Significance
Iron is essential for microbial growth, and minimizing iron acquisition by pathogens is a cornerstone of the nutritional immunity response. Nevertheless, strategies by which intracellular pathogens circumvent these defenses are poorly understood. Herein, we demonstrate that the highly conserved Legionella effector protein MavN is the conduit for iron piracy by the bacterium. We map the in-membrane topology of MavN and pinpoint mutations that render the protein incompetent for sustaining Legionella intracellular proliferation. MavN iron transport activity was reconstituted in vitro and mutations identified as critical for growth were similarly detrimental to iron uptake. Our findings illuminate mechanistic details of iron accrual by Legionella and illustrate a scheme by which opportunistic bacteria can procure a limited but crucial nutrient.
Keywords: Legionella, iron, MavN, transport, vacuole
Abstract
Legionella pneumophila causes a potentially fatal form of pneumonia by replicating within macrophages in the Legionella-containing vacuole (LCV). Bacterial survival and proliferation within the LCV rely on hundreds of secreted effector proteins comprising high functional redundancy. The vacuolar membrane-localized MavN, hypothesized to support iron transport, is unique among effectors because loss-of-function mutations result in severe intracellular growth defects. We show here an iron starvation response by L. pneumophila after infection of macrophages that was prematurely induced in the absence of MavN, consistent with MavN granting access to limiting cellular iron stores. MavN cysteine accessibilities to a membrane-impermeant label were determined during macrophage infections, revealing a topological pattern supporting multipass membrane transporter models. Mutations to several highly conserved residues that can take part in metal recognition and transport resulted in defective intracellular growth. Purified MavN and mutant derivatives were directly tested for transporter activity after heterologous purification and liposome reconstitution. Proteoliposomes harboring MavN exhibited robust transport of Fe2+, with the severity of defect of most mutants closely mimicking the magnitude of defects during intracellular growth. Surprisingly, MavN was equivalently proficient at transporting Fe2+, Mn2+, Co2+, or Zn2+. Consequently, flooding infected cells with either Mn2+ or Zn2+ allowed collaboration with iron to enhance intracellular growth of L. pneumophila ΔmavN strains, indicating a clear role for MavN in transporting each of these ions. These findings reveal that MavN is a transition-metal-ion transporter that plays a critical role in response to iron limitation during Legionella infection.
The gram-negative bacterium Legionella pneumophila grows within alveolar macrophages during pneumonic disease (1). Infection of the lungs results from aspiration of aerosolized or ingested water supplies contaminated with bacteria growing in biofilms or within environmental amoebae, the natural host of the microorganism (2, 3). After internalization by phagocytes, the bacterium avoids entry into the host endolysosomal network by establishing the Legionella-containing vacuole (LCV), a membrane-bound compartment that is tightly associated with components of the host cell endoplasmic reticulum (4, 5). Once the LCV is established, bacteria grow within the vacuole until the host cell lyses, liberating bacteria for another round of intracellular replication, ∼18 to 24 h after initial contact.
Intracellular growth within the LCV strictly requires the Icm/Dot type IV secretion system (T4SS) (6, 7). An impressive cadre of bacterial proteins is translocated through this system into host cells, with typical clinical isolates having 300 or more different effector proteins extruded into the host cell (8, 9). The activities of a large number of these proteins have been described, including hijacking host anterograde membrane traffic proteins, modulating endocytic processes, or interfering with host protein synthesis (10–23). In contrast to the absolute requirement for T4SS function, single effector deletions are strikingly competent for intracellular replication. Such capability alludes to pervasive genetic redundancy in this system, presumably due to the accumulation of genes necessary to support encounters with diverse amoebal hosts in the environment (24). An exception to this rule is the T4SS substrate MavN (lpg2815) that localizes circumferentially about the LCV (25). Deletion mutants lacking MavN exhibit growth arrest after approximately one bacterial division, although these mutants show no growth defect in broth cultures. Mutants lacking MavN also construct LCVs that are morphologically indistinguishable from wild-type (WT) organisms, indicating that the protein plays no role in interfacing with the host secretory pathway (25, 26).
MavN is one of only 7 highly conserved “core effectors” within the Legionella clade, out of almost 6,000 such proteins identified, arguing it fulfills a critical role during replication in a large number of hosts (8). Previous work supported a role in iron uptake by LCVs. MavN transcription is regulated by iron availability and its promoter contains a ferric uptake regulator (Fur)-binding element (Fur box) (25, 26). Furthermore, flooding macrophages with either Fe2+ or Fe3+ ions partially bypasses a requirement for MavN function, consistent with the inability of the mutant to acquire iron under normal homeostatic conditions of the host cytoplasm (25). Particularly low levels of iron are available for exploitation by intracellular pathogens in the host cytosol, as the metal is found in trace quantities within labile iron stores or tightly associated with cytoplasmic iron-binding proteins in the insoluble ferric (Fe3+) form (27). Therefore, to propagate themselves, intravacuolar pathogens must solve two problems: first, identify accessible iron stores within the host cytoplasm, and then convey the nutrient into an insulated, membrane-bound compartment. In the case of many intravacuolar pathogens, iron is acquired via the endocytic pathway, permitting access to transferrin-bound iron (28, 29). In contrast, the L. pneumophila vacuole is sequestered from the endocytic path. Therefore, the well-characterized routes of iron acquisition during growth in culture, using either the inner-membrane FeoB transporter (30) or siderophores (31, 32), are predicted to be insufficient to support intracellular growth in all cell types. Given MavN regulation by iron levels, its localization about the vacuole, and its predicted multipass integral membrane topology, this effector is hypothesized to be a transporter for piracy of iron from host cells (25, 26).
In this report, we test the model that MavN is an iron transporter by topological mapping, reconstituting transport activity with purified protein, and interrogating the importance of conserved residues in both transport activity and Legionella growth. This approach revealed defective intracellular proliferation resulting from impaired iron uptake by MavN and uncovered a role for the protein in transporting multiple divalent cations into replication vacuoles.
Results
An LCV-Activated Iron Starvation Response That Is Exacerbated by the Absence of MavN.
MavN is localized about the Legionella-containing vacuole and predicted to be a multipass integral membrane protein based on amino acid sequence. Transcription of the mavN gene is repressed by iron, raising the possibility that the protein only functions under conditions of iron starvation (25, 26).
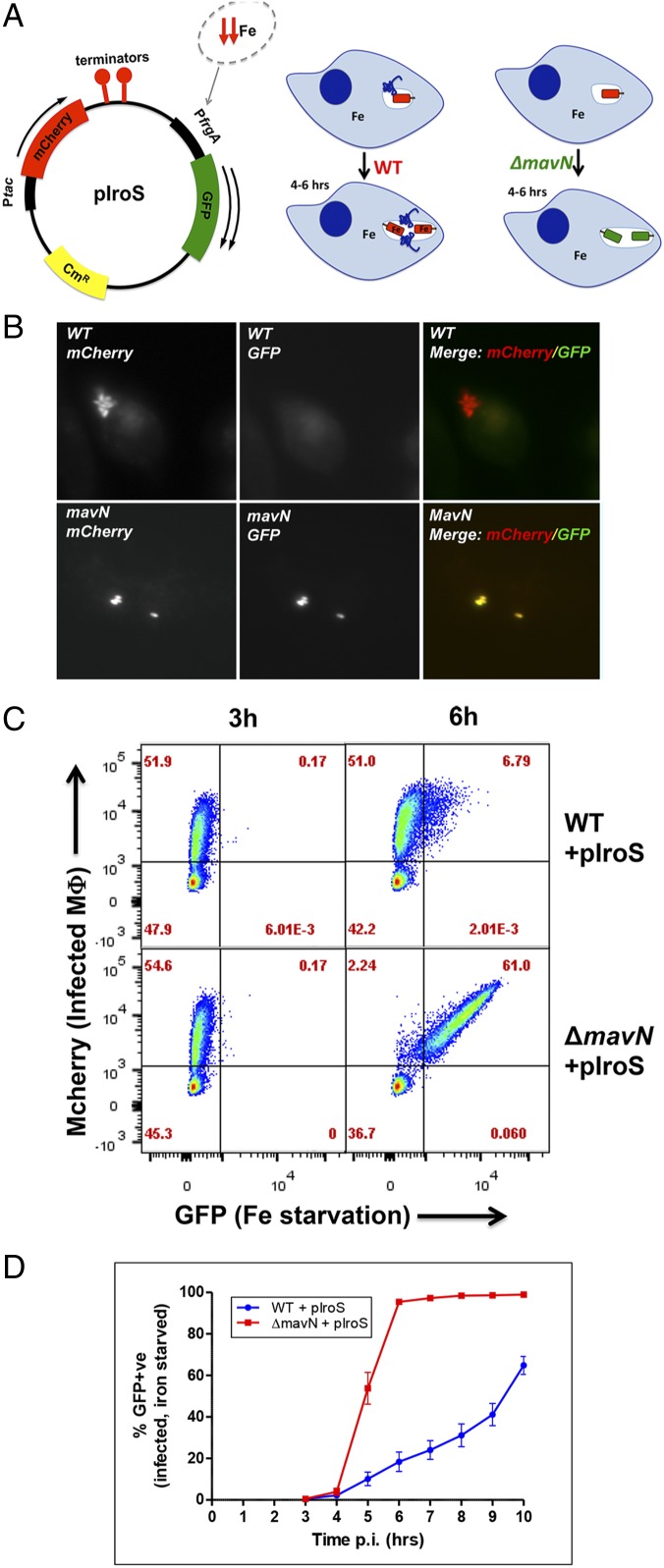
To determine if iron-regulated genes are produced throughout the L. pneumophila infection cycle to promote iron access, we generated a fluorescent reporter for assessing bacterial iron starvation (Fig. 1A). WT and ΔmavN strains were constructed in which GFP was placed under control of the iron-repressible frgA promoter in a previously described vector (33) with mCherry expression controlled by the Ptac promoter to allow bacterial detection (Fig. 1A). Analysis of bone marrow-derived macrophages (BMDMs) 10 h post infection (hpi) showed robust expression of the frgA-GFP construction in the ΔmavN strain, while the WT was largely repressed (Fig. 1B). Flow cytometric analysis of infected macrophages 6 h after challenge with bacteria indicated that the majority of ΔmavN-infected macrophages harbored bacteria undergoing iron starvation, while iron starvation was observed in ∼10-fold fewer WT-infected macrophages (6.8 vs. 61%, respectively; Fig. 1C). This disparity reveals that there is sufficient mavN expression at early time points to prevent iron starvation. Even in WT bacterial populations undergoing iron starvation in BMDMs at 6 hpi there were lower GFP levels relative to the ΔmavN strain, consistent with basal expression of MavN having the functional consequence of delaying and down-modulating the iron starvation response (Fig. 1C). To measure iron starvation kinetics in the two strain backgrounds, triplicate infections were collected at 1-h time points, starting at 3 hpi, and analyzed by fluorescence-activated cell sorting (FACS). By 5 hpi, a majority of the ΔmavN strain-infected macrophages showed detectable GFP, with the entire population evincing iron starvation 1 h later (Fig. 1D). In contrast, the percentage of GFP-positive WT strain-infected macrophages increased slowly over time (Fig. 1D), corresponding to iron starvation that occurs as a consequence of continued replication and increased demands on the intravacuolar iron supplies. We conclude that the expression of MavN allows bacteria to establish infection without experiencing iron starvation during the time period in which the replication compartment is being established.
Fig. 1.
MavN function suppresses the Legionella iron starvation response. (A) A fluorescent reporter allows quantification of the intracellular iron starvation response. The pIroS plasmid, derived from pXDC94 (33), enables detection of iron starvation by derepression of the frgA promoter. Bacteria undergoing starvation response appear GFP+. (B) In the presence of MavN, large replication centers exist that exhibit no iron starvation response. Shown are BMDMs challenged with the noted L. pneumophila strains harboring pIroS in the presence of isopropyl β-d-1-thiogalactopyranoside for 10 h, fixed, and processed for fluorescence microscopy to detect mCherry (under Ptac control) and GFP (repressed by iron). (C) By 6 hpi, the majority of BMDMs harboring the ΔmavN strain exhibited an iron starvation response. BMDMs were challenged for the indicated times with the noted strains harboring pIroS, fixed, and processed for FACS analysis. Displayed are the percentage of total cells found in the noted gates. (D) L. pneumophila ΔmavN strain exhibits an iron starvation response shortly after infection. BMDMs were challenged with the noted L. pneumophila strains, fixed at various times after infection, and processed for FACS analysis to determine the fraction of BMDMs harboring bacteria that fire the frgA promoter. Experiments displayed in C and D were performed on separate days. Data shown are means ± SE.
Topology of MavN Predicts a Multipass Membrane Transporter.
As reported previously (25), a hydrophobicity algorithm predicts that MavN has 8 transmembrane (TM) regions when inserted into the LCV [TOPCONS (34)]. We also confirmed experimentally that the MavN amino terminus is present in the host cell cytosol (25). To further validate this 8-TM model, we devised a strategy to selectively label cytosol-exposed regions during intracellular growth using the transmembrane-substituted cysteine accessibility method (TM-SCAM) (35, 36). Cysteine (Cys) replacements were made at various residues throughout the FLAG-MavN coding sequence, introduced into L. pneumophila, and used to challenge phorbol ester-transformed U937 cells (7). After plasma membrane disruption, cytosol accessibility of the single-Cys derivatives was assayed by exposing intact LCVs to the bilayer-impermeant sulfhydryl-reactive maleimide-PEG2-biotin (MP2B), followed by immunoprecipitation with anti-FLAG and immunoblotting with anti-biotin (Fig. 2 A and B). To this end, a cysteineless (Cys0) mutant of FLAG-MavN was constructed by mutating all 3 endogenous Cys residues to Ala (C101A/C301A/C509A). Single-Cys mutations were introduced on this Cys0 background and ΔmavN strains harboring plasmids with each of the mutations were analyzed. The Cys0 strain replicated relatively efficiently in BMDMs, with yields approaching that of WT after 72-h incubation with macrophages (Fig. 2C; P ∼ 1).
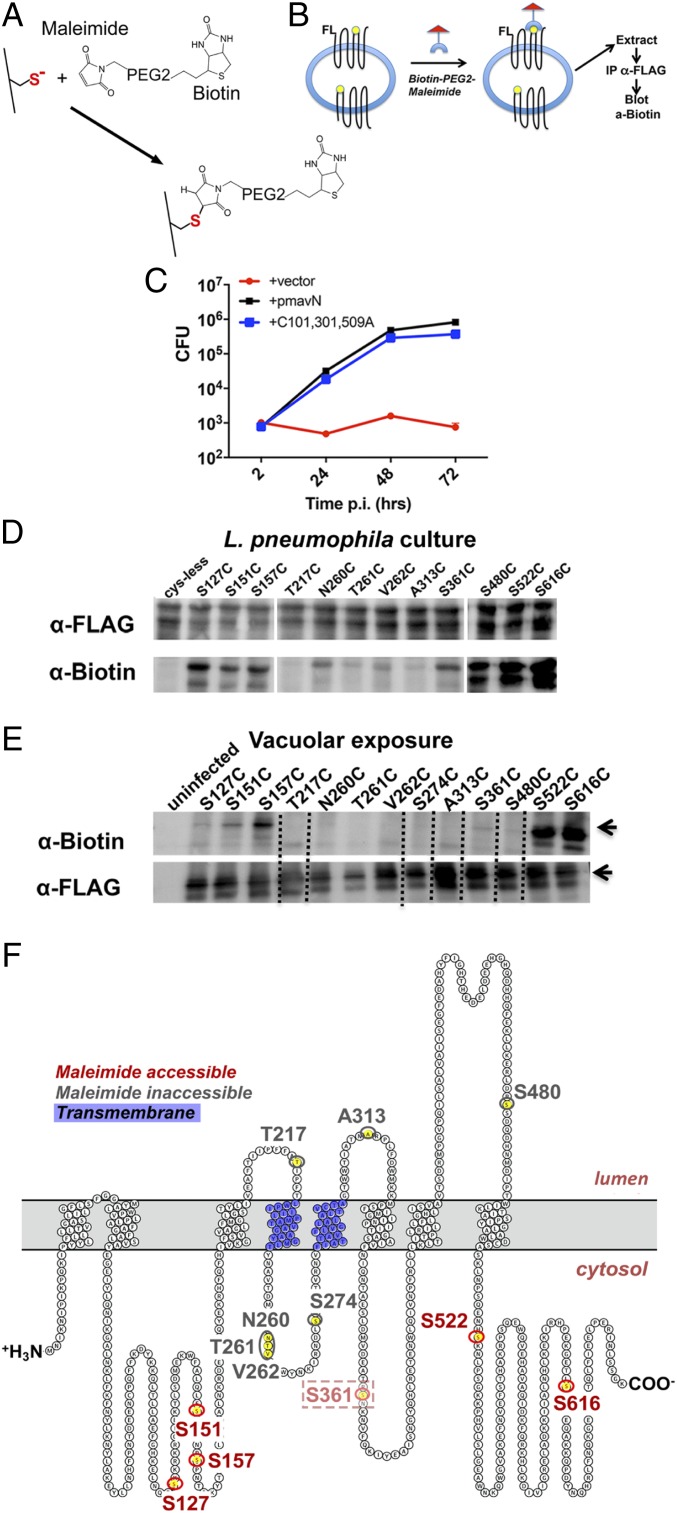
Fig. 2.
MavN topology determined by the scanning cysteine accessibility method. (A) Maleimide-PEG2-biotin covalently links to exposed sulfhydryls to effect biotin labeling. (B) Disrupted macrophages permit access of label to FLAG-MavN on the LCV surface as detected by immunoprecipitation (IP) with anti-FLAG (MavN) followed by blotting with anti-biotin. (C) BMDMs were challenged with the indicated L. pneumophila ΔmavN strains harboring a plasmid copy of either WT mavN or the mavN Cys0 derivative harboring the indicated Cys substitution mutants (mavN*) at an MOI of 0.01. At 72 hpi, the infected macrophages were lysed and plated for bacterial CFUs. Fold increase in CFUs relative to 2 hpi is plotted. Data are representative of triplicate infections. (D) Identification of Cys substitutions accessible to MP2B during L. pneumophila growth in broth. Extracts of the indicated ΔmavN strains harboring plasmids with the indicated substitution mutations were incubated with MP2B, blocked, immunoprecipitated with anti-FLAG, fractionated on 7.5% SDS/PAGE, and probed with either anti-FLAG or anti-biotin to detect derivatives with reactive (accessible) Cys substitutions. (E) U937 macrophages were challenged at an MOI of 5 with the ΔmavN strain harboring plasmids with the indicated substitution mutations. At 6 hpi, infected cells were mechanically lysed under conditions that preserve intact Legionella-containing vacuoles (Materials and Methods) (37). The lysate was then incubated with MP2B to label exposed Cys prior to protein isolation and extraction from vacuolar membranes. Proteins were immunoprecipitated with anti-FLAG, separated on 7.5% SDS/PAGE, and probed with either anti-FLAG or anti-biotin. Dotted lines represent predicted transmembrane domains. (F) MavN protein prediction based on TOPCONS. Shown are map positions of Cys substitution mutations. Red: residues reactive with MP2B; charcoal: nonreactive residues.
The reactivity of the maleimide reagent was then tested against FLAG-MavN derivatives having the single-Cys substitutions in the Cys0 background. Each of the mutant derivatives was present at high steady-state levels during growth of L. pneumophila in broth culture, indicating that the mutations did not destabilize the derivatives prior to translocation (Fig. 2D, α-FLAG). To determine accessibility of the proteins to sulfhydryl-reactive reagents during broth growth, Triton X-100 extracts were prepared, incubated with MP2B, immunoprecipitated with anti-FLAG, and probed on immunoblots with anti-biotin (Fig. 2D and SI Appendix, Fig. S1). Surprisingly, many of the Cys1 mutants exhibited low accessibility to MP2B during broth growth, with one (T217C) showing no detectable reaction with the reagent (Fig. 2D). MavN may be in a conformation that prevents accessibility of the reagent to some of the Cys1 substitutions or the protein is bound by chaperones that prevent exposure of the Cys residues during bacterial growth in culture. This nonuniform labeling in the absence of membrane insertion complicates the analysis, but a sufficiently diverse group of derivatives displayed reactivity to enable further topological analysis after LCV insertion during intracellular growth.
To identify cytosol-exposed regions of MavN, L. pneumophila harboring Cys1 derivatives were incubated with BMDMs for 6 h to allow LCV establishment. BMDMs were then disrupted by Dounce homogenization using conditions known to preserve the LCV integrity (37) and the crude lysates were incubated with MP2B. After solubilization with Triton X-100 to extract FLAG-MavN from LCVs, samples were immunoprecipitated with anti-FLAG, fractionated, and then immunoprobed for FLAG-MavN and biotinylated MavN (Fig. 2E). All Cys1 FLAG-MavN derivatives were efficiently inserted and extracted from the LCV, implying they were in a well-folded, WT-like conformation (Fig. 2E). When the Cys1 derivatives were probed with MP2B, 3 noncontiguous regions of the protein, separated by even numbers of ∼20-residue hydrophobic stretches, were found to have maleimide-reactive Cys1 substitutions (Fig. 2E). All biotinylated residues are predicted to be on the cytosolic side of the LCV membrane, with the carboxyl-terminal 150 residues having 2 particularly reactive Cys substitutions (S522C, S616C; Fig. 2 E and F). The reactivity pattern of these residues agrees with the previously described 8-TM model. It should be noted that one of these residues, S361C, showed detectable reactivity, with MP2B, but this was low relative to the other reactive residues (Fig. 2 E and F, designated as faint red). The protein also showed low reactivity when lysed L. pneumophila was probed with MP2B, consistent with the S361C substitution being relatively poorly accessible even under complete lysis conditions. This pattern is also consistent with 3 Cys1 mutants (T217C, A313C, S480C) that are nonreactive and placed in the LCV lumen by the 8-TM model. The identification of 4 nonreactive Cys1 substitution mutations predicted to be cytosolically exposed within a 35-amino acid loop between Y234 and V279 (Fig. 2 E and F, flanked by blue residues) is likely due to the 234- to 279-aa loop being masked by interdomain interactions or this segment forming a partial reentrant moiety in the membrane that is resistant to labeling. These 3 residues show strikingly low reactivity when completely lysed L. pneumophila is treated with MP2B (Fig. 2D, α-biotin), implying the probe is physically occluded from these residues.
Identification of MavN Residues Required for Function during Intracellular Growth.
To initiate structure–function analyses of MavN with the goal of connecting intracellular growth to in vitro iron transport, a series of directed mavN point mutations was designed based on sequence conservation, focusing on residues that typically coordinate first-row transition-metal ions. These mutations, constructed in an otherwise WT gene, were introduced on plasmids into a ΔmavN strain and assayed for intracellular growth (Fig. 3). The information derived from this approach was then used to prioritize mutants for analysis in a reconstituted transport assay.
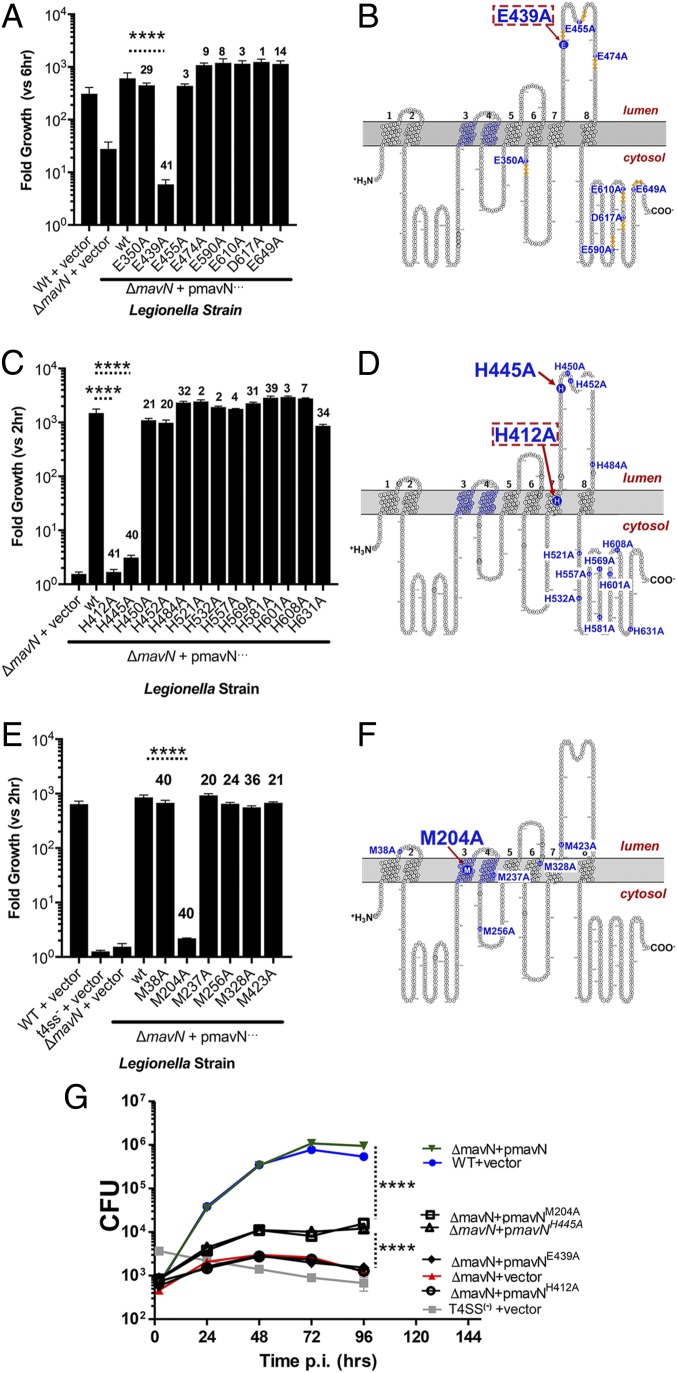
Fig. 3.
Putative iron-coordinating residues are critical for MavN function during intracellular growth. (A, C, and E) BMDMs were challenged with the indicated strains of Legionella at an MOI of 0.01. At 72 hpi, intracellular bacterial growth was determined based on either measurement of bacterial luminescence (A) or enumeration of bacterial CFUs from infected cell lysates (C and E) and is plotted as the fold increase relative to either 6 hpi (A) or 2 hpi (C and E). The numbers above each bar represent the number of Legionella species for which the residue of interest is conserved [from a total of 41 species examined (8)]. (B, D, and F) Map position of each of the mutations analyzed in the graph directly to the left of each map. Large font: substitutions resulting in a defect in intracellular growth; red dashed boxes: substitutions showing no ability to support intracellular growth. (G) Substitution mutations can be separated into two classes based on magnitude of defects. BMDMs were challenged with the indicated L. pneumophila strains at an MOI of 0.01. At the indicated time points, cells were lysed and bacterial CFUs were enumerated (Materials and Methods). Data are from triplicate infections. For statistics, an unpaired, two-tailed Student’s t test was performed on logarithmically transformed data of mutants relative to the ΔmavN/pmavN+ strain in each panel. ****P < 0.0001. Data shown are means ± SE.
We have previously determined that the E439A substitution, now mapped to the luminal side of the LCV in the loop that extends from amino acids 420 to 490, interferes with intracellular growth (25). This residue is at the amino terminus of the tetrapeptide EGFE, which corresponds to an EXXE motif associated with iron-binding regions of fungal iron transporters, ferritin, and bacterial iron sensors (38–41). There are several of these motifs in MavN, particularly in poorly conserved residues in the carboxyl-terminal cytosolic domain (Fig. 3B). When single E→A or D→A substitutions were introduced into each of these motifs and the resultant MavN mutants were produced in the ΔmavN strain, only the previously characterized E439A substitution resulted in a growth defect (Fig. 3A), and this phenotype was indistinguishable from that of a ΔmavN strain (Fig. 3G). Notably, E439 was the only absolutely conserved residue among this group of targeted MavN residues in the Legionella clade. Among the 41 Legionellaceae species sequenced, including amoeba-associated organisms not known to cause human disease, all 41 MavN orthologs have the conserved E439 residue (8) (SI Appendix, Dataset S1). In contrast, with the exception of E350 located in a cytosol-accessible loop (present in 29/41 isolates), the others were poorly conserved, with a distinct minority having the L. pneumophila Philadelphia-1 residue present (Fig. 3A and SI Appendix, Dataset S1). The predicted luminal location of E439 comports with a role in iron binding or in mediating important contacts in at least one of the states in the transport pathway (Fig. 2F).
In addition to the EXXE motif, the luminal 420-to-490 loop is enriched in His residues that have been associated with a number of iron-binding proteins and transporters (42, 43). We demonstrated previously that a MavN mutant with a triple H→A substitution (H445A/H450A/H452A) impaired intracellular L. pneumophila growth (25). To identify the specific His residues involved in supporting L. pneumophila intracellular growth, single H→A changes were made including the only His predicted to be in a transmembrane segment as well as all residues C-terminal to the site of the triple mutant (Fig. 3 C and D). The single H445A mutation reproduced the defect observed in the original triple mutant, although there was detectable growth relative to a ΔmavN strain (Fig. 3G). H445 is well-conserved, with 40/41 Legionellaceae MavN orthologs having this residue. No other single-His mutations in the carboxyl-terminal hydrophilic region caused defective growth (Fig. 3C). In contrast, the TM-localized H412A mutation elicited growth defects that were indistinguishable from a ΔmavN strain—and even stronger than the H445A mutation—arguing that this residue alters metal transport across the membrane (Fig. 3G). H412 is highly conserved, with all sequenced Legionellaceae MavN orthologs having this residue, while most of the nonessential His residues were weakly conserved, with the exception of H581 (SI Appendix, Dataset S1).
A single Met residue has been demonstrated to afford Nramp1 selectivity for transition metals such as Fe, Co, Mn, and Zn (44). In addition, Met residues are proposed to play important roles in Cu transport by bacterial metal efflux pumps (45). MavN has 4 Met residues predicted to be within (M204, M237, M328) or directly abutting TM regions (M38). Two of these are well-conserved (M38 and M204, found in 40/41 MavN orthologs) (Fig. 3E). Alanine substitutions were introduced for each of these 4 Met residues as well as 2 other Met residues predicted to be located on opposite sides of the membrane (M256, M423) (Fig. 3F). Only Ala substitutions in the highly conserved M204 evoked a considerable growth defect (Fig. 3E), with the mutant protein retaining some residual function (Fig. 3G). Interestingly, in no case, based on the 8-TM model, were we able to identify predicted cytoplasmic residues that, when mutated, depressed growth. In summary, analysis of these mutants pointed to MavN being an iron transporter, in which case the chemistry of the important residues could be reconciled in the context of its metal-ion transport function. To gain direct evidence for such an activity, MavN was purified and tested for transport activity.
Purification of Functional MavN from Pichia pastoris.
To obtain biochemical support for a transport function, we sought to reconstitute the activity of MavN in a proteoliposome-based assay with purified protein. Confoundingly, initial attempts to express MavN in Escherichia coli yielded minuscule quantities with no discernable transport activity after liposome incorporation. Integral membrane proteins often present such an obstacle, but this can often be circumvented by testing expression of suitable orthologs that are dissimilar in amino acid sequence. Database mining for MavN orthologs revealed that the protein is present only in Legionellaeae, perhaps owing to its specialized function in the LCV, and that the sequence variation was quite narrow (SI Appendix, Dataset S1). Although MavN is translated by the bacterium, the protein is inserted in membranes derived from its host, either amoebae and macrophages, and thus we reasoned that a eukaryotic expression host could provide essential folding machinery and an amenable bilayer environment. Pichia pastoris proved compatible for reproducibly producing milligram quantities of highly pure MavN (yields of 1 to 2 mg/10 g cells) (Fig. 4A). The molecular identity of MavN was confirmed by tryptic digestion and mass spectrometry (SI Appendix, Fig. S2). Detergent compatibility of MavN was then assessed, focusing on mild nonionic detergents. Alkyl polyethylene glycol (PEG) detergents were found to produce the best chromatographic profile (Fig. 4 A and B). However, MavN eluted from a Superdex 200 Increase column at a molecular mass larger than the predicted 75-kDa protein (elution volume 11.5 mL, Fig. 4A).
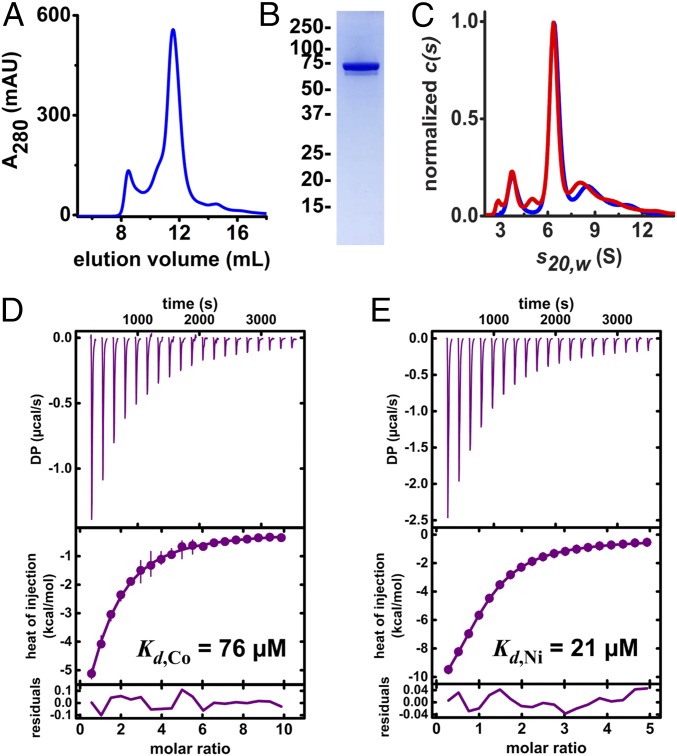
Fig. 4.
Characterization of recombinant MavN. MavN was heterologously expressed in Pichia and isolated via affinity chromatography. (A) Size-exclusion chromatogram of purified MavN solubilized in 1 mM C12E6. (B) Representative SDS/PAGE and Coomassie stain of SEC-purified MavN. (C) Normalized sedimentation velocity absorbance (blue) and interference (red) c(s) profiles for C12E8-solubilized MavN indicating the presence of a dominant species at 6.34S. The contribution of the free detergent to the interference c(s) distribution is not shown. (D and E) Isothermal titration calorimetry of MavN binding to Co2+ and Ni2+, respectively. MavN (∼50 μM) was titrated with (D) 2.5 mM CoCl2 or (E) 1.25 mM NiCl2. Isotherms were fit to a single-site binding model to extract thermodynamic parameters. DP, differential power.
Sedimentation velocity analytical ultracentrifugation was used to characterize the oligomeric state of the purified MavN. The absorbance c(s) profiles revealed the presence of a dominant species at 6.34S accounting for ∼62% of the sedimenting signal (Fig. 4C). Membrane protein analysis in GUSSI of the absorbance and interference c(s) profiles for this species predicted that a species sedimenting at 6.34 ± 0.06 S will have a protein mass contribution of 148 ± 30 kDa and a detergent contribution of 73 ± 9 C12E8. These data support the conclusion that the dominant species observed is a detergent-solubilized MavN dimer.
Substrate binding is an essential prerequisite for transport. So, to evaluate whether MavN binds transition-metal ions, the detergent-solubilized protein was subjected to isothermal titration calorimetry. MavN was titrated with either CoCl2 or NiCl2 (Fig. 4 D and E), which are surrogates for oxygen-sensitive Fe2+. Isotherms were well-fit by a single-site model, implying a 1:1 stoichiometry, namely that each MavN protomer binds one metal ion. The MavN dissociation constants, Kd, for Co2+ and Ni+2 were found to be 76 and 21 μM, respectively, affinity values that are typical for substrates of transporters.
MavN Transports Iron and Other Transition-Metal Ions.
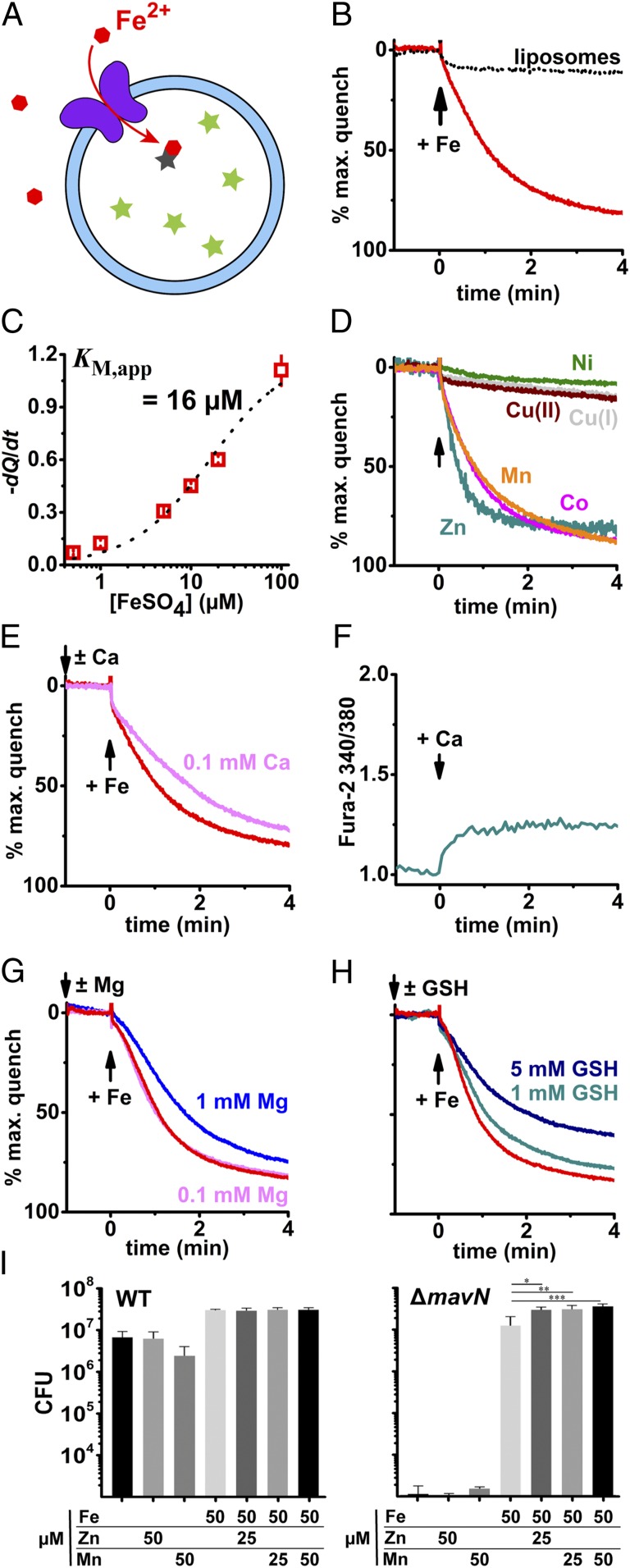
We next sought confirmation that MavN is a bona fide iron transporter. It is worthwhile to note that despite the universal biological importance of controlled iron flux through membranes, until recently there was no reported robust proteoliposome-reconstituted iron transport assay. We recently developed such an assay for the mitochondrial iron importer mitoferrin-1 (46), and used the same assay to verify that MavN can transport iron in a reconstituted system (Fig. 5A). MavN was incorporated into proteoliposomes and metal-ion uptake was assessed by fluorescence quenching of encapsulated calcein. MavN efficiently transported Fe2+ (Fig. 5B) with an apparent KM of 16 μM (Fig. 5C). Additionally, the first-row transition metals Mn2+, Co2+, and Zn2+ could serve as substrates for MavN (Fig. 5D). Conversely, MavN did not discernably transport Ni2+, Cu+, or Cu2+ (Fig. 5D). To probe MavN selectivity for transition versus alkali divalent metals, we competed MavN iron transport with a 10- or 100-fold greater quantity of calcium or magnesium (Fig. 5 E and G). Addition of 0.1 mM calcium mildly slowed MavN-mediated iron uptake whereas magnesium at 0.1 mM had no effect; boosting magnesium to 1 mM, however, diminished iron transport similar to 0.1 mM calcium. Though calcium competed with MavN iron transport, calcium is not a transported substrate, as demonstrated by the fluorescent ratiometric sensor Fura-2 (Fig. 5F). These data reveal that MavN effectively discriminates in favor of transition metals and against divalent alkali ions.
Fig. 5.
MavN transports iron and other transition metals. MavN was reconstituted into defined proteoliposomes by hydrophobic chromatography. (A) Fluorescence quenching of intraliposomal calcein (250 μM) was monitored to assess metal uptake; measurements were taken at room temperature. (B) Proteoliposome uptake of ferrous iron mediated by MavN. FeSO4 (10 μM) was added at t = 0 (black arrow). The black dotted trace shows protein-free liposomes similarly exposed to iron. (C) Concentration dependence of MavN iron transport. Depicted in red are initial quenching rates as determined by linear fits of the first 60 s after FeSO4 addition; error bars are SD from 3 or 4 independent measurements. Quenching rate points were fit to the Michaelis–Menten equation (black dotted line). (D) MavN transports manganese, cobalt, and zinc but neither copper nor nickel. Metal ions were added at t = 0 (black arrow) up to 10 μM MnCl2, CoCl2, NiCl2, CuCl, CuCl2, or ZnCl2. (E) Competition of MavN iron transport with calcium. CaCl2 (0.1 mM) was added 1 min prior to 10 μM FeSO4. (F) MavN does not transport calcium. Fura-2 (50 μM) was incorporated into MavN proteoliposomes and fluorescence was monitored to assess calcium uptake. CaCl2 (0.1 mM) was added at t = 0. (G) Competition of MavN iron transport with magnesium. MgCl2 (0.1 or 1 mM) was added 1 min prior to 10 μM FeSO4. (H) Glutathione slows iron uptake mediated by MavN. Reduced glutathione (GSH; 1 or 5 mM) was added 1 min prior to 10 μM FeSO4. (I) Growth of the ΔmavN strain is stimulated by Zn2+ or Mn2+ in the presence of iron. BMDMs were challenged with either WT or ΔmavN Legionella strains at an MOI of 0.01 and incubated for 96 h. At 96 hpi, bacterial CFUs were determined (Materials and Methods) to determine total yield. Metal concentrations are noted (FeSO4, MnSO4, ZnSO4). Data shown are means ± SE. P values were determined by Tukey’s multiple comparisons test. *P < 0.011, **P < 0.0067, ***P < 0.0007.
Iron is not available in the cytosol as free ions but rather is chaperoned by proteins, stored in ferritins, and sequestered by low–molecular-weight ligands, the last dubbed the labile iron pool (LIP). Glutathione (GSH) has been proposed to be the dominant LIP ligand (47) and is present at 10−3 to 10−2 M in the cytosol, raising the possibility that MavN could receive substrate from a GSH–iron complex. As depicted in Fig. 5H, we supplemented GSH in iron uptake assays with the hypothesis that if MavN receives iron from the GSH-bound pool, then addition of the ligand would accelerate transport. GSH addition proved inhibitory, however, indicating the tripeptide acts as a competing ligand for iron rather than facilitating substrate recognition by MavN.
The efficacy of MavN in vitro for transporting Mn2+, Co2+, and Zn2+ argues that the protein transports these metals also within host cells. Our previous work is inconsistent with this role, as only iron addition stimulated intracellular growth of the ΔmavN strain (25). If iron concentrations within the vacuole were too low to allow growth under conditions in which other metals were added, however, then this would have abrogated stimulation of intracellular growth by another substrate. Assuming that iron is limiting requires that it be added in conjunction with other substrates to test for bypass of growth defects. Therefore, excess iron was added along with the simultaneous addition of other transition metals and intracellular growth of the L. pneumophila ΔmavN strain was measured over 96 h after challenging BMDMs. The addition of Mn2+ or Zn2+ had no effect on intracellular growth of the L. pneumophila WT in the presence or absence of iron, indicating that the WT strain had sufficient access to these metals in the presence of intact MavN (Fig. 5 I, Left). In contrast, Mn2+ or Zn2+ added simultaneously with iron flooding clearly stimulated growth of the ΔmavN strain in BMDMs compared with addition of iron alone (Fig. 5 I, Right; P < 0.0007 for 50 μM MnSO4). Therefore, the ΔmavN strain is defective for access to Mn2+ and Zn2+ during intravacuolar growth, demonstrating these two metals are substrates for the protein. We attempted the identical experiments with Co2+, but addition of this metal was quite toxic to BMDMs, preventing us from directly evaluating whether it is a MavN substrate.
Proliferation-Defective MavN Mutants Are Impaired for Iron Transport.
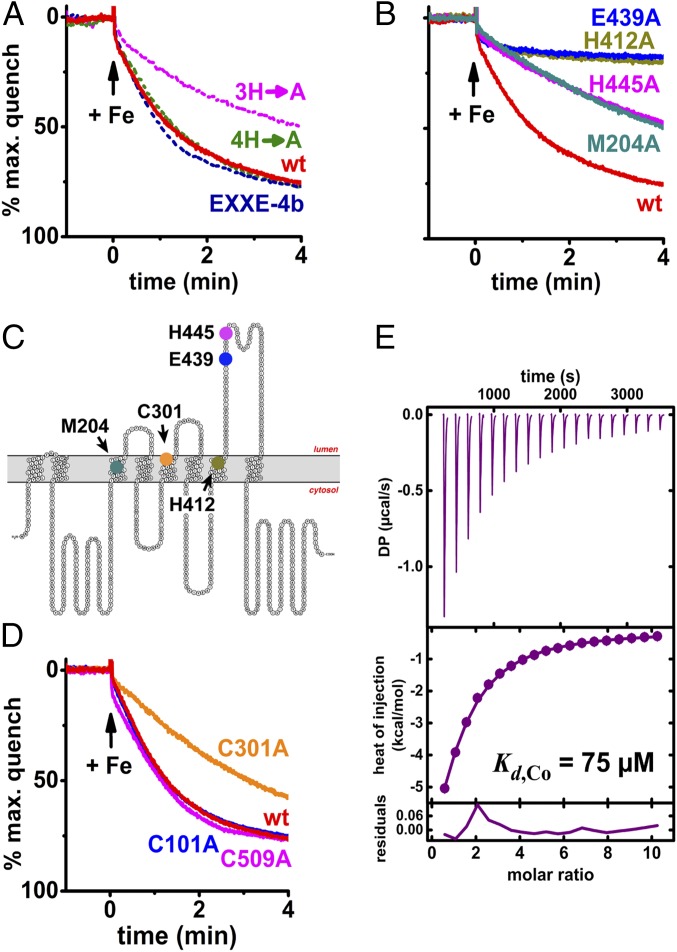
Having established that MavN can transport iron in vitro, we sought to ascertain whether the proliferation-defective mutants we previously identified (Fig. 3) (25) were similarly deficient for transporting iron. Notably, the defective mutants could be divided into two groups: (i) those that caused growth defects indistinguishable from the ΔmavN strain and (ii) those that supported intermediate levels of intracellular growth. These MavN derivatives were produced and purified from Pichia for proteoliposome incorporation. One of the mutants with 3 mutations, 3H→A (H445A/H450A/H452A), displayed an intracellular growth defect whereas the quadruple mutant, 4H→A (H460A/H462A/H465A/H466A) and the EXXE-4B hexamutant (453EDELDEE459→453AAALAAA459) were indistinguishable from WT (25). Iron uptake of the MavN variants in the reconstituted system recapitulated their cognate proliferation phenotypes in the cell-based assay (Fig. 3). The 3H→A mutant showed a partial defect in metal transport in the liposome-reconstituted assay, retaining only 40% of activity in a 4-min measurement (Fig. 6A). The 4H→A and EXXE-4B mutations, however, had no effect on iron uptake and were indistinguishable from the WT protein. Therefore, loss of MavN function in the reconstituted system reproduced loss of function during intracellular growth.
Fig. 6.
Proliferation-defective MavN mutants are impaired for iron uptake. (A) MavN mutant transport activities in vitro correlate with their capacities to support Legionella proliferation as described previously (25). Proteoliposome uptake assays were performed as in Fig. 5. (B) MavN point mutant activities in vitro similarly recapitulate Legionella growth defects depicted in Fig. 3. (C) Transport-defective point mutants mapped onto MavN topology. (D) Effects of Cys ablation on MavN transport activity in vitro. (E) Binding isotherm of MavN C301A titrated with 2.5 mM CoCl2.
Four MavN point mutants caused growth defects during growth in macrophages, with strains harboring the H412A and E439A alleles being indistinguishable from a ΔmavN strain, while strains containing M204A and H445A showed intermediate phenotypes (Fig. 3G). In the reconstituted system, the activities of the H445A and 3H→A variants showed identical partial defects (Fig. 6 A and B), indicating the phenotype of the 3H→A mutation can be entirely reproduced by substitution of the single highly conserved H445 residue. Similarly, the M204A MavN derivative retained only partial transport activity (Fig. 6B), mirroring the moderate growth defect of Legionella harboring the same derivative during challenge of BMDMs (Fig. 3G). Most notable were the defects observed with strains containing the H412A and E439A mutants. These derivatives were entirely incapable of fostering intracellular growth (Fig. 3G) and also demonstrated undetectable iron transport activity, mimicking that of the protein-free control (Figs. 5A and 6B). As noted above, these two residues are 100% conserved in all MavN orthologs within Legionellaceae species, highlighting critical roles for H412 and E439 in metal transport (SI Appendix, Dataset S1) (8). In short, the same 4 single-residue substitutions identified here as critical for intracellular growth also showed transport defects in the reconstitution assay.
Lastly, in one case, we uncovered a discordance between the effect of a mutation on intracellular growth and iron transport. Substitution of all 3 native Cys residues in MavN with Ala resulted in intracellular growth similar to that of WT (Fig. 2C). In the transport assay, however, the Cys0 mutant transport activity was impaired to nearly the extent as the M204A and H445A mutants. Subsequently, individual Cys→Ala mutants were tested in the proteoliposome-reconstituted transport assay and only C301A exhibited deficient iron uptake (Fig. 6D) while both C101A and C509A were indistinguishable from WT. Calorimetry of metal binding by the C301A mutant, by titrating with CoCl2, indicated no discernible defect in substrate recognition (Fig. 6E). This functional discrepancy argues that insertion in the native LCV may mask a subtle conformational defect in this particular mutant, or that overproduction by L. pneumophila may bypass this defect during intracellular growth assays (Fig. 2C).
The impaired MavN mutants were also tested for transport of transition metals other than iron. The mutations that resulted in close to null phenotypes for iron transport, H412A and E439A, were found to have negligible transport activity for all metals tested (SI Appendix, Fig. S3 A and B). Mutant M204A, as with iron, retained partial uptake activity for the metals tested (SI Appendix, Fig. S3C). The partial-function C301A and H445A mutations, however, resulted in some degree of metal specificity (SI Appendix, Fig. S3 D and E). C301A, although defective relative to WT, appeared to be more efficient at transporting Zn2+ than other metals (SI Appendix, Fig. S4). The H445A mutant, which showed similar defects in iron and Co2+ transport, displayed only a small defect for transport of Zn2+ and a transport efficiency for Mn2+ that approached that of WT (SI Appendix, Fig. S4). Therefore, mutations eliciting defects in iron transport may retain considerable activity for other selected transition metals.
Discussion
In this report, we have established that the Icm/Dot translocated effector MavN (lpg2815) is a transition-metal transporter that allows piracy of ferrous iron, and potentially other metals, from the host cell cytoplasm. By using a labeling method (TM-SCAM) based on cysteine accessibility, we compiled a tentative map of the MavN native topology within the LCV. The map revealed several noncontiguous regions of the protein to be localized to the cytosol-accessible LCV surface (Fig. 2). Extensive mutagenesis of typical iron-coordinating residues uncovered two classes of defects from single-residue substitutions: mutations that eliminate intracellular bacterial growth or retain partial function (Fig. 3). Both classes affected residues that were highly conserved, with the absolutely essential side chains being conserved in all MavN orthologs from sequenced Legionella species (48). We heterologously produced, purified, and reconstituted MavN in vitro to validate its proposed iron transport function in primary macrophages (25) and found striking correlation between mutant MavN transport activities and the effect of the same mutations on Legionella intracellular proliferation, with the magnitude of intracellular growth defect mirroring the severity of the transport defect.
The translocated effector repertoire of L. pneumophila encompasses substantial functional redundancy (49), with only 7 core effectors found in all members of the Legionella clade (8). MavN is one of these 7 and, conspicuously, single deletions of only 2 of these—MavN and LegA3—attenuate Legionella intracellular growth. Absence of MavN causes a severe intracellular growth defect in all tested host cells, indicating its importance in supporting replication within the LCV (25, 26). The use of an iron transporter to procure this crucial element is unsurprising, although the lack of functional redundancy is remarkable. Four of seven core effectors are predicted integral membrane proteins (IMPs) and L. pneumophila strain 130b comprises 56 predicted multipass IMPs (50) out of roughly 300 effectors. Perhaps secretion of transmembrane proteins by the T4SS is inefficient or otherwise energetically costly, thus diminishing demand for redundancy of IMP effectors. Alternatively, unlike other effectors, there may be no selective pressure to acquire other proteins that could replace MavN function. One explanation for the massive expansion of Icm/Dot substrates is that the pathogen is a generalist and must be able to grow in multiple unrelated hosts in the environment (24). This requires interaction with host proteins that potentially diverge extensively by sequence. To ensure growth in disparate hosts, the bacterium has acquired genes that increase fitness for one host, without losing genes that provide it with a fitness advantage in other hosts, resulting in gene expansion. Perhaps MavN interacts with an evolutionarily conserved protein or binds directly with iron and a conserved chaperone molecule. Nevertheless, the presence of >50 IMP effectors implies the organism has multiple pathways for other nutrient acquisition such as phosphate or the alkali divalent metals magnesium and calcium.
MavN can employ iron, manganese, and zinc as transport substrates, although expression of the mavN gene is tightly regulated by available iron via the Fur transcriptional regulator (25, 26). We provide evidence that within the Legionella-containing vacuole, MavN allows access of the bacterium to cytoplasmic pools of these nutrients. Loss of MavN can be compensated maximally by flooding culture medium with excess iron in combination with either Mn2+ or Zn2+, indicating that the inability to acquire these metals is the primary cause of the intracellular growth defect exhibited in ΔmavN strains. The chelatable (labile) iron and manganese intracellular pools are typically in the 10−5 M range, permitting MavN to directly uptake the metal ions from the cytosol based on the apparent KM of iron uptake. MavN discriminates against copper and may prevent intoxication by the metal. Notably, L. pneumophila employs the detoxification pump CopA, a PIB-type ATPase, to efflux copper, but this protective mechanism is superfluous for intracellular growth (51).
MavN has no discernible sequence similarity to any known transporters. Based on primary sequence, LCV-inserted MavN is predicted to encompass 8 transmembrane regions (Fig. 2F). The TM-SCAM labelings are largely congruous with this model, placing residues predicted to be in three short loops of 8 to 20 amino acids as well as a 70-amino acid domain that is critical for function within the lumen of the LCV. Predicted as cytosolically exposed is the carboxyl-terminal 150-amino acid region that we found is readily accessible to maleimide labeling (Fig. 2F). The TM-SCAM experiments identified two other cytosol-accessible regions flanked by transmembrane domains, consistent with the prediction of the topology model. Unexpected was the inability to label a region predicted to generate a 35-amino acid loop facing the cytosol (Fig. 2F). We hypothesize that this 35-amino acid loop faces the cytosol and is resistant to labeling because it is protected in some fashion, either through inter- or intramolecular interactions with other parts of the protein, or by looping back to form a partial reentrant structure in the membrane, preventing accessibility of the maleimide labeling reagent. That there is one other large loop and the long carboxyl-terminal segment of the protein on the same side of the membrane increases the possibility of such interactions (Fig. 2).
Our mutational analyses identified residues critical for both metal transport and intracellular growth that mapped to two predicted TM regions (TM3 and TM7) and within a 70-amino acid span located in the LCV lumen (between TM7 and TM8; Fig. 3). We hypothesize that these residues are involved in transporting metal ions through the membrane and chaperoning the substrates inside the LCV through coordination by the luminal domain. The directed approach, however, failed to identify any cytoplasmically localized residues that alter function even though 13 potential metal-binding residues (Cys, His, Asp, Glu) were mutated in the 150-amino acid carboxyl-terminal domain (Fig. 3). The interaction of metals with the cytoplasmic face is likely to be of weak affinity, facilitating transfer to higher-affinity TM and luminal residues that can receive and sequester the ions. Accordingly, the abundance of potential metal-coordinating residues suggests that the carboxyl-terminal domain could act as a sponge for metal ions, locally concentrating substrate for LCV uptake. By this formulation, the substitution of single residues would likely be undetectable in our assays. More perplexing is that the C301A mutation in MavN caused a defect in iron uptake into proteoliposomes but promoted proficient intracellular growth of Legionella (Fig. 6D). Based on its partial defect in iron transport, the C301A mutation would be predicted to generate a phenotype similar to the H445A or M204A mutations that show partial loss of function (Fig. 3). This phenomenon may be attributable to the mutant retaining substrate-binding capability (Fig. 6E) while the transport defect arises from inefficient release of metals into the lumen. Therefore, factors contained in the LCV lumen, and absent from the in vitro liposome assay, could be involved in releasing metal ions from MavN, and the factors’ presence could suppress a proliferation defect. Such factors exist, as Legionella is known to synthesize both siderophores and pyomelanin, which have high affinity for iron (26, 52, 53).
In summary, the conserved effector MavN plays an indispensable role in L. pneumophila growth by directly acting as a conduit for the trafficking of transition-metal ions into the LCV. Although it has long been considered likely that intravacuolar bacterial pathogens insert microbial nutrient transporters into the vacuolar membrane, demonstration of their existence has remained elusive. For this reason, MavN now provides a paradigm for microbial acquisition of essential growth factors from the host cell cytosol. Of particular interest is the critical role that this protein plays in the piracy of iron, a key target of host nutritional immunity. As such, MavN is now clearly established as the key vacuolar component in competing with the host to access essential transition metals and facilitate intracellular growth in the presence of limiting resources.
Materials and Methods
Materials.
EZ-Link-maleimide-PEG2-biotin (MP2B), calcein, Fura-2, and yeast and E. coli medium components were obtained from Thermo Fisher Scientific. Yeast nitrogen base was from RPI. Anti–FLAG-M2 antibody, Triton X-100, and Chelex and Amberlite resins were from Sigma-Aldrich. FLAG resin (anti-DYKDDDK G1 affinity resin) was purchased from GenScript. IgG fraction mouse monoclonal anti-biotin was from Jackson ImmunoResearch. N-dodecyl-β-d-maltopyranoside (DDM) and hexa- and octaethylene glycol monododecyl ethers (C12E6, C12E8) were purchased from Anatrace. Lipids were purchased from Avanti Polar Lipids. Zeocin was obtained from Invitrogen. Talon cobalt metal affinity resin was from Clontech. Other chemicals were obtained from Sigma-Aldrich.
Bacterial and Mammalian Cell Culture.
L. pneumophila strains LP02 [WT thyA (7)] and isogenic LP02 ΔmavN were grown on CYE solid agar and AYE broth as described (25). Bone marrow-derived macrophages were from the permissive A/J mouse, and isolated and cultured as described (5). U937 cells were phorbol ester-transformed and cultured for L. pneumophila challenge following previous protocols (7).
Intracellular Growth Assays.
For challenge of BMDMs, postexponentially grown bacteria were incubated with macrophages at a multiplicity of infection (MOI) of 0.05, and viable bacteria were enumerated by lysing cells at the indicated time points and plating for colony-forming units (CFUs) on CYE medium. To visualize intracellular growth microscopically, BMDMs were seeded on sterile glass coverslips in flat-bottom 24-well plates and challenged with postexponentially grown bacteria at an MOI of 0.5. Macrophages were fixed with 4% paraformaldehyde in PBS for 15 min, washed three times with PBS, and visualized microscopically for mCherry and GFP fluorescence.
To determine the role of metals in supporting intracellular growth, BMDMs from A/J mice were seeded in a 24-well plate and challenged with bacteria at an MOI of 0.05. At 2 hpi, BMDMs were washed with PBS to remove extracellular bacteria. Medium was supplemented with the metals at the indicated final concentration and added to cells after the final wash. At 96 hpi, macrophages were lysed with saponin, the lysate and media were pooled, and CFUs were determined by culturing on CYE solid medium. Significance was determined by Tukey’s multiple comparisons.
Construction of Reporters and Mutations.
The pIroS reporter plasmid was constructed from the parental plasmid pXDC94 that allows simultaneous expression of Ptac-controlled mCherry and GFP regulated by a promoter of interest [kind gift of Howard Shuman, Columbia University Medical Center, New York, NY (33)]. To analyze the kinetics of iron starvation, the promoter for the L. pneumophila frgA gene, which is controlled by the Fur repressor and up-regulated by iron starvation, was placed upstream of the gfp gene (31). To this end, the fragment encoding the Fur box and promoter region upstream of frgA was amplified to generate XmaI/KpnI ends, digested by the two enzymes, and inserted into the cognate sites upstream of gfp. To isolate substitution mutations, overlapping primer pairs containing the desired mutations were first used introduced into mavN in the pDTI133 pUC18-based plasmid (25), according to the site-directed mutagenesis protocol (Stratagene). Substitution mutations in the mavN coding sequence were then moved into the pJB908 plasmid (54), to allow expression in L. pneumophila, by digesting with SacI/BamHI.
Transmembrane-Substituted Cysteine Accessibility Method.
To identify accessible residues during broth growth, L. pneumophila strains were grown overnight in 200 mL AYE broth (7), centrifuged, and washed with 50 mM Tris⋅HCl (pH 7.5). After washing, the cells were pelleted at 6,000 × g for 15 min and snap-frozen in liquid N2. The pellets were resuspended in 2 mL ice-cold homogenization buffer (20 mM Hepes⋅KOH, pH 7.2, 250 mM sucrose, 5 mM EGTA, cOmplete protease inhibitor cocktail; PIC), incubated at 4 °C for 15 min with 150 μg/mL lysozyme, and lysed by one freeze–thaw cycle in liquid N2 followed by 10-min incubation with 1% Triton X-100. The extract was clarified by centrifugation at 16,000 × g for 30 min at 4 °C. The supernatant was preincubated with 5 mM TCEP [Tris(2-carboxyethyl)phosphine hydrochloride] at room temperature for 20 min to reduce disulfides, and labeling was initiated with 1.33 mM maleimide-PEG2-biotin. The labeling reaction was allowed to proceed overnight at 4 °C with end-over-end rotation. To quench the reaction, β-mercaptoethanol (β-ME) was added to 1% final concentration (1% of total solution; total solution volume 550 μL) and extracts were processed for immunoprecipitation.
To identify MavN Cys residues accessible to MP2B after localization on the Legionella-containing vacuole, 4 × 106 phorbol ester-transformed U937 cells were challenged with L. pneumophila strains at an MOI of 5.0 as described (7). At 6 hpi, U937 cells were washed to remove noninternalized bacteria, lifted, and washed with PBS. The cells were then suspended in 800 μL homogenization buffer supplemented with PIC, disrupted by 2 passages in a Dounce homogenizer to maintain LCV integrity (37), and preincubated with 5 mM TCEP prior to treatment with 1.33 mM MP2B. After overnight incubation at 4 °C with end-over-end rotation, the cells were treated with β-ME (1% of total; total volume 550 μL) for 5 min to quench the reaction and washed twice with 1 mL FLAG buffer (50 mM Tris⋅HCl, pH 7.4, 150 mM NaCl, PIC) and pelleted. The cell pellets were resuspended in 550 μL FLAG buffer supplemented with 1% Triton X-100 and incubated for 20 min at room temperature with end-over-end rotation. The samples were then subjected to centrifugation at 16,000 × g for 20 min at 4 °C and 500 μL of the detergent-solubilized membrane proteins in the supernatant was collected for further processing.
Immunoprecipitation and Immunoblotting.
Cleared lysates were added to 40 μL FLAG resin which had been prewashed three times in lysis buffer and incubated overnight at 4 °C with end-over-end rotation. The following day the resin was pelleted at 6,000 × g for 1 min, and the supernatant was aspirated and washed three times with FLAG buffer containing 0.5% Triton X-100. The resin was resuspended in 100 μL FLAG buffer and transferred to a microspin column. The columns were then centrifuged at 6,000 × g for 1 min and bound proteins were removed by incubation with 75 μL elution buffer (150 μg/mL 3×FLAG peptide, 0.5% Triton X-100, FLAG buffer) for 1 h at room temperature with end-over-end rotation. Columns were again centrifuged at 6,000 × g for 1 min and eluate was collected.
For immunoblotting analysis of immunoprecipitates, the eluates were mixed with equal volumes of 2× Laemmli buffer containing β-ME and resolved on 7.5% SDS/PAGE. The fractionated proteins were transferred to polyvinylidene difluoride membranes (GE Healthcare) and probed with affinity-purified mouse anti-FLAG M2 or mouse anti-biotin antibodies. Blots were then incubated with Near-Infrared Fluorescent conjugated secondary antibodies (LI-COR) and imaged with the LI-COR Odyssey CLX Imaging System.
Fluorescence-Activated Cell Sorting Analysis of Iron Starvation Reporter.
Phorbol ester-transformed U937 cells were plated at 4 × 106 per well. Twenty-four hours after plating, cells were challenged with L. pneumophila strains harboring the pIroS plasmid. For measuring mCherry expression (indicating infectivity) and GFP expression (indicating iron starvation), infections were performed at an MOI of 3 for the noted times. Cells were harvested with ice-cold PBS and fixed with 4% paraformaldehyde for 15 min. Cells were washed twice and analyzed by Becton Dickinson LSR II flow cytometer. Data were processed using FlowJo software.
Protein Expression in P. pastoris and Purification.
The cDNA encoding MavN from L. pneumophila, strain Philadelphia-1, was subcloned into expression plasmid pPICZ A (Invitrogen) with a carboxyl-terminal decahistidine tag; a 3C protease site was included for proteolytic tag removal. Mutageneses were performed via PCR and confirmed by DNA sequencing. PmeI-linearized plasmids were electroporated into Pichia strain SMD1163H (Invitrogen) and transformants were selected on YPDS plates supplemented with 400 μg/mL Zeocin. Individual Pichia colonies were screened for MavN expression by Western blotting against the affinity tags. Pichia cultures were grown in buffered minimal glycerol medium at 30 °C to an OD600 of ∼25, pelleted by centrifugation at 3,000 × g for 5 min, resuspended in buffered minimal methanol medium supplemented with 1 mM ZnSO4, and shaken for 24 h at 25 °C with two tranches of methanol, up to 0.6%, roughly 12 h apart. Induced Pichia were again pelleted and flash-frozen in liquid N2.
MavN and mutants were purified from cell powder produced by cryomilling Pichia (three cycles, 3 min each, 25 rpm) in liquid N2-cooled stainless steel-ball milling jars (Retsch). All subsequent steps were performed at 4 °C. Powder was resuspended (10 g up to 50 mL) in extraction buffer (50 mM Hepes⋅NaOH, pH 7.9, 150 mM NaCl, 5 mM β-ME, 2 mM TCEP, protease inhibitors, 0.5 μg/mL DNase) then DDM added to 3% and stirred for 2 h. Extracts were clarified by centrifuging at 38,000 × g for 30 min, pH was adjusted to ∼7.4 with 1 N NaOH, and extracts were batch-bound to Talon resin for 2 h. Resin was collected by gravity flow in a polypropylene column and MavN exchanged to C12E6 by washing with 10 bed volumes wash buffer (20 mM Mops⋅NaOH, pH 7.4, 140 mM NaCl, 1 mM C12E6, 10 mM imidazole, 5 mM β-ME, 2 mM TCEP). The column was plugged, 1 bed volume wash buffer was added, C12E6 was raised to 2 mM, and house-made 3C protease was added to cleave MavN from the affinity tag. After rocking the resin slurry overnight, imidazole was added to 30 mM and MavN was eluted from the column. Affinity-purified MavN was concentrated with a 50-kDa MWCO centrifugal concentrator and subjected to size-exclusion chromatography (SEC) over Superdex 200 Increase 10/300 GL equilibrated in SEC buffer (20 mM Mops⋅NaOH, pH 7.4, 140 mM NaCl, 2 mM TCEP, 1 mM C12E6). MavN was finally concentrated with a 100-kDa MWCO concentrator to ∼2 mg/mL for downstream use.
Peptide Mass Fingerprinting.
Purified recombinant MavN was run on SDS/PAGE under reducing conditions. The gel was stained with Coomassie brilliant blue, destained, and thoroughly washed in deionized water. The MavN gel band (∼10 μg protein) was excised, protein was digested with LysC/trypsin, and extract was applied to a C4 reverse-phase column run at a flow rate of 300 nL/min on an UltiMate 3000 HPLC (Thermo Dionex) connected to an Orbitrap Elite mass spectrometer (Thermo Scientific). LC/MS/MS data were searched against the house-built database containing the MavN sequence and the whole Swiss-Prot database (https://www.uniprot.org). The false discovery rate (FDR) of the searches is less than 1%.
Sedimentation Velocity Analytical Ultracentrifugation.
Sedimentation velocity experiments were performed at 50,000 rpm and 10 °C on a Beckman Coulter ProteomeLab XL-I analytical ultracentrifuge and An50-Ti rotor following standard protocols (55). Three samples of MavN, at concentrations corresponding to an A280 of 1.0 in 140 mM KCl, 20 mM Hepes⋅KOH (pH 7.4), 2 mM TCEP, 2 mM C12E8 detergent, were loaded in 12-mm two-channel centerpiece cells and sedimentation data were collected using the absorbance (280 nm) and interference (655 nm) optical detection systems. Time-corrected (56) data were analyzed in SEDFIT 16.1 (57) in terms of a continuous c(s) distribution of sedimenting species using an s range of 0 to 15 with a linear resolution of 300 and a maximum entropy regularization confidence interval of 0.68. Interference data were analyzed using a bimodal fictional ratio to account for the presence of free detergent at ∼0.3S. The solution density, solution viscosity, protein extinction coefficient, and protein partial specific volume were calculated based on their composition in SEDNTERP (58). The protein refractive index increment was calculated based on its composition in SEDFIT (59). A partial specific volume of 0.973 cm3⋅g−1 (60) and refractive index increment of 0.1352 cm3⋅g−1 (61) were used for C12E8.
Absorbance and interference c(s) distributions were analyzed simultaneously using the membrane protein calculation module in GUSSI 1.4.1 (62) to obtain the protein and detergent contributions to the sedimenting complex. The fitted f/fo method, based on the work of Ebel and coworkers (63, 64), was utilized. Sedimentation coefficients were corrected to standard conditions in water at 20 °C, s20,w utilizing the partial specific volume for the major protein–detergent complex identified in the analysis.
Isothermal Titration Calorimetry.
MavN was freshly purified, concentrated, and then dialyzed at 4 °C for 20 to 24 h against Chelex-treated size-exclusion buffer (20 mM Mops⋅NaOH, pH 7.4, 140 mM NaCl, 0.5 mM TCEP, 1 mM C12E6). Titrations were performed at 10 °C on a MicroCal iTC200. Protein solutions were degassed for 15 min prior to placement into the calorimeter, and dialysis buffer was degassed for 30 min before addition of CoCl2 or NiCl2. Thermograms were analyzed using the software NITPIC, SEDPHAT, and GUSSI (65, 66).
Proteoliposome Preparation.
MavN proteoliposomes were prepared via hydrophobic chromatography (46); protein-free liposomes were prepared in the same manner, except with C12E6-containing SEC buffer replacing MavN. Lipids in chloroform were dried with argon to a film and then further vacuum-dried overnight. Lipid composition by mass was 50:35:5:5:5 1-palmitoyl-2-oleoylphosphatidylcholine (POPC):1-palmitoyl-2-oleoylphosphatidylethanolamine (POPE):1-palmitoyl-2-oleoylphosphatidylglycerol (POPG):1-palmitoyl-2-oleoylphosphatidylserine (POPS):soy phosphatidylinositol (PI). All buffers were treated with Chelex resin to remove trace divalent ions. Lipid films were resuspended to 20 mg/mL in 10 mM Mops (pH 7.4), layered with argon, bath-sonicated to large unilamellar vesicles, and NaCl added up to 140 mM. Ten sequential additions of 10% Triton X-100 were added to destabilize the liposomes and the final lipid:detergent ratio was 10:6 by mass. To the destabilized liposomes, freshly purified MavN was added (10 mg lipid/50 μg protein), and the suspension was brought up to a total volume of 700 μL with inside buffer (10 mM Mops, pH 7.4, 140 mM NaCl) and rocked at 4 °C for 30 min. The proteoliposome suspension was then passed 15 times over 550 mg Amberlite XAD-2 at room temperature, diluted to 6.5 mg/mL lipid with inside buffer, and flash-frozen in liquid N2. Calcein (250 μM) or Fura-2 (50 μM) was incorporated by freezing–thawing three times; liposomes were then extruded through a 400-nm filter and free dye was removed by passing the vesicles over Sepharose CL-2B equilibrated in inside buffer.
Proteoliposome Uptake Assays.
Fluorescence quenching of calcein (λex 495 nm, λem 515 nm) and Fura-2 (λex 340/380 nm, λem 510 nm) was monitored using a Cary Varian Eclipse spectrofluorometer at room temperature. (Proteo)Liposomes were diluted ∼30-fold to 20 μM lipid in Chelex-treated assay buffer (10 mM Mops⋅KOH, pH 7.4, 140 mM KCl). Metal stock solutions were made to 100 mM Me2+—FeSO4 in 20 mM H2SO4; CuCl in 1.1 M HCl; CoCl2, CuCl2, MnCl2, NiCl2, and ZnCl2 in water—each experimental day. For iron uptake, assays were supplemented with 1 mM sorbitol to scavenge radical oxygen species. Maximal dye quenching was determined by addition of the ionophores pyrithione (for iron, cobalt, nickel, and zinc) or calcimycin (for copper and manganese) to liposomes to equilibrate internal and external divalent ions.
Supplementary Material
Acknowledgments
This work was supported by the intramural program of the Eunice Kennedy Shriver National Institute of Child Health and Human Development (NICHD); National Institutes of Health (NIH) (Project ZIA HD008928); NIH/National Institute of Allergy and Infectious Diseases (NIAID) Training Grants T32 AI007329 (to D.T.I.) and T32GM07310 (to E.L.); as well as NIAID Grant R21AI115261 (to R.R.I.). We thank Stephen Kwok and Allen Parmelee from the Tufts University Flow Cytometry Core for their technical assistance in sorting experiments. We thank Matthias Machner (NICHD) and Gisela Storz (NICHD) for comments on the manuscript, and Yan Li of the National Institute of Neurological Diseases and Stroke for mass spectrometric analysis of purified MavN.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1902806116/-/DCSupplemental.
References
- 1.Horwitz M. A., Cell-mediated immunity in Legionnaires’ disease. J. Clin. Invest. 71, 1686–1697 (1983). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rowbotham T. J., Isolation of Legionella pneumophila from clinical specimens via amoebae, and the interaction of those and other isolates with amoebae. J. Clin. Pathol. 36, 978–986 (1983). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Thomas V., et al. , Amoebae in domestic water systems: Resistance to disinfection treatments and implication in Legionella persistence. J. Appl. Microbiol. 97, 950–963 (2004). [DOI] [PubMed] [Google Scholar]
- 4.Isberg R. R., O’Connor T. J., Heidtman M., The Legionella pneumophila replication vacuole: Making a cosy niche inside host cells. Nat. Rev. Microbiol. 7, 13–24 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Swanson M. S., Isberg R. R., Association of Legionella pneumophila with the macrophage endoplasmic reticulum. Infect. Immun. 63, 3609–3620 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Marra A., Blander S. J., Horwitz M. A., Shuman H. A., Identification of a Legionella pneumophila locus required for intracellular multiplication in human macrophages. Proc. Natl. Acad. Sci. U.S.A. 89, 9607–9611 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Berger K. H., Isberg R. R., Two distinct defects in intracellular growth complemented by a single genetic locus in Legionella pneumophila. Mol. Microbiol. 7, 7–19 (1993). [DOI] [PubMed] [Google Scholar]
- 8.Burstein D., et al. , Genomic analysis of 38 Legionella species identifies large and diverse effector repertoires. Nat. Genet. 48, 167–175 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Huang L., et al. , The E Block motif is associated with Legionella pneumophila translocated substrates. Cell Microbiol. 13, 227–245 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kagan J. C., Stein M. P., Pypaert M., Roy C. R., Legionella subvert the functions of Rab1 and Sec22b to create a replicative organelle. J. Exp. Med. 199, 1201–1211 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nagai H., Kagan J. C., Zhu X., Kahn R. A., Roy C. R., A bacterial guanine nucleotide exchange factor activates ARF on Legionella phagosomes. Science 295, 679–682 (2002). [DOI] [PubMed] [Google Scholar]
- 12.Kagan J. C., Murata T., Roy C. R., Analysis of Rab1 recruitment to vacuoles containing Legionella pneumophila. Methods Enzymol. 403, 71–81 (2005). [DOI] [PubMed] [Google Scholar]
- 13.Murata T., et al. , The Legionella pneumophila effector protein DrrA is a Rab1 guanine nucleotide-exchange factor. Nat. Cell Biol. 8, 971–977 (2006). [DOI] [PubMed] [Google Scholar]
- 14.Ingmundson A., Delprato A., Lambright D. G., Roy C. R., Legionella pneumophila proteins that regulate Rab1 membrane cycling. Nature 450, 365–369 (2007). [DOI] [PubMed] [Google Scholar]
- 15.Mukherjee S., et al. , Modulation of Rab GTPase function by a protein phosphocholine transferase. Nature 477, 103–106 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Choy A., et al. , The Legionella effector RavZ inhibits host autophagy through irreversible Atg8 deconjugation. Science 338, 1072–1076 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tan Y., Luo Z. Q., Legionella pneumophila SidD is a deAMPylase that modifies Rab1. Nature 475, 506–509 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tan Y., Arnold R. J., Luo Z. Q., Legionella pneumophila regulates the small GTPase Rab1 activity by reversible phosphorylcholination. Proc. Natl. Acad. Sci. U.S.A. 108, 21212–21217 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fontana M. F., et al. , Secreted bacterial effectors that inhibit host protein synthesis are critical for induction of the innate immune response to virulent Legionella pneumophila. PLoS Pathog. 7, e1001289 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shen X., et al. , Targeting eEF1A by a Legionella pneumophila effector leads to inhibition of protein synthesis and induction of host stress response. Cell Microbiol. 11, 911–926 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Belyi Y., Tabakova I., Stahl M., Aktories K., Lgt: A family of cytotoxic glucosyltransferases produced by Legionella pneumophila. J. Bacteriol. 190, 3026–3035 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Machner M. P., Isberg R. R., Targeting of host Rab GTPase function by the intravacuolar pathogen Legionella pneumophila. Dev. Cell 11, 47–56 (2006). [DOI] [PubMed] [Google Scholar]
- 23.Machner M. P., Isberg R. R., A bifunctional bacterial protein links GDI displacement to Rab1 activation. Science 318, 974–977 (2007). [DOI] [PubMed] [Google Scholar]
- 24.O’Connor T. J., Adepoju Y., Boyd D., Isberg R. R., Minimization of the Legionella pneumophila genome reveals chromosomal regions involved in host range expansion. Proc. Natl. Acad. Sci. U.S.A. 108, 14733–14740 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Isaac D. T., Laguna R. K., Valtz N., Isberg R. R., MavN is a Legionella pneumophila vacuole-associated protein required for efficient iron acquisition during intracellular growth. Proc. Natl. Acad. Sci. U.S.A. 112, E5208–E5217 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Portier E., et al. , IroT/mavN, a new iron-regulated gene involved in Legionella pneumophila virulence against amoebae and macrophages. Environ. Microbiol. 17, 1338–1350 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Philpott C. C., Ryu M. S., Frey A., Patel S., Cytosolic iron chaperones: Proteins delivering iron cofactors in the cytosol of mammalian cells. J. Biol. Chem. 292, 12764–12771 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Clemens D. L., Horwitz M. A., The Mycobacterium tuberculosis phagosome interacts with early endosomes and is accessible to exogenously administered transferrin. J. Exp. Med. 184, 1349–1355 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sturgill-Koszycki S., Schaible U. E., Russell D. G., Mycobacterium-containing phagosomes are accessible to early endosomes and reflect a transitional state in normal phagosome biogenesis. EMBO J. 15, 6960–6968 (1996). [PMC free article] [PubMed] [Google Scholar]
- 30.Robey M., Cianciotto N. P., Legionella pneumophila feoAB promotes ferrous iron uptake and intracellular infection. Infect. Immun. 70, 5659–5669 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hickey E. K., Cianciotto N. P., An iron- and Fur-repressed Legionella pneumophila gene that promotes intracellular infection and encodes a protein with similarity to the Escherichia coli aerobactin synthetases. Infect. Immun. 65, 133–143 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cianciotto N. P., Iron acquisition by Legionella pneumophila. Biometals 20, 323–331 (2007). [DOI] [PubMed] [Google Scholar]
- 33.Hovel-Miner G., Faucher S. P., Charpentier X., Shuman H. A., ArgR-regulated genes are derepressed in the Legionella-containing vacuole. J. Bacteriol. 192, 4504–4516 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tsirigos K. D., Peters C., Shu N., Käll L., Elofsson A., The TOPCONS web server for consensus prediction of membrane protein topology and signal peptides. Nucleic Acids Res. 43, W401–W407 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bogdanov M., Zhang W., Xie J., Dowhan W., Transmembrane protein topology mapping by the substituted cysteine accessibility method (SCAM(TM)): Application to lipid-specific membrane protein topogenesis. Methods 36, 148–171 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Karlin A., Akabas M. H., Substituted-cysteine accessibility method. Methods Enzymol. 293, 123–145 (1998). [DOI] [PubMed] [Google Scholar]
- 37.Derré I., Isberg R. R., Legionella pneumophila replication vacuole formation involves rapid recruitment of proteins of the early secretory system. Infect. Immun. 72, 3048–3053 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Stearman R., Yuan D. S., Yamaguchi-Iwai Y., Klausner R. D., Dancis A., A permease-oxidase complex involved in high-affinity iron uptake in yeast. Science 271, 1552–1557 (1996). [DOI] [PubMed] [Google Scholar]
- 39.Ramanan N., Wang Y., A high-affinity iron permease essential for Candida albicans virulence. Science 288, 1062–1064 (2000). [DOI] [PubMed] [Google Scholar]
- 40.Trikha J., Theil E. C., Allewell N. M., High resolution crystal structures of amphibian red-cell L ferritin: Potential roles for structural plasticity and solvation in function. J. Mol. Biol. 248, 949–967 (1995). [DOI] [PubMed] [Google Scholar]
- 41.Wösten M. M., Kox L. F., Chamnongpol S., Soncini F. C., Groisman E. A., A signal transduction system that responds to extracellular iron. Cell 103, 113–125 (2000). [DOI] [PubMed] [Google Scholar]
- 42.Grossoehme N. E., Akilesh S., Guerinot M. L., Wilcox D. E., Metal-binding thermodynamics of the histidine-rich sequence from the metal-transport protein IRT1 of Arabidopsis thaliana. Inorg. Chem. 45, 8500–8508 (2006). [DOI] [PubMed] [Google Scholar]
- 43.Lu C. H., Lin Y. F., Lin J. J., Yu C. S., Prediction of metal ion-binding sites in proteins using the fragment transformation method. PLoS One 7, e39252 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bozzi A. T., et al. , Conserved methionine dictates substrate preference in Nramp-family divalent metal transporters. Proc. Natl. Acad. Sci. U.S.A. 113, 10310–10315 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Long F., et al. , Crystal structures of the CusA efflux pump suggest methionine-mediated metal transport. Nature 467, 484–488 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Christenson E. T., Gallegos A. S., Banerjee A., In vitro reconstitution, functional dissection, and mutational analysis of metal ion transport by mitoferrin-1. J. Biol. Chem. 293, 3819–3828 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hider R. C., Kong X. L., Glutathione: A key component of the cytoplasmic labile iron pool. Biometals 24, 1179–1187 (2011). [DOI] [PubMed] [Google Scholar]
- 48.Burstein D., et al. , Genome-scale identification of Legionella pneumophila effectors using a machine learning approach. PLoS Pathog. 5, e1000508 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.O’Connor T. J., Boyd D., Dorer M. S., Isberg R. R., Aggravating genetic interactions allow a solution to redundancy in a bacterial pathogen. Science 338, 1440–1444 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dolezal P., et al. , Legionella pneumophila secretes a mitochondrial carrier protein during infection. PLoS Pathog. 8, e1002459 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kim E. H., Charpentier X., Torres-Urquidy O., McEvoy M. M., Rensing C., The metal efflux island of Legionella pneumophila is not required for survival in macrophages and amoebas. FEMS Microbiol. Lett. 301, 164–170 (2009). [DOI] [PubMed] [Google Scholar]
- 52.Cianciotto N. P., An update on iron acquisition by Legionella pneumophila: New pathways for siderophore uptake and ferric iron reduction. Future Microbiol. 10, 841–851 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zheng H., Chatfield C. H., Liles M. R., Cianciotto N. P., Secreted pyomelanin of Legionella pneumophila promotes bacterial iron uptake and growth under iron-limiting conditions. Infect. Immun. 81, 4182–4191 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sexton J. A., et al. , The Legionella pneumophila PilT homologue DotB exhibits ATPase activity that is critical for intracellular growth. J. Bacteriol. 186, 1658–1666 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhao H., Brautigam C. A., Ghirlando R., Schuck P., Overview of current methods in sedimentation velocity and sedimentation equilibrium analytical ultracentrifugation. Curr. Protoc. Protein Sci. 71, 20.12.1–20.12.49 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhao H., et al. , Recorded scan times can limit the accuracy of sedimentation coefficients in analytical ultracentrifugation. Anal. Biochem. 437, 104–108 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Schuck P., Size-distribution analysis of macromolecules by sedimentation velocity ultracentrifugation and Lamm equation modeling. Biophys. J. 78, 1606–1619 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Cole J. L., Lary J. W., Moody T. P., Laue T. M., Analytical ultracentrifugation: Sedimentation velocity and sedimentation equilibrium. Methods Cell Biol. 84, 143–179 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhao H., Brown P. H., Schuck P., On the distribution of protein refractive index increments. Biophys. J. 100, 2309–2317 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.le Maire M., Champeil P., Moller J. V., Interaction of membrane proteins and lipids with solubilizing detergents. Biochim. Biophys. Acta 1508, 86–111 (2000). [DOI] [PubMed] [Google Scholar]
- 61.Corti M., Miner C., Degiorgio V., Cloud point transition in nonionic micellar solutions. J. Phys. Chem. 88, 309–317 (1984). [Google Scholar]
- 62.Brautigam C. A., Calculations and publication-quality illustrations for analytical ultracentrifugation data. Methods Enzymol. 562, 109–133 (2015). [DOI] [PubMed] [Google Scholar]
- 63.Salvay A. G., Santamaria M., le Maire M., Ebel C., Analytical ultracentrifugation sedimentation velocity for the characterization of detergent-solubilized membrane proteins Ca++-ATPase and ExbB. J. Biol. Phys. 33, 399–419 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.le Maire M., et al. , Gel chromatography and analytical ultracentrifugation to determine the extent of detergent binding and aggregation, and Stokes radius of membrane proteins using sarcoplasmic reticulum Ca2+-ATPase as an example. Nat. Protoc. 3, 1782–1795 (2008). [DOI] [PubMed] [Google Scholar]
- 65.Keller S., et al. , High-precision isothermal titration calorimetry with automated peak-shape analysis. Anal. Chem. 84, 5066–5073 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zhao H., Piszczek G., Schuck P., SEDPHAT—A platform for global ITC analysis and global multi-method analysis of molecular interactions. Methods 76, 137–148 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.