the evolution of cancer. Yet, the clinical consequences of such ITH on risk prediction remain unclear. Here we show ITH-driven variance on patient stratification and argue that the level of ITH of individual genes should be considered when developing single sector-based prognostic multigene tests (MGTs) in non–small-cell lung cancer (NSCLC). Single-cell molecular analysis of enriched, patient-derived circulating tumor cells (CTCs) further revealed predictive biomarkers for metastatic risk. Through systematic analysis of genes implicated in multiple steps of the metastatic spectrum, we demonstrate that the refined signatures achieve superior accuracy in identifying patients with early-stage disease at high risk of recurrence of NSCLC.
Keywords: microfluidics, circulating biomarkers, tumor heterogeneity
Abstract
Despite pronounced genomic and transcriptomic heterogeneity in non–small-cell lung cancer (NSCLC) not only between tumors, but also within a tumor, validation of clinically relevant gene signatures for prognostication has relied upon single-tissue samples, including 2 commercially available multigene tests (MGTs). Here we report an unanticipated impact of intratumor heterogeneity (ITH) on risk prediction of recurrence in NSCLC, underscoring the need for a better genomic strategy to refine prognostication. By leveraging label-free, inertial-focusing microfluidic approaches in retrieving circulating tumor cells (CTCs) at single-cell resolution, we further identified specific gene signatures with distinct expression profiles in CTCs from patients with differing metastatic potential. Notably, a refined prognostic risk model that reconciles the level of ITH and CTC-derived gene expression data outperformed the initial classifier in predicting recurrence-free survival (RFS). We propose tailored approaches to providing reliable risk estimates while accounting for ITH-driven variance in NSCLC.
Emerging multiregion sequencing data provide clear evidence of genomic intratumor heterogeneity (ITH) in largely smoking-dominated Caucasian lung cancers (1–3). Recently, we observed a complex genomic landscape with variegated copy number landscape and early diversification in never-smoker Asian non–small-cell lung cancer (NSCLC), despite low mutation burden (4). The clinical consequences of such ITH at multiple molecular levels are also becoming apparent in other cancer types, suggesting that ITH-driven variance may result in patient misclassification (5–8). Nevertheless, no current multigene test (MGT) had factored in ITH for feature selection (7), including 2 gene expression-based MGTs for lung cancer patients (9, 10).
By applying a prognostic multigene classifier to multiregion profiling data, we first delineated transcriptomic ITH and examined the extent to which NSCLC patient stratification was confounded by ITH. The classifier, termed tumor matrisome index (TMi), has been validated for its predictive value in prognosis and adjuvant chemotherapy response in more than 2,000 patients with early-stage NSCLC (11). In essence, TMi is computed based on the expression level of 29 matrisome genes, primarily encoding noncore matrisome proteins including extracellular matrix (ECM) regulators (MMP12, MMP1, ADAMTS5), ECM-affiliated proteins (GREM1, SFTPC, SFTPA2, SFTPD, FCN3), secreted factors (S100A2, CXCL13, WIF1, CHRDL1, CXCL2, IL6, HHIP, S100A12), and other ECM-related components (LPL, CPB2, MAMDC2, CD36), as well as core matrisome molecules including collagens (COL11A1, COL10A1, COL6A6) and ECM glycoproteins (SPP1, CTHRC1, TNNC1, ABI3BP, PCOLCE2), all of which were found to be more differentially expressed in NSCLC compared with matched tumor-free tissues (11).
Here, we found that, even though TMi remained a valid prognostic predictor, a significant number of TMi genes displayed substantial ITH and contributed to discordant classifications within the same tumor (having both TMilow and TMihigh sectors), suggesting the need to reconstruct gene signature based on the level of ITH and interpatient heterogeneity (IPH) of actual genes themselves, as recently proposed for breast cancer MGTs (7). We hypothesized that the observed aberrant matrisomal expression pattern accompanying tumor progression in the course of primary tumor invasion might also prove useful and thus be reflected at later steps of metastasis (12), as during circulation. Accordingly, we assessed circulating tumor cells (CTCs), in addition to multiregion primary tumor tissues, to address intratumoral phenotypic variation of prognostic TMi signatures in this work.
This approach was further motivated by recent single-cell sequencing studies suggesting that spatiotemporally heterogeneous CTCs could provide a comprehensive window into metastatic disease at the genomic (13–15) and transcriptomic level (16–18) across various malignancies. Although a single tumor biopsy may not always be representative of the entire tumor harboring spatially segregated clones (19), the spatial and temporal variation of CTCs may recapitulate gene expression and pathways found in primary and metastatic cancer. We further employed single-cell, and not bulk-cell, analysis to rule out possible leukocyte contamination (20–22), which is particularly pronounced in transcriptomic studies when activated leukocytes concurrently overexpress cancer-associated genes, as well as epithelial–mesenchymal transition (EMT) and stem cell markers, given their mesenchymal and hematopoietic nature, complicating expression analysis of CTC-specific transcripts (20, 21). Single-cell analysis further allows evaluation whether cell-to-cell variation in expression of prognostic matrisome signatures would differ in patients based on clinical features and disease status.
Despite the apparent limitation of bulk CTC analysis, however, a generalized workflow for isolation and molecular characterization of single CTCs is lacking as a result of the extreme rarity of detectable and intact CTCs and the associated technical challenges (23). Our group recently developed an integrated ClearCell FX and microfluidic platform workflow to 1) measure full-length mRNA transcriptome from single patient-derived CTCs (24) and 2) detect dominant mutations found in matched primary tumors (25). Uncompromised genetic integrity of ClearCell FX enriched CTCs were evidenced by high-quality sequencing performance metrics in both studies, demonstrating the feasibility of incorporating label-free, marker-independent microfluidic technology for downstream molecular analyses and functional studies. Recent single-cell sequencing studies conducted at different external laboratories further confirmed that the DNA extracted from ClearCell FX-enriched CTCs isolated by DEPArray technology or micromanipulator subjected to whole-genome amplification (WGA) was of high quality and suitable for sequencing, showing the robustness of the ClearCell FX system (26, 27).
Here we employed the same microfluidic approaches to develop an integrative workflow for single-cell gene expression analysis of patient-derived CTCs (SI Appendix, Fig. S1). Single-cell transcriptomic analysis of 61 circulating tumor cells (CTCs) identified specific gene signatures that distinguished metastatic from nonmetastatic NSCLC, providing metastasis-associated biomarkers that could potentially serve as predictors of cancer recurrence (28). Through systematic in silico validation in a total of 2,748 patient-derived samples, we further show that a newly developed risk model comprising exclusively single-CTC-derived signatures, specifically tailored to the level of ITH, has robust prognostic ability in predicting tissue-based recurrence-free survival (RFS), and argue that such approaches may supersede previous attempts in identifying patients with early-stage disease at high risk of NSCLC recurrence.
Results
ITH-Driven Patient Misclassification.
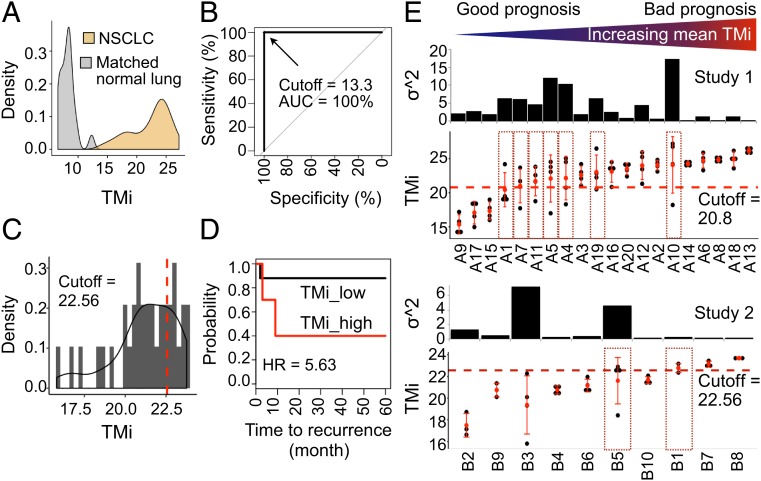
To examine the impact of ITH on risk predictions, we analyzed multiregion gene expression profiles derived from surgical specimens (3 or 4 regions per tumor) from 2 recently published studies (Methods), denoted as study 1 and 2 in this work, using prognostic TMi gene panel (SI Appendix, Fig. S2). As samples were annotated with disease status (tumor or normal) and recurrence-free survival in study 1 (SI Appendix, Table S1) and study 2 (SI Appendix, Table S2), respectively, we first examined diagnostic and prognostic accuracy of TMi. TMi achieved an excellent diagnostic accuracy in differentiating normal from tumor samples in study 1, consisting of 80 regions from 20 early-stage NSCLC tumors and 20 matched normal lung tissues (Fig. 1A), in which sensitivity, specificity, and the area under the receiver operating characteristic (ROC) curve (AUC) were all 100% (Fig. 1B). To test the prognostic performance of TMi, we next stratified all 35 sectors from 10 NSCLC patients from study 2 into TMilow and TMihigh groups based on the optimal cutoff index (Fig. 1C) for recurrence-free survival (RFS) analyses, as previously described (11). In this small patient cohort, tumors predicted as being recurrent by the model had significantly worse survival outcomes, demonstrating a robust predictive value of the TMi for RFS predictions (Fig. 1D). Despite the small sample size, we further assessed the TMi at the patient level by utilizing the highest index for each patient, and observed that 1 of 6 (16.7%) TMilow patients and 2 of 4 (50%) TMihigh patients had recurrence, suggesting that the worse scored sector is sufficient to impact on an adverse outcome (SI Appendix, Fig. S3).
Fig. 1.
ITH-driven patient misclassification in lung cancers. (A) Density distribution of TMi in NSCLC (n = 80) and matched normal lung (n = 20) from study 1. (B) ROC curves using the best TMi cutoff value. (C) Gaussian kernel density distribution of TMi in tumor sectors (n = 35) from study 2. (D) Kaplan–Meier survival curves using the optimal cutoff value (95% CI = 1.4 to 22.7; log-rank P = 0.00628). (E) TMi distribution and the variance of TMi (σ2). The universal cutoff value and the optimal cutoff value were used for patient stratification in study 1 (Top) and study 2 (Bottom), respectively. Dotted red boxes represent discordant tumor samples with TMilow and TMihigh sectors. Patients are ordered by increasing mean TMi.
Having validated the clinical utility of TMi at the tumor sector level, we next computed the level of ITH of each matrisome gene by fitting a linear mixed-effects model (29). A marked ITH in matrisome expression was found in both studies; among the 29 TMi genes analyzed, 7 genes (ADAMTS8, CD36, COL6A6, FCN3, IL6, SFTPD, and WIF1) and 8 genes (ABI3BP, ADAMTS8, COL6A6, CPB2, FCN3, HHIP, LPL, and OGN) displayed greater ITH than IPH in study 1 and study 2, respectively (SI Appendix, Fig. S4). By grouping genes based on the level of ITH as previously described (7), we found that a high proportion of genes (34.5% and 41.4%) exhibited moderate (0.4 to 0.6) to high (0.6 to 1.0) ITH (SI Appendix, Table S3).
Indeed, when we scored each tumor sector according to TMi and applied a universal cutoff index (11) for risk stratification in Affymetrix GPL 570-profiled study 1, 35% of patients (7 of 20) showed discordant tumor samples, having TMilow and TMihigh sectors (Fig. 1 E, Top, dotted red boxes). Similarly, a number of patients in study 2 exhibited significantly high variance of TMi (σ2), with 20% of patients (2 of 10) who could be misclassified if the earlier prognostic cutoff value of 22.56 had been applied (Fig. 1 E, Bottom, dotted red boxes). Of note, the optimal, not universal, cutoff was used in study 2, as samples were assayed with a non-GPL570 platform (11). Although the expression values of these matrisome genes were generally correlated between different sectors within the same tumors, the correlations were weak in a number of patients, including patient 3 from study 2 (SI Appendix, Fig. S5), indicating a substantial level of ITH in tumor microenvironment. Adding to the earlier observations in breast cancer (5, 7, 30), these data highlight the need to consider ITH as a determinant for construction of prognostic gene signatures for lung cancer.
Microfluidic Enrichment of Patient-Derived CTCs at the Single-Cell Level.
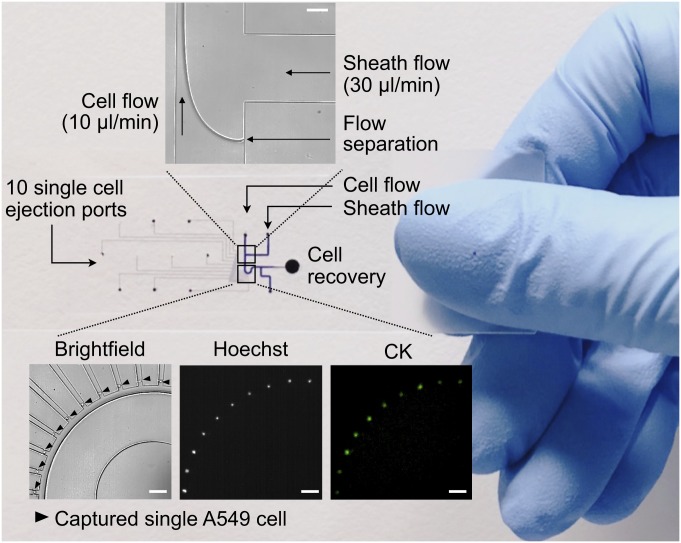
We next examined if cancer cells in primary tumor selected for distant metastasis would further require distinct patterns of matrisome gene expression during circulation and thus could serve as predictors for metastasis or recurrence. Using a cell mechanics-based microfluidic device, we previously demonstrated a size-based separation of single lung CTCs, representative of T790M/L858R mutations found in matched bulk NSCLC tumors, to 100% purity with CD45+ depletion method (25). After having validated the device performance (Fig. 2) and the integrity of mRNA transcripts upon 1% paraformaldehyde (PFA) fixation (31–34) with A549 lung cancer cells (SI Appendix, Fig. S6), we isolated and assessed a total of 61 CTCs from 20 Asian patients with NSCLC, in which the number of analyzed CTCs was less than 5 per 7.5 mL peripheral blood (SI Appendix, Table S4). Such low CTC yield is attributed to high-purity isolation required for single-cell genomic analyses and is comparable to previous studies regarding the number of QC-passed lung CTCs (15, 25, 35).
Fig. 2.
Microfluidic enrichment for single-cell analysis. Inertial focusing, label-free capture of single cancer cells using a microfluidic device (25). (Top) Hydrodynamic focusing of cell flow (A549 lung adenocarcinoma) by sheath flow (glycerol). (Bottom) Bright-field and immunofluorescent images of isolated single A549 cells. (Scale bar, 100 μm.)
Initial screening of 3 candidate housekeeping genes in 14 patient-derived CTCs revealed heterogeneous expression of ACTB (SI Appendix, Fig. S7), which was thus excluded from subsequent normalization. Among the 29 TMi genes that were analyzed, 15 (51.7%) had detectable expression, from which a final multigene panel was established (SI Appendix, Fig. S8 and Table S5). Highly sensitive PCR-based multiplex preamplification protocol was developed and validated through melting curve analysis (SI Appendix, Fig. S9). To probe heterogeneity within the CTC population, we further isolated and analyzed 24 single cells of 7 different lung cancer cell lines. Expression levels of 3 matrisome genes that were up-regulated in primary NSCLC tumors relative to normal lung tissues (11) varied the most, whereas that of down-regulated genes in tumors had the least variability in CTCs compared with cancer cell lines (SI Appendix, Fig. S10).
Distinct Matrisome Profiles in Patients with and without Distant Metastasis.
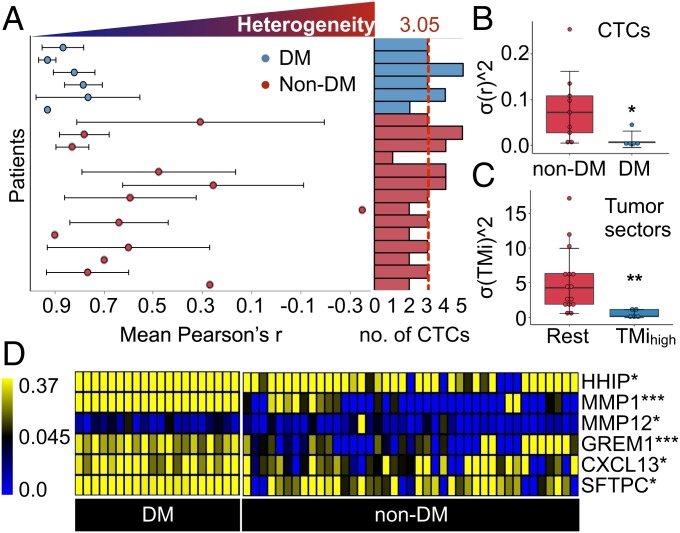
As a result of sample collection and processing from 2 different laboratories, batch effects were observed (SI Appendix, Fig. S11) and removed by using informatics approaches (SI Appendix, SI Materials and Methods) (36). Batch effects should not be exclusive to high-throughput omics data and are also critical to address for low-dimensional qPCR measurements (37). Having processed the expression data of CTCs, we next computed Pearson correlation coefficients (r) between TMi gene-expression levels for each patient to assess the degree of cell-to-cell heterogeneity (Fig. 3A). Interestingly, we found a significantly higher variance (σr2) in TMi expression in CTCs from nonmetastatic disease compared with metastatic (M) disease (Fig. 3B). This is consistent with our earlier observation in NSCLC tumor specimens (study 1), in which high-risk patients (highest quartile) displayed a considerably lower degree of within-patient variance of TMi than the rest of patients (Fig. 3C). A recent study has in fact observed a substantial level of ITH in EMT scores in patients with NSCLC harboring low EMT-scored sectors (38). Altogether these data indicate that there may be an environmental trigger that stabilizes EMT-related processes in advanced cancers. We postulate that quantification of matrisomal ITH may predict early metastasis following surgical resection.
Fig. 3.
Potential predictors of distant metastasis or recurrence. (A) Heterogeneity in 15-gene matrisome expression (±SD) measured by mean Pearson correlation coefficient (r) across all CTCs detected within the same patient with (blue) or without (red) distant metastases (DM). Each clustered bar (Right) represents the number of analyzed CTCs. The vertical red dashed line represents the mean number of analyzed CTCs. (B and C) Intrapatient variability in matrisome gene expression in (B) liquid biopsies and (C) tumor tissues (***P < 0.001 and **P < 0.01, Wilcoxon rank-sum test). (D) Heat map comparing expression profiles of selected matrisome genes between DM and non-DM patient groups (***P < 0.001, **P < 0.01, and *P < 0.05, Wilcoxon rank-sum test).
Transcriptomic characterization of single CTCs identified distinct matrisome profiles in NSCLC with differing metastatic potential (Fig. 3D and SI Appendix, Fig. S12), suggesting potential contribution of CTC-autonomous ECM gene expression to cancer dissemination, as recently proposed (39). Most CTCs expressed high levels of alveolar type II epithelial cell marker (SFTPC), reflecting the common tissue-specific origin for NSCLC. Of 9 genes previously up-regulated in primary tumors compared with tumor-free tissues (11), 4 genes (MMP1, MMP12, GREM1, CXCL13) were highly expressed in CTCs of metastatic NSCLC (SI Appendix, Fig. S13), exhibiting a conserved expression pattern between tumor specimens and liquid biopsies. Accordingly, we hypothesized that these genes implicated in multiple steps of the metastatic spectrum might serve as clinically applicable biomarkers for refining tissue-based prognostication. Taking multiregion profiling data and single-CTC-derived gene expression data into consideration (SI Appendix, Fig. S14), we suggest a prognostic index tailored to the level of ITH, termed as the MMP index (MMPi), consisting of 2-gene MMP signature (Methods).
Identification of Patients at High Risk of Recurrence.
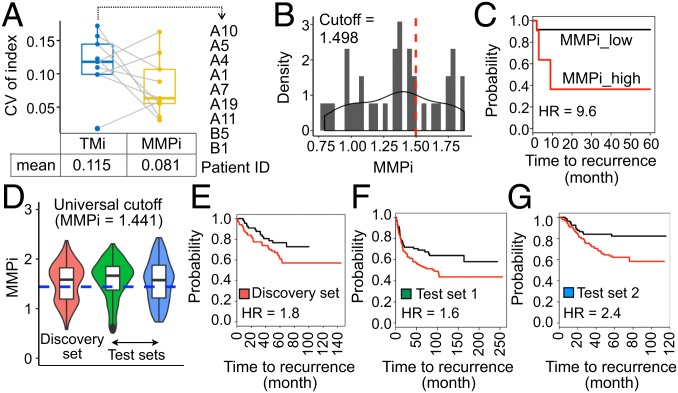
By using the earlier multiregion profiling data (study 1 and study 2), we first computed the coefficient of variation (CV) of the index for patient samples previously identified to have discordant tumor sectors by TMi to assess if the refined metrics would address the issue of ITH-driven discordance in tumor classification. The mean CV of the initial (TMi) and refined (MMPi) model were 0.115 and 0.081, respectively, implying a 30% reduction in the variability of the index in these samples (Fig. 4A). Although 6 of 30 (20%) remained discordant, it accounted for a smaller proportion of patients than TMi when predefined cutoff values were applied to both studies as previously described (SI Appendix, Fig. S15). Importantly, compared with TMi (Fig. 2D), MMPi demonstrated superior performance in classifying tumors with markedly different RFS outcomes at tumor sector and patient levels with optimal cutoffs in our discovery cohort (Fig. 4 B and C and SI Appendix, Fig. S16), highlighting the improved accuracy of the refined index in predicting NSCLC recurrence.
Fig. 4.
Refined prognostication in NSCLC patients with MMPi. (A) CV of the index in discordant samples previously identified with TMi. Patient IDs and the mean CV for each prognostic index are stated. (B) Gaussian kernel density distribution of MMPi in tumor sectors (n = 35) from study 2. (C) Kaplan–Meier survival curves using the optimal cutoff value (95% CI = 2.0 to 46.8; log-rank P = 0.00062). (D) Violin plot depicting MMPi distribution in datasets probed with the same profiling platform and the universal cutoff value (blue dotted line). (E–G) Kaplan–Meier survival curves using (E) GSE50081 (95% CI = 0.99 to 3.3; log-rank P = 0.049; n = 177), (F) GSE30219 (95% CI = 1.0 to 2.4; log-rank P = 0.0345; n = 278), and (G) GSE31210 (95% CI = 1.3–4.3; log-rank P = 0.00343; n = 226).
A robust prognostic performance of MMPi was confirmed in multiple independent validation cohorts comprising a total of 2,748 patients with NSCLC (SI Appendix, Fig. S17). The HR varies from 1.71 to 3.78 in 9 (of 12) datasets for overall survival (OS) analyses (SI Appendix, Table S6) and from 1.7 to 4.08 in 5 (of 6) datasets for RFS analyses (SI Appendix, Table S7). Having comprehensive clinical features, TCGA lung adenocarcinomas (LUADs) were used to perform multivariate Cox regression analysis and revealed MMPi as a biomarker independently associated with mortality (SI Appendix, Table S8). Given that the patient classification was done using different cutoffs, which might make clinical translation of our findings rather difficult, we next tested the potential of a common, or universal, cutoff index for all patients.
The Universal MMPi Cutoff for Patient Stratification.
To avoid the effect of profiling platform on scoring, we examined 3 datasets (GSE50081, GSE30219, GSE31210) that were annotated with RFS outcomes and probed with the same profiling platform (Affymetrix GPL570). Two studies (GSE50081, GSE31210) comprised exclusively early-stage (stage I/II) carcinomas, whereas the remaining set (GSE30219) included all 4 stages of cancer. The universal cutoff value was determined as the optimal cutoff value in the discovery set (MMPi = 1.441), and was tested on the other 2 independent test sets (Fig. 4D). Patients stratified according to this fixed, universal cutoff index exhibited significantly different RFS outcomes in both analyzed datasets (Fig. 4 E–G), demonstrating clinical applicability of MMPi in identifying recurrence-prone lung cancers in patients with early-stage NSCLC.
Although the universal cutoff was identified with Affymetrix GPL570, we finally applied it to the earlier study 2 cohort, which probed genes with Affymetrix GeneChip Human Gene 1.0 ST arrays, to assess its clinical applicability in a different profiling platform. Kaplan–Meier survival analyses revealed that 2 of 21 (9.5%) MMPilow sectors and 7 of 14 (50%) MMPihigh sectors had recurrence, and no MMPilow and 3 of 6 (50%) MMPihigh patients had recurrence at the patient level with the universal cutoff value of 1.441 (SI Appendix, Fig. S18). Altogether, these data reinforce and highlight the wide clinical applicability of the present scoring metrics and the predefined cutoff value for better prognostication of recurrence risk in NSCLC.
Discussion
Single-cell analyses of CTCs have revealed clinically useful copy number variations (15, 35) and point mutations (40) while resolving the degree of heterogeneity in lung cancer. However, these findings are pertinent only to epithelial marker-expressing CTCs, missing out on dedifferentiated EpCAM− or mesenchymal/EMT-like CTCs, all of which have been inextricably linked to disease progression and treatment response (41–43). By relying upon a label-free approach, we found that metastatic potential of an NSCLC tumor lies in the profile of its heterogeneity in matrisome expression, which is in turn reflected in the populations of CTCs. In line with the findings supporting a nonexclusive hypothesis of EMT’s contribution to CTC phenotype (44), TMihigh cells in primary tumor may be functionally equipped with key properties required for their survival in bloodstream and metastatic niche formation, particularly given the close association between matrisome and EMT (11, 45). Repetitive observation of CTCs expressing mesenchymal attributes correlated with appearance of metastases in recent clinical studies (44), and the role of their heterogeneity in organ-specific metastases (42, 46, 47) further points toward these aggressive cells as major constituents of putative metastatic founders.
However, it is now apparent that the organs of future metastasis, called premetastatic niches (PMNs), are not just passive receivers of CTCs, but are actively modulated by the tumor-secreted factors or tumor-shed extracellular vesicles (e.g., exosomes) prior to the occurrence of metastasis (48). The well-established regulators of this stepwise progression of PMN are MMPs released by cells in the primary tumor and nonresident cells (e.g., bone marrow-derived cells [BMDCs], stromal fibroblasts, and endothelial cells) recruited at the local PMN site (49–51). The enzymatic activity of MMPs indeed have direct functional impact on vasculature integrity, in which biologically active ECM fragments (e.g., chemoattractant collagen IV peptides) released during ECM degradation promote the recruitment of BMDCs and CTCs to the PMN site (52). Here, we observed the cell-autonomous expression of ECM-modulating genes, specifically MMP1 and MMP12, in metastatic CTCs, providing a potentially new cellular player in remodeling of the ECM at the PMN site. Collectively, our experimental data suggest that MMPihigh CTCs may be an active source of the PMN formation carrying their own “soil” (53), highlighting the significance of tumor stromal signaling during the PMN evolution (54).
Matrisomal abnormalities represent a promising biomarker for prognostication and prediction of immunotherapy response (45). TMi profiles further reflected sex, but not racial, differences (SI Appendix, Fig. S19) previously associated with the prevalence and prognosis of NSCLC (SI Appendix, Fig. S20). Given the presence of such confounding factors, multivariate regression models were fitted and revealed TMi (11) and MMPi (SI Appendix, Table S8) as independent predictors of recurrence and mortality. We further posit that other bodily fluids such as epithelial-lining fluid (ELF) could serve as an alternative preoperative source to tissue biopsy, providing a noninvasive microsampling probe to examine prognostic TMi signature. Our preliminary data confirm the high classification accuracy achieved by TMi in differentiating benign nodules from malignant cancers using ELF samples (SI Appendix, Fig. S21), supporting the increasingly recognized clinical value of biochemical substances in ELF, including tumor markers and tumor-derived nucleic acids, as diagnostic biomarkers of primary lung adenocarcinoma (55). Benign nodules further remained as a nonconfounding variable even in the classification of other lung diseases, such as chronic obstructive pulmonary disease (COPD) and interstitial lung diseases (ILD), validating the robustness of TMi performance (SI Appendix, Fig. S22).
Nevertheless, unlike TMi metrics, for which the clinical utility remains robust for samples with missing expression data in a few genes, MMPi would require the entire gene set given the small number of genes used to construct the assay. Future assessments of whether the proposed metrics could be directly applied to FFPE specimens following surgical resection and quantified with conventional RT-qPCR are warranted to facilitate its incorporation into routine clinical practice.
Methods
Expression Datasets.
Raw data of multisector gene expression profiles from study 1 (GSE33532) were acquired from the National Center for Biotechnology Information (NCBI) Gene Expression Omnibus (GEO) repository through the GEOquery package (56) in R. Preprocessing of data, such as background correction and adjustment, was performed with Robust Multiarray Average (RMA) through the affy package (57). Probes having higher mean expression across the samples were collapsed to the genes. Detailed description, including data preprocessing techniques and clinical information, of study 2 profiling data can be found in the original work (38). For TCGA data processing, the TCGA-Assembler package (58) in R was used to extract normalized RPKM count values. Genes with RPKM counts in at least 20% of the total number of samples were included for subsequent processing using the edgeR package (59), and were normalized with the Trimmed Mean of M-values (TMM) method. GEO datasets were acquired for raw expression profiles as described earlier or processed (normalized) data directly from the NCBI GEO.
Computation of ITH and Prognostic Indices.
The lme4 package (29) in R was used to compute the level of ITH of each matrisome gene through linear mixed-effects analyses as previously described (7). TMi of each patient was computed by using 29 matrisome genes as previously described (11). MMPi was computed by using the same Cox regression coefficient as follows: MMPi = (0.1102 * MMP12 expression) + (0.07096 * MMP1 expression). The optimal cutoff index for survival analyses was defined as the most significant split using the log-rank test, and determined by using a web-based Cutoff Finder algorithm (http://molpath.charite.de/cutoff) as previously described (11).
CTC Enrichment and Single-Cell Isolation.
Informed consent for use of blood samples for CTC analysis in this paper was obtained through protocols approved by the SingHealth Centralized Institutional Review Board. Whole blood samples (7.5 mL) collected from recruited patients with NSCLC were enriched by using the ClearCell FX System according to the manufacturer’s manual (Biolidics). Enriched samples were fixed with 1% PFA before staining with anti-human CD45-PE (eBioscience) and Hoechst 33342, trihydrochloride, and trihydrate (Life Technologies). Preparation of the sample involves adding the enriched, stained cells to a 1-mL syringe and coupling it to the microfluidic device (25). The device was mounted on a microscope (Olympus BX61), and CTCs were selected based on the detection of immunofluorescence (CD45−) by the user. The same principle was used to negatively deplete WBCs (CD45+) in the capture chambers. The cell flow to sheath flow rates were set at constant conditions of 10 μL/min and 30 µL/min, respectively, and achieved with 2 syringe pumps (Chemyx Fusion 200 Classic). The same parameters were used for lung cancer cell lines. The microfluidic device was calibrated by using a high-speed camera (Photron Fastcam 1024PCI), ensuring the cell flow width reached a maximum of 25 μm in the main channel to facilitate cell propulsion in a single file using hydrodynamic focusing. Glycerol 65% (Thermo Fisher Scientific) was used for the sheath buffer. The basic design of the microchannel device consists of 10 chambers that block additional cells from entering once occupied, allowing the capture and isolation of 10 individual cells in the channel.
Single-Cell Lysis and Reverse Transcription.
Recovered single CTCs or cancer cell lines were transferred to 0.2-mL PCR tubes and subjected to RNA extraction using Ambion Single Cell Lysis Kit according to the manufacturer’s specifications (Life Technologies). In each lysed sample, 2.5 μM oligo (dT) primers and 0.5 mM dNTP Mix (Life Technologies) were added, incubated at 65 °C for 5 min, and subsequently cooled on ice for at least 1 min. First-strand buffer (1×), 5 mM DTT, 10 U RNaseOUT Recombinant RNase Inhibitor, and 50 U SuperScript III RT (Life Technologies) were added to a final volume of 20 μL. The following thermal setting was applied to the final RT product on a Veriti 96-well thermal cycler (Applied Biosystems): 25 °C for 5 min, 55 °C for 60 min, and 85 °C for 5 min. cDNA was stored at −20 °C.
Target-Specific Preamplification.
Multigene primer mix (1 μM) was prepared by adding the following components to a nuclease-free (NF) 0.2-mL centrifuge tube: 1 μL of 100 μM forward gene primer, 1 μL of 100 μM reverse gene primer, and NF water up to 100 μL. cDNA template (10 μL) generated from single cells was preamplified in a total volume of 20 μL containing 1× PCRBIO Ultra Mix (PCR Biosystems), 100 nM of each primer, and NF water. The following thermal setting was applied on the PCR cycler: 95 °C for 10 min followed by 25 cycles of amplification (95 °C for 20 s, 60 °C for 1 min, and 72 °C for 20 s) and a final additional incubation at 72 °C for 7 min. Amplified target amplicons were purified before being subjected to purification using Agencourt AMpure XP beads at a 1:1.5 ratio following the manufacturer’s manual (Beckman Coulter), with the final elution in 60 μL of NF water before quantification.
Real-Time Quantitative PCR.
SYBR Green I detection chemistry on a Bio-Rad CFX96 Real-Time PCR Detection System (BioRad Laboratories) was used to carry out qPCR in real time. Diluted RT product (1 μL) was added to a final volume of 10 μL containing 300 nM of each primer (Integrated DNA Technologies), 1× FastStart SYBR Green Master mix (Roche), and NF water. Melting curve analyses were performed to confirm a single peak for primer specificities. The following thermal setting was applied on the RT-qPCR cycler: 95 °C for 10 min followed by 40 cycles of amplification (95 °C for 20 s, 55 °C or 60 °C for 30 s, and 72 °C for 20 s) and a final additional incubation at 72 °C for 7 min. Expression data were normalized to 2 housekeeping genes (GADPH and UBB) with the following equation: relative expression = 2−(Cq[gene of interest] − mean Cq[housekeeping genes]). Each experiment was performed in duplicate.
Data and Code Availability.
Validation datasets used in this study are available at NCBI GEO under the accession codes GSE31210, GSE42127, GSE30219, GSE11969, GSE50081, GSE3141, GSE37745, GSE41271, GSE68465, GSE26939, and GSE19188. Our single cell expression data and an R script for performing PCA can be found in Figshare (https://doi.org/10.6084/m9.figshare.9202241.v1).
Details on cell culture, primer design, multiplex gene panel, and bioinformatics are described in SI Appendix, SI Materials and Methods.
Supplementary Material
Acknowledgments
This work was conceived and carried out at the MechanoBioEngineering Laboratory at the Department of Biomedical Engineering, National University of Singapore (NUS). We acknowledge support provided by the Institute for Health Innovation and Technology (iHealthtech) at NUS. We thank Dr. Won-Chul Lee and Dr. Jianjun Zhang at the MD Anderson Cancer Center for providing multiregion profiling data and Dr. Goh Kah Yee at National Cancer Centre Singapore and Mr. Terence Cheng at Institute of Molecular and Cell Biology for providing lung cancer cell lines. W.-T.L. is supported by the National Medical Research Council (NMRC/CSA/040/2012 and NMRC/CSA-INV/0025/2017). S.B.L. acknowledges support provided by the NUS Graduate School for Integrative Sciences and Engineering, Mogam Science Scholarship Foundation, and Daewoong Foundation.
Footnotes
Conflict of interest statement: C.T.L. serves as an advisor of Biolidics. S.B.L. and C.T.L. have filed a patent for the TMi assay. S.J.T. has filed a patent for a single-cell microfluidic device presented in this work. A.A.S.B., S.J.T., W.-T.L., and C.T.L. are shareholders of Biolidics. The remaining authors declare no competing interest.
This article is a PNAS Direct Submission.
Data deposition: The single cell expression data and an R script for performing PCA have been deposited on Figshare (DOI: 10.6084/m9.figshare.9202241.v1).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1907904116/-/DCSupplemental.
References
- 1.de Bruin E. C., et al. , Spatial and temporal diversity in genomic instability processes defines lung cancer evolution. Science 346, 251–256 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jamal-Hanjani M., et al. ; TRACERx Consortium , Tracking the evolution of non-small-cell lung cancer. N. Engl. J. Med. 376, 2109–2121 (2017). [DOI] [PubMed] [Google Scholar]
- 3.Zhang J., et al. , Intratumor heterogeneity in localized lung adenocarcinomas delineated by multiregion sequencing. Science 346, 256–259 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nahar R., et al. , Elucidating the genomic architecture of Asian EGFR-mutant lung adenocarcinoma through multi-region exome sequencing. Nat. Commun. 9, 216 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barry W. T., et al. , Intratumor heterogeneity and precision of microarray-based predictors of breast cancer biology and clinical outcome. J. Clin. Oncol. 28, 2198–2206 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gerlinger M., et al. , Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. N. Engl. J. Med. 366, 883–892 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gyanchandani R., et al. , Intratumor heterogeneity affects gene expression profile test prognostic risk stratification in early breast cancer. Clin. Cancer Res. 22, 5362–5369 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gulati S., et al. , Systematic evaluation of the prognostic impact and intratumour heterogeneity of clear cell renal cell carcinoma biomarkers. Eur. Urol. 66, 936–948 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bueno R., et al. , Validation of a molecular and pathological model for five-year mortality risk in patients with early stage lung adenocarcinoma. J. Thorac. Oncol. 10, 67–73 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kratz J. R., et al. , A practical molecular assay to predict survival in resected non-squamous, non-small-cell lung cancer: Development and international validation studies. Lancet 379, 823–832 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lim S. B., Tan S. J., Lim W. T., Lim C. T., An extracellular matrix-related prognostic and predictive indicator for early-stage non-small cell lung cancer. Nat. Commun. 8, 1734 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lambert A. W., Pattabiraman D. R., Weinberg R. A., Emerging biological principles of metastasis. Cell 168, 670–691 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lohr J. G., et al. , Whole-exome sequencing of circulating tumor cells provides a window into metastatic prostate cancer. Nat. Biotechnol. 32, 479–484 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lohr J. G., et al. , Genetic interrogation of circulating multiple myeloma cells at single-cell resolution. Sci. Transl. Med. 8, 363ra147 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ni X., et al. , Reproducible copy number variation patterns among single circulating tumor cells of lung cancer patients. Proc. Natl. Acad. Sci. U.S.A. 110, 21083–21088 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Aceto N., et al. , Circulating tumor cell clusters are oligoclonal precursors of breast cancer metastasis. Cell 158, 1110–1122 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Miyamoto D. T., et al. , RNA-seq of single prostate CTCs implicates noncanonical Wnt signaling in antiandrogen resistance. Science 349, 1351–1356 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ramsköld D., et al. , Full-length mRNA-Seq from single-cell levels of RNA and individual circulating tumor cells. Nat. Biotechnol. 30, 777–782 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Alizadeh A. A., et al. , Toward understanding and exploiting tumor heterogeneity. Nat. Med. 21, 846–853 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Aktas B., et al. , Stem cell and epithelial-mesenchymal transition markers are frequently overexpressed in circulating tumor cells of metastatic breast cancer patients. Breast Cancer Res. 11, R46 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Blassl C., et al. , Gene expression profiling of single circulating tumor cells in ovarian cancer–Establishment of a multi-marker gene panel. Mol. Oncol. 10, 1030–1042 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sieuwerts A. M., et al. , Molecular characterization of circulating tumor cells in large quantities of contaminating leukocytes by a multiplex real-time PCR. Breast Cancer Res. Treat. 118, 455–468 (2009). [DOI] [PubMed] [Google Scholar]
- 23.Alix-Panabières C., Pantel K., Challenges in circulating tumour cell research. Nat. Rev. Cancer 14, 623–631 (2014). [DOI] [PubMed] [Google Scholar]
- 24.Ramalingam N., et al. , Abstract 2923: Label-free enrichment and integrated full-length mRNA transcriptome analysis of single live circulating tumor cells from breast cancer patients. Cancer Res. 77 (suppl. 13), 2923 (2017). [Google Scholar]
- 25.Yeo T., et al. , Microfluidic enrichment for the single cell analysis of circulating tumor cells. Sci. Rep. 6, 22076 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yin J., et al. , Characterization of circulating tumor cells in breast cancer patients by spiral microfluidics. Cell Biol. Toxicol. 35, 59–66 (2019). [DOI] [PubMed] [Google Scholar]
- 27.Mohammad S., et al. , ClearCell FX, a Marker-Independent Process for Enriching Viable Circulating Tumour Cells (CTCs) from Melanoma Patients’ Blood (NCRI Cancer Conference, 2016). [Google Scholar]
- 28.Tian X., et al. , Recurrence-associated gene signature optimizes recurrence-free survival prediction of colorectal cancer. Mol. Oncol. 11, 1544–1560 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bates D., Mächler M., Bolker B., Walker S., Fitting linear mixed-effects models using lme4. J. Stat. Soft. 67, 48 (2015). [Google Scholar]
- 30.Drury S., Salter J., Baehner F. L., Shak S., Dowsett M., Feasibility of using tissue microarray cores of paraffin-embedded breast cancer tissue for measurement of gene expression: A proof-of-concept study. J. Clin. Pathol. 63, 513–517 (2010). [DOI] [PubMed] [Google Scholar]
- 31.Machado L., et al. , In situ fixation redefines quiescence and early activation of skeletal muscle stem cells. Cell Rep. 21, 1982–1993 (2017). [DOI] [PubMed] [Google Scholar]
- 32.Pechhold S., et al. , Transcriptional analysis of intracytoplasmically stained, FACS-purified cells by high-throughput, quantitative nuclease protection. Nat. Biotechnol. 27, 1038–1042 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Russell J. N., Clements J. E., Gama L., Quantitation of gene expression in formaldehyde-fixed and fluorescence-activated sorted cells. PLoS One 8, e73849 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Calzone F. J., Britten R. J., Davidson E. H., Mapping of gene transcripts by nuclease protection assays and cDNA primer extension. Methods Enzymol. 152, 611–632 (1987). [DOI] [PubMed] [Google Scholar]
- 35.Carter L., et al. , Molecular analysis of circulating tumor cells identifies distinct copy-number profiles in patients with chemosensitive and chemorefractory small-cell lung cancer. Nat. Med. 23, 114–119 (2017). [DOI] [PubMed] [Google Scholar]
- 36.Lim S. B., Single-cell analysis of circulating tumor cells. Figshare. 10.6084/m9.figshare.9202241.v1. Deposited 1 August 2019. [DOI]
- 37.Leek J. T., et al. , Tackling the widespread and critical impact of batch effects in high-throughput data. Nat. Rev. Genet. 11, 733–739 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lee W. C., et al. , Multiregion gene expression profiling reveals heterogeneity in molecular subtypes and immunotherapy response signatures in lung cancer. Mod. Pathol. 31, 947–955 (2018). [DOI] [PubMed] [Google Scholar]
- 39.Ting D. T., et al. , Single-cell RNA sequencing identifies extracellular matrix gene expression by pancreatic circulating tumor cells. Cell Rep. 8, 1905–1918 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Park S. M., et al. , Molecular profiling of single circulating tumor cells from lung cancer patients. Proc. Natl. Acad. Sci. U.S.A. 113, E8379–E8386 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yu M., et al. , Circulating breast tumor cells exhibit dynamic changes in epithelial and mesenchymal composition. Science 339, 580–584 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang L., et al. , The identification and characterization of breast cancer CTCs competent for brain metastasis. Sci. Transl. Med. 5, 180ra48 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Satelli A., et al. , EMT circulating tumor cells detected by cell-surface vimentin are associated with prostate cancer progression. Oncotarget 8, 49329–49337 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Francart M. E., et al. , Epithelial-mesenchymal plasticity and circulating tumor cells: Travel companions to metastases. Dev. Dyn. 247, 432–450 (2018). [DOI] [PubMed] [Google Scholar]
- 45.Bin Lim S., et al. , Pan-cancer analysis connects tumor matrisome to immune response. NPJ Precis. Oncol. 3, 15 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Alix-Panabières C., Riethdorf S., Pantel K., Circulating tumor cells and bone marrow micrometastasis. Clin. Cancer Res. 14, 5013–5021 (2008). [DOI] [PubMed] [Google Scholar]
- 47.Boral D., et al. , Molecular characterization of breast cancer CTCs associated with brain metastasis. Nat. Commun. 8, 196 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Peinado H., et al. , Pre-metastatic niches: Organ-specific homes for metastases. Nat. Rev. Cancer 17, 302–317 (2017). [DOI] [PubMed] [Google Scholar]
- 49.Gupta G. P., et al. , Mediators of vascular remodelling co-opted for sequential steps in lung metastasis. Nature 446, 765–770 (2007). [DOI] [PubMed] [Google Scholar]
- 50.Hiratsuka S., et al. , MMP9 induction by vascular endothelial growth factor receptor-1 is involved in lung-specific metastasis. Cancer Cell 2, 289–300 (2002). [DOI] [PubMed] [Google Scholar]
- 51.Kaplan R. N., et al. , VEGFR1-positive haematopoietic bone marrow progenitors initiate the pre-metastatic niche. Nature 438, 820–827 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Erler J. T., et al. , Hypoxia-induced lysyl oxidase is a critical mediator of bone marrow cell recruitment to form the premetastatic niche. Cancer Cell 15, 35–44 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Nowell P. C., The clonal evolution of tumor cell populations. Science 194, 23–28 (1976). [DOI] [PubMed] [Google Scholar]
- 54.Zhang X. H., et al. , Selection of bone metastasis seeds by mesenchymal signals in the primary tumor stroma. Cell 154, 1060–1073 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Uchida A., et al. , Napsin A levels in epithelial lining fluid as a diagnostic biomarker of primary lung adenocarcinoma. BMC Pulm. Med. 17, 195 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Davis S., Meltzer P. S., GEOquery: A bridge between the gene expression omnibus (GEO) and bioconductor. Bioinformatics 23, 1846–1847 (2007). [DOI] [PubMed] [Google Scholar]
- 57.Gautier L., Cope L., Bolstad B. M., Irizarry R. A., Affy–Analysis of Affymetrix GeneChip data at the probe level. Bioinformatics 20, 307–315 (2004). [DOI] [PubMed] [Google Scholar]
- 58.Zhu Y., Qiu P., Ji Y., TCGA-assembler: Open-source software for retrieving and processing TCGA data. Nat. Methods 11, 599–600 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Robinson M. D., McCarthy D. J., Smyth G. K., edgeR: A bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Validation datasets used in this study are available at NCBI GEO under the accession codes GSE31210, GSE42127, GSE30219, GSE11969, GSE50081, GSE3141, GSE37745, GSE41271, GSE68465, GSE26939, and GSE19188. Our single cell expression data and an R script for performing PCA can be found in Figshare (https://doi.org/10.6084/m9.figshare.9202241.v1).
Details on cell culture, primer design, multiplex gene panel, and bioinformatics are described in SI Appendix, SI Materials and Methods.