Abstract
Purpose/Objectives:
Deep learning is an emerging technique that allows us to capture imaging information beyond the visually recognizable level of a human being. For its anatomical characteristics and location, on-board target verification for radiation delivery to pancreatic tumors is a challenging task. Our goal was to employ deep neural network to localize the pancreatic tumor target on kV X-ray images acquired using on-board imager (OBI) for image-guided radiation therapy (IGRT).
Materials/Methods:
The network is setup up in such a way that the input is either a digitally reconstructed radiograph (DRR) image or a monoscopic X-ray projection image acquired by the OBI from a given direction, and the output is the location of the planning target volume (PTV) target in the projection image. To produce sufficient amount of training X-ray images reflecting vast possible clinical scenarios of anatomy distribution, a series of changes were introduced to the planning CT (pCT) images, including deformation, rotation and translation, to simulate different inter- and intra-fractional variations. After model training, the accuracy of the model was evaluated by retrospectively studying patients who underwent pancreatic cancer radiotherapy. Statistical analysis using mean absolute differences (MADs) and Lin’s concordance correlation coefficient were used to assess the accuracy of the predicted target positions.
Results:
MADs between the model-predicted and the actual positions were found to be less than 2.60 mm in anterior-posterior, lateral, and oblique directions for both two axes in the detector plane. For comparison studies with and without fiducials, MADs are less than 2.49 mm. For all cases, Lin’s concordance correlation coefficients between the predicted and actual positions were found to be better than 93%, demonstrating the success of the proposed deep learning for IGRT.
Conclusions:
We demonstrated that markerless pancreatic tumor target localization is achievable with high accuracy by using a deep learning technique approach.
SUMMARY
Accurate tumor localization is of great significance for the success of radiotherapy directed to the pancreas. For hypofractionated regimens, tumor localization is commonly achieved by relying on the use of implanted fiducials. In this study, we present a novel deep learning (DL)-based strategy to localize the pancreatic tumor in projection X-ray images for image-guided radiation therapy without the necessity for fiducials.
INTRODUCTION
Radiotherapy is an important treatment modality in the management of pancreatic cancer [1, 2], which is one of the leading causes of cancer death worldwide [3]. Recent advances in conformal radiotherapy techniques such as VMAT and gated VMAT [4, 5] have substantially improved our ability to achieve highly conformal dose distributions. However, for patients to truly benefit from the technical advancements, image guidance that ensures a safe and accurate delivery of the planned dose distribution [6–9] must be in place.
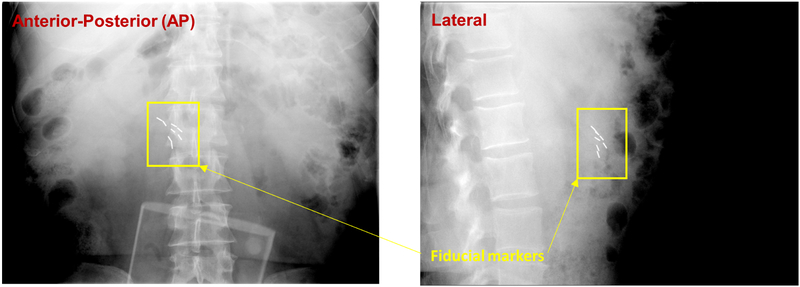
Significant effort has been devoted in developing image guidance strategies for accurate beam targeting over the last decade, including the use of stereoscopic or monoscopic kV X-ray imaging [10–13], hybrid kV and MV imaging [14, 15], cone-beam computed tomography (CBCT) [16], on-board magnetic resonance imaging (MRI) [17], ultrasound imaging [18], etc. For many practical reasons, the kV X-ray imaging is perhaps the most commonly used technique, but it offers insufficient soft-tissue contrast, making it difficult to accurately localize soft-tissue targets. Thus, metallic fiducials are often implanted into the tumor target or adjacent normal tissue to facilitate the patient setup and real-time tumor monitoring [19–21] (Figure 1). The placement of fiducials into the pancreas is an invasive procedure and requires availability of interventional radiologist or other specialist with expertise on this task, prolonging treatment initiation. Moreover, the presence of fiducials within the target may cause metal artifacts during treatment simulation scanning, not rarely obscuring certain regions of interest during target delineation [22].
Figure 1.
Implanted gold fiducials used for daily positioning and real-time tracking in pancreatic image-guided radiotherapy.
Deep learning utilizes hierarchical artificial neural networks with layers of interconnected neurons to carry out the process of machine learning. A deep learning-based image analysis of monoscopic or stereoscopic kV X-ray image(s) taken before or during treatment delivery can provide us information about the position of tumor target that is usually invisible without implanted fiducials. In this study, we assessed the utility of a patient-specific region-based convolutional neural network (PRCNN) for tumor localization and tracking. To mitigate the nuance associated with the clinical implementation of deep learning algorithms in training data collection, we propose a novel personalized PRCNN model training strategy based on synthetically generated digitally reconstructed radiographs (DRRs) without relying on the use of a vast amount of clinical kV X-ray images.
METHODS AND MATERIALS
Deep Learning for IGRT
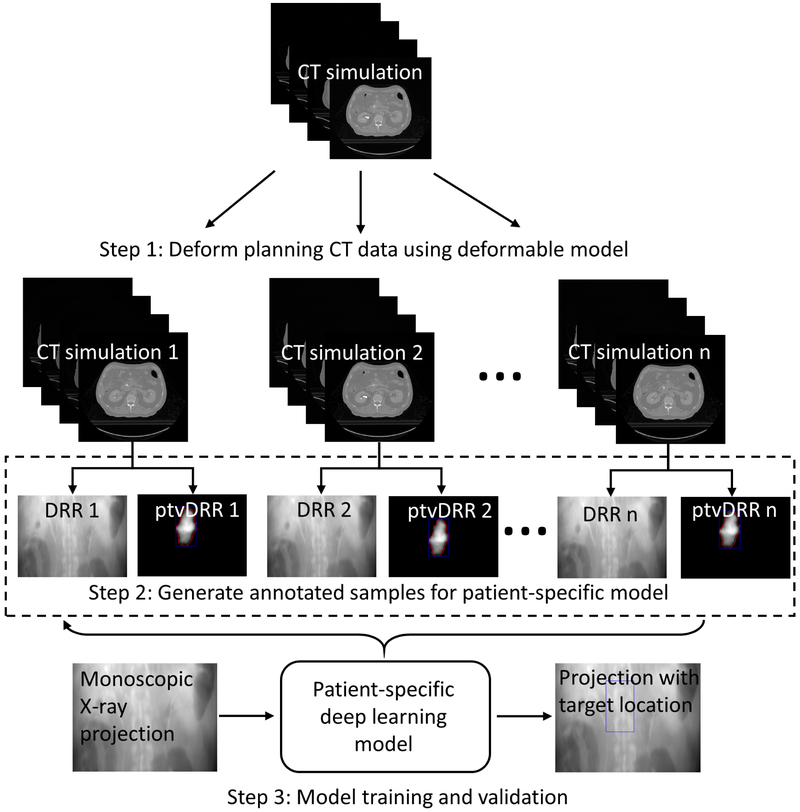
A deep learning model learns multiple levels of representations that correspond to different levels of abstraction from the input images to perform predictions. For a case such as target localization, a large amount of annotated training datasets is needed to train the deep learning model and validate the accuracy of the model. Figure 2 shows the workflow of the proposed target localization process for pancreatic IGRT. The first step was to generate training datasets of kV projection X-ray images reflecting differences in patient anatomy. To obtain realistic training data, we used a robust deformable model described by a motion vector field (MVF) to deform the planning CT (pCT) images of the patients. Secondly, after deformation/rotation/translation processes were completed, the pCT was projected in the geometry of the OBI system to generate DRRs. The planning target volume (PTV) delineated in the pCT was also projected to generate a PTV-only radiograph. Bounding box of the PTV-only radiograph was calculated and embedded to the corresponding DRR as an annotation. Finally, we used the annotated DRRs to train a patient-specific target localization neural network and used it for subsequent pancreatic tumor target localization. Validation tests using independent simulated kV projection images were then performed.
Figure 2.
Overall flowchart of the proposed deep learning-based treatment target localization method. Abbreviation: CT = computed tomography, DRR = digitally reconstructed radiograph, ptvDRR = planning target volume (PTV)-only digitally reconstructed radiograph.
Generation of Training Datasets
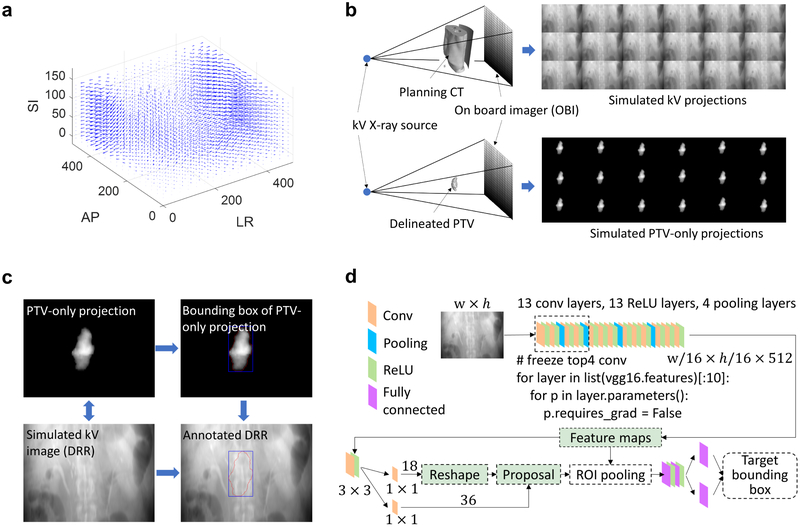
Training of a deep learning model requires a large number of annotated datasets and this often presents a bottleneck problem in establishing a deep learning model. We generated annotated projection images from the patient’s pCT. In our institution, a 4DCT is generally acquired for radiation treatments directed to the pancreas, and the exhale phase is chosen for treatment planning of gated dose deliveries. To this end, we introduced a series of changes (deformations, translations, and rotations) to the pCT to simulate different inter- and intra-fractional variations. The MVF used to deform the pCT was extracted by registering the pCT phase with other phases of the patient’s 4D-CT. For a pCT dataset with a size of 512 × 512 × 180, the corresponding MVF for our deformable model was a 4D matrix with a size of 512 × 512 × 180 × 3, where the three 512 × 512 × 180 matrices in the 4D matrix contain displacements along the x-axis, y-axis and z-axis, respectively. An example of the MVF is shown in Figure 3a. By registering the pCT to each phase of the 4D-CT, we extracted 9 MVFs. In order to increase the number of training data samples, we randomly selected two consecutive MVFs (extracted by two consecutive phase-resolved CT data with respect to the pCT) to generate a new MVF’ as follows:
where MVFi is one of the MVF in the 9 MVFs set, and MVF is a uniformly distributed random number in the interval (0,1). In this way, a total of 80 MVFs were generated and applied to deform the pCT data to create realistic deformations. The deformed image datasets were then divided into two parts, where 64 deformed CT datasets were used for model training and the other 16 deformed CT datasets used for model testing. For each of the deformed pCT data, we further augmented the data by incorporating rotation and random translation. Specifically, each of the deformed pCT data was rotated around the superior-interior (SI) direction from −3° to 3° with 1.5° intervals, and randomly shifted 5 times. With this method, a series of 1600 and 400 high-quality 3D CT data that incorporate inter- and intra-fractional variations of tumor motion for pancreatic cancer were generated for model training and testing, respectively.
Figure 3.
(a) Example of motion vector field extracted by registering the phaseresolved 4D-CT with respect to the pCT. (b) Generation of the simulated kV projection using deformation, rotation and translation incorporated pCT images with geometry consistent with the OBI system on a realistic treatment linac. (c) Data annotation for the patient-specific deep learning model. (d) Network architecture of the deep learning model used for localizing the pancreatic cancer treatment target. Abbreviation: AP = anterior-posterior, LR = left-right, SI = superior-inferior, PTV = planning target volume.
Subsequently, DRRs along the directions of anterior-posterior (AP), left-right (LR), and an oblique degree (135°) were produced by projecting each of the above pCT datasets using a forward X-ray projection model in the OBI system (Figure 3b). Namely, the X-ray source to the flat-panel detector distance is 1500 mm and the X-ray source to the isocenter distance is 1000 mm. The detector pixel and array sizes were 0.388 mm and 768 × 1024, respectively. The projection calculation was implemented using CUDA C programming language to take advantage of graphics processing unit acceleration (Nvidia GeForce GTX Titan X, Santa Clara, CA). The bounding box of the projected PTV was mapped to the corresponding DRR as an annotation (Figure 3c). The DRRs and the corresponding bounding box information (top-left corner, width, height) were then used to train the deep learning model.
Deep Learning Model
With the annotated datasets, we trained a deep learning model to localize the PTV for IGRT. Conventional target detection model using region-based convolutional neural network (ConvNet) can achieve nearly real-time detection for a given region proposal [23]. However, the calculation of the proposals is computationally intensive and presents a bottleneck for real-time detection of targets of interest. Here we trained a region-proposed network (RPN) to provide proposals for the region-based detection network [24], with the two networks sharing all the image feature hierarchies with each other, as illustrated in Figure 3d. The framework jointly generates region proposals and refines its spatial locations. The projection image was first passed through a feature extractor to generate high-level feature hierarchies. We used VGG16 ConvNet [25] which has 13 convolutional layers, 13 ReLU layers and 4 pooling layers as the feature extractor and it was pre-trained to take advantage of transfer learning. We also froze the top 4 convolutional layers of the VGG16 ConvNet to reduce the number of training weights and to accelerate training. Based on the feature hierarchies, the fully convolutional RPN was constructed by adding a few additional convolutional layers, specifically, a 3 × 3 convolutional layer followed by two siblings 1 × 1 convolutional layers. By providing the annotation dataset, the RPN selected 2000 region proposals from 12000 initial anchors during training, each with a score. For each proposal, a position sensitive region of interest pooling layer extracted a fixed length feature vector from the feature hierarchies, which were then fed into fully connected layers for multi-task loss calculation (i.e., the summation of a classification loss and a regression loss).
With the above framework, the RPN and the region-based detection network share the ConvNet and allow real-time accurate target detection. Here we used 10 epochs to train the model and the learning rate was set to 0.0001, and all training samples were randomly permuted. For efficient model training, all of the annotated DRRs were resized to 384 × 512 and then cropped into the size of 380 × 450.
Validation of the PTV Localization Model
In this institutional review board (IRB)-approved study, the trained target localization model was validated by retrospectively analyzing a cohort of 2400 DRR datasets for two pancreatic SBRT cases (see Supplemental data for five additional cases). For each patient, we retrieved pCT images, PTV contours and 4D-CT images. A set of 1600 synthetic DRRs was generated for each of the three directions (AP, LR, and an oblique direction 135°) and used to train a patient specific model, which was then tested using 400 independent DRRs in the corresponding direction by comparing the predicted target position to the annotation of the DRRs. The training and testing datasets were generated using different MVFs results from different phases of the 4D CT data. Thus, the training and testing datasets do not overlap. Statistical analysis using Lin’s concordance correlation coefficient (ρc) [26] [27] was performed to assess the results along with the difference between the annotated position and model predicted position of the PTV. ρc is defined as:
with μx and μy the mean values for the predicted positions and the annotated positions of the 400 testing samples, respectively. σx and σy are the corresponding standard deviations. ρ is the correlation coefficient between the predicted and annotated positions.
To demonstrate the advantage of the proposed method, we compared the performance of the proposed target localization method with and without the presence of fiducials. To obtain CT images without fiducials, we identified the implanted fiducials in the patient’s pCT data, and then replaced them with soft tissue. The pCT images with fiducials were used to generate DRRs. For both cases (with and without fiducials), the deep learning models were trained and tested using the same number of samples and the same hyperparameters.
RESULTS
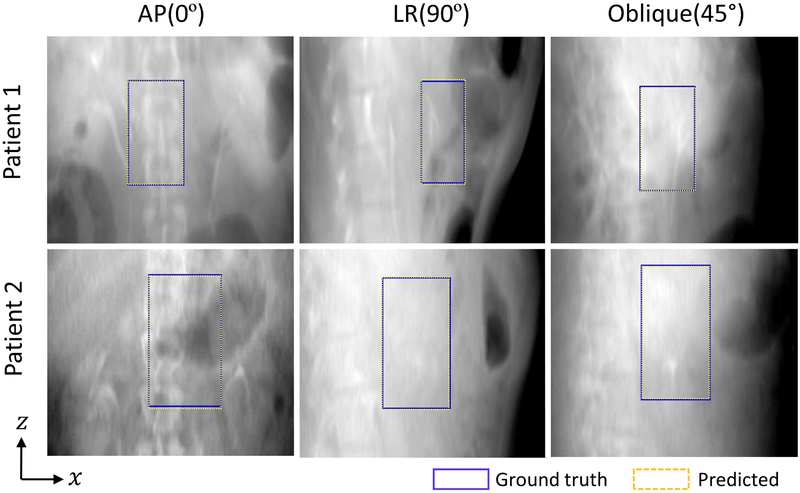
We observed that the predicted PTV bounding box positions match the known positions in the detector plane with significant precision. Figure 4 shows some examples of PTV bounding boxes (dashed yellow lines) derived from the proposed approach together with the known bounding box positions (blue lines) for the patients in AP, LR and oblique directions. The known positions of the target bounding boxes were obtained by projecting the PTV onto the corresponding DRR planes. To facilitate visualization, the patients’ DRRs after MVF deformations, rotations and translations are presented in the figure as the background.
Figure 4.
Examples of the treatment target bounding boxes derived from the patient-specific deep models (yellow dashed box) and their corresponding annotations (blue box), overlaid on top of the patients’ DRRs. The first, second and third columns show the results in AP, LR and oblique directions, respectively.
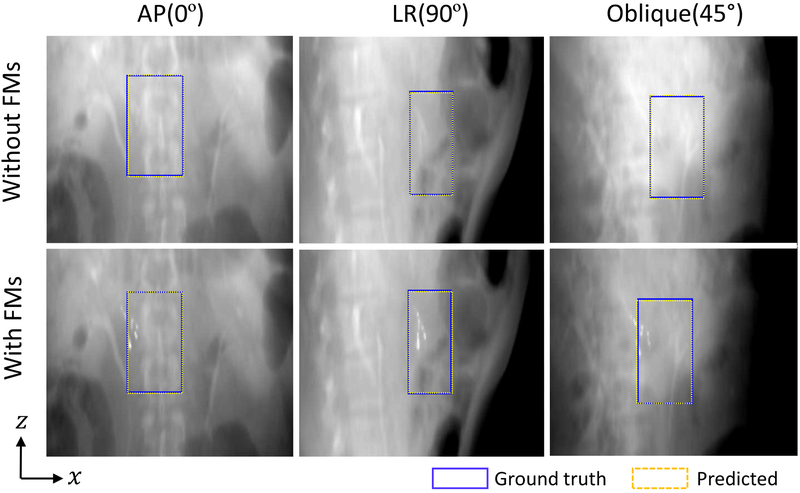
Target localization in AP, LR and oblique directions with and without fiducials can be appreciated in Figure 5. The first row shows the results of target bounding box localization in the absence of fiducials (i.e., the fiducials in pCT data were identified and replaced with the adjacent soft tissue). The results with fiducials are illustrated in the second row. For both cases, the predicted target bounding boxes agree with the ground truths very well, suggesting the proposed method can provide accurate target positions without implanted fiducials. We also measured the positions of the fiducials and found that the results also support our conclusion.
Figure 5.
Examples of the target bounding boxes derived from the deep learning model (yellow dashed box) and their corresponding annotations (blue box) with and without fiducials. In both cases, the predicted target positions agree with the known positions better than 2.6 mm in all three directions, showing the advantage of the proposed target localization method.
A summary of the quantitative evaluations using Lin’s concordance correlation coefficients and MADs between the predictions and the annotated positions of the top-left corner of the PTV bounding boxes is shown in Table 1. Results for five additional cases are included in Supplemental data. The predicted and annotated positions agree with each other well, with observed ρc values greater than 0.93 for all three directions and for target localization with and without fiducials, suggesting a high level of accuracy of the proposed deep learning model, with the MADs being less than 2.6 mm across all cases and in all directions.
Table 1.
Mean absolute differences and Lin’s concordance correlation coefficients between the predicted and annotated PTV positions in anterior-posterior, left-right, and oblique directions. Data are shown as means±standard deviations.
| Index | Anterior-posterior | Left-right | Oblique | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| MADx (mm) |
ρc | MADz (mm) |
ρc | MADx (mm) |
ρc | MADz (mm) |
ρc | MADx (mm) |
ρc | MADz (mm) |
ρc | |
| 1 | 1.95±0.75 | 0.94 | 2.55±1.28 | 0.95 | 0.46±0.48 | 0.99 | 0.97±0.64 | 0.98 | 0.74±0.64 | 0.98 | 1.49±1.14 | 0.97 |
| 2 | 1.49±1.53 | 0.95 | 2.41±1.86 | 0.94 | 0.60±2.21 | 0.94 | 0.38±1.31 | 0.95 | 1.02±0.72 | 0.98 | 2.25±1.44 | 0.95 |
| wo/ FMs | 1.36±0.65 | 0.97 | 1.41±1.48 | 0.97 | 0.34±0.41 | 0.99 | 1.32±0.92 | 0.98 | 0.68±0.68 | 0.98 | 1.31±0.89 | 0.98 |
| w/ FMs | 1.33±1.15 | 0.96 | 2.49±1.75 | 0.94 | 0.51±0.64 | 0.98 | 1.57±1.21 | 0.97 | 0.83±1.38 | 0.95 | 1.47±1.81 | 0.94 |
The deep learning-based prediction model was found to be computationally efficient once the training of the model was performed. In a PC platform (Intel Core i7–6700K, RAM 32 GB) with GPU (Nvidia GeForce GTX Titan X, Memory 12 GB), the training took about two hours for a given direction. The execution of the prediction model took less than 200 ms (the time could be further reduced to ~100 ms with the most recent GPU with Turing™ architecture), which is well suited for real-time tracking of the tumor target position during the radiation dose delivery or during other interventional procedures.
DISCUSSION
Organ motion is an important limiting factor for the maximum exploitation of modern radiation therapy [10, 16]. Adverse influence of organ motion is even more relevant in hypofractionated regimens or SBRT due to the high doses per fraction. For accurate beam targeting, current IGRT relies heavily on the use of implanted fiducials for online/offline target localization, which is invasive, costly, and not free of potential adverse effects. In this work, we made use of the enormous potential of deep learning to investigate a novel markerless target localization strategy by effective extraction of deeper layers of image features embedded in online projection X-ray images via a novel deep learning model. With the proposed model, pancreatic cancer target tracking was achieved with high accuracy and celerity, demonstrating enormous potential for clinical applications.
It is important to emphasize that, different from population-based prognostic studies where hundreds or thousands human subjects are required, this study investigates a patient-specific model to localize treatment target. While a large number of annotated training datasets are still needed, a unique feature of the patient specific deep learning modelling is that it alleviates the demand for a large number of patients. To avoid overfitting, a thumb of rule in establishing a patient specific predictive model is that the training datasets should be large enough to cover the clinical scenarios of anatomic changes for the given patient. For this purpose, for each patient, we deformed, translated and rotated the planning CT data to mimic different scenarios of the patient anatomy and generated DRRs in each direction with the corresponding PTV contour and bounding box as the annotation. In other words, although only two human subjects were investigated, the training for each patient in each direction involved 1600 annotated training datasets, and the testing was performed by analyzing a cohort of 400 independent DRRs for the patient in a specific direction (a specific model was trained for each specific direction of each patient). Hence, the datasets for training and testing the deep learning model are abundant.
While the choice of a patient-specific model provides a useful solution to carry out deep learning by focusing the algorithm on target geometries most relevant to the patient under treatment, the approach may have two potential pitfalls. First, it is possible that patient anatomy changes outside the MVF generated from the 4DCT. In general, a deep learning model is limited to the data distribution used for model training. To mitigate the problem, efforts should be made to ensure that the completeness of distribution of training datasets by considering all possible scenarios before proceeding to model training. Secondly, the planning 4DCT may not be perfect as breathing irregularities and 4DCT image artifacts may occur, which would adversely influence the accuracy of the resultant predictive model. To enhance the robustness of the model against the abnormalities, artifacts removal techniques such as temporal domain regularization may be applied so that the obtained MVF presents a reliable description of the organ motion. A patient with severe breathing irregularities may not be a good candidate for the approach. In this case, the use of multiple scans may improve the robustness of the model.
Incorporating deep learning into radiotherapy is being actively studied for improved clinical decision making and outcome prediction [28–30]. This study presented a direct application of deep learning to IGRT. The novelties of the proposed approach include: (1) a patient-specific deep learning approach for nearly real-time localization of the pancreas target without relying on implanted fiducials; (2) a novel model training data generation scheme; and (3) a model training procedure based on the use of synthetic DRRs derived from the motion-incorporated pCT images. The use of synthetic DRRs mitigates the need for a vast amount of X-ray images for the model training process, making the proposed approach clinically practical. While this study is focused on target localization in monoscopic X-ray imaging, the approach can be easily extended to the stereoscopic X-ray image guidance or other imaging modalities such as cone beam CT.
A natural question is how to implement the deep learning-based model into RT workflow to benefit patient care. We foresee that the approach can be utilized in two ways: guiding patient setup process and monitoring target position during the dose delivery process. The achievable MADs of this work are better than 2.6mm, which is comparable with the IGRT data reported in the literature with the use of implanted fiducials [31, 32]. Similar to current fiducial-based IGRT, a 3 mm margin to the ITV seems to be appropriate when AP/LAT kV images and the proposed deep learning model are used for patient setup. We note that our deep learning model is capable of extracting the PTV position in 100–200 ms after an onboard kV image is acquired, which not only makes it possible to rapidly setup the patient, but also opens the possibility of nearly real-time monitoring of target position at selected gantry angle(s) during treatment delivery. Currently, an angle specific deep learning model needs to be trained in order to obtain the target position at the angle, which is similar to the image registration-based target tracking (e.g., the image guidance system implemented in CyberKnife® from Accuray Inc., Sunnyvale, CA). It is thus practically sensible to monitor the target at a few pre-defined angles when the model is used for target tracking. Finally, we emphasize that the calculation time of 100–200 ms can be improved with further optimization of hardware and software. According to AAPM Task Group 76 [33], the overall system latency, which include the times of image acquisition (~20 ms), image processing, and the linac beam-on/off triggering latency (~6ms) [34], should be less than 500 ms. Thus, the computing time here is acceptable for nearly real-time target monitoring applications.
In conclusion, this study brings up an accurate markerless pancreatic target localization technique based on deep learning. The strategy provides a clinically sensible solution for pancreatic IGRT. The approach is quite general and can be extended to many types of cancers [35]. Future clinical implementation of the technique with thorough validation can provide radiation oncology discipline an important new tool to aid on patient setup and treatment delivery.
Supplementary Material
Acknowledgement:
This work was partially supported by NIH (1R01 CA176553 and R01CA227713) and a Faculty Research Award from Google Inc. We gratefully acknowledge the support of NVIDIA Corporation for the GPU donation. We also would like to thank Prof. Lei Dong for his useful discussion.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflicts of interest: Lei Xing and Daniel Chang serve as the principal investigators of a master research agreement between Stanford University and Varian Medical System. Albert C Koong holds stocks of Aravive, Inc.
REFERENCES
- 1.Vincent A, et al. , Pancreatic cancer. The lancet, 2011. 378(9791): p. 607–620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wang F and Kumar P, The role of radiotherapy in management of pancreatic cancer. Journal of gastrointestinal oncology, 2011. 2(3): p. 157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Siegel RL, Miller KD, and Jemal A, Cancer statistics, 2017. CA: a cancer journal for clinicians, 2017. 67(1): p. 7–30. [DOI] [PubMed] [Google Scholar]
- 4.Veldeman L, et al. , Evidence behind use of intensity-modulated radiotherapy: a systematic review of comparative clinical studies. The lancet oncology, 2008. 9(4): p. 367–375. [DOI] [PubMed] [Google Scholar]
- 5.Staffurth J, A review of the clinical evidence for intensity-modulated radiotherapy. Clinical oncology, 2010. 22(8): p. 643–657. [DOI] [PubMed] [Google Scholar]
- 6.Dawson LA and Sharpe MB, Image-guided radiotherapy: rationale, benefits, and limitations. The lancet oncology, 2006. 7(10): p. 848–858. [DOI] [PubMed] [Google Scholar]
- 7.Xing L, et al. , Overview of image-guided radiation therapy. Medical Dosimetry, 2006. 31(2): p. 91–112. [DOI] [PubMed] [Google Scholar]
- 8.Verellen D, et al. , Innovations in image-guided radiotherapy. Nature Reviews Cancer, 2007. 7(12): p. 949. [DOI] [PubMed] [Google Scholar]
- 9.Jaffray DA, Image-guided radiotherapy: from current concept to future perspectives.Nature Reviews Clinical Oncology, 2012. 9(12): p. 688. [DOI] [PubMed] [Google Scholar]
- 10.Shirato H, et al. , Real-time tumour-tracking radiotherapy. The Lancet, 1999. 353(9161): p. 1331–1332. [DOI] [PubMed] [Google Scholar]
- 11.Shirato H, et al. , Physical aspects of a real-time tumor-tracking system for gated radiotherapy. International Journal of Radiation Oncology* Biology* Physics, 2000. 48(4):p. 1187–1195. [DOI] [PubMed] [Google Scholar]
- 12.Zhang X, et al. , Tracking tumor boundary in MV-EPID images without implanted markers: A feasibility study. Medical physics, 2015. 42(5): p. 2510–2523. [DOI] [PubMed] [Google Scholar]
- 13.Campbell WG, Miften M, and Jones BL, Automated target tracking in kilovoltage images using dynamic templates of fiducial marker clusters. Medical physics, 2017. 44(2): p. 364–374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu W, et al. , Real-time 3D internal marker tracking during arc radiotherapy by the use of combined MV–kV imaging. Physics in Medicine & Biology, 2008. 53(24): p. 7197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wiersma R, Mao W, and Xing L, Combined kV and MV imaging for real-time tracking of implanted fiducial markers a. Medical physics, 2008. 35(4): p. 1191–1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jaffray DA, et al. , Flat-panel cone-beam computed tomography for image-guided radiation therapy. International Journal of Radiation Oncology* Biology* Physics, 2002. 53(5): p. 1337–1349. [DOI] [PubMed] [Google Scholar]
- 17.Acharya S, et al. , Online magnetic resonance image guided adaptive radiation therapy: first clinical applications. International Journal of Radiation Oncology* Biology* Physics, 2016. 94(2): p. 394–403. [DOI] [PubMed] [Google Scholar]
- 18.Omari EA, et al. , Preliminary results on the feasibility of using ultrasound to monitor intrafractional motion during radiation therapy for pancreatic cancer. Medical physics, 2016. 43(9): p. 5252–5260. [DOI] [PubMed] [Google Scholar]
- 19.Khashab MA, et al. , Comparative analysis of traditional and coiled fiducials implanted during EUS for pancreatic cancer patients receiving stereotactic body radiation therapy. Gastrointestinal endoscopy, 2012. 76(5): p. 962–971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Van der Horst A, et al. , Interfractional position variation of pancreatic tumors quantified using intratumoral fiducial markers and daily cone beam computed tomography. International Journal of Radiation Oncology* Biology* Physics, 2013. 87(1): p. 202–208. [DOI] [PubMed] [Google Scholar]
- 21.Majumder S, et al. , Endoscopic Ultrasound–Guided Pancreatic Fiducial Placement: How Important Is Ideal Fiducial Geometry? Pancreas, 2013. 42(4): p. 692–695. [DOI] [PubMed] [Google Scholar]
- 22.Gurney-Champion OJ, et al. , Visibility and artifacts of gold fiducial markers used for image guided radiation therapy of pancreatic cancer on MRI. Medical physics, 2015. 42(5): p. 2638–2647. [DOI] [PubMed] [Google Scholar]
- 23.Girshick R Fast r-cnn. in Proceedings of the IEEE international conference on computer vision. 2015.
- 24.Ren S, et al. , Faster R-CNN: towards real-time object detection with region proposal networks. IEEE Transactions on Pattern Analysis & Machine Intelligence, 2017(6): p. 1137–1149. [DOI] [PubMed] [Google Scholar]
- 25.Simonyan K and Zisserman A, Very deep convolutional networks for large-scale image recognition. arXiv preprint arXiv:1409.1556, 2014. [Google Scholar]
- 26.Lawrence I and Lin K, A concordance correlation coefficient to evaluate reproducibility. Biometrics, 1989: p. 255–268. [PubMed] [Google Scholar]
- 27.Hamilton DG, McKenzie DP, and Perkins AE, Comparison between electromagnetic transponders and radiographic imaging for prostate localization: A pelvic phantom study with rotations and translations. Journal of applied clinical medical physics, 2017. 18(5): p. 43–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cha KH, et al. , Bladder cancer treatment response assessment in CT using radiomics with deep-learning. Scientific reports, 2017. 7(1): p. 8738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ibragimov B, et al. , Development of deep neural network for individualized hepatobiliary toxicity prediction after liver SBRT. Medical physics, 2018. 45(10): p. 4763–4774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Luo Y, et al. , Development of a Fully Cross-Validated Bayesian Network Approach for Local Control Prediction in Lung Cancer. IEEE Transactions on Radiation and Plasma Medical Sciences, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jayachandran P, et al. , Interfractional uncertainty in the treatment of pancreatic cancer with radiation. International Journal of Radiation Oncology* Biology* Physics, 2010. 76(2): p. 603–607. [DOI] [PubMed] [Google Scholar]
- 32.Whitfield G, et al. , Quantifying motion for pancreatic radiotherapy margin calculation. Radiotherapy and Oncology, 2012. 103(3): p. 360–366. [DOI] [PubMed] [Google Scholar]
- 33.Keall PJ, et al. , The management of respiratory motion in radiation oncology report of AAPM Task Group 76 a. Medical physics, 2006. 33(10): p. 3874–3900. [DOI] [PubMed] [Google Scholar]
- 34.Shepard AJ, et al. , Characterization of clinical linear accelerator triggering latency for motion management system development. Medical physics, 2018. 45(11): p. 4816–4821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhao W, et al. , Visualizing the Invisible in Prostate Radiation Therapy: Markerless Prostate Target Localization Via a Deep Learning Model and Monoscopic Kv Projection XRay Image. International Journal of Radiation Oncology• Biology• Physics, 2018. 102(3): p. S128–S129. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.