Abstract
Mantle cell lymphoma (MCL) is a comparatively rare non-Hodgkin’s lymphoma characterised by overexpression of cyclin D1. Many patients present with or progress to advanced stage disease within 3 years. MCL is considered an incurable disease with median survival between 3 and 4 years. We have investigated the role(s) of CCN1 (CYR61) and cell cycle regulators in progressive MCL. We have used the human MCL cell lines REC1 < G519 < JVM2 as a model for disease aggression. The magnitude of CCN1 expression in human MCL cells is REC1 > G519 > JVM2 cells by RQ-PCR, depicting a decrease in CCN1 expression with disease progression. Investigation of CCN1 isoform expression by western blotting showed that whilst expression of full-length CCN1 was barely altered in the cell lines, expression of truncated forms (18–20 and 28–30 kDa) decreased with disease progression. We have then demonstrated that cyclin D1 and cyclin dependent kinase inhibitors (p21CIP1and p27KIP1) are also involved in disease progression. Cyclin D1 was highly expressed in REC1 cells (OD: 1.0), reduced to one fifth in G519 cells (OD: 0.2) and not detected by western blotting in JVM2 cells. p27KIP1 followed a similar profile of expression as cyclin D1. Conversely, p21CIP1 was absent in the REC1 cells and showed increasing expression in G519 and JVM2 cells. Subcellular localization detected p21CIP1/ p27KIP1 primarily within the cytoplasm and absent from the nucleus, consistent with altered roles in treatment resistance. Dysregulation of the CCN1 truncated forms are associated with MCL progression. In conjunction with reduced expression of cyclin D1 and increased expression of p21, this molecular signature may depict aggressive disease and treatment resistance.
Keywords: CCN1, Cyclin D1, CYR61, MCL, p21CIP1, p27KIP1
Introduction
Mantle cell lymphoma (MCL) is a distinct subset of B cell Non Hodgkinʼs Lymphoma (NHL) characterised by overexpression of cyclin D1 as a result of t(11;14)(q13;q32) chromosomal translocation (Pérez-Galán et al. 2011). This translocation juxtaposes the cyclin D1 gene on chromosome 11q13 with the immunoglobulin heavy chain gene on 14q32 which leads to cyclin D1 overexpression and the dysregulation of the cell cycle (O’Connor 2007; Zucca and Bertoni 2013). The clinical course of this disease is highly variable and ranges from indolent classic morphology to aggressive variants with blastoid or pleomorphic morphology (Royo et al. 2012). MCL is characterised by almost inevitable relapses; many patients present with or progress to advanced stage disease within 3 years due to increasing resistance to chemotherapy and other agents (Dreyling et al. 2018; Liu et al. 2015). MCL is broadly considered an incurable disease with the median survival of patients being 3–4 years.
The REC1, G519, JVM2 cell line model for MCL progression
In this study, we have used three human MCL cell lines REC1, Granta 519 (G519) and JVM2 as a model for MCL disease progression. The 2016 revision of World Health Organisation classification of lymphoid neoplasms describes the variants of MCL in addition to the characteristic Cyclin D1 rearrangent (i) 2 types of clinically indolent forms with either Immunoglobulin heavy chain variable region gene (IGHV) unmutated or minimally mutated and usually with expression of SOX11 typically involving lymph nodes or extranodal sites, (ii) acquisition of additional molecular or cytogenetic abnormalities leading to an aggressive blastoid or pleomorphic phenotype and (iii) leukaemic non-nodal MCL develops from IGHV mutated and SOX11 negative B cells with usual bone marrow and / or spleen involvement where abnormalities in TP53 contributes to enhance aggressive nature (Swerdlow et al. 2016). REC1 cells display unmutated IGHV with expression of SOX11 (Beà et al. 2013) consistent with the classic indolent forms of MCL. GRANTA 519 (G 519) cells display blastoid phenotype with SOX11 positivity and amplification of BCL2 gene leading to Bcl2 overexpression enhancing cell survival associated with the aggressive blastoid variant forms (Queirós et al. 2016; Rudolph et al. 2004). JVM2 cells are IGHV unmutated and SOX11 negative, expressing low levels of cyclinD1 with increased expression of cyclin D2, BCL2 positive (Tucker et al. 2006) and is considered a blastoid variant (Camps et al. 2006) consistent with the aggressive blastoid variant, SOX11 negativity overlaps with the aggressive leukaemic non-nodal MCL forms. In addition, the cell lines show increasing resistance to lenalidomide in the order of REC1 < G519 < JVM2 (Zhang et al. 2008) with G519 and JVM2 cells also showing increased resistance to Ibrutinib (Balsas et al. 2017). REC1 cells are observed as early stage/ indolent MCL showing sensitivity to conventional therapy in contrast to G519 and JVM2 cell lines which behave consistently with aggressive stages (Nordgren et al. 2012; Rauert-Wunderlich et al. 2016).
Cyclin D1 and cell cycle progression
Overexpression of cyclin D1 (CCND1) as a result of t(11;14) chromosomal translocation is the hallmark feature of mantle cell lymphoma (Cassaday et al. 2015). Cyclin D1 plays a central role in the cell cycle regulation by binding to either cyclin-dependent kinase 4 (CDK4) or CDK6. The CCND1-CDK4 or CCND1-CDK6 complex phosphorylates the retinoblastoma protein (pRb) which leads to degradation of CCND1 suppressor effect on cell cycle progression. This process leads to release of the E2F family of transcription factors and then S-phase entry (Cassaday et al. 2015). E2F transcription factor regulates genes that encode DNA replication and cell cycle control (Nevins 2001). In MCL, CCND1 overexpression leads to hyperphosphorylate pRb and accumulation of E2F which facilitates the G1/S transition and uncontrolled cell proliferation (Cassaday et al. 2015). CCND1 is upregulated in almost all MCL patients, however cyclin D1 alone is insufficient to promote MCL. Additional oncogenic aberrations have been implicated in the generation of MCL, including, c-Myc overexpression, lack of the ataxia telangiectasia mutated (ATM) gene or p53 dysregulation (Müller et al. 2013). Many secondary genetic events are involved in MCL lymphomagenesis and include inactivation of the DNA damage response pathways, activation of cell-survival pathways, and suppression of apoptosis. Additional oncogenic aberrations are likely to contribute to the development of MCL involving cell proliferation, survival, and interactions with microenvironment (Jares et al. 2012; Müller et al. 2013). Mutations within stem cell signalling pathways have been identified that may contribute to the pathogenesis of this disease. For example, Notch1 mutations are found in 12% of MCL and are associated with poor survival, suppression of the Notch pathway in MCL, decreased cell proliferation and increased apoptosis (Kridel et al. 2012). Similarly, deregulation of the Wnt canonical pathway was found in MCL by inactivation of phospho-GSK3B suggesting that the Wnt pathway may also contribute to pathogenesis (Gelebart et al. 2008). Further investigations are required to develop novel, effective therapeutic agents for MCL.
Many studies have indicated that activation or upregulation of cell cycle regulators, p21, p27 and cyclin D1 are induced through CCN1 signalling (Sawai et al. 2007; Tong et al. 2004; Xie et al. 2004a, b), whilst the functional effect or output appears to be cell type specific. In 2014, (Saglam et al. 2014) have found that induction of cyclin D1 expression in grade ductal carcinoma in situ (DCIS) can occur through CCN1 signalling leading to cell cycle progression. CCN1 protein promoted cell cycle arrest by increased cell senescence at G0/G1 phase through activation notch-1-p21 pathway and reduced proliferation of human trophoblast cells (Kipkeew et al. 2016). Similarly, CCN1 signalling induced accumulation of p53 and p21 driving cell senescence leading to suppression of lung cancer cell proliferation (Jim Leu et al. 2013).
CCN1 (CYR61)
CCN1, a matricellular protein is involved in stem cell signalling within the haematopoietic microenvironment (McCallum and Irvine 2009; Wells et al. 2015) and has been associated with a number of haematological malignancies including acute myeloid leukaemia (AML) and multiple myeloma (MM) (Crawford and Irvine 2016). CCN1, also known as CYR61, belongs to CCN family of proteins that are comprised of a signal peptide (SP), Insulin like growth factor binding domain (IGFBP), Von Willebrand type C repeat (VWC), a hinge region leading to the Thrombospondin type I domain (TSP-1) and cysteine rich carboxyl terminal (CT) (Planque and Perbal 2003).
CCN1 has been shown to be involved in a diverse array of cellular processes including regulation of cell migration, cell adhesion, proliferation, differentiation, apoptosis and angiogenesis through direct binding to cell surface receptors such as integrins and heparan sulfate proteoglycans (HSPGs) (Jandova et al. 2012; Kireeva et al. 1996). CCN1 is a ligand for integrins and acts through direct binding to integrins (via VWC, TSP-1 and CT domains) and HSPGs in order to enhance specific functions (Leu et al. 2004). Increasing evidence has shown that CCN1 promotes cell adhesion by binding to integrin α6β1-HSPG co-receptors in fibroblasts (Todorovicç et al. 2005), αMβ2 in murine macrophages (Bai et al. 2010) αDβ2 in macrophage foam cells (Yakubenko et al. 2006), α‖bβ3 in activated platelets (Jedsadayanmata et al. 1999). CCN1 may interact with other growth factors such as bone morphogenetic proteins (BMP), transforming growth factor β (TGF-β) and vascular endothelial growth factor (VEGF) which need more investigation (Lau 2011). Recently, it has been shown that ALK5 suppression prevents TGF-β-induced CCN1 expression in human dermal fibroblasts (Thompson et al. 2014). More importantly, CCN1 is a transcriptional target of TGF-β and may potentiate an autocrine regulatory mechanism in tumourigenesis (Bartholin et al. 2007) and is similarly a target of the Wnt B catenin pathway as demonstrated in mesenchymal stem cell differentiation to osteoblasts and hepatocellular carcinoma (Si et al. 2006; Li et al. 2012). Constitutive activation of the Wnt Signalling pathway was also identified in MCL (Gelebart et al. 2008), with MCL initiating cells (MCL-ICs) displaying activation of the Wnt signalling pathway (Samaniego et al. 2014). Targeting of Wnt signalling specifically reduces the growth of MCL-ICs (Mathur et al. 2015).
CCN1 can be observed in cancer as “a double-edged sword” (Lau 2011) where altered CCN1 expression in various cancers may induce or suppress tumour growth (Feng et al. 2008; Holloway et al. 2005). CCN1 plays unique roles in different cancers: it is a tumour promoter in cancers of the breast (Menendez et al. 2005), prostate (Sun et al. 2008), pancreatic (Maity et al. 2014), gastric carcinogenesis (Cheng et al. 2014), ovarian carcinoma (Gery et al. 2005), colorectal (Jeong et al. 2014), myeloma (Roodman 2014) and acute myeloid leukaemia (AML) (Niu et al. 2014) due to either overexpression of CCN1 or truncated isoforms. Paradoxically, CCN1 is a tumour suppressor in melanoma (Dobroff et al. 2009), non-small cell lung cancer (Tong et al. 2001) and endometrial adenocarcinoma (Chien et al. 2004). Full-length CCN proteins can play an anti-proliferative role, while truncated isoforms may induce tumour proliferation (Planque and Perbal 2003). For example, when CCN1 is cleaved by plasmin, this releases a truncated protein of CCN1 (28 kDa) supporting endothelial cell migration in breast carcinoma cells (Pendurthi et al. 2005). In 2013, (Choi et al. 2013) identified truncated CCN1 in the vitreous fluid secretome of proliferative diabetic retinopathy patients rather than full-length protein. Furthermore, a truncated CCN1 protein which includes complete or partial length forms of the two-modules form IGFBP-VWC is generated by proteolytic cleavage of matrix metalloproteinase MMPs (Choi et al. 2013). In addition to proteolysis generating production of a CCN1 truncated isoform, it may also be generated by alternative mRNA splicing (Perbal 2009).
Increasing evidence suggests that CCN1 plays important roles in tumour development, including migration, survival, proliferation and metastasis (Sun et al. 2008). In AML, CCN1 induces tumour survival through activation of the Ras/Raf/MEK/ERK pathway by up regulating c-Myc and Bcl-xL and by down regulating Bax (Niu et al. 2014). Similarly, CCN1 confers human breast cancer resistance to chemotherapeutic agent-induced apoptosis and suppresses apoptosis by activating the integrins/NF-kb/XIAP signalling pathway (Lin et al. 2004). CCN1 has been found in sites of bone remodelling, involvement enhancing osteoblast differentiation while suppressing osteoclast formation (Crockett et al. 2007; Si et al. 2006).
Furthermore, CCN1 has been implicated in Wnt3A-induced osteoblast differentiation of mesenchymal stem cells (Si et al. 2006). In vivo, (Johnson et al. 2014) have reported that overexpression of CCN1 which is produced by mesenchymal cells in bone marrow microenvironment of patients with monoclonal gammopathy of undetermined significance (MGUS), asymptomatic myeloma (AMM), and multiple myeloma (MM) suppressed tumour cell growth and reduced MGUS progression to MM. However, CCN1 may enhance myeloma cell viability through supporting survival of the INA-6 myeloma cell line lacking interleukin-6(IL-6) (Dotterweich et al. 2014).
Many studies have indicated that activation or upregulation of cell cycle regulators, p21, p27 and cyclin D1 induced through CCN1 signalling (Sawai et al. 2007; Tong et al. 2004; Xie et al. 2004a, b). In 2014, (Saglam et al. 2014) have found that induction of cyclin D1 expression in grade ductal carcinoma in situ (DCIS) can occur through CCN1 signalling leading to cell cycle progression. However, CCN1 protein promoted cell cycle arrest by increased senescence-cell at G0/G1 phase through activation notch-1-p21 pathway and reduced cell proliferation of human trophoblast cells (Kipkeew et al. 2016). Moreover, CCN1 signalling induced accumulation of p53 and p21 driving cell senescence leading to suppression of lung cancer cell proliferation (Jim Leu et al. 2013).
p21
p21CIP1 (p21) belongs to the CIP/KIP family of proteins that regulate cell cycle progression. The CIP/KIP family comprises three members; p21CIP1 (Cdk Interacting Protein 1), p27KIP1 (Kinase Inhibitory Protein 1) and p57KIP2 (Kinase Inhibitory Protein 2) (Lu and Hunter 2010; Bretones et al. 2015) which bind and inhibit most cyclin-CDK complexes. The CIP/KIP family of proteins, particularly p21, plays an essential role in cell cycle control, halting the transition from G1 phase to S phase (Pérez-Sayáns et al. 2013). Two pathways regulate p21; a p53-dependent pathway (in response to DNA damage which activates p53 leading to upregulation of p21 and repression of cell growth in G1 phase with potential DNA repair or stimulation of programmed cell death) and a p53-independent pathway, in which cellular growth factors regulate p21 expression (Brennan et al. 2002; Ciccarelli et al. 2005). p21 binds to CCND1-CDK4 or -CDK6 complex and inhibits the kinase activity of CDKs in response to many stimuli leading to regulation G1/S progression, at the restriction point. This process leads to repression of phosphorylation of pRb protein which in turn prevents the expression of E2F factors and blocks G1/S transition (Zhang and Yan 2012).
In addition to negative regulation of the cell cycle, p21 can regulate gene transcription; p21 suppresses E2F transcription factor through a pathway independent of CDKs or Rb (Perkins 2002). p21 is involved in controlling cellular growth by suppressing of E2F1 transcription factor via Wnt4 expression and Notch 1 activation (Devgan et al. 2005) and p21 stimulates NFkB-mediated transcription by activation of p300 and CBP (Perkins 2002). Studies have identified additional roles for p21, in cancer it is a tumour suppressor through cell cycle arrest and blocking DNA synthesis by binding to proliferating cell nuclear antigen (PCNA) (Waga et al. 1994) or as oncogenic factor, promoting carcinogenesis and tumour development (Roninson 2002). The expression of p21 varies in different cancers, it is down regulated in small-cell lung (Komiya et al. 1997), colorectal (Zirbes et al. 2000), cervical (Lu et al. 1998a), and head and neck cancer (Kapranos et al. 2000) that are associated with tumour progression. In contrast, it is upregulated in prostate (Baretton et al. 1999), ovarian (Ferrandina et al. 2000), breast, oesophageal squamous cell carcinomas (Queiroz et al. 2010; Nemes et al. 2005), and in brain tumours (Roninson 2002). The function of p21 depends on subcellular localization which may be nuclear, cytoplasmic or mitochondrial (Picolo and Crispi 2012). p21 regulates cell proliferation and differentiation by localizing in the nucleus (Abbas and Dutta 2009), whilst p21 enables resistance to DNA damage by inhibiting proteins essential for apoptosis by localising to the cytoplasm or mitochondria, enhancing tumourigenesis (de Renty et al. 2014; Sohn et al. 2006).
p27
In G1 phase of cell cycle, reviewed in (Hirama and Koeffler 1995) p27 binds to CCNE-CDK2 complex and inhibits the catalytic activity of CDK2 resulting in cell cycle arrest at the restriction point in response to DNA damage or anti-mitogenic signals. As a result, this prevents the phosphorylation of pRb which leads to block in the transcription of genes required for G1/S progression (Toyoshima and Hunter 1994). Beyond the restriction point, cell cycle proceeds independent of mitogenic signals (Coats et al. 1996).
Interestingly, p27KIP1 and p21CIP1 have an important role in promotion of assembly of CCND-CDK4/6 complexes (LaBaer et al. 1997). This interaction leads to sequestration of p27 in CCND-CDK4 complex which blocks inhibition of the CCNE-CDK2 complex (Perez-Roger et al. 1999). Binding of p27KIP1 with CCND-CDK4 complex suppresses the kinase activity of CDK4 (Ray et al. 2009). Expression of p27KIP1 is reduced in several types of cancer associated with poor prognosis including breast (He et al. 2012), prostate (Roy et al. 2008), lung and colon cancer (Timmerbeul et al. 2006). In contrast, p27KIP1 is overexpressed in hepatocellular carcinoma (HCC) which is associated with longer disease free survival (Qin and Ng 2001). In MCL, Quintanilla-Martinez et al. (2003) suggested overexpression of cyclin D1 contributed to a change in p27KIP1 levels leading to inhibition of cellular growth.
In addition to cell cycle regulation, there is some evidence that p27KIP1 has important roles in apoptosis, transcriptional activation, and migration depending on its localization. In the nucleus, it has an essential role to inhibit cell growth and is considered as a tumour suppressor (Jeannot et al. 2015). Phosphorylation of specific sites on p27KIP1 leads to its export into the cytoplasm where it can act as a tumour promotor as reviewed in (Besson et al. 2008). Many studies have shown that cytoplasmic p27KIP1 in cancer including melanoma (Chen et al. 2011; Denicourt et al. 2007), ovarian carcinoma (Duncan et al. 2010), renal cell carcinoma (Kruck et al. 2012), osteosarcoma (Li et al. 2016) and acute myelogenous leukaemia (Min et al. 2004) is associated with cell migration, high tumour grade and metastasis, poor prognosis and survival. Moreover, cytoplasmic p27KIP1 contributed to treatment resistance mediated suppression of apoptosis in Her2+ breast cancer cells (Zhao et al. 2014).
This study investigated the expression of CCN1 and cell cycle regulators Cyclin D1, p21 and p27 in progressive MCL.
Materials and methods
Cell culture
Human Mantle Cell Lymphoma cell lines REC1, G519 and JVM2 were purchased from the Deutsche Sammlung von Mikroorganismen and Zellkulturen (DSMZ-Germany). All cell lines were cultured in Roswell Park Memorial Institute (RPMI) 1640 (Gibco) supplemented with 10% FBS (Gibco) and were incubated at 37 °C in a humidified atmosphere of 5% CO2. Cells were passaged twice weekly to maintain log phase. Experiments were conducted within 10 passages from cell recovery and cells were seeded at 2 × 105 cells per ml for experimental procedures. Cells were counted using a haemocytometer and viability assessed using the trypan blue exclusion assay.
Normal peripheral blood (nPB) samples were obtained from Normal blood donors (n = 5) via NHS Blood and Transplant Bristol (NHSBT, Bristol, UK) with material transfer agreement and approval for use via Plymouth University ethics committee. Lymphocytes were extracted using Lymphoprep™ (Axis Shield, UK) using manufacturer’s instructions and cells harvested in Trizol® reagent (Fisher Scientific, UK) for RNA analyses.
Protein extraction and quantification
Total cell lysates for three cell lines (REC1, G519 and JVM2) were prepared in Radioimmune Precipitation Assay buffer (RIPA): Tris 50 mM, NaCl 150 mM, Triton X-100 1%, Na-Deoxycholate 0.5%, SDS 0.1% supplemented with Complete™ protease inhibitor (Roche UK) and stored at −20 °C until required.
Subcellular fractions
Nuclear, mitochondria and cytoplasmic proteins from REC1, G519 and JVM2 cell lines were extracted using the Cell Fraction Kit-Standard (Abcam, UK) according to manufacturer instructions with the following modifications: reduced cell number to 1 × 106/ml incubated with 500 μl of buffer A (1X) and samples were incubated for 8 min to extract clean mitochondrial fractions. Buffer A (1X) was prepared using 5 ml 2X buffer A stock, 4900 μl dH2O and 100 μl PI 100X (Halt Protease Inhibitor Single-Use Cocktail EDTA-Free, ThermoScientific). All fractions were stored at −80 °C until required.
Protein was quantified using the micro BCA™ Assay Kit following the supplier instructions (Pierce, ThermoScientific, UK). The subcellular fractions for nuclear, cytoplasmic and mitochondrial extracts were run simultaneously on one gel (NuPage 10 well 10% Bis Tris gel (Invitrogen, UK)) to enable direct comparison. Experiments were conducted in triplicate (n = 3).
Western blotting
Protein samples (10 μg) were loaded on a 10% NuPAGE® Bis-Tris Gels (Invitrogen UK) and transferred onto a PVDF membrane (Millipore, UK). Membranes were blocked with 2.5% skimmed milk for 30 min. Membranes were then incubated overnight at 4 °C with primary antibody (rabbit anti-p21CIP1 (2947S) (1:1000, Cell Signalling Technology), rabbit anti-p27KIP1 (3688 s) (1:1000, New England Biolabs), mouse cyclin D1 (1:1000, Cell Signalling Technology), rabbit anti-CCN1 (ab24448, Lot # GR26258–7) (1:2000, Abcam), anti-GAPDH (ab9485) (1:2500, Abcam), mouse anti-COX IV (ab33985) (1:1000, Abcam), rabbit anti Histone H3 (9717 s)(1:1000, New England Biolabs)). Membrane was washed with PBS-T and incubated with an appropriate secondary antibody conjugated to HRP. Bands were detected using chemiluminesence (Thermo Scientific SuperSignal®West Dura Extended Duration Substrate) and viewed using the Image Quant LAS4000.
RNA extraction and cDNA synthesis
Total RNA was extracted using Trizol® reagent (Fisher Scientific, UK). Trizol (1 ml for 1 × 106 cells) was pipetted up and down several times until the samples were homogenised. 200 μl of chloroform was added per ml of Trizol®, shaken vigorously for 15 s and incubated at room temperature for 2–3 min. Samples were centrifuged at 13000 rpm for 15 min at 4 °C. The upper layer was decanted to new tubes and 500 μl isopropanol added and incubated at room temperature for 10 min. Samples were centrifuged at 13000 rpm for 15 min at 4 °C. Supernatant was discarded and the white RNA pellet washed three times with 70% ethanol. RNase free water was used to re-suspend the pellet. Samples were stored at −80 °C until required. RNA quantity and purity was measured using a nanodrop spectrophotometer using A260/A280 ratios (Thermo-Fisher scientific, Waltham, MA, USA). cDNA was synthesised using 2 μg of total RNA in 20 μl volume using the High Capacity RNA to cDNA kit (Applied Biosystems) according to the manufacturer’s instructions. cDNA samples were stored at −20 °C until required. Negative template controls consisting of reaction without cDNA were run for all experiments.
RQ-PCR
The quantitative real-time PCR was performed by using StepOne Software v2.3 analytical software (Thermo-cycler 96 well plate Real time PCR, Applied Biosystems, US) in a volume of 12.5 μl containing 1 μl cDNA (100 ng), 0.625 μl ppp, 6.25 μl 2xMM (Prime time Master Mix, Integrated DNA technologies). The PCR reaction conditions were 95 °C for 3 min, followed by 40 cycles comprising denaturation at 95 °C for 5 s, annealing at 60 °C for 30 s. Predesigned assay reagents with FAM/ TAMRA fluorescence were used for CCN1 (Hs00155479_m1), GAPDH ((PT.39a.22214836) from Integrated DNA Technologies), p21 (00355782-M1), p27 (01597588-M1) and cyclin D1 ((00765553-M1) from Applied Biosystems). GAPDH used as an endogenous control, gene expression levels were reported using the ∆∆CT method with experimental samples run in triplicate and independent replicates of RNA completed in triplicate for reporting.
Statistics
Statistical analysis was performed on data from at least 3 independent experiments. For RQ-PCR, samples were run in triplicate and then independent replicates performed in triplicate for publication. Students t-test was performed to identify significance between samples and where p < 0.05 was deemed significant.
Results
CCN1 expression is inversely correlated with MCL aggressiveness
In order to assess the potential role(s) of CCN1 in MCL, we have investigated CCN1 expression using RQ-PCR and Western blotting. REC1, G519, JVM2 human MCL cell lines were used as a model for MCL disease progression in the order from indolent to aggressive disease REC1 < G519 < JVM2 respectively.
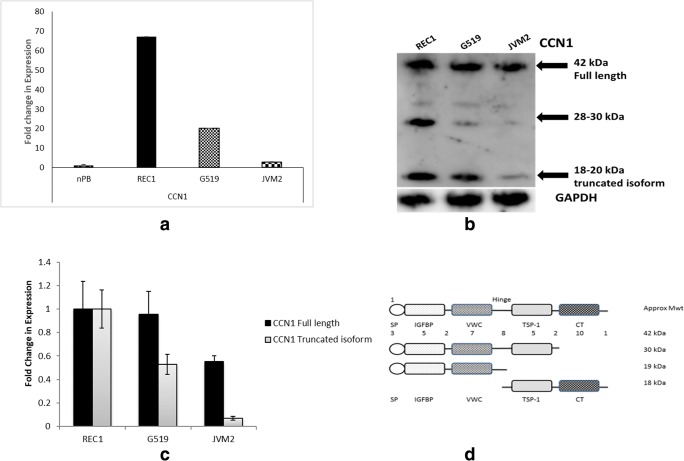
RQ-PCR for CCN1 expression shows an inverse relationship with disease aggression. Normal peripheral blood (nPB) lymphocytes from donors were also assessed for CCN1 expression. CCN1 was barely detected in nPB (CT = 34.9 ± 0.75) and therefore set to a fold change of 1 to asses relative expression within the MCL cell lines. CCN1 expression was high in REC1 cells (Fold change 67.18 ± 0.14) and sequentially decreased in progressive G519 cells (Fold change 20.1 ± 0.22) and JVM2 cells (Fold change 2.86 ± 0.52) (Fig. 1a).
Fig. 1.
CCN1 expression in MCL progression. a RQ-PCR screen of MCL cell lines for CCN1 expression in normal peripheral blood (nPB), low-aggression phenotype (REC1) and in aggressive disease (G519 and JVM2). b Western blot showing CCN1 expression for the MCL cell lines, REC1, G519 and JVM2 c Optical densitometry for CCN1 band pattern by Western Blotting. d CCN1 domain structure of full length and potential truncated CCN1 proteins adapted from Choi et al. 2013. GAPDH was used as a loading control, data generated were normalised against GAPDH control (n = 3 independent samples) and (nPB, n = 5 independent samples)
Investigation of CCN1 total protein expression using western blot analysis showed CCN1 protein expression at the following approximate molecular weights; 42 kDa consistent with expression of full length CCN1, 28-30 kDa and 18-20 kDa consistent with expression of truncated proteins. Expression of full–length CCN1 barely altered through the cell lines however, expression of the truncated form (18-20 kDa) was high in REC1 cells (OD:1.0) reduced in G519 cells (OD:0.5) and barely detected in JVM2 cells (Fig. 1b and c). Reports from a previous study (Choi et al. 2013) suggests the 28-30 kDa moiety could comprise the SP, IGFBP, VWC and TSP-1 domains whilst the 18–20 kDa moiety could be either the SP, IGFBP, VWC fragment or TSP-1 and CT fragment, or potentially a mix of both (Fig. 1d).
Cyclin D1 is downregulated in progressive MCL
We have investigated cyclin D1 expression using RQ-PCR and Western blotting using total protein extracts from the three MCL cell lines; REC1, G519 and JVM2. RQ-PCR shows cyclin D1 expression is high in REC1 cells and decreased in G519 and JVM2 cells consistent with deregulation of cyclin D1 in aggressive disease (Fig. 2a). Fold changes in cyclin D1 expression were 10.1, 4.6 and 1.0 for REC1, G519 and JVM2 respectively.
Fig. 2.
Cyclin D1, p21 and p27 expression in MCL progression. RQ-PCR of cyclin D1 (a), p21 (d) and p27 (g) in MCL for REC1, G519 and JVM2 cells. Western blotting for expression of total protein for cyclin D1 (b), p21 (e) and p27 (h) in MCL for REC1, G519 and JVM2 cells. Optical densitometry for western blotting of cyclin D1 (c), p21 (f) and p27 (i). Densitometry was performed on banding and data generated normalised against GAPDH loading control (n = 3 independent replicates)
Cyclin D1 total protein expression mirrored that of the gene expression where cyclin D1 was highly expressed in the Rec1 cells (OD: 1.0), reduced to one fifth in the G519 cells (OD: 0.2) and not detected by western blotting in the JVM2 cell line (Fig. 2b and c).
High expression of p21CIP1 and down expression of p27KIP1 in aggressive MCL
Cyclin dependent kinase inhibitors, p21CIP1 and p27KIP1, were also investigated for an involvement in disease progression.
RQ-PCR shows increasing expression of p21 with disease progression whilst expression levels of p27 are decreased with disease progression in G519 and JVM2 cells (Fig. 2d). Total cell lysates showed that p21CIP1 was not detected in REC1 cells but had increasing expression in G519 (OD: 0.6) and JVM2 cells (OD: 1.0) (Fig. 2e and f). Whilst RQ-PCR and western blotting show that p27KIP1 expression was high in the REC1 and decreased with disease progression in G519 and JVM2 cells (Fig. 2g–i). Total cell lysates showed that p27KIP1 was highly expressed in REC1 (OD: 1.5), decreased in G519 (OD: 1.0) and not detected in JVM2 cells (Fig. 2h and i).
Altered subcellular localization of p21CIP1 and p27KIP1 portray resistance in progressive MCL
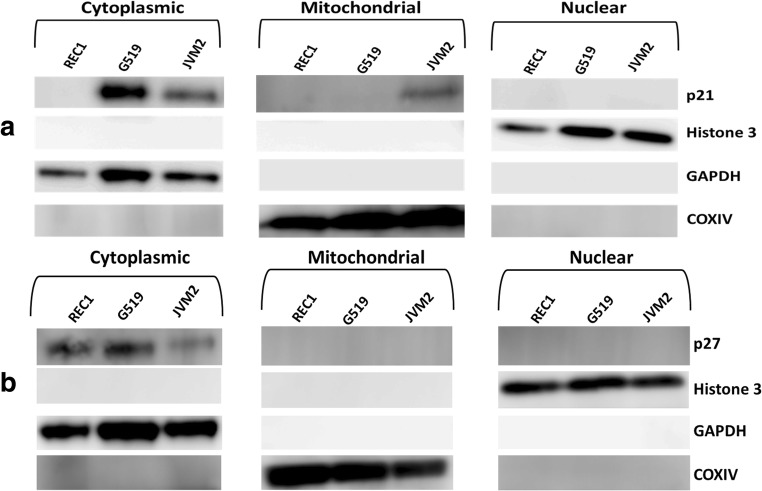
To investigate the subcellular localization of p21CIP1 and p27KIP1 in MCL, we extracted cytoplasmic, mitochondrial and nuclear protein from cells and performed western blotting.
Expression of p21CIP1 was not detected in any fraction for early stage REC1 cells. For the progressive G519 and JVM2 cells, p21CIP1 was primarily expressed in the cytoplasm and was not detected in the nucleus (Fig. 3a). Expression of p21CIP1 was detected in the mitochondrial fraction for JVM2 cells. Blotting for GAPDH (cytoplasmic), COX IV (Mitochondrial) and Histone 3 (Nuclear) markers were used as controls for each fraction.
Fig. 3.
Subcellular localisation of p21 and p27 in MCL. a Western blot for p21 cellular content for cytoplasmic, mitochondrial and nuclear fractions for REC1, G519 and JVM2 cells. b Western blot for p27 cellular content for cytoplasmic, mitochondrial and nuclear fractions for REC1, G519 and JVM2 cells. GAPDH, Histone 3 and COXIV were used as a loading control and to ensure clean fractions free from cross contamination during the fractionation process were obtained (n = 3 independent samples)
For p27KIP1, expression was only detected in the cytoplasmic fraction and not in mitochondrial or nuclear fractions (Fig. 3b). GAPDH (cytoplasmic), COX IV (Mitochondrial) and Histone 3 (Nuclear) markers were used as loading controls and to ensure clean fractions were obtained for each, without cross contamination during the fractionation process.
Discussion
MCL is considered an aggressive disease and has one of the worst outcomes among B cell lymphomas due to the dysregulation of the DNA damage response pathways accompanied with abnormal cell survival mechanisms suppressing apoptosis (Campo and Rule 2015; Moros et al. 2014). MCL characterised by overexpression of cyclin D1 as a result of t(11;14) chromosomal translocation that leads to dysregulation of the cell cycle (O’Connor 2007; Zucca and Bertoni 2013). Furthermore, MCL is characterised by clinical course where variants are subdivided into indolent form (classic morphology) and aggressive (blastoid or pleomorphic appearance) (Kridel et al. 2012). The heterogeneous biology and aggressive behaviour of MCL present a challenge for designing standard therapies (Smith 2011) and therefore require further investigation to identify more effective treatment strategies.
In this study, we have investigated the role(s) of CCN1 in MCL and investigated cell cycle regulation using expression of cyclin D1, p21CIP1 and p27KIP1 in the cell line model representing disease progression in MCL where the magnitude of aggressive behaviour is in the order REC1 < G519 < JVM2. CCN1 dysregulation was identified in MCL progression where CCN1 was highly expressed in the REC1 cells and reduced in the aggressive G519 and JVM2 cells. CCN1 expression decreases with disease progression. We found that the lower expression of CCN1 in aggressive MCL cell lines (G519 and JVM2) is additional risk factor for disease progression. However, many studies have indicated that the high expression of CCN1 is implicated in disease progression, tumorigenesis and invasion of hepatocellular carcinoma (HCC) (Li et al. 2012), breast cancer (O'Kelly et al. 2008), prostatic carcinoma (D'Antonio et al. 2010), gliomas (Xie et al. 2004b) and gastric cancer (Lin et al. 2007).
Further investigation demonstrated that whilst full-length CCN1 remains relatively constant within the cell lines, the 18–20 kDa truncated form is decreased within aggressive G519 and JVM2 cells. Previous reports identify that this truncated form could potentially be a fragment consisting of the SP-IGFBP-VWC domains or the TSP-1-CT domains (Choi et al. 2013) inferring that either transcriptional control or post-translational control of CCN1 (via degradation by MMP2 / MMP14 for example (Choi et al. 2013)) is lost within aggressive MCL. In 2006, Planque et al. 2006, have found that full length of CCN3 inhibited cell growth whilst a truncated isoform induced “morphological transformation of chicken embryo fibroblasts” suggesting a role in oncogenic activity. The truncated isoform of CCN3 translocated to the nucleus of cancer cell lines supporting the role of the carboxyl terminal being involved in transcriptional regulation. Increasing evidence indicates the Nuclear Localisation Signal (NLS), a short peptide motif mediating nuclear localisation of proteins, is located in the CT module of CCN3 (lysine-rich PTDKKGKKCLRTKKSLKA) (Planque 2006). Perbal (1999) found nuclear localisation of the truncated isoform of CCN3 (NOV) protein (31/32 kDa) in the nucleus of 143 and HeLa cells. It is suggested the truncated isoform may be deficient of N-terminus to have a role in the gene expression of target cells since antibodies against the C-terminus of CCN3 / NOV were used (KKGKKCLRTKKS). CCN1 (CYR61) and CCN3 (NOV) are members in the CCN family of matricellular proteins sharing 40%–60% amino acid homology which may infer potential for the CCN1 truncated protein to have nuclear localisation and role(s) in gene regulation (Perbal 1999; Planque and Perbal 2003). Supporting this, the CCN3 NLS region (PTDKKGKKCLRTKKSLKA)(Planque 2006) is highly conserved in the CCN1 CT domain (KKGKKCSKTKKS). This may suggest that the CCN1 truncated form (18–20 k Da) may translocate to the nucleus of MCL cell lines. This is consistent with CCN1 found in the nucleus of bladder smooth muscle cells (Chen and Du 2007; Tamura et al. 2001).
Full length CCN proteins can play an anti-proliferative role, whilst truncated isoforms may induce tumour proliferation (Planque and Perbal 2003). The truncated isoform appears to have altered biological function(s) owing to CCN1 partition into the soluble phase (28 kDa), diffusing freely within tissue and may act as an antagonist towards the full-length CCN1 form within the insoluble matrix (42 kDa) (Pendurthi et al. 2005). For example, CCN1 is cleaved by plasmin and releases a truncated form of CCN1 (28 kDa) which may support endothelial cell migration in breast carcinoma (Pendurthi et al. 2005). In 2013, (Choi et al. 2013), found the CCN1 truncated form (11-23 kDa) expressed in diabetic retinopathy patients instead of the full-length 42 kDa protein. It is also postulated that a truncated isoform of CCN1 can arise due to alternative mRNA splicing (Perbal 2009).
In this study, Cyclin D1 was also deregulated in MCL and our findings are consistent with previous reports of cyclin D1 down regulation and disease progression (Peng et al. 1998). Interestingly, Saglam et al. (2014) have demonstrated that the up regulation of cyclin D1 and p53 are activated by the CCN1 pathway in high-grade ductal carcinoma in situ (DCIS). Consistent with these findings, we showed a positive association between CCN1 and cyclin D1 expression in all MCL cell lines. In other solid tumours, where cyclin D1 is overexpressed; breast, liver, lung, and brain cancer (Gillett et al. 1996; Hall and Peters 1996; Molenaar et al. 2008) requires consistent signalling from the extracellular matrix and growth factors (Assoian and Klein 2008).
MCL progression involving down regulation of cyclin D1 and the up regulation of p21CIP1 may contribute to treatment resistance (Abukhdeir and Park 2008). We have shown that p21CIP1 levels increase with disease progression. This is consistent with other studies that report overexpression of p21CIP1 was correlated with tumour progression; in breast (Ceccarelli et al. 2001) and ovarian carcinoma (Ferrandina et al. 2000) and in brain tumours (Jung et al. 1995).
We have shown that p27KIP1 levels decrease with disease progression also consistent with findings from Izban et al. (2000), where p27KIP1 was overexpressed in early stage disease (typical MCL) and down regulated in aggressive stage (blastoid variants) where it was associated with a high proliferation rate of blastoid MCL. Conversely, in 1998 (Quintanilla-Martinez et al. 1998) showed expression of p27KIP1 was inversely associated with the proliferation rate of MCL cells; undetected in typical MCL cells (classic disease) associated with low proliferation rate but was overexpressed in the blastic variant of MCL cells (aggressive disease) with higher proliferation rate.
P27KIP1 and p21CIP1 have important roles in promotion of assembly of CCND-CDK4/6 complexes (LaBaer et al. 1997). This interaction leads to sequestration of p27KIP1 in CCND-CDK4 complex which blocks inhibition of the CCNE-CDK2 complex (Perez-Roger et al. 1999). Furthermore, investigation of p21CIP1 expression in MCL progression showed down regulation at early stage disease and overexpression at advanced stage that mirrors its roles in MCL progression.
More importantly, in cancer, the tumour suppressor function of p21CIP1 and p27KIP1 depends on their nuclear localization (Jeannot et al. 2015; Romanov et al. 2012). Many studies have found that phosphorylation of specific sites of p27KIP1 and p21CIP1 lead to their export into the cytoplasm where they can act as a tumour promotors and induce drug resistance (Ohkoshi et al. 2015; Zhao et al. 2014). In this study, p21CIP1 and p27KIP1 were found in the cytoplasmic fractions and absent in nuclear fractions of all three cell lines (REC1, G519 and JVM2). This suggests in MCL, p21CIP1 and p27KIP1 have lost their tumour suppressor roles and acquired tumour promoter roles by localisation to the cytoplasm. Whilst mutation of p21CIP1 has not been investigated here, mutation of the p21CIP1 gene frequently occurs in cancer cells leading to inactivation of p21CIP1 with loss of function to block the cell cycle, even when overexpressed (Lu et al. 1998b; Lukas et al. 1997). While regulation of p27KIP1 is different from other cell cycle inhibitors, p27KIP1 gene mutation is rare (Garrett-Engele et al. 2007).
CCN1 likely plays key role(s) in haematopoiesis and in B cell development through modulation of stem cell signalling pathways, TGF β, BMP, Notch, Wnt-β catenin (McCallum and Irvine 2009; Wells et al. 2015). CCN1 roles within haematological malignancy show CCN1 promotes survival and inhibits apoptosis in AML (Niu et al. 2014) and overexpression of CCN1 in multiple myeloma (MM) postponed tumour growth and suppressed bone destruction (Johnson et al. 2014). CCN1 signalling involves many stem cell pathways active within the bone marrow microenvironment where haematopoiesis ensues; CCN1 can activate the Wnt-β catenin-TCF4 signalling pathway in glioma cell (Xie et al. 2004a) and induces Wnt3A osteoblast differentiation of mesenchymal stem cells (Si et al. 2006). CCN1 in some cancers plays important roles in enhancing apoptosis, suppressing tumour growth, such as non-small-cell lung cancer (NSCLC) cell lines through activating the β-catenin-c-myc-p53-p21 signalling pathway (Tong et al. 2004). Moreover, CCN1 enhances pancreatic cancer cell motility in vitro and cell tumorgenic growth in vivo by regulating sonic-Hedgehog through integrin-Notch-signalling pathway (Haque et al. 2012).
In conclusion, CCN1 expression appears to be regulated in MCL, where reduced expression of the truncated forms (18–20 and 28–30 kDa) is associated with aggressive disease. In combination with reduced expression of cyclin D1 and increased expression of p21, this molecular signature may depict aggressive disease and treatment resistance. Further investigation will ascertain CCN1 role(s) in MCL progression.
Acknowledgements
We would like to thank the Iraqi Government for funding this work.
Abbreviations
- MCL
Mantle cell lymphoma
- NHL
Non Hodgkin’s Lymphoma
- CCND1
Cyclin D1
- CDK4 or 6
Cyclin-dependent kinase 4 (CDK4) or cyclin-dependent kinase 6
- pRb
Retinoblastoma protein
- ATM
Ataxia telangiectasia mutated
- CYR61
Cysteine-rich protein 61
- AML
Acute myeloid leukaemia
- SP
Signal peptide
- IGFBP
Insulin like growth factor binding domain
- VWC
Von Willebrand type C repeat
- TSP-1
Thrombospondin type 1 domain
- CT
Cysteine rich carboxyl terminal
- HSPGs
Heparan sulfate proteoglycans
- BMP
Bone morphogenetic protein
- TGF-β
Transforming growth factor β
- VEGF
Vascular endothelial growth factor
- MMPs
Matrix metalloproteinases
- MM
Multiple myeloma
- OSC
Oesophageal squamous carcinoma
- MEK/ERK
Mitogen-activated protein kinase/extracellular-signal-regulated kinase
- OAFs
Osteoclast activating factors
- OBIs
Osteoblast inhibitors
- IL-6
Interleukin-6
- p21CIP1
(Cdk Interacting Protein 1)
- p27KIP1
(Kinase Inhibitory Protein 1)
- p57KIP2
(Kinase Inhibitory Protein 2)
- TNBC
Triple negative breast carcinoma
- FOXO3a
Forkhead box O3
- PCNA
Proliferating cell nuclear antigen
- OSCC
Oral squamous cell carcinoma
- HCC
Hepatocellular carcinoma
- NLS
Nuclear Localisation Signal
- DCIS
High-grade ductal carcinoma in situ
- NSCLC
Non-small-cell lung cancer
References
- Abbas T, Dutta A. p21 in cancer: intricate networks and multiple activities. Nat Rev Cancer. 2009;9(6):400–414. doi: 10.1038/nrc2657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abukhdeir AM, Park BH. P21 and p27: roles in carcinogenesis and drug resistance. Expert Rev Mol Med. 2008;10:e19. doi: 10.1017/S1462399408000744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Assoian RK, Klein EA. Growth control by intracellular tension and extracellular stiffness. Trends Cell Biol. 2008;18(7):347–352. doi: 10.1016/j.tcb.2008.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai T, Chen C-C, Lau LF. Matricellular protein CCN1 activates a proinflammatory genetic program in murine macrophages. J Immunol. 2010;184(6):3223–3232. doi: 10.4049/jimmunol.0902792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balsas P, Esteve-Arenys A, Roldán J, Jiménez L, Rodríguez V, Valero JG, Chamorro-Jorganes A, de la Bellacasa RP, Teixidó J, Matas-Céspedes A, Moros A, Martínez A, Campo E, Sáez-Borderías A, Borrell JI, Pérez-Galán P, Colomer D, Roué G. Activity of the novel BCR kinase inhibitor IQS019 in preclinical models of B-cell non-Hodgkin lymphoma. J Hematol Oncol. 2017;10(1):80. doi: 10.1186/s13045-017-0447-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baretton G, Klenk U, Diebold J, Schmeller N, Löhrs U. Proliferation-and apoptosis-associated factors in advanced prostatic carcinomas before and after androgen deprivation therapy: prognostic significance of p21/WAF1/CIP1 expression. Br J Cancer. 1999;80(3–4):546–555. doi: 10.1038/sj.bjc.6690390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartholin L, Wessner LL, Chirgwin JM, Guise TA. The human Cyr61 gene is a transcriptional target of transforming growth factor beta in cancer cells. Cancer Lett. 2007;246(1):230–236. doi: 10.1016/j.canlet.2006.02.019. [DOI] [PubMed] [Google Scholar]
- Beà S, Valdés-Mas R, Navarro A, Salaverria I, Martín-Garcia D, Jares P, Giné E, Pinyol M, Royo C, Nadeu F, Conde L, Juan M, Clot G, Vizàn P, Croce LD, Puente DA, López-Guerra M, Moros A, Roue G, Aymerich M, Villamor N, Colomo L, Martínez A, Valera A, Martín-Subero JI, Amador V, Hernàndez L, Rozman M, Enjuanes A, Forcada P, Muntañola A, Hartmann EM, Calasanz MJ, Rosenwald A, Ott G, Hernàndez-Rivas JM, Klapper W, Siebert R, Wiestner A, Wilson WH, Colomer D, López-Guillermo A, López-Otin C, Puente XS, Campo E. Landscape of somatic mutations and clonal evolution in mantle cell lymphoma. Proc Natl Acad Sci U S A. 2013;110(45):18250–18255. doi: 10.1073/pnas.1314608110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Besson A, Dowdy SF, Roberts JM. CDK inhibitors: cell cycle regulators and beyond. Dev Cell. 2008;14(2):159–169. doi: 10.1016/j.devcel.2008.01.013. [DOI] [PubMed] [Google Scholar]
- Brennan P, Palacios-Callender M, Umar T, Tant S, Langdon J. Expression of type 2 nitric oxide synthase and p21 in oral squamous cell carcinoma. Int J Oral Maxillofac Surg. 2002;31(2):200–205. doi: 10.1054/ijom.2001.0214. [DOI] [PubMed] [Google Scholar]
- Bretones G, Delgado MD, León J. Myc and cell cycle control. BBA Gene Regulatory Mechanisms. 2015;1849(5):506–516. doi: 10.1016/j.bbagrm.2014.03.013. [DOI] [PubMed] [Google Scholar]
- Campo E, Rule S. Mantle cell lymphoma: evolving management strategies. Blood. 2015;125(1):48–55. doi: 10.1182/blood-2014-05-521898. [DOI] [PubMed] [Google Scholar]
- Camps J, Salaverria I, Garcia MJ, Prat E, Beà S, Pole JC, Hernández L, Del Rey J, Cigudosa JC, Bernués M. Genomic imbalances and patterns of karyotypic variability in mantle-cell lymphoma cell lines. Leuk Res. 2006;30(8):923–934. doi: 10.1016/j.leukres.2005.11.013. [DOI] [PubMed] [Google Scholar]
- Cassaday RD, Goy A, Advani S, Chawla P, Nachankar R, Gandhi M, Gopal AK. A phase II, single-arm, open-label, multicenter study to evaluate the efficacy and safety of P276-00, a cyclin-dependent kinase inhibitor, in patients with relapsed or refractory mantle cell lymphoma. Clin Lymphoma Myeloma Leuk. 2015;15(7):392–397. doi: 10.1016/j.clml.2015.02.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ceccarelli C, Santini D, Chieco P, Lanciotti C, Taffurelli M, Paladini G, Marrano D. Quantitative p21WAF-1/p53 immunohistochemical analysis defines groups of primary invasive breast carcinomas with different prognostic indicators. Int J Cancer. 2001;95(2):128–134. doi: 10.1002/1097-0215(20010320)95:2<128::aid-ijc1022>3.0.co;2-d. [DOI] [PubMed] [Google Scholar]
- Chen Y, Du X. Functional properties and intracellular signaling of CCN1/Cyr61. J Cell Biochem. 2007;100:1337–1345. doi: 10.1002/jcb.21194. [DOI] [PubMed] [Google Scholar]
- Chen G, Cheng Y, Zhang Z, Martinka M, Li G. Prognostic significance of cytoplasmic p27 expression in human melanoma. Cancer Epidemiol Biomark Prev. 2011;20(10):2212–2221. doi: 10.1158/1055-9965.EPI-11-0472. [DOI] [PubMed] [Google Scholar]
- Cheng T-Y, Wu M-S, Hua K-T, Kuo M-L, Lin M-T. Cyr61/CTGF/Nov family proteins in gastric carcinogenesis. World J Gastroenterol. 2014;20(7):1694–1700. doi: 10.3748/wjg.v20.i7.1694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chien W, Kumagai T, Miller CW, Desmond JC, Frank JM, Said JW, Koeffler HP. Cyr61 suppresses growth of human endometrial cancer cells. J Biol Chem. 2004;279(51):53087–53096. doi: 10.1074/jbc.M410254200. [DOI] [PubMed] [Google Scholar]
- Choi J, Lin A, Shrier E, Lau LF, Grant MB, Chaqour B. Degradome products of the matricellular protein CCN1 as modulators of pathological angiogenesis in the retina. J Biol Chem. 2013;288(32):23075–23089. doi: 10.1074/jbc.M113.475418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciccarelli C, Marampon F, Scoglio A, Mauro A, Giacinti C, De Cesaris P, Zani BM. p21WAF1 expression induced by MEK/ERK pathway activation or inhibition correlates with growth arrest, myogenic differentiation and onco-phenotype reversal in rhabdomyosarcoma cells. Mol Cancer. 2005;4(1):41. doi: 10.1186/1476-4598-4-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coats S, Flanagan WM, Nourse J, Roberts JM. Requirement of p27Kip1 for restriction point control of the fibroblast cell cycle. Science. 1996;272(5263):877–880. doi: 10.1126/science.272.5263.877. [DOI] [PubMed] [Google Scholar]
- Crawford LJ, Irvine AE. The role of the CCN family of proteins in blood cancers. J Cell Commun Signal. 2016;10(3):197–205. doi: 10.1007/s12079-016-0342-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crockett JC, Schütze N, Tosh D, Jatzke S, Duthie A, Jakob F, Rogers MJ. The matricellular protein CYR61 inhibits osteoclastogenesis by a mechanism independent of αvβ3 and αvβ5. Endocrinology. 2007;148(12):5761–5768. doi: 10.1210/en.2007-0473. [DOI] [PubMed] [Google Scholar]
- D'Antonio KB, Toubaji A, Albadine R, Mondul AM, Platz EA, Netto GJ, Getzenberg RH. Extracellular matrix associated protein CYR61 is linked to prostate cancer development. J Urol. 2010;183(4):1604–1610. doi: 10.1016/j.juro.2009.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Renty C, DePamphilis ML, Ullah Z. Cytoplasmic localization of p21 protects trophoblast giant cells from DNA damage induced apoptosis. PLoS One. 2014;9(5):e97434. doi: 10.1371/journal.pone.0097434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denicourt C, Saenz CC, Datnow B, Cui X-S, Dowdy SF. Relocalized p27Kip1 tumor suppressor functions as a cytoplasmic metastatic oncogene in melanoma. Cancer Res. 2007;67(19):9238–9243. doi: 10.1158/0008-5472.CAN-07-1375. [DOI] [PubMed] [Google Scholar]
- Devgan V, Mammucari C, Millar SE, Brisken C, Dotto GP. p21WAF1/Cip1 is a negative transcriptional regulator of Wnt4 expression downstream of Notch1 activation. Genes Dev. 2005;19(12):1485–1495. doi: 10.1101/gad.341405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobroff AS, Wang H, Melnikova VO, Villares GJ, Zigler M, Huang L, Bar-Eli M. Silencing cAMP-response element-binding protein (CREB) identifies CYR61 as a tumor suppressor gene in melanoma. J Biol Chem. 2009;284(38):26194–26206. doi: 10.1074/jbc.M109.019836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dotterweich J, Ebert R, Kraus S, Tower RJ, Jakob F, Schütze N. Mesenchymal stem cell contact promotes CCN1 splicing and transcription in myeloma cells. Cell Commun Signal. 2014;12(1):36. doi: 10.1186/1478-811X-12-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dreyling M, Aurer I, Cortelazzo S, Hermine O, Hess G, Jerkeman M, Le Gouill S, Ribrag V, Trněný M, Visco C, Walewski J, Zaja F, Zinzani PL. Treatment for patients with relapsed/refractory mantle cell lymphoma: European-based recommendations. Leuk Lymphoma. 2018;59(8):1814–1828. doi: 10.1080/10428194.2017.1403602. [DOI] [PubMed] [Google Scholar]
- Duncan TJ, Al-Attar A, Rolland P, Harper S, Spendlove I, Durrant LG. Cytoplasmic p27 expression is an independent prognostic factor in ovarian cancer. Int J Gynecol Pathol. 2010;29(1):8–18. doi: 10.1097/PGP.0b013e3181b64ec3. [DOI] [PubMed] [Google Scholar]
- Feng P, Wang B, Ren EC. Cyr61/CCN1 is a tumor suppressor in human hepatocellular carcinoma and involved in DNA damage response. Int J Biochem Cell Biol. 2008;40(1):98–109. doi: 10.1016/j.biocel.2007.06.020. [DOI] [PubMed] [Google Scholar]
- Ferrandina G, Stoler A, Fagotti A, Fanfani F, Sacco R, De Pasqua A, Mancuso S, Scambia G. p21WAF1/CIP1 protein expression in primary ovarian cancer. Int J Oncol. 2000;17(6):1231–1236. doi: 10.3892/ijo.17.6.1231. [DOI] [PubMed] [Google Scholar]
- Garrett-Engele CM, Tasch MA, Hwang HC, Fero ML, Perlmutter RM, Clurman BE, Roberts JM. A mechanism misregulating p27 in tumors discovered in a functional genomic screen. PLoS Genet. 2007;3(12):e219. doi: 10.1371/journal.pgen.0030219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gelebart P, Anand M, Armanious H, Peters AC, Bard JD, Amin HM, Lai R. Constitutive activation of the Wnt canonical pathway in mantle cell lymphoma. Blood. 2008;112(13):5171–5179. doi: 10.1182/blood-2008-02-139212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gery S, Xie D, Yin D, Gabra H, Miller C, Wang H, Scott D, William SY, Popoviciu ML, Said JW. Ovarian carcinomas: CCN genes are aberrantly expressed and CCN1 promotes proliferation of these cells. Clin Cancer Res. 2005;11(20):7243–7254. doi: 10.1158/1078-0432.CCR-05-0231. [DOI] [PubMed] [Google Scholar]
- Gillett C, Smith P, Gregory W, Richards M, Millis R, Peters G, Barnes D. Cyclin D1 and prognosis in human breast cancer. Int J Cancer. 1996;69(2):92–99. doi: 10.1002/(SICI)1097-0215(19960422)69:2<92::AID-IJC4>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- Hall M, Peters G. Genetic alterations of cyclins, cyclin-dependent kinases, and Cdk inhibitors in human cancer. Adv Cancer Res. 1996;68:67–108. doi: 10.1016/s0065-230x(08)60352-8. [DOI] [PubMed] [Google Scholar]
- Haque I, De A, Majumder M, Mehta S, McGregor D, Banerjee SK, Van Veldhuizen P, Banerjee S. The matricellular protein CCN1/Cyr61 is a critical regulator of Sonic Hedgehog in pancreatic carcinogenesis. J Biol Chem. 2012;287(46):38569–38579. doi: 10.1074/jbc.M112.389064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He W, Wang X, Chen L, Guan X. A crosstalk imbalance between p27kip1 and its interacting molecules enhances breast carcinogenesis. Cancer Biother Radiopharm. 2012;27(7):399–402. doi: 10.1089/cbr.2010.0802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirama T, Koeffler HP. Role of the cyclin-dependent kinase inhibitors in the development of cancer. Blood. 1995;86(3):841–854. [PubMed] [Google Scholar]
- Holloway SE, Beck AW, Girard L, Jaber MR, Barnett CC, Brekken RA, Fleming JB. Increased expression of Cyr61 (CCN1) identified in peritoneal metastases from human pancreatic cancer. J Am Coll Surg. 2005;200(3):371–377. doi: 10.1016/j.jamcollsurg.2004.10.005. [DOI] [PubMed] [Google Scholar]
- Izban KF, Alkan S, Singleton TP, Hsi ED. Multiparameter immunohistochemical analysis of the cell cycle proteins cyclin D1, Ki-67, p21WAF1, p27KIP1, and p53 in mantle cell lymphoma. Arch Pathol Lab Med. 2000;124(10):1457–1462. doi: 10.5858/2000-124-1457-MIAOTC. [DOI] [PubMed] [Google Scholar]
- Jandova J, Beyer TE, Meuillet EJ, Watts GS. The matrix protein CCN1/CYR61 is required for αVβ5-mediated cancer cell migration. Cell Biochem Funct. 2012;30(8):687–695. doi: 10.1002/cbf.2853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jares P, Colomer D, Campo E. Molecular pathogenesis of mantle cell lymphoma. J Clin Invest. 2012;122(10):3416–3423. doi: 10.1172/JCI61272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeannot P, Callot C, Baer R, Duquesnes N, Guerra C, Guillermet-Guibert J, Bachs O, Besson A. Loss of p27Kip1 promotes metaplasia in the pancreas via the regulation of Sox9 expression. Oncotarget. 2015;6(34):35880. doi: 10.18632/oncotarget.5770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jedsadayanmata A, Chen C-C, Kireeva ML, Lau LF, Lam SC-T. Activation-dependent adhesion of human platelets to Cyr61 and Fisp12/mouse connective tissue growth factor is mediated through integrin αIIbβ3. J Biol Chem. 1999;274(34):24321–24327. doi: 10.1074/jbc.274.34.24321. [DOI] [PubMed] [Google Scholar]
- Jeong D, Heo S, Ahn TS, Lee S, Park S, Kim H, Park D, Bae SB, Lee SS, Lee MS. Cyr61 expression is associated with prognosis in patients with colorectal cancer. BMC Cancer. 2014;14(1):164. doi: 10.1186/1471-2407-14-164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jim Leu SJ, Sung JS, Chen MY, Chen CW, Cheng JY, Wang TY, Wang JJ. The matricellular protein CCN1 suppresses lung cancer cell growth by inducing senescence via the p53/p21 pathway. J Cell Biochem. 2013;114(9):2082–2093. doi: 10.1002/jcb.24557. [DOI] [PubMed] [Google Scholar]
- Johnson SK, Stewart JP, Bam R, Qu P, Barlogie B, van Rhee F, Shaughnessy JD, Jr, Epstein J, Yaccoby S. CYR61/CCN1 overexpression in the myeloma microenvironment is associated with superior survival and reduced bone disease. Blood. 2014;124(13):2051–2060. doi: 10.1182/blood-2014-02-555813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung J-M, Bruner JM, Ruan S, Langford LA, Kyritsis AP, Kobayashi T, Levin VA, Zhang W. Increased levels of p21WAF1/Cip1 in human brain tumors. Oncogene. 1995;11(10):2021–2028. [PubMed] [Google Scholar]
- Kapranos N, Stathopoulos G, Manolopoulos L, Kokka E, Papadimitriou C, Bibas A, Yiotakis J, Adamopoulos G. p53, p21 and p27 protein expression in head and neck cancer and their prognostic value. Anticancer Res. 2000;21(1B):521–528. [PubMed] [Google Scholar]
- Kipkeew F, Kirsch M, Klein D, Wuelling M, Winterhager E, Gellhaus A. CCN1 (CYR61) and CCN3 (NOV) signaling drives human trophoblast cells into senescence and stimulates migration properties. Cell Adhes Migr. 2016;10(1–2):163–178. doi: 10.1080/19336918.2016.1139265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kireeva ML, Mo F-E, Yang GP, Lau LF. Cyr61, a product of a growth factor-inducible immediate-early gene, promotes cell proliferation, migration, and adhesion. Mol Cell Biol. 1996;16(4):1326–1334. doi: 10.1128/mcb.16.4.1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komiya T, Hosono Y, Hirashima T, Masuda N, Yasumitsu T, Nakagawa K, Kikui M, Ohno A, Fukuoka M, Kawase I. p21 expression as a predictor for favorable prognosis in squamous cell carcinoma of the lung. Clin Cancer Res. 1997;3(10):1831–1835. [PubMed] [Google Scholar]
- Kridel R, Meissner B, Rogic S, Boyle M, Telenius A, Woolcock B, Gunawardana J, Jenkins C, Cochrane C, Ben-Neriah S. Whole transcriptome sequencing reveals recurrent NOTCH1 mutations in mantle cell lymphoma. Blood. 2012;119(9):1963–1971. doi: 10.1182/blood-2011-11-391474. [DOI] [PubMed] [Google Scholar]
- Kruck S, Merseburger AS, Hennenlotter J, Scharpf M, Eyrich C, Amend B, Sievert KD, Stenzl A, Bedke J. High cytoplasmic expression of p27Kip1 is associated with a worse cancer-specific survival in clear cell renal cell carcinoma. BJU Int. 2012;109(10):1565–1570. doi: 10.1111/j.1464-410X.2011.10649.x. [DOI] [PubMed] [Google Scholar]
- LaBaer J, Garrett MD, Stevenson LF, Slingerland JM, Sandhu C, Chou HS, Fattaey A, Harlow E. New functional activities for the p21 family of CDK inhibitors. Genes Dev. 1997;11(7):847–862. doi: 10.1101/gad.11.7.847. [DOI] [PubMed] [Google Scholar]
- Lau LF. CCN1/CYR61: the very model of a modern matricellular protein. Cell Mol Life Sci. 2011;68(19):3149–3163. doi: 10.1007/s00018-011-0778-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leu S-J, Chen N, Chen C-C, Todorović V, Bai T, Juric V, Liu Y, Yan G, Lam SC-T, Lau LF. Targeted mutagenesis of the angiogenic protein CCN1 (CYR61) selective inactivation of integrin α6β1-heparan sulfate proteoglycan coreceptor-mediated cellular functions. J Biol Chem. 2004;279(42):44177–44187. doi: 10.1074/jbc.M407850200. [DOI] [PubMed] [Google Scholar]
- Li Z-Q, Ding W, Sun S-J, Li J, Pan J, Zhao C, Wu W-R, Si W-K. Cyr61/CCN1 is regulated by Wnt/β-catenin signaling and plays an important role in the progression of hepatocellular carcinoma (Cyr61 is regulated by Wnt and plays a role in HCC) PLoS One. 2012;7(4):e35754. doi: 10.1371/journal.pone.0035754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Nakka M, Kelly AJ, Lau CC, Krailo M, Barkauskas DA, Hicks JM, Man T-K. p27 is a candidate prognostic biomarker and metastatic promoter in osteosarcoma. Cancer Res. 2016;76(13):4002–4011. doi: 10.1158/0008-5472.CAN-15-3189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin M-T, Chang C-C, Chen S-T, Chang H-L, Su J-L, Chau Y-P, Kuo M-L. Cyr61 expression confers resistance to apoptosis in breast cancer MCF-7 cells by a mechanism of NF-κB-dependent XIAP up-regulation. J Biol Chem. 2004;279(23):24015–24023. doi: 10.1074/jbc.M402305200. [DOI] [PubMed] [Google Scholar]
- Lin M-T, Chang C-C, Lin B-R, Yang H-Y, Chu C-Y, Wu M-H, Kuo M-L. Elevated expression of Cyr61 enhances peritoneal dissemination of gastric cancer cells through integrin α2β1. J Biol Chem. 2007;282(47):34594–34604. doi: 10.1074/jbc.M706600200. [DOI] [PubMed] [Google Scholar]
- Liu Y, Zhang X, Zhong JF. Current approaches and advance in mantle cell lymphoma treatment. Stem Cell Investig. 2015;2:18. doi: 10.3978/j.issn.2306-9759.2015.09.02. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Z, Hunter T. Ubiquitylation and proteasomal degradation of the p21Cip1, p27Kip1 and p57Kip2 CDK inhibitors. Cell Cycle. 2010;9(12):2342–2352. doi: 10.4161/cc.9.12.11988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu X, Toki T, Konishi I, Nikaido T, Fujii S. Expression of p21WAF1/CIP1 in adenocarcinoma of the uterine cervix. Cancer. 1998;82(12):2409–2417. [PubMed] [Google Scholar]
- Lu Y, Yamagishi N, Yagi T, Takebe H. Mutated p21 (WAF1/CIP1/SDI1) lacking CDK-inhibitory activity fails to prevent apoptosis in human colorectal carcinoma cells. Oncogene. 1998;16(6):705–712. doi: 10.1038/sj.onc.1201585. [DOI] [PubMed] [Google Scholar]
- Lukas J, Groshen S, Saffari B, Niu N, Reles A, Wen W-H, Felix J, Jones LA, Hall FL, Press MF. WAF1/Cip1 gene polymorphism and expression in carcinomas of the breast, ovary, and endometrium. Am J Pathol. 1997;150(1):167–175. [PMC free article] [PubMed] [Google Scholar]
- Maity G, Mehta S, Haque I, Dhar K, Sarkar S, Banerjee SK, Banerjee S. Pancreatic tumor cell secreted CCN1/Cyr61 promotes endothelial cell migration and aberrant neovascularization. Sci Rep. 2014;4:4995. doi: 10.1038/srep04995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathur R, Sehgal L, Braun FK, Berkova Z, Romaguerra J, Wang M, Rodriguez MA, Fayad L, Neelapu SS, Samaniego F. Targeting Wnt pathway in mantle cell lymphoma-initiating cells. J Hematol Oncol. 2015;8(1):63. doi: 10.1186/s13045-015-0161-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCallum L, Irvine A. CCN3–a key regulator of the hematopoietic compartment. Blood Rev. 2009;23(2):79–85. doi: 10.1016/j.blre.2008.07.002. [DOI] [PubMed] [Google Scholar]
- Menendez JA, Vellon L, Mehmi I, Teng PK, Griggs DW, Lupu R. A novel CYR61-triggered ‘CYR61-αvβ3 integrin loop’regulates breast cancer cell survival and chemosensitivity through activation of ERK1/ERK2 MAPK signaling pathway. Oncogene. 2005;24(5):761–779. doi: 10.1038/sj.onc.1208238. [DOI] [PubMed] [Google Scholar]
- Min YH, Cheong J-W, Kim JY, Eom JI, Lee ST, Hahn JS, Ko YW, Lee MH. Cytoplasmic mislocalization of p27Kip1 protein is associated with constitutive phosphorylation of Akt or protein kinase B and poor prognosis in acute myelogenous leukemia. Cancer Res. 2004;64(15):5225–5231. doi: 10.1158/0008-5472.CAN-04-0174. [DOI] [PubMed] [Google Scholar]
- Molenaar JJ, Ebus ME, Koster J, van Sluis P, van Noesel CJ, Versteeg R, Caron HN. Cyclin D1 and CDK4 activity contribute to the undifferentiated phenotype in neuroblastoma. Cancer Res. 2008;68(8):2599–2609. doi: 10.1158/0008-5472.CAN-07-5032. [DOI] [PubMed] [Google Scholar]
- Moros A, Bustany S, Cahu J, Saborit-Villarroya I, Martínez A, Colomer D, Sola B, Roué G. Antitumoral activity of lenalidomide in in vitro and in vivo models of mantle cell lymphoma involves the destabilization of cyclin D1/p27KIP1 complexes. Clin Cancer Res. 2014;20(2):393–403. doi: 10.1158/1078-0432.CCR-13-1569. [DOI] [PubMed] [Google Scholar]
- Müller A, Zang C, Chumduri C, Dörken B, Daniel PT, Scholz CW. Concurrent inhibition of PI3K and mTORC1/mTORC2 overcomes resistance to rapamycin induced apoptosis by down-regulation of Mcl-1 in mantle cell lymphoma. Int J Cancer. 2013;133(8):1813–1824. doi: 10.1002/ijc.28206. [DOI] [PubMed] [Google Scholar]
- Nemes JA, Nemes Z, Márton IJ. p21WAF1/CIP1 expression is a marker of poor prognosis in oral squamous cell carcinoma. J Oral Pathol Med. 2005;34(5):274–279. doi: 10.1111/j.1600-0714.2005.00310.x. [DOI] [PubMed] [Google Scholar]
- Nevins JR. The Rb/E2F pathway and cancer. Hum Mol Genet. 2001;10(7):699–703. doi: 10.1093/hmg/10.7.699. [DOI] [PubMed] [Google Scholar]
- Niu C-C, Zhao C, Yang Z, Zhang X-L, Pan J, Si W-K. Inhibiting CCN1 blocks AML cell growth by disrupting the MEK/ERK pathway. Cancer Cell Int. 2014;14(1):74–74. doi: 10.1186/s12935-014-0074-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nordgren TM, Hegde GV, Joshi SS. Ritonavir exhibits limited efficacy as a single agent in treating aggressive mantle cell lymphoma. J Cancer Sci Ther. 2012;4(4):61–68. [Google Scholar]
- O’Connor OA. Mantle cell lymphoma: identifying novel molecular targets in growth and survival pathways. ASH Education Program Book. 2007;2007(1):270–276. doi: 10.1182/asheducation-2007.1.270. [DOI] [PubMed] [Google Scholar]
- Ohkoshi S, Yano M, Matsuda Y. Oncogenic role of p21 in hepatocarcinogenesis suggests a new treatment strategy. World J Gastroenterol. 2015;21(42):12150–12156. doi: 10.3748/wjg.v21.i42.12150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Kelly J, Chung A, Lemp N, Chumakova K, Yin D, Wang H-J, Said J, Gui D, Miller CW, Karlan BY. Functional domains of CCN1 (Cyr61) regulate breast cancer progression. Int J Oncol. 2008;33(1):59–67. [PubMed] [Google Scholar]
- Pendurthi UR, Tran TT, Post M, Rao LVM. Proteolysis of CCN1 by plasmin: functional implications. Cancer Res. 2005;65(21):9705–9711. doi: 10.1158/0008-5472.CAN-05-0982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng S-Y, Chou S-P, Hsu H-C. Association of downregulation of cyclin D1 and of overexpression of cyclin E with p53 mutation, high tumor grade and poor prognosis in hepatocellular carcinoma. J Hepatol. 1998;29(2):281–289. doi: 10.1016/s0168-8278(98)80014-7. [DOI] [PubMed] [Google Scholar]
- Perbal B. Nuclear localisation of NOVH protein: a potential role for NOV in the regulation of gene expression. Mol Pathol. 1999;52(2):84–91. doi: 10.1136/mp.52.2.84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perbal B. Alternative splicing of CCN mRNAs…. It has been upon us. J Cell Commun Signal. 2009;3(2):153–157. doi: 10.1007/s12079-009-0051-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez-Galán P, Dreyling M, Wiestner A. Mantle cell lymphoma: biology, pathogenesis, and the molecular basis of treatment in the genomic era. Blood. 2011;117(1):26–38. doi: 10.1182/blood-2010-04-189977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Roger I, Kim SH, Griffiths B, Sewing A, Land H. Cyclins D1 and D2 mediate Myc-induced proliferation via sequestration of p27Kip1 and p21Cip1. EMBO J. 1999;18(19):5310–5320. doi: 10.1093/emboj/18.19.5310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez-Sayáns M, Suárez-Peñaranda JM, Gayoso-Diz P, Barros-Angueira F, Gándara-Rey JM, García-García A. The role of p21Waf1/CIP1 as a Cip/Kip type cell-cycle regulator in oral squamous cell carcinoma (review) Med Oral Patol Oral Cir Bucal. 2013;18(2):e219. doi: 10.4317/medoral.18213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perkins ND. Not just a CDK inhibitor: regulation of transcription by p21WAF1/CIP1/SDI1. Cell Cycle. 2002;1(1):35–37. [PubMed] [Google Scholar]
- Piccolo MT, Crispi S. The dual role played by p21 may influence the apoptotic or anti-apoptotic fate in cancer. J Cancer Res Updates. 2012;1(2):189–202. [Google Scholar]
- Planque N. Nuclear trafficking of secreted factors and cell-surface receptors: new pathways to regulate cell proliferation and differentiation, and involvement in cancers. Cell Commun Signal. 2006;4(1):7. doi: 10.1186/1478-811X-4-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Planque N, Perbal B. A structural approach to the role of CCN (CYR61/CTGF/NOV) proteins in tumourigenesis. Cancer Cell Int. 2003;3(1):15. doi: 10.1186/1475-2867-3-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Planque N, Long Li C, Saule S, Bleau AM, Perbal B. Nuclear addressing provides a clue for the transforming activity of amino-truncated CCN3 proteins. J Cell Biochem. 2006;99(1):105–116. doi: 10.1002/jcb.20887. [DOI] [PubMed] [Google Scholar]
- Qin L-F, Ng I O-l. Expression of p27 KIP1 and p21 WAF1/CIP1 in primary hepatocellular carcinoma: clinicopathologic correlation and survival analysis. Hum Pathol. 2001;32(8):778–785. doi: 10.1053/hupa.2001.27105. [DOI] [PubMed] [Google Scholar]
- Queirós AC, Beekman R, Vilarrasa-Blasi R, Duran-Ferrer M, Clot G, Merkel A, Raineri E, Russiñol N, Castellano G, Beà S, Navarro A, Kulis M, Verdaguer-Dot N, Jares P, Enjuanes A, Calasanz MJ, Bergmann A, Vater I, Salaverría I, vane de Werken HJG, Wilson WH, Datta A, Flicek P, Royo R, Martens J, Giné E, Lopez-Guillermo A, Stunnenberg HG, Klapper W, Pott C, Heath S, Gut IG, Siebert R, Campo E, Martín-Subero JI. Decoding the DNA Methylome of mantle cell lymphoma in the light of the entire B cell lineage. Cancer Cell. 2016;30(5):806–821. doi: 10.1016/j.ccell.2016.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Queiroz AB, Focchi G, Dobo C, Gomes TS, Ribeiro DA, Oshima CT. Expression of P27, P21WAF/Cip1, and P16INK4a in normal oral epithelium, oral squamous papilloma, and oral squamous cell carcinoma. Anticancer Res. 2010;30(7):2799–2803. [PubMed] [Google Scholar]
- Quintanilla-Martinez L, Thieblemont C, Fend F, Kumar S, Pinyol M, Campo E, Jaffe ES, Raffeld M. Mantle cell lymphomas lack expression of p27 Kip1, a cyclin-dependent kinase inhibitor. Am J Pathol. 1998;153(1):175–182. doi: 10.1016/S0002-9440(10)65558-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quintanilla-Martinez L, Davies-Hill T, Fend F, Calzada-Wack J, Sorbara L, Campo E, Jaffe ES, Raffeld M. Sequestration of p27 Kip1 protein by cyclin D1 in typical and blastic variants of mantle cell lymphoma (MCL): implications for pathogenesis. Blood-New York. 2003;101(8):3181–3187. doi: 10.1182/blood-2002-01-0263. [DOI] [PubMed] [Google Scholar]
- Rauert-Wunderlich H, Rudelius M, Ott G, Rosenwald A. Targeting protein kinase C in mantle cell lymphoma. Br J Haematol. 2016;173(3):394–403. doi: 10.1111/bjh.13973. [DOI] [PubMed] [Google Scholar]
- Ray A, James MK, Larochelle S, Fisher RP, Blain SW. p27Kip1 inhibits cyclin D-cyclin-dependent kinase 4 by two independent modes. Mol Cell Biol. 2009;29(4):986–999. doi: 10.1128/MCB.00898-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romanov V, Pospelov V, Pospelova T. Cyclin-dependent kinase inhibitor p21Waf1: contemporary view on its role in senescence and oncogenesis. Biochem Mosc. 2012;77(6):575–584. doi: 10.1134/S000629791206003X. [DOI] [PubMed] [Google Scholar]
- Roninson IB. Oncogenic functions of tumour suppressor p21 Waf1/Cip1/Sdi1: association with cell senescence and tumour-promoting activities of stromal fibroblasts. Cancer Lett. 2002;179(1):1–14. doi: 10.1016/s0304-3835(01)00847-3. [DOI] [PubMed] [Google Scholar]
- Roodman GD. CCN1: a sticky issue in myeloma. Blood. 2014;124(13):2006–2008. doi: 10.1182/blood-2014-08-592303. [DOI] [PubMed] [Google Scholar]
- Roy S, Singh RP, Agarwal C, Siriwardana S, Sclafani RA, Agarwal R. Downregulation of both p21/Cip1 and p27/Kip1 produces a more aggressive prostate cancer phenotype. Cell Cycle. 2008;7(12):1828–1835. doi: 10.4161/cc.7.12.6024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Royo C, Navarro A, Clot G, Salaverria I, Giné E, Jares P, Colomer D, Wiestner A, Wilson WH, Vegliante MC, Fernandez V, Hartmann EM, Trim N, Erber WN, Swerdlow SH, Klapper W, Dyer MJ, Vargas-Pabón M, Ott G, Rosenwald A, Siebert R, López-Guillermo A, Campo E, Beà S. Non-nodal type of mantle cell lymphoma isa specific biological and clinical subgroup of the disease. Leukemia. 2012;26(8):1895–1898. doi: 10.1038/leu.2012.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudolph C, Steinemann D, Von Neuhoff N, Gadzicki D, Ripperger T, Drexler HG, Mrasek K, Liehr T, Claussen U, Emura M, Schrock E, Schlegelberger B. Molecular cytogenetic characterization of the mantle cell lymphoma cell line GRANTA-519. Cancer Genet Cytogenet. 2004;153(2):144–150. doi: 10.1016/j.cancergencyto.2004.01.006. [DOI] [PubMed] [Google Scholar]
- Saglam O, Dai F, Husain S, Zhan Y, Toruner G, Haines GK. Matricellular protein CCN1 (CYR61) expression is associated with high-grade ductal carcinoma in situ. Hum Pathol. 2014;45(6):1269–1275. doi: 10.1016/j.humpath.2014.02.007. [DOI] [PubMed] [Google Scholar]
- Samaniego F, Sehgal L, Braun FK, Berkova Z, Romaguera JE, Wang M, Rodriguez A, Neelapu SS, Mathur R. Molecular signatures of tumor-initiating cells unveil wnt pathway as a therapeutic target in mantle cell lymphoma. Blood. 2014;2014(124):2148. [Google Scholar]
- Sawai K, Mukoyama M, Mori K, Kasahara M, Koshikawa M, Yokoi H, Yoshioka T, Ogawa Y, Sugawara A, Nishiyama H. Expression of CCN1 (CYR61) in developing, normal, and diseased human kidney. Am J Physiol Renal Physiol. 2007;293(4):F1363–F1372. doi: 10.1152/ajprenal.00205.2007. [DOI] [PubMed] [Google Scholar]
- Si W, Kang Q, Luu HH, Park JK, Luo Q, Song W-X, Jiang W, Luo X, Li X, Yin H. CCN1/Cyr61 is regulated by the canonical Wnt signal and plays an important role in Wnt3A-induced osteoblast differentiation of mesenchymal stem cells. Mol Cell Biol. 2006;26(8):2955–2964. doi: 10.1128/MCB.26.8.2955-2964.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith MR. Should there be a standard therapy for mantle cell lymphoma? Future Oncol. 2011;7(2):227–237. doi: 10.2217/fon.10.189. [DOI] [PubMed] [Google Scholar]
- Sohn D, Essmann F, Schulze-Osthoff K, Jänicke RU. p21 blocks irradiation-induced apoptosis downstream of mitochondria by inhibition of cyclin-dependent kinase–mediated caspase-9 activation. Cancer Res. 2006;66(23):11254–11262. doi: 10.1158/0008-5472.CAN-06-1569. [DOI] [PubMed] [Google Scholar]
- Sun Z, Wang Y, Cai Z, Chen P, Tong X, Xie D. Involvement of Cyr61 in growth, migration, and metastasis of prostate cancer cells. Br J Cancer. 2008;99(10):1656–1667. doi: 10.1038/sj.bjc.6604712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swerdlow SH, Campo E, Pileri SA, Harris NL, Stein H, Siebert R, Advani R, Ghielmini M, Salles GA, Zelenetz AD, Jaffe ES. The 2016 revision of the World Health Organization classification of lymphoid neoplasms. Blood. 2016;127(20):2375–2390. doi: 10.1182/blood-2016-01-643569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura I, Rosenbloom J, Macarak E, Chaqour B. Regulation of Cyr61 gene expression by mechanical stretch through multiple signaling pathways. Am J Physiol Cell Physiol. 2001;281:C1524–C1532. doi: 10.1152/ajpcell.2001.281.5.C1524. [DOI] [PubMed] [Google Scholar]
- Thompson K, Murphy-Marshman H, Leask A. ALK5 inhibition blocks TGFβ-induced CCN1 expression in human foreskin fibroblasts. J Cell Commun Signal. 2014;8(1):59–63. doi: 10.1007/s12079-014-0229-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timmerbeul I, Garrett-Engele CM, Kossatz U, Chen X, Firpo E, Grünwald V, Kamino K, Wilkens L, Lehmann U, Buer J. Testing the importance of p27 degradation by the SCFskp2 pathway in murine models of lung and colon cancer. Proc Natl Acad Sci. 2006;103(38):14009–14014. doi: 10.1073/pnas.0606316103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Todorovicç V, Chen C-C, Hay N, Lau LF. The matrix protein CCN1 (CYR61) induces apoptosis in fibroblasts. J Cell Biol. 2005;171(3):559–568. doi: 10.1083/jcb.200504015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong X, Xie D, O'Kelly J, Miller CW, Muller-Tidow C, Koeffler HP. Cyr61, a member of CCN family, is a tumor suppressor in non-small cell lung cancer. J Biol Chem. 2001;276(50):47709–47714. doi: 10.1074/jbc.M107878200. [DOI] [PubMed] [Google Scholar]
- Tong X, O'Kelly J, Xie D, Mori A, Lemp N, McKenna R, Miller CW, Koeffler HP. Cyr61 suppresses the growth of non-small-cell lung cancer cells via the β-catenin–c-myc–p53 pathway. Oncogene. 2004;23(28):4847–4855. doi: 10.1038/sj.onc.1207628. [DOI] [PubMed] [Google Scholar]
- Toyoshima H, Hunter T. p27, a novel inhibitor of G1 cyclin-Cdk protein kinase activity, is related to p21. Cell. 1994;78(1):67–74. doi: 10.1016/0092-8674(94)90573-8. [DOI] [PubMed] [Google Scholar]
- Tucker CA, Bebb G, Klasa RJ, Chhanabhai M, Lestou V, Horsman DE, Gascoyne RD, Wiestner A, Masin D, Bally M. Four human t (11; 14)(q13; q32)-containing cell lines having classic and variant features of mantle cell lymphoma. Leuk Res. 2006;30(4):449–457. doi: 10.1016/j.leukres.2005.08.016. [DOI] [PubMed] [Google Scholar]
- Waga S, Hannon GJ, Beach D, Stillman B. The p21 inhibitor of cyclin-dependent kinases controls DNA replication by interaction with PCNA. Nature. 1994;369(6481):574–578. doi: 10.1038/369574a0. [DOI] [PubMed] [Google Scholar]
- Wells J, Howlett M, Cheung L, Kees UR. The role of CCN family genes in haematological malignancies. J Cell commun Signal. 2015;9(3):267–278. doi: 10.1007/s12079-015-0296-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie D, Yin D, Tong X, O’Kelly J, Mori A, Miller C, Black K, Gui D, Said JW, Koeffler HP. Cyr61 is overexpressed in gliomas and involved in integrin-linked kinase-mediated Akt and β-catenin-TCF/Lef signaling pathways. Cancer Res. 2004;64(6):1987–1996. doi: 10.1158/0008-5472.can-03-0666. [DOI] [PubMed] [Google Scholar]
- Xie D, Yin D, Wang H-J, Liu G-T, Elashoff R, Black K, Koeffler HP. Levels of expression of CYR61 and CTGF are prognostic for tumor progression and survival of individuals with gliomas. Clin Cancer Res. 2004;10(6):2072–2081. doi: 10.1158/1078-0432.ccr-0659-03. [DOI] [PubMed] [Google Scholar]
- Yakubenko VP, Yadav SP, Ugarova TP. Integrin α D β 2, an adhesion receptor up-regulated on macrophage foam cells, exhibits multiligand-binding properties. Blood. 2006;107(4):1643–1650. doi: 10.1182/blood-2005-06-2509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Yan B. Cell cycle regulation by carboxylated multiwalled carbon nanotubes through p53-independent induction of p21 under the control of the BMP signaling pathway. Chem Res Toxicol. 2012;25(6):1212–1221. doi: 10.1021/tx300059m. [DOI] [PubMed] [Google Scholar]
- Zhang L, Schafer P, Muller G, Stirling D, Bartlett B. The ratio of cyclin D1/p21kip baseline gene expression and SPARC gene expression can be potential predictors of non-Hodgkin's lymphoma (NHL) patient response to lenalidomide therapy. J Clin Oncol. 2008;26(15_suppl):22150–22150. [Google Scholar]
- Zhao H, Faltermeier CM, Mendelsohn L, Porter PL, Clurman BE, Roberts JM. Mislocalization of p27 to the cytoplasm of breast cancer cells confers resistance to anti-HER2 targeted therapy. Oncotarget. 2014;5(24):12704. doi: 10.18632/oncotarget.2871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zirbes TK, Baldus SE, Moenig SP, Nolden S, Kunze D, Shafizadeh ST, Schneider PM, Thiele J, Hoelscher AH, Dienes HP. Prognostic impact of p21/waf1/cip1 in colorectal cancer. Int J Cancer. 2000;89(1):14–18. doi: 10.1002/(sici)1097-0215(20000120)89:1<14::aid-ijc3>3.0.co;2-l. [DOI] [PubMed] [Google Scholar]
- Zucca E, Bertoni F. Toward new treatments for mantle-cell lymphoma? N Engl J Med. 2013;369(6):571–572. doi: 10.1056/NEJMe1307596. [DOI] [PubMed] [Google Scholar]