Significance
We describe the convergence of autophagy with YAP/TAZ, 2 highly related transcriptional regulators playing essential roles in cell plasticity for tissue regeneration and cancer. We show that a key event in autophagy, the fusion of autophagosomal vesicles with lysosomes, is transcriptionally regulated by YAP/TAZ. At least in part, this is mediated by the YAP/TAZ target Armus, a protein of the RAB-GAP family. Consistently with YAP/TAZ being patterned by mechanical signals, we found that cellular mechanics, dictated by the physical properties of the ECM, potently regulate autophagy efficiency. The connection described here is crucial for YAP-mediated reprogramming of normal cells into stem-like cells and of benign tumor cells in cancer stem cells. Thus, nuclear reprogramming must go hand in hand with autophagy-mediated cytoplasmic restructuring.
Keywords: mechanotransduction, YAP/TAZ, autophagy, cell plasticity
Abstract
Autophagy, besides ensuring energy metabolism and organelle renewal, is crucial for the biology of adult normal and cancer stem cells. However, it remains incompletely understood how autophagy connects to stemness factors and the nature of the microenvironmental signals that pattern autophagy in different cell types. Here we advance in these directions by reporting that YAP/TAZ transcriptionally control autophagy, being critical for autophagosomal degradation into autolysosomes. YAP/TAZ are downstream effectors of cellular mechanotransduction and indeed we found that cell mechanics, dictated by the physical property of the ECM and cytoskeletal tension, profoundly impact on autophagic flux in a YAP/TAZ-mediated manner. Functionally, by using pancreatic and mammary organoid cultures, we found that YAP/TAZ-regulated autophagy is essential in normal cells for YAP/TAZ-mediated dedifferentiation and acquisition of self-renewing properties. In tumor cells, the YAP/TAZ–autophagy connection is key to sustain transformed traits and for acquisition of a cancer stem cell state by otherwise more benign cells. Mechanistically, YAP/TAZ promote autophagic flux by directly promoting the expression of Armus, a RAB7-GAP required for autophagosome turnover and whose add-back rescues autophagy in YAP/TAZ-depleted cells. These findings expand the influence of YAP/TAZ mechanotransduction to the control of autophagy and, vice versa, the role of autophagy in YAP/TAZ biology, and suggest a mechanism to coordinate transcriptional rewiring with cytoplasmic restructuring during cell reprogramming.
Autophagy is a fundamental process in cell and tissue homeostasis, preserving nutrient metabolism and ensuring organellar quality control (1). In its most classical depiction (i.e., macroautophagy), this process involves sequestration of portions of cytoplasms within double-membrane vesicles (autophagosomes) that fuses with lysosomes to generate autolysosomes, where the autophagic cargo gets degraded. These functions typically protect against a host of diseases, from cancer to neurodegeneration and aging (2, 3); in other conditions, however, autophagy is causal for pathological cell states, most notably tumor progression (4). Autophagy is also required for the fitness of adult somatic stem cells (SCs), allowing either self-renewal or differentiation, as such accompanying tissue needs (5–7). In the context of cancer, autophagy can foster the generation of tumor cells with attributes of cancer SCs (CSCs) (5). The connection between autophagy and stemness is, however, complicated by the intrinsic fate plasticity of normal and tumor cells alike, as, depending on microenvironmental inputs, cells can reversibly switch from a nonstem to a stem state (8, 9). Still unclear in these scenarios remain the mechanisms by which autophagy connects to stemness factors in different somatic lineages, and how the autophagic flux can be controlled spatiotemporally by extrinsic signals.
Here we report the convergence of autophagy with another field of burgeoning interest, YAP/TAZ biology. YAP/TAZ are transcriptional coactivators playing essential roles in tissue regeneration by promoting either dedifferentiation and acquisition of stem/progenitor states by more differentiated cell types, or promoting expansion of resident SCs (10, 11). Moreover, YAP/TAZ are central mediators of tumorigenesis whose activation leads to acquisition of malignant properties in most solid malignancies (12). One of the most appealing features of YAP/TAZ is their regulation by the cell’s structural features, such as polarity, shape, and cytoskeletal organization. In turn, these depend on a cell’s location within the 3D architecture of tissues, including the attachment to other cells and to the surrounding extracellular matrix (ECM) (13, 14). Here we found that YAP/TAZ transcriptionally control a cardinal step in autophagy, the generation of the autolysosomes from autophagosomes, offering a means to connect autophagy to the main players of cell plasticity and tumorigenesis; this connection also allows harmonizing autophagic flux with overarching microenvironmental signals mechanically informing cells about their shape, location, and neighborhood relationships.
Results
YAP/TAZ Control Autophagic Flux by Regulating Autophagosome Degradation.
This work was initiated by a serendipitous discovery: In the context of other studies (15, 16), we were investigating whether YAP/TAZ inactivation affected cell viability. We first checked for apoptosis, but did not detect it. We then expected induction of autophagy, as an alternative means to cell consumption in YAP/TAZ-depleted cells. Surprisingly, we found the opposite, namely, that loss of YAP/TAZ in fact potently impaired autophagy. We found this unexpected result attractive as it hinted to a previously underappreciated positive link between autophagy and YAP/TAZ biology.
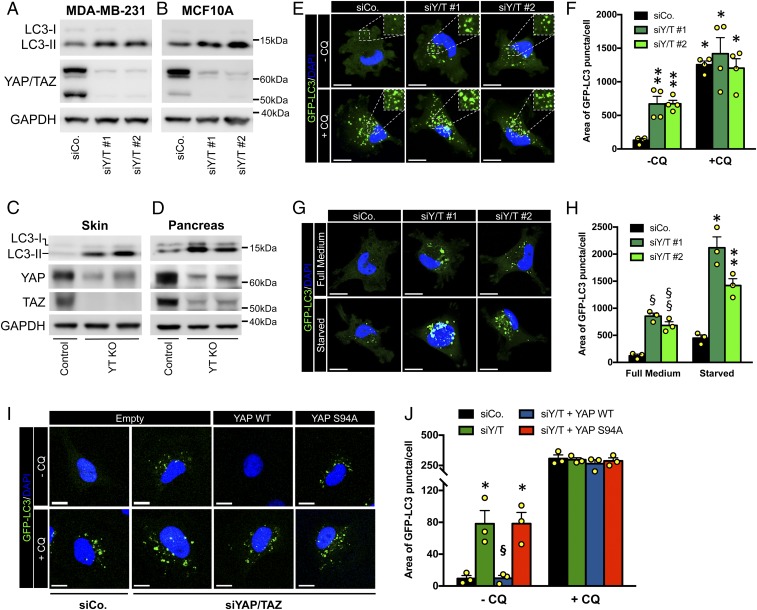
By immunoblot of lysates from control- and YAP/TAZ-depleted MDA-MB-231, a most established triple negative breast cancer (TNBC) cellular model system for studying YAP/TAZ addictions (16), we found that YAP/TAZ inactivation increased the levels of LC3-II, the lipidated form of LC3, a marker of autophagosome formation (17) (Fig. 1A). The generality of this finding was confirmed by extending our analysis to nontransformed mammary epithelial cells, MCF10A (Fig. 1B), and to colon cancer HCT116 cells (SI Appendix, Fig. S1A), all with consistent results. We further extended these conclusions in vivo, by monitoring LC3-II levels in lysates from skin and pancreatic explants of YAP/TAZ knockout mice: By immunoblotting with anti-LC3, loss of YAP/TAZ potently increased LC3 lipidation (Fig. 1 C and D).
Fig. 1.
YAP/TAZ control autophagic flux by regulating autophagosome degradation. (A and B) Immunoblot analysis for YAP/TAZ and LC3 in MDA-MB-231 (A) and MCF10A (B) transfected with control siRNA (siCo.) or 2 independent YAP/TAZ siRNA mixes (siY/T #1 or siY/T #2) for 48 h. The cleaved LC3 peptide and its phosphatidylethanolamine conjugated form are indicated as LC3-I and LC3-II, respectively. GAPDH serves as loading control. (C and D) R26CAG-CreER; Yapfl/fl; Tazfl/fl mice carrying an ubiquitous inducible Cre recombinase were treated with tamoxifen to obtain the knockout of both YAP and TAZ floxed alleles in vivo (Yap−/−; Taz−/−; i.e., Y/T KO). Tamoxifen-treated Yapfl/fl; Tazfl/fl littermate mice (without CRE) were used as control. See also SI Appendix, Methods. Immunoblot analysis of lysates from skin (C) and pancreatic biopsies (D) shows increased levels of LC3-II in YAP/TAZ knockout mice, compared to their controls (Top). Knockout of YAP and TAZ was confirmed by Western blot (Middle). Residual YAP and TAZ proteins remain in these blots due to cells escaping recombination. GAPDH serves as loading control (Bottom). (E and F) MDA-MB-231 cells stably expressing GFP-LC3 construct (MDA-GFP-LC3) were transfected with control siRNA (siCo) or 2 independent YAP/TAZ siRNA mixes (siY/T #1, siY/T #2) for 48 h. Cells were treated with medium (−CQ) or CQ 50 μM (+CQ) for the last 4 h. E shows representative confocal images. The Insets (2×) show higher magnification of the GFP-LC3 puncta. DAPI (blue) is a nuclear counterstain. (Scale bar, 20 μm.) (F) Quantification of GFP-LC3 puncta accumulation, measured as area of GFP-LC3 puncta per cell. Bars represent mean + SEM from 4 independent experiments. (*P ≤ 0.0001, **P < 0.05, compared to −CQ siCo; 2-way ANOVA). See SI Appendix, Methods for detailed GFP-LC3 quantification method. (G and H) MDA-GFP-LC3 cells transfected as in E, were cultured in full nutrient medium (full medium) or subjected to nutrient starvation in Hank's balanced salt solution (HBSS; starved) for the last 4 h. (G) Panels are representative confocal images. (H) Quantification of GFP-LC3 puncta accumulation as in F. Bars represent mean + SEM from 3 independent experiments (§P < 0.05, §§P < 0.01, compared to full medium siCo; *P < 0.0001, **P < 0.001 compared to starved siCo; 2-way ANOVA). (I and J) MII cells stably expressing GFP-LC3 construct (MII-GFP-LC3) were infected with an empty lentiviral vector (empty) or with the indicated siRNA-insensitive doxycycline-inducible lentiviral Flag-tagged YAP constructs. Cells were transfected with either control (siCo) or YAP/TAZ siRNAs (siYAP/TAZ), treated with doxycycline and analyzed 48 h after siRNA transfection. Cells were concomitantly treated with or without CQ for the last 4 h. (I) Panels are representative confocal images. (Scale bar, 20 μm.) (J) Quantification of GFP-LC3 puncta as in F. Bars represent mean + SEM (*P < 0.01 compared to −CQ empty-infected siCo, §P < 0.01 compared to −CQ empty-infected siYAP/TAZ; 1-way ANOVA). See also SI Appendix, Fig. S1G for validation of YAP/TAZ transcriptional activity in I and J.
To further confirm these data, we monitored autophagy through fluorescence microscopy in MDA-MB-231 cells stably expressing GFP-LC3 (MDA-GFP-LC3), by following the GFP-positive punctate structures that represent autophagosomes (17). As shown in Fig. 1 E and F, YAP/TAZ knockdown increased the area of GFP-LC3 puncta per cell; similar results were obtained with the Ras-transformed MCF10A-T1k (MII) cell line, stably expressing GFP-LC3 (MII-GFP-LC3) (SI Appendix, Fig. S1 B and C).
Next, we asked if the increased GFP-LC3 puncta observed upon YAP/TAZ knockdown reflects an increased autophagosome formation or, in fact, a blocked autophagosome turnover. To discriminate between these possibilities, the effects of YAP/TAZ knockdown were quantified in the presence of chloroquine (CQ), an inhibitor of autophagosome degradation after the fusion of autophagosomes with lysosomes. The rationale of these experiments is the following: If YAP/TAZ inhibit autophagosome formation, then YAP/TAZ inactivation plus CQ treatment should synergize with each other at inducing autophagosome accumulation by affecting both initiation and degradation. If instead, YAP/TAZ promote autophagosome degradation, then CQ treatment should be epistatic to YAP/TAZ inactivation, with no difference detected between control and YAP/TAZ-depleted cells after CQ treatment. This second scenario was the one experimentally validated, as we found that the area of GFP-LC3 puncta per cell increased to a similar extent in CQ-treated control and YAP/TAZ-depleted cells, consistently with YAP/TAZ being involved in lysosomal degradation of the autophagosomes (Fig. 1 E and F and SI Appendix, Fig. S1 B and C).
To validate these findings, we monitored the autophagic flux by scoring the number of LC3 endogenous puncta in the absence and presence of bafilomycin A1 (BafA1), a second independent inhibitor of autophagosome degradation (18). Consistently with previous results, the number of LC3 endogenous puncta significantly increased in YAP/TAZ-depleted MDA-MB-231 compared to control siRNA-treated cells, while BafA1 induced autophagosome accumulation to the same extent in both control and YAP/TAZ-depleted cells (SI Appendix, Fig. S1 D and E). Similar conclusions were derived from Western blot analysis of endogenous LC3 from cell lysates treated with CQ and BafA1 (SI Appendix, Fig. S1F).
To complement the above conclusions, we used starvation as a means to increase autophagic flux from its initial steps. Indeed, if YAP/TAZ act at the lysosomal degradation step, their knockdown should still elicit autophagosome accumulation under nutrient starvation conditions. This conclusion was validated in Fig. 1 G and H: by monitoring GFP-LC3 dots in nutrient-deprived cells, YAP/TAZ depletion still induced autophagosome accumulation, just at higher levels when compared to YAP/TAZ depletion alone. The accumulation of GFP-LC3 puncta induced by YAP/TAZ knockdown was rescued by siRNA-insensitive YAP wild type, but not transcriptionally deficient YAP mutant (YAP S94A) (Fig. 1 I and J and SI Appendix, Fig. S1G).
Finally, we examined the role of YAP/TAZ in early stages of autophagy and phagophore formation by following membrane accumulation of the WD repeat phosphoinositide interacting protein 2 (WIPI2) (18). No significant increase in the number of WIPI2 puncta was observed in YAP/TAZ-depleted cells compared to control siRNA-treated ones (SI Appendix, Fig. S1 H and I; see starvation [Starv] as positive control of autophagy induction).
We conclude from this collective set of evidence that YAP/TAZ are critical for autophagosomal degradation under both steady-state and induced autophagic settings, and that this relies on YAP/TAZ-dependent transcriptional regulation of autophagic flux.
YAP/TAZ Regulate the Fusion of Autophagosomes with Lysosomes.
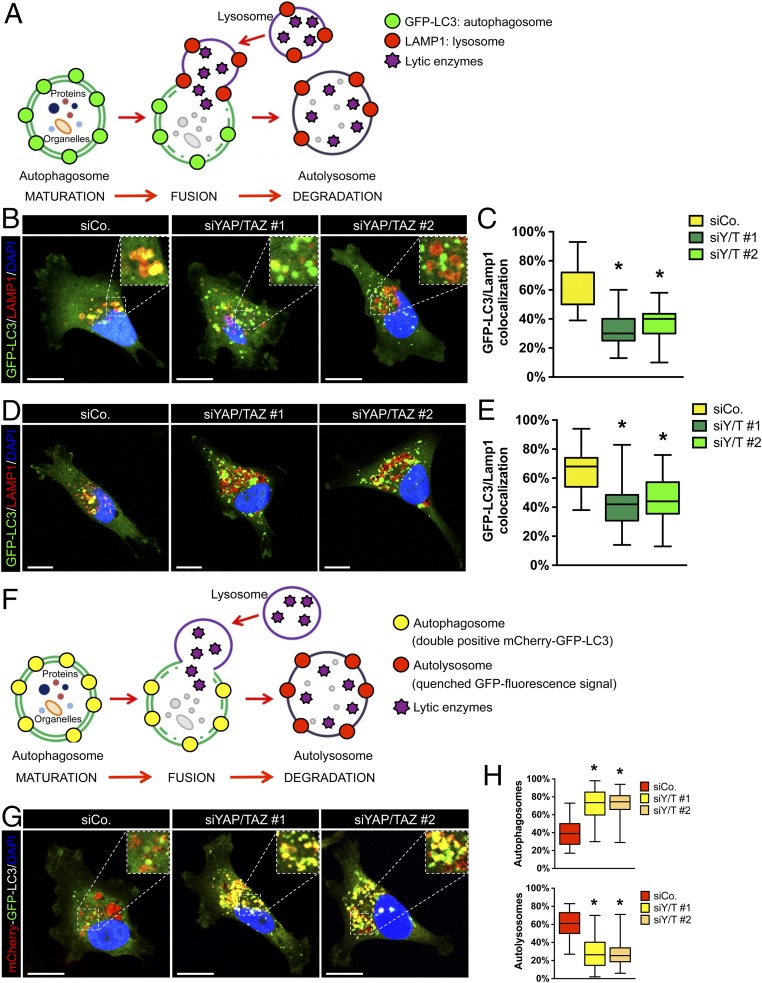
To test more directly the involvement of YAP/TAZ in autolysosome biogenesis, we examined the colocalization of the GFP-LC3 puncta with the lysosome-associated protein 1 (LAMP1), a marker of late endosomes and lysosomal compartments (Fig. 2A). In control MDA-GFP-LC3 cells, LAMP1 (red in Fig. 2 A–C) colocalizes with the GFP-LC3 puncta, indicating successful formation of autolysosomes (yellow) (Fig. 2 A–C). YAP/TAZ are critical for this fusion, as revealed by GFP-LC3 puncta and LAMP1-positive vesicles remaining as separate entities in YAP/TAZ knockdown cells (Fig. 2 A–C). Similar results were also observed in starved cells (Fig. 2 D and E), indicating the relevance of YAP/TAZ for autolysosome formation both in basal and starvation-induced autophagy (Fig. 2 A–E).
Fig. 2.
YAP/TAZ regulate the fusion of autophagosomes with lysosomes. (A) Schematic of autophagosome degradation during autophagic flux. Mature autophagosomes (stained by GFP-LC3; green) containing cytoplasmic materials fuse with lysosomes (stained by LAMP-1; red) to become autolysosomes. (B and C) MDA-GFP-LC3 transfected with control siRNA (siCo) or 2 independent YAP/TAZ siRNAs mixes (siY/T #1 and siY/T #2) were stained for the lysosomal membrane protein LAMP-1. Panels are representative confocal images of the colocalization between GFP-LC3 puncta (green) and LAMP1-positive vesicles (red) (B), and the corresponding quantification, scored as the ratio of the double-positive vesicles (yellow) to the total of GFP-LC3 puncta (green) per cell (C). DAPI (blue) is a nuclear counterstain. The Inset magnification is 2.5×. (Scale bar, 20 μm.) (D and E) MDA-GFP-LC3 transfected as previously described (B and C) were starved in HBSS for 4 h and stained for LAMP-1 to evaluate the colocalization between GFP-LC3 puncta (green) and LAMP-1 positive vesicles (red). Panel shows representative confocal images of GFP-LC3 (green) and LAMP-1 staining (red) (D) and the corresponding quantification of the colocalization between GFP-LC3 puncta (green) and LAMP-1 positive vesicles (red) (E). (F) Schematics of differently labeled autophagosomes (mCherry+/GFP+) and autolysosomes (mCherry+/GFP−) in MDA-mCherry-GFP-LC3. (G and H) Cells were transfected with control siRNA (siCo) or 2 independent YAP/TAZ siRNAs mixes (siY/T #1 and siY/T #2). (G) Representative confocal images of MDA-mCherry-GFP-LC3 cells treated as above. DAPI (blue) is a nuclear counterstain. The Inset magnification is 2×. (Scale bar, 20 μm.) (H) Quantification of the percentage of autophagosomes and autolysosomes scored, respectively, as the ratio of GFP+/mCherry+ puncta (autophagosomes) and GFP-/mCherry+ puncta (autolysosomes) to the total number of mCherry positive puncta (autophagosomes and autolysosomes). (C, E, and H) At least 30 cells from 3 independent experiments were scored for each condition. Data are presented as box and whisker plots: the box extends from the 25th to the 75th percentile; the line within the box represents the median; whiskers extend to show the highest and lowest values (*P < 0.0001 compared to siCo; 1-way ANOVA). (G and H) Number of autophagosomes ± SEM: siCo = 10.44 ± 2.31, siYAP/TAZ #1 = 21.92 ± 3.10, siYAP/TAZ #2 = 22.06 ± 3.14; number of autolysosomes ± SEM: siCo = 15.15 ± 2.96, siYAP/TAZ #1 = 6.65 ± 0.93, siYAP/TAZ #2 = 6.12 ± 0.95.
We confirmed that YAP/TAZ inhibition interferes with autolysosome formation by means of an independent experimental setup, that is, by using a tandem-tagged mCherry-GFP-LC3 probe. In autophagosome vesicles, both GFP and mCherry retain their fluorescence, while fluorescence of GFP, but not mCherry, is progressively quenched after fusion with lysosomes (due to the low pH of the autolysosome lumen) (Fig. 2F). In line with prior results, YAP/TAZ depletion in MDA-MB-231 cells stably expressing the mCherry-GFP-LC3 construct (MDA-mCherry-GFP-LC3), caused an increased rate of vesicles double positive for mCherry and GFP, i.e., blocked at the autophagosome stage (Fig. 2 F–H).
YAP/TAZ Mechanotransduction Regulates Autophagy.
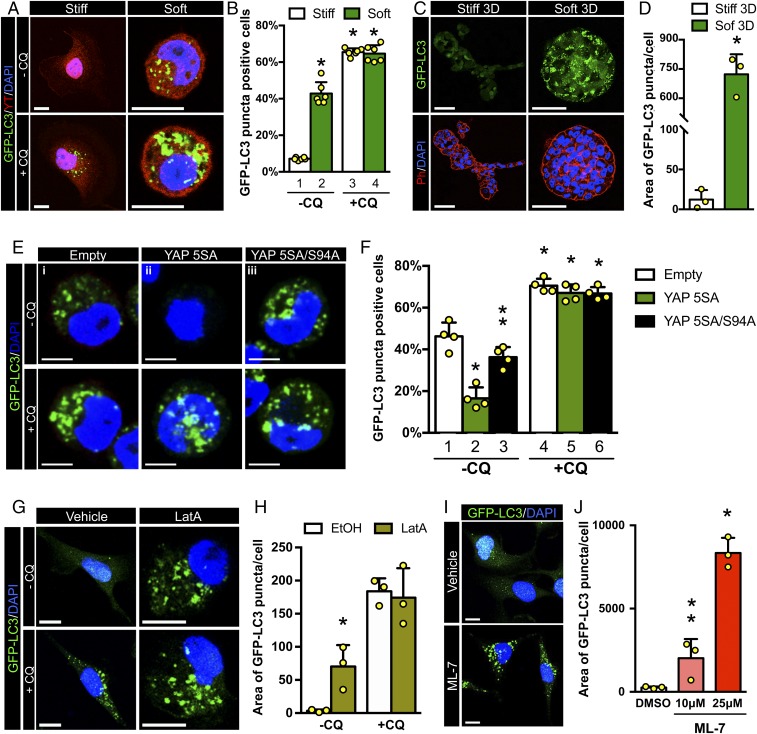
The results presented so far indicate that YAP/TAZ control autophagic flux favoring fusion of autophagosomes with lysosomes. But how is this regulated by environmental signals? YAP/TAZ activity is overarchingly induced by the mechanical inputs that cells receive from their attachment to a more or less compliant ECM or from neighboring cells. These physical cues in turn impact on cytoskeletal organization and cell shape (13, 14). Yet, little is known about the links between cellular mechanotransduction and autophagy. To test the relevance of ECM physical attributes for the regulation of autophagic flux, we plated MII-GFP-LC3 cells on soft vs. stiff fibronectin-coated acrylamide hydrogels (19, 20). Placing cells on soft hydrogels, where YAP/TAZ are inactive (SI Appendix, Fig. S1J), greatly increases the number of cells accumulating GFP-LC3 puncta (Fig. 3 A, Top and Fig. 3B, compare lanes 1 vs. 2). However, CQ treatment erases any difference in autophagosome accumulation between cells plated on stiff and soft ECM (Fig. 3 A, Bottom and Fig. 3B, compare lanes 3 and 4). Thus, autophagy is mechanically regulated, with low mechanical signaling ostensibly slowing down the autophagic flux at the level of autophagosome degradation recapitulating the effects of YAP/TAZ knockdown.
Fig. 3.
YAP/TAZ mechanotransduction regulates autophagy. (A) Representative confocal images of GFP-LC3 (green) and YAP/TAZ (red) in MII-GFP-LC3 cells plated on stiff (40 kPa) vs. soft (2.0 kPa) fibronectin-coated acrylamide hydrogels for 24 h. Cells were treated with vehicle (−CQ) or with CQ (+CQ) for the last 4 h. DAPI (blue) is a nuclear counterstain. (Scale bar, 20 μm.) (B) Percentage of MII-GFP-LC3 cells accumulating GFP-LC3 puncta as in A. Bars represent mean + SD from 6 different experiments (*P < 0.0001 compared to −CQ stiff; 2-way ANOVA). Cells with more than 10 GFP-LC3 puncta were scored as positive. (C) MII-GFP-LC3 were embedded as a single cell in a 3D matrix formed of Matrigel supplemented with different doses of collagen-I. Soft matrix contained 0.25 mg/mL collagen-I (soft 3D), whereas stiff matrix contained 1 mg/mL collagen-I (stiff 3D). After 5 d, cells were fixed, stained with phalloidin (red) to visualize the morphology of multicellular structures, and analyzed for the presence of GFP-LC3 puncta (green). (Scale bar, 50 μm.) (D) Quantification of GFP-LC3 puncta induced of cells experiencing a soft 3D ECM and plated as in C. At least 30 multicellular structures (>1,500 cells) from 3 independent experiments were scored for each condition; bars represent mean + SD (*P < 0.001, compared to stiff 3D; 2-tailed Student’s t test). (E and F) MII-GFP-LC3 cells infected with either an empty lentiviral vector (empty) or with the indicated doxycycline-inducible lentiviral YAP constructs, were plated on soft fibronectin-coated acrylamide hydrogels in the presence of doxycycline for 24 h and treated with medium (−CQ) or with CQ (+CQ) for the last 4 h. (E) Representative confocal images showing GFP-LC3 puncta (green). (F) Bars show accumulating GFP-LC3 puncta, and represent mean + SD from 4 independent experiments (*P < 0.0001 compared to −CQ empty infected, **P < 0.001 compared to −CQ YAP 5SA-infected cells; 2-way ANOVA). (G) Representative confocal images of MII-GFP-LC3 cells treated for 20 h with ethanol (EtOH) or 0.4 μM LatA. Cells were treated without (−CQ) or with CQ (+CQ) for the last 4 h; GFP-LC3 (green). (Scale bar, 20 μm.) (H) Quantification of GFP-LC3 puncta of MII-GFP-LC3 cells treated as in G. Bars represent mean + SD from 3 independent experiments (*P < 0.05 compared to −CQ EtOH-treated cells; 2-way ANOVA). (I) Representative confocal images of MII-GFP-LC3 cells treated for 20 h with DMSO or ML-7 (10 μM, 25 μM); GFP-LC3 puncta (green). (Scale bar, 20 μm.) (J) Quantification of GFP-LC3 puncta of MII-GFP-LC3 cells treated as in I. Bars represent mean + SD from 3 independent experiments (*P < 0.0001; **P < 0.05 compared to DMSO; 1-way ANOVA).
To further validate the causal connections between mechanical inputs and regulation of autophagy, we expanded the above conclusions on a 3D model system, culturing MII-GFP-LC3 cells in a Matrigel supplemented with low vs. high doses of collagen I (0.25 mg/mL and 1 mg/mL, respectively) to obtain a soft or stiff 3D ECM, as previously described (21). MII-GFP-LC3 growing within the stiffer 3D ECM formed tubule-like structures with no evidence of GFP-LC3 puncta (Fig. 3 C and D). In contrast, cells growing within the softer 3D ECM formed smaller acinus-like spheres displaying accumulation of GFP-LC3 puncta, similarly to what was observed above for cells plated on soft 2D hydrogels (Fig. 3 C and D).
To prove that the impairment of the autophagic flux induced by a soft ECM depends on the inhibition of YAP/TAZ, we investigated whether raising YAP/TAZ activity in cells experiencing low mechanics is sufficient to prevent autophagosome accumulation. Indeed, transduction of a doxy-inducible lentiviral vector expressing an activated form of YAP (YAP5SA) prevented accumulation of GFP-LC3 puncta in MII-GFP-LC3 cells cultured on soft hydrogels (Fig. 3E, compare frames i and ii; Fig. 3F, compare lanes 1 and 2). In contrast, the transcriptionally inactive YAP5SA/S94A mutant was uneffective (Fig. 3E, compare frames i and iii; Fig. 3F compare lanes 1 and 3). Again, upon CQ treatment we observed a dominant increased level in the percentage of GFP-LC3 puncta positive cells irrespective of any YAP activity (Fig. 3 E and F). Taken together, these data confirm that cell mechanics control autophagic flux through the regulation of YAP/TAZ transcriptional activity.
The physical features of the ECM control YAP/TAZ activity by changing actomyosin contractility and F-actin organization (13). We thus inhibited cellular mechanotransduction intracellularly by impairing either F-actin integrity, through treatment with latrunculin A (LatA), or actomyosin contractility, using ML-7, a drug inhibiting the myosin light chain kinase. As shown in Fig. 3 G and H, LatA-treated cells accumulated GFP-LC3 positive autophagosomes compared to mechanocompetent EtOH-treated control cells. Similarly, we observed a dose-dependent accumulation of the GFP-LC3 puncta with ML-7, when compared to DMSO-treated cells (Fig. 3 I and J). This accumulation occurs at the level of autolysosome dynamics, as CQ treatment did not show any significant difference at inducing GFP-LC3 puncta between EtOH- and LatA-treated cells (Fig. 3 G and H). Taken together, these data confirm that cell mechanics control autophagic flux through the regulation of YAP/TAZ transcriptional activity.
YAP/TAZ Require Autophagy to Sustain Cell Phenotypic Plasticity.
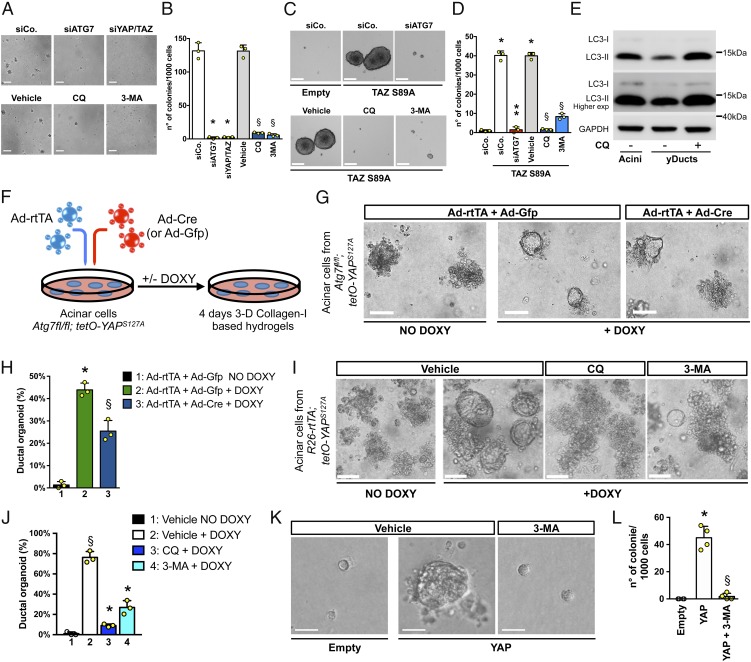
We then aimed to determine the role of autophagic flux regulation for some key aspects of YAP/TAZ biology. A classic tumorigenic attribute endowed by high levels of YAP/TAZ is the ability to overcome the widespread requirement cell–ECM adhesion to support cell proliferation (22). We found that impairing autophagic flux by depletion of the essential autophagy gene ATG7 (1) reduced anchorage-independent growth of MDA-MB-231 cells, as such phenocopying the effects of YAP/TAZ inactivation (Fig. 4 A and B). Consistent results were observed by blocking autophagy with other independent tools, that is, by either low doses of CQ or 3-methyladenine (3-MA, blocking the autophagy-initiation complex) (Fig. 4 A and B). These results establish a parallel between anchorage-independent growth relying on a high-level of YAP/TAZ, and YAP/TAZ-mediated sustainment of proficient autophagic flux.
Fig. 4.
YAP/TAZ require efficient autophagy flux to sustain their biological responses. (A and B) Impairing autophagic flux inhibits anchorage-independent growth. Representative pictures (A) and quantification (B) of colonies formed by MDA-MB-231 cells transfected with control siRNA (siCo) or with siRNAs targeting either ATG7 (siATG7) or YAP/TAZ (siYAP/TAZ) and then plated in soft-agar conditions. For pharmacological inhibition of autophagy, MDA-MB-231 were plated for soft-agar assay and treated with medium (vehicle) or with 2 independent autophagy inhibitors (CQ: 25 μM; 3-MA: 10 μM). (Scale bar, 200 μm.) Data are presented as mean + SD of 3 independent experiments. (*P < 0.0001 compared to siCo; §P < 0.0001 compared to vehicle; 1-way ANOVA). (C and D) Impairing autophagic flux inhibits YAP/TAZ-induced mammosphere formation. TAZ S89A-overexpressing MII cells (TAZ S89A), either transfected with the indicated siRNAs or treated with the autophagy inhibitors, were tested for mammosphere formation. MII cells transduced with empty vector (empty) were used as negative control of mammosphere growth. Data are mean + SD of 3 independent experiments. (*P < 0.0001 compared to empty siCo; **P < 0.0001 compared to TAZ S89A siCo; §P < 0.0001 compared to TAZ S89A vehicle; 1-way ANOVA). (E) Immunoblot analysis for LC3 protein of lysates obtained from acinar cells (acini) and YAP-induced pancreatic organoids (yDuct) treated with normal medium (−CQ) or with CQ 50 μM (+CQ) for 4 h before harvesting. (F) Schematic of the experiment performed with acinar cells isolated from Atg7fl/fl; tetO-YAPS127A mice. Pancreatic acini were coinfected with an adenoviral vector expressing rtTA (Ad-rtTA), to allow a doxycyline-inducible YAP expression, in combination with either a Cre- (Ad-Cre) or GFP-encoding adenoviral vector (Ad-Gfp), to obtain Atg7−/− or Atg7fl/fl cells, respectively. (G) Bright field images of pancreatic acini treated as indicated and seeded in collagen-I based hydrogels. Atg7fl/fl acini lacking exogenous YAP expression (e.g., Ad-rtTA + Ad-Gfp NO DOXY) were used as negative control of pancreatic reprogramming. (Scale bar, 50 μm.) (H) The ability to form ductal organoids was scored as percentage of acinar colonies converting to ductal structures. Data are mean + SD (n = 3 independent replicates) from 1 of 3 experiments, providing similar results (*P < 0.0001, lane 2 vs. lane 1; §P < 0.01 lane 3 vs. lane 2; 1-way ANOVA). (I and J) Pancreatic acini from R26-rtTA; tetO-YAPS127A mice seeded in collagen-I based hydrogels were cultured in the presence of DOXY, to induce the expression of the transgenic YAP, and treated either with medium (vehicle) or with 2 independent autophagy inhibitors CQ (25 μM) or 3-MA (10 μM). Acini lacking exogenous YAP expression (NO DOXY) were used as negative control of pancreatic reprogramming. See also SI Appendix, Fig. S2C for schematic of the experiment. Bright field images (I) and quantification (J) of yDuct organoids obtained upon YAP-dependent reprogramming of acinar cells treated as described above. (Scale bars, 50 μm.) Data are mean + SD of 3 independent experiments. (§P < 0.0001 compared to lane 1; *P < 0.0001 compared to lane 2; 1-way ANOVA). (K and L) FACS-purified LD cells were transduced with an rtTA-encoding lentivirus in combination with an empty vector (empty) or an inducible lentiviral YAP construct (YAP). Cells were cultured in presence of doxycycline and treated either with medium (vehicle) or 3-MA 10 μM (3-MA) as indicated. Representative images (K) and quantification (L) of yMaSC colonies formed by the indicated cells 15 d after plating in 5% Matrigel cultures. See also SI Appendix, Fig. S2D for representative FACS plots illustrating LD cells sorting procedure and SI Appendix, Fig. S2E for schematic of the experiment. Data are mean + SD of 2 independent experiments with 2 technical replicates each (*P < 0.0001, YAP compared to empty; §P < 0.0001, YAP+3MA compared to YAP; 1-way ANOVA). See SI Appendix, Methods for additional information on YAP-induced reprogramming experiments.
To address more directly whether autophagy is intrinsic to YAP/TAZ biological responses, we focused on YAP/TAZ-induced cell-fate plasticity. For example, it has been previously shown that raising the levels of either YAP or TAZ converts MII cells into cells with CSC-associated immunological traits (SI Appendix, Fig. S2A) and confers CSC properties to more differentiated and otherwise benign tumor cells (23, 24). To investigate whether autophagic flux is relevant for these responses, we performed mammosphere assays with otherwise benign MII tumor cells transduced with a cDNA enconding for an activated version of TAZ (MII-TAZ S89A). As shown in Fig. 4 C and D, control MII cells (MII-empty) are, per se, poorly able to grow as mammosphere, but acquire this capacity upon overexpression of TAZ S89A (MII-TAZ S89A). Inhibition of autophagy, by depletion of ATG7 or treatment with autophagy inhibitors (CQ or 3-MA), severely reduced TAZ-induced mammosphere formation (Fig. 4 C and D). Taken together, these data indicate that YAP/TAZ require autophagy to sustain transformed traits and CSC-inducing properties in cancer cells.
YAP/TAZ can also promote plasticity of terminally differentiated epithelial cells of different tissues, converting them into their corresponding tissue-specific progenitor/SCs (25). We asked whether YAP/TAZ-dependent regulation of autophagy is required for these reprogramming events. For example, in the pancreas, YAP/TAZ are critical for the earliest transformation event of normal acinar cells, namely their “metaplasia” that converts them into ductal-like progenitors from which pancreatic ductal adenocarcinoma (PDAC) eventually emerges. At least in part, this cell fate transition can be dissected in vitro using pancreatic organoids: Expression of YAP in acini explanted from transgenic R26-rtTA; tetO-YAPS127A mice and treated with doxycycline (DOXY) leads to acinar reprogramming into ductal cells that can be propagated as cyst-like organoids (yDucts) (25). This YAP-reprogramming step requires a YAP-induced rise in autophagic flux. Indeed, we found that YAP expression greatly increases autophagosome clearance in acinar cells (Fig. 4E). More remarkably, genetic inactivation of autophagy impairs YAP-mediated reprogramming of acinar cells. We showed this using acini isolated from Atg7fl/fl; tetO-YAPS127A mice, treated ex vivo with adenoviral vectors expressing rtTA (Ad-rtTA) and Cre to obtain YAP-expressing Atg7−/− cells (Fig. 4F and SI Appendix, Fig. S2B). These acini display reduced ability to convert into ductal cells (Fig. 4 G and H). Similar results were obtained after pharmacological inhibition of autophagy by CQ or 3-MA in acini explanted from R26-rtTA; tetO-YAPS127A transgenics (SI Appendix, Fig. S2C), leading to severe reduction in the number of yDucts when compared to the control (vehicle + DOXY) condition (Fig. 4 I and J).
We finally extended these conclusions to primary mammary gland cells. As previously reported, introduction of YAP or TAZ in terminally differentiated luminal mammary gland cells (LD, FACS-sorted as EpCAMhighCD49flowCD61− fraction), turn them into YAP-induced mammary SCs (yMaSCs) (SI Appendix, Fig. S2D). The latter are indistinguishable from the tissue resident mammary gland SCs (25) and indeed formed solid colonies when plated at clonogenic density in 3D 5% Matrigel cultures (Fig. 4 K and L and SI Appendix, Fig. S2E). However, 3-MA treatment completely blocks the appearance of yMaSCs (Fig. 4 K and L), indicating that YAP/TAZ also require an efficient autophagy flux to induce the reprogramming of normal mammary gland cells into a MaSC-like state.
YAP/TAZ Control Autophagic Flux through Their Direct Target Armus.
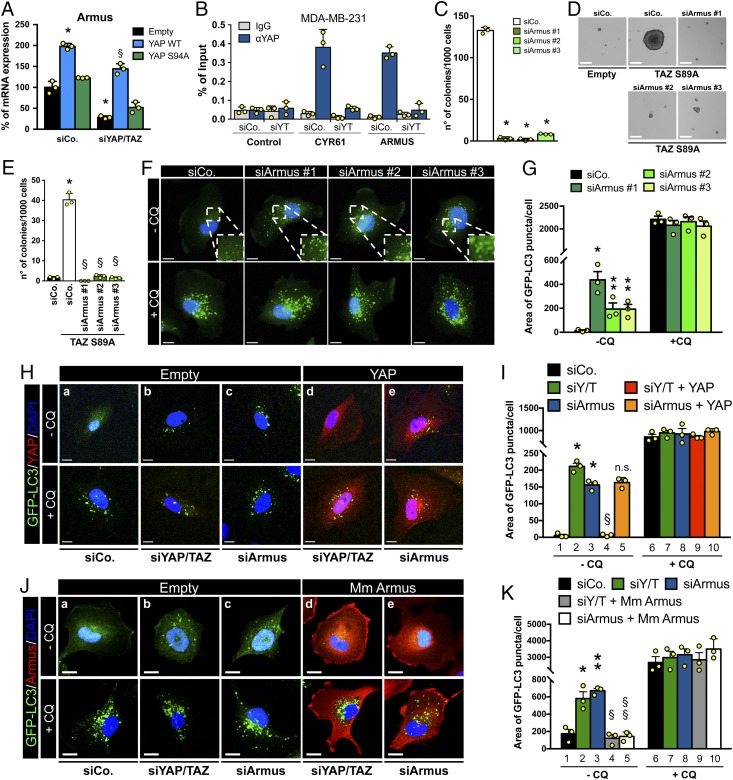
To study how YAP/TAZ might promote autophagic flux, we searched among YAP/TAZ direct target genes. We previously reported a chromatin “YAP/TAZ interactome map” obtained by the intersection of YAP/TAZ ChIP-seq datasets (i.e., YAP/TAZ bound enhancers) with Hi-C datasets, as such identifying a putative list of YAP/TAZ direct target genes (20). Searching this list for genes known to be involved in autophagy regulation, we identified as candidate effectors 4 members of the Tre2–Bub2–Cdc16 domain-containing family (TBC1D): Armus/TBC1D2A, TBC1D7, TBC1D9, and TBC1D10A (26). These genes are RAB GTPase activating proteins (RAB-GAPs; negative regulators of RAB GTPases) known to interact with LC3 proteins and, thus, coordinating trafficking and fusion events of intracellular vesicles in autophagic pathway (27). By RT-qPCR, however, only Armus, a RAB7-GAP known to regulate the fusion of autophagosomes with lysosomes (28, 29), was validated as a YAP/TAZ-regulated gene, at least in the MDA-MB-231 cellular model system. Indeed, we found that Armus mRNA levels were down-regulated upon YAP/TAZ knockdown (Fig. 5A and SI Appendix, Fig. S3 A and B) and rescued by add-back of wild-type YAP, but not transcriptionally inactive YAP (Fig. 5A).
Fig. 5.
YAP/TAZ control autophagic flux through their direct target Armus. (A) MII-GFP-LC3, infected with an empty vector (empty) or with the indicated siRNA-insensitive doxycycline-inducible lentiviral YAP constructs, were transfected with control (siCo) or YAP/TAZ-targeting siRNAs (siYAP/TAZ). Cells were treated with doxycycline, harvested 48 h after siRNA transfection, and analyzed by RT-qPCR for Armus mRNA levels. Data were normalized to empty-infected cells transfected with siCo (black bar). (*P ≤ 0.0001 siCo + YAP WT compared to empty siCo; §P ≤ 0.0001 siYAP/TAZ + YAP WT compared to empty siYAP/TAZ; 2-way ANOVA). See also SI Appendix, Fig. S3 A and B. (B) Validation by ChIP-qPCR in MDA-MB-231 cells of the YAP/TAZ-binding site on the Armus-associated enhancer (see also SI Appendix, Fig. S3 C and D). CYR61 promoter is positive control, HBB promoter is negative control (control). Data from 3 replicates (mean + SD) are shown normalized to the percent input (1% of starting chromatin used as input). (C) Armus depletion impairs anchorage-independent growth. Quantification of colonies formed by MDA-MB-231 transfected with siCo or with 3 independent Armus siRNAs (siArmus #1, #2, and #3) plated in soft-agar assays. (*P < 0.0001 compared to siCo; 1-way ANOVA). See also SI Appendix, Fig. S3G for the validation of siRNAs. (D and E) Armus is required for YAP/TAZ-induced mammosphere formation. TAZ S89A-overexpressing MII cells (TAZ S89A) were transfected with the indicated siRNAs and tested for mammosphere formation. MII cells transduced with empty vector (empty) are negative control. (*P < 0.0001 TAZ S89A siCo compared to empty siCo; §P < 0.0001 TAZ S89A siArmus compared to TAZ S89A siCo; 1-way ANOVA). (F) Representative confocal images of MII-GFP-LC3 transfected as in C. The Insets (2×) show higher magnification of the GFP-LC3 puncta. (G) Quantification of GFP-LC3 puncta of MII-GFP-LC3 cells treated as in F, measured as area of GFP-LC3 puncta per cell. (*P < 0.001, **P < 0.05 compared to −CQ siCo; 2-tailed Student’s t test). (H and I) MII-GFP-LC3, infected with an empty vector (empty) or with a siRNA-insensitive doxycycline-inducible lentiviral Flag-tagged YAP constructs (YAP), were transfected with siCo, YAP/TAZ siRNAs mix (siYAP/TAZ), or Armus siRNA (siArmus), and treated with doxycycline. Representative confocal images (YAP in red) (H) and quantification (I) of GFP-LC3 puncta of MII-GFP-LC3 cells treated as above, measured as area of GFP-LC3 puncta per cell. (*P < 0.0001 compared to lane 1; §P < 0.0001 compared to lane 2; n.s., not significant compared to lane 3; 1-way ANOVA). (J) MII-GFP-LC3, infected with an empty vector (empty) or with a doxycycline-inducible vector encoding a siRNA-insensitive HA-tagged mouse Armus construct (Mm Armus, red), were transfected with the indicated siRNAs. (K) Quantification of GFP-LC3 puncta of MII-GFP-LC3 cells treated as in J. (*P < 0.01, **P < 0.001 compared to lane 1; §P < 0.001 compared to lane 2; §§P < 0.0001 compared to lane 3; 1-way ANOVA). (F–K) Cells were treated with medium (−CQ) or CQ 50 μM (+CQ) for the last 4 h and analyzed 48 h after siRNA transfection. (F, H, and J) GFP-LC3 (green); DAPI (blue) is a nuclear counterstain. (Scale bar, 20 μm.) (A, C, E, G, I, and K) Bars represent mean (n = 3) + SD.
We first validated by ChIP-qPCR the association of YAP/TAZ with the enhancer element of Armus predicted by the YAP/TAZ “interactome map” (15, 16, 20) (SI Appendix, Fig. S3C). This binding occurred in both MDA-MB-231 (Fig. 5B) and MII cells (SI Appendix, Fig. S3D). Consistently with Armus being a YAP/TAZ target, we found its mRNA levels are regulated by cellular mechanotransduction, being suppressed in cells experiencing a soft ECM or treated with inhibitors of the F-actin cytoskeleton (SI Appendix, Fig. S3 E and F).
Functionally, upon knockdown with independent siRNAs, Armus is essential for anchorage-independent growth of MDA-MB-231 cells (Fig. 5C), phenocopying at least in part the requirement of YAP/TAZ; more crucially, Armus is up-regulated upon TAZ activation (SI Appendix, Fig. S2A) and also required downstream of TAZ for induction of CSCs in MII cells (Fig. 5 D and E).
Having established the transcriptional and functional connections between YAP/TAZ and Armus, we next addressed if YAP/TAZ control autophagy through Armus. First, depletion of Armus by 3 independent siRNAs led to autophagosome accumulation, as shown by the increase of the area occupied by GFP-LC3 puncta per cell (Fig. 5 F and G). CQ treatment leveled autophagosome accumulation in all experimental conditions, without any additive effects when compared to Armus knockdown alone, in keeping with the model that Armus regulates autophagosome turnover, rather than initiation (Fig. 5 F and G). Then, we investigated if Armus is epistatic to YAP/TAZ in the regulation of autophagy. We found that experimentally raising YAP activity, although sufficient to rescue YAP/TAZ depletion (Fig. 5H, compare frames a/b with d; Fig. 5I, compare lanes 1/3 with 4), was unable to rescue autophagic flux impaired in Armus knockdown cells (Fig. 5H, compare frames a/c with e; Fig. 5I, compare lanes 1/3 with 5). Thus, Armus acts downstream of YAP. Consistently, adding back Armus through lentiviral delivery in fact restored autophagosome degradation in both Armus-depleted cells (Fig. 5J, compare frames a/c with e; Fig. 5K, compare lanes 1/3 with 5) and YAP/TAZ-depleted cells (Fig. 5J, compare frames a/b with d; Fig. 5K, compare lanes 1/2 with 4). Thus, transcriptional regulation of Armus is a mechanism by which YAP/TAZ regulate autophagosome turnover.
Discussion
The findings presented here describe an unexpected convergence of 2 areas of intense investigation, autophagy and the activities of YAP/TAZ. The paradigm identified here holds a number of implications, suggesting the involvement of YAP/TAZ in the biological effects of autophagy and, vice versa, of autophagy in YAP/TAZ responses. If broadly applicable, this would expand manifolds the influence of YAP/TAZ mechanotransduction to realms of biology that are well established for autophagy but so far unsuspected in the YAP/TAZ field. This includes neurodegeneration, aging, and a host of human genetic disorders associated with defective autophagy (2, 3). In the other direction, the present findings also invite the adoption of a wider interpretative lens for the potent biology of YAP/TAZ: most of our understanding of how YAP/TAZ control SCs, tissue regeneration, and tumorigenesis is very partial, limited to transcriptional regulation of genes involved in cell proliferation (13); the contribution of YAP/TAZ-mediated increased autophagic flux may be in fact instrumental for a number of these responses. As proof of principle of this concept, here we found the involvement of autophagy in the context of YAP/TAZ-mediated induction of stemness properties, that is, conversion of otherwise benign breast tumor cells into CSCs, which indeed others have shown to rely on an increased autophagic flux to sustain their CSC-related traits (reviewed in ref. 5) and of YAP-driven reprogramming of normal differentiated cells into their corresponding tissue-specific stem cells. Such requirements may reflect the need to accommodate the higher metabolic demands associated with a transition from a quiescent cell state to one endowed with proliferative potential (5). However, since reprogramming can in fact be uncoupled from proliferation (25), and given the unrestricted availability of nutrients in the experimental systems here adopted, we favor a different interpretation: We propose that YAP/TAZ-driven autophagy represents a form of “cytoplasmic” reprogramming, a checkpoint ensuring that YAP/TAZ transcriptional rewiring in the nucleus goes hand in hand with the need of cytoplasmic and in fact whole-cell renovation and restructuring. It is tempting to speculate that the inner composition and structural organization of the cell’s constituents (organelles, cytoskeleton, vesicles) may represent a currently poorly appreciated form of “epigenetic memory,” a roadblock to normally unwarranted cell fate changes that helps in preserving cell differentiation.
Over the last decades a large body of knowledge has accumulated on the mechanisms and players of autophagy. Linking YAP/TAZ activity to a key step in autophagic flux, the autophagosome–autolysosome fusion, advances on the less understood means by which autophagy can be spatially and temporally patterned. Mechanical signals can indeed target individual cells with exquisite specificity depending on the tensional force applied to each individual cell. The positive connection between YAP/TAZ and autophagy has been previously noticed, although through a proposed indirect mechanism involving YAP acting upstream of myosin-light chain II (MLC2) expression (30). Our findings instead suggest a different mechanistic interpretation, as we envision autophagy as a downstream event of YAP/TAZ mechanotransduction: in our model, changes in ECM physicality serve as upstream inputs to control YAP/TAZ-mediated transcriptional activation of members of the TBC1D family, such as Armus. As such, here we highlight a previously neglected role of YAP/TAZ in autophagic flux and provide functional and epistatic validation of Armus acting downstream of YAP/TAZ for regulation of autophagic flux. Given the potential functional overlap of this otherwise poorly characterized family of proteins, it is plausible that, in other cellular contexts YAP/TAZ may control autophagy through regulation of Armus and other members of this family (26, 27). At the same time, by no mean do our data exclude other mechanisms that may exist for YAP or TAZ to regulate autophagy. It is possible that different cell types or microenvironmental conditions may favor a role of YAP/TAZ-mediated transcription in promoting both autophagy initiation and autophagosome turnover, or rather tip the balance toward 1 of the 2 steps. Also intriguing is the possibility that in cells infected with viral RNA/DNA, YAP/TAZ may also regulate autophagy in a transcriptionally independent manner by associating with TBK1 in the cytoplasm (31, 32).
Autophagy is an appealing therapeutic target in a number of diseases, but also a particularly challenging one, given the broad relevance of autophagy for normal homeostasis. In contrast, YAP/TAZ are crucial in cancer and to accommodate tissue needs after injury, but remarkably ostensibly dispensable for homeostasis (12). Thus, the emerging connections here between autophagy and the YAP/TAZ signaling ecosystems imply that targeting YAP/TAZ mechanotransduction may offer new insights to block or fuel autophagy in various contexts, and do so by either affecting ECM rigidity, cell mechanics, or YAP/TAZ transcription itself.
Methods
Cell Cultures and Reagents.
MDA-MB-231 cells (from ICLC) were cultured in DMEM/F12 (Thermo Fisher) supplemented with 10% FBS, glutamine, and antibiotics. MCF-10A and MII cells were cultured in DMEM/F12 with 5% horse serum (HS), glutamine, and antibiotics, freshly supplemented with insulin, EGF, hydrocortisone, and cholera toxin (23, 33). HEK293 (ATCC), and HCT116 (ICLC) and HEK293GP (Takara) cells were cultured in DMEM (Thermo Fisher) supplemented with 10% FBS, glutamine, and antibiotics. All cells were checked routinely for absence of mycoplasma contaminations. To generate MDA-MB-231-GFP-LC3 and MDA-MB-231-mCherry-GFP-LC3 stable cell lines, MDA-MB-231 cells were respectively transduced with pBABE-puro-GFP-LC3 or pBABE-puro-mCherry-GFP-LC3 and selected with puromycin. To generate MII-GFP-LC3 cells, MII cells were transduced with pBABE-blasti-GFP-LC3 and selected with blasticidin.
Mice.
Animal experiments were performed adhering to our institutional guidelines (University of Padua) as approved by the Organismo Preposto al Benessere Degli Animali (OPBA) and the Italian Ministry of Health.
Supplementary Material
Acknowledgments
We thank D. J. Pan, F. Camargo, M. Sandri, D. Saur, J. Siveke, and P. Bonaldo for gifts of mice and I. Dikic and L. Naldini for plasmids. This work is supported by Fondazione “Associazione Italiana per la Ricerca sul Cancro” (AIRC) under 5 per Mille 2019, ID 22759 program, by an AIRC PI-Grant, by a “Framework per l’Attrazione e il Rafforzamento delle Eccellenze per la Ricerca in Italia” (FARE) Grant from Ministero dell’Istruzione, dell'Università e della Ricerca (MIUR), and by Epigenetics Flagship project Consiglio Nazionale delle Ricerche (CNR) - MIUR grants (to S.P.). This work was also funded by a “Fondazione Cassa di Risparmio di Padova e Rovigo” (CARIPARO) starting grant (to T.P.), by “Progetti di Rilevante Interesse Nazionale” (PRIN) 2017 grants (to S.P., L.A., and T.P.), and by “Progetti di Ricerca Dipartimentali/Investimento Strategico di Dipartimento” (PRID/SID) (to L.A.). Q.Z. was supported by a fellowship from Marie Skłodowska-Curie Innovative Training Network – “Biochemical and Mechanochemical Signalling in Polarized Cells” (BIOPOL) Consortium. This project has received funding from the European Program Horizon 2020-European Research Council research and innovation programme “De Novo Generation of Somatic Stem Cells” (DENOVOSTEM) Grant 670126 (to S.P.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. G.K. is a guest editor invited by the Editorial Board.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1908228116/-/DCSupplemental.
References
- 1.Mizushima N., Komatsu M., Autophagy: Renovation of cells and tissues. Cell 147, 728–741 (2011). [DOI] [PubMed] [Google Scholar]
- 2.Levine B., Kroemer G., Biological functions of autophagy genes: A disease perspective. Cell 176, 11–42 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dikic I., Elazar Z., Mechanism and medical implications of mammalian autophagy. Nat. Rev. Mol. Cell Biol. 19, 349–364 (2018). [DOI] [PubMed] [Google Scholar]
- 4.Galluzzi L., et al. , Autophagy in malignant transformation and cancer progression. EMBO J. 34, 856–880 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boya P., Codogno P., Rodriguez-Muela N., Autophagy in stem cells: Repair, remodelling and metabolic reprogramming. Development 145, dev146506 (2018). [DOI] [PubMed] [Google Scholar]
- 6.García-Prat L., Sousa-Victor P., Muñoz-Cánoves P., Proteostatic and metabolic control of stemness. Cell Stem Cell 20, 593–608 (2017). [DOI] [PubMed] [Google Scholar]
- 7.Salemi S., Yousefi S., Constantinescu M. A., Fey M. F., Simon H.-U., Autophagy is required for self-renewal and differentiation of adult human stem cells. Cell Res. 22, 432–435 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rybstein M. D., Bravo-San Pedro J. M., Kroemer G., Galluzzi L., The autophagic network and cancer. Nat. Cell Biol. 20, 243–251 (2018). [DOI] [PubMed] [Google Scholar]
- 9.Lin B., et al. , Modulating cell fate as a therapeutic strategy. Cell Stem Cell 23, 329–341 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Panciera T., Azzolin L., Cordenonsi M., Piccolo S., Mechanobiology of YAP and TAZ in physiology and disease. Nat. Rev. Mol. Cell Biol. 18, 758–770 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Moya I. M., Halder G., Hippo-YAP/TAZ signalling in organ regeneration and regenerative medicine. Nat. Rev. Mol. Cell Biol. 20, 211–226 (2019). [DOI] [PubMed] [Google Scholar]
- 12.Zanconato F., Cordenonsi M., Piccolo S., YAP/TAZ at the roots of cancer. Cancer Cell 29, 783–803 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Totaro A., Panciera T., Piccolo S., YAP/TAZ upstream signals and downstream responses. Nat. Cell Biol. 20, 888–899 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Humphrey J. D., Dufresne E. R., Schwartz M. A., Mechanotransduction and extracellular matrix homeostasis. Nat. Rev. Mol. Cell Biol. 15, 802–812 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zanconato F., et al. , Genome-wide association between YAP/TAZ/TEAD and AP-1 at enhancers drives oncogenic growth. Nat. Cell Biol. 17, 1218–1227 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zanconato F., et al. , Transcriptional addiction in cancer cells is mediated by YAP/TAZ through BRD4. Nat. Med. 24, 1599–1610 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mizushima N., Yoshimori T., Levine B., Methods in mammalian autophagy research. Cell 140, 313–326 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Klionsky D. J., et al. , Guidelines for the use and interpretation of assays for monitoring autophagy (3rd edition). Autophagy 12, 1–222 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dupont S., et al. , Role of YAP/TAZ in mechanotransduction. Nature 474, 179–183 (2011). [DOI] [PubMed] [Google Scholar]
- 20.Totaro A., et al. , YAP/TAZ link cell mechanics to Notch signalling to control epidermal stem cell fate. Nat. Commun. 8, 15206 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Aragona M., et al. , A mechanical checkpoint controls multicellular growth through YAP/TAZ regulation by actin-processing factors. Cell 154, 1047–1059 (2013). [DOI] [PubMed] [Google Scholar]
- 22.Brusatin G., Panciera T., Gandin A., Citron A., Piccolo S., Biomaterials and engineered microenvironments to control YAP/TAZ-dependent cell behaviour. Nat. Mater. 17, 1063–1075 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cordenonsi M., et al. , The Hippo transducer TAZ confers cancer stem cell-related traits on breast cancer cells. Cell 147, 759–772 (2011). [DOI] [PubMed] [Google Scholar]
- 24.Lamar J. M., et al. , The Hippo pathway target, YAP, promotes metastasis through its TEAD-interaction domain. Proc. Natl. Acad. Sci. U.S.A. 109, E2441–E2450 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Panciera T., et al. , Induction of expandable tissue-specific stem/progenitor cells through transient expression of YAP/TAZ. Cell Stem Cell 19, 725–737 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Frasa M. A. M., Koessmeier K. T., Ahmadian M. R., Braga V. M. M., Illuminating the functional and structural repertoire of human TBC/RABGAPs. Nat. Rev. Mol. Cell Biol. 13, 67–73 (2012). [DOI] [PubMed] [Google Scholar]
- 27.Kern A., Dikic I., Behl C., The integration of autophagy and cellular trafficking pathways via RAB GAPs. Autophagy 11, 2393–2397 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Carroll B., et al. , The TBC/RabGAP Armus coordinates Rac1 and Rab7 functions during autophagy. Dev. Cell 25, 15–28 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Toyofuku T., Morimoto K., Sasawatari S., Kumanogoh A., Leucine-rich repeat kinase 1 regulates autophagy through turning on TBC1D2-dependent Rab7 inactivation. Mol. Cell. Biol. 35, 3044–3058 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pavel M., et al. , Contact inhibition controls cell survival and proliferation via YAP/TAZ-autophagy axis. Nat. Commun. 9, 2961 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pilli M., et al. , TBK-1 promotes autophagy-mediated antimicrobial defense by controlling autophagosome maturation. Immunity 37, 223–234 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang Q., et al. , Hippo signalling governs cytosolic nucleic acid sensing through YAP/TAZ-mediated TBK1 blockade. Nat. Cell Biol. 19, 362–374 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Santner S. J., et al. , Malignant MCF10CA1 cell lines derived from premalignant human breast epithelial MCF10AT cells. Breast Cancer Res. Treat. 65, 101–110 (2001). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.