Abstract
In our previous study, the expression profile of tight junction (TJ) protein claudins (CLDNs) in human osteosarcoma (OS) cells was examined, and the data found the CLDN10 was high expressed in OS cells versus fetal osteoblast cells. Hence, we aim to determine the impacts and the molecular mechanisms of CLDN10 in the metastatic phenotype of OS. The exact expression profiles of CLDN10 and phosphorylated Janus kinase 1 (JAK1) in noncancerous bone tissues and OS tissues were detected via a western blotting and immunohistochemistry method. The OS cells with CLDN10 or JAK1 silencing was established via an RNA interference (RNAi) method, and an osteoblast cell line stably expressing CLDN10 was established via cell transfection. Then, the transfection effects and activation states of JAK1/ signal transducer and activator of transcription1 (Stat1) pathway in OS and osteoblast cells were detected via a western blotting assay. Moreover, the metastatic ability of osteoblast cells and OS cells in vitro were evaluated by means of a cell counting kit-8 (CCK8) assay, colony formation assay in soft agar, transwell assay and wound-healing experiment. The present data revealed that CLDN10 and phospho-JAK1 were up-regulated in OS tissues compared with noncancerous bone tissues. Genetic loss of CLDN10 or JAK1 inhibited the activation of the Stat1 and the malignant phenotype in OS cells. To sum up, our study suggested the CLDN10 enhanced the metastatic phenotype of OS cells via the activation of the JAK1/Stat1 signaling pathway.
Keywords: Claudin-10, Osteosarcoma, Janus kinase 1, Signal transducer and activator of transcription1, Metastasis
Background
To date, the issue of constraining the early metastasis has been suggested to be a blockage in the therapy of OS (Li et al. 2011). Currently, it has been received more and more attentions that distant metastasis of tumor is frequently coupled with the aberrations in tight junction (TJ) proteins (González-Mariscal et al. 2008; Tsukita et al. 1999). Claudins (CLDNs) were found to be the major constituent proteins of TJs, and the abnormalities of CLDNs can lead to the destructions in structure and function of TJs, which are prerequisite for invasion and distant metastasis of tumor cells (Oliveira and Morgado-Diaz 2007). Furthermore, recent studies have confirmed that the tumor cells commonly display the alterations in CLDNs expression (Tabaries and Siegel 2017). For instance, CLDN1 was found to be a suppressor for the metastasis of lung adenocarcinoma and the loss of CLDN1 indicated a poor prognosis patient in lung adenocarcinoma patients (Chao et al. 2009). Moreover, CLDN7 was indicated to be low expressed in head and neck cancer cells and the reduced CLDN7 enhanced the cell invasion and migration in these tumor (Usami et al. 2006). Briefly, these researches showed the CLDNs protein have the opportunity to be set as the diagnostic markers of human tumors.
The distinct expression profiles of CLDNs in diverse malignancies displayed an opportunity to be utilized in early diagnosis and therapy of these tumors. Additionally, accumulating evidence reveals that CLDNs were involved in the regulation of cell proliferation, differentiation and motility through participating in the transduction of intracellular/extracellular signals (Merino-Gracia et al. 2016; Jeansonne et al. 2003; Hamazaki et al. 2002). For instance, CLDN3 was revealed to increase the malignant potential of colorectal cancer cells as modulators of epidermal growth factor receptor signaling (de Souza et al. 2013). CLDN7 was found to promote the epithelial-mesenchymal transition (EMT) process through recruiting the epithelial cell adhesion molecule (EpCAM) in human colorectal cancer (Philip et al. 2015). Furthermore, a recent study demonstrated that enhanced CLDN18 expression activated extracellular signal-regulated kinase (ERK)1/2 to contribute to the malignant potential of bile duct cancer (Takasawa et al. 2017). Given these associations of CLDN proteins and various human cancer types, these proteins may have the potential to be established as novel targets in the diagnosis and therapeutic of certain tumor types. Previously, we found that CLDN10 was high expressed in OS cells versus fetal osteoblast cells. However, at present, there is no report on the impact of CLDN10 in the tumorigenesis of OS; therefore, we aim to determine the exact roles and molecular mechanisms of CLDN10 in OS malignancy and to recognize innovative targets for OS therapy.
Materials and methods
Cell culture
Human fetal-osteoblast cell line (hFOB.1.19), human kidney 293 T (HKT 293 T) cell line, and human OS cell lines (U2OS, 143B, MG63, Saos2) were obtained from the Shanghai Cell Bank of the Chinese Academy of Sciences. These cell lines were cultured in Dulbecco’s modified Eagle’s medium (DMEM) (cat. no. 12491–015, Thermo Fisher Scientific, Inc., Waltham, MA, USA) containing 10% fetal bovine serum (FBS) in a humidified incubator at 37 °C, 5% CO2, and 95% humidity.
Antibodies
Rabbit anti-human phosphor-Stat1 (cat. no. 7649), phosphor-JAK1(cat. no. 66245), Stat1 (cat. no. 14994) and JAK1 (cat. no. 3344) antibodies were purchased from Cell Signaling Technology, Inc. (Danvers, MA, USA). Mouse anti-human β-actin (cat. no. ab8226) and Rabbit anti-human CLDN10 (cat. no. ab24792) were purchased from Abcam (Cambridge, MA, USA).
Western blotting
Total protein extraction from cell culture was performed according to protocols from protein extraction kits (cat. no.310003, Best Bio, Shanghai, China). Then, the protein concentration was quantified using an Enhanced BCA Protein Assay Kit (Pierce Chemical Co., Dallas, TX, USA). Total protein (30 μg) from each sample was detached SDS-PAGE electrophoresis, and transferred into nitrocellulose membrane (EMD Millipore, Billerica, MA, USA). Subsequently, the membrane was incubated in blocking buffer (5% no-fat milk in TBST) and probed with primary antibody at a dilution of 1:1000 at 4 °C overnight. Following this, the membranes were rinsed 3 times with PBS and incubated with horseradish peroxidase (HRP)-conjugated secondary antibodies (1:1000; Santa Cruz Biotechnology, Inc., Dallas, TX, USA). Immunoreactive bands were visualized using chemiluminescence detection system (GE, Fairfield, Connecticut, USA) and evaluated with Image Lab 6.0.1 software (Bio-Rad Laboratories).
Cell counting Kit-8 (CCK-8) assay
The CCK-8 assay was performed following the manufacturer’s protocols (CCK-8; Dojindo, Kumamoto, Japan). Cells were seeded into 96-well plates at a density of 1 × 103 cells/well in triplicate, and cultured overnight to allow for adherence. The final optical density (OD) was measured every 12 h for 4 days at a wavelength of 450 nm to estimate cell proliferation ability in different groups.
Colony formation assays
The experimental process was performed as described formerly (Zhang et al. 2015).
Wound-healing assay
Cells were cultured in DMEM (cat. no. 12491–015, Thermo Fisher Scientific, Inc., Waltham, MA, USA) containing10% FBS with a 70% convergence on 24-well plastic dishes and the cell monolayer was scratched with a 100-μl pipette tip. The wounds were imaged under a light microscope (magnification ×200; E100; Nikon Instruments, Inc., Japan) at the identical location at 0, 12 and 24 h after scratching.
Matrigel chamber method
For the invasion assay, 5 × 104 cells/well were planked in the top chamber on Matrigel-coated (BD Biosciences) membranes. Cells were seeded in medium without FBS in the top chamber, and medium with chemotactic factors obtained from the cell culture was supplemented to the lower chamber to stimulate invasion. Cells were incubated for 6 h in a cell incubator at 37 °C before they were stained with Diff-Quik Kit (Sysmex, Kobe, Japan) and counted under a light microscope (magnification ×200; E100; Nikon Instruments, Japan).
RNA interference (RNAi) method
The frozen glycerol bacterial stocks of pGCSIL-scramble, pGCSIL-green fluorescent protein (GFP), pGCSIL-CLDN10-RNAi, pGCSIL-JAK1-RNAi, VSVG expression plasmid and virion-packaging elements (pHelper 1.0) were obtained from Nanjing KeyGen Biotech Co, Ltd. (Nanjing, China). HEK 293 T cells (0.2 × 107) in 6-well dishes were cultured in DMEM with 10% FBS for 24 h at 37 °C incubator to attain 70–80% confluence. A total of four plasmids, containing 10 μg pGCSIL-JAK1-RNAi, pGCSIL-CLDN10-RNAi or pGCSIL-scramble, 5 μg packaging vector pHelper 1.0, 5 μg pGCSIL- GFP plasmid and 5 μg VSVG expression plasmid were supplemented to 950 μl Opti-MEM (cat. no. 31985062, Thermo Fisher Scientific, Inc., Waltham, MA, USA) to a final volume of 1.0 ml. The cell transfection was performed following the manufacturer’s protocol of Lipofectamine 2000 (cat. no. 11668027, Thermo Fisher Scientific, Inc., Waltham, MA, USA) and the lentiviral particles were collected 48 h after transfection.
Patients and tissue samples
21 cases of histologically noncancerous sections were obtained from osteoarthritis patients with an average age of 47 years who were treated at the Qilu Hospital of Shandong University between October 2006 and September 2011.
27 cases of tumor sections were obtained from OS patients with an average age of 24 years who were treated at the Qilu Hospital of Shandong University between June 2007 and May 2012. The OS patient was selected based on the subsequent standards: no prior malignant disease and no histories of radiotherapy or chemotherapy. The grading and staging of these tumors were according to the American Joint Committee on Cancer (AJCC) tumor node metastasis (TNM) staging system. The details of the clinicopathological characteristics were summarized in Table 1.
Table 1.
Expression of CLDN10 and clinicopathological characteristics in OS patients
| Item | N | CLDN10 (+) | CLDN10 (−) | P |
|---|---|---|---|---|
| Tumor tissue | 27 | 17 | 10 | <0.001* |
| Normal | 21 | 6 | 15 | |
| Age (years) | ||||
| ≤19 | 15 | 9 | 6 | 0.762 |
| >19 | 12 | 8 | 4 | |
| Gender | ||||
| Male | 16 | 10 | 6 | 0.893 |
| Female | 11 | 7 | 4 | |
| Stage | ||||
| IA- IIA | 10 | 7 | 3 | 0.426 |
| IIB- III | 17 | 10 | 7 | |
| Response to chemotherapy | ||||
| Poor | 9 | 6 | 3 | 0.242 |
| Good | 7 | 4 | 3 | |
| NA (n = 8) | 11 | |||
| Pulmonary metastasis | ||||
| + | 21 | 16 | 5 | <0.001* |
| – | 6 | 1 | 5 | |
NA not available
*Statistical significance was found with the Chi-square test/Chi-Square Goodness-of-Fit Test
Immunohistochemistry
The detailed expression profiles of CLDN10 and phospho-JAK1 protein in sections from 21 noncancerous bone tissues and 27 OS patients were explored via a immunohistochemistry method as described formerly (Zhang et al. 2015), and the primary antibodies utilized were rabbit anti-human CLDN10 and phospho-JAK1. The expression localization at the cell membrane was considered as positive for CLDN10 and the expression localization of phospho-JAK1 located at the cell nucleus was considered as positive. The staining and scoring of these protein expression were measured semi-quantitatively as previously described (Yang et al. 2008).
Cell transfection
The pNSE-IRES-EGFP1-C1/CLDN10 plasmid and empty vector plasimid pNSE-IRES2-EGFP1-C1 (+) were obtained from KeyGEN BioTECH Company. The plasmid (Five micrograms) was transfected into cells according to the protocol of SuperFect Transfection Reagent (TaKaRa, Japan). The monoclonal population was obtained and expanded via G418 (Sigma, St. Louis, Missouri, USA)-resistant screening.
Statistical methods
All the data are expressed as the mean ± standard deviation of at least 3 rounds of experimental data. Data were analyzed using Student’s t test or ANOVA with Dunnett’s multiple comparisons test. The prognostic implication of CLDN10 expression was detected by the χ2 test/χ2 Goodness-of-Fit Test. *P < 0.05 and **P < 0.01 was considered statistically significant.
Results
Expression of CLDN10 is upregulated in the OS cell lines and tissues
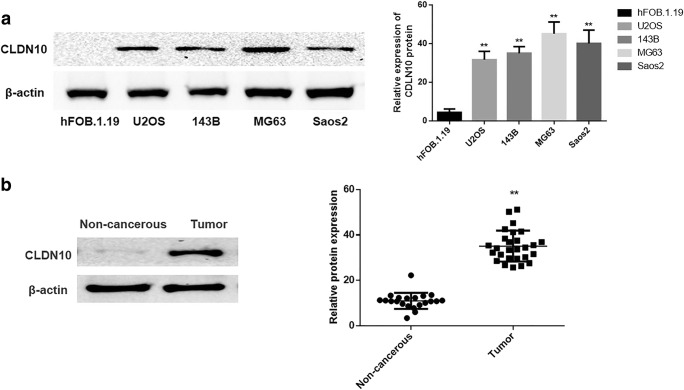
The expression level of CLDN10 in human fetal-osteoblast cell line (hFOB.1.19) and OS cell lines (U2OS, 143B, MG63 and Saos2) were explored via western blotting method. The results demonstrated that the protein expression level of CLDN10 was low in human fetal-osteoblast cell line hFOB.1.19 but highly expressed in OS cell lines (Fig. 1a).
Fig. 1.
Expressions of CLDN10 in the OS cell lines and tissues. a The protein expression of CLDN10 in osteoblast and OS cell lines (left), and the corresponding statistical analysis of protein expression (right). Note: normalized with β-actin, **P < 0.01, versus osteoblast cell line b The expression profiles of CLDN10 protein in primary osteosarcoma tissues and noncancerous sections (left), and the relative CLDN10 protein expression levels are expressed as dots (right). Note: normalized with β-actin, **P < 0.01, versus noncancerous bone sections
The alterations in the expression profiles of CLDN10 between OS tissues and non-cancerous bone tissues were also detected via semiquantitative immunoblotting analysis. As seen in Fig. 1b, the CLDN10 expression was significantly up-regulated in the OS tissues versus in noncancerous bone tissues (P = 0.0003).
Expression of CLDN10 and phospho-JAK1 in human OS tissues
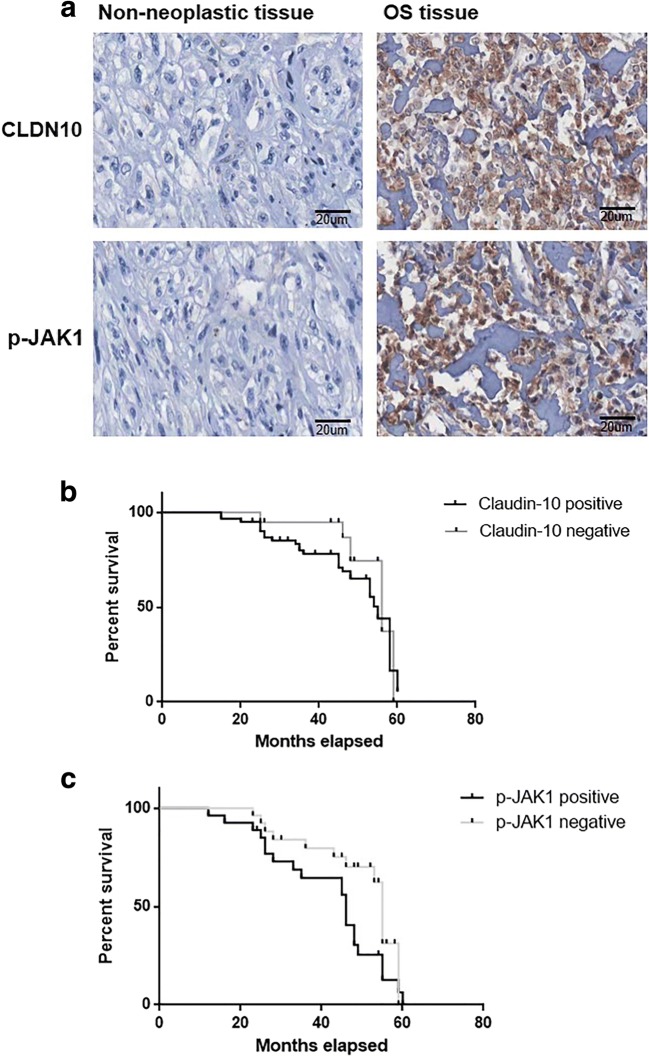
The expression profiles of CLDN10 and phospho-JAK1 in the 27 cases of OS tissues and 21 cases of non-cancerous bone tissues were investigated via an immunohistochemistry method. As depicted in Fig. 2a, the expression of CLDN10 was found to be mainly located at the cell membrane, and the expression of CLDN10 was highly expressed in 80.9% (17/27) of OS tissues and 28.5% (6/21) of noncancerous bone tissues (Table 1). In addition, the expression of CLDN10 was revealed to not be notably associated with patient age (P = 0.762), gender (P = 0.893), TNM stage (P = 0.426), response to chemotherapy (P = 0.242), but associated with the pulmonary metastasis (P = 0.0003) in OS patients.
Fig. 2.
Expression levels of CLDN10 and phospho-JAK1 in human primary OS patients a Detection of CLDN10 and phospho-JAK1 in human primary OS tissues and bone tissues. b Association between CLDN10 and survival time were analyzed via Kaplan-Meier Survival Curves and the Log-Rank Test c Association between phospho-JAK1 and survival time in OS patients
The phospho-JAK1 expression was investigated in the OS tissues and noncancerous bone sections (Fig. 2). As displayed in Table 2, the expression of phospho-Stat1 was highly expressed in 55.5% (15/27) of OS tissues and 33.3% (7/21) of noncancerous bone tissues. The results demonstrated that the phospho-JAK1 expression was increased (P = 0.0079) in human OS tissues versus noncancerous bone sections (Table 2). Furthermore, the expression of phospho-JAK1 was associated with TNM stage (P = 0.0007) and the pulmonary metastasis (P = 0.0003) of patients with OS (Table 2).
Table 2.
Expression of phospho-JAK1 and clinicopathological characteristics in OS patients
| Item | N | phospho-JAK1 (+) | phospho-JAK1 (−) | P |
|---|---|---|---|---|
| Tumor tissue | 27 | 15 | 12 | <0.01* |
| Non-neoplastic | 21 | 7 | 14 | |
| Age (years) | ||||
| ≤19 | 15 | 8 | 7 | 0.862 |
| >19 | 12 | 7 | 5 | |
| Gender | ||||
| Male | 16 | 9 | 7 | 0.792 |
| Female | 11 | 6 | 5 | |
| Stage | ||||
| IA- IIA | 10 | 8 | 3 | <0.001* |
| IIB- III | 17 | 7 | 10 | |
| Response to chemotherapy | ||||
| Poor | 9 | 4 | 5 | 0.143 |
| Good | 7 | 3 | 4 | |
| NA (n = 8) | 11 | |||
| Pulmonary metastasis | ||||
| + | 21 | 13 | 7 | <0.001* |
| – | 6 | 2 | 4 | |
NA not available
*Statistical significance was found with the Chi-square test/Chi-Square Goodness-of-Fit Test
Association with survival and clinical outcomes
The associations among CLDN10 and phospho-JAK1 with survival time of OS patients were determined via Kaplan-Meier Survival Curves and the Log-Rank Tests. As shown in Fig. 2b, the median survival of patients with tumors that were positive for CLDN10 protein (34.46 months) had a notably shorter survival time compared with those patients with tumors were negative for CLDN10 protein (45.54 months) (Chi square = 7.218, P = 0.0071). Besides, as shown in Fig. 2c, the median survival of patients with tumors that were positive for phospho-JAK1 protein (33.26 months) had a notably shorter survival time compared with those patients with tumors were negative for phospho-JAK1 protein (46.63 months) (Chi square = 8.176, P = 0.0021).
The loss of CLDN10 inhibited the proliferation and metastasis of OS cell line
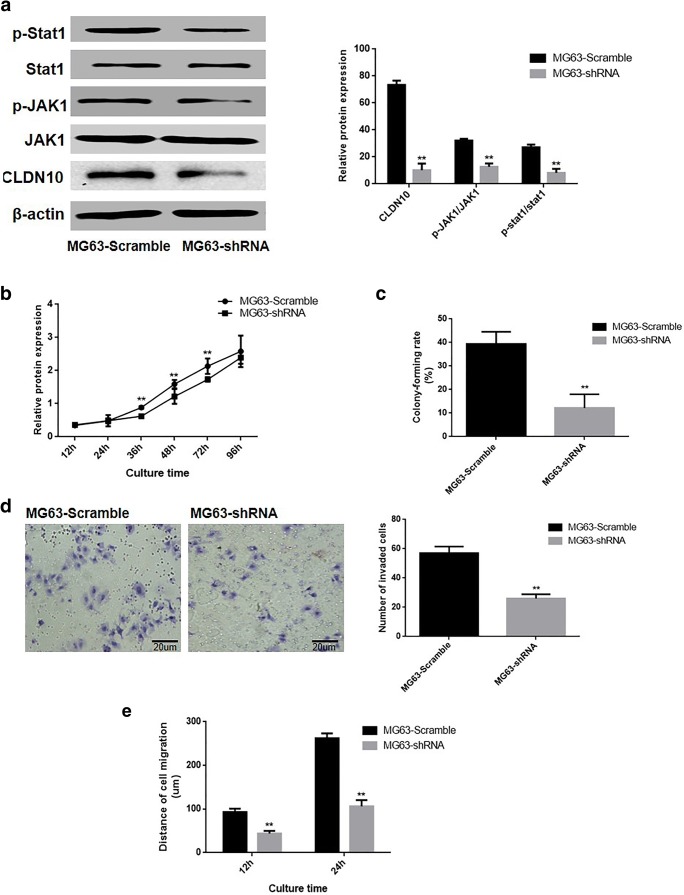
The effect of CLDN10 knockdown on proliferation, invasion and migration ability was determined in osteoblast cells. A pGCSIL-scramble plasmid and a pGCSIL-CLDN10-RNAi plasmid were transfected into an OS cell line, MG63. The western blotting assay in Fig. 3a demonstrated that the ratio of phosphorylation JAK1 (P = 0.0027) and Stat1 (P = 0.0003) were notably reduced in the MG63 cells that silence CLDN10.
Fig. 3.
The loss of CLDN10 inhibited the proliferation and metastasis of OS cell line. a The effects of CLDN10 knockout and the activation of the JAK1/Stat1 signaling pathway (left), and the corresponding statistical analysis of protein expression (right). Note: normalized with β-actin, **P < 0.01, versus scramble group. b Growth curve of MG63 cells. c The abilities of MG63 cells to form colonies. d The impact of CLDN10 knockout on the invasive ability of MG63 cells in vitro (left), and the corresponding statistical analysis (right). e The impact of CLDN10 knockout on the migration ability of MG63 cells in vitro. **P < 0.01, versus the scramble group
Effects of CLDN10 knockdown on malignant phenotype were analyzed in MG63 cells. The Cell Counting kit-8 results in Fig. 3b depicted that the proliferation rate of MG63-CLDN10-shRNA cells was notably reduced than the scramble groups (P = 0.653). The data derived from 2D monolayer culture (Fig. 2e) revealed that the number of colonies formed in CLDN10-silencing cells was significantly lower than the scramble group (P = 0.0002). The results of the transwell chamber assay and wound-healing assay demonstrated that the numbers of invasive MG63 cells were notably increased (P = 0.0024) in CLDN10 silencing cells (Fig. 3d). Results also displayed that the migration distances of MG63 cells in the CLDN10-RNAi group were significant shorter than in scramble group at 12 h (P = 0.0014) and 24 h (P = 0.0016) (Fig. 3e).
Impact of JAK1/Stat1 signaling pathway activation on the malignant phenotype of OS cells
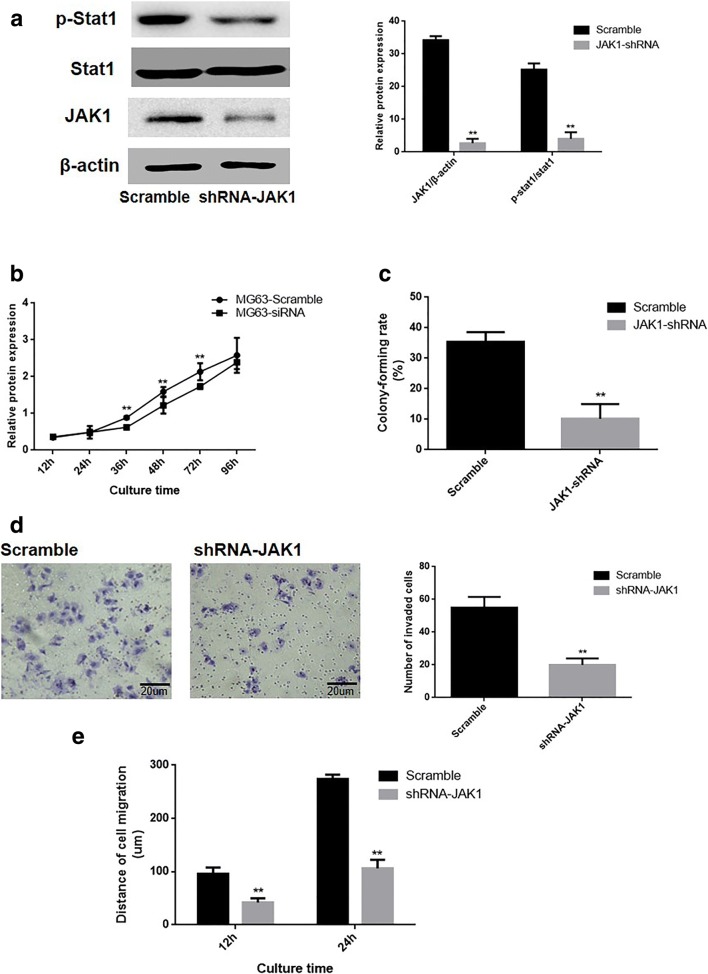
This study elucidated the effects of JAK1/Stat1 signaling pathway on the malignant phenotype in MG63 cells. The pGCSIL-scramble or pGCSIL-JAK1-RNAi plasmids were transfected into MG63 cells, and western blotting data revealed that genetic silencing of JAK1 inhibited the activation of the Stat1 in OS cells (Fig. 4a). The results obtained from the CCK8 assay (Fig. 4b) and colony formation assays (Fig. 4c) demonstrated that proliferation rate (P = 0.0016) and the number of colonies formed of JAK1-RNAi cells (P = 0.0004) were significantly reduced versus the scramble group Furthermore, The data of transwell chamber assay and wound-healing assay displayed that the number of invasive MG63 cells were significantly reduced (P = 0.0012) following JAK1 silencing (Fig. 4d). Additionally, the migration distances of JAK1-RNAi cells were notably reduced than the scramble group after 12 and 24 h (P = 0.0001; P = 0.0002, respectively; Fig. 4e).
Fig. 4.
Impact of JAK1 knockout on the malignant phenotype of OS cells. a The impacts of JAK1 knockout on the activation of the Stat1 in OS cells (left), and the corresponding statistical analysis of protein expression (right). Note: normalized with β-actin, **P < 0.01, versus scramble group. b Growth curve of MG63 cells. c The abilities of MG63 cells to form colonies. d The impact of JAK1 knockout on the invasive ability of cells in vitro (left), and the corresponding statistical analysis (right). e The migration ability of the MG63 cell line in vitro. **P < 0.01, versus the scramble group
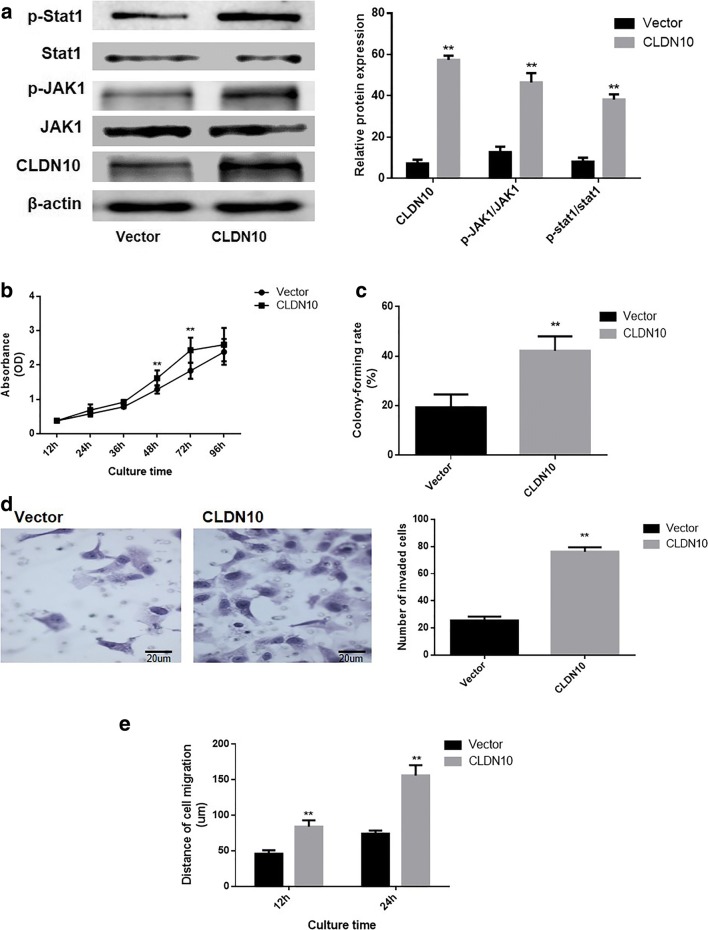
Impact of CLDN10 overexpression on the malignant phenotype of osteoblast cells
The impact of CLDN10 overexpression on malignant phenotype were also analyzed in osteoblast cells. A pNSE-IRES-EGFP1-C1/CLDN10 plasmid was transfected into an osteoblast cell line, hFOB.1.19. The data from western blotting (Fig. 5a) showed that the ratio of phosphorylation JAK1 (P = 0.0003) and phosphorylation Stat1 (P = 0.0001) were significantly increased in the hFOB.1.19 cells with CLDN10 overexpression. The results obtained from the CCK8 assay (Fig. 5b) and colony formation assays (Fig. 5c) demonstrated that proliferation rate (P = 0.0011) and the number of colonies formed of cells with CLDN10 overexpression (P = 0.0007) were significantly increased than the vector group. Furthermore, it was also determined that the number of invasive hFOB.1.19 cells were significantly increased (P = 0.0006) following CLDN10 overexpression (Fig. 5d). Additionally, the migration distances of hFOB.1.19 cells with CLDN10 overexpression were significantly increased than the vector group after 12 and 24 h (P = 0.0003; P = 0.0002, respectively; Fig. 5e).
Fig. 5.
Impact of CLDN10 overexpression on the proliferation and metastasis of osteoblast cells. a The expression of CLDN10 and the activation of the JAK1/Stat1 signaling pathway (left), and the corresponding statistical analysis of protein expression (right). Note: normalized with β-actin, **P < 0.01, compared with vector group. b Growth curve of osteoblast cells. c The abilities of osteoblast cells to form colonies. d The impact of CLDN10 on the invasive ability of osteoblast cells in vitro (left), and the corresponding statistical analysis (right). e The migration ability of osteoblast cells in vitro. **P < 0.01, versus the vector group
Discussion
The current understanding of the biological functions of CLDN10, a member of the CLDN protein family, is mostly restricted to the function in cell barriers and connections (Koval 2013). Previously, we assumed that the increase in CLDN10 was involved in the OS tumorigenesis. To confirm this theory, an OS cell line with a CLDN10 knockout was established, and our data showed that the knockout of CLDN10 notably suppressed the proliferation and motility of OS cells. Most of the research indicated that the CLDNs protein were often decreased in human various tumors. The expression level of CLDN1 was decreased and the re-gain of CLDN1 suppressed the invasive ability in pancreatic cancer cells (Hewitt et al. 2006; Morin 2005). However, a number of the latest researches have indicated that the CLDNs protein expression in certain tumors was strongly involved with the metastatic abilities of tumor cells (Ouban and Ahmed 2010; Zavala-Zendejas et al. 2011). These observations revealed that the diverse CLDNs have specific role on the biological behaviors of certain tumor (D'Souza et al. 2005, 2007; Li et al. 2013). One possible reason contributing to this phenomenon was that CLDNs may have different function in certain cells and depend on specific interacting molecules (Micke et al. 2014). Studies have indicated CLDN1 overexpression induced the cell migration and invasion in human liver cells via c-Abl-ERK signaling pathway (Suh et al. 2017). In contrast, several results were recently observed that the CLDN18 expression can lead to a malignant phenotype of bile duct cancer together with EGFR/ERK signaling (Takasawa et al. 2017). However, the impacts and its specific molecular mechanisms of CLDN10 in OS and remain to be elucidated.
Currently, few studies have determined the functional association between bone carcinogenesis and the CLDN10 protein. This study revealed that CLDN10 expression was overexpressed in OS tissues, and this overexpression in CLDN10 protein significantly promoted the proliferation and motility of OS cells. Previously, gene chip analysis was conducted to screen for and analyze CLDN10 target genes and associated signaling pathways, and the Kyoto Encyclopedia of Genes and Genomes analysis suggested that the overexpression of CLDN10 enhanced JAK1/Stat1 signaling (data not shown). In this study, the effect of CLDN10 on the JAK1/Stat1 signaling pathway in OS was examined and it was revealed that CLDN10 significantly promoted the JAK1/Stat1 signaling pathway. Besides, JAK1-RNAi assays were used to silence the expression of JAK1 and the results demonstrated that JAK1 silencing resulted in inhibition of the Stat1 signaling pathway and metastatic abilities in OS cells. Currently, a limited number of researches have revealed that JAK1/Stat1 signaling was associated with the impacts of CLDNs in human tumorigenesis. Notably, our results validated that the loss of CLDN10 contributes to a suppression of metastatic ability via the JAK1/Stat1 signaling pathway in OS cells. Besides, in osteoblast cells, the CLDN10 overexpression resulted in an activation of JAK1/Stat1 signaling pathway and enhanced the metastasis phenotype. To date, studies have certificated that CLDN proteins can interact with various cellular signal transduction proteins, and related with the regulation a range of cell biological behaviors (18). Hence, we assumed that CLDN10 may bind with the specific cell signal-conducting molecule and participate in the regulation of the JAK1/Stat1 signaling pathway in OS. However, the exact mechanism of how the signal is transduced from membrane-anchored CLDNs to JAK1 proteins located in the cell interior remains to be clarified.
Conclusion
Currently, the specific molecular mechanisms of CLDNs and their impacts in OS remains unclear. Findings in the present study confirmed that CLDN10 affects the Stat1 signaling pathway via JAK1, ultimately enhancing the malignant phenotype of OS cells.
Acknowledgements
We would like to thank American Journal Experts (AJE) for English language editing. This manuscript was edited for English language by American Journal Experts (AJE).
Abbreviations
- CCK8
Cell counting kit-8
- CLDNs
Claudins
- TJ
Tight junction
- IHC
Immunohistochemical analysis
- RNAi
RNA interference
- OS
Osteosarcoma
- JAK1
Janus kinase 1
- Stat1
Signal transducer and activator of transcription1
Author contributions
XZ, XW and AW performed the experiments and analyzed the data. MZ and QL contributed to the conception and design of the study. TL revised the manuscript critically for important intellectual content. All authors read and approved the final manuscript.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional and/or national research committee and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards. Ethics approval (approval no. SDU06384) was approved by the Ethics Committee of Central Hospital of Zibo, Affiliated with Shandong University. This article does not contain any studies with animals performed by any of the authors. The informed consent for participation was obtained from all patients and their parents who participated in this study in an appropriate method.
Consent for publication
Written informed consent was obtained from all patients and their parents who participated in this study.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Xiaowei Zhang, Email: zxw90@foxmail.com.
Xianbin Wang, Email: wangxianbin1111@163.com.
Aiyu Wang, Email: 18678185317@163.com.
Qian Li, Email: liqianjz@163.com.
Ming Zhou, Email: zmzxyy@126.com.
Tao Li, Phone: 05332360685, Email: taolichina@126.com.
References
- Chao Y-C, Pan S-H, Yang S-C, Yu S-L, Che T-F, Lin C-W, Tsai M-S, Chang G-C, Wu C-H, Wu Y-Y. Claudin-1 is a metastasis suppressor and correlates with clinical outcome in lung adenocarcinoma. Am J Respir Crit Care Med. 2009;179(2):123–133. doi: 10.1164/rccm.200803-456OC. [DOI] [PubMed] [Google Scholar]
- de Souza WF, Fortunato-Miranda N, Robbs BK, de Araujo WM, de-Freitas-Junior JC, Bastos LG, Viola JP, Morgado-Diaz JA. Claudin-3 overexpression increases the malignant potential of colorectal cancer cells: roles of ERK1/2 and PI3K-Akt as modulators of EGFR signaling. PLoS One. 2013;8(9):e74994. doi: 10.1371/journal.pone.0074994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Souza T, Agarwal R, Morin PJ. Phosphorylation of claudin-3 at threonine 192 by cAMP-dependent protein kinase regulates tight junction barrier function in ovarian cancer cells. J Biol Chem. 2005;280(28):26233–26240. doi: 10.1074/jbc.M502003200. [DOI] [PubMed] [Google Scholar]
- D'Souza T, Indig FE, Morin PJ. Phosphorylation of claudin-4 by PKCε regulates tight junction barrier function in ovarian cancer cells. Exp Cell Res. 2007;313(15):3364–3375. doi: 10.1016/j.yexcr.2007.06.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- González-Mariscal L, Tapia R, Chamorro D. Crosstalk of tight junction components with signaling pathways. Biochim Biophys Acta Biomembr. 2008;1778(3):729–756. doi: 10.1016/j.bbamem.2007.08.018. [DOI] [PubMed] [Google Scholar]
- Hamazaki Y, Itoh M, Sasaki H, Furuse M, Tsukita S. Multi-PDZ domain protein 1 (MUPP1) is concentrated at tight junctions through its possible interaction with claudin-1 and junctional adhesion molecule. J Biol Chem. 2002;277(1):455–461. doi: 10.1074/jbc.M109005200. [DOI] [PubMed] [Google Scholar]
- Hewitt KJ, Agarwal R, Morin PJ. The claudin gene family: expression in normal and neoplastic tissues. BMC Cancer. 2006;6(1):186. doi: 10.1186/1471-2407-6-186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeansonne B, Lu Q, Goodenough DA, Chen YH. Claudin-8 interacts with multi-PDZ domain protein 1 (MUPP1) and reduces paracellular conductance in epithelial cells. Cell Mol Biol. 2003;49(1):13–21. [PubMed] [Google Scholar]
- Koval M. Claudin heterogeneity and control of lung tight junctions. Annu Rev Physiol. 2013;75:551–567. doi: 10.1146/annurev-physiol-030212-183809. [DOI] [PubMed] [Google Scholar]
- Li X, Ashana AO, Moretti VM, Lackman RD. The relation of tumour necrosis and survival in patients with osteosarcoma. Int Orthop. 2011;35(12):1847–1853. doi: 10.1007/s00264-011-1209-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Li Y, Qiu H, Wang Y. Downregulation of claudin-7 potentiates cellular proliferation and invasion in endometrial cancer. Oncol Lett. 2013;6(1):101–105. doi: 10.3892/ol.2013.1330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merino-Gracia J, Costas-Insua C, Canales MA, Rodriguez-Crespo I. Insights into the C-terminal peptide binding specificity of the PDZ domain of neuronal nitric-oxide synthase: CHARACTERIZATION OF THE INTERACTION WITH THE TIGHT JUNCTION PROTEIN CLAUDIN-3. J Biol Chem. 2016;291(22):11581–11595. doi: 10.1074/jbc.M116.724427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Micke P, Mattsson JS, Edlund K, Lohr M, Jirstrom K, Berglund A, Botling J, Rahnenfuehrer J, Marincevic M, Ponten F, Ekman S, Hengstler J, Woll S, Sahin U, Tureci O. Aberrantly activated claudin 6 and 18.2 as potential therapy targets in non-small-cell lung cancer. Int J Cancer. 2014;135(9):2206–2214. doi: 10.1002/ijc.28857. [DOI] [PubMed] [Google Scholar]
- Morin PJ. Claudin proteins in human cancer: promising new targets for diagnosis and therapy. Cancer Res. 2005;65(21):9603–9606. doi: 10.1158/0008-5472.CAN-05-2782. [DOI] [PubMed] [Google Scholar]
- Oliveira SS, Morgado-Diaz JA. Claudins: multifunctional players in epithelial tight junctions and their role in cancer. Cell Mol Life Sci. 2007;64(1):17–28. doi: 10.1007/s00018-006-6314-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouban A, Ahmed AA (2010) Claudins in human cancer: a review Histology and histopathology 25:83–90. 10.14670/HH-25.83 [DOI] [PubMed]
- Philip R, Heiler S, Mu W, Buchler MW, Zoller M, Thuma F. Claudin-7 promotes the epithelial-mesenchymal transition in human colorectal cancer. Oncotarget. 2015;6(4):2046–2063. doi: 10.18632/oncotarget.2858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suh Y, Yoon CH, Kim RK, Lim EJ, Oh YS, Hwang SG, An S, Yoon G, Gye MC, Yi JM, Kim MJ, Lee SJ. Claudin-1 induces epithelial-mesenchymal transition through activation of the c-Abl-ERK signaling pathway in human liver cells. Oncogene. 2017;36(8):1167–1168. doi: 10.1038/onc.2016.294. [DOI] [PubMed] [Google Scholar]
- Tabaries S, Siegel PM. The role of claudins in cancer metastasis. Oncogene. 2017;36(9):1176–1190. doi: 10.1038/onc.2016.289. [DOI] [PubMed] [Google Scholar]
- Takasawa K, Takasawa A, Osanai M, Aoyama T, Ono Y, Kono T, Hirohashi Y, Murata M, Sawada N. Claudin-18 coupled with EGFR/ERK signaling contributes to the malignant potentials of bile duct cancer. Cancer Lett. 2017;403:66–73. doi: 10.1016/j.canlet.2017.05.033. [DOI] [PubMed] [Google Scholar]
- Tsukita S, Furuse M, Itoh M. Structural and signalling molecules come together at tight junctions. Curr Opin Cell Biol. 1999;11(5):628–633. doi: 10.1016/S0955-0674(99)00016-2. [DOI] [PubMed] [Google Scholar]
- Usami Y, Chiba H, Nakayama F, Ueda J, Matsuda Y, Sawada N, Komori T, Ito A, Yokozaki H. Reduced expression of claudin-7 correlates with invasion and metastasis in squamous cell carcinoma of the esophagus. Hum Pathol. 2006;37(5):569–577. doi: 10.1016/j.humpath.2005.12.018. [DOI] [PubMed] [Google Scholar]
- Yang Y, Yang H, McNutt MA, Xiong F, Nie X, Li L, Zhou R. LAPTM4B overexpression is an independent prognostic marker in ovarian carcinoma. Oncol Rep. 2008;20(5):1077–1083. [PubMed] [Google Scholar]
- Zavala-Zendejas VE, Torres-Martinez AC, Salas-Morales B, Fortoul TI, Montano LF, Rendon-Huerta EP. Claudin-6, 7, or 9 overexpression in the human gastric adenocarcinoma cell line AGS increases its invasiveness, migration, and proliferation rate. Cancer Investig. 2011;29(1):1–11. doi: 10.3109/07357907.2010.512594. [DOI] [PubMed] [Google Scholar]
- Zhang X, Ruan Y, Li Y, Lin D, Quan C. Tight junction protein claudin-6 inhibits growth and induces the apoptosis of cervical carcinoma cells in vitro and in vivo. Med Oncol. 2015;32(5):148. doi: 10.1007/s12032-015-0600-4. [DOI] [PubMed] [Google Scholar]