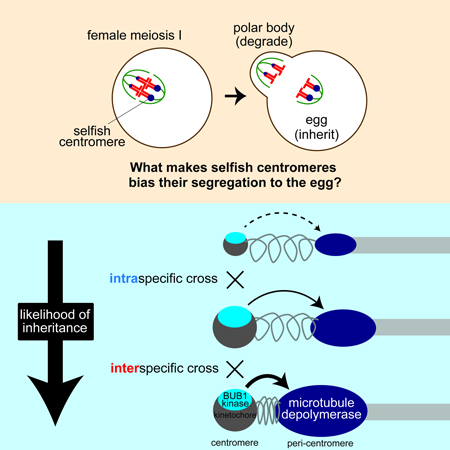
SUMMARY
Asymmetric division in female meiosis creates selective pressure favoring selfish centromeres that bias their transmission to the egg. This centromere drive can explain the paradoxical rapid evolution of both centromere DNA and centromere-binding proteins despite conserved centromere function. Here, we define a molecular pathway linking expanded centromeres to histone phosphorylation and recruitment of microtubule destabilizing factors, leading to detachment of selfish centromeres from spindle microtubules that would direct them to the polar body. Exploiting centromere divergence between species, we show that selfish centromeres in two hybrid mouse models use the same molecular pathway but modulate it differently to enrich destabilizing factors. Our results indicate that increasing microtubule destabilizing activity is a general strategy for drive in both models, but centromeres have evolved distinct mechanisms to increase that activity. Furthermore, we show that drive depends on slowing meiotic progression, suggesting that selfish centromeres can be suppressed by regulating meiotic timing.
Keywords: meiotic drive, meiosis, mouse, oocyte, chromosome segregation, centromere
eTOC
The enrichment of microtubule-destabilizing activity on selfish centromeres provides a mechanistic basis for centromere drive
Graphical Abstract
INTRODUCTION
Genomes are vulnerable to selfish genetic elements, which increase in frequency by forming additional copies of themselves (e.g., transposons) or distorting transmission ratios during meiosis (i.e., meiotic drive), and are neutral or harmful to the host (McLaughlin and Malik, 2017). In female meiosis, selfish elements violate Mendel’s Law of Segregation by preferentially segregating to the egg, which increases their transmission to the progeny (Chmátal et al., 2017; Pardo-Manuel de Villena and Sapienza, 2001) (Figure 1A). Because centromeres direct chromosome segregation, they are the genetic elements with the best opportunity to cheat the segregation process. Meiotic drive of selfish centromeres, or centromere drive, can explain the “centromere paradox”: rapid evolution of both centromere DNA sequences and genes encoding centromere-binding proteins despite highly conserved centromere function in chromosome segregation (Henikoff et al., 2001). The centromere drive theory is based on the idea that natural selection favors centromere DNA sequences that act selfishly in female meiosis. Fitness costs associated with drive, for example due to deleterious alleles linked to driving centromeres, would also select for alleles of centromere-binding proteins that suppress drive. Thus, centromere DNA sequences and centromere proteins continually evolve in conflict with each other, analogous to a molecular arms race between viruses and the immune system. Supporting this theory, centromeres with expanded satellite repeats drive in monkeyflowers and in mice (Fishman and Saunders, 2008; Iwata-Otsubo et al., 2017; Wu et al., 2018). However, the theory raises several fundamental questions: how do centromeres cheat at a molecular level, linking from centromere expansion to selfish behavior, and how can centromere drive be suppressed?
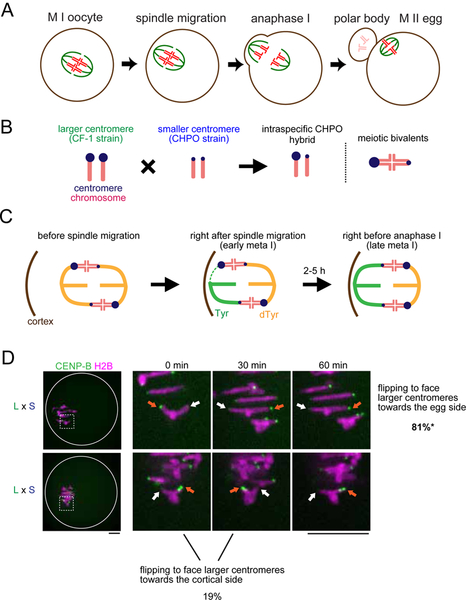
Figure 1. Biased flipping underlies the biased orientation of selfish centromeres towards the egg pole.
(A) Schematic of female meiosis. The meiosis I (MI) spindle initially forms in the center of the oocyte and later migrates towards the cortex and orients perpendicular to the cortex, followed by the highly asymmetric cell division. Selfish elements cheat by preferentially orienting to the egg side of the spindle. (B) Schematic of the intraspecific CHPO hybrid system for centromere drive. A Mus musculus strain with larger (L) centromeres, CF-1, is crossed to a strain with smaller (S) centromeres, CHPO. In the hybrid offspring, chromosomes with larger and smaller centromeres are paired in meiotic bivalents. (C) Schematic showing spindle asymmetry and biased orientation of larger centromeres in the intraspecific CHPO hybrid, based on previous results (Akera et al., 2017). Initial MT attachments are established when the spindle is still in the center and symmetric. Hybrid bivalents are off-center on the spindle, with the larger centromere closer to the pole, indicating that larger and smaller centromeres interact differentially with spindle MTs. Bivalent orientation on the spindle is unbiased right after spindle migration (early meta I), but the attachment of larger centromeres to the cortical side of the spindle is especially unstable, leading to detachment and flipping to establish biased orientation (late meta I). (D) CF-1 x CHPO (L x S) hybrid oocytes expressing CENP-B-mCherry and H2B-EGFP were imaged live after spindle migration. Time-lapse images show examples of flipping events to face larger centromeres towards the egg (top) or cortical (bottom, 0 – 30 min) side. Images are maximum intensity z-projections showing all chromosomes (left), or optical slices magnified to show flipping events (timelapse). Orange and white arrows indicate larger and smaller centromeres, respectively. Scale bar, 10 μm. Percentages indicate the frequency of each case (n = 21 flipping events from 48 cells). *P < 0.05, indicating significant deviation from 50%. Two out of four flipping events that faced larger centromeres towards the cortical side were subsequently reversed (bottom, 30 – 60 min), demonstrating the difficulty for larger centromeres to remain attached to the cortical side.
To address these questions, we need a system to examine cell biological mechanisms of centromere drive. We previously established a Mus musculus hybrid between a standard laboratory strain with larger centromeres (either CF-1 or C57BL/6J) and a wild-derived strain from an isolated population with smaller centromeres (CHPO) (Figure 1B, also see STAR METHOD “Mice” section). In this intraspecific hybrid system (hereafter refer to as CHPO hybrid), larger centromeres have more centromeric minor satellite repetitive sequence, more CENP-A nucleosomes which specify the site of kinetochore assembly, and more kinetochore proteins (e.g. CENP-C and HEC1) relative to smaller centromeres (Chmátal et al., 2014; Iwata-Otsubo et al., 2017). In meiosis I in the CHPO hybrid, there are six bivalents in which larger and smaller centromeres of homologous chromosomes are paired (Figure 1B), and seven trivalents in which a Robertsonian fusion chromosome from CHPO pairs with two homologous chromosomes (Chmátal et al., 2014). We focused our analyses on the bivalents, which preferentially orient with larger centromeres towards the egg side of the spindle (Iwata-Otsubo et al., 2017), which will segregate to the egg (Figure 1C). We use this biased orientation on the spindle as a readout for centromere drive because trivalents mis-segregate frequently and cause subfertility (Bint et al., 2011; Daniel, 2002; Pacchierotti et al., 1995), making it technically difficult to analyze inheritance in this system.
Our previous results suggest that larger centromeres detach from the cortical side of the spindle to re-orient towards the egg side (Akera et al., 2017) (Figure 1C). The findings raise the question of why larger centromeres, which build larger kinetochores, are more susceptible to detachment (Lampson and Black, 2017). Moreover, it is unclear whether findings in one hybrid model system represent a general strategy for selfish centromeres to cheat. The large divergence in centromere DNA sequences between mouse species (Kipling et al., 1995; Narayanswami et al., 1992; Wong et al., 1990) suggests that genetic conflict between centromere DNA and centromere-binding proteins has generated different evolutionary trajectories in different species and potentially different mechanisms of centromere drive. In this study, we uncovered molecular mechanisms and evolutionary strategies of meiotic cheating by selfish centromeres, exploiting both intraspecific variation and interspecific divergence in combination with cell biological analyses and experimental manipulation of centromeres. We establish a molecular pathway linking expanded centromeres to microtubule (MT)-destabilizing activity at peri-centromeres and show that this activity makes centromeres selfish. Moreover, we show that centromeres from different mouse species have evolved distinct strategies to enrich destabilizing activity. Finally, our findings suggest that rapid progression through meiosis I can be a mechanism to suppress drive.
RESULTS
BUB1 links selfish centromeres and higher MT-destabilizing activity
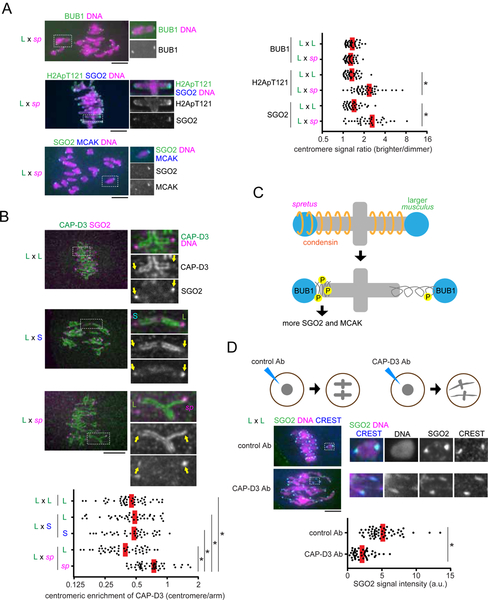
To confirm that bivalents in CHPO hybrid oocytes preferentially re-orient to direct larger centromeres towards the egg side during metaphase I, we imaged these flipping events live. Since larger centromeres have 6- to 10-fold more minor satellite repeats, we can distinguish larger and smaller centromeres by expressing fluorescently-tagged CENP-B protein, which binds minor satellite (Masumoto et al., 1989). We find that flipping events are biased to detach larger centromeres from the cortical side and re-orient them towards the egg side (81%, Figure 1D). Consistent with this result, we previously showed that the orientation of larger centromeres is initially unbiased, but later becomes biased towards the egg side of the spindle before anaphase I (Figure 1C). We also showed that larger centromeres form more unstable MT attachments compared to smaller centromeres, particularly with the cortical side of the spindle (Figure 1C, early meta I) (Akera et al., 2017). These findings suggest that selfish larger centromeres with larger kinetochores detach more readily from the spindle. To uncover the underlying mechanisms, we examined factors that destabilize MTs at centromeres to correct erroneous attachments: MCAK (mitotic centromere associated kinesin), which is a member of the kinesin-13 family, and the chromosome passenger complex (CPC) composed of Survivin, Borealin, INCENP, and Aurora B kinase (Carmena et al., 2012; Godek et al., 2015; Lampson and Grishchuk, 2017). By analyzing the bivalents in CHPO hybrid oocytes, we found asymmetry in MCAK, Survivin, and phosphorylated INCENP (Salimian et al., 2011) as a marker of active Aurora B kinase (Figures 2A and S1A). We did not observe such asymmetry in control oocytes in which centromeres of homologous chromosomes should be the same. Using CENP-B to label minor satellite, we found that larger centromeres have more of these MT-destabilizing factors compared to smaller centromeres (Figure 2B), similar to previous results for kinetochore proteins (Iwata-Otsubo et al., 2017). These observations suggest that selfish centromeres enrich MT-destabilizing activity to detach MTs and re-orient on the spindle.
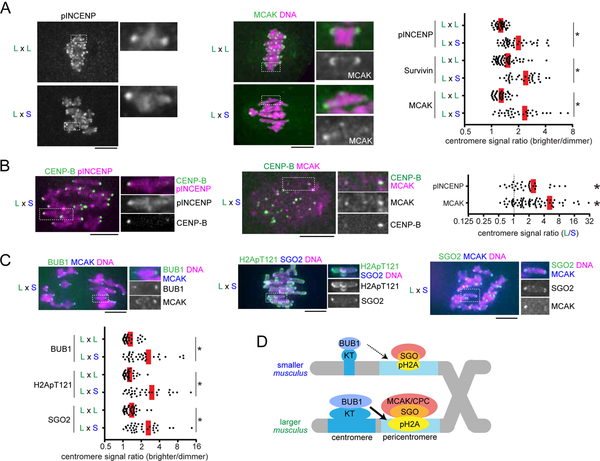
Figure 2. Selfish centromeres enrich more MT-destabilizing factors through the BUB1 pathway.
(A and C) CF-1 x CHPO (L x S) hybrid oocytes, or CF-1 x CF-1 (L x L) as controls, were fixed at metaphase I and stained for phosphorylated INCENP, Survivin, MCAK, BUB1, H2ApT121, or SGO2. Graph shows centromere signal ratios, calculated as the brighter divided by the dimmer signal for each bivalent (n > 32 for each condition); red line, mean; *P < 0.001. (B) CF-1 x CHPO hybrid oocytes expressing CENP-B-EGFP were stained for pINCENP or MCAK. Graph shows centromere signal ratios, calculated as the CF-1 centromere divided by the CHPO centromere signal for each bivalent. Each dot represents a single bivalent (n > 31 for each condition); red line, mean; *P < 0.001, indicating significant deviation from 1. Images (A-C) are maximum intensity z-projections showing all chromosomes (left), or optical slices magnified to show single bivalents (right); scale bars, 10 μm. (D) Model of the amplified BUB1 pathway in larger centromeres compared to smaller centromeres in the intraspecific CHPO hybrid.
MT-destabilizing factors localize to peri-centromeres, which are nearby but distinct chromosome regions from centromeres (Watanabe, 2012). Further, the amount of peri-centromeric repetitive major satellite DNA is similar between larger and smaller centromeres in the CHPO hybrid (Iwata-Otsubo et al., 2017). Therefore, it was unclear how larger centromeres can enrich more destabilizing activity. Shugoshin serves as a scaffold for both CPC and MCAK and is recruited to peri-centromeres by directly binding histone H2A threonine 121 phosphorylation marks (H2A pT121) (Watanabe, 2012). This histone phosphorylation is catalyzed by BUB1 kinase, which localizes at kinetochores (Kawashima et al., 2010). We hypothesized that larger centromeres recruit higher levels of BUB1 kinase relative to smaller centromeres, which would induce the asymmetric localization of Shugoshin and MT-destabilizing factors. Indeed, we found asymmetry in BUB1, H2A pT121, and SGO2, the major Shugoshin paralog in mouse oocytes (Lee et al., 2008), across the bivalents in CHPO hybrid oocytes but not in control oocytes (Figures 2C and S1B). MCAK is enriched more on larger centromeres relative to smaller centromeres (Figure 2B), and BUB1 and SGO2 are enriched on the centromere with more MCAK (Figure 2C). Therefore, BUB1 and SGO2 are also enriched on larger centromeres (Figure 2D). Similarly, since H2ApT121 is enriched on the centromere with more SGO2 (Figure 2C), H2ApT121 is also higher on larger centromeres (Figure 2D). Together, these results indicate that BUB1 kinase is the molecular link between larger kinetochores and MT-destabilizing factors at peri-centromeres (Figure 2D).
Asymmetry in MT destabilizing activity is essential for centromere drive
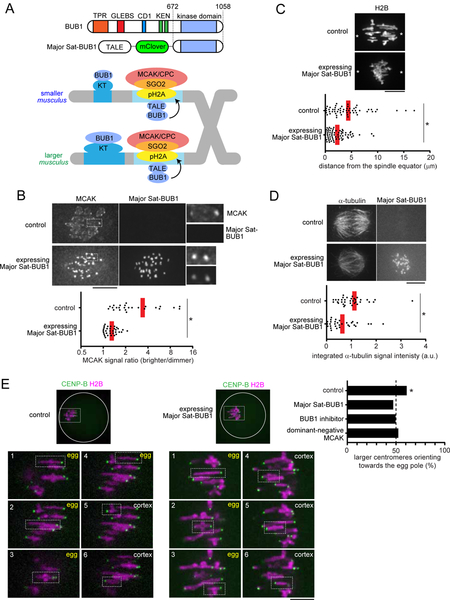
To test the significance of the BUB1 pathway and MT-destabilizing activity for centromere drive, we developed an approach to experimentally equalize destabilizing activity between larger and smaller centromeres. We took advantage of the peri-centromeric repetitive major satellite DNA, which is similar between larger and smaller centromeres (Iwata-Otsubo et al., 2017), by genetically fusing the kinase domain of BUB1 to a TALE construct that targets major satellite (hereafter, Major Sat-BUB1) (Miyanari et al., 2013) (Figure 3A). Expressing this construct in hybrid oocytes increased MCAK and CPC levels on both sides of the bivalent and canceled the asymmetry (Figures 3B and S2). To determine the functional consequences of BUB1 targeting, we first examined the position of hybrid bivalents, which are off-center on the spindle at metaphase I in control hybrid oocytes, with larger centromeres closer to the pole (Figure 1C), indicating functional differences in MT interactions between larger and smaller centromeres (Chmátal et al., 2014). Bivalents were positioned close to the equator in hybrid oocytes expressing Major Sat-BUB1 (Figure 3C), which strongly suggests that centromere functions are equalized by this manipulation. Second, we confirmed that increasing MT-destabilizing factors at centromeres through BUB1 targeting indeed makes MTs more unstable based on cold-stable kinetochore-MT fibers (Rieder, 1981) (Figure 3D). Finally, we measured the orientation of hybrid bivalents on the spindle, using CENP-B to distinguish larger and smaller centromeres. We found that larger centromeres are no longer biased towards the egg pole in oocytes expressing Major Sat-BUB1, demonstrating that the asymmetry in destabilizing activity is essential for centromere drive (Figure 3E).
Figure 3. Asymmetry in MT destabilizing activity is essential for centromere drive.
(A) Schematic of the strategy to equalize MT-destabilizing activity between larger and smaller centromeres by targeting BUB1 to major satellite; TPR, tetratricopeptide repeat domain; GLEBS, BUB3 binding domain; CD1, conserved domain 1;KEN, KEN box (Vleugel et al., 2015). (B) CF-1 x CHPO (L x S) oocytes expressing a TALE targeting major satellite fused to the fluorescent protein mClover and to BUB1 lacking the N-terminal kinetochore-targeting domain (Major Sat-BUB1). Cells were fixed at metaphase I and stained for MCAK. Graph shows centromere signal ratios, calculated as the brighter divided by the dimmer signal for each bivalent. Each dot represents a single bivalent (n > 25 for each condition); red line, mean. (C) CF-1 x CHPO oocytes expressing Major Sat-BUB1 and H2B-EGFP were imaged live at metaphase I. Asterisks indicate the position of spindle poles determined by differential interference contrast imaging. Graph shows the distance between the spindle equator and the crossover position of each bivalent (n > 60 bivalents for each condition). (D) CF-1 x CF-1 oocytes expressing Major Sat-BUB1 were analyzed for cold-stable MTs at metaphase I. Graph shows integrated α-tubulin signal intensity in the spindle (n > 32 spindles for each condition). (E) CF-1 x CHPO oocytes expressing CENP-B-mCherry and H2B-EGFP were imaged live shortly before anaphase I onset (within 30 min). Oocytes also expressed Major Sat-BUB1 or a dominant-negative MCAK mutant, RAMFLhyp, or were treated with a BUB1 inhibitor, BAY-1816032, as indicated. The fraction of bivalents with the larger centromere oriented towards the egg pole was quantified; n = 272 bivalents for control, 110 for Major Sat-BUB1, 115 for BAY-1816032, and 126 for RAMFLhyp. Images show the most common configuration for conditions with or without biased orientation (control or Major Sat-BUB1), with each of the six bivalents labeled to indicate the orientation of the larger centromere towards the egg or cortex. Images (B-E) are maximum intensity z-projections or optical slices magnified to show single bivalents. *P < 0.005, indicating significant deviation from 50% in (E).
As a complementary approach, we inhibited the BUB1 pathway using a BUB1 inhibitor, BAY-1816032 (Siemeister et al., 2019). This manipulation significantly reduced H2ApT121, SGO2 and MCAK levels (Figure S3) and canceled biased orientation of larger centromeres towards the egg pole (Figure 3E). Further, we directly perturbed MT-destabilizing activity by expressing a dominant-negative MCAK construct, RAMFLhyp (Illingworth et al., 2010; Wordeman et al., 2007). This construct is a motor-dead, full-length MCAK designed to deplete endogenous MCAK from centromeres by competing for centromere binding (Wordeman et al., 2007). Expressing this mutant MCAK disrupted biased orientation, demonstrating the significance of destabilizing activity for centromere drive (Figure 3E). Together, these results indicate that selfish centromeres exploit BUB1 signaling to recruit MT destabilizers to win the competition in female meiosis.
Centromeres in an interspecific hybrid exhibit asymmetry in destabilizers governed by condensin
Our experiments with the intraspecific CHPO hybrid system revealed molecular mechanisms of drive, linking selfish centromeres to an amplified BUB1 pathway and recruitment of MT-destabilizing factors to the peri-centromere. To determine whether these findings represent general properties of driving centromeres in mouse, we exploited the large divergence in centromere DNA sequences between mouse species (Wong et al., 1990) to identify a second hybrid model for centromere drive. We selected Mus musculus and Mus spretus because centromere DNA has diverged between the two species with spretus centromeres having substantially more minor satellite and less major satellite repeats compared to musculus centromeres (Miyanari et al., 2013; Narayanswami et al., 1992; Wong et al., 1990). We crossed SPRET/EiJ (Mus spretus) with CF-1 or C57BL/6J (Mus musculus with larger centromeres relative to CHPO) to produce an interspecific hybrid (hereafter, spretus hybrid) (Figure 4A). We used CF-1 to mate with CHPO in the intraspecific cross because they efficiently produce hybrid offspring, but we primarily used C57BL/6J as the musculus strain in the spretus cross because of difficulties mating spretus with CF-1.
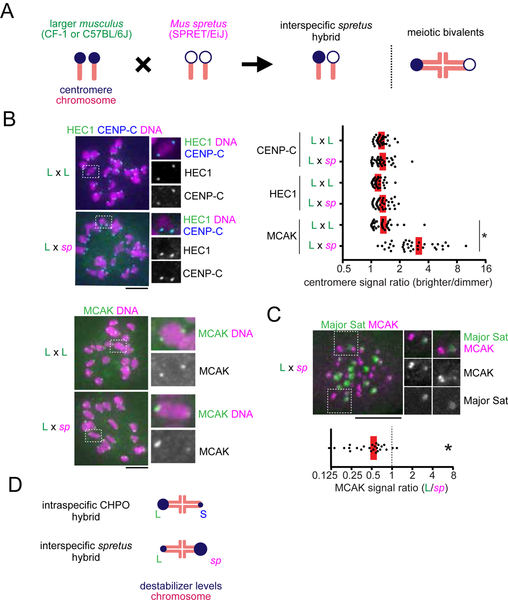
Figure 4. Centromeres in an interspecific hybrid exhibit asymmetry in destabilizers but not in kinetochore size.
(A) Schematic of the interspecific spretus hybrid system. A Mus musculus strain with larger centromeres (L, CF-1 or C57BL/6J) is crossed to a Mus spretus strain (sp, SPRET/EiJ). In the hybrid offspring, chromosomes with musculus and spretus centromeres are paired in meiotic bivalents. (B) C57BL/6J x SPRET/EiJ (L x sp) hybrid oocytes, or C57BL/6J x C57BL/6J (L x L) as controls, were fixed at metaphase I and stained for the indicated centromere proteins. Graph shows centromere signal ratios, calculated as the brighter divided by the dimmer signal for each bivalent (n > 36 bivalents for each condition). (C) C57BL/6J x SPRET/EiJ (L x sp) oocytes expressing Major Sat. TALE-mClover were stained for MCAK. Graph shows centromere signal ratios, calculated as the C57BL/6J centromere divided by the spretus centromere signal for each bivalent (n = 24 bivalents). Images (B, C) are maximum intensity z-projections showing all chromosomes (left), or optical slices magnified to show single bivalents (right); scale bars, 10 μm. In the graphs, each dot represents a single bivalent; red line, mean; *P < 0.001, indicating significant deviation from 1 in (C). (D) Schematic of relative MT destabilizer levels in both intraspecific and interspecific hybrid models.
We first measured centromere protein levels in spretus hybrid oocytes. Both the inner kinetochore protein CENP-C that binds to CENP-A nucleosomes and the outer kinetochore protein HEC1 that binds to MTs were similar across the hybrid bivalents (Figure 4B). In contrast, MCAK showed significant asymmetry across the bivalents in the spretus hybrid, but not in control musculus oocytes (Figure 4B). The CPC localized all over the chromosomes in this hybrid without obvious enrichment on centromeres (Figure S4), probably due to higher levels of cohesin complex on chromosome arms (Sodek et al., 2017), which contributes to CPC recruitment through the Haspin kinase pathway (Goto et al., 2017). Therefore, we focused on MCAK as a MT-destabilizing factor in the spretus hybrid. Since musculus centromeres have more peri-centromeric repetitive major satellite DNA, we can distinguish musculus and spretus centromeres by expressing a fluorescently-tagged TALE construct that recognizes major satellite (Miyanari et al., 2013). Using this approach, we found that the larger musculus centromeres, which recruited more MCAK in the intraspecific CHPO hybrid, have less MCAK compared to spretus centromeres in the interspecific hybrid (Figures 2B, 4C, and 4D). These data indicate that spretus centromeres in the interspecific hybrid enrich destabilizing activity independent of kinetochore size, in contrast to the intraspecific CHPO hybrid where increased destabilizing activity is associated with larger kinetochores.
To understand how spretus centromeres enrich MT-destabilizing factors at peri-centromeres, we examined the BUB1 pathway in the spretus hybrid. Consistent with other kinetochore proteins, BUB1 localization was similar across the hybrid bivalents (Figure 5A). In contrast, the scaffold for MT-destabilizing factors, SGO2, showed significant asymmetry across the bivalents in a consistent orientation with MCAK (Figure 5A). Furthermore, H2ApT121 was enriched on spretus centromeres with more SGO2 (Figure 5A), raising the question of how spretus centromeres acquire more H2A phosphorylation without recruiting more BUB1 kinase, especially since BUB1 is the dominant kinase for H2ApT121 (El Yakoubi et al., 2017). One possibility is that differences in centromere geometry between musculus and spretus centromeres impact the accessibility of kinetochore-localized BUB1 kinase to peri-centromeric chromatin, thereby modulating H2A phosphorylation. The condensin complex regulates centromere geometry by preventing deformation of both centromeric and peri-centromeric chromatin during mitosis and meiosis (Houlard et al., 2015; Lee et al., 2011; Oliveira et al., 2005; Samoshkin et al., 2009). Therefore, we examined condensin localization in spretus hybrid oocytes. The CAP-D3 subunit of condensin II, the major condensin complex in mouse oocytes (Houlard et al., 2015), localized all along the chromosome axis, including spretus centromeres, but was reduced on musculus centromeres, leading to asymmetric centromere localization of condensin on hybrid bivalents (Figure 5B, L x sp). CAP-D3 was also partially excluded from centromeres in control musculus oocytes and in the intraspecific CHPO hybrid, with no detectable differences between larger and smaller musculus centromeres (Figure 5B, L x S and L x L). These results suggest that condensin induces SGO2 and MCAK asymmetry by modulating centromere geometry in the spretus hybrid (Figure 5C), but centromere asymmetry in the CHPO hybrid is not due to condensin differences.
Figure 5. Condensin governs the asymmetry in MT-destabilizing factors in the interspecific spretus hybrid.
(A) C57BL/6J x SPRET/EiJ (L x sp) hybrid oocytes, or C57BL/6J x C57BL/6J (L x L) as controls, were fixed at metaphase I and stained for BUB1, H2ApT121, or SGO2. Graph shows centromere signal ratios, calculated as the brighter divided by the dimmer signal for each bivalent. Each dot represents a single bivalent (n > 36 for each condition). (B) C57BL/6J x SPRET/EiJ (L x sp) hybrid oocytes, or CF-1 x CHPO (L x S) and CF-1 x CF-1 (L x L) as controls, were fixed at metaphase I and stained for CAP-D3 and SGO2. Graph shows centromeric enrichment of CAP-D3, calculated as the centromeric signal divided by the chromosome arm signal for each half-bivalent. Each dot represents a single centromere (n > 40 for each condition). (C) Model for how SGO2 and MCAK recruitment depends on condensin in the spretus hybrid. (D) CF-1 x CF-1 (L x L) oocytes microinjected with control IgG or anti-CAP-D3 antibody were fixed at metaphase I and stained for SGO2 and CREST. Graph shows centromeric SGO2 signal intensity. Each dot represents a single centromere (n > 46 for each condition). Images (A, B, D) are maximum intensity z-projections showing all chromosomes (left), or optical slices magnified to show single bivalents (right); red line, mean; *P < 0.001; scale bars, 10 μm.
To directly test the significance of condensin for SGO2 enrichment, we inhibited condensin II function by microinjecting CAP-D3 antibody. Condensin II inhibition caused centromere stretching (Lee et al., 2011) and significantly reduced SGO2 levels (Figure 5D). Together, our results indicate that both hybrid systems use the same molecular pathway but modulate it differently to enrich MT-destabilizing factors at peri-centromeres. In the intraspecific CHPO hybrid, larger musculus centromeres assemble larger kinetochores to amplify the BUB1 pathway by recruiting more BUB1 kinase. In contrast, spretus centromeres modulate centromere geometry, independent of kinetochore size, to amplify the BUB1 pathway in the spretus hybrid.
Winning centromeres in one hybrid become losers in another hybrid based on the relative destabilizing activity
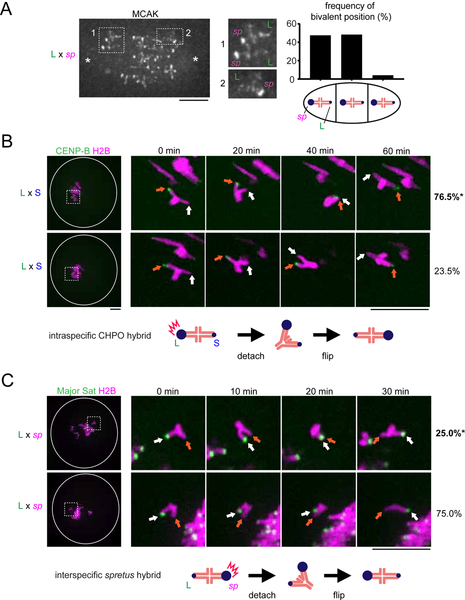
We performed three assays to test the functional consequences of asymmetry in MT-destabilizing factors. First, based on the findings from the CHPO hybrid, we predicted that spretus hybrid bivalents would be positioned off-center on the spindle, with centromeres with higher destabilizing activity closer to the pole. Indeed, we found that spretus centromeres with more MCAK are closer to the pole (Figure 6A). Second, we predicted that centromeres with more MCAK should initiate flipping events by detaching MTs first. To test this prediction, we tracked the flipping process by live imaging in both hybrid models to determine which centromere moved towards the opposite pole first to initiate flipping, indicating that it detached first. In the intraspecific CHPO hybrid, we found that larger musculus centromeres initiated 76% of flipping events (Figure 6B), but the same larger musculus centromeres initiated only 25% of flipping events when paired with spretus centromeres in the interspecific hybrid (Figure 6C). These results are consistent with relative MCAK levels: spretus > larger musculus > smaller musculus (Figures 2B and 4C).
Figure 6. Relative MCAK levels on centromeres predict their destabilizing activity.
(A) C57BL/6J x SPRET/EiJ (L x sp) oocytes were fixed at metaphase I and stained for MCAK. Images are maximum intensity z-projections showing all chromosomes or optical slices magnified to show two bivalents closer to the left pole (1) or a single bivalent closer to the right pole (2). Schematic shows bivalent positions as equidistant between the two poles (middle) or off-center with either the spretus centromere or the larger musculus centromere closer to the pole. The frequency of each case is plotted (n = 120 bivalents). (B) CF-1 x CHPO (L x S) oocytes expressing CENP-B-mCherry and H2B-EGFP were imaged live. Time-lapse images show examples of flipping events, which were analyzed to determine the frequency of either the larger (orange arrows) or smaller (white arrows) musculus centromere moving first to initiate flipping (top and bottom panels respectively). Percentages on the right indicate frequency of each case (n = 45 flipping events from 61 cells). (C) CF-1 x SPRET/EiJ and C57BL/6J x SPRET/EiJ (L x sp) oocytes expressing Major Sat. TALE-mClover and H2B-mCherry were imaged live and analyzed to determine whether the larger musculus (white arrows) or spretus (orange arrows) centromere initiates flipping (top and bottom panels respectively) (n = 27 flipping events from 20 cells). Images (B, C) are maximum intensity z-projections showing all chromosomes (left), or optical slices magnified to show flipping events (timelapse). White circle indicates the cell outline. Schematics show the more frequent flipping events, with relative MCAK levels indicated by the size of the blue circle. Scale bars, 10 μm; *P < 0.05, indicating significant deviation from 50%.
Third, if higher destabilizing activity is a general property of driving centromeres, the prediction is that larger musculus centromeres, which win in the CHPO hybrid by preferentially orienting towards the egg side of the spindle, would be losers in the spretus hybrid. We found that the orientation of spretus hybrid bivalents was unbiased just before anaphase I (Figure 7B, control), but spretus hybrid oocytes do not delay anaphase onset (Sebestova et al., 2012), in sharp contrast to the CHPO hybrid (Akera et al., 2017) (Figure 7A). Timing is important because the spindle initially forms in the center of the oocyte, and later migrates towards the cortex (Almonacid et al., 2014; Azoury et al., 2011; Holubcová et al., 2013). CDC42 signaling from the cortex regulates MT tyrosination, which generates asymmetry between the two sides of the spindle after spindle migration (Akera et al., 2017; Dehapiot et al., 2013) (Figures 1C, 7A, and S5). Biased orientation arises from biased flipping while the spindle is positioned close to the cortex and asymmetric (Figures 1C and 1D). Consistent with this idea, the orientation of CHPO hybrid bivalents is initially unbiased at the earlier stage right after spindle migration, but anaphase I is delayed 2 – 5 hours, which provides time for the flipping events (Akera et al., 2017) (Figure 1C). In contrast, spretus hybrid oocytes progress to anaphase I immediately after spindle migration.
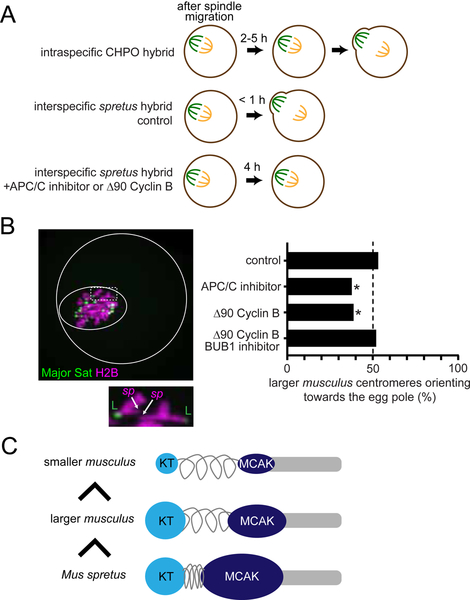
Figure 7. Relative MT-destabilizing activity determines the direction of centromere drive.
(A) Schematics of meiotic progression. Expression of non-degradable Δ90 Cyclin B or treatment with ProTAME, an APC/C inhibitor, delays anaphase I onset in the spretus hybrid to at least 4 hours, comparable to the CHPO hybrid. (B) CF-1 x SPRET/EiJ and C57BL/6J x SPRET/EiJ oocytes expressing Major Sat. TALE-mClover and H2B-mCherry were imaged live either shortly before anaphase I (control) or 2–4 hours after spindle migration. Oocytes also expressed Δ90 Cyclin B or were treated with ProTAME or the BUB1 inhibitor BAY-1816032 as indicated. Images are a maximum intensity z-projection of the whole oocyte (top) and an optical slice magnified to show two bivalents (bottom). Solid and dashed white circles indicate the outline of the cell and the spindle, respectively. Graph shows the fraction of bivalents with the larger musculus centromere oriented towards the egg pole; n = 295 bivalents for control, 135 for ProTAME, 134 for Δ90 Cyclin B, and 150 for Δ90 Cyclin B + BAY-1816032. *P < 0.01, indicating significant deviation from 50%. Scale bars, 10 μm. (C) Schematic showing that the direction of centromere drive correlates with MT-destabilizer levels. Musculus centromeres enrich destabilizing activity by increasing kinetochore size, whereas spretus centromeres do so by modulating centromere geometry.
These observations suggest that centromere drive might depend on slowing meiotic progression so that selfish centromeres have time to flip after the spindle has acquired asymmetry. Therefore, we experimentally delayed anaphase in spretus hybrid oocytes either using an Anaphase Promoting Complex/Cyclosome (APC/C) inhibitor, ProTAME (Zeng et al., 2010), or by expressing non-degradable Cyclin B (Δ90 Cyclin B) (Madgwick et al., 2004; Schindler and Schultz, 2009) (Figure 7A). Delaying anaphase by either manipulation induced biased orientation 2–4 hours after spindle migration, with larger musculus centromeres preferentially oriented towards the cortical side of the spindle, which will direct them to the polar body (Figure 7B). These results demonstrate that relative destabilizing activity defines the directionality of centromere drive and that centromere drive depends on slowing meiotic progression. Finally, BUB1 inhibitor treatment canceled biased orientation in the spretus hybrid (Figures 7B and S6), as in the CHPO hybrid (Figure 3E), showing that centromere drive in both systems depends on the BUB1 pathway.
DISCUSSION
Our findings reveal both molecular and evolutionary strategies of meiotic cheating by selfish centromeres in mouse. Our results are consistent with the centromere drive theory, which proposes that selfish centromeres expand centromeric satellite repeats to win the competition in female meiosis (Henikoff et al., 2001). We find that more expanded centromeres win in both hybrids: larger musculus centromeres win against smaller musculus centromeres but lose against spretus centromeres, which have even more minor satellite DNA (Iwata-Otsubo et al., 2017; Miyanari et al., 2013; Wong et al., 1990) (Figure 7B). However, it has been unclear what activity at centromeres leads to selfish behavior. Centromeres incorporate both MT-binding activity at kinetochores and counteracting MT-destabilizing activities, which promote re-orientation of incorrect attachments to prevent segregation errors (Heald and Khodjakov, 2015). We show that selfish centromeres exploit the same destabilizing activity to bias their segregation to the egg. Multiple lines of evidence support this conclusion. First, we observed higher levels of MT-destabilizing factors at selfish centromeres in both intraspecific and interspecific hybrids (Figures 2 and 4). Second, both equalizing and diminishing destabilizers across hybrid bivalents prevented drive (Figures 3 and 7). Third, selfish centromeres initiated flipping events by detaching MTs (Figure 6). Fourth, relative levels of MT-destabilizing factors determine the direction of centromere drive, converting winners in one hybrid to losers in another hybrid (Figure 7).
Our finding that MT-destabilizing activity underlies non-Mendelian segregation is consistent with the cell biology of chromosome segregation in mouse oocytes (Kitajima, 2018). Initial MT attachments are established when the spindle is still symmetric and therefore lacks spatial cues to guide selfish centromeres (Kitajima et al., 2011), which must selectively detach to flip towards the egg pole after the spindle has migrated and acquired asymmetry. This process implies some destabilizing activity that acts specifically on the cortical side of the spindle, which is more tyrosinated (Akera et al., 2017). We propose MCAK as this activity because it preferentially destabilizes tyrosinated MTs (Peris et al., 2009; Sirajuddin et al., 2014) and is recruited at higher levels to selfish centromeres in both hybrid models. Also, MCAK localizes to centromeres only at late metaphase I (Illingworth et al., 2010), which matches the timing of flipping to orient selfish centromeres towards the egg pole. Based on this model, selfish centromeres with more MCAK destabilize MTs preferentially on the cortical side enriched in tyrosinated MTs. Once selfish centromeres detach and flip to the egg side, MT attachments are relatively more stable because MTs are less tyrosinated on the egg side, preventing further flipping. This model mechanistically links spindle asymmetry and centromere asymmetry and explains the source of biased directionality in flipping (Figure 1D). There may be additional mechanisms for centromeres to bias their transmission in female meiosis, as suggested by a different musculus hybrid model in which biased re-orientation before spindle migration leads to biased segregation (Wu et al., 2018), in contrast to our findings.
We show that the BUB1 pathway links expanded centromeres to recruitment of MT-destabilizing factors at the peri-centromere. In the intraspecific CHPO hybrid, larger kinetochores lead to more BUB1 kinase and histone phosphorylation, which recruits Shugoshin and MT-destabilizing factors (Figure 2). By experimentally equalizing destabilizing activity through BUB1 targeting to major satellite sequences, we demonstrate the significance of this pathway for centromere drive (Figure 3). In contrast, spretus centromeres amplify the BUB1 pathway by recruiting condensin, which likely increases the accessibility of kinetochore-localized BUB1 kinase to peri-centromeric histone H2A (Figure 5). These results suggest that genetic conflict between centromere DNA and centromere-binding proteins has played out differently in different species, leading to distinct mechanisms to enrich destabilizing activity through the BUB1 pathway, either by increasing the amount of BUB1 kinase or its accessibility to the substrate (Figure 7C). Evolution of centromeres to increase destabilizing activity would be constrained, however, since excessive destabilizing activity is detrimental during mitosis and meiosis by preventing centromeres from establishing stable MT attachments to faithfully segregate chromosomes (Liu et al., 2009).
The core of the centromere drive theory is the idea that suppression of drive provides selective pressure for evolution of centromere proteins (Henikoff et al., 2001). Although this idea has been influential to explain the paradoxical rapid evolution of centromere proteins, it has been difficult to directly test without some understanding of the cell biological basis of centromere drive. Our results provide the first step towards a mechanistic model for the selective pressure. We show that flipping events to face selfish centromeres towards the egg pole take time, and rapid progression through meiosis I prevents drive (Figures 7A and 7B). Meiotic progression is controlled by the spindle assembly checkpoint, which delays anaphase until all chromosomes are attached to spindle MTs (Joglekar, 2016). This checkpoint is weaker in oocytes compared to somatic cells, which is counter-intuitive because of the risk of producing aneuploid eggs (Nagaoka et al., 2011; Shao et al., 2013). We propose that the weakened checkpoint may be a consequence of adaptive evolution to suppress centromere drive. Multiple mechanisms could weaken the spindle assembly checkpoint, for example dampening the signaling cascade at centromeres or strengthening APC/C activity. Moreover, a large cytoplasmic volume, which is a general feature of female meiosis, is directly linked to the weakened checkpoint (Kyogoku and Kitajima, 2017; Lane and Jones, 2017). Identifying genes with signatures of rapid evolution may provide further insights into how genomes have evolved to suppress drive, through either a weak checkpoint or other mechanisms. One intriguing candidate is the rapidly evolving kinetochore protein KNL1 (Tromer et al., 2015), which recruits BUB1 and other checkpoint components to kinetochores. By modulating both MT-destabilizing and spindle checkpoint activity, KNL1 may have evolved to suppress centromere drive, although its recently discovered role in neurodevelopment offers an alternative explanation (Cheerambathur et al., 2019; Zhao et al., 2019). Another potential suppressor is the condensin complex, which is rapidly evolving in multiple lineages (Beck and Llopart, 2015; King et al., 2018) and modulates destabilizing activity through centromere geometry (Samoshkin et al., 2009). Our cell biological studies of centromere drive, combined with molecular evolution analysis, will lead to a deeper understanding of the molecular arms race between selfish elements and the rest of the genome.
STAR METHODS
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Michael A. Lampson (lampson@sas.upenn.edu).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Mice
Mouse strains were purchased from the Jackson Laboratory (ZALENDE/EiJ, stock #001392 corresponds to CHPO; C57BL/6J, stock# 000664; SPRET/EiJ, stock# 001146) and from Envigo (NSA, stock# 033 corresponds to CF-1). CHPO males were crossed CF-1 females and SPRET males were crossed to CF-1 or C57BL/6J females to generate hybrids. The CHPO strain contains seven Robertsonian fusions (Rb(1.3), Rb(4.6), Rb(5.15), Rb(11.13), Rb(8.12), Rb(9.14), and Rb(16.17)), each of which pairs with two CF-1 chromosomes in CF-1 x CHPO hybrid meiosis I to form a trivalent (Chmátal et al., 2014). We included only bivalents (chromosome 2, 7, 10, 18, 19, X) in our analyses to avoid complications of trivalents. All mice used in this study were 8–14 week-old females. All animal experiments were approved by the Institutional Animal Care and Use Committee and were consistent with the National Institutes of Health guidelines.
METHOD DETAILS
Oocyte collection and culture
Female mice were hormonally primed with 5U of Pregnant Mare Serum Gonadotropin (PMSG, Calbiochem, cat# 367222) 44–48 h prior to oocyte collection. Germinal vesicle (GV)-intact oocytes were collected in bicarbonate-free minimal essential medium with polyvinylpyrrolidone and Hepes (MEM-PVP) (Stein and Schindler, 2011), denuded from cumulus cells, and cultured in Chatot-Ziomek-Bavister (CZB) (Chatot et al., 1989) medium covered with mineral oil (Sigma, cat# M5310) in a humidified atmosphere of 5% CO2 in air at 37°C. During collection, meiotic resumption was inhibited by addition of 2.5 μM milrinone. Milrinone was subsequently washed out to allow meiotic resumption. Oocytes were checked for GVBD (germinal vesicle breakdown) 1.5 h after milrinone washout, and those that did not enter GVBD stage were removed from the culture. BUB1 inhibitor, BAY-1816032, was added to the medium at a final concentration of 50 μM, 3 h after GVBD, to disrupt the BUB1 pathway.
Oocyte microinjection
GV oocytes were microinjected with ~5 pl of cRNAs or antibodies in MEM-PVP containing milrinone at room temperature (RT) with a micromanipulator TransferMan NK 2 (Eppendorf) and picoinjector (Medical Systems Corp.). After the injection, oocytes were kept in milrinone for 16 h to allow protein expression. cRNAs used for microinjections were H2B-mCherry (human histone H2B with mCherry at the C-terminus) at 400 ng/μl, TALE-mClover (TALE construct that recognize Major satellite repeats fused to mClover and 3 tandem Halo tag at the C-terminus) at 1000 ng/μl, H2B-Egfp (human histone H2B with EGFP at the C-terminus) at 600 ng/μl, Cenpb-mCherry (mouse CENP-B with mCherry at the C-terminus) at 1300 ng/μl, Major Sat-Bub1 (TALE construct that recognize Major satellite repeats fused to mClover and the kinase domain of mouse BUB1 (a.a. 672–1058) at the C-terminus) at 100 ng/μl, RAMFLhyp (a dominant negative Chinese hamster MCAK construct carrying three point mutations in the motor domain and five Aurora B phosphorylation sites mutated to alanine)(Wordeman et al., 2007) at 500 ng/μl, and Δ90 Cyclin B-EGFP (human cyclin B1 lacking the first 90 a.a. with EGFP at the C-terminus) at 700 ng/μl (Schindler and Schultz, 2009). cRNAs were synthesized using the T7 mScript™ Standard mRNA Production System (CELL SCRIPT). Control rabbit IgG (Cell Signaling Technology) and rabbit anti-mouse CAP-D3 (a gift from Tatsuya Hirano) antibodies were used for microinjections at a concentration of 1 mg/ml. We analyzed oocytes with stretched centromeres, as shown previously (Lee et al., 2011).
Live imaging
Oocytes were placed into 2 μl drops of CZB media covered with mineral oil in a glass-bottom tissue culture dish (FluoroDish FD35–100) in a heated environmental chamber with a stage top incubator (Incubator BL and Heating Insert P; PeCon GmBH) to maintain 5% CO2 in air and 37°C. Confocal images were collected with a microscope (DMI4000 B; Leica) equipped with a 63× 1.3 NA glycerol-immersion objective lens, an xy piezo Z stage (Applied Scientific Instrumentation), a spinning disk confocal scanner (Yokogawa Corporation of America), an electron multiplier charge-coupled device camera (ImageEM C9100–13; Hamamatsu Photonics), and an LMM5 laser merge module with 488- and 593-nm diode lasers (Spectral Applied Research) controlled by MetaMorph software (Molecular Devices). Confocal images were collected as z-stacks at 1 μm intervals to visualize the entire meiotic spindle.
Oocyte immunocytochemistry
MI oocytes at different times after GVBD were cultured in CZB media. Oocytes were fixed in freshly prepared 2% paraformaldehyde in PBS with 0.1% Triton X-100, pH 7.4, for 20 min at RT, permeabilized in PBS with 0.1% Triton X-100 for 15 min at RT, placed in blocking solution (PBS containing 0.3% BSA and 0.01% Tween-20) overnight at 4 °C, incubated 1 h with primary antibodies in blocking solution, washed 3 times for 15 min, incubated 1 h with secondary antibodies, washed 3 times for 15 min, and mounted in Vectashield (Vector, cat# H-1400) with bisbenzimide (Hoechst 33342, Sigma-Aldrich) to visualize chromosomes. For Figure S5, 0.05% glutaraldehyde was added to the fixative to better preserve spindle MTs (Schuh and Ellenberg, 2007). The primary antibodies used for this study were rat anti-tyrosinated α-tubulin (1:1000, Serotec, YL1/2), rabbit anti-β-tubulin (9F3) monoclonal conjugated to Alexa Fluor 488 (1:50 dilution; Cell Signaling Technology), mouse anti-α-tubulin (1:500, Sigma, DM1A), CREST human autoantibody against centromere (1:100, Immunovision), rabbit anti-human p-INCENP (Salimian et al., 2011) (1:200), rabbit anti-human Survivin (1:500, Cell signaling, 71G4B7), rabbit anti-human MCAK (1:1000, a gift from Duane Compton), mouse anti-mouse BUB1 (1:100, a gift from Yoshinori Watanabe), mouse anti-mouse SGO2 (1:500, a gift from Yoshinori Watanabe), rabbit anti-histone H2AT120ph (1:2500, Active motif, 39391), mouse anti-mouse HEC1 (1:100, Santa Cruz, C-11), rabbit anti-mouse CENP-C (1:2000, a gift from Yoshinori Watanabe), rabbit anti-mouse CAP-D3 (1:500, a gift from Tatsuya Hirano). Secondary antibodies were Alexa Fluor 488–conjugated donkey anti-mouse or donkey anti-rabbit or Alexa Fluor 594–conjugated donkey anti-rat, donkey anti-rabbit, donkey anti-mouse or goat anti-human (1:500, Invitrogen). Confocal images were collected as z-stacks at 1 μm intervals to visualize the entire meiotic spindle, using the spinning disc confocal microscope described above.
To quantify centromere signal ratios, optical slices containing centromeres from the same bivalent were added to produce a sum projection using Fiji/ImageJ. Ellipses were drawn around the centromeres, and signal intensity was integrated over each ellipse after subtracting background, obtained by near the centromeres. Ratios were obtained for each bivalent by dividing the intensity of the brighter centromere by that of the dimmer centromere unless otherwise specified in the figure legend. Relative centromeric enrichment of condensin on the chromosomes (Figure 5B) was calculated in the same way except that the centromeric signals were divided by the signals on the chromosome arm, obtained by measuring the average intensity of a region on the arm.
Biased orientation assay
GV oocytes from CF-1 x CHPO (CHPO hybrid) or SPRET/EiJ x CF-1 and SPRET/EiJ x C57BL/6J (spretus hybrid) were collected and microinjected with cRNAs encoding CENP-B-mCh and H2B-EGFP (CHPO hybrid) or Major Satellite-mClover and H2B-mCh (spretus hybrid). Oocytes were induced for meiotic resumption by washing out milrinone. Live imaging was performed as described above, starting 10 h (CHPO hybrid) or 5 h (spretus hybrid, control) after GVBD to capture the time just before anaphase onset. Images were taken every 30 min. Spretus hybrid oocytes arrested in metaphase I by 1 μM ProTAME or expressing Δ90 Cyclin B were imaged at 10 h after GVBD. The position of the spindle near the cortex was confirmed by differential interference contrast images, and the fraction of bivalents with the larger musculus centromere (CF-1 or C57BL/6J) oriented towards the egg was quantified, using CENP-B to distinguish CF-1 centromeres from CHPO or Major Satellite to distinguish spretus centromeres from CF-1 or C57BL/6J.
Flipping assay
Oocytes from CF-1 x CHPO (CHPO hybrid) or SPRET/EiJ x CF-1 and SPRET/EiJ x C57BL/6J (spretus hybrid) were imaged live as in the biased orientation assay except for starting 7 h (CHPO hybrid) or 4.5 h (spretus hybrid) after GVBD and taking images every 10 or 20 min. To measure the frequency of each centromere initiating the flipping, we only analyzed flipping events in which we captured the intermediate state (only one of the two centromeres moving towards the opposite pole). To measure biased flipping, oocytes with the spindle completely migrated towards the cortex (distance between the cortex and the center of the spindle < 25 μm) were analyzed. In the CHPO hybrid, we analyzed 6 bivalents in each oocyte because other chromosomes form trivalents (see “Mice” section). If 3 are initially facing in each direction (i.e., no bias), then one flipping event gives 4 facing one way and 2 the other way, which is sufficient for the 60/40 bias observed in this system (Iwata-Otsubo et al., 2017) (Figure 3E).
Cold-stable MT assay
Oocytes were placed into ice cold MEM-PVP for 6 min before fixation and stained for α-tubulin. Confocal images were collected to visualize the entire meiotic spindle, using the spinning disc confocal microscope described above. To calculate tubulin signal intensity, ellipses were drawn around the spindle, and α-tubulin intensity was integrated over each ellipse in optical slices containing the spindle, after subtracting background.
Statistical analysis
Data points are pooled from at least two independent experiments unless specified in the figure legend. The following statistical methods were used: unpaired t test in Figures 2A, 2C, 3B, 3C, 3D, 4B, 5A, 5B, 5D, S2A, S2B, S3, and S6; chi square test for goodness of fit for deviations from the expected 50:50 ratio in Figures 1D, 3E, 6B, 6C, 7B, and for deviations from 1 in Figures 2B and 4C.
QUANTIFICATION AND STATISTICAL ANALYSIS
Data points are pooled from at least two independent experiments unless specified in the figure legend. The following statistical methods were used: unpaired t test in Figures 2A, 2C, 3B, 3C, 3D, 4B, 5A, 5B, 5D, S2A, S2B, S3, and S6; chi square test for goodness of fit for deviations from the expected 50:50 ratio in Figures 1D, 3E, 6B, 6C, 7B, and for deviations from 1 in Figures 2B and 4C., The exact value of n, what n represents and definition of center can be found in the figure legends for each experiment. Statistical tests were performed using GraphPad Prism, and a P value of less than 0.05 was judged as statistically significant.
DATA AND CODE AVAILABILITY
This study did not generate datasets or code that require deposition in a public repository. Raw data is available from the authors upon request.
Supplementary Material
Figure S1. BUB1, H2ApT121, and SGO2 are symmetric across bivalents in control oocytes. Related to Figure 2. (A) CF-1 x CHPO (L x S) hybrid oocytes, or CF-1 x CF-1 (L x L) as controls, were fixed at metaphase I and stained for Survivin. (B) CF-1 x CF-1 (L x L) oocytes were fixed at metaphase I and stained for BUB1, H2ApT121, or SGO2. Images (A and B) are maximum intensity z-projections showing all chromosomes (left), or optical slices magnified to show single bivalents (right). Quantification is shown in Figure 2.
Figure S2. BUB1 targeting cancels asymmetry in both MCAK and CPC. Related to Figure 3. (A and B) CF-1 x CHPO oocytes expressing Major Sat-BUB1 were fixed at metaphase I and stained for MCAK (A) or pINCENP (B). Images are maximum intensity z-projections showing all chromosomes (left) or optical slices magnified to show single bivalents (right). Scale bar, 10 μm. Graphs show centromere signal intensities or centromere signal ratios, calculated as the brighter divided by the dimmer signal for each bivalent. Each dot represents a single centromere (A and B, bottom, n > 53 for each condition) or a single bivalent (B, top, n > 51 for each condition). Red line, mean; *P < 0.001.
Figure S3. Bub1 inhibition disrupts the BUB1 pathway in CHPO hybrid oocytes. Related to Figure 3. CF-1 x CHPO (L x S) oocytes treated with a BUB1 inhibitor, BAY-1816032, were fixed at metaphase I and stained for H2ApT121, SGO2, or MCAK. Images are maximum intensity z-projections showing all chromosomes (left), or optical slices magnified to show single bivalents (right); scale bars, 10 μm. Graph shows centromere signal intensities. Each dot represents a single centromere (n > 56 for each condition); red line, mean; *P < 0.001.
Figure S4. CPC is spread across the chromosomes in the interspecific spretus hybrid. Related to Figure 4. C57BL/6J x SPRET/EiJ (L x sp) oocytes were fixed at metaphase I and stained for pINCENP and CREST. Images are maximum intensity z-projections showing all chromosomes (left) or an optical slice magnified to show a single bivalent (right). Scale bar, 10 μm.
Figure S5. Spindle asymmetry in tyrosinated MTs in the interspecific spretus hybrid. Related to Figure 7. C57BL/6J x SPRET/EiJ (L x sp) oocytes were fixed at metaphase I and stained for tyrosinated α-tubulin and β-tubulin. Images are maximum intensity z-projections showing the whole oocyte (left) or a magnified view of the spindle (right). Dashed line, cortex; scale bars, 10 μm.
Figure S6. Bub1 inhibition disrupts the BUB1 pathway in spretus hybrid oocytes. Related to Figure 7. C57BL/6J x SPRET/EiJ (L x sp) oocytes treated with a BUB1 inhibitor, BAY-1816032, were fixed at metaphase I and stained for H2ApT121, SGO2, or MCAK. Images are maximum intensity z-projections showing all chromosomes (left), or optical slices magnified to show single bivalents (right); scale bars, 10 μm. Graph shows centromere signal intensities. Each dot represents a single centromere (n > 72 for each condition); red line, mean; *P < 0.001.
High microtubule-destabilizing activity makes mouse centromeres selfish in meiosis
Amplified BUB1 signaling enriches destabilizing activity on selfish centromeres
Selfish centromeres can modulate the BUB1 pathway by different mechanisms
Rapid progression through meiosis I can suppress centromere drive
Acknowledgements
We thank B. E. Black, R. M. Schultz, and M. T. Levine for comments on the manuscript, T. Hirano for the CAP-D3 antibody, D. A. Compton for the MCAK antibody, Y. Watanabe for the CENP-C, SGO2, and BUB1 antibodies, L. Wordeman for the RAMFLhyp construct, and M. E. Torres-Padilla and Y. Miyanari for the TALE constructs. The research was supported by NIH grant GM122475 (M.A.L.) and by a research fellowship from Uehara Memorial Foundation (T.A.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
DECLARATION OF INTERESTS
The authors declare no competing interests.
References
- Akera T, Chmátal L, Trimm E, Yang K, Aonbangkhen C, Chenoweth DM, Janke C, Schultz RM, and Lampson MA (2017). Spindle asymmetry drives non-Mendelian chromosome segregation. Science 358, 668–672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almonacid M, Terret M-E, and Verlhac M-H (2014). Actin-based spindle positioning: new insights from female gametes. J. Cell Sci. 127, 477–483. [DOI] [PubMed] [Google Scholar]
- Azoury J, Lee KW, Georget V, Hikal P, and Verlhac M-H (2011). Symmetry breaking in mouse oocytes requires transient F-actin meshwork destabilization. Development 138, 2903–2908. [DOI] [PubMed] [Google Scholar]
- Beck EA, and Llopart A (2015). Widespread Positive Selection Drives Differentiation of Centromeric Proteins in the Drosophila melanogaster subgroup. Sci. Rep 5, 17197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bint SM, Ogilvie CM, Flinter FA, Khalaf Y, and Scriven PN (2011). Meiotic segregation of Robertsonian translocations ascertained in cleavage-stage embryos--implications for preimplantation genetic diagnosis. Hum. Reprod 26, 1575–1584. [DOI] [PubMed] [Google Scholar]
- Carmena M, Wheelock M, Funabiki H, and Earnshaw WC (2012). The chromosomal passenger complex (CPC): from easy rider to the godfather of mitosis. Nat. Rev. Mol. Cell Biol. 13, 789–803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatot CL, Ziomek CA, Bavister BD, Lewis JL, and Torres I (1989). An improved culture medium supports development of random-bred 1-cell mouse embryos in vitro. J. Reprod. Fertil 86, 679–688. [DOI] [PubMed] [Google Scholar]
- Cheerambathur DK, Prevo B, Chow T-L, Hattersley N, Wang S, Zhao Z, Kim T, Gerson-Gurwitz A, Oegema K, Green R, et al. (2019). The Kinetochore-Microtubule Coupling Machinery Is Repurposed in Sensory Nervous System Morphogenesis. Dev. Cell 48, 864–872.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chmátal L, Gabriel SI, Mitsainas GP, Martínez-Vargas J, Ventura J, Searle JB, Schultz RM, and Lampson MA (2014). Centromere strength provides the cell biological basis for meiotic drive and karyotype evolution in mice. Curr. Biol 24, 2295–2300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chmátal L, Schultz RM, Black BE, and Lampson MA (2017). Cell Biology of Cheating-Transmission of Centromeres and Other Selfish Elements Through Asymmetric Meiosis In Centromeres and Kinetochores, (Springer, Cham: ), pp. 377–396. [DOI] [PubMed] [Google Scholar]
- Daniel A (2002). Distortion of female meiotic segregation and reduced male fertility in human Robertsonian translocations: consistent with the centromere model of co-evolving centromere DNA/centromeric histone (CENP-A). Am. J. Med. Genet 111, 450–452. [DOI] [PubMed] [Google Scholar]
- Dehapiot B, Carrière V, Carroll J, and Halet G (2013). Polarized Cdc42 activation promotes polar body protrusion and asymmetric division in mouse oocytes. Dev. Biol 377, 202–212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fishman L, and Saunders A (2008). Centromere-associated female meiotic drive entails male fitness costs in monkeyflowers. Science 322, 1559–1562. [DOI] [PubMed] [Google Scholar]
- Godek KM, Kabeche L, and Compton DA (2015). Regulation of kinetochore–microtubule attachments through homeostatic control during mitosis. Nat. Rev. Mol. Cell Biol. 16, 57–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goto Y, Yamagishi Y, Shintomi-Kawamura M, Abe M, Tanno Y, and Watanabe Y (2017). Pds5 Regulates Sister-Chromatid Cohesion and Chromosome Bi-orientation through a Conserved Protein Interaction Module. Curr. Biol 27, 1005–1012. [DOI] [PubMed] [Google Scholar]
- Heald R, and Khodjakov A (2015). Thirty years of search and capture: The complex simplicity of mitotic spindle assembly. J. Cell Biol. 211, 1103–1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henikoff S, Ahmad K, and Malik HS (2001). The centromere paradox: stable inheritance with rapidly evolving DNA. Science 293, 1098–1102. [DOI] [PubMed] [Google Scholar]
- Holubcová Z, Howard G, and Schuh M (2013). Vesicles modulate an actin network for asymmetric spindle positioning. Nat. Cell Biol. 15, 937–947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houlard M, Godwin J, Metson J, Lee J, Hirano T, and Nasmyth K (2015). Condensin confers the longitudinal rigidity of chromosomes. Nat. Cell Biol. 17, 771–781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Illingworth C, Pirmadjid N, Serhal P, Howe K, and Fitzharris G (2010). MCAK regulates chromosome alignment but is not necessary for preventing aneuploidy in mouse oocyte meiosis I. Development 137, 2133–2138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwata-Otsubo A, Dawicki-McKenna JM, Akera T, Falk SJ, Chmátal L, Yang K, Sullivan BA, Schultz RM, Lampson MA, and Black BE (2017). Expanded Satellite Repeats Amplify a Discrete CENP-A Nucleosome Assembly Site on Chromosomes that Drive in Female Meiosis. Curr. Biol 27, 2365–2373.e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joglekar AP (2016). A Cell Biological Perspective on Past, Present and Future Investigations of the Spindle Assembly Checkpoint. Biology (Basel). 5, 44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawashima SA, Yamagishi Y, Honda T, Ishiguro K, and Watanabe Y (2010). Phosphorylation of H2A by Bub1 prevents chromosomal instability through localizing shugoshin. Science 327, 172–177. [DOI] [PubMed] [Google Scholar]
- King T, Leonard CJ, Cooper JC, Nguyen S, Joyce E, and Phadnis N (2018). No Recurrent losses and rapid evolution of the condensin II complex in insects. BioRxiv. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kipling D, Mitchell AR, Masumoto H, Wilson HE, Nicol L, and Cooke HJ (1995). CENP-B binds a novel centromeric sequence in the Asian mouse Mus caroli. Mol. Cell. Biol 15, 4009–4020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitajima TS (2018). Mechanisms of kinetochore-microtubule attachment errors in mammalian oocytes. Dev. Growth Differ. 60, 33–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitajima TS, Ohsugi M, and Ellenberg J (2011). Complete kinetochore tracking reveals error-prone homologous chromosome biorientation in mammalian oocytes. Cell 146, 568–581. [DOI] [PubMed] [Google Scholar]
- Kyogoku H, and Kitajima TS (2017). Large Cytoplasm Is Linked to the Error-Prone Nature of Oocytes. Dev. Cell 41, 287–298.e4. [DOI] [PubMed] [Google Scholar]
- Lampson M, and Grishchuk E (2017). Mechanisms to Avoid and Correct Erroneous Kinetochore-Microtubule Attachments. Biology (Basel). 6, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lampson MA, and Black BE (2017). Cellular and Molecular Mechanisms of Centromere Drive. Cold Spring Harb. Symp. Quant. Biol 82, 249–257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lane SIR, and Jones KT (2017). Chromosome biorientation and APC activity remain uncoupled in oocytes with reduced volume. J. Cell Biol. 216, 3949–3957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J, Kitajima TS, Tanno Y, Yoshida K, Morita T, Miyano T, Miyake M, and Watanabe Y (2008). Unified mode of centromeric protection by shugoshin in mammalian oocytes and somatic cells. Nat. Cell Biol. 10, 42–52. [DOI] [PubMed] [Google Scholar]
- Lee J, Ogushi S, Saitou M, and Hirano T (2011). Condensins I and II are essential for construction of bivalent chromosomes in mouse oocytes. Mol. Biol. Cell 22, 3465–3477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu D, Vader G, Vromans MJM, Lampson MA, and Lens SMA (2009). Sensing chromosome biorientation by spatial separation of aurora B kinase from kinetochore substrates. Science 323, 1350–1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madgwick S, Nixon VL, Chang H-Y, Herbert M, Levasseur M, and Jones KT (2004). Maintenance of sister chromatid attachment in mouse eggs through maturation-promoting factor activity. Dev. Biol 275, 68–81. [DOI] [PubMed] [Google Scholar]
- Masumoto H, Masukata H, Muro Y, Nozaki N, and Okazaki T (1989). A human centromere antigen (CENP-B) interacts with a short specific sequence in alphoid DNA, a human centromeric satellite. J. Cell Biol. 109, 1963–1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLaughlin RN, and Malik HS (2017). Genetic conflicts: the usual suspects and beyond. J. Exp. Biol 220, 6–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyanari Y, Ziegler-Birling C, and Torres-Padilla M-E (2013). Live visualization of chromatin dynamics with fluorescent TALEs. Nat. Struct. Mol. Biol 20, 1321–1324. [DOI] [PubMed] [Google Scholar]
- Nagaoka SI, Hodges CA, Albertini DF, and Hunt PA (2011). Oocyte-specific differences in cell-cycle control create an innate susceptibility to meiotic errors. Curr. Biol 21, 651–657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narayanswami S, Doggett NA, Clark LM, Hildebrand CE, Weier HU, and Hamkalo BA (1992). Cytological and molecular characterization of centromeres in Mus domesticus and Mus spretus. Mamm. Genome 2, 186–194. [DOI] [PubMed] [Google Scholar]
- Oliveira RA, Coelho PA, and Sunkel CE (2005). The condensin I subunit Barren/CAP-H is essential for the structural integrity of centromeric heterochromatin during mitosis. Mol. Cell. Biol 25, 8971–8984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pacchierotti F, Tiveron C, Mailhes JB, and Davisson MT (1995). Susceptibility to vinblastine-induced aneuploidy and preferential chromosome segregation during meiosis I in Robertsonian heterozygous mice. Teratog. Carcinog. Mutagen 15, 217–230. [DOI] [PubMed] [Google Scholar]
- Pardo-Manuel de Villena F, and Sapienza C (2001). Nonrandom segregation during meiosis: the unfairness of females. Mamm. Genome 12, 331–339. [DOI] [PubMed] [Google Scholar]
- Peris L, Wagenbach M, Lafanechère L, Brocard J, Moore AT, Kozielski F, Job D, Wordeman L, and Andrieux A (2009). Motor-dependent microtubule disassembly driven by tubulin tyrosination. J. Cell Biol. 185, 1159–1166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rieder CL (1981). The structure of the cold-stable kinetochore fiber in metaphase PtK1 cells. Chromosoma 84, 145–158. [DOI] [PubMed] [Google Scholar]
- Salimian KJ, Ballister ER, Smoak EM, Wood S, Panchenko T, Lampson MA, and Black BE (2011). Feedback control in sensing chromosome biorientation by the Aurora B kinase. Curr. Biol 21, 1158–1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samoshkin A, Arnaoutov A, Jansen LET, Ouspenski I, Dye L, Karpova T, McNally J, Dasso M, Cleveland DW, and Strunnikov A (2009). Human condensin function is essential for centromeric chromatin assembly and proper sister kinetochore orientation. PLoS One 4, e6831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindler K, and Schultz RM (2009). CDC14B acts through FZR1 (CDH1) to prevent meiotic maturation of mouse oocytes. Biol. Reprod 80, 795–803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuh M, and Ellenberg J (2007). Self-organization of MTOCs replaces centrosome function during acentrosomal spindle assembly in live mouse oocytes. Cell 130, 484–498. [DOI] [PubMed] [Google Scholar]
- Sebestova J, Danylevska A, Novakova L, Kubelka M, and Anger M (2012). Lack of response to unaligned chromosomes in mammalian female gametes. Cell Cycle 11, 3011–3018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao H, Li R, Ma C, Chen E, and Liu XJ (2013). Xenopus oocyte meiosis lacks spindle assembly checkpoint control. J. Cell Biol. 201, 191–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siemeister G, Mengel A, Fernández-Montalván AE, Bone W, Schröder J, Zitzmann-Kolbe S, Briem H, Prechtl S, Holton SJ, Mönning U, et al. (2019). Inhibition of BUB1 Kinase by BAY 1816032 Sensitizes Tumor Cells toward Taxanes, ATR, and PARP Inhibitors In Vitro and In Vivo. Clin. Cancer Res. 25, 1404–1414. [DOI] [PubMed] [Google Scholar]
- Sirajuddin M, Rice LM, and Vale RD (2014). Regulation of microtubule motors by tubulin isotypes and post-translational modifications. Nat. Cell Biol. 16, 335–344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sodek M, Kovacovicova K, and Anger M (2017). True Nondisjunction of Whole Bivalents in Oocytes with Attachment and Congression Defects. Cytogenet. Genome Res. 151, 10–17. [DOI] [PubMed] [Google Scholar]
- Stein P, and Schindler K (2011). Mouse oocyte microinjection, maturation and ploidy assessment. J. Vis. Exp [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tromer E, Snel B, and Kops GJPL (2015). Widespread Recurrent Patterns of Rapid Repeat Evolution in the Kinetochore Scaffold KNL1. Genome Biol. Evol 7, 2383–2393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vleugel M, Hoek TA, Tromer E, Sliedrecht T, Groenewold V, Omerzu M, and Kops GJPL (2015). Dissecting the roles of human BUB1 in the spindle assembly checkpoint. J. Cell Sci. 128, 2975–2982. [DOI] [PubMed] [Google Scholar]
- Watanabe Y (2012). Geometry and force behind kinetochore orientation: lessons from meiosis. Nat. Rev. Mol. Cell Biol. 13, 370–382. [DOI] [PubMed] [Google Scholar]
- Wong AKC, Biddle FG, and Rattner JB (1990). The chromosomal distribution of the major and minor satellite is not conserved in the genus Mus. Chromosoma 99, 190–195. [DOI] [PubMed] [Google Scholar]
- Wordeman L, Wagenbach M, and von Dassow G (2007). MCAK facilitates chromosome movement by promoting kinetochore microtubule turnover. J. Cell Biol. 179, 869–879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu T, Lane SIR, Morgan SL, and Jones KT (2018). Spindle tubulin and MTOC asymmetries may explain meiotic drive in oocytes. Nat. Commun 9, 2952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El Yakoubi W, Buffin E, Cladière D, Gryaznova Y, Berenguer I, Touati SA, Gómez R, Suja JA, van Deursen JM, and Wassmann K (2017). Mps1 kinase-dependent Sgo2 centromere localisation mediates cohesin protection in mouse oocyte meiosis I. Nat. Commun 8, 694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng X, Sigoillot F, Gaur S, Choi S, Pfaff KL, Oh D-C, Hathaway N, Dimova N, Cuny GD, and King RW (2010). Pharmacologic inhibition of the anaphase-promoting complex induces a spindle checkpoint-dependent mitotic arrest in the absence of spindle damage. Cancer Cell 18, 382–395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao G, Oztan A, Ye Y, and Schwarz TL (2019). Kinetochore Proteins Have a Post-Mitotic Function in Neurodevelopment. Dev. Cell 48, 873–882.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. BUB1, H2ApT121, and SGO2 are symmetric across bivalents in control oocytes. Related to Figure 2. (A) CF-1 x CHPO (L x S) hybrid oocytes, or CF-1 x CF-1 (L x L) as controls, were fixed at metaphase I and stained for Survivin. (B) CF-1 x CF-1 (L x L) oocytes were fixed at metaphase I and stained for BUB1, H2ApT121, or SGO2. Images (A and B) are maximum intensity z-projections showing all chromosomes (left), or optical slices magnified to show single bivalents (right). Quantification is shown in Figure 2.
Figure S2. BUB1 targeting cancels asymmetry in both MCAK and CPC. Related to Figure 3. (A and B) CF-1 x CHPO oocytes expressing Major Sat-BUB1 were fixed at metaphase I and stained for MCAK (A) or pINCENP (B). Images are maximum intensity z-projections showing all chromosomes (left) or optical slices magnified to show single bivalents (right). Scale bar, 10 μm. Graphs show centromere signal intensities or centromere signal ratios, calculated as the brighter divided by the dimmer signal for each bivalent. Each dot represents a single centromere (A and B, bottom, n > 53 for each condition) or a single bivalent (B, top, n > 51 for each condition). Red line, mean; *P < 0.001.
Figure S3. Bub1 inhibition disrupts the BUB1 pathway in CHPO hybrid oocytes. Related to Figure 3. CF-1 x CHPO (L x S) oocytes treated with a BUB1 inhibitor, BAY-1816032, were fixed at metaphase I and stained for H2ApT121, SGO2, or MCAK. Images are maximum intensity z-projections showing all chromosomes (left), or optical slices magnified to show single bivalents (right); scale bars, 10 μm. Graph shows centromere signal intensities. Each dot represents a single centromere (n > 56 for each condition); red line, mean; *P < 0.001.
Figure S4. CPC is spread across the chromosomes in the interspecific spretus hybrid. Related to Figure 4. C57BL/6J x SPRET/EiJ (L x sp) oocytes were fixed at metaphase I and stained for pINCENP and CREST. Images are maximum intensity z-projections showing all chromosomes (left) or an optical slice magnified to show a single bivalent (right). Scale bar, 10 μm.
Figure S5. Spindle asymmetry in tyrosinated MTs in the interspecific spretus hybrid. Related to Figure 7. C57BL/6J x SPRET/EiJ (L x sp) oocytes were fixed at metaphase I and stained for tyrosinated α-tubulin and β-tubulin. Images are maximum intensity z-projections showing the whole oocyte (left) or a magnified view of the spindle (right). Dashed line, cortex; scale bars, 10 μm.
Figure S6. Bub1 inhibition disrupts the BUB1 pathway in spretus hybrid oocytes. Related to Figure 7. C57BL/6J x SPRET/EiJ (L x sp) oocytes treated with a BUB1 inhibitor, BAY-1816032, were fixed at metaphase I and stained for H2ApT121, SGO2, or MCAK. Images are maximum intensity z-projections showing all chromosomes (left), or optical slices magnified to show single bivalents (right); scale bars, 10 μm. Graph shows centromere signal intensities. Each dot represents a single centromere (n > 72 for each condition); red line, mean; *P < 0.001.
Data Availability Statement
This study did not generate datasets or code that require deposition in a public repository. Raw data is available from the authors upon request.